Abstract
Single-cell technologies are offering unparalleled insight into complex biology, revealing the behavior of rare cell populations that are masked in bulk population analyses. One current limitation of single-cell approaches is that lineage relationships are typically lost as a result of cell processing. We recently established a method, CellTagging, permitting the parallel capture of lineage information and cell identity via a combinatorial cell indexing approach. CellTagging integrates with high-throughput single-cell RNA sequencing, where sequential rounds of cell labeling enable the construction of multi-level lineage trees. Here, we provide a detailed protocol to (i) generate complex plasmid and lentivirus CellTag libraries for labeling of cells; (ii) sequentially CellTag cells over the course of a biological process; (iii) profile single-cell transcriptomes via high-throughput droplet-based platforms; and (iv) generate a CellTag expression matrix, followed by clone calling and lineage reconstruction. This lentiviral-labeling approach can be deployed in any organism or in vitro culture system that is amenable to viral transduction to simultaneously profile lineage and identity at single-cell resolution.
Introduction
Enabled by recent advances in single-cell technology, many features of cell identity and state can be assayed across numerous individual cells, supporting the curation of high-resolution cell atlases1–3. Since its introduction in the last decade4, single-cell RNA sequencing (scRNA-seq) has seen wide adoption for single-cell resolution analyses. Early scRNA-seq methods were relatively low throughput5–7, until higher-capacity microfluidic technologies enabled huge gains in cell capture rate8–10. These methods are now moving beyond the requirement for physical separation of individual cells, enabling further improvements in capture rates and cost reductions11,12. Beyond high-throughput scRNA-seq, single-cell measurement of chromatin accessibility is now possible13,14, even in concert with transcriptome capture15. Computational methods are also emerging to integrate these multi-omic datasets16,17. Together, this technological progress has enabled population heterogeneity to be deconstructed, revealing rare cell types and states across a range of biological systems. However, the application of these technologies can be limited as cell harvest generally requires tissue disruption, resulting in the loss of crucial spatial, temporal and lineage information.
Reconstruction of lineage relationships at single-cell resolution
The construction of lineage hierarchies reveals valuable information about cell potential, identity and behavior. Several computational approaches have been developed to reconstruct differentiation trajectories, inferring lineage relationships. In this respect, Monocle18,19 was an early leader, using dimensionality reduction via independent component analysis to project cells in a two-dimensional space. A minimum spanning tree algorithm is applied to ‘join-the-dots’ between transcriptionally similar cells, mapping the longest path through the data to create a pseudo-temporal cell fate trajectory. Many comparable methods adopt a similar strategy to Monocle20–24, while other approaches such as k-nearest neighbor graphs25,26 and degree of RNA splicing27 rely on connecting cell clusters28,29 to reconstruct differentiation trajectories. However, these methods often produce conflicting differentiation trees from identical input data30. Furthermore, misleading branches in the trajectory can arise as a result of overfitting31, an error where a computational model performs very well with training data but performs poorly on new datasets.
The direct experimental connection of progenitors to their progeny enables true lineage trees to be reconstructed across a biological process, independent of or complementary to the aforementioned computational methods for trajectory reconstruction. To enable capture of ‘ground truth’ lineage information at single-cell resolution, several elegant approaches have recently emerged. Fundamentally, lineage tracing is based on the unique labeling of individual cells, either via the exploitation of naturally occurring somatic mutations32–34 or experimentally induced heritable marks35. Traditionally, experimental methods have relied on virus-36,37 or transposon-mediated38 delivery of heritable DNA barcodes or the introduction of CRISPR–Cas9-induced stochastic mutations39–41 into genomic DNA to uniquely mark cells. Although these approaches were initially not compatible with scRNA-seq, recent adaptations have enabled readout of cell labels in the transcriptome, supporting the parallel capture of lineage and identity42–48. With CRISPR–Cas9-based technologies44–46, cell labels are progressively mutable; thus, lineages can be reconstructed from tracking sequential mutations. In contrast, although virus-based cell labeling is easier to deploy without complex genetic manipulation, the barcodes are not mutable42,43. Therefore, while virus-based approaches have supported clonal analysis, it was not possible to reconstruct lineage maps using these strategies.
Parallel reconstruction of lineage and identity with CellTagging
We have established a tractable cellular barcoding technology, CellTagging, that can be easily deployed across a broad range of biological systems, enabling high-resolution, high-throughput lineage reconstruction without the requirement for complex genome engineering strategies42,49. To label cells with a heritable barcode, permitting their subsequent identification, we transduce fibroblasts with lentivirus carrying GFP and an SV40 polyadenylation signal sequence, where GFP expression is driven by a minimal cytomegalovirus (CMV) promoter. This design, leveraging the pSMAL backbone, was first used in van Galen et al.50 to transduce human hematopoietic stem cells efficiently. Within the 3′ untranslated region (UTR) of GFP, we engineered an 8-bp random index sequence that allows for the generation of abundant barcodes expressed as polyadenylated mRNA, thereby allowing both lineage information and cell identity to be captured in parallel, using high-throughput scRNA-seq platforms. In the complex libraries generated via this approach, each lentivirus can carry one of up to 65,536 unique CellTags. Starting cell populations are transduced with this library at a multiplicity of infection >1 to ensure the delivery of multiple CellTags into each cell (Fig. 1a). This combinatorial barcoding method results in the unique labeling of cells with permanent and heritable CellTag ‘signatures’. Crucially, to enable the reconstruction of lineage trees, unlike other viral approaches, we create indexed libraries to enable the sequential labeling of cells to map lineage relationships (Fig. 1b–d). This flexible labeling approach enabled us to longitudinally track lineage and identity during cell fate conversion, elucidating the molecular regulatory mechanisms underpinning defined trajectories and enabling identification of factors to enhance reprogramming42.
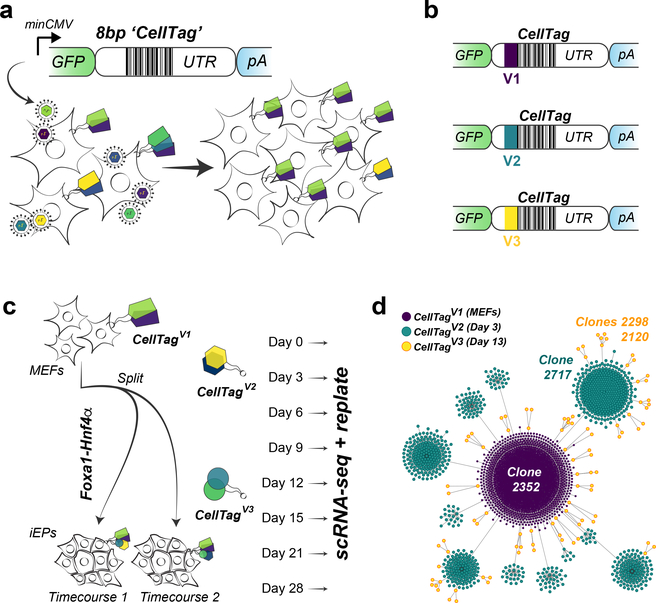
Fig. 1 |. The CellTag workflow for parallel capture of lineage and identity.
a, Schematic of the CellTagging workflow. A lentiviral construct is engineered with an 8-bp random ‘CellTag’ barcode in the 3′ UTR of GFP, followed by an SV40 polyadenylation signal. Cells are transduced with this lentiviral library (produced via transfection of HEK293T cells with the complex plasmid library) so that each cell expresses ~3–4 CellTags, resulting in a unique, heritable signature, enabling clonally related cells to be tracked over the course of an experiment. b, Design of CellTag constructs for multiplexing: a short, 6-bp index sequence is inserted in front of the variable CellTag region, allowing different rounds of CellTagging to be demultiplexed and lineage trees to be subsequently constructed. c, Schematic of experimental approach: reprogramming of MEFs to iEPs via retroviral delivery of Foxa1 and Hnf4α. Cells were transduced as fibroblasts with CellTagV1, and again at 3 d (with CellTagV2) and 13 d (with CellTagV3) after initiation of reprogramming. A fraction of cells were recovered for scRNA-seq every 3–7 d, and the remainder were replated. d, Reconstruction and visualization of lineages via force-directed graphing. Each node represents an individual cell, and edges represent clonal relationships between cells: purple, CellTagV1 clones; blue, CellTagV2 clones; yellow, CellTagV3 clones. n = 2,199 cells.
In this protocol, we provide detailed, stepwise directions on how to perform sequential CellTagging on in vitro cultured cells, using direct reprogramming of fibroblasts to induced endoderm progenitors (iEPs) as an example (Fig. 1c). CellTagging can be deployed in any organism or in vitro culture system that is amenable to viral transduction, enabling the investigation of lineage and cell identity at single-cell resolution, across a range of biological questions.
Comparison with other lineage-tracing strategies
Prospective lineage tracing has traditionally relied on cell labeling using reporter genes such as GFP or β-galactosidase, allowing cells to be followed over time51,52. However, these approaches require sparse labeling to ensure that independent cells and their progeny can be tracked, limiting their throughput. New sequencing technologies ushered in rapid advances in tracing capabilities, where high-complexity DNA barcode libraries were initially used to uniquely label cells, permitting highly parallel cell tracing36. Subsequent sequencing-based approaches have incorporated Cre-mediated recombination to generate unique genetic barcode combinations, enabling large-scale clonal analyses in whole animals53. Altogether, these strategies have generally been limited by a requirement for DNA-based barcode sequencing, neglecting the cell transcriptome and hence assessment of cell identity. More recently, sequencing-based methods, have evolved in concert with high-throughput scRNA-seq, where barcodes introduced using lentivirus are expressed as RNA and captured within the single-cell transcriptome43. This approach has supported the parallel capture of both clonal and cellular identity information. However, the DNA- and RNA-based prospective tracking approaches discussed thus far support only clonal analysis; the barcodes introduced are not mutable, and therefore lineage relationships cannot be mapped. To expand on these strategies, we integrated short index sequences immediately upstream of the CellTag sequence, permitting sequential rounds of cell labeling (Figs. 1 and 2) and lineage tree reconstruction42. Furthermore, unlike previous approaches where additional PCR steps are needed to capture cell barcodes, no additional steps in single-cell library preparation are required for CellTag recovery, adding to the benefits of this technology.
