Abstract
Although knowledge of the coordination chemistry and metal-withholding function of the innate immune protein human calprotectin (hCP) has broadened in recent years, understanding of its Ca2+-binding properties in solution remains incomplete. In particular, the molecular basis by which Ca2+ binding affects structure and enhances the functional properties of this remarkable transition-metal-sequestering protein has remained enigmatic. To achieve a molecular picture of how Ca2+ binding triggers hCP oligomerization, increases protease stability, and enhances antimicrobial activity, we implemented a new integrated mass spectrometry (MS)-based approach that can be readily generalized to study other protein-metal and protein-ligand interactions. Three MS-based methods, HDX-MS, PLIMSTEX and native MS, provided a comprehensive analysis of Ca2+ binding and oligomerization to hCP without modifying the protein in any way. Integration of these methods allowed us to (i) observe the four regions of hCP that serve as Ca2+ binding sites, (ii) determine the binding stoichiometry to be four Ca2+ per CP heterodimer and eight Ca2+ per CP heterotetramer, (iii) establish the protein-to-Ca2+ molar ratio that causes the dimer-to-tetramer transition, and (iv) calculate the binding affinities associated with the four Ca2+-binding sites per heterodimer. These quantitative results support a model in which hCP exists in its heterodimeric form and is at most half-bound to Ca2+ in the cytoplasm of resting cells. With release into the extracellular space, hCP encounters elevated Ca2+ concentrations and binds more Ca2+ ions, forming a heterotetramer that is poised to compete with microbial pathogens for essential metal nutrients.
Graphical Abstract
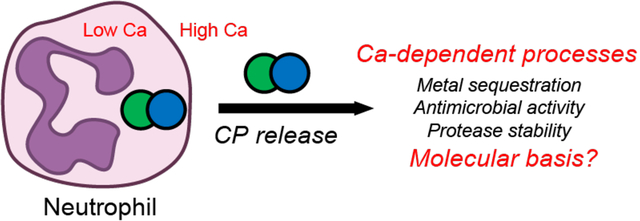
Introduction
Calprotectin (CP, S100A8/S100A9 oligomer, MRP8/MRP14 oligomer) is a Ca2+- and transition-metal-ion-binding protein and an abundant component of the innate immune system.1 CP functions as a high-affinity metal-ion chelator to compete with microbial pathogens for available transition metal ions that are essential nutrients, including Mn2+, Fe2+, Ni2+ and Zn2+.1–8 This nutrient-starvation strategy is one aspect of innate immunity, a front-line, non-specific defense strategy against invading pathogens to prevent their growth and ultimately infection.
In the current model for the metal-sequestering function of CP, white blood cells (i.e., neutrophils) and epithelial cells produce and release CP into the extracellular space in response to microbial invasion.1 In this milieu, the Ca2+ ions (≈ 2 mM)9 modulate the metal-withholding function of the protein. CP binds Ca2+ ions and transition metal ions at independent sites (vide infra). Many previous studies demonstrated that the Ca2+-bound form of CP exhibits enhanced transition metal ion affinities,3, 10–12 antimicrobial activity,2–3, 10 and resistance to serine proteases13 compared to the apo protein. Although the metal-sequestering function of this fascinating and complex system depends on an interplay of metal binding, conformational change, and self-association, the overall picture for these processes has remained enigmatic. We do not have a molecular model for how CP binds Ca2+ ions and how this binding affects the protein’s structure, dynamics, and biological function in metal-withholding and beyond. To address this lack of understanding, we implemented a novel, integrated mass spectrometry (MS)-based approach to unravel the complexities of Ca2+ binding to human CP (hCP).
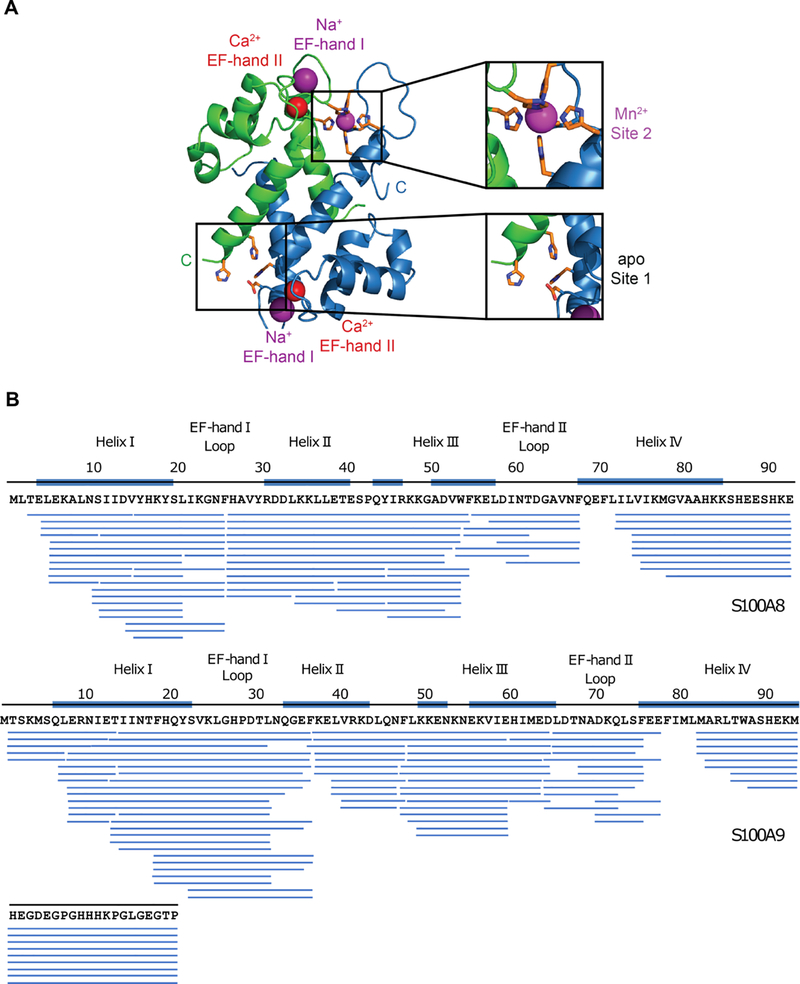
We chose Ca2+ binding to hCP because it is important in disease, and the majority of reported coordination chemistry studies of CP focused on this ortholog. hCP is a heterooligomer of two S100 proteins, S100A8 (α subunit, 10.8 kDa) and S100A9 (β subunit, 13.2 kDa).14 Each S100 subunit has two Ca2+-coordination sites: a C-terminal “canonical” EF-hand and an N-terminal “non-canonical” EF-hand.15 Apo hCP is a S100A8/S100A9 heterodimer, and Ca2+ binding to the EF-hands causes self-association of two heterodimers to form a heterotetramer10, 16–17 by a process that is not understood. Each S100A8/S100A9 heterodimer harbors two sites for coordinating transition metal ions (Figure 1A). These sites, at the heterodimer interface, include two motifs: His3Asp (site 1) and a His6 (site 2).3,10, 18–19 The His3Asp site sequesters Zn2+, and the functionally versatile His6 site withholds Mn2+, Fe2+, Ni2+ and Zn2+ from microbes.1 CP can also sequester copper, but the details of this coordination chemistry are unclear.20
Figure 1.
hCP structure and sequence coverage map of pepsin-digested CP-Ser. (A) Crystal structure of Mn2+-, Ca2+-, and Na+-bound CP-Ser (PDB 4XJK).18 A heterodimer unit taken from the heterotetramer is shown. S100A8 is green and S100A is blue. (B) Sequence coverage map of pepsin-digested S100A8(C42S) (top), and S100A9(C3S) (bottom) of CP-Ser. Each blue bar represents a peptide identified in a tandem mass spectrometry mapping experiment. Secondary structure elements are indicated by blue lines on top of the sequence. The Ca2+-binding loops are indicated by text only.
Our early studies of CP-Ser, a S100A8(C42S)/S100A9(C3S) variant of hCP, showed that Ca2+ ions enhance the Zn2+ affinities of both the His3Asp and His6 sites,10 and our subsequent work demonstrated that Ca2+ also enhances the binding affinities for Mn2+, Fe2+ and Ni2+.3, 11–12 Moreover, studies of Mn2+- and Fe2+-bound to the His6 site of CP-Ser indicated that Ca2+ binding does not affect the coordination geometry or the electronic structure of the His6 site.18, 21 In contrast, studies of Fe2+ and Ni2+ bound to the His3Asp site of CP-Ser showed that the coordination environment of this site is perturbed by Ca2+ binding.12, 21 Magnetic circular dichroism spectroscopy demonstrated that the Fe2+-His3Asp site is 5-coordinate in the absence and presence of bound Ca2+, but that the ligand-field strength decreases upon Ca2+ binding.21 Crystallographic evaluation of Ni2+-bound CP-Ser in the absence and presence of Ca2+ revealed that the coordination number of Ni2+ changed from 6 to 4 at the His3Asp site as a result of Ca2+ binding.12 Although these observations are informative, they do little to advance our understanding of how Ca2+ ions modulate the conformation and transition-metal binding of CP.
