Abstract
Biodegradation is simply the metabolism of anthropogenic, or otherwise unwanted, chemicals in our environment, typically by microorganisms. The metabolism of compounds commonly found in living things is limited to several thousand metabolites whereas ∼100 million chemical substances have been devised by chemical synthesis, and ∼100 000 are used commercially. Since most of those compounds are not natively found in living things, and some are toxic or carcinogenic, the question arises as to whether there is some organism somewhere with the enzymes that can biodegrade them. Repeatedly, anthropogenic chemicals have been denoted ‘non-biodegradable,’ only to find they are reactive with one or more enzyme(s). Enzyme reactivity has been organized into categories of functional group transformations. The discovery of new functional group transformations has continually expanded our knowledge of enzymes and biodegradation. This expansion of new-chemical biodegradation is driven by the evolution and spread of newly evolved enzymes. This review describes the biodegradation of widespread commercial chemicals with a focus on four classes: polyaromatic, polychlorinated, polyfluorinated, and polymeric compounds. Polyaromatic hydrocarbons include some of the most carcinogenic compounds known. Polychlorinated compounds include polychlorinated biphenyls (PCBs) and many pesticides of the twentieth century. Polyfluorinated compounds are a major focus of bioremediation efforts today. Polymers are clogging landfills, killing aquatic species in the oceans and increasingly found in our bodies. All of these classes of compounds, each thought at one time to be non-biodegradable, have been shown to react with natural enzymes. The known limits of enzyme catalysis, and hence biodegradation, are continuing to expand.
Keywords: bacteria, biodegradation, enzyme, fluorinated, halogenated, polymers
Overarching theme
The question of whether any and all synthetic chemicals might be metabolized has long been the subject of speculation, but it will never be answered because chemical synthesis is essentially endless. The field continually advances by examining chemicals thought to be non-metabolizable, isolating organisms and their corresponding enzymes that break them down, and thus extending the known limits of enzyme catalysis.
In his 1951 textbook, Ernest Gale asserted, ‘….somewhere or other some organism exists which can, under suitable conditions, oxidize any substance which is theoretically capable of being oxidized' [1]. In another textbook published in 1999 [2], Martin Alexander wrote, ‘…only some of the countless reactions possible of organic molecules have been exploited in the processes necessary for metabolism. The resulting anabolic, and presumably catabolic, processes are thus few in number.' More recently, the idea of metabolic limitations, or the lack thereof, has been further elaborated as the ‘Microbial Infallibility Hypothesis' [3]. The Infallibility Hypothesis suggests that all chemicals now or in the future are metabolizable by at least one living thing, but this is formally not a hypothesis as it is not amenable to experimental proof or falsification. We cannot know a priori whether any chemical devised by humans will be metabolizable, or not, because the numbers are too vast. Approximately 108 molecules have now been synthesized and to date the number of theoretically possible small organic molecules is reported at 1060 [4,5], which is greater than the number of atoms on Earth [6]. A microbial ‘census' enumerated prokaryotes on Earth at >1030 [7] and most prokaryotes contain >103 enzymes. The number of possible enzymes consisting of 300 amino acids is 20300, greater than the number of atoms in the universe [8]. We can never expect to categorize all combinations of enzymes and chemical substances, we can only extend our knowledge of what is possible.
Defining the limits of biodegradation and enzyme catalysis
Chemicals of concern have driven studies of biodegradability
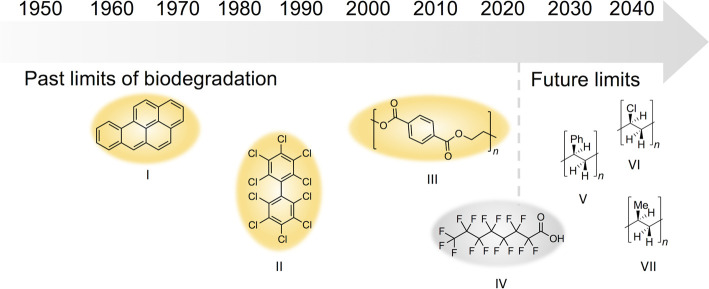
Biodegradation is a field of study focused principally on the metabolism of anthropogenic chemicals by microorganisms, with the term bioremediation typically reserved for the application of microbes, plants, and enzymes in environmental cleanup. Biodegradation research and bioremediation practice have been driven by the chemicals of concern during different decades. Discoveries of the carcinogenicity of polycyclic aromatic compounds like benzo[a]pyrene in the 1960's (Figure 1) drove investigations into the biodegradability of these compounds. Growing knowledge that polychlorinated biphenyls (PCBs) impacted human and ecosystem health in the 1970's incentivized research and led to the finding that decades-old deposits of PCBs were slowly biodegrading. Today's major pollution concerns are polymers and perfluorinated compounds, and these classes are now the foci of biodegradation research.
Figure 1. Timeline of past, present, and possible future discoveries of microbes and enzymes capable of breaking down natural and anthropogenic compounds including benzo[a]pyrene (I), perchloro-1,1′-biphenyl (a PCB) (II), and polyethylene terephthalate (III).
One major challenge currently facing our society are the ‘forever chemicals' such as perfluorooctanoic acid (IV) poised at the current ‘limit' of biodegradation. As the field advances, our understanding will also expand to the biodegradation of recalcitrant polymers such as polystyrene (V), polyvinyl chloride (VI), polypropylene (VII) and yet-to-be-synthesized chemicals.
This review does not seek to cover biodegradation metabolism and enzymes exhaustively. A search in article title for one biodegradative enzyme, urease, yielded 2800 papers indexed on PubMed as of July 1, 2020. In light of the scale, this review has focused on several of the major classes of compounds studied for biodegradation and initially considered hard to degrade or to be completely non-biodegradable. Additionally, through studying the limits of biodegradation, the field has contributed greatly to our knowledge of enzyme catalysis and evolution.
The emergence of enzyme and biodegradation research
Enzymes were recognized as biological catalysts in 1833 and have traditionally been named for the functional group undergoing reaction along with the letters ‘ase' added at the end; for example, amidase [9]. Functional groups are defined here as stable configurations of atoms, typically 2–7, with distinct chemical properties and undergoing defined chemical transformations. A standardized functional group reaction classification was established by 1964 that recognized their reactions as being: (1) oxidation-reduction; (2) group transfer; (3) hydrolysis; (4) lyase reaction; (5) isomerization; or (6) ligation. In 2018, a new class, (7) translocases, was added to include transport proteins [10]. The number of functional groups undergoing one or more reactions in the initial incarnation of the Enzyme Commission (EC) nomenclature system was ∼30. The 712 enzymes classified by the EC in 1964 reacted largely with compounds derived from biological systems: major cellular metabolites proteins, lipids, carbohydrates, and nucleic acids. Accordingly, initial enzymes classified by the EC catalyzed reactions with the most prevalent biological functional groups; for example: amides, amines, alcohols, aldehydes, carboxylates.
The twentieth century saw the rise of the petrochemical industries, leading to an enormous expansion in synthetic consumer products. Presently, ∼105 chemical compounds [11] are found in these products, introducing both new functional groups and their combinations into the biosphere. Industrialization and urbanization led to concentrated pockets of unnatural chemicals and concerns about their effects on biological systems. Mammalian xenobiotic metabolism and human epidemiological studies were carried out to study health effects of industrial chemicals. Biodegradation and bioremediation also arose as important fields of study during that time.
Database and prediction tools define current limits of biodegradation knowledge
To promote understanding and applications of biodegradation and bioremediation, the University of Minnesota Biocatalysis/Biodegradation Database (BBD) went online in 1995 [12] to detail known enzymatic functional group reaction types. We quickly realized it would not be feasible to compile enzyme data for all chemicals. The majority of commercial chemicals have never been studied at the level of enzymatic biodegradation. To highlight the burden for assessing new chemicals, the United States Environmental Protection Agency received 36 000 chemicals for proposed commercial use between 1999 and 2004 alone [13].
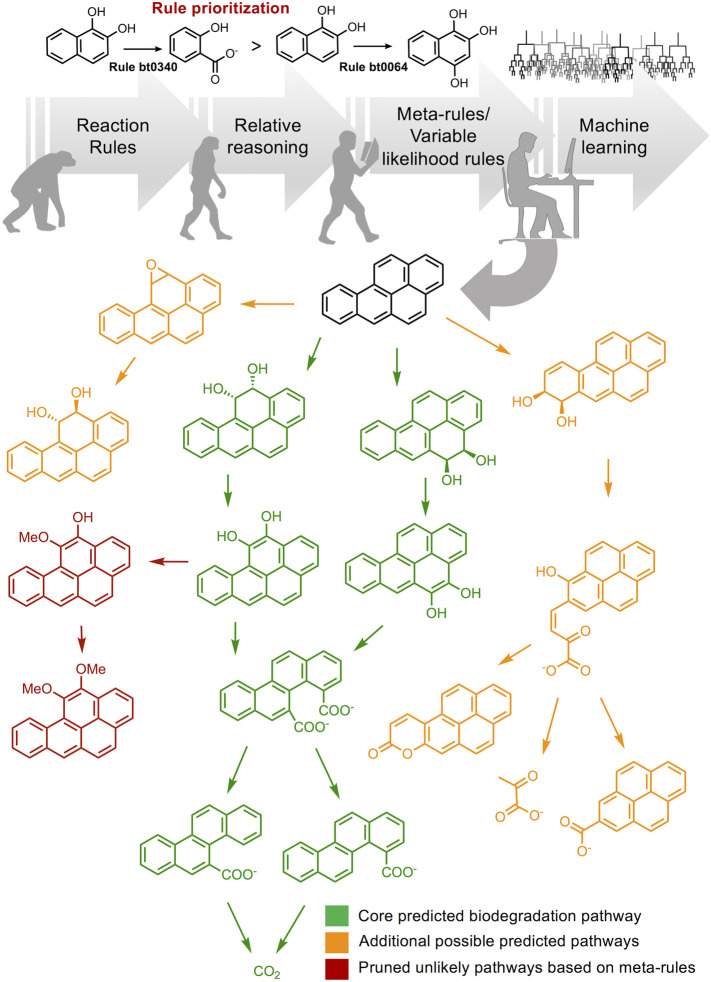
To meet the needs of the research community to consider the biodegradation of chemicals comprehensively, the existing knowledge of enzymatic functional group transformation was further developed to predict biodegradation pathways for new chemicals [14]. The in silico prediction tool, now known as the Eawag-BBD Pathway Prediction System (PPS), is based on computational reaction rules (Figure 2). Each rule describes a reaction for one of sixty functional groups undergoing on average, one of four types of reactions. The application of 240 functional group reaction rules alone produced many feasible and unlikely pathways. The latter include things like futile cycles that would be selected against in naturally occurring bacteria. To retain the plausible pathways and constrain others, higher order rules that reined in the functional group rules were extracted from existing pathways in the BBD database [15]. These rules on top of rules, denoted as meta- and variable likelihood rules, only added an additional 130 rules but greatly decreased extraneous, implausible pathways and made the prediction tool much more user friendly [16].
