Abstract
A rapid flow cytometric antimicrobial susceptibility test for bacteria isolated from companion animals – the FASTvet assay, developed by FASTinov®, was evaluated. Bacterial strains isolated from different biological samples of companion animals with infectious diseases in progress were obtained from several veterinary clinical laboratories across the country. A total of 115 strains, comprising 65 Gram-negative and 50 Gram positive isolates, were incubated with 13 antimicrobial drugs (ampicillin, amoxicillin-clavulanic acid, piperacillin-tazobactam, cefpodoxime, imipenem, enrofloxacin, gentamicin, amikacin for Gram-negative; penicillin, cefoxitin, enrofloxacin, vancomycin and ampicillin for Gram-positive) at breakpoint concentrations following CLSI protocol (CLSI Vet 01, 2018) for 1 h and analyzed by flow cytometry. The overall categorical agreement was 95.6% in case of Gram-negative and of 96.7% in Gram-positive isolates when compared to microdilution. FASTvet kits contribute to reduce the turnaround time (2 vs. 24 h) with early determination of the antimicrobial susceptibility profile. The correct and rapid choice of the target antibiotic therapy, will have a positive impact on animal care, contributing for preventing antimicrobial resistance. In conclusion, FASTinov® vet kits showed an excellent performance, both for Gram-negative and Gram-positive isolates encouraging us to enlarge the sample size and planning multicentric studies.
Keywords: antimicrobial resistance, antimicrobial susceptibility test, veterinary, flow cytometry, one health concept
Introduction
The availability of effective antimicrobials has enabled considerable advancements in modern medical and veterinary practice that would have been unachievable without them. Antimicrobial drugs are a precious resource daily used all over the world to treat infections both in humans and animals. Increased antimicrobial resistance due to unnecessary use of antibiotics is a major obstacle to keep these drugs useful for as long as possible. Microbial multidrug resistance is emerging worldwide at an alarming rate and is now recognized as a major health threat (Infectious Diseases Society of America, 2011).
There is a growing concern regarding both public health and animal welfare about the consequences of antimicrobial resistance (AMR) in bacteria from animal sources. The appropriate use of antimicrobial drugs (AMDs) in veterinary medicine is one of the key areas of European Union (EU) policy objectives to combat AMR. Various initiatives have been started by many national, European and international bodies to promote a wise use of AMDs both in human and veterinary medicine. The importance of accurate and rapid laboratorial diagnostics regarding Antimicrobial Susceptibility Testing (AST) as the basis for a rational prescription of antimicrobials to treat an infection has been advocated by numerous international and national guidelines and regulatory agencies. AST is considered as one of the most important factors governing the adequate selection of antimicrobials, both for human and veterinary use. The primary objective is the target selection of the most appropriate antibiotic, the second involves a better control of public health hazards and the third objective is the provision of valid epidemiological AST surveillance data.
The early start of effective antibiotic therapy during the course of an infectious disease reduces the probability of pathogens developing AMR (Deresinski, 2007). Particularly in sepsis cases and bloodstream infections, early and targeted antimicrobial therapy is also crucial to decrease mortality rates (Kumar et al., 2006). Empirical antimicrobial therapy is often started as soon as possible, especially on sepsis management without waiting for AST results (Cohen, 2009; Dellinger et al., 2013). In most of such cases, therapy involves the selection of broad-spectrum antibiotics or a combination of antibiotics which promotes the emergence of resistance (Micek et al., 2010). Early assessment of AST is therefore essential for statement of targeted antimicrobial therapy in order to avoid the escalating rates of resistance and to decrease mortality. At present, in routine clinical microbiology laboratories, ASTs is usually assessed by automated systems such as Vitek2 (BioMérieux), MicroScan Walkaway (Beckman Coulter) or Phoenix (Becton Dickinson) (Mittman et al., 2009; Duggal et al., 2012; Gherardi et al., 2012); or semi-automated method like Sensititre (Thermofisher) (Rene, 2010); in alternative, manual tests such as disc diffusion, broth dilution or Etest can be used (Jenkins and Schuetz, 2012; Van Belkum et al., 2013). While a diversity of tests can be performed, all of them are based upon the study of the growth ability of microorganisms which takes at least 24 h to test results.