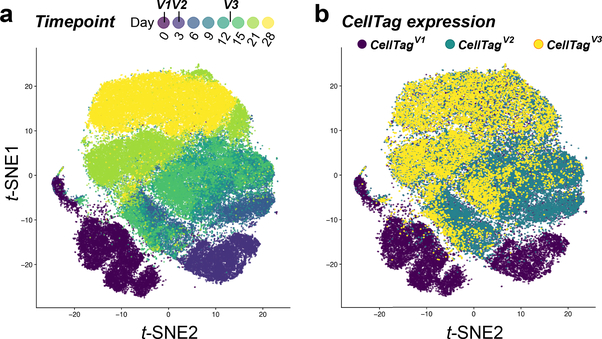
Fig. 2 |. CellTagging applied to a cell reprogramming time course.
a, A t-SNE plot of a 28-d fibroblast-to-iEP reprogramming time course experiment, with time point information projected onto the plot (n = 85,010 cells). b, Overlay of CellTag expression, broken down by CellTag library version, onto the t-SNE plot in a. Before the initiation of reprogramming, fibroblasts were transduced with the CellTagV1 library. Three days after initiation of reprogramming, cells were transduced with the CellTagV2 library. Finally, 13 d after the start of the reprogramming process, cells were transduced with the CellTagV3 library. Following this scheme, CellTag expression is detected in 99% of cells, with almost 70% of cells expressing two or more CellTags to support confident cell tracking.
Virus-independent cell labeling strategies also form a valuable component of the current lineage-tracing toolkit. For example, CRISPR–Cas9 barcode editing has recently been coupled with scRNA-seq in zebrafish44–46 and mice54,55. In these methods, genetic barcodes in a multi-copy transgenic reporter are edited via injection of Cas9 protein or RNA, along with a single-guide RNA (sgRNA) targeting the transgenic reporter, which is expressed in the cell transcriptome. Cumulative edits allow lineages to be reconstructed, although Cas9 degradation can restrict the temporal window of lineage tracing. Furthermore, the number of distinct barcodes that can be generated is limited, and all integration site edits must be recovered to build a full picture of cell lineage, which can be a problem considering dropout in scRNA-seq. Dropout describes a scenario where a transcript is expressed in a cell but is undetected in its mRNA profile. scGESTALT (single-cell genome editing of synthetic target arrays for lineage tracing) overcomes some of these limitations; employing nine editable sites, in tandem, a combination of injection and transgenic-driver Cas9/sgRNA expression coupled with increased barcode diversity enables extended lineage recording45,56. However, the lineage barcodes are recovered from a low percentage of cells, relative to CellTagging. In addition, some lineage records can be erased by large deletions spanning the multiple CRISPR sites that are in tandem. Furthermore, the same edits can be introduced into independent cells, due to saturation of Cas9 editing. These limitations restrict longer-term tracking and the generation of more complex lineage trees. This latter point is already being addressed by generating edits from multiple independent sites54, and by generating a self-targeting form of CRISPR–Cas957,58, where ‘homing’ increases complexity so that scarring can occur over a longer period of time55. As a CRISPR–Cas9 alternative, transposon-based TracerSeq, exploits the Tol2 transposase to randomly integrate unique and heritable labels into individual cell genomes. Asynchronous insertion over successive cell divisions permits lineage tree reconstruction in zebrafish development, avoiding repeat editing from which CRISPR–Cas9-based approaches can suffer47.
While CRISPR–Cas9-based strategies offer many benefits for whole-organism lineage tracing, they do require considerable transgenesis, which may not be easily deployed in some systems. Here, CellTagging is valuable in that it can be easily applied to any cell type that is amenable to lentiviral transduction. In addition, the timing of sequential tagging to build lineage trees is extremely flexible. Moreover, unlike previous cellular barcoding approaches, no additional PCR steps are required to capture CellTags. Together, this offers considerable advantages over emerging gene-editing technologies to track cells, particularly in cell types where gene editing is challenging, or the generation of transgenic lines is not feasible. Furthermore, CellTagging enables accurate tracking of almost 70% of labeled cells, with a low false-positive rate. These advantages position CellTagging as a tractable option for lineage reconstruction in targeted organs/cell types in vivo, in vitro cell culture, and transplant of tagged cells as GFP expression is maintained over long periods of time in vivo49,59.
Limitations of CellTagging
There are several considerations to make before selecting CellTagging as a tool to track clonally related cells and reconstruct lineage relationships. The base pSMAL construct has been used to generate lentivirus to transduce difficult-to-infect cells such as human hematopoietic stem cells50 and myeloid precursors60 at a high multiplicity of infection (MOI). Indeed, we have successfully CellTagged and traced a range of cells, including mouse fibroblasts, B cells, macrophages and human embryonic kidney cells42,49. Furthermore, we have previously transduced mouse iEPs, followed by their transplant into a mouse model and successful tracking for 7 d after engraftment49. However, CellTagging may be incompatible with some cell types that are not amenable to efficient viral transduction. Related to this point, cell types in which expression of lentiviral genes is heavily silenced are not good candidates for CellTagging, although multiple CellTag integrations may overcome partial silencing. Future improvements on this cell tracking tool to guard against silencing via the use of alternate promoters61,62 or insulator sequences63–65 will address this current limitation for some systems. In addition, recovery of CellTag information from genomic DNA will provide a potential alternative. The impact of silencing is cell-type dependent: in our fibroblast to iEP reprogramming time course, although CellTag expression becomes weaker over time, we did not observe considerable silencing42. We are also currently developing improved computational methods to infer clonal relationships upon partial silencing of CellTag signatures. Experimentally, a further enhancement would consist of mutable barcodes to avoid multiple rounds of viral transduction. Finally, because this approach is based on scRNA-seq, cells are dissociated for analysis, resulting in loss of spatial information. Future improvements could incorporate in situ sequencing of cell barcodes to recover valuable spatial and phenotypic data.
Experimental design
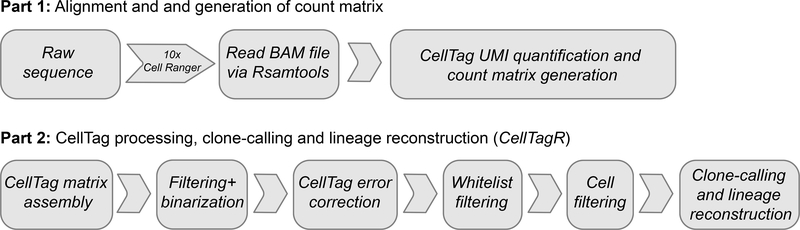
The CellTagging protocol is divided into three main experimental parts and one analytical part: (i) generation of complex plasmid and lentivirus CellTag libraries for labeling of cells; (ii) sequential CellTagging of cells over the course of a biological process; (iii) single-cell transcriptome profiling; and (iv) generation and filtering of the CellTag expression matrix, followed by clone calling and lineage reconstruction. To aid the design of CellTag-based experiments across a range of different cell types, we have developed a simulation-based calculator, available at http://celltag.org/ (also see Software below).
Construction of complex CellTag libraries
The construction of complex CellTag libraries is based on the introduction of a random 8-bp barcode into the pSMAL lentiviral plasmid backbone (Fig. 1a) via restriction-free cloning. CellTags are positioned within the 3′ UTR of GFP, followed by an SV40 polyadenylation signal sequence. A 6-bp index sequence is inserted immediately upstream of the CellTag region to enable sequential rounds of labeling and lineage reconstruction (Figs. 1 and 2). High levels of GFP-CellTag expression are driven by a minimal CMV promoter, resulting in abundant, indexed and polyadenylated transcripts that are captured as part of standard scRNA-seq pipelines. In the complex lentivirus libraries generated via this approach, each viral particle can carry one of up to 65,536 unique CellTags. Three pooled CellTag libraries (V1, V2 and V3), each containing a unique index sequence, are available via Addgene (https://www.addgene.org/pooled-library/morris-lab-celltag/). Depending on the user’s goal, these libraries can be modified to contain additional index sequences or longer barcodes. In addition, defined CellTags can be used for sample multiplexing, as we have previously demonstrated49. It is crucial to maintain the complexity of these pooled libraries via a liquid culture amplification approach, followed by sequencing-based assessment of CellTag diversity. This latter step also generates a ‘whitelist’ of CellTags that can be used to correct PCR and sequencing errors in subsequent analytical steps to increase the specificity and sensitivity of clone-calling (see Generation of CellTag expression matrix, clone-calling and lineage reconstruction). Our computational stochastic simulation at http://celltag.org/ demonstrates that it is essential to maintain a high library complexity, to reduce the chances of two unrelated cells becoming labeled with the same combination of CellTags, particularly when labeling a large number of cells. Vesicular stomatitis virus G (VSV-G) pseudotyped lentiviral particles are then produced via transfection of 293T cells, followed by determination of titer. High-titer virus is used for infection of cells at an optimal MOI to ensure integration of multiple unique CellTags per cell, to increase tracking confidence. Through our simulation, supporting our previous experimental results42, we found that to avoid the same CellTag signatures labeling independent cells, an MOI of ≥3 is recommended.
Sequential CellTag labeling, cell harvesting and replating
In this section of the experimental protocol, cells are transduced with complex CellTag lentivirus libraries. This step is highly dependent on the biological system under investigation. It is critical that cells are labeled with multiple CellTags to mark cells with a unique combinatorial tag signature, where we aim for cells to express three unique CellTags on average. In downstream analyses, cells expressing fewer than two unique CellTags are filtered out to ensure high-confidence clone-calling and lineage reconstruction. In this protocol, we outline the sequential CellTagging of mouse embryonic fibroblasts as they directly reprogram into iEPs over a 4-week time course experiment. This system is extremely amenable to multiple rounds of lentiviral transduction, with little impact on cell physiology or reprogramming efficiency42,49. In this example, we outline the initial CellTagging of cells as fibroblasts, followed by second and third rounds of labeling, 72 h and 13 d after reprogramming initiation, respectively (Fig. 1b). This scheme was designed to capture early and late cell fate decisions in the reprogramming process and, again, is highly dependent on the system under study.