Two recent X-ray structures of metal-bound CP-Ser, obtained with crystallization conditions of 10 equivalents of Ca2+ per homodimer,5, 18 show a Ca2+ stoichiometry of < 4 per heterodimer. In the case of Mn2+-, Ca2+- and Na+-bound CP-Ser, the protein crystallized as a heterotetramer.18 Although anomalous electron density corresponding to bound Ca2+ was observed in the four C-terminal EF-hands of the heterotetramer, no evidence for bound Ca2+ was found in any of the four N-terminal EF-hands (Figure 1A).18 Na+ ions, abundant in the crystallization solution, were modeled into the N-terminal EF-hands. These results are consistent with the notion that the C-terminal EF-hands bind Ca2+ with higher affinity than the N-terminal EF-hands and demonstrate that 10 equivalents of Ca2+ are insufficient for Ca2+ binding at the N-terminal EF-hands, at least under the crystallization conditions. The results, however, do not provide an answer to the Ca2+ binding stoichiometry of CP-Ser or its behavior in solution, motivating the research described here.
Compared to investigating transition-metal binding to hCP, molecular-level insight into how Ca2+ interacts with hCP is difficult to achieve. Ca2+ is spectroscopically silent, each heterodimer has four different Ca2+-binding sites, and Ca2+-dependent self-association of heterodimers to form heterotetramers presents an additional variable that must be considered. As a result, methods routinely applied to study transition-metal binding to hCP – ranging from competition titrations for apparent binding affinities to advanced spectroscopic methods for the electronic structure determination of transition-metal-bound species – are poorly suited for evaluating Ca2+ binding.
Therefore, we implemented a combination of MS-based structural proteomics approaches that allow not only direct observation of Ca2+ binding to hCP but also assessment of oligomerization, and determination of apparent binding affinities. The approaches are (i) hydrogen/deuterium exchange MS (HDX-MS), (ii) protein ligand interactions in solution by mass spectrometry, titration and hydrogen/deuterium exchange (PLIMSTEX), and (iii) native mass spectrometry (native-MS). Importantly, the approaches apply to the native protein and do not require a surrogate for Ca2+.
HDX-MS measures relative rates of exchange of backbone amide hydrogen atoms with deuterium, permitting measurement of the changes in hydrogen-bonding and solvent accessibility accompanying both Ca2+-binding and self-association in solution. This approach facilitates identification of Ca2+-binding sites and protein-protein interfaces.22–25
PLIMSTEX provides an indirect readout of ligand binding in solution whereby the protein is titrated with a ligand (Ca2+) and its conformation is monitored by HDX at each titration point. These measurements can pertain to the entire protein or to regional (peptide) levels by following fragments formed by proteolytic digestion. In either case, the resulting data are fit to models to obtain apparent binding affinities (i.e., association (Ka) or dissociation (Kd) constants). This approach is particularly appropriate for proteins like CP that coordinate ligands with several stoichiometries.26–28
Lastly, employing native-MS to monitor ligand-binding titrations allows observation of protein oligomers resulting from ligand complexation. The titration is performed in solution, and the use of a volatile supporting electrolyte (ammonium acetate, NH4OAc) enables direct MS detection of the various oligomers with and without a bound ligand (in this case, Ca2+). This titration affords information on molecular weight, stoichiometry, and relative stabilities of protein-ligand complexes.29–30
Prior MS studies using HDX kinetics and PLIMSTEX for the Ca2+-binding proteins calmodulin31 and using HDX kinetics for troponin C27 and DREAM (Downstream Regulatory Element Antagonist Modulator)32 provided insight on conformational changes upon Ca2+ binding for systems that are less complex than CP. Unlike these prior studies, here we integrate data from HDX kinetics, PLIMSTEX, and native MS. This new, combined approach provides details about the apo heterodimer and the Ca2+-bound heterotetramer, uncovers the Ca2+-binding stoichiometry, and enables expanded modeling to give stepwise binding affinities. The combination, more than any single approach, delivers a detailed picture of how Ca2+ binding affects the structure and function of hCP. In particular, the experimental results accompanied by equilibrium modeling show pathways for Ca2+ binding and the tetramerization and provide equilibrium constants for the various steps. The approach should be useful for future investigations of CP and its role in infectious disease2, 6, 33 and other pathologies that include Alzheimer’s disease34 and cancer,35–36 and for other intricate metal-binding proteins.
Results and Discussion
HDX Kinetics Identify the Ca2+-binding Sites
We studied the hCP variant CP-Ser, a S100A8(C42S)/S100A9(C3S) oligomer, obtained from recombinant protein expression.10 CP-Ser allows experiments to be conducted without the complications associated with disulfide bonding (e.g., no need for a reducing agent in the HDX or other measurement). This Cys-null variant has been employed extensively in metal-binding and antimicrobial activity studies, and all available data indicate that it displays the same Ca2+-dependent heterotetramerization and transition-metal-binding properties as hCP.10, 37
We first measured HDX kinetics for CP-Ser at the peptide level in the absence and presence of excess Ca2+ ions. By examining the S100A8 and S100A9 peptide fragments obtained following pepsin digestion, we identified peptide fragments corresponding to each of the four EF-hand domains and the tetramer interface. A total of 65 (40 unique – duplicates with different charge states are not counted) and 85 (55 unique) peptides were identified in a mapping experiment for the S100A8 and S100A9 subunits, respectively, with nearly complete coverage (≈ 95%) for both the subunits (Figure 1B). We identified multiple overlapping peptides containing the Ca2+-binding sites of each subunit. A comparison of HDX for Ca2+-free and Ca2+-bound CP-Ser revealed protection (reduced HDX) upon Ca2+ binding for nearly the entire S100A8 subunit, except for regions represented by peptides 22–26 and 40–52. Likewise, protection of nearly the entire S100A9 subunit was observed, except regions 48–60 and 83–114 of the Ca2+-bound state. Protection increases in the presence of Ca2+ for both the S100A8 and S100A9 N- and C-terminal EF-hands and for the neighboring regions in each protein subunit (Figures S1 and S2).
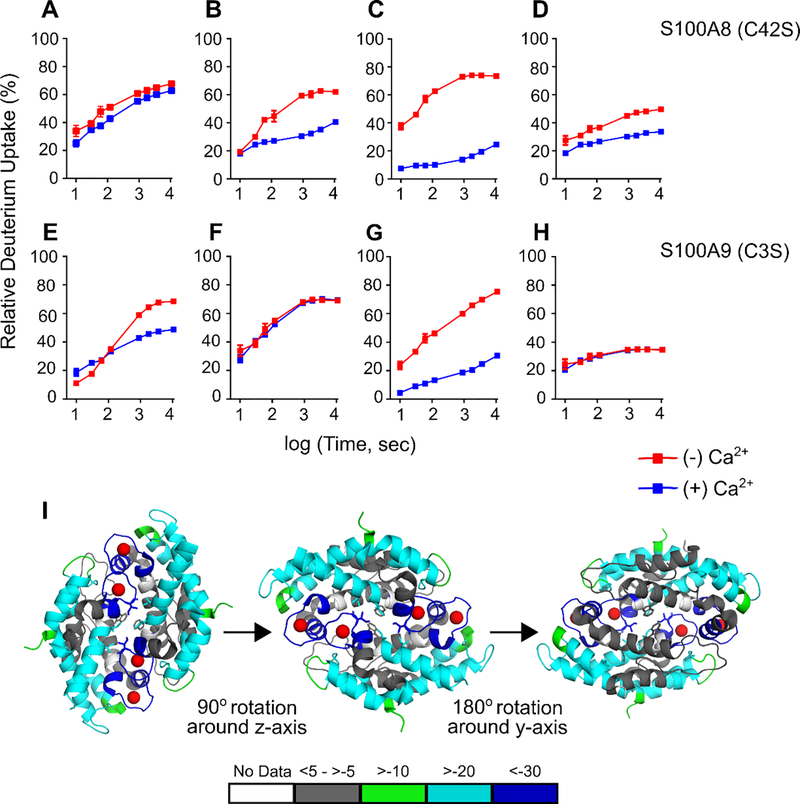
Upon Ca2+ binding, greater reduction in HDX is observed at the C-terminal EF-hands than at the N-terminal EF-hands; the extent of HDX decreases by as much as 59% at the C-terminal EF-hand of each subunit, indicating strong protection and a clear conformational change (Figures 2, S1 and S2). For example, peptides 16–26 and 27–34 (Figure 2A, B) from the N-terminal EF-hand of S100A8 exhibit smaller differences in HDX between bound and unbound than does peptide 55–68 from the C-terminal EF-hand of S100A8 (Figure 2C). Similarly, for S100A9, the C-terminal EF-hand region 65–77 (Figure 2G) shows greater protection to HDX compared to the N-terminal EF-hand peptide 22–36 (Figure 2E). These results are consistent with stronger Ca2+ binding at the canonical C-terminal EF-hands. These sites have greater negative charge and more oxygen donors compared to the non-canonical N-terminal EF-hands.15, 18 We also observed reduction in deuterium uptake upon Ca2+ binding for peptides adjoining the C-terminal Ca2+-binding site for S100A8 (Figure 2D), whereas regions of CP-Ser not involved in Ca2+ binding showed nearly invariant HDX behavior (Figure 2F).
