Abstract
Background
Glioma is one of the leading causes of cancer-related deaths. This study aimed to investigate the function and mechanism of long noncoding RNA (lncRNA) LINC00173 in the regulation of glioma progression.
Methods
LINC00173 expression was measured using qRT-PCR. Survival rate was analyzed through Kaplan–Meier method. CCK8, colony formation and EdU assays were performed to measure cell proliferation while transwell was used to determine cell migration and invasion. Luciferase reporter assay was conducted to test RNA interaction.
Results
LINC00173 expression was elevated in glioma tissues and cells. LINC00173 high expression predicted poor prognosis. Loss of LINC00173 inhibited proliferation, migration and invasion. LINC00173 interacted with miR-765 to enhance NUTF2 expression. MiR-765 expression was negatively correlated with LINC00173 and NUTF2 in glioma tissues. NUTF2 level was increased in glioma tissues. NUTF2 overexpression rescued the potential of proliferation, migration and invasion in LINC00173-silenced cells.
Conclusion
Our research demonstrated that LINC00173 promotes glioma progression through targeting miR-765/NUTF2 axis.
Keywords: LINC00173, miR-765, NUTF2, glioma
Introduction
Glioma initiates from glial cells and contains several types, such as astrocytoma and oligodendroglioma.1 Over a quarter of brain tumors are glioma, which causes a large number of cancer-related deaths every year around the world.1 The current clinically therapeutic strategies are surgery combined with chemotherapy and radiotherapy.2 However, the prognosis of glioma patients remains not well post therapy.3 Hence, it is urgently required to discover new molecular mechanism for glioma therapy.
Both long noncoding RNA (lncRNA) and microRNA (miRNA) belong to noncoding RNAs, which have no protein-coding ability. LncRNA is characterized with more than 200 nucleotides while miRNA is about 22 nucleotides in length.4 LncRNA and miRNA are involved in various cellular processes, including cell division, invasion and survival.5 Dysregulation of lncRNA or miRNA usually causes tumor initiation and progression.6,7 For example, lncRNA LINC00152 upregulation promotes gastric cancer growth and metastasis.8 LncRNA SNHG6 overexpression facilitates lung cancer cell proliferation and metastasis.9 MiR-340-5p dysregulation promotes tumorigenesis of esophageal squamous cell carcinoma.10 In addition, miR-126 suppresses colon cancer cell survival and induces apoptosis.11 Besides, lncRNA has been identified as potential competing endogenous RNA (ceRNA) for miRNA to function in cancer.12 The potential roles underlying lncRNA and miRNA still require much investigation. And the relationship between lncRNA and miRNA also needs to be defined.
LINC00173 is an oncogene in lung cancer and breast cancer.13,14 The function of LINC00173 in glioma is unclear. In the current study, we found that LINC00173 was upregulated in glioma tissues. LINC00173 high expression was associated with a low survival rate. LINC00173 depletion suppressed proliferation, migration and invasion of glioma cells. LINC00173 was discovered to sponge miR-765 to elevate NUTF2 expression. Taken together, our findings supported that LINC00173 plays essential oncogenic roles in glioma through activating miR-765/NUTF2 pathway.
Materials and Methods
Clinical Samples
Thirty-seven glioma tissues and normal tissues were collected from Lishui City People’s Hospital. Patients received no radiotherapy or chemotherapy before surgery. All tissues were stored in liquid nitrogen. Association between LINC00173 expression and clinical characteristics in glioma tissues was analyzed in Table 1. Written informed consent was obtained from every patient. This study was approved by the Ethics Committee of Lishui City People’s Hospital (No. 201705150324). And the experiments were conducted in accordance with the Declaration of Helsinki.
Table 1.
