Abstract
SARS-CoV-2-specific memory T cells will likely prove critical for long-term immune protection against COVID-19. Here, we systematically mapped the functional and phenotypic landscape of SARS-CoV-2-specific T cell responses in unexposed individuals, exposed family members, and individuals with acute or convalescent COVID-19. Acute-phase SARS-CoV-2-specific T cells displayed a highly activated cytotoxic phenotype that correlated with various clinical markers of disease severity, whereas convalescent-phase SARS-CoV-2-specific T cells were polyfunctional and displayed a stem-like memory phenotype. Importantly, SARS-CoV-2-specific T cells were detectable in antibody-seronegative exposed family members and convalescent individuals with a history of asymptomatic and mild COVID-19. Our collective dataset shows that SARS-CoV-2 elicits broadly directed and functionally replete memory T cell responses, suggesting that natural exposure or infection may prevent recurrent episodes of severe COVID-19.
Graphical Abstract
Highlights
-
•
Acute-phase SARS-CoV-2-specific T cells display an activated cytotoxic phenotype
-
•
Convalescent-phase SARS-CoV-2-specific T cells generate broad responses
-
•
Polyfunctional SARS-CoV-2-specific T cells also occur in seronegative individuals
Sekine et al. provide a functional and phenotypic map of SARS-CoV-2-specific T cells across the full spectrum of exposure, infection, and COVID-19 severity. They observe that SARS-CoV-2 elicits broadly directed and functionally replete memory T cells that may protect against recurrent episodes of severe COVID-19.
Introduction
The world changed in December 2019 with the emergence of a new zoonotic pathogen, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes a variety of clinical syndromes collectively termed coronavirus disease 2019 (COVID-19). At present, there is no vaccine against SARS-CoV-2, and the excessive inflammation associated with severe COVID-19 can lead to respiratory failure, septic shock, and ultimately, death (Guan et al., 2020; Wolfel et al., 2020; Wu and McGoogan, 2020). The overall mortality rate is 0.5%–3.5% (Guan et al., 2020; Wolfel et al., 2020; Wu and McGoogan, 2020). However, most people seem to be affected less severely and remain asymptomatic or develop only mild symptoms during COVID-19 (He et al., 2020b; Wei et al., 2020; Yang et al., 2020). It will therefore be critical for public health reasons to determine whether people with milder forms of COVID-19 develop robust immunity against SARS-CoV-2.
Global efforts are currently underway to map the determinants of immune protection against SARS-CoV-2. Recent data have shown that SARS-CoV-2 infection generates near-complete protection against rechallenge in rhesus macaques (Chandrashekar et al., 2020), and similarly, there is limited evidence of reinfection in humans with previously documented COVID-19 (Kirkcaldy et al., 2020). Further work is therefore required to define the mechanisms that underlie these observations and evaluate the durability of protective immune responses elicited by primary infection with SARS-CoV-2. Most correlative studies of immune protection against SARS-CoV-2 have focused on induction of neutralizing antibodies (Hotez et al., 2020; Robbiani et al., 2020; Seydoux et al., 2020; Wang et al., 2020). However, antibody responses are not detectable in all patients, especially those with less severe forms of COVID-19 (Long et al., 2020; Mallapaty, 2020; Woloshin et al., 2020). Previous work has also shown that memory B cell responses tend to be short lived after infection with SARS-CoV-1 (Channappanavar et al., 2014; Tang et al., 2011). In contrast, memory T cell responses can persist for many years (Le Bert et al., 2020; Tang et al., 2011; Yang et al., 2006) and, in mice, protect against lethal challenge with SARS-CoV-1 (Channappanavar et al., 2014).
SARS-CoV-2-specific T cells have been identified in humans (Grifoni et al., 2020; Ni et al., 2020). It has nonetheless remained unclear to what extent various features of the T cell immune response associate with serostatus and the clinical course of COVID-19. To address this knowledge gap, we characterized SARS-CoV-2-specific CD4+ and CD8+ T cells in outcome-defined cohorts of donors (total n = 206) from Sweden, which has experienced a more open spread of COVID-19 than many other countries in Europe (Habib, 2020).
Results
T Cell Perturbations in COVID-19
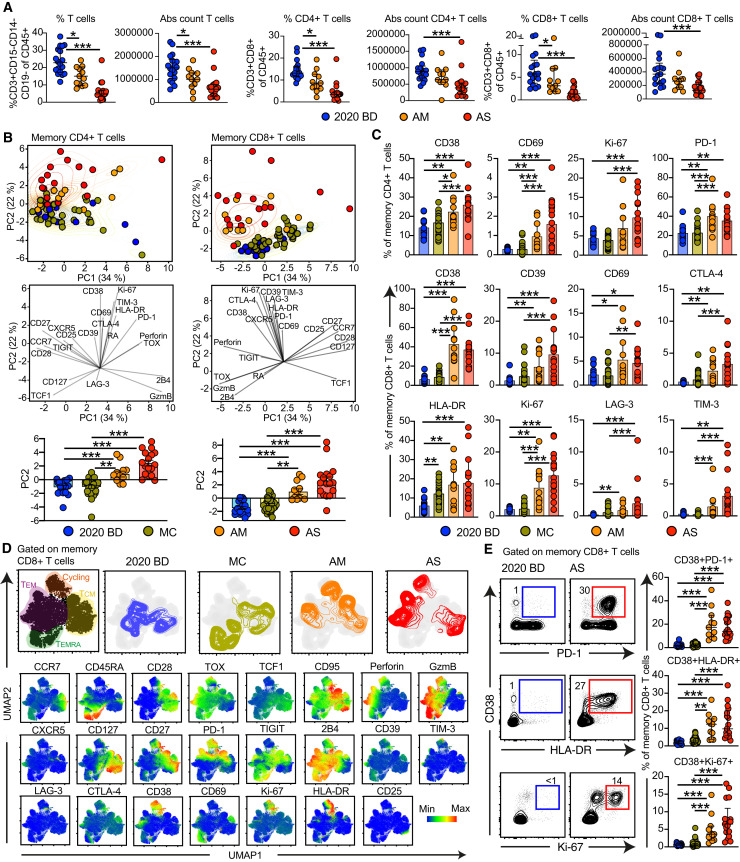
Our preliminary analyses showed that the absolute numbers and relative frequencies of CD4+ and CD8+ T cells were unphysiologically low in patients with acute moderate or severe COVID-19 (Figures 1 A, S1 A, and S1B). This finding has been reported previously (He et al., 2020a; Liu et al., 2020). We then used a 29-color flow cytometry panel to assess the phenotypic landscape of these immune perturbations in direct comparisons with healthy blood donors and individuals who had recovered from mild COVID-19 acquired early during the pandemic (February–March 2020). Cohort demographics are described in the STAR Methods section. None of these variables were associated with disease severity. The following parameters were measured in each sample: viability, C-C chemokine receptor type 7 (CCR7), cluster of differentiation 3 (CD3), CD4, CD8, CD14, CD19, CD25, CD27, CD28, CD38, CD39, CD45RA, CD69, CD95, CD127, cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), C-X-C chemokine receptor type 5 (CXCR5), granzyme B, human leukocyte antigen (HLA)-DR, Ki-67, lymphocyte activation gene 3 (LAG-3), programmed cell death protein 1 (PD-1), perforin, T cell factor 1 (TCF1), T cell immunoreceptor with immunoglobulin (Ig) and ITIM domains (TIGIT), T cell Ig and mucin domain-containing protein 3 (TIM-3), thymocyte selection-associated high-mobility group box factor (TOX), and natural killer cell receptor 2B4. Unbiased principal component analysis (PCA) revealed clear segregation between memory T cells from patients with acute moderate or severe COVID-19 and memory T cells from convalescent individuals and healthy blood donors (Figure 1B), driven largely by expression of CD38, CD69, Ki-67, and PD-1 in the CD4+ compartment and expression of CD38, CD39, CD69, CTLA-4, HLA-DR, Ki-67, LAG-3, and TIM-3 in the CD8+ compartment (Figures 1B, 1C, and S1C).
Figure 1.
T Cell Perturbations in COVID-19
(A) Dot plots summarizing the absolute counts and relative frequencies of CD3+ (left), CD4+ (center), and CD8+ T cells (right) in healthy blood donors from 2020 (2020 BD ) and patients with acute moderate (AM ) or acute severe COVID-19 (AS). Each dot represents one donor. Data are shown as median ± IQR. ∗p < 0.05, ∗∗∗p < 0.001. Kruskal-Wallis rank-sum test with Dunn’s post hoc test for multiple comparisons.
(B) Top: PCA plots showing the distribution and segregation of memory CD4+ and CD8+ T cells by group. Each dot represents one donor. Memory cells were defined by exclusion of naive cells (CCR7+ CD45RA+ CD95−). Center: PCA plots showing the corresponding trajectories of key markers that influenced the group-defined segregation of memory CD4+ and CD8+ T cells. Bottom: dot plots showing the group-defined distribution of markers in PC2. Each dot represents one donor. MC, individuals in the convalescent phase after mild COVID-19. ∗∗p < 0.01, ∗∗∗p < 0.001. Kruskal-Wallis rank-sum test with Dunn’s post hoc test for multiple comparisons.
(C) Dot plots summarizing the expression frequencies of activation/cycling markers among memory CD4+ (top) and CD8+ T cells (bottom) by group. Each dot represents one donor. Data are shown as median ± IQR. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Kruskal-Wallis rank-sum test with Dunn’s post hoc test for multiple comparisons.
(D) Top: UMAP plots showing the clustering of memory CD8+ T cells by group in relation to all memory CD8+ T cells (left). Bottom: UMAP plots showing the expression of individual markers (n = 3 donors per group). UMAP plots were based on all markers distinguished in the bottom row.
(E) Left: representative flow cytometry plots showing the expression of activation/cycling markers among memory CD8+ T cells by group. Numbers indicate percentages in the drawn gates. Right: dot plots showing the expression frequencies of activation/cycling markers among memory CD8+ T cells by group. Each dot represents one donor. Data are shown as median ± IQR. Key as in (B) and (C). ∗∗p < 0.01, ∗∗∗p < 0.001. Kruskal-Wallis rank-sum test with Dunn’s post hoc test for multiple comparisons.
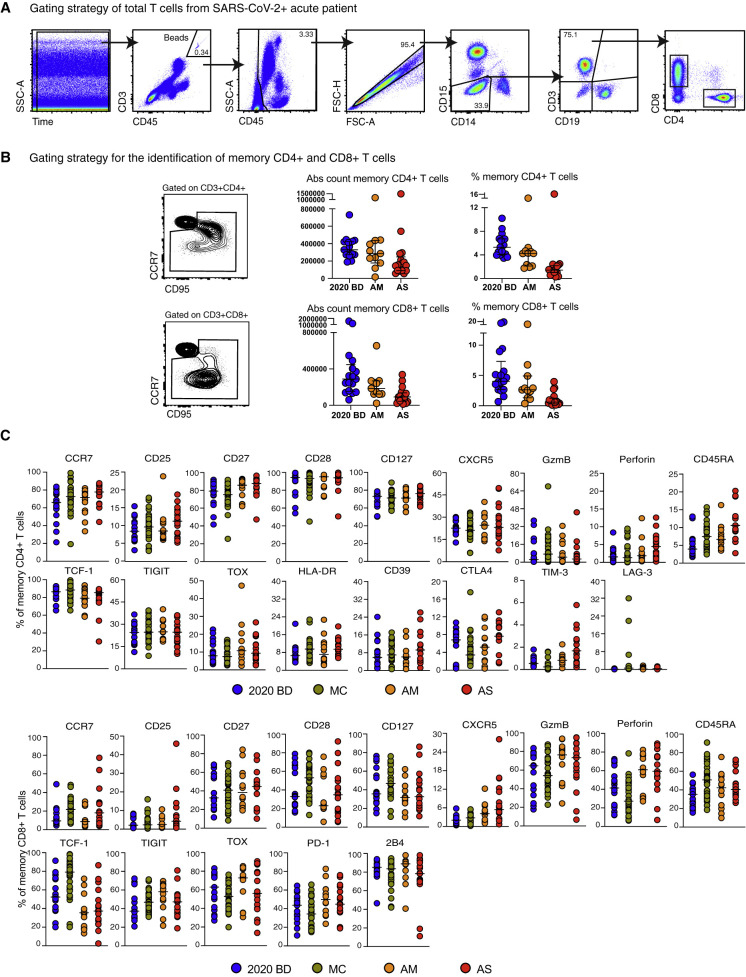
Figure S1.
