Abstract
Background
Severe, symptomatic aortic stenosis (AS) is associated with poor prognoses. However, early detection of AS is difficult because of the long asymptomatic period experienced by many patients, during which screening tools are ineffective. The aim of this study was to develop and validate a deep learning–based algorithm, combining a multilayer perceptron and convolutional neural network, for detecting significant AS using ECGs.
Methods and Results
This retrospective cohort study included adult patients who had undergone both ECG and echocardiography. A deep learning–based algorithm was developed using 39 371 ECGs. Internal validation of the algorithm was performed with 6453 ECGs from one hospital, and external validation was performed with 10 865 ECGs from another hospital. The end point was significant AS (beyond moderate). We used demographic information, features, and 500‐Hz, 12‐lead ECG raw data as predictive variables. In addition, we identified which region had the most significant effect on the decision‐making of the algorithm using a sensitivity map. During internal and external validation, the areas under the receiver operating characteristic curve of the deep learning–based algorithm using 12‐lead ECG for detecting significant AS were 0.884 (95% CI, 0.880–0.887) and 0.861 (95% CI, 0.858–0.863), respectively; those using a single‐lead ECG signal were 0.845 (95% CI, 0.841–0.848) and 0.821 (95% CI, 0.816–0.825), respectively. The sensitivity map showed the algorithm focused on the T wave of the precordial lead to determine the presence of significant AS.
Conclusions
The deep learning–based algorithm demonstrated high accuracy for significant AS detection using both 12‐lead and single‐lead ECGs.
Keywords: aortic valve stenosis, deep learning, electrocardiography
Subject Categories: Valvular Heart Disease, Electrophysiology, Information Technology
Clinical Perspective
What Is New?
We developed a deep learning–based algorithm, combining a multilayer perceptron and convolutional neural network, for detecting significant aortic stenosis using ECGs.
The developed algorithm achieved performance as a potentially reliable screening tool for detecting significant aortic stenosis.
We used a sensitivity map to visualize the region of the ECG that was used for discrimination by the convolutional neural network–based algorithm.
What Are the Clinical Implications?
Reliable ECG screening to detect aortic stenosis may prove important because the majority of patients with AS are asymptomatic, and early diagnosis is essential for preventing irreversible disease progression and mortality.
Nonstandard Abbreviations and Acronyms.
2D 2‐dimensional
AS aortic stenosis
AUC area under the receiver operating characteristic curve
CNN convolutional neural network
MLP multilayer perceptron
The burden of valvular heart disease is increasing owing to prolonged life expectancy.1 Aortic stenosis (AS) is the most common of these diseases in developed countries.2 The typical course of AS involves a long asymptomatic period—many patients with severe AS are asymptomatic.3, 4 Once symptoms begin, mortality increases.5 Without surgery, 40% to 50% of patients with classic symptoms die within 1 year.5, 6 Good outcomes generally result from careful follow‐up in asymptomatic individuals and urgent aortic valve replacement in symptomatic individuals.3, 7, 8 Screening is important to avoid irreversible disease progression and preventable death; however, there are no suitable screening tools for asymptomatic patients.
Diagnostic methods for AS include ECG, chest radiography, and echocardiography.9, 10 ECG and chest radiography lack sensitivity and specificity.10 In AS patients, an ECG usually demonstrates left ventricular hypertrophy.11 In addition, a left or right bundle‐branch block may be identified in up to 10% of patients. Chest radiography results typically appear normal in the early stages of the disease, but signs of left ventricular hypertrophy and congestive heart failure eventually develop.10, 11 Echocardiography is used to confirm an AS diagnosis and to determine severity.9 However, echocardiography is an expensive, time‐consuming, and less accessible among screening tools. As such, echocardiography is conducted for patients suspected of having severe symptomatic AS rather than for asymptomatic patients.9
To develop a reliable screening method based on ECG, we used a deep learning–based algorithm combining a multilayer perceptron (MLP) and convolutional neural network (CNN). Deep learning has shown high accuracy and applicability in computer vision, speech recognition, and signal processing.12 Deep learning has also been applied in several medical domains, such as detecting retinopathy and cardiac arrest, diagnosing left systolic dysfunction, and predicting the occurrence of atrial fibrillation using ECG.13, 14, 15, 16 In this study, we developed and validated an algorithm based on deep learning for detecting AS using 12‐lead ECG. Furthermore, we evaluated the performance of the algorithm in detecting AS using single‐lead ECG and visualized the algorithm's decision‐making using a sensitivity map.
Methods
The data that support the findings of this study are available from the corresponding author on reasonable request.
Study Design and Population
This multicenter retrospective cohort study involved data from 2 hospitals (labeled A and B) to develop and validate an MLP‐ and CNN‐based algorithm for detecting AS. Hospital A is a cardiovascular teaching hospital, and hospital B is a community general hospital. The study participants were adult patients (aged ≥18 years) who underwent both ECG and echocardiography within 4 weeks. In other words, we included patients who underwent electro‐ and echocardiography during the study period and those in whom the difference between the ECG and echocardiogram dates was <4 weeks. We excluded patients for whom demographic, ECG, or echocardiogram information was missing. As shown in Figure 1, patients who were treated at hospital A (October 2016–March 2019) were randomly split into algorithm derivation (80%) and internal validation (20%) data sets. The derivation data set was used to develop the algorithm based on deep learning. In the derivation data set, we used several ECGs within 4 weeks of the echocardiography date. Using this method, we amplified and created an ECG data set sufficient for developing a deep learning–based algorithm. We then evaluated the accuracy of the algorithm using the internal validation data. Furthermore, we used the hospital B data as an external validation data set (March 2017–March 2019) to verify that the algorithm was applicable across centers. Because the purpose of the validation data was to assess the accuracy of the algorithm, we used only 1 ECG from each patient—the most recent before their echocardiography—for the internal and external validation data sets. This study complied with the Declaration of Helsinki. The institutional review boards of Sejong General Hospital (2019‐0356) and Mediplex Sejong Hospital (2019‐064) approved this study protocol and waived the need for informed consent, given impracticality and minimal harm.