Figure 2.
HDX studies of CP-Ser (5 μM) in the absence and presence of excess Ca2+ ions (250 μM). (A-H) Representative HDX kinetic plots for peptides from the CP-Ser subunits S100A8(C42S) (A-D) and S100A9(C3S) (E-H) in the absence (red) and presence (blue) of Ca2+ ions. Kinetic plots for peptides from EF-hands (A, B, C, E, and G) and other regions (D, F, H) are shown. Peptides sequences and associated charge states are: (A) YHKYSLIKGNF (S100A8(C42S) 16–26 (+2)), (B) HAVYRDDL (S100A8(C42S) 27–34 (+2)), (C) FKELDINTDGAVNF (S100A8(C24S) 55–68 (+2)), (D) VIKMGVAAHKKSHEESHKE (S100A8(C42S) 75–93 (+3)), (E) SVKLGHPDTLNQGEF (S100A9(C3S) 23–37 (+2)), (F) FLKKENKNEKVIE (S100A9 (C3S) 48–60 (+2)), (G) LDTNADKQLSFEE (S100A9(C3S) 66–78 (+2)), and (H) TWASHEKMHEGDEGPGHHHKPGLGEGTP (S100A9(C3S) 87–114 (+3), respectively. (I) Differential HDX uptake in % is mapped onto the crystal structure of the Ca2+-bound CP-Ser tetramer (PDB: 4XJK). Different views of the differential HDX map rotated 90° around z-axis from and 180° around the y-axis from are shown. Colored bars represent averaged HDX differences between apo and holo CP-Ser across all seven time points in duplicate. The extent of HDX is represented by colored regions in the map, gray regions mark no or little change with green, cyan and blue representing small, medium and large changes in deuterium uptake in the holo-state, respectively.
HDX indicates that Ca2+ binding has a negligible effect on the C-terminal tail of S100A9, defined as residues 96–114; this region is essential for metal sequestration by CP. In addition to containing two histidine residues that complete the His6 coordination sphere at site 2, the tail changes conformation upon coordination of a divalent metal ion at the His6 site and thereby effectively encapsulates the bound metal ion.18 We identified multiple peptides from the tail region of S100A9 (Figures 1B, S4), one of which covers residues 87–114. This peptide and others from this region of S100A9, display comparable HDX in the absence and presence of Ca2+ (Figure 2H), indicating that the enhanced transition-metal affinities that occur at the His6 site upon Ca2+ binding do not result from a Ca2+-dependent conformational change in the tail region. Because short, overlapping peptides from the tail region of S100A9 were not identified (lost during chromatography or lack of proteolysis), it remains possible that large changes in HDX with the addition of Ca2+ occur within this region but were not observed. In such a scenario, signals from residues strongly affected by Ca2+ would be diluted by those that change little in this 28-residue long peptide and would be missed.
To obtain a global picture of how Ca2+ binding affects HDX across the protein, we calculated the averaged deuterium uptake differences between CP-Ser with and without Ca2+ bound. We mapped these differences onto the crystal structure of the Ca2+-, Na+- and Mn2+-bound CP-Ser heterotetramer (PDB: 4XJK)18 as a model (Figure 2I). That structure contains Ca2+ ions bound at the C-terminal EF-hand domains, Na+ ions bound in the N-terminal EF-hand domains, and a Mn2+ ion bound at the His6 site, but only the Ca2+ ions are shown in the HDX model (Figure 2I). The structural map presented here shows that the CP-Ser heterotetramer exhibits significant protection in HDX at the Ca2+-binding sites, the heterotetramer interface, and at several regions remote from the Ca2+-binding sites for the S100A8 and S100A9 subunits. Because Ca2+ binding causes apo CP heterodimers to self-associate and form heterotetramers,10, 16–17 these results indicate that heterotetramer formation observed following Ca2+ addition decreases the extent of HDX.
The crystal structure shows the tetramer interface is primarily comprised of hydrophobic residues (Ile60, Ile73 and Ile76) from S100A8 and tryptophan (W88) from the S100A9 subunit. HDX interrogates the tetramer interface in solution and shows a gain in protection for regions represented by the S100A8 peptides containing Ile 60 (also in a Ca2+-binding site), Ile73, and Ile76 (Figures 2C, 2D, S1) but not for the tryptophan (W88)-containing peptide at the N-terminus of S100A9 (Figures 2H, S2). Any effect of W88 is presumably diluted by the large number of unaffected residues contained in that peptide, as discussed above.
PLIMSTEX Reports on the CP-Ser Heterodimer
The HDX kinetics provided a foundation for the application of PLIMSTEX to further interrogate the four Ca2+-binding events and determine the Ca2+-binding affinities of the EF-hand domains in CP-Ser (Figures 3 and 4). In a PLIMSTEX experiment, an appropriate exchange time from the HDX kinetics is chosen, and exchange at specific regions of the protein (e.g., the Ca2+-binding sites) is monitored in a titration format (see Experimental Methods), permitting determination of the binding affinity at each site. We titrated CP-Ser with increasing amounts of Ca2+ and identified the peptides that underwent significantly decreasing HDX with increasing [Ca2+], particularly peptides corresponding to the Ca2+-binding sites (Figure 3A,C,E,G) and peptides from the S100A8 that form the tetramer interface (Figure 3C,D). For instance, the peptide (55–68) from S100A8 contains both a Ca2+-binding site and residues that contribute to the tetramer interface (Figure 3C). In contrast, one peptide (87–113) from the S100A9 subunit that contains interfacial W88 does not change (Figure 3H), because of its large size (vide supra). We also found peptides corresponding to other protein regions that showed negligible change in HDX (Figure 3B,F); these peptides can be considered to be controls.
Figure 3.
Representative plots of model-free PLIMSTEX data for the titration of 4 μM CP-Ser with Ca2+ ions. Data for S100A8(C42S) (A-D) and S100A9(C3S) (E-H) are shown. Peptide sequences and associated charge states are: (A) HAVYRDDL (S100A8(C42S) 27–34 (+2)), (B) TESPQYIRKKGADVW (S100A8(C42S) 40–54 (+2)), (C) FKELDINTDGAVNF (S100A8(C24S) 55–68 (+2)), (D) VIKMGVAAHKKSHEESHKE (S100A8(C42S) 75–93 (+3)), (E) SVKLGHPDTLNQGEF (S100A9(C3S) 23–37 (+2)), (F) FLKKENKNEKVIE (S100A9(C3S) 48–60 (+2)),(G)LDTNADKQLSFEE(S100A9(C3S)66–78(+2)), and(H) TWASHEKMHEGDEGPGHHHKPGLGEGTP (S100A9(C3S) 87–114 (+3), respectively. PLIMSTEX data for peptides from the EF-hands (A, C, E, and G) and other regions (B, D, F, H) are shown.
Figure 4.
PLIMSTEX data for the titration of 4 μM CP-Ser with Ca2+ ions fit to the NLLS algorithm using a 1:4 sequential binding model with no consideration of tetramerization. The data were acquired for four EF-hand peptides in the following charge states: HAVYRDDL (S100A8(C42S) 27–34 (+2)) (blue circles), FKELDINTDGAVNF (S100A8(C42S) 55–68 (+2)) (green squares), SVKLGHPDTLNQGEF (S100A9(C3S) 23–37 (+2)) (red diamonds), LDTNADKQLSFEE (S100A9(C3S) 66–78 (+2)) (black triangles), respectively.
For modeling Ca2+ binding to CP-Ser, we considered the four different Ca2+ sites in the heterodimer and the Ca2+-dependent tetramerization event. To begin, we utilized PLIMSTEX data from four peptides that originate from the four Ca2+-binding EF-hands of the S100A8 and S100A9 subunits (Figure 3A,C,E,G) and fit the data to a 1:4 sequential binding model that did not consider tetramerization (see Experimental Methods and Supporting Discussion in the SI). The calculated curves for the four peptides fit well the experimental data as judged from both visual inspection and the statistical parameters (resRMS of 0.92 and R2 of 0.997) (Figure 4).
