Abstract
Background
Monogenic diseases are individually rare but collectively common, and are likely underdiagnosed.
Objectives
Estimate the prevalence of monogenic cardiovascular diseases (MCVD) and potentially missed diagnoses in a cardiovascular cohort.
Methods
Exomes from 8,574 individuals referred for cardiac catheterization were analyzed. Pathogenic/likely pathogenic (P/LP) variants associated with MCVD (cardiomyopathies, arrhythmias, connective tissue disorders, and familial hypercholesterolemia) were identified. Electronic health records (EHR) were reviewed for individuals harboring P/LP variants who were predicted to develop disease (G+). G+ individuals who did not have a documented relevant diagnosis were classified into groups of whether they may represent missed diagnoses (unknown, unlikely, possible, probable, definite), based on relevant diagnostic criteria/features for that disease.
Results
We identified 159 P/LP variants; 2,361 individuals harbored at least one P/LP variant, of whom 389 G+ individuals (4.5% of total cohort) were predicted to have at least one MCVD. EHR review of 342 G+ individuals predicted to have one MCVD with sufficient EHR data revealed that 52 had been given the relevant clinical diagnosis. The remaining 290 individuals were classified as potentially having a MCVD as follows: 193 unlikely (66.6%), 50 possible (17.2%), 30 probable (10.3%), 17 definite (5.9%). Grouping possible, probable, definite and known diagnoses, 149 were considered to have an MCVD. Novel MCVD pathogenic variants were identified in 16 individuals.
Conclusions
Overall, 149 individuals (1.7% of cohort) have MCVDs, but only 35% were diagnosed. These patients represents a “missed opportunity” which could be addressed by greater use of genetic testing of patients seen by cardiologists.
Keywords: Monogenic diseases, Genetics, Exome Sequencing, Amyloidosis
Condensed abstract:
We sought to determine the prevalence of monogenic cardiovascular diseases (MCVD) and estimate the number of potentially missed diagnoses in a cardiovascular cohort. Exomes from 8,574 individuals referred to the Duke University cardiac catheterization lab (CATHGEN) were analyzed and 389 individuals (4.5% of total population) were predicted to have at least one MCVD. Electronic health record (EHR) review of 342 individuals predicted to have one MCVD with sufficient EHR data revealed 149 individuals (1.7% of entire cohort) have MCVDs, however, only 35% had been clinically diagnosed. These patients represents a “missed opportunity” which could be addressed by genetic screening.
Background
Monogenic, or Mendelian, diseases represent the extreme end of the genotype-phenotype dyad in which a rare, deleterious mutation in a single gene leads to disease. Although individually rare, in aggregate monogenic diseases are common, with an estimated prevalence of 7–8% in the United States (1). Additionally, monogenic diseases are often associated with severe clinical phenotypes that have high morbidity and mortality rates (1). Cardiovascular monogenic diseases are no exception and encompass a wide range of phenotypes that ultimately lead to coronary artery disease (2), heart failure (3), aortic dissection (4), and/or malignant ventricular arrhythmias (3,5–7). Recent advances have led to an increasing number of effective treatments that are able to alter disease progression and improve outcomes (2,8–10). To achieve the benefits of these treatments, early recognition, diagnosis of the disease and family screening is imperative.
The increasing accessibility of whole exome sequencing has allowed for a thorough estimation of the genotypic prevalence of pathogenic variants that may lead to a monogenic disease (11,12). Several studies have demonstrated a genotypic prevalence of pathogenic variants greater than the estimated phenotypic prevalence of the disease caused by the pathogenic variants (3,11,13), which could be attributed to variable penetrance of the genetic variants, but may imply that some of these diseases are under-recognized in the clinical setting. Therefore, using a cohort of individuals consecutively referred to the cardiac catheterization lab (CATHeterization GENetics [CATHGEN]), we sought to determine the prevalence of disease- causing variants associated with monogenic cardiovascular diseases and estimate the number of potentially missed diagnoses.
Methods.
Study Population
The CATHGEN biorepository was used for this study; details of the cohort have been described previously (14). The cohort is composed of 9,334 individuals who were consecutively referred to the Duke University Hospital cardiac catheterization lab between 2001 and 2010. Arterial blood samples were obtained at the time of catheterization; whole blood was stored in EDTA tubes. All participants provided informed consent and the study was approved by the Duke University Institutional Review Board (IRB).
Whole Exome Sequencing
Whole exome sequencing was performed by the Regeneron Genetics Center® on DNA extracted from whole blood using the Illumina Hiseq 2500 platform. Full sequencing and genotype filtering details can be found in the supplementary methods.
Identification of pathogenic/likely pathogenic variants
Monogenic cardiovascular disease (MCVD) genes were identified by reviewing clinical genetic testing panels for cardiomyopathies (including hypertrophic, familial and transthyretin cardiomyopathies), arrhythmias (including long QT syndrome, Brugada syndrome), connective tissue disorders/aortopathies (including Marfan syndrome, Ehlers-Danlos syndrome, Loeys-Dietz syndrome), and familial hypercholesterolemia (FH) (Supplementary Table 1), and included genes for congenital heart disease and skeletal myopathies. All observed variants within these genes in CATHGEN were initially assessed for pathogenicity using the ClinVar database(15), matching on chromosome, position, reference and alternate alleles (ClinVar data downloads available online: ftp://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh38/archive_2.0/2019/clinvar_20190923.vcf.gz). All candidate pathogenic or likely pathogenic (P/LP) variants were then classified using the American College of Genetics and Genomics/Association for Molecular Pathology (ACMG/AMP) 2015 guidelines for variant classification(16). For the PP3 (computational and predictive data) criterion found in the guidelines(16), variants identified as P/LP according to ClinVar were annotated for functional consequences using CADD v1.5(17), MutationTaster2(18), and the Ensembl Variant Effect Predictor (VEP)(19). The SIFT(20), PolyPhen-2(21), and dbscSNV(22) predictions were also generated using VEP. Further details can be found in the supplementary methods.