Figure 1.
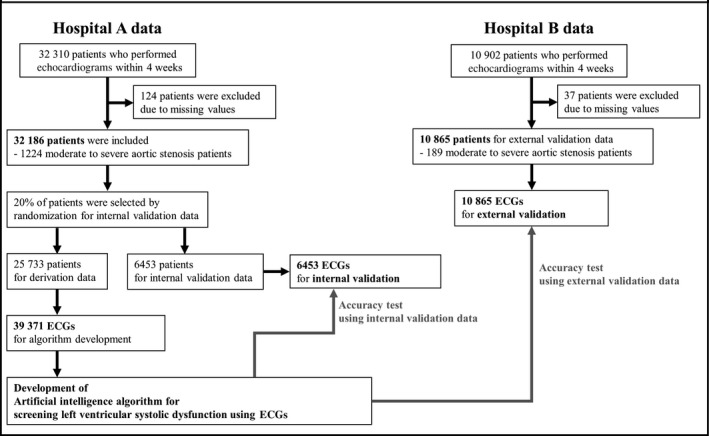
Study flowchart.
End Point and Predictive Variables
The primary end point of this study was significant (beyond moderate) AS, defined as an aortic valve area ≤1.5 cm2 or a mean pressure gradient ≥20 mm Hg, confirmed by echocardiography.17 We used each patient's demographic information and ECG as the predictive variables. We used 4 variables—age, sex, weight, and height—as the demographic information. As shown in Figure 2, we used the ECG data in 2 ways. First, we used features of the ECG, such as heart rate, presence of atrial fibrillation or flutter, QT interval, corrected QT interval (QTc), QRS duration, R‐wave axis, and T‐wave axis, to develop an algorithm with the demographic information. Second, we used the raw ECG data. In the raw data of each 12‐lead ECG, there were 5000 numbers for each lead, recorded over 10 seconds (500 Hz)—60 000 numbers in total. We used 8 seconds of ECG data by excluding the first and last 1‐second periods because more artifacts were contained within these ranges. Consequently, we created 2‐dimensional (2D) data of 12×4000 from each ECG to develop and validate the algorithm.
Figure 2.
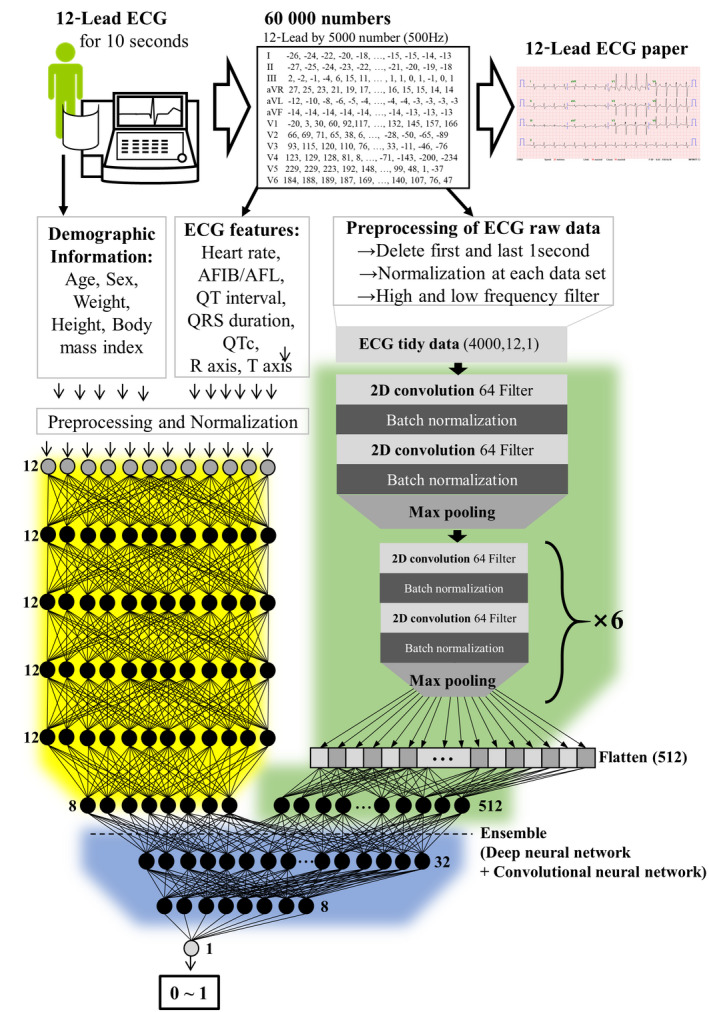
ECG data and artificial intelligence algorithm. AFIB indicates atrial fibrillation; AFL, atrial flutter; QTc, corrected QT interval; and 2D, 2‐dimensional.
Algorithm Development
As shown in Figure 2, the algorithm was developed using 3 deep learning methods. First, we developed an MLP with 6 hidden layers, 81 nodes, and batch‐normalization layers (Figure 2,yellowarea) to detect AS; the input comprised the 12 patient features (age, sex, weight, height, body mass index, heart rate, presence of atrial fibrillation of atrial flutter, QT interval, QTc, QRS duration, R‐wave axis, and T‐wave axis), and the output was a prediction between 0 and 1.18, 19 Second, we developed a CNN with 2D convolutional, max‐pooling, flattened, and batch‐normalization layers (Figure 2,greenarea) to detect AS; the input was the raw ECG data (sampled at 500 Hz, or 500 samples per second for 12‐lead data), and the output was a prediction between 0 and 1.20, 21, 22, 23 We confirmed the architecture of the algorithm and the hyperparameters for training using a grid search. Each experiment was conducted 100 times, and we chose the smallest layer unless a statistically significant difference was found (P <0.01). The best algorithm was selected using the binary cross entropy and mean absolute error as the loss function and metric, respectively. The dimensions of the original ECG data (2D, 5000×12) and the input data (2D, 4000×12) for the algorithm were the same. Because a flattening layer was included, a process to decrease the number of dimensions was applied using the deep learning algorithm. Numerous studies on deep learning applied to an ECG have used a flattening layer at the end of the architecture to obtain generalization for the classification tasks.