Next, using the PLIMSTEX data, we tested four different models incorporating tetramerization in the model-fitting (Figure S3). Because a number of other Ca2+-binding proteins (e.g., calmodulin) show cooperative Ca2+ binding, we incorporated a combination of tetramerization and cooperativity for up to eight Ca2+ ions bound to the CP-Ser heterotetramer into these four models. All four models fit the data well, resulting in our inability to discriminate between models and suggesting that PLIMSTEX modeling at this stage is insufficiently constrained to determine the affinities for Ca2+ binding and tetramerization (Figure S3, Table S1, and Supporting Discussion in the SI). Moreover, for each of the four models, the inflection in the titration plot occurs at approximately two equivalents of Ca2+ per heterodimer (Figure 4). This result contrasts with results of prior biophysical studies indicating that, in solution, full tetramerization requires approximately 20 equivalents of Ca2+.11, 13> Thus, the PLIMSTEX data must primarily pertain to the CP-Ser heterodimer. Also, the observations from PLIMSTEX suggest that changes in protein conformation are largely complete when two (rather than four) equivalents of Ca2+ are added, reinforcing the notion that the information obtained from PLIMSTEX may be incomplete. It is possible, however, that binding of the second Ca2+ ion already induces conformational propagation in other EF-hands that bind the third and fourth Ca2+ ions (i.e., minimal conformational changes occur upon additional Ca2+ binding).32 To draw a comprehensive picture of Ca2+ binding to CP-Ser, we reasoned that the PLIMSTEX modeling needed to be constrained by the binding stoichiometry and the equivalents of Ca2+ that causes the heterodimer-to-heterotetramer transition.
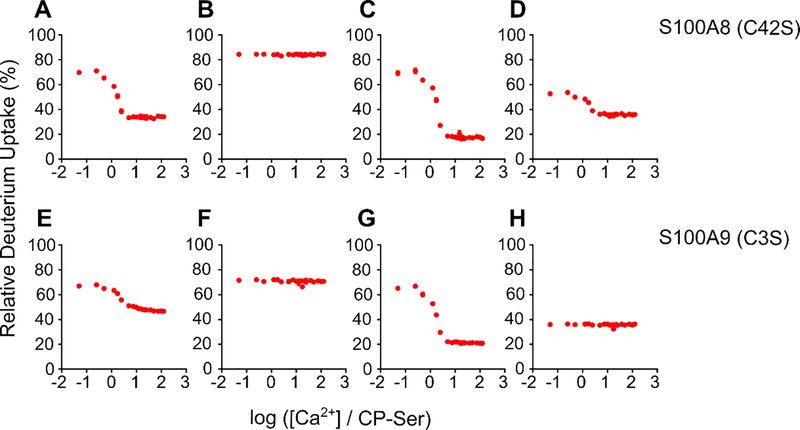
Sharp-break PLIMSTEX Curves Show that the CP-Ser Heterodimer Binds Four Ca2+ Ions
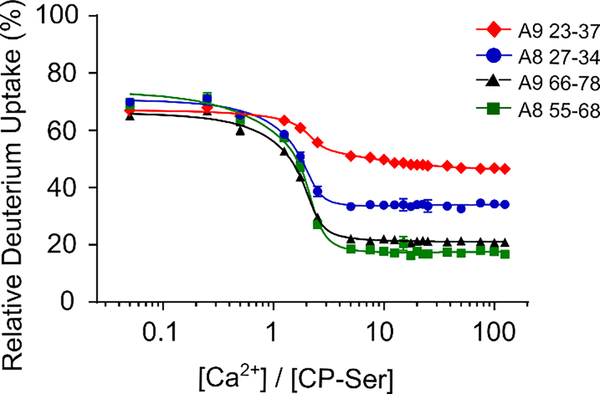
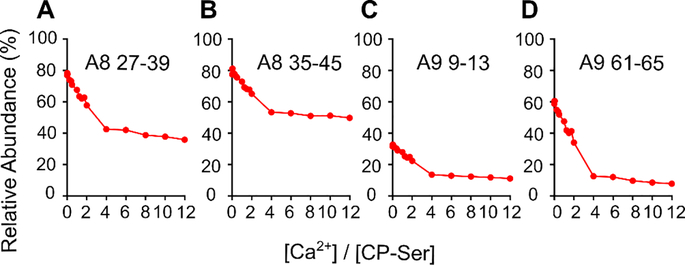
To constrain the PLIMSTEX fitting by the Ca2+-binding stoichiometry, we obtained sharp-break PLIMSTEX26 curves by titrating CP-Ser at concentrations that likely exceed the Kd values (Figure 5). We titrated 40 μM CP-Ser – a concentration 10-fold greater than the CP-Ser concentration used in the PLIMSTEX experiment described above – with up to 12 equivalents (480 μM) of Ca2+. Almost 90% of the peptides directly from or adjoining the Ca2+-binding sites exhibited a near-sharp break at a 4:1 Ca2+:CP-Ser heterodimer molar ratio (Figure 5). In contrast, peptides corresponding to the N-terminal region ( residues 1–7), and middle region (residues 48–60) of the S100A8 subunit, and C-terminus (83–114) of the S100A9 subunit showed negligible change in HDX (Figure S4), consistent with the behavior expected for regions that do not bind Ca2+. At least two peptides, WFKELDINT (52–62 (+2)) from S100A8 and IINTFHQY (14–21 (+2)) from A9 showed breaks at lower Ca2+:CP-Ser stoichiometry, indicating that a few regions of the protein complete their conformational changes when binding less than four Ca2+ ions (Figure S4B). With these exceptions, most of the results indicate that the heterodimer binds up to four Ca2+ ions and, by corollary, the tetramer binds up to eight Ca2+ ions. This analysis is consistent with previous MS studies that reported an 8:1 stoichiometry for Ca2+ binding to hCP.16–17
Figure 5.
Sharp-break PLIMSTEX curves obtained for the S100A8(C42S) (A, B) and S100A9(C3S) (C, D) subunits, respectively. The data were acquired for the four peptides in the following charge states: (A) HAVYRDDLKKLLE (S100A8(C42S) 27–39 (+2)), (B) KKLLETESPQY (S100A8(C42S) 35–45 (+2)), (C) ERNIE (S100A9(C3S) 9–13 (+1)), (D) HIMED (S100A9(C3S) 61–65 (+1)).
Native-MS Reveals the Ca2+ Equivalents Required for Dimer-to-Tetramer Transition
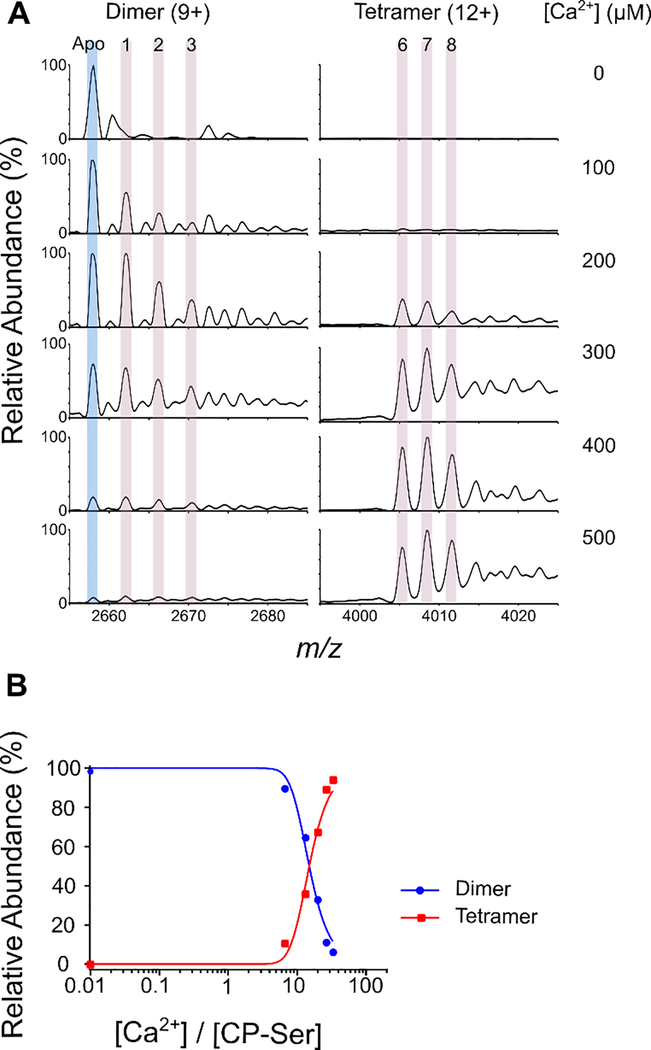
To add a further constraint for the modeling of Ca2+ binding to CP-Ser, we chose a direct method that simultaneously monitors the heterodimer and heterotetramer and the numbers of Ca2+ ions bound to each (Figure 6). For this, we acquired native mass spectra of 15 μM CP-Ser in the presence of 0–500 μM Ca2+. We observed that the abundance of the heterodimer decreased and the abundance of the heterotetramer increased as more Ca2+ was added (Figure 6A). Moreover, peaks representing up to eight Ca2+ ions bound to the heterotetramer occurred at >200 μM Ca2+, in agreement with the 8:1 binding stoichiometry for the CP-Ser heterotetramer deduced from the sharp-break PLIMSTEX titration (Figure 5). Importantly, the native-MS experiments also demonstrate that the transition of CP-Ser from the heterodimer to the heterotetramer occurs between 13–20 equivalents (200–300 μM) of added Ca2+, and the results suggest that the heterodimer bearing four Ca2+ prefers to be in the tetramer state (Figure 6A and Supporting Information). The observation of incomplete Ca2+ binding in the heterotetramer (i.e., 6 and 7 Ca2+ species) is likely due to loss of the metal ion in the ionization process. A calculation (Supporting Information) starting with 15 μM CP-Ser shows that the probability of having one molecule of CP-Ser heterodimer in a 10-nm ESI droplet is ≈ 0.04 and an upper bound on the fraction of two or more unassociated heterodimers in the droplet being counted in the heterotetramer signal is at most 4%. This analysis supports that the overall signals for heterodimer and heterotetramer in native MS represent the species in solution and are not artifacts of solvent evaporation in the ESI process. In this calculation, the droplet radius in nanospray is taken to be approximately 10 nm after a single droplet fission.38–40
Figure 6.
