Abstract
Eating disorders and substance use disorders frequently co-occur. Twin studies reveal shared genetic risk between eating disorders and substance use, with the strongest associations between symptoms of bulimia nervosa (BN) and problem alcohol use, mainly abuse and dependence (twin-based genetic correlation [rg]=0.23–0.53). Analytic advances facilitate the computation of genetic correlations using summary statistics from existing genome-wide association studies (GWAS). We investigated shared genetic risk between eating disorder and substance use and disorder phenotypes using GWAS data. Four eating disorder phenotypes (anorexia nervosa [AN], AN with binge-eating, AN without binge-eating, and a BN factor score), and eight substance use-related phenotypes (drinks per week, alcohol use disorder [AUD], smoking initiation, current smoking, cigarettes per day, nicotine dependence, cannabis initiation, and cannabis use disorder) from eight studies were included. Total sample sizes per phenotype ranged from ~2,400 to ~537,000 individuals. We used linkage disequilibrium score regression to calculate single nucleotide polymorphism-based genetic correlations between eating disorder and substance use-related phenotypes. Significant positive genetic associations emerged between AUD and AN (rg=0.18; false discovery rate q=0.0006), cannabis initiation and AN (rg=0.23; q<0.0001), and cannabis initiation and AN with binge-eating (rg=0.27; q=0.0016). Conversely, significant negative genetic correlations were observed between three non-diagnostic smoking phenotypes (smoking initiation, smoking cessation, and cigarettes per day) and AN without binge-eating (rgs=−0.19 to −0.23; qs<0.04). The observed patterns of association between different eating disorder and substance use-related phenotypes highlights the potentially complex and substance-specific relationships between these behaviors associated with significant public health burden.
Keywords: comorbidity, eating disorders, genetic correlation, genome-wide association, linkage disequilibrium score regression, substance use disorders
A well-established phenotypic association exists between eating disorder and substance use phenotypes, with evidence for specific relations between particular types of eating disorders and substance use disorders. The prevalence of an alcohol use disorder (AUD) is greater among individuals with bulimia nervosa (BN) and binge-eating disorder (BED) than individuals with anorexia nervosa (AN) or healthy controls (Gadalla and Piran, 2007, Root et al., 2010). Similarly, individuals with BN or BED are at increased risk for smoking, nicotine dependence (ND) (Solmi et al., 2016, Wiederman and Pryor, 1996), and cannabis use (Krug et al., 2008, Wiederman and Pryor, 1996) compared with individuals with AN or healthy controls, though these results are not consistent (Root et al., 2010). Importantly, women with the binge-eating/purging subtype of AN report a higher prevalence of AUD, smoking, ND, and cannabis use than women with the restricting subtype of AN (Anzengruber et al., 2006, Krug et al., 2008, Root et al., 2010). Thus, binge eating—a transdiagnostic symptom defined as eating a large amount of food in a short period of time while experiencing loss of control—may be a key component of the observed association.
However, prior research has only partially addressed whether binge eating is the critical eating disorder symptom in the comorbidity, especially across different milestones of substance use (i.e., initiation through dependence) and across a variety of substances (i.e., alcohol, nicotine, and cannabis). It is crucial to elucidate shared etiological mechanisms for these associations because of the increased morbidity and mortality associated with comorbid presentations (Duncan et al., 2006, Franko et al., 2013) and because improvements in one disorder may exacerbate (or weaken) symptoms of the other disorder (Center on Addiction and Substance Abuse, 2003). Refining our understanding of these associations could improve prevention and treatment approaches for these debilitating disorders, their comorbidity, and their sequelae.
Accumulating findings from twin studies implicate shared genetic factors between eating disorder and substance use-related phenotypes. The strongest reported association is between BN symptoms, including binge eating, and problem alcohol use (Munn-Chernoff and Baker, 2016), with a genetic correlation (twin-based rg) ranging from 0.23 to 0.53 (Baker et al., 2010, Baker et al., 2017, Munn-Chernoff et al., 2013, Munn-Chernoff et al., 2015, Slane et al., 2012, Trace et al., 2013). Although there has been less focus on the genetic associations between BN symptoms and regular smoking and BN symptoms and illicit drug use disorder, twin-based rgs of 0.35 and approximately 0.38, respectively, have been reported (Baker et al., 2007, Baker et al., 2010). A paucity of information exists regarding whether less problematic aspects of substance use exhibit a significant rg with eating disorder phenotypes. Genetic factors influencing this comorbidity may only come into play once an individual has progressed to problematic alcohol use, as genetic effects are more prominent in problem substance use, such as abuse and dependence than with the initiation and general use of substances (Heath et al., 1997, Rhee et al., 2003, True et al., 1997, van den Bree et al., 1998). No study has comprehensively examined a range of eating disorder and substance use-related phenotypes to determine whether the rg varies with different aspects of substance use and whether the rg varies depending on the eating disorder and substance examined.
Recent advances in genomic methods allow for an assessment of rg using existing genome-wide association study (GWAS) summary statistics. Unlike twin studies, these genome-wide methods allow for use of unrelated cases and controls, which typically yield large sample sizes (i.e., in the tens to hundreds of thousands). One such method is linkage disequilibrium score regression (LDSC; Bulik-Sullivan et al., 2015a, Bulik-Sullivan et al., 2015b), which estimates single-nucleotide polymorphism (SNP)-based heritability and rg between phenotypes. Of particular relevance to low prevalence phenotypes, such as AN, estimation of SNP-based rg does not require both phenotypes to be measured in the same individual, meaning that independent studies that assess only one phenotype can be jointly examined.
Thus, the aim of the current study was to estimate SNP-based rgs between eating disorder and substance use-related phenotypes based upon summary statistics from the largest published eating disorder GWAS and existing GWAS encompassing a range of substance use-related phenotypes. This study examines shared SNP-based genetic risk between eating disorder and multiple substance use-related phenotypes (i.e., alcohol, nicotine, and cannabis), using robust data from twin studies to shape our expectations. First, we hypothesize that the strongest SNP-based rg will be between eating disorder phenotypes that have binge eating as a core symptom and alcohol use phenotypes (Munn-Chernoff and Baker, 2016). Second, we hypothesize that for binge-eating-related phenotypes, the SNP-based rg will be lowest when examining typical alcohol consumption and highest when assessing AUD (Munn-Chernoff and Baker, 2016). Because we have less information from twin studies about genetic associations between eating disorders and smoking and cannabis use-related phenotypes, we do not forward any hypotheses for these substances. Findings from this study could yield important information about this clinically challenging pattern of comorbidity (Gregorowski et al., 2013), ultimately suggesting biologically informed prevention efforts and improved treatments for patients presenting with these dual diagnoses.
Method
Participants
We included summary statistics from two existing GWAS of eating disorder phenotypes where particiants were primarily of European ancestry (Wade et al., 2013, Watson et al., in press) and six existing GWAS of substance use-related phenotypes using data only from individuals of European ancestry (Demontis et al., 2019, Hancock et al., 2017, Kranzler et al., 2019, Liu et al., 2019, Pasman et al., 2018, Walters et al., 2018). The eating disorder phenotypes (Table 1) included a diagnosis of AN (which was further parsed into AN with binge-eating or AN without binge-eating), and a BN factor score derived from the Eating Disorder Examination (EDE; Fairburn and Cooper, 1993). The EDE is a well-established, structured clinical interview used to determine eating disorder diagnoses. We did not examine BN or BED diagnoses because there are currently no published GWAS of either disorder; thus, the EDE-BN factor score represents the closest to a GWAS of BN available. Substance use-related phenotypes ranged from typical use (e.g., drinks per week, smoking initiation, and cannabis initiation) to abuse/dependence (i.e., AUD, ND, and cannabis use disorder [CUD]). Table 2 provides individual study details.
Table 1.