Patterns of disease inheritance and the associated phenotypes were determined using the ClinVar (15) submitted interpretations, evidence, and citations for each variant, and the Online Mendelian Inheritance in Man® database (OMIM®)(23); details for each gene and variant are in Supplementary Tables 1 and 2. Individuals not predicted to develop disease (i.e., individuals who are heterozygous for autosomal recessive diseases) were excluded, however, female carriers of X-linked recessive diseases were of interest, because they may have milder phenotypes due to skewed X inactivation(24) but they are described separately. Electronic health record (EHR) review was done on individuals harboring P/LP variants and predicted to develop the relevant MCVD up until the last point of follow up. EHR review included semi-automated examination of administrative data (i.e. laboratory data), structured data from cardiac imaging and electrocardiograms, and manual review of clinic and inpatient notes. EHR review thus determined if individuals who carried P/LP variants were given the relevant clinical diagnosis, as well as enabled determination of diagnostic criteria/expression of disease in those not diagnosed (“potentially missed diagnoses”).
Classification of “Potentially Missed Diagnoses”
Individuals predicted to develop one MCVD and not clinically diagnosed with the respective MCVD were classified into the following five groups depending on the presence of clinical diagnostic criteria or expression of relevant disease phenotypes determined through EHR review, (i.e., “potentially missed diagnosis”): unlikely (i.e., did not have phenotypic expression/diagnostic criteria of the relevant MCVD), possible (i.e. had some features of the MCVD), probable, definite (i.e. met diagnostic criteria for the MCVD), or unknown/insufficient information. The relevant diagnostic criteria for each phenotype was used for classification when available, however, not all MCVD have well-established diagnostic criteria. In those cases, a classification system was developed for the respective phenotype using data published in the literature, and individuals were classified according to their most typical mode of presentation. For MCVDs that included the presence of a deleterious variant as part of the diagnostic criteria (i.e. Marfan syndrome, and Arrhythmogenic right ventricular cardiomyopathy), that criterion was utilized as part of our classification for those diseases. Full details of the classification scheme for each phenotype can be found in the Supplementary Methods. A panel of board-certified cardiologists with expertise in cardiomyopathies, electrophysiology, cardiac imaging and inherited cardiovascular diseases (A.S., C.H., R.M., J.D., A.W., S.S.) developed and came to a consensus on the classification criteria used. Individuals who had at least two clinical encounters identifiable in the EHR were classified and, depending on the MCVD, had to satisfy additional requirements (e.g., presence of electrocardiograms for arrhythmia phenotypes, presence of cardiac imaging for cardiomyopathies, etc.). Individuals with fewer than two clinical encounters were not further analyzed and were not included in the denominator for potentially missed diagnoses. The classification scheme was used to primarily account for variable mutation penetrance and, less likely, for potential false positive variant calls. Individuals carrying two P/LP variants and at risk of two predicted MCVDs are described separately and were not included in the group of potentially missed diagnoses because of the potential overlap of the manifestations of the two diseases. Individuals that harbored the Sodium Voltage-Gated Channel Alpha Subunit 5 (SCN5A) p.Arg225Trp and p.Asp1274Asn mutations (N=2) were also described separately, because the mutations are associated with multiple phenotypes (Long QT syndrome and Brugada Syndrome).
Identification and Classification of Novel Null Variants
Variants that were not found in the ClinVar database and were absent from the dbSNP(25), gnomAD (https://gnomad.broadinstitute.org/) (26), NHLBI Exome Sequencing Project(27) and 1000Genomes(28) databases were considered novel. Novel frameshift, nonsense, start loss, or canonical ±1 or 2 splice mutations predicted to lead to nonsense-mediated decay were considered novel null variants. Full details on the variant selection can be found in the supplementary methods. Individuals found to harbor the variants of interest underwent EHR review. Only variants present in individuals with clear clinical phenotypes related to the gene (i.e., given the relevant clinical diagnosis) were assessed for pathogenicity according to the ACMG/AMP guidelines(16).
Results
Study population and whole exome sequencing quality control.
A total of 8,783 participants had whole exome sequencing performed. Of these, 209 were removed during QC for meeting one or more of the following criteria: low DNA quality (N=96), mismatch between reported and genotypic sex (N=53), high heterozygosity (N=50), ancestry outliers (N=3), or an unusually high number of relationships detected with other samples (N=1), leaving 8,574 exomes. The final analysis set of 8,574 samples had an average of 94.8% of targeted bases with at least 20× coverage and an average of >99.5% of genotypes called across the targeted regions.
