Abstract
During the wound healing process, human mesenchymal stem cells (hMSCs) are recruited to the injury where they regulate inflammation and initiate healing and tissue regeneration. To aid in healing, synthetic cell-laden hydrogel scaffolds are being designed to deliver additional hMSCs to wounds to enhance or restart the healing process. These scaffolds are being designed to mimic the native tissue environments, which includes physical cues, such as scaffold stiffness. In this work, we focus on how the initial scaffold stiffness hMSCs are encapsulated in changes cell-mediated remodeling and degradation and motility of the encapsulated cells. To do this, we encapsulate hMSCs in a well-defined synthetic hydrogel scaffold that recapitulates aspects of the native extracellular matrix (ECM). We then characterize cell-mediated degradation in the pericellular region as a function of the initial microenvironmental stiffness. Our hydrogel consists of a 4-arm poly(ethylene glycol) (PEG) end-functionalized with norbornene which is chemically cross-linked with a matrix metalloproteinase (MMP) degradable peptide sequence. This peptide sequence is cleaved by hMSC-secreted MMPs. The hydrogel elastic modulus is varied from 80 to 2400 Pa by changing the concentration of the peptide cross-linker. We use multiple particle tracking microrheology (MPT) to characterize the spatio-temporal cell-mediated degradation in the pericellular region. In MPT, fluorescently labeled particles are embedded in the material and their Brownian motion is measured. We measure an increase in cell-mediated degradation and remodeling as the post-encapsulation time increases. MPT also measures changes in the degradation profile in the pericellular region as hydrogel stiffness is increased. We hypothesize that the change in the degradation profile is due to a change in the amount and type of molecules secreted by hMSCs. We also measure a significant decrease in cell speed as hydrogel stiffness increases due to the increased physical barrier that needs to be degraded to enable motility. These measurements increase our understanding of the rheological changes in the pericellular region in different physical microenvironments which could lead to better design of implantable biomaterials for cell delivery to wounded areas.
Keywords: poly(ethylene glycol)-peptide hydrogel, multiple particle tracking microrheology (MPT), matrix metalloproteinases (MMPs), cell motility, degradation profile
Graphical Abstract
Introduction
Human mesenchymal stem cells (hMSCs) play an essential role during the wound healing process. During healing, hMSCs are recruited to the injury to regulate inflammation, progress the wound to the proliferative phase and coordinate tissue formation. hMSCs can also specify lineage by differentiating into several cell types, including osteoblasts, chondrocytes and adipocytes.1-6 Due to these properties, hMSCs are encapsulated into synthetic scaffolds with well-defined microenvironments that can be implanted in a wound site to enhance or restart healing in chronic wounds.7-11 These scaffolds will mimic native tissue stiffness and be designed to deliver additional hMSCs to the wound, necessitating cell migration within and, eventually, out of the scaffold to the native tissue.12-22
To enable migration to the wounded areas, hMSCs degrade and re-engineer their microenvironment, creating micron-sized channels to migrate through by secreting matrix metalloproteinases (MMPs), proteolytic enzymes, and exerting cytoskeletal tension on the network. 1,5,12-14,20,23 During this process, hMSCs receive physical cues from the scaffold. These physical cues affect basic cellular processes, but cells are also changing these cues through scaffold degradation. This complex process depends on the stiffness of the initial environment. In this work, we focus on the effect of scaffold stiffness, a physical cue, on cell-mediated degradation and motility in a synthetic hydrogel scaffold. This scaffold recapitulates critical aspects of the native extracellular matrix (ECM) to enable cell migration and isolates the role of physical cues in this process.12-22,24
Cell-responsive synthetic hydrogel scaffolds have been widely used as a powerful tool to study basic cellular functions and cell-material interactions in both 2D and 3D.12-22,24,25 These hydrogels are degradable and biocompatible, enabling high cell viability, and also enable control over physical and chemical cues presented to cells.14,17,19,20,26 The design of these materials draws inspiration from cellular and developmental biology and regenerative medicine, slowly increasing the scaffold complexity to recreate in vivo environments in vitro.12,13,19,25,27-31 The ECM is a network of different collagens, elastin and glycoproteins which surrounds cells in all tissues and provides structural support to the tissue.5,24,32-34 Physical and chemical cues from the ECM regulate cellular functions such as differentiation, migration and proliferation.14,33-35 Physical cues include matrix elasticity, which varies in the body from soft tissues, such as the brain at 100 Pa to bone which is 2–4 GPa.33 ECM composition is continuously changing during tissue development, wound healing and pathological conditions such as cancer progression and fibrotic disease. Therefore, it is important to investigate how microenvironmental stiffness alters cellular behavior to better understand cellular responses during wound healing and disease progression.
