Abstract
Antisense oligonucleotides represent a novel therapeutic platform for the discovery of medicines that have the potential to treat most neurodegenerative diseases. Antisense drugs are currently in development for the treatment of amyotrophic lateral sclerosis, Huntington’s disease, and Alzheimer’s disease, and multiple research programs are underway for additional neurodegenerative diseases. One antisense drug, nusinersen, has been approved for the treatment of spinal muscular atrophy. Importantly, nusinersen improves disease symptoms when administered to symptomatic patients rather than just slowing the progression of the disease. In addition to the benefit to spinal muscular atrophy patients, there are discoveries from nusinersen that can be applied to other neurological diseases, including method of delivery, doses, tolerability of intrathecally delivered antisense drugs, and the biodistribution of intrathecal dosed antisense drugs. Based in part on the early success of nusinersen, antisense drugs hold great promise as a therapeutic platform for the treatment of neurological diseases.
Keywords: amyotrophic lateral sclerosis, antisense oligonucleotides, Huntington’s disease, RNA, spinal muscular atrophy
INTRODUCTION
The genetic basis for most of the common inherited neurodegenerative diseases was determined over the past 25 years, and subsequently, a wealth of information on the biochemical, molecular, and neural pathways impacted by these genetic changes has been uncovered. In addition to inherited neurodegenerative diseases, there has been enormous progress in our understanding of the mechanisms of pathology in sporadic neurodegenerative diseases such as Alzheimer’s disease. Although there are several promising therapies currently in clinical trials, except for spinal muscular atrophy (SMA), there are no available treatments that significantly modify the course of these diseases. As it is estimated that Alzheimer’s and other neurodegenerative diseases will consume a large part of our health care dollars, it is imperative that more effective therapies be identified.
A key challenge presented by these discoveries from a drug-discovery perspective is that most identified genes are not readily targeted with current small-molecule therapeutics. For example, the genetic change identified in Huntington’s disease (HD) is a polyglutamine expansion in the amino terminus of the huntingtin protein that is thought to result in a dominant gain of function (MacDonald et al. 1993, Munoz-Sanjuan & Bates 2011). Huntingtin is a large (>300 kDa) protein that lacks a characterized enzymatic function and, as such, has not been amenable to direct targeting with small-molecule drugs. Although antibody-based (Yu & Watts 2013) and conformation-correcting (Eisele et al. 2015) therapeutics are being developed to enhance the removal or spread of protein species that are implicated in causing some instances of neurodegenerative disease, candidate genes for other dominantly inherited neurodegenerative disease are not obvious targets for small-molecule or antibody drugs. Similarly, studies into the mechanisms of recessively inherited genes have yielded a limited number of candidate proteins for developing conventional small-molecule-based drugs.
RNA represents a unique target for developing therapeutics. There are several advantages in using RNA as a therapeutic target such as broad applicability to most RNAs in the cell, including noncoding RNAs, direct translation of genetic discoveries to drug discovery programs, and the speed and efficiency of the drug-discovery process. Although there has been some encouraging progress in the identification of small-molecule drugs that modulate RNA function (Naryshkin et al. 2014, Palacino et al. 2015), antisense oligonucleotides (ASOs) offer a more direct approach. The first concerted studies to evaluate antisense drugs as possible therapeutics for neurodegenerative diseases began approximately 15 years ago. The initial focus was on inherited neurodegenerative diseases with a clearly defined therapeutic target. Given the unknowns about the distribution of ASOs in the central nervous system (CNS), these studies focused on diseases that primarily produced pathology in more accessible tissue from the cerebral spinal fluid (CSF) (e.g., spinal cord) (Cartegni & Krainer 2003, Hua et al. 2007, Smith et al. 2006). Once proof of concept was established, the goal was to expand first to inherited diseases that produced pathology in the brain and then to sporadic diseases with more target risk. The strategy has been successful, producing one approved drug, six drugs in clinical development, and numerous drug discovery programs. In this review, we highlight some of the more advanced programs as examples of the potential of antisense technology for treating neurodegenerative and neurological diseases.
Antisense Therapeutics
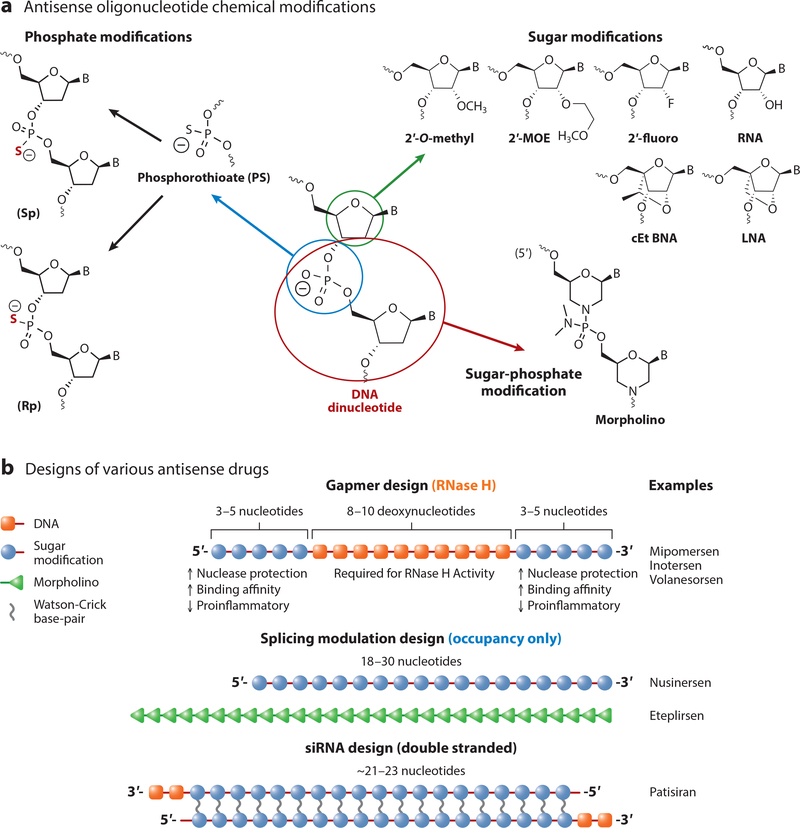
ASOs are synthetic oligonucleotide or oligonucleotide analogs usually between 12 and 30 nucleotides in length that are designed to bind to RNA by Watson-Crick base pairing. ASOs can be designed to bind to protein coding RNAs (messenger RNAs) as well as noncoding RNAs such as microRNAs or large noncoding RNAs. After binding to the targeted RNA, the antisense drug can modulate the function of the targeted RNA by several different mechanisms (Bennett & Swayze 2010, Crooke et al. 2018). The molecular mechanisms through which an ASO modulates RNA function are dependent on the chemical modifications, the position the modifications are incorporated into the oligonucleotide, and where on the target RNA the oligonucleotide binds. A detailed discussion of different antisense mechanisms and chemical modifications can be found in several recent reviews (Bennett et al. 2017, Bennett & Swayze 2010, Crooke 2017, Crooke et al. 2018). Briefly, ASOs generally incorporate sugar modifications to enhance binding affinity to the target RNA, increase metabolic stability, and decrease adverse effects and phosphate modifications; which enhance metabolic stability and increase protein binding to enhance tissue distribution and cellular uptake (Figure 1). The morpholino modification increases metabolic stability but decreases protein binding (Bennett et al. 2017). The most widely used modifications for CNS applications are oligonucleotides containing both phosphorothioate- and 2′-O-methoxyethyl (MOE)- modifications, either as uniform 2′-MOE modified for splicing applications or as chimeric 2′-MOE/DNA modifications (gapmer), for the RNase H mechanism (Figure 1).
Figure 1.
Common chemical modifications and designs used for ASO therapeutics. Common chemical modifications include 2′-sugar modifications such as 2′-O-methyl, 2′-O- MOE), and 2′fluoro; 2′,4′-bridged sugar modifications such as LNA and cEt BNA; the phosphate modification phosphorothioate, which can be synthesized as the (Sp) and (Rp) isomers; and the sugar-phosphate replacement morpholino. ASOs can be designed as single-stranded ASOs that utilize the RNase H mechanism (gapmer), uniformly modified to promote splicing, or as double-stranded designs that work through the RISC pathway (siRNA). Abbreviations: ASO, antisense oligonucleotide; cEt BNA, constrained ethyl bicyclic nucleic acid; fluoro, fluorine; LNA, locked nucleic acid; MOE, methoxyethyl; RISC, RNA-induced silencing complex; siRNA, small interfering RNA.
