Highlights
-
•
Influenza A virus-like particles (VLPs) were produced using recombinant insect cells.
-
•
VLPs were produced using insect cells as host cells without using a baculovirus.
-
•
A secretory form of VLPs consists of hemagglutinin and matrix protein 1.
-
•
The VLP productivity is comparable to that of the baculovirus–insect cell system.
Keywords: Insect cell culture, Recombinant protein production, Virus-like particles, Influenza virus, High Five cells
Abstract
Virus-like particles (VLPs) are hollow nanoparticles composed of recombinant viral surface proteins without a virus genome. In the present study, we investigated the production of influenza VLPs using recombinant insect cells. DNA fragments encoding influenza A virus hemagglutinin (HA) and matrix protein 1 (M1) were cloned with the Drosophila BiP signal sequence in plasmid vectors containing a blasticidin and a neomycin resistance gene, respectively. After Trichoplusia ni BTI-TN-5B1-4 (High Five) cells were co-transfected with a pair of constructed plasmid vectors, stably transformed cells were established via incubation with blasticidin and G418. Western blot analyses showed that recombinant High Five cells secreted HA and M1 proteins into the culture supernatant. Immunoprecipitation of the culture supernatant with an anti-HA antibody and transmission electron microscopy suggested that secreted HA and M1 proteins were in a particulate structure with a morphology similar to that of an influenza virus. Hemagglutination assay indicated that expressed HA molecules retained hemagglutination activity. In a shake-flask culture, recombinant cells achieved a high HA yield (≈ 10 μg/ml) comparable to the yields obtained using the baculovirus–insect cell system. Recombinant insect cells may serve as excellent platforms for the efficient production of influenza VLPs for use as safe and effective vaccines and diagnostic antigens.
1. Introduction
Vaccination is the most effective way to prevent influenza, an acute respiratory infection that causes several hundred-thousands of deaths in people worldwide yearly. Current influenza vaccines manufactured using embryonated hens’ eggs holds some bottlenecks including long manufacturing time and difficulty in scaling up the process [1,2]. To overcome such bottlenecks, some influenza vaccines have recently been produced using cultured mammalian cells such as MDCK and Vero cells for virus propagation [1,2]. Cell culture-based vaccine manufacture offers advantages such as short production time and easy scale-up, but the vaccines are still manufactured from infectious viruses, which is a potential safety concern. Recombinant subunit vaccines represent an attractive alternative to cell-culture based vaccines. Subunit vaccines are safe because they are produced without using viruses, but reportedly they often suffer from low immunogenicity [[3], [4], [5]]. Virus-like particles (VLPs) can present viral antigens in an authentic conformation densely on the particle surface and therefore induce strong humoral and cellular immune responses [[3], [4], [5], [6], [7]]. Among various expression systems including bacteria, yeast, plant, and mammalian cell systems [[7], [8], [9], [10]], the baculovirus–insect cell system has been extensively used for the production of a wide variety of VLPs [[11], [12], [13], [14]]. A human papillomavirus-like particle vaccine, Cervarix, which has been approved for the prevention of cervical cancers, has been manufactured using the baculovirus–insect cell system, demonstrating that insect cells are a practical and excellent platform for the production of novel recombinant protein vaccines such as VLPs [13,[15], [16], [17]]. However, since the baculovirus infection leads to the death of the host insect cells, continuous production is virtually impossible in the baculovirus–insect cell system. In addition, in the production of VLPs, progeny recombinant baculoviruses become contaminants that are difficult to separate from VLPs [5,12,13].
Recombinant insect cells have recently emerged as promising alternative platforms to the baculovirus–insect cell system [6,18,19]. Once a recombinant insect cell line is established, target proteins can be produced simply by culturing the cells. Mammalian cells are commonly used as host cells for the stable expression of recombinant therapeutic proteins. On the other hand, insect cells are suitable host cells for the production of large quantities of recombinant proteins derived from mammalian viruses, since the cytotoxicity of mammalian virus proteins against insect cells may be lower than that against mammalian cells [20,21]. Furthermore, insect cells can be easily cultivated to a high cell density in suspension with a serum-free or animal-derived component-free medium. In a previous study, a secreted form of Japanese encephalitis VLPs was successfully produced by recombinant lepidopteran insect cells transformed using a plasmid vector encoding the virus prM/E gene [20,21]. In the present study, we investigated the production and characterization of influenza VLPs from recombinant insect cells stably co-expressing structural proteins of influenza A viruses, hemagglutinin (HA) and matrix protein 1 (M1). HA is the major protective antigen of an influenza A virus inducing neutralizing antibodies in vivo, while M1 is another structural protein of the influenza virus. Reportedly, the expression of HA and M1 leads to the formation of VLPs, which are as immunologically efficient as conventional vaccines, in the baculovirus–insect cell system [22,23]. The results obtained in the present study demonstrated that a secretory form of influenza VLPs consisting of HA and M1 proteins was successfully produced by recombinant insect cells. A considerably high HA yield (≈ 10 μg/ml), which was comparable to the yield obtained by the baculovirus–insect cell system, was attained in a shake-flask culture of recombinant insect cells.