Quantification and Characterization of CD4+ and CD8+ T Cells in COVID-19, Related to Figure 1
(A) Flow cytometric gating strategy for the identification and quantification of CD4+ and CD8+ T cells. (B) Left: flow cytometric gating strategy for the identification and quantification of memory CD4+ and CD8+ T cells. Right: dot plots summarizing the absolute numbers and relative frequencies of memory CD4+ and CD8+ T cells by group. Each dot represents one donor. Data are shown as median ± IQR. 2020 BD: healthy blood donors from 2020 (n = 18). AM: patients with acute moderate COVID-19 (n = 11). AS: patients with acute severe COVID-19 (n = 17). (C) Dot plots summarizing the expression frequencies of phenotypic markers among memory CD4+ and CD8+ T cells by group. Each dot represents one donor. Bars indicate median values. 2020 BD: healthy blood donors from 2020 (n = 18). MC: individuals in the convalescent phase after mild COVID-19 (n = 31). AM: patients with acute moderate COVID-19 (n = 11). AS: patients with acute severe COVID-19 (n = 17).
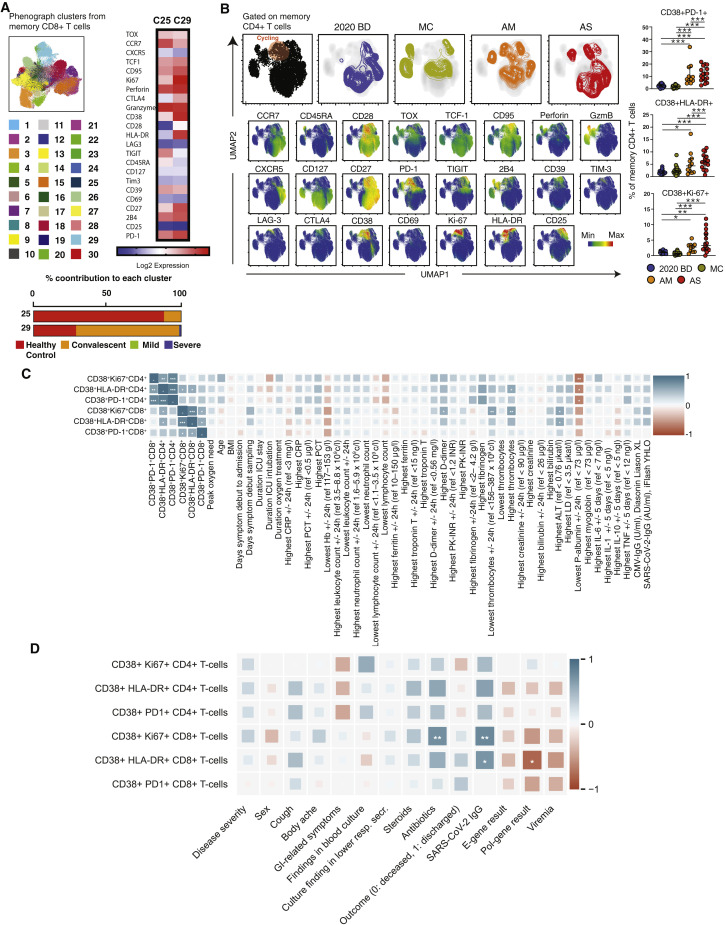
To extend these findings, we concatenated all memory CD4+ T cells and all memory CD8+ T cells from healthy blood donors, convalescent individuals, and patients with acute moderate or severe COVID-19. Phenotypically related cells were identified using the clustering algorithm PhenoGraph, and marker expression patterns were visualized using the dimensionality reduction algorithm Uniform Manifold Approximation and Projection (UMAP). Distinct topographical clusters were apparent in each group (Figures 1D, S2 A, and S2B). In particular, memory CD8+ T cells from patients with acute moderate or severe COVID-19 expressed a distinct cluster of markers associated with activation and the cell cycle, including CD38, HLA-DR, Ki-67, and PD-1 (Figures 1D and S2A). A similar pattern was observed among memory CD4+ T cells from patients with acute moderate or severe COVID-19 (Figure S2B). These findings were confirmed via manual gating of the flow cytometry data (Figure 1E). Correlative analyses further demonstrated that the activated/cycling phenotype was strongly associated with SARS-CoV-2-specific IgG levels and various clinical parameters, including age, hemoglobin concentration, platelet count, and plasma levels of alanine aminotransferase, albumin, D-dimer, fibrinogen, and myoglobin (Figures S2C and S2D), but less strongly associated with plasma levels of various inflammatory markers, including interleukin (IL)-1β, IL-10, and tumor necrosis factor (TNF) (Table S1).
Figure S2.
Phenograph and UMAP Clustering of Memory CD4+ and CD8+ T Cells with Correlative Analyses of Immune Activation Phenotypes versus Clinical Parameters in Acute COVID-19, Related to Figure 1
(A) Phenograph plots showing the clustering of memory CD8+ T cells and heatmap highlighting clusters 20 and 29. (B) Top left: UMAP plots showing the clustering of memory CD4+ T cells by group in relation to all memory CD4+ T cells (left). Bottom left: UMAP plots showing the expression of individual markers (n = 3 donors per group). Right: dot plots summarizing the expression frequencies of activation/cycling markers among memory CD4+ T cells by group. Each dot represents one donor. Data are shown as median ± IQR. 2020 BD: healthy blood donors from 2020 (n = 18). MC: individuals in the convalescent phase after mild COVID-19 (n = 31). AM: patients with acute moderate COVID-19 (n = 11). AS: patients with acute severe COVID-19 (n = 17). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Kruskal-Wallis rank-sum test with Dunn’s post hoc test for multiple comparisons. (C) and (D) Heatmaps summarizing the pairwise correlations between phenotypically defined subpopulations of memory CD4+ or CD8+ T cells and various clinical parameters in patients with acute COVID-19. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Spearman rank correlation.
Phenotypic Characteristics of SARS-CoV-2-Specific T Cells in Acute and Convalescent COVID-19
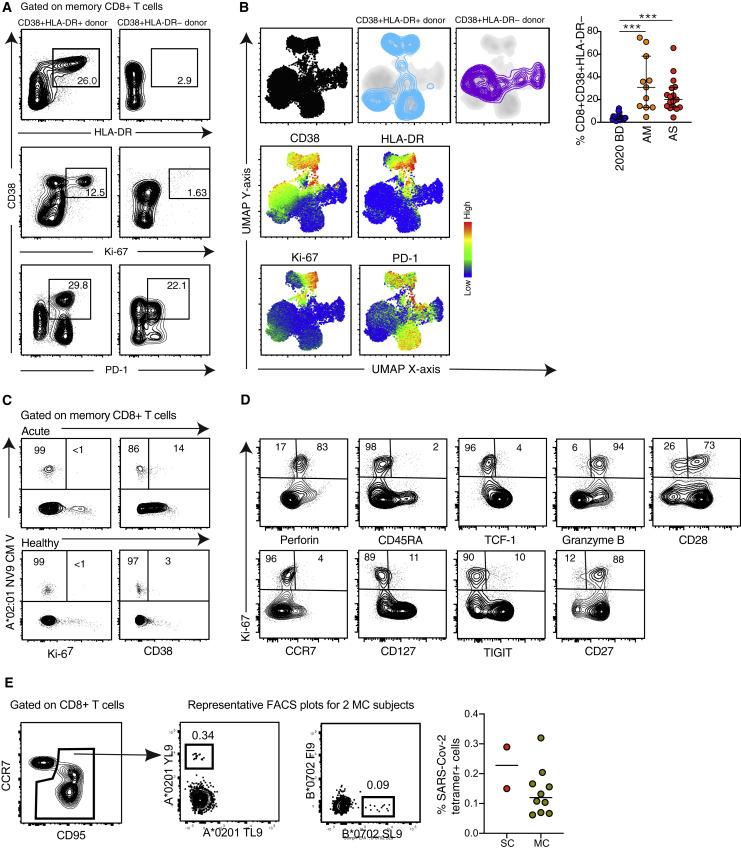
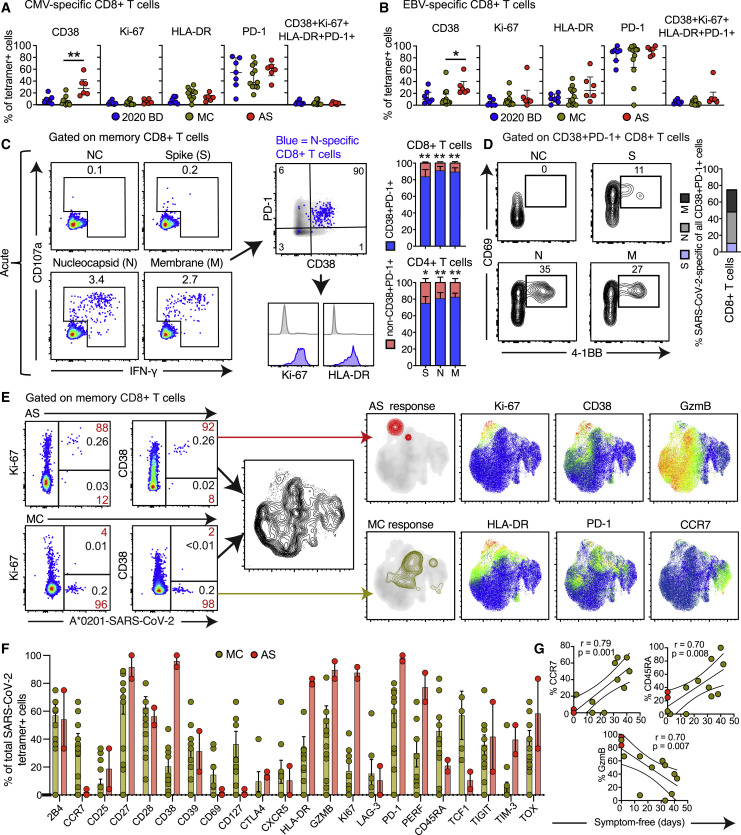
Unphysiologically high expression frequencies of CD38, potentially driven by a highly inflammatory environment, were observed consistently among memory CD8+ T cells from patients with acute moderate or severe COVID-19 (Figures S3 A and S3B). In line with these data, we found that CD8+ T cells specific for cytomegalovirus (CMV) or Epstein-Barr virus (EBV) more commonly expressed CD38, but not HLA-DR, Ki-67, or PD-1, in patients with acute moderate or severe COVID-19 compared with convalescent individuals and healthy blood donors, indicating limited bystander activation and proliferation during the early phase of infection with SARS-CoV-2 (Figures 2 A, 2B, and S3C). Of note, actively proliferating CD8+ T cells, defined by expression of Ki-67, exhibited a predominant CCR7− CD27+ CD28+ CD45RA− CD127− phenotype in patients with acute moderate or severe COVID-19 (Figure S3D), as reported previously in the context of vaccination and other viral infections (Buggert et al., 2018; Miller et al., 2008). On the basis of these findings, we used overlapping peptides spanning the immunogenic domains of the SARS-CoV-2 spike, membrane, and nucleocapsid proteins to stimulate peripheral blood mononuclear cells (PBMCs) from patients with acute moderate or severe COVID-19. A vast majority of responding CD4+ and CD8+ T cells displayed an activated/cycling (CD38+ HLA-DR+ Ki67+ PD-1+) phenotype (Figure 2C). These results were confirmed using an activation-induced marker (AIM) assay to measure upregulation of CD69 and 4-1BB (CD137), suggesting that most CD38+ PD-1+ CD8+ T cells were specific for SARS-CoV-2 (Figure 2D).