One type of material commonly used for cell encapsulation is a synthetic hydrogel scaffold based on a poly(ethylene glycol) (PEG) backbone. PEG is a bio-inert macromolecule and resistant to protein absorption. It has hydrophilic properties enabling high water absorption, which is essential for cell survival.15,16,25,36-38 Due to these advantages, we use a PEG-peptide hydrogel scaffold for 3D hMSC encapsulation. In our hydrogel, PEG is end-functionalized with norbornene and is chemically cross-linked with an MMP-degradable peptide sequence through radical-mediated step-growth photopolymerization.20,39 This peptide sequence is cleaved by hMSC-secreted MMPs.13,14,19,20,25,29,39-41 Therefore, this scaffold can be remodeled by 3D encapsulated hMSCs to enable motility and other basic processes. The physical microenvironment can also be varied by changing the concentration of the MMP-degradable peptide cross-linker. In this work, we vary physical cues initially presented to encapsulated hMSCs in the pericellular region to characterize cell-mediated degradation and motility.
Cells respond to the stiffness of their microenvironment through a process called mechanotransduction.24,33-35,42 In this process, cells apply force to deform their surrounding matrix. This mechanical force is then converted to biochemical signals through focal adhesion (FA) proteins. A FA is a complex protein that connects the intracellular actin-myosin cytoskeleton to the extracellular adhesion ligands in the native ECM.24,33-35,42 FAs are connected to ECM adhesion ligands through integrins, which are a family of transmembrane proteins. Force applied to the matrix changes the composition or structure of the FAs and provides signals to the cell.33-35,42 Through this process, the cell not only applies force that enables basic processes but modifies the physical microenvironment and, subsequently, the signal they are receiving. This complex evolution of physical cues leads to changes in basic cellular processes and an increased need to understand these interactions for the design of implantable scaffolds that can inform and manipulate cellular processes.
Previous research has shown that physical properties of the ECM affect cell lineage specification, morphology and migration in 2D and 3D scaffolds.14,24,34,43 Engler et al. cultured hMSCs on 2D substrates with different elasticities to mimic different tissues. They measured hMSCs differentiated into different lineages based on the stiffness of the substrate.24 Changes in cellular elongation has also been studied in systems where cells are encapsulated in 3D in hydrogels.14,43 Kyburz et al. and Dikovsky et al. measured a decrease in cellular elongation as stiffness is increased.14,43 Kyburz et al. also showed that stiffness of a hydrogel scaffold changes cellular migration. They measured a decrease in cellular speed and percentage of cells migrating with an increase in hydrogel stiffness.14 From these studies, it is clear that physical cues change hMSC motility and other basic processes. This change in hMSC motility also suggests that there will be a change in hMSC-mediated degradation in the pericellular region.
To measure hMSC-mediated degradation and re-engineering in the pericellular region, we use multiple particle tracking microrheology (MPT). MPT measures the Brownian motion of fluorescent particles embedded in the hydrogel. Material rheological properties are then calculated from this Brownian motion using the Generalized Stokes-Einstein Relation.44-52 We use MPT due to several advantages that make it ideal to characterize pericellular degradation. MPT is sensitive in the low moduli range which enables characterization of the weak gel network during critical phase transitions, i.e. gel to sol. This technique rapidly acquires data, ~ 30 s, enabling data collection of temporally evolving materials, such as during cell-mediated degradation, at a quasi steady-state. MPT also enables spatial characterization of the cell microenvironment. This technique uses video microscopy to capture particle movement enabling the rheological properties in the pericellular region to be quantified spatially.47-54
MPT was first used by Schultz et al. to measure dynamic cell-mediated re-engineering in the pericellular region caused by cytoskeleton tension and cell-secreted MMPs.20 In this work, they measure that hMSCs create a degradation profile where the greatest degradation is far from the cell center and immediately around the hMSC material remains in a gel phase, a reverse reaction-diffusion degradation profile.20 In our previous work, we investigated the role of cytoskeleton tension and cell-secreted MMPs in matrix degradation by inhibiting cytoskeleton tension.39 There was no change in the degradation profile around cells that could not exert tension on the scaffold suggesting that tension is playing a minimal role in matrix degradation and remodeling.39 From this result, we hypothesized that tissue inhibitors of metalloproteinases (TIMPs), a molecule secreted by hMSCs, inactivate MMP activity around the cell to create a microenvironment where the material remains stiff to enable attachment and spreading.5,55 We characterize the role of TIMPs in scaffold remodeling by inhibiting these molecules and measuring changes in the degradation profile.41,56 We measure that cells change their degradation strategy and create profiles where the scaffold is degraded around the cell and cross-link density increases as distance from the cell center increases, a reaction-diffusion degradation profile. In all of these experiments, the stiffness of the hydrogels was kept constant with cells experiencing the same physical cues.