2. Materials and methods
2.1. Insect cells and culture medium
A lepidopteran insect cell line Trichoplusia ni BTI-TN-5B1-4 (High Five; Thermo Fisher Scientific, Waltham, MA, USA) was used as the host cell in the present study. The cells were maintained at 27 °C in T-flasks with a serum-free medium called Express Five SFM (Thermo Fisher Scientific) supplemented with 2.41 g/L of l-glutamine and 10 mg/L of gentamicin sulfate. Cell density was determined by microscopically counting the number of cells with a Countess II automated cell counter (Thermo Fisher Scientific), while cell viability was judged by trypan blue dye exclusion.
2.2. Plasmid construction
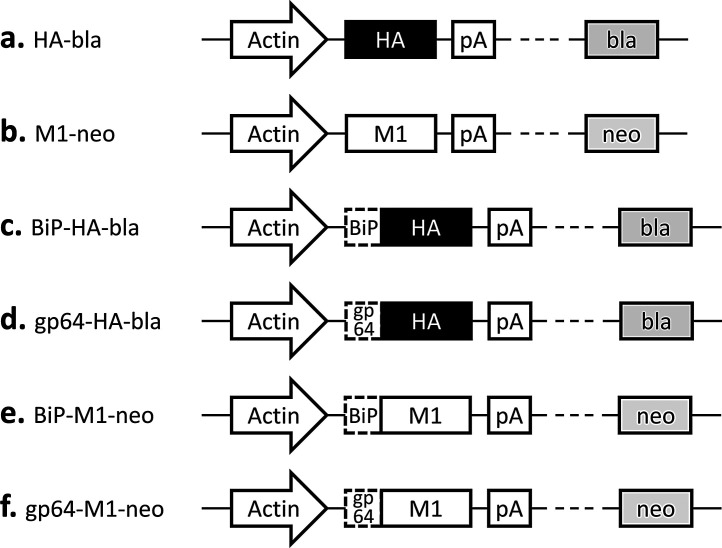
The cDNA fragments encoding the influenza A virus HA (A/New Caledonia/20/1999 (H1N1); GenBank AY289929.1) and M1 (A/Puerto Rico/8/1934 (H1N1); GenBank CY009445.1) were PCR-cloned into plasmid vectors pIHAbla and pIHAneo, respectively (Fig. 1 a, b). The pIHAbla and pIHAneo contain the Bombyx mori actin promoter downstream of the B. mori nucleopolyhedrovirus (BmNPV) IE-1 transactivator and the BmNPV HR3 enhancer for high-level expression in insect cells [24]. The pIHAbla and pIHAneo also carry a blasticidin and a neomycin resistance gene, respectively, for use as a selectable marker. Plasmid vectors containing the HA or M1 gene downstream of either the Drosophila immunoglobulin heavy chain binding protein (BiP) signal sequence [24] or the Autographa californica NPV (AcNPV) gp64 signal sequence [25] were also constructed (Fig. 1 c–f). All the DNA sequences were codon-optimized for lepidopteran insect cells.
Fig. 1.
Schematic representation of the plasmid vectors constructed. Actin, Bombyx mori cytoplasmic actin promotor; HA, influenza A virus hemagglutinin (HA) gene; M1, influenza A virus matrix protein 1 (M1) gene; BiP, Drosophila BiP signal sequence; gp64, Autographa californica nucleopolyhedrovirus (NPV) gp64 signal sequence; pA, OpIE2 polyadenylation sequence from Orgyia pseudotsugata NPV; bla, blasticidinr; neo, neomycinr.
2.3. Transient expression
High Five cells in the exponential growth phase were inoculated into 35-mm culture dishes with 2 ml of fresh medium at a cell density of 2.0 × 105 cells/cm3 1 h before transfection. The cells were transfected with 1 μg of plasmid vectors for the HA and M1 genes at a ratio (w/w) of 1:1 using 3 μl of FuGENE 6 transfection reagent (Promega, Madison, WI, USA). Three days after transfection, the culture supernatant was collected to evaluate the production of HA and M1 proteins.
2.4. Stable transformation
High Five cells in the exponential growth phase were inoculated into 35-mm dishes with 2 ml of medium at a cell density of 2.0 × 105 cells/cm3 1 h before transfection. The cells were transfected with 1 μg of plasmids for the HA and M1 genes at a ratio (w/w) of 1:1 using 3 μl of FuGENE 6. Three days after transfection, the cells were removed from the dishes and inoculated into 100-mm culture dishes with 10 ml of fresh medium at a cell density of 0.8 × 105 cell/cm3. After 24 h of incubation, blasticidin (InvivoGen, San Diego, CA, USA) and G418 (InvivoGen) were added at concentrations of 22 and 280 μg/ml, respectively, to allow the selection of stably transformed cells. The selective medium was replaced every 3 days until colonies of blasticidin- and neomycin-resistant cells were formed. Cells were isolated from each colony into a 96-well plate with 100 μl of medium containing blasticidin and G418. After cells had grown to confluency, the cells were expanded gradually in 35-mm dishes, T25-flasks, and T75-flasks. Expanded cells were incubated with the medium without blasticidin and G418.