The input data for ECG were composed of a 2D array (12 x 4000) of numbers. To make the input of 2D ECG data, we rearranged the data in the following order: V1, V2, V3, V4, V6, aVL, lead 1, −aVR, lead 2, aVF, and lead 3. In this manner, the data were rearranged in the order of the axis angle. Consequently, the data in similar rows were arranged with similar angles. Because ECG was recorded over time, data in similar columns contained information from similar times. The CNN and pooling layer are famous architectures for learning the 2D image data because the architectures are suitable for filtering the spatial locality of the 2D data and extracting the features from the relationship between data, which is closed location. Image data and ECG input data have similar characteristics in that similar information is arranged in similar locations, and thus the CNN has shown high accuracy in many studies of deep learning algorithms for ECG raw data. We tested several different arrangements of the ECG input data with grid searches and confirmed the arrangement that showed the best performance.
The CNN network architecture consisted of 7 residual blocks with 2 CNN layers per block. The proposed deep learning–based algorithm was developed using an ensemble method combining the MLP and CNN algorithms (Figure 2,bluearea), for which the input is the raw ECG data and 12 features, and the output is a prediction between 0 and 1.24 TensorFlow (Google Brain Team) was the back end.25 We used the Adam optimizer with the default parameters β1=0.9 and β2=0.999 and a mini batch size of 32. We initialized the learning rate at 1×10−3 and reduced it by a factor of 10 when the developmentally set loss stopped improving for each consecutive epoch. We chose the model that achieved the lowest error on the derivation data set.
The hyperparameters of the algorithm architecture and the optimization algorithm were selected using a grid search. We searched the number of convolutional layers, the size and number of the convolutional filters, and the use of a dropout layer and a batch‐normalization layer. When we added a residual block of CNN >7, there was no significant increase in accuracy. Consequently, we selected the final CNN algorithm with a residual block of 7. The number of filters in each convolutional layer was selected by a grid search. Experiments were performed to confirm the number of nodes in the MLP layer. We chose the smallest node unless there was a statistical significantly difference (P<0.001). Because the dropout and batch‐normalization layers added to the accuracy, we selected the layers in the final architecture.
To evaluate the performance of the algorithm when using 1 ECG lead, we developed the algorithm using 1 ECG lead and validated the same ECG lead. For example, we developed an algorithm using raw data from lead 1 and validated the algorithm using raw data from lead 1. We then developed an algorithm using raw data of lead 2 and validated the algorithm using raw data from lead 2. In the same manner, we developed and validated the algorithm using each ECG lead (lead 1, lead 2, lead 3, aVF, aVR, aVL, V1, V2, V3, V4, V5, and V6). We developed an additional deep learning–based algorithm for which we used 4000 numbers from each single lead in the derivation data set as input information. The single‐lead algorithm was developed as an ensemble method, combining the MLP (age, sex, weight, height, heart rate, presence of atrial fibrillation or flutter, QT interval, QTc, and QRS duration) and CNN (raw data of each single lead). To enhance the performance of the single‐lead algorithm, we used a short‐time Fourier transformation that separated the signal into different frequency components, which helped the generalizability of the model.26 Because the short‐time Fourier transformation did not enhance the performance of the 12‐lead algorithm, it was left out of that design.
We also developed additional algorithms based on conventional machine learning models to compare with the ensemble method. For this, we used logistic regression, random forest, simple neural network (1 hidden layer), and support vector machine methods developed with the glm, randomForest, nnet, and e1071 packages, respectively, in R (R Development Core Team).27 These machine learning methods showed better performance than traditional methods in several medical domains in previous studies.28, 29
Visualizing Developed Algorithms Based on Deep Learning
To understand the model and make a comparison with existing medical knowledge, it was important to identify which region had a significant effect on the decision of the algorithm based on the CNN. In this work, we used a sensitivity map as a saliency method. The map was computed utilizing the first‐order gradients of the classifier probabilities with respect to the input signals. If the probability of a classifier was sensitive to a specific region of the signal, the region would be considered significant in the model. In this study, we used a gradient‐class activation map as a sensitivity map and guided gradient back‐propagation methods.30 The sensitivity map showed the region of importance of the first convolutional layer in the CNN part. Because the number of filters of the first convolutional layer was 64, the sensitivity map described the region of importance for determining the presence of AS as grade 64. We visualized grade 0 as black and grade 64 as yellow.
Algorithm Evaluation
After developing the prediction algorithms, we input the feature data and ECG raw signal of each patient in the validation data into the developed algorithms. Each deep learning–based algorithm calculated the probability of significant AS in the range from 0 (non‐AS) to 1 (AS). To confirm the performance of the developed deep learning–based algorithms, we compared the probability calculated by the algorithm with the presence of AS in the validation data set.
Statistical Analysis
We used the area under the receiver operating characteristic curve (AUC) as a comparative metric.31 A 2‐sided P<0.05 was considered significant for all tests. We evaluated the 95% CI using bootstrapping (resampling 10 000 times with replacement).32 For the bootstrapping, we applied 50% random sampling with replacement using all validation data. We conducted the bootstrapping 10 000 times and confirmed each confidence interval. All statistical analyses were performed using R. Because the purpose of this algorithm was screening for AS in the general population for transferring to confirmative diagnosis using echocardiography, we computed the specificity and accuracy when the sensitivity was 0.8 in each predictive algorithm. We also evaluated the 95% CI for each result. Because the purpose of the proposed algorithm is the screening of AS using an ECG, we also confirmed the specificity, negative predictive value, and positive predictive value at an operating point with high sensitivity (90%).