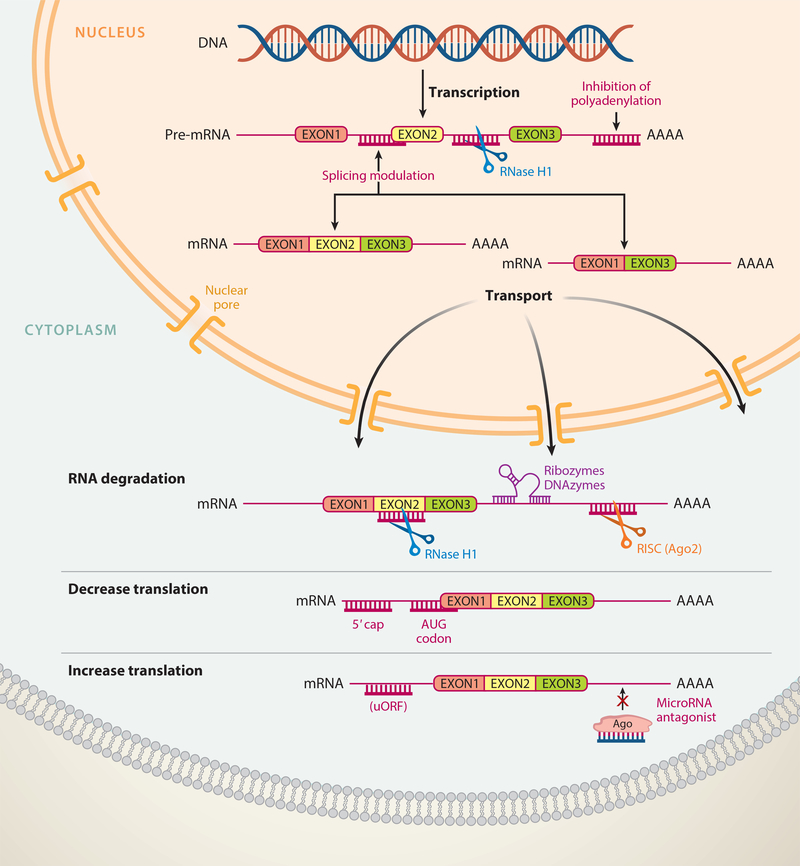
There are multiple mechanisms by which ASOs can interfere with RNA function (Figure 2). ASOs can be designed to promote degradation of the bound RNA by endogenous nucleases such as RNase H1 or, alternatively, argonaute 2 (Ago2), which is the active enzyme in the RNA-induced silencing complex (RISC). Oligonucleotides that work through RNase H1 generally have between 8 and 10 consecutive DNA nucleotides to support binding and cleavage by RNase H1 (Figure 1), while ASOs that work through Ago2 are generally delivered to the cell as double-stranded RNA or modified RNA oligonucleotides (Figure 1). Once in the cell, the double-stranded molecule interacts with RISC, followed by the selective binding of one strand to Ago2 to form a ribonuclear protein complex (Meister & Tuschl 2004). ASOs can also be designed to modulate the processing of the RNA molecule, such as by modulating RNA splicing (Kole et al. 2012) or polyadenylation site selection (Vickers et al. 2001) or by nonsense-mediated mRNA decay (Nomakuchi et al. 2016). Alternatively, ASOs can be designed to disrupt RNA structures that inhibit translation, block upstream AUG codons (upstream open reading frames or uORFs), or bind to microRNAs so as to block their interaction with messenger RNA (mRNA), all of which can increase protein translation (Esau et al. 2006; Liang et al. 2016, 2017). Each of these antisense mechanisms has been documented to work in cell culture, and most have been documented to occur in animals. The choice of which method to exploit depends on the specific molecular changes desired.
Figure 2.
Mechanisms of action for ASOs. ASOs can modulate RNA function by a variety of mechanisms, including degradation of the pre-mRNA in the nucleus or mature RNA in the cytoplasm by RNase H1, and degradation of RNA in the cytoplasm by the RISC complex (Ago2) or ribozymes or DNAzymes. ASOs can also modulate RNA functiona by nondegradative mechanisms such as splicing or polyadenylation modulation in the nucleus and decrease or increase protein translation in the cytoplasm. Abbreviations: Ago2, argonaute 2; ASO, antisense oligonucleotide; mRNA, messenger RNA; pre-mRNA, precursor mRNA; RISC, RNA-induced silencing complex; uORF, upstream open reading frame.
ASO Distribution and Pharmacokinetics in the Central Nervous System
Chemical modifications have a major impact on the pharmacokinetic properties of ASO drugs (Geary et al. 2015). Single-stranded phosphorothioate-modified oligodeoxynucleotides (ODNs) as well as phosphorothioate-modified chimeric DNA/2′-sugar oligonucleotides (Figure 1) distribute broadly when administered systemically, with the highest concentrations found in kidney, liver, spleen, and lymphatic tissues (Geary et al. 2001, 2015). As mentioned above, morpholino-modified ASOs have limited protein binding and are rapidly cleared in the urine. The same is true for double-stranded ASOs, unless they are conjugated to specific targeting ligands or formulated in nanoparticulate formulations (Geary et al. 2015). Although there are reports of phosphorothioate modified ODNs crossing the blood-brain barrier (BBB) (Banks et al. 2001), in our experience, the amount that crosses an intact BBB is not sufficient for pharmacological activity. Injection of single-stranded phosphorothioate-modified and 2′-MOE-modified ASOs into the CSF results in rapid distribution throughout the spinal cord and into most regions of the brain (Butler et al. 2000, Finkel et al. 2016, Kordasiewicz et al. 2012, Passini et al. 2011, Rigo et al. 2014, Smith et al. 2006). Approximately 80% or more of an intrathecal injected dose appears in systemic circulation, peaking 4–6 h after injection (Finkel et al. 2016, Rigo et al. 2014), which is consistent with clearance out of the CSF by normal CSF turnover pathways. The remaining 20% of the injected dose remains in brain tissue. The tissue half-life for 2′-MOE-modified oligonucleotides ranges from 3 weeks to 6 months in CNS tissues of mice and nonhuman primates. Depending on the chemistry and chemical design, the effects of single-stranded phosphorothioate ASOs on gene expression can last from 6 weeks to more than 6 months following a single injection, supporting infrequent administration of the antisense drug (Hua et al. 2010, Kordasiewicz et al. 2012, Rigo et al. 2014).
Following intrathecal administration, the highest drug concentrations are found in the lumbar spinal cord (near the site of injection), with slightly lower concentrations found in other regions of the spinal cord and cortical regions of the brain (DeVos et al. 2017, Finkel et al. 2016, McCampbell et al. 2018, Rigo et al. 2014). In the brain there is a concentration gradient, with the highest drug concentrations found in the cortex, hippocampus, and Purkinje cells of the cerebellum, and the lowest drug concentrations are found in deeper brain structures such as the striatum and in the granular layer of the cerebellum. The distribution is similar following intracerebral ventricle administration, although higher drug concentrations are found in the tissue surrounding the ventricle. Similar distribution patterns have been observed in mice, rats, cynomolgus and Rhesus monkeys, dogs, pigs, and human subjects.
NEURODEGENERATIVE DISEASES
As discussed above, there is an urgent need to identify more effective therapies for neurological diseases, including neurodegenerative diseases. Based on the experience with nusinersen, antisense technology has the potential to create transformative medicines for the treatment of a variety of neurodegenerative diseases. This section briefly reviews nusinersen and several ASO drugs currently in clinical trials.
Spinal Muscular Atrophy
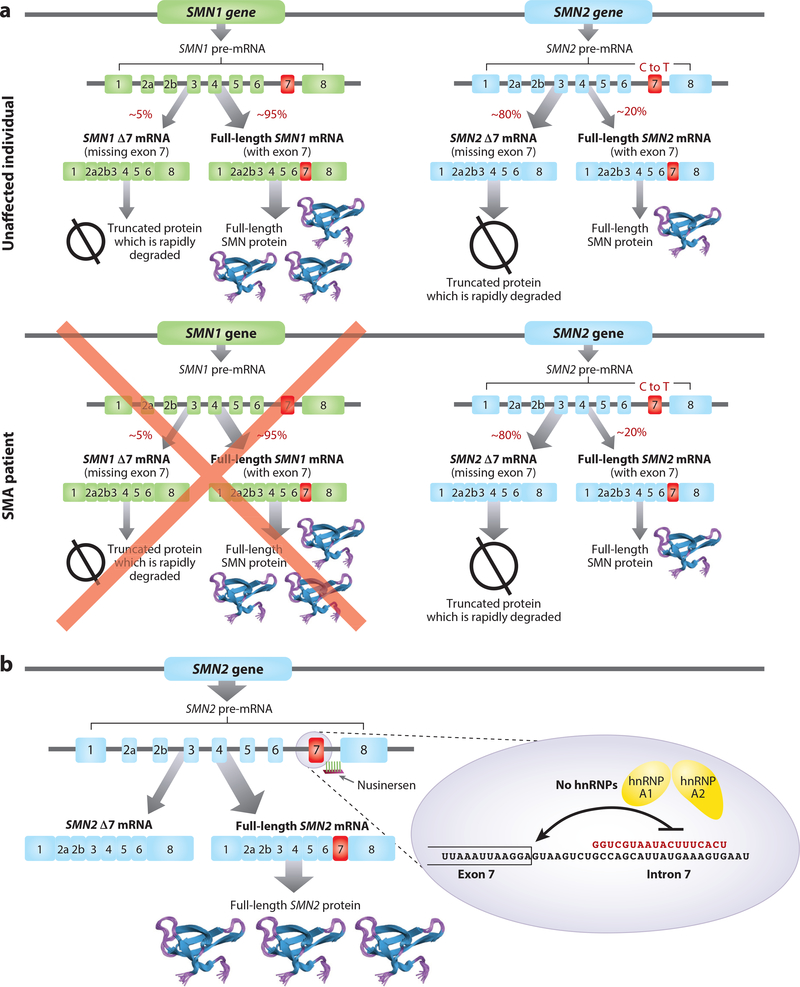
SMA is a rare autosomal recessive neuromuscular disease caused by deletions or point mutations in the telomeric Survival Motor Neuron 1 (SMN1) gene (Figure 3) (Lefebvre et al. 1995, Sugarman et al. 2012, Sumner 2007). The disease presents as a spectrum of phenotypes, with the most severe form, type I SMA, resulting in mortality shortly after birth. Children with type I SMA present with generalized muscle weakness and hypotonia in the first few months of life. These children are never able to sit unaided and usually die by 2 years of age without nutritional and respiratory support (Cobben et al. 2008, Finkel et al. 2014b, Rudnik-Schoneborn et al. 2009). Type II SMA children are able to sit or stand unaided but unable to walk unaided, with survival expected into adulthood (Kaufmann et al. 2011, 2012; Oskoui & Kaufmann 2008). Type III SMA patients are able to sit and walk unaided and have a normal life span. However, as they age, type III patients gradually lose muscle function and become dependent on wheelchairs. Type IV or adult-onset SMA patients have a normal life expectancy but develop weakness over time due to a loss of muscle function.