2.5. Production, concentration, and purification of VLPs
Recombinant cells in the exponential growth phase were collected and suspended at a density of 2.0 × 105 cells/cm3 in fresh medium. Fifty ml of cell suspension was transferred into a 200-ml screw-capped Erlenmeyer flask. Cells in the flask were cultivated at 27 °C on a rotary shaker at 90 rpm. After 5 days of culture, the supernatant was harvested and clarified by centrifugation. The supernatant was precipitated with 10% (w/w) polyethylene glycol (PEG; molecular mass, approximately 6000 Da) and 1.9% (w/w) NaCl. After 2-h incubation, the precipitate was collected by centrifugation for 20 min at 10,000 rpm and 4 °C. The pellet was then resuspended in phosphate-buffered saline (PBS; pH 7.4), applied to a 10 to 40% (w/w) continuous sucrose density gradient in TNE buffer (10 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA, pH 8.0) in 2.2-ml tubes, and centrifuged at 55,000 rpm at 4 °C for 1.5 h using an S55S swing rotor (Koki Holdings, Tokyo, Japan) in a Himac CS100GX micro-ultracentrifuge (Koki Holdings). Following centrifugation, fractions were collected from the bottom of the tube. The fractions containing both HA and M1 proteins were pooled and stored at 4 °C for subsequent analyses.
2.6. Immunocytochemistry
Cells in the exponential growth phase were inoculated into 35-mm dishes at a density of 2.0 × 105 cells/cm3 and incubated at 27 °C overnight. After aspirating the culture medium, cells were washed with PBS and fixed with 200 μl of PBS containing 4% paraformaldehyde for 10 min. After washing with ice-cold PBS twice, 200 μl of PBS containing 100 μg/ml digitonin (Merck, Darmstadt, Germany) was added to the dishes for cell membrane permeabilization, and the dishes were then incubated for 10 min. After washing with ice-cold PBS three times, blocking solution (PBS containing 0.1% Tween 20, 1% bovine serum albumin (BSA), and 22.5 mg/ml glycine) was added to the dishes, and they were incubated for 30 min. After washing with PBS twice, mouse anti-M1 monoclonal antibody (Abcam, Cambridge, UK) and rabbit anti-HA polyclonal antibody (Sino Biological, Beijing, China) diluted with PBS containing 0.1% Tween 20 and 1% BSA were added, and the dishes were incubated for 1 h under humid conditions. After washing with PBS three times, Alexa Fluor 488-conjugated anti-mouse IgG H&L (Abcam) and Alexa Fluor 647-conjugated anti-rabbit IgG H&L (Abcam) diluted with PBS containing 0.1% Tween 20 and 1% BSA were added, and the dishes were incubated for 1 h under dark conditions. After being washed with PBS three times, the cells were mounted using SlowFade Diamond antifade mountant (Thermo Fisher Scientific). Processed cells were observed via an FV1000-KDM confocal laser microscope (Olympus, Tokyo, Japan).
2.7. Immunoprecipitation
Culture supernatant of recombinant cells was mixed with either rabbit anti-HA polyclonal antibody (Sino Biological) or rabbit anti-M1 polyclonal antibody (Sino Biological) in microtubes that were then incubated for 1 h. After protein G PLUS-agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were added, the microtubes were rotated for 1 h. Bead-antigen complexes were then pelleted by centrifugation, and the pellet was washed 3 times with PBS. The pellet was resuspended in electrophoresis sample buffer, and the supernatant was analyzed via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting.
2.8. Western blotting
Samples were subjected to SDS-PAGE using a 12.5% gel under reducing conditions, followed by transfer to a poly(vinylidene difluoride) (PVDF) membrane using an iBlot 2 gel transfer device (Thermo Fisher Scientific). Immunoreactive proteins were detected with either mouse anti-HA monoclonal antibody (Abcam) or mouse anti-M1 monoclonal antibody (Abcam) and alkaline phosphatase-conjugated anti-mouse IgG (H + L) polyclonal antibody (Promega). The proteins were then stained using a BCIP/NBT color development substrate (Promega) following the manufacturer’s protocol.
2.9. Transmission electron microscopy (TEM)
The culture supernatant was subjected to ultracentrifugation for 6 h at 35,200 rpm and 4 °C. The resultant pellet was resuspended in PBS, which was fractionated by sucrose density-gradient centrifugation, as described above. Fractions containing both HA and M1 proteins were then collected and added onto an elastic carbon-coated copper grid (Okenshoji, Tokyo, Japan). After being stained with 2% phosphotungstic acid, the sample was observed using a JEM-2100 F transmission electron microscope (JEOL, Tokyo, Japan) at 200 kV and 100–200 k-fold magnification.
In the case of TEM with gold nanoparticle probes, the sample was placed onto an elastic carbon-coated copper grid, which was washed with Tris-buffered saline (TBS) and blocking solution (TBS containing 1% BSA) and then incubated for 1 h. Mouse anti-HA monoclonal antibody (Abcam) diluted with the blocking solution was then loaded, and the grid again was incubated for 1 h. After washing with TBS, 15-nm gold nanoparticle-conjugated anti-mouse IgG (H + L) (Cytodiagnostics, Ontario, Canada) diluted with the blocking solution was added to the grid, which again was incubated for 1 h. After washing twice with TBS, the grid was fixed with 1% glutaraldehyde, washed by distilled water, and then stained with 2% phosphotungstic acid. The sample was observed using the transmission electron microscope, as described above.
2.10. Hemagglutination assay
A series of 2-fold dilutions of culture supernatant precipitated with PEG was prepared on a 96-well plate. After 50 μl of PBS containing 1% chicken red blood cells (RBCs; Nippon Bio-test Laboratories, Saitama, Japan) was added, the plate was incubated at 25 °C for 30 min. The plate was monitored visually to detect agglutination of RBCs in the wells.