Figure S3.
Immune Activation Patterns in Acute COVID-19, Related to Figure 2
(A) Representative flow cytometry plots showing the expression of activation/cycling markers among memory CD8+ T cells in patients with acute severe COVID-19. Numbers indicate percentages in the drawn gates. (B) Top left: UMAP plots showing the clustering of memory CD8+ T cells by phenotype in relation to all memory CD8+ T cells (left). Bottom left: UMAP plots showing the expression of individual markers (n = 1 donor per group). Right: dot plot summarizing the frequencies of CD38+ HLA-DR− memory CD8+ T cells by group. Each dot represents one donor. Data are shown as median ± IQR. 2020 BD: healthy blood donors from 2020 (n = 18). AM: patients with acute moderate COVID-19 (n = 11). AS: patients with acute severe COVID-19 (n = 17). ∗∗∗p < 0.001. Kruskal-Wallis rank-sum test with Dunn’s post hoc test for multiple comparisons. (C) Representative flow cytometry plots showing the expression of activation/cycling markers among CMV-specific memory CD8+ T cells in a healthy control and a patient with acute severe COVID-19. Numbers indicate percentages in the drawn gates. (D) Representative flow cytometry plots showing the phenotype of Ki-67+ memory CD8+ T cells in a patient with acute severe COVID-19. Numbers indicate percentages in the drawn gates. (E) Left: representative flow cytometry plots showing SARS-CoV-2-specific tetramer+ CD8+ T cells in two donors from group MC. Right: dot plot summarizing the frequencies of SARS-CoV-2-specific tetramer+ CD8+ T cells in donors from groups MC (n = 10) and SC (n = 2).
Figure 2.
Phenotypic Characteristics of SARS-CoV-2-Specific T Cells in Acute and Convalescent COVID-19
(A and B) Dot plots summarizing the expression frequencies of activation/cycling markers among tetramer+ CMV-specific (A) or EBV-specific CD8+ T cells (B) by group. Each dot represents one specificity in one donor. Data are shown as median ± IQR. ∗p < 0.05, ∗∗p < 0.01. Kruskal-Wallis rank-sum test with Dunn’s post hoc test for multiple comparisons.
(C) Representative flow cytometry plots (left) and bar graphs (right) showing the expression of activation/cycling markers among CD107a+ and/or IFN-γ+ SARS-CoV-2-specific CD4+ and CD8+ T cells (n = 6 donors). Numbers indicate percentages in the drawn gates. Data are shown as median ± IQR. NC, negative control. ∗p < 0.05, ∗∗p < 0.01. Paired t test or Wilcoxon signed-rank test.
(D) Representative flow cytometry plots (left) and bar graph (right) showing the upregulation of CD69 and 4-1BB (AIM assay) among CD38+ PD-1+ SARS-CoV-2-specific CD8+ T cells (n = 6 donors). Numbers indicate percentages in the drawn gates. S, spike; M, membrane; N, nucleocapsid.
(E) Left: representative flow cytometry plots showing the expression of activation/cycling markers among tetramer+ SARS-CoV-2-specific CD8+ T cells by group (red) and by total frequency (black). Center: UMAP plot showing the clustering of memory CD8+ T cells. Right: UMAP plots showing the clustering of tetramer+ SARS-CoV-2-specific CD8+ T cells by group and the expression of individual markers (n = 2 donors).
(F) Dot plots summarizing the expression frequencies of all quantified markers among tetramer+ SARS-CoV-2-specific CD8+ T cells by group. Each dot represents combined specificities in one donor. Data are shown as median ± IQR.
(G) Bivariate plots showing the pairwise correlations between symptom-free days and the expression frequencies of CCR7, CD45RA, or granzyme B (GzmB). Each dot represents combined specificities in one donor. Key as in (F). Spearman rank correlation.
In further experiments, we used HLA class I tetramers as probes to detect CD8+ T cells specific for the predicted optimal epitopes from SARS-CoV-2 (Figure S3E; Table S2). A vast majority of tetramer+ CD8+ T cells in the acute phase of infection, but not during convalescence, displayed an activated/cycling phenotype (Figure 2E). In general, early SARS-CoV-2-specific CD8+ T cell populations were characterized by expression of immune activation molecules (CD38, HLA-DR, and Ki-67), inhibitory receptors (PD-1 and TIM-3), and cytotoxic molecules (granzyme B and perforin), whereas convalescent-phase SARS-CoV-2-specific CD8+ T cell populations were skewed toward an early differentiated memory (CCR7+ CD127+ CD45RA−/+ TCF1+) phenotype (Figure 2F). Importantly, the expression frequencies of CCR7 and CD45RA among SARS-CoV-2-specific CD8+ T cells were positively correlated with the number of symptom-free days after infection (CCR7: r = 0.79, p = 0.001; CD45RA: r = 0.70, p = 0.008), whereas the expression frequency of granzyme B among SARS-CoV-2-specific CD8+ T cells was inversely correlated with the number of symptom-free days after infection (r = 0.70, p = 0.007) (Figure 2G). Time from exposure was therefore associated with emergence of stem-like memory SARS-CoV-2-specific CD8+ T cells.
Functional Characteristics of SARS-CoV-2-Specific T Cells in Convalescent COVID-19
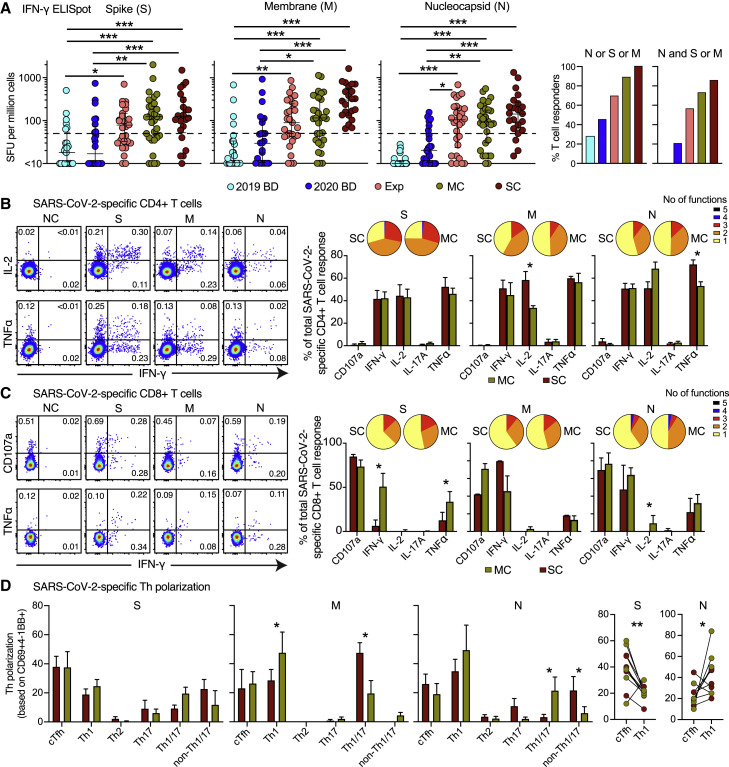
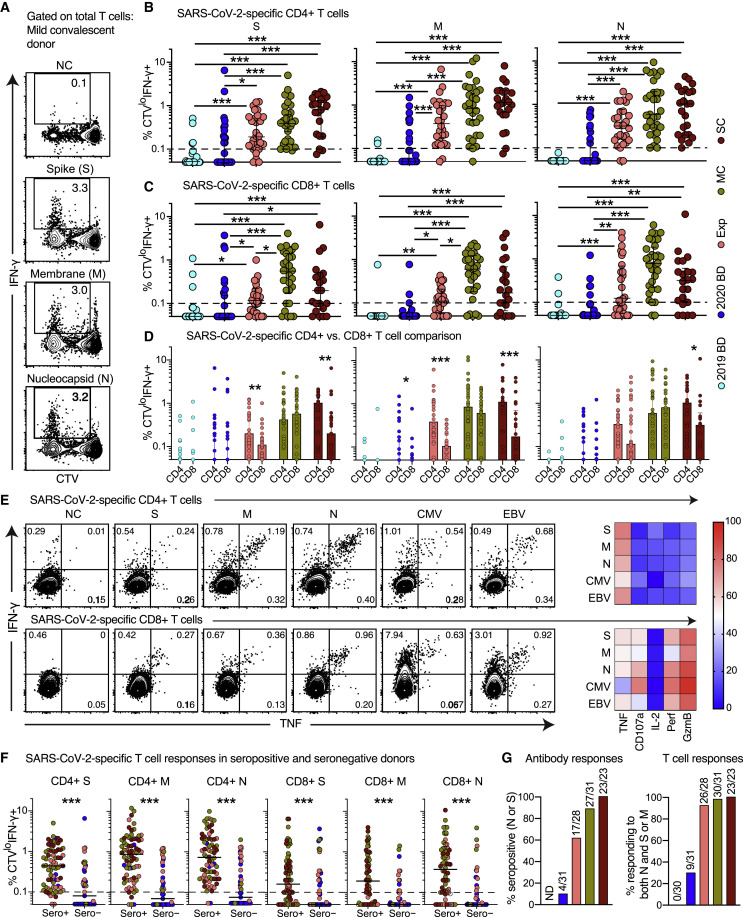
On the basis of these observations, we quantified functional SARS-CoV-2-specific memory T cell responses across five distinct cohorts, including healthy individuals who donated blood before or during the pandemic, family members who shared a household with convalescent individuals and were exposed at the time of symptomatic disease, and individuals in the convalescent phase after mild or severe COVID-19. We detected potentially cross-reactive T cell responses directed against the spike and/or membrane proteins in 28% of healthy individuals who donated blood before the pandemic, consistent with previous reports (Grifoni et al., 2020; Le Bert et al., 2020), but nucleocapsid reactivity was notably absent in this cohort (Figures 3 A, S4 A, and S4B). The highest response frequencies against any of these three proteins were observed in convalescent individuals who experienced severe COVID-19 (100%). Progressively lower response frequencies were observed in convalescent individuals with a history of mild COVID-19 (87%), exposed family members (67%), and healthy individuals who donated blood during the pandemic (46%) (Figure 3A).
Figure 3.
Functional Characteristics of SARS-CoV-2-Specific T Cells in Convalescent COVID-19
(A) Left: dot plots summarizing the frequencies of IFN-γ-producing cells responding to overlapping peptides spanning the immunogenic domains of the SARS-CoV-2 S, M, and N proteins by group (ELISpot assays). Each dot represents one donor. The dotted line indicates the cutoff for positive responses. Right: bar graph showing the frequencies of IFN-γ-producing cells responding to the internal (N) and surface antigens (M and/or S) of SARS-CoV-2 by group (ELISpot assays). 2019 BD, healthy blood donors from 2019; Exp, exposed family members; SC, individuals in the convalescent phase after severe COVID-19; SFU, spot-forming unit. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Kruskal-Wallis rank-sum test with Dunn’s post hoc test for multiple comparisons.