Figure 2. Evolution of biodegradation prediction tools from reaction-rule based software (Eawag-BBD) towards the integration of relative reasoning and meta-rules to limit the combinatorial explosion of possibilities [15].
The latest software (e.g. enviPath) incorporates probabilistic machine learning models to further prune the search space and predict additional likely pathways for biodegradation [18]. Possible degradation pathways for benzo[a]pyrene are collated from the Eawag-BBD, PPS, and enviPath. At the top (Rule prioritization), rule bt0340 in the Eawag-BBD is the prioritized rule for dihydroxylation of polyaromatics overriding rule bt0064 for formation of hydroxybenzenoid derivatives.
In the years since their initial development, computational methods for biodegradation prediction have undergone major updates including the use of probabilistic models to encode biotransformation rules. Wicker and Fenner pioneered a machine learning approach using multi-label classifiers to calculate probability scores for microbial biotransformations [17]. Probability thresholds from these classifiers were then used to further prune the combinatorial nature of predicted chemical degradation pathways. These models were integrated in a more flexible tool built on the Eawag-BBD known as enviPath, which also allows users to include and analyze their own data [18]. A number of other metabolic pathway prediction tools including PathPred, CATALOGIC, and BNICE have been developed and are primarily still built on biotransformation logic resembling the Eawag-BBD [19–21]. Indeed, the Eawag-BBD remains the largest manually curated and publicly available collection of biotransformation rules specifically tailored towards contaminants in the environment. A key limitation of existing pathway prediction tools is the inability to quantitatively model the half-lives of chemicals. As a first step towards this ambitious goal, the Eawag-Soil package was published with over 4700 half-life values for the transformations of pesticides and other xenobiotics in soil [22]. Improved availability of chemical half-life data through data packages such as Eawag-Soil will advance progress towards quantitatively predicting the persistence and fate of contaminants in the environment.
New biodegradation limits: new enzymes acting on new chemicals
New enzymes can evolve rapidly
Increasingly, industry is seeking to make molecules that biodegrade after their useful lifetime. Even before environmental concerns were of high priority in industry, most industrial chemicals were found to be biodegradable because microbes are inherently efficient in biodegradation. This is, in part, due to their large numbers (109/gm in a typical topsoil) and rapid rates of evolution due to extraordinary generation times as short as 9.8 minutes [23]. Those facts are coupled with the exceptional capacity of enzyme catalysis provided by amino acid side chains, metals, and organic cofactors. Enzymes were traditionally thought to be highly specific catalysts [24], but more recent studies have called attention to enzyme promiscuity and its role in the evolution of new catalytic functions [25,26]. The term ‘enzyme promiscuity' highlights observations that many enzymes have low activities with substrates typically not present in cells and thus are generally irrelevant to cell physiology, but new anthropogenic substances can act to select for and thus promulgate these off-target enzyme activities. Indeed, multiple selective events can lead to the evolution of new metabolic pathways for the complete biodegradation of anthropogenic chemicals [27]. For example, bacterial melamine metabolism combines a highly specific melamine deaminase producing ammeline with a guanine deaminase that serves the cell by deaminating guanine. The guanine deaminase also does double-duty to deaminate ammeline to ammelide, filling a gap in the pathway [28]. The concept of catalytic promiscuity has also been central to developments in directed laboratory evolution for generating robust industrial enzyme catalysts. Directed evolution typically starts with a modest non-physiological reactivity of an enzyme being boosted through rounds of experimental selection to achieve activity levels approaching the kcat/KM values of naturally occurring enzymes [29]. The techniques making that possible were the subject of Frances Arnold's Nobel Prize lecture in Chemistry in 2018 [30]. Indeed, ‘directed evolution' is a keyword in over 3700 PubMed articles and these techniques have been effective to shift or enhance the natural biocatalytic capabilities of enzymes to act on desired substrates including xenobiotics [31].
Biodegradative enzymes quickly evolve to ‘average' enzyme activity
In directed evolution in the laboratory, the practitioner typically takes an enzyme with a low promiscuous activity and carries out mutation and selection to evolve a higher activity toward a new substrate of interest [8]. In some cases, an entirely new biocatalytic reaction has been evolved, for example a new enzyme that makes carbon-to-silicon bonds [32]. The improvement in catalytic efficiency of these enzymes is often expressed as an increase in the steady-state kinetic parameter, kcat/KM. This is directly relevant to the limits of biodegradation as many new synthetic chemicals are introduced into commerce annually and efficient biodegradation will require that high catalytic rates evolve quickly in new enzymes.
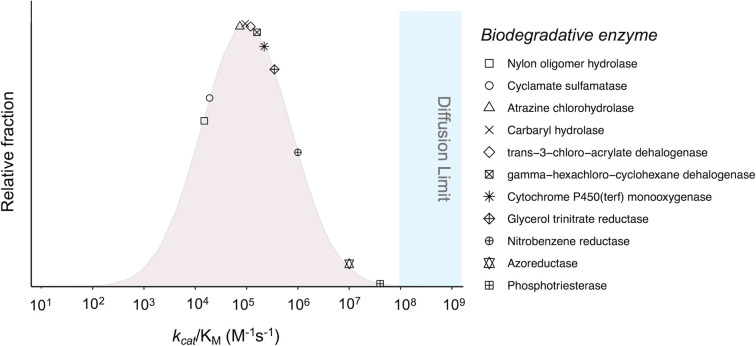
Evolution in nature has been shown to proceed in a similar manner as in the laboratory and low promiscuous activities are selected for to generate new enzymes [27]. The important question is how rapidly does natural evolution occur to biodegrade emerging environmental pollutants, and we will use here the same metric used in directed evolution experiments: kcat/KM. Enzymes clearly demonstrated to be newly evolved must be compared with the set of well-studied enzymes, most of which are common to many forms of life and are thus ancient in an evolutionary sense. The BRENDA enzyme database contains such a set of kcat/KM parameters for >3500 enzymes with their preferred substrates. This data was recently compiled and analyzed [33,34] and forms the basis for the comparisons made here. The BRENDA dataset showed median kinetic values for kcat and KM of 10 s−1 and 100 µM, respectively, yielding a combined kcat/KM of 1 × 105 M−1 s−1. The plotted values across all enzyme show a Gaussian distribution (Figure 3) with most falling in the range of 104–106 M−1 s−1 [33,34]. Moreover, <1% of enzymes operate in the diffusion-controlled reaction regime of >109 M−1 s−1, which was an unexpected finding since many enzyme examples in biochemistry textbooks have been selected for their exceptional catalytic rates, but these are not representative of enzymes at large.
Figure 3. Distribution of kcat/KM enzyme values, comparing all enzymes in the BRENDA database to enzymes for which there is evidence of their recent evolution to specifically react with new anthropogenic chemicals.
The pink curve represents the nearly Gaussian distribution of kcat/KM values for 4451 enzymes as analyzed by Davidi et al. [32]. The symbols each represent an enzyme isolated specifically for its activity against an anthropogenic chemical and derived from a microorganism evolved to biodegrade the chemical. Details of each enzyme, its source, and steady-state kinetic parameters can be found in Supplementary Table S1.
Interestingly, many enzymes that we believe to have recently evolved to biodegrade new chemicals have a kcat/KM in the same range, ∼104–4 × 107 M−1 s−1, with a similar median (Figure 3 and Supplementary Table S1). The survey is inherently limited because we restricted the biodegradative enzymes to those for which there is reasonable evidence supporting a recent evolution in response to new chemical inputs into the environment. For example, recent evolutionary origins are strongly suggested for atrazine chlorohydrolase (AtzA). After atrazine was introduced as a new commercial herbicide in 1959 [35], it was considered poorly biodegradable but that was followed after some decades with a sudden emergence of rapid biodegradation in many soils linked to the presence of the atzA gene [36,37]. Rapid atrazine biodegradation was further linked to the spread of the atzA gene on transposable DNA segments found on broad host-range plasmids [38] supported by strong AtzA sequence conservation (>98%) amongst enzyme variants from six continents [39]. Moreover, AtzA is highly specific; it reacts with atrazine and more slowly with an atrazine analog containing a fluorine in place of chlorine. It specifically catalyzes hydrolytic dehalogenation, a reactivity obtained principally via two point mutations that switch its chemospecificity from deamination to dehalogenation [40]. AtzA was found to be 98% identical in amino acid sequence to a deaminase enzyme, TriA [41]. The deaminase, that had very low dehalogenase activity, was mutated in the laboratory and evolved into a dehalogenase, recapitulating the natural evolutionary pathway [42]. The naturally evolved enzyme shows a respectable kcat/KM of 7.4 × 104 M−1 s−1 (Figure 3 and Supplementary Table S1). In total, these combined observations demonstrate that new enzymatic activities directed against anthropogenic chemicals can arise very quickly, show high substrate selectivity, and substantial catalytic efficiency.
A similar set of observations implicate trans-3-chloroacrylate dehalogenase as a recently evolved enzyme. The first enzyme identified with this reactivity was derived from Pseudomonas cichorii 170 that was isolated from soil repeatedly treated with the commercial nematocide (+/−)-1,3-dichloropropene [43]. The biodegradative pathway for racemic 1,3-dichloropropene produces both cis- and trans-3-chloroacrylate as intermediates. Distinct dehalogenase enzymes react with each. Each enzyme has a kcat/KM of >1 × 105 M−1 s−1 against its respective haloacrylate substrate. The stereospecific enzyme reactive with the trans-intermediate has been more exhaustively studied and is included here (Figure 3). The dehalogenase X-ray structure has been solved and it is a member of a major tautomerase superfamily [44]. Indeed, the enzyme 4-oxalocrotonate that catalyzes the isomerization of 2-oxo-4-hexenedioate to 2-oxo-3-hexenedioate also catalyzes hydrolytic dehalogenation of trans-3-chloroacrylate, albeit at a very modest kcat/KM of 1.1 × 10−2 M−1 s−1. Poelarends et al. introduced two point mutations that increased the kcat/KM for trans-3-chloroacrylate by two orders of magnitude [45]. These mutations were chosen based on the knowledge that both reactions proceed through a proposed enolate intermediate.