Native MS of CP-Ser in the absence and presence of Ca2+ ions. (A) Native mass spectra of 15 μM CP-Ser in the presence of 0–500 μM Ca2+ ions, zoomed in to show the most abundant charge states of dimer (9+) and tetramer (12+) species. The apo CP-Ser peak is highlighted in light blue and peaks for Ca2+-bound species are highlighted in pink. (B) Plots of CP-Ser species distribution at different [Ca2+] represented for dimer (blue circles) and tetramer (red squares). Signal intensities for each CP-Ser species are corrected for the fraction of the total dimer subunit for each [Ca2+].
An accurate estimate of the equivalents of Ca2+ ions required for the heterodimer-to-heterotetramer transition comes from the relative abundance of the CP-Ser heterodimer and heterotetramer species, determined from the corresponding native-MS peak intensities and plotted vs. the [Ca2+]/[CP-Ser] ratio ( Figure 6B). A 1:1 ratio of heterodimer and heterotetramer occurs for 15 equivalents of Ca2+ (225 μM). This value is reminiscent of the number of Ca2+ equivalents required to fully enhance the transition-metal-ion affinities of hCP.3, 11 We also note that both the CP-Ser and Ca2+ ion concentrations used in this experiment are physiologically relevant. Levels of CP in tissue abscess fluid can exceed 1 mg/mL (≈ 40 μM heterodimer), and the total extracellular Ca2+ concentration is ≈ 2000 μM. 9, 41
Combination of PLIMSTEX, Native-MS, and Sharp-break PLIMSTEX Allows Assessment of Ca2+-binding Affinities
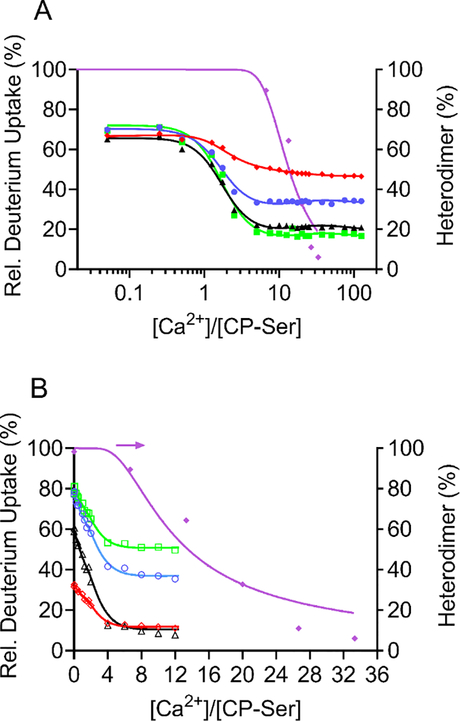
The results from HDX and PLIMSTEX (at 4 μM CP-Ser) delineate the Ca2+-binding behavior of the CP-Ser heterodimer, whereas those from native MS at a higher concentration (15 μM CP-Ser) and from sharp-break PLIMSTEX (40 μM CP-Ser) report the Ca2+ equivalents required for the dimer-to-tetramer transition. Together, these data allow us to constrain the modeling and incorporate up to four independently binding Ca2+ ions for the heterodimer and eight for the heterotetramer (Figure 7 and Supporting Discussion in SI). The best fit of the data with the model (referred to as Model K in SI and described above) provides the Ca2+-binding affinities associated with the four EF-hands of CP-Ser, a tetramerization constant, and an overall Adair binding constant (β) (Table 1). The overall Adair binding constant, β = 2.7 × 1045 M−9, is the product of the binding constants for the four Ca2+ ions for each heterodimer and the tetramerization constant (see Supporting Discussion in SI for details).
Figure 7.
Data fitted with NLLS to a model incorporating four independent Ca2+ (1–4 Ca2+) in the dimer and 8 Ca2+ in the tetramer. (A) The data are PLIMSTEX (4 μM CP-Ser) and native MS (15 μM CP-Ser) and represent the binding sites A9(C3S) 23–37 (+2) (red diamonds), A8(C42S) 27–34 (+2) (blue circles), A9(C3S) 66–78 (+2) (black triangles), A8(C42S) 55–68 (+2) (green squares), and native-MS data (purple diamonds). (B) The data are sharp-break PLIMSTEX (40 μM CP-Ser) and native-MS (15 μM CP-Ser). The data are from the Ca2+-binding sites A9(C3S) 9–13 (+1) (red diamonds), A8(C42S) 27–39 (+2) (blue circles), A9(C3S) 61–65 (+1) (black triangles), A8(C42S) 35–45 (+2) (green squares), and native-MS data (purple diamonds).
Table 1.
Calculated dissociation constant values from experimental data and model fit.a
| Dissociation Constant | Value (M−1) | Propagated Errorb |
|---|---|---|
| K1 | 9.4 × 10−7 | 0.74 × 10−7 |
| K2 | 9.4 × 10−7 | 0.74 × 10−7 |
| K3 | 4.9 × 10−5 | 0.28 × 10−5 |
| K4 | 4.3 × 10−1 | 0.06 × 10−1 |
| Ktetramer | 1.1 × 10−12 (β = 2.7 × 1045M−9) | 0.006 × 10−12 |
Dissociation constants were determined from the data presented in Figure 7.
First order approximation of the uncertainty in the fit parameters as propagated through the least squares fitting from the standard deviations of the replicate HDX measurements.
Because the PLIMSTEX data are similar for the four EF-hands and do not allow clear assignment of the order of Ca2+ binding, we did not use those data to assign the four dissociation constants (K1-K4) to specific EF-hands. Our modeling shows that two EF-hand domains coordinate Ca2+ micromolar affinity whereas the other two EF-hands coordinate Ca2+ with substantially lower affinity. We associate K1 and K2 with the C-terminal EF-hands and K3 and K4 with the N-terminal EF-hands, in accord with the X-ray crystallography18 and with the HDX kinetics (vide supra). Further, we speculate that the S100A9 and S100A8 non-canonical EF-hands bind the third and the fourth Ca2+ ions, respectively. The S100A8 noncanonical EF-hand (LIKGNFHAVY), lacking amino acids that tightly bind Ca2+, shows asharp change in conformation when CP-Ser binds two Ca2+ ions and then shows no further change with increasing Ca2+ equivalents (Figure 4). Once this EF-hand binds the fourth Ca2+ ion, the protein transitions to the heterotetramer. The S100A9 noncanonical EF-hand (GHPDTLNQGE) changes its conformation more slowly (Figure 4), and we assign this site as the third to bind Ca2+. Indeed, this EF-hand has more side chains that strongly bind Ca2+ than does the S100A8 noncanonical EF-hand. The character of the PLIMSTEX curves make it difficult to judge whether the binding is cooperative; thus, the four binding events were modeled to be independent (see Supporting Discussion in SI).
To test whether the model provides equivalent results for a more limited data set, we modeled the PLIMSTEX data at 4 μM CP-Ser in combination with the native-MS titration at 15 μM CP-Ser. These results (Table S4, Figures S5–S8, and Supporting Discussion) provide comparable estimates of K1 and K2 and of Ktetramer but underestimate the role of the heterodimer with three bound Ca2+ ions by yielding a smaller K3 (compare Table 1 and Table S4) and a larger K4. These comparisons underscore the importance of incorporating measurements at a higher protein concentration than 4 μM and show that a more complete picture is provided by modeling the combination of three titrations at 4, 15, and 40 μM, as described above.
Although this work provides the first examination of the Ca2+-binding affinities of CP, Ca2+ affinities of other S100 proteins have been reported. Those prior studies indicate that C-terminal canonical EF-hands generally have Kd values in the range of 1–50 μM.15, 42–44 The Kd values determined here are ≈ 1 μM, at the strong-binding end of this range. Reported Kd values for N-terminal EF-hands fall in the range of 100–500 μM.15, 42–44 Our modeling indicates that one of the N-terminal hands (assigned as S100A9, see above) has a Kd value of 50 μM whereas the other (S100A8) is remarkably low at 0.4 M. We speculate that the low binding affinity for the fourth Ca2+ ion is likely offset by the high propensity to tetramerize once the heterodimer is loaded, even at a trace level, with a fourth Ca2+ ion.