Figure 3.
Mechanism of nusinersen action. (a) Humans express two SMN genes, SMN1 and SMN2, that differ by a few nucleotides due to a duplication in chromosome 5. A cytosine to thymine transition in exon 7 of SMN2 weakens the RNA splicing signal such that most transcripts derived from SMN2 skip exon 7, creating a truncated protein that is rapidly degraded. SMA patients have a deletion mutation in the SMN1 gene, resulting in an SMN protein deficiency. (b) Nusinersen binds to a site in intron 7 of the SMN2 premRNA, displacing the splicing suppressor proteins hnRNP A1/A2 and allowing U1 small nuclear ribonucleoproteins to bind to the 5′-splice site, promoting inclusion of SMN2 exon 2 into the mRNA. Abbreviations: mRNA, messenger RNA; premRNA, precursor mRNA; SMA, spinal muscular atrophy.
The chromosomal region where the SMN1 gene lies, 5q13, underwent a duplication during primate evolution, resulting in a nearly identical SMN2 gene located centromeric to the SMN1 gene (Figure 3) (Rochette et al. 2001). The SMN2 gene differs from SMN1 by 5–13 nucleotides (Lefebvre et al. 1995, Lorson et al. 1999, Monani et al. 1999). The most important difference between SMN1 and SMN2 is a C to T transition in exon 7, which inactivates a splicing enhancer and simultaneously introduces a splicing silencer, resulting in the skipping of exon 7 in most SMN2 transcripts (Cartegni & Krainer 2002, Cartegni et al. 2006, Kashima & Manley 2003, Kashima et al. 2007, Lorson et al. 1999). Because the SMN1 and SMN2 genes lie in an unstable chromosomal region, there is variation in the number of copies of both genes in the human population, including individuals with more than two copies of SMN2 without any known deleterious effect. Patients with type 1 SMA typically have two SMN2 gene copies, those with type 2 SMA have three SMN2 gene copies, and patients with type 3 SMA have three to four SMN2 gene copies. These observations support the idea that SMN2 copy number is a robust disease modifier (Prior et al. 2009; Rudnik-Schoneborn et al. 2009; Wirth et al. 1999, 2006).
Nusinersen (Spinraza) is an antisense drug designed to increase the expression of SMN protein by modulating splicing of the SMN2 precursor messenger RNA (premRNA) to make a full-length mature messenger RNA (mRNA) from the SMN2 premRNA. Nusinersen was identified following an extensive screen of uniform 2′-MOE-modified ASOs. Several sites within exon 7, intron 6, and intron 7 enhanced exon 7 inclusion when targeted with ASOs (Hua et al. 2007, 2008, 2012). The most effective ASO target site was within intron 7, approximately 10 nucleotides downstream of the 5′-splice site (Hua et al. 2008). The same regulatory region was also independently identified by another laboratory and termed ISS-N1, for intronic splicing silencer N1 (Singh et al. 2006). Nusinersen produced dose-dependent hSMN2 exon 7 inclusion in peripheral tissues in transgenic mice expressing the human SMN2 gene when administered systemically and in CNS tissues when administered directly into CSF (Hua et al. 2008, 2010, 2011; Rigo et al. 2014). In neonatal mice but not older mice, systemic dosing with nusinersen resulted in SMN2 exon 7 inclusion in spinal cord and brain tissue, consistent with the postnatal formation of an intact BBB. In adult mice, a single administration into the CSF by injection into the lateral ventricle had long-lasting effects, with enhanced exon 7 inclusion observed between 6 months and 1 year (Hua et al. 2010, Rigo et al. 2014). In mouse models of SMA, nusinersen was shown to prolong survival, prevent motor neuron and muscle pathology, and increase muscle strength when injected into the CSF (Hua et al. 2010, Passini et al. 2011, Sahashi et al. 2012). Systemic dosing produced remarkable increases in survival in severe SMA mouse models (Hua et al. 2011, 2015), likely due to the prevention of cardiac, gastrointestinal, and other systemic pathologies known to occur in severe SMA mouse models (Bevan et al. 2010, Heier et al. 2010, Hua et al. 2011). These systemic pathologies are not common features of SMA, so it is not clear at present if systemic treatment is warranted in SMA patients. Interestingly, systemic dosing in neonate mice can rescue motor neuron and neuromuscular junction phenotypes even when the fraction of nusinersen that crosses the immature BBB is effectively neutralized with an ASO that targets nusinersen and antagonizes the action of nusinersen in the CNS (Hua et al. 2015). This observation suggests that motor neuron health can be indirectly affected by nusinersen effects in peripheral tissues, perhaps via a neurotrophic factor(s), in addition to its direct effects when administered to the CNS.
Nusinersen was advanced into clinical development based on the consistent improvements in pathology and muscle function in multiple SMA mouse models. Toxicology and pharmacokinetic studies were performed following administration of nusinersen by lumbar puncture in juvenile cynomolgus monkeys in 3- and 12-month studies, which supported its advancement into clinical trials. The initial phase 1 clinical study was an open-label study in which increasing doses of nusinersen were administered by lumbar puncture to type 2 and type 3 SMA children (Chiriboga et al. 2016). The drug was well tolerated, and encouraging signs of activity were observed. The phase 1 study was followed by two different open-label phase 2 studies, one in the same population as the phase 1 study and a second in infantile-onset SMA infants (consistent with type 1 SMA). A loading regime was utilized in these studies to more quickly achieve steady-state drug concentrations in tissues. Results from the phase 2 study in children with type 2/3 SMA again demonstrated no safety or tolerability concerns with multiple doses, feasible intrathecal dosing, and dose-related CSF and plasma pharmacokinetics that were consistent with predictions from nonclinical studies of the extended drug half-life. In addition, the initial observations of dose- and time-dependent improvement in motor function scores were replicated, and improvements utilizing the new end points of the upper limb module and 6-min walk test were also evident (Darras et al. Neurology, In press). In the infantile-onset study, intrathecal-administered nusinersen was well tolerated in these infants, and changes in clinical efficacy on measures of motor function (CHOP-INTEND), incremental motor milestone achievement, muscle electrophysiology, and ventilation-free survival data were promising compared to published natural history cohorts (Finkel et al. 2014a, 2016).
A third phase 2 open-label study was initiated once the safety and tolerability of nusinersen were established in infants and children with SMA (NURTURE, NCT02386553); the study was designed to evaluate the effects of nusinersen in infants genetically diagnosed with SMA (most likely to develop type 1 or type 2 SMA), with treatment initiated before symptom onset. A key objective of the study is to determine if nusinersen can delay disease onset and/or result in a milder form of the disease. Subjects had to receive their first dose of the drug at or before 6 weeks of age and were required to have a baseline ulnar compound muscle action potential (CMAP) amplitude value ≥1 mV. This trial is currently ongoing; however, interim data from the NUTURE study are encouraging and support early treatment.
Based on safety and tolerability of the drug and the lumbar puncture procedure, as well as encouraging clinical activity, two phase 3 clinical studies were initiated, one in infantile-onset SMA and the second in childhood-onset SMA (Finkel et al. 2017, Mercuri et al. 2018). In general, the inclusion/exclusion criteria were the same as in the infantile-onset phase 2 study with the exception that enrollment was limited to infants with 2 copies of SMN2 (i.e., most likely to develop type I SMA). A sham procedure control was selected, as it was determined that placebo injections would represent an unfavorable risk/benefit in this patient population. To mitigate the ethical concerns of a controlled study, the study was designed with an interim analysis to be conducted when at least 80 infants had reached 6 months of study duration, using the last available data for each infant in the analysis. In August 2016, the planned interim analysis was conducted, which demonstrated a significantly greater proportion of nusinersen-treated motor milestone responders using part 2 of the Hammersmith Infant Neurological Examination (HINE) versus sham procedures (41% versus 0%; p < 0.001), in addition to a favorable survival benefit. Following the interim analysis, the study was ended early out of ethical consideration for the sham control group. Nusinersen demonstrated a favorable safety profile, with commonly reported adverse events consistent with those expected in infants with SMA (Finkel et al. 2017).
The design of the global phase 3 study in children with later-onset SMA (CHERISH, NCT02292537) was based on a detailed analysis of the data from the phase 1 and phase 2 studies in children. This global phase 3 randomized, double-blind, sham-procedure-controlled study to assess the clinical efficacy, safety, tolerability, and pharmacokinetics of nusinersen in patients with later-onset SMA also contained a prespecified interim analysis to be conducted when all subjects had completed their 6-month or later evaluation and more than 50 subjects had completed the 15-month assessment (which was the end of study assessment), using the last data available for each subject. As was the case for ENDEAR, the CHERISH interim analysis was also successful, and in November 2016 it was announced that results from the prespecified analysis of the primary end point demonstrated a clinically meaningful difference (Mercuri et al. 2018).