Although sensitivity maps indicate specific regions, we could not confirm the relationship between the decision of the algorithm and qualitative features that physicians used. To overcome this limitation, we used 2 methods. First, we used variable importance in the MLP part of the developed deep learning algorithm; the features included demographic and ECG parameters. Second, we studied the degree to which the convolutional part of the developed algorithm learned the features understood by physicians. To determine the correlation between the process of the CNN part of the developed algorithm and qualitative features of ECG, we tested 6 features: heart rate, presence of atrial fibrillation or flutter, QT interval, QRS duration, R‐wave axis, and T‐wave axis. The performance of the linear regressors was evaluated using the coefficient of determination. If the encoded features contained enough information to regress on the qualitative features, the regressor would predict the qualitative features correctly and have a high value for the coefficient of determination.
Results
In total, 43 212 patients were eligible to be included in this study (Figure 1). We excluded 161 patients who were missing values. The study included 43 051 patients, of whom 1413 had significant AS. The baseline characteristics of the study participants are shown in Table. Algorithms based on deep learning and conventional machine learning were developed using a derivation data set of 39 371 ECGs (12‐lead) from 25 733 patients. An additional deep learning–based algorithm was developed using the single‐lead data from the same data set. The performance of the algorithm was then verified using 6453 ECGs from the 6453 patients in the internal validation data set from hospital A and 10 865 ECGs from the 10 865 patients in the external validation data set from hospital B. We provided the developed deep learning algorithm to other researchers in Table S1. The H5 file can be used with Python to validate the algorithm and better understand its architecture.
Table 1.
Baseline Characteristics
| Characteristic | Hospital A (Derivation and Internal Validation Data) | Hospital B (External Validation Data) | P Value‡ | ||||
|---|---|---|---|---|---|---|---|
| Non‐AS | AS | P Value† | Non‐AS | AS | P Value† | ||
| Study participants, n (%) | 30 962 (96.2%) | 1224 (3.8) | 10 676 (98.3%) | 189 (1.7) | <0.001 | ||
| Participant characteristics | |||||||
| Age, y | 60.21 (15.27) | 71.64 (12.12) | <0.001 | 58.01 (15.26) | 73.19 (12.33) | <0.001 | <0.001 |
| Male, n (%) | 15 695 (50.7) | 480 (39.2) | <0.001 | 5368 (50.3) | 67 (35.4) | <0.001 | 0.684 |
| Weight, Kg | 64.75 (12.30) | 59.84 (10.97) | <0.001 | 66.01 (13.34) | 59.79 (11.22) | <0.001 | <0.001 |
| Height, cm | 162.29 (9.32) | 157.58 (9.09) | <0.001 | 163.12 (9.49) | 157.26 (9.58) | <0.001 | <0.001 |
| BMI, kg/m2 | 24.48 (3.55) | 24.01 (3.37) | <0.001 | 24.69 (3.80) | 24.12 (3.72) | 0.040 | <0.001 |
| Heart rate, bpm | 75.23 (18.88) | 76.59 (20.28) | 0.014 | 72.54 (16.05) | 76.56 (20.27) | 0.001 | <0.001 |
| Echocardiographic findings | |||||||
| LVSD, mm | 30.57 (7.49) | 30.62 (8.05) | 0.821 | 30.71 (6.24) | 30.84 (7.21) | 0.777 | 0.087 |
| LVDD, mm | 47.95 (6.28) | 48.23 (7.07) | 0.133 | 48.66 (5.24) | 48.82 (6.24) | 0.680 | <0.001 |
| Septum, mm | 10.01 (1.82) | 11.51 (1.91) | <0.001 | 9.45 (1.81) | 10.94 (1.97) | <0.001 | <0.001 |
| PWT, mm | 9.60 (1.60) | 10.94 (1.60) | <0.001 | 9.15 (1.53) | 10.46 (1.52) | <0.001 | <0.001 |
| Aorta, mm | 31.97 (4.22) | 31.82 (4.35) | 0.234 | 30.49 (3.91) | 29.95 (3.80) | 0.061 | <0.001 |
| LAD, mm | 39.99 (8.36) | 47.51 (10.65) | <0.001 | 37.40 (6.79) | 44.12 (8.39) | <0.001 | <0.001 |
| E, cm/s | 64.44 (21.05) | 78.21 (32.53) | <0.001 | 66.71 (18.83) | 81.62 (27.83) | <0.001 | <0.001 |
| A, cm/s | 67.37 (16.50) | 75.70 (17.96) | <0.001 | 68.20 (16.24) | 78.24 (17.14) | <0.001 | <0.001 |
| DT, ms | 198.61 (57.64) | 247.75 (96.42) | <0.001 | 214.48 (50.61) | 247.69 (86.10) | <0.001 | <0.001 |
| E′, cm/s | 6.66 (2.60) | 4.72 (1.57) | <0.001 | 6.79 (2.52) | 4.71 (1.61) | <0.001 | <0.001 |
| A′, cm/s | 8.65 (2.14) | 7.38 (2.12) | <0.001 | 8.51 (2.01) | 7.64 (2.37) | <0.001 | <0.001 |
| E/E′ | 10.75 (5.05) | 17.58 (7.98) | <0.001 | 10.85 (4.65) | 18.53 (8.17) | <0.001 | 0.990 |
| TRPG | 21.62 (7.74) | 28.21 (10.53) | <0.001 | 20.95 (6.95) | 28.07 (10.60) | <0.001 | <0.001 |
| PA pressure, mmHg | 25.46 (9.04) | 32.99 (12.25) | <0.001 | 24.19 (7.65) | 31.93 (11.96) | <0.001 | <0.001 |
| LVMI, g/m2 | 100.15 (30.28) | 129.54 (36.60) | <0.001 | 94.25 (26.48) | 123.48 (34.98) | <0.001 | <0.001 |