Pathogenic/likely pathogenic variants
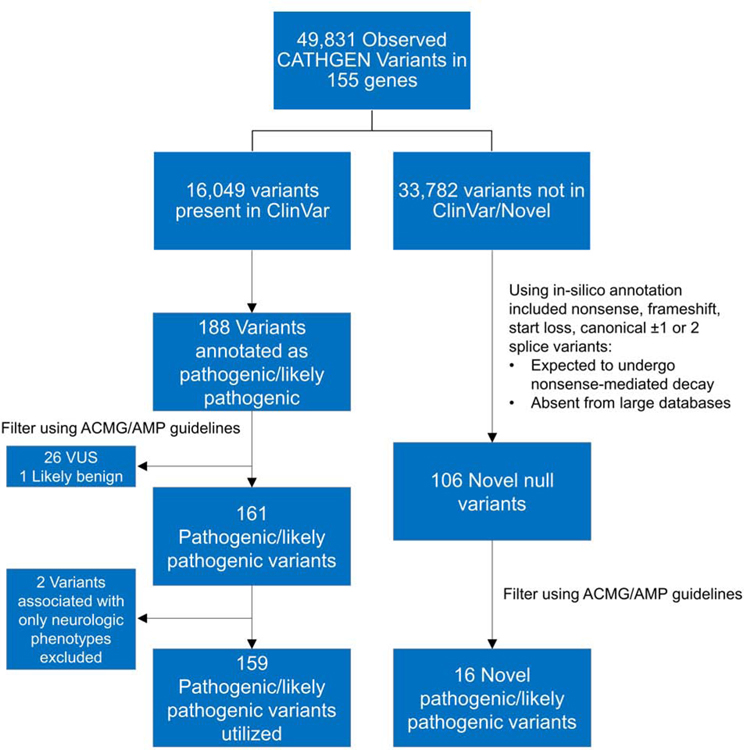
Using clinical genetic testing panels, we identified 155 genes causing or potentially causing MCVD to analyze in this study. In these 155 genes, 49,831 variants were identified in the 8,574 individuals, and of these, 16,049 variants were present in the ClinVar database. A total of 188 variants (0.4% of all 49,831 identified variants) present in 2,506 individuals were annotated as germline P/LP and were thus carried forward for further classification using the ACMG/AMP guidelines. Of these 188 variants, 161 variants were deemed P/LP after further annotation using the ACMG/AMP guidelines (Supplementary Table 2). Although they met criteria for pathogenicity, the Sodium Voltage-Gated Channel Beta Subunit 1 (SCN1B) p.Cys121Trp and Lamin A/C (LMNA) p.Arg298Cys variants were excluded because they are only associated with neurologic phenotypes (Generalized Epilepsy with Febrile Seizures-Plus and Charcot-Marie Tooth Disease, respectively), leaving 159 P/LP variants (Figure 1; Supplementary Table 2). The gene GAA (glucosidase alpha, acid) had the greatest number of unique P/LP variants (N=21) (Figure 2) and the HFE (homeostatic iron regulator) p.His63Asp variant had the highest minor allele frequency (MAF=0.13).
Figure 1. Identification and selection of pathogenic/likely pathogenic variants in the CATHGEN cohort.
In total, 49,831 variants were identified in 155 genes and 159 P/LP variants were utilized after annotation using the ACMG/AMP guidelines (Left). ACMG/AMP classification of the novel null variants in individuals with a clear clinical phenotype revealed 16 novel pathogenic variants (Right).
Figure 2. Unique pathogenic/likely pathogenic variants identified in each gene.
The bars represent the number of P/LP variants identified in each gene, and they were colored according to the respective phenotype the gene is associated with.
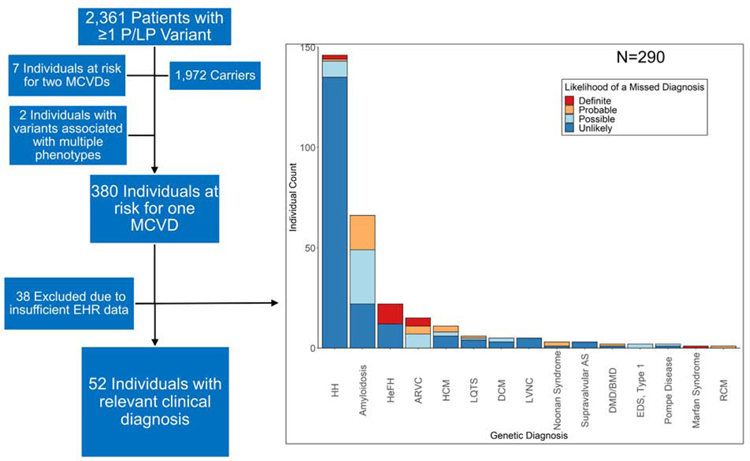
Not taking inheritance pattern into account, a total of 2,361 individuals harbored at least one P/LP variant, and the vast majority (N=1,847; 78.2% of the 2,361 that harbored ≥1 P/LP variant) of them were heterozygous for the HFE p.His63Asp mutation. After filtering on the inheritance pattern of the disease (i.e. excluding individuals heterozygous for an autosomal recessive disease), 389 (4.5% of the total cohort of 8,574) harbored P/LP variants and were predicted to be at risk for the respective disease based on the inheritance pattern. Of these, 7 were at risk for two MCVDs and 2 harbored variants associated with multiple phenotypes (Central Illustration).
Central Illustration. Individuals diagnosed and potentially missed diagnoses.
Flow chart (left) illustrating the number of individuals harboring ≥1 P/LP variant, the number of individuals predicted to develop ≥1 MCVDs, and the number of individuals with the relevant clinical diagnosis. Barplot (right) illustrating the proportions of undiagnosed patients with their likelihood of a missed diagnosis. Abbreviations: P/LP: Pathogenic/Likely Pathogenic; MCVD: Monogenic Cardiovascular Disease; HH: Hereditary Hemochromatosis; HeFH: Heterozygous Familial Hypercholesterolemia; AS: Aortic Stenosis; DMD/BMD: Duchenne/Becker Muscular Dystrophy; EDS: Ehler-Danlos Syndrome
Of the 380 individuals predicted to develop one MCVD, the most prevalent disease based on genotype was hereditary hemochromatosis (N=164, 43.1% of the 380 individuals predicted to develop one MCVD; gene mutated: HFE), followed by familial transthyretin amyloidosis (N=70, 18.4%; gene mutated: TTR), cardiomyopathies (N=54, 14.2%: hypertrophic [N=30; genes mutated: MYH7, MYBPC3, TNNI3, and TNNT2], dilated [N=22; genes mutated: TTN, LMNA, MYH7, TNNT2, and BAG3], and restrictive [N=2; genes mutated: DES and FLNC] cardiomyopathies) and heterozygous FH (N=30, 7.9%; gene mutated: LDLR).