Based on the striking results from the phase 3 study in infantile-onset SMA (ENDEAR), in conjunction with the overwhelming evidence of a highly favorable benefit-risk profile in multiple SMA populations, the US Food and Drug Administration (FDA) approved nusinersen for the treatment of pediatric and adult patients with all types of SMA approximately three months after the initial filing, which is among the fastest approvals by the FDA. Nusinersen was approved almost five years to the day following the first patient being exposed to the drug, which reflects a thoughtful and well-executed development plan (Xu et al. 2017). Market authorization for nusinersen has been approved in more than 40 countries throughout the world. Nusinersin is the first approved drug for the treatment of SMA and the first intrathecally dosed antisense drug. Nusinersen demonstrated that neurodegenerative diseases are treatable and that it is possible to improve disease symptoms rather than only slow down disease progression. As expected, results from multiple clinical studies with nusinersen demonstrate greater efficacy if treatment is initiated early in the disease process and support newborn screening for this disease.
Amyotrophic Lateral Sclerosis
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease affecting motor neurons in the cortex and spinal cord. The loss of motor neurons results in progressive muscle weakness and ultimately death within 3 to 5 years of symptom onset, usually due to respiratory failure. Riluzole and edaravone have been approved for the treatment of ALS, based on modest increases in survival, with other therapies focused on managing patient symptoms. For most patients, the cause of their disease is currently unknown, although approximately 10–15% of ALS cases are familial (Hardiman et al. 2017). There are over 20 known genes that have been documented to cause ALS, with dominant mutations in chromosome 9 open reading frame number 72 (C9orf72) and superoxide dismutase 1 (SOD1) accounting for approximately 40% of cases (Hardiman et al. 2017, Volk et al. 2018). There is not a common cellular pathway affected by the mutations, suggesting that dysfunction in several pathways may lead to clinical ALS. The familial forms of ALS are an attractive opportunity for antisense therapeutics, as target risk is minimized because therapies aimed at lowering the mutant protein or RNA are clearly on target. Currently, there are antisense therapies in development targeting SOD1 and C9orf72 transcripts.
SOD1
The first gene to be identified that causes familial ALS was SOD1, which accounts for approximately 20% of familial ALS cases (Rosen et al. 1993). More than 100 different mutations in the SOD1 gene have been associated with ALS (Andersen & Al-Chalabi 2011). The exact mechanism by which mutations in SOD1 cause disease is still not understood, but most data suggest that it is due to a toxic gain of function that may impact neuronal function and survival (Bruijn et al. 2004). Numerous preclinical studies demonstrated that decreasing the expression of the toxic protein increases survival in transgenic mice and rats (Bruijn & Cleveland 1996, Foust et al. 2013, Raoul et al. 2005, Smith et al. 2006). Based on data showing that reducing SOD1 expression in rodent models of ALS delayed disease onset and increased survival, an ASO was developed that targets SOD1 mRNA via an RNase H1 mechanism of action (Smith et al. 2006). ISIS 333611, a 2′-MOE gapmer, produced a dose-dependent lowering of SOD1 mRNA and protein in transgenic rats expressing human mutant SOD1G93A RNA (Smith et al. 2006). The decrease in SOD1 expression was well tolerated and delayed disease onset in the transgenic rats. Based on the encouraging preclinical data, ISIS 333611 was advanced into clinical development to become the first experimental antisense drug administered intrathecally into patients for the treatment of a neurodegenerative disease (Miller et al. 2013). When this trial was started, it was anticipated that the antisense drug would be given as a continuous infusion using implantable pumps. Therefore, to model a single dose, subjects were infused intrathecally with increasing doses of the drug over 12 h using external pumps. No drug-related safety issues were identified in the study. Although the data were encouraging, advances in antisense therapies for other neurological diseases, including SMA, identified more effective designs for CNS-administered antisense drugs that improved potency and tolerability. In addition, preclinical work in mice and nonhuman primates demonstrated that the delivery of antisense drugs by intrathecal bolus injection was more effective than continuous infusion (Rigo et al. 2014). Therefore, additional work on ISIS 333611 was halted, and a new drug discovery project was initiated to identify a more potent backup drug.
BIIB067 (IONIS-SOD1Rx) was identified following a screen of 2′-MOE gapmer oligonucleotides in cell culture and hSOD1G93A transgenic mice and rats (McCampbell et al. 2018). BIIB067, which targets a different region on the SOD1 premRNA than ISIS 333611, was approximately six-fold more potent than ISIS 333611 in cultured cells and three- to four-fold more potent for inhibition of SOD1 mRNA expression in transgenic rodents. BIIB067 was also more effective in delaying disease progression and increasing survival in transgenic rodents compared to ISIS 333611. Interestingly, administration of BIIB067 to symptomatic hSOD1G93A mice reversed CMAP decrements in the mice, suggesting a partial recovery of motor neuron function (McCampbell et al. 2018). Intrathecal injection of BIIB067 into cynomolgus monkeys, which has a single mismatch to cynomolgus monkey SOD1, produced a dose-dependent reduction of SOD1 mRNA and protein in CNS tissues and CSF. A phase 1b/2a single- and multiple-ascending-dose clinical study of BIIB067 in familial ALS with a mutation in SOD1 is currently in progress (NCT02623699).
C9orf72
A hexanucleotide expansion (GGGGCC) in the noncoding region of the C9orf72 gene was recently identified as the most common cause of familial ALS and frontotemporal dementia, accounting for 40% of familial cases of ALS (DeJesus-Hernandez et al. 2011, Majounie et al. 2012, Renton et al. 2011). The C9orf72 gene codes for a protein with homology to DENN proteins, likely functioning as a guanine nucleotide exchange factor for small GTPases involved in membrane trafficking (Farg et al. 2017, Levine et al. 2013). There are two transcription initiation sites for the C9orf72 gene, resulting in alternative first exons (exons 1A and 1B) (DeJesus-Hernandez et al. 2011). The hexanucleotide expansion occurs within the first intron corresponding to transcripts that initiate with the 1A exon (transcription variant 2). The number of hexanucleotide repeats is variable and can range from 2 to 24 hexanucleotide repeats in unaffected controls and from the mid-thirties to several thousand repeats in cells from C9orf72 ALS patient samples (Van Mossevelde et al. 2017). At least three different potential mechanisms have been described for how the hexanucleotide expansion causes neurodegeneration (Balendra & Isaacs 2018, DeJesus-Hernandez et al. 2011, Renton et al. 2011). Because the repeat occurs in a noncoding region of the transcribed RNA, one proposed mechanism is a toxic gain of function for the RNA in which the repetitive sequence binds to and depletes RNA-binding proteins (Donnelly et al. 2013, Lagier-Tourenne et al. 2013, Sareen et al. 2013), similar to the mechanism of myotonic dystrophy (Cooper et al. 2009, Wojciechowska & Krzyzosiak 2011). A second proposed mechanism is that the toxic dipeptides are translated from the repeat sequence via an unconventional repeat-associated nonATG (RAN) translation (Ash et al. 2013, Gendron et al. 2013, Kwon et al. 2014), which was first described for spinocerebellar ataxia (SCA)8 and myotonic dystrophy type 1 (Zu et al. 2011). Five different poly-dipeptides may be translated from the sense and antisense transcripts, with poly(glycine-arginine) and poly(proline-arginine) demonstrating the greatest degree of toxicity to cultured cells and in fly models when over expressed (Kwon et al. 2014, Mizielinska et al. 2014). The third proposed mechanism is decreased production of the C9orf72 protein. As mentioned above, there are two transcription initiation sites: one upstream from the hexanucleotide repeat producing the 1A exon that produces transcript variant 2 and one originating at the 1B exon producing transcript variant 1. The repeat expansion decreases transcription initiation from the downstream initiation site (transcript variant 1), resulting in approximately a 50% decrease in total C9orf72 protein (Balendra & Isaacs 2018, DeJesus-Hernandez et al. 2011). Deletion of the C9orf72 gene in mice does not result in detectable neurodegeneration (Jiang et al. 2016, O’Rourke et al. 2016); therefore, the loss of C9orf72 protein by itself does not explain the mechanism of neurodegeneration. However, it cannot be ruled out that the loss of C9orf72 protein may exacerbate the toxicity of the hexanucleotide-expanded RNA or poly-dipeptides.
