Abstract
The recent events of outbreaks related to different respiratory viruses in the past few years, exponentiated by the pandemic caused by the coronavirus disease 2019 (COVID-19), reported worldwide caused by SARS-CoV-2, raised a concern and increased the search for more information on viruses-based diseases. The detection of the virus with high specificity and sensitivity plays an important role for an accurate diagnosis. Despite the many efforts to identify the SARS-CoV-2, the diagnosis still relays on expensive and time-consuming analysis. A fast and reliable alternative is the use of low-cost biosensor for in loco detection. This review gathers important contributions in the biosensor area regarding the most current respiratory viruses, presents the advances in the assembly of the devices and figures of merit. All information is useful for further biosensor development for the detection of respiratory viruses, such as for the new coronavirus.
Keywords: Biosensors, Respiratory virus, Detection, Diagnosis
Graphical abstract
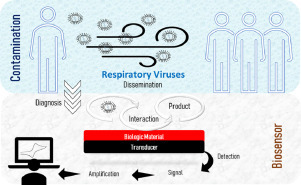
1. Introduction
At the end of 2019, the whole world was affected by a new pandemic, respiratory virus, named Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), which causes the coronavirus disease 2019 (COVID-19), reported initially in Wuhan, in China. According to the data provided from the Centers for Disease Control and Prevention (CDC) and the World Health Organization (WHO), respiratory diseases outbreaks have become more constant over the past few years, as the Severe Acute Respiratory Syndrome (SARS) in 2002, H5N1 Influenza (avian FLU) in 2004, H1N1 Influenza (swine FLU) in 2009, the Middle East Respiratory Syndrome (MERS) in 2012, H7N9 Influenza in 2013, the Enterovirus D68 in 2014, and more [1,2].
The determination of the sequence of virus genome, proteins structure and the host immunologic response is important to understand the virus pathogenicity, transmission, and infectivity, which facilitates the development of efficient vaccines and therapies. Moreover, these studies open novel possibilities to different methodologies for early detection, like biosensors, an easy-to-use device that can rapidly detect the disease even in asymptomatic conditions with high reliability and low cost.
One effective way, reported at media briefing from WHO Director-General to slow down the virus dissemination during the 2019 Covid-19 pandemic, was the social isolation, recommendation adopted worldwide [3]. The most effective way to this date, to detect the virus is the use of real-time quantitative polymerase chain reaction (RT-qPCR) [4], while antibody-based techniques (IgG/IgM) are being introduced as supplemental tools [5]. So far, expensive, time-consuming detection systems with the need for skilled labor personal are being used and evaluated. Moreover, during the social isolation, the individual need to break out from the confinement to perform the medical test, which increases the risk to enter into contact with the virus in the process. Therefore, the development of biosensors is important because the device can be operated in loco and it is ready-to-use by any personal.
This review focus on the most current contributions of biosensors designed for respiratory virus detection. The world scenario with the COVID-19 pandemic and the concerns regarding the latest and worrying outbreaks makes this survey of great importance for researchers planning to develop strategies for fast diagnosis. Some papers developing biosensors for SARS-CoV-2 determination have already been published, which proves that the biological material is already at hand to researches for further studies regarding this subject.
2. Biosensors
Biosensors are analytical devices that convert biological reactions into measurable signals. The biological material such as enzymes, tissues, microorganisms, antibodies, cell receptors, or a biomimetic component, is immobilized over a transducer, and interacts with the analyte in the solution, producing a biochemical response (Fig. 1 ). The transducer, in turn, converts this biochemical response into a quantifiable signal measured by the digital detector module [6,7]. The main types of transducing systems are the electrochemical, optical, and piezoelectric. Electrochemical biosensors monitor alterations in charge distribution over the transducer surface, based on potentiometric [8,9], amperometric [10], [11], [12] or impedimetric [13], [14], [15] transduction principles. Optical biosensors are versatile tools for analytical purposes because it provides multiplexed detection within a single device. These devices focus on the measurement of optical properties and characteristics of the transducer surface when occurring the interaction of the analyte with the recognition element. [16], [17], [18], [19]. Piezoelectric biosensors employ transducers that resonate when an external alternating electrical field is applied. They are based on the measurement of changes in the resonance frequency caused by the mass of the crystal and the immobilized biological material. According to the corresponding variation of electrical signal upon contact with the analyte, the difference in mass can be assessed. Among the many applications, these biosensors have been used in a wide in medicine to detect targets in biological systems [20], [21], [22].
Fig. 1.
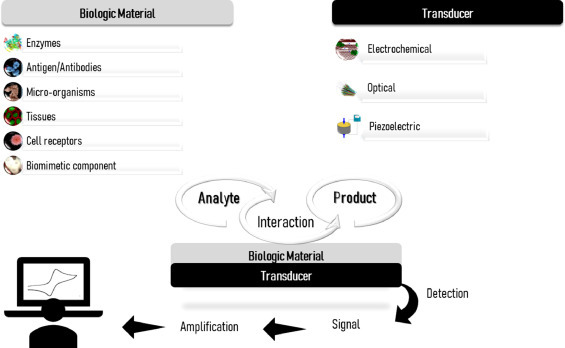
A general illustration of a biosensor.
The fast detection of biological pathogens plays a crucial role in the prevention of disease spread, infections, and pathologies [23]. Biosensors have found immense applications in medical diagnostics, and it offers more specific, sensitive, fast, and reproducible results as compared to the conventional techniques like biochemical assays and immunoassays [24].
Moreover, biosensors are been increasingly applied in clinical analysis due to their portability and point of care testing, which can analyze real biological samples in routine clinical use [25]. The incorporation of nanotechnology in the design of biosensors has improved the detection of biological specimens, as the preparation of biointerfaces of self-assembled monolayers (SAMs), improving biocompatibility and resistance against nonspecific adsorption [26], [27], [28], [29].
Point-of-care testing (POCT) is one of the most important applications of biosensors for infection diagnosis. POCT measurement is the practice of performing a diagnostic test near the patient to provide rapid results, providing appropriate, convenient care to patients, and more effective treatment of rapidly progressing infections. Moreover, these devices can be used without expensive instrumentation [30,31]. Accurate and early diagnoses play a crucial role in identifying the actual cause and nature of any disease. Currently, the focus shifted toward the early detection of COVID-19 disease. Saliva has a pivotal role in non-invasive salivary diagnostics that provide a convenient and cost-effective POCT platform for fast detection and may be an attempt to improve the chances of survival of patients from COVID-19 disease [32,33].
Biosensors can be further classified according to the analytes or reactions that they monitor as immunosensors (antibody–antigen interaction), enzymatic biosensors (enzyme–target analyte interaction) DNA biosensor (hybridization) and whole-cells biosensor.
2.1. Immunosensors
Immunosensors are biological sensors based on the specific interaction between antibodies and antigens. The lymphocyte B produces the antibody upon the host contact with an antigen and performs clonal expansion and differentiation. After, the antigens are eliminated, followed by the apoptosis of effector lymphocytes and remaining of memory B cells [34]. It has been developed for continuous monitoring of analytes through point-of-care devices, which provide low cost, full automation, portability, fast response, high sensitivity, accuracy, and precision [35]. The application of immunosensors in clinical diagnosis and monitoring of diseases has been emphasized in recent works and it is mainly reported for the detection of biomarkers [36,37] hormones [38], [39], [40], pathogenic bacteria [41], [42], [43], [44], [45], viruses [46], [47], [48], [49] and toxins [50], [51], [52], [53].
2.2. Enzymatic biosensors
Enzymatic biosensors exploit the catalytic property of enzymes, biorecognition molecules that possess high chemical specificity and have provided excellent selectivity for their targeted substrate. In enzymatic sensing, the device is combined with a transducer, which reacts selectively with its analyte, generating an electrochemical [54], optical [55] or piezoelectric [56] signal. This signal correlates to the concentration of the analyte in the sample [57], [58], [59]. Since enzyme electrodes devices offer several distinct advantages, they are used in many point-of-care and clinical applications for a broad range of analytes [60], [61], [62], [63].
2.3. Genosensors
Deoxyribonucleic acid (DNA) biosensors, or genosensors, have been exploited for their inherent physicochemical stability and suitability to provide practical ways to identify and diagnose various diseases. DNA is the carrier of genetic information, and it is distinct in any living organism, virus, or pathogen. Therefore, through their specific nucleic acid sequences, the DNA biosensor can be able to discriminate different organisms and diagnose various diseases and human pathogens. The principle of detection of a DNA biosensor relies on the immobilization of an immobilized DNA or RNA strand (probe), on the surface of a physical transducer, to detect its complementary (target) sequence via hybridization. The duplex formation can be detected following the association of an appropriate hybridization indicator, or through other changes accrued from the binding event [64], [65], [66]. Through the efforts of researchers, many new genosensors have emerged in recent years into clinical applications to detect various diseases and pathogens [67], [68], [69], [70].
2.4. Whole-cells biosensors
Whole cells can be used as recognition elements. A variety of surface antigens presented on the cell envelopes, including proteins, glycoproteins, lipopolysaccharides, and peptidoglycan, can act as targets for biorecognition. The development of biosensors for whole microorganisms is challenging because it requires the detection of analytes that are much larger (micrometer scale) than typical molecular analytes, such as proteins (nanometer scale). Many surface epitopes can lead to nonspecific interactions with the sensor surface. Nevertheless, organisms used to develop whole-cell biosensors are generally experimentally modified to incorporate transducer capacity or increase their sensitivity [71], [72], [73], [74], [75]. The whole cell-based biosensor is increasingly being reported in the literature, and these reports have shown high selectivity, sensitivity, and great potential for their use in biomedical diagnostics [76], [77], [78], [79].
3. Virus
As intracellular parasites, viruses use the cellular machineries for completion of their replication cycle. The basic structure of virus is comprised of genome (DNA or RNA), protein capsid (for nucleic acid protection) and in some a lipidic envelope that covers the capsid. [80,81].
The first reported studies on viruses began at the end of the XIX and the beginning of the XX century, through experiments with tobacco mosaic, in 1882, and with the action of bacteriophages in Shigella culture, in 1915 [82,83]. The first Influenza virus (FLU) isolated in laboratory was reported in 1933 [84]. It is a respiratory virus, associated with the 1918–1919 pandemic Spanish FLU that resulted in the death of about a quarter of the entire population, and is still the cause of seasonal flu in several countries [85]. From this and previous works, several types of researches on cell culture during the 40 s and 50 s alongside with the bacteriophage studies of Hershey and Chase in 1952, increased the interest regarding viruses, culminating at the beginning of the modern virology [86], [87], [88].
A virus replication only occurs in intracellular space, a crucial step for virus cycle. Fig. 2 represents a generic enveloped RNA virus cycle, the most common among respiratory viruses. In general, the replication steps consist firstly in the attachment between a viral protein and the receptor of the target-cell surface. After the attachment, the virus penetrates the membrane cell to cytoplasm. Right after, an uncoating step is responsible for the release of viral nucleic acid into the host cell and allows the synthesis of viral proteins and genome. Then, the viral particles are assembled according to the viral symmetry. With the new viral particles assembled, the maturation step leaves the virus infectious, increasing the viral tropism. Finally, the virus is released by different mechanisms, like cell lysis, budding, or exocytosis [89,90].
Fig. 2.
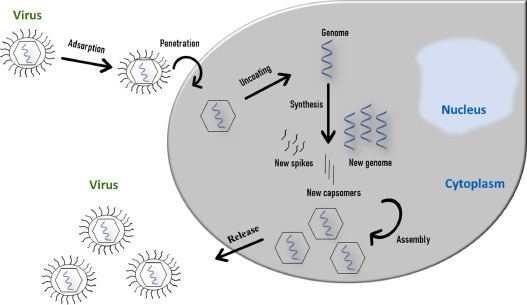
RNA virus replication in an intracellular space.
3.1. Respiratory viruses
In upper and lower airways in the respiratory tract, there are physical barriers composed by epithelial cells and mucus besides the alveolar macrophages in the lungs [91]. This set of protection seeks to safeguard the immune response of the host body in several ways, like the inhibition of interferon, latent infections, or the presence of fusion proteins in the viral capsid [90]. Besides, viruses can mutate and might result in more tropism and virulence, thus increasing the evasion from the immunologic system, aggravating the clinical condition of the host [92,93].
The most worrying respiratory viruses in public health: respiratory syncytial virus (RSV), coronavirus (MERS and SARS-CoV), and FLU [2], are based in RNA, in which the mutation process is more accentuated [94]. This mutation comes from the low fidelity of RNA polymerase, resulting in the incorporation of nucleotides errors during the replication of the material genetic with more frequency (10−3 to 10−4 nucleotides) compared to DNA polymerase (10−8 to 10−11 nucleotides), which is present in eukaryotic cells and some classes of viruses, as adenoviruses [94], [95], [96], [97]. The RNA viruses are responsible for countless cases of illness from these acute respiratory tract infections resulting in deaths worldwide, as summarized in Table 1 .
Table 1.
Data of the deadliest RNA viruses around the world.
| Virus | Cases | Deaths | Reference |
|---|---|---|---|
| Influenza (seasonal)a | 3 – 5 million | 290,000 – 650,000 | [98] |
| RSVa | ~30 million | > 100,000 | [99] |
| MERSb | 2494 | 858 | [100] |
| SARSc | 8096 | 774 | [101] |
These are annual estimates.
Data obtained from September 2012 – February 2019.
Data obtained from November 2002 – July 2003.
Despite the shorter evaluated period, SARS and MERS shows a high mortality rate. With annual estimates, unfortunately, Influenza and RSV are still the deadliest among the respiratory viruses. The other viruses, although data are not shown in the table, can also lead to death, even in a small proportion or in a local scenario, as it is the case for some reports regarding human adenovirus [102], human metapneumovirus [103] and rhinovirus [104].
The gold standard test to diagnose respiratory virus disease is based on cell culture, upon the evaluation of the cytopathic effect and hemadsorption caused by the virus [105] . However, molecular assays based on nucleic acid have also been employed [106], [107], [108], [109], [110] as the respiratory virus multiplex PCR systems for respiratory virus detection, with high accuracy for early diagnosis [111]. However, the need for a diagnosis with high accuracy and precision, associated with fast analysis, low cost, in an easy-to-handle device, is still not observed [112,113]. Therefore, alternatives as the use of biosensors have been under evaluation to supply these demands [114,115]. The next topics surveyed the most recent contributions related to biosensors applied to the determination of respiratory viruses. This compilation is important to disseminate information to researches interested in the development of such devices.
4. Viral respiratory biosensors
4.1. Influenza
Influenza virus is a member of the Orthomyxoviridae family, with (-)ssRNA nucleic acid, and the species A and B are the most common associated with human infection and disease [116]. The Influenza virus exhaust path consists of the alteration in the hemagglutinin antigens (1–18) and the neuraminidase (1–11), responsible for the attachment and penetration in the host cell, respectively [117,118]. Upon that, it is possible to distinguish the virus A subtypes. As for the seasonal specie B, it differs basically in the HA1 antigen between the two strains, B/Yamagata/16/88, and B/Victoria/2/87, and the classification is based on this difference [119,120].