Demographics of the individuals at risk for MCVD can be found in Table 1. The majority were male (63.5%) and were of European ancestry (73.3%), followed by African ancestry (26.2%). The average age of the individuals at enrollment was 59.8 years and the most common primary indication for referral to the cardiac catherization lab was ischemic heart disease (61.4%). The median follow-up time since the time of enrollment was 7.5 (IQR: 1.6, 11.8) years.
Table 1.
Demographics of individuals harboring a pathogenic/likely pathogenic variant and predicted to develop a cardiovascular monogenic disease based on mode of inheritance.
| Total (N=389) | Missing | Cardiomyopathy (N=54) | Arrhythmia* (N=41) | Connective Tissue Disease (N=5) | Other† (N=289) | |
|---|---|---|---|---|---|---|
| Age at enrollment (Mean[SD]) | 59.8 (12.5) | 0 | 57.7 (11.2) | 57.3 (8.9) | 51.2 (14.4) | 60.7 (13.0) |
| Age at last follow up (Mean [SD]) | 67.1 (12.7) | 0 | 65.4 (10.5) | 65.7 (10.3) | 58.6 (11.9) | 67.8 (13.4) |
| Years of follow up since enrollment (Median [IQR]) | 7.5 (1.6, 11.8) | 0 | 8.0 (2.2, 11.7) | 10.3 (3.4, 11.8) | 9.4 (3.4, 11.0) | 6.6 (1.3, 11.7) |
| Sex (Male) (n [%]) | 247 (63.5%) | 0 | 35 (64.8%) | 23 (56.1%) | 5 (100.0%) | 184 (63.7%) |
| Coronary Artery Disease (n [%]) | 213 (57.4%) | 18 | 15 (27.8%) | 20 (48.8%) | 2 (40.0%) | 176 (60.9%) |
| Primary Indication for Procedure (n [%]) | 0 | |||||
| Ischemic Heart Disease | 239 (61.4%) | 15 (27.8%) | 25 (61.0%) | 3 (60.0%) | 196 (67.8%) | |
| Valvular Heart Disease | 11 (2.8%) | 2 (3.7%) | 1 (2.4%) | 0 (0.0%) | 8 (2.8%) | |
| Congenital Heart Disease | 1 (0.2%) | 0 (0.0%) | 1 (2.4%) | 0 (0.0%) | 0 (0.0%) | |
| Other | 138 (35.4%) | 37 (68.5%) | 14 (34.1%) | 2 (40.0%) | 85 (29.4%) | |
| Ejection Fraction (Mean [SD]) | 53.6 (15.2) | 20 | 47.5 (19.9) | 55.5 (11.0) | 39.9 (18.6) | 54.7 (14.4) |
| Ancestry (n [%]) | 0 | |||||
| European (EUR) | 285 (73.3%) | 49 (90.7%) | 29 (70.7%) | 3 (60.0%) | 204 (70.6%) | |
| African (AFR) | 102 (26.2%) | 5 (9.3%) | 12 (29.3%) | 2 (40.0%) | 83 (28.7%) | |
| Admixed American (AMR) | 2 (0.5%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 2 (0.7%) |
Included individuals harboring the SCN5A p.Arg225Trp and p.Asp1274Asn mutations
Included individuals with 2 predicted phenotypes
Electronic health record review
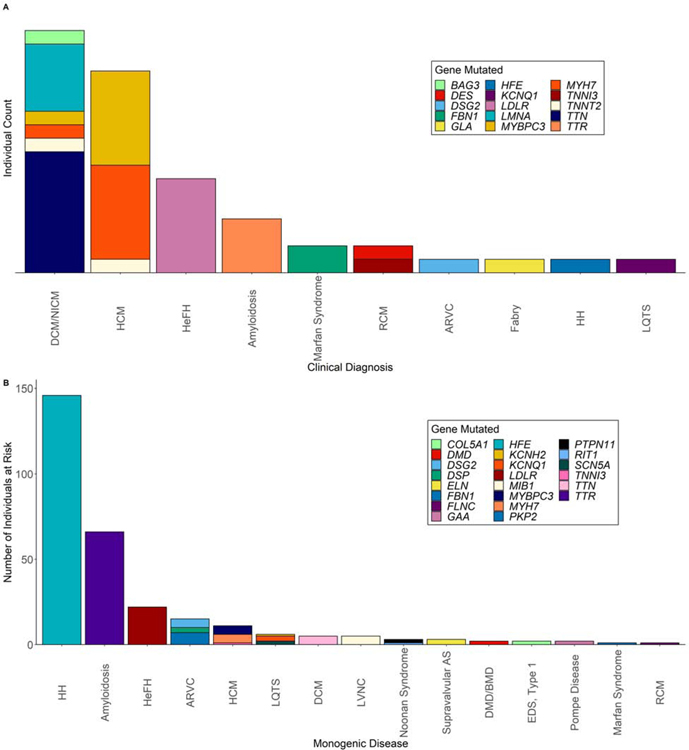
Of the 380 individuals predicted to develop one MCVD, 342 had sufficient information in the EHR to determine diagnosis and expression of disease. Only 52 out of 342 (15.2%) individuals had been given the relevant clinical diagnosis (Central Illustration, Figure 3A). Hypertrophic and dilated cardiomyopathies were the two diseases that had been most commonly clinically diagnosed (Table 2; Central Illustration, Figure 3A). Of note, our criteria did not classify diagnosis of an idiopathic cardiomyopathy (instead of familial dilated cardiomyopathy) as a “potentially missed diagnosis” since patients would have been appropriately treated regardless of the clinical recognition of the genetic cause (except for cascade screening). Despite this, the clinically documented etiology was determined to be idiopathic in all 17 of the dilated cardiomyopathy cases, and genetic testing had not been performed on any of them.