| AVA, cm2 | 1.90 (0.39) | 1.06 (0.34) | <0.001 | 1.74 (0.20) | 1.13 (0.34) | <0.001 | 0.017 |
| Mean PG, mmHg | 7.44 (3.87) | 32.05 (19.17) | <0.001 | 11.04 (3.88) | 32.06 (19.36) | <0.001 | <0.001 |
| EF, % | 57.44 (9.91) | 55.88 (10.09) | <0.001 | 63.51 (9.68) | 60.90 (12.27) | <0.001 | <0.001 |
| Electrocardiographic findings | |||||||
| AF, n (%) | 3920 (12.7) | 324 (26.5) | <0.001 | 742 (7.0) | 38 (20.1) | <0.001 | <0.001 |
| QT interval, ms | 400.44 (45.18) | 414.16 (54.30) | <0.001 | 400.92 (40.27) | 407.83 (52.17) | 0.020 | 0.880 |
| QTc | 440.72 (36.33) | 458.17 (40.40) | <0.001 | 434.89 (34.43) | 452.24 (44.91) | <0.001 | <0.001 |
| QRS duration, ms | 97.25 (18.57) | 100.67 (22.62) | <0.001 | 96.35 (16.50) | 99.78 (21.12) | 0.005 | <0.001 |
| R axis, angle | 38.03 (45.97) | 36.80 (46.38) | 0.358 | 37.94 (40.44) | 34.16 (43.94) | 0.203 | 0.831 |
| T‐wave peak, mV | 0.23 (0.30) | 0.25 (0.37) | 0.021 | 0.24 (0.25) | 0.27 (0.26) | 0.102 | 0.007 |
| T‐wave inversion, n | 4223 (13.6%) | 188 (15.4%) | 0.094 | 1177 (11.0%) | 23 (12.2%) | 0.704 | <0.001 |
| T axis, angle | 47.87 (54.77) | 78.09 (77.64) | <0.001 | 43.22 (44.03) | 76.23 (74.61) | <0.001 | <0.001 |
A indicates late diastolic mitral inflow velocity, A′ late diastolic mitral annular tissue velocity; AF, atrial fibrillation or atrial flutter; AS, aortic stenosis; AVA, aortic valve area; BMI, body mass index; DT, deceleration time; E, early diastolic mitral inflow velocity; E′, early diastolic mitral annular tissue velocity; EF, ejection fraction; LAD, left atrial dimension; LVDD, left ventricular diastolic dimension; LVMI, left ventricular mass index; LVSD, left ventricular systolic dimension; PA, pulmonary artery; PG, pressure gradient; PWT, posterior wall thickness; QTc, corrected QT interval; and TRPG, tricuspid regurgitation peak gradient.
The alternative hypothesis for this P value was that there was a difference between the AS and non‐AS data groups for each variable.
The alternative hypothesis for this P value was that there is a difference between hospital A (derivation and internal validation data group) and hospital B (external validation group) for each variable.
As shown in Figure 3, during internal validation, the AUC of the ensemble algorithm combining CNN and MLP was 0.884 (95% CI, 0.880–0.887)—significantly greater than that of the CNN (0.825; 95% CI, 0.821–0.829), MLP (0.800; 95% CI, 0.792–0.808), and other machine learning algorithms. In external validation, the AUC of the ensemble algorithm combining CNN and MLP was 0.861 (95% CI, 0.858–0.863)—significantly greater than that of the CNN (0.816; 95% CI, 0.812–0.819), MLP (0.807; 95% CI, 0.800–0.815), and other machine learning algorithms. As shown in the Tables S2 and S3, at the highly sensitive operating point, the negative predictive value was >99%.
Figure 3.
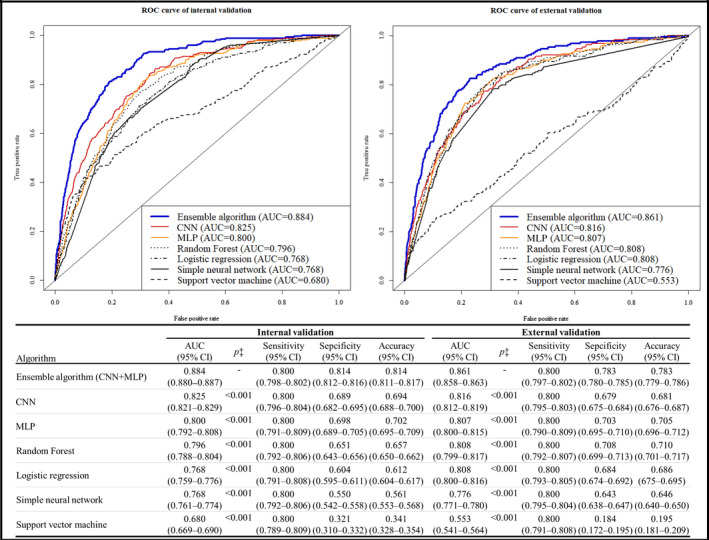
Performance of artificial intelligence algorithms for detecting aortic stenosis. AUC indicates area under the receiver operating characteristic curve; CNN, convolutional neural network; MLP, multilayer perceptron; and ROC, receiver operating characteristic.
The AUCs of the single‐lead ensemble algorithm during internal and external validation using lead 2 were 0.845 (95% CI, 0.841–0.848) and 0.821 (95% CI, 0.816–0.825), respectively; the results of the ensemble algorithms using other single leads are shown in Table S4.
As shown in Figure 4, we used a sensitivity map to visualize the ECG region used by the algorithm to identify AS. The map shows that the proposed algorithm focused on the T wave of the precordial lead to determine the presence of significant AS.
Figure 4.

Sensitivity map for confirming the region associated with prediction of aortic stenosis (AS).
The sensitivity map showed the convolutional neural network's (CNN's) region of algorithm attention for determining the presence of AS. The most important region is in yellow, and the least important region is in black. Because the number of filters for the first convolutional layer was 64, the sensitivity map described the region of importance for determining the presence of AS as grade 64. We visualized grade 0 as black and grade 64 as yellow. The sensitivity map showed the initial area of T wave in V2–V5 as the most important region used by the developed CNN algorithm for the decision.
As shown in Table S1, T‐wave axis, age, and QTc were the most importance variables in the MLP part of the developed algorithm. T‐wave axis and QT interval had high correlations with the model‐derived features of the CNN part (AUC: 0.695 and 0.566, respectively).