Overall, the Kd values for Ca2+ binding determined in this work support the working model postulated for the CP-Ser.1 In particular, these results indicate that hCP is only partially Ca2+-bound at the C-terminal EF-hands in the cytoplasm where Ca2+ concentrations are low (nanomolar range). It is likely that a mixture of species (apo, 1 and 2 Ca2+ dimers) exists in this milieu. With release into the extracellular space, three EF-hands readily bind Ca2+, but the fourth site is only weakly populated. The two low-affinity Kd values are consistent with the non-canonical N-terminal EF-hands of S100A8 and S100A9. The PLIMSTEX data indicate substantial conformational changes upon binding of two Ca2+. This behavior suggests that Ca2+ binding at the C-terminal EF-hands induces conformational changes at the N-terminal EF-hands to poise the protein to bind additional Ca2+ ions once the Ca2+ concentration becomes high, as is the case for Ca2+ binding to DREAM.32
Given the weak binding affinity of the fourth Ca2+ ion, the binding interface between the two heterodimers is likely to be the deciding factor in the tetramerization of CP-Ser. The best model fit gives a tetramerization Kd value of 1.1 x 10−12 M (Table 1), indicative of a strong binding interface between the two heterodimers. When the heterodimer has bound one or two Ca2+ ions, which occurs at low Ca2+-to-protein ratios, no tetramerization occurs, and the protein is in a “resting state”. Once the protein is released to extracellular space, it binds a third Ca2+ and begins to bind a fourth Ca2+ ion. The driving force for tetramerization of the dimer with 4 Ca2+ is so favorable that it offsets the weak affinity for a fourth Ca2+ ion and “pulls” the equilibrium toward product to form the heterotetramer binding 8 Ca2+ ions, preparing the protein to play its role in innate immunity by sequestering transition metal ions.
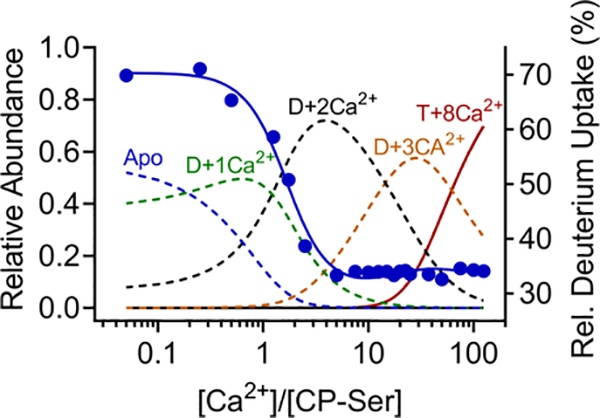
The working model described above indicates that the Ca2+ affinities of hCP are tuned such that the heterodimeric protein becomes heterotetrameric when it encounters higher extracellular Ca2+ levels. An informative way to view this behavior is the variation in fractional species from the model (Figures 8 and S10). The fractional-species variation shows clearly that the concentration of the heterodimers containing two and three bound Ca2+ ions decreases as the heterotetramer appears and becomes ultimately the dominant species. This working model has support from prior evaluations of the protein under conditions of low and high Ca2+ concentrations;3, 10–12 however, until now, this view has lacked convincing quantitative support.
Figure 8.
Fractional-species variations as a function of the relative concentration of Ca2+ showing the concomitant fit of the 4 μM protein PLIMSTEX data for A8 27–34. The model does show a dimer plus 4Ca2+, but its abundance is too low to be seen on this graph. The fractional species also for the apo protein does not start at zero because there was a small amount of adventitious Ca2+ in the sample solution, and that is an input to the model. This is also seen in Figures S7 and S10.
Summary and Outlook
The present study provides comprehensive, quantitative molecular information of the Ca2+-binding properties of CP and its metal-sequestering function in solution. In particular, the use of an integrated MS approach allowed us to bring new understanding to a complicated system that involves multiple Ca2+-binding events and self-association. The resulting quantitative insights support and guide our development of the working model for how CP sequesters transition metal ions in the extracellular space. To the best of our knowledge, this work constitutes the first report of the affinity between S100 oligomers.
The current working model of how Ca2+ ions modulate the structure and function of CP starts with the protein existing primarily as a Ca2+-free heterodimer when stored in the cytoplasm of immune and epithelial cells where the resting Ca2+ concentration is low (≈ 100 nM).9 When CP is released into the extracellular space, it encounters higher Ca2+ concentrations (≈ 2 mM)9 and binds Ca2+, facilitating production of a heterotetramer, increasing the transition-metal-ion affinities of the His3Asp and His6 sites, enhancing antimicrobial activity, and affording protease resistance.1, 13 This model has been largely based on transition-metal-binding studies demonstrating that excess Ca2+ ions enhance the transition-metal affinities of the protein and has been corroborated by functional studies.1, 3, 10–13 Although there is compelling evidence from prior studies that Ca2+-induced changes in CP are essential for function and also impact its fate in the biological milieu, the molecular basis for these effects has remained elusive. Moreover, there is no reported crystallographic or solution structure of the apo CP heterodimer. As a result, we have no ability to link structural snapshots to achieve depictions of conformational change.
The integration of three MS-based approaches enables a further evaluation of the hypothesis that Ca2+ ions modulate hCP function in vivo and to (i) confirm the four regions that serve as Ca2+ binding sites, (ii) determine the binding stoichiometry for the CP-Ser to be four Ca2+ per dimer and eight Ca2+ per tetramer, (iii) establish the protein-to-Ca2+ molar ratio that causes the dimer-to-tetramer transition, and (iv) determine the Ca2+-binding affinities associated with four binding sites (Table 1) and the self-association constant for the Ca2+-bound protein. Taken together, the results are consistent with the view that local Ca2+ levels in the biological milieu modulate hCP structure, metal-sequestering function, and fate. In particular, our modeling indicates that the third and fourth EF-hands of CP will be populated with Ca2+ only at high Ca2+ concentrations when the heterotetramer has formed, supporting the idea that intracellular CP is a heterodimer in resting cells. With regard to the fate of the protein, prior work from some of us indicates that self-association modulates the lifetime of CP in biological milieu.13, 45 The decrease in HDX for the Ca2+-bound heterotetramer relative to the apo heterodimer demonstrates that the protein adopts a less dynamic structure upon Ca2+ binding, providing a new structural insight into the marked protease resistance observed for the former species. Furthermore, the number of bound Ca2+ ions required for tetramerization suggests that serine proteases degrade apo CP as well as CP heterodimers that have several Ca2+-binding stoichiometries.
Despite the success of the integrated workflow developed here, one limitation is the inability to ascertain whether cooperative Ca2+ binding occurs (Supporting Discussion in SI). Given that cooperativity is a hallmark of EF-hand proteins, it is possible that the current experimental design precluded observation of cooperativity, and further studies along these lines are certainly warranted.
In closing, we expect that this integrated MS methodology to be applicable to other complicated protein-ligand systems where other approaches afford incomplete pictures. We look forward to extending this approach to evaluate transition-metal binding by hCP and other S100 proteins. We anticipate that the resulting insights from such endeavors will enhance our appreciation of the molecular mechanisms of S100 proteins at the host-pathogen interface and in associated pathologies such as Alzheimer’s disease and cancer.
Experimental Methods
Materials
All chemicals, proteases and solvents were purchased from Sigma Aldrich (St. Louis, MO) unless otherwise stated. Deuterium oxide was from Cambridge Isotope Laboratories Inc. (Andover, MA). All buffers and metal solutions were prepared using Milli-Q water (18.2 MΩ•cm, 0.22-μm filter, Millipore). All mobile chromatography phases for protein purification were prepared with Ultrol grade HEPES (free acid, Calbiochem) and TraceSELECT NaCl (Fluka), and aqueous TraceSELECT NaOH (Sigma) was used for pH adjustments. All buffers were stored in polypropylene bottles. The highest available purity of calcium chloride (99.99%) was purchased from Alfa Aesar. Stock solutions of Ca2+ (1 M, 100 mL) were prepared by using Milli-Q water and acid-washed volumetric glassware, and the solutions were transferred to and stored in polypropylene tubes. Protein concentrations were determined by using their optical absorbance at 280 nm (ε280 = 18,450 cm−1M−1 for the CP-Ser heterodimer10) by using a Nanodrop (Thermo Scientific). All concentrations of CP-Ser and equivalents of Ca2+ are reported in terms of the heterodimer.
