Abstract
Background
As fat and obesity play a vital role in the pathophysiology of metabolic dysfunction-associated fatty liver disease (MAFLD), this study aims to investigate the association between the fat mass and obesity-associated gene (FTO) and MAFLD.
Methods
Six SNPs (rs6499640, rs1421085, rs8050136, rs3751812, rs9939609 and rs9930506) within FTO were genotyped for 741 MAFLD patients (median age, 69.98; interquartile range, 66.55–75.93) and 825 healthy people (median age, 69.94; interquartile range, 66.39–75.64). Allele and genotype frequencies, pairwise linkage disequilibrium (LD) and haplotype analysis were calculated.
Results
BMI, waist circumference, systolic blood pressure, diastolic blood pressure, fasting plasma glucose, triglyceride, alanine transaminase, glutamyl transpeptidase and the prevalence of diabetes were found to be higher in the MAFLD individuals comparing to the control ones (P < 0.05). For rs1421085, the C allele frequency was remarkably higher in MAFLD after Bonferroni correction (OR [95% CI] =1.353 [1.095–1.671]; Pcorr =0.030), and a significantly different genotype result was observed in log-additive model (OR [95% CI] =1.369 [1.108–1.691]; Pcorr =0.024). For rs8050136, significantly increased A allele frequency was observed in MAFLD (OR [95% CI] =1.371 [1.109–1.695]; Pcorr =0.024), and A-allele carriers showed increased MAFLD risk (OR [95% CI] =1.393 [1.103–1.759]; Pcorr =0.030). For rs3751812, the T allele frequency was remarkably higher in MAFLD (OR [95% CI] =1.369 [1.108–1.691]; Pcorr =0.024), and T-allele carriers demonstrated high MAFLD risk (OR [95% CI] =1.392 [1.103–1.756]; Pcorr =0.030). For rs9939609, A allele frequency was also remarkably high in MAFLD (OR [95% CI] =1.369 [1.108–1.691]; Pcorr =0.024), and A-allele carriers were more susceptible to MAFLD (OR [95% CI] =[1.103–1.756]; Pcorr =0.030). A strong LD was found among rs1421085, rs8050136, rs3751812 and rs9939609 (r2 >0.8), and individuals with C-A-T-A haplotype had an elevated MAFLD risk (P =0.005).
Conclusion
The case-control study indicated that C variant of rs1421085, A variant of rs8050136, T variant of rs3751812 and A variant of rs9939609 are associated with elevated MAFLD risk in the older Chinese Han population.
Keywords: MAFLD, FTO, polymorphism, genetic association, susceptibility
Background
Metabolic dysfunction-associated fatty liver disease (MAFLD), also known as non-alcoholic fatty liver disease (NAFLD), is widely recognized as the most common cause of chronic liver disease worldwide, affecting over one-quarter of the global population.1,2 The overall prevalence of MAFLD in Asia is estimated to be 29.6% up till now.2 Due to great changes in lifestyles and dietary habits, MAFLD has become a great social health burden in China, with prevalence of 18.2% in 2000–2006, 20.0% in 2007–2009, and 20.9% in 2010–2013.3,4 A recent meta-analysis reported that the national prevalence of MAFLD in China had reached up to 29.1%.4 MAFLD can manifest as non-alcoholic fatty liver or non-alcoholic steatohepatitis (NASH) and may lead to liver cirrhosis, hepatic failure and hepatocellular carcinoma.5,6
Both genetic and environmental factors can influence the pathogenesis of MAFLD, and genetic factors are believed to play a primary role in MAFLD occurrence. Increasing numbers of susceptibility genes for MAFLD have been reported recently, such as the patatin-like phospholipase domain-containing protein 3 (PNPLA3),7,8 transmembrane 6 superfamily 2 (TM6SF2),9 and adiponectin-encoding (ADIPOQ)10 among others. MAFLD patients are characterized by fatty liver without excessive alcohol consumption. Fat and obesity are related to the pathophysiology of MAFLD. A landmark study published in 2015 suggested that the single nucleotide variant rs1421085 of the fat mass and obesity-associated gene (FTO) has been shown to work as a super-enhancer in adipocyte regulating IRX3 and IRX5 genes, which involved in adipocyte browning and thermogenesis.11 The locus modulates the enhancer activity, and plays a dominant role in the body mass index (BMI), and possibly in obesity-based MAFLD as well.