Eating disorder-related phenotype descriptions.
| Phenotype | Definitions |
|---|---|
| Anorexia nervosa (AN)a | Diagnostic criteria included: 1. BMI less than minimally expected 2. Intense fear of gaining weight 3. Weight or shape disturbance, undue influence of weight or shape, or denial of the seriousness of the disorder |
| AN with binge-eatingb | Individuals with AN who also engaged in binge eating episodes, defined as eating a large amount of food in a short period of time while having a sense of loss of control over the eating episode. The binge eating episodes must have occurred at least twice a week for three months. |
| AN without binge-eatingb | Individuals with AN who did not engage in binge eating episodes. |
| Bulimia nervosa (BN)c factor | Derived from a factor analysis that included the following items: 1. Reporting self-induced vomiting to control body weight 2. Reporting suffering from or being treated for binge eating 3. Reporting suffering from or being treated for bulimia |
Note:
A fourth diagnostic criterion for AN includes amenorrhea. However, amenorrhea was excluded as a required criterion for cases in the Psychiatric Genomics Consortium datasets since it is no longer a diagnostic criterion in the DSM-5.
The DSM and ICD include two subtypes of anorexia nervosa (AN)—a binge-eating/purging subtype and a restricting subtype. Although it would have been ideal to examine differences between the AN binge-eating/purging subtype and AN restricting subtype, this was not possible with current Psychiatric Genomics Consortium data. However, there was sufficient information about presence or absence of binge eating, which resulted in creating the AN with binge-eating and AN without binge-eating subtypes.
Bulimia nervosa is defined as: 1) recurrent episodes of binge eating; 2) recurrent inappropriate compensatory behaviors (e.g., self-induced vomiting, laxative use) to prevent weight gain; 3) the binge eating and inappropriate compensatory behaviors occurring an average of twice a week for three months; 4) having undue influence of body weight and shape; and 5) disturbance not occurring during AN.
Table 2.
Details of samples included in analyses.
| Study | Sample/Consortium | Phenotype(s) | Definition | Sample Size (cases / controls if binary) | Number of SNPs in summary statistics file |
|---|---|---|---|---|---|
| Eating Disorder Phenotype | |||||
| Watson et al. (in press) | PGC-ED | 1. Anorexia nervosa 2. Anorexia nervosa with binge-eating 3. Anorexia nervosa without binge-eating |
DSM-III-R, DSM-IV, ICD-8, ICD-9, ICD-10, or self-reported anorexia nervosa | 16,992 / 55,525 2,381 / 10,249 2,262 / 10,254 |
8,219,102 8,982,440 8,671,192 |
| Wade et al. (2013) | Australian Twin Registry | Bulimia nervosa factor | Eating Disorder Examination | 151 / 2,291 | 6,150,213 |
| Substance Use-Related Phenotype | |||||
| Kranzler et al. (2019) | MVP | Alcohol use disorder | ICD-9 or ICD-10 | 34,658 / 167,346 | 6,895,251 |
| Walters et al. (2018) | PGC-SUD | Alcohol dependence | DSM-IV | 8,485 / 20,272 | 9,271,145 |
| Liu et al. (2019) | GSCAN | 1. Drinks per week* 2. Smoking initiation 3. Current smokinga 4. Cigarettes per day* |
Average number of drinks each week Ever vs. never regular smoker Current vs. former smokers Average number of cigarettes smoked per day |
537,349 311,629 / 321,173 92,573 / 220,248 263,954 |
11,916,707 11,733,344 12,197,133 12,003,613 |
| Hancock et al. (2017) | 14 consortia | Nicotine dependence** | Mild (FTND score 0–3), Moderate (FTND score 4–6), or Severe (FTND score 7–10) | 14,184 (Mild) 9,206 (Moderate) 5,287 (Severe) |
10,622,668 |
| Pasman et al. (2018) | ICC UK Biobank |
Cannabis initiation | Lifetime cannabis use | 43,380 / 118,702 | 11,733,371 |
| Demontis et al. (2019) | iPSYCH | Cannabis use disorder | ICD-10 | 2,387 / 48,985 | 8,969,939 |
Note: SNPs=single nucleotide polymorphisms; PGC-ED=Eating Disorders Working Group of the Psychiatric Genomics Consortium; DSM=Diagnostic and Statistical Manual; ICD=International Classification of Diseases; PGC-SUD=Substance Use Disorders Working Group of the Psychiatric Genomics Consortium; MVP=Million Veteran Program; GSCAN=GWAS & Sequencing Consortium of Alcohol and Nicotine use; FTND=Fagerstrӧm Test of Nicotine Dependence; ICC=International Cannabis Consortium; iPSYCH=Lundbeck Foundation Initiative for Integrative Psychiatric Research.
Treated as a continuous phenotype.
Treated as an ordinal phenotype.
In Lui et al. (2019), the phenotype is labeled as “smoking cessation”. It was renamed as “current smoking” to reflect the coding scheme and for ease in comparing across all smoking phenotypes.
Statistical Analysis
We used LDSC (Bulik-Sullivan et al., 2015a, Bulik-Sullivan et al., 2015b) to evaluate SNP-based rg between samples. This method uses the linkage disequilibrium (LD) structure of the genome to estimate the distribution of effect sizes for individual SNPs as a function of their LD score. Under a polygenic model, causal SNPs are likely to be overrepresented in higher LD score bins (i.e., including additional SNPs in high LD) such that associations with SNPs in these LD bins will make stronger contributions to the phenotype under study. This polygenic distribution of effect sizes across LD score bins provides an estimate of SNP-based heritability, i.e., the proportion of phenotypic variance that is attributable to the aggregate effects of genome-wide SNPs. The correlation between distributions of effect sizes across LD bins between two phenotypes then provides an estimate of SNP-based rg.
Genetic correlations range from −1 to +1, where the sign indicates that the same genetic factors are contributing to variation in the target traits in opposite or same directions, respectively. The LDSC intercept for the genetic covariance provides evidence about sample overlap across two traits. SNPs (MAF>0.01) found in the HapMap3 EUR population were used to calculate LD scores. We used the false discovery rate (FDR; Benjamini and Hochberg, 1995) to correct for multiple testing (n=66 tests; q<0.05). Finally, post-hoc analyses examined whether significant differences between two rgs existed, using the jackknife procedure implemented through LDSC (Bulik-Sullivan et al., 2015b).
For significant rgs detected in LDSC where both the individual eating disorder and substance use-related phenotypes each had ~10 or more significant GWAS SNPs (Zhu et al., 2018), bidirectional Mendelian randomization (MR) analyses (Smith and Ebrahim, 2003) were conducted using Generalised Summary-data-based Mendelian Randomisation (GSMR; Zhu et al., 2018) to preliminarily investigate potentially causal relationships between liability to these phenotypes. As GSMR requires a reference sample with individual genotypes to account for LD, we used the 1000 Genomes Phase 3 European ancestries sample as our reference panel (1000 Genomes Project Consortium et al., 2015). SNPs with evidence of horizontal pleiotropy were excluded (using the HEIDI-Outlier method; default p-value threshold=0.01). We only included genome-wide significant SNPs (p<5x10−8) as possible instruments, and SNPs were clumped to ensure independence (i.e., amongst a set of genome-wide significant SNPs correlated at r2>0.05 within a 1-Mb window, only the SNP with the lowest p-value was retained). As MR analyses are sensitive to sample overlap, we scanned the published papers to determine which samples were included in the discovery GWAS, and any known samples in common across the two discovery GWAS were excluded from the eating disorder GWAS and summary statistics were regenerated for MR analyses.
Results
The overall SNP-based heritability for the eating disorder phenotypes ranged from 0.1992 to 0.3911, whereas the corresponding heritabilities for the substance use-related phenotypes ranged from 0.0273 to 0.3548 (Supplemental Table 1). Figure 1 and Supplemental Table 1 show the rgs between all four eating disorder phenotypes and eight substance use-related phenotypes. Broadly speaking, there were significant rgs across substance use-related phenotypes, ranging from 0.21 (AUD and cigarettes per day) to 0.70 (drinks per week and AUD). Cannabis initiation was not significantly genetically correlated with cigarettes per day or ND. For the remainder of the results, we focus on previously unexplored associations of interest in this study—correlations between eating disorder and substance use-related phenotypes. For these associations, the genetic covariance intercepts ranged from −0.0252 (standard error [SE]=0.007; AN and cannabis initiation) to 0.0113 (SE=0.0072; AN and CUD), indicating some sample overlap (or low-level confounding) existed (Yengo et al., 2018), although the LDSC approach parses this overlap from the rg estimation.
Figure 1. Genetic correlations between eating disorder subtypes and substance use-related phenotypes.