Figure 3. Genes mutated in individuals clinically diagnosed and individuals at risk of developing a monogenic disease but not diagnosed.
(A) Bar plot illustrating the 52 individuals who were given the relevant clinical diagnosis and the respective gene that was mutated. (B) Bar plot illustrating the 290 individuals at risk for developing a MCVD, and the respective gene that was mutated. Abbreviations as in Central Illustration.
Table 2.
Individuals predicted to develop one monogenic disease, the number clinically diagnosed and the likelihood of a missed diagnosis
| Predicted Phenotype based on Genotype | Total | Clinically Diagnosed | Likelihood of Missed Diagnosis | Unknown/Insufficient Information | |||
|---|---|---|---|---|---|---|---|
| Unlikely | Possible | Probable | Definite | ||||
| Hereditary Hemochromatosis | 164 | 1 | 135 | 8 | 1 | 2 | 17 |
| Amyloidosis | 70 | 4 | 22 | 27 | 17 | 0 | 0 |
| Hypertrophic Cardiomyopathy (HCM) | 30 | 17* | 6 | 2 | 3 | 0 | 2 |
| Heterozygous Familial Hypercholesterolemia | 30 | 7 | 12 | 0 | 0 | 10 | 1 |
| Dilated Cardiomyopathy (DCM) | 22 | 17† | 3 | 2 | 0 | 0 | 0 |
| Autosomal Dominant Long QT Syndrome | 20 | 1 | 4 | 1 | 1 | 0 | 13 |
| Arrhythmogenic Right Ventricular Cardiomyopathy | 16 | 1 | 0 | 7 | 4 | 4 | 0 |
| Noonan Syndrome | 3 | 0 | 1 | 0 | 2 | 0 | 0 |
| Brugada Syndrome | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| Left Ventricular Noncompaction | 5 | 0 | 5 | 0 | 0 | 0 | 0 |
| Marfan syndrome | 3 | 2 | 0 | 0 | 0 | 1 | 0 |
| Supravalvular Aortic Stenosis | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| Duchenne/Becker Muscular Dystrophy‡ | 2 | 0 | 1 | 0 | 1 | 0 | 0 |
| Pompe Disease | 2 | 0 | 1 | 1 | 0 | 0 | 0 |
| Ehler-Danlos Syndrome, Type 1 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| Restrictive Cardiomyopathy (RCM) | 2 | 1 | 0 | 0 | 1 | 0 | 0 |
| Fabry Disease | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Autosomal Recessive Long QT Syndrome | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| MYH7 Congenital Myopathy | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| Total | 380 | 52 | 193 | 50 | 30 | 17 | 38 |
15 were diagnosed with HCM, 1 with DCM, and 1 with RCM
11 were diagnosed with DCM and 6 with Nonischemic Cardiomyopathy (NICM)
Male hemizygotes only
The remaining 290 out of 342 individuals who were predicted to develop one MCVD and had not been clinically diagnosed were then classified into four groups through EHR review based on the likelihood of a “potentially missed diagnosis” as follows: 193 unlikely (66.6%), 50 possible (17.2%), 30 probable (10.3%), 17 definite (5.9%) (Figure 3B;Table 2). Thus, grouping individuals that had a phenotypic expression of the MCVD (possible, probable, and definite) and those that were diagnosed, 149 out of the 342 (43.6%) of the individuals carrying P/LP variants had phenotypic expression of the relevant MCVD. Thus, restated at the level of the total cohort: (1) 389 individuals (4.5% of total cohort of 8,574 individuals) harbored P/LP variants and were predicted to be at risk for ≥1 MCVD; (2) 149 out of 8,574 (1.7%) of these individuals harboring P/LP variants had diagnostic criteria/phenotypic expression of the relevant MCVD; and (3) 52 of the 149 (35%) individuals had been given the relevant clinical diagnosis. In absolute numbers, familial transthyretin (TTR) amyloidosis was the most commonly missed disease (27 possible and 17 probable; Table 2) and accounted for approximately 45% of the potentially missed diagnoses. Details of all 380 individuals that were predicted to develop one MCVD with the culprit mutation they harbored, if they were diagnosed and the likelihood of a missed diagnosis can be found in Supplemental Table 3.
We also examined female mutation carriers of X-linked recessive diseases, who were not included in the prior 389 participants predicted to develop at least one MCVD, since they can have milder disease manifestation due to skewed X chromosome inactivation (24). Only one such individual was identified and did have expression of disease: a carrier of a truncating dystrophin (DMD) mutation (p.Glu1536Ter) diagnosed clinically with idiopathic dilated cardiomyopathy with a left ventricular ejection fraction of 20%, without an alternative etiology, and had no documented evidence of skeletal myopathy.
Individuals with more than one predicted phenotype
Surprisingly, we identified seven individuals who harbored two P/LP variants and were predicted to develop the two relevant MCVD based on inheritance pattern (Supplemental Table 4). Only four of the seven individuals had been given the relevant clinical diagnosis of one of the two MCVDs, and two had evidence of phenotypic expression for the second MCVD (Patients No. 381 and 384; Supplemental Table 4). Phenotypic features of the individuals were described in detail in Supplemental Table 4.