Association Between LINC00173 Expression and Clinical Characteristics in Glioma Tissues
| Features | Low (n=19) | High (n=18) | P-value |
|---|---|---|---|
| Age (years) | 0.714 | ||
| ≤60 | 13 | 14 | |
| >60 | 6 | 4 | |
| Gender | 0.743 | ||
| Male | 12 | 10 | |
| Female | 7 | 8 | |
| Grade | 0.022 | ||
| I–II | 14 | 6 | |
| III–IV | 5 | 12 | |
| Tumor size (cm) | 0.038 | ||
| ≤4 | 16 | 9 | |
| >4 | 3 | 9 |
Cell Culture and Treatment
The normal human astrocyte (NHA) and glioma cell lines were purchased from Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured using PMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen). shRNAs against LINC00160, miR-629-3p mimics, miR-629-3p inhibitors and negative controls were obtained from GenePharma and transfected into glioma cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Efficiency was validated using qRT-PCR after 48 h.
qRT-PCR
Total RNA was extracted from tissues and cell lines using TRIzol (Invitrogen, Carlsbad, CA). PrimeScript RT reagent kit (RR047A, Takara Holdings Inc., Tokyo, Japan) was used to generate cDNA from RNA template. qPCR was completed through SYBR Premix EX Taq™ II (Takara, Japan). GAPDH was the normalized control. Relative expression was calculated through the 2−ΔΔCt method.
Luciferase Reporter Assay
The fragment of LINC00173 or NUTF2 containing indicated miR-765 binding site was constructed into pMIR-REPORT vector. For luciferase reporter assay, glioma cells were transfected with REPORT vector and miR-765 mimics. After 48 h, the luciferase reporter activity was measured through the Dual-Luciferase Reporter Assay System (Promega, Madison, WI).
Western Blot Assay
Cells were lyzed using Radioimmunoprecipitation buffer (Beyotime, Shanghai, China). Protein concentration was determined by a BCA Protein Assay Kit (Thermo Fisher Scientific, MA). Then, proteins were separated using 10% SDS-PAGE and transferred onto PVDF membranes. After blockage using 5% BSA for 2 h, the membrane was incubated the primary antibodies at 4°C overnight. After washed 3 times using PBST, the membranes were incubated with horseradish peroxidase-labeled second antibody, followed by detection through the enhanced chemiluminescence reagent (EMD Millipore, USA).
CCK8 and Colony Forming Assays
Proliferation was measured using CCK8 and colony formation assay. CCK8 assay was performed using the CCK-8 reagent (Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions and absorbance was determined at 450 nm using a microplate reader (BioTek, Winooski, VT). For colony formation assay, 500 cells were seeded into 6-well plates and cultured for 14 days. Then the cells were fixed with methanol and stained with 0.1% crystal violet for 10 minutes.
EdU Assay
Cells were plated into 96-well plates and incubated with EdU (100 μL) at 37°C for 4 h, followed by detection using FACS.
Transwell Migration and Invasion Assays
Transwell plates (Corning, NY) were used to measure migration and invasion according to the manufacturer’s instructions. In brief, cells were suspended into 200 μL serum-free medium and seeded into the upper chamber while the lower chamber was filled with 600 µL of complete medium. After incubation for 24, cells in the lower chamber were fixed with methanol and stained with 0.1% crystal violet for 10 minutes. Migrated and invaded cells were counted through a light microscope.
Statistical Analysis
GraphPad Prism 6.0 (GraphPad, CA, USA) was used to analyze results. Data were presented as means±standard deviation (SD). Significant differences were analyzed using Student’s t-test or one-way ANOVA. Survival rate was analyzed by the Kaplan–Meier method and Log rank test. P<0.05 was considered to be significant.
Results
LINC00173 Expression is Elevated in Glioma
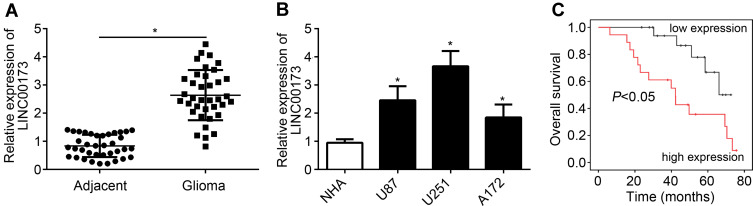
The expression of LINC00173 was firstly analyzed. Through qRT-PCR, we found that LINC00173 level was elevated in glioma tissues compared with normal tissues (Figure 1A). Besides, we found that LINC00173 was also upregulated in glioma cell lines compared to NHA cells (Figure 1B). Then, according to the median value of LINC00173, glioma tissues were classified into two groups. After analysis, we found that LINC00173 high expression correlated with poor prognosis (Figure 1C).