2.11. Enzyme-linked immunosorbent assay (ELISA)
The HA concentration in the culture supernatant was measured by sandwich ELISA. VLPs purified as describe above were used as a standard. The concentration of HA in the reference standard solution was determined on the total protein concentration, which was based on the results of a bicinchoninic acid assay, and the purity of the HA, which was established using silver-stained gel. Mouse anti-HA monoclonal antibody (Abcam) was diluted with Na2CO3/NaHCO3 buffer and allowed to adsorb onto 96-well plates. Each well of the plates was incubated in succession with either a sample or a standard, rabbit hyperimmune serum against the purified VLPs (Cosmo Bio, Tokyo, Japan), and horseradish peroxidase-conjugated goat anti-rabbit IgG (Promega). The ELISA POD substrate TMB kit (Nacalai Tesque, Kyoto, Japan) was used for detection according to the manufacturer’s protocol. The absorbance was measured with a microplate reader (EnSpire; PerkinElmer Japan, Yokohama, Japan) using a 450 nm test wavelength and a 650 nm reference wavelength. The difference between the two absorbances was converted to the HA concentration by interpolating the value on a standard curve prepared simultaneously.
3. Results and discussion
3.1. Effect of signal sequence
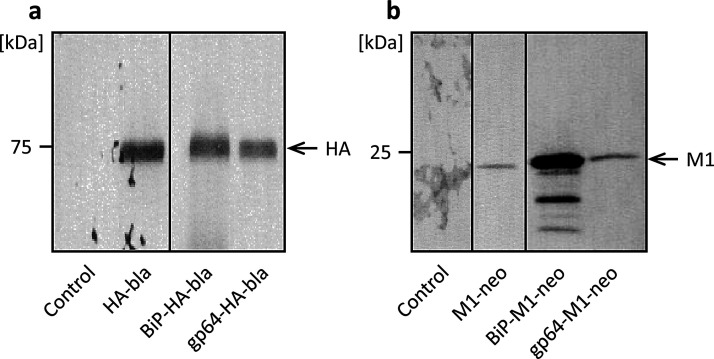
Signal sequences located at the N-terminus of nascent proteins are involved in the transport of proteins to or through cell membranes and the secretion of proteins from the cell. Use of insect- or baculovirus-derived signal sequences affects recombinant protein production in High Five cells [21,26,27]. Hence, we first performed transient expression experiments to investigate the effect of signal sequence on the expressions of the HA and M1 proteins of influenza A viruses. We constructed plasmid vectors carrying the HA or M1 gene downstream of either the Drosophila BiP signal sequence [24] or the AcNPV gp64 signal sequence [25] (Fig. 1 c–d), along with plasmids that had no signal sequence (Fig. 1 a, b). High Five cells were transfected with the respective plasmids, and the culture supernatants were analyzed by western blotting. Specific protein bands were detected at electrophoretic mobilities of approximately 75 and 25 kDa, which coincided with the molecular weights of HA and M1 proteins, respectively, of influenza A viruses with or without a signal sequence (Fig. 2 a, b). These results suggest that cells transfected with the HA and the M1 genes of influenza A viruses expressed and secreted the HA and the M1 proteins, respectively. A comparable level of HA expression was observed regardless of whether the BiP or gp64 signal sequence was used, but a significantly higher level of M1 expression was obtained with the BiP signal sequence than with the gp64 signal sequence. Therefore, we employed the BiP signal sequence for the generation of stably transformed High Five cells expressing both HA and M1 in subsequent investigations.
Fig. 2.
Western blot analysis of culture supernatant from the transient expression of the HA gene (a) and the M1 gene (b) with different signal sequences in Trichoplusia ni BTI-TN-5B1-4 (High Five) cells. Control, untransfected cells.
3.2. Establishment of stably transformed insect cells
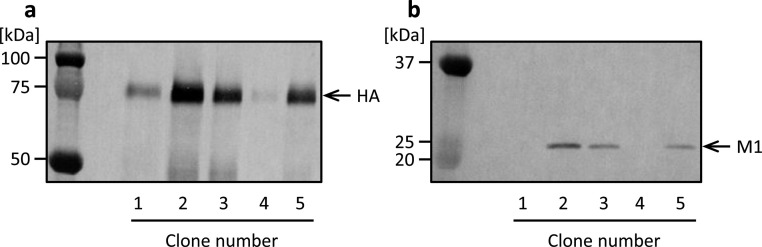
We constructed stably transformed High Five cells by co-transfection using the plasmid vectors BiP-HA-bla and BiP-M1-neo (Fig. 1 c, e). Incubation with blasticidin and G418 over 2 weeks after co-transfection allowed us to obtain cells resistant to the antibiotics. Culture supernatants of several resistant cell clones were analyzed by western blotting. HA- and M1-specific bands were detected at electronic mobilities of approximately 75 and 25 kDa, respectively (Fig. 3 a, b). A highly productive clone of both HA and M1 was obtained among the resultant cell clones (Fig. 3, clone 2). These observations suggest that established cells secreted both the HA and M1 proteins into the culture medium and that simply by culturing in the presence of two antibiotics, recombinant cells into which both the HA and M1 genes were integrated in the genome were efficiently obtained in the present study. Highly productive cells derived from clone 2 were used for the following analyses.
Fig. 3.
Western blot analysis of culture supernatant of High Five cells stably transformed with plasmid vectors BiP-HA-bla and BiP-M1-neo. Culture supernatants in static culture were analyzed using anti-HA (a) and anti-M1 (b) antibodies.