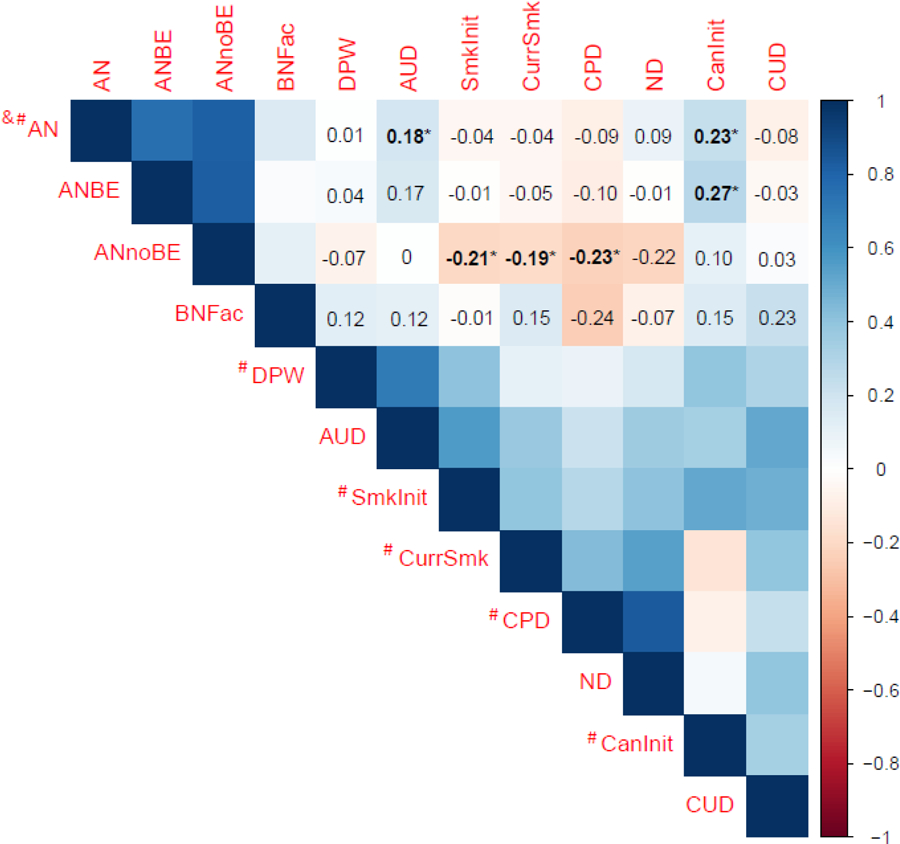
AN=anorexia nervosa; ANBE=anorexia nervosa with binge-eating; ANnoBE=anorexia nervosa without binge-eating; BNFac=bulimia nervosa factor score; DPW=drinks per week; AUD=alcohol use disorder; SmkInit=smoking initiation; CurrSmk=current smoking; CPD=cigarettes per day; ND=nicotine dependence; CanInit=cannabis initiation; CUD=cannabis use disorder. # indicates known or potential sample overlap with UK Biobank; & indicates known sample overlap with iPSYCH. Bolded and * values denote significant genetic correlations after correcting for multiple comparisons using False Discovery Rate (n tests=66; q<0.05).
Significant positive rgs were observed for alcohol use- and cannabis use-related phenotypes. First, the rg was significiant between AN and AUD (rg=0.18; SE=0.05; q=0.0006), but not between AN and drinks per week (rg=0.01; SE=0.03; q=0.91), suggesting that genetic factors that increase risk for AN also increase risk for AUD, but little evidence exists for shared risk between AN and typical alcohol consumption. These two correlations significantly differed from each other (z-score=3.51, p=0.0005). Intriguingly, there was a significant difference in rgs for AN and AUD versus AN without binge-eating and AUD (z-score=2.28, p=0.02), but not for AN and AUD versus AN with binge-eating and AUD (z-score=0.23, p=0.82). No significant association between the BN factor, which included items pertaining to both binge eating and compensatory behaviors, and either alcohol use-related phenotype was observed.
Second, the significant rg between AN and cannabis initiation was 0.23 (SE=0.04, q<0.0001), and the significant rg between AN with binge-eating and cannabis initiation was 0.27 (SE=0.08, q=0.0017), indicating that genetic factors that increase the risk for AN also increase the risk for cannabis initiation. However, cannabis initiation was not significantly correlated with the BN factor (rg=0.15, SE=0.18, q=0.57) or with AN without binge-eating (rg=0.10, SE=0.08, q=0.31). No significant associations were observed between any eating disorder phenotype and CUD (rgs=−0.08–0.23; SEs=0.01; qs<0.57). Post-hoc analyses revealed significant differences in the rgs for AN and cannabis initiation versus AN and CUD (z-score=2.70, p=0.01). However, the rg between AN with binge-eating and cannabis initiation, while significant, was statistically different from the rg between AN with binge-eating and CUD.
Conversely, for smoking phenotypes, significant correlations were only observed for the AN without binge-eating subtype. Smoking initiation (rg=−0.21, SE=0.06, q=0.0006), current smoking (referred to as smoking cessation in Liu et al., 2019)1 (rg=−0.19, SE=0.08, q=0.03), and cigarettes per day (rg=−0.23, SE=0.07, q=0.003) were significantly and negatively associated with AN without binge-eating. Although the correlation between ND and AN without binge-eating was in the same direction as the other smoking phenotypes, it was not significant (rg=−0.22, SE=0.12, q=0.14). The rgs comparing AN diagnosis and AN without binge-eating with each of the three non-diagnostic smoking traits all differed significantly from each other (z-scores ranged from −3.22 to −2.11; p-values<0.04).
Because the AN GWAS and cannabis initiation GWAS each identified eight significant loci (Pasman et al., 2018, Watson et al., in press), and the two studies comprising the AUD sample identified 10 significant loci (Kranzler et al., 2019, Walters et al., 2018), we also conducted exploratory follow-up MR analyses for AN-AUD and AN-cannabis initiation to examine whether there might be evidence of a causal relationship, given their significant rg. We used summary statistics from a subset of the AN GWAS that did not include the UK Biobank cohort for the AN-cannabis initiation analysis, as this cohort overlapped with the cannabis initiation sample (AN subset without the UK Biobank cohort, Ncases=16,224, Ncontrols=52,460). We did not obtain an estimate for the cannabis inititiation to AN direction of effect because only four SNPs were available after clumping, all of which were excluded for potential pleiotropy by the HEIDI-outlier analysis. There was no evidence of a causal relationship for any comparison (p>0.05; see Supplemental Table 2).
Discussion
Using existing GWAS data, we investigated genetic associations between liabilities to four eating disorder and eight substance use-related phenotypes spanning initiation and typical use to dependence. We found specific patterns of association between the two diagnostic categories for eating disorders, suggesting that differences between AN and BN, and between AN subtypes, with substance use-related phenotypes may point toward substance-specific genetic relationships. Additionally, there may be some degree of symptom overlap contributing to these associations.
Three main patterns emerged. First, in line with prior twin studies, we observed a positive rg between problem alcohol use (i.e., AUD) and AN diagnosis. Second, we estimated positive rgs between cannabis initiation and AN diagnosis, as well as cannabis initiation and the AN with binge-eating subtype. This is a novel finding not previously examined in twin research. The positive genetic associations suggest that some genetic loci are influencing these traits in the same direction. Second, negative genetic correlations emerged between the three smoking phenotypes from the GWAS & Sequencing Consortium of Alcohol and Nicotine use (GSCAN) cohort and AN without binge-eating, but not with the other three eating disorder phenotypes. These negative genetic correlations indicate that some of the loci influencing the liability to these eating disorder and smoking phenotypes are shared, but are affecting the liability to these traits in opposite directions. Indeed, genetic correlations cannot identify specific loci or underlying mechanisms that contribute to the shared risk. Nevertheless, the results provide initial evidence for differential genetic associations between the liability to varying eating disorder and substance use-related phenotypes.