Recently, several technologies have been pointed out as more rapid alternatives for ASTs such as whole-genome sequencing, mass spectrometry, fluorescence-activated cell sorting and microarrays particularly useful regarding the detection of known mechanisms of resistance (Van Belkum et al., 2013). However, such approaches use very expensive instruments and procedures, resulting in high analysis costs. In addition, they can only detect previously known mechanisms of resistance. In clinical practice, human or veterinary, faster AST methods are required in order to get an effective treatment, applying the most suitable antibiotic as soon as possible, ideally within the same day. We validated a novel methodology of AST involving the use of flow cytometry (FC), using microbial isolates obtained from companion animals with infectious diseases in progress. This is a disruptive approach for rapid AST. FC allows single cell analysis and is a very fast and accurate method (Andrade et al., 2018). This study was conducted in collaboration with FASTinov®, a spin-off company of Porto University, Portugal from June 2018 to June 2019, the company responsible for FASTvet kit manufacturing.
Materials and Methods
Bacterial Strains
A total of 65 Gram-negative bacilli (40 E. coli, 10 Klebsiella pneumoniae, 5 Proteus mirabilis, 8 Salmonella spp., 2 Pasteurella multocida), 30 Staphylococcus spp. (10 S. aureus, 10 S. epidermidis and 10 S. pseudintermedius) and 20 Enterococcus spp. (14 E. faecalis and 6 E. faecium) isolated from various species of companion animals with infectious diseases in progress and from different biological samples (skin soft tissue, cyst content, infected wounds) were made available by several microbiological veterinary laboratories across the country. In addition, type strains belonging to American Type Culture Collection (ATCC): Escherichia coli 35218, E. coli 25922; P. aeruginosa 27853; Staphylococcus aureus 29213, S. aureus 43300, Enterococcus faecalis 29212, E. faecalis 51299 and E. faecium 700221 were also included in this study.
FASTvet Panels
Two AST panels were previously developed and tested by FASTinov® in a microplate format: the FASTvet gramneg (for Gram-negative bacilli) and the FASTvet grampos (for Staphylococcus and Enterococcus). Antibiotics (see Table 1), at drug breakpoint concentrations adequate to the origin of the bacterial strain (see Table 2), were selected according the CLSI vet protocol from 2018 (CLSI 5th Edition, 2018). A fluorescent probe is also present in each well. Distinct probes, such as nucleic acid staining or membrane depolarization status, previously optimized, were selected in order to reveal distinct cell lesions produced by antimicrobials.
TABLE 1.
Susceptibility phenotypes evaluated using reference methods according CLSI, comparison with flow cytometry assay.
| BMD |
FASTinov versus BMD |
|||||||||
| Antimicrobial | n | MIC intervale μg/ml | S | I | R | mE | ME | VME | CA | |
| Gram negative bacilli | Ampicillin | 65 | 1–≥64 | 15 | – | 50 | 0 | 0 | 0 | 100% |
| Amoxicillin-clavulanic acid | 65 | 0.06/4–≥32/4 | 16 | 1 | 48 | 2 | 3 | 0 | 92.3% | |
| Piperacillin-tazobactam | 65 | ≤0.125/4–≥64/4 | 56 | 1 | 8 | 4 | 0 | 0 | 93.9% | |
| Cefpodoxime | 65 | 0.06–≥64 | 52 | 1 | 12 | 0 | 0 | 0 | 100% | |
| Imipenem | 65 | 0.25–≥64 | 60 | 3 | 2 | 3 | 1 | 0 | 93.9% | |
| Enrofloxacin | 65 | ≤0.06–≥32 | 46 | 2 | 17 | 2 | 1 | 0 | 95.4% | |
| Gentamicin | 65 | ≤0.125–≥32 | 56 | 2 | 7 | 3 | 1 | 0 | 93.9% | |
| Amikacin | 65 | 0.06–64 | 58 | 2 | 5 | 3 | 0 | 0 | 95.4% | |
| Overall | 520 | 359 | 12 | 149 | 17 | 6 | 0 | 95.6% | ||
| Staphylococcus spp. | Penicillin | 30 | ≤0.06–36 | 6 | – | 24 | 0 | 1 | 0 | 96.7% |
| Cefoxitin | 30 | 0.25–8 | 23 | – | 7 | 1 | 1 | 0 | 93.3% | |
| Enrofloxacin | 30 | ≤0.06–≥32 | 28 | – | 2 | 1 | 0 | 0 | 96.7% | |
| Vancomycin | 30 | 0.06–1 | 30 | – | 2 | 0 | 0 | 93.3% | ||
| Enterococcus spp. | Ampicillin | 20 | 1–8 | 16 | 2 | 2 | 0 | 0 | 0 | 100% |
| Vancomycin | 20 | 0.5–8 | 16 | 2 | 2 | 0 | 0 | 0 | 100% | |
| Overall | 180 | 135 | 6 | 39 | 4 | 2 | 0 | 96.7% | ||
S-susceptible; I-intermediate and Resistant. Categorical agreement (CA) and number of errors: mE-minor errors; ME- major errors and VME-very major errors.