The Influenza virus is responsible for countless deaths around the world, especially at the end of the pandemic in 1910, and only in the 30 s, researchers acquired useful information regarding the virus. It is estimated that the Influenza virus, especially the subtype H1N1, has infected about 500 million people [117].
The virus isolation at 1933 [84], alongside with Louis Pasteur research on a vaccine against rabies, in 1885 [121], were important developments for the soviet development of the first known attenuated vaccine against the influenza virus A [122]. However, only in the 40 s, an inactivated vaccine was approved. Although safer, it was less efficient [81]. It contained viral particles from both Influenza A and B, for population distribution [123].
The majority of biosensor development found in the literature are designed to Influenza virus detection, as attested by the high number of reviews regarding this subject [124], [125], [126], [127], [128]. Table 2 contains data based on biosensor research directed to Influenza virus detection in the past five years. It is the most studied virus among the other respiratory viruses, and some reports stand out for low limit of detection, simplicity, fastness, and cutting-edge technology.
Table 2.
Influenza virus biosensors data.
| Detection technique | Substrate | Immobilized material / Analyte | Monitored compound | Working Range | LOD | Sample | Ref. |
|---|---|---|---|---|---|---|---|
| Electrochemical – Chronoamperometry and DPV | Dual carbon SPE | H1N1 antibody/ HA protein of the H1N1 | Methylene Blue | 25.0 – 500 pM | 9.40 pM | Commercial H1N1 | [129] |
| H5N1 antibody/ HA protein of the H5N1 | 8.30 pM | Commercial H5N1 | |||||
| Electrochemical – Amperometry | Glass wafer/PDMS | H1N1 antibody/ H1N1 antigen (sandwich ELISA-like assay) | TMB | 1.00 pg.mL−1 – 10.0 ng.mL−1 | 1.00 pg.mL−1 | Commercial ELISA kits | [130] |
| H5N1 antibody/ H5N1 antigen (sandwich ELISA-like assay) | |||||||
| H7N9 antibody/ H7N9 antigen (sandwich ELISA-like assay) | |||||||
| Electrochemical – OSWV | Gold electrode | His6—H1 HA/ anti-H1N1 | Potencial of peak redox Cu(II)/Cu (I) current | 1.00 × 109- 1.00 × 108 sera dilution | 1.00 × 109 – 1.00 × 108 sera dilution | Vaccinated mice sera | [131] |
| Optical – SPR | Gold sensor | H5N1 antigen/ H5N1 antibody | Label-free detection | Not determined | 193.3 ng.mL−1 | – | [132] |
| Electrochemical - EIS | Gold SPE | ssDNA of the H1N1/ ss-cDNA of theH1N1 | Label-free detection | 0.000 ng.mL−1 – 166,7 ng.mL−1 | 0.667 ng.mL−1 | Nasal swab and oropharyngeal samples | [133] |
| Electrochemical – Amperometry | Gold electrode | DNA tetrahedral probe of the H7N9/ ssDNA of H7N9 | TMB | 1.00 pM – 2.50 nM | 0.750 pM | Throat swab samples | [134] |
| Electrochemical – EIS | Rolled-up gold nanomembrane microelectrodes | ssDNA of the H1N1/ ssDNA of the H1N1 | Label-free detection | 20.0 aM – 2.00 pM | 20 aM | Commercil samples | [135] |
| Optical – UV–vis spectrometer | PDA vesicles | H5N1 antibody/ HA of the H5N1 | PDA color change | 5.40 × 10−4 – 13.5 copies.μL−1 | 0.530 copies.µL−1 | Tracheal swabs collected from wild birds | [136] |
| Electrochemical – DPV | Super-hydrophobic paper / conductive carbon paste | H1N1 antibody/ H1N1 antigen | Label-free detection | 10.0 – 1 × 104 PFU.mL−1 | 113 PFU.mL−1 | Saliva from a healthy person | [137] |
| Optical – Scanner and ImageJ | Gold paper electrode | H1N1 antibody/ H1N1 antigen (sandwich ELISA-like assay) | TMB | 0.000 – 1 × 104 PFU.mL−1 | 1.34 PFU.mL−1 2.27 PFU.mL−1 |
Commercial sample Spike saliva |
[138] |
| Electrochemical – EIS | Label-free | 0.000 – 1 × 104 PFU.mL−1 | 3.30 PFU.mL−1 4.70 PFU.mL−1 |
Commercial sample | [138] | ||
| Spike saliva | |||||||
| Electrochemical – EIS | Gold electrode | Glycan / H3N2 virus | Label-free | 8.00 aM – 0.800 nM | 5.00 aM | Intact, but inactivated H3N2 particles | [139] |
| BioScan nanomechanical cantilever system | Nanomechanical cantilever | 3′SL-PAA polymer / H5N3 surface glycoprotein | Label-free | 10.06 – 10.08 vp.mL−1 | 10.06 vp.mL−1 | Commercial H5N3 | [141] |
| Piezoelectric – SPM | Lead zirconate titanate piezoelectric disk | 3′SL-PAA polymer / H5N3 surface glycoprotein | Label-free | 10.05 – 10.07 vp.mL−1 | 10.05 vp.mL−1 (100 µm thick) | Commercial H5N3 | [142] |
| 10.04 vp.mL−1 (10 µm thick) | |||||||
| Electrochemical – EIS | BDD | anti-M1 H1N1 protein/ M1 protein of H1N1 | Label-free | 5.00 – 10.0 PFU.mL−1 | 1.00 fg.mL−1 | Saliva buffer | [143] |
| Electrochemical – EIS | ITO/glass electrode | H1N1 antibody/ H1N1 antigen | Label-free | 10.0 – 10.04 PFU.mL−1 | 26.0 PFU.mL−1 (PBS) | Saliva sample | [144] |
| 33.0 PFU.mL−1 (Saliva) | |||||||
| Multiplex RT-PCR-electrochemical genosensor - Voltmeter | Gold electrode | ssDNA of the H1N1 PCR products/ H1N1 antigen | Ferrocene | 10.04 TCID50 to 10.0° TCID50 | 10.0 TCID50.mL−1 (H1N1) | Pediatric pneumonia patients samples | [145] |
| ssDNA of the H3N2 PCR products/ H3N2 antigen | 100 TCID50.mL−1 (H3N2) | ||||||
| Electrochemical – DPV | ITO/glass electrode | ssDNA of the H1N1/ mini-HA protein | Potassium ferrocyanide/ferricyanide | 10.0 – 10.04 PFU.mL−1 | 3.70 PFU.mL−1 | Commercial H1N1 | [146] |
| Electrochemiluminescence | Gold electrode | HA of the H1N1/ anti-H1N1 | Immunoliposome encapsulating tris (2,2′- bipyridyl) ruthenium (II) complex | 2.70 × 102 – 2.70 × 103 PFU.mL−1 | Not determined | Commercial H1N1 | [147] |
| Electrochemical – EIS | Gold electrode in PDMS microchannels | H1N1 antibody/ H1N1 antigen | Label-free | 1.00 – 10.04 PFU.mL−1 | 0.500 PFU.mL−1 | Commercial H1N1 | [148] |
| Optical – IM-SPR | Gold chip | H7N9 antibody/ Attenuated reassorted H7N9 antigen | Label-free | 2.30 × 102 – 2.30 × 105 copies.mL−1 | 402 copies.mL−1 | Nasal mucosa from flu-like syndrome patients | [149] |
| 144 copies.mL−1 | Commercial H7N9 | ||||||
| Electrochemical – DPV | SPCE | H5N1 DNA aptamer/ anti-H5N1 (sandwich ELISA-like assay) | Electrocatalytic reaction of the surface ALP with APP | 100 fM –10.0 pM | 100 fM | Diluted human serum samples spiked | [150] |
| Electrochemical – CV | Gold electrode | Multi-functional DNA 3WJ/ HA protein of H5N1 | Fe3+/2+ of hemin | 1.00 pM – 100 nM | 1.00 pM | Chicken serum | [151] |
| Optical – UV–vis spectra | 96-well microplate | H5N1 antibody/ H5N1 antigen (sandwich ELISA-like assay) | TMB | 0.100 – 4.00 ng.mL−1 | 0.040 ng.mL−1 | Commercial H5N1 | [152] |
| Optical – SPR | Gold chip | H5N1 aptamer/ H5N1 whole virus (sandwich ELISA-like assay) | Aptamer-AuNPs | 1.00 × 104 – 8.00 × 104 EID50.mL−1 | 200 EID50.mL−1 | H5N1-infected feces samples | [153] |
| Electrochemical – OSWV | Gold electrode | His6—H5 HA/ anti-H5N1 | Cu(II) ions redox current decreasing | 4.00 – 100 pg.mL−1 | 2.40 pg.mL−1 | Hen sera from individuals vaccinated and non-vaccinated | [154] |
| Electrochemical – OSWV | Gold electrode | ssDNA of H5N1/ RNA of the H5N1 | Fe3+ | 3.00 × 103 – 3.00 × 105 copies.mL−1 | 3.00 copies.mL−1 | Biological sample | [155] |
| Optical – Fluorescence | Ag@SiO2 NPs | H5N1 aptamer/ Recombinant HA protein of the H5N1 | Thiazole orange | 2.00 – 100 ng.mL−1 | 2.00 ng.mL−1 | Commercial H5N1 | [156] |
| 3.50 ng.mL−1 | Human serum | ||||||
| Electrochemical – EIS | BDD | 2 – 4 mers peptide dendrimer/ H1N1 and H3N2 antigens | Label-free | 6.00 – 400 PFU,sample−1 for H3N2 and 3.00 – 400 PFU.sample−1 for H1N1 | 0.330 PFU.sample−1 for H1N1 and 0.91 PFU.sample−1 for H3N2 | Commercials HxNx | [157] |
| Electrochemical – IT method | SPCE | H1N1 antibody/ H1N1 antigen | TMB | 4.00 – 64.0 HA unit | 0.430 HA unit | Chick embryo allantoic saliva simulated sample | [158] |
| Electrochemical – CV | Tungsten rods | Anti-AIV NP aptamer/ AIV NP | Methylene blue | 2.00 – 12.0 nM | 1.13 nM | Negative oral and cloacal swabs from chicken | [159] |
| Electrochemical - modified and tailored MOSFET | Portable TFT | AIV antibody/ Nucleoprotein of the AIV | Label-free | 10.01 – 10.06 EID50 mL−1 | 10.02 EID50.mL−1 | Ducks and Mallards swab samples | [160] |
| Electrochemical – LSV | Gold electrode | H7 antibody / AIV H7 (sandwich ELISA-like assay) | AgNPs | 1.60 × 10−3 – 16.0 ng.mL−1 | 1.60 pg.mL−1 | Commercial AIV H7 | [161] |
| Optical – NNLFA | Sample pads | AIV antibody/ AIV nucleoproteins from H5N2 (sandwich ELISA-like assay) AIV antibody/ AIV nucleoproteins from H5N6 (sandwich ELISA-like assay) |
Ca2+ enhanced | 10.0°.5 to 10.04 EID50 mL−1 10.02.5 to 10.05 EID50 mL−1 |
10.02 EID50.mL−1 10.03.5 EID50.mL−1 |
Oropharyngeal swabs and cloacal swabs from ducks experimentally infected | [162] |
| Electrical - Magnetoresistance | GMR sensor | AIV antibody/ AIV antigen (sandwich ELISA-like assay) | Binding magnetic nanoparticles onto the GMR causes change in the resistance | 1.00 × 103 – 1.00 × 105 TCID50.mL−1 | 1.50 × 102 TCID50.mL−1 | AIVs obtained through colaboration | [163] |
| Electrochemical – DPV | Carbon DEP-chip | H1N1 antigen/ Aptamer-AuNPs | AuNPs | 0.400 – 100 µg.mL−1 | 0.51 μg.L − 1 | Human serum | [164] |
| Optical – Reflectance measurements | SiO2-based IO nanostructures | H1N1 antibody/ H1N1 antigen | Label-free detection | 10.03 – 10.05 PFU | Not determined | Commercial H1N1 | [165] |
| Optical – LSPR | ITO | DNA 3WJ/ HA protein from H5N1 | Label-free | 1.00 pM – 100 nM | 1.00 pM | Chicken serum | [166] |
| Electrochemical – IPA | SPCE | Biotin and fluorescein-labelled H5 PCR amplicons | TMB | 1.00 – 15.0 mM | 7.43 µM | Commercial H5N1 | [167] |
| Volumetric assay | MWCNT-IDE | H1N1 aptamer / H1N1 antigen (sandwich ELISA-like assay) | AuNPs | 0.010 – 100 pM | 10.0 fM (using aptamer for sandwich) | Commercial H1N1 | [168] |
| MWCNT- IDE | H1N1 antibody / H1N1 antigen (sandwich ELISA-like assay) | AuNPs | 1.00 pM (using antibody for sandwich) | ||||
| Optical – SPR | Gold optical fiber | H6 antibody/ H6 antigen | Label-free | 10.05 – 10.09 EID50.mL−1 | 5.14 × 105 EID50.mL−1 | Tracheal samples from chickens | [169] |
| Optical – IE | Silicon wafer | H5N1 antibody / H5N1 antigen | Label-free detection | Not determined | Not determined | Commercial H5N1 | [170] |
3′SL-PAA = synthetic sialylglycoconjugates based on a polymer matrix; 3WJ = 3 way-junction; AgNPs = silver nanoparticles; Ag@SiO2 NPs = silver@silicon dioxide nanoparticles; AIV = Avian influenza viruses; ALP = alcaline phosphatase; APP = 4-amino phenyl phosphate; AuNPs = gold nanoparticles; BDD = boron-doped diamond; DEP = disposable three-electrode screen-printed; DPV = differential pulse voltammetry; EIS = impedance spectroscopy; GMR = giant magnetoresistance; His6-H5 = histidine-tagged hemagglutinin; IDE = interdigitated dielectrode; IE = imaging ellipsometry; IM-SPR = intensity-modulated surface plasmon resonance; IO = inverse opal; IPA = intermittent pulse amperometry; ITO = indium tin oxide; LSPR = localized surface plasmon resonance; LSV = linear sweep voltammetry; MOSFET = metal−oxide semiconductor field-effect; MWCNT = multiwalled carbon nanotube; NNLPA = NIR-to-NIR lateral flow immunoassay;.
OSWV = Osteryoung square-wave voltammetric; PDA = polydiacetylene; PDMS = polydimethylsiloxane; SiO2 = silicon dioxide; SPCE = screen-printed carbon electrode; SPE = screen-printed electrode; SPM = scanning probe microscopy; SPR = surface plasmon resonance; ss-cDNA = single stranded-cDNA; TFT = thin film transistor; TMB = 3,3,5,5-tetramethylbenzidine;.
In a fast analysis, high sensitivity and selectivity are mandatory. Veerapandian [129] developed an electrochemical biosensor using a carbon screen-printed electrode modified with graphene oxide nanosheets followed by methylene blue adsorption, chitosan, protein-A from S. aureus, and monoclonal antibodies (H5N1 and H1N1) immobilized through drop-casting. Despite the many steps, a simple device can be assembled, with the possibility to detect two different subtypes simultaneously with a limit of detection below nM (9.4 pM for H1N1 and 8.3 for H5N1). The authors also informed the fastness of the analysis, with the detection time below 1 min, far better characteristics than the traditional methods. This work presents a biosensor with all the desirable features required for this device as high sensitivity, specificity, low cost, fastness, besides the simultaneous determination of two different virus subtypes in one measurement using small-volume samples (50 to 100 μL).