An important consideration for this part of the experimental design is the rate at which cells divide; for example, in fast-dividing cell populations, large clones of cells and lineages can be reconstructed. In contrast, labeling post-mitotic cells will not yield any clonal information as CellTagging relies on the inheritance of CellTags and their detection in progeny to identify clonally related cells. Our simulations indicate that rapidly dividing cells require earlier sequencing to reliably detect rare clones—alternatively, more cells can be sequenced at later time points to obtain the same resolution. In addition to this, the number of starting cells is an important consideration, where the initial number of cells should be minimized to facilitate capture of a high percentage of labeled cells, promoting detection of clonally related cells. To aid the design of CellTagging experiments, we provide an experimental simulator at http://celltag.org/, taking into account these design considerations, along with a detailed troubleshooting guide.
Following CellTagging, cells are then cultured, portions are collected and methanol fixed for later single-cell sequencing, and the remaining cells are replated to allow clones to continue expanding. We opt for methanol fixation66 in these longer time course experiments, enabling cells to be stored and processed together for single-cell profiling. One benefit of this ‘cell-banking’ approach is that it allows for the outcome of the experiment to be assessed (for example, by monitoring successful reprogramming) via more cost-effective means before committing to relatively expensive scRNA-seq. Alternatively, for investigation of a simple lineage bifurcation, for example, cells can be CellTagged, followed by culture and harvest at a single later time point. This strategy is often sufficient to capture cells in many transcriptional states across an unsynchronized biological process.
Cell hydration and scRNA-seq
After completion of CellTagging, culture, and methanol fixation, cells are rehydrated and processed on either Drop-seq8 or 10x Genomics Chromium single-cell10 platforms, where we have successfully called clones from data acquired using both of these strategies. We expect that other high-throughput scRNA-seq modalities, such as InDrops9, will work well with CellTagging. One important consideration here is the method of library preparation where 3′ enrichment of transcripts is compatible with the location of the CellTag motif within 200 nt of the polyadenylation sequence. The position of the CellTag could easily be moved to the 5′ portion of GFP to support barcode capture from library preparation strategies designed to capture the 5′ end of transcripts. Another critical consideration is the cell capture rate of the single-cell platform of choice. For example, although Drop-seq is more cost effective, typically only 5% of cells are recovered during library preparation. In contrast, 60% of cells loaded onto the 10x Chromium device are captured, enabling smaller pools of cells to be cultured and maximizing clone detection to support lineage reconstruction.
Experimental controls
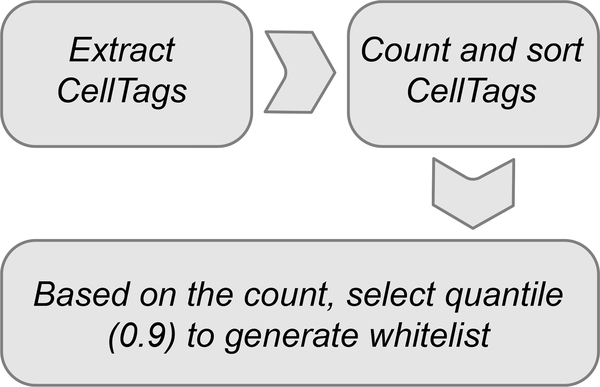
Non-CellTagged controls should also be included in single-cell profiling to test the impact of cell labeling on the biological process of interest, via assessment of potential perturbations on cellular physiology at the transcriptome level. These controls are particularly important where the impact of multiple rounds of viral integration on the cells being investigated is unknown. We have typically profiled one time point, at the end of the experiment, to assess this. To evaluate the efficacy of CellTagging to uniquely label cells, we also recommend labeling two independent biological replicates of cell populations of interest with the same CellTag library. If the library is sufficiently complex, there should be minimal overlap of CellTag signatures between the independent replicates (Fig. 3). If there is substantial overlap, this could indicate that the complexity of your CellTag library is insufficient, leading to false-positive clone calls arising from independent cells that are not uniquely labeled. In terms of quality control, it is important to also assess whether any significant CellTag silencing occurs across the course of an experiment. This can be evaluated by comparing the proportions of cells passing the cell tracking threshold of expressing two or more unique CellTags per cell across the course of an experiment. From our direct reprogramming experiments, we find that CellTag expression diminishes over 4 weeks but is silenced in only 10% of cells42. In these assessments, it is also important to gauge whether CellTags are specifically silenced in any subpopulations of cells under investigation.
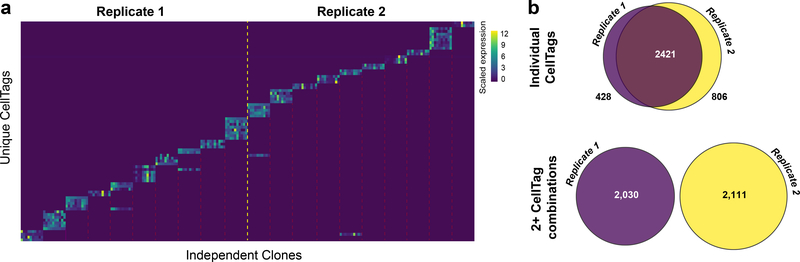
Fig. 3 |. CellTag signatures uniquely label cells across independent biological replicates.
a, Heatmap showing scaled expression of individual CellTags in 20 major clones (n = 10 representative cells per clone) from two independent biological replicates tagged with the same CellTag library. The dashed yellow line marks the separation between the two time courses. Dashed red lines mark separation between independent clones. Although some CellTags are shared between these independent biological replicates, the combined CellTag signatures are unique. b, Top: overlap of individual CellTags detected in two independent biological replicates (replicate 1: n = 8,535 cells; replicate 2: n = 11,997 cells) tagged with the same CellTag library preparation. Bottom: there is no overlap of CellTag signatures between the two replicates.
Generation of CellTag expression matrix, clone-calling and lineage reconstruction
Following the sequencing of single-cell libraries, a CellTag expression matrix is generated alongside the standard digital expression matrix representing each single-cell transcriptome. The CellTag expression matrix is then filtered and corrected to support high-confidence clone calling, according to the following steps. (i) Closely related CellTags (i.e., Levenshtein edit distance ≤2) are collapsed on a cell-by-cell basis to correct for sequencing and PCR errors. (ii) CellTags reported by less than two independent transcripts are filtered out. (iii) ‘Whitelisting’ is performed to remove PCR and sequencing artifacts that are not corrected in the previous step. This whitelisting consists of filtering out CellTags that are not detected from sequencing of the original complex CellTag library. (iv) Cells with >20 CellTags (likely to correspond to cell multiplets) and less than two unique CellTags per cell are filtered out. (v) Clone calling is performed where Jaccard coefficient scores were calculated to assess the similarity of CellTag expression signatures in all cells in a pairwise manner, thereby identifying clonally related cells. These steps form the basis of our CellTagR pipeline (https://github.com/morris-lab/CellTagR; Supplementary Manual 1) and together increase the sensitivity and specificity of clone calling (Fig. 4). We define clones as groups of three or more related cells. To reconstruct lineage relationships, cells are assembled into sub-clusters according to clone identity, and then sub-clusters are connected to each other to build lineages of related cells. We use a force-directed graph-drawing algorithm to visualize these relationships between cells.
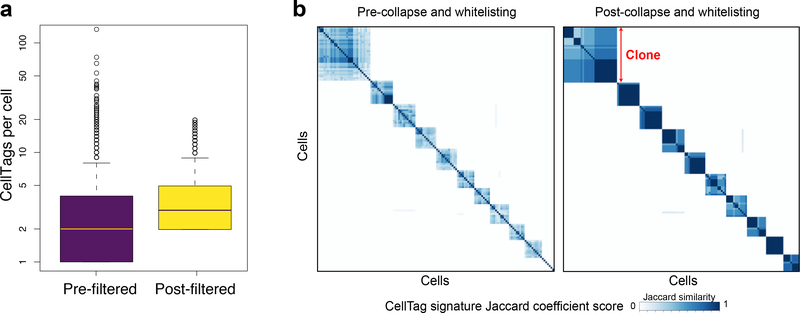
Fig. 4 |. CellTag filtering and error correction.
a, Mean CellTags per cell before and after CellTag pipeline filtering (n = 20,532 cells). The box plots show the median, first and third quantile and error bar with outliers. b, Pairwise correlation scores (Jaccard similarity) and hierarchical clustering of 10 major clones arising from this tag-and-trace experiment. Hierarchical clustering (Ward’s method (Ward.D2)) is based on each cell’s Jaccard correlation relationships with other cells, where each defined ‘block’ of cells represents a clone. Left panel: scoring and clustering of pairwise correlations, before whitelisting and filtering. Right panel: after whitelisting and filtering, pairwise correlations are stronger, and more cells are detected within each clone (n = 869 cells).
Materials
Biological materials
Stellar chemically competent Escherichia coli (Takara Bio, cat. no. 636766)
293T Human embryonic kidney cells (ATCC, cat. no. CRL-3216)
Mouse embryonic fibroblasts (MEFs) derived67 from E13.5 C57BL/6J mouse embryos (The Jackson Laboratory, cat. no. 000664) !CAUTION Our cell lines tested negative for mycoplasma contamination. The cell lines used in your research should be regularly checked to ensure they are authentic and are not infected with mycoplasma.