Figure 1.
LINC00173 expression is elevated in glioma. (A) The level of LINC00173 in glioma tissues was measured. (B) The expression of LINC00173 in glioma cell lines and NHAs. (C) Association between overall survival and LINC00173 expression in glioma patients. *P<0.05.
LINC00173 Enhanced Glioma Cell Proliferation, Migration and Invasion
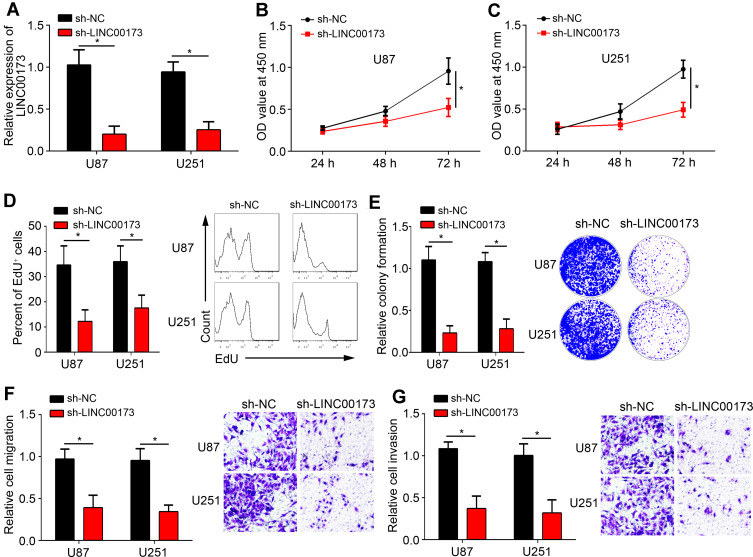
To explore the function of LINC00173, U87 and U251 cells were chosen. After transfection of sh-LINC00173, LINC00173 expression was significantly downregulated (Figure 2A). Through CCK8 assay, we observed that LINC00173 knockdown suppressed the proliferation capacity of glioma cells (Figure 2B and C), which was validated by EdU and colony formation assays (Figure 2D and E). Afterwards, transwell assay was performed. Results indicated that LINC00173 loss inhibited migration and invasion of glioma cells (Figure 2F and G). Thus, LINC00173 exerted oncogenic roles by affecting proliferation, migration and invasion.
Figure 2.
LINC00173 enhanced glioma cell proliferation, migration and invasion. (A) qRT-PCR analysis of LINC00173 expression in U87 and U251 cells. (B–E) Proliferation ability was measured using CCK8, EdU and colony formation assays. (F and G) Migration and invasion capacity was evaluated after LINC00173 knockdown in glioma cells. *P<0.05.