TABLE 2.
Antimicrobial concentrations included on FASTvet (breakpoint values according CLSI).
| Antimicrobial | FASTvet panel concentrations (μg/ml) |
|||||
| Phenotype | S | I | R | S (urine) | R (urine) | |
| Gram negative bacilli | Ampicillin | ≤0.25 | 0.5 | >0.5 | ≤8 | >8 |
| Amoxicillin-clavulanic acid | ≤0.25/0.12 | 0.5/0.25 | >0.5/0.25 | ≤8/4 | >8/4 | |
| Piperacillin-tazobactam* | ≤16/4 | 64/4 | >64/4 | |||
| Cefpodoxime | ≤2 | 4 | >4 | |||
| Imipenem* Enterobacterales | ≤1 | 2 | >2 | |||
| Imipenem* Pseudomonas spp. | ≤2 | 4 | >4 | |||
| Enrofloxacin Enterobacterales | ≤0.12 | 0.25 | >0.25 | |||
| Enrofloxacin Pseudomonas spp. | ≤0.5 | 2 | >2 | |||
| Gentamicin | ≤2 | 4 | >4 | |||
| Amikacin | ≤4 | 8 | >8 | |||
| Staphylococcus spp. | Penicillin* | ≤0.12 | >0.12 | |||
| Cefoxitin* | ≤4 | >4 | ||||
| Enrofloxacin | ≤0.5 | 2 | >2 | |||
| Vancomycin* S. aureus | ≤2 | 8 | >8 | |||
| Enterococcus spp. | Vancomycin* S. coagulase neg | ≤4 | 16 | >16 | ||
| Ampicillin* | ≤8 | >8 | ||||
| Vancomycin* | ≤4 | 16 | >16 | |||
*Human breakpoints. According to flow cytometry analysis when the cells show lesions with the lowest concentration tested, the strain is considered S (susceptible); if no lesion is shown with the lower concentration and but only with the second concentration, the strain is considered I (intermediate); if even with the highest concentration no effect is seen, the strain is considered R (resistant). The category of I is not always present.
Inoculation and Incubation of the AST Panels
Pure colonies isolated in solid media, were inoculated in Brain-Heart broth cation adjusted (Sigma) and incubated at 37°C shaking until turbidity (aprox. 1 h), in order to obtain an exponential growth phase culture (Faria-Ramos et al., 2013); after a 2 centrifugation step, a microbial suspension was obtained and adjusted to the optimized concentration of cells and the FASTvet gramneg or FASTvet grampos panels inoculated accordingly to the microorganism. FASTvet panels were then incubated during 1 h, protected from light, at 37°C, shaking 540 rpm/min. Repeatability and reproducibility were determined by testing 10 strains in triplicate, from independent inoculums, which MIC values are on-scale, according ISO 20776-2 (ISO 20776-2, 2016).
Flow Cytometric Analysis
The FASTvet panels were analyzed in a CytoFLEX flow cytometer (Beckman Coulter) using previously optimized templates. The number of cells on the “gate”- zone of analysis; the complexity of the cells (measured by the side scatter); size of the cells (measured by the forward scatter) and the intensity of fluorescence of the cells are the 4 main parameters analyzed. Treated cells are always compared with non-treated cells for each tested microbial strain. A dedicated software, the BioFAST®, developed for AST for human health, was adapted to this veterinary assay. Antibiotic susceptibility was determined by the use of an algorithm in the BioFAST software (FASTinov®, Porto, Portugal) that determines the ratio between the mean fluorescence intensity (MFI) of treated bacteria compared with control cells (non-treated bacteria); this ratio was calculated through ROC curves and defined as the staining index (SI). For each antibiotic and bacterial combination, cut-off values were incorporated into the BioFAST software for antimicrobial susceptibility testing.