With trans-3-chloroacrylate dehalogenase and atrazine chlorohydrolase both, structural and mechanistic studies have revealed adaptive pathways by which common cellular deamination and isomerization catalysts can undergo minimal mutational changes to assume new cellular functions as dechlorinating enzymes. That activity can then spread in ecosystems when there is widespread environmental use of the chemical. It is likely that enzymes capable of reacting with atrazine and trans-3-chloroacrylate arose previously in enzyme evolutionary history but only became widespread when human commercial activities provided the appropriate selective pressure. The enzymes in Figure 3 show significant activity, in the range of 104–108 M−1 s−1, with anthropogenic compounds believed to have existed only within the last century, a very short time period on an evolutionary time scale.
New enzymes act on new functional groups, further expanding limits of biodegradation
Newly evolved enzymes allow bacteria to acquire carbon, nitrogen, phosphorus or sulfur from additional chemicals by functional group transformations liberating those elements in assimilable forms. The functional groups undergoing enzymatic reactions for the anthropogenic chemicals depicted in Supplementary Table S1 are organohalides, phosphotriesters, carbamates, esters, sulfamates, azo, nitro, amide and alkyl chemical groups. Many of these functional groups are uncommon in biology but are found in industrial, agricultural, and consumer products. As previously mentioned, there are ∼30 common functional groups (alcohols, aldehydes, carboxylates, etc.) that combine to make most intermediary metabolites. The Eawag-BBD catalogues the biodegradative metabolism of ∼30 more distinct functional groups [16]. The Chemical Abstracts Service Registry adds 15 000 new chemical entries daily [46]. Some of those new chemicals contain functional groups not represented in the Eawag-BBD rule base and so complete predictions for their biodegradation cannot be made. Supplementary Table S2 compiles information on additional functional groups for which enzymatic transformations have been incompletely studied. The metabolism of compounds containing those functional group can provide carbon, nitrogen, phosphorus, or sulfur to support microbial growth, making it very likely that they are biodegradable. The compilation in Supplementary Table S2 calls out a need for continuing biodegradation studies to fill knowledge gaps and inform new prediction capabilities that will extend to newly synthesized chemicals containing those functional groups.
Case studies: biodegradation limits expanded in studies of polyfunctionalized compounds
Polycyclic aromatic hydrocarbons
Polyfunctionalized molecules are often slow to biodegrade, even when composed of the same functional group, for example, polycyclic aromatic hydrocarbons (PAHs) are compounds containing multiple aromatic rings. PAHs are abundant in coal, petroleum, and as by-products from combustion of organic materials [47]. PAH ring systems are highly stable thermodynamically and it has been proposed that a significant fraction of carbon in the universe exists in this form [48].
Initial studies on the metabolism of polycyclic aromatic hydrocarbons (PAHs) were carried out using rats, decades before comparable studies with bacteria. In the 1930's, Eric Boyland, a founder of the molecular toxicology field, showed certain PAHs were exquisitely carcinogenic to rats [49]. Over the next 5 decades, Boyland and co-workers carried out extensive studies to elucidate the metabolic fate of these chemicals in mammals, which proved to be the key to understanding their carcinogenicity [50]. PAH metabolism in mammals was shown to be initiated by a membrane-bound heme-containing monooxygenase that came to be known as cytochrome P450 [51]. A soluble counterpart P450 was later identified in bacteria in 1968 that functioned in the biodegradative pathway for a non-aromatic natural product, camphor [52]. The mammalian metabolism of PAHs produced epoxides and trans-dihydrodiol metabolites (Figure 4A) that were further functionalized to make them more water soluble to promote their excretion in the animal's urine [53].
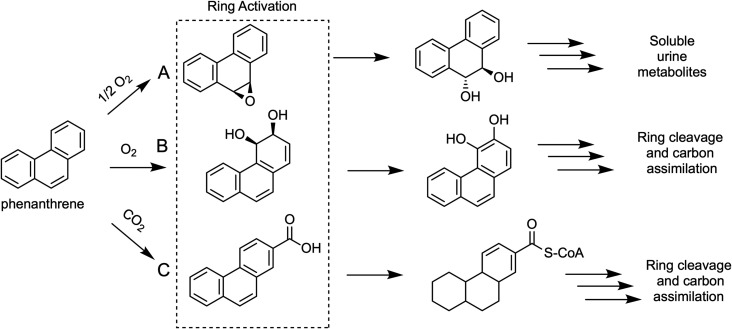
Figure 4. Differential activation of a representative polycyclic aromatic hydrocarbon (PAH) under aerobic (A & B) and anaerobic conditions (C) showing the key ring activation mechanism by each of the three major types of metabolic pathways.
(A) shows ring activation by a cytochrome P450 monooxygenase that breaks the aromaticity of the central ring to make an epoxide intermediate shown at the top in the dotted box. Further reactions serve to make the molecule more water soluble for excretion and so this pathway is common in eukaryotes, including humans [66]. (B) shows ring activation by a dioxygenase enzyme to produce a dihydrodiol intermediate in the middle of the dotted box [67]. Further oxidations lead to cleavage of the aromatic ring and ultimate assimilation of the carbon by biodegrading bacteria. (C) shows initial ring activation by a carboxylase enzyme to make a carboxylated PAH at the bottom in the dotted box. The carboxyl group is further functionalized with coenzyme A and then reductases reduce the aromatic ring to allow hydrolases, dehydrogenases, and ring cleavage enzymes to degrade the structure and allow carbon assimilation [60].
The bacterial metabolism of several PAHs was reported in 1958 by Fernley and Evans [54]. In early studies, it was presumed that microorganisms produced trans-dihydrodiols based on the mammalian paradigm. However, in the 1970's, bacteria were demonstrated to oxidize PAHs via dioxygenase enzymes producing cis-dihydrodiols with naphthalene [55], benzo[a]anthracene and benzo[a]pyrene [56] (Figure 4B). Recently, the type of dioxygenases that oxidize PAHs (Rieske oxygenases) have been found to be very common throughout the microbial world [57] and their substrate range extends beyond aromatic compounds. In some bacteria that are particularly active in degrading diverse PAH structures, both Rieske dioxygenase and cytochrome P450 monooxygenases participate in the biodegradation via pathways that closely resemble the predicted biodegradation pathway for benzo[a]pyrene represented in Figure 2 [58]. The cytochrome P450 monooxygenase transforms PAHs to epoxide metabolites as shown in Figure 4A. The set of biodegradative enzymes needed to completely mineralize multi-ring PAHs such as benzo[a]pyrene is extensive and not highly common in bacteria. As shown in Figure 2, four enzymatic reactions are required to remove just one carbon atom from the 20 carbon atoms in benzo[a]pyrene.
It was once believed PAHs were only biodegraded in aerobic environments, but exciting recent advances have been made in demonstrating mechanisms by which anaerobic bacteria biodegrade these compounds [59]. A critical research question in anaerobic systems remained: how are highly resonance-stabilized PAHs activated for metabolism in the absence of the reactive oxygen species generated at the active sites of cytochrome P450 and Riseke oxygenases? The key initiating activation is catalyzed by carboxylases as suggested by metabolite studies [60]. This difficult carboxylation biochemistry may be carried out at least in part by enzymes from a large family of reversible UbiD-like (de)carboxylases that have a unique prenylated-FMN cofactor and were recently characterized for carrying out a wide range of reactions [61].
Subsequent to carboxylation, the carboxyl group is esterified to coenzyme A and then goes through ring reduction (Figure 4C). The aromatic ring reduction is truly remarkable biochemistry mimicking a chemical reaction known as the Birch reduction which uses sodium metal in ammonia to achieve the very low redox potential necessary to overcome the highly resonance-stabilized benzene ring. Two distinct benzoyl-CoA reductase enzyme systems (BCRs) have evolved that reach to the known biological limits of negative redox potential at −622 mV [62,63]. BCR class I contain three Fe4S4 clusters and use ATP to help drive the reaction [62]. Class II BCRs contain Fe4S4 clusters and carry out the reduction at a tungsten center in an ATP-independent reaction [63]. It is not clear which, or if both types, participate in the anaerobic biodegradation of multi-ring aromatic systems. These studies are at the limits of current knowledge of PAH biodegradation.
In nature, aromatic hydrocarbons are often functionalized, most commonly with alkyl substituents. As such, there are millions of distinct aromatic ring compounds. Showing complete pathways for even a small fraction of these compounds is beyond the scope of the present review and readers are referred to the metabolic databases such as enviPath [18], KEGG [64], or MetaCyc [65] for details on all of the reactions and metabolites in known pathways.
Polychlorinated compounds
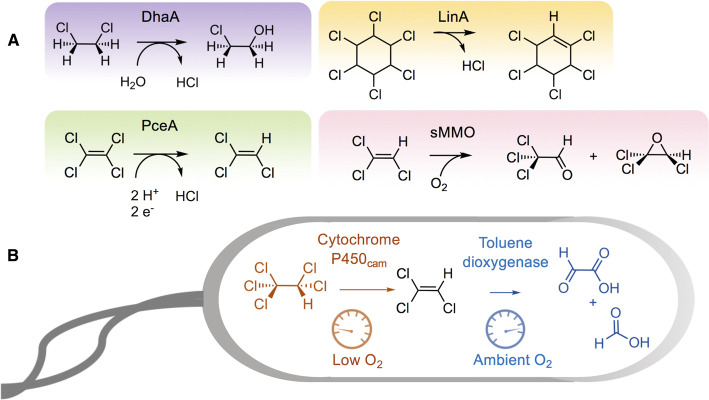
While enzymes for the biodegradation of PAHs were being elucidated in the 1970's, insights into the microbial metabolism of polychlorinated compounds came in the succeeding decades (Figure 1). In the early to mid-twentieth century, many polychlorinated compounds were used commercially because they resisted biodegradation and that was considered a desirable property, for example, with long-lasting pesticides such as chlordane. Subsequently, many commercial polychlorinated compounds were found to be toxic to animals, including humans, and so the biodegradation of chlorinated compounds was studied extensively [66]. Also during that period, chemists discovered a wide variety of chlorinated natural products, which likely are produced as biotoxins against competing organisms [67].