Regardless of their classification, antibodies present a unique and specific interaction with the correspondent antigen with a low equilibrium dissociation constant. The use of immunosensors stands out in numbers for Influenza virus detection, as can be seen in Table 2, including the simultaneous determination of different subtypes. An attractive aspect of the immunosensor is the possibility of the development of two different devices for the same purpose. It is possible to immobilize either the antibody or the antigen. Han [130] developed a biosensor for simultaneous detection of H1N1, H5N1, and H7N9 virus using a triple-arrayed three-electrode chip over a polydimethylsiloxane (PDMS) with a limit of detection as low as 1 pg.mL−1. In their work, the antibodies were immobilized over the transducer, and the virus antigens were detected. Fig. 3 depicts the sensor used by Han.
Fig. 3.
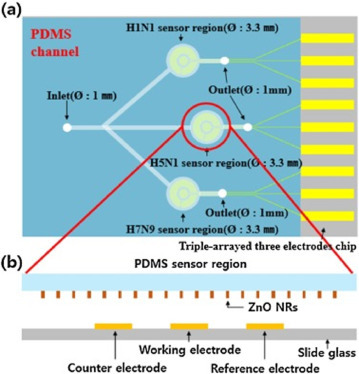
A schematic diagram of the designed triple-arrayed three-electrode immunosensor chip consisting of a top PDMS channel layer and a bottom glass substrate. (a) Top view of immunosensor chip with three different sensing regions for H1N1, H5N1, and H7N9, respectively. (b) Cross sectional view of H5N1 sensor region with ZnO NRs grown on the surface of PDMS and three electrodes aligned to the sensing chamber. Republished from [130]; permission conveyed through Copyright Clearance Center, Inc, License Number 4855350166819.
On the other hand, Miluka [131] immobilized the antigen, recombinant His-tagged HA subtype H1N1- monomer from the H1N1 pdm09 influenza virus for the detection of anti-hemagglutinin antibodies against the swine virus H1N1. The antigen was immobilized over a dipyrromethene-Cu(II)-modified gold electrode, and the anti-hemagglutinin H1 antibodies from mice sera were determined. Wong [132] reported a biosensor for H5N1 based on the H5N1 virus protein-modified gold electrode for the detection of H5N1 antibodies. The authors of these two works claimed that their devices present a lower limit of detection than commercial systems.
Another type of sensor growing considered attention is the genosensor, also applied to Influenza virus detection. An advantage of using RNA or DNA is that the classic biosensor assembly is dependent on the hybridization of single strands, a reversible process, meaning that the transducer surface can be regenerated. Genosensors are also known for their excellent limit of detection. Ravina [133] reported a biosensor based on a gold screen-printed electrode modified with 5′-amine labeled 22-mer ssDNA of hemagglutinin gene to the detection of the complementary DNA strand through electrochemical impedance spectroscopy (EIS). A limit of detection of 4 pg was obtained. Dong [134] developed an electrochemical genosensor for H7N9 with a limit of detection of 0.75 fM. However, Medina-Sánchez [135] has found an outstanding limit of detection of 20 aM through EIS without any amplification, introducing a new electrode with tubular geometry.
Electrochemical transducers are the basis of most of the devices studied for Influenza virus determination, but optical transducers also provide high efficiency and sensitivity. He [136] has found a remarkable limit of detection of 0.53 copies.mL−1 for H5N1 detection in a 20 min experiment using a colorimetric transducer based on the chromism and fluorescence properties of polydiacetylene vesicles modified through covalent binding to HA of H5 monoclonal antibody.
The variety of different electrodes, transducers, and detection techniques for Influenza virus detection, encouraged the development of innovative systems. Jang was the first to develop a low-cost, miniaturized paper-based [137], followed by a flow-based paper [138] immunosensor for electrochemical and colorimetric detection of Influenza H1N1 virus. The evolution from simple paper to flow-paper promoted a decrease in the limit of detection from 113 PFU.mL−1 to 3.3 PFU.mL−1 (electrochemical) and 1.34 PFU.mL−1 (colorimetric). Hushegyi [139] developed a glycan biosensor for the Influenza virus. Glycan are complex organic structures present in the surface of viruses and host cells, as for Influenza viruses and human cells [140]. Tkac used a polycrystalline gold electrode modified with a mixture of two thiols in a self-assembled monolayer (SAM) to immobilize a glycan (a 2,3-sialyllactose derivative). This biosensor was used to detect two Influenza virus subtypes, H1N1 and H5N1, through the interaction between the immobilized glycan and the virus glycoprotein hemagglutinin. This was the first detection at aM range (140 aM for H5N1 and 14 fM for H1N1) using an impedimetric glycan biosensor. A similar approach was used by Erofeev and Gorelkin [141,142], a syalylglycopolymer constituted of an H5N3 glycoprotein modified-polyacrylamide. The striking difference is the detection methodologies. In the first moment, they used a standard cantilever from atomic force microscope as the transducer. The measurement was based on the cantilever deflection induced by lateral intermolecular forces in a flow system upon contact with the virus. As it used a complex optical system, it was not classified by the authors for home appliances. Therefore, in the second moment, they switched into a piezoelectric transducer using a lead zirconate titanate piezoelectric disk. By keeping the flowing system and the same syalylglycopolymer, they achieved a sensitive, selective, label-free device that can be applied for commercial use.
Real biological samples are usually based on blood, but fluids as urine and saliva can also be used. Another advantage of biosensors for Influenza virus detection is the possibility of using less invasive fluids obtained from mouth, throat, or nose to get viral biological material. Nidworski [143] developed an impedimetric immunosensor for the Influenza virus based on a boron-doped diamond (BDD) electrode functionalized with polyclonal anti-M1 antibodies. Real samples based on throat and nasal swabs were treated to release the M1 protein from the virus. The sensor showed high sensitivity, selectivity, and rapid analysis (5 min).
4.2. Coronaviruses
The first studies of the coronavirus were performed in the late 1960s by distinct researcher groups. They have isolated, independently, different strains of a new virus in cell culture, which presented an unusual ether-sensitive property. Tyrrell demonstrated through electron microscopy that this new group of viruses was also morphological identical, with a crown-like appearance (so the name corona). Years after the understanding of the first coronavirus strains – B814, 229E, and OC43, and due to the virus study and its presence in many animals, the coronavirus was divided into three distinct groups: the first one containing 229E; the second one containing OC43; and the third one containing aviary virus, all with classification based on the genome and specific antigens [171].
In this century, three significant outbreaks have already changed the world scenario regarding the Coronaviridae family. At the end of 2002 and the beginning of 2003, in southern China, a new type of coronavirus was discovered. Named severe acute respiratory syndrome coronavirus (SARS-CoV), it promoted an outbreak with near a thousand deaths and more than eight thousand infected [172]. In 2012, a second outbreak was attributed to the middle east respiratory syndrome coronavirus (MERS-CoV) [173], with origin in the south of Asia and the middle east of Africa. Around 35% of the patients died, a higher mortality than SARS. At the end of 2019, the new pandemic spread worldwide from China was due to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes COVID-19 disease.
These viruses’ transmission to humans is believed to occur via animals as natural virus reservoirs, especially bats, civets, and camels [172]. The human contact with the animal environment, their secretions, and their meat may be the principal transmission mode. The reason for the high infection rate reported for SARS-CoV-2 is because the virus is easily transmitted via respiratory droplet, through aerial droplets and contact. SARS-CoV-2 has a higher propagation rate when compared to other coronaviruses. It is believed that it has a high viral load right in the beginning of the infection, which can lead to an inter-human transmission ever since. In the other hand, SARS-CoV has different moments of high viral load, higher at the end of the disease, which justify its lower transmission in the beginning of the infection [174]. It is considered that humans are transient or terminal host regarding inter-human infection by the MERS virus, and there are, until now, no supporting evidence that the transmission can occur [175].
The coronaviruses are developed in the host cellular cytoplasm, promoting cell destruction [171]. The diseases can be asymptomatic, specially MERS. Some unspecific symptoms are fever, cough, loss of air, and, in severe cases, severe respiratory problems such as respiratory insufficiency and associated comorbidities [173,176].
Despite the lethality of SARS-CoV, SARS-CoV-2, and MERS, it was not found in the literature many publications regarding coronavirus biosensor development compared to the growing number of studies for the Influenza virus. A partial explanation may come from the fact that Influenza virus has many harmful subtypes, and the morbidity and mortality are reported annually worldwide, which demands the fast diagnosis with reliability found in biosensors. The more significant events related to coronaviruses were the SARS-CoV and MERS outbreaks, which were controlled. Therefore, fewer efforts are found regarding these two specifics strains. However, the emergence of the new SARS-CoV-2 enhanced the concern, and the research for coronavirus became vital. Within months it was published two papers reporting biosensor development for detection of SARS-CoV-2, so far. These are the first steps for a real device that can be used to identify infected people and hasten the treatment. Table 3 contains data of these two papers alongside other reports regarding SARS, and MERS since the first outbreak, in 2002.
Table 3.
Coronavirus biosensors data.
| Detection technique | Substrate | Immobilized material / Analyte | Monitored compound | Working Range | LOD | Sample | Ref. |
|---|---|---|---|---|---|---|---|
| Optical – LSPR | PMMA optical fiber | Anti-SARS-CoV N protein / SARS-CoV N protein (sandwich ELISA-like assay) | Fluorophore DyLight™ 649-modified secondary antibody | 0.100 pg.mL−1 – 1.00 ng.mL−1 | 1.00 pg.mL−1 | Human serum from healthy donor | [177] |
| Optical – Confocal laser scanning microscopy | Glass chip | SARS-CoV N protein/ QDs-conjugated RNA aptamer | Fluorescence intensy of the QDs | 0.1 – 50 pg.mL−1 | 0.100 pg.mL−1 | Synthetic RNA aptamer | [178] |
| Optical – SPR | Gold-micropatterned chip | GBP-E-SCVme (SARS-CoV) fusion proteins / anti-SCVme | Label-free detection | Not determined | 0.200 µg.mL−1 | Rabbit anti-SCVme | [179] |
| Colorimetric assay | Multiplexed Paper | MERS-CoV DNA/ acpcPNA-AgNPs | Color change of AgNPs aggregation | 20.0 – 1.00 × 103 nM | 1.53 nM | Synthetic DNA oligonucleotides | [180] |
| Electrochemical – SWV | Array of carbon electrode | MERS-CoV and HumanCoV proteins/ Antibody for each virus | Reduction peak current of ferro/ferricyanide redox couple | 0.010 –1.00 × 104 ng.mL−1 | 0.400 pg.mL−1 | Spiked nasal samples (HumasCoV) | [181] |
| 0.001 – 100 ng.mL−1 | 1.00 pg.mL−1 | Spiked nasal samples (MERS-CoV) | |||||
| Optical – LSPR | Two-dimensional gold nanoisland | SARS-CoV 2 cDNA/ SARS-CoV 2 nucleic acid | Label-free detection | 0.100 pM – 1.00 μM | 0.220 pM | Synthetic oligonucleotide | [182] |
| Electrochemical – Semiconductor analyzer | Graphene FET | SARS-CoV 2 spike antibody/ SARS-CoV 2 spike protein | Label-free detection | 1.00 fg.mL−1 – 10.0 pg.mL−1 | 1.00 fg.mL−1 | PBS buffer | [183] |
| 16.0 – 1.60 × 104 PFU.mL−1 | 1.6 × 101 PFU.mL−1 | Cells culture | |||||
| 10.0 – 1.00 × 105 copies.mL−1 | 242 copies.mL−1 | Nasopharyngeal swab specimens from COVID-19 patients | |||||
| Piezoeletric | PQC sensor | SARS-CoV NG-8 aptamer/ SARS-CoV helicase protein | Magnetic bead enrichment | 0.050 – 1.00 µg.mL−1 | 3.50 ng.mL−1 | Human serum | [184] |
| Optical – SPR | Gold chip | SARS-CoV oligonucleotide probe / SARS-CoV PCR product | Label-free detection | 1.00 nM – 1 µM | 2.00 nM | Throat swab specimens | [185] |
acpcPNA = pyrrolidinyl peptide nucleic acid; FET = Field-effect transistor; GBP-E = Gold binding polypeptides-enhanced green fluorescent protein; QD = Quantum-dots; PMMA = Polymethyl methacrylate; PPT = Plasmonic photothermal; PQC = Piezoelectric quartz crystal.
It was observed for the biosensors developed for the Influenza virus that electrochemical systems were the majority among the papers found. For coronavirus detection, only six optical systems were found as the main sensor type with one work using a piezoelectric system for SARS-CoV-1 determination, one work using an electrochemical system for MERS determination and one electrochemical system for SARS-CoV-2 determination. The efficient and innovative devices developed for the Influenza virus and presented in this paper can be used as models to the determination of these viruses, since the biological material are essentially the same, such as antibodies, antigens, DNA, PCR products, and more.
Huang [177] developed a fiber-optic biosensor to detect the nucleocapsid (N) protein, a specific SARS-CoV antigen as a faster and more sensitive alternative to the RT-qPCR and serological tests. The same limit of detection (0.1 pg.mL−1) was also achieved two years after by Roh [178] using quantum dots-conjugated RNA aptamer immobilized over a designed chip to recognize the same N protein from SARS-CoV, and it is, to date, the lowest limit of detection found for a coronavirus through biosensor devices. Seeking an efficient biomolecule immobilization, Park [179] developed a surface plasmon-ressonance (SPR) biosensor for SARS-CoV based on the use of gold binding polypeptide (GBP). GBP was fused to enhanced green fluorescent protein (GBP-E) and to SARS-CoV membrane envelope (SCVme), the latter that can bind to anti-SCVme antibodies. This interesting system presents high specificity to gold substrates without losing the biomolecule activity. A representation of the work of Park can be seen in Fig. 4 .
Fig. 4.
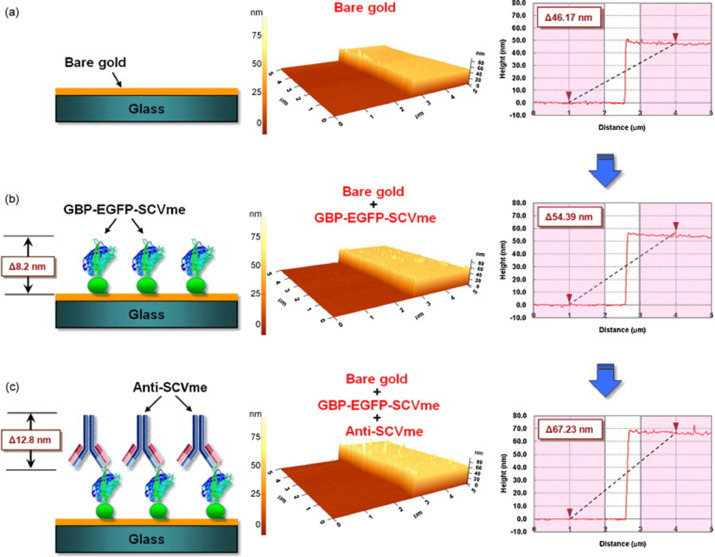
AFM images of the sequential binding of GBP-E-SCVme and anti-SCVme on the gold-micropatterned surface. (a) Bare gold surface, (b) binding of the GBP-E-SCVme fusion proteins onto the gold surface, and (c) subsequent binding of the anti-SCVme antibodies on the GBP-E-SCVme layer. Left, schematic diagrams for the successive binding of GBP-E-SCVme and anti-SCVme on the gold micropatterns; middle, three-dimensional topological images; right, the cross-sectional contours of samples a–c, sequentially (these are average height differences of the individual scan lines from each area). Republished from [179]; permission conveyed through Copyright Clearance Center, Inc, License Number 4855350410966.