FTO encodes a nuclear protein of 506 amino acids and shares sequence motifs with the Fe (II)- and 2-oxoglutarate-dependent oxygenases, making it able to alter DNA methylation and regulate gene transcription.12 Genome-wide association studies (GWAS) repeatedly indicated that variants within the introns of FTO were related to an increased risk for obesity in humans.13–15 Experimental research has demonstrated that FTO gene expression is upregulated in the liver of MAFLD rats and the overexpression of FTO can characterize MAFLD via increasing oxidative stress and lipid overaccumulation in human hepatocyte cells.16 Additionally, it was reported that FTO and the forkhead transcription factor O1 gene (FoxO1) cooperatively contribute to the disturbance of lipid metabolism and the interactions between FTO and FoxO1 are closely related to the pathogenesis of MAFLD.17
Six single nucleotide polymorphisms (SNPs) within FTO (rs6499640, rs1421085, rs8050136, rs3751812, rs9939609 and rs9930506) have been shown to reach genome-wide significance for obesity from previous GWAS studies.13–15,18 Given the predictive potential of FTO, we hypothesized that FTO could influence the occurrence of MAFLD. We performed an association study in a Chinese Han population to in-depth investigate the relationship between FTO and MAFLD.
Methods
Subjects
The study was performed according to the guidelines of the Helsinki Declaration in 1975, revised in 2000. A standard protocol was designed by Shanghai Innovation Center of TCM Health Service. This study was approved by the Ethics Committee of Shanghai University of Traditional Chinese Medicine. A total of 1566 participants took part in this case-control study from April 2017 to July 2018. All the participants signed the informed consent voluntarily. They met the following criteria: Permanent residents of Zhangjiang area of Pudong District, Shanghai; Chinese Han population with no blood relationship to each other; all medical examination information, biochemical and specimen collection information were complete and reliable. Subjects who abused alcohol (< 140 g/week in male and < 70 g/week in female), or were carriers of hepatitis B or C, or had a history of drug-induced liver disease or autoimmune liver disease were excluded. The MAFLD was diagnosed according to the guidelines for the management of MAFLD of the Chinese Medical Association in 2010.19 The color ultrasound system was employed by two experienced radiologists to evaluate fatty liver. Based on the criteria above, 741 MAFLD patients with median (interquartile range) age being 69.98 (66.55–75.93) and 825 gender-matched healthy controls from the same geographic areas with median (interquartile range) age being 69.94 (66.39–75.64) recruited in this study.
The baseline information such as age, gender, alcohol consumption, current smoking and medical history were collected by questionnaire. Subjects were required to wear light clothing without hats and shoes when measuring height and bodyweight, to the nearest 0.1cm and 0.1kg by electronic measurement instrument (Shengyuan, Zhengzhou, China). Body mass index (BMI) was calculated according to the formula bodyweight (kg)/height2 (m2). Blood pressure was measured by electronic sphygmomanometers (Biospace, Cheonan, South Korea). Blood samples were obtained from the antecubital vein in the morning after an overnight fasting period. Fasting plasma glucose (FPG), total cholesterol (TC), triglyceride (TG), low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), alanine transaminase (ALT), aspartate transaminase (AST) and gamma-glutamyl transferase (GGT) were measured using an automatic biochemistry analyzer (Hitachi, Tokyo, Japan).
DNA Extraction, SNP Selection and Genotyping
5mL of peripheral venous blood were collected from each subject. Then, the genomic DNA was extracted using the standard phenol-chloroform method for genotyping. SNPs were selected based on positive reported variants for obesity from previous GWAS studies.13–15,18 Six SNPs (rs6499640, rs1421085, rs8050136, rs3751812, rs9939609 and rs9930506) were genotyped in this study. Among them, all these six SNPs were successfully genotyped by matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry using the MassARRAY® Analyzer 4 platform (Sequenom, San Diego, CA). The probes and primers were designed using Mysequenom online software with the accompanying Assay Design Suite v2.0. The standard polymerase chain reaction (PCR) was performed in a total volume of 5 μL containing 10 ng of genomic DNA. Detailed information regarding the primers and PCR reaction conditions is available on request.
Statistical Analysis
R programming language (Lucent Technologies, New Jersey, USA) was adopted to analyze baseline characteristics. The allele and genotype distributions were analyzed using SHEsis (http://analysis.bio-x.cn/myAnalysis.php) (Bio-X Institutes, Shanghai, China).20 The data normalization was performed by R as well. Variables with normal distribution are presented as mean ± standard deviation and then compared using two-sided Student’s T-tests. Skewed variables are presented as median (interquartile range) and then compared using Wilcoxon’s rank-sum test. Categorical variables are expressed as counts or percentages and were compared using Pearson’s χ2 tests. Hardy-Weinberg equilibrium (HWE) was assessed by Pearson’s χ2 tests. Odds ratios (OR) and their 95% confidence intervals (CIs) were also assessed. The criteria for r2 were set at 0.8 for defining a strong pairwise linkage disequilibrium (LD) using Haploview 4.221 (Broad Institute, Cambridge, MA, USA) and Haplotype analysis conducted via SHEsis.22 For the analyses of the six SNPs, we perform Bonferroni correction (Pcoor =P-value*6) to prevent an inflation of type I error. Pcoor <0.05 was identified as statistical significance. The power of the sample size was calculated by G*power software version 3.1.23
Results
Demographics of Study Subjects
The overall clinical and laboratory characteristics of the 741 MAFLD patients and the 825 controls are listed in Table 1. Age or gender differences between MAFLD and control groups were neglectable. BMI, waist circumference (WC), systolic blood pressure, diastolic blood pressure, FPG, TG, ALT, GGT and the prevalence of diabetes were found to be higher in the MAFLD group than the control group (P <0.05). HDL-C was lower in the MAFLD group than in the control group (P <0.05).