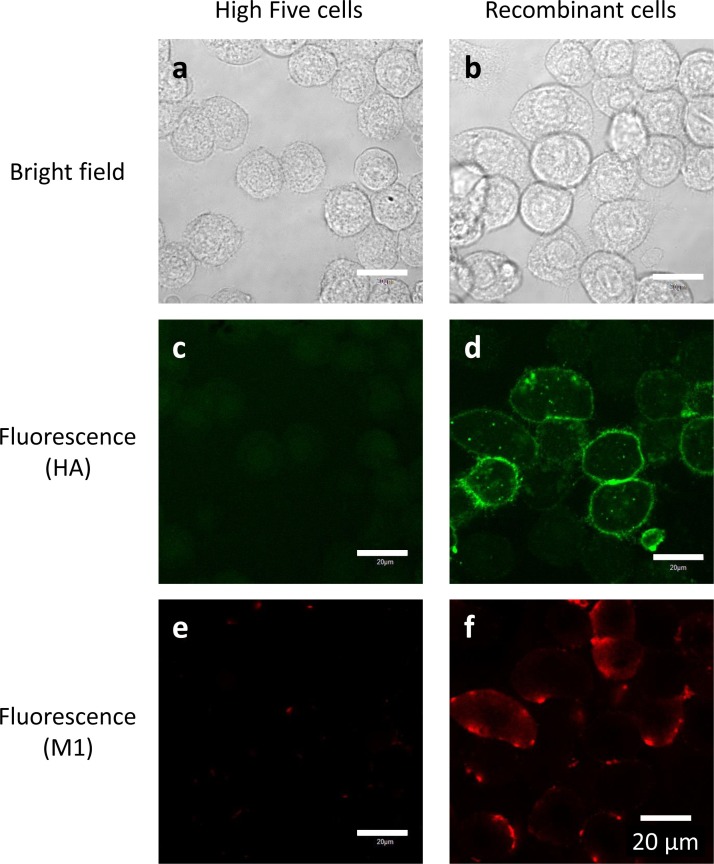
The envelope of influenza A viruses consists of a lipid bilayer that includes transmembrane protein HA that protrudes outward and M1 that lies underneath the membrane. For virus assembly and budding, all virus components must be transported to the assembly site at the cell membrane where they interact with each other in infected cells [[28], [29], [30]]. HA is transported to the budding site, which is a specific region on the cell membrane that contains lipid rafts. M1 then binds to the cytoplasmic tails of HA. Reportedly, influenza VLPs are produced via a similar mechanism in host cells [31]. Therefore, we used a confocal laser microscope to observe the localization of HA and M1 proteins within recombinant High Five cells. Immunocytochemical staining showed that neither HA nor M1 was detected in untransfected High Five cells (Fig. 4 c, e). By contrast, in recombinant cells, HA proteins were observed mostly on the outer side of the cell membrane (Fig. 4d), while M1 molecules existed near the inner side of the cell membrane in the cytoplasm (Fig. 4f). These observations indicate that expressed HA and M1 proteins are appropriately transported for assembly and budding of VLPs in recombinant insect cells.
Fig. 4.
Immunocytochemical staining of recombinant High Five cells (b, d, f) and untransfected cells (a, c, e). HA proteins were stained green with Alexa Fluor 647 (c, d), and M1 proteins were stained red with Alexa Fluor 488 (e, f). White bars indicate 20 μm.
3.3. Characterization of VLPs
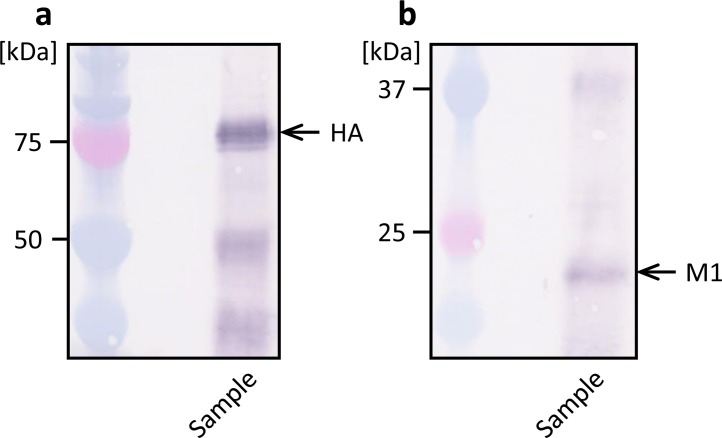
Next, the culture supernatant of recombinant High Five cells was immunoprecipitated with anti-HA antibody, and the precipitate was analyzed by western blotting (Fig. 5 ). Both M1 and HA were detected in the precipitate prepared with anti-HA, which suggests that a complex of HA and M1 molecules was present in the culture supernatant. In contrast, when the culture supernatant was immunoprecipitated with anti-M1 antibody, only a small amount of M1 was observed in the precipitate, but HA was not detected (data not shown).
Fig. 5.
Western blot analysis of culture supernatant immunoprecipitated with rabbit anti-HA polyclonal antibody. Mouse anti-HA monoclonal antibody (a) and mouse anti-M1 monoclonal antibody (b) was used to detect HA and M1, respectively.