(B) and (C) Left: representative flow cytometry plots showing the functional profiles of SARS-CoV-2-specific CD4+ (B) and CD8+ T cells (C) from a convalescent individual (group MC). Numbers indicate percentages in the drawn gates. Right: bar graphs and pie charts summarizing the distribution of individual functions among SARS-CoV-2-specific CD4+ (B) and CD8+ T cells (C) from convalescent individuals in groups MC (n = 12) or SC (n = 14). Data are shown as median ± IQR. Key as in (A). ∗p < 0.05. Unpaired t test or Mann-Whitney U test.
(D) Left: bar graphs summarizing the functional polarization of SARS-CoV-2-specific CD4+ T cells from convalescent individuals in groups MC (n = 5) and SC (n = 6). Subsets were defined as CXCR5+ (cTfh), CCR4− CCR6− CXCR3+ CXCR5− (Th1), CCR4+ CCR6− CXCR3− CXCR5− (Th2), CCR4− CCR6+ CXCR3− CXCR5− (Th17), CCR4− CCR6+ CXCR3+ CXCR5− (Th1/17), and CCR4− CCR6− CXCR3− CXCR5− (non-Th1/2/17). Data are shown as median ± IQR. ∗p < 0.05. Unpaired t test or Mann-Whitney U test. Right: line graph comparing cTfh versus Th1 polarization by specificity in convalescent individuals from groups MC and SC. Each dot represents one donor. Key as in (A). ∗p < 0.05, ∗∗p < 0.01. Paired t test.
Figure S4.
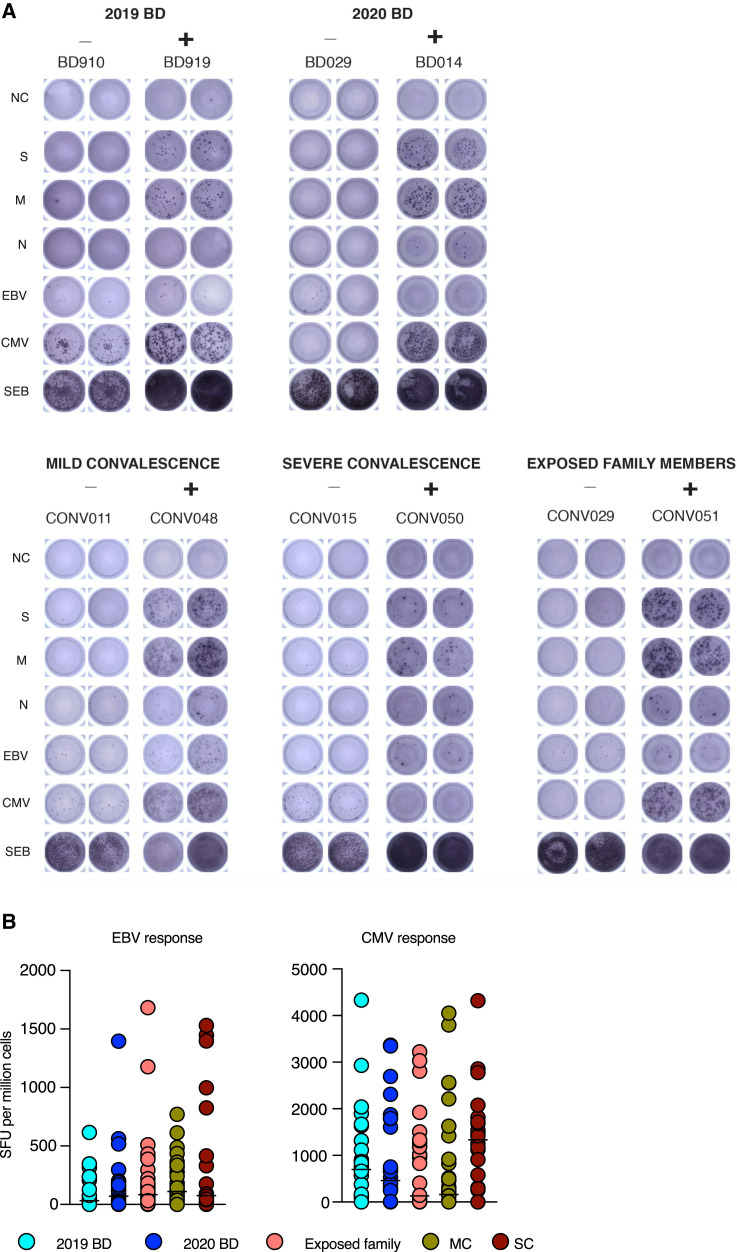
Quantification of Functional T Cell Reactivity in COVID-19, Related to Figure 3
(A) Representative images showing the detection of IFN-γ-producing cells responding to overlapping peptides spanning the immunogenic domains of the SARS-CoV-2 spike (S), membrane (M), and nucleocapsid proteins (N) by group (ELISpot assays). NC: negative control. EBV: Epstein-Barr virus. CMV: cytomegalovirus. SEB: staphylococcal enterotoxin B. (B) Dot plots summarizing the frequencies of IFN-γ-producing cells responding to optimal peptide epitopes derived from EBV BZLF1 and EBNA-1 (left) or CMV pp65 (right) by group (ELISpot assays). Each dot represents the mean of combined specificities in one donor. Bars indicate median values. No significant differences were detected among groups for any specificity. 2019 BD: healthy blood donors from 2019 (n = 25). 2020 BD: healthy blood donors from 2020 (n = 24). Exp: exposed family members (n = 30). MC: individuals in the convalescent phase after mild COVID-19 (n = 31). SC: individuals in the convalescent phase after severe COVID-19 (n = 22). SFU: spot-forming unit.
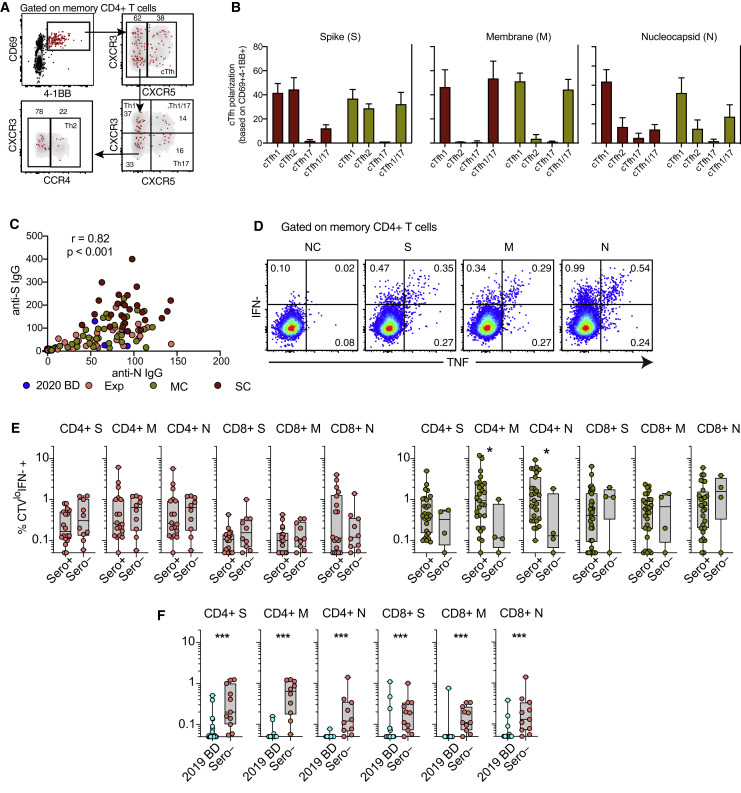
To assess the functional capabilities of SARS-CoV-2-specific memory CD4+ and CD8+ T cells in convalescent individuals, we stimulated PBMCs with the overlapping spike, membrane, and nucleocapsid peptide sets and measured a surrogate marker of degranulation (CD107a) along with production of interferon (IFN)-γ, IL-2, and TNF (Figures 3B and 3C). SARS-CoV-2-specific CD4+ T cells predominantly expressed IFN-γ, IL-2, and TNF (Figure 3B), whereas SARS-CoV-2-specific CD8+ T cells predominantly expressed IFN-γ and mobilized CD107a (Figure 3C). We then used the AIM assay to determine the functional polarization of SARS-CoV-2-specific CD4+ T cells. Interestingly, spike-specific CD4+ T cells were skewed toward a circulating T follicular helper (cTfh) profile, suggesting a key role in the generation of potent antibody responses, whereas membrane-specific and nucleocapsid-specific CD4+ T cells were skewed toward a Th1 or a Th1/Th17 profile (Figures 3D, S5 A, and S5B).
Figure S5.
Functional Polarization of SARS-CoV-2-Specific Memory CD4+ T Cells, Antibody Correlations, and Comparative Analyses of SARS-CoV-2-Specific CD4+ and CD8+ T Cell Responses versus Serostatus in COVID-19, Related to Figures 3 and 4
(A) Representative flow cytometry plots showing the identification of memory CD4+ T cells responding to overlapping peptides spanning the immunogenic domains of the SARS-CoV-2 nucleocapsid protein by subset (AIM assay). Subsets were defined as CXCR5+ (cTfh), CCR4− CCR6− CXCR3+ CXCR5− (Th1), CCR4+ CCR6− CXCR3− CXCR5− (Th2), CCR4− CCR6+ CXCR3− CXCR5− (Th17), CCR4− CCR6+ CXCR3+ CXCR5− (Th1/17), and CCR4− CCR6− CXCR3− CXCR5− (non-Th1/2/17). (B) Bar graphs summarizing the functional polarization of memory CXCR5+ (cTfh) CD4+ T cells responding to overlapping peptides spanning the immunogenic domains of the SARS-CoV-2 spike (S), membrane (M), and nucleocapsid proteins (N). Data are shown as median ± IQR. Key as in C. (C) Correlation between anti-spike (S) and anti-nucleocapsid (N) IgG levels. Each dot represents one donor. 2020 BD: healthy blood donors from 2020 (n = 31). Exp: exposed family members (n = 28). MC: individuals in the convalescent phase after mild COVID-19 (n = 31). SC: individuals in the convalescent phase after severe COVID-19 (n = 23). Spearman rank correlation. (D) Representative flow cytometry plots showing functional SARS-CoV-2-specific memory CD4+ T cell responses in a seronegative convalescent donor (group MC). Numbers indicate percentages in the drawn gates. NC: negative control. S: spike. M: membrane. N: nucleocapsid. (E) Dot plots summarizing SARS-CoV-2-specific CD4+ and CD8+ T cell responses versus serostatus in exposed family members (left) and individuals in the convalescent phase after mild COVID-19 (right). Each dot represents one donor. Data are shown as median ± IQR. S: spike. M: membrane. N: nucleocapsid. ∗p < 0.05. Mann-Whitney U test. (F) Dot plots summarizing SARS-CoV-2-specific CD4+ and CD8+ T cell responses in exposed seronegative family members and unexposed healthy blood donors (group 2019 BD). Each dot represents one donor. Data are shown as median ± IQR. ∗p < 0.05. Mann-Whitney U test.
Antibody Responses and Proliferative Capabilities of SARS-CoV-2-Specific T Cells in Convalescent COVID-19
In a final series of experiments, we assessed the recall capabilities of SARS-CoV-2-specific CD4+ and CD8+ T cells in convalescent individuals, exposed family members, and healthy blood donors. Proliferative responses were identified by tracking the progressive dilution of a cytoplasmic dye (CellTrace Violet [CTV]) after stimulation with the overlapping spike, membrane, and nucleocapsid peptide sets, and functional responses to the same antigens were evaluated 5 days later by measuring the production of IFN-γ (Blom et al., 2013; Buggert et al., 2014). Anamnestic responses in the CD4+ and CD8+ T cell compartments, quantified as a function of CTVlow IFN-γ+ events (Figure 4 A), were detected in most convalescent individuals (mild COVID-19 [MC] = 96%, severe COVID-19 [SC] = 100%) and exposed family members (92%) (Figures 4B and 4C). SARS-CoV-2-specific CD4+ T cell responses were proportionately larger overall than the corresponding SARS-CoV-2-specific CD8+ T cell responses (exposed family members = 1.8-fold, MC = 1.4-fold, SC = 1.8-fold) (Figure 4D). In addition, most IFN-γ+ SARS-CoV-2-specific CD4+ T cells produced TNF, and most IFN-γ+ SARS-CoV-2-specific CD8+ T cells expressed granzyme B and perforin (Figure 4E).