Reagents
Ambion nuclease-free water (Thermo Fisher Scientific, cat. no. AM9937)
Gibco DMEM (Thermo Fisher Scientific, cat. no. 11965084)
Gibco FBS (Thermo Fisher Scientific, cat. no. 10438026)
Gibco DPBS, magnesium- and calcium-free (Thermo Fisher Scientific, cat. no. 14190136)
Gibco penicillin–streptomycin (100×; Thermo Fisher Scientific, cat. no. 15140122)
Gibco β-mercaptoethanol (Thermo Fisher Scientific, cat. no. 21985023)
Qiagen Plasmid Plus Mega Kit (Qiagen, cat. no. 12981)
2× Kapa HiFi Hotstart Readymix (Roche, cat. no. KK2601)
Agencourt Ampure XP beads (Beckman Coulter, cat. no. A63880)
High Sensitivity D5000 reagents (Agilent Technologies, cat. no. 5067–5593)
X-tremeGENE 9 DNA Transfection reagent (Sigma-Aldrich, cat. no. 6365779001)
Protamine sulfate (Sigma-Aldrich, cat. no. P3369)
TrypLE Express enzyme (Thermo Fisher Scientific, cat. no. 12604013)
Gelatin solution (2% (vol/vol); Sigma-Aldrich, cat. no. G1393)
Methanol (Thermo Fisher Scientific, cat. no. A4521)
Chromium Single Cell 3′ Library & Gel Bead Kit v3 (10x Genomics, cat. no. PN-1000075)
Chromium Single Cell B Chip Kit (10x Genomics, cat. no. PN-1000074)
Chromium i7 Multiplex Kit (10x Genomics, cat. no. PN-120262)
DpnI (New England BioLabs, cat. no. R0176S)
Phusion High-Fidelity DNA Polymerase (New England BioLabs, cat. no. M0530S)
dNTP mix (Clontech, cat. no. 639125)
SOC medium (super optimal broth with catabolite repression) (Thermo Fisher Scientific, cat. no. 15544034)
Lysogeny broth (LB)/agar tablets (Sigma-Aldrich, cat. no. L7025)
Ampicillin solution (100 mg/ml; Sigma-Aldrich, cat. no. A5354)
BSA powder (Sigma-Aldrich, cat. no. A8806)
DTT solution (1 M; Sigma-Aldrich, cat. no. 43816)
SSC buffer (20×; Sigma-Aldrich, cat. no. S6639)
NxGen RNAse inhibitor (Lucigen, cat. no. 30281–1)
gBlock Gene Fragments (500 ng) and oligos (Table 1) ordered from IDT ▲CRITICAL Oligos are synthesized 100 nM scale, with HPLC purification. gBlocks and oligos should be resuspended in low-EDTA TE buffer (10 mM Tris, 0.1 mM EDTA, pH 8.0) and stored at −20 °C.
Table 1 |.
DNA oligonucleotide sequences
| Oligo name | Sequences | Purpose | Associated steps |
|---|---|---|---|
| CellTag-V1 gBlock | 5′-ACCTGAGCACCCAGTCCGCCCTGAGCAAAGACCCCAACGAGAAGCGCGATCACATGGTCCTGCTGGAGTTCGTGACCGCCGCCGGGATCACTCTCGGCATGGACGAGCTGTACAAGTAAACCGGTNNNNNNNNGAATTCGATGACAGGCGCAGCTTCCGAGGGATTTGAGATCCAGACATGATAAGATACATTGATGAGTTTGGACAAACCAAAACTAGAATGCAGTGAAAAAAATGCCTTATTTGTGAAATTTGTGA-3′ | Generation of complex CellTag libraries | 1–5 |
| Forward CellTag sequencing primer | 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT CATGGACGAGCTGTACAAGTAA-3′ | Assessment of CellTag library complexity | 19–31 |
| Reverse CellTag sequencing primer | 5′-CAAGCAGAAGACGGCATACGAGATACAGTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGTGCAGGGGAAAGAATAGTAGAC-3′ | Assessment of CellTag library complexity | 19–31 |
Plasmids
pCMV-VSV-G (Addgene, Plasmid ID 8454)
pCMV-dR8.2 dvpr (Addgene, Plasmid ID 8455)
Pooled CellTag libraries
Libraries are described at https://www.addgene.org/pooled-library/morris-lab-celltag/. These libraries are delivered as suspended DNA (10 ng/μl in a volume of 10 μl) in a microcentrifuge tube on blue ice. For best results, minimize freeze–thaws.
Pooled CellTag library V1, V2 and V3 (Addgene, cat. nos. 115643, 115644 and 115645)
Equipment
High Sensitivity D5000 Screen Tape (Agilent Technologies, cat. no. 5067–5592)
High Sensitivity D5000 Reagents (Agilent Technologies, cat. no. 5067–5593)
Microcentrifuge tubes (1.5 ml)
PCR tubes (0.2 ml)
Tubes (15 and 50 ml)
Sterile serological pipettes (5 and 10 ml)
Syringes (10 ml)
Syringe filters (0.45 μm; Millipore, cat. no. SLHVM33RS)
Vacuum filtration system (250 ml)
Tissue culture dishes (100 mm)
Veriti 96-Well Fast Thermal Cycler (Thermo Fisher Scientific, cat. no. 4375305)
Benchtop centrifuge for 1.5-ml tubes
Water bath
Heat block
Digital vortex mixer
Hemocytometers (Incyto, cat. no. DHC-F015)
Shaker incubator
Bacterial plate incubator
Nanodrop spectrophotometer (Thermo Fisher Scientific, cat. no. ND-2000)
10x Genomics Chromium Controller and Accessories (10x Genomics, cat. no.120263)
Software
10x Cell Ranger Analysis Pipeline (https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest)
CellTagR Analysis Pipeline (https://github.com/morris-lab/CellTagR; Supplementary Manual 1 and Supplementary Software 1)
R (https://www.r-project.org/, R version ≥3.5.0)
RStudio (https://www.rstudio.com/)
CellTag Simulator (http://celltag.org; Supplementary Software 2): This clonal simulation tool allows the user to explore potential false-positive rates and the evolution of clone sizes over time under different experimental conditions. Further instructions on setting parameters and performing a simulation can be found in Box 1.
Box 1 |. Using the CellTag simulator.
The simulation takes the following parameters.
N: starting cell population
L: the library complexity
μ: the average MOI. MOI used in the simulation is sampled from a Poisson distribution with mean of μ
pr: the rate of passaging
f: the fraction of cells retained in each passage
gr: the average rate of cell division
Ts: the time of sequencing
s: the number of cells sequenced
Using the above parameters, the simulation initializes a cell population N and randomly tags each cell with a CellTag from library L with the MOI of transfection sampled from a Poisson distribution with a mean of a given average MOI, μ. Here, we define a duplicate as a group of two or more cells expressing the same combination of CellTags. Considering that viral transduction is a stochastic process, the simulator runs a specified number of times (1,000 is the default setting) and the average number of duplicates is found. Next, the cells are simulated to proliferate at the growth rate gr, from which a subset of cells is maintained at the passage frequency pr until the time of sequencing. A subset of the population is sampled at each passage, based on a binomial distribution with the input fraction of cells to keep. The simulation terminates at the time of sequencing, and s cells to be sequenced are drawn at random out of the entire population. The ‘sequenced’ cells, s, containing more than one unique CellTag are used to calculate the metrics, including the number of clones, the average size of a clone, and the average size of a clone above some threshold clone size specified by the user.
Reagent setup
MEF/293T culture medium
For 500 ml of MEF/293T medium, combine 444.5 ml of DMEM with 50 ml of FBS (10% (vol/vol)), 5 ml of penicillin–streptomycin (1% (vol/vol)) and 500 μl of β-mercaptoethanol (0.1% (vol/vol)). Complete MEF medium can be stored at 2–8 °C for up to 4 weeks.
LB/agar plates
Add one LB/agar tablet to an autoclavable container and bring the volume to 50 ml with deionized water. Autoclave for 20 min at 121 °C. Cool to 50 °C in a temperature-controlled water bath. Add 0.50 ml of 100-mg/ml ampicillin. Pour onto four 100 mm × 15 mm sterile plates. Poured plates can be stored at 2–8 °C for up to 4 weeks.
Procedure
Generation of complex CellTag libraries ● Timing 2 d
▲CRITICAL These first steps (1–5) are optional should the user wish to modify the existing CellTag design. In these steps, using restriction-free cloning, random CellTags are inserted into the pSMAL backbone to generate a complex plasmid library. The CellTag-V1 gBlock sequence can be modified to include different index sequences to support more than three rounds of CellTagging. In addition, the length of the CellTag sequence can be changed to modify the library complexity. If obtaining pooled libraries from Addgene, begin at Step 6 (Amplification of pooled CellTag libraries). We have made three indexed pooled libraries available to support CellTagging of cells in three rounds to reconstruct lineages.
-
1Add the following to a PCR tube and mix by pipetting.
Reagent Amount 5× PCR buffer 4 μl 10 mM dNTP mix 0.4 μl 5 ng CellTag-V1 gBlock x μl 100 ng pSMAL destination plasmid x μl Phusion polymerase 0.2 μl Water To 20 μl -
2Run the following PCR program.
Temperature Time No. of cycles 98 °C 30 s 1 98 °C 8 s 15 60 °C 20 s 72 °C 140 s 72 °C 5 min 1 4 °C Forever -
3
DpnI-treat PCR product by adding 20 units of DpnI directly to the reaction mix, mix by pipetting and incubate for 2 h at 37 °C, followed by 20 min at 80 °C.
-
4
Thaw 100 μl of Stellar competent cells on ice, in a 1.5-ml microcentrifuge tube.
-
5
Add 10 μl of DpnI-treated PCR product to the competent cells and mix by pipetting. Proceed to the amplification steps (8–18) to generate your pooled CellTag library.
Amplification of pooled CellTag libraries ● Timing 2 d
-
6
Thaw Stellar chemically competent cells in an ice bath in a 1.5-ml microcentrifuge tube.
▲CRITICAL STEP Here we use high-efficiency competent cells to maintain library complexity. Users should be aware that lentivirus plasmids are prone to recombination. In our experience, we have not detected any issues with recombination, which can be investigated via a diagnostic digest of the CellTag library.
-
7
Add 10–50 ng of pooled CellTag DNA library to the competent cells and mix by pipetting.
▲CRITICAL STEP CellTag libraries are available from Addgene: https://www.addgene.org/pooledlibrary/morris-lab-celltag/ (including CellTag V1, V2, and V3).
-
8
Incubate the transformation mixture on ice for 30 min.
-
9
Heat shock the transformation mixture in the tube for 60 s on a heat block at 42 °C.