Discussion
In this study, we developed a deep learning–based algorithm, combining MLP and CNN, for detecting AS using 12‐lead and single‐lead ECGs. In addition, we developed an ensemble algorithm using single‐lead ECG that showed reasonable performance. We then visualized the CNN part of developed ensemble algorithm to determine the regions and characteristics of the ECG used for detecting AS.
Developing a reliable screening tool for detecting significant AS is important because the majority of patients with severe AS are asymptomatic, and early diagnosis is essential for preventing irreversible disease progression and mortality.3 AS is one of the most common valvular diseases in developed countries, and its prevalence is projected to increase over the next decade with an aging population.2 If significant AS could be detected using a conventional 12‐lead ECG or a single‐lead device, patients could be referred for echocardiography and early diagnosis. However, no reliable screening tools exist currently. Electrocardiography and chest radiography lack sensitivity and specificity, and echocardiography and exercise tests are expensive, time‐consuming, and inaccessible.9, 10
To address this need, we developed a deep learning–based algorithm as a reliable AS‐screening tool. Deep learning includes feature learning, which is a set of methods that allow the creation of a model that uses raw data for automatic identification of the features and relationships needed to perform a task.12 As the learning process evolves automatically, the model becomes increasingly effective at identifying intricate structures in high‐dimensional data without information loss and requires little engineering by humans.12 Consequently, it can be applied quickly and easily to many tasks and can extract meaningful information from the data without human bias. In the MLP algorithm, we used the value selected by the physician; therefore, the performance of the algorithm was limited by manual feature extraction. In the CNN algorithm, we used the raw ECG data; thus, the complete information of the ECG raw signal was used. Although more computing power and data storage were required to process and use the raw signal for the CNN, we were able to uncover new information from the ECG and use the features of ECG itself over human bias. Therefore, we were able to make more accurate algorithms using CNN.
The most important aspect of deep learning is its ability to use various types of data, such as images, 2D data, and waveforms. In this study, we used not only variables from domain knowledge (age, sex, weight, height, heart rate, presence of atrial fibrillation or flutter, QT interval, QTc, QRS duration, R‐wave axis, and T‐wave axis) but also ECG raw data (2D numerical data, 12×4000). Similar to our use of ECG patterns for the diagnosis of AS, Attia et al14, 16 developed an algorithm based on CNN for screening cardiac contractile dysfunction and predicting the occurrence of atrial fibrillation during sinus rhythm using 12‐lead ECGs and demonstrated its feasibility. However, deep learning is often criticized for the unreliability of its outcomes because of the unpredictability of the process. Consequently, we used a sensitivity map to visualize the region of the ECG that was used for decision‐making by the CNN‐based algorithm. To the best of our knowledge, this study is the first to develop a deep learning–based algorithm for detecting AS and to visualize the ECG region that the algorithm used for decision‐making.
In this study, a sensitivity map showed that the CNN part of the developed ensemble algorithm focused on the T wave of the right precordial lead (V1–4) to determine the presence of significant AS. Furthermore, the variable importance of the MLP part and correlation results of the convolutional part also showed that the T‐wave axis and QT interval were both important factors for determining the presence of AS. As shown in Table, there were significant differences between QT interval and T‐wave axis in the AS and non‐AS data groups. The T‐wave peak in ECGs from the AS data group showed higher values than those in the non‐AS data group. In addition, a T‐wave inversion occurred less frequently in the ECGs of the AS data group. Russo et al33 also described that hypertrophy in response to systolic ventricular overload prolongs ventricular activation time, which in turn may cause reversal of repolarization from endocardium to epicardium and thus the inversion of the T wave (also see Xin et al33, 34). And Greve et al35 confirmed that T‐wave inversion in leads V4 through V6 reflects peak aortic jet velocity better than ST‐segment depression. T‐wave inversion was independently predictive of poor prognosis in patients with asymptomatic AS in previous studies.36, 37 In addition, AS was correlated with S‐wave amplitude and T‐wave strain patterns of the right precordial lead in previous studies. Vranic38 showed that S wave changes in right precordial leads can predict increases in the pressure gradient and critical narrowing of the aortic valve area, and Xiao et al39 confirmed that right precordial Q waves help to distinguish anterior myocardial infarction from AS.38, 39 Taniguchi et al40 showed associations of ST‐segment elevations in the right precordial lead with different clinical outcomes in AS patients. Although the T wave represents the repolarization of ventricles, it is also related to the QRS complex and ST segment, which correspond to the depolarization and interval, respectively. Owing to statistical and technical limitations, we do not yet know the exact meaning of the T wave in AS. Nevertheless, this study confirmed that the T wave of the right precordial lead is important for discriminating AS and that there were the differences between the ECGs in the AS and non‐AS data groups. We need to conduct additional experiments regarding the shape and slope of the T wave, which remains a subject of our next study.
The purpose of the developed algorithm was screening for AS. The developed algorithm achieved an AUC of ≈0.861 to 0.884. The performance of the model is greater than that of other common screening tests, such as mammography used for breast cancer screening (AUC=0.78; positive predictive value: 3–12%) and fecal occult blood testing for the detection of colorectal neoplasia (AUC=0.71; overall sensitivity: 29%).41, 42 As shown in Tables S2 and S3, at the highly sensitive operating point, the developed algorithm performs well as a potential screening tool for ruling out AS, with a negative predictive value >99%. Although the performance of the developed algorithm remains unsatisfactory, the possibility of applying deep learning to the field of electrocardiography is shown in this study.
This study has several limitations. First, because the study was conducted in only 2 hospitals in Korea, it is necessary to validate the model with patients in other countries. Because an algorithm based on deep learning can overfit the training data, it is important to confirm its accuracy in other situations. Second, “advanced ECG” has powerful features such as spatial QRS‐T angle, spatial ventricular gradient, azimuths, and elevations of the QRS, especially of the T wave.43 However, those values could not be calculated automatically with precision, so we could not adopt those features. Because the data set is too large to take those values manually, we could use only 7 features in the MLP algorithm. Third, we need to further explore the decision process of the algorithm based on deep learning (MLP and CNN).44, 45 For example, additional experiments are required to advance our understanding of the deep learning process and thus determine which characteristics of the precordial T wave influence the algorithm's decisions. This will be our next area of study.