Figure 4.
Antibody Responses and Proliferation Capabilities of SARS-CoV-2-Specific T Cells in Convalescent COVID-19
(A) Representative flow cytometry plots showing the proliferation (CTV−) and functionality (IFN-γ+) of SARS-CoV-2-specific T cells from a convalescent individual (group MC) after stimulation with overlapping peptides spanning the immunogenic domains of the SARS-CoV-2 S, M, and N proteins. Numbers indicate percentages in the drawn gates.
(B) and (C) Dot plots summarizing the frequencies of CTV− IFN-γ+ SARS-CoV-2-specific CD4+ (B) and CD8+ T cells (C) by group and specificity. Each dot represents one donor. The dotted line indicates the cutoff for positive responses. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Kruskal-Wallis rank-sum test with Dunn’s post hoc test for multiple comparisons.
(D) Dot plots comparing the frequencies of CTV− IFN-γ+ SARS-CoV-2-specific CD4+ versus CD8+ T cells by group and specificity. Each dot represents one donor. Data are shown as median ± IQR. Key as in (B) and (C). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Paired t test or Wilcoxon signed-rank test.
(E) Left: representative flow cytometry plots showing the production of IFN-γ and TNF among CTV− virus-specific CD4+ (top) and CD8+ T cells (bottom) from a convalescent individual (group MC). Numbers indicate percentages in the drawn gates. Right: heatmaps summarizing the functional profiles of CTV− IFN-γ+ virus-specific CD4+ (top) and CD8+ T cells (bottom). Data are shown as mean frequencies (key).
(F) Dot plots summarizing the frequencies of CTV− IFN-γ+ SARS-CoV-2-specific CD4+ and CD8+ T cells by group, serostatus, and specificity. Each dot represents one donor. The dotted line indicates the cutoff for positive responses. Key as in (B) and (C). ∗∗∗p < 0.001. Mann-Whitney U test.
(G) Left: bar graph summarizing percent seropositivity by group. Right: bar graph summarizing the percentage of individuals in each group with detectable T cell responses directed against the internal (N) and surface antigens (M and/or S) of SARS-CoV-2.
Serological evaluations revealed a strong positive correlation between IgG responses directed against the spike protein of SARS-CoV-2 and IgG responses directed against the nucleocapsid protein of SARS-CoV-2 (r = 0.82, p < 0.001) (Figure S5C). Moreover, SARS-CoV-2-specific CD4+ and CD8+ T cell responses were present in seronegative individuals, albeit at lower frequencies compared with seropositive individuals (41% versus 99%, respectively) (Figure 4F). These discordant responses were nonetheless pronounced in some convalescent individuals with a history of mild COVID-19 (3 of 31), exposed family members (9 of 28), and healthy individuals who donated blood during the pandemic (5 of 31) (Figures 4F and S5D), often targeting the internal (nucleocapsid) and surface antigens (spike and/or membrane) of SARS-CoV-2 (Figure 4G). Higher frequencies of SARS-CoV-2-specific T cells were also found in exposed seronegative family members compared with unexposed donors (Figure S5E). Potent memory T cell responses were therefore elicited in the absence or presence of circulating antibodies, consistent with a non-redundant role as key determinants of immune protection against COVID-19 (Chandrashekar et al., 2020).
Discussion
We are currently facing the biggest global health emergency in decades, namely the devastating outbreak of COVID-19. In the absence of a protective vaccine, it will be critical to determine whether exposed and/or infected people, especially those with asymptomatic or very mild forms of the disease who likely act inadvertently as major transmitters, develop robust adaptive immunity against SARS-CoV-2 (Long et al., 2020).
In this study, we used a systematic approach to map cellular and humoral immune responses against SARS-CoV-2 in patients with acute moderate or severe COVID-19, individuals in the convalescent phase after mild or severe COVID-19, exposed family members, and healthy individuals who donated blood before (2019) or during the pandemic (2020). Individuals in the convalescent phase after mild COVID-19 were traced after returning to Sweden from endemic areas (mostly northern Italy). These donors exhibited robust memory T cell responses months after infection, even in the absence of detectable circulating antibodies specific for SARS-CoV-2, that may contribute to protection against severe COVID-19.
We found that T cell activation, characterized by expression of CD38, was a hallmark of acute COVID-19. Similar findings have been reported previously in the absence of specificity data (Huang et al., 2020; Thevarajan et al., 2020; Wilk et al., 2020). Many of these T cells also expressed HLA-DR, Ki-67, and PD-1, indicating a combined activation/cycling phenotype, which correlated with early SARS-CoV-2-specific IgG levels and, to a lesser extent, plasma levels of various inflammatory markers. Our data also showed that many activated/cycling T cells in the acute phase were functionally replete and specific for SARS-CoV-2. Equivalent functional profiles have been observed early after immunization with successful vaccines (Blom et al., 2013; Miller et al., 2008; Precopio et al., 2007). Accordingly, the expression of multiple inhibitory receptors, including PD-1, likely indicates early activation rather than exhaustion (Zheng et al., 2020a, 2020b).
Virus-specific memory T cells have been shown to persist for many years after infection with SARS-CoV-1 (Le Bert et al., 2020; Tang et al., 2011; Yang et al., 2006). In line with these observations, we found that SARS-CoV-2-specific T cells acquired an early differentiated memory (CCR7+ CD127+ CD45RA−/+ TCF1+) phenotype in the convalescent phase, as reported previously in the context of other viral infections and successful vaccines (Blom et al., 2013; Demkowicz et al., 1996; Fuertes Marraco et al., 2015; Precopio et al., 2007). This phenotype has been associated with stem-like properties (Betts et al., 2006; Blom et al., 2013; Demkowicz et al., 1996; Fuertes Marraco et al., 2015; Precopio et al., 2007). Accordingly, we found that SARS-CoV-2-specific T cells generated anamnestic responses to cognate antigens in the convalescent phase, characterized by extensive proliferation and polyfunctionality. Of particular note, we detected similar memory T cell responses directed against the internal (nucleocapsid) and surface proteins (membrane and/or spike) in some individuals lacking detectable circulating antibodies specific for SARS-CoV-2. Indeed, almost twice as many healthy individuals who donated blood during the pandemic had memory T cell responses versus antibody responses, implying that seroprevalence as an indicator may underestimate the extent of adaptive immune responses against SARS-CoV-2.
Our study was cross-sectional in nature and limited in terms of clinical follow-up and overall donor numbers in each outcome-defined group. It therefore remains to be determined whether robust memory T cell responses in the absence of detectable circulating antibodies can protect against severe forms of COVID-19. This scenario has nonetheless been inferred from previous studies of MERS and SARS-CoV-1 (Channappanavar et al., 2014; Li et al., 2008; Zhao et al., 2016, 2017), both of which have been shown to induce potent memory T cell responses that persist for years, in contrast to the corresponding antibody responses (Alshukairi et al., 2016; Shin et al., 2019; Tang et al., 2011). Antibody responses have also been shown to wane after infection with SARS-CoV-2 (Ibarrondo et al., 2020; Long et al., 2020), suggesting that transient humoral immunity is a general feature of coronavirus infections (Callow et al., 1990). However, the fact that memory B cells (Juno et al., 2020) and memory T cells are generated in response to SARS-CoV-2 suggests that natural infection may elicit protection from severe COVID-19. In line with these observations, none of the convalescent individuals in this study, including those with previous mild disease, experienced further episodes of COVID-19.
Of note, we detected cross-reactive T cell responses directed against the spike and/or membrane proteins of SARS-CoV-2 in 28% of unexposed healthy blood donors, consistent with a high degree of pre-existing immunity in the general population (Braun et al., 2020; Grifoni et al., 2020; Le Bert et al., 2020). Moreover, these particular data were derived from cryopreserved samples, so this figure might be considered as a lower bound estimate of overall prevalence (Owen et al., 2007). In this context, our findings most likely reflect widespread exposure to seasonal coronaviruses, which could shape the subsequent immune response to SARS-CoV-2. As such, it remains likely that a fraction of the anamnestic SARS-CoV-2-specific T cell response was initially induced by seasonal coronaviruses in seronegative individuals (Mateus et al., 2020). It is also tempting to speculate that such responses may provide at least partial protection against SARS-CoV-2, given that pre-existing T cell immunity has been associated with beneficial outcomes after challenge with the pandemic influenza virus strain H1N1 (Sridhar et al., 2013; Wilkinson et al., 2012).
Collectively, our data provide a functional and phenotypic map of SARS-CoV-2-specific T cell immunity across the full spectrum of exposure, infection, and disease. The observation that many individuals with asymptomatic or mild COVID-19 had highly durable and functionally replete memory T cell responses, not uncommonly in the absence of detectable humoral responses, further suggests that natural exposure or infection could prevent recurrent episodes of severe COVID-19.