-
10
Chill the tube on ice for 1 min.
-
11
Add SOC medium to the tube, bringing the volume to 1,000 μl.
-
12
Incubate the transformation/SOC mixture at 37 °C while shaking (~250 rpm) for 1 h.
-
13
Using 5 μl of the transformation/SOC mixture, prepare serial dilutions from 1:10 to 1:1,000 in LB and plate 50 μl of each dilution onto one LB/agar plate (with 50 μg/ml ampicillin) per dilution (a total of three plates).
-
14
Incubate the plates overnight at 37 °C and add the remaining transformation/SOC mixture from Step 13 to 500 ml of LB (with 100 mg/ml of ampicillin) in a 1-liter flask.
-
15
Incubate the flask overnight while shaking at 37 °C.
-
16
After overnight incubation, count the number of colonies on the plates to calculate the number of c.f.u.
▲CRITICAL STEP To maintain the complexity of the CellTag library, 100–200 c.f.u. are required per unique CellTag in the library.
-
17
Harvest the cells from the overnight liquid culture and purify the library using Qiagen Megaprep columns (from the Qiagen Plasmid Plus Mega Kit) following the manufacturer’s protocol and elute the library in 1 ml of EB buffer.
-
18
Measure the concentration of the purified library using a Nanodrop spectrophotometer.
∎ PAUSE POINT Pooled libraries can be stored long term at –20 °C.
Assessment of CellTag library complexity via sequencing ● Timing 1 d + sequencing
-
19Add the following to one PCR tube for each library to be assessed. Mix by pipetting.
-
20Run the following PCR program.
Temperature Time No. of cycles 95 °C 3 min 1 95 °C 20 s 12 65 °C 15 s 72 °C 20 s 72 °C 1 min 1 4 °C Forever -
21
Equilibrate Ampure XP beads to room temperature (RT, 15–25 °C) and mix thoroughly by vortexing.
-
22
Add 30 μl of Ampure beads to each tube of sample from Step 20. This is a 0.6× beads-to-sample ratio. Pipette mix 15 times and incubate at RT for 15 min.
-
23
Place the tube strip in a magnetic separator until the solution is clear and then remove and discard the supernatant.
-
24
Add 200 μl of 80% (vol/vol) ethanol to the pellet and let stand for 30 s.
-
25
Remove and discard the ethanol wash. Repeat Steps 24 and 25 for a total of two ethanol washes.
-
26
Centrifuge the PCR tube briefly, remove any remaining supernatant and discard. Air dry the pellet for 2 min at RT.
-
27
Add 10 μl nuclease-free water to the pellet, pipette mix and incubate for 2 min at RT to elute the DNA.
-
28
Place the tube strip in a magnetic separator until the solution is clear and then remove and transfer the supernatant to a new PCR tube.
-
29
Using 1 μl of the purified DNA sample as input, run an Agilent Tapestation High Sensitivity D5000 Screen Tape according to the manufacturer’s instructions with high sensitivity D5000 reagents.
-
30
Sequence the prepared library on an Illumina MiSeq. This provides sufficient read depth to assess the complexity of at least three pooled CellTag libraries.
-
31
Generate the whitelist for the sequenced pooled CellTag library following the steps outlined in Fig. 5. Table 2 shows the number of whitelisted CellTags contained in our Addgene libraries, for comparison. Detailed tutorials and script resources are provided in Supplementary Manual 1 and Supplementary Software 1 and can also be found at https://github.com/morris-lab/CellTagR. ? TROUBLESHOOTING
Fig. 5 |.
Flowchart of the steps required for CellTag whitelist generation from raw sequence data.
Table 2 |.
Number of whitelisted CellTags contained in Addgene libraries
| Addgene library | No. of unique CellTags in each pooled library |
|---|---|
| CellTag-V1 | 19,973 CellTags |
| CellTag-V2 | 4,934 CellTags |
| CellTag-V3 | 5,737 CellTags |
Production of CellTag lentivirus ● Timing 6 d
!CAUTION Follow biosafety level 2 precautions for Steps 34–53.
-
32
Prepare maxipreps of the two lentivirus packaging plasmids: pCMV-VSV-G and pCMV-dR8.2 dvpr.
-
33
Day 0: seed 293T cells on a 100-mm dish at 50–60% confluency (~5 million cells) per plate in MEF/ 293T culture medium.
-
34
Day 1: transfect 293T cells. First change medium ~2 h before transfection.
-
35Prepare two 1.5-ml Eppendorf tubes.
- To tube 1 add 15 μl of X-tremeGENE 9 DNA Transfection Reagent directly to 185 μl of DMEM.
- To tube 2, bringing the volume to 200 μl with DMEM, add the following.
Plasmids Amount CellTag library (Step 18) 2 μg pCMV-VSV-G (Step 32) 200 ng pCMV-dR8.2 dvpr (Step 32) 2 μg -
36
Add the DMEM + DNA liquid mixture (tube 2) into the DMEM + X-tremeGENE 9 tube (tube 1) dropwise and mix by pipetting.
-
37
Incubate at RT for 15 min.
-
38
Add the DMEM + DNA + X-tremeGENE 9 mixture dropwise to the plate of 293T cells.
-
39
Gently slide the plate back and forth and side to side to evenly distribute the transfection reagents.
-
40
Incubate overnight at 37 °C.
-
41
Day 2: aspirate medium and add fresh medium to transfected 293T cells.
-
42
Day 3: first virus collection. Collect the cell supernatant from the transfected 293T cells using a 10-ml syringe. Immediately and gently add fresh medium to the cells and return them to the incubator.
-
43
Filter the cell supernatant through a low-protein-binding 0.45-μm syringe filter to remove cell debris. Collect the supernatant in a 15-ml tube. See below for storage details.
-
44
Day 4: second virus collection. Collect the cell supernatant from the transfected 293T cells using a 10-ml syringe and dispose of the culture plate.
-
45
Filter the supernatant through a low-protein-binding 0.45-μm syringe filter to remove cell debris. Collect the supernatant in a 15-ml tube.
▲CRITICAL STEP Virus is ideally used as fresh as possible to achieve the required high transduction efficiencies.
∎PAUSE POINT CellTag virus can be stored for several days at 2–8 °C or long term at −80 °C, although it is best used the day it is collected.
-
46
The CMV promoter in the CellTag construct drives expression of GFP to support accurate viral titering. Titer the virus according to the fluorescence titer assay from Addgene (https://www.addgene.org/protocols/fluorescence-titering-assay/). Higher-accuracy assessment of viral titer can be performed via flow cytometry. Alternatively, our imaging-based titration software can be used. Code and tutorials can be found here: https://github.com/morris-lab/CellTag-Titration.
Transduction of cells with CellTag lentivirus ● Timing 3 d
-
47
Day 1: Plate MEFs. Prepare a 0.1% (vol/vol) gelatin coating solution, diluting a 2% (vol/vol) stock using DPBS and filtering through a 0.22-μm syringe filter.
-
48
Coat a 6-well plate with 0.1% (vol/vol) gelatin solution and incubate for 30 min at RT.
-
49
Aspirate the gelatin solution and wash three times with DPBS. For the last wash, leave the DPBS in the well to prevent the well from drying out until the cells are ready to be plated.
-
50
After aspirating DPBS, plate MEFs at a density of 50,000 cells per well in a 6-well plate in MEF/293T culture medium. The cells will adhere to the plate.
▲CRITICAL STEP The starting cell population size is extremely important. To maximize the number and size of clones that can be detected and traced, we recommend keeping the starting number of cells to be CellTagged relatively small. In addition, the downstream choice of platform for single-cell capture should be considered when determining the starting cell number. For example, higher-efficiency cell capture platforms require fewer cells to be loaded, thus supporting the culture of smaller population sizes in earlier stages. These parameters can be explored using the CellTag Simulator at http://celltag.org/.
-
51
Day 2: Transduce cells plated in Step 47. Add 5 μg/ml protamine sulfate to viral supernatant generated in Steps 32–46 to enhance transduction efficiency. Polybrene (1–10 μg/ml) can be used as an alternative to protamine sulfate.
-
52
Remove medium from the cells plated in Step 47 and replace it with the viral supernatant. Incubate cells overnight at 37 °C.
▲CRITICAL STEP We transduce MEFs at an MOI of ~3–4, resulting in each cell expressing a unique combination of CellTags. This increases the confidence of downstream clone calling. With an MOI of ~3, we find that ~70% of MEFs express two or more unique CellTags. We use fresh viral supernatant for this step. However, the medium used to culture HEK293 cells can be incompatible for some cell types and applications. In these instances, we recommend the concentration of CellTag viral particles via ultracentrifugation68 and resuspension in an appropriate medium.
-
53
Day 3: Replace the cell culture medium with fresh medium.
At this stage, GFP-positive cells should begin to be visible. At 48 h after transduction, almost all cells should be GFP positive (Fig. 6a).
In initial experiments, we recommend a ‘trial run’ to assess cell response to CellTagging and any potential viral silencing. For our own transduction and culture of MEFs over a 10-week period, we observed that CellTag expression becomes weaker but is not fully silenced (Fig. 6b). This can be assessed visually by fluorescence microscopy, by flow cytometry or via single-cell sequencing as outlined from Step 69 onward. ? TROUBLESHOOTING
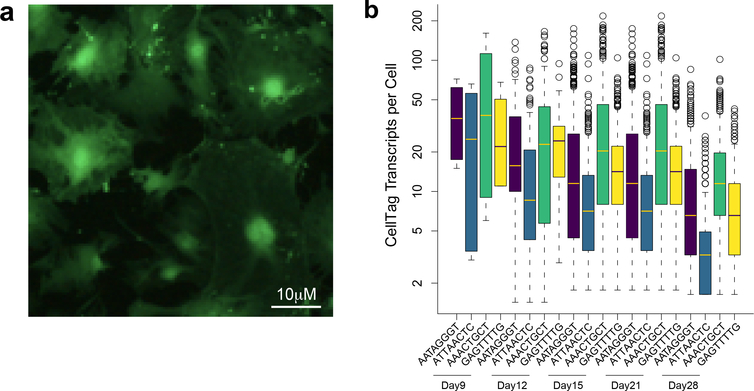
Fig. 6 |. Expression of CellTags.
a, Visualization of GFP-CellTag expression in MEFs 48 h after transduction. b, Expression levels of individual CellTags per cell over 3 weeks in a representative clone labeled by four unique CellTags (n = 1,998 cells). Expression diminishes over time but is not completely silenced.