Table 1.
Baseline Characteristics of Study Subjects
| Variables | MAFLD (n = 741) | Control (n = 825) | P |
|---|---|---|---|
| Age (years) | 69.98 (66.55–75.93) | 69.94 (66.39–75.64) | 0.691 |
| Male, n (%) | 296 (39.95%) | 351 (42.55%) | 0.297 |
| BMI (kg/m2) | 25.90 (24.08–27.99) | 24.24 (22.03–26.33) | < 0.001 |
| WC (cm) | 96 (92–100) | 93 (90–97) | < 0.001 |
| SBP (mmHg) | 145.08 ± 21.37 | 138.27 ± 24.62 | < 0.001 |
| DBP (mmHg) | 84 (76–90) | 80 (74.00–89.25) | < 0.001 |
| FPG (mmol/L) | 6 (5.50–7.00) | 5.8 (5.20–6.50) | < 0.001 |
| TC (mmol/L) | 5.14 (4.498–5.73) | 5.09 (4.44–5.75) | 0.421 |
| TG (mmol/L) | 2.01 ± 1.19 | 1.74 ± 1.13 | < 0.001 |
| LDL-C (mmol/L) | 3.21 (2.66–3.72) | 3.15 (2.60–3.72) | 0.210 |
| HDL-C (mmol/L) | 1.12 (0.97–1.28) | 1.17 (1.02–1.37) | < 0.001 |
| ALT (U/L) | 19.00 (14.00–27.00) | 18.00 (13.00–24.00) | < 0.001 |
| AST (U/L) | 22.00 (19.00–26.00) | 22 (19.00–26.25) | 0.612 |
| GGT (U/L) | 26.00 (20.00–36.00) | 23.00 (17.00–33.00) | < 0.001 |
| Diabetes | 189 (25.51%) | 165 (20.00%) | 0.009 |
Notes: Variables with normal distribution were presented as mean ± standard deviation. Skewed variables were presented as median (interquartile range).
Abbreviations: BMI, body mass index; WC, waist circumference; SBP, systolic blood pressure; DBP, diastolic blood pressure; FPG, fasting plasma glucose; TC, total cholesterol; TG, triglyceride; LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; ALT, alanine transaminase; AST, aspartate transaminase; GGT, glutamyl transpeptidase.
Single SNP Site Associations
The detailed information of six SNPs are listed in Table 2. For all six SNPs, there was no significant deviation from HWE in either the case or control groups (P >0.05). The allele frequencies of rs1421085, rs8050136, rs3751812 and rs9939609 were significantly different between MAFLD and controls (Pcorr <0.05). The C allele frequency of rs1421085 was remarkably higher in MAFLD patients (OR=1.353; 95% CI=1.095–1.671; P =0.005; Pcorr =0.030). The occurrence of A allele of rs8050136 was significantly increased in MAFLD group (OR=1.371; 95% CI=1.109–1.695; P =0.004; Pcorr =0.024). In rs3751812, the T allele frequency was remarkably higher in MAFLD (OR=1.369; 95% CI=1.108–1.691; P =0.004; Pcorr =0.024). And the A allele frequency of rs9939609 was also significantly increased in MAFLD (OR=1.369; 95% CI=1.108–1.691; P =0.004; Pcorr =0.024).
Table 2.
