Abstract
Nowadays, automatic disease detection has become a crucial issue in medical science due to rapid population growth. An automatic disease detection framework assists doctors in the diagnosis of disease and provides exact, consistent, and fast results and reduces the death rate. Coronavirus (COVID-19) has become one of the most severe and acute diseases in recent times and has spread globally. Therefore, an automated detection system, as the fastest diagnostic option, should be implemented to impede COVID-19 from spreading. This paper aims to introduce a deep learning technique based on the combination of a convolutional neural network (CNN) and long short-term memory (LSTM) to diagnose COVID-19 automatically from X-ray images. In this system, CNN is used for deep feature extraction and LSTM is used for detection using the extracted feature. A collection of 4575 X-ray images, including 1525 images of COVID-19, were used as a dataset in this system. The experimental results show that our proposed system achieved an accuracy of 99.4%, AUC of 99.9%, specificity of 99.2%, sensitivity of 99.3%, and F1-score of 98.9%. The system achieved desired results on the currently available dataset, which can be further improved when more COVID-19 images become available. The proposed system can help doctors to diagnose and treat COVID-19 patients easily.
Keywords: Coronavirus, COVID-19, Deep learning, Chest X-ray, Convolutional neural network, Long short-term memory
1. Introduction
The coronavirus epidemic that has spread across the world has placed all sectors on lockdown. According to the World Health Organization's latest estimates, as of July 9th, 2020, more than twelve million people have been infected with close to 552,050 deaths [1]. Health systems have reached the point of failure, even in developed countries, due to the shortage of intensive care units (ICUs; COVID-19 patients with worse conditions are admitted into ICUs). The strain that began to spread in Wuhan, China was identified from two different coronaviruses, severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) [2]. The symptoms of COVID-19 can range from cold to fever, shortness of breath, and acute respiratory syndrome [3]. In comparison to SARS, the kidneys and liver are affected by coronavirus as well as the respiratory system [4].
Coronavirus detection at an early stage plays a vital role in controlling COVID-19 due to its high transmissibility. The diagnosis of coronavirus by gene sequencing for respiratory or blood samples should be confirmed as the main pointer for reverse transcription-polymerase chain reaction (RT-PCR), according to the guidelines of the Chinese government [5]. The process of RT-PCR takes 4–6 hours to get results, which takes a long time compared to COVID-19's rapid spread rate. RT-PCR test kits are in huge shortage, in addition to being inefficient [6]. As a result, many infected patients cannot be detected in time and tend to unknowingly infect others. With the detection of this disease at an early stage, the prevalence of COVID-19 disease will decrease [7]. To alleviate the inefficiency and scarcity of current COVID-19 tests, a lot of effort has been made to look for alternative test methods. Another visualization method is to diagnose COVID-19 infections using radiological images such as X-rays or computed tomography (CT). Earlier works have shown that anomalies can be found in COVID-19 patients in chest CT scans in the shape of ground-glass opacities [8]. The researchers have claimed that a system based on chest CT scans can be an important method for diagnosis and quantifying of COVID-19 cases [9].
Many researchers have demonstrated various approaches to detect COVID-19 utilizing X-ray images. Recently, computer vision [10], machine learning [[11], [12], [13]], and deep learning [14,15] have been used to automatically diagnose several diverse ailments in the human body, which ensures smart healthcare [16,17]. The deep learning method is used as a feature extractor that enhances classification accuracies [18]. The detection of tumor regions in the lungs, X-ray bone suppression, diabetic retinopathy, prostate segmentation, skin lesions, and the presence of the myocardium in coronary CT scans are examples of the contributions [19,20] of deep learning.
Therefore, this paper aims to propose a deep learning based system that combines the CNN and LSTM networks to automatically detect COVID-19 from X-ray images. In the proposed system, CNN is used for feature extraction and LSTM is used to classify COVID-19 based on those features. The LSTM network has an internal memory that is capable of learning from imperative experiences with long-term states. In fully connected networks, the layers are fully connected and the nodes between layers are connectionless and process only one input. In the case of LSTM, the nodes are connected from a directed graph along a temporal sequence that is considered an input with a specific order [21]. Hence, the 2-D CNN and LSTM layout feature combination improves classification greatly. The dataset used for this paper was collected from multiple sources and a preprocessing was performed to reduce the noise.
In the following, the contributions of this research are summarized.
-
a)
Developing a combined deep CNN-LSTM network to automatically assist the early diagnosis of patients with COVID-19 efficiently.
-
b)
To detect COVID-19 using chest X-rays using a dataset formed comprising 4575 images.
-
c)
A detailed experimental analysis is provided in terms of accuracy, sensitivity, specificity, F1-score, a confusion matrix, and AUC using receiver operating characteristic (ROC) to measure the performance of the proposed system.
The paper is organized as follows: A review of recent scholarly works related to this study is described in Section 2. A description of the proposed system, including dataset collection and preparation, is presented in Section 3. The experimental results and comparative performance of the proposed deep learning system are provided in Section 4. The discussion is given in Section 5. Section 6 concludes the paper.
2. Related works
To address the COVID-19 epidemic, researchers have developed deep learning techniques to diagnose COVID-19 based on clinical images, CT scans, and X-rays of the chest. This review describes the recently developed systems that have applied deep learning techniques in the field of COVID-19 detection.
Rahimzadeh et al. [22] developed a concatenated CNN based on Xception and ResNet50V2 models that classified COVID-19 cases using chest X-rays. The developed system used a data set that contained 180 images of COVID-19 patients, 6054 images of pneumonia patients, and 8851 images of normal people. 633 images were selected for each of the eight training phases. The experimental outcome obtained 99.56% accuracy and 80.53% recall for COVID-19 cases. Alqudah et al. [23] used artificial intelligence techniques to develop a system that detected COVID-19 from chest X-rays. The images used were classified with different machine learning techniques, such as support vector machine (SVM), CNN, and random forest (RF). The system obtained 95.2% accuracy, 100% specificity, and 93.3% sensitivity. Loey et al. [24] introduced a generative adversarial network (GAN) with deep learning to diagnose COVID-19 from chest X-rays. The scheme used the three pre-trained models AlexNet, GoogleNet, and RestNet18. The collected data included 69 images of COVID-19 cases, 79 images of pneumonia bacterial cases, 79 images of pneumonia virus cases, and 79 images of normal cases. GoogleNet was selected as a main deep learning technique with 80.6% test accuracy in the four classes scenario, AlexNet with 85.2% test accuracy in the three classes scenario, and GoogleNet with 99.9% test accuracy in the two classes scenario.
Ucar et al. [25] proposed a COVID-19 detection system based on deep architecture that utilized X-ray images. The data set included 76 images of COVID-19 cases, 4290 images of pneumonia cases, and 1583 images of normal cases. The scheme achieved 98.3% accuracy for COVID-19 cases. Apostolopoulos et al. [26] introduced a transfer learning strategy with CNN that could automatically diagnose COVID-19 cases by extracting essential features from chest X-rays. The system used the five CNN variants VGG19, Inception, MobileNet, Xception, and Inception-ResNetV2 to classify COVID-19 images. The data set included 224 images of COVID-19 patients, 700 images of pneumonia patients, and 504 images of normal patients. The data set was split using the concept of 10-fold cross-validation for training and evaluation purposes. VGG19 was selected as a main deep learning model with 93.48% accuracy, 92.85% specificity, and 98.75% sensitivity in the developed system. Bandyopadhyay et al. [27] proposed a novel model that used the LSTM-GRU to automatically classify confirmed, released, negative, and death cases of COVID-19. The developed scheme achieved 87% accuracy for the confirmed cases, 67.8% accuracy for negative cases, 62% accuracy for deceased cases, and 40.5% accuracy for released cases. Khan et al. [28] presented a deep learning network to automatically predict COVID-19 cases from chest X-rays. The data set included 284 images of COVID-19 cases, 330 images of pneumonia bacterial cases, 327 images of pneumonia viral cases, and 310 images of normal cases. Overall, the proposed system obtained 89.5% accuracy, 97% precision, and 100% recall for COVID-19 cases.