RNase H1–dependent ASOs have been shown to reduce RNA foci and RAN dipeptides in both cellular and mouse models of C9orf72 ALS (Donnelly et al. 2013, Gendron et al. 2017, Jiang et al. 2016, Lagier-Tourenne et al. 2013, Sareen et al. 2013). Given the theoretical concerns about the reduction of C9orf72 protein, ASOs were evaluated for their effects on transcript variants 1 and 2. ASOs designed to bind to the C9orf72 premRNA 5′ to the repeat selectively promoted the loss of the repeat-containing transcript variant 2 without affecting the levels of the transcript variant 1 and total C9orf72 protein (Lagier-Tourenne et al. 2013). In contrast, ASOs designed to bind 3′ to the repeat reduced both transcripts and total C9orf72 protein. Reduction of the repeat-containing transcripts in induced pluripotent stem cells decreased RNA foci, decreased dipeptide production, reversed transcriptional changes, and enhanced sensitivity to glutamate toxicity (Donnelly et al. 2013, Lagier-Tourenne et al. 2013, Sareen et al. 2013). These results were extended to mouse models expressing the human C9orf72 gene with the hexanucleotide expansion (Jiang et al. 2016). Mice expressing the human C9orf72 gene with 450 GGGGCC repeats have both RNA foci and aggregates of RAN dipeptide repeats in cortical and hippocampal neural cells but no loss of motor neurons or motor deficits. The mice do manifest some cognitive and behavioral deficits such as in spatial learning, the marble-burying test, and the elevated plus maze (Jiang et al. 2016). Treatment with an ASO that selectively reduced the repeat-containing transcript variant 2 decreased the number of neural cells with RNA foci and dipeptide aggregates and attenuated the cognitive function and anxiety dysfunction observed in the mice. Based on these findings, an ASO selectively targeting the repeat-containing C9orf72 transcript has started clinical trials in ALS patients documented to have the C9orf72 hexanucleotide expansion (NCT03626012).
Ataxin 2
Ataxin 2 is an RNA-binding protein found in RNA-containing granules, including stress granules, P bodies, and neuronal RNP granules. Ataxin 2 has a polyglutamine tract encoded by up to 23 cytosine-adenine-guanine (CAG) repeats. CAG repeat lengths of 34 or larger are associated with the severe neurodegenerative disease spinocerebellar ataxia 2 (SCA2) (Scoles & Pulst 2018). Intermediate-size expansions between 27 and 32 CAG repeats are associated with an increased risk of ALS (Elden et al. 2010, Gispert et al. 2012). Decreasing the expression of ataxin 2 using genetic methods or ASOs reduced neurodegeneration introduced in yeast, flies, and mice by increased expression of mutated TDP-43 (Becker et al. 2017, Elden et al. 2010). As TDP-43 protein mislocalization to cytoplasmic aggregates is a common pathological feature for both sporadic and familial forms of ALS (Neumann et al. 2006), reduction of ataxin 2 by ASOs may be broadly beneficial as a therapy for most forms of ALS. In addition, SCA2 patients should also benefit from an ataxin 2 antisense drug (Scoles et al. 2017). Based on these findings, efforts are underway to identify and develop an antisense drug targeting ataxin 2 for ALS and SCA2.
Huntington’s Disease
HD is an autosomal-dominant neurodegenerative disorder that commonly affects adults in mid-life, although there are juvenile variants of the disease. The disease is characterized by progressive motor decline, cognitive decline, and psychiatric symptoms, accompanied by brain atrophy. The disease is also caused by a CAG repeat expansion in the first exon of the huntingtin (HTT) gene (MacDonald et al. 1993). The mechanisms by which the CAG repeat expansion causes HD have been extensively studied but are still not well understood, although most published data suggest that the disease is caused by a toxic gain of function for the huntingtin protein (Zuccato et al. 2010).
The identification of therapies that reduce the expression of the huntingtin protein (huntingtin lowering) has been an active area of research for the past 15 years. Most strategies focus on the reduction of huntingtin RNA and the subsequent decrease in the production of huntingtin protein using viral-delivered small hairpin RNAs or microRNAs, synthetic small interfering RNAs (siRNAs), or single-stranded ASOs that work through the RNase H1 mechanism (Boudreau et al. 2009, Carroll et al. 2011, DiFiglia et al. 2007, Evers et al. 2018, Grondin et al. 2015, Harper et al. 2005, Hu et al. 2012, Keiser et al. 2016, Kordasiewicz et al. 2012, McBride et al. 2011, Ostergaard et al. 2013, Pfister et al. 2009, Southwell et al. 2018, Yu et al. 2012). In addition, synthetic ASOs targeting the CAG repeats to selectively block translation of the mutant huntingtin transcript have been described (Evers et al. 2011, Gagnon et al. 2010, Hu et al. 2009). All approaches have been documented to work in cell culture assays, and most have been shown to produce benefits in various mouse models of HD, with a 40–50% reduction of mutant huntingtin being sufficient for therapeutic benefit. Short-term (2-week infusion) administration of a 2′-MOE gapmer ASO targeting huntingtin reduced huntingtin protein for up to 4 months following the termination of dosing and produced a motor function benefit for more than 9 months, suggesting that huntingtin does not need to be continually suppressed (Kordasiewicz et al. 2012).
In most cases, the siRNAs or viral-delivered siRNAs/microRNAs were administered into the parenchyma of the striatum and were shown to knock down the huntingtin RNA in tissue adjacent to the site of injection, with limited effects in more distal tissue (DiFiglia et al. 2007, Evers et al. 2018, Grondin et al. 2015, McBride et al. 2011). Single-stranded ASOs delivered into the CSF diffuse more broadly throughout the brain, with cortical and hippocampal regions showing the best response, although some activity is observed in deeper brain structures such as the striatum. Studies in mouse models of HD demonstrate that the selective removal of mutant huntingtin from cortical neurons partially reversed motor and behavioral deficits; selective removal from striatal neurons had minimal benefits, and removal from both cortical and striatal neurons ameliorated all deficits (Estrada-Sanchez et al. 2015, Wang et al. 2014).
The most advanced strategy for reducing mutant huntingtin expression is the use of single-stranded ASOs that work through the RNase H1 mechanism; three different drugs are currently in clinical trials (NCT02519036, NCT03225846, and NCT03225833). Two different therapeutic strategies are being pursued: nonallele selective, which will result in reductions in both wild-type and mutant huntingtin proteins (Kordasiewicz et al. 2012, Southwell et al. 2018), and allele-selective, which targets polymorphisms that are linked to the expanded CAG repeat (Carroll et al. 2011; Ostergaard et al. 2013; Skotte et al. 2014; Southwell et al. 2014, 2018). The advantage of the nonallele-selective approach is that the therapeutic agent could be used in all HD patients, while the allele-selective approach would only treat a subset of HD patients, with a drug directed to the more prevalent polymorphism treating approximately 40% of HD patients of European heritage (Kay et al. 2015). Thus, several allele-selective drugs would be needed to treat the entire HD population. The main concern with total huntingtin–lowering approaches is that huntingtin may be required for normal neural function. Complete deletion of huntingtin is embryonically lethal in mice (Duyao et al. 1995, Zeitlin et al. 1995). Inducible knockout of huntingtin in adult mice did not produce neurodegeneration or behavioral effects but did result in acute pancreatitis (Wang et al. 2016). A second study performed conditional knockout of the huntingtin gene in adult hemizygous mice (e.g., they had 50% of normal huntingtin throughout development), which resulted in behavioral deficits, reactive gliosis, and bilateral thalamic calcification (Dietrich et al. 2017). Additional studies are needed to reconcile these results. ASOs targeting huntingtin produce predictable dose-dependent knockdown of the target in brain tissue, achieving a maximal reduction of approximately 80%. In addition, several studies using a variety of huntingtin-lowering strategies in rodents and nonhuman primates failed to identify a toxicity related to the lowering of total huntingtin protein (Boudreau et al. 2009, DiFiglia et al. 2007, Kordasiewicz et al. 2012, McBride et al. 2011).
ISIS 443139 is a 5–10-5 2′-MOE gapmer, with both phosphodiester and phosphorothioate linkages in the backbone, that targets an exonic sequence in the huntingtin RNA, and it has advanced into clinical studies. The initial clinical study was a randomized, double-blind, multiple-ascending-dose study in early manifest HD patients (Tabrizi et al., manuscript submitted). ISIS 443139 (RG6042) was delivered by bolus intrathecal injection every month for 3 months (4 injections) to 46 subjects, followed by a 4-month untreated follow-up period. The primary objectives of the study were safety and tolerability, with the secondary objective being characterization of the CSF pharmacokinetics of ISIS 443139. Five dose cohorts were investigated: 10 mg, 30 mg, 60 mg, 90 mg, and 120 mg. The drug was well tolerated at all dose levels, with adverse effects being generally mild and not related to the study drug. Drug exposure in the CSF was dose-related and appeared to plateau at the higher doses. A dose-dependent reduction in mutant huntingtin protein was detected in the CSF, documenting target engagement, which also appeared to plateau at the higher doses. Based on the preclinical modeling, the mean 40% reduction in CSF mutant huntingtin is predicted to produce a mean 60% reduction of mutant huntingtin protein in cortical tissue. As expected, based on the short-term nature of the study and limited number of subjects in each dose cohort (4–12), there were no group-wise differences in clinical outcomes. Based on these encouraging results, an open-label extension study was started in which subjects participating in the phase 1 study are eligible to participate (NCT03342053). In addition, a pivotal phase 3 study is being planned.
Wave Life Sciences has started clinical studies of two allele-selective antisense drugs, which target two different polymorphisms linked to the CAG expansion. There is limited publicly available information on the drugs, including the chemistry and design of the ASOs.