The Basic Information of SNPs and Allele Frequencies in MAFLD Patients and Controls
| SNP | Allele Frequency | Genotype Frequency | HWE | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF | OR (95% CI) | P | Pcorr | P | Pcorr | |||||||
| rs6499640 | G/A | G/G | A/G | A/A | ||||||||
| Case | 1235/247 | 0.17 | 513 | 209 | 19 | 0.916 | ||||||
| Control | 1383/267 | 0.16 | 1.036 (0.857–1.252) | 0.715 | >1.000 | 576 | 231 | 18 | 0.875 | >1.000 | 0.652 | |
| rs1421085 | T/C | T/T | T/C | C/C | ||||||||
| Case | 1268/214 | 0.14 | 541 | 186 | 14 | 0.911 | ||||||
| Control | 1467/183 | 0.11 | 1.353 (1.095–1.671) | 0.005 | 0.03 | 649 | 169 | 7 | 0.014 | >1.000 | 0.539 | |
| rs8050136 | C/A | C/C | C/A | A/A | ||||||||
| Case | 1269/213 | 0.14 | 541 | 187 | 13 | 0.788 | ||||||
| Control | 1470/180 | 0.11 | 1.371 (1.109–1.695) | 0.004 | 0.024 | 652 | 166 | 7 | 0.012 | >1.000 | 0.6 | |
| rs3751812 | G/T | G/G | G/T | T/T | ||||||||
| Case | 1267/215 | 0.15 | 539 | 189 | 13 | 0.744 | ||||||
| Control | 1468/182 | 0.11 | 1.369 (1.108–1.691) | 0.004 | 0.024 | 650 | 168 | 7 | 0.011 | >1.000 | 0.559 | |
| rs9939609 | T/A | T/T | T/A | A/A | ||||||||
| Case | 1267/215 | 0.15 | 539 | 189 | 13 | 0.744 | ||||||
| Control | 1468/182 | 0.11 | 1.369 (1.108–1.691) | 0.004 | 0.024 | 650 | 168 | 7 | 0.011 | >1.000 | 0.559 | |
| rs9930506 | A/G | A/A | A/G | G/G | ||||||||
| Case | 1190/292 | 0.2 | 482 | 226 | 33 | 0.616 | ||||||
| Control | 1370/280 | 0.17 | 1.201 (1.001–1.439) | 0.048 | 0.288 | 564 | 242 | 19 | 0.043 | >1.000 | 0.501 | |
Abbreviations: SNP, single nucleotide polymorphisms; MAF, minor allele frequency; HWE, Hardy–Weinberg equilibrium; OR, odds ratio; CI, confidence interval; Pcorr, P value after Bonferroni correction.
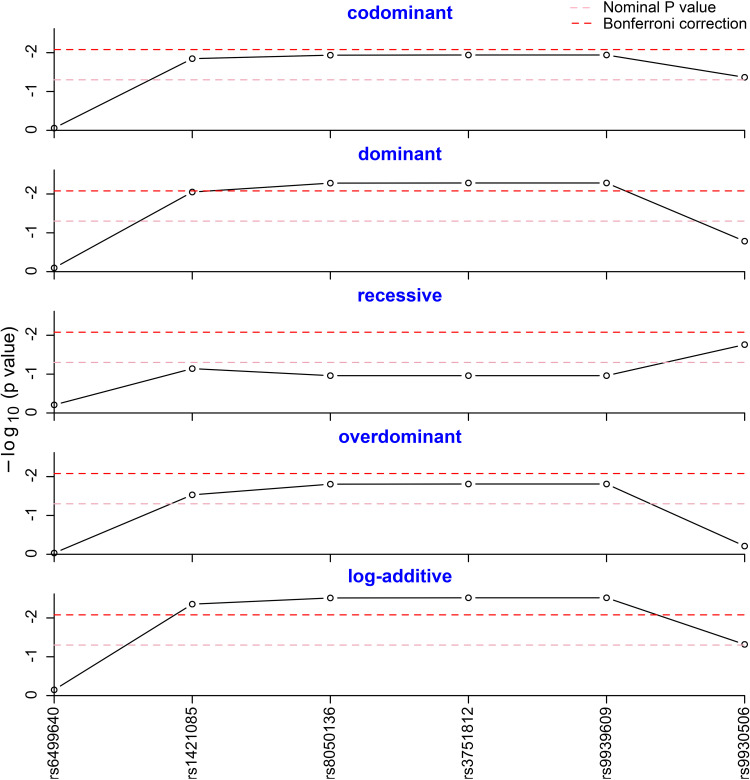
Five different genetic models (codominant, dominant, recessive, overdominant and log-additive) were tested to assess the association of genotype frequencies with MAFLD. As shown in Figure 1, the dominant model of rs8050136, rs3751812 and rs9939609 was statistically different after Bonferroni correction, as well as the log-additive model of rs1421085, rs8050136, rs3751812 and rs9939609 (Pcorr <0.05). As described in Table 3, a significantly different result was observed in log-additive model of rs1421085 (OR=1.369; 95% CI=1.108–1.691; P =0.004; Pcorr =0.024). For rs8050136, A-allele carriers were found to have significantly increased risk of MAFLD with an OR of 1.393 (Pcorr =0.030, 95% CI =1.103–1.759), compared to the carriers of homozygous CC genotype. For rs3751812, T-allele carriers were observed to strikingly add MAFLD risk with an OR of 1.392 (Pcorr =0.030, 95% CI = 1.103–1.756). For rs9939609, individuals with T-allele were also detected to be more susceptible to MAFLD, compared to the individuals of homozygous TT genotype with an OR of 1.392 (Pcorr =0.030, 95% CI = 1.103–1.756). Given the effect sizes of 0.1, we obtained the power above 95% at p = 0.05. The SNP genetic models without significant differences can be found in Supplementary Table 1.