Chlorinated compounds are enzymatically biodegraded by several different mechanisms and specific examples are represented in Figure 5A. They are: (1) hydrolytic (DhaA), (2) beta-eliminative (LinA, a lyase reaction), (3) reductive (PceA) and (4) oxygenative (sMMO) mechanisms [66]. For example, 1,2-dichloroethane is a high-volume commercial chemical that is dechlorinated via hydrolase enzymes (e.g. DhaA) [68]. An alkyl carbon atom bearing a single chlorine atom is highly susceptible to nucleophilic displacement, making it readily biodegradable by a wide range of bacteria [69]. Highly chlorinated compounds are almost invariably more resistant to biodegradation and were found to be persistent in many environments [70]. In those cases where they do biodegrade, reductive dechlorination often initiates their catabolism. The underlying biochemistry has now been studied in some detail. Reduced biological metallo- cofactors such as Fe-heme, Co-cobalamins, and Ni-F430, in the absence of an associated enzyme, were shown to be capable of catalyzing reductive cleavage of carbon to chlorine bonds [71]. Previously, coenzyme F430 alone had been shown to catalyze reductive dechlorination of chloromethanes [72]. Enzymes containing those cofactors may carry out non-physiological [73] or energy-linked reductive dehalogenation [74]. In contrast, less chlorinated olefinic and aromatic compounds are often biodegraded rapidly by oxygenase-catalyzed reactions [75–78]. The complementarity of reductive reactions for polyhalogenated compounds and oxygenative degradation after partial halogen displacement led to the concept of cycling between those reactions to carry out complete dehalogenation (Figure 5B). A recombinant bacterium was shown to catalyze anaerobic reduction and elimination of chloride from pentachloroethane to yield trichloroethylene that was oxidized to completely non-chlorinated end-products aerobically [79]. This study highlights that nature must also draw on multiple enzymes and mechanisms to biodegrade multiply halogenated compounds.
Figure 5. Chlorinated compounds are biodegraded by different mechanisms and complete biodegradation of highly chlorinated compounds may require multiple mechanisms.
(A) Four mechanisms of dehalogenation are illustrated by four different enzymatic examples. The examples (left to right, top to bottom) are haloalkane dehalogenase (DhaA), lindane dehalogenase (LinA), perchloroethylene reductive dehalogenase (PceA), and soluble methane monooxygenase (sMMO). (B) Biodegradation of pentachloroethane by a recombinant bacterium combines reductive and eliminative dechlorination at low oxygen tension and oxygenative biodegradation at ambient oxygen tension [70]. This mimics an ideal situation in the environment in which highly chlorinated compounds can be biodegraded by reductive dehalogenation and subsequent oxygenative dehalogenation either by different or the same microorganism(s), as some anaerobes are known to express oxygenases.
Energy-linked organohalide respiration was initially discovered from careful analytical chemical analysis of polychlorinated biphenyl (PCB)-laden sediments where highly chlorinated PCBs were giving rise to less-chlorinated products [80]. The process was clearly shown to be microbiological in subsequent laboratory studies with mixed cultures of bacteria [81]. Since then, a range of chlorinated aliphatic and aromatic compounds have been shown to undergo reduction as part of bacterial processes yielding ATP [74]. These reductases may be membrane-bound or soluble; the latter have been chosen for mechanistic studies because of the greater facility with which they could be studied and crystallized.
To date, two soluble reductive dehalogenases have had X-ray structures solved. They derive from the strict anaerobe Sulfurospirillum multivorans that grows with tetrachloroethene as the terminal electron acceptor [82] and from Nitratireductor pacificus strain pht-3BT [83] that was isolated from a deep-sea pyrene-degrading bacterial consortium. The latter enzyme is a non-respiratory reductive dehalogenase [84]. Both enzymes contain iron sulfur clusters and a corrinoid cofactor lacking an axial base ligand, which is atypical of most enzyme-associated corrinoids. Earlier non-enzymatic studies had shown that corrinoids lacking an axial base showed significantly higher rates of reductive dehalogenation [85] and nature seems to have adopted this strategy. Both structure-based enzyme studies suggest that a carbon-centered radical may be an intermediate along the reaction pathway. However, the mechanistic proposals differ in that Bommer et al. propose an electron transfer into the carbon-to-halogen bond with a possible carbon-to-cobalt bond forming, whereas Payne et al. propose cobalt forming a direct bond with the halogen during catalysis. Carbon-to-cobalt bonds are known in enzymes and model reactions with corrinoids [86] but the proposed halogen-cobalt interaction was novel. The latter was further supported by electron paramagnetic resonance (EPR) spectroscopy [83]. Beyond these two examples, there are >100 phylogenetically distinct orthologous groups of reductive dehalogenases, and multiple reductive dehalogenases per genome are common in some classes of bacteria such as the Dehalococcoidia [87,88]. The reductive dehalogenases are considered to be evolutionarily ancient, with a likely expansion in genomes due to the recent anthropogenic release of chlorinated industrial chemicals.
Perfluorinated compounds
While the biodegradation of perchlorinated compounds is now reasonably well-defined, perfluorinated compounds were developed commercially at a later date and are exceptionally challenging to degrade due to their extreme inertness. The carbon-to-fluorine bond is considered amongst the strongest known, with a bond dissociation energy of up to 130 kcal per mol. Despite this, compounds containing single carbon-to-fluorine bonds have been shown to react with enzymes. For example, fluoroacetate is a potent toxin produced by some plants and Streptomyces spp. [89]. A selective pressure for detoxification and carbon acquisition from fluoroacetate likely led to the evolution of a specific hydrolytic defluorinating enzyme that transforms fluoroacetate to glycolate via a direct displacement mechanism [90]. Fluoroacetate was the first among a handful of fluorinated natural products to have been reported and they are mainly limited to fluorinated amino acids, fatty acids, and nucleoside derivatives [89]. Although no fluoroaromatic natural products have been isolated to date, monofluorinated aromatics are known to be biodegradable [91–93]. In organochemical reactions, fluoroaromatics are known to undergo nucleophilic aromatic substitution reactions more readily than their chlorinated counterparts [94]. In light of that, it is not surprising that these compounds are readily accessible to enzymatic degradation.
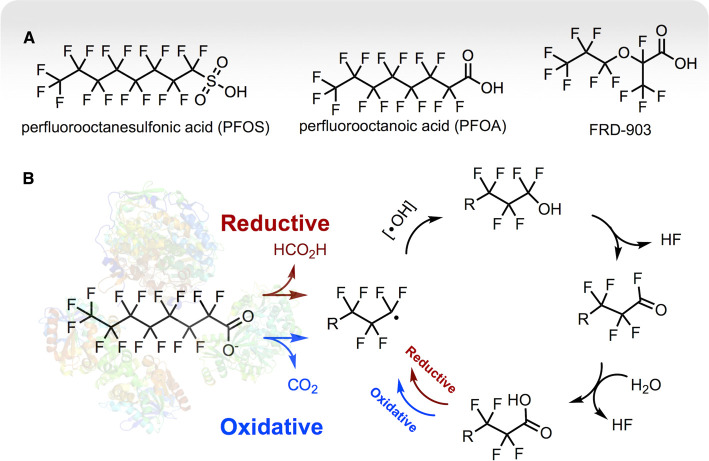
Over the last century, industries have presented a much more imposing challenge to biological systems by synthesizing a diverse collection of perfluoroalkyl sulfonic and carboxylic acids. Principal among those are perfluorooctane sulfonic acid (PFOS), perfluorooctanoic acid (PFOA), and a hexafluoropropylene oxide dimer acid (FRD-903), the latter being a more recently introduced substitute for the other two (Figure 6A). These and more than 4000 related highly fluorinated compounds have been produced and used extensively in household products, fire suppression, metal-plating, electronics, and other industrial applications [95]. As such, they have become widespread in the environment. More importantly, numerous human health and ecosystem effects are now attributed to exposure to PFOS and PFOA [96]. FRD-903 is newer (Figure 6A), and it is not yet clear if this represents a safer alternative. Concerns have been compounded by the prevailing notion that many of these compounds persist and may be completely non-biodegradable.
Figure 6. Perfluorinated compound biodegradation is currently under intense investigation.
Specific biodegradative enzymes are yet to be identified and the proposed mechanism for defluorination is based on chemical studies. (A) shows important commercial perfluorinated compounds. (B) shows a proposed mechanism by which a representative perfluorocarboxylate (PFOA) could be completely degraded by one or more enzymes via consecutive one-carbon chain shortening cycles. Although enzyme(s) responsible for catalyzing PFOA degradation have not yet been characterized, this proposal is consistent with studies on Acidimicrobium sp. strain A6 [97] and abiotic chemical degradation reactions. The key intermediate is a carbon-centered radical that could be generated via a reductive or oxidative enzymatic process. Recombination of the radical with a yet unknown oxygen species (hydroxyl radical only represents the proper electron balance) could generate a difluorohydroxy species that could undergo spontaneous displacement of two fluorine atoms. That would generate a new fluorocarboxylate that is one carbon shorter, and then the cycle can repeat, leading to complete degradation. R = one or more CFx groups.
There have been numerous reports of PFOS or PFOA biodegradation in sediments or microbial consortia but studies based on substrate disappearance only are prone to artifact. Ideal biodegradation studies: (1) document compound purity, (2) identify reaction products, (3) show a kinetic course of biodegradation, and (4) show the specific participation of an identified microorganism or enzyme. In this context, a recent study by Huang and Jaffé that addressed those issues comprehensively has attracted significant attention [97]. The paper describes the biodegradation of PFOS and PFOA by a bacterium, Acidimicrobium sp. strain A6, both as an isolate and in combination with other uncharacterized microbes in an enrichment culture. Both the isolate and enrichment culture showed biodegradation of both PFOA and PFOS with the corresponding formation of fluoride.
Additionally, time course analysis of PFOA and PFOS biodegradation showed C7, C6, C5, and C4 perfluorinated carboxylic acid breakdown products accumulating in the enrichment cultures along with fluoride. PFOA showed an initial drop during the first one-third of the time course and the corresponding C7 perfluorinated acid formed and plateaued. Shorter chain perfluoro acids (C6, C5, C4) also increased and the C4 acid started declining at the longest time point, suggesting continued degradation of the carbon chain in the enrichment culture. These observations are consistent with a biological mechanism by which the chain is shortened from the carboxylic acid end, one carbon at a time, with the concomitant liberation of fluorine atoms as fluoride (Figure 6B).