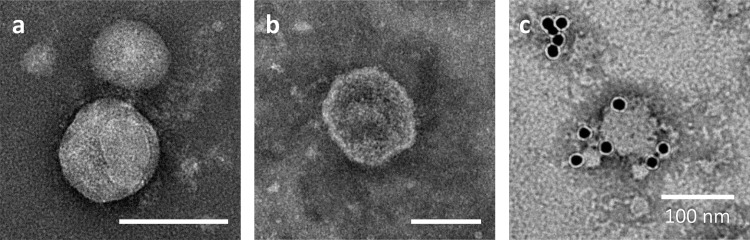
We further examined the culture supernatant of recombinant High Five cells by negative staining and TEM. Influenza virus particles are often spherical and approximately 100 nm in diameter [28,29]. Transmembrane protein HA forms the outer spikes that appear on the viral envelope. Transmission electron micrographs of the culture supernatant precipitated with PEG and fractionated by sucrose density-gradient centrifugation show particles with a spike-like structure (Fig. 6 a) and a lipid bilayer (Fig. 6b). The particle diameter was approximately 80–200 nm, which coincides with those of influenza virions and influenza VLPs produced in different expression systems such as a mammalian cell expression system and the baculovirus–insect cell system [9,31]. Fig. 6c depicts the surface of a particle labelled with an anti-HA monoclonal antibody and a secondary antibody conjugated to a 15-nm gold nanoparticle, which demonstrates the presence of HA molecules on the particle surface. Immunoprecipitation analysis and TEM suggest that HA and M1 proteins were produced in a particulate form and that HA was embedded in the surface of the particles while M1 was present inside the particles.
Fig. 6.
Negative stain transmission electron micrographs of culture supernatant precipitated with polyethylene glycol (PEG) and fractionated by sucrose density-gradient centrifugation. (c) HA was immunostained with an anti-HA monoclonal antibody followed by a secondary antibody conjugated to a 15-nm gold nanoparticle. White bars indicate 100 nm.
3.4. Qualitative and quantitative analyses of HA
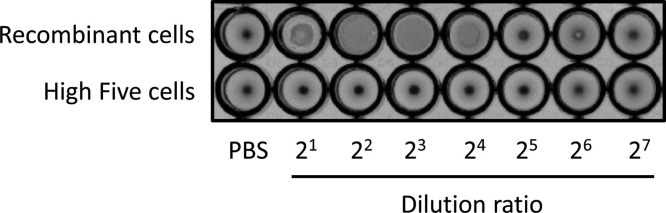
Influenza HA protein is characterized and detected by adsorption onto the membrane of RBCs, which causes them to clump together. To examine the functionality of HA on VLPs, we assayed the hemagglutination activity of culture supernatants using 1% chicken RBCs. In the case of the supernatant of recombinant High Five cells, RBCs were dispersed as a clump, and an agglutination of RBCs was observed in the wells with dilution ratios that ranged from 2 to 16-folds (Fig. 7 ). By contrast, the supernatant of untransfected High Five cells did not show RBC agglutination. These results indicate that HA proteins expressed by recombinant insect cells were embedded in the lipid bilayer of VLPs in a native orientation and then exhibited hemagglutination activity.
Fig. 7.
Hemagglutination assay of culture supernatant precipitated with PEG. Phosphate-buffered saline (PBS) was included as a negative control.
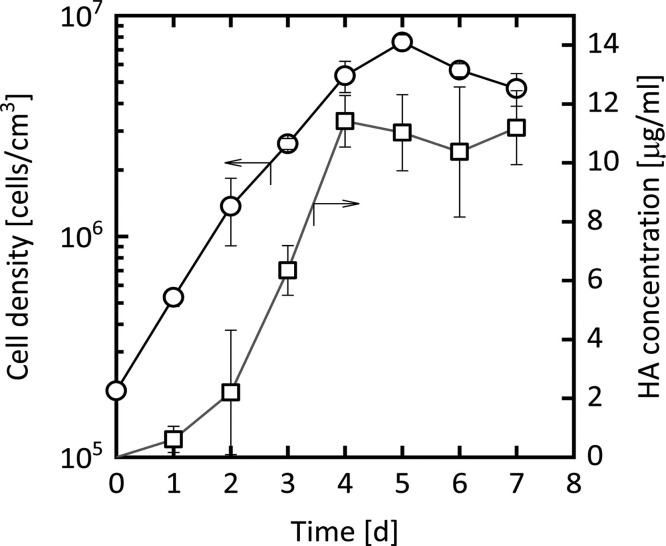
Finally, we evaluated the productivity of HA in a shake-flask culture of recombinant High Five cells via sandwich ELISA. As shown in Fig. 8 , the HA concentration increased with cell growth until day 4. On day 4 the yield of HA reached approximately 11 μg/ml, which is comparable to that obtained by the baculovirus–insect cell system [32]. A considerably high productivity of biologically functional HA was attained with recombinant insect cells in the present study.
Fig. 8.
Secretory production of HA protein by recombinant cells in shake-flask culture. Shown are the changes over time in the density of viable cells (circles) and in the concentration of HA in the culture supernatant (squares). Each plot represents the mean ± S.D. obtained from three different determinations.