Cell harvest and replating for clonal expansion ● Timing 4 weeks; system dependent
-
54
Harvest transduced MEFs for single-cell sequencing. Aspirate medium from the culture plate.
-
55
Wash the plate once with DPBS.
-
56
Add 0.5 ml of TrypLE Express per well of the 6-well plate and incubate for 5 min at 37 °C to dissociate cells from the plate.
-
57
Add 4 ml of MEF/293T medium to neutralize TrypLE Express.
-
58
Collect the cell suspension in a 15-ml tube and centrifuge at 200g for 5 min at RT.
-
59
Aspirate the supernatant from the tube and discard, without disturbing the cell pellet.
-
60
Resuspend cells in 5 ml of MEF/293T medium and carefully count cells using a hemocytometer.
-
61
Prepare a portion of cells for methanol fixation (adapted from Alles et al.66) by transferring a minimum of 10,000 cells into a 1.5-ml Eppendorf tube. Reserve the remaining cells on ice.
-
62
Centrifuge the cells to be methanol fixed at 300g for 5 min at RT.
-
63
Aspirate and discard the supernatant. Resuspend the cell pellet in 1 ml of ice-cold DPBS.
-
64
Centrifuge the cells at 300g for 5 min at RT.
-
65
Aspirate and discard the supernatant and resuspend the cell pellet in 200 μl of ice-cold DPBS.
-
66
Add 800 μl of methanol (pre-chilled to −20 °C) dropwise while gently mixing the cell suspension using a vortex on a low-speed setting.
-
67
Place the cell suspension on ice for 15 min.
For longitudinal analyses, we recommend methanol fixation of cells at each time point, capturing cells and performing library preparation in one batch.
For 10x Genomics-based single-cell processing, we methanol fix a minimum of 10,000 cells per sample. Ideally, 25,000 cells are fixed to yield ~10,000 single-cell transcriptomes per sample. For Drop-Seq, we fix a minimum of 150,000 cells per sample.
∎PAUSE POINT Cell suspensions can be stored long term at −80 °C. Using this methanol fixation approach, we have recovered high-quality single-cell transcriptomes from cells stored for up to 12 months.
-
68
Replate the remaining cells for continued clonal expansion, according to Step 47.
For replated MEFs, we CellTagged cells with pooled CellTag-V2 library 5 d after CellTag-V1 transduction and pooled CellTag-V3 library 10 d after V2 CellTagging. At each cell collection (at intervals of 3–7 d), we recovered and fixed 10,000 cells for single-cell profiling, replating the remaining cells.
▲CRITICAL STEP Analysis of clonal expansion can be achieved with a single round of CellTagging. For more complex lineage reconstruction to support trajectory analysis, additional rounds of CellTagging with V2 and V3 pooled libraries are required. This strategy leverages the unique index sequences preceding each variable CellTag region, supporting demultiplexing and reconstruction of lineage relationships.
Single-cell library preparation ● Timing 2 d
-
69
Following harvest of all samples in the study, rehydrate methanol-fixed cells by first placing them on ice for 15 min to equilibrate them to 4 °C.
-
70
Centrifuge cells at 300g for 5 min at RT and then remove and discard the supernatant.
-
71
Resuspend cells in 0.04% (wt/vol) BSA + 1 mM DTT + 0.2 U/μl RNase inhibitor in 3× SSC (diluted from the 20× stock with nuclease-free water) buffer to obtain a suspension of 700–1,200 cells per μl.
-
72
Process cells for single-cell capture, library preparation and sequencing using 10x Genomics or Drop-Seq platforms, according to standard protocols (https://support.10xgenomics.com/single-cell-gene-expression).
We aim to sequence the cells to a depth of ≥30,000 reads per cell. Overall, if cell population sizes are maintained relatively small while the proportions of cells sequenced is high, the number and size of clones detected are increased.
▲CRITICAL STEP 10x Genomics Chromium 3′ Reagent Kits were used for scRNA-seq. Do not use Chromium 5′ Reagent Kits as we estimate that the fragmentation step in this library preparation results in the loss of CellTag motifs from ~50% of GFP transcripts. We have successfully used Chromium 3′ V2 and V3 Reagent Kits, from both cells and nuclei, for library preparation and recovery of CellTag information.
∎PAUSE POINT Libraries can be stored long term at −20 °C. If storing for >1 month, the library should be requantified to assess possible degradation.
Single-cell analysis, clone calling and lineage reconstruction ● Timing 3 d for preliminary analysis
-
73
Refer to Fig. 7 for an overview of analytical Steps 73–79. Process raw reads using Cell Ranger: https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger. Starting from the aligned data (BAM) from the 10x Cell Ranger pipeline, using Rsamtools, the BAM file is read line by line to identify cell barcodes, the CellTag motif and unique molecular identifiers (UMIs). From this information, CellTag UMI counts are quantified and a count matrix is constructed.
-
74
Following alignment, process CellTags using our R-based package, ‘CellTagR’ (Supplementary Manual 1 and Supplementary Software 1), and a walkthrough at https://github.com/morris-lab/CellTagR, supporting recovery of CellTag expression from FASTQ and BAM files. First, process the raw CellTag UMI count matrices to remove CellTags represented by only one transcript and then binarize the matrix.
-
75
Apply error correction to collapse similar barcodes on a cell-by-cell basis, resolving any CellTag errors arising during PCR amplification or sequencing.
For this step we use Starcode69, which calculates the Levenshtein distances between DNA sequences to collapse similar CellTags, increasing the sensitivity and specificity of downstream clone calling.
-
76
Filter the matrix to include only those CellTags confirmed to exist in the library, from whitelisting in Step 31.
-
77
Filter the matrix to exclude cells expressing <2, and >20 CellTags.
-
78
Using the filtered CellTag matrix, perform Jaccard analysis to identify clonally related cells by calculating the similarity between CellTag signatures. Using hierarchical clustering, generate lists of cells with their associated clone IDs.
-
79
Connect clones across different rounds of CellTagging to reconstruct lineage relationships: assemble cells into sub-clusters according to clone identity and then connect sub-clusters to each other to build lineages of related cells. Visualize these lineages by force-directed graphing.
▲CRITICAL STEP For Step 78, we use a Jaccard score threshold of 0.7. Based on species mixing of CellTagged mouse and human cells, we found that this cutoff produces a low false-positive rate42. We performed a sensitivity analysis and found that lowering this threshold can increase the likelihood that unrelated cells will be falsely identified as being clonally related. Increasing this threshold can lead to cells being dropped from a clone, or clones being split into two sub-clones. This latter instance can arise if one CellTag comprising a clonal signature becomes prone to dropout or is silenced over time.
Our datasets on clonal and lineage dynamics during reprogramming can be explored at http://celltag.org/. Our CellTag calculator (Supplementary Software 2) is also available via this website to simulate expected clone sizes, aiding experimental design and troubleshooting.
Our raw data is available from GEO: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE99915.
Fig. 7 |. Processing CellTag data.
Flowchart of the steps required for alignment of raw sequence data to quantify CellTag UMIs and generate a transcriptome digital expression matrix (part 1). Flowchart of the steps involved in data processing using CellTagR package to call clones and reconstruct lineages (part 2).
Troubleshooting
To aid the design of CellTagging experiments, we provide an experimental simulator at http://celltag.org/ (Supplementary Software 2), along with a detailed troubleshooting guide. Extensive troubleshooting for 10x Genomics scRNA-seq library preparation is provided here: https://support.10xgenomics.com/single-cell-gene-expression. In this section, we highlight several steps in the CellTagging protocol where troubleshooting may be required.
Low complexity of CellTag libraries (Steps 6–31)
After pooled CellTag library amplification, plasmid preparations are sequenced to assess their complexity, i.e., how many unique CellTags are present in the library. For example, our existing CellTag-V1 pooled library contains ~2 × 104 unique CellTags, based on our whitelisting analysis (Step 31). If the pooled library contains an order of magnitude fewer CellTags than expected, it indicates that plasmid amplification was not optimal. In this instance, optimize the transformation of chemically competent cells and screen the numbers of colonies formed from the liquid culture. Following transduction and sequencing of cells, we detect almost 75% of whitelisted CellTags on average (Table 3). It is important to use sufficiently complex libraries for CellTagging in order to ensure that unrelated cells are not labeled with the same CellTag combinations by chance.
Table 3 |.
Expected library complexity at the beginning and end of the protocol
| CellTag library | No. of unique CellTags in library | No. of unique CellTags detected after sequencing (percentage) | No. of cells sequenced |
|---|---|---|---|
| CellTag-V1 | 19,973 | 12,933 (65) | 37,612 |
| CellTag-V2 | 4,934 | 4,487 (91) | 32,176 |
| CellTag-V3 | 5,737 | 3,655 (64) | 10,212 |
Cells not transduced with CellTags at high efficiency (Steps 47–53)
CellTag lentiviral transduction efficiency is initially assessed via visualization of GFP expression. If fewer than 70% of cells are GFP positive 48 h after transduction, this step needs to be refined to maximize the proportion of cells passing the tracking threshold (defined as the expression of two or more CellTags per cell). This can be achieved by increasing viral titer via optimization of pooled library transfection into 293T cells. In addition, using freshly prepared virus, or avoiding multiple freeze–thaw cycles of stored virus, may improve transduction efficiencies. Alternatively, virus can be spin concentrated68 or cells can be ‘spinfected’70 to enhance transduction efficiency. Finally, alternative additives such as polybrene may be used to maximize viral transduction.