Alzheimer’s Disease
Alzheimer’s disease is the most prevalent of the neurodegenerative diseases, with over 5 million people affected in the United States and more than 20 million worldwide. The disease is characterized by progressive cognitive decline, leading to complete dependency and ultimately death. Alzheimer’s disease has characteristic neuropathology with diffuse extracellular amyloid plaques containing β-amyloid and intracellular neurofibrillary tangles largely composed of the tau protein. With newer in vivo imaging techniques and CSF measurements of β-amyloid and tau in a genetically predisposed patient population, the temporal relationship between these two pathologies has become better defined (Fleisher et al. 2015, McDade & Bateman 2018, McDade et al. 2018, Quiroz et al. 2018). The results show that cortical β-amyloid deposits occur 15 years or more before the clinical onset of symptoms, while increases in CSF β-amyloid peptides occur 5 or more years prior to β-amyloid deposition. Neurofibrillary tangles of tau protein are temporally more closely aligned to the onset of clinical symptoms. Targeting the production, disaggregation, and clearance of β-amyloid has been an active area of drug discovery and research over the past 20 years but has yielded limited success (Murphy 2018). Possible explanations include that treatments are initiated too late, trial designs are inadequate, the wrong form of β-amyloid is being targeted, or the β-amyloid hypothesis is incorrect. Despite the limited success to date, β-amyloid-targeting therapies remain an active area of investigation.
Tau is a microtubule-associated protein encoded by the Microtubule-Associated Protein Tau (MAPT) gene located on chromosome 17 and is expressed primarily in neurons. Intracellular tau can be hyperphosphorylated and can form toxic oligomers and aggregates, which are visualized as neurofibrillary tangles in certain neurodegenerative diseases and are referred to as tauopathies. The most common tauopathy is Alzheimer’s disease, but other examples include frontotemporal dementias due to mutations in MAPT, progressive supranuclear palsy, corticobasal degeneration, and Pick’s disease. Interestingly, even though tau is an intracellular protein, it has been shown that oligomerized tau can spread from cell to cell, causing tau aggregation in the newly infected cell (de Calignon et al. 2012, Kaufman et al. 2016, Wu et al. 2016). Antibody-based therapies to reduce the cell-to-cell spread of tau are currently in clinical trials.
ASOs represent another strategy to reduce the intracellular load of tau, reducing the formation and spread of intracellular neurofibrillary tangles. Similar to the experience with huntingtin-targeting ASOs, a 2′-MOE-modified gapmer ASO targeting tau RNA produced a dose-dependent reduction in tau expression, with effects lasting longer than four months (DeVos et al. 2013). Reducing tau expression was well tolerated in normal mice and protected against picrotoxin-induced seizures. These studies were extended to a mouse model of human tauopathy. Treatment with a tau-targeting RNase H ASO prevented and reversed tau aggregates in the PS19 transgenic mice that express the human P301S mutation (DeVos et al. 2017). The tau ASO also reduced tau seeding activity in brain lysates from treated mice. A single administration of the tau ASO prevented hippocampal volume and neuron loss and prolonged survival in the mice as well as reversed nesting deficits. Intrathecal dosing of cynomolgus monkeys with a tau ASO produced dose- and time-dependent reductions of tau mRNA and protein in the spinal cord, cortex, hippocampus, and CSF (DeVos et al. 2017). The reduction of tau in the CSF was important to observe as it could translate to a pharmacodynamic biomarker in clinical studies. After the completion of toxicology studies, a phase 1 clinical trial of a MAPT-targeting ASO has been initiated in mild Alzheimer’s disease patients (NCT03186989).
CONCLUSIONS
The examples discussed above highlight the broad potential of antisense therapies for neurodegenerative diseases and other neurological diseases. It is still early in the application of this technology, so caution is warranted; however, there is strong foundational science driving the development of ASO drugs for the treatment of neurological diseases. In addition to the projects discussed above, there are active antisense drug–discovery programs underway for additional diseases, including Parkinson’s disease, various SCAs, prion disease, ALS, Alzheimer’s disease, Angelman syndrome, and infantile seizure disorders. It is unlikely that all these projects will yield drugs, but if a few are successful, antisense drugs will be an important treatment option available to neurologists.
Currently, ASOs are dosed by an invasive procedure, e.g., lumbar puncture; therefore, applications of ASO technology have been focused on the more severe, rapidly progressive diseases. Although lumbar puncture procedures are commonly done in the clinical setting, the procedure is more technically demanding than other drug administration routes, and it will put additional pressure on the health care system to deliver these novel therapeutics. Developing more convenient methods of delivering antisense drugs, such as the use of subcutaneously accessed intrathecal catheters, should broaden the use of ASOs for neurological diseases (Strauss et al. 2018). In the longer term, advances in ASO chemistry to enhance potency, conjugation of targeting ligands to the ASO, and novel formulations are predicted to result in more effective antisense drugs. Finally, the holy grail will be to identify methods to safely and efficiently deliver ASOs across the BBB, allowing systemic dosing of antisense drugs.
In summary, ASO technology has made remarkable advances in the past 10 years, with a drug approval and multiple potential therapeutic agents for neurodegenerative diseases in development. The profound clinical benefit of nusinersen in SMA patients validates the platform. Further validation should occur over the next few years based on the large pipeline of drugs in clinical development. We may be at the cusp of finally addressing the therapeutic needs of patients who suffer from these devastating neurodegenerative diseases.
ACKNOWLEDGMENTS
We thank Yimin Hua, Richard Smith, Timothy Miller, Holly Kordasiewicz, and Frank Rigo for discussions and contributions to the work cited in this review.
DISCLOSURE STATEMENT
C.F.B. is an employee of Ionis Pharmaceuticals Inc. and receives salary and stock options from the company. D.W.C. is a consultant for Ionis Pharmaceuticals. A.R.K., through his employer, Cold Spring Harbor Laboratory, receives royalty income from Ionis Pharmaceuticals tied to sales of the spinal muscular atrophy drug Spinraza, which is marketed and sold by Biogen. A.R.K. is a consultant, advisor, and director of Stoke Therapeutics and receives compensation and stock from the company.
LITERATURE CITED
- Andersen PM, Al-Chalabi A. 2011. Clinical genetics of amyotrophic lateral sclerosis: What do we really know? Nat. Rev. Neurol. 7:603–15 [DOI] [PubMed] [Google Scholar]
- Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin WL, et al. 2013. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77:639–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balendra R, Isaacs AM. 2018. C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat. Rev. Neurol. 14:544–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks WA, Farr SA, Butt W, Kumar VB, Franko MW, Morley JE. 2001. Delivery across the blood-brain barrier of antisense directed against amyloid β: reversal of learning and memory deficits in mice overexpressing amyloid precursor protein. J. Pharmacol. Exp. Ther. 297:1113–21 [PubMed] [Google Scholar]
- Becker LA, Huang B, Bieri G, Ma R, Knowles DA, et al. 2017. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 544:367–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CF, Baker BF, Pham N, Swayze EE, Geary RS. 2017. Pharmacology of antisense drugs. Annu. Rev. Pharmacol. Toxicol. 57:81–105 [DOI] [PubMed] [Google Scholar]
- Bennett CF, Swayze EE. 2010. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 50:259–93 [DOI] [PubMed] [Google Scholar]
- Bevan AK, Hutchinson KR, Foust KD, Braun L, McGovern VL, et al. 2010. Early heart failure in the SMNΔ7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery. Hum. Mol. Genet. 19:3895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau RL, McBride JL, Martins I, Shen S, Xing Y, et al. 2009. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol. Ther. 17:1053–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruijn LI, Cleveland DW. 1996. Mechanisms of selective motor neuron death in ALS: insights from transgenic mouse models of motor neuron disease. Neuropathol. Appl. Neurobiol. 22:373–87 [DOI] [PubMed] [Google Scholar]
- Bruijn LI, Miller TM, Cleveland DW. 2004. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu. Rev. Neurosci. 27:723–49 [DOI] [PubMed] [Google Scholar]
- Butler M, Crooke R, Graham M, Lougheed M, Murray S, et al. 2000. Distribution of phosphorothioate oligodeoxynucleotides in class A scavenger receptor knockout mice. J. Pharm. Exp. Ther. 292:489–96 [PubMed] [Google Scholar]
- Carroll JB, Warby SC, Southwell AL, Doty CN, Greenlee S, et al. 2011. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene/allele-specific silencing of mutant huntingtin. Mol. Ther. 19:2178–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Hastings ML, Calarco JA, de Stanchina E, Krainer AR. 2006. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 78:63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR. 2002. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 30:377–84 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR. 2003. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat. Structural Biol. 10:120–25 [DOI] [PubMed] [Google Scholar]
- Chiriboga CA, Swoboda KJ, Darras BT, Iannaccone ST, Montes J, et al. 2016. Results from a phase 1 study of nusinersen (ISIS-SMNRx) in children with spinal muscular atrophy. Neurology 86:890–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobben JM, Lemmink HH, Snoeck I, Barth PA, van der Lee JH, de Visser M. 2008. Survival in SMA type I: a prospective analysis of 34 consecutive cases. Neuromuscul. Disord. 18:541–44 [DOI] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. 2009. RNA and disease. Cell 136:777–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke ST. 2017. Molecular mechanisms of antisense oligonucleotides. Nucleic Acid Ther. 27:70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke ST, Witztum JL, Bennett CF, Baker BF. 2018. RNA-targeted therapeutics. Cell Metab. 27:714–39 [DOI] [PubMed] [Google Scholar]