Conclusions
An algorithm based on MLP and CNN had accurate performance to detect significant AS using both 12‐lead and single‐lead ECGs.
Disclosures
None.
Supporting information
Tables S1–S4
J Am Heart Assoc. 2020;9:e014717 DOI: 10.1161/JAHA.119.014717
See Editorial by Gladding et al.
References
- 1. Kodali SK, Velagapudi P, Hahn RT, Abbott D, Leon MB. Valvular heart disease in patients ≥80 years of age. J Am Coll Cardiol. 2018;71:2058–2072. [DOI] [PubMed] [Google Scholar]
- 2. Carabello BA, Paulus WJ. Aortic stenosis. Lancet. 2009;373:956–966. [DOI] [PubMed] [Google Scholar]
- 3. Lancellotti P, Magne J, Dulgheru R, Clavel M‐A, Donal E, Vannan MA, Chambers J, Rosenhek R, Habib G, Lloyd G, et al. Outcomes of patients with asymptomatic aortic stenosis followed up in heart valve clinics. JAMA Cardiol. 2018;3:1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bonow RO. Exercise hemodynamics and risk assessment in asymptomatic aortic stenosis. Circulation. 2012;126:803–805. [DOI] [PubMed] [Google Scholar]
- 5. Leon MB, Smith CR, Mack M, Miller DC, Moses JW, Svensson LG, Tuzcu EM, Webb JG, Fontana GP, Makkar RR, et al. Transcatheter aortic‐valve implantation for aortic stenosis in patients who cannot undergo surgery. N Engl J Med. 2010;363:1597–1607. [DOI] [PubMed] [Google Scholar]
- 6. Ben‐Dor I, Pichard AD, Gonzalez MA, Weissman G, Li Y, Goldstein SA, Okubagzi P, Syed AI, Maluenda G, Collins SD, et al. Correlates and causes of death in patients with severe symptomatic aortic stenosis who are not eligible to participate in a clinical trial of transcatheter aortic valve implantation. Circulation. 2010;122:S37–S42. [DOI] [PubMed] [Google Scholar]
- 7. Maes F, Lerakis S, Barbosa Ribeiro H, Gilard M, Cavalcante JL, Makkar R, Herrmann HC, Windecker S, Enriquez‐Sarano M, Cheema AN, et al. Outcomes from transcatheter aortic valve replacement in patients with low‐flow, low‐gradient aortic stenosis and left ventricular ejection fraction less than 30%. JAMA Cardiol. 2019;4:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baumgartner H, Falk V, Bax JJ, De Bonis M, Hamm C, Holm PJ, Iung B, Lancellotti P, Lansac E, Rodriguez Muñoz D, et al. 2017 ESC/EACTS guidelines for the management of valvular heart disease. Eur Heart J. 2017;38:2739–2791. [DOI] [PubMed] [Google Scholar]
- 9. Saikrishnan N, Kumar G, Sawaya FJ, Lerakis S, Yoganathan AP. Accurate assessment of aortic stenosis. Circulation. 2014;129:244–253. [DOI] [PubMed] [Google Scholar]
- 10. Maganti K, Rigolin VH, Sarano ME, Bonow RO. Valvular heart disease: diagnosis and management. Mayo Clin Proc. 2010;85:483–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alley W, Mahler S. Chapter 54: valvular emergency In: Tintinalli JE, Stapczynski JS, Ma OJ, Yealy DM, Meckler GD, Cline D, eds. Tintinalli's Emergency Medicine: A Comprehensive Study Guide. New York, NY: McGraw Hill Education; 2016;373–380. [Google Scholar]
- 12. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. [DOI] [PubMed] [Google Scholar]
- 13. Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;304:649–656. [DOI] [PubMed] [Google Scholar]
- 14. Attia ZI, Kapa S, Lopez‐Jimenez F, McKie PM, Ladewig DJ, Satam G, Pellikka PA, Enriquez‐Sarano M, Noseworthy PA, Munger TM, et al. Screening for cardiac contractile dysfunction using an artificial intelligence–enabled electrocardiogram. Nat Med. 2019;25:70–74. [DOI] [PubMed] [Google Scholar]
- 15. Kwon J‐M, Lee Y, Lee Y, Lee S, Park J. An algorithm based on deep learning for predicting in‐hospital cardiac arrest. J Am Heart Assoc. 2018;7:e008678 DOI: 10.1161/JAHA.118.008678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Attia ZI, Noseworthy PA, Lopez‐Jimenez F, Asirvatham SJ, Deshmukh AJ, Gersh BJ, Carter RE, Yao X, Rabinstein AA, Erickson BJ, et al. An artificial intelligence‐enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet. 2019;394:861–867. [DOI] [PubMed] [Google Scholar]
- 17. Bonow RO, Brown AS, Gillam LD, Kapadia SR, Kavinsky CJ, Lindman BR, Mack MJ, Thourani VH. ACC/AATS/AHA/ASE/EACTS/HVS/SCA/SCAI/SCCT/SCMR/STS 2017 appropriate use criteria for the treatment of patients with severe aortic stenosis. J Am Coll Cardiol. 2017;70:2566–2598. [DOI] [PubMed] [Google Scholar]
- 18. Bengio Y, Courville A, Vincent P. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell. 2013;35:1798–1828. [DOI] [PubMed] [Google Scholar]
- 19. Schmidhuber J. Deep learning in neural networks: an overview. Neural Networks. 2015;61:85–117. [DOI] [PubMed] [Google Scholar]
- 20. Hannun AY, Rajpurkar P, Haghpanahi M, Tison GH, Bourn C, Turakhia MP, Ng AY. Cardiologist‐level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med. 2019;25:65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Proc IEEE Int Conf Comput Vis. 2016;1:770–778. [Google Scholar]
- 22. LeCun Y, Boser B, Denker JS, Howard RE, Habbard W, Jackel LD. Handwritten digit recognition with a back‐propagation network. Adv Neural Inf Process Syst. 1990;1:396–404. [Google Scholar]
- 23. LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient‐based learning applied to document recognition. Proc IEEE. 1998;1:2278–2324. [Google Scholar]
- 24. Dietterich TG. Ensemble methods in machine learning. In: Multiple Classifier System. London, UK: Springer; 2000;1–15. [Google Scholar]
- 25. Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, et al. TensorFlow: a system for large‐scale machine learning TensorFlow: a system for large‐scale machine learning. 12th USENIX Symp Oper Syst Des Implement (OSDI ‘16). 2016;265–284.