Kumar et al. [29] introduced a deep learning methodology to classify COVID-19 infected patients using chest X-rays. The scheme used nine pre-trained models for feature extraction and SVM for classification. The two data sets contained 158 X-ray images of both COVID-19 and non-COVID-19 patients. The combined ResNet50 and SVM model was statistically superior to other models, with 95.38% accuracy and 95.52% F1-score. Horry et al. [30] developed a system based on pre-trained models to detect COVID-19 from chest X-rays. The proposed system used Xception, VGG, ResNet, and Inception for classification. The dataset used in the system includes 115 images of COVID-19 cases, 322 images of pneumonia cases, and 60361 images of normal cases. Both the VGG16 and VGG19 classifiers had roughly 80% recall and precision. Hemdan et al. [31] introduced a deep learning technique to detect COVID-19 from X-ray images. The framework included the seven pre-trained models VGG19, MobileNetV2, InceptionV3, ResNetV2, DenseNet201, Xception, and InceptionResNetV2. In the developed system, 50 X-ray images were used, with 25 images from COVID-19 patients and 25 images from non-COVID-19 patients. Among the tested classifiers, VGG19 and DenseNet201 achieved the highest values of 90% accuracy and 83% precision.
Singh et al. [32] used VGG16, a deep transfer learning architecture, to detect COVID-19 from CT scans. The extracted features were selected using principal component analysis (PCA) and classified by four different classifiers. The best result achieved 95.7% accuracy, 95.8% precision, and 95.3% F1-score with a bagging ensemble method and SVM classifier. Ahuja et al. [33] introduced a transfer learning technique to diagnose COVID-19 symptoms using a three phase approach. The augmented images were applied to several pre-trained models to localize the abnormality of the CT scans, and the ResNet18 architecture obtained 99.4% accuracy in test cases. Fong et al. [34] presented a deep learning-based case study that utilized composite Monte-Carlo (CMC) and fuzzy rule induction to address the limited data for forecasting methods.
3. Methods and materials
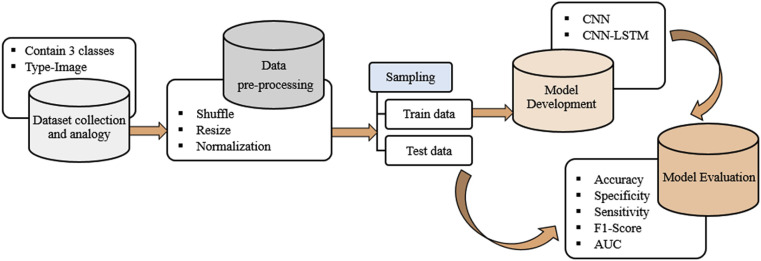
Fig. 1 illustrated the overall system for the detection of COVID-19 consisting of several phases. Raw X-ray images were first passed through the preprocessing pipeline. Data resizing, shuffling, and normalization were done in the preprocessing pipeline. The preprocessed data set was then partitioned into a training set and testing set, and we trained the CNN and CNN-LSTM architecture using the training data. After each epoch, the training accuracy and loss were determined. At the same time, using 5-fold cross-validation, validation accuracy and loss were also obtained. The performance of the proposed system was measured with the following evaluation metrics: confusion matrix, accuracy, AUC using ROC, specificity, sensitivity, and F1-score.
Fig. 1.
The overall system architecture of the proposed COVID-19 detection system.
3.1. Dataset collection and description
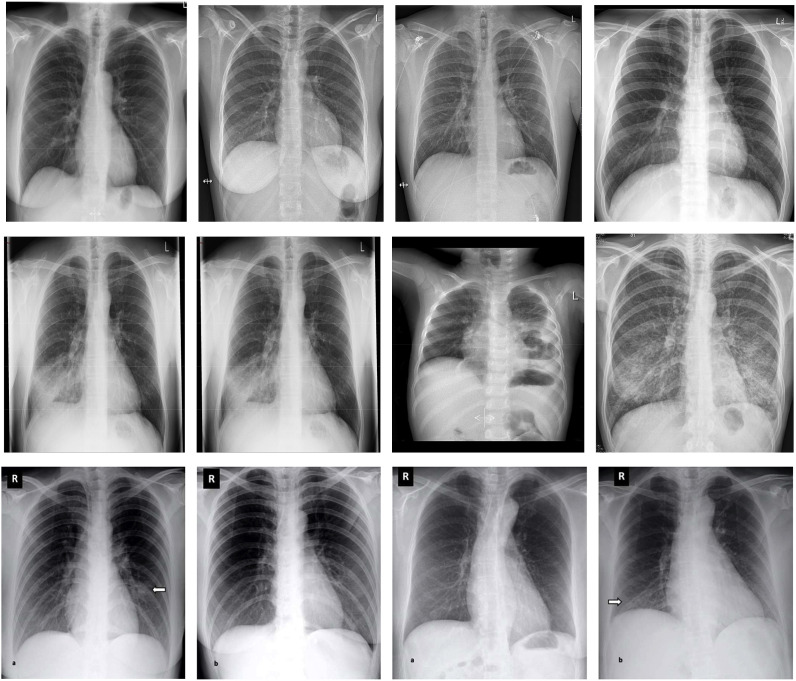
As the emergence of COVID-19 is very recent, none of the large repositories contain any COVID-19 labeled data, thereby requiring us to rely on different sources of images of normal, pneumonia, and COVID-19 cases. First, 613 X-ray images of COVID-19 cases were collected from the following websites: GitHub [35,36], Radiopaedia [37], The Cancer Imaging Archive (TCIA) [38], and the Italian Society of Radiology (SIRM) [39]. Then, instead of data being independently augmented, a dataset containing 912 already augmented images was collected from Mendeley [40]. Finally, 1525 images of pneumonia cases and 1525 X-ray images of normal cases were collected from the Kaggle repository [41] and NIH dataset [42]. The main objective of the dataset selection was to make it available to the public so that it is accessible and extensible to researchers. The use of this dataset in further studies may also enable more efficient diagnoses of COVID-19 patients. We resized the images to ones with a resolution of 224 × 224 pixels. The number of X-ray images of each set was partitioned in Table 1 . The visualization of X-ray images of each class is shown in Fig. 2 .
Table 1.
The partitioning description of used dataset.
| Data/Cases | COVID-19 | Normal | Pneumonia | Overall |
|---|---|---|---|---|
| Training | 1220 | 1220 | 1220 | 3660 |
| Testing | 305 | 305 | 305 | 915 |
| Overall | 1525 | 1525 | 1525 | 4575 |
Fig. 2.
The images in the first, second, and third rows show 4 sample images of COVID-19 cases, pneumonia cases, and normal cases respectively.
3.2. Development of combined network
The proposed architecture was developed with a combination of a convolutional neural network (CNN) and a long short-term memory (LSTM) network, which are briefly described as follows.