All MERS biosensors found were developed as nanoparticle-based devices. An explanation may be that by the time of MERS outbreak, the use of nanoparticles and quantum dots for the most diverse systems were sedimented in the scientific community with easiness in the fabrication, stability, and many nanostructure options. Teengam [180] presented a biosensor for MERS detection as an innovative optical system. It was used a pyrrolidinyl peptide nucleic acid (PNA) immobilized over a paper-based analytical device (PAD) to detect synthetic oligonucleotides with a sequence corresponding to MERS. The paper-type colorimetric biosensor response occurs through aggregation/de-aggregation of negatively charged silver nanoparticles upon addition with positively charged PNA followed by complementary DNA. The results analysis does not require a computer interface, as the responses can be observed in the naked eye. Layqah has recently [181] developed the first electrochemical immunosensor for MERS detection. The biosensor consists of an array of eight gold nanoparticles modified-carbon electrodes containing MERS antigen immobilized through glutaraldehyde cross-linking immobilization technique. The measurement of the specific antibody was performed through square wave voltammetry in a solution containing ferrocyanide/ferricyanide, and a limit of detection of 0.4 pg.mL−1 was achieved.
As far as we know, Qiu and coworkers [182] presented the development of the first SARS-CoV-2 biosensor. Using the knowledge acquired from RT-PCR from SARS-CoV and SARS-CoV-2, they selected oligonucleotides containing specific sequences for both the diseases and their thiol-complementary DNA receptor. The hybridization event was analyzed through a dual-functional plasmonic system, integrating a plasmonic photothermal (PPT) effect and localized surface plasmon resonance (LSPR) transduction on a single chip. The innovative dual sensor achieved a low limit of detection of 0.22 pM and the use of a label-free system, with perspective to point-of-care use. With just a few days after the work of Qiu, Seo [183] presented a label-free field-effect transistor-based biosensor using a specific spike antibody for the detection of SARS-CoV-2 antigen protein. The sensor was evaluated in the universal transport medium, used to suspend nasopharyngeal swabs and in clinical samples, and the results were satisfactory. Despite the fact that one system is a genosensor, in which the virus can be detected right after the infection, both systems present some drawbacks as complexity and noise signals. Work still needs to be done to develop low-cost and miniaturized devices that can be accessible to the population in their own homes, avoiding agglomeration in health clinics and hospitals.
The biosensors developed for coronavirus presented excellent sensitivity, selectivity, and innovative assembly by taking advantage of the nanotechnology. The recent COVID-19 pandemic already boosted the development of the first biologic sensors, which can imply in revisiting the study of older coronaviruses as SARS and MERS. It is expected more research on these viruses, as simultaneous determination, miniaturization, and development of point-of-care devices for the population.
4.3. Respiratory syncytial virus
The respiratory syncytial virus (RSV) was firstly isolated from chimpanzees in 1956 [186]. It presents an (-)ssRNA genome [187] and belongs to the Paramyxoviridae family, which is composed of other viruses of public health interest, as the measles virus and the Newcastle disease virus [188]. It is one of the major causes of respiratory diseases around the world with a lethality compared to the Influenza virus, mainly in children under five years old [189]. In this group of children RSV can lead to bronchiolitis and pneumonia, the former presenting in almost 50% of the cases [186,190].
There is currently no vaccine against RSV [187]. An attempt was performed in 1967 to develop an inactive vaccine, but it was not effective [191]. This current scenario was responsible for more than 59,000 deaths of children under 5 years old in 2015 [192] with estimative of deaths between 66,000 and 234,000 children, teenagers, and elders [191]. Just in Brazil, RSV was responsible for 2366 new cases from 2007 to 2012 with more deaths even than the deaths correlated with the Influenza virus [189].
It is interesting to evaluate the discrepancies in the biosensor developments in the past few years. There are currently only vaccines for some Influenza virus subtypes, but none for the coronaviruses presented here and RSV. Therefore, the previous determination of these viruses in the contaminated people is of utmost importance for healthcare and to prevent new epidemics or even worse as SARS-CoV-2 pandemic. However, even with all of these concerns, few studies were reported with the development of biosensors for RSV detection. Table 4 contains the data of the few papers found in the literature. Only three optical systems and two electrochemical systems were found. Nevertheless, regarding the other viruses approaches, RSV biosensors present more heterogenicity than the others.
Table 4.
Respiratory syncytial virus biosensors data.
| Detection technique | Substrate | Immobilized material / Analyte | Monitored compound | Working Range | LOD | Sample | Ref. |
|---|---|---|---|---|---|---|---|
| Optical – SPR | Gold chip | RSV oligonucleotide probe / RSV PCR product | Label-free detection | 1.00 nM – 1.00 µM | 3.00 nM | Throat swab specimens | [185] |
| Optical – Microarray scanner | Gold-plated tungsten filaments | RSV DNA hairpin structures (molecular beacon style)/ RNA from cell culture | Label-free detection | Not determined | 11.9 PFU | RNA target extrated from cell culture | [193] |
| Electrochemical – Amperometric through SPE | Polystyrene slide | Anti-RSV/ RSV antigen (sandwich ELISA-like assay) | TMB | Not determined | 1:5000 (RSV Ag dilution) | Respiratory secretion clinical samples | [194] |
| Electrochemical – Amperometric | Gold chip | aMB with SA-HRP/ RSV DNA | TMB | 100 pM – 100 nM | 11.0 pM | RSV DNA in 10% human serum | [195] |
| aMB with SA-HRP/ RSV miRNA let-7a | 20.0 pM – 100 nM | 3.40 pM | |||||
| Optical – Absorbance | Plate/Gold nanoparticle layer | Anti-RSV / RSV antigen | TMB | 0.050 – 30.0 pg.mL−1 | 0.010 pg.mL−1 | Human serum from healthy donor | [197] |
aMB = Allosteric molecular beacons; SA-HRP = Streptavidin aptamer-horseradish peroxidase; TMB = 3,3,5,5-Tetramethylbenzidine;.
With the purpose to detect viral particles, Peres [193] developed an optic genosensor based in gold filament substrate. With a detection limit of 11.9 PFU, this is one of the most sensitive biosensors for RSV detection. Rochelet [194] developed an immunosensor based on a polymer-modified screen-printed electrode. With a low limit of detection of 1:5000 RSV Ag dilution, the device showed efficiency compared to a standard serological assay. Moreover, the device also presented low cost, and fast response time (25 min), features not related to the ELISA or the RT-PCR assays.
Cai [195] developed a genosensor based on an interesting allosteric molecular beacon (aMB), as depicted in the Fig. 5 . The aMB used in their work, upon contact with an RSV specific DNA target, has its stem opened and formed a streptavidin aptamer that was coupled to a streptavidin-horseradish peroxidase enzyme (SA-HRP). The electrochemical reduction signal of 3,3′,5,5′-tetramethylbenzidine (TMB) formed by the enzymatic reaction in the hydrogen peroxide presence was correlated to RSV concentration. With a limit of detection of 11.0 pM using a small sample volume (4.0 μL), this biosensor presented the lowest LOD among the studies found for RSV detection. The aMB strategy provides excellent sensitivity, which was also observed for the Influenza virus [196], and it is a new alternative for genosensor development.
Fig. 5.
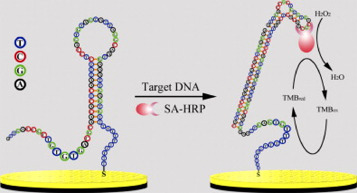
The principle of the RSV-aMB E-sensor for RSV DNA detection. Republished from [195]; permission conveyed through Copyright Clearance Center, Inc, License Number 4855350615984.
Shi [185] developed a genosensor based on gene gold chips through surface plasmon resonance that was able to detect simultaneously nine respiratory viruses: influenza virus A and B, H1N1, RSV, parainfluenza virus 1–3 (PIV1, 2, 3), adenovirus, and SARS-CoV-1. The biosensor presents high sensitivity and selectivity. Moreover, it represents an important step for further studies in the simultaneous determination of several viruses, since the associated illnesses cause similar symptoms. The possibility to discard and to confirm diseases with reliability is the ultimate goal in the frontline in the fight against respiratory viruses.
4.4. Other respiratory viruses
Despite the high incidence rates and mortality related to the viruses previously described in this paper, other respiratory viruses present lower virulence, but they are responsible for complications in humans and overload in health systems, like human adenovirus (AdV), human bocavirus (HBoV) and human Rhinovirus (HRV) [198], [199], [200].
Probably because of their low virulence, there are just a few biosensors reporting their determination. Regardless, the development of such device is of the most importance because the symptoms that these viruses cause are the same of the other viruses reported so far. The use of these specific biosensors by the population may be the first step to rule out other deadly viruses and, thus, to decrease the swelling in health services.
From the few papers found, it is worth mention the work of Ostroff [201], the only biosensor developed for human rhinovirus (HRV) determination, using an optically coated silicon surface modified with HRV polyclonal antibodies. The system was able to detect the virus-specific non-structural protein 3C protease. The biosensor presented high sensitivity (picomolar range) and fast response 28 min.
One significant advantage of biosensors is the possibility of simultaneous determinations of several analytes. Alongside the many requirements for a biosensor, the development of a device that could analyze and discriminate several respiratory viruses in one single analysis is a remarkable achievement. The works of Jin [202], Shi [185], and Jenison [203] were performed regarding human adenovirus. The work of Shi was already discussed for RSV because this device was able to determine nine different respiratory viruses, including human adenovirus. Jin developed an electrochemical biosensor based on gold chips modified with carbon nanotubes to determine the virus through surface plasmon resonance. The work of Jenison went through the same way, with a ten-minute assay for the detection of seven respiratory viruses, including rhinovirus, as depicted in Fig. 6 .
Fig. 6.
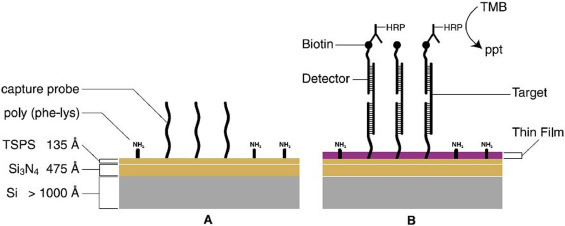
Schematic representation of the thin film biosensor. (A) Unreacted thin film biosensor surface with covalently attached capture probe. The surface coating, silicon nitride (Si3N4), appears gold in white light. (B) Surface reacting with target sequence to produce thin film. Target immobilization triggers reactions that enzymatically transduce the formation of hybrids on the surface into molecular thin films causing a change in color from gold to purple. Republished from [203]; permission conveyed through Copyright Clearance Center, Inc, License Number 4876701161684.
A great number of papers are addressed to the development of biosensors, but still, few devices are available in commerce as the glucose biosensor or using commercial detection systems. One good example of respiratory virus detection is the work of Owens [204], in which it was developed a label-free optical biosensor for the determination of HRV using the Corning EpicⓇ system, as depicted in Fig. 7 . This work, alongside other studies presented here, shows an excellent perspective for commercial devices.
Fig. 7.
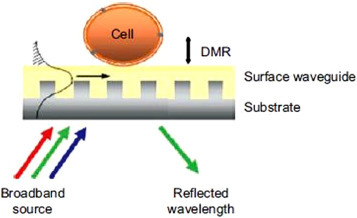
A schematic view of the EpicⓇ biosensor. When the glass substrate is illuminated with broadband light, only a ‘single’ wavelength that is resonant with the waveguide grating structure is strongly reflected. The EpicⓇ system measures the wavelength reflected by the sensor which is determined by the optical properties of the sensing zone within approximately 150 nm of the sensor. The magnitude of this wavelength shift is proportional to the amount of DMR. Republished from [204]; permission conveyed through Copyright Clearance Center, Inc, License Number 4876710020488.
There are other important respiratory viruses, but it was not found any biosensors reports, such as the human bocavirus, which proves that much work can still be performed to achieve early diagnosis and availability of commercial devices for these viruses determination [198].
Conclusion
The diseases caused by respiratory viruses are a matter of public health, with more than a million new cases and hundreds of deaths reported annually. Moreover, viruses can evolve with time, and humans are continually susceptible to new deadly forms, as it is the case of the COVID-19 pandemic that is plaguing the world. The current scenario relies upon the use of specific diagnosis, but with time-consuming analysis, high cost of the whole procedure, with the possibility of false-positives and false-negatives results, and it is not accessible to the entire population. The concept of point-of-care devices has been growing considered attention because of its advantages, such as the high sensitivity, selectivity, reproducibility, low-cost, small sample requirement, allied to a miniaturized device that can be used with simple handling and easy operation.
Biosensors satisfy these requirements, and they have been developed for many purposes, including in the health area. This review exploited the most recent development in biosensors regarding the respiratory viruses, with a stress on the Influenza virus, the coronaviruses, and the respiratory syncytial virus. The majority of papers found in the literature approached the Influenza virus. This number of studies considers the existence of many subtypes (H1N1, H5N1, H7N9, H3N2, and others), the annual recurrence, and the high number of deaths. SARS-CoV and MERS present valuable, but fewer studies when compared to the Influenza virus. Despite their lethality, these viruses were controlled after their respective outbreaks, which seems not to justify many efforts in biosensor development. Unfortunately, biosensors for RSV determination are the scarcest. With annual reports of new cases and deaths, just like Influenza, but especially of young children, it was expected more efforts regarding its determination through the point of care devices.
Researches have been using different approaches and innovative systems to develop their sensors. With the advent of nanotechnology, high sensitivity is being reached, with the detections in the attomolar range, better than the traditional methods. Optical and electrochemical are the central choices for transducing, as the specific nucleic acids, and antigen/antibodies are the usual choices for biological material to be immobilized. Previous serological assays and the genome study of the viruses are important because allows researchers to use this information and the virus's biological material to the development of immunosensor and genosensors.
The use of commercial electrodes such as screen-printed, and systems that can detect more than one respiratory virus in a simultaneous determination, provide some perspective. There is not available any device, as the glucose biosensor, regarding the determination of the viruses reported in this review. Nevertheless, with just months since the beginning of the COVID-19 pandemic, at least two biosensors were developed for SARS-CoV-2 determination, which can be the first step for further studies to evolve from the proof of concept to the point of care device.