Figure 1.
Genotype frequency analysis in five genetic models. The genotype frequencies of rs8050136, rs3751812 and rs9939609 were statistically different in the dominant model after Bonferroni correction. The genotype frequencies of rs1421085, rs8050136, rs3751812 and rs9939609 were statistically different in the log-additive model after Bonferroni correction.
Table 3.
Genotype Distributions in MAFLD Patients and Controls
| SNP | Model | Case | Control | OR (95% CI) | P | Pcorr | |
|---|---|---|---|---|---|---|---|
| rs1421085 | Log-additive | – | 741 | 825 | 1.364 (1.101–1.690) | 0.004 | 0.024 |
| rs8050136 | Dominant | C/C | 541 | 652 | 1 | 0.005 | 0.030 |
| C/A-A/A | 200 | 173 | 1.393 (1.103–1.759) | ||||
| Log-additive | – | 741 | 825 | 1.384 (1.115–1.718) | 0.003 | 0.018 | |
| rs3751812 | Dominant | G/G | 539 | 650 | 1 | 0.005 | 0.030 |
| G/T-T/T | 202 | 175 | 1.392 (1.103–1.756) | ||||
| Log-additive | – | 741 | 825 | 1.383 (1.115–1.715) | 0.003 | 0.018 | |
| rs9939609 | Dominant | T/T | 539 | 650 | 1 | 0.005 | 0.030 |
| T/A-A/A | 202 | 175 | 1.392 (1.103–1.756) | ||||
| Log-additive | – | 741 | 825 | 1.383 (1.115–1.715) | 0.003 | 0.018 | |
Note: The table above was the genetic model whose results are positive for each locus analysis.
Abbreviation: Pcorr, P value after Bonferroni correction.
Haplotype Analysis
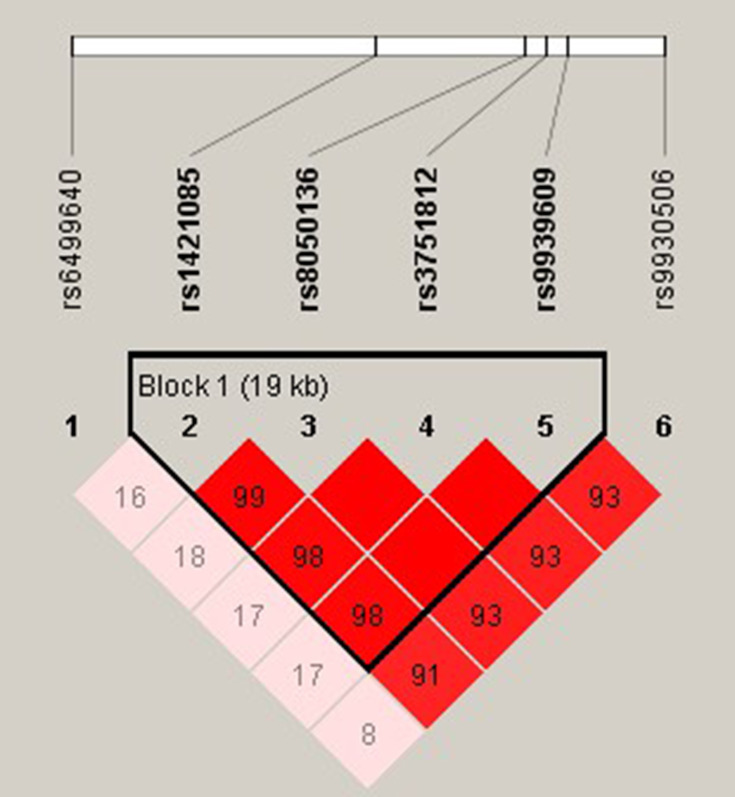
The r2 were calculated for all random combinations of SNPs. We identified rs1421085-rs8050136-rs3751812-rs9939609 as a strong LD block within FTO gene with Haploview analysis (Figure 2). Haplotypes in our study with an estimated frequency below 3% in both cases and controls were excluded from further analysis. We found that the haplotype of TCGT (rs1421085-rs8050136-rs3751812-rs9939609) can appreciably lower the risk of MAFLD (P =0.002; OR = 0.728; 95% CI =0.590–0.898) and that individuals with CATA haplotype had an elevated MAFLD risk (P =0.005; OR =1.348; 95% CI 1.089–1.667) (Table 4).