A low percentage of profiled cells pass the CellTag expression threshold for cell tracking
After sequencing, data are processed through the CellTagR pipeline, where the average number of unique CellTags labeling each cell is calculated, along with the proportion of cells passing the two or more CellTag threshold to support clone calling. A low proportion of cells passing this threshold could indicate the following: (i) as above, cells are not transduced at a sufficient multiplicity of infection; (ii) the CellTag library used to label the cells is not sufficiently complex; or (iii) CellTags are silenced in the biological system of interest. This latter issue can be investigated by periodically assessing the average number of CellTags expressed per cell over a time course experiment. In our iEP reprogramming system, we initially assessed CellTag expression every 2 weeks over a 10-week period.
Overabundance of clonally related cells
Over the duration of an experiment, clones should gradually appear and increase in size, or in some cases decrease in size, over time. In the early time points of an experiment, days in the case of our CellTagged MEF expansion, the cell population should not be dominated by any individual clones. If overabundance of a particular clone is observed after processing of sequence data via CellTagR, this suggests an excessive false-positive clone-calling rate. This result would suggest that the CellTag library used to label the cells is insufficiently complex, leading to many unrelated cells being tagged with the same CellTag combinations.
Too few clones or small clones identified
The CellTag simulator can assist in setting expectations for the number and size of clones detected. If the number of clones called is lower than expected, or clones contain fewer cells than expected, this can impede informative lineage reconstruction. Sequencing only a small proportion of the labeled population can lead to this issue, which in turn decreases the probability that clonally related cells will be captured for analysis. This situation can be addressed by: (i) initiating the experiment with a smaller pool of cells, maximizing the proportion of cells analyzed; (ii) using a high-capture rate single-cell analysis platform; or (iii) in cases where clones contain too few cells, providing additional time for cells to divide. Finally, depending on the biological system under study, there may be limited cell division, in which case fewer and smaller clones will be called.
Lineage collisions are detected
Following construction of lineage trees, in a small proportion (~4%) of called clones, we have observed that clones labeled in the second and third rounds of CellTagging arise from different ancestors42. This can indicate two issues. (i) CellTag libraries lack complexity and do not uniquely label independent cells. This is addressed in the previous points, above. (ii) The clone-calling threshold (based on Jaccard score) is stringent and can result in clones being ‘split’ into sub-clones. This is caused by CellTag dropout or failure to correct more pronounced CellTag sequence errors. In this instance, after identification of such ‘collisions’, where a clone called from later labeling is derived from two seemingly independent clones, the CellTag signatures of these clones should be visually inspected to make a determination of whether the clones should be collapsed. Using this approach, we reduced the collision rate to ~1%42.
Timing
Steps 1–5, generation of complex CellTag libraries: 2 d
Steps 6–18, amplification of pooled CellTag libraries: 2 d
Steps 19–31, assessment of CellTag library complexity via sequencing: 1 d + sequencing
Steps 32–46, production of CellTag lentivirus: 6 d
Steps 47–53, transduction of cells with CellTag lentivirus: 3 d
Steps 54–68, cell harvest and replating for clonal expansion: 4 weeks; system dependent
Steps 69–72, single-cell library preparation: 2 d
Steps 73–79, downstream data processing: 3 d for preliminary analysis
Anticipated results
The protocol we have described here enables the user to label cells with CellTags in iterative rounds to enable clonal and lineage relationships to be defined. As CellTags are expressed as polyadenylated mRNA, both lineage information and cell identity can be captured in parallel, using high-throughput scRNA-seq. Associations between lineage and cell identity can then be investigated across the course of a biological process. Throughout the CellTagging protocol, there are several stages at which the success of the experiment can be assessed.
Evaluation of successful CellTagging
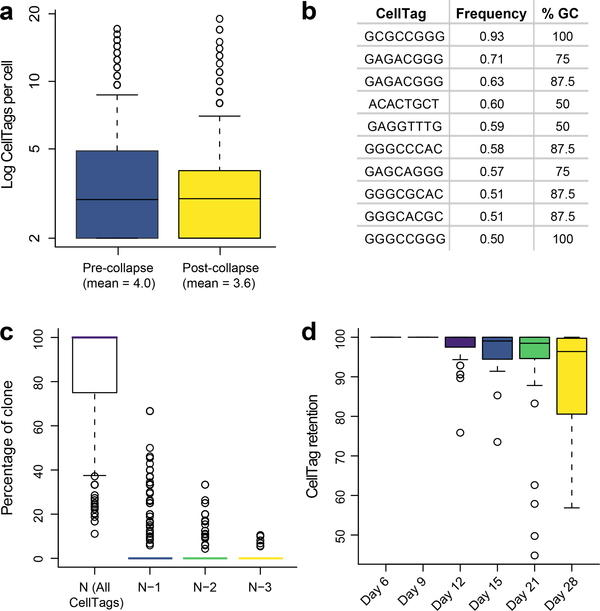
CellTagging efficiency can initially be assessed simply via visualization of GFP expression. A majority of cells expressing GFP within 48 h after viral transduction indicates that CellTag expression will be detected in each single-cell transcriptome. Beyond this initial assessment, processing of sequencing results via the CellTagR pipeline provides more detailed information. For example, we find that CellTags are collapsed in 61% of cells, with a mean of 0.4 CellTags corrected per cell (Fig. 8a). The ten most frequently collapsed CellTags contain a high average GC content of 80% (Fig. 8b); errors likely arise due to the relative difficulty of amplifying and sequencing GC-rich regions. In terms of CellTag detection, in almost 90% of cells possessing clonal relatives, we detect all CellTags comprising a complete signature, with only a mean 6% of cells missing one CellTag from the full signature (Fig. 8c). Partial silencing of viral gene expression accounts for some of this CellTag dropout, although 89% of the expected CellTag expression is retained 28 d after initial transduction (Fig. 8d). Provided that the CellTag library generated by the user is sufficiently complex, a large proportion of cells should be uniquely labeled with distinct CellTag combinations. For example, in fibroblasts, 60–70% of cells should pass the two or more unique CellTags per cell threshold to support clone calling.
Fig. 8 |. Expected CellTag expression metrics.
a, Mean number of CellTags detected per cell (n = 19,581 cells) before and after CellTag error correction via collapsing. b, Sequences and %GC content of the 10 most frequently collapsed CellTags. c, Analysis of CellTag signatures across n = 339 clones, 4,130 cells. 84.6% (±1.3% s.e.m.) of cells have full CellTag signature (N), with 6.1% (±0.75% s.e.m.) of cells missing one CellTag (N−1) of the full signature. d, Percentage of cells within a clone retaining a full CellTag expression signature, measured over 22 days. The full signature for each clone is defined by aggregating all CellTags associated with the cells of a specified clone. Singleton CellTags, likely arising from uncorrected errors or cell lysis, are removed from the signature.
Clone calling
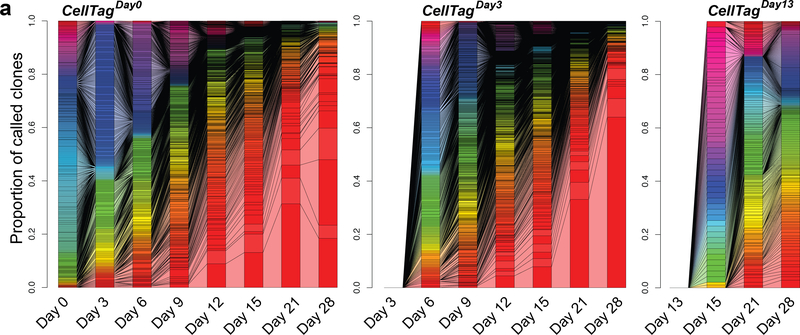
Following initial data processing via CellTagR, many clonal relationships between cells should be identified. Defining a clone as a group of three or more cells sharing significant overlap of their CellTag signature, the user should expect to see small clones of cells appearing at the beginning of the experiment, gradually increasing in size as the experiment progresses. We assess these clonal dynamics via connected bar graphs, providing a simple visualization of clone expansion and contraction over time (Fig. 9). While this is highly dependent on the system under study and the sampling rate, in our reprogramming system we frequently observe the collapse of initial clones, which we presume to be non-reprogrammed cells senescing after a period of growth. We also often observe the dominance of the population by one, or a handful, of clones. Our viral integration analysis did not point to this being a transduction artifact; rather, this reflects the normal growth and expansion of our reprogrammed cells42. The largest clones at the end of the experiment are derived from the earliest rounds of CellTagging, whereas later rounds give rise to smaller clones that have had less time to expand. This timing should also be taken into consideration in any experimental design and analysis.
Fig. 9 |. Clonal dynamics over a reprogramming time course experiment.
Connected bar plots showing individual clones as a proportion of all clones called at each time point for a MEF-to-iEP reprogramming time course, for each round of CellTagging (n = 14,088 cells across 1,120 clones). Connected bars denote clonal expansion and growth over time.
Lineage tree construction
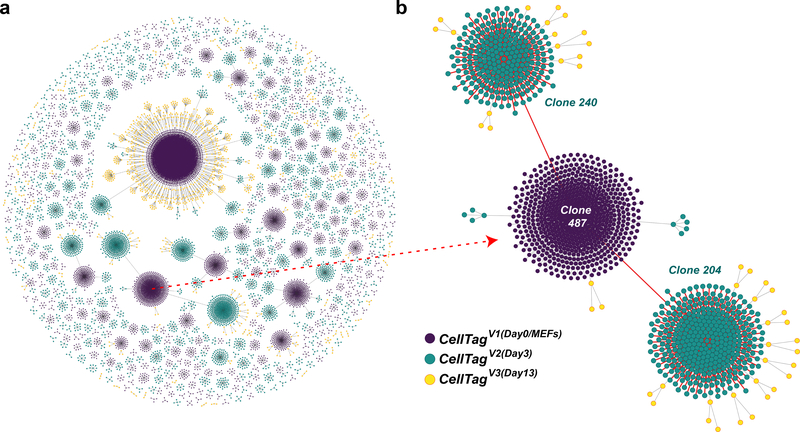
To reconstruct lineage relationships, cells are assembled into sub-clusters according to clone identity, and then sub-clusters are connected to each other to build lineages of related cells, visualized by force-directed graphing. By inspecting the lineages, there should be few lineage collisions, i.e., clonally related cells called from later rounds of labeling that are derived from independent ancestors. Few lineage collisions indicate a high-quality experiment where independent cells have been labeled with unique CellTag signatures. To produce more complete lineages including later rounds of CellTagged cells, we relaxed the definition of these later clones to consist of two or more related cells. Given efficient CellTagging, clonal expansion and a high proportion of the labeled cells captured, many lineages of varying size should be observed (Fig. 10). From this point, the single-cell transcriptome data, captured in parallel with this lineage information, can be used to explore cell identity. For example, specific lineages can be projected into low-dimensional space using a variety of methods17,18,71,72, differential expression analyses can be performed, and lineage restriction at various time points can be assessed, to list just a few possible approaches to support further exploration of the data.