Consortia
The members of the Karolinska COVID-19 Study Group are Mira Akber, Soo Aleman, Lena Berglin, Helena Bergsten, Niklas K. Björkström, Susanna Brighenti, Demi Brownlie, Marcus Buggert, Marta Butrym, Benedict Chambers, Puran Chen, Martin Cornillet Jeannin, Jonathan Grip, Angelica Cuapio Gomez, Lena Dillner, Jean-Baptiste Gorin, Isabel Diaz Lozano, Majda Dzidic, Johanna Emgård, Lars I. Eriksson, Malin Flodström Tullberg, Anna Färnert, Hedvig Glans, Sara Gredmark Russ, Alvaro Haroun-Izquierdo, Elizabeth Henriksson, Laura Hertwig, Habiba Kamal, Tobias Kamann, Jonas Klingstrom, Efthymia Kokkinou, Egle Kvedaraite, Hans-Gustaf Ljunggren, Sian Llewellyn-Lacey, Marco Loreti, Magalini Lourda, Kimia Maleki, Karl-Johan Malmberg, Nicole Marquardt, Christopher Maucourant, Jakob Michaelsson, Jenny Mjösberg, Kirsten Moll, Jagadees Muva, Johan Mårtensson, Pontus Nauclér, Anna Norrby-Teglund, Annika Olsson, Laura Palma Medina, Tiphaine Parrot, Björn Persson, André Perez-Potti, Lena Radler, Emma Ringqvist, Olga Rivera-Ballesteros, Olav Rooyackers, Johan Sandberg, John Tyler Sandberg, Takuya Sekine, Ebba Sohlberg, Tea Soini, Kristoffer Strålin, Anders Sönnerborg, Mattias Svensson, Janne Tynell, Renata Varnaite, Andreas Von Kries, Christian Unge, and David Wulliman.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| 1G1 (PE) [anti-CCR4] | BD Biosciences | 551120 (RRID:AB_394054) |
| 11A9 (PE-Cy7) [anti-CCR6] | BD Biosciences | 560620 (RRID:AB_1727440) |
| RPA-T8 or UCHT1 (BUV805) [anti-CD3] | BD Biosciences | 565515 (RRID:AB_2739277) |
| RPA-T8 (BUV395) [anti-CD8] | BD Biosciences | 563795 (RRID:AB_2722501) |
| M-A251 (PE-Cy5) [anti-CD25] | BD Biosciences | 560988 (RRID:AB_2033955) |
| CD28.2 (BUV563) [anti-CD28] | BD Biosciences | 564438 (RRID:AB_2738808) |
| HIT2 (BUV496) [anti-CD38] | BD Biosciences | 564657 (RRID:AB_2744376) |
| FN50 (BV750) [anti-CD69] | BD Biosciences | 563835 (RRID:AB_2738442) |
| FN50 (BUV737) [anti-CD69] | BD Biosciences | 564439 (RRID:AB_2722502) |
| DX2 (BB630) [anti-CD95] | BD Biosciences | Custom conjugate |
| H4A3 (PE-CF594) [anti-CD107a] | BD Biosciences | 562628 (RRID:AB_2737686) |
| BNI3 (BB755) [anti-CTLA-4] | BD Biosciences | 560938 (RRID:AB_2033942) |
| RF8B2 (APC-R700) [anti-CXCR5] | BD Biosciences | 565191 (RRID:AB_2739103) |
| GB11 (BB790) [anti-granzyme B] | BD Biosciences | Custom conjugate |
| G46-6 (BUV615) [anti-HLA-DR] | BD Biosciences | 565073 (RRID:AB_2722500) |
| MQ1-17H12 (APC-R700) [anti-IL-2] | BD Biosciences | 565136 (RRID:AB_2739079) |
| B56 (BB660) [anti-Ki-67] | BD Biosciences | Custom conjugate |
| T47-530 (BUV661) [anti-LAG-3] | BD Biosciences | Custom conjugate |
| δG9 (BB700) [anti-perforin] | BD Biosciences | Custom conjugate |
| 741182 (BUV737) [anti-TIGIT] | BD Biosciences | Custom conjugate |
| C1.7 (PE/Dazzle 594) [anti-2B4] | BioLegend | 393506 (RRID:AB_2734464) |
| G043H7 (APC-Cy7) [anti-CCR7] | BioLegend | 353211 (RRID:AB_10915272) |
| M5E2 (BV510) [anti-CD14] | BioLegend | 301842 (RRID:AB_2561946) |
| HIB19 (BV510) [anti-CD19] | BioLegend | 302242 (RRID:AB_2561668) |
| O323 (BV785) [anti-CD27] | BioLegend | 302832 (RRID:AB_2562674) |
| A1 (BV711) [anti-CD39] | BioLegend | 328227 (RRID:AB_2632893) |
| HI100 (BV421) [anti-CD45RA] | BioLegend | 304129 (RRID:AB_10900421) |
| HI100 (BV570) [anti-CD45RA] | BioLegend | 304131 (RRID:AB_10897946) |
| A019D5 (BV605) [anti-CD127] | BioLegend | 351334 (RRID:AB_2562022) |
| G025H7 (AF647) [anti-CXCR3] | BioLegend | 353712 (RRID:AB_10962948) |
| 4S.B3 (BV785) [anti-IFN-γ] | BioLegend | 502542 (RRID:AB_2563882) |
| EH12.2H7 (PE-Cy7) [anti-PD-1] | BioLegend | 367415 (RRID:AB_2616743) |
| F38-2E2 (BV650) [anti-TIM-3] | BioLegend | 345027 (RRID:AB_2565828) |
| Mab11 (BV650) [anti-TNF] | BioLegend | 502937 (RRID:AB_2561355) |
| 4B41 (BV421) [anti-4-1BB] | BioLegend | 309820 (RRID:AB_2563830) |
| C63D9 (AF488) [anti-TCF1] | Cell Signaling | 6444 (RRID:AB_2797627) |
| REA473 (A647) [anti-TOX] | Miltenyi Biotec | 130-107-838 (RRID:AB_2654224) |
| S3.5 (PE-Cy5.5) [anti-CD4] | Thermo Fisher Scientific | MHCD0418 (RRID:AB_10376013) |
| eBio64DEC17 (PE) [anti-IL-17A] | Thermo Fisher Scientific | 12-7179-42 (RRID:AB_1724136) |
| Purified anti-CD28/CD49d | BD Biosciences | 347690 (RRID:AB_647457) |
| Ultra-LEAF OKT3 Purified [anti-CD3] | BioLegend | 317326 (RRID: AB_11150592) |
| LIVE/DEAD Fixable Aqua Dead Cell Stain Kit | Thermo Fisher Scientific | Cat#L34957 |
| Biological Samples | ||
| Acute moderate blood samples | Karolinska Hospital | N/A |
| Acute severe blood samples | Karolinska Hospital | N/A |
| Mild convalescent blood samples | Karolinska Hospital | N/A |
| Severe convalescent blood samples | Karolinska Hospital | N/A |
| Exposed family member blood samples | Karolinska Hospital | N/A |
| 2020 blood donor samples | Karolinska Universitetslaboratoriet | N/A |
| 2019 blood donor samples | Karolinska Universitetslaboratoriet | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Synthetic CMV peptides | Peptides & Elephants GmbH | https://www.peptides.de/ |
| Synthetic EBV peptides | Peptides & Elephants GmbH | https://www.peptides.de/ |
| Synthetic overlapping SARS-CoV-2 peptides (Prot_M, N, S) | Miltenyi Biotec | https://www.miltenyibiotec.com/SE-en/ |
| Optimal SARS-CoV-2 peptides | Peptides & Elephants GmbH | https://www.peptides.de/ |
| Foxp3/TF Staining Buffer Set | Invitrogen | Cat#00-5523-00 |
| Staphylococcal Enterotoxin B (SEB) | Sigma-Aldrich | Cat#S4881 |
| Brefeldin A | BioLegend | Cat#420601 |
| BD GolgiStop (with Monensin) | BD Biosciences | Cat#554724 |
| BCIP/NBT Substrate | Mabtech | Cat#3650-10 |
| BL21(DE3)pLysS Competent Cells | Novagen | Cat#69451 |
| Triton X-100 Buffer | Sigma-Aldrich | Cat#X100 |
| 8M Urea Buffer | Sigma-Aldrich | Cat#51457 |
| Cysteamine | Sigma-Aldrich | Cat#M9768 |
| D-(+)-Biotin | Sigma-Aldrich | Cat#2031 |
| Critical Commercial Assays | ||
| iFLASH Anti-SARS-CoV-2 IgG Kit | YHLO | Cat#20210217 |
| LIAISON SARS-CoV-2 ELISA IgG Kit | DiaSorin | Cat#311450 |
| Human IFN-γ ELISpot Kit | Mabtech | Cat#3420-2A |
| BD Multitest 6-color TBNK reagent with BD Trucount tubes | BD Biosciences | Cat#337166 |
| CellTrace Violet Cell Proliferation Kit | Thermo Fisher Scientific | Cat#C34571 |
| Software and Algorithms | ||
| NetMHCpan EL 4.1 | DTU Bioinformatics | http://www.cbs.dtu.dk/services/NetMHCpan-4.1/ |
| FlowJo 10.6.1 | FlowJo | https://www.flowjo.com/ |
| UMAP plugin 2.2 | FlowJo | https://www.flowjo.com/ |
| GraphPad Prism 8.4.2 | GraphPad Software | https://www.graphpad.com/ |
| PhenoGraph 0.2.1 | PhenoGraph | https://omictools.com/phenograph-tool |
| RStudio | RStudio | https://rstudio.com/ |
| Python (scikit-learn 0.22.1) | Python | https://www.python.org/ |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Marcus Buggert (marcus.buggert@ki.se)
Material Availability
HLA class I tetramers can be generated and shared on a collaborative basis.
Data and Code Availability
The published article includes all data generated during this study. All codes are freely available at source.
Experimental Model and Subject Details
Human subjects
Maximal disease severity was assessed using the NIH Ordinal Scale and Sequential Organ Failure Assessment (SOFA) (Beigel et al., 2020; Singer et al., 2016). The NIH Ordinal Scale was defined as follows: (1) not hospitalized with no limitation of activities; (2) not hospitalized with limitation of activities and/or home oxygen requirement; (3) hospitalized but not requiring supplemental oxygen and no longer requiring ongoing medical care; (4) hospitalized and not requiring supplemental oxygen but requiring ongoing medical care; (5) hospitalized requiring supplemental oxygen; (6) hospitalized requiring non-invasive ventilation or the use of high-flow oxygen devices; (7) hospitalized receiving invasive mechanical ventilation or extracorporeal membrane oxygenation; and (8) death.
Donors were assigned to one of seven groups for the purposes of this study. AS: patients with acute severe disease requiring hospitalization in the high dependency or intensive care unit, with low-flow oxygen support (< 10 L/min), high-flow oxygen support, or invasive mechanical ventilation (n = 17). These patients had a median NIH Ordinal Scale score of 7 (IQR 6–7) and a median SOFA score of 6 (IQR 3–6) at the time of sampling 12–17 days after disease onset (47% were viremic, and 82% were antibody-seropositive for SARS-CoV-2). AM: patients with acute moderate disease requiring hospitalization and low-flow oxygen support (0–3 L/min; n = 10). These patients had a median NIH Ordinal Scale score of 5 (IQR 5–5) and a median SOFA score of 1 (IQR 1–1) at the time of sampling 11–14 days after disease onset (40% were viremic, and 50% were antibody-seropositive for SARS-CoV-2). SC: individuals in the convalescent phase after severe disease (n = 26). Samples were collected 42–58 days after disease onset, corresponding to 3–21 days after resolution of symptoms (100% were antibody-seropositive for SARS-CoV-2). MC: individuals in the convalescent phase after mild disease (n = 40). Samples were collected 49–64 days after disease onset, corresponding to 25–53 days after resolution of symptoms (85% were antibody-seropositive for SARS-CoV-2). Exp: family members who shared a household with donors in groups MC or SC (n = 30). These individuals were exposed at the time of symptomatic disease (21% remained asymptomatic, and 63% were antibody-seropositive for SARS-CoV-2). 2020 BD: individuals who donated blood at the Karolinska University Hospital in May 2020 (during the pandemic; n = 55). 2019 BD: individuals who donated blood at the Karolinska University Hospital between July and September 2019 (before the pandemic; n = 28).
PBMCs were isolated from heparin-coated tubes (groups AS, AM, SC, MC, Exp, and 2020 BD) or citrate-anticoagulated buffy coats (group 2019 BD). A separate serum separator tube was collected from each donor. All donors provided written informed consent in accordance with the Declaration of Helsinki. The study was approved by the Swedish Ethical Review Authority. Donor characteristics are summarized in Table S3, and immunological assay breakdowns are summarized in Table S4.
Method Details
Flow cytometry
PBMCs were isolated from venous blood samples via standard density gradient centrifugation and used immediately (groups AS, AM, SC, MC, Exp, and 2020 BD) or after cryopreservation in liquid nitrogen (group 2019 BD). Cells were washed in phosphate-buffered saline (PBS) supplemented with 2% fetal bovine serum (FBS) and 2 μM EDTA (FACS buffer) and stained with PE-conjugated or BV421-conjugated HLA class I tetramers and/or anti-CCR7–APC-Cy7 (clone G043H7; BioLegend) for 10 min at 37°C. Other surface markers were detected via the subsequent addition of directly conjugated antibodies at pre-titrated concentrations for 20 min at room temperature, and viable cells were identified by exclusion using a LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Thermo Fisher Scientific). Cells were then washed again in FACS buffer and fixed/permeabilized using a FoxP3 / Transcription Factor Staining Buffer Set (eBioscience). Intracellular markers were detected via the addition of directly conjugated antibodies at pre-titrated concentrations for 1 hr at 4°C. Stained cells were fixed in PBS containing 1% paraformaldehyde (PFA; Biotium) and stored at 4°C. Samples were acquired using a FACSymphony A5 (BD Biosciences). Data were analyzed with FlowJo software version 10.6.1 (FlowJo LLC). Gating strategies were described previously (Buggert et al., 2014, 2018). Core gates included singlet isolation (FSC-H versus FSC-A), live CD3 selection (CD3 versus Aqua, CD14, and CD19), lymphocyte enrichment (SSC-A versus FSC-A), CD4 or CD8 selection (CD4 versus CD8). Detailed gating strategies for individual markers are depicted in the relevant figures.