4. Conclusions
In the present study, we investigated the production of influenza VLPs in recombinant High Five cells. The cDNA fragments encoding the HA and M1 proteins of influenza A viruses were separately cloned with the Drosophila BiP signal sequence into the plasmid vectors pIHAbla and pIHAneo, respectively. After co-transfection with a set of resultant plasmids, High Five cells were incubated with blasticidin and G418, and cells resistant to the antibiotics were efficiently obtained. The resultant recombinant High Five cells secreted the HA and M1 proteins of influenza A viruses in the culture supernatant. Immunoprecipitation and TEM analyses of the culture supernatant showed that secreted HA and M1 molecules formed VLPs with a morphology equivalent to authentic influenza virus particles. Hemagglutination assay indicated that HA proteins within the VLPs retained hemagglutination activity. A significantly high yield of HA (≈ 10 μg/ml) was achieved with recombinant cells in a shake-flask culture. These results suggest that recombinant insect cells may serve as promising platforms for the production of functional influenza VLPs for use as next-generation vaccines and diagnostic antigens. Approaches demonstrated in the present study could also be used to develop and produce VLPs from other enveloped viruses including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as well as from influenza A viruses of different subtypes.
Recombinant insect cells expressing both the HA and M1 proteins were efficiently established in the present study, but the approaches employed were based on random integration of foreign genes into the genome of host cells. Targeted integration of genes in the genome of insect cells, such as a knock-in mediated using the genome editing technology CRISPR-Cas9, should be investigated in order to more efficiently generate highly productive recombinant cells with a high level of genetic stability. Further characterizations of the VLPs including glycosylation profiling and stability should be examined in future study. Investigations using animal experimentation are also needed in order to examine the immunogenicity of VLPs produced by recombinant insect cells, which includes the induction of neutralizing antibody and cellular immune responses.
CRediT authorship contribution statement
Takuya Matsuda: Investigation, Writing - original draft. Toshikazu Tanijima: Investigation. Akito Hirose: Investigation. Kyoko Masumi-Koizumi: Investigation, Methodology, Validation. Tomohisa Katsuda: Validation. Hideki Yamaji: Conceptualization, Methodology, Writing - review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgement
This research was partially supported by a Grant-in-Aid for Scientific Research (B) (15H04195) from the Japan Society for the Promotion of Science and by the Japan Agency for Medical Research and Development (AMED) under Grant Number JP19ae0101054. The authors thank Mr. H. Iwamoto, Mr. T. Ogawa (Institute of Pathology, Kyodo Byori, Inc.), and Dr. K. Morita (Department of Chemical Science and Engineering, Graduate School of Engineering, Kobe University) for their kind support in TEM.
References
- 1.Feng S.Z., Jiao P.R., Qi W.B., Fan H.Y., Liao M. Development and strategies of cell-culture technology for influenza vaccine. Appl. Microbiol. Biotechnol. 2011;89:893–902. doi: 10.1007/s00253-010-2973-9. [DOI] [PubMed] [Google Scholar]
- 2.Milián E., Kamen A.A. Current and emerging cell culture manufacturing technologies for influenza vaccines. Biomed. Res. Int. 2015;2015:504831. doi: 10.1155/2015/504831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noad R., Roy P. Virus-like particles as immunogens. Trends Microbiol. 2003;11:438–444. doi: 10.1016/s0966-842x(03)00208-7. [DOI] [PubMed] [Google Scholar]
- 4.Roy P., Noad R. Virus-like particles as a vaccine delivery system: myths and facts. Hum. Vaccin. 2008;4:5–8. doi: 10.4161/hv.4.1.5559. [DOI] [PubMed] [Google Scholar]
- 5.Kushnir N., Streatfield S.J., Yusibov V. Virus-like particles as a highly efficient vaccine platform: diversity of targets and production systems and advances in clinical development. Vaccine. 2012;31:58–83. doi: 10.1016/j.vaccine.2012.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamaji H. Suitability and perspectives on using recombinant insect cells for the production of virus-like particles. Appl. Microbiol. Biotechnol. 2014;98:1963–1970. doi: 10.1007/s00253-013-5474-9. [DOI] [PubMed] [Google Scholar]
- 7.Fuenmayor J., Gòdia F., Cervera L. Production of virus-like particles for vaccines. N. Biotechnol. 2017;39:174–180. doi: 10.1016/j.nbt.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C.Y., Yeh Y.C., Yang Y.C., Chou C., Liu M.T., Wu H.S., Chan J.T., Hsiao P.W. Mammalian expression of virus-like particles for advanced mimicry of authentic influenza virus. PLoS One. 2010;5:e9784. doi: 10.1371/journal.pone.0009784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmeisser F., Adamo J.E., Blumberg B., Friedman R., Muller J., Soto J., Weir J.P. Production and characterization of mammalian virus-like particles from modified vaccinia virus Ankara vectors expressing influenza H5N1 hemagglutinin and neuraminidase. Vaccine. 2012;30:3413–3422. doi: 10.1016/j.vaccine.2012.03.033. [DOI] [PubMed] [Google Scholar]