LINC00173 Worked as the Sponge for miR-765
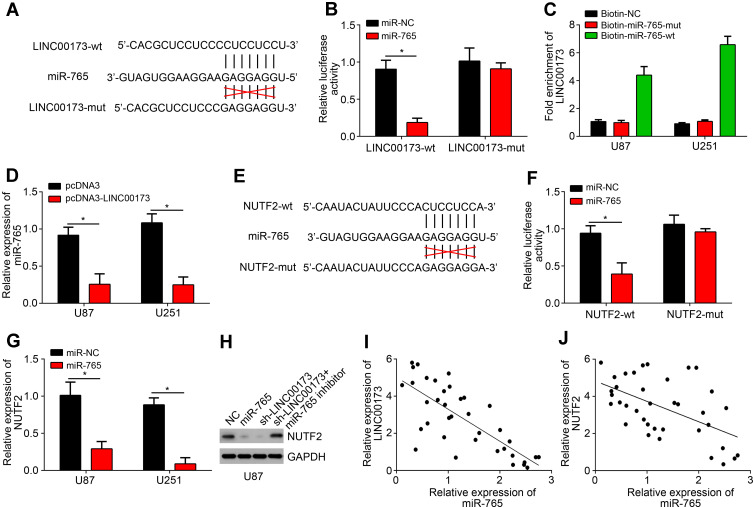
LINC00173 has been found to serve as ceRNA for several miRNAs, such as miR-490 and miR-218.13,14 To determine the mechanism of LINC00173 in glioma, we also performed bioinformatics analysis using miRDB. We identified that miR-765 was the most potential candidate because it scored the highest. To validate it, we constructed luciferase reporters (Figure 3A), followed by luciferase reporter assay. Results showed that miR-765 only suppressed the activity of LINC00173-wt (Figure 3B), supporting their direct interaction. Pulldown assay further demonstrated their interaction (Figure 3C). qRT-PCR found that LINC00173 overexpression suppressed the level of miR-765 (Figure 3D). Next, bioinformatics analysis using miRDB and TargetSan implied that NUTF2 is the most potential target of miR-765. The corresponding luciferase reporters were further constructed (Figure 3E). Luciferase reporter assay also demonstrated the interaction between NUTF2 and miR-765 (Figure 3F). Besides, NUTF2 expression was suppressed by miR-765 mimics (Figure 3G). Moreover, NUTF2 level was decreased after LINC00173 knockdown while miR-765 inhibitors reversed it (Figure 3H). Finally, we found that miR-765 level was negatively correlated with LINC00173 or NUTF2 in glioma tissues (Figure 3I and J).
Figure 3.
LINC00173 worked as the sponge for miR-765. (A) Bioinformatics analysis indicated the binding sites between LINC00173 and miR-765. (B) U87 cells were transfected with miR-765 mimics or negative controls (miR-NC) and luciferase reporter (LINC00173-wt or LINC00173-mut). Then relative luciferase activity was determined. (C) RNA pulldown assay using biotin-labeled miRNAs. (D) Relative expression of miR-765 after LINC00173 knockdown. (E) Bioinformatics analysis indicated the binding sites between miR-765 and NUTF2. (F) U87 cells were transfected with miR-765 mimics or negative controls (miR-NC) and luciferase reporter (NUTF2-wt or NUTF2-mut). Then relative luciferase activity was determined. (G) qRT-PCR analysis for NUTF2 expression. (H) Western blotting analysis for NUTF2 protein level. (I and J) Correlation analyses of LINC00173, miR-765 and NUTF2 in glioma tissues using Pearson’s correlation coefficient. *P<0.05.
LINC00173 Promoted Glioma Progression Through miR-765/NUTF2 Pathway
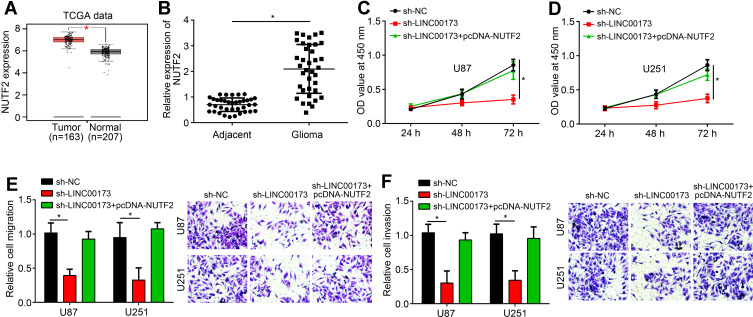
We noticed that NUTF2 expression was upregulated in glioma tissues (Figure 4A and B), suggesting an oncogenic role. To investigate whether LINC00173 regulates glioma progression through miR-765/NUTF2, we restored the expression of NUTF2 in sh-LINC00173 transfected cells. CCK8 and transwell assays demonstrated that NUTF2 restoration successfully rescued the capacities of proliferation, migration and invasion in glioma cells transfected with sh-LINC00173 (Figure 4C–F). Therefore, LINC00173 contributes to glioma progression through miR-765/NUTF2 pathway.