Based on findings from twin studies, we hypothesized that: 1) the strongest SNP-based rg would be between eating disorder phenotypes that have binge eating as a core symptom and alcohol use phenotypes; and 2) a significant positive genetic correlation between eating disorder phenotypes with binge eating as a key symptom and AUD would emerge. In line with these hypotheses, we found a signifant genetic association between AUD and AN diagnosis, but not between typical alcohol consumption (i.e., drinks per week) and AN. No twin study has examined genetic association between AN and alcohol use-related phenotypes, and previous studies (Walters et al., 2018, Watson et al., in press) using LDSC have not reported significant rgs between these traits. That we found a significant association most likely reflects the larger AN sample size in our study (from 3,495 cases and 10,982 controls to 16,992 cases and 55,525 controls), as well as combining two large existing GWAS of AUD, emphasizing the importance of increasing sample sizes for GWAS.
Intriguingly, although we did not detect a significant rg for AN with binge-eating or the BN factor score with AUD, the point estimate for the rg between AUD and AN with binge eating was similar to that for AUD and AN diagnosis (0.17 vs. 0.18, respectively) and higher than AUD and AN without BE (0.01). Sample sizes for these AN subtypes were smaller than for AN diagnosis; however, the two subtypes included approximately equal numbers of cases and controls. Indeed, binge eating was assessed in such a way that we were unable to tease apart purging behaviors, and AN diagosis is heterogenous even within subytpes. Thus, binge eating may be one plausible key component of the observed genetic association. For example, binge eating has been shown to activate brain reward circuitry in a similar manner to substances (Kaye et al., 2013, Volkow et al., 2013), and administration of Naltrexone, an opioid antagonist approved by the U.S. Food and Drug Administration for the treatment of AUD (Kranzler and Soyka, 2018), has been shown to reduce the frequency of binge-eating episodes among individuals with an eating disorder (Jonas and Gold, 1988, Stancil et al., in press). Thus, our findings highlight the importance of expanding GWAS to include BN and binge-eating disorder, where a core symptom of both disorders is binge eating, to elucidate whether binge eating is a critical eating disorder symptom in the comorbidity with AUD and to home in on the relevant shared mechanisms.
The significant genetic associations between cannabis initiation and AN are novel, yet consistent with the negative genetic association between cannabis use and BMI, and with observational (Pasman et al., 2018) and experimental (Di Marzo and Matias, 2005, Volkow et al., 2017) studies regarding the role of endocannabinoids in appetite regulation, energy expenditure, stress, and reward. One of the principal psychoactive agents of cannabis, delta-9-tetrahydrocannabinol (THC), a partial agonist of the endogenous cannabinoid 1 (CB1) receptor, is presumed to be orexigenic and may acutely increase appetite and food intake, contributing to its potential role as an appetite stimulant in patients with an anorexia or cachexia syndrome (Reuter and Martin, 2016) due to a disease (e.g., HIV AIDS) or in response to treatment (e.g., chemotherapy). An antagonist of the CB1 receptor was previously tested as a highly promising anti-obesity medication (Rimonabant, SR141716). Further, the endocannabinoid anandamide has been shown to be elevated in individuals with acute AN (Monteleone and Maj, 2013), indicating disruption in food-related reward and eating behavior regulation. Animal and human studies have also provided initial evidence for the therapeutic effectiveness of cannabinoid agonists in treating eating disorders (Andries et al., 2014, Avraham et al., 2017). It is also likely that individuals with high genetic liability to AN are less likely to experiment with a substance that has a documented hyperphagia component. Thus, there is evidence of a complex biological relationship between cannabis use and eating disorders, as well as BMI. Nonetheless, the preliminary MR analyses did not provide persuasive support for causal relationships, in either direction, between genetic liability to cannabis initiation and AN. The small number of SNPs used as instruments, due to the low number of independent genome-wide significant loci and exclusion of overlapping SNPs that were not ambiguous and palindromic, may have significantly limited power to make causal inferences. Nonetheless, in the absence of strong causal evidence, several hypotheses regarding the negative genetic correlation between cannabis initiation and AN are possible.
Finally, the significant negative rgs between three smoking phenotypes—smoking initiation, current smoking, and cigarettes per day—and AN without binge-eating are intriguing, suggesting that increased genetic liability to AN without binge-eating is associated with decreased genetic liability to multiple smoking behaviors. Phenotypic studies are inconsistent about the association between the restricting subtype of AN and smoking. Some studies suggest that individuals with restricting AN have a higher prevalence of various smoking phenotypes than controls (Krug et al., 2008), whereas other studies indicate no significant difference between the two groups (Anzengruber et al., 2006). A recent meta-analysis did not find differences in the odds of lifetime smoking between individuals with AN and healthy controls (Solmi et al., 2016), yet the authors did not assess differences by AN subtype. Individuals with AN may smoke as a way to control or lose weight (White, 2011), and temporary weight gain does occur with smoking cessation (Filozof et al., 2004). However, a positive phenotypic correlation need not be accompanied by a rg in the same direction (or genetic contributors to the phenotypic association at all). Still, there is plausible support for the negative genetic correlation. Although not significant, a negative genetic correlation between smoking and AN has been reported (Bulik-Sullivan et al., 2015a, Watson et al., in press). Notably, our study includes individuals from these earlier reports and extends findings by including larger sample sizes for both AN and smoking phenotypes. Even though it is not evident from the existing literature that these opposing directions of effect directly relate to a shared predisposition versus a causal process, there is speculative support for loci related to nicotine addiction that might also influence decreased liability to food intake (Mineur et al., 2011). Unfortunately, there are no twin studies of AN or AN-like traits and smoking with which to compare findings. Such speculations should be reviewed as one of several possible mechanisms that link smoking to AN, as AN is a complex multi-faceted disorder that extends well-beyond reduced food intake.
Another explanation for the negative genetic association is that it is due to a third, underlying variable influencing both AN without binge-eating and smoking. In the largest GWAS of smoking phenotypes, positive genetic correlations were observed between smoking initiation and cigarettes per day with multiple cardiometabolic traits, including type 2 diabetes and fasting glucose (Liu et al., 2019). These same metabolic traits were negatively genetically correlated with AN (Duncan et al., 2017, Watson et al., in press). Thus, the patterns of rgs point to metabolic, rather than psychiatric, factors in influencing the apparent genetic association between smoking phenotypes and AN. However, the associations could also reflect adoption of unhealthy lifestyles that promote obesity and are correlated with smoking. In addition, the rgs between smoking and BMI, as well as AN and BMI, could reflect underlying disinhibitory pathways, as variants associated with BMI show enrichment in the central nervous system (Goodarzi, 2018). The current approach is not designed to disentangle these putative etiological mechanisms, but our findings do encourage careful study of the specific relationships between eating and substance use disorders.
Substance use and substance use disorders are partially distinct, and although excessive substance use is a necessary component of it, substance use disorders relate to psychological and physiological impairment related to excess use and aspects of loss of control over the behavior. Consistent with our findings for alcohol, accumulating evidence suggests that genetic liability to other psychiatric traits (e.g., schizophrenia) is strongly correlated with liability to substance use disorders (e.g., AUD) but not substance use (e.g., alcohol consumption). Genetic liability to alcohol use has also been correlated with liability to psychiatric traits (e.g., major depression) in opposite directions depending on level of involvement (Kranzler et al., 2019). However, we did not find similar elevations in rgs when contrasting ever smoking and ND, nor comparing cannabis initiation to CUD. It is possible that the lack of genetic overlap between AN and ND, as well as AN and CUD, is related to the relatively modest sample size of those discovery GWAS. A similar non-significant rg was noted for AUD when the Walters et al. (2018) alcohol dependence GWAS was used as the sole source of summary statistics for problem drinking in the current study. Still, there are several other explanations for this divergence in findings. For instance, for tobacco, the highly addictive nature of nicotine may result in convergence in genomic effects on earlier and later stages of smoking (i.e., a much larger proportion of those who ever smoke become dependent compared with the proportion of those who drink alcohol and develop AUD). For cannabis, given its lower addictive potential, we might have expected stronger associations with CUD than cannabis initiation. In addition to the considerably smaller sample size of the CUD GWAS, the association with cannabis initiation could also be attributed to the small number of cohorts in that discovery GWAS that included individuals with a high likelihood of CUD. It is also possible that the relationship between AN and cannabis use is distinct and that earlier, but not later stages of cannabis use are genetically related to liability to AN. Future studies should consider the multi-stage nature of substance use and misuse when examining cross-trait correlations.