- 26. Pandit SV. ECG baseline drift removal through STFT. In: Proceedings of 18th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE; 1:1405–1406. [Google Scholar]
- 27. Kuhn M, Johnson K. Applied Predictive Modeling. New York, NY: Springer; 2013. [Google Scholar]
- 28. Mortazavi BJ, Downing NS, Bucholz EM, Dharmarajan K, Manhapra A, Li SX, Negahban SN, Krumholz HM. Analysis of machine learning techniques for heart failure readmissions. Circ Cardiovasc Qual Outcomes. 2016;9:629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frizzell JD, Liang L, Schulte PJ, Yancy CW, Heidenreich PA, Hernandez AF, Bhatt DL, Fonarow GC, Laskey WK. Prediction of 30‐day all‐cause readmissions in patients hospitalized for heart failure. JAMA Cardiol. 2017;2:204. [DOI] [PubMed] [Google Scholar]
- 30. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad‐CAM: visual explanations from deep networks via gradient‐based localization. Proc IEEE Int Conf Comput Vis. 2017;1:618–626. [Google Scholar]
- 31. Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006;27:861–874. [Google Scholar]
- 32. Carpenter J, Bithell J. Bootstrap confidence intervals: when, which, what? A practical guide for medical statisticians. Stat Med. 2000;19:1141–1164. [DOI] [PubMed] [Google Scholar]
- 33. Russo R, Rizzoli G, Stritoni P, Seminara G, Rubino M, Brumana T. T‐wave changes in patients with hemodynamic evidence of systolic or diastolic overload of the left ventricle: a retrospective study on 168 patients with isolated chronic aortic valve disease. Int J Cardiol. 1987;14:137–143. [DOI] [PubMed] [Google Scholar]
- 34. Xin D, Zheng W, Xuyu J, Yuemin S, Wenjuan Z, Bo B, Xuefang Y, Canliang H. Changes of the electrocardiographic strain pattern in patients with aortic stenosis and its underling mechanisms. Heart. 2011;97:A239–A240. [Google Scholar]
- 35. Greve AM, Gerdts E, Boman K, Gohlke‐Baerwolf C, Rossebø AB, Hammer‐Hansen S, Køber L, Willenheimer R, Wachtell K. Differences in cardiovascular risk profile between electrocardiographic hypertrophy versus strain in asymptomatic patients with aortic stenosis (from SEAS Data). Am J Cardiol. 2011;108:541–547. [DOI] [PubMed] [Google Scholar]
- 36. Greve AM, Boman K, Gohlke‐Baerwolf C, Kesäniemi YA, Nienaber C, Ray S, Egstrup K, Rossebø AB, Devereux RB, Køber L, et al. Clinical implications of electrocardiographic left ventricular strain and hypertrophy in asymptomatic patients with aortic stenosis. Circulation. 2012;125:346–353. [DOI] [PubMed] [Google Scholar]
- 37. Hering D, Piper C, Horstkotte D. Influence of atypical symptoms and electrocardiographic signs of left ventricular hypertrophy or ST‐segment/T‐wave abnormalities on the natural history of otherwise asymptomatic adults with moderate to severe aortic stenosis: preliminary communication. J Heart Valve Dis. 2004;13:182–187. [PubMed] [Google Scholar]
- 38. Vranic II. Electrocardiographic appearance of aortic stenosis before and after aortic valve replacement. Ann Noninvasive Electrocardiol. 2017;22:e12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xiao HB, Ramzy IS, Bowker TJ, Dancy M. Electrocardiographic associations of right precordial Q waves help to distinguish anterior myocardial infarction from aortic stenosis. Int J Cardiol. 2002;82:159–166. [DOI] [PubMed] [Google Scholar]
- 40. Taniguchi T, Shiomi H, Kosuge M, Morimoto T, Nakatsuma K, Nishiga M, Sasa T, Saito N, Kimura T. Prognostic significance of ST‐segment elevation in leads V1–2 in patients with severe aortic stenosis. Circ J. 2016;80:526–534. [DOI] [PubMed] [Google Scholar]
- 41. Pisano ED, Gatsonis C, Hendrick E, Yaffe M, Baum JK, Acharyya S, Conant EF, Fajardo LL, Bassett L, D'Orsi C, et al. Diagnostic performance of digital versus film mammography for breast‐cancer screening. N Engl J Med. 2005;353:1773–1783. [DOI] [PubMed] [Google Scholar]
- 42. Haug U, Kuntz KM, Knudsen AB, Hundt S, Brenner H. Sensitivity of immunochemical faecal occult blood testing for detecting left‐ vs right‐sided colorectal neoplasia. Br J Cancer. 2011;104:1779–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schlegel TT, Kulecz WB, Feiveson AH, Greco EC, DePalma JL, Starc V, Vrtovec B, Rahman MA, Bungo MW, Hayat MJ, et al. Accuracy of advanced versus strictly conventional 12‐lead ECG for detection and screening of coronary artery disease, left ventricular hypertrophy and left ventricular systolic dysfunction. BMC Cardiovasc Disord. 2010;10:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen X, Duan Y, Houthooft R, Schulman J, Sutskever I, Abbeel P. InfoGAN: interpretable representation learning by information maximizing generative adversarial nets. Neural Inf Process Syst. 2016;1:2172–2180. [Google Scholar]
- 45. Fong RC, Vedaldi A. Interpretable explanations of black boxes by meaningful perturbation. Proc IEEE Int Conf Comput Vis. 2017;1:3449–3457. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S4