Individuals harboring a variant associated with multiple phenotypes
Two individuals harbored a P/LP variant that had been associated with multiple MCVD phenotypes (long QT syndrome and Brugada Syndrome). The individual (Patient no. 387; Supplemental Table 4) harboring the SCN5A p.Asp1274Asn mutation had a QTc of 520 on her most recent electrocardiogram in the absence of an alternate QTc prolonging etiologies. Phenotypic features of the individuals were described in detail in Supplemental Table 4.
Novel null/truncating variants.
A total of 106 novel null variants in 45 genes were identified (Figure 1; Supplemental Table 5); 185 individuals were found to harbor one of the null variants and nine individuals harbored two. Of these 194 individuals, 176 had sufficient EHR data and did not harbor a culprit P/LP variant: 17 had been given the relevant clinical diagnosis and one had clear expression of the relevant disease on EHR review (Supplemental Table 5). Given the uncertainty of these variants, we performed pathogenicity assessment on variants in the aforementioned 18 individuals. ACMG classification of the 18 novel null variants in these individuals (each individual harboring one) with a clear clinical phenotype revealed 16 pathogenic variants in Titin (TTN; 10 unique variants in 10 individuals), Filamin C (FLNC; 3 variants), Myosin Binding Protein C (MYBPC3; 1 variant), LDL Receptor (LDLR; 1 variant) and DMD (1 variant), and two variants of undetermined significance (VUS) in Myosin Heavy Chain 7 (MYH7). The most common MCVD associated with novel null variants was dilated cardiomyopathy/nonischemic cardiomyopathy (N=14) with the majority of individuals (N=10) harboring novel TTN truncating variants.
Discussion
In this study we evaluated the burden of MCVDs in a cardiovascular cohort, and importantly, with availability of phenotyping data through EHR and imaging data determined the burden of missed diagnoses, which is unique to our study. We found that 4.5% of the 8,574 individuals studied in this cohort enriched for cardiovascular disease were at risk of a MCVD based on carriage of a P/LP variant. Further, 149 individuals (1.7% of the cohort) both carried a P/LP variant and had evidence of the disease, but only 35% (52/149 individuals) had previously been given a clinical diagnosis of the relevant disease. Unfortunately, this meant 97 individuals both carried a P/LP variant and had diagnostic criteria/clinical expression of disease but had not been given the diagnosis despite being referred for an invasive cardiac procedure. These patients thus constitute a “potentially missed diagnosis”. Thus, our results support greater use of genetic testing in patients being evaluated for cardiovascular disorders, with genetic testing guided by clinical, ECG and echocardiographic data, but with consideration of genetic screening even in the absence of clear diagnostic criteria for an MCVD given the high percentage of missed diagnoses in patients who had some expressed phenotypes of the disease.
Our study highlights the under-recognition of MCVD which are important to diagnose for several reasons. First, many of the MCVDs identified have evidence-based treatment modalities available(8–10) or are clinically actionable(29), e.g., hereditary hemochromatosis(9) and familial transthyretin amyloidosis(8). Second, several of the MCVDs described herein, e.g., ARVC, HCM, and LQTS, lead to arrhythmogenic phenotypes where sudden death is a relatively common presentation, so early identification of these individuals can increase survival (3,5,6). Third, identifying individuals harboring pathogenic mutations is important for cascade screening of family members at risk of inheriting the pathogenic mutations. In addition, the under- recognition of these diseases underscores the importance of including monogenic diseases in the treating physician’s differential diagnosis, despite their individual rarity. One of the most common potentially missed diagnoses in our study was familial TTR amyloidosis and it serves as an example where a genome-first approach in patient care may potentially improve outcomes. A previous study (30) has shown increasing temporal trends of mortality resulting from cardiac amyloidosis. The underlying reason, particularly in the 3–4% of African Americans (>1 million allele carriers) with a pathogenic p.Val142Ile mutation may be due to a clinical under-diagnosis of the disease. As an early diagnosis predicts response to therapy(8), there is a need to utilize tools (e.g., genetic testing) that can facilitate identification of occult TTR amyloidosis. Because of the aforementioned reasons, at the time of writing this manuscript, we are in the process of returning these genetic results to the participants.
We have also identified novel deleterious variants that were associated with a clear clinical phenotype and were considered pathogenic using the ACMG/AMP guidelines. The most common phenotype associated with these variants was dilated cardiomyopathy/nonischemic cardiomyopathy, with TTN truncating variants being the most common novel variants identified. This is in line with the robust evidence implicating TTN truncating variants as a common cause of dilated cardiomyopathy with an age-dependent but variable penetrance (31). Identification of these novel variants in our study adds to the body of knowledge available to geneticists to aid in cardiovascular genetics evaluations. We also provide data that potentially support the reclassification of some variants currently classified as P/LP in the ClinVar database, which is used clinically in the care of cardiovascular genetics patients. Using the ACMG/AMP guidelines, 26 of these variants were determined to be VUSs and one was likely benign, and none of the individuals with sufficient EHR data harboring these variants had an associated diagnosis to support their pathogenicity (Supplemental Table 2). These data also will assist clinicians in cardiovascular genetic evaluations. Of note, a significant portion of the individuals did not show phenotypic features or fulfill diagnostic criteria (i.e. unlikely missed diagnoses), which is likely due to the incomplete penetrance of some of the variants identified, which are in genes that have known incomplete penetrance even in families harboring the same variant (e.g. hereditary hemochromatosis, TTR amyloidosis, HCM).