This is the largest and most comprehensive assessment of shared genetic risk between eating disorder and substance use-related phenotypes to date, using existing GWAS data from large cohorts (up to ~537,000 individuals per phenotype). We were able to separately assess approximate AN subtypes (i.e., with binge-eating vs. without binge-eating) to evaluate the extent to which binge eating, in the context of AN, may share genetic risk with substance use-related phenotypes. Using these large datasets—many of which are publically available—allows for the rapid development of scientific knowledge regarding the underlying etiology of psychiatric disorder and substance use comorbidity. Nevertheless, some limitations exist. First, sample sizes for the BN factor score and CUD GWAS were relatively small compared with the other GWAS, resulting in large standard errors and low power. Second, we were unable to uniformally examine sex differences in these rgs. Since the prevalence of eating disorders is higher in women than men, and the prevalence of substance use disorders is higher in men than women (American Psychiatric Association, 2013), it will be important to explore possible sex differences in genetic associations as the GWAS data become available. Notably, we previously did not find evidence for sex differences in the rg between binge eating and problem alcohol use (Munn-Chernoff et al., 2013). Finally, SNP coverage was limited in the earlier GWAS of the BN factor score because that study used older genotyping platforms and imputation panels that included fewer SNPs than current imputation panels. The Eating Disorders and Substance Use Disorders Working Groups of the Psychiatric Genomics Consortium (PGC) are continuously adding samples and releasing data freezes with incrementally larger sample sizes, while collecting information on multiple substances (e.g., opioids). Thus, in coming years, the statistical power is expected to increase for AN (including the with and without binge-eating subtypes), BN, and BED, as well as AUD, ND, and CUD, from within and outside the PGC. This will allow for a more refined assessment of specific eating disorder symptoms, including binge eating, in relation to substance use-related phenotypes.
In conclusion, findings from this study suggest that the underlying etiology between eating disorder and substance use-related phenotypes is not consistent across traits or levels of substance involvement, extending results from twin studies to a genome-wide SNP approach. Despite the typically high co-occurrence of alcohol, tobacco, and cannabis use, and their genetic overlap (Pasman et al., 2018), the differential patterns seen between the eating disorder and substance use-related phenotypes highlights the uniqueness and complexity of their shared etiology. Additional research using contemporary genomic methods such as cross-disorder association studies could identify the specific loci contributing to this comorbidity. Once loci are identified, additional research that combines polygenic risk scores with measured environmental constructs could enhance the prediction, prevention, and treatment of co-occurring eating disorder and substance use-related traits.
Supplementary Material
Acknowledgements
Grant support for individual authors can be found in Supplementary Table 3. This study included summary statistics of a genetic study on cannabis use (Pasman et al. [2018] Nature Neuroscience). We would like to acknowledge all participating groups of the International Cannabis Consortium, and in particular, the members of the working group including Joelle Pasman, Karin Verweij, Nathan Gillespie, Eske Derks, and Jacqueline Vink. Pasman et al. (2018) included data from the UK Biobank resource under application numbers 9905, 16406, and 25331.
Eating Disorders Working Group of the Psychiatric Genomics Consortium (PGC-ED)
We thank all study volunteers, study coordinators, and research staff who enabled this study. ANGI: The Anorexia Nervosa Genetics Initiative was an initiative of the Klarman Family Foundation. Additional support was offered by the National Institute of Mental Health. We acknowledge support from the North Carolina Translational and Clinical Sciences Institute (NC TraCS) and the Carolina Data Warehouse. PGC: We are deeply indebted to the investigators who comprise the PGC, and to the hundreds of thousands of individuals who have shared their life experiences with PGC investigators and the contributing studies. We are grateful to the Children’s Hospital of Philadelphia (CHOP), the Price Foundation Collaborative Group (PFCG), Genetic Consortium for Anorexia Nervosa (GCAN), Wellcome Trust Case-Control Consortium-3 (WTCCC-3), the Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH), the QSkin Sun and Health Study, Riksät (Swedish National Quality Register for Eating Disorders), the Stockholm Center for Eating Disorders (SCÄ), LifeGene, the UK Biobank, and all PGC-ED members for their support in providing individual samples used in this study. We thank SURFsara (http://www.surf.nl) for support in using the Lisa Compute Cluster. We thank Max Lam, Institute of Mental Health, Singapore, for Ricopili consultation. This study also represents independent research partly funded by the English National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the English Department of Health and Social Care. High performance computing facilities were funded with capital equipment grants from the GSTT Charity (TR130505) and Maudsley Charity (980). Research reported in this publication was supported by the National Institute of Mental Health of the US National Institutes of Health under Award Number U01MH109514. The content is solely the responsibility of the authors and does not necessarily represent the official views of the US National Institutes of Health.
Substance Use Disorders Working Group of the Psychiatric Genomics Consortium (PGC-SUD)
The PGC-SUD receives support from the National Institute on Drug Abuse and the National Institute of Mental Health via MH109532. We gratefully acknowledge prior support from the National Institute on Alcohol Abuse and Alcoholism. Statistical analyses for the PGC were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org) hosted by SURFsara and financially supported by the Netherlands Scientific Organization (NWO 480–05-003) along with a supplement from the Dutch Brain Foundation and the VU University Amsterdam. Cohort specific acknowledgements may be found in Walters et al. (2018) Nature Neuroscience.
Footnotes
Competing Financial Interests
The authors report the following potential competing interests. O. Andreassen received a speaker’s honorarium from Lundbeck. G. Breen received grant funding and consultancy fees in preclinical genetics from Eli Lilly, consultancy fees from Otsuka and has received honoraria from Illumina. C. Bulik served on Shire Scientific Advisory Boards; she receives author royalties from Pearson. D. Degortes served as a speaker and on advisory boards, and has received consultancy fees for participation in research from various pharmaceutical industry companies including: AstraZeneca, Boehringer, Bristol Myers Squibb, Eli Lilly, Genesis Pharma, GlaxoSmithKline, Janssen, Lundbeck, Organon, Sanofi, UniPharma, and Wyeth; he has received unrestricted grants from Lilly and AstraZeneca as director of the Sleep Research Unit of Eginition Hospital (National and Kapodistrian University of Athens, Greece). J. Hudson has received grant support from Shire and Sunovion, and has received consulting fees from DiaMentis, Shire, and Sunovion. A. Kaplan is a member of the Shire Canadian BED Advisory Board and is on the steering committee for the Shire B/educated Educational Symposium: June 15–16, 2018. J. Kennedy served as an unpaid member of the scientific advisory board of AssurexHealth Inc. M. Landén declares that, over the past 36 months, he has received lecture honoraria from Lundbeck and served as scientific consultant for EPID Research Oy. No other equity ownership, profit-sharing agreements, royalties, or patent. P. Sullivan is on the Lundbeck advisory committee and is a Lundbeck grant recipient; he has served on the scientific advisory board for Pfizer, has received a consultation fee from Element Genomics, and a speaker reimbursement fee from Roche. J. Treasure has received an honorarium for participation in an EAP meeting and has received royalties from several books from Routledge, Wiley, and Oxford University press. T. Werge has acted as a lecturer and scientific advisor to H. Lundbeck A/S. L. Bierut, A. Goate, J. Rice, J.-C. Wang, and the spouse of N. Saccone are listed as inventors on Issued US Patent 8080,371, “Markers for Addiction” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. N. Wodarz has received funding from the German Research Foundation (DFG) and Federal Ministry of Education and Research Germany (BMBF); he has received speaker’s honoraria and travel funds from Janssen-Cilag and Indivior. He took part in industry-sponsored multicenter randomized trials by D&A Pharma and Lundbeck. M. Ridinger received compensation from Lundbeck Switzerland and Lundbeck institute for advisory boards and expert meetings, and from Lundbeck and Lilly Suisse for workshops and presentations. K. Mann received honoraria from Lundbeck, Pfizer, Novartis, and AbbVie. K. Mann also received Honoraria (Advisory Board) from Lundbeck and Pfizer and speaker fees from Janssen Cilag. H. Kranzler has been an advisory board member, consultant, or continuing medical education speaker for Indivior, Lundbeck, and Otsuka. He is a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was sponsored in the past three years by AbbVie, Alkermes, Amygdala Neurosciences, Arbor Pharmaceuticals, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, and Pfizer. H. Kranzler and J. Gelernter are named as inventors on PCT patent application #15/878,640, entitled “Genotype-guided dosing of opioid agonists,” filed 24 January 2018. D.-S. Choi is a scientific advisory member of Peptron Inc. M. Frye has received grant support from Assurex Health, Mayo Foundation, Myriad, NIAAA, National Institute of Mental Health (NIMH), and Pfizer; he has been a consultant for Intra-Cellular Therapies, Inc., Janssen, Mitsubishi Tanabe Pharma Corporation, Myriad, Neuralstem Inc., Otsuka American Pharmaceutical, Sunovion, and Teva Pharmaceuticals. H. de Wit has received support from Insys Therapeutics and Indivior for studies unrelated to this project, and she has consulted for Marinus and Jazz Pharmaceuticals, also unrelated to this project. T. Wall has previously received funds from ABMRF. J. Nurnberger is an investigator for Janssen and Assurex. M. Nöthen has received honoraria from the Lundbeck Foundation and the Robert Bosch Stiftung for membership on advisory boards. N. Scherbaum has received honoraria from Abbvie, Sanofi-Aventis, Reckitt Benckiser, Indivior, Lundbeck, and Janssen-Cilag for advisory board membership and the preparation of lectures, manuscripts, and educational materials. Since 2013, N. Scherbaum has also participated in clinical trials financed by Reckitt Benckiser and Indivior. W. Gäbel has received symposia support from Janssen-Cilag GmbH, Neuss, Lilly Deutschland GmbH, Bad Homburg, and Servier, Munich, and is a member of the Faculty of the Lundbeck International Neuroscience Foundation (LINF), Denmark. J. Kaprio has provided consultations on nicotine dependence for Pfizer (Finland) 2012–2015. In the past three years, L. Degenhardt has received investigator-initiated untied educational grants for studies of opioid medications in Australia from Indivior, Mundipharma, and Seqirus. B. Neale is a member of the scientific advisory board for Deep Genomics and has consulted for Camp4 Therapeutics Corporation, Merck & Co., and Avanir Pharmaceuticals, Inc. A. Agrawal previously received peer-reviewed funding and travel reimbursement from ABMRF for unrelated research. All other authors have no conflicts of interest to disclose.