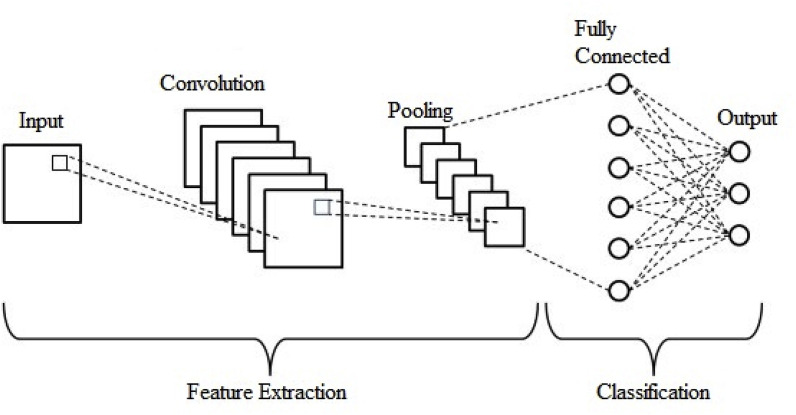
3.2.1. Convolutional neural network
A particular type of multilayer perceptron is a CNN, but a simple neural network cannot learn complex features, unlike a deep learning architecture. CNNs have shown excellent performance in many applications [43,44], such as image classification, object detection, and medical image analysis. The main idea behind a CNN is that it can obtain local features from high layer inputs and transfer them to lower layers for more complex features. A CNN comprises convolutional, pooling, and fully connected (FC) layers. A typical CNN architecture along with these layers is depicted in Fig. 3 .
Fig. 3.
A typical architecture of the convolutional neural network.
The convolutional layer encompasses a set of kernels [45] for determining a tensor of feature maps. These kernels convolve an entire input using “stride(s)” so that the dimensions of an output volume become integers [46]. The dimensions of an input volume decrease after the convolutional layer is used to execute the striding process. Therefore, zero padding [47] is required to pad an input volume with zeros and maintain the dimensions of an input volume with low-level features. The operation of the convolutional layer is given as:
| (1) |
where I refers to the input matrix, K denotes a 2D filter of size m × n, and F represents the output of a 2D feature map. The operation of the convolutional layer is denoted by I*K. To increase nonlinearity in feature maps, the rectified linear unit (ReLU) layer is used [48]. ReLU computes activation by keeping the threshold input at zero. It is mathematically expressed as follows:
| f(x) = max(0, x) | (2) |
The pooling layer [49] performs a downsampling of a given input dimension to reduce the number of parameters. Max pooling is the most common method, which produces the maximum value in an input region. The FC layer [50] is used as a classifier that makes a decision on the basis of features obtained from the convolutional and pooling layers.
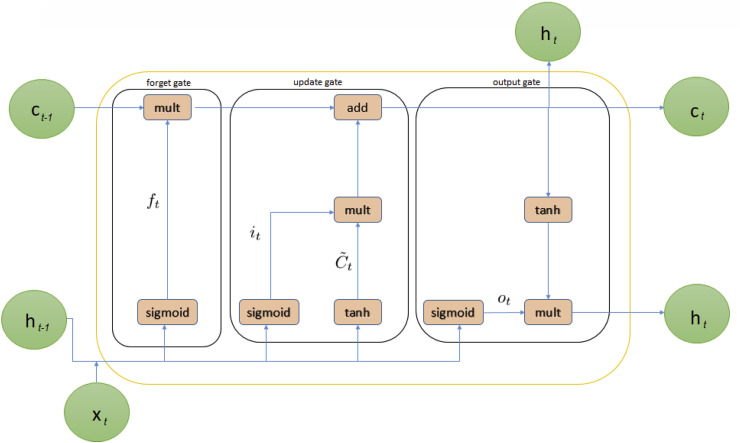
3.2.2. Long short-term memory
Long short-term memory is an improvement of recurrent neural networks (RNNs). LSTM proposes memory blocks instead of conventional RNN units in solving the vanishing and exploding gradient problem [51]. It then adds a cell state to save long-term states, which is its main difference from RNNs. An LSTM network can remember and connect previous information to data obtained in the present [52]. LSTM is combined with three gates, such as an input gate, a “forget” gate, and an output gate, where refers to the current input; and denote the new and previous cell states, respectively; and h t and are the current and previous outputs, respectively. The internal structure of LSTM is shown in Fig. 4 .
Fig. 4.
The internal structure of Long short-term memory.
The principle of the input gate of LSTM is shown in the following forms.
| (3) |
| (4) |
| (5) |
where (3) is used to pass and through a sigmoid layer to decide on which portion of information should be added. Subsequently, (4) is employed to obtain new information after and are passed through the tanh layer. The current moment information, , and long-term memory information into are combined in (5), where Wi refers to a sigmoid output, and refers to a tanh output. Here, Wi denotes weight matrices, and bi represents the input gate bias of LSTM. The LSTM's forget gate then allows the selective passage of information using a sigmoid layer and a dot product. The decision about whether to forget related information from a previous cell with a certain probability is executed using (6), in which Wf refers to the weight matrix, bf is the offset, and σ is the sigmoid function.
| (6) |
The LSTM's output gate determines the states that are required for continuation by the and inputs following (7) and (8). The final output is obtained and multiplied by the state decision vectors that pass new information, Ct, through the tanh layer.
| (7) |
| (8) |
where Wo and bo are the output gate's weighted matrices and LSTM bias, respectively.
3.2.3. Combined CNN-LSTM network
In this study, a combined method was developed to automatically detect COVID-19 cases using three types of X-ray images. The structure of this architecture was designed by combining CNN and LSTM networks, where the CNN is used to extract complex features from images, and LSTM is used as a classifier.
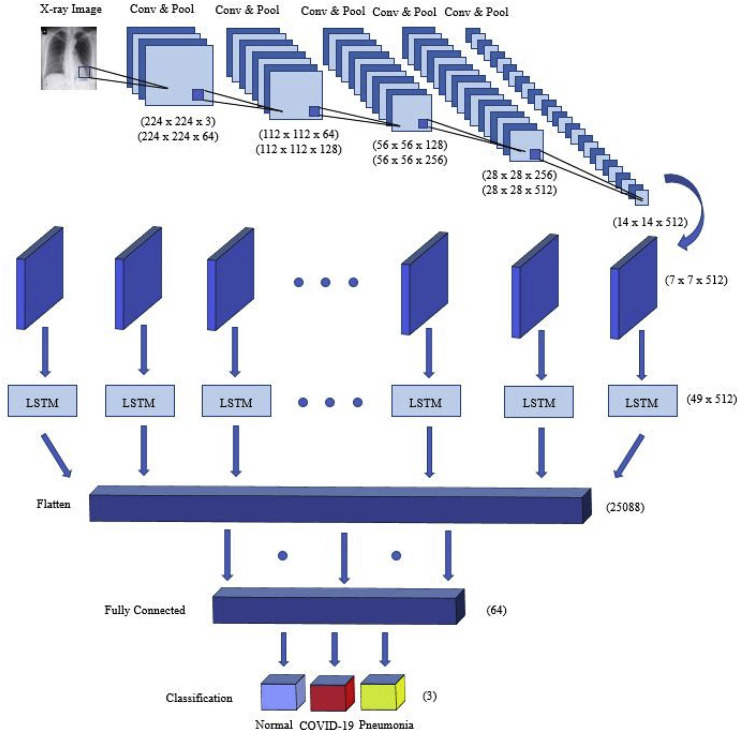
Fig. 5 illustrates the proposed hybrid network for COVID-19 detection. The network has 20 layers: 12 convolutional layers, five pooling layers, one FC layer, one LSTM layer, and one output layer with the softmax function. Each convolution block is combined with two or three 2D CNNs and one pooling layer, followed by a dropout layer characterized by a 25% dropout rate. The convolutional layer with a size of 3 × 3 kernels is used for feature extraction that is activated by the ReLU function. The max-pooling layer with a size of 2 × 2 kernels is used to reduce the dimensions of an input image. In the last part of the architecture, the function map is transferred to the LSTM layer to extract time information. After the convolutional block, the output shape is found to be (none, 7, 7, 512).
Fig. 5.
An illustration of the proposed hybrid network for COVID-19 detection.