Fig. 10 |. Lineages reconstructed from a reprogramming time course.
a, Force-directed graph of clonally related cells and lineages reconstructed from a reprogramming time course (1,031 clones, 12,932 cells). All lineages and clone distributions can be interactively explored using our companion website, CellTag Viz (http://www.celltag.org/). b, A detailed example of a lineage tree, where we follow a CellTagV1-labeled clone (cells labeled as MEFs, at day 0) and its descendants. Each node represents an individual cell, and edges represent clonal relationships between cells. Purple, CellTagV1 clones; blue, CellTagV2 clones (cells labeled at day 3, after reprogramming initiation); yellow, CellTagV3 clones (cells labeled at day 13, after reprogramming initiation).
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
CellTagging of fibroblast to iEP lineage reprogramming42 data are available via GEO: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE99915. The clones and lineages reconstructed from this dataset can be interactively explored via http://celltag.org/, along with our simulator to support CellTag experimental design. CellTagging constructs are available from Addgene: https://www.addgene.org/pooled-library/morris-lab-celltag/. Updates to this protocol will be provided at https://www.protocols.io/view/single-cell-mapping-of-lineage-and-identity-via-ce-yxifxke.
Code availability
Our R package, CellTagR, code and analysis tutorials are available via GitHub: https://github.com/morris-lab/CellTagR.
Supplementary Material
Acknowledgements
We thank members of the Morris laboratory for critical discussions and J. Dick (University of Toronto) for the gift of the pSMAL backbone. This work was funded by National Institutes of Health grants R01-GM126112, R21-HG009750 and P30-DK052574; Silicon Valley Community Foundation, Chan Zuckerberg Initiative Grants HCA-A-1704-01646 and HCA2-A-1708-02799; The Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital grant MI-II-2016-544. S.A.M. is supported by a Vallee Scholar Award; B.A.B. is supported by NIH-T32HG000045-18; and K.K. is supported by a Japan Society for the Promotion of Science Postdoctoral Fellowship.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41596-019-0247-2.
Peer review information Nature Protocols thanks Jennifer Adair, James Gagnon and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Regev A et al. The human cell atlas. Elife 6, 27041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han X et al. Mapping the mouse cell atlas by microwell-seq. Cell 172, 1091–1107.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Tabula Muris Consortium. et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang F et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Picelli S et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Ramsköld D et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Streets AM et al. Microfluidic single-cell whole-transcriptome sequencing. Proc. Natl Acad. Sci. USA 111, 7048–7053 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macosko EZZ et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein AMM et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng GXY et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg AB et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao J et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cusanovich DA et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174, 1309–1324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lareau CA et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat. Biotechnol. 37, 916–924 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao J et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361, 1380–1385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welch JD et al. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell 177, 1873–1887.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu X et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo M, Bao EL, Wagner M, Whitsett JA & Xu Y SLICE: determining cell differentiation and lineage based on single cell entropy. Nucleic Acids Res. 45, e54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji Z & Ji H TSCAN: Pseudo-time reconstruction and evaluation in single-cell RNA-seq analysis. Nucleic Acids Res. 44, e117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin J et al. Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell 17, 360–372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marco E et al. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc. Natl Acad. Sci. USA 111, 5643–5650 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Street K et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Setty M et al. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat. Biotechnol. 34, 637–645 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bendall SC et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 157, 714–725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.La Manno G et al. RNA velocity of single cells. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grün D et al. De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell 19, 266–277 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Schlitzer A, Chakarov S, Ginhoux F & Poidinger M Mpath maps multi-branching single-cell trajectories revealing progenitor cell progression during development. Nat. Commun. 7, 11988 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinreb C, Wolock S, Tusi BK, Socolovsky M & Klein AM Fundamental limits on dynamic inference from single-cell snapshots. Proc. Natl Acad. Sci. Usa. 115, E2467–E2476 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giecold G, Marco E, Garcia SP, Trippa L & Yuan G-C Robust lineage reconstruction from high-dimensional single-cell data. Nucleic Acids Res. 44, e122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lodato MA et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 350, 94–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leung ML et al. Single-cell DNA sequencing reveals a late-dissemination model in metastatic colorectal cancer. Genome Res. 27, 1287–1299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ludwig LS et al. Lineage tracing in humans enabled by mitochondrial mutations and single-cell genomics. Cell 176, 1325–1339.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kester L & van Oudenaarden A Single-cell transcriptomics meets lineage tracing. Cell Stem Cell 23, 166–179 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Lu R, Neff NF, Quake SR & Weissman IL Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat. Biotechnol. 29, 928–933 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porter SN, Baker LC, Mittelman D & Porteus MH Lentiviral and targeted cellular barcoding reveals ongoing clonal dynamics of cell lines in vitro and in vivo. Genome Biol. 15, R75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun J et al. Clonal dynamics of native haematopoiesis. Nature 514, 322–327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKenna A et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frieda KL et al. Synthetic recording and in situ readout of lineage information in single cells. Nature 541, 107–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt ST, Zimmerman SM, Wang J, Kim SK & Quake SR Quantitative analysis of synthetic cell lineage tracing using nuclease barcoding. ACS Synth. Biol. 6, 936–942 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biddy BA et al. Single-cell mapping of lineage and identity in direct reprogramming. Nature 564, 219–224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao Z et al. A single-cell roadmap of lineage bifurcation in human ESC models of embryonic brain development. Cell Stem Cell 20, 120–134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spanjaard B et al. Simultaneous lineage tracing and cell-type identification using CRISPR–Cas9-induced genetic scars. Nat. Biotechnol. 36, 469–473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raj B et al. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol. 36, 442–450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alemany A, Florescu M, Baron CS, Peterson-Maduro J & van Oudenaarden A Whole-organism clone tracing using single-cell sequencing. Nature 556, 108–112 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Wagner DE et al. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinreb C, Rodriguez-Fraticelli A, Camargo FD & Klein AM Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science eaaw3381 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo C et al. CellTag indexing: genetic barcode-based sample multiplexing for single-cell genomics. Genome Biol. 20, 90 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Galen P et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature 510, 268–72 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Turner DL & Cepko CL A common progenitor for neurons and glia persists in rat retina late in development. Nature 328, 131–136 (1987). [DOI] [PubMed] [Google Scholar]
- 52.Frank E & Sanes JR Lineage of neurons and glia in chick dorsal root ganglia: analysis in vivo with a recombinant retrovirus. Development 111, 895–908 (1991). [DOI] [PubMed] [Google Scholar]
- 53.Pei W et al. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548, 456–460 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan MM et al. Molecular recording of mammalian embryogenesis. Nature 1, 77–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalhor R et al. Developmental barcoding of whole mouse via homing CRISPR. Science 361, eaat9804 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raj B, Gagnon JA & Schier AF Large-scale reconstruction of cell lineages using single-cell readout of transcriptomes and CRISPR–Cas9 barcodes by scGESTALT. Nat. Protoc 13, 2685–2713 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalhor R, Mali P & Church GM Rapidly evolving homing CRISPR barcodes. Nat. Methods 14, 195–200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perli SD, Cui CH & Lu TK Continuous genetic recording with self-targeting CRISPR-Cas in human cells. Science 353, aag0511 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Morris SA et al. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell 158, 889–902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doulatov S et al. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell 13, 459–470 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kita-Matsuo H et al. Lentiviral vectors and protocols for creation of stable hESC lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLoS ONE 4, e5046 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hong S et al. Functional analysis of various promoters in lentiviral vectors at different stages of in vitro differentiation of mouse embryonic stem cells. Mol. Ther. 15, 1630–1639 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramezani A & Hawley RG Strategies to insulate lentiviral vector-expressed transgenes. Methods Mol. Biol. 614, 77–100 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benabdellah K, Gutierrez-Guerrero A, Cobo M, Muñoz P & Martín F A chimeric HS4-SAR insulator (IS2) that prevents silencing and enhances expression of lentiviral vectors in pluripotent stem cells. PLoS ONE 9, e84268 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pfaff N et al. A ubiquitous chromatin opening element prevents transgene silencing in pluripotent stem cells and their differentiated progeny. Stem Cells 31, 488–499 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Alles J et al. Cell fixation and preservation for droplet-based single-cell transcriptomics. BMC Biol. 15, 44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garfield AS Derivation of primary mouse embryonic fibroblast (PMEF) cultures. Methods Mol. Biol. 633, 19–27 (2010). [DOI] [PubMed] [Google Scholar]
- 68.Ichim CV & Wells RA Generation of high-titer viral preparations by concentration using successive rounds of ultracentrifugation. J. Transl. Med 9, 137 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zorita E, Cuscó P & Filion GJ Starcode: sequence clustering based on all-pairs search. Bioinformatics 31, 1913–1919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berggren WT, Lutz M & Modesto V General spinfection protocol in StemBook (ed The Stem Cell Community) (Harvard Stem Cell Institute, 2008). [PubMed] [Google Scholar]
- 71.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Becht E et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CellTagging of fibroblast to iEP lineage reprogramming42 data are available via GEO: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE99915. The clones and lineages reconstructed from this dataset can be interactively explored via http://celltag.org/, along with our simulator to support CellTag experimental design. CellTagging constructs are available from Addgene: https://www.addgene.org/pooled-library/morris-lab-celltag/. Updates to this protocol will be provided at https://www.protocols.io/view/single-cell-mapping-of-lineage-and-identity-via-ce-yxifxke.