Protein Preparation
The hCP variant CP-Ser, a S100A8(C42S)/(S100A9(C3S) oligomer, was used for all the experiments in this study. CP-Ser was prepared as the apo heterodimer and handled as previously described.10, 37
Hydrogen-Deuterium Exchange (HDX)
The HDX experiment and data analysis procedures were similar to previously described protocols.32 Briefly, CP-Ser (50 μM) was equilibrated in the absence or presence of 2.5 mM Ca2+ for at least 1 h at 25°C in 11 mM HEPES, 11 mM NaCl, pH 7.0 in H2O. A 2-μL aliquot of the solution was then diluted ten-fold with buffer prepared in D2O (11 mM HEPES, 11 mM NaCl, pH = 7.0) (or H2O buffer for an undeuterated control sample) to initiate the HDX reaction. After various exchange times (10, 30, 60, 120, 900, 1800, 3600 and 10800 s), the HDX was quenched by adding 30 μL of ice-cold quench solution (3 M urea in 1% (v/v) aqueous trifluoroacetic acid), which lowered the solution pH to 2.5. Peptide identification was accomplished through a combination of accurate mass analysis and LC-MS/MS by using MassMatrix (version 2.4.2) for identification.46 Raw HDX mass spectra were submitted to HDX Workbench for calculation and data visualization.47 The HDX data were acquired in LTQ-FTICR mass spectrometer (Thermo Scientific) and presented here as an average of duplicate measurements for each exchange time.
PLIMSTEX
CP-Ser was incubated with increasing concentrations of Ca2+ for 1 h at 25°C. The samples were then diluted with an excess (ten-fold) amount of buffer containing D2O. The final protein and ligand concentrations for the PLIMSTEX experiments were 4 μM CP-Ser and 0–500 μM Ca2+ for the normal (not sharp-break) PLIMSTEX titration, and 40 μM CP-Ser and 0–480 μM Ca2+ for the sharp-break PLIMSTEX experiment. A total of 21 (0, 0.2, 1, 2, 5, 7, 10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 150, 200, 300, 400, 500 μM) and 16 (0, 1, 2, 10, 16, 20, 40, 50, 60, 70, 80, 160, 240, 320, 400, 480 μM) Ca2+ concentrations were used for the normal PLIMSTEX titrations and the sharp-break PLIMSTEX titrations, respectively. CP-Ser underwent HDX for 30 min at 25 °C and was submitted to on-line pepsin digestion and desalting prior to analysis with an LTQ-FTICR mass spectrometer (Thermo Scientific, Santa Clara, CA). The buffer conditions and instrumental conditions were similar to those described in the HDX experimental section except for the sharp-break PLIMSTEX where an aliquot of 500 μM stock protein in 75 mM HEPES, 100 mM NaCl, pH 7.5 was incubated with the ligand for at least 1 h at 25 °C. From this stock, 2 μL protein stock was diluted 1:10 in D2O buffer for HDX reactions. The final concentration of CP-Ser injected onto the mass spectrometer was kept at low levels by diluting each sample to a final volume of 500 μL (25-fold dilution) volume with quench solution (1.9 M urea prepared in 1% (v/v) aqueous TFA) for the sharp-break experiment. The mass spectra were searched by using HDX Workbench47 to calculate deuterium uptake percentages. Each PLIMSTEX experiment was performed in duplicate, and the data presented here were an average of two measurements.
Native Mass Spectrometry
CP-Ser was buffer exchanged a total of five times in 200 mM NH4OAc solution, pH 7, by using a 10K MWCO filter (15,000 × g, 10 min, 4 °C). After the fifth round of buffer exchange, the concentration of CP-Ser was determined and the protein solution was diluted to 250 μM with 200 mM ammonium acetate solution and aliquots of this solution were stored at −80 °C until the day of the experiment. At that time, protein solutions (3 μL, 250 μM) were diluted to a final volume of 50 μL by using 200 mM ammonium acetate, pH 7.0. To the protein solutions were added different volumes of 1 mM Ca2+ to give final concentrations of 0, 100, 200, 300, 400 and 500 μM. The solutions were then incubated at room temperature for at least 1 h. Each mixture was then loaded onto nano-spray emitter tips ES480 (Thermo Scientific, Waltham, MA) and were analyzed by using the Exactive plus EMR instrument (Thermo Scientific, Waltham, MA). The native mass spectra were acquired from 500–20,000 m/z at a source temperature of 55 °C, CID 100 volts, CE 100 volts, and spray voltage of 1.2–1.5 kV at a mass resolving power of 17,500 (8,750 for the apo-sample with no Ca2+). From the native mass spectra obtained for each Ca2+ concentration, the fraction heterodimer and heterotetramer were calculated from signal intensities of each species at each Ca2+ concentration, and the relative abundances (%) were plotted as a function of the Ca2+ concentration to obtain a titration curve. These native-MS titration data for the heterodimer were incorporated in the model fitting dataset as a separate peptide (see Supporting Discussion in the SI) together with PLIMSTEX data for four peptides from the EF-hands. The data presented is the signal averaged over 5–10 minutes in a Thermo Exactive EMR instrument for each sample in an experiment.
PLIMSTEX Data Analysis and Binding Affinity Calculation
The HDX kinetic experiment described above provided a guide for the PLIMSTEX experiment.48 From the HDX kinetics, the exchange time at which the difference in deuterium uptake between the apo- and the holo sample was relatively large and constant was chosen to be the time (30 min) for PLIMSTEX experiment. Titration of CP-Ser with increasing concentrations of Ca2+ (as shown in Figure 4 and 7) was followed by measuring the extent of deuterium uptake as a function of the Ca2+ concentration. The change in deuterium uptake was plotted as a function of the ligand-to-protein ratio to obtain a PLIMSTEX titration curve. The data for four peptides from the EF-hands were then fit to a mathematical model to obtain the binding affinities for Ca2+ binding to CP-Ser by using a nonlinear least squares (NLLS) regression analysis, as described previously.48–49 Several different mathematical models were fit to the PLIMSTEX and native-MS data to arrive at a best-fit model (see Supporting Discussion of the SI).
Supplementary Material
Acknowledgements
This work was supported by the NIH National Institute for General Medical Sciences grants 2P41GM103422 and R24GM136766 to M.L.G. and R01GM118695 to E.M.N.
Footnotes
Supporting Information
Figures S1-S11, Tables S1-S6, and Supporting Discussion on mass spectrometry analyses and modeling. Additional models were suggested by one of the reviewers, and the results of their implementation are described in the Supporting Discussion of the SI (referred to as Models L, M, and N). This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zygiel EM; Nolan EM, Transition metal sequestration by the host-defense protein calprotectin. Annu. Rev. Biochem. 2018, 87, 621–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corbin BD; Seeley EH; Raab A; Feldmann J; Miller MR; Torres VJ; Anderson KL; Dattilo BM; Dunman PM; Gerads R; Caprioli RM; Nacken W; Chazin WJ; Skaar EP, Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 2008, 319 (5865), 962–965. [DOI] [PubMed] [Google Scholar]
- 3.Nakashige TG; Zhang B; Krebs C; Nolan EM, Human calprotectin is an iron-sequestering host-defense protein. Nat. Chem. Biol. 2015, 11 (10), 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zygiel EM; Nelson CE; Brewer LK; Oglesby-Sherrouse AG; Nolan EM, The human innate immune protein calprotectin induces iron starvation responses in Pseudomonas aeruginosa. J. Biol. Chem. 2019, 294 (10), 3549–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakashige TG; Zygiel EM; Drennan CL; Nolan EM, Nickel sequestration by the host-defense protein human calprotectin. J. Am. Chem. Soc. 2017, 139 (26), 8828–8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu JZ; Jellbauer S; Poe AJ; Ton V; Pesciaroli M; Kehl-Fie TE; Restrepo NA; Hosking MP; Edwards RA; Battistoni A; Pasquali P; Lane TE; Chazin WJ; Vogl T; Roth J; Skaar EP; Raffatellu M, Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 2012, 11 (3), 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hood MI; Mortensen BL; Moore JL; Zhang Y; Kehl-Fie TE; Sugitani N; Chazin WJ; Caprioli RM; Skaar EP, Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog. 2012, 8 (12), e1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakashige TG; Stephan JR; Cunden LS; Brophy MB; Wommack AJ; Keegan BC; Shearer JM; Nolan EM, The hexahistidine motif of host-defense protein human calprotectin contributes to zinc withholding and its functional versatility. J. Am. Chem. Soc. 2016, 138 (37), 12243–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brini M; Ottolini D; Cali T; Carafoli E, Calcium in health and disease. Met.Ions Life Sci. 2013, 13, 81–137. [DOI] [PubMed] [Google Scholar]