Using the reshape method, the input size of the LSTM layer has become (49, 512). The summary of the proposed architecture is shown in Table 2 . After analyzing the time characteristics, the architecture sorts the X-ray images through a fully connected layer to predict whether they belong under any of the three categories (COVID-19, pneumonia, and normal).
Table 2.
The full summary of CNN-LSTM network.
| Layer | Type | Kernel Size | Stride | Kernel | Input Size |
|---|---|---|---|---|---|
| 1 | Convolution2D | 3 × 3 | 1 | 64 | 224 × 224 × 3 |
| 2 | Convolution2D | 3 × 3 | 1 | 64 | 224 × 224 × 64 |
| 3 | Pool | 2 × 2 | 2 | – | 224 × 224 × 64 |
| 4 | Convolution2D | 3 × 3 | 1 | 128 | 112 × 112 × 64 |
| 5 | Convolution2D | 3 × 3 | 1 | 128 | 112 × 112 × 128 |
| 6 | Pool | 2 × 2 | 2 | – | 112 × 112 × 128 |
| 7 | Convolution2D | 3 × 3 | 1 | 256 | 56 × 56 × 128 |
| 8 | Convolution2D | 3 × 3 | 1 | 256 | 56 × 56 × 256 |
| 9 | Pool | 2 × 2 | 2 | – | 56 × 56 × 256 |
| 10 | Convolution2D | 3 × 3 | 1 | 512 | 28 × 28 × 256 |
| 11 | Convolution2D | 3 × 3 | 1 | 512 | 28 × 28 × 512 |
| 12 | Convolution2D | 3 × 3 | 1 | 512 | 28 × 28 × 512 |
| 13 | Pool | 2 × 2 | 2 | – | 28 × 28 × 512 |
| 14 | Convolution2D | 3 × 3 | 1 | 512 | 14 × 14 × 512 |
| 15 | Convolution2D | 3 × 3 | 1 | 512 | 14 × 14 × 512 |
| 16 | Convolution2D | 3 × 3 | 1 | 512 | 14 × 14 × 512 |
| 17 | Pool | 2 × 2 | 2 | – | 14 × 14 × 512 |
| 18 | LSTM | – | – | – | 49 × 512 |
| 19 | FC | – | – | 64 | 25,088 |
| 20 | Output | – | – | 3 | 64 |
3.3. Performance evaluation metrics
The following metrics are used to measure the performance of the proposed system: TP denotes the correctly predicted COVID-19 cases, FP denotes the normal or pneumonia cases that are misclassified as COVID-19 by the proposed system, TN denotes the normal or pneumonia cases that are correctly classified, and FN denotes the COVID-19 cases that are misclassified as normal or pneumonia cases.
| (9) |
| (10) |
| (11) |
| (12) |
4. Experimental results analysis
4.1. Experimental setup
In the experiment, the dataset was split into 80% and 20% for training and testing, respectively. The results were obtained using 5-fold cross-validation technique. The proposed network consists of 12 convolutional layers, as described in Table 2, the learning rate is 0.0001, and the maximum epoch number is 125, as determined experimentally. The CNN and CNN-LSTM networks were implemented using Python and the Keras package with TensorFlow2 on an Intel(R) Core(TM) i7-2.2 GHz processor. In addition, the experiments were executed using the graphical processing unit (GPU) NVIDIA GTX 1050 Ti with 4 GB and 16 GB RAM, respectively.
4.2. Results analysis
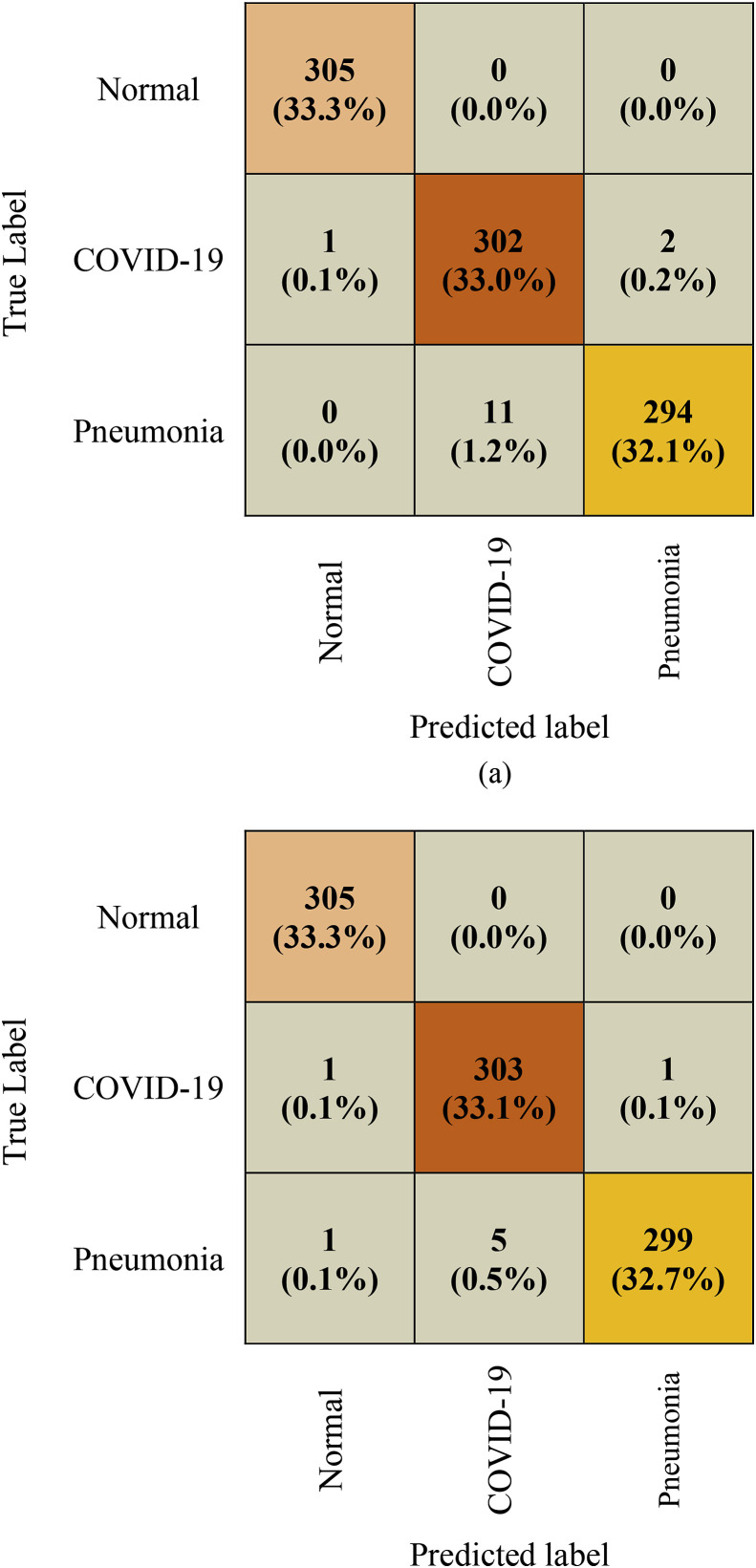
Fig. 6 depicts the confusion matrix of the test phase of the competitive CNN and proposed CNN-LSTM architecture for COVID-19 disease classification. Among the 915 images, 14 were misclassified by the CNN architecture, with three images for COVID-19. Meanwhile, only eight images were misclassified by the proposed CNN-LSTM architecture, including two images for COVID-19. It was found that the proposed CNN-LSTM network outperforms the competitive CNN network as it has better and consistent true positive and true negative values and lesser false negative and false positive values. Therefore, the proposed system can efficiently classify COVID-19 cases.
Fig. 6.
Confusion matrix of the proposed COVID-19 detection system. (a) CNN (b) CNN- LSTM.