The available prototype provides a stand-alone application with limited licensing support, interacting with flow cytometers using a commonly supported file format, which most cytometers export.
Reference AST Method
All tested strains were analyzed through reference methods, either microdilution or disk diffusion tests, according to CLSI AST vet document (CLSI 5th Edition, 2018) and classified as susceptible (S), intermediate (I) or resistant (R). Recommended QC were included.
Data Analysis and Comparison With Reference AST Methods
Taking into consideration the classification of the AST phenotypes determined by reference methods, the cut-off values for the cytometric assay were calculated using ROC curves and included in the BioFAST vet software. The phenotype determined using the FASTvet assay was therefore recorded and compared with the obtained with reference methods. In case of discrepancies, the susceptibility test was repeated by both methods. Categorical agreement was calculated and errors were classified as very major errors (VMEs; a false susceptible), major errors (ME; a false resistant) and minor errors (mEs; any false result involving an intermediate result) according to International Organization for Standardization [ISO] (20776), 2006 part 2.
Statistical Analysis
The percentage agreement (PA) was calculated in order to evaluate repeatability and reproducibility.
Results
A total of 115 microbial isolates were included in the present study, including 65 Gram-negative isolates and 50 Gram-positive strains. The overall categorical agreement (CA) between the new cytometric AST assay and the reference method was 95.6% (range between 92.3–100%) for FASTvet gramneg and 96.7% (range between 93.3–100%) on FASTgrampos. CA and errors distribution is detailed on Table 1, no VM errors were found.
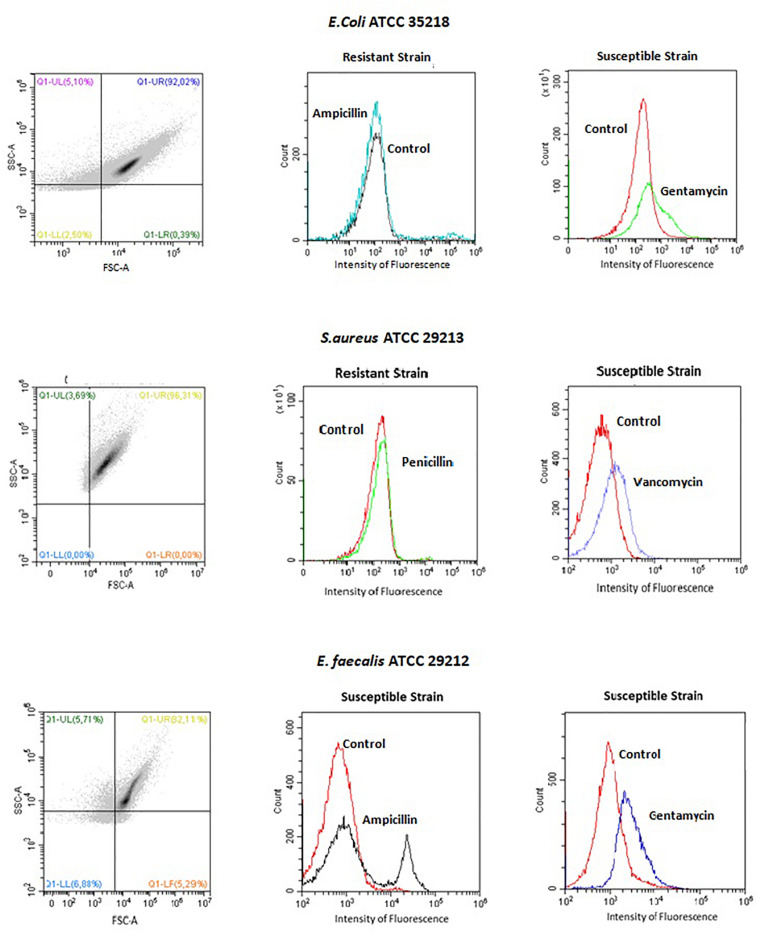
Typical dot plots and histogram are shown in Figure 1, as examples. An increase of the intensity of fluorescence (moving to the right) of the drug-treated microbial population comparing with non-treated cells (control), means that the cells depolarized, corresponding to a susceptible phenotype. In such strains, a decrease of the number of cells (cell counts) is common. Resistant phenotypes, reveal similar pattern to the control (no changes on fluorescence or reduction on cell number was seen). An intermediate phenotype is detected whenever an increase of the intensity of fluorescence is observed only following incubation with the highest concentration of the drug (the lowest concentrations produce no effect). Regarding reproducibility, 96.6% was obtained (PA = 0.966) and the repeatability was 100% (PA = 1).