- 10.Buffin S., Peubez I., Barrière F., Nicolaï M.C., Tapia T., Dhir V., Forma E., Sève N., Legastelois I. Influenza A and B virus-like particles produced in mammalian cells are highly immunogenic and induce functional antibodies. Vaccine. 2019;37:6857–6867. doi: 10.1016/j.vaccine.2019.09.057. [DOI] [PubMed] [Google Scholar]
- 11.van Oers M.M. Vaccines for viral and parasitic diseases produced with baculovirus vectors. Adv. Virus Res. 2006;68:193–253. doi: 10.1016/S0065-3527(06)68006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vicente T., Roldão A., Peixoto C., Carrondo M.J.T., Alves P.M. Large-scale production and purification of VLP-based vaccines. J. Invertebr. Pathol. 2011;107:S42–S48. doi: 10.1016/j.jip.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandes F., Teixeira A.P., Carinhas N., Carrondo M.J.T., Alves P.M. Insect cells as a production platform of complex virus-like particles. Expert Rev. Vaccines. 2013;12:225–236. doi: 10.1586/erv.12.153. [DOI] [PubMed] [Google Scholar]
- 14.Lai C.C., Cheng Y.C., Chen P.W., Lin T.H., Tzeng T.T., Lu C.C., Lee M.S., Hu A.Y.C. Process development for pandemic influenza VLP vaccine production using a baculovirus expression system. J. Biol. Eng. 2019;13:78. doi: 10.1186/s13036-019-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox M.M.J. Recombinant protein vaccines produced in insect cells. Vaccine. 2012;30:1759–1766. doi: 10.1016/j.vaccine.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felberbaum R.S. The baculovirus expression vector system: a commercial manufacturing platform for viral vaccines and gene therapy vectors. Biotechnol. J. 2015;10:702–714. doi: 10.1002/biot.201400438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Oers M.M., Pijlman G.P., Valk J.M. Thirty years of baculovirus–insect cell protein expression: from dark horse to mainstream technology. J. Gen. Virol. 2015;96:6–23. doi: 10.1099/vir.0.067108-0. [DOI] [PubMed] [Google Scholar]
- 18.Douris V., Swevers L., Labropoulou V., Andronopoulou E., Georgoussi Z., Iatrou K. Stably transformed insect cell lines: tools for expression of secreted and membrane-anchored proteins and high-throughput screening platforms for drug and insecticide discovery. Adv. Virus Res. 2006;68:113–156. doi: 10.1016/S0065-3527(06)68004-4. [DOI] [PubMed] [Google Scholar]
- 19.Yamaji H. Production of antibody in insect cells. In: Al-Rubeai M., editor. vol. 7. Springer Science+Business Media; Dordrecht, The Netherland: 2011. pp. 53–76. (Antibody Expression and Production, Cell Engineering). [Google Scholar]
- 20.Yamaji H., Nakamura M., Kuwahara M., Takahashi Y., Katsuda T., Konishi E. Efficient production of Japanese encephalitis virus-like particles by recombinant lepidopteran insect cells. Appl. Microbiol. Biotechnol. 2013;97:1071–1079. doi: 10.1007/s00253-012-4371-y. [DOI] [PubMed] [Google Scholar]
- 21.Yamaji H., Konishi E. Production of Japanese encephalitis virus-like particles in insect cells. Bioengineered. 2013;4:438–442. doi: 10.4161/bioe.24514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang S.M., Song J.M., Quan F.S., Compans R.W. Influenza vaccines based on virus-like particles. Virus Res. 2009;143:140–146. doi: 10.1016/j.virusres.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krammer F., Grabherr R. Alternative influenza vaccines made by insect cells. Trends Mol. Med. 2010;16:313–320. doi: 10.1016/j.molmed.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Yamaji H., Manebe T., Watakabe K., Muraoka M., Fujii I., Fukuda H. Production of functional antibody Fab fragment by recombinant insect cells. Biochem. Eng. J. 2008;41:203–209. [Google Scholar]
- 25.Jarvis D.L., Garcia A., Jr. Biosynthesis and processing of the Autographa californica nuclear polyhedrosis virus gp64 protein. Virology. 1994;205:300–313. doi: 10.1006/viro.1994.1646. [DOI] [PubMed] [Google Scholar]
- 26.Sonoda H., Kumada Y., Katsuda T., Yamaji H. Production of single-chain Fv-Fc fusion protein in stably transformed insect cells. Biochem. Eng. J. 2012;67:77–82. [Google Scholar]
- 27.Yamaji H., Segawa M., Nakamura M., Katsuda T., Kuwahara M., Konishi E. Production of Japanese encephalitis virus-like particles using the baculovirus–insect cell system. J. Biosci. Bioeng. 2012;114:657–662. doi: 10.1016/j.jbiosc.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 28.Nayak D.P., Hui E.K.W., Barman S. Assembly and budding of influenza virus. Virus Res. 2004;106:147–165. doi: 10.1016/j.virusres.2004.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nayak D.P., Balogun R.A., Yamada H., Zhou Z.H., Barman S. Influenza virus morphogenesis and budding. Virus Res. 2009;143:147–161. doi: 10.1016/j.virusres.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossman J.S., Lamb R.A. Influenza virus assembly and budding. Virology. 2011;411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei H.J., Chang W., Lin S.C., Liu W.C., Chang D.K., Chong P., Wu S.C. Fabrication of influenza virus-like particles using M2 fusion proteins for imaging single viruses and designing vaccines. Vaccine. 2011;29:7163–7172. doi: 10.1016/j.vaccine.2011.05.077. [DOI] [PubMed] [Google Scholar]
- 32.Krammer F., Schinko T., Palmberger D., Tauer C., Messner P., Grabherr R. Trichoplusia ni cells (High FiveTM) are highly efficient for the production of influenza A virus-like particles: a comparison of two insect cell lines as production platforms for influenza vaccines. Mol. Biotechnol. 2010;45:226–234. doi: 10.1007/s12033-010-9268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]