CRediT authorship contribution statement
Brayan Viana Ribeiro: Conceptualization, Investigation, Writing - original draft. Taís Aparecida Reis Cordeiro: Writing - original draft. Guilherme Ramos Oliveira e Freitas: Methodology, Writing - original draft. Lucas Franco Ferreira: Methodology, Writing - review & editing. Diego Leoni Franco: Conceptualization, Methodology, Investigation, Writing - original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are grateful for the technical support from the Federal University of Uberlândia and Federal University of Vales do Jequitinhonha e Mucuri. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors
References
- 1.Centers for Disease Control and Prevention, Initial page, CDC. (2020). https://www.cdc.gov/(accessed March 28, 2020).
- 2.World Health Organization (WHO), Disease Outbreaks by Year, WHO. (2020). https://www.who.int/csr/don/archive/year/en/(accessed April 1, 2020).
- 3.World Health Organization (WHO) WHO; 2020. WHO Director-General's Opening Remarks at the Media Briefing on COVID-19 - 16 March 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19—16-march-2020 (accessed March 28, 2020) [Google Scholar]
- 4.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., Van Der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus ( 2019-nCoV ) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiang J., Yan M., Li H., Liu T., Lin C., Huang S., Shen C. Evaluation of enzyme-linked immunoassay and colloidal gold- immunochromatographic assay kit for detection of novel coronavirus ( SARS-Cov-2 ) causing an outbreak of pneumonia ( COVID-19 ) MedRxiv. 2020:1–13. doi: 10.1101/2020.02.27.20028787. [DOI] [Google Scholar]
- 6.Goode J.A., Rushworth J.V.H., Millner P.A. Biosensor regeneration: a review of common techniques and outcomes. Langmuir. 2015;31:6267–6276. doi: 10.1021/la503533g. [DOI] [PubMed] [Google Scholar]
- 7.Yang F., Ma Y., Stanciu S.G., Wu A. In: Nanobiosensors From Des. to Appl. Wu A., Khan W.S., editors. Wiley‐VCH; 2020. Transduction process‐based classification of biosensors. [DOI] [Google Scholar]
- 8.Saeedfar K., yook Heng L., Rezayi M. Fabricating long shelf life potentiometric urea biosensors using modified MWCNTs on screen printed electrodes. Sens. Lett. 2017;15:97–103. doi: 10.1166/sl.2017.3780. [DOI] [Google Scholar]
- 9.Ding J., Qin W. Recent advances in potentiometric biosensors. TrAC Trends Anal. Chem. 2020;124 doi: 10.1016/j.trac.2019.115803. [DOI] [Google Scholar]
- 10.German N., Ramanavicius A., Ramanaviciene A. Amperometric glucose biosensor based on electrochemically deposited gold nanoparticles covered by polypyrrole. Electroanalysis. 2017;29:1267–1277. doi: 10.1002/elan.201600680. [DOI] [Google Scholar]
- 11.Labban N., Hughes L., Wayu M., Pollock J., Leopold M. MON-182 adaptable amperometric biosensor platforms for the diagnosis of endocrine disorders. J. Endocr. Soc. 2019;3 doi: 10.1210/js.2019-MON-182. [DOI] [Google Scholar]
- 12.Lima A.B., Ferreira L.F., Barbosa S.L., de S. Gil E., da Silva R.A.B., dos Santos W.T.P. Selective determination of verapamil in pharmaceutics and urine using a boron‐doped diamond electrode coupled to flow injection analysis with multiple‐pulse amperometric detection. Electroanalysis. 2018;30:1880–1885. doi: 10.1002/elan.201800206. [DOI] [Google Scholar]
- 13.Aydın E.B. Highly sensitive impedimetric immunosensor for determination of interleukin 6 as a cancer biomarker by using conjugated polymer containing epoxy side groups modified disposable ITO electrode. Talanta. 2020;215 doi: 10.1016/j.talanta.2020.120909. [DOI] [PubMed] [Google Scholar]
- 14.Chakraborty A., Tibarewala D.N., Barui A. Bioelectron. Med. Devices. Woodhead Publishing; 2019. Impedance-based biosensors; pp. 97–122. [DOI] [Google Scholar]
- 15.Cordeiro T.A.R., Gonçalves M.V.C., Franco D.L., Reis A.B., Martins H.R., Ferreira L.F. Label-free electrochemical impedance immunosensor based on modified screen-printed gold electrodes for the diagnosis of canine visceral leishmaniasis. Talanta. 2019;195:327–332. doi: 10.1016/j.talanta.2018.11.087. [DOI] [PubMed] [Google Scholar]
- 16.Soni A., Surana R.K., Jha S.K. Smartphone based optical biosensor for the detection of urea in saliva. Sens. Actuat. B Chem. 2018;269:346–353. doi: 10.1016/j.snb.2018.04.108. [DOI] [Google Scholar]
- 17.Masson J.-.F. Surface plasmon resonance clinical biosensors for medical diagnostics. ACS Sens. 2017;2:16–30. doi: 10.1021/acssensors.6b00763. [DOI] [PubMed] [Google Scholar]
- 18.Garzón V., Pinacho D.G., Bustos R.-.H., Garzón G., Bustamante S. Optical biosensors for therapeutic drug monitoring. Biosensors. 2019;9:132. doi: 10.3390/bios9040132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soler M., Lechuga L.M. Boosting cancer immunotherapies with optical biosensor nanotechnologies. Eur. Med. J. 2019;4:124–132. [Google Scholar]
- 20.Chorsi M.T., Curry E.J., Chorsi H.T., Das R., Baroody J., Purohit P.K., Ilies H., Nguyen T.D. Piezoelectric biomaterials for sensors and actuators. Adv. Mater. 2019;31 doi: 10.1002/adma.201802084. [DOI] [PubMed] [Google Scholar]
- 21.Jeffree A.I., Karman S., Ibrahim S., Karim M.S.A., Rozali S. In: RITA 2018. Majeed A.P.P.A., Mat-Jizat J.A., Hassan M.H.A., Taha Z., Choi H.L., Kim J., editors. Springer; Singapore: 2019. Biosensors approach for lung cancer diagnosis—a review; pp. 425–435. [DOI] [Google Scholar]
- 22.Pohanka M. Piezoelectric biosensor for the determination of tumor necrosis factor alpha. Talanta. 2018;178:970–973. doi: 10.1016/j.talanta.2017.10.031. [DOI] [PubMed] [Google Scholar]
- 23.Ricci F., Adornetto G., Palleschi G. A review of experimental aspects of electrochemical immunosensors. Electrochim. Acta. 2012;84:74–83. doi: 10.1016/j.electacta.2012.06.033. [DOI] [Google Scholar]
- 24.Saylan Y., Yilmaz F., Özgür E., Derazshamshir A., Yavuz H., Denizli A. Molecular imprinting of macromolecules for sensor applications. Sensors. 2017;17:898. doi: 10.3390/s17040898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Justino C.I.L., Duarte A.C., Rocha-Santos T.A.P. In: Adv. Clin. Chem. Makowski G.S., editor. Elsevier; 2016. Immunosensors in clinical laboratory diagnostics; pp. 65–108. [DOI] [PubMed] [Google Scholar]
- 26.Huang C.-.J. In: Chem. Gas, Biosens. Internet Things Relat. Appl. Mitsubayashi K., Niwa O., Ueno Y., editors. Elsevier; 2019. Advanced surface modification technologies for biosensors; pp. 65–86. [DOI] [Google Scholar]
- 27.Lan W.-.C., Huang T.-.S., Cho Y.-.C., Huang Y.-.T., Walinski C.J., Chiang P.-.C., Rusilin M., Pai F.-.T., Huang C.-.C., Huang M.-.S. The potential of a nanostructured titanium oxide layer with self-assembled monolayers for biomedical applications: surface properties and biomechanical behaviors. Appl. Sci. 2020;10:590. doi: 10.3390/app10020590. [DOI] [Google Scholar]
- 28.Acosta S., Quintanilla L., Alonso M., Aparicio C., Rodríguez-Cabello J.C. Recombinant AMP/polypeptide self-assembled monolayers with synergistic antimicrobial properties for bacterial strains of medical relevance. ACS Biomater. Sci. Eng. 2019;5:4708–4716. doi: 10.1021/acsbiomaterials.9b00247. [DOI] [PubMed] [Google Scholar]
- 29.Lv H., Ye L., Liu Q., Li S., Li T., Huang N., Gao Y., Fan L., Du W. S‐S‐PEG‐COOH self‐assembled monolayer on gold surface enabled a combined assay for serological EBV antibody isotypes. Proteomics Clin. Appl. 2018;13 doi: 10.1002/prca.201800067. [DOI] [PubMed] [Google Scholar]
- 30.Baryeh K., Takalkar S., Lund M., Liu G. In: Med. Biosens. Point Care Appl. Narayan R.J., editor. Woodhead Publishing; 2017. Introduction to medical biosensors for point of care applications; pp. 3–25. [DOI] [Google Scholar]
- 31.Brazaca L.C., Ribovski L., Janegitz B.C., Zucolotto V. In: Med. Biosens. Point Care Appl. Narayan R.J., editor. Woodhead Publishing; 2017. Nanostructured materials and nanoparticles for point of care (POC) medical biosensors; pp. 229–254. [DOI] [Google Scholar]
- 32.Sabino-Silva R., Jardim A.C.G., Siqueira W.L. Coronavirus COVID-19 impacts to dentistry and potential salivary diagnosis. Clin. Oral Investig. 2020;24:1619–1621. doi: 10.1007/s00784-020-03248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen T., Bang D.D., Wolff A. 2019 Novel Coronavirus disease (COVID-19): paving the road for rapid detection and point-of-care diagnostics. Micromachines (Basel) 2020;11:306. doi: 10.3390/mi11030306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbas A.K., Lichtman A.H., Pillai S. 8th Editio. Elsevier; 2015. Cellular and Molecular Immunology. [Google Scholar]
- 35.Bahadır E.B., Sezgintürk M.K. Applications of electrochemical immunosensors for early clinical diagnostics. Talanta. 2015;132:162–174. doi: 10.1016/j.talanta.2014.08.063. [DOI] [PubMed] [Google Scholar]
- 36.Li F., Li Y., Feng J., Dong Y., Wang P., Chen L., Chen Z., Liu H., Wei Q. Ultrasensitive amperometric immunosensor for PSA detection based on Cu2O@CeO2-Au nanocomposites as integrated triple signal amplification strategy. Biosens. Bioelectron. 2017;87:630–637. doi: 10.1016/j.bios.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 37.Tuteja S.K., Chen R., Kukkar M., Song C.K., Mutreja R., Singh S., Paul A.K., Lee H., Kim K.-.H., Deep A., Suri C.R. A label-free electrochemical immunosensor for the detection of cardiac marker using graphene quantum dots (GQDs) Biosens. Bioelectron. 2016;86:548–556. doi: 10.1016/j.bios.2016.07.052. [DOI] [PubMed] [Google Scholar]
- 38.Castiello F.R., Tabrizian M. Gold nanoparticle amplification strategies for multiplex SPRi-based immunosensing of human pancreatic islet hormones. Analyst. 2019;144:2541–2549. doi: 10.1039/C9AN00140A. [DOI] [PubMed] [Google Scholar]
- 39.Kelch J., Delaney A., Kelleher F., Baker P., Iwuoha E., Dempsey E. Impedimetric and electrochemical evaluation of a new redox active steroid derivative for hormone immunosensing. Biosens. Bioelectron. 2020;150 doi: 10.1016/j.bios.2019.111876. [DOI] [PubMed] [Google Scholar]
- 40.Li N., Chow A.M., Ganesh H.V.S., Ratnam M., Brown I.R., Kerman K. Diazonium-modified screen-printed electrodes for immunosensing growth hormone in blood samples. Biosensors. 2019;9:88. doi: 10.3390/bios9030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jampasa S., Lae-ngee P., Patarakul K., Ngamrojanavanich N., Chailapakul O., Rodthongkum N. Electrochemical immunosensor based on gold-labeled monoclonal anti-LipL32 for leptospirosis diagnosis. Biosens. Bioelectron. 2019;142 doi: 10.1016/j.bios.2019.111539. [DOI] [PubMed] [Google Scholar]
- 42.Juronen D., Kuusk A., Kivirand K., Rinken A., Rinken T. Immunosensing system for rapid multiplex detection of mastitis-causing pathogens in milk. Talanta. 2018;178:949–954. doi: 10.1016/j.talanta.2017.10.043. [DOI] [PubMed] [Google Scholar]
- 43.Pandey C.M., Tiwari I., Singh V.N., Sood K.N., Sumana G., Malhotra B.D. Highly sensitive electrochemical immunosensor based on graphene-wrapped copper oxide-cysteine hierarchical structure for detection of pathogenic bacteria. Sens. Actuat. B Chem. 2017;238:1060–1069. doi: 10.1016/j.snb.2016.07.121. [DOI] [Google Scholar]
- 44.Verdoodt N., Basso C.R., Rossi B.F., Pedrosa V.A. Development of a rapid and sensitive immunosensor for the detection of bacteria. Food Chem. 2017;221:1792–1796. doi: 10.1016/j.foodchem.2016.10.102. [DOI] [PubMed] [Google Scholar]
- 45.Rodrigues L.P., Ferreira D.C., Ferreira L.F., de S.G.C., Cuadros-Orellana O., Brito-Madurro A.G., de Oliveira R.J., Jr. O.A., Madurro J.M. Electropolymerization of hydroxyphenylacetic acid isomers and the development of a bioelectrode for the diagnosis of bacterial meningitis. J. Appl. Electrochem. 2015;45:1277–1285. doi: 10.1007/s10800-015-0892-2. [DOI] [Google Scholar]
- 46.Sun Y., Xu L., Zhang F., Song Z., Hu Y., Ji Y., Shen J., Li B., Lu H., Yang H. A promising magnetic SERS immunosensor for sensitive detection of avian influenza virus. Biosens. Bioelectron. 2017;89(Part 2):906–912. doi: 10.1016/j.bios.2016.09.100. [DOI] [PubMed] [Google Scholar]
- 47.Zhang H., Miller B.L. Immunosensor-based label-free and multiplex detection of influenza viruses: state of the art. Biosens. Bioelectron. 2019;141 doi: 10.1016/j.bios.2019.111476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.da C. Santos C., Santos P.C.M., Rocha K.L.S., Thomasini R.L., de Oliveira D.B., Franco D.L., Ferreira L.F. A new tool for dengue virus diagnosis: optimization and detection of anti-NS1 antibodies in serum samples by impedimetric transducers. Microchem. J. 2020;154 doi: 10.1016/j.microc.2019.104544. [DOI] [Google Scholar]