Figure 4.
LINC00173 promoted glioma progression through miR-765/NUTF2 pathway. (A and B) NUTF2 expression in glioma tissues and normal tissues according to TCGA data (using GEPIA tool) and qRT-PCR. (C and D) Proliferation was measured by CCK8 assay. (E and F) migration and invasion potential was determined by transwell assay. *P<0.05.
Discussion
As the most malignant brain tumor, glioma leads to a huge number of deaths. Patients with glioma display a poor prognosis. Therefore, it is of great significance to reveal the mechanism underlying glioma progression. In this study, we found that LINC00173 was upregulated in glioma tissues and cells. LINC00173 overexpression predicted a poor prognosis. Moreover, LINC00173 knockdown suppressed the proliferation, migration and invasion of glioma cells. LINC00173 was also found to inhibit miR-765 and promote NUTF2 expression. Summarily, our research discovered that LINC00173 is an important oncogenic lncRNA in glioma.
The potential roles of lncRNA in glioma have been researched for a long time. Many lncRNAs have been identified to participate in glioma development. For example, lncRNA NCK1-AS1 enhances growth and metastasis of glioma through targeting miR-138-2-3p to activate β-catenin signaling.2 LncRNA CCAT2 contributes to glioma progression by activating VEGFA pathway.15 LncRNA LINC00467 upregulation promotes glioma development through repressing P53 level.16 Previous study showed that LINC00173 downregulation promotes non-small cell lung cancer cell growth and survival.17 However, another study showed that Linc00173 enhances chemoresistance and facilitates tumor progression in small cell lung cancer.13 Besides, LINC00173 contributes to breast cancer development.14 Yet, how LINC00173 works in glioma remains undermined. In our study, we found that LINC00173 was upregulated in glioma tissues. LINC00173 knockdown inhibited the proliferation, migration and invasion of glioma cells. Therefore, our data discovered that LINC00173 is a new oncogene in glioma for the first time.
LncRNA has been found to serve as miRNA sponge in tumor cells. For instance, lncRNA TTN-AS1 sponges miR-140-5p to promote breast cancer metastasis.18 LncRNA CDKN2B-AS1 sponges miR-324-5p to regulate cell-cycle progression in laryngeal squamous cell cancer.19 Previous studies also revealed LINC00173 was a sponge for some miRNAs, such as miR-490-3p and miR-218.13,14 In our study, we did not observe LINC00173 sponges above miRNAs. However, through bioinformatics, we identified LINC00173 targeted miR-765 in glioma. We demonstrated their direct interaction and found that LINC00173 overexpression inhibited miR-765 expression. MiR-765 has important roles in cancers. MiR-765 was found to suppress tongue squamous cell carcinoma development.20 MiR-765 also promotes myeloma and osteosarcoma progression.21,22 Besides, miR-765 plays oncogenic or anti-cancer roles in gastric cancer and breast cancer.23,24 Its role in glioma remains unclear. Our results suggested that miR-765 was a tumor suppressor in glioma.
LncRNA-miRNA-mRNA regulatory axis is widely observed in cancer. For example, LINC00703/miR-181a/KLF6 axis suppresses the development of gastric cancer.25 LINC00312/miR-9/CDH1 axis was found to promote breast cancer progression.26 Through bioinformatics, we found that miR-765 targeted NUTF2 in glioma. Moreover, we showed that NUTF2 expression was regulated by LINC00173/miR-765 axis. The function of NUTF2 in cancer is nearly unknown. In our work, we found that NUTF2 expression was upregulated in glioma tissues compared to normal tissues. Moreover, we found that NUTF2 overexpression promoted the proliferation, migration and invasion of glioma cells, indicating NUTF2 was an oncogene in glioma.