Figure 2.
Linkage disequilibrium plot between SNPs. The number in each square is r2*100 between two SNPs. As shown in the picture above rs1421085-rs8050136-rs3751812-rs9939609 was identified as a strong block.
Table 4.
Frequencies of Estimated Haplotypes and Test Statistics Between the MAFLD and Control
| Haplotype | Case | Control | OR (95% CI) | P |
|---|---|---|---|---|
| C A T A | 210 | 180 | 1.348 (1.089–1.667) | 0.005 |
| T C G T | 1263 | 1465 | 0.728 (0.590–0.898) | 0.002 |
Note: Haplotypes with a frequency under 3% were excluded.
Discussion
The role of gene FTO in MAFLD within Chinese Han population was evaluated in the present study. Rs1421085, rs8050136, rs3751812 and rs9939609 within FTO were found significantly associated with MAFLD based on our case-control study in 1566 older Chinese Han subjects. These four FTO SNPs demonstrated significant MAF differences between MAFLD patients and controls. The C variant of rs1421085, A variant of rs8050136, T variant of rs3751812, A variant of rs9939609 increased MAFLD risk. To the best of our knowledge, this is the first genetic association study to investigate the relationship between these six SNPs within FTO gene and MAFLD in an elderly Chinese population.
Although numerous studies have confirmed that FTO polymorphisms are closely related to the risk of obesity, only very few studies have explored the genetic discrepancies of FTO between MAFLD and non-MAFLD subjects. A Spanish study found that three SNPs (rs8050136, rs9939609 and rs9940128) within the FTO gene were associated with fatty liver disease in HIV-infected patients and suggested that variations within FTO may be predictors of fatty liver disease in HIV-infected patients, independent of metabolic factors.24 This study supported our finding that rs8050136 and rs9939609 were associated with MAFLD. Additionally, individuals with the rs9939609 AA genotype were found to present higher likelihoods of liver steatosis (>10% fatty hepatocytes) among HIV/HCV-coinfected European white patients in a cross-sectional study.25 Behaviorally, Harbon et al found the relationship between rs1421085 within FTO and greater fat and refined starch intakes in 133 overweight/obese Caucasians.26 Besides, FTO has been reported to play a role in individual’s preference for high-fat foods,27 caused by reduced satiety responsiveness,28 leading to increased consumption of highly palatable food. Another notable study pointed out that excess caloric consumption may be a leading risk factor for MAFLD.1 In supporting these evidences, our results may explain the increased the risk of MAFLD which may be due to an individual’s excessive dietary fat, from the perspective of genetic variance.
The molecular pathogenesis of MAFLD is complex and multifactorial, with oxidative stress and insulin resistance as major pathophysiological mechanisms. Genetic factors influence MAFLD development as they hold the key to better understanding the pathogenesis and developing new and better treatments.29 A recent landmark study showed that a variant within FTO (rs1421085 T>C) led to the derepression of a potent preadipocyte enhancer and a doubling of IRX3 and IRX5 expression during early adipocyte differentiation by disrupting the conserved sequence of the ARID5B repressor. This led the autonomous development of cells from energy-consuming adipocytes to energy-storing white one, reducing mitochondrial heat generation by 5 times and increasing lipid storage which have been reported to be leading risk factors for MAFLD.11 Interestingly, FTO is highly expressed in the hypothalamus. And individuals with rs9939609 AA genotype have impaired satiety in the central nervous system, which increases food intake and increases the risk of obesity.30 These evidences together with our genetic association study all support the hypothesis that FTO influences the occurrence of MAFLD and might be a new significant therapeutic target for MAFLD treatment.
Several studies focused on the age-related association of FTO polymorphisms and food cravings. It has been found that food cravings declined with age, but this age effect was different across variants of FTO rs9939609, while TT homozygotes showed the typical age-related decline in food cravings, as well as no such decline among A-carriers.31 Besides, the FTO genotype have effect on the relationship between age and emotional eating, with age-related decline in this behavior only present in the A-carriers.32 In our study, allele A of rs9939609 was associated with higher MAFLD risk in the old. Our finding may result in that A-carriers have higher emotional to eating which has been proven that subjects homozygous for the FTO rs9939609 A allele have dysregulated circulating levels of the orexigenic hormone acyl-ghrelin, a key appetite-regulating hormone.33
The results of the LD analysis indicated that rs1421085-rs8050136-rs3751812-rs9939609 were in strong LD and that the frequencies of haplotypes with different combinations of the four polymorphisms significantly differed in the MAFLD and control groups. The individual haplotype CATA was a genetic risk factor for MAFLD, while the haplotype TCGT was a genetic protective factor for MAFLD. To the best of our knowledge, this is the first time that rs1421085, rs8050136, rs3751812 and rs9939609 have been shown to form a haplotype block associated with MAFLD.