Our results support the heightened use of sequencing of known genes for MCVD in patients referred for evaluation of cardiovascular disorders, which involves filtering for rare, deleterious variants in the coding regions of the genome. Although data are contradictory (32), there is a growing body of evidence that polygenic risk scores (PRS), which are composed primarily of common variants, may have clinical utility in common complex diseases (33). In fact, these PRS may also have utility in MCVD including LQTS (34) and DCM (35), and while more data are necessary, one could envision a time in the future where we would simultaneously screen for rare and common variation. Regardless, the clinical utility for identification of monogenic diseases with a culprit P/LP variant is well-established, with clear implications for patients and family members.
Other studies have been published on the burden of monogenic diseases in totality(1), where they have shown a similar burden of under-recognition of monogenic diseases. For example, a similar study (36) focused on renal and genitourinary monogenic disorders in a sample of healthy adults, found that 1.4% of the individuals had clinically actionable genotypes. However, health record data was unavailable for the vast majority of the participants (36), limiting determination of missed diagnoses.
Limitations
While our study includes a large cardiovascular disease-enriched cohort and could have significant clinical implications, it is important to note some limitations. The absolute number of missed diagnoses is relatively low, however given the morbid and mortal consequences of these MCVD, and the low risk of genetic testing with declining associated costs, even this small absolute number is worth preventing. Although next-generation whole exome sequencing is highly accurate, there are some uncertainty in areas of the genome that can lead to false positive variant calls, particularly for insertions and deletions(16), and false negative calls in regions with high GC content or low depth of coverage (37). In order to minimize the number of false positives, we used stringent variant filtering to balance sensitivity and specificity. Also, copy number variants were not analyzed. Our phenotyping was limited by the EHR data that was available for each individual and may therefore lead to an underestimate of true disease penetrance. In addition, some MCVD diagnoses may have been documented outside of the Duke Health system EHR, which may therefore lead to an overestimate of potentially missed diagnoses. Further, our data likely underestimates the true prevalence of all MCVDs, particularly because VUSs may be pathogenic, but there is currently not enough data to support their pathogenicity. Also, pathogenic variants undergo reclassification on a regular basis(38), so a lack of a clinical phenotype observed in some of the individuals may simply may be due to the culprit variant being benign and may lead to an overestimate of genotype-positive individuals. To overcome this potential limitation, we did, however, carefully classify variants using the most current ACMG/AMP guidelines to minimize the number of false positives.
Conclusions
In this cohort of 8,574 patients referred for cardiac catheterization, whole exome sequencing identified P/LP variants associated with monogenic cardiovascular disease in 4.5% of the cohort. Additionally, up to 149 (1.7% of entire cohort) patients harbor both a P/LP variant and have features suggesting phenotypic expression of the relevant disease, however only 35% of those patients had been given a clinical diagnosis. This highlights the under-recognition of monogenic diseases in the clinical setting and represents a “missed opportunity” which could be addressed by genetic screening.
Supplementary Material
Perspectives:
Competency in Patient Care:
Monogenic (Mendelian) cardiovascular diseases, though individually rare, are relatively common in aggregate but undetected in over half of affected patients. More systematic exome sequencing of patients referred for evaluation of cardiovascular disorders could have important implications for long-term clinical surveillance, cascade screening of potentially affected family members, and implementation of evidence-based therapies.
Translational Outlook:
Additional studies are needed to determine whether a genotype-first approach can improved clinical outcomes in patients carrying pathogenic variants associated with monogenic cardiovascular disease.
Acknowledgments:
We gratefully recognize the CATHGEN participants for taking part in this study and Regeneron Pharmaceuticals for performing the whole exome sequencing.
Sources of Funding: This work was supported by National Heart, Lung and Blood Institute grant 5P01-HL036587 and Duke Forge Award - Duke CTSA grant UL1TR002553.
Abbreviations list:
- EHR
Electronic Health Record
- ACMG/AMP
American College of Medical Genetics and Genomics/Association for Molecular Pathology
- EDTA
Ethylenediaminetetraacetic acid
- DNA
Deoxyribonucleic acid
- NHLBI
National Heart, Lung, and Blood Institute
- HCM
Hypertrophic Cardiomyopathy
- LQTS
Long QT Syndrome
- DCM
Dilated Cardiomyopathy
- ARVC
Arrhythmogenic Right Ventricular Cardiomyopathy
- LVNC
Left Ventricular Noncompaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: Aris Baras: Employee and stockholder of Regeneron Pharmaceuticals. James Daubert: Honoraria for: Events Committee, Data Safety Monitoring Board, Consulting, Advisory Boards or for lectures from Abbott, Biosense, Biotronik, Boston Scientific, Microport, Farapulse, Medtronic and Vytronus. Research grants from: Abbott and Medtronic. Sreekanth Vemulapalli: Grants / Contracts: National Institutes of Health, Patient Centered Outcomes Research Institute, Food and Drug Administration (NEST), Society of Thoracic Surgeons, American College of Cardiology, Boston Scientific, Abbott Vascular. Consulting / Adboard: HeartFlow, Baylabs (Caption Health), Janssen, Boston Scientific. Andrew Wang: Advisory board/consulting: Cytokinetics; Educational grant (to institution): MyoKardia Inc; Research grants (to institution): MyoKardia Inc, Cytokinetics. Svati H. Shah: Primary investigator of research sponsored agreement: Verily Life Sciences; Primary investigator of research sponsored agreement: AstraZeneca. The other authors report no relevant conflicts.