In conclusion, our study showed that LINC00173 acted as a sponge for miR-765 to promote NUTF2 expression. And LINC00173/miR-765/NUTF2 axis plays a critical function in promoting glioma progression.
Funding Statement
This work was supported by Zhejiang Province Analytical Testing and Experimental Animal Program (LGD19H090001) and Zhejiang Province Welfare Technology Applied Research Project (2017C37111).
Disclosure
All authors declare no conflicts of interest in this work.
References
- 1.Ostrom QT, Cioffi G, Gittleman H, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012-2016. Neuro Oncol. 2019;21(Suppl 5):v1–v100. doi: 10.1093/neuonc/noz150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang L, Li X, Ye H, et al. Long non-coding RNA NCK1-AS1 promotes the tumorigenesis of glioma through sponging microRNA-138-2-3p and activating the TRIM24/Wnt/beta-catenin axis. J Exp Clin Cancer Res. 2020;39(1):63. doi: 10.1186/s13046-020-01567-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Lei C, Liu P, et al. Progress and prospects of recurrent glioma: a recent scientometric analysis of the web of science in 2019. World Neurosurg. 2020;134:e387–e399. doi: 10.1016/j.wneu.2019.10.078 [DOI] [PubMed] [Google Scholar]
- 4.Sun B, Meng M, Wei J, Wang S. Long noncoding RNA PVT1 contributes to vascular endothelial cell proliferation via inhibition of miR-190a-5p in diagnostic biomarker evaluation of chronic heart failure. Exp Ther Med. 2020;19(5):3348–3354. doi: 10.3892/etm.2020.8599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng S, Yao J, Chen Y, et al. Functional role of reprogramming-related long noncoding RNA (lincRNA-ROR) in glioma. J Mol Neurosci. 2015;56(3):623–630. doi: 10.1007/s12031-014-0488-z [DOI] [PubMed] [Google Scholar]
- 6.Zhang D, Zhou H, Liu J, Mao J. Long noncoding RNA ASB16-AS1 promotes proliferation, migration, and invasion in glioma cells. Biomed Res Int. 2019;2019:5437531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun L, Zhao M, Wang Y, et al. Neuroprotective effects of miR-27a against traumatic brain injury via suppressing FoxO3a-mediated neuronal autophagy. Biochem Biophys Res Commun. 2017;482(4):1141–1147. doi: 10.1016/j.bbrc.2016.12.001 [DOI] [PubMed] [Google Scholar]
- 8.Shi Y, Sun H. Down-regulation of lncRNA LINC00152 suppresses gastric cancer cell migration and invasion through inhibition of the ERK/MAPK signaling pathway. Onco Targets Ther. 2020;13:2115–2124. doi: 10.2147/OTT.S217452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li K, Jiang Y, Xiang X, et al. Long non-coding RNA SNHG6 promotes the growth and invasion of non-small cell lung cancer by downregulating miR-101-3p. Thorac Cancer. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Gu M, Ju Y, Zhou J. PIK3C3 acts as a tumor suppressor in esophageal squamous cell carcinoma and was regulated by MiR-340-5p. Med Sci Monit. 2020;26:e920642. doi: 10.12659/MSM.923909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei L, Chen Z, Cheng N, et al. MicroRNA-126 inhibit viability of colorectal cancer cell by repressing mTOR induced apoptosis and autophagy. Onco Targets Ther. 2020;13:2459–2468. doi: 10.2147/OTT.S238348 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Chen Y, Shen Z, Zhi Y, et al. Long non-coding RNA ROR promotes radioresistance in hepatocellular carcinoma cells by acting as a ceRNA for microRNA-145 to regulate RAD18 expression. Arch Biochem Biophys. 2018;645:117–125. doi: 10.1016/j.abb.2018.03.018 [DOI] [PubMed] [Google Scholar]