Overall, despite the limitation of insufficient coverage of SNPs in the candidate gene, further genetic studies with more comprehensive SNPs covering FTO and a larger sample size are needed to further evaluate or conform the role of the FTO gene in MAFLD. While the Ultrasonography used to detect MAFLD in our study was a good parameter for measuring fatty liver, it might cause concern that it is not a good enough gold standard compared to histology. But histology specimens are hard to obtain, and a meta-analysis compared these two methods and demonstrated that Ultrasonography is reliable and accurate detection of moderate-severe fatty liver with low cost, safety, and accessibility.34 In addition, this study did not assess the food intake of the subjects which should be taken into consideration in the future works.
Conclusions
In summary, our results demonstrate that the A variant of rs8050136, T variant of rs3751812 and A variant of rs9939609 within FTO gene are associated with elevated MAFLD in the older Chinese Han population. These findings indicate that it is worth exploring the role of FTO in MAFLD, and further functional investigations are needed to elucidate its molecular mechanisms.
Acknowledgments
The writing of this article was supported by Shanghai Innovation Center of TCM Health Service of Shanghai University of Traditional Chinese Medicine. We thank all the partners and staff who helped us in the process of this study.
Funding Statement
This work is supported by Three-year action plan for Shanghai (project number: ZY (2018-2020)-CCCX-2001-01), National Natural Science Foundation of China (81973730, 81603411), 2020 Shanghai Innovation Center Development (2020 Science and Technology 01-01-30).
Abbreviations
BMI, body mass index; CI, confidence intervals; FTO, fat mass and obesity-associated gene; GWAS, genome-wide association studies; HWE, Hardy–Weinberg equilibrium; LD, linkage disequilibrium; MAF, minor allele frequency; MAFLD, metabolic dysfunction-associated fatty liver disease; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; OR, odds ratios; SNP, single nucleotide polymorphism; WC, waist circumference.
Ethics Approval and Consent to Participate
The study was approved by the Ethics Committee of Shanghai University of Traditional Chinese Medicine. Written informed consent was obtained from all subjects voluntarily.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64(1):73–84. doi: 10.1002/hep.28431 [DOI] [PubMed] [Google Scholar]
- 2.Wong SW, Chan WK. Epidemiology of non-alcoholic fatty liver disease in Asia. Indian J Gastroenterol. 2020;39(1):1–8. doi: 10.1007/s12664-020-01018-x [DOI] [PubMed] [Google Scholar]
- 3.Li Z, Xue J, Chen P, Chen L, Yan S, Liu L. Prevalence of nonalcoholic fatty liver disease in mainland of China: a meta-analysis of published studies. J Gastroenterol Hepatol. 2014;29(1):42–51. doi: 10.1111/jgh.12428 [DOI] [PubMed] [Google Scholar]
- 4.Fan JG, Kim SU, Wong VW. New trends on obesity and NAFLD in Asia. J Hepatol. 2017;67(4):862–873. doi: 10.1016/j.jhep.2017.06.003 [DOI] [PubMed] [Google Scholar]
- 5.Estes C, Razavi H, Loomba R, Younossi Z, Sanyal AJ. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalasani N, Younossi Z, Lavine JE, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology. 2012;55(6):2005–2023. doi: 10.1002/hep.25762 [DOI] [PubMed] [Google Scholar]
- 7.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40(12):1461–1465. doi: 10.1038/ng.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Xing C, Cohen JC, Hobbs HH. Genetic variant in PNPLA3 is associated with nonalcoholic fatty liver disease in China. Hepatology. 2012;55(1):327–328. doi: 10.1002/hep.24659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong VW, Wong GL, Tse CH, Chan HL. Prevalence of the TM6SF2 variant and non-alcoholic fatty liver disease in Chinese. J Hepatol. 2014;61(3):708–709. doi: 10.1016/j.jhep.2014.04.047 [DOI] [PubMed] [Google Scholar]
- 10.Gupta AC, Misra R, Sakhuja P, Singh Y, Basir SF, Sarin SK. Association of adiponectin gene functional polymorphisms (−11377C/G and +45T/G) with nonalcoholic fatty liver disease. Gene. 2012;496(1):63–67. doi: 10.1016/j.gene.2011.12.023 [DOI] [PubMed] [Google Scholar]
- 11.Claussnitzer M, Dankel SN, Kim KH, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373(10):895–907. doi: 10.1056/NEJMoa1502214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerken T, Girard CA, Tung YC, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–1472. doi: 10.1126/science.1151710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39(6):724–726. doi: 10.1038/ng2048 [DOI] [PubMed] [Google Scholar]