In Liu et al. (2019), the phenotype is noted as “smoking cessation”, where current smokers were coded as 2 and former smokers were coded as 1. Because the comparison group is “current smokers”, we have renamed this phenotype as “current smoking” for clarification and ease of interpretation across all smoking phenotypes.
References
- 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR (2015). A global reference for human genetic variation. Nature 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andries A, Frystyk J, Flyvbjerg A, Stoving RK (2014). Dronabinol in severe, enduring anorexia nervosa: A randomized controlled trial. Int J Eat Disord 47:18–23. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association (2013). Diagnostic and Statistical Manual of Mental Disorders. American Psychiatric Publishing: Arlington, Virginia. [Google Scholar]
- Anzengruber D, Klump KL, Thornton L, Brandt H, Crawford S, Fichter MM, Halmi KA, Johnson C, Kaplan AS, LaVia M, Mitchell J, Strober M, Woodside DB, Rotondo A, Berrettini WH, Kaye WH, Bulik CM (2006). Smoking in eating disorders. Eat Behav 7:291–299. [DOI] [PubMed] [Google Scholar]
- Avraham Y, Paturski I, Magen I, Vorobiev L, Berry EM (2017). 2-Arachidonoylglycerol as a possible treatment for anorexia nervosa in animal model in mice. Brain Res 1670:185–190. [DOI] [PubMed] [Google Scholar]
- Baker JH, Mazzeo SE, Kendler KS (2007). Association between broadly defined bulimia nervosa and drug use disorders: Common genetic and environmental influences. Int J Eat Disord 40:673–678. [DOI] [PubMed] [Google Scholar]
- Baker JH, Mitchell KS, Neale MC, Kendler KS (2010). Eating disorder symptomatology and substance use disorders: Prevalence and shared risk in a population based twin sample. Int J Eat Disord 43:648–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JH, Munn-Chernoff MA, Lichtenstein P, Larsson H, Maes H, Kendler KS (2017). Shared familial risk between bulimic symptoms and alcohol involvement during adolescence. J Abnorm Psychol 126:506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y and Hochberg Y (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Statist Soc, Series B, 57:449–518. [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, ReproGen Consortium, Psychiatric Genomics Consortium, Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3, Duncan L, Perry JR, Patterson N, Robinson EB, Daly MJ, Price AL, Neale BM (2015a). An atlas of genetic correlations across human diseases and traits. Nat Genet 47:1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium, Patterson N, Daly MJ, Price AL, Neale BM (2015b). LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center on Addiction and Substance Abuse (2003). Food for thought: Substance abuse and eating disorders. Columbia University (ed). The National Center on Addiction and Substance Abuse at Columbia University: New York, NY: pp 1–83. [Google Scholar]
- Demontis D, Rajagopal VM, Thorgeirsson TE, Als TD, Grove J, Leppala K, Gudbjartsson DF, Pallesen J, Hjorthoj C, Reginsson GW, Tyrfingsson T, Runarsdottir V, Qvist P, Christensen JH, Bybjerg-Grauholm J, Baekvad-Hansen M, Huckins LM, Stahl EA, Timmermann A, Agerbo E, Hougaard DM, Werge T, Mors O, Mortensen PB, Nordentoft M, Daly MJ, Stefansson H, Stefansson K, Nyegaard M, Borglum AD (2019). Genome-wide association study implicates CHRNA2 in cannabis use disorder. Nat Neurosci 22:1066–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marzo V and Matias I (2005). Endocannabinoid control of food intake and energy balance. Nat Neurosci 8:585–589. [DOI] [PubMed] [Google Scholar]
- Duncan AE, Neuman RJ, Kramer JR, Kuperman S, Hesselbrock VM, Bucholz KK (2006). Lifetime psychiatric comorbidity of alcohol dependence and bulimia nervosa in women. Drug Alcohol Depend 84:122–132. [DOI] [PubMed] [Google Scholar]
- Duncan L, Yilmaz Z, Gaspar H, Walters R, Goldstein J, Anttila V, Bulik-Sullivan B, Ripke S, Eating Disorders Working Group of the Psychiatric Genomics Consortium, Thornton L, Hinney A, Daly M, Sullivan PF, Zeggini E, Breen G, Bulik CM (2017). Significant locus and metabolic genetic correlations revealed in genome-wide association study of anorexia nervosa. Am J Psychiatry 174:850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairburn CG and Cooper Z (1993). The Eating Disorder Examination In: Binge Eating: Nature, Assessment and Treatment. Fairburn CG, Wilson GT (eds). Guilford Press: New York: pp. 317–359. [Google Scholar]
- Filozof C, Fernandez Pinilla MC, Fernandez-Cruz A (2004). Smoking cessation and weight gain. Obes Rev 5:95–103. [DOI] [PubMed] [Google Scholar]
- Franko DL, Keshaviah A, Eddy KT, Krishna M, Davis MC, Keel PK, Herzog DB (2013). A longitudinal investigation of mortality in anorexia nervosa and bulimia nervosa. Am J Psychiatry 170:917–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadalla T and Piran N (2007). Co-occurrence of eating disorders and alcohol use disorders in women: A meta analysis. Arch Womens Ment Health 10:133–140. [DOI] [PubMed] [Google Scholar]
- Goodarzi MO (2018). Genetics of obesity: What genetic association studies have taught us about the biology of obesity and its complications. Lancet Diabetes Endocrinol 6:223–236. [DOI] [PubMed] [Google Scholar]
- Gregorowski C, Seedat S, Jordaan GP (2013). A clinical approach to the assessment and management of co-morbid eating disorders and substance use disorders. BMC Psychiatry 13, 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock DB, Guo Y, Reginsson GW, Gaddis NC, Lutz SM, Sherva R, Loukola A, Minica CC, Markunas CA, Han Y, Young KA, Gudbjartsson DF, Gu F, McNeil DW, Qaiser B, Glasheen C, Olson S, Landi MT, Madden PAF, Farrer LA, Vink J, Saccone NL, Neale MC, Kranzler HR, McKay J, Hung RJ, Amos CI, Marazita ML, Boomsma DI, Baker TB, Gelernter J, Kaprio J, Caporaso NE, Thorgeirsson TE, Hokanson JE, Bierut LJ, Stefansson K, Johnson EO (2017). Genome-wide association study across European and African American ancestries identifies a SNP in DNMT3B contributing to nicotine dependence. Mol Psychiatry 23:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG (1997). Genetic and environmental contributions to alcohol dependence risk in a national twin sample: Consistency of findings in women and men. Psychol Med 27:1381–1396. [DOI] [PubMed] [Google Scholar]
- Jonas JM and Gold MS (1988). The use of opiate antagonists in treating bulimia: A study of low-dose versus high-dose naltrexone. Psychiatry Res 24:195–199. [DOI] [PubMed] [Google Scholar]