While purified enzymes catalyzing defluorination of perfluorinated compounds have not been reported to date, insights can be derived from the whole-cell studies described above and in the context of chemical degradation experiments that use strong oxidants and reductants. Perfluorinated compounds are highly inert to nucleophilic displacement reactions, but persulfate and nickel-iron reagents have been shown to catalyze PFOA oxidation and reduction reactions, respectively, that lead to fluoride release and chain shortening [98–100]. In those studies, it is generally agreed that both oxidation and reduction reactions proceed through a carbon-centered radical intermediate (Figure 6B). The radical is proposed to be quenched with an oxygen atom derived from water to produce the difluoro alcohol that would undergo spontaneous and rapid gem-elimination of HF to produce an acyl-fluoride that would hydrolyze rapidly to the carboxylic acid. These chemical mechanisms, like the Acidimicrobium sp. strain A6 cultures [97], release fluoride and produce a series of shorter chain perfluorinated carboxylic acids. A non-biological study using an electrical plasma-based system that generates both oxidizing and reducing species showed the formation of formate during the course of PFOA removal from water [101]. It is proposed that a reductive mechanism would produce formate and an oxidative process would produce carbon dioxide [98,99,101]. Differentiating between formate or carbon dioxide end products is difficult in a whole-cell biological system with thousands of enzymes present but should be addressable in future in vitro studies with defined enzymes.
Polymer biodegradation
Nature has made and biodegraded polymers for millions of years. Natural polymers can be recycled on short time scales but also, as in the case of lignin, support trees more than one hundred meters tall that live for thousands of years [102]. The durability of lignin is due partly to its heterogeneous structure, derived from various biological radical coupling reactions [103]. Lignin biosynthesis is thought to have evolved 400–450 million years ago [104], providing microbes sufficient time to evolve biodegradative enzymes that recycle carbon from lignin into the shared biosphere.
In contrast, the beginnings of synthetic polymers go back merely two hundred years [105]. The earliest contrivances were semi-synthetic. Polymers such as cellulose acetate were generated by chemical modification of the naturally occurring cellulose polymers. Bakelite was the first polymer, or plastic, built up completely starting with synthetic non-polymer precursors, phenols and formaldehyde. Bakelite was first prepared and molded for consumer goods in 1910 [105]. Over the ensuing 100 years, plastic types and quantities increased enormously. Major polymers in use today are polyethylene (PE), polypropylene (PP), polystyrene (PS), polyvinyl chloride (PVC), polyurethanes (PUR), and polyethylene terephthalate (PET) (Figure 1). Today, up to 8% of total fossil fuel resources are used annually to supply the carbon and energy for producing synthetic polymers [106].
Until recently, research and development of synthetic polymers was largely focused on their properties and performance pertaining to intended applications, with little thought of their biodegradability or recyclability. This has changed to some degree. While performance is still important for commercial applicability, the pervasiveness and consequences of polymers in the environment are changing public and political perceptions. For example, the Great Pacific Garbage Patch is estimated to contain more than 1.8 trillion pieces of plastic. If it continues to grow at its current pace, the mass of ocean plastics is estimated to exceed that of fish by the year 2050 [107,108]. Recently, concerns have been heightened by the analytical documentation and purported health effects of micro- and nano-plastic particles in the bodies of humans [109]. These developments have brought increasing focus on the biodegradation and recycling of synthetic polymers.
It is important to investigate plastic biodegradation for purposes of environmental remediation and guidance on designing new polymers, but many investigations have been faulted for a lack of scientific rigor [110]. The biodegradation of low molecular mass chemicals is typically demonstrated with mass balances, substrate disappearance and commensurate product formation, and the involvement of specific enzymes or genes. These criteria are difficult to achieve with plastics biodegradation studies and so rarely met in published accounts. Instead more indirect evidence of biodegradation may be shown, for example, microscopic images of microbes inhabiting surfaces, increases in microbial biomass over time, or loss of plastic over time [110–112]. These are not immune to artifacts, however. Microbes are well-documented to grow on chemicals commonly used as plasticizers in polymer formulations, and these plasticizers are not covalently linked to the polymer matrix. PVC plastics can contain up 50% plasticizer, of which phthalate esters are most common [113]. Phthalates are highly biodegradable [114]. Bisphenol A (BPA) is used in polyester-styrene and polycarbonate plastics and is also known to be biodegradable by a number of bacteria [115]. These comments are not meant to imply that polymers such as low-density polyethylene are non-biodegradable, given that nature makes an impressive array of oxygenolytic enzymes for degrading natural polymers [111]. However, this present review will focus on polymer degradation studies that have met the standard of mass balances and enzyme delineation.
The most detailed studies of biodegradation for a major synthetic polymer have been conducted with polyethylene terephthalate (PET) (see structure III in Figure 1). Recently, a bacterium was isolated in pure culture that was capable of growing on PET as a sole source of carbon and energy [116]. The bacterium, Ideonella sakaiensis 201-F6, was part of a consortium that almost completely degraded a PET film in 6 weeks. Sequencing the bacterium's genome revealed the presence of enzymes that degraded PET to mono(2-hydroxyethyl) terephthalic acid that was further transformed to terephthalate and ethylene glycol. The metabolism of terephthalate [117] and ethylene glycol [118] has been demonstrated in a large number of divergent bacteria.
The PET-digesting enzyme, denoted PETase, that is secreted by I. sakaiensis was studied in mechanistic detail and its structure was solved to 0.92 Angstrom resolution [119]. PETase shows a classical α/β-hydrolase fold and >50% sequence identity with cutinase enzymes known to hydrolyze the plant leaf-surface polymer, cutin. To accommodate the polymers, both enzymes contain an active site in a cleft that binds the respective polymer strands. PETase is distinctive in showing a cleft that is almost three-fold wider than the cleft in the cutinase from Thermobifida fusca, for which an X-ray structure is also available [120]. The PETase active site is characterized by a catalytic triad with a serine nucleophile in relay with an aspartate and a histidine, an arrangement common in α/β-hydrolases. The wild-type enzyme, and double mutant with higher activity, were shown to have activity in biodegrading PET and a commercially relevant PET replacement polymer, polyethylene-2,5-furandicarboxylate (PEF) [119].
Most recently, Tournier et al. reported on the results of a comprehensive study for using a PETase enzyme to degrade commercial PET, recycle the terephthalic acid to produce new PET, and thus show that PETase can potentially serve in a use-reuse circular system [121]. To accomplish that, extensive enzyme engineering was carried out to increase enzymatic activity and melting temperature. The former was carried out by modeling to identify eleven residues for saturation mutagenesis and subsequent combinatorial testing of best variants. To increase protein stability, a disulfide bond was engineered into the structure, creating a PETase with a melting temperature of 94.5°C. The engineered PETase was tested in a 150 L pilot scale-up to mimic an industrial process. The pilot scale proved effective when using a volume of PETase enzyme that would cost 4% of the value of the PET being treated. This suggested that an enzyme-based recycling process using the engineered PETase would be feasible in terms of cost. In the process that was demonstrated, terephthalic acid could be recovered and used with commercial ethylene glycol to regenerate new PET. This makes sense economically as terephthalic acid represents 86.3% of the mass of the PET being recycled and it can be crystallized, whereas ethylene glycol is difficult to recover.
Polymer biodegradation represents one frontier in the effort to handle the recalcitrant compounds that humans have continually devised for safety, health and convenience. Beyond PET, there are signs of progress on the biodegradation of other polymers, for example polystyrene (see structure V in Figure 1) [122]. In this example, superworms were observed to ingest macroscopic pieces of polystyrene. Isolates from the worm gut microbiota were cultured and a Psuedomonas strain was identified as a polystyrene-degrading bacterium. Gene expression and enzyme inhibition studies implicated the involvement a serine hydrolase enzyme in polystyrene degradation, although it is unclear that any bonds in polystyrene are susceptible to hydrolysis. In the long term, the sheer magnitude of polymer production will require multiple solutions beyond individual enzymes. These solutions include shifting to more biodegradable polymers [123,124], mechanical reformulation, and recycling [108]. Recycling will take different forms, from direct reuse to chemical and biological breakdown and resynthesis. The latter work on biorecycling is already underway [121] and advancing to biotechnology start-up stage.
Prospects and conclusions
Biodegradation is an ever-expanding field. With 15 000 new chemical entities being added to databases daily [46], enzyme-based studies on individual chemicals can never keep up. As it has in the past, efforts will focus on the largest volume and most disruptive chemicals from an ecosystem and human health perspective. Additionally, fundamental knowledge of the leading edges of enzyme reactions, coupled with more powerful computational prediction tools will help fill the gaps in our ability to keep pace with new chemical synthesis and usage. Furthermore, advances in next-generation sequencing and other -omics technologies will allow for better prediction and manipulation of biodegradation in natural and built environments such as wastewater treatment plants.
Abbreviations
- BBD
Biocatalysis/Biodegradation Database
- BCRs
benzoyl-CoA reductase enzyme systems
- EC
Enzyme Commission
- EPR
electron paramagnetic resonance
- PAH
polycyclic aromatic hydrocarbon
- PCB
polychlorinated biphenyl
- PFOA
perfluorooctanoic acid
- PPS
Pathway Prediction System
Open Access
Open access for this article was enabled by the participation of University of Minnesota in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work has been supported by funding from the MnDRIVE Initiative at the University of Minnesota and by the USDA National Institute of Food and Agriculture, Agricultural and Food Research Initiative Competitive Program, Ecosystem Services and Agro-Ecosystem Management, grant no. 2019-67019-29403. S.L.R. was supported by a National Science Foundation (NSF) Graduate Research Fellowship (grant no. 00039202).