References
- 1.Chong JX, Buckingham KJ, Jhangiani SN et al. The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. Am J Hum Genet 2015;97:199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luirink IK, Wiegman A, Kusters DM et al. 20-Year Follow-up of Statins in Children with Familial Hypercholesterolemia. N Engl J Med 2019;381:1547–1556. [DOI] [PubMed] [Google Scholar]
- 3.Teekakirikul P, Kelly MA, Rehm HL, Lakdawala NK, Funke BH. Inherited cardiomyopathies: molecular genetics and clinical genetic testing in the postgenomic era. J Mol Diagn 2013;15:158–70. [DOI] [PubMed] [Google Scholar]
- 4.Paterick TE, Humphries JA, Ammar KA et al. Aortopathies: etiologies, genetics, differential diagnosis, prognosis and management. Am J Med 2013;126:670–8. [DOI] [PubMed] [Google Scholar]
- 5.Khera AV, Mason-Suares H, Brockman D et al. Rare Genetic Variants Associated With Sudden Cardiac Death in Adults. J Am Coll Cardiol 2019. [DOI] [PMC free article] [PubMed]
- 6.Modell SM, Lehmann MH. The long QT syndrome family of cardiac ion channelopathies: a HuGE review. Genet Med 2006;8:143–55. [DOI] [PubMed] [Google Scholar]
- 7.Marcus FI, McKenna WJ, Sherrill D et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the task force criteria. Circulation 2010;121:1533–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maurer MS, Schwartz JH, Gundapaneni B et al. Tafamidis Treatment for Patients with Transthyretin Amyloid Cardiomyopathy. N Engl J Med 2018;379:1007–1016. [DOI] [PubMed] [Google Scholar]
- 9.Bacon BR, Adams PC, Kowdley KV, Powell LW, Tavill AS, American Association for the Study of Liver D. Diagnosis and management of hemochromatosis: 2011 practice guideline by the American Association for the Study of Liver Diseases. Hepatology 2011;54:328–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biegstraaten M, Arngrimsson R, Barbey F et al. Recommendations for initiation and cessation of enzyme replacement therapy in patients with Fabry disease: the European Fabry Working Group consensus document. Orphanet J Rare Dis 2015;10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bick AG, Flannick J, Ito K et al. Burden of rare sarcomere gene variants in the Framingham and Jackson Heart Study cohorts. Am J Hum Genet 2012;91:513–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lek M, Karczewski KJ, Minikel EV et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semsarian C, Ingles J, Maron MS, Maron BJ. New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol 2015;65:1249–1254. [DOI] [PubMed] [Google Scholar]
- 14.Kraus WE, Granger CB, Sketch MH Jr. et al. A Guide for a Cardiovascular Genomics Biorepository: the CATHGEN Experience. J Cardiovasc Transl Res 2015;8:449–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landrum MJ, Lee JM, Riley GR et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 2014;42:D980–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards S, Aziz N, Bale S et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 2019;47:D886–D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014;11:361–2. [DOI] [PubMed] [Google Scholar]
- 19.McLaren W, Gil L, Hunt SE et al. The Ensembl Variant Effect Predictor. Genome Biol 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 2012;40:W452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adzhubei IA, Schmidt S, Peshkin L et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jian X, Boerwinkle E, Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res 2014;42:13534–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 2005;33:D514–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van den Veyver IB. Skewed X inactivation in X-linked disorders. Semin Reprod Med 2001;19:183–91. [DOI] [PubMed] [Google Scholar]
- 25.Sherry ST, Ward MH, Kholodov M et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001;29:308–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karczewski KJ, Francioli LC, Tiao G et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein- coding genes. bioRxiv 2019:531210.
- 27.Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP). Seattle, WA, 2019. [Google Scholar]
- 28.Genomes Project C, Auton A, Brooks LD et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalia SS, Adelman K, Bale SJ et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med 2017;19:249–255. [DOI] [PubMed] [Google Scholar]
- 30.Alexander KM, Orav J, Singh A et al. Geographic Disparities in Reported US Amyloidosis Mortality From 1979 to 2015: Potential Underdetection of Cardiac Amyloidosis. JAMA Cardiol 2018;3:865–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ware JS, Cook SA. Role of titin in cardiomyopathy: from DNA variants to patient stratification. Nat Rev Cardiol 2018;15:241–252. [DOI] [PubMed] [Google Scholar]
- 32.Mosley JD, Gupta DK, Tan J et al. Predictive Accuracy of a Polygenic Risk Score Compared With a Clinical Risk Score for Incident Coronary Heart Disease. JAMA 2020;323:627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marston NA, Kamanu FK, Nordio F et al. Predicting Benefit From Evolocumab Therapy in Patients With Atherosclerotic Disease Using a Genetic Risk Score: Results From the FOURIER Trial. Circulation 2020;141:616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lahrouchi N, Tadros R, Crotti L et al. Transethnic Genome-Wide Association Study Provides Insights in the Genetic Architecture and Heritability of Long QT Syndrome. Circulation 2020. [DOI] [PMC free article] [PubMed]
- 35.Pirruccello JP, Bick A, Wang M et al. Analysis of cardiac magnetic resonance imaging in 36,000 individuals yields genetic insights into dilated cardiomyopathy. Nat Commun 2020;11:2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasouly HM, Groopman EE, Heyman-Kantor R et al. The Burden of Candidate Pathogenic Variants for Kidney and Genitourinary Disorders Emerging From Exome Sequencing. Ann Intern Med 2019;170:11–21. [DOI] [PubMed] [Google Scholar]
- 37.Goldfeder RL, Priest JR, Zook JM et al. Medical implications of technical accuracy in genome sequencing. Genome Med 2016;8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrison SM, Rehm HL. Is ‘likely pathogenic’ really 90% likely? Reclassification data in ClinVar. Genome Med 2019;11:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.