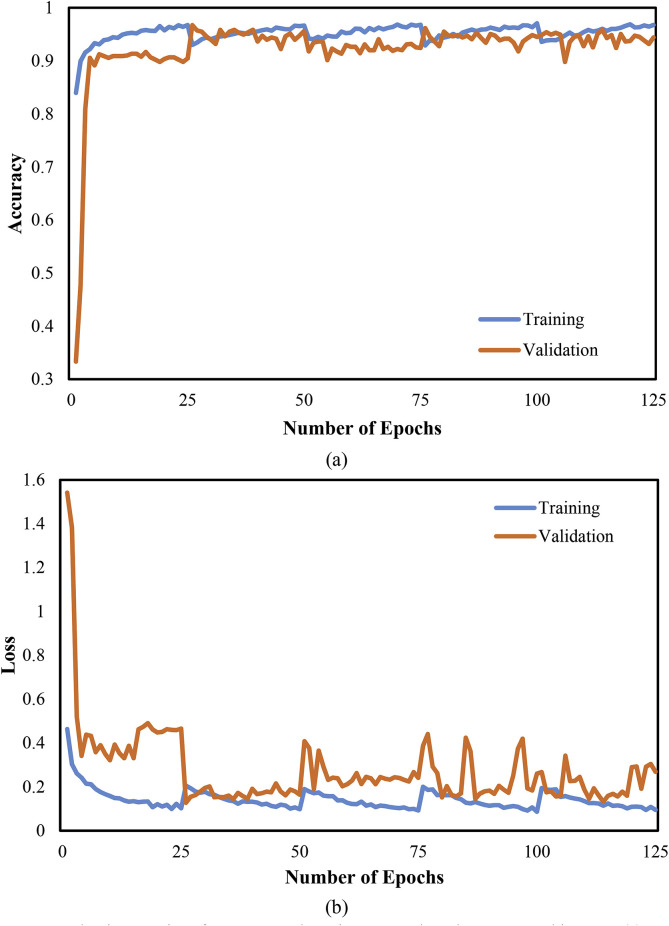
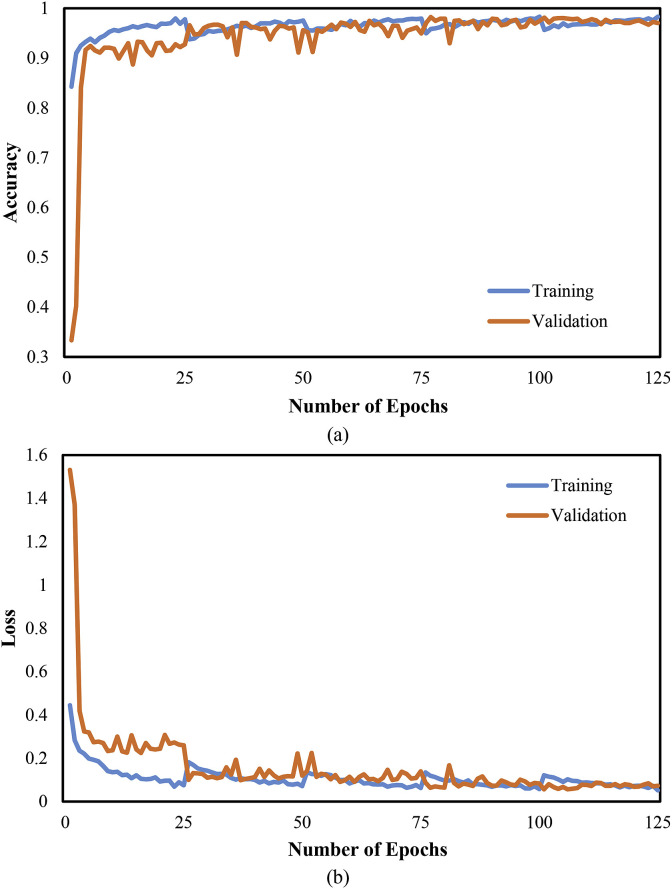
Moreover, Fig. 7 graphically the performance evaluation of the CNN classifier with accuracy and cross-entropy (loss) in the training and validation phase. The training and validation accuracy is 96.7% and 94.4%, respectively, at epoch 125. Similarly, the training and validation loss is 0.09 and 0.26, respectively, for the CNN architecture. Further, Fig. 8 depicts the performance evaluation of the CNN-LSTM classifier graphically with accuracy and cross-entropy (loss) in the training and validation phase. The obtained training and validation accuracy is 98.3% and 97.0%, respectively, at epoch 125. Similarly, the training and validation loss is 0.05 and 0.07, respectively, for the CNN-LSTM architecture. Better scores of training and validation accuracy were achieved using the CNN-LSTM architecture as compared with the CNN architecture.
Fig. 7.
Evaluation metrics of COVID-19 detection system based on CNN architecture. (a) Accuracy (b) Loss.
Fig. 8.
Evaluation metrics of COVID-19 detection system based on CNN-LSTM architecture (a) Accuracy (b) Loss.
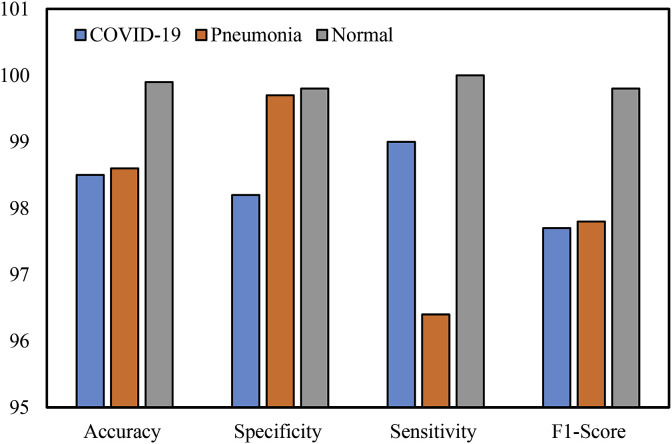
The overall accuracy, specificity, sensitivity, and F1-score for each case of CNN architecture are summarized in Table 3 and visually shown in Fig. 9 . The CNN network achieved 98.2% specificity, 99.0% sensitivity, and 97.7% F1-score for the COVID-19 cases. For the pneumonia classification, it recorded 99.7% specificity, 96.4% sensitivity, and 97.8% F1-score. In the normal cases, it obtained 99.8% specificity, 100% sensitivity, and 99.8% F1-score. While the highest specificity, sensitivity, and F1-score were obtained in the normal cases, the lower values of sensitivity were found in the pneumonia cases.
Table 3.
Performance of the CNN network.
| Class | Accuracy (%) | Specificity (%) | Sensitivity (%) | F1-Score (%) |
|---|---|---|---|---|
| COVID-19 | 98.5 | 98.2 | 99.0 | 97.7 |
| Pneumonia | 98.6 | 99.7 | 96.4 | 97.8 |
| Normal | 99.9 | 99.8 | 100.0 | 99.8 |
Fig. 9.
The graphical representation of the results of the CNN network.
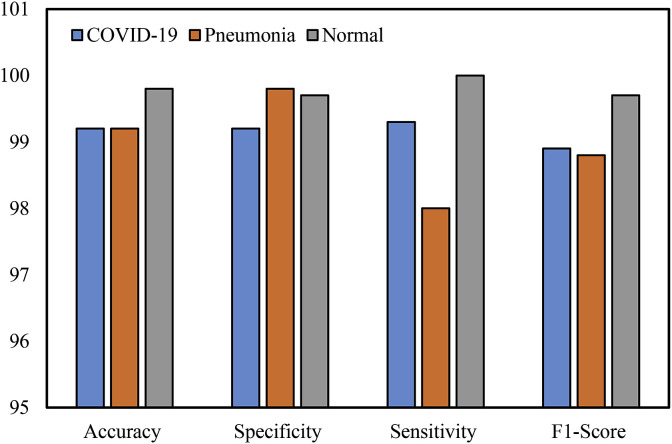
Furthermore, Table 4 and Fig. 10 shows the performance metrics of each class of the developed CNN-LSTM network. The COVID-19 category was classified with good sensitivity, specificity, and F1-score (99.3%, 99.2%, and 98.9%). The value of sensitivity (99.3%) means that the sum of the false negatives is low while the specificity value (99.2%) means that the sum of the true negatives is high. For the pneumonia classification, it obtained 99.8% specificity, 98.0% sensitivity, and 98.8% F1-score. For the normal cases, it obtained 99.7% specificity, 100% sensitivity, and 99.7% F1-score. While the maximum sensitivity and F1-score were achieved in the normal cases, the lower values of sensitivity were obtained in the pneumonia cases.