- Kaye WH, Wierenga CE, Bailer UF, Simmons AN, Wagner A, Bischoff-Grethe A (2013). Does a shared neurobiology for foods and drugs of abuse contribute to extremes of food ingestion in anorexia and bulimia nervosa? Biol Psychiatry 73, 836–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzler HR and Soyka M (2018). Diagnosis and pharmacotherapy of alcohol use disorder: A review. JAMA 320:815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, Tsao PS, Klarin D, Baras A, Reid J, Overton J, Rader DJ, Cheng Z, Tate JP, Becker WC, Concato J, Xu K, Polimanti R, Zhao H, Gelernter J (2019). Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun 10:1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug I, Treasure J, Anderluh M, Bellodi L, Cellini E, di BM, Granero R, Karwautz A, Nacmias B, Penelo E, Ricca V, Sorbi S, Tchanturia K, Wagner G, Collier D, Fernandez-Aranda F (2008). Present and lifetime comorbidity of tobacco, alcohol and drug use in eating disorders: A European multicenter study. Drug Alcohol Depend 97:169–179. [DOI] [PubMed] [Google Scholar]
- Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J, McGuire D, Tian C, Zhan X, 23andMe Research Team, HUNT All-In Psychiatry, Choquet H, Docherty AR, Faul JD, Foerster JR, Fritsche LG, Gabrielsen ME, Gordon SD, Haessler J, Hottenga JJ, Huang H, Jang SK, Jansen PR, Ling Y, Magi R, Matoba N, McMahon G, Mulas A, Orru V, Palviainen T, Pandit A, Reginsson GW, Skogholt AH, Smith JA, Taylor AE, Turman C, Willemsen G, Young H, Young KA, Zajac GJM, Zhao W, Zhou W, Bjornsdottir G, Boardman JD, Boehnke M, Boomsma DI, Chen C, Cucca F, Davies GE, Eaton CB, Ehringer MA, Esko T, Fiorillo E, Gillespie NA, Gudbjartsson DF, Haller T, Harris KM, Heath AC, Hewitt JK, Hickie IB, Hokanson JE, Hopfer CJ, Hunter DJ, Iacono WG, Johnson EO, Kamatani Y, Kardia SLR, Keller MC, Kellis M, Kooperberg C, Kraft P, Krauter KS, Laakso M, Lind PA, Loukola A, Lutz SM, Madden PAF, Martin NG, McGue M, McQueen MB, Medland SE, Metspalu A, Mohlke KL, Nielsen JB, Okada Y, Peters U, Polderman TJC, Posthuma D, Reiner AP, Rice JP, Rimm E, Rose RJ, Runarsdottir V, Stallings MC, Stancakova A, Stefansson H, Thai KK, Tindle HA, Tyrfingsson T, Wall TL, Weir DR, Weisner C, Whitfield JB, Winsvold BS, Yin J, Zuccolo L, Bierut LJ, Hveem K, Lee JJ, Munafo MR, Saccone NL, Willer CJ, Cornelis MC, David SP, Hinds DA, Jorgenson E, Kaprio J, Stitzel JA, Stefansson K, Thorgeirsson TE, Abecasis G, Liu DJ, Vrieze S (2019). Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet 51:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineur YS, Abizaid A, Rao Y, Salas R, DiLeone RJ, Gundisch D, Diano S, De Biasi M, Horvath TL, Gao XB, Picciotto MR (2011). Nicotine decreases food intake through activation of POMC neurons. Science 332:1330–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone P and Maj M (2013). Dysfunctions of leptin, ghrelin, BDNF and endocannabinoids in eating disorders: Beyond the homeostatic control of food intake. Psychoneuroendocrinology 38, 312–330. [DOI] [PubMed] [Google Scholar]
- Munn-Chernoff MA and Baker JH (2016). A primer on the genetics of comorbid eating disorders and substance use disorders. Eur Eat Disord Rev 24:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn-Chernoff MA, Duncan AE, Grant JD, Wade TD, Agrawal A, Bucholz KK, Madden PAF, Martin NG, Heath AC (2013). A twin study of the association between alcohol dependence, binge eating, and compensatory behaviors. J Stud Alcohol Drugs 74, 664–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn-Chernoff MA, Grant JD, Agrawal A, Sartor CE, Werner KB, Bucholz KK, Madden PA, Heath AC, Duncan AE (2015). Genetic overlap between alcohol use disorder and bulimic behaviors in European American and African American women. Drug Alcohol Depend 153:335–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasman JA, Verweij KJH, Gerring Z, Stringer S, Sanchez-Roige S, Treur JL, Abdellaoui A, Nivard MG, Baselmans BML, Ong JS, Ip HF, van der Zee MD, Bartels M, Day FR, Fontanillas P, Elson SL, 23andMe Research Team, de Wit H, Davis LK, MacKillop J, Substance Use Disorders Working Group of the Psychiatric Genomics Consortium, International Cannabis Consoritum, Derringer JL, Branje SJT, Hartman CA, Heath AC, van Lier PAC, Madden PAF, Magi R, Meeus W, Montgomery GW, Oldehinkel AJ, Pausova Z, Ramos-Quiroga JA, Paus T, Ribases M, Kaprio J, Boks MPM, Bell JT, Spector TD, Gelernter J, Boomsma DI, Martin NG, MacGregor S, Perry JRB, Palmer AA, Posthuma D, Munafo MR, Gillespie NA, Derks EM, Vink JM (2018). GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat Neurosci 21:1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter SE and Martin JH (2016). Pharmacokinetics of cannabis in cancer cachexia-anorexia syndrome. Clin Pharmacokinet 55:807–812. [DOI] [PubMed] [Google Scholar]
- Rhee SH, Hewitt JK, Young SE, Corley RP, Crowley TJ, Stallings MC (2003). Genetic and environmental influences on substance initiation, use, and problem use in adolescents. Arch Gen Psychiatry 60:1256–1264. [DOI] [PubMed] [Google Scholar]
- Root TL, Pisetsky EM, Thornton L, Lichtenstein P, Pedersen NL, Bulik CM (2010). Patterns of co-morbidity of eating disorders and substance use in Swedish females. Psychol Med 40:105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slane JD, Burt SA, Klump KL (2012). Bulimic behaviors and alcohol use: Shared genetic influences. Behav Genet 42:603–613. [DOI] [PubMed] [Google Scholar]
- Smith GD and Ebrahim S (2003). ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 32:1–22. [DOI] [PubMed] [Google Scholar]
- Solmi M, Veronese N, Sergi G, Luchini C, Favaro A, Santonastaso P, Vancampfort D, Correll CU, Ussher M, Thapa-Chhetri N, Fornaro M, Stubbs B (2016). The association between smoking prevalence and eating disorders: A systematic review and meta-analysis. Addiction 111, 1914–1922. [DOI] [PubMed] [Google Scholar]
- Stancil SL, Adelman W, Dietz A, Abdel-Rahman S (in press). Naltrexone reduces binge eating and purging in adolescents in an eating disorder program. J Child Adolesc Psychopharmacol. doi: 10.1089/cap.2019.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trace SE, Thornton LM, Baker JH, Root TL, Janson LE, Lichtenstein P, Pedersen NL, Bulik CM (2013). A behavioral-genetic investigation of bulimia nervosa and its relationship with alcohol use disorder. Psychiatry Res 208:232–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True WR, Heath AC, Scherrer JF, Waterman B, Goldberg J, Lin N, Eisen SA, Lyons MJ, Tsuang MT (1997). Genetic and environmental contributions to smoking. Addiction 92:1277–1287. [PubMed] [Google Scholar]