- Darras BT Chiriboga CA, Iannaccone ST, Swoboda KJ, Montes J, Mignon L, Xia S, Bennett CF, Bishop KM, Shefner JM, Green AM Sun S, Bhan I, Gheuens S, Schneider E, Farwell W and De Vivo DC (2019) Nusinersen in later-onset spinal muscular atrophy: long-term results from the phase 1/2 studies. Neurology, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Calignon A, Polydoro M, Suarez-Calvet M, William C, Adamowicz DH, et al. 2012. Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron 73:685–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, et al. 2011. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72:245–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVos SL, Goncharoff DK, Chen G, Kebodeaux CS, Yamada K, et al. 2013. Antisense reduction of tau in adult mice protects against seizures. J. Neurosci. 33:12887–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVos SL, Miller RL, Schoch KM, Holmes BB, Kebodeaux CS, et al. 2017. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci. Transl. Med. 9:eaag0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich P, Johnson IM, Alli S, Dragatsis I. 2017. Elimination of huntingtin in the adult mouse leads to progressive behavioral deficits, bilateral thalamic calcification, and altered brain iron homeostasis. PLOS Genet. 13:e1006846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, et al. 2007. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. PNAS 104:17204–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, et al. 2013. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80:415–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyao MP, Auerbach AB, Ryan A, Persichetti F, Barnes GT, et al. 1995. Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science 269:407–10 [DOI] [PubMed] [Google Scholar]
- Eisele YS, Monteiro C, Fearns C, Encalada SE, Wiseman RL, et al. 2015. Targeting protein aggregation for the treatment of degenerative diseases. Nat. Rev. Drug Discov. 14:759–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, et al. 2010. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466:1069–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, et al. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3:87–98 [DOI] [PubMed] [Google Scholar]
- Estrada-Sanchez AM, Burroughs CL, Cavaliere S, Barton SJ, Chen S, et al. 2015. Cortical efferents lacking mutant huntingtin improve striatal neuronal activity and behavior in a conditional mouse model of Huntington’s disease. J. Neurosci. 35:4440–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers MM, Miniarikova J, Juhas S, Valles A, Bohuslavova B, et al. 2018. AAV5-miHTT gene therapy demonstrates broad distribution and strong human mutant huntingtin lowering in a Huntington’s disease minipig model. Mol. Ther. 26:2163–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers MM, Pepers BA, van Deutekom JC, Mulders SA, den Dunnen JT, et al. 2011. Targeting several CAG expansion diseases by a single antisense oligonucleotide. PLOS ONE 6:e24308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RAK, et al. 2017. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum. Mol. Genet. 26:4093–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, et al. 2016. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet 388:3017–26 [DOI] [PubMed] [Google Scholar]
- Finkel RS, McDermott MP, Kaufmann P, Darras BT, Chung WK, et al. 2014a. Observational study of spinal muscular atrophy type I and implications for clinical trials. Neurology 83:810–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel RS, McDermott MP, Kaufmann P, Darras BT, Chung WK, et al. 2014b. Observational study of spinal muscular atrophy type 1 and implications for clinical trials. Neurology 83:974–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel RS, Mercuri E, Darras BT, Connolly AM, Kuntz NL, et al. 2017. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N. Engl. J. Med. 377:1723–32 [DOI] [PubMed] [Google Scholar]
- Fleisher AS, Chen K, Quiroz YT, Jakimovich LJ, Gutierrez Gomez M, et al. 2015. Associations between biomarkers and age in the presenilin 1 E280A autosomal dominant Alzheimer disease kindred: a cross-sectional study. JAMA Neurol. 72:316–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust KD, Salazar DL, Likhite S, Ferraiuolo L, Ditsworth D, et al. 2013. Therapeutic AAV9-mediated suppression of mutant SOD1 slows disease progression and extends survival in models of inherited ALS. Mol. Ther. 21:2148–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Pendergraff HM, Deleavey GF, Swayze EE, Potier P, et al. 2010. Allele-selective inhibition of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochemistry 49:10166–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary RS, Norris D, Yu R, Bennett CF. 2015. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 87:46–51 [DOI] [PubMed] [Google Scholar]
- Geary RS, Watanabe TA, Truong L, Freier S, Lesnik EA, et al. 2001. Pharmacokinetic properties of 2′-O-(2-methoxyethyl)-modified oligonucleotide analogs in rats. J. Pharmacol. Exp. Ther. 296:890–97 [PubMed] [Google Scholar]
- Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, et al. 2013. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 126:829–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Chew J, Stankowski JN, Hayes LR, Zhang YJ, et al. 2017. Poly(GP) proteins are a useful pharmacodynamic marker for C9ORF72-associated amyotrophic lateral sclerosis. Sci. Transl. Med. 9:eaai7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gispert S, Kurz A, Waibel S, Bauer P, Liepelt I, et al. 2012. The modulation of amyotrophic lateral sclerosis risk by ataxin-2 intermediate polyglutamine expansions is a specific effect. Neurobiol. Dis. 45:356–61 [DOI] [PubMed] [Google Scholar]
- Grondin R, Ge P, Chen Q, Sutherland JE, Zhang Z, et al. 2015. Onset time and durability of huntingtin suppression in rhesus putamen after direct infusion of antihuntingtin siRNA. Mol. Ther. Nucleic Acids 4:e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardiman O, Al-Chalabi A, Chio A, Corr EM, Logroscino G, et al. 2017. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers 3:17071. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, et al. 2005. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. PNAS 102:5820–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heier CR, Satta R, Lutz C, DiDonato CJ. 2010. Arrhythmia and cardiac defects are a feature of spinal muscular atrophy model mice. Hum. Mol. Genet. 19:3906–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Liu J, Yu D, Chu Y, Corey DR. 2012. Mechanism of allele-selective inhibition of huntingtin expression by duplex RNAs that target CAG repeats: function through the RNAi pathway. Nucleic Acids Res. 40:11270–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Matsui M, Gagnon KT, Schwartz JC, Gabillet S, et al. 2009. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol. 27:478–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Bennett CF, Baker BF, Vickers TA, Krainer AR. 2006. Enhancement of alternative exon 7 inclusion in SMN2 by antisense 2’-MOE oligonucleotides targeting the exon. PLOS In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Liu YH, Sahashi K, Rigo F, Bennett CF, Krainer AR. 2015. Motor neuron cell-nonautonomous rescue of spinal muscular atrophy phenotypes in mild and severe transgenic mouse models. Genes Dev. 29:288–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Sahashi K, Hung G, Rigo F, Passini MA, et al. 2010. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 24:1634–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Sahashi K, Rigo F, Hung G, Bennett CF, Krainer AR. 2012. Correction of RNA splicing with antisense oligonucleotides as therapeutic strategy for a neurodegenerative disease In Chembiomolecular Science: At the Frontier of Chemistry and Biology, ed. Shibasaki M, lino M, Osada H, pp. 301–14. Tokyo: Springer [Google Scholar]
- Hua Y, Sahashi K, Rigo F, Hung G, Horev G, et al. 2011. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 478:123–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR. 2007. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLOS Biol. 5:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR. 2008. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 82:834–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Zhu Q, Gendron TF, Saberi S, McAlonis-Downes M, et al. 2016. Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron 90:535–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima T, Manley JL. 2003. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 34:460–63 [DOI] [PubMed] [Google Scholar]
- Kashima T, Rao N, David CJ, Manley JL. 2007. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum. Mol. Genet. 16:3149–59 [DOI] [PubMed] [Google Scholar]
- Kaufman SK, Sanders DW, Thomas TL, Ruchinskas AJ, Vaquer-Alicea J, et al. 2016. Tau prion strains dictate patterns of cell pathology, progression rate, and regional vulnerability in vivo. Neuron 92:796–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann P, McDermott MP, Darras BT, Finkel R, Kang P, et al. 2011. Observational study of spinal muscular atrophy type 2 and 3: functional outcomes over 1 year. Arch. Neurol. 68:779–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann P, McDermott MP, Darras BT, Finkel RS, Sproule DM, et al. 2012. Prospective cohort study of spinal muscular atrophy types 2 and 3. Neurology 79:1889–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay C, Collins JA, Skotte NH, Southwell AL, Warby SC, et al. 2015. Huntingtin haplotypes provide prioritized target panels for allele-specific silencing in Huntington disease patients of European ancestry. Mol. Ther. 23:1759–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser MS, Kordasiewicz HB, McBride JL. 2016. Gene suppression strategies for dominantly inherited neurodegenerative diseases: lessons from Huntington’s disease and spinocerebellar ataxia. Hum. Mol. Genet. 25:R53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole R, Krainer AR, Altman S. 2012. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 11:125–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, et al. 2012. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron 74:1031–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, et al. 2014. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345:1139–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, et al. 2013. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. PNAS 110:E4530–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, et al. 1995. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80:155–65 [DOI] [PubMed] [Google Scholar]
- Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. 2013. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics 29:499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Shen W, Sun H, Migawa MT, Vickers TA, Crooke ST. 2016. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat. Biotechnol. 34:875–80 [DOI] [PubMed] [Google Scholar]
- Liang XH, Sun H, Shen W, Wang S, Yao J, et al. 2017. Antisense oligonucleotides targeting translation inhibitory elements in 5′ UTRs can selectively increase protein levels. Nucleic Acids Res. 45:9528–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B. 1999. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. PNAS 96:6307–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, et al. 1993. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72:971–83 [DOI] [PubMed] [Google Scholar]