FIGURE 1.
Dot plot and histograms of a typical flow cytometry analysis of one Gram negative bacilli (E. coli ATCC 35218) and two Gram positive cocci (S. aureus ATCC 29213 and E. faecalis ATCC 29212). Note an increase on the intensity of fluorescence on the susceptible strains.
The time required to obtain a final AST, using FASTvet panel was 2 h, in contrast with the minimum of 24 h required by the current standard AST methods, thus resulting in a gain in time to result of 22 h in average.
Discussion
The purpose of this study was to evaluate the performance of FASTvet (flow cytometry antimicrobial susceptibility test for veterinary) and to determine whether this procedure could be used routinely to reduce AST turnaround time in veterinary medicine. This will allow the early choice of targeted antibiotic therapy, by the veterinary surgeon whenever managing an animal with an infectious disease in progress. This methodology was first applied to human health and, attending to the many similitudes of microorganisms/drugs required, we proposed it could be transferred, with similar advantages, to veterinary field. The One Health Concept stresses in particular, the problem of antimicrobial resistance that is global and affects both human and animals (American Veterinary Medical Association, 2008; Andrade et al., 2018). Among the various alternatives and contingencies considered to prevent the emergence of resistance to antimicrobials, a fast and accurate AST is urgently needed. This study demonstrates the potential utility of FASTvet as a useful AST tool to assist the veterinary surgeon in clinical useful time in the selection of targeted antimicrobials (Andrade et al., 2018) and supports further investigation and validation on this test methodology. A quick and accurate clinical treatment would prevent the dissemination of multidrug-resistant bacteria.
Pure colonies were used in this study and, around 1 h subculture in a broth was needed especially for β-lactamic drugs since they only act when cells reach the exponential growth phase. However, such a FC assay could be started not only with colonies but also directly from positive blood cultures (Costa-de-Oliveira et al., 2017) or in urine (data not shown) as there are enough cells on exponential growth phase. This will avoid the sub-culture step fast-tracking the answer to a maximum of 2 h instead of 2 days.
A limitation of this technology is that, like all other phenotypic AST tests, it cannot be performed directly from polymicrobial products or mixed cultures. In such instances, molecular tests could detect a resistance gene, e.g., mecA in a nasal swab meaning the presence of a MRSA. Nevertheless, only some genes could be searched for detection of resistance and its absence does not guarantee susceptibility. Another limitation is the fact that this test does not provide minimum inhibitory concentrations (MIC values), as only breakpoint concentrations were included. Flow cytometry assay could provide MIC values but we privileged the speed (more concentrations will increase the time of analysis) and few clinical situations require that information. By contrary, quantitative values could be very relevant for epidemiological purpose but, in that case, it would be better to calculate epidemiological cut-off values (ECV) than MIC.
With the present study, we demonstrate that AST results for veterinary antimicrobials could be obtained within the same working day, about 24 h earlier than results obtained with the usual standard diagnostic procedure. The performance of the FASTvet kit agrees with the ISO recommendations for AST. Those results are promising and lead us to increase the sample size. An external validation, in dedicated veterinary labs, is being planned. This new and disruptive technology could change the clinical diagnostic paradigm in relation to AST in veterinary medicine.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
FA wrote the manuscript, collected the biological samples, and designed the study. RG and IM-O executed the experimental work. AR wrote the manuscript and designed the study. CP-V wrote the manuscript and designed the study and coordinated the research. All authors contributed to the article and approved the submitted version.
Conflict of Interest
RG, IM-O, AD, and CP-V were employed by FASTinov. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This article was supported by National Funds through FCT – Fundação para a Ciência e a Tecnologia, I.P., within CINTESIS, R&D Unit (reference UIDB/4255/2020).