Peptides
Peptides corresponding to known optimal epitopes derived from CMV (pp65) and EBV (BZLF1 and EBNA-1), overlapping peptides spanning the immunogenic domains of the SARS-CoV-2 spike (Prot_S), membrane (Prot_M), and nucleocapsid proteins (Prot_N), and optimal peptides for the manufacture of HLA class I tetramers were synthesized at > 95% purity. Lyophilized peptides were reconstituted at a stock concentration of 10 mg/mL in DMSO and further diluted to 100 μg/mL in PBS.
Epitope prediction
Peptides were selected from full-length SARS-CoV-2 sequences spanning 82 different strains from 13 countries (National Center for Biotechnology Information). The predicted binding affinities of conserved 9-mer peptides for HLA-A∗0201 and HLA-B∗0702 were determined using NetMHCpan version 4.1 (Reynisson et al., 2020). Binders were defined by a threshold IC50 value of 500 nM. Strong binders were defined by a % Rank < 0.5, and weak binders were defined by a % Rank < 2 (Table S2). A total of 13 strong binders were identified for tetramer generation (Table S2).
Tetramers
HLA class I tetramers were generated as described previously (Price et al., 2005). Briefly, biotin-tagged HLA-A∗0201 and HLA-B∗0702 heavy chains were expressed under the control of a T7 promoter as insoluble inclusion bodies in Escherichia coli strain BL21(DE3)pLysS (Novagen). IPTG-induced Escherichia coli were lyzed via repeated freeze/thaw cycles to release inclusion bodies that were subsequently purified by washing in 0.5% Triton X-100 buffer (Sigma-Aldrich). Heavy chain and β2 m inclusion body preparations were denatured separately in 8 M urea buffer (Sigma-Aldrich) and mixed at a 1:1 molar ratio. Each monomeric protein was refolded in 2-mercaptoethylamine/cystamine redox buffer (Sigma-Aldrich) with the appropriate synthetic peptide (Peptides & Elephants GmbH). Refolded monomers were purified via anion exchange after buffer replacement (10 mM Tris, pH 8.1). Purified monomers were biotinylated using d-biotin and BirA (Sigma-Aldrich). Excess biotin was removed via gel filtration. Biotinylated monomers were then tetramerized via the addition of fluorochrome-conjugated streptavidin at a 4:1 molar ratio, respectively. The following specificities were used in this study: CMV A∗0201 NV9 (NLVPMVATV), EBV A∗0201 GL9 (GLCTLVAML), SARS-CoV-2 A∗0201 AV9 (ALSKGVHFV), SARS-CoV-2 A∗0201 HI9 (HLVDFQVTI), SARS-CoV-2 A∗0201 KV9 (KLLEQWNLV), SARS-CoV-2 A∗0201 LL9 (LLLDRLNQL), SARS-CoV-2 A∗0201 LLY (LLYDANYFL), SARS-CoV-2 A∗0201 SV9 (SLVKPSFYV), SARS-CoV-2 A∗0201 TL9 (TLDSKTQSL), SARS-CoV-2 A∗0201 VL9 (VLNDILSRL), SARS-CoV-2 A∗0201 YL9 (YLQPRTFLL), SARS-CoV-2 B∗0702 FI9 (FPRGQGVPI), SARS-CoV-2 B∗0702 KT9 (KPRQKRTAT), SARS-CoV-2 B∗0702 SA9 (SPRRARSVA), and SARS-CoV-2 B∗0702 SL9 (SPRWYFYYL).
Functional assay
PBMCs were resuspended in complete medium (RPMI 1640 supplemented with 10% FBS, 1% L-glutamine, and 1% penicillin/streptomycin) at 1 × 107 cells/mL and cultured at 1 × 106 cells/well in 96-well V-bottom plates (Corning) with the relevant peptides (each at 0.5 μg/mL) for 30 min prior to the addition of anti-CD28/CD49d (3 μL/mL; clone L293/L25; BD Biosciences), brefeldin A (1 μL/mL; Sigma-Aldrich), monensin (0.7 μL/mL; BD Biosciences), and anti-CD107a–PE-CF594 (clone H4A3; BD Biosciences). Negative control wells lacked peptides, and positive control wells included staphylococcal enterotoxin B (SEB; 0.5 μg/mL; Sigma-Aldrich) or plate-bound anti-CD3 (1 μg/mL; clone OKT3; BioLegend). Cells were analyzed by flow cytometry after incubation for 8 hr at 37°C.
Proliferation assay
PBMCs were labeled with CTV (0.5 μM; Thermo Fisher Scientific), resuspended in complete medium at 1 × 107 cells/mL, and cultured at 1 × 106 cells/well in 96-well U-bottom plates (Corning) with the relevant peptides (each at 1 μg/mL) in the presence of anti-CD28/CD49d (3 μL/mL; clone L293/L25; BD Biosciences) and IL-2 (10 IU/mL; PeproTech). Negative control wells lacked peptides, and positive control wells included SEB (0.5 μg/mL; Sigma-Aldrich) or plate-bound anti-CD3 (1 μg/mL; clone OKT3; BioLegend). Functional assays were performed as described above after incubation for 5 days at 37°C.
AIM assay
PBMCs were resuspended in complete medium at 1 × 107 cells/mL and cultured at 1 × 106 cells/well in 96-well U-bottom plates (Corning) with the relevant peptides (each at 1 μg/mL) in the presence of anti-CD28/CD49d (3 μL/mL; clone L293/L25; BD Biosciences). Cells were analyzed by flow cytometry after incubation for 24 hr at 37°C. The following directly conjugated monoclonal antibodies were used to detect activation markers: anti-CD69–BUV737 (clone FN50; BD Biosciences) and anti-4-1BB–BV421 (clone 4B41; BioLegend).
Trucount
Absolute counts were obtained using Multitest 6-color TBNK reagent with Trucount tubes (BD Biosciences). Samples were fixed with 2% PFA for 2 hr prior to acquisition. Absolute CD3+ cell counts were calculated using the following formula: (# CD3+ events acquired x total # beads x 1000) / (# beads acquired x volume of whole blood stained in μL). CD4+ and CD8+ cell counts were computed from the respective frequencies relative to CD3+ cells.
Principal component analysis
Analyses were performed using scikit-learn version 0.22.1 in Python. Data were normalized using sklearn.preprocessing.StandardScaler in the same package to generate z-scores for PCA.
UMAP
FCS 3.0 data files were imported into FlowJo software version 10.6.0 (FlowJo LLC). All samples were compensated electronically. Dimensionality reduction was performed using the FlowJo plugin UMAP version 2.2 (FlowJo LLC). The downsample version 3.0.0 plugin and concatenation tool was used to visualize multiparametric data from up to a total of 120,000 CD8+ T cells (n = 3 donors per group). Representative donors were selected from the 50th percentile for all markers. The following parameters were used in these analyses: metric = euclidean, nearest neighbors = 30, and minimum distance = 0.5. Clusters of phenotypically related cells were detected using PhenoGraph version 0.2.1. The following markers were included in the cluster analysis: CCR7, CD27, CD28, CD38, CD39, CD45RA, CD95, CD127, CTLA-4, CXCR5, granzyme B, Ki-67, LAG-3, PD-1, perforin, TCF1, TIGIT, TIM-3, TOX, and 2B4. Plots were generated using Prism version 8.2.0 (GraphPad Software Inc.).
ELISpot assay
PBMCs were rested overnight in complete medium and seeded at 2 × 105 cells/well in MultiScreen HTS Filter Plates (Merck Millipore) pre-coated with anti-IFN-γ (15 μg/mL; clone 1-D1K; Mabtech). Test wells were supplemented with overlapping peptides spanning Prot_S, Prot_M, and Prot_N (each at 2 μg/mL; Miltenyi Biotec) or peptides corresponding to known optimal epitopes derived from CMV (pp65) or EBV (BZLF1 and EBNA-1) (each at 2 μg/mL; Peptides & Elephants GmbH). Negative control wells lacked peptides, and positive control wells included SEB (0.5 μg/mL; Sigma-Aldrich). Assays were incubated for 24 hr at 37°C. Plates were then washed six times with PBS (Sigma-Aldrich) and incubated for 2 hr at room temperature with biotinylated anti-IFN-γ (1 μg/mL; clone mAb-7B6-1; Mabtech). After six further washes, a 1:1,000 dilution of alkaline phosphatase-conjugated streptavidin (Mabtech) was added for 1 hr at room temperature. Plates were then washed a further six times and developed for 20 min with BCIP/NBT Substrate (Mabtech). All assays were performed in duplicate. Mean values from duplicate wells were used for data representation. Spots were counted using an automated ELISpot Reader System (Autoimmun Diagnostika GmbH).
Serology
SARS-CoV-2-specific antibodies were detected in serum using both the iFLASH Anti-SARS-CoV-2 IgG chemiluminescent microparticle immunoassay against the nucleocapsid and spike proteins (Shenzhen YHLO Biotech Co. Ltd.) as well as the LIAISON SARS-CoV-2 IgG fully automated indirect chemiluminescent immunoassay against the S1 and S2 (spike) proteins (DiaSorin). These assays produced highly concordant results and have been shown to perform adequately as diagnostic tools (Plebani et al., 2020). An individual was considered seropositive if one of the two methods generated a positive result. All assays were performed by trained employees at the clinical laboratory according to standard procedures.
Quantification and Statistical Analysis
Statistical analyses were performed using R studio or Prism version 7.0 (GraphPad Software Inc.). Differences between unmatched groups were compared using an unpaired t test, the Mann-Whitney U test, or the Kruskal-Wallis rank-sum test with Dunn’s post hoc test for multiple comparisons, and differences between matched groups were compared using a paired t test or the Wilcoxon signed-rank test. Correlations were assessed using the Spearman rank correlation. Non-parametric tests were used if the data were not distributed normally according to the Shapiro-Wilk normality test. Phenotypic relationships within multivariate datasets were visualized using FlowJo software version 10.6.1 (FlowJo LLC).
Acknowledgments
We express our gratitude to all donors, health care personnel, study coordinators, administrators, and laboratory managers involved in this work. M.B. was supported by the Swedish Research Council, the Karolinska Institutet, the Swedish Society for Medical Research, the Jeansson Stiftelser, the Åke Wibergs Stiftelse, the Swedish Society of Medicine, the Swedish Cancer Society, the Swedish Childhood Cancer Fund, the Magnus Bergvalls Stiftelse, the Hedlunds Stiftelse, the Lars Hiertas Stiftelse, the Swedish Physicians against AIDS Foundation, the Jonas Söderquist Stiftelse, and the Clas Groschinskys Minnesfond. H.-G.L. and the Karolinska COVID-19 Study Group were supported by the Alice och Knut Wallenbergs Stiftelse and Nordstjernan AB. D.A.P. was supported by a Wellcome Trust Senior Investigator Award (100326/Z/12/Z).
Author Contributions
H.-G.L., S.A., and M.B. conceived the project. T.S., A.P.-P., O.R.-B., J.-B.G., S.L.-L., S.M., D.J.W., T.K., J.E., T.P., D.A.P., and M.B. designed and performed experiments. T.S., A.P.-P., O.R.-B., J.-B.G., and M.B. analyzed data. K.S., A.O., H.K., G.B., E.F., O.R., L.I.E., J.-I.H., A.S., T.A., J.A., M.N., J.K., S.G.-R., N.K.B., J.K.S., D.A.P., H.-G.L., S.A., and M.B. provided critical resources. D.A.P. and M.B. supervised experiments. T.S., A.P.-P., O.R.-B., and M.B. drafted the manuscript. D.A.P. and M.B. edited the manuscript. All authors contributed intellectually and approved the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: August 14, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.08.017.