- 13.Zeng F, Wang Q, Wang S, et al. Linc00173 promotes chemoresistance and progression of small cell lung cancer by sponging miR-218 to regulate Etk expression. Oncogene. 2020;39(2):293–307. doi: 10.1038/s41388-019-0984-2 [DOI] [PubMed] [Google Scholar]
- 14.Fan H, Yuan J, Li X, et al. LncRNA LINC00173 enhances triple-negative breast cancer progression by suppressing miR-490-3p expression. Biomed Pharmacother. 2020;125:109987. doi: 10.1016/j.biopha.2020.109987 [DOI] [PubMed] [Google Scholar]
- 15.Sun SL, Shu YG, Tao MY. LncRNA CCAT2 promotes angiogenesis in glioma through activation of VEGFA signalling by sponging miR-424. Mol Cell Biochem. 2020;468(1–2):69–82. doi: 10.1007/s11010-020-03712-y [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Jiang X, Wu Z, et al. Long noncoding RNA LINC00467 promotes glioma progression through inhibiting P53 expression via binding to DNMT1. J Cancer. 2020;11(10):2935–2944. doi: 10.7150/jca.41942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Q, Tang Y, Tang C, et al. Diminished LINC00173 expression induced miR-182-5p accumulation promotes cell proliferation, migration and apoptosis inhibition via AGER/NF-kappaB pathway in non-small-cell lung cancer. Am J Transl Res. 2019;11(7):4248–4262. [PMC free article] [PubMed] [Google Scholar]
- 18.Xue J, Zhang Z, Li X, Ren Q, Wang Q. Long non-coding RNA TTN-AS1 promotes breast cancer cell migration and invasion via sponging miR-140-5p. Oncol Lett. 2020;19(2):1255–1260. doi: 10.3892/ol.2019.11222 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Liu F, Xiao Y, Ma L, Wang J. Regulating of cell cycle progression by the lncRNA CDKN2B-AS1/miR-324-5p/ROCK1 axis in laryngeal squamous cell cancer. Int J Biol Markers. 2020;35(1):47–56. doi: 10.1177/1724600819898489 [DOI] [PubMed] [Google Scholar]
- 20.Ding J, Yang C, Yang S. LINC00511 interacts with miR-765 and modulates tongue squamous cell carcinoma progression by targeting LAMC2. J Oral Pathol Med. 2018;47(5):468–476. doi: 10.1111/jop.12677 [DOI] [PubMed] [Google Scholar]
- 21.Long S, Long S, He H, Chen G. MicroRNA-765 is preregulated in multiple myeloma and serves an oncogenic role by directly targeting SOX6. Exp Ther Med. 2019;17(6):4741–4747. doi: 10.3892/etm.2019.7473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lv DB, Zhang JY, Gao K, et al. MicroRNA-765 targets MTUS1 to promote the progression of osteosarcoma via mediating ERK/EMT pathway. Eur Rev Med Pharmacol Sci. 2019;23(11):4618–4628. doi: 10.26355/eurrev_201906_18040 [DOI] [PubMed] [Google Scholar]
- 23.Jiao Y, Yuan C, Wu H, Li X, Yu J. Oncogenic microRNA-765 promotes the growth and metastasis of breast carcinoma by directly targeting ING4. J Cell Biochem. 2019. [DOI] [PubMed] [Google Scholar]
- 24.Yuan L, Ma T, Liu W, et al. LINC00994 promoted invasion and proliferation of gastric cancer cell via regulating miR-765-3p. Am J Transl Res. 2019;11(10):6641–6649. [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H, Peng M, Li Y, Zhu R, Li X, Qian Z. LINC00703 acts as a tumor suppressor via regulating miR-181a/KLF6 axis in gastric cancer. J Gastric Cancer. 2019;19(4):460–472. doi: 10.5230/jgc.2019.19.e43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Qiu F, Huang L, et al. Long noncoding RNA LINC00312 regulates breast cancer progression through the miR9/CDH1 axis. Mol Med Rep. 2020;21(3):1296–1303. doi: 10.3892/mmr.2019.10895 [DOI] [PubMed] [Google Scholar]