References
- American Veterinary Medical Association (2008). One Health: A New Professional Imperative. One Health Initiative Task Force: Final Report. Schaumburg, IL: The American Veterinary Medical Association, 1–71. Available online at: https://www.avma.org/KB/Resources/Reports/Documents/onehealth_final.pdf [Google Scholar]
- Andrade F. F., Pina-Vaz C., Rodrigues A. G. (2018). Antimicrobial resistance: a one health concept perspective analysis∗. Infect. Dis. Diag. 1–9. 10.29011/2577-1515.100027 [DOI] [Google Scholar]
- CLSI Vet 01 (2018). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals, 5th Edn Wayne, PA: CLSI [Google Scholar]
- Cohen J. (2009). Sepsis. Medicine 37 562–565. [Google Scholar]
- Costa-de-Oliveira S., Teixeira-Santos R., Silva A. P., Pinho E., Mergulhão P., Silva-Dias A., et al. (2017). Potential impact of flow cytometry antimicrobial susceptibility testing on the clinical management of gram-negative bacteremia using the FASTinov® kit. Front. Microbiol. 8:2455. 10.3389/fmicb.2017.02455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellinger R. P., Levy M. M., Rhodes A., Annane D., Gerlach H., Opal S. M., et al. (2013). Surviving sepsis campaign: International guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 39 165–228. 10.1007/s00134-012-2769-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deresinski S. (2007). Principles of antibiotic therapy in severe infections: optimizing the therapeutic approach by use of laboratory and clinical Data. Clin. Infect. Dis. 45(Suppl 3), S177–S1783. 10.1086/519472 [DOI] [PubMed] [Google Scholar]
- Duggal S., Gaind R., Tandon N., Deb M., Chugh T., Das (2012). Comparison of an automated system with conventional identification and antimicrobial susceptibility testing. ISRN Microbiol. 2012:107203. 10.5402/2012/107203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria-Ramos I., Espinar M. J., Rocha R., Santos-Antunes J., Rodrigues A. G., Cantón R., et al. (2013). A novel flow cytometric assay for rapid detection of extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 19 E8–E15. 10.1111/j.1469-0691.2012.03986.x [DOI] [PubMed] [Google Scholar]
- Gherardi G., Angeletti S., Panitti M., Pompilio A., Di Bonaventura G., Crea F., et al. (2012). Comparative evaluation of the Vitek-2 Compact and Phoenix systems for rapid identification and antibiotic susceptibility testing directly from blood cultures of Gram-negative and Gram-positive isolates. Diagnostic Microbiol. Infect. Dis. 72 20–31. 10.1016/j.diagmicrobio.2011.09.015 [DOI] [PubMed] [Google Scholar]
- Infectious Diseases Society of America (2011). Combating antimicrobial resistance: policy recommendations to save lives. Clin. Infect. Dis. 52 S397–S428. 10.1093/cid/cir154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Organization for Standardization [ISO] (20776) (2006). Clinical Laboratory Testing and in Vitro Diagnostic Test Systems: Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices. Geneva: ISO, Part 2. [Google Scholar]
- Jenkins S. G., Schuetz A. N. (2012). Current concepts in laboratory testing to guide antimicrobial therapy. Mayo Clin. Proc. 87 290–308. 10.1016/j.mayocp.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Roberts D., Wood K. E., Light B., Parrillo J. E., Sharma S., et al. (2006). Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 34 1589–1596. 10.1097/01.CCM.0000217961.75225.E9 [DOI] [PubMed] [Google Scholar]
- Micek S. T., Welch E. C., Khan J., Pervez M., Doherty J. A., Reichley R. M., et al. (2010). Empiric combination antibiotic therapy is associated with improved outcome against sepsis due to gram-negative bacteria: a retrospective analysis. Antimicrob. Agents Chemother. 54 1742–1748. 10.1128/AAC.01365-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittman S. A., Huard R. C., Della-Latta P., Whittier S. (2009). Comparison of BD Phoenix to Vitek 2, MicroScan MICroSTREP, and etest for antimicrobial susceptibility testing of Streptococcus pneumoniae. J. Clin. Microbiol. 47 3557–3561. 10.1128/JCM.01137-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rene S. H. (2010). MIC Determination by Broth Dilution using Sensititre. Geneva: World Health Organization Laboratory. [Google Scholar]
- Van Belkum A., Durand G., Peyret M., Chatellier S., Zambardi G., Schrenzel J., et al. (2013). Rapid clinical bacteriology and its future impact. Ann. Laboratory Med. 33 14–27. 10.3343/alm.2013.33.1.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.