- 10.Brophy MB; Hayden JA; Nolan EM, Calcium ion gradients modulate the zinc affinity and antibacterial activity of human calprotectin. J. Am. Chem. Soc. 2012, 134 (43), 18089–18100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayden JA; Brophy MB; Cunden LS; Nolan EM, High-affinity manganese coordination by human calprotectin is calcium-dependent and requires the histidine-rich site formed at the dimer interface. J. Am. Chem. Soc. 2013, 135 (2), 775–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakashige TG; Bowman SEJ; Zygiel EM; Drennan CL; Nolan EM, Biophysical examination of the calcium-modulated nickel-binding properties of human calprotectin reveals conformational change in the EF-hand domains and His3Asp site. Biochemistry 2018, 57 (28), 4155–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephan JR; Nolan EM, Calcium-induced tetramerization and zinc chelation shield human calprotectin from degradation by host and bacterial extracellular proteases. Chem. Sci. 2016, 7 (3), 1962–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter MJ; Chazin WJ, High level expression and dimer characterization of the S100 EF-hand proteins, migration inhibitory factor-related proteins 8 and 14. J. Biol. Chem. 1998, 273 (20), 12427–12435. [DOI] [PubMed] [Google Scholar]
- 15.Gifford JL; Walsh MP; Vogel HJ, Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 2007, 405, 199–221. [DOI] [PubMed] [Google Scholar]
- 16.Vogl T; Roth J; Sorg C; Hillenkamp F; Strupat K, Calcium-induced noncovalently linked tetramers of MRP8 and MRP14 detected by ultraviolet matrix-assisted laser desorption/ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 1999, 10 (11), 1124–1130. [DOI] [PubMed] [Google Scholar]
- 17.Strupat K; Rogniaux H; Van Dorsselaer A; Roth J; Vogl T, Calcium-induced noncovalently linked tetramers of MRP8 and MRP14 are confirmed by electrospray ionization-mass analysis. J. Am. Soc. Mass Spectrom. 2000, 11 (9), 780–788. [DOI] [PubMed] [Google Scholar]
- 18.Gagnon DM; Brophy MB; Bowman SE; Stich TA; Drennan CL; Britt RD; Nolan EM, Manganese binding properties of human calprotectin under conditions of high and low calcium: X-ray crystallographic and advanced electron paramagnetic resonance spectroscopic analysis. J. Am. Chem. Soc. 2015, 137 (8), 3004–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korndörfer IP; Brueckner F; Skerra A, The crystal structure of the human (S100A8/S100A9)2 heterotetramer, calprotectin, illustrates how conformational changes of interacting alpha-helices can determine specific association of two EF-hand proteins. J. Mol. Biol. 2007, 370 (5), 887–898. [DOI] [PubMed] [Google Scholar]
- 20.Besold AN; Gilston BA; Radin JN; Ramsoomair C; Culbertson EM; Li CX; Cormack BP; Chazin WJ; Kehl-Fie TE; Culotta VC, The role of calprotectin in withholding zinc and copper from Candida albicans. Infect. Immun. 2017, 86 (2), e00779–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker TM; Nakashige TG; Nolan EM; Neidig ML, Magnetic circular dichroism studies of iron(ii) binding to human calprotectin. Chem. Sci. 2017, 8 (2), 1369–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z; Smith DL, Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993, 2 (4), 522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iacob RE; Krystek SR; Huang RY; Wei H; Tao L; Lin Z; Morin PE; Doyle ML; Tymiak AA; Engen JR; Chen G, Hydrogen/deuterium exchange mass spectrometry applied to IL-23 interaction characteristics: potential impact for therapeutics. Expert Rev. Proteomics 2015, 12 (2), 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J; Rodnin MV; Ladokhin AS; Gross ML, Hydrogen-deuterium exchange and mass spectrometry reveal the ph-dependent conformational changes of diphtheria toxin t domain. Biochemistry 2014, 53 (43), 6849–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konermann L; Vahidi S; Sowole MA, Mass spectrometry methods for studying structure and dynamics of biological macromolecules. Anal. Chem. 2014, 86 (1), 213–232. [DOI] [PubMed] [Google Scholar]
- 26.Zhu MM; Rempel DL; Du Z; Gross ML, Quantification of protein-ligand interactions by mass spectrometry, titration, and H/D exchange: PLIMSTEX. J. Am. Chem. Soc. 2003, 125 (18), 5252–5253. [DOI] [PubMed] [Google Scholar]
- 27.Huang RY; Rempel DL; Gross ML, HD exchange and PLIMSTEX determine the affinities and order of binding of Ca2+ with troponin C. Biochemistry 2011, 50 (24), 5426–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H; Rempel DL; Giblin D; Frieden C; Gross ML, Peptide-level interactions between proteins and small-molecule drug candidates by two hydrogen-deuterium exchange MS-based methods: the example of apolipoprotein E3. Anal. Chem. 2017, 89 (20), 10687–10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosati S; Yang Y; Barendregt A; Heck AJ, Detailed mass analysis of structural heterogeneity in monoclonal antibodies using native mass spectrometry. Nat. Protoc. 2014, 9 (4), 967–976. [DOI] [PubMed] [Google Scholar]
- 30.Thompson NJ; Rosati S; Heck AJ, Performing native mass spectrometry analysis on therapeutic antibodies. Methods 2014, 65 (1), 11–17. [DOI] [PubMed] [Google Scholar]
- 31.Sperry JB; Huang RY; Zhu MM; Rempel DL; Gross ML, Hydrophobic peptides affect binding of calmodulin and Ca as explored by H/D amide exchange and mass spectrometry. Int. J. Mass Spectrom. 2011, 302 (1–3), 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J; Li J; Craig TA; Kumar R; Gross ML, Hydrogen-deuterium exchange mass spectrometry reveals calcium binding properties and allosteric regulation of downstream regulatory element antagonist modulator (DREAM). Biochemistry 2017, 56 (28), 3523–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kehl-Fie TE; Skaar EP, Nutritional immunity beyond iron: a role for manganese and zinc. Curr. Opin. Chem. Biol. 2010, 14 (2), 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HJ; Savelieff MG; Kang J; Brophy MB; Nakashige TG; Lee SJC; Nolan EM; Lim MH, Calprotectin influences the aggregation of metal-free and metal-bound amyloid-beta by direct interaction. Metallomics 2018, 10 (8), 1116–1127. [DOI] [PubMed] [Google Scholar]
- 35.Tibble J; Sigthorsson G; Foster R; Sherwood R; Fagerhol M; Bjarnason I, Faecal calprotectin and faecal occult blood tests in the diagnosis of colorectal carcinoma and adenoma. Gut 2001, 49 (3), 402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turvill J; Aghahoseini A; Sivarajasingham N; Abbas K; Choudhry M; Polyzois K; Lasithiotakis K; Volanaki D; Kim B; Langlands F; Andrew H; Roos J; Mellen S; Turnock D; Jones A, Faecal calprotectin in patients with suspected colorectal cancer: a diagnostic accuracy study. Br. J. Gen. Pract. 2016, 66 (648), e499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hadley RC; Nolan EM, Preparation and iron redox speciation study of the Fe(II)-binding antimicrobial protein calprotectin. Methods Mol. Biol. 2019, 1929, 397–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juraschek R; Dulcks T; Karas M, Nanoelectrospray– more than just a minimized-flow electrospray ionization source. J. Am. Soc. Mass Spectrom. 1999, 10 (4), 300–308. [DOI] [PubMed] [Google Scholar]
- 39.Kebarle P; Tang L, From ions in solution to ions in the gas phase –the mechanism of electrospray mass spectrometry. Anal. Chem. 1993, 65 (22), 972A–986A. [Google Scholar]
- 40.Thomson B. A. a. I. J. V., Field induced ion evaporation from liquid surfaces at atmospheric pressure. J. Chem. Phys. 1979, 71 (11), 4451–4463. [Google Scholar]
- 41.Johne B; Fagerhol MK; Lyberg T; Prydz H; Brandtzaeg P; Naess -Andresen CF; Dale I, Functional and clinical aspects of the myelomonocyte protein calprotectin. J. Clin. Pathol: Mol. Pathol 1997, 50 (3), 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donato R, S-100proteins. Cell Calcium 1986, 7, 123–145. [DOI] [PubMed] [Google Scholar]
- 43.Santamaria-Kisiel L; Rintala-Dempsey AC; Shaw GS, Calcium-dependent and independent interactions of the S100 protein family. Biochem. J. 2006, 396 (2), 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garrett SC; Hodgson L; Rybin A; Toutchkine A; Hahn KM; Lawrence DS; Bresnick AR, A biosensor of S100A4 metastasis factor activation: inhibitor screening and cellular activation dynamics. Biochemistry 2008, 47, 986–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stephan JR; Yu F; Costello RM; Bleier BS; Nolan EM, Oxidative post-translational modifications accelerate proteolytic degradation of calprotectin. J. Am. Chem. Soc. 2018, 140 (50), 17444–17455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu H; Freitas MA, MassMatrix: a database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics 2009, 9 (6), 1548–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pascal BD; Willis S; Lauer JL; Landgraf RR; West GM; Marciano D; Novick S; Goswami D; Chalmers MJ; Griffin PR, HDX workbench: software for the analysis of H/D exchange MS data. J. Am. Soc. Mass Spectrom. 2012, 23 (9), 1512–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu MM; Rempel DL; Gross ML, Modeling datafrom titration, amide H/D exchange, and mass spectrometry to obtain protein-ligand binding constants. J. Am. Soc. Mass Spectrom. 2004, 15 (3), 388–97. [DOI] [PubMed] [Google Scholar]
- 49.Zhang YR, D. L.; Gross ML, Hydrogen exchange mass spectrometry for the analysis of ligand binding and protein aggregation. In Hydrogen exchange mass spectrometry of proteins, Weis DD, Ed. John Wiley & Sons: 2016; pp 185–207. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.