- 49.Kaushik A., Yndart A., Kumar S., Jayant R.D., Vashist A., Brown A.N., Li C.-.Z., Nair M. A sensitive electrochemical immunosensor for label-free detection of Zika-virus protein. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-28035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng H.-.P., Chuang H.-.S. Rapid and sensitive nano-immunosensors for botulinum. ACS Sens. 2019;4:1754–1760. doi: 10.1021/acssensors.9b00644. [DOI] [PubMed] [Google Scholar]
- 51.Ozoemena O.C., Maphumulo T., Shai J.L., Ozoemena K.I. Electrospun carbon nanofibers as an electrochemical immunosensing platform for vibrio cholerae toxin: aging effect of the redox probe. ACS Omega. 2020;5:5762–5771. doi: 10.1021/acsomega.9b03820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu L., Gunasekaran S. Dual-channel ITO-microfluidic electrochemical immunosensor for simultaneous detection of two mycotoxins. Talanta. 2019;194:709–716. doi: 10.1016/j.talanta.2018.10.091. [DOI] [PubMed] [Google Scholar]
- 53.Solanki P.R., Singh J., Rupavali B., Tiwari S., Malhotra B.D. Bismuth oxide nanorods based immunosensor for mycotoxin detection. Mater. Sci. Eng. C. 2017;70(Part 1):564–571. doi: 10.1016/j.msec.2016.09.027. [DOI] [PubMed] [Google Scholar]
- 54.Liu A., Lang Q., Liang B., Shi J. Sensitive detection of maltose and glucose based on dual enzyme-displayed bacteria electrochemical biosensor. Biosens. Bioelectron. 2017;87:25–30. doi: 10.1016/j.bios.2016.07.050. [DOI] [PubMed] [Google Scholar]
- 55.Tabrizi M.A., Ferré-Borrull J., Marsa L.F. An optical biosensor for the determination of cathepsin B as a cancer-associated enzyme using nanoporous anodic alumina modified with human serum albumin-thionine. Microchim. Acta. 2020;187:230. doi: 10.1007/s00604-020-4188-9. [DOI] [PubMed] [Google Scholar]
- 56.Zhao T., Fu Y., He H., Dong C., Zhang L., Zeng H., Xing L., Xue X. Self-powered gustation electronic skin for mimicking taste buds based on piezoelectric–enzymatic reaction coupling process. Nanotechnology. 2018;29 doi: 10.1088/1361-6528/aaa2b9. [DOI] [PubMed] [Google Scholar]
- 57.Bollella P., Gorton L. Enzyme based amperometric biosensors. Curr. Opin. Electrochem. 2018;10:157–173. doi: 10.1016/j.coelec.2018.06.003. [DOI] [Google Scholar]
- 58.Mehrotra P. Biosensors and their applications – A review. J. Oral Biol. Craniofacial Res. 2016;6:153–159. doi: 10.1016/j.jobcr.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoo Y.J., Feng Y., Kim Y.-.H., Yagonia C.F.J. 1st Edit. Springer; Dordrecht: 2017. Fundamentals of Enzyme Engineering. [DOI] [Google Scholar]
- 60.P. P., S. V. A numerical modelling of an amperometric-enzymatic based uric acid biosensor for GOUT arthritis diseases. Inf. Med. Unlocked. 2018;12:143–147. doi: 10.1016/j.imu.2018.03.001. [DOI] [Google Scholar]
- 61.Fraser L.A., Kinghorn A.B., Dirkzwager R.M., Liang S., Cheung Y.-.W., Lim B., Shiu S.C.-C., Tang M.S.L., Andrew D., Manitta J., Richards J.S., Tanner J.A. A portable microfluidic Aptamer-Tethered Enzyme Capture (APTEC) biosensor for malaria diagnosis. Biosens. Bioelectron. 2018;100:591–596. doi: 10.1016/j.bios.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 62.El Harrad L., Bourais I., Mohammadi H., Amine A. Recent advances in electrochemical biosensors based on enzyme inhibition for clinical and pharmaceutical applications. Sensors. 2018;18:164. doi: 10.3390/s18010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L., Tianjiao M., Zhao D., Jia H., An S., Yang X., Wang H., Zhang Y. An enzyme-free electrochemical biosensor based on well monodisperse Au nanorods for ultra-sensitive detection of telomerase activity. Biosens. Bioelectron. 2020;148 doi: 10.1016/j.bios.2019.111834. [DOI] [PubMed] [Google Scholar]
- 64.Rasheed P.A., Sandhyarani N. Electrochemical DNA sensors based on the use of gold nanoparticles: a review on recent developments. Microchim. Acta. 2017;184:981–1000. doi: 10.1007/s00604-017-2143-1. [DOI] [Google Scholar]
- 65.Rashid J.I.A., Yusof N.A. The strategies of DNA immobilization and hybridization detection mechanism in the construction of electrochemical DNA sensor: a review. Sens. Bio-Sensing Res. 2017;16:19–31. doi: 10.1016/j.sbsr.2017.09.001. [DOI] [Google Scholar]
- 66.Vasdev K. DNA biosensors-a review. J. Bioeng. Biomed. Sci. 2017;7 doi: 10.4172/2155-9538.1000222. [DOI] [Google Scholar]
- 67.Kaur G., Paliwal A., Tomar M., Gupta V. Detection of Neisseria meningitidis using surface plasmon resonance based DNA biosensor. Biosens. Bioelectron. 2016;78:106–110. doi: 10.1016/j.bios.2015.11.025. [DOI] [PubMed] [Google Scholar]
- 68.Saeed A.A., Sánchez J.L.A., O'Sullivan C.K., Abbas M.N. DNA biosensors based on gold nanoparticles-modified graphene oxide for the detection of breast cancer biomarkers for early diagnosis. Bioelectrochemistry. 2017;118:91–99. doi: 10.1016/j.bioelechem.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 69.Khan M., Khan A.R., Shin J.-.H., Park S.-.Y. A liquid-crystal-based DNA biosensor for pathogen detection. Sci. Rep. 2016;6:22676. doi: 10.1038/srep22676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cui M., Wang Y., Wang H., Wu Y., Luo X. A label-free electrochemical DNA biosensor for breast cancer marker BRCA1 based on self-assembled antifouling peptide monolayer. Sens. Actuat. B Chem. 2017;244:742–749. doi: 10.1016/j.snb.2017.01.060. [DOI] [Google Scholar]
- 71.Ahmed A., Rushworth J.V., Hirst N.A., Millner P.A. Biosensors for whole-cell bacterial detection. Clin. Microbiol. Rev. 2014;27:631–646. doi: 10.1128/CMR.00120-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu Y., Xie X., Duan Y., Wang L., Cheng Z., Cheng J. A review of impedance measurements of whole cells. Biosens. Bioelectron. 2016;77:824–836. doi: 10.1016/j.bios.2015.10.027. [DOI] [PubMed] [Google Scholar]
- 73.Roggo C., der Meer J.R. Miniaturized and integrated whole cell living bacterial sensors in field applicable autonomous devices. Curr. Opin. Biotechnol. 2017;45:24–33. doi: 10.1016/j.copbio.2016.11.023. [DOI] [PubMed] [Google Scholar]
- 74.Brutesco C., Prévéral S., Escoffier C., Descamps E.C.T., Prudent E., Cayron J., Dumas L., Ricquebourg M., Adryanczyk-Perrier G., de Groot A., Garcia D., Rodrigue A., Pignol D., Ginet N. Bacterial host and reporter gene optimization for genetically encoded whole cell biosensors. Environ. Sci. Pollut. Res. Int. 2017;24:52–65. doi: 10.1007/s11356-016-6952-2. [DOI] [PubMed] [Google Scholar]
- 75.Gui Q., Lawson T., Shan S., Yan L., Liu Y. The application of whole cell-based biosensors for use in environmental analysis and in medical diagnostics. Sensors. 2017;17:1623. doi: 10.3390/s17071623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Riangrungroj P., Bever C.S., Hammock B.D., Polizzi K.M. A label-free optical whole-cell Escherichia coli biosensor for the detection of pyrethroid insecticide exposure. Sci. Rep. 2019;9:12466. doi: 10.1038/s41598-019-48907-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kylilis N., Riangrungroj P., Lai H.-.E., Salema V., Fernández L.Á., Stan G.-B.V., Freemont P.S., Polizzi K.M. Whole-cell biosensor with tunable limit of detection enables low-cost agglutination assays for medical diagnostic applications. ACS Sens. 2019;4:370–378. doi: 10.1021/acssensors.8b01163. [DOI] [PubMed] [Google Scholar]
- 78.Inda M.E., Mimee M., Lu T.K. Cell-based biosensors for immunology, inflammation, and allergy. J. Allergy Clin. Immunol. 2019;144:645–647. doi: 10.1016/j.jaci.2019.07.024. [DOI] [PubMed] [Google Scholar]
- 79.Goers L., Ainsworth C., Goey C.H., Kontoravdi C., Freemont P.S., Polizzi K.M. Whole‐cell Escherichia coli lactate biosensor for monitoring mammalian cell cultures during biopharmaceutical production. Biotechnol. Bioeng. 2017;114:1290–1300. doi: 10.1002/bit.26254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wessner D.R. The origins of viruses. Scitable by Nat. Educ. 2010 https://www.nature.com/scitable/topicpage/the-origins-of-viruses-14398218/ (accessed April 1, 2020) [Google Scholar]
- 81.Tortora G.J., Funke B.R., Case C.L., Vírus . In: Microbiologia. 8th Edit. House Laser., editor. Artmed, Porto Alegre; 2006. Viróides e Prions; pp. 376–408. [Google Scholar]
- 82.Gooding G.V., Jr. In: Plant Viruses. 2nd Edit. Van Regenmortel M.H.V., Fraenkel-Conrat H., editors. Springer; Boston, MA: 1986. Tobacco mosaic virus epidemiology and control; pp. 133–152. [DOI] [Google Scholar]
- 83.Summers W.C. Félix Hubert d'Herelle (1873–1949): history of a scientific mind. Bacteriophage. 2016;6 doi: 10.1080/21597081.2016.1270090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kilbourne E.D. Pandora’ s box and the history of the respiratory viruses : a case study of serendipity in research. Hist. Philos. Life Sci. 1992;14:299–308. https://www.jstor.org/stable/23331665 [PubMed] [Google Scholar]
- 85.Saunders-hastings P.R., Krewski D. Reviewing the history of pandemic influenza: understanding patterns of emergence and transmission. Pathogens. 2016;5:66. doi: 10.3390/pathogens5040066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hershey A.D., Chase M. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 1952;36:39–56. doi: 10.1085/jgp.36.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Enders J.F., Weller T.H., Robbins F.C. Cultivation of the lansing strain of poliomyelitis virus in cultures of various human embryonic tissues. Science. 1949;109:85–87. doi: 10.1126/science.109.2822.85. [DOI] [PubMed] [Google Scholar]
- 88.C.J. Burrell, C.R. Howard, F.A. Murphy, History and impact of virology, in: N.J. MacLachlan, E.J. Dubovi (Eds.), Fenner White's Med. Virol., 5th Editio, Academic Press, 2017: pp. 3–14. doi:10.1016/B978-0-12-375156-0.00001-1.
- 89.Louten J. : Essent. Hum. Virol. 1st Edit. Academic Press; 2016. Virus replication pp. 49–70. [DOI] [Google Scholar]
- 90.Manjarrez-Zavala E., Rosete-Olvera D.P., Gutiérrez-González L.H., Ocadiz-Delgado R., Cabello-Gutiérrez C. In: Respir. Dis. Infect. - A New Insight. 1st Edit. Mahboub B.H., editor. IntechOpen; 2013. Pathogenesis of viral respiratory infection. [DOI] [Google Scholar]
- 91.Sato S., Kiyono H. The mucosal immune system of the respiratory tract. Curr. Opin. Virol. 2012;2:225–232. doi: 10.1016/j.coviro.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 92.Yu Z., Cheng K., Sun W., Zhang X., Xia X., Gao Y. PB2 and HA mutations increase the virulence of highly pathogenic H5N5 clade 2.3.4.4 avian influenza virus in mice, Arch. Virol. 2018;163:401–410. doi: 10.1007/s00705-017-3631-7. [DOI] [PubMed] [Google Scholar]
- 93.Hulswit R.J.G., de Haan C.A.M., Bosch B.-.J. In: Adv. Virus Res. Ziebuhr J., editor. Elsevier; The Netherlands: 2016. Coronavirus spike protein and tropism changes; pp. 29–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cann A.J. Princ. Mol. Virol. 5th Editio. Elsevier; 2012. Genome; pp. 55–101. [DOI] [Google Scholar]
- 95.Baltimore D. Expression of animal virus genomes. Bacteriol. Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.World Health Organization (WHO) Water Recreation and Disease: Plausibility of Associated Infections: Acute Effects, Sequelae and Mortality. World Health Organization; 2005. Viruses; pp. 192–229. [Google Scholar]
- 97.Balasuriya U.B.R., Barratt-Boyes S., Beer M., Bird B., Brownlie J., Coffey L.L., Cullen J.M., Delhon G.A., Donis R.O., Gardner I., Gilkerson J., Golde W.T., Hartley C., Heidner H., Herden C., Kirkland P. In: Fenner and White's Medical Virology. 5th Edit. MacLachlan N.J., Dubovi E.J., editors. Academic Press; 2017. Adenoviridae; pp. 217–227. [DOI] [Google Scholar]
- 98.World Health Organization (WHO), Influenza (Seasonal), WHO. (2018). https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (accessed January 4, 2020).
- 99.PATH, The global respiratory syncytial virus burden, (2017) 1–2. https://path.azureedge.net/media/documents/CVIA_RSV_disease_burden_fs.pdf (accessed January 4, 2020).
- 100.World Health Organization (WHO), Middle East respiratory syndrome coronavirus (MERS-CoV), WHO. (2019). https://www.who.int/emergencies/mers-cov/en/ (accessed January 4, 2020).
- 101.World Health Organization (WHO), Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003, WHO. (2003). https://www.who.int/csr/sars/country/table2004_04_21/en/ (accessed January 4, 2020).