References
- 1.Gale E.F. (1951) The Chemical Activities of Bacteria, 3rd edn, Academic Press, New York, U.S.A. [Google Scholar]
- 2.Alexander M. (1999) Biodegradation and Bioremediation, 2nd edn, Academic Press, New York, U.S.A. [Google Scholar]
- 3.Ramakrishnan B. (2012) Microbial diversity and degradation of pollutants. J. Bioremed. Biodeg. 3, e128 10.4172/2155-6199.1000e128 [DOI] [Google Scholar]
- 4.Gómez-Bombarelli R., Wei J.N., Duvenaud D., Hernández-Lobato J.M., Sánchez- Lengeling B., Sheberla D. et al. (2018) Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 4, 268–276 10.1021/acscentsci.7b00572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemonick S. (2020) Artificial intelligence leaps into chemical space. Chem. Eng. News 98, 30–35 10.1021/cen-09813-cover [DOI] [Google Scholar]
- 6.Weisenberger D. (2020) How many atoms are there in the world? Accessed 20 April 2020. Available from: https://education.jlab.org/qa/mathatom_05.html
- 7.Whitman W.B., Coleman D.C. and Wiebe W.J. (1998) Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. U.S.A. 95, 6578–6583 10.1073/pnas.95.12.6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romero P.A. and Arnold F.H. (2009) Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell. Biol. 10, 866–876 10.1038/nrm2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tipton K. and Boyce S. (2000) History of the enzyme nomenclature system. Bioinformatics 16, 34–40 10.1093/bioinformatics/16.1.34 [DOI] [PubMed] [Google Scholar]
- 10.ExplorEnz - The Enzyme Database https://www.enzyme-database.org/news.php. Accessed 20 April 2020
- 11.Erickson B.E. (2017) How many chemicals are in use today. Chem. Eng. News 95, 23–24 10.1021/cen-09509-govpol [DOI] [Google Scholar]
- 12.Ellis L.B.M. and Wackett L.P. (1997) Microbial biocatalysis and biodegradation informatics. Nat. Biotechnol. 15, 1406–1406 10.1038/nbt1297-1406 [DOI] [PubMed] [Google Scholar]
- 13.Krimsky S. (2017) The unsteady state and inertia of chemical regulation under the US toxic substances control act. PLoS Biol. 15, 1–10 10.1371/journal.pbio.2002404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou B.K., Wackett L.P. and Ellis L.B. (2003) Microbial pathway prediction: a functional group approach. J. Chem. Inf. Comput. Sci. 43, 1051–1057 10.1021/ci034018f [DOI] [PubMed] [Google Scholar]
- 15.Fenner K., Gao J., Kramer S., Ellis L. and Wackett L.P. (2008) Data-driven extraction of relative reasoning rules to limit combinatorial explosion in biodegradation pathway prediction. Bioinformatics 24, 2079–2085 10.1093/bioinformatics/btn378 [DOI] [PubMed] [Google Scholar]
- 16.Gao J.F., Ellis L.B.M. and Wackett L.P. (2011) The University of Minnesota pathway prediction system: multi-level prediction and visualization. Nucleic Acids Res. 39, W406–W411 10.1093/nar/gkr200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wicker J., Fenner K., Ellis L.B.M., Wackett L.P. and Kramer S. (2010) Predicting biodegradation products and pathways: a hybrid knowledge-based and machine learning based approach. Bioinformatics 26, 814–821 10.1093/bioinformatics/btq024 [DOI] [PubMed] [Google Scholar]
- 18.Wicker J., Lorsbach T., Gütlein M., Schmid E., Latino D., Kramer S. et al. (2016) enviPath-The environmental contaminant biotransformation pathway resource. Nucleic Acids Res. 44, D502–D508 10.1093/nar/gkv1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moriya Y., Shigemizu D., Hattori M., Tokimatsu T., Kotera M., Goto S. et al. (2010) PathPred: an enzyme-catalyzed metabolic pathway prediction server. Nucleic Acids Res. 38, W138–W143 10.1093/nar/gkq318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finley S.D., Broadbelt L.J. and Hatzimanikatis V. (2009) Computational framework for predictive biodegradation. Biotechnol. Bioeng. 104, 1086–1097 10.1002/bit.22489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dimitrova N.H., Dermen I.A., Todorova N.D., Vasilev K.G., Dimitrov S.D., Mekenyan O.G. et al. (2017) CATALOGIC 301c model: validation and improvement. SAR QSAR Environ. Res. 28, 511–524 10.1080/1062936X.2017.1343255 [DOI] [PubMed] [Google Scholar]
- 22.Latino D.A., Wicker J., Gütlein M., Schmid E., Kramer S. and Fenner K. (2017) Eawag-Soil in enviPath: a new resource for exploring regulatory pesticide soil biodegradation pathways and half-life data. Environ. Sci. Process Impacts 19, 449–464 10.1039/c6em00697c [DOI] [PubMed] [Google Scholar]
- 23.Gibson B., Wilson D.J., Feil E. and Eyre-Walker A. (2018) The distribution of bacterial doubling times in the wild. Proc. Biol. Sci. 285, 20180789 10.1098/rspb.2018.0789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koshland D.E. (1978) Role of flexibility of the specificity, control and evolution of enzymes. FEBS Lett. 62, E47–E52 10.1016/0014-5793(76)80853-8 [DOI] [PubMed] [Google Scholar]
- 25.Copley S.D. (2015) An evolutionary biochemist's perspective on promiscuity. Trends Biochem. Sci. 40, 72–78 10.1016/j.tibs.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Copley S.D. (2017) Shining a light on enzyme promiscuity. Curr. Opin. Struct. Biol. 47, 167–175 10.1016/j.sbi.2017.11.001 [DOI] [PubMed] [Google Scholar]
- 27.Copley S.D. (2009) Evolution of efficient pathways for degradation of anthropogenic chemicals. Nat. Chem. Biol. 5, 559–566 10.1038/nchembio.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seffernick J.L., Dodge A.G., Sadowsky M.J., Bumpus J.A. and Wackett L.P. (2010) Bacterial ammeline metabolism by guanine deaminase. J. Bacteriol. 192, 1106–1112 10.1128/JB.01243-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obexer R., Godina A., Garrabou X., Mittl P.R., Baker D., Griffiths A.D. et al. (2017) Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase. Nat. Chem. 9, 50–56 10.1038/nchem.2596 [DOI] [PubMed] [Google Scholar]
- 30.Arnold F.H. (2019) Innovation by evolution: bringing new chemistry to life (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 58, 14420–14426 10.1002/anie.201907729 [DOI] [PubMed] [Google Scholar]
- 31.Nagata Y., Ohtsubo Y. and Tsuda M. (2015) Properties and biotechnological applications of natural and engineered haloalkane dehalogenases. Appl. Microbiol. Biotechnol. 99, 9865–9881 10.1007/s00253-015-6954-x [DOI] [PubMed] [Google Scholar]
- 32.Kan S.J., Lewis R.D., Chen K. and Arnold F.H. (2016) Directed evolution of cytochrome c for carbon-silicon bond formation: bringing silicon to life. Science 354, 1048–1051 10.1126/science.aah6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bar-Even A., Noor E., Savir Y., Liebermeister W., Davidi D., Tawfik D.S. et al. (2011) The moderately efficient enzyme: evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry 50, 4402–4410 10.1021/bi2002289 [DOI] [PubMed] [Google Scholar]
- 34.Davidi D., Longo L.M., Jabłońska J., Milo R. and Tawfik D.S. (2018) A bird's-eye view of enzyme evolution: chemical, physicochemical, and physiological considerations. Chem. Rev. 118, 8786–8797 10.1021/acs.chemrev.8b00039 [DOI] [PubMed] [Google Scholar]
- 35.LeBaron H.M., McFarland J. and Burnside O. (2008) The Triazine Herbicides, Elsevier, Amsterdam [Google Scholar]
- 36.Udiković-Kolić N., Scott C. and Martin-Laurent F. (2011) Evolution of atrazine-degrading capabilities in the environment. Appl. Microbiol. Biotechnol. 96, 1175–1189 10.1007/s00253-012-4495-0 [DOI] [PubMed] [Google Scholar]
- 37.Wackett L.P. (2003) Evolution of new enzymes and pathways: Soil microbes adapt to s-triazine herbicides In Pesticide Decontamination and Detoxification (Gan J.J., Zhu P.C., Aust S. D. and Lemley A.T. eds.), pp. 37–48, American Chemical Society Symposium Series No, Washington, D.C [Google Scholar]
- 38.Martinez B., Tomkins J., Wackett L.P., Wing R. and Sadowsky M.J. (2001) Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. ADP. J. Bacteriol. 183, 5684–5697 10.1128/JB.183.19.5684-5697.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Souza M.L., Seffernick J., Martinez B., Sadowsky M.J. and Wackett L.P. (1998) The atrazine catabolism genes atzABC are widespread and highly conserved. J. Bacteriol. 180, 1951–1954 10.1128/JB.180.7.1951-1954.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raillard S.-A., Krebber Y., Chen J.E., Ness E., Bermudez R., Trinidad R. et al. (2001) Novel enzyme activities and functional plasticity revealed by recombining homologous enzymes. Chem. Biol. 8, 891–898 10.1016/S1074-5521(01)00061-8 [DOI] [PubMed] [Google Scholar]
- 41.Seffernick J.L., deSouza M.L., Sadowsky M.J. and Wackett L.P. (2001) Melamine deaminase and atrazine chlorohydrolase: 98% identical but functionally different. J. Bacteriol. 183, 2405–2410 10.1128/JB.183.8.2405-2410.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noor S., Taylor M.C., Russell R.J., Jermiin L.S., Jackson C.J., Oakeshott J.G. et al. (2012) Intramolecular epistasis and the evolution of a new enzymatic function. PLoS ONE 7, 11 10.1371/journal.pone.0039822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poelarends G.J., Wilkens M., Larkin M.J., van Elsas J.D. and Janssen D.B. (1998) Degradation of 1,3-dichloropropene by Pseudomonas cichorii 170. Appl. Environ. Microbiol. 64, 2931–2936 10.1128/AEM.64.8.2931-2936.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poelarends G.J. and Whitman C.P. (2004) Evolution of enzymatic activity in the tautomerase superfamily: mechanistic and structural studies of the 1,3-dichloropropene catabolic enzymes. Bioorg. Chem. 32, 376–392 10.1016/j.bioorg.2004.05.006 [DOI] [PubMed] [Google Scholar]