Table 4.
Performance of the CNN-LSTM network.
| Class | Accuracy (%) | Specificity (%) | Sensitivity (%) | F1-score (%) |
|---|---|---|---|---|
| COVID-19 | 99.2 | 99.2 | 99.3 | 98.9 |
| Pneumonia | 99.2 | 99.8 | 98.0 | 98.8 |
| Normal | 99.8 | 99.7 | 100.0 | 99.7 |
Fig. 10.
The graphical representation of the results of the CNN-LSTM network.
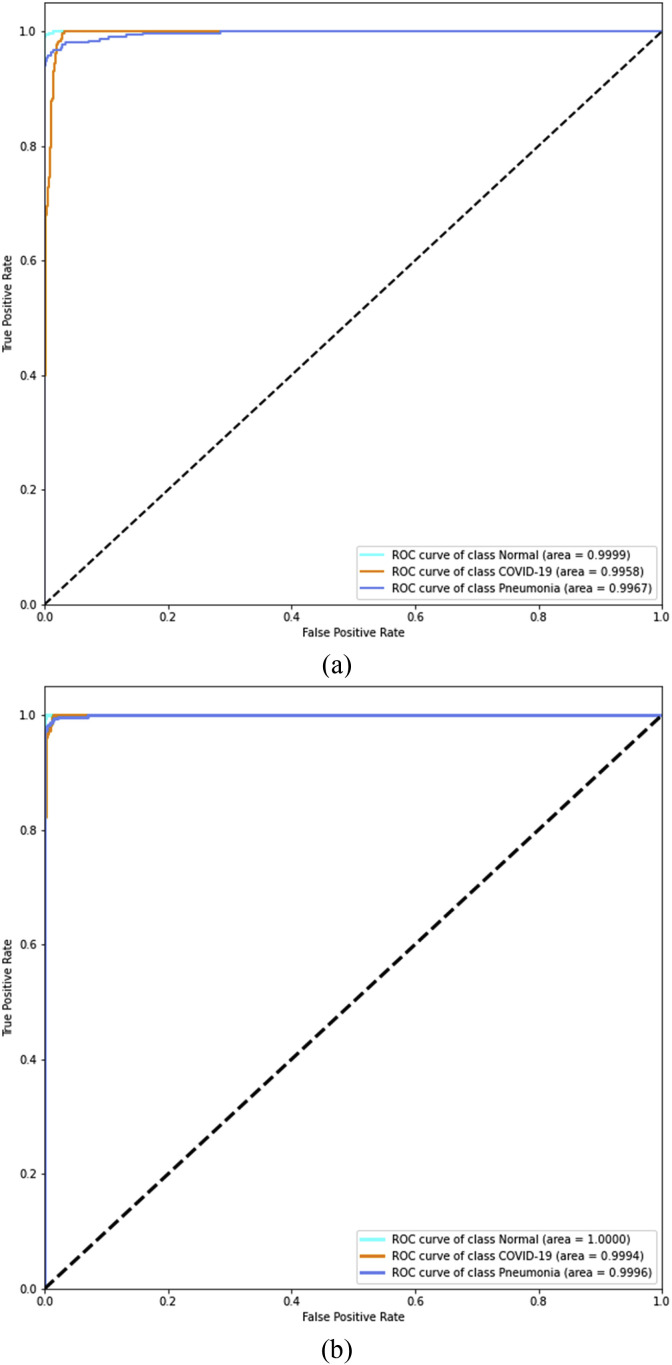
Furthermore, the ROC curves are added between the true positive rate (TPR) and the false positive rate (FPR) to compare the overall performance in Fig. 11 . The area under the ROC curve (AUC) was calculated to be 99.8% and 99.9% for the CNN and the CNN-LSTM architectures, respectively, clearly showing that the proposed network obtained a higher value as compared to the CNN architecture.
Fig. 11.
ROC analysis of the developed system (a) CNN (b) CNN-LSTM.
From the experimental findings, it is evident that the CNN architecture achieved 95.3% AUC, 98.2% specificity, 99.0% sensitivity, and 97.7% F1-score after experimental verification for the COVID-19-infected cases. Comparing the outcomes, the proposed CNN-LSTM network obtained an overall 99.9% AUC, 99.2% specificity, 99.3% sensitivity, and 98.9% F1-score, respectively, for the COVID-19 cases. The main purpose of this research is to achieve good results in detecting COVID-19 cases and not detecting false COVID-19 cases. The experimental results revealed that the proposed CNN-LSTM architecture outperforms the competitive CNN network.
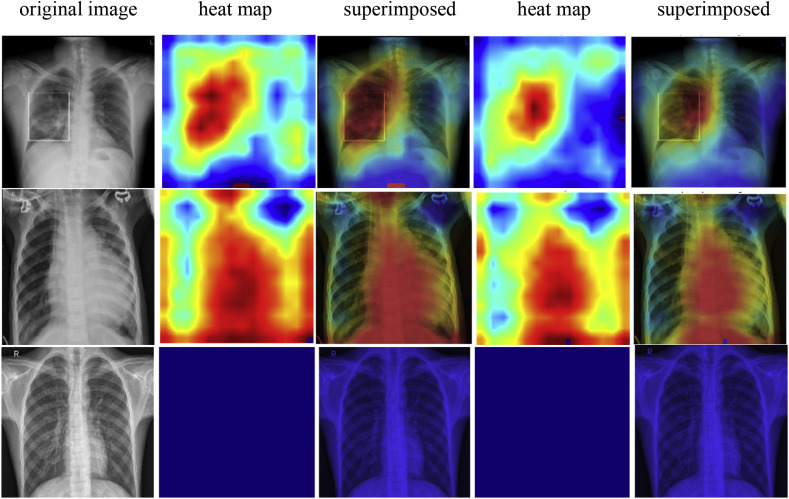
Finally, Gradient-weighted Class Activation Mapping (Grad-CAM) refers to the heat map used for the visual explanation of our experiment using the gradients of a target concept. A coarse localization map highlights the important regions in the image for prediction after passing into the final layer. In Fig. 12 , the heat map of the classified test examples is shown for COVID-19, pneumonia, and normal cases both for CNN and CNN-LSTM architecture.
Fig. 12.
The first, second, and third rows show COVID-19 samples, pneumonia samples and normal samples respectively. Besides, the first column refers to the original image; the second and third column refers to heat map and superimposed image for CNN; fourth and fifth column for CNN-LSTM.