- van den Bree MB, Johnson EO, Neale MC, Pickens RW (1998). Genetic and environmental influences on drug use and abuse/dependence in male and female twins. Drug Alcohol Depend 52:231–241. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Hampson AJ, Baler RD (2017). Don’t worry, be happy: Endocannabinoids and cannabis at the intersection of stress and reward. Annu Rev Pharmacol Toxicol 57:285–308. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Tomasi D, Baler RD (2013). The addictive dimensionality of obesity. Biol Psychiatry 73:811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade TD, Gordon S, Medland S, Bulik CM, Heath AC, Montgomery GW, Martin NG (2013). Genetic variants associated with disordered eating. Int J Eat Disord 46:594–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, Aliev F, Bacanu SA, Batzler A, Bertelsen S, Biernacka JM, Bigdeli TB, Chen LS, Clarke TK, Chou YL, Degenhardt F, Docherty AR, Edwards AC, Fontanillas P, Foo JC, Fox L, Frank J, Giegling I, Gordon S, Hack LM, Hartmann AM, Hartz SM, Heilmann-Heimbach S, Herms S, Hodgkinson C, Hoffmann P, Jan Hottenga J, Kennedy MA, Alanne-Kinnunen M, Konte B, Lahti J, Lahti-Pulkkinen M, Lai D, Ligthart L, Loukola A, Maher BS, Mbarek H, McIntosh AM, McQueen MB, Meyers JL, Milaneschi Y, Palviainen T, Pearson JF, Peterson RE, Ripatti S, Ryu E, Saccone NL, Salvatore JE, Sanchez-Roige S, Schwandt M, Sherva R, Streit F, Strohmaier J, Thomas N, Wang JC, Webb BT, Wedow R, Wetherill L, Wills AG, 23andMe Research Team,Boardman JD, Chen D, Choi DS, Copeland WE, Culverhouse RC, Dahmen N, Degenhardt L, Domingue BW, Elson SL, Frye MA, Gabel W, Hayward C, Ising M, Keyes M, Kiefer F, Kramer J, Kuperman S, Lucae S, Lynskey MT, Maier W, Mann K, Mannisto S, Muller-Myhsok B, Murray AD, Nurnberger JI, Palotie A, Preuss U, Raikkonen K, Reynolds MD, Ridinger M, Scherbaum N, Schuckit MA, Soyka M, Treutlein J, Witt S, Wodarz N, Zill P, Adkins DE, Boden JM, Boomsma DI, Bierut LJ, Brown SA, Bucholz KK, Cichon S, Costello EJ, de Wit H, Diazgranados N, Dick DM, Eriksson JG, Farrer LA, Foroud TM, Gillespie NA, Goate AM, Goldman D, Grucza RA, Hancock DB, Harris KM, Heath AC, Hesselbrock V, Hewitt JK, Hopfer CJ, Horwood J, Iacono W, Johnson EO, Kaprio JA, Karpyak VM, Kendler KS, Kranzler HR, Krauter K, Lichtenstein P, Lind PA, McGue M, MacKillop J, Madden PAF, Maes HH, Magnusson P, Martin NG, Medland SE, Montgomery GW, Nelson EC, Nothen MM, Palmer AA, Pedersen NL, Penninx B, Porjesz B, Rice JP, Rietschel M, Riley BP, Rose R, Rujescu D, Shen PH, Silberg J, Stallings MC, Tarter RE, Vanyukov MM, Vrieze S, Wall TL, Whitfield JB, Zhao H, Neale BM, Gelernter J, Edenberg HJ, Agrawal A (2018). Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci 21:1656–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson HJ, Yilmaz Z, Thornton LM, Hubel C, Coleman JRI, Gaspar HA, Bryois J, Hinney A, Leppa VM, Mattheisen M, Medland SE, Ripke S, Yao S, Giusti-Rodriguez P, Anorexia Nervosa Genetics Initiative, Hanscombe KB, Purves KL, Eating Disorders Working Group of the Psychiatric Genomics Consortium, Adan RAH, Alfredsson L, Ando T, Andreassen OA, Baker JH, Berrettini WH, Boehm I, Boni C, Perica VB, Buehren K, Burghardt R, Cassina M, Cichon S, Clementi M, Cone RD, Courtet P, Crow S, Crowley JJ, Danner UN, Davis OSP, de Zwaan M, Dedoussis G, Degortes D, DeSocio JE, Dick DM, Dikeos D, Dina C, Dmitrzak-Weglarz M, Docampo E, Duncan LE, Egberts K, Ehrlich S, Escaramis G, Esko T, Estivill X, Farmer A, Favaro A, Fernandez-Aranda F, Fichter MM, Fischer K, Focker M, Foretova L, Forstner AJ, Forzan M, Franklin CS, Gallinger S, Giegling I, Giuranna J, Gonidakis F, Gorwood P, Mayora MG, Guillaume S, Guo Y, Hakonarson H, Hatzikotoulas K, Hauser J, Hebebrand J, Helder SG, Herms S, Herpertz-Dahlmann B, Herzog W, Huckins LM, Hudson JI, Imgart H, Inoko H, Janout V, Jimenez-Murcia S, Julia A, Kalsi G, Kaminska D, Kaprio J, Karhunen L, Karwautz A, Kas MJH, Kennedy JL, Keski-Rahkonen A, Kiezebrink K, Kim YR, Klareskog L, Klump KL, Knudsen GPS, La Via MC, Le Hellard S, Levitan RD, Li D, Lilenfeld L, Lin BD, Lissowska J, Luykx J, Magistretti PJ, Maj M, Mannik K, Marsal S, Marshall CR, Mattingsdal M, McDevitt S, McGuffin P, Metspalu A, Meulenbelt I, Micali N, Mitchell K, Monteleone AM, Monteleone P, Munn-Chernoff MA, Nacmias B, Navratilova M, Ntalla I, O’Toole JK, Ophoff RA, Padyukov L, Palotie A, Pantel J, Papezova H, Pinto D, Rabionet R, Raevuori A, Ramoz N, Reichborn-Kjennerud T, Ricca V, Ripatti S, Ritschel F, Roberts M, Rotondo A, Rujescu D, Rybakowski F, Santonastaso P, Scherag A, Scherer SW, Schmidt U, Schork NJ, Schosser A, Seitz J, Slachtova L, Slagboom PE, Slof-Op’t Landt MCT, Slopien A, Sorbi S, Swiatkowska B, Szatkiewicz JP, Tachmazidou I, Tenconi E, Tortorella A, Tozzi F, Treasure J, Tsitsika A, Tyszkiewicz-Nwafor M, Tziouvas K, van Elburg AA, van Furth EF, Wagner G, Walton E, Widen E, Zeggini E, Zerwas S, Zipfel S, Bergen AW, Boden JM, Brandt H, Crawford S, Halmi KA, Horwood LJ, Johnson C, Kaplan AS, Kaye WH, Mitchell JE, Olsen CM, Pearson JF, Pedersen NL, Strober M, Werge T, Whiteman DC, Woodside DB, Stuber GD, Gordon S, Grove J, Henders AK, Jureus A, Kirk KM, Larsen JT, Parker R, Petersen L, Jordan J, Kennedy M, Montgomery GW, Wade TD, Birgegard A, Lichtenstein P, Norring C, Landen M, Martin NG, Mortensen PB, Sullivan PF, Breen G, Bulik CM (in press). Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat Genet doi: 10.1038/s41588-019-0439-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA (2011). Smoking for weight control and its associations with eating disorder symptomatology. Compr Psychiatry 53:403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederman MW and Pryor T (1996). Substance use among women with eating disorders. Int J Eat Disord 20:163–168. [DOI] [PubMed] [Google Scholar]
- Yengo L, Yang J, Visscher PM (2018). Expectation of the intercept from bivariate LD score regression in the presence of population stratification. bioRxiv, https://doi.org.10.1101/310565. [Google Scholar]
- Zhu Z, Zheng Z, Zhang F, Wu Y, Trzaskowski M, Maier R, Robinson MR, McGrath JJ, Visscher PM, Wray NR, Yang J (2018). Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun 9, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