- 102.Rozwadowski F., Caulcrick-Grimes M., Mchugh L., Haldeman A., Fulton T., Killerby M., Schneider E., Lu X., Sakthivel S.K., Bhatnagar J., Rabeneck D.B., Zaki S., Watson J. Fatalities associated with human adenovirus type 7 at a substance abuse rehabilitation facility — New Jersey, 2017. Morb. Mortal. Wkly. Rep. 2018;67:371–372. doi: 10.15585/mmwr.mm6712a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schildgen V., Van Den Hoogen B., Fouchier R., Tripp R.A., Alvarez R., Manoha C., Williams J., Schildgen O. Human metapneumovirus: lessons learned over the first decade. Clin. Microbiol. Rev. 2011;24:734–754. doi: 10.1128/CMR.00015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hung I.F.N., Zhang A.J., To K.K.W., Chan J.F.W., Zhu S.H.S., Zhang R., Chan T., Chan K., Yuen K. Unexpectedly higher morbidity and mortality of hospitalized elderly patients associated with rhinovirus compared with influenza virus respiratory tract infection. Int. J. Mol. Sci. 2017;18:259. doi: 10.3390/ijms18020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Das S., Dunbar S., Tang Y. Laboratory diagnosis of respiratory tract infections in children – the state of the art. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Júnior R.B.M., Carney S., Goldemberg D., Bonine L., Spano L.C., Siqueira M., Checon R.E. Detection of respiratory viruses by real-time polymerase chain reaction in outpatients with acute respiratory infection. Mem. Inst. Oswaldo Cruz. 2014;109:716–721. doi: 10.1590/0074-0276140046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mahony J.B. Detection of respiratory viruses by molecular methods. Clin. Microbiol. Rev. 2008;21:716–747. doi: 10.1128/CMR.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Olofsson S., Brittain-long R., Andersson L.M., Westin J., Lindh M. PCR for detection of respiratory viruses: seasonal variations of virus infections. Expert Rev. Anti. Infect. Ther. 2011;9:615–626. doi: 10.1586/eri.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lin C., Hwang D., Chiu N., Weng L., Liu H., Mu J.-.J., Liu C.-.P., Chi H. Increased detection of viruses in children with respiratory tract infection using PCR. Int. J. Environ. Res. Public Health. 2020;17:564. doi: 10.3390/ijerph17020564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brennan-Krohn T. Making sense of respiratory viral panel results. Am. Soc. Microbiol. 2020 https://asm.org/Articles/2020/March/Making-Sense-of-Respiratory-Viral-Panel-Results (accessed April 4, 2020) [Google Scholar]
- 111.Huang H.-.S., Tsai C.-.L., Chang J., Hsu T.-.C., Lin S., Lee C.-.C. Multiplex PCR system for the rapid diagnosis of respiratory virus infection : systematic review and meta-analysis. Clin. Microbiol. Infect. 2018;24:1055–1063. doi: 10.1016/j.cmi.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Malhotra B.D., Chaubey A. Biosensors for clinical diagnostics industry. Sens. Actuat. B Chem. Chem. 2003;91:117–127. doi: 10.1016/S0925-4005(03)00075-3. [DOI] [Google Scholar]
- 113.Janegitz B.C., Cancino J., Zucolotto V. Disposable biosensors for clinical diagnosis. J. Nanosci. Nanotechnol. 2014;14:378–389. doi: 10.1166/jnn.2014.9234. [DOI] [PubMed] [Google Scholar]
- 114.Pejcic B., De Marco R., Parkinson G. The role of biosensors in the detection of emerging infectious diseases. Analyst. 2006;131:1079–1090. doi: 10.1039/B603402K. [DOI] [PubMed] [Google Scholar]
- 115.Guliy O.I., Zaitsev B.D., Larionova O.S., Borodina I.A. Virus detection methods and biosensor technologies. Biophysics (Oxf). 2019;64:890–897. doi: 10.1134/S0006350919060095. [DOI] [Google Scholar]
- 116.The editors of encyclopædia Britannica, Influenza, Encycl. Br. (2020). https://www.britannica.com/science/influenza.
- 117.Hamborsky J., Kroger A., Charles, editors. Epidemiology and Prevention of Vaccine-Preventable Diseases. 13th Edit. Public Health Foundation; Washington D.C.: 2015. Centers for disease control and prevention, influenza; pp. 187–208.https://www.cdc.gov/vaccines/pubs/pinkbook/front-matter.html [Google Scholar]
- 118.Yang J.M., Kim K.R., Kim C.S. Biosensor for rapid and sensitive detection of influenza virus. Biotechnol. Bioprocess Eng. 2018;23:371–382. doi: 10.1007/s12257-018-0220-x. [DOI] [Google Scholar]
- 119.Rota P.A., Wallis T.R., Harmon M.W., Rota J.S., Kendal A.P., Nerome K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990;175:59–68. doi: 10.1016/0042-6822(90)90186-U. [DOI] [PubMed] [Google Scholar]
- 120.Centers for Disease Control and Prevention, Types of influenza viruses, CDC. (2019). https://www.cdc.gov/flu/about/viruses/types.htm(accessed April 8, 2020).
- 121.Rappuoli R. Inner workings: 1885, the first rabies vaccination in humans. PNAS. 2014;111:12273. doi: 10.1073/pnas.1414226111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Palese P. Influenza virus vaccines: past and future. Nat. Microbiol. 2018 https://naturemicrobiologycommunity.nature.com/users/79539-peter-palese/posts/29264-influenza-virus-vaccines-past-and-future (accessed April 6, 2020) [Google Scholar]
- 123.Hannoun C. The evolving history of influenza viruses and influenza vaccines. Expert Rev. Vaccines. 2013;12:1085–1094. doi: 10.1586/14760584.2013.824709. [DOI] [PubMed] [Google Scholar]
- 124.El Adnani Z., Mcharfi M., Sfaira M., Benzakour M., Benjelloun A.T., Touhami M.E. DFT theoretical study of 7-R-3methylquinoxalin-2(1H)-thiones (RH; CH3; Cl) as corrosion inhibitors in hydrochloric acid. Corros. Sci. 2013;68:223–230. doi: 10.1016/j.corsci.2012.11.020. [DOI] [Google Scholar]
- 125.Dziąbowska K., Czaczyk E., Nidzworski D. Detection methods of human and animal influenza virus-current trends. Biosensors. 2018;8:94. doi: 10.3390/bios8040094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Diouani M.F., Helali S., Hafaid I., Hassen W.M., Snoussi M.A., Ghram A., Jaffrezic-Renault N., Abdelghani A. Miniaturized biosensor for avian influenza virus detection. Mater. Sci. Eng. C. 2008;28:580–583. doi: 10.1016/j.msec.2007.10.043. [DOI] [Google Scholar]
- 127.Ravina A.Dalal, Mohan H., Prasad M., Pundir C.S. Detection methods for influenza A H1N1 virus with special reference to biosensors: a review. Biosci. Rep. 2020;40 doi: 10.1042/BSR20193852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang J.M., Kim K.R., Kim C.S. Biosensor for rapid and sensitive detection of influenza virus. Biotechnol. Bioprocess Eng. 2018;23:371–382. doi: 10.1007/s12257-018-0220-x. [DOI] [Google Scholar]
- 129.Veerapandian M., Hunter R., Neethirajan S. Dual immunosensor based on methylene blue-electroadsorbed graphene oxide for rapid detection of the in fl uenza A virus antigen. Talanta. 2016;155:250–257. doi: 10.1016/j.talanta.2016.04.047. [DOI] [PubMed] [Google Scholar]
- 130.Han J.-.H., Lee D., Chew C.H.C., Kim T., Pak J.J. A multi-virus detectable microfluidic electrochemical immunosensor for simultaneous detection of H1N1, H5N1, and H7N9 virus using ZnO nanorods for sensitivity enhancement. Sens. Actuat. B Chem. 2016;228:36–42. doi: 10.1016/j.snb.2015.07.068. [DOI] [Google Scholar]
- 131.Mikula E., Silva C.E., Kopera E., Zdanowski K., Radecki J., Radecka H. Highly sensitive electrochemical biosensor based on redox - active monolayer for detection of anti-hemagglutinin antibodies against swine-origin influenza virus H1N1 in sera of vaccinated mice. BMC Vet. Res. 2018;14:1–9. doi: 10.1186/s12917-018-1668-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wong C.L., Chua M., Mittman H., Choo L.X., Lim H.Q., Olivo M. A phase-intensity surface plasmon resonance biosensor for avian influenza A (H5N1) detection. Sensors. 2017;17:2363. doi: 10.3390/s17102363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ravina, Mohan H., Gill P.S., Kumar A. Hemagglutinin gene based biosensor for early detection of swine flu (H1N1) infection in human. Int. J. Biol. Macromol. 2019;130:720–726. doi: 10.1016/j.ijbiomac.2019.02.149. [DOI] [PubMed] [Google Scholar]
- 134.Dong S., Zhao R., Zhu J., Lu X., Li Y., Qiu S., Jia L., Jiao X., Song S., Fan C., Hao R., Song H. Electrochemical DNA biosensor based on a tetrahedral nanostructure probe for the detection of avian influenza A (H7N9) virus. Appl. Mater. Interfaces. 2015;7:8834–8842. doi: 10.1021/acsami.5b01438. [DOI] [PubMed] [Google Scholar]
- 135.Medina-sánchez M., Ibarlucea B., Pérez N., Karnaushenko D.D., Weiz S.M., Baraban L., Cuniberti G., Schmidt O.G. High-performance 3D tubular nanomembrane sensor for DNA detection. Nano Lett. 2016;16:4288–4296. doi: 10.1021/acs.nanolett.6b01337. [DOI] [PubMed] [Google Scholar]
- 136.Jiang L., Luo J., Dong W., Wang C., Jin W., Xia Y., Wang H., Ding H., Jiang L., He H. Development and evaluation of a polydiacetylene based biosensor for the detection of H5 influenza virus. J. Virol. Methods. 2015;219:38–45. doi: 10.1016/j.jviromet.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 137.Devarakonda S., Singh R., Bhardwaj J., Jang J. Cost-effective and handmade paper-based immunosensing device for electrochemical detection of influenza virus. Sensors. 2017;11:2597. doi: 10.3390/s17112597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bhardwaj J., Sharma A., Jang J. Vertical flow-based paper immunosensor for rapid electrochemical and colorimetric detection of influenza virus using a different pore size sample pad. Biosens. Bioelectron. 2018;126:36–43. doi: 10.1016/j.bios.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 139.Hushegyi A., Pihíková D., Bertok T., Adam V., Kizek R., Tkac J. Ultrasensitive detection of influenza viruses with a glycan-based impedimetric biosensor. Biosens. Bioelectron. 2016;79:644–649. doi: 10.1016/j.bios.2015.12.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Raman R., Tharakaraman K., Sasisekharan V., Sasisekharan R. Glycan-protein interactions in viral pathogenesis. Curr. Opin. Struct. Biol. 2016;40:153–162. doi: 10.1016/j.sbi.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gorelkin P.V., Erofeev A.S., Kiselev G.A., Kolesov D.V., Dubrovin E.V., Yaminsky I.V. Synthetic sialylglycopolymer receptor for virus detection using cantilever-based sensors. Analyst. 2015;140:6131–6137. doi: 10.1039/C5AN01102G. [DOI] [PubMed] [Google Scholar]
- 142.Erofeev A.S., Gorelkin P.V., Kolesov D.V., Kiselev G.A., Dubrovin E.V., Yaminsky I.V. Label-free sensitive detection of influenza virus using PZT discs with a synthetic sialylglycopolymer receptor layer. R. Soc. Open Sci. 2019;6 doi: 10.1098/rsos.190255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nidzworski D., Siuzdak K., Niedziałkowski P., Bogdanowicz R., Sobaszek M., Ryl J., Weiher P., Sawczak M., Wnuk E., Goddard W.A., III, Jaramillo-Botero A., Ossowski T. A rapid-response ultrasensitive biosensor for influenza virus detection using antibody modified boron-doped diamond. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-15806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Joshi S.R., Sharma A., Kim G., Jang J. Low cost synthesis of reduced graphene oxide using biopolymer for influenza virus sensor. Mater. Sci. Eng. C. 2020;108 doi: 10.1016/j.msec.2019.110465. [DOI] [PubMed] [Google Scholar]
- 145.Xu L., Jiang X., Zhu Y., Duan Y., Huang T., Huang Z., Liu C., Xu B., Xie Z. A multiplex asymmetric reverse transcription-PCR assay combined with an electrochemical DNA sensor for simultaneously detecting and subtyping influenza a viruses. Front. Microbiol. 2018;27:1405. doi: 10.3389/fmicb.2018.01405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bhardwaj J., Chaudhary N., Kim H., Jang J. Subtyping of influenza A H1N1 virus using a label-free electrochemical biosensor based on the DNA aptamer targeting the stem region of HA protein. Anal. Chim. Acta. 2019;1064:94–103. doi: 10.1016/j.aca.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 147.Katayama Y., Ohgi T., Mitoma Y., Hifumi E., Egashira N. Detection of influenza virus by a biosensor based on the method combining electrochemiluminescence on binary SAMs modified Au electrode with an immunoliposome encapsulating Ru ( II ) complex. Anal. Bioanal. Chem. 2016;408:5963–5971. doi: 10.1007/s00216-016-9587-8. [DOI] [PubMed] [Google Scholar]
- 148.Singh R., Hong S., Jang J. Label-free detection of influenza viruses using a reduced graphene oxide-based electrochemical immunosensor integrated with a microfluidic platform. Sci. Rep. 2017;7 doi: 10.1038/srep42771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chang Y., Wang W., Hong Y., Yuan R., Chen K., Huang Y.-.W., Lu P.-.L., Chen Y.-.H., Chen Y.-M.A., Su L.-.C., Wang S.-.F. Simple strategy for rapid and sensitive detection of avian influenza A H7N9 virus based on intensity-modulated SPR biosensor and new generated antibody. Anal. Chem. 2018;90:1861–1869. doi: 10.1021/acs.analchem.7b03934. [DOI] [PubMed] [Google Scholar]
- 150.Diba F.S., Kim S., Lee H.J. Amperometric bioaffinity sensing platform for avian influenza virus proteins with aptamer modified gold nanoparticles on carbon chips. Biosens. Bioelectron. 2015;72:355–361. doi: 10.1016/j.bios.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 151.Lee T., Park S.Y., Jang H., Kim G.-.H., Lee Y., Park C., Mohammadniaei M., Lee M.-.H., Min J. Fabrication of electrochemical biosensor consisted of multi functional DNA structure/porous au nanoparticle for avian influenza virus (H5N1) in chicken serum. Mater. Sci. Eng. C. 2019;99:511–519. doi: 10.1016/j.msec.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 152.Lin C., Guo Y., Zhao M., Sun M., Luo F., Guo L., Qiu B., Lin Z., Chen G. Highly sensitive colorimetric immunosensor for influenza virus H5N1 based on enzyme-encapsulated liposome. Anal. Chim. Acta. 2017;963:112–118. doi: 10.1016/j.aca.2017.01.031. [DOI] [PubMed] [Google Scholar]
- 153.Nguyen V., Bin Seo H., Kim B.C., Kim S.K., Song C.-.S., Gu M.B. Highly sensitive sandwich-type SPR based detection of whole H5Nx viruses using a pair of aptamers. Biosens. Bioelectron. 2016;86:293–300. doi: 10.1016/j.bios.2016.06.064. [DOI] [PubMed] [Google Scholar]
- 154.Jarocka U., Sawicka R., Stachyra A., Góra-Sochacka A., Sirko A., Zagórski-Ostoja W., Sączyńska V., Porębska A., Dehaen W., Radecki J., Radecka H. A biosensor based on electroactive dipyrromethene-Cu(II) layer deposited onto gold electrodes for the detection of antibodies against avian influenza virus type H5N1 in hen sera. Anal. Bioanal. Chem. 2015;407:7807–7814. doi: 10.1007/s00216-015-8949-y. [DOI] [PubMed] [Google Scholar]
- 155.Malecka K., Świętoń E., Verwilst P., Stachyra A., Sirko A., Dehaen W., Radecki J., Radecka H. Ultrasensitive electrochemical genosensor for direct detection of specific RNA sequences derived from avian influenza viruses present in biological samples. Acta Biochim Pol. 2019;66:299–304. doi: 10.18388/abp.2019_2774. [DOI] [PubMed] [Google Scholar]