5. Discussions
By analyzing the results, it is demonstrated that a combination of CNN and LSTM has significant effects on the detection of COVID-19 based on the automatic extraction of features from X-ray images. The proposed system could distinguish COVID-19 from pneumonia and normal cases with high accuracy. A comparison between existing systems and our proposed system, in terms of accuracy and computational time, is shown in Table 5 . From Table 5, it is found that some of the proposed systems [22,28,31,31,[53], [54], [55]], and [56] obtained a slightly lower accuracy in range of 80.6%–92.3%. The moderately highest accuracy of 93.5%, 95.2%, 95.4%, 98.3% and 98.3% are found in [23,26,29,57], and [25] respectively. The system developed in [58] obtained an overall accuracy of 98.08% considering the multi-classes. Moreover, a comparison between existing systems in terms of computational time depicted that the system developed in [23] required 6.3s to classify 21 test images [25], needed 2277.6s for 8997 training images [31], took 2641.0s and 4.0s for training and testing of 40 and 10 images respectively, and [56] consumed 79184.3s and 262.0s for training and testing of 4449 and 1638 images respectively. In our experiment, the CNN architecture required 18950.0s and 114.0s for training and testing 3660 and 915 images respectively. Our proposed CNN-LSTM architecture provides good performance and it is also faster than the CNN approach, taking 18372.0s and 113.0s for training and testing which is proportionally faster than other studies. Overall, the result of our proposed system is superior compared to other existing systems.
Table 5.
Comparison of the proposed system with existing systems in terms of accuracy and computational time.
| Author | Architecture | Accuracy (%) | Accuracy (COVID-19) (%) | Training (s) | Testing (s) |
|---|---|---|---|---|---|
| Rahimzadeh et al. [22] | Xception + ResNet50V2 | 91.4 | 99.6 | – | – |
| Alqudah et al. [23] | AOCT-NET | 95.2 | – | – | 6.3 |
| Loey et al. [53] | GoogleNet | 80.6 | 100.0 | – | – |
| Ucar et al. [25] | COVIDiagnosis-Net | 98.3 | 100.0 | 2277.6 | – |
| Apostolopoulos et al. [26] | VGG19 | 93.5 | – | – | – |
| Khan et al. [28] | CoroNet (Xception) | 89.5 | 96.6 | – | – |
| Kumar et al. [29] | ResNet50 + SVM | 95.4 | – | – | – |
| Hemdan et al. [31] | VGG19 | 90.0 | – | 2641.0 | 4.0 |
| Li et al. [54] | DenseNet | 88.9 | 79.2 | – | – |
| Wang et al. [55] | Tailored CNN | 92.3 | 80.0 | – | – |
| Asnaoui et al. [56] | Incpetion_ResNet_V2 | 92.2 | – | 79184.3 | 262.0 |
| Chowdhury et al. [57] | Sgdm-SqueezeNet | 98.3 | 96.7 | – | – |
| Ozturk et al. [58] | DarkCovidNet | 98.08 | – | – | – |
| Farooq et al. [59] | ResNet50 | 96.2 | 100.0 | – | – |
| Proposed System | CNN-LSTM | 99.4 | 99.2 | 18372.0 | 113.0 |
6. Conclusion
As COVID-19 cases are increasing daily, many countries are facing resource shortages. Hence, it is necessary to identify every single positive case during this health emergency. We introduced a deep CNN-LSTM network for the detection of novel COVID-19 from X-ray images. Here, CNN is used as a feature extractor and the LSTM network as a classifier for the detection of coronavirus. The performance of the proposed system is improved by combining extracted features with LSTM that differentiate COVID-19 cases from others. The developed system obtained an accuracy of 99.4%, AUC of 99.9%, specificity of 99.2%, sensitivity of 99.3%, and F1-score of 98.9%. The proposed CNN-LSTM and competitive CNN architecture are applied both on the same dataset. Our extensive experimental results revealed that our proposed architecture outperforms a competitive CNN network. In this global COVID-19 pandemic, we hope that the proposed system would be able to develop a tool for COVID-19 patients and reduce the workload of the medical diagnosis for COVID-19.
The proposed system has some limitations. Firstly, the sample size is relatively small that needs to be increased to test the generalizability of the developed system. This would be overcomed if more images are found in the coming days. Secondly, it only focuses on the posterior-anterior (PA) view of X-rays, hence it cannot differentiate other views of X-rays such as anterior-posterior (AP), lateral, etc. Thirdly, COVID-19 images comprising multiple disease symptoms cannot be efficiently classified. Finally, the performance of our proposed system was not compared with radiologists. Hence, a comparison of the proposed system with radiologists would be part of a future study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors wish to acknowledge the Department of Computer Science and Engineering (CSE), Khulna University of Engineering & Technology (KUET) for facilitating the work.
References
- 1.About worldometer COVID-19 data - Worldometer. https://www.worldometers.info/coronavirus/
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Everything about the corona virus - Medicine and health. https://medicine-and-mental-health.xyz/archieves/4510
- 4.Culp W.C. Coronavirus disease 2019. A & A Pract. 2020;14 doi: 10.1213/xaa.0000000000001218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;2019 doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jibril S.I.A.M.L., Islam Md M., Sharif U.S. Predictive data mining models for novel coronavirus (COVID-19) infected patients recovery. SN Comput Sci. 2020;1:206. doi: 10.1007/s42979-020-00216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y., Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. AJR Am J Roentgenol. 2020;1–7 doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- 9.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR, Radiology. 2020. 200432. [DOI] [PMC free article] [PubMed]
- 10.Thevenot J., Lopez M.B., Hadid A. A survey on computer vision for assistive medical diagnosis from faces. IEEE J Biomed Health Inf. 2018;22:1497–1511. doi: 10.1109/JBHI.2017.2754861. [DOI] [PubMed] [Google Scholar]
- 11.Islam M.M., Iqbal H., Haque M.R., Hasan M.K. 2017 IEEE Reg. 10 Humanit. Technol. Conf. IEEE; 2017. Prediction of breast cancer using support vector machine and K-Nearest neighbors; pp. 226–229. [DOI] [Google Scholar]
- 12.Haque M.R., Islam M.M., Iqbal H., Reza M.S., Hasan M.K. 2018 Int. Conf. Comput. Commun. Chem. Mater. Electron. Eng. IEEE; 2018. Performance evaluation of random forests and artificial neural networks for the classification of liver disorder; pp. 1–5. [DOI] [Google Scholar]
- 13.Hasan M.K., Islam M.M., Hashem M.M.A. 2016 5th Int. Conf. Informatics, Electron. Vis. IEEE; 2016. Mathematical model development to detect breast cancer using multigene genetic programming; pp. 574–579. [DOI] [Google Scholar]
- 14.Islam Ayon S., Milon Islam M. Diabetes prediction: a deep learning approach. Int J Inf Eng Electron Bus. 2019;11:21–27. doi: 10.5815/ijieeb.2019.02.03. [DOI] [Google Scholar]
- 15.Ayon S.I., Islam M.M., Hossain M.R. Coronary artery heart disease prediction: a comparative study of computational intelligence techniques. IETE J Res. 2020:1–20. doi: 10.1080/03772063.2020.1713916. 0. [DOI] [Google Scholar]
- 16.Rahaman A., Islam M., Islam M., Sadi M., Nooruddin S. Developing IoT based smart health monitoring systems: a review. Rev Intelligence Artif. 2019;33:435–440. doi: 10.18280/ria.330605. [DOI] [Google Scholar]
- 17.Islam M.M., Rahaman A., Islam M.R. Development of smart healthcare monitoring system in IoT environment. SN Comput Sci. 2020;1:185. doi: 10.1007/s42979-020-00195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang X. Proc. - 2009 2nd IEEE Int. Conf. Comput. Sci. Inf. Technol. ICCSIT. vol. 2009. 2009. Feature extraction for image recognition and computer vision; pp. 1–15. [DOI] [Google Scholar]