Contributor Information
Karolinska COVID-19 Study Group:
Mira Akber, Lena Berglin, Helena Bergsten, Susanna Brighenti, Demi Brownlie, Marta Butrym, Benedict Chambers, Puran Chen, Martin Cornillet Jeannin, Jonathan Grip, Angelica Cuapio Gomez, Lena Dillner, Isabel Diaz Lozano, Majda Dzidic, Malin Flodström Tullberg, Anna Färnert, Hedvig Glans, Alvaro Haroun-Izquierdo, Elizabeth Henriksson, Laura Hertwig, Sadaf Kalsum, Efthymia Kokkinou, Egle Kvedaraite, Marco Loreti, Magalini Lourda, Kimia Maleki, Karl-Johan Malmberg, Nicole Marquardt, Christopher Maucourant, Jakob Michaelsson, Jenny Mjösberg, Kirsten Moll, Jagadees Muva, Johan Mårtensson, Pontus Nauclér, Anna Norrby-Teglund, Laura Palma Medina, Björn Persson, Lena Radler, Emma Ringqvist, John Tyler Sandberg, Ebba Sohlberg, Tea Soini, Mattias Svensson, Janne Tynell, Renata Varnaite, Andreas Von Kries, and Christian Unge
Supplemental Information
Correlative Analyses of Immune Activation Phenotypes versus Soluble Factor Measurements in Acute COVID-19, Related to Figure 1 Pairwise correlations between phenotypically defined subpopulations of memory CD4+ and CD8+ T cells and the relative concentrations of various soluble factors in the peripheral blood of patients with acute COVID-19. ∗p < 0.05, ∗∗p < 0.01. Spearman rank correlation.
References
- Alshukairi A.N., Khalid I., Ahmed W.A., Dada A.M., Bayumi D.T., Malic L.S., Althawadi S., Ignacio K., Alsalmi H.S., Al-Abdely H.M. Antibody Response and Disease Severity in Healthcare Worker MERS Survivors. Emerg. Infect. Dis. 2016;22:1113–1115. doi: 10.3201/eid2206.160010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S. Remdesivir for the Treatment of Covid-19— Preliminary Report. N Engl J Med. 2020 doi: 10.1056/NEJMoa2007764. Published online May 22, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts M.R., Nason M.C., West S.M., De Rosa S.C., Migueles S.A., Abraham J., Lederman M.M., Benito J.M., Goepfert P.A., Connors M. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom K., Braun M., Ivarsson M.A., Gonzalez V.D., Falconer K., Moll M., Ljunggren H.G., Michaëlsson J., Sandberg J.K. Temporal dynamics of the primary human T cell response to yellow fever virus 17D as it matures from an effector- to a memory-type response. J. Immunol. 2013;190:2150–2158. doi: 10.4049/jimmunol.1202234. [DOI] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020 doi: 10.1038/s41586-020-2598-9. Published online July 29, 2020. [DOI] [PubMed] [Google Scholar]
- Buggert M., Norström M.M., Salemi M., Hecht F.M., Karlsson A.C. Functional avidity and IL-2/perforin production is linked to the emergence of mutations within HLA-B∗5701-restricted epitopes and HIV-1 disease progression. J. Immunol. 2014;192:4685–4696. doi: 10.4049/jimmunol.1302253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggert M., Nguyen S., Salgado-Montes de Oca G., Bengsch B., Darko S., Ransier A., Roberts E.R., Del Alcazar D., Brody I.B., Vella L.A. Identification and characterization of HIV-specific resident memory CD8+ T cells in human lymphoid tissue. Sci. Immunol. 2018;3 doi: 10.1126/sciimmunol.aar4526. eaar4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow K.A., Parry H.F., Sergeant M., Tyrrell D.A. The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 1990;105:435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fett C., Zhao J., Meyerholz D.K., Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J. Virol. 2014;88:11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demkowicz W.E., Jr., Littaua R.A., Wang J., Ennis F.A. Human cytotoxic T-cell memory: long-lived responses to vaccinia virus. J. Virol. 1996;70:2627–2631. doi: 10.1128/jvi.70.4.2627-2631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes Marraco S.A., Soneson C., Cagnon L., Gannon P.O., Allard M., Abed Maillard S., Montandon N., Rufer N., Waldvogel S., Delorenzi M., Speiser D.E. Long-lasting stem cell-like memory CD8+ T cells with a naïve-like profile upon yellow fever vaccination. Sci. Transl. Med. 2015;7:282ra48. doi: 10.1126/scitranslmed.aaa3700. [DOI] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., China Medical Treatment Expert Group for Covid-19 Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib H. Has Sweden’s controversial covid-19 strategy been successful? BMJ. 2020;369:m2376. doi: 10.1136/bmj.m2376. [DOI] [PubMed] [Google Scholar]
- He R., Lu Z., Zhang L., Fan T., Xiong R., Shen X., Feng H., Meng H., Lin W., Jiang W., Geng Q. The clinical course and its correlated immune status in COVID-19 pneumonia. J. Clin. Virol. 2020;127:104361. doi: 10.1016/j.jcv.2020.104361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Hotez P.J., Corry D.B., Strych U., Bottazzi M.E. COVID-19 vaccines: neutralizing antibodies and the alum advantage. Nat. Rev. Immunol. 2020;20:399–400. doi: 10.1038/s41577-020-0358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C., Hausner M.A., Ferbas K.G., Tobin N.H., Aldrovandi G.M., Yang O.O. Rapid Decay of Anti-SARS-CoV-2 Antibodies in Persons with Mild Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2025179. Published online July 21, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juno J.A., Tan H.X., Lee W.S., Reynaldi A., Kelly H.G., Wragg K., Esterbauer R., Kent H.E., Batten C.J., Mordant F.L. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0995-0. Published online July 13, 2020. [DOI] [PubMed] [Google Scholar]
- Kirkcaldy R.D., King B.A., Brooks J.T. COVID-19 and Postinfection Immunity: Limited Evidence, Many Remaining Questions. JAMA. 2020 doi: 10.1001/jama.2020.7869. Published online May 11, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020 doi: 10.1038/s41586-020-2550-z. Published online July 15, 2020. [DOI] [PubMed] [Google Scholar]
- Li C.K., Wu H., Yan H., Ma S., Wang L., Zhang M., Tang X., Temperton N.J., Weiss R.A., Brenchley J.M. T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Li S., Liu J., Liang B., Wang X., Wang H., Li W., Tong Q., Yi J., Zhao L. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., Hu J.L., Xu W., Zhang Y., Lv F.J. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Mallapaty S. Will antibody tests for the coronavirus really change everything? Nature. 2020;580:571–572. doi: 10.1038/d41586-020-01115-z. [DOI] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020 doi: 10.1126/science.abd3871. Published online August 4, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.D., van der Most R.G., Akondy R.S., Glidewell J.T., Albott S., Masopust D., Murali-Krishna K., Mahar P.L., Edupuganti S., Lalor S. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28:710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H., Wei P., Ge J., Gou M., Li X. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity. 2020;52:971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen R.E., Sinclair E., Emu B., Heitman J.W., Hirschkorn D.F., Epling C.L., Tan Q.X., Custer B., Harris J.M., Jacobson M.A. Loss of T cell responses following long-term cryopreservation. J. Immunol. Methods. 2007;326:93–115. doi: 10.1016/j.jim.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plebani M., Padoan A., Negrini D., Carpinteri B., Sciacovelli L. Diagnostic performances and thresholds: The key to harmonization in serological SARS-CoV-2 assays? Clin. Chim. Acta. 2020;509:1–7. doi: 10.1016/j.cca.2020.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Precopio M.L., Betts M.R., Parrino J., Price D.A., Gostick E., Ambrozak D.R., Asher T.E., Douek D.C., Harari A., Pantaleo G. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8+ T cell responses. J. Exp. Med. 2007;204:1405–1416. doi: 10.1084/jem.20062363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D.A., Brenchley J.M., Ruff L.E., Betts M.R., Hill B.J., Roederer M., Koup R.A., Migueles S.A., Gostick E., Wooldridge L. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S. Convergent Antibody Responses to SARS-CoV-2 Infection in Convalescent Individuals. bioRxiv. 2020 doi: 10.1101/2020.05.13.092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux E., Homad L.J., MacCamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., Akins N.R., Stuart A.B., Wan Y.H., Feng J. Characterization of neutralizing antibodies from a SARS-CoV-2 infected individual. bioRxiv. 2020 doi: 10.1101/2020.05.12.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.S., Kim Y., Kim G., Lee J.Y., Jeong I., Joh J.S., Kim H., Chang E., Sim S.Y., Park J.S., Lim D.G. Immune Responses to Middle East Respiratory Syndrome Coronavirus During the Acute and Convalescent Phases of Human Infection. Clin. Infect. Dis. 2019;68:984–992. doi: 10.1093/cid/ciy595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Deustchman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.D., Coopersmith C.M. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar S., Begom S., Bermingham A., Hoschler K., Adamson W., Carman W., Bean T., Barclay W., Deeks J.J., Lalvani A. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 2013;19:1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- Tang F., Quan Y., Xin Z.T., Wrammert J., Ma M.J., Lv H., Wang T.B., Yang H., Richardus J.H., Liu W., Cao W.C. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W.E., Li Z., Chiew C.J., Yong S.E., Toh M.P., Lee V.J. Presymptomatic Transmission of SARS-CoV-2 — Singapore, January 23-March 16, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:411–415. doi: 10.15585/mmwr.mm6914e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martinez-Colon G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson T.M., Li C.K., Chui C.S., Huang A.K., Perkins M., Liebner J.C., Lambkin-Williams R., Gilbert A., Oxford J., Nicholas B. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 2012;18:274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Woloshin S., Patel N., Kesselheim A.S. False Negative Tests for SARS-CoV-2 Infection — Challenges and Implications. N. Engl. J. Med. 2020;383:e38. doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. Published online February 24, 2020. [DOI] [PubMed] [Google Scholar]
- Yang L.T., Peng H., Zhu Z.L., Li G., Huang Z.T., Zhao Z.X., Koup R.A., Bailer R.T., Wu C.Y. Long-lived effector/central memory T-cell responses to severe acute respiratory syndrome coronavirus (SARS-CoV) S antigen in recovered SARS patients. Clin. Immunol. 2006;120:171–178. doi: 10.1016/j.clim.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Gui X., Xiong Y. Comparison of Clinical Characteristics of Patients with Asymptomatic vs Symptomatic Coronavirus Disease 2019 in Wuhan, China. JAMA Netw. Open. 2020;3:e2010182. doi: 10.1001/jamanetworkopen.2020.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., Agnihothram S., Baric R.S., David C.S., Perlman S. Airway Memory CD4+ T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Alshukairi A.N., Baharoon S.A., Ahmed W.A., Bokhari A.A., Nehdi A.M., Layqah L.A., Alghamdi M.G., Al Gethamy M.M., Dada A.M. Recovery from the Middle East respiratory syndrome is associated with antibody and T-cell responses. Sci. Immunol. 2017;2 doi: 10.1126/sciimmunol.aan5393. eaan5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H.Y., Zhang M., Yang C.X., Zhang N., Wang X.C., Yang X.P., Dong X.Q., Zheng Y.T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlative Analyses of Immune Activation Phenotypes versus Soluble Factor Measurements in Acute COVID-19, Related to Figure 1 Pairwise correlations between phenotypically defined subpopulations of memory CD4+ and CD8+ T cells and the relative concentrations of various soluble factors in the peripheral blood of patients with acute COVID-19. ∗p < 0.05, ∗∗p < 0.01. Spearman rank correlation.
Data Availability Statement
The published article includes all data generated during this study. All codes are freely available at source.