- Majounie E, Renton AE, Mok K, Dopper EG, Waite A, et al. 2012. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 11:323–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JL, Pitzer MR, Boudreau RL, Dufour B, Hobbs T, et al. 2011. Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington’s disease. Mol. Ther. 19:2152–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCampbell A, Cole T, Wegener AJ, Tomassy GS, Setnicka A, et al. 2018. Antisense oligonucleotides extend survival and reverse decrement in muscle response in ALS models. J. Clin. Investig. 128:3558–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade E, Bateman RJ. 2018. Tau positron emission tomography in autosomal dominant Alzheimer disease: small windows, big picture. JAMA Neurol. 75:536–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade E, Wang G, Gordon BA, Hassenstab J, Benzinger TLS, et al. 2018. Longitudinal cognitive and biomarker changes in dominantly inherited Alzheimer disease. Neurology 91:e1295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Tuschl T. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431:343–49 [DOI] [PubMed] [Google Scholar]
- Mercuri E, Darras BT, Chiriboga CA, Day JW, Campbell C, et al. 2018. Nusinersen versus sham control in later-onset spinal muscular atrophy. N. Engl. J. Med. 378:625–35 [DOI] [PubMed] [Google Scholar]
- Miller TM, Pestronk A, David W, Rothstein J, Simpson E, et al. 2013. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol. 12:435–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, et al. 2014. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345:1192–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, et al. 1999. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 8:1177–83 [DOI] [PubMed] [Google Scholar]
- Munoz-Sanjuan I, Bates GP. 2011. The importance of integrating basic and clinical research toward the development of new therapies for Huntington disease. J. Clin. Investig. 121:476–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP. 2018. Amyloid-beta solubility in the treatment of Alzheimer’s disease. N. Engl. J. Med. 378:391–92 [DOI] [PubMed] [Google Scholar]
- Naryshkin NA, Weetall M, Dakka A, Narasimhan J, Zhao X, et al. 2014. Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science 345:688–93 [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, et al. 2006. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130–33 [DOI] [PubMed] [Google Scholar]
- Nomakuchi TT, Rigo F, Aznarez I, Krainer AR. 2016. Antisense oligonucleotide-directed inhibition of nonsense-mediated mRNA decay. Nat. Biotechnol. 34:164–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke JG, Bogdanik L, Yanez A, Lall D, Wolf AJ, et al. 2016. C9orf72 is required for proper macrophage and microglial function in mice. Science 351:1324–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskoui M, Kaufmann P. 2008. Spinal muscular atrophy. Neurotherapeutics 5:499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard ME, Southwell AL, Kordasiewicz H, Watt AT, Skotte NH, et al. 2013. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 41:9634–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacino J, Swalley SE, Song C, Cheung AK, Shu L, et al. 2015. SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat. Chem. Biol. 11:511–17 [DOI] [PubMed] [Google Scholar]
- Passini MA, Bu J, Richards AM, Kinnecom C, Sardi SP, et al. 2011. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 3:72ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister EL, Kennington L, Straubhaar J, Wagh S, Liu W, et al. 2009. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington’s disease patients. Curr. Biol. 19:774–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior TW, Krainer AR, Hua Y, Swoboda KJ, Snyder PC, et al. 2009. A positive modifier of spinal muscular atrophy in the SMN2 gene. Am. J. Hum. Genet. 85:408–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz YT, Sperling RA, Norton DJ, Baena A, Arboleda-Velasquez JF, et al. 2018. Association between amyloid and tau accumulation in young adults with autosomal dominant Alzheimer disease. JAMA Neurol. 75:548–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoul C, Abbas-Terki T, Bensadoun JC, Guillot S, Haase G, et al. 2005. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat. Med. 11:423–28 [DOI] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, et al. 2011. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72:257–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigo F, Chun SJ, Norris DA, Hung G, Lee S, et al. 2014. Pharmacology of a central nervous system delivered 2′-O-methoxyethyl-modified survival of motor neuron splicing oligonucleotide in mice and nonhuman primates. J. Pharmacol. Exp. Ther. 350:46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochette CF, Gilbert N, Simard LR. 2001. SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum. Genet. 108:255–66 [DOI] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, et al. 1993. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362:59–62 [DOI] [PubMed] [Google Scholar]
- Rudnik-Schoneborn S, Berg C, Zerres K, Betzler C, Grimm T, et al. 2009. Genotype-phenotype studies in infantile spinal muscular atrophy (SMA) type I in Germany: implications for clinical trials and genetic counselling. Clin. Genet. 76:168–78 [DOI] [PubMed] [Google Scholar]
- Sahashi K, Hua Y, Ling KK, Hung G, Rigo F, et al. 2012. TSUNAMI: an antisense method to phenocopy splicing-associated diseases in animals. Genes Dev. 26:1874–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D, O’Rourke JG, Meera P, Muhammad AK, Grant S, et al. 2013. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci. Transl. Med. 5:208ra149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoles DR, Meera P, Schneider MD, Paul S, Dansithong W, et al. 2017. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature 544:362–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoles DR, Pulst SM. 2018. Spinocerebellar ataxia type 2. Adv. Exp. Med. Biol. 1049:175–95 [DOI] [PubMed] [Google Scholar]
- Singh NK, Singh NN, Androphy EJ, Singh RN. 2006. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 26:1333–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotte NH, Southwell AL, Ostergaard ME, Carroll JB, Warby SC, et al. 2014. Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: providing a therapeutic option for all Huntington disease patients. PLOS ONE 9:e107434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RA, Miller TM, Yamanaka K, Monia BP, Condon TP, et al. 2006. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Investig. 116:2290–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell AL, Kordasiewicz HB, Langbehn D, Skotte NH, Parsons MP, et al. 2018. Huntingtin suppression restores cognitive function in a mouse model of Huntington’s disease. Sci. Transl. Med. 10:eear3959. [DOI] [PubMed] [Google Scholar]
- Southwell AL, Skotte NH, Kordasiewicz HB, Ostergaard ME, Watt AT, et al. 2014. In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol. Ther. 22:2093–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss KA, Carson VJ, Brigatti KW, Young M, Robinson DL, et al. 2018. Preliminary safety and tolerability of a novel subcutaneous intrathecal catheter system for repeated outpatient dosing of nusinersen to children and adults with spinal muscular atrophy. J. Pediatr. Orthop. 38:e610–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugarman EA, Nagan N, Zhu H, Akmaev VR, Zhou Z, et al. 2012. Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of >72,400 specimens. Eur. J. Hum. Genet. 20:27–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner CJ. 2007. Molecular mechanisms of spinal muscular atrophy. J. Child Neurol. 22:979–89 [DOI] [PubMed] [Google Scholar]
- Tabrizi SJ, Leavitt BR, Landwehrmeyer B, Wild EJ, Saft C, Blair NF, Craufurd D, Priller J, Rickards H, Rosser A, Kordasiewicz HB, Czech C, Swayze E, Norris DA, Baumann T, Gerlach I, Schobel SA, Paz E, Bennett CF, Lane RM Inhibition of mutant huntingtin expression in a randomized trial of patients with Huntington’s disease. Submitted to New England J. of Med. [Google Scholar]
- Van Mossevelde S, van der Zee J, Cruts M, Van Broeckhoven C. 2017. Relationship between C9orf72 repeat size and clinical phenotype. Curr. Opin. Genet. Dev. 44:117–24 [DOI] [PubMed] [Google Scholar]
- Vickers TA, Wyatt JR, Burckin T, Bennett CF, Freier SM. 2001. Fully modified 2′-MOE oligonucleotides redirect polyadenylation. Nucleic Acids Res. 29:1293–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk AE, Weishaupt JH, Andersen PM, Ludolph AC, Kubisch C. 2018. Current knowledge and recent insights into the genetic basis of amyotrophic lateral sclerosis. Med. Genet. 30:252–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Liu X, Gaertig MA, Li S, Li XJ. 2016. Ablation of huntingtin in adult neurons is nondeleterious but its depletion in young mice causes acute pancreatitis. PNAS 113:3359–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Gray M, Lu XH, Cantle JP, Holley SM, et al. 2014. Neuronal targets for reducing mutant huntingtin expression to ameliorate disease in a mouse model of Huntington’s disease. Nat. Med. 20:536–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth B, Brichta L, Schrank B, Lochmuller H, Blick S, et al. 2006. Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum. Genet. 119:422–28 [DOI] [PubMed] [Google Scholar]
- Wirth B, Herz M, Wetter A, Moskau S, Hahnen E, et al. 1999. Quantitative analysis of survival motor neuron copies: identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype-phenotype correlation, and implications for genetic counseling. Am. J. Hum. Genet. 64:1340–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowska M, Krzyzosiak WJ. 2011. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet. 20:3811–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JW, Hussaini SA, Bastille IM, Rodriguez GA, Mrejeru A, et al. 2016. Neuronal activity enhances tau propagation and tau pathology in vivo. Nat. Neurosci. 19:1085–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Irony I, Bryan WW, Dunn B. 2017. Development of gene therapies—lessons from nusinersen. Gene Ther. 24:527–28 [DOI] [PubMed] [Google Scholar]
- Yu D, Pendergraff H, Liu J, Kordasiewicz HB, Cleveland DW, et al. 2012. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell 150:895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YJ, Watts RJ. 2013. Developing therapeutic antibodies for neurodegenerative disease. Neurotherapeutics 10:459–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. 1995. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat. Genet. 11:155–63 [DOI] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, et al. 2011. Non-ATG-initiated translation directed by microsatellite expansions. PNAS 108:260–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Valenza M, Cattaneo E. 2010. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol. Rev. 90:905–81 [DOI] [PubMed] [Google Scholar]