- 15.Scuteri A, Sanna S, Chen WM, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115. doi: 10.1371/journal.pgen.0030115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo J, Ren W, Li A, et al. Fat mass and obesity-associated gene enhances oxidative stress and lipogenesis in nonalcoholic fatty liver disease. Dig Dis Sci. 2013;58(4):1004–1009. doi: 10.1007/s10620-012-2516-6 [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Li S, Li J, et al. Expression and significance of fat mass and obesity associated gene and forkhead transcription factor O1 in non-alcoholic fatty liver disease. Chin Med J (Engl). 2014;127(21):3771–3776. [PubMed] [Google Scholar]
- 18.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41(1):18–24. doi: 10.1038/ng.274 [DOI] [PubMed] [Google Scholar]
- 19.Fan JG, Jia JD, Li YM, et al. Guidelines for the diagnosis and management of nonalcoholic fatty liver disease: update 2010: (published in Chinese on Chinese Journal of Hepatology 2010; 18: 163–166). J Dig Dis. 2011;12(1):38–44. doi: 10.1111/j.1751-2980.2010.00476.x [DOI] [PubMed] [Google Scholar]
- 20.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97–98. doi: 10.1038/sj.cr.7290272 [DOI] [PubMed] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 22.Li Z, Zhang Z, He Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. 2009;19(4):519–523. [DOI] [PubMed] [Google Scholar]
- 23.Faul F, Erdfelder E, Buchner A, Lang AG. Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav Res Methods. 2009;41(4):1149–1160. doi: 10.3758/BRM.41.4.1149 [DOI] [PubMed] [Google Scholar]
- 24.Nunez-Torres R, Macias J, Rivero-Juarez A, et al. Fat mass and obesity-associated gene variations are related to fatty liver disease in HIV-infected patients. HIV Med. 2017;18(8):546–554. doi: 10.1111/hiv.12489 [DOI] [PubMed] [Google Scholar]
- 25.Pineda-Tenor D, Berenguer J, Jimenez-Sousa MA, et al. FTO rs9939609 polymorphism is associated with metabolic disturbances and response to HCV therapy in HIV/HCV-coinfected patients. BMC Med. 2014;12:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harbron J, van der Merwe L, Zaahl MG, Kotze MJ, Senekal M. Fat mass and obesity-associated (FTO) gene polymorphisms are associated with physical activity, food intake, eating behaviors, psychological health, and modeled change in body mass index in overweight/obese Caucasian adults. Nutrients. 2014;6(8):3130–3152. doi: 10.3390/nu6083130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CNA. An obesity-associated FTO gene variant and increased energy intake in children. N Engl J Med. 2008;359(24):2558–2566. doi: 10.1056/NEJMoa0803839 [DOI] [PubMed] [Google Scholar]
- 28.Wardle J, Carnell S, Haworth CMA, Farooqi IS, O’Rahilly S, Plomin R. Obesity associated genetic variation in FTO is associated with diminished satiety. J Clin Endocrinol Metab. 2008;93(9):3640–3643. doi: 10.1210/jc.2008-0472 [DOI] [PubMed] [Google Scholar]
- 29.Severson TJ, Besur S, Bonkovsky HL. Genetic factors that affect nonalcoholic fatty liver disease: a systematic clinical review. World J Gastroenterol. 2016;22(29):6742–6756. doi: 10.3748/wjg.v22.i29.6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melhorn SJ, Askren MK, Chung WK, et al. FTO genotype impacts food intake and corticolimbic activation. Am J Clin Nutr. 2018;107(2):145–154. doi: 10.1093/ajcn/nqx029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dang LC, Samanez-Larkin GR, Smith CT, et al. FTO affects food cravings and interacts with age to influence age-related decline in food cravings. Physiol Behav. 2018;192:188–193. doi: 10.1016/j.physbeh.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abdella HM, El Farssi HO, Broom DR, Hadden DA, Dalton CF; Eating Behaviours and Food Cravings. Influence of age, sex, BMI and FTO genotype. Nutrients. 2019;11(2):377. doi: 10.3390/nu11020377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karra E, O’Daly OG, Choudhury AI, et al. A link between FTO, ghrelin, and impaired brain food-cue responsivity. J Clin Invest. 2013;123(8):3539–3551. doi: 10.1172/JCI44403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernaez R, Lazo M, Bonekamp S, et al. Diagnostic accuracy and reliability of ultrasonography for the detection of fatty liver: a meta-analysis. Hepatology. 2011;54(3):1082–1090. doi: 10.1002/hep.24452 [DOI] [PMC free article] [PubMed] [Google Scholar]