- 45.Poelarends G.J., Almrud J.J., Serrano H., Darty J.E., Johnson W.H. Jr, Hackert M.L. et al. (2006) Evolution of enzymatic activity in the tautomerase superfamily: mechanistic and structural consequences of the L8R mutation in 4-oxalocrotonate tautomerase. Biochemistry 45, 7700–7708 10.1021/bi0600603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.CAS registry (2020) https://www.cas.org/support/documentation/chemical-substances
- 47.Alegbeleye O.O., Opeolu B.O. and Jackson V.A. (2017) Polycyclic aromatic hydrocarbons: a critical review of environmental occurrence and bioremediation. Environ. Manag. 60, 758–783 10.1007/s00267-017-0896-2 [DOI] [PubMed] [Google Scholar]
- 48.Zhao L., Xu B., Ablikim U., Lu W., Ahmed M., Evseev M.M. et al. (2019) Gas-phase synthesis of triphenylene. ChemPhysChem 20, 791–797 10.1002/cphc.201801154 [DOI] [PubMed] [Google Scholar]
- 49.Boyland E. and Warren F.L. (1937) The induction of tumours by methylcholanthrene in two strains of mice. J. Path. Bact. 45, 171–177 10.1002/path.1700450116 [DOI] [Google Scholar]
- 50.Ewa B. and Danuta M.Š (2017) Polycyclic aromatic hydrocarbons and PAH-related DNA adducts. J. Appl. Genet. 58, 321–330 10.1007/s13353-016-0380-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brodie B.B., Axelrod J., Cooper J.R., Gaudette L., La Du B.N., Mitoma C. et al. (1955) Detoxication of drugs and other foreign compounds by liver microsomes. Science 121, 603–604 10.1126/science.121.3147.603 [DOI] [PubMed] [Google Scholar]
- 52.Katagiri M., Ganguli B.N. and Gunsalus I.C. (1968) A soluble cytochrome P-450 functional in methylene hydroxylation. J. Biol. Chem. 243, 3543–3546 PMID: [PubMed] [Google Scholar]
- 53.Booth J. and Boyland E. (1949) Metabolism of polycyclic compounds; formation of 1,2-dihydroxy-1,2-dihydronaphthalenes. Biochem. J. 44, 361–365 10.1042/bj0440361 [DOI] [PubMed] [Google Scholar]
- 54.Fernley H.N. and Evans W.C. (1958) Oxidative metabolism of polycyclic hydrocarbons by soil Pseudomonads. Nature. 182, 373–375 10.1038/182373a0 [DOI] [PubMed] [Google Scholar]
- 55.Jeffrey A.M., Yeh H.J., Jerina D.M., Patel T.R., Davey J.F. and Gibson D.T. (1975) Initial reactions in the oxidation of naphthalene by Pseudomonas putida. Biochemistry 14, 575–584 10.1021/bi00674a018 [DOI] [PubMed] [Google Scholar]
- 56.Gibson D.T., Mahadevan V., Jerina D.M., Yogi H. and Yeh H.J. (1975) Oxidation of the carcinogens benzo[a]pyrene and benzo[a]anthracene to dihydrodiols by a bacterium. Science 189, 295–297 10.1126/science.1145203 [DOI] [PubMed] [Google Scholar]
- 57.Ramaswamy S. (2011) Naphthalene 1,2-Dioxygenase. Encyclopedia of Inorganic and Bioinorganic Chemistry
- 58.Moody J.D., Freeman J.P., Fu P.P. and Cerniglia C.E. (2004) Degradation of benzo[a]pyrene by Mycobacterium vanbaalenii PYR-1. Appl. Environ. Microbiol. 70, 340–345 10.1128/aem.70.1.340-345.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rabus R., Boll M., Heider J., Meckenstock R.U., Buckel W., Einsle O. et al. (2016) Anaerobic microbial degradation of hydrocarbons: from enzymatic reactions to the environment. J. Mol. Microbiol. Biotechnol. 26, 25–28 10.1159/000443997 [DOI] [PubMed] [Google Scholar]
- 60.Himmelberg A.M., Brüls T., Farmani Z., Weyrauch P., Barthel G., Schrader W. et al. (2018) Anaerobic degradation of phenanthrene by a sulfate-reducin enrichment culture. Environ. Microbiol. 20, 3589–3600 10.1111/1462-2920.14335 [DOI] [PubMed] [Google Scholar]
- 61.Batyrova K.A., Khusnutdinova A.N., Wang P.H., Di Leo R., Flick R., Edwards E.A. et al. (2020) Biocatalytic in vitro and in vivo FMN prenylation and (de)carboxylase activation. ACS Chem. Biol. 15, 1874–1882 10.1021/acschembio.0c00136 [DOI] [PubMed] [Google Scholar]
- 62.Weyrauch P., Heker I., Zaytsev A.V., von Hagen C.A., Arnold M.E., Golding B.T. et al. (2020) The 5,6,7,8-tetrahydro-2-naphthoyl-CoA reductase reaction in the anaerobic degradation of naphthalene and identification of downstream metabolites. Appl. Environ. Microbiol. 86, e00996–e02020 10.1128/AEM.00996-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huwiler S.G., Löffler C., Anselmann S.E.L., Stärk H.J., von Bergen M., Flechsler J. et al. (2019) One-megadalton metalloenzyme complex in Geobacter metallireducens involved in benzene ring reduction beyond the biological redox window. Proc. Natl. Acad. Sci U.S.A. 116, 2259–2264 10.1073/pnas.1819636116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kanehisa M. and Goto S. (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caspi R., Billington R., Keseler I.M., Kothari A., Krummenacker M., Midford P.E. et al. (2020) The MetaCyc database of metabolic pathways and enzymes - a 2019 update. Nucleic Acids Res. 48, D445–D453 10.1093/nar/gkz862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wackett L.P. (1994) Dehalogenation in environmental biotechnology. Curr. Opin. Biotechnol. 5, 260–265 10.1016/0958-1669(94)90027-2 [DOI] [PubMed] [Google Scholar]
- 67.Gribble G.W. (2015) Biological activity of recently discovered halogenated marine natural products. Mar. Drugs 13, 4044–4136 10.3390/md13074044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Janssen D.B., van der Ploeg J.R. and Pries F. (1994) Genetics and biochemistry of 1,2- dichloroethane degradation. Biodegradation 5, 249–257 10.1007/BF00696463 [DOI] [PubMed] [Google Scholar]
- 69.McDonald I.R., Warner K.L., McAnulla C., Woodall C.A., Oremland R.S. and Murrell J.C. (2002) A review of bacterial methyl halide degradation: biochemistry, genetics and molecular ecology. Environ. Microbiol. 4, 193–203 10.1046/j.1462-2920.2002.00290.x [DOI] [PubMed] [Google Scholar]
- 70.Frantz B., Aldrich T. and Chakrabarty A.M. (1987) Microbial degradation of synthetic recalcitrant compounds. Biotechnol. Adv. 5, 85–99 10.1016/0734-9750(87)90005-x [DOI] [PubMed] [Google Scholar]
- 71.Gantzer C.J. and Wackett L.P. (1991) Reductive dechlorination catalyzed by bacterial transition-metal coenzymes. Environ. Sci. Technol. 25, 715–722 10.1021/es00016a017 [DOI] [Google Scholar]
- 72.Krone U.E., Laufer K., Thauer R.K. and Hogenkamp H.P. (1989) Coenzyme F430 as a possible catalyst for the reductive dehalogenation of chlorinated C1 hydrocarbons in methanogenic bacteria. Biochemistry 28, 10061–10065 10.1021/bi00452a027 [DOI] [PubMed] [Google Scholar]
- 73.Li S. and Wackett L.P. (1993) Reductive dehalogenation by cytochrome P450CAM: substrate binding and catalysis. Biochemistry 32, 9355–9361 10.1021/bi00087a014 [DOI] [PubMed] [Google Scholar]
- 74.Fincker M. and Spormann A.M. (2017) Biochemistry of catabolic reductive dehalogenation. Annu. Rev. Biochem. 86, 357–386 10.1146/annurev-biochem-061516-044829 [DOI] [PubMed] [Google Scholar]
- 75.van Hylckama Vlieg J.E. and Janssen D.B. (2001) Formation and detoxification of reactive intermediates in the metabolism of chlorinated ethenes. J. Biotechnol. 85, 81–102 10.1016/s0168-1656(00)00364-3 [DOI] [PubMed] [Google Scholar]
- 76.Parales R.E. and Haddock J.D. (2004) Biocatalytic degradation of pollutants. Curr. Opin. Biotechnol. 15, 374–379 10.1016/j.copbio.2004.06.003 [DOI] [PubMed] [Google Scholar]
- 77.Fox B., Borneman J.G., Wackett L.P. and Lipscomb J.D. (1990) Haloalkene oxidation by the soluble methane monooxygenase from Methylosinus trichosporium OB3b: mechanistic and environmental implications. Biochemistry 29, 6419–6427 10.1021/bi00479a013 [DOI] [PubMed] [Google Scholar]
- 78.Oldenhuis R., Vink R.L., Janssen D.B. and Witholt B. (1989) Degradation of chlorinated aliphatic hydrocarbons by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase. Appl. Environ. Microbiol. 55, 2819–2826 10.1128/AEM.55.11.2819-2826.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wackett L.P., Sadowsky M.J., Newman L.M., Hur H.-G. and Li S. (1994) Metabolism of polyhalogenated compounds by a genetically engineered bacterium. Nature 368, 627–629 10.1038/368627a0 [DOI] [PubMed] [Google Scholar]
- 80.Brown J.F. Jr, Bedard D.L., Brennan M.J., Carnahan J.C., Feng H. and Wagner R.E. (1987) Polychlorinated biphenyl dechlorination in aquatic sediments. Science 236, 709–712 10.1126/science.236.4802.709 [DOI] [PubMed] [Google Scholar]
- 81.Abramowicz D.A. (1995) Aerobic and anaerobic PCB biodegradation in the environment. Environ. Health Perspect. 103, 97–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bommer M., Kunze C., Fesseler J., Schubert T., Diekert G. and Dobbek H. (2014) Structural basis for organohalide respiration. Science 346, 455–458 10.1126/science.1258118 [DOI] [PubMed] [Google Scholar]
- 83.Payne K.A., Quezada C.P., Fisher K., Dunstan M.S., Collins F.A., Sjuts H. et al. (2015) Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation. Nature 517, 513–516 10.1038/nature13901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Collins F.A., Fisher K., Payne K.A.P., Gaytan Mondragon S., Rigby S.E.J. and Leys D. (2018) NADP(H)-driven organohalide reduction by a nonrespiratory reductive dehalogenase. Biochemistry 57, 3493–3502 10.1021/acs.biochem.8b00255 [DOI] [PubMed] [Google Scholar]
- 85.Marks T.S., Allpress J.D. and Maule A. (1989) Dehalogenation of lindane by a variety of porphyrins and corrins. Appl. Environ. Microbiol. 55, 1258–1261 10.1128/AEM.55.5.1258-1261.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Giedyk M., Goliszewska K. and Gryko D. (2015) Vitamin B12-catalysed reactions. Chem. Soc. Rev. 44, 3391–3404 10.1039/c5cs00165j [DOI] [PubMed] [Google Scholar]