- 19.Lakshmanaprabu S.K., Mohanty S.N., Shankar K., Arunkumar N., Ramirez G. Optimal deep learning model for classification of lung cancer on CT images. Future Generat Comput Syst. 2019;92:374–382. doi: 10.1016/j.future.2018.10.009. [DOI] [Google Scholar]
- 20.Chen Y., Gou X., Feng X., Liu Y., Qin G., Feng Q., Yang W., Chen W. Bone suppression of chest radiographs with cascaded convolutional networks in wavelet domain. IEEE Access. 2019;7:8346–8357. doi: 10.1109/ACCESS.2018.2890300. [DOI] [Google Scholar]
- 21.Wang Y., Wu Q., Dey N., Fong S., Ashour A.S. Deep back propagation–long short-term memory network based upper-limb sEMG signal classification for automated rehabilitation. Biocybern Biomed Eng. 2020;40:987–1001. doi: 10.1016/j.bbe.2020.05.003. [DOI] [Google Scholar]
- 22.Rahimzadeh M., Attar A. A new modified deep convolutional neural network for detecting COVID-19 from X-ray images. 2020. http://arxiv.org/abs/2004.08052 [DOI] [PMC free article] [PubMed]
- 23.Alqudah A.M., Qazan S., Alquran H.H., Alquran H., Qasmieh I.A., Alqudah A. Covid-2019 detection using X-ray images and artificial intelligence hybrid systems. 2020. [DOI]
- 24.Loey M., Smarandache F., Khalifa N.E.M. Within the lack of chest COVID-19 X-ray dataset: a novel detection model based on GAN and deep transfer learning. Symmetry (Basel) 2020;12:651. doi: 10.3390/sym12040651. [DOI] [Google Scholar]
- 25.Ucar F., Korkmaz D. COVIDiagnosis-Net: deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med Hypotheses. 2020;140:109761. doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020:1–8. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bandyopadhyay S.K., Dutta S. Death and Release; MedRxiv: 2020. Machine learning approach for confirmation of COVID-19 cases: positive, negative. 2020.03.25.20043505. [DOI] [Google Scholar]
- 28.Khan A.I., Shah J.L., Bhat M. CoroNet: a deep neural network for detection and diagnosis of covid-19 from chest X-ray images. 2020. http://arxiv.org/abs/2004.04931 [DOI] [PMC free article] [PubMed]
- 29.Kumar P., Kumari S. Detection of coronavirus disease (COVID-19 ) based on deep features. 2020. https://Www.Preprints.Org/Manuscript/202003.0300/V1 9. [DOI]
- 30.Horry M.J., Chakraborty S., Paul M., Ulhaq A., Pradhan B. 2007. X-ray image based COVID-19 detection using pre-trained deep learning models. [Google Scholar]
- 31.Hemdan E.E.-D., Shouman M.A., Karar M.E. COVIDX-net: a framework of deep learning classifiers to diagnose COVID-19 in X-ray images. 2020. http://arxiv.org/abs/2003.11055
- 32.Singh M., Bansal S. Transfer learning based ensemble support vector machine model for automated COVID-19 detection using lung computerized tomography scan data. 2020. https://assets.researchsquare.com/files/rs-32493/v1_stamped.pdf [DOI] [PMC free article] [PubMed]
- 33.Ahuja S., Panigrahi B.K., Dey N., Rajinikanth V., Gandhi T.K. Deep transfer learning - based automated detection of COVID-19 from lung CT scan slices. 2020. [DOI] [PMC free article] [PubMed]
- 34.Fong S.J., Li G., Dey N., Crespo R.G., Herrera-Viedma E. Composite Monte Carlo decision making under high uncertainty of novel coronavirus epidemic using hybridized deep learning and fuzzy rule induction. Appl Soft Comput J. 2020;93 doi: 10.1016/j.asoc.2020.106282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen J.P., Morrison P., Dao L. COVID-19 image data collection. 2020. http://arxiv.org/abs/2003.11597
- 36.COVID-19 chest X-ray. https://github.com/agchung
- 37.Covid-19. 2020. https://radiopaedia.org/
- 38.Welcome to the cancer imaging archive - the cancer imaging archive (TCIA) https://www.cancerimagingarchive.net/
- 39.COVID-19 database | SIRM. https://www.sirm.org/en/category/articles/covid-19-database/
- 40.Mendeley data - augmented COVID-19 X-ray images dataset. https://data.mendeley.com/datasets/2fxz4px6d8/4
- 41.Kaggle chest x-ray repository. https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia
- 42.NIH chest X-rays | Kaggle. https://www.kaggle.com/nih-chest-xrays/data?select=Data_Entry_2017.csv
- 43.Galvez R.L., Bandala A.A., Dadios E.P., Vicerra R.R.P., Maningo J.M.Z. IEEE Reg. 10 Annu. Int. Conf. Proceedings/TENCON. Institute of Electrical and Electronics Engineers Inc.; 2019. Object detection using convolutional neural networks; pp. 2023–2027. [DOI] [Google Scholar]
- 44.Tajbakhsh N., Shin J.Y., Gurudu S.R., Hurst R.T., Kendall C.B., Gotway M.B., Liang J. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imag. 2016;35:1299–1312. doi: 10.1109/TMI.2016.2535302. [DOI] [PubMed] [Google Scholar]
- 45.Hasan A.M., Jalab H.A., Meziane F., Kahtan H., Al-Ahmad A.S. Combining deep and handcrafted image features for MRI brain scan classification. IEEE Access. 2019;7:79959–79967. doi: 10.1109/ACCESS.2019.2922691. [DOI] [Google Scholar]
- 46.Gu J., Wang Z., Kuen J., Ma L., Shahroudy A., Shuai B., Liu T., Wang X., Wang G., Cai J., Chen T. Recent advances in convolutional neural networks. Pattern Recogn. 2018 doi: 10.1016/j.patcog.2017.10.013. [DOI] [Google Scholar]
- 47.Kutlu Avcı. A novel method for classifying liver and brain tumors using convolutional neural networks, discrete wavelet transform and long short-term memory networks. Sensors. 2019;19:1992. doi: 10.3390/s19091992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh D., Kumar V., Vaishali, Kaur M. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution-based convolutional neural networks. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 2020 doi: 10.1007/s10096-020-03901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lundervold A.S., Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys. 2019;29:102–127. doi: 10.1016/j.zemedi.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 50.Chang P., Grinband J., Weinberg B.D., Bardis M., Khy M., Cadena G., Su M.Y., Cha S., Filippi C.G., Bota D., Baldi P., Poisson X.L.M., Jain X.R., Chow X.D. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. Am J Neuroradiol. 2018;39:1201–1207. doi: 10.3174/ajnr.A5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hochreiter S. The vanishing gradient problem during learning recurrent neural nets and problem solutions. Int J Uncertain Fuzziness Knowledge-Based Syst. 1998;6:107–116. doi: 10.1142/S0218488598000094. [DOI] [Google Scholar]
- 52.Chen G. A gentle tutorial of recurrent neural network with error backpropagation. 2016. http://arxiv.org/abs/1610.02583 1–10.
- 53.Loey M., Smarandache F., Khalifa N.E.M. Within the lack of chest COVID-19 X-ray dataset: a novel detection model based on GAN and deep transfer learning. Symmetry. 2020;12:651. doi: 10.3390/SYM12040651. 12 (2020) 651. [DOI] [Google Scholar]
- 54.Li X., Li C., Zhu D. 2020. COVID-MobileXpert: on-device COVID-19 screening using snapshots of chest X-ray. [Google Scholar]
- 55.Wang L., Wong A. 2020. COVID-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.El Asnaoui K., Chawki Y. Using X-ray images and deep learning for automated detection of coronavirus disease. J Biomol Struct Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1767212. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chowdhury M.E.H., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Bin Mahbub Z., Islam K.R., Khan M.S., Iqbal A., Al-Emadi N., Reaz M.B.I. 2020. Can AI help in screening Viral and COVID-19 pneumonia. [Google Scholar]
- 58.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farooq M., Hafeez A. COVID-ResNet: a deep learning framework for screening of COVID19 from radiographs. 2020. http://arxiv.org/abs/2003.14395