- 156.Pang Y., Rong Z., Wang J., Xiao R., Wang S. A fluorescent aptasensor for H5N1 influenza virus detection based-on the CORE-shell nanoparticles metal-enhanced fluorescence (MEF) Biosens. Bioelectron. 2015;66:527–532. doi: 10.1016/j.bios.2014.10.052. [DOI] [PubMed] [Google Scholar]
- 157.Matsubara T., Ujie M., Yamamoto T., Einaga Y., Daidoji T., Nakaya T., Sato T. Avian influenza virus detection by optimized peptide termination on a boron-doped diamond electrode. Sensors. 2020;5:431–439. doi: 10.1021/acssensors.9b02126. [DOI] [PubMed] [Google Scholar]
- 158.Zhang H., Zhang S., Liu N. Prevention and control of emergent infectious disease with high specific antigen sensor. Artif. Cells, Nanomedicine, Biotechnol. 2017;45:1298–1303. doi: 10.3109/21691401.2016.1161638. [DOI] [PubMed] [Google Scholar]
- 159.Lee I., Kim S.-.E., Lee J., Woo D.H., Lee S., Pyo H., Song C.-.S., Lee J. A self-calibrating electrochemical aptasensing platform: correcting external interference errors for the reliable and stable detection of avian influenza viruses. Biosens. Bioelectron. 2020;152 doi: 10.1016/j.bios.2020.112010. [DOI] [PubMed] [Google Scholar]
- 160.Choi J., Jeun M., Yuk S., Park S., Choi J., Lee D., Shin H., Kim H., Cho I., Kim S.K., Lee S., Song C.S., Lee K.H. Fully packaged portable thin film biosensor for the direct detection of highly pathogenic viruses from on-site samples. ACS Nano. 2019;13:812–820. doi: 10.1021/acsnano.8b08298. [DOI] [PubMed] [Google Scholar]
- 161.Huang J., Xie Z., Xie Z., Luo S., Xie L., Huang L., Fan Q., Zhang Y., Wang S., Zeng T. Silver nanoparticles coated graphene electrochemical sensor for the ultrasensitive analysis of avian influenza virus H7. Anal. Chim. Acta. 2016;913:121–127. doi: 10.1016/j.aca.2016.01.050. [DOI] [PubMed] [Google Scholar]
- 162.Kim J., Kwon J.H., Jang J., Lee H., Kim S., Hahn Y.K., Kim S.K., Lee K.H., Lee S., Pyo H., Song C.-.S., Lee J. Rapid and background-free detection of avian influenza virus in opaque sample using NIR-to-NIR upconversion nanoparticle-based lateral flow immunoassay platform. Biosens. Bioelectron. 2018;112:209–215. doi: 10.1016/j.bios.2018.04.047. [DOI] [PubMed] [Google Scholar]
- 163.Krishna V.D., Wu K., Perez A.M., Wang J.-.P. Giant magnetoresistance-based biosensor for detection of influenza a virus. Front. Microbiol. 2016;7:400. doi: 10.3389/fmicb.2016.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kushwaha A., Takamura Y., Nishigaki K., Biyani M. Competitive non-SELEX for the selective and rapid enrichment of DNA aptamers and its use in electrochemical aptasensor. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-43187-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lee W.S., Kang T., Kim S.-.H., Jeong J. An Antibody-immobilized silica inverse opal nanostructure for label-free optical biosensors. Sensors. 2018;18:307. doi: 10.3390/s18010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lee T., Kim G.H., Kim S.M., Hong K., Kim Y., Park C., Sohn H., Min J. Label-free localized surface plasmon resonance biosensor composed of multi-functional DNA 3 way junction on hollow Au spike-like nanoparticles (HAuSN) for avian influenza virus detection. Colloids Surf. B Biointerfaces. 2019;182 doi: 10.1016/j.colsurfb.2019.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Low S.S., Tan M.T.T., Loh H.-.S., Khiew P.S., Chiu W.S. Facile hydrothermal growth graphene/ZnO nanocomposite for development of enhanced biosensor. Anal. Chim. Acta. 2016;903:131–141. doi: 10.1016/j.aca.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 168.Wang F., Gopinath S.C., Lakshmipriya T. Aptamer-antibody complementation on multiwalled carbon nanotube-gold transduced dielectrode surfaces to detect pandemic swine influenza virus. Int. J. Nanomed. 2019;14:8469–8481. doi: 10.2147/IJN.S219976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhao X., Tsao Y.-.C., Lee F.-.J., Tsai W.-.H., Wang C.-.H., Chuang T.-.L., Wu M.-.S., Lin C.-.W. Optical fiber sensor based on surface plasmon resonance for rapiddetection of avian influenza virus subtype H6: initial studies. J. Virol. Methods. 2016;233:15–22. doi: 10.1016/j.jviromet.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 170.Qi C., Tian X.-.S., Chen S., Yan J.-.H., Cao Z., Tian K.-.G., Gao G.F., Jin G. Detection of avian influenza virus subtype H5 using a biosensor based on imaging ellipsometry. Biosens. Bioelectron. 2010;25:1530–1534. doi: 10.1016/j.bios.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 171.Kahn J.S., McIntosh K. History and recent advances in coronavirus discovery. Pediatr. Infect. Dis. J. 2005;24:S223–S227. doi: 10.1097/01.inf.0000188166.17324.60. [DOI] [PubMed] [Google Scholar]
- 172.Gong S., Bao L. The battle against SARS and MERS coronaviruses: reservoirs and Animal models. Anim. Model. Exp. Med. 2018;1:125–133. doi: 10.1002/ame2.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.World Health Organization (WHO) WHO; 2019. Middle East respiratory Syndrome Coronavirus (MERS-CoV)https://www.who.int/emergencies/mers-cov/en/ (accessed January 4, 2020) [Google Scholar]
- 174.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.-.L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Killerby M.E., Biggs H.M., Midgley C.M., Gerber S.I., Watson J.T. Middle east respiratory syndrome coronavirus transmission. Emerg. Infect. Dis. 2020;26:191–198. doi: 10.3201/eid2602.190697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.World Health Organization (WHO), Infectious diseases of potential risk for travellers, in: G. Poumerol, A. Wilder-Smith (Eds.), Int. Travel Heal., WHO, Geneva, Switzerland, 2012: pp. 5581. https://www.who.int/publications/i/item/9789241580472.
- 177.Huang J.C., Chang Y.-.F., Chen K.-.H., Su L.-.C., Lee C.-.W., Chen C.-.C., Chen Y.-M.A., Chou C. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in human serum using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens. Bioelectron. 2009;25:320–325. doi: 10.1016/j.bios.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Roh C., Jo S.K. Quantitative and sensitive detection of SARS coronavirus nucleocapsid protein using quantum dots-conjugated RNA aptamer on chip. J. Chem. Technol. Biotechnol. 2011;86:1475–1479. doi: 10.1002/jctb.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Park T.J., Hyun M.S., Lee H.J., Lee S.Y., Ko S. A self-assembled fusion protein-based surface plasmon resonance biosensor for rapid diagnosis of severe acute respiratory syndrome. Talanta. 2009;79:295–301. doi: 10.1016/j.talanta.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Teengam P., Siangproh W., Tuantranont A., Vilaivan T., Chailapakul O., Henry C.S. Multiplex paper-based colorimetric DNA sensor using pyrrolidinyl peptide nucleic acid-induced AgNPs aggregation for detecting MERS-CoV, MTB and HPV oligonucleotides. Anal. Chem. 2017;89:5428–5435. doi: 10.1021/acs.analchem.7b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Layqah L.A., Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim. Acta. 2019;186 doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 183.Seo G., Lee G., Kim M.J., Baek S.-.H., Choi M., Ku K.B., Lee C.-.S., Jun S., Park D., Kim H.G., Kim S.-.J., Lee J.-.O., Kim B.T., Park E.C., Il Kim S. Rapid Detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 184.Albano D.R.B., Shum K., Tanner J.A., Fung Y.S. 17th Int. Meet. Chem. Sensors - IMCS. AMA; Viena, Austria: 2018. Piezoelectric quartz crystal aptamer biosensor for detection and quantification of SARS CoV helicase protein; pp. 211–213. [DOI] [Google Scholar]
- 185.Shi L., Sun Q., He J., Xu H., Liu C., Zhao C., Xu Y., Wu C., Xiang J., Gu D., Long J., Lan H. Development of SPR biosensor for simultaneous detection of multiplex respiratory viruses. Biomed. Mater. Eng. 2015;26:S2207–S2216. doi: 10.3233/BME-151526. [DOI] [PubMed] [Google Scholar]
- 186.Collins P.L., Graham B.S. Viral and host factors in human respiratory syncytial virus pathogenesis. J. Virol. 2008;82:2040–2055. doi: 10.1128/JVI.01625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Collins P.L., Fearns R., Graham B.S. In: Challenges and Opportunities for Respiratory Syncytial Virus Vaccines. Anderson L.J., Graham B.S., editors. Springer; Berlin: 2013. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease; pp. 3–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.MacLachlan N.J., Dubovi E.J., editors. Fenner's Veterinary Virology. 5th Editio. Academic Press; 2017. Paramyxoviridae and Pneumoviridae; pp. 327–356. [DOI] [Google Scholar]
- 189.Alonso W.J., Tamerius J., Freitas A.R.R. Respiratory syncytial virus causes more hospitalizations and deaths in equatorial Brazil than influenza (including during the 2009 pandemic) An. Acad. Bras. Cienc. 2020;92 doi: 10.1590/0001-3765202020180584. [DOI] [PubMed] [Google Scholar]
- 190.Murphy B.R., Prince G.A., Collins P.L., Coelingh K.V.W., Olmsted R.A., Spriggs M.K., Parrott R.H., Kim H.-.W., Brandt C.D., Chanock R.M. Current approaches to the development of vaccines effective against parainfluenza and respiratory syncytial viruses. Virus Res. 1988;11:1–15. doi: 10.1016/0168-1702(88)90063-9. [DOI] [PubMed] [Google Scholar]
- 191.Acosta P.L., Caballero M.T., Polack F.P. Brief history and characterization of enhanced respiratory syncytial virus disease. Clin. Vaccine Immunol. 2016;23:189–195. doi: 10.1128/CVI.00609-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Shi T., McAllister D.A., O'Brien K.L., Simoes E.A.F., Madhi S.A., Gessner B.D., Polack F.P., Balsells E., Acacio S., Aguayo C., Alassani I., Ali A., Antonio M., Awasthi S., Awori J.O., Azziz-Baumgartner E., Baggett H.C., Baillie V.L., Balmaseda A., Barahona A., Basnet S., Bassat Q., Basualdo W., Bigogo G., Bont L., Breiman R.F., Brooks W.A., Broor S., Bruce N., Bruden D., Buchy P., Campbell S., Carosone-Link P., Chadha M., Chipeta J., Chou M., Clara W., Cohen C., de Cuellar E., Dang D.-.A., Dash-yandag B., Deloria-Knoll M., Dherani M., Eap T., Ebruke B.E., Echavarria M., de C.C., Emediato F.L., Fasce R.A., Feikin D.R., Feng L., Gentile A., Gordon A., Goswami D., Goyet S., Groome M., Halasa N., Hirve S., Homaira N., Howie S.R.C., Jara J., Jround I., Kartasasmita C.B., Khuri-Bulos N., Kotloff K.L., Krishnan A., Libster R., Lopez O., Lucero M.G., Lucion F., Lupisan S.P., Marcone D.N., McCracken J.P., Mejia M., Moisi J.C., Montgomery J.M., Moore D.P., Moraleda C., Moyes J., Munywoki P., Mutyara K., Nicol M.P., Nokes D.J., Nymadawa P., da C. Oliveira M.T., Oshitani H., Pandey N., Paranhos-Baccalà G., MD L.N.P., Picot V.S., Rahman M., Rakoto-Andrianarivelo M., Rasmussen Z.A., Rath B.A., Robinson A., Romero C., Russomando G., Salimi V., Sawatwong P., Scheltema N., Schweiger B., Scott J.A.G., Seidenberg P., Shen K., Singleton R., Sotomayor V., Strand T.A., Sutanto A., Sylla M., Tapia M.D., Thamthitiwat S., Thomas E.D., Tokarz R., Turner C., Venter M., Waicharoen S., Wang J., Watthanaworawit W., Yoshida L.-.M., Yu H., Zar H.J., Campbell H., Nair H. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet. 2017;390:946–958. doi: 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Perez J.W., Haselton F.R., Wright D.W. Viral detection using DNA functionalized gold filaments. Analyst. 2009;134:1548–1553. doi: 10.1039/B904191E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Rochelet M., Solanas S., Grossiord C., Maréchal P., Résa C., Vienney F., Barranger C., Joannes M. A thin layer-based amperometric enzyme immunoassay for the rapid and sensitive diagnosis of respiratory syncytial virus infections. Talanta. 2012;100:139–144. doi: 10.1016/j.talanta.2012.07.088. [DOI] [PubMed] [Google Scholar]
- 195.Cai Z., Song Y., Wu Y., Zhu Z., Yang C.J., Chen X. An electrochemical sensor based on label-free functional allosteric molecular beacons for detection target DNA/miRNA. Biosens. Bioelectron. 2013;41:783–788. doi: 10.1016/j.bios.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 196.Lee C.-.C., Liao Y.-.C., Lai Y.-.H., Chuang M.-.C. Recognition of dual targets by a molecular beacon-based sensor: subtyping of influenza a virus. Anal. Chem. 2015;87:5410–5416. doi: 10.1021/acs.analchem.5b00810. [DOI] [PubMed] [Google Scholar]
- 197.Zhan L., Yang T., Li C.M., Wu W.B., Huang C.Z. Sensitive immunosensor for respiratory syncytial virus based on dual signal amplification of gold nanopaticle layer-modified plates and catalyzed reporter deposition An enhanced immunoassay was designed and utilized for respiratory syncytial virus ( RSV ) Sens. Actuat. B Chem. 2018;255(Part 2):1291–1297. doi: 10.1016/j.snb.2017.08.110. [DOI] [Google Scholar]
- 198.Schildgen O. Human bocavirus: lessons learned to date. Pathogens. 2013;2:1–12. doi: 10.3390/pathogens2010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nichols W.G., Campbell A.J.P., Boeckh M. Respiratory viruses other than influenza virus: impact and therapeutic advances. Clin. Microbiol. Rev. 2008;21:274–290. doi: 10.1128/CMR.00045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Leotte J., Trombetta H., Faggion H.Z., Almeida B.M., Nogueira M.B., Vidal L.R., Raboni S.M. Impact and seasonality of human rhinovirus infection in hospitalized patients for two consecutive years. J. Pediatr. (Rio. J). 2017;93:294–300. doi: 10.1016/j.jped.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ostroff R., Ettinger A., La H., Rihanek M., Zalman L., Meador J., III, Patick A.K., Worland S., Polisky B. Rapid multiserotype detection of human rhinoviruses on optically coated silicon surfaces. J. Clin. Virol. 2001;21:105–117. doi: 10.1016/S1386-6532(01)00150-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Jin X.-.H., Ishii A., Aoki K., Ishida S., Mukasa K., Ohno S. Detection of human adenovirus hexon antigen using carbon nanotube sensors. J. Virol. Methods. 2011;171:405–407. doi: 10.1016/j.jviromet.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 203.Jenison R., Rihanek M., Polisky B. Use of a thin film biosensor for rapid visual detection of PCR products in a multiplex format. Biosens. Bioelectron. 2001;16:757–763. doi: 10.1016/S0956-5663(01)00217-2. [DOI] [PubMed] [Google Scholar]
- 204.Owens R.M., Wang C.Q., You J.A., Jiambutr J., Xu A.S.L., Marala R.B., Jin M.M. Real-time quantitation of viral replication and inhibitor potency using a label-free optical biosensor. J. Recept. Signal Transduct. 2009;29:195–201. doi: 10.1080/10799890903079919. [DOI] [PubMed] [Google Scholar]