- 87.Molenda O., Puentes Jácome L.A., Cao X., Nesbø C.L., Tang S., Morson N. et al. (2020) Insights into origins and function of the unexplored majority of the reductive dehalogenase gene family as a result of genome assembly and ortholog group classification. Environ. Sci. Process Impacts 22, 663–678 10.1039/c9em00605b [DOI] [PubMed] [Google Scholar]
- 88.Hug L.A. and Edwards E.A. (2013) Diversity of reductive dehalogenase genes from environmental samples and enrichment cultures identified with degenerate primer PCR screens. Front. Microbiol. 4, 341 10.3389/fmicb.2013.00341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Walker M.C. and Chang M.C. (2014) Natural and engineered biosynthesis of fluorinated natural products. Chem. Soc. Rev. 43, 6527–6536 10.1039/c4cs00027g [DOI] [PubMed] [Google Scholar]
- 90.Au K. and Walsh C.T. (1984) Stereochemical studies on a plasmid-coded fluoroacetate halidohydrolase. Bioorg. Chem. 12, 197–205 10.1016/0045-2068(84)90003-8 [DOI] [Google Scholar]
- 91.Zhao Z.Q., Zheng T.C., Zhang W.J., Shen X.L., Lv L. and Li Y.M. (2019) Degradation of 3-fluoroaniline by Rhizobium sp. JF-3. Biodegradation 30, 433–445 10.1007/s10532-019-09885-8 [DOI] [PubMed] [Google Scholar]
- 92.Tiedt O., Mergelsberg M., Boll K., Müller M., Adrian L., Jehmlich N. et al. (2016) ATP-dependent C-F bond cleavage allows the complete degradation of 4-fluoroaromatics without oxygen. mBio 7, e00990-16 10.1128/mBio.00990-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kiel M. and Engesser K.H. (2015) The biodegradation vs. biotransformation of fluorosubstituted aromatics. Appl. Microbiol. Biotechnol. 99, 7433–7464 10.1007/s00253-015-6817-5 [DOI] [PubMed] [Google Scholar]
- 94.March J. (1992) Advanced Organic Chemistry, 2nd edn, McGraw-Hill Publishers, New York [Google Scholar]
- 95.Organisation for Economic Co-operation and Development. (2019) https://www.oecd.org/chemicalsafety/portal-perfluorinated-chemicals/aboutpfass/
- 96.Sunderland E.M., Hu X.C., Dassuncao C., Tokranov A.K., Wagner C.C. and Allen J.G. (2019) A review of the pathways of human exposure to poly- and perfluoroalkyl substances (PFASs) and present understanding of health effects. J. Expo. Sci. Environ. Epidemiol. 29, 131–147 10.1038/s41370-018-0094-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang S. and Jaffé P.R. (2019) Defluorination of perfluorooctanoic acid (PFOA) and perfluorooctane sulfonate (PFOS) by Acidimicrobium sp. strain A6. Environ. Sci. Technol. 53, 11410–11419 10.1021/acs.est.9b04047 [DOI] [PubMed] [Google Scholar]
- 98.Bentel M.J., Yu Y., Xu L., Li Z., Wong B.M., Men Y. et al. (2019) Defluorination of Per- and polyfluoroalkyl substances (PFASs) with hydrated electrons: structural dependence and implications to PFAS remediation and management. Environ. Sci. Technol. 53, 3718–3728 10.1021/acs.est.8b06648 [DOI] [PubMed] [Google Scholar]
- 99.Zenobio J.E., Modiri-Gharehveran M., de Perre C., Vecitis C.D. and Lee L.S. (2020) Reductive transformation of perfluorooctanesulfonate by NiFe-activated carbon. J. Hazard. Mater. 397, 122782 10.1016/j.jhazmat.2020.122782 [DOI] [PubMed] [Google Scholar]
- 100.Yang L., He L., Xue J., Ma Y., Xie Z., Wu L. et al. (2020) Persulfate-based degradation of perfluorooctanoic acid (PFOA) and perfluorooctane sulfonate (PFOS) in aqueous solution: review on influences, mechanisms and prospective. J. Hazard. Mater. 5, 122405 10.1016/j.jhazmat.2020.122405 [DOI] [PubMed] [Google Scholar]
- 101.Singh R.K., Fernando S., Baygi S.F., Multari N., Thagard S.M. and Holsen T.M. (2019) Breakdown products from perfluorinated alkyl substances (PFAS) degradation in a plasma- based water treatment process. Environ. Sci. Tech. 53, 2731–2738 10.1021/acs.est.8b07031 [DOI] [PubMed] [Google Scholar]
- 102.Howell E. (2013) What is the world's tallest tree? Web page https://www.livescience.com/28729-tallest-tree-in-world.html. Accessed 6 June, 2020
- 103.Sangha A.K., Parks J.M., Standaert R.F., Ziebell A., Davis M. and Smith J.C. (2012) Radical coupling reactions in lignin synthesis: a density functional theory study. J. Phys. Chem. B. 116, 4760–4768 10.1021/jp2122449 [DOI] [PubMed] [Google Scholar]
- 104.Janusz G., Pawlik A., Sulej J., Swiderska-Burek U., Jarosz-Wilkolazka A. and Paszczynski A. (2017) Lignin degradation: microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 41, 941–962 10.1093/femsre/fux049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Feldman D. (2008) Polymer history. Design. Monomers Polymers 11, 1–15 10.1163/156855508X292383 [DOI] [Google Scholar]
- 106.Andrady A.L. (2015) Plastic Products. In Plastics and Environmental Sustainability, pp. 83–119, John Wiley & Sons, Hoboken, New Jersey [Google Scholar]
- 107.Lebreton L., Slat B., Ferrari F., Sainte-Rose B., Aitken J., Marthouse R. et al. (2018) Evidence that the Great Pacific Garbage Patch is rapidly accumulating plastic. Sci. Rep. 8, 4666 10.1038/s41598-018-22939-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Johnson A.F. and Mantus E. (2020) Closing the Loop on the Plastics Dilemma: Proceedings of A Workshop in Brief, National Academies Press; 10.17226/25647 [DOI] [Google Scholar]
- 109.Stapleton P.A. (2019) Toxicological considerations of nano-sized plastics. AIMS Environ. Sci. 6, 367–378 10.3934/environsci.2019.5.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zumstein M.T., Narayan R., Kohler H.E., McNeill K. and Sander M. (2019) Dos and do nots when assessing the biodegradation of plastics. Environ. Sci. Technol. 53, 9967–9969 10.1021/acs.est.9b04513 [DOI] [PubMed] [Google Scholar]
- 111.Montazer Z., Habibi Najafi M.B. and Levin D.B. (2020) Challenges with verifying microbial degradation of polyethylene. Polymers 12, 123 10.3390/polym12010123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Syranidou E., Karkanorachaki K., Amorotti F., Repouskou E., Kroll K., Kolvenbach B. et al. (2017) Development of tailored indigenous marine consortia for the degradation of naturally weathered polyethylene films. PLoS ONE 12, e0183984 10.1371/journal.pone.0183984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lowell Center for Sustainable Production (2011) Phthalates and their alternatives: health and environmental concerns. Technical Briefing
- 114.Meeker J.D., Sathyanarayana S. and Swan S.H. (2009) Phthalates and other additives in plastics: human exposure and associated health outcomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 2097–2113 10.1098/rstb.2008.0268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang W.W., Yin K. and Chen L.X. (2013) Bacteria-mediated bisphenol A degradation. Appl. Microbiol. Biotechnol. 97, 5681–5689 10.1007/s00253-013-4949-z [DOI] [PubMed] [Google Scholar]
- 116.Yoshida S., Hiraga K., Takehana T., Taniguchi I., Yamaji H., Maeda Y. et al. (2016) A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351, 1196–1199 10.1126/science.aad6359 [DOI] [PubMed] [Google Scholar]
- 117.Burke B., Cain S.M., Fitzgerald M., Stephens S., Turnbull M., and Mittapalli S. Terephthalate metabolism. Phthalate Family Map http://eawag-bbd.ethz.ch/pth/pth_map.html
- 118. https://www.genome.jp/kegg-bin/show_pathway?map00630+C01380 Ethylene glycol metabolism. Glyoxylate and dicarboxylate pathways, KEGG.
- 119.Austin H.P., Allen M.D., Donohoe B.S., Rorrer N.A., Kearns F.L., Silveira R.L. et al. (2018) Characterization and engineering of a plastic- degrading aromatic polyesterase. Proc. Natl Acad. Sci. U.S.A. 115, E4350–E4357 10.1073/pnas.1718804115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Roth C., Wei R., Oeser T., Then J., Föllner C., Zimmermann W. et al. (2014) Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca. Appl. Microbiol. Biotechnol. 98, 7815–7823 10.1007/s00253-014-5672-0 [DOI] [PubMed] [Google Scholar]
- 121.Tournier V., Topham C.M., Gilles A., David B., Folgoas C., Moya-Leclair E. et al. (2020) An engineered PET depolymerase to break down and recycle plastic bottles. Nature 580, 216–219 10.1038/s41586-020-2149-4 [DOI] [PubMed] [Google Scholar]
- 122.Kim H.R., Lee H.M., Yu H.C., Jeon E., Lee S., Li J. et al. (2020) Biodegradation of polystyrene by Pseudomonas sp. isolated from the gut of superworms (larvae of Zophobas atratus). Environ. Sci. Technol. 54, 6987–6996 10.1021/acs.est.0c01495 [DOI] [PubMed] [Google Scholar]
- 123.Terzopoulou Z., Papadopoulos L., Zamboulis A., Papageorgiou D.G., Papageorgiou G.Z. and Bikiaris D.N. (2020) Tuning the properties of furandicarboxylic acid-based polyesters with copolymerization: a review. Polymers 12, 1209 10.3390/polym12061209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nazrin A., Sapuan S.M., Zuhri M.Y.M., Ilyas R.A., Syafiq R. and Sherwani S.F.K. (2020) Nanocellulose reinforced thermoplastic starch (TPS), polylactic acid (PLA), and polybutylene succinate (PBS) for food packaging applications. Front. Chem. 8, 213 10.3389/fchem.2020.00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jacob J., Schmoldt A. and Grimmer G. (1982) Influence of monooxygenase inducers on the metabolic profile of phenanthrene in rat-liver microsomes. Toxicology 25, 333–343 10.1016/0300-483X(82)90111-1 [DOI] [PubMed] [Google Scholar]
- 126.Gibson D.T. (1999) Beijerinckia sp strain B1: a strain by any other name. J. Ind. Microbiol. Biotechnol. 23, 284–293 10.1038/sj.jim.2900715 [DOI] [PubMed] [Google Scholar]