Here, we use MPT to measure dynamic cell-material interactions in PEG-peptide hydrogels that present different initial physical microenvironments to 3D encapsulated hMSCs over 6 days. Hydrogel elasticity is changed by changing the concentration of the peptide cross-linker. The initial hydrogel elastic moduli, , varies from 80 to 2400 Pa. As we increase scaffold elasticity, we measure changes in cellular degradation profiles in the pericellular region. We hypothesize that these changes are due to a change in the amount of MMPs secreted by the cells at each stiffness in response to increased amount of cross-links that need to be degraded to create microchannels for migration. By increasing the cross-link density, we also measure a statistically significant decrease in cellular speed due to the increased physical barrier presented by stiffer materials. These measurements enhance our understanding of the strategy cells use to degrade their pericellular region and the microenvironments they engineer to initiate motility in response to hydrogel stiffness. This knowledge will lead to better design of pericellular regions in biomaterials that recapitulate physical aspects of native tissue with the goal of creating scaffolds that mimic these environments to deliver hMSCs to wounded areas.
Materials and Methods
hMSC Culture
Bone marrow derived hMSCs are purchased from Lonza in passage 2. hMSCs are cultured in a 150 cm2 tissue culture treated Petri dish (Sigma-Aldrich) with 35–45 mL of growth medium. Growth medium contains low-glucose Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies), 0.5 μg mL−1 Fungizone (Life Technologies), 50 U mL−1 Penicillin/streptomycin (Life Technologies), 10% fetal bovine serum (FBS, VWR) and 1 ng mL−1 recombinant human fibroblast growth factor (hFGF, PeproTech). hMSCs are incubated at 37°C and 5% CO2 with growth medium changed every 3–4 days. In all experiments, cells are passaged at ~ 90 % confluency. For all experiments, hMSCs from passage 2–6 are used.
Hydrogel Fabrication
The hydrogel precursor solution used for 3D cell encapsulation is composed of a four-arm star PEG end-functionalized with norbornene (PEG-N, 3 mM, Mn = 20 000 g mol−1, Sigma-Aldrich) backbone that is chemically reacted with thiol residues in an MMP-degradable peptide sequence, KCGPQG↓IWGQCK (Mn = 1 305 g mol−1, Bachem). This peptide is cleaved by hMSC-secreted MMPs and enables cellular remodeling which creates microenvironments for cells to spread and migrate.14,20,39,40,57 To change the microenvironmental stiffness, different concentrations of the MMP-degradable peptide sequence are used. For all experiments, the concentration of PEG-N is kept constant and the concentration of the cross-linker is changed, which changes the ratio of thiol:ene (R) in the scaffold. Re-engineering and remodeling of the pericellular region around encapsulated hMSCs is measured for R = 0.55, 0.65, 0.7, 0.75, 0.85 and 1, which correspond to peptide concentrations of 3.3, 3.9, 4.2, 4.5, 5.1 and 6 mM and corresponding weight percentages of 0.43, 0.51, 0.55, 0.59, 0.67 and 0.78 wt%. Information about changes in the MMP-degradable peptide weight percentage, which is equivalent to the overall gel weight percent change, can be found in the Supplemental Information, Table S1.
An adhesion ligand, CRGDS (1 mM, Mn = 594 g mol−1, American Peptide, Inc.), is tethered to the hydrogel to enable cellular attachment to the scaffold through integrin binding on the cell surface. A highly water soluble photoinitiator, lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) (1.7 mM) is added to the scaffold precursor solution to initiate a thiol:ene step-growth photopolymerization. LAP is synthesized using a previously published protocol.58 This hydrogel has been widely used for 3D cell encapsulation and enables cellular migration and maintains high cell viability.14,17,19,20,54,58 All materials are dissolved in 1× phosphate buffered saline (1× PBS, Life Technologies) to the desired concentration. Fluorescently labeled 1 μm carboxylated polystyrene probe particles (0.2% solids per volume, 2a = 0.97 ± 0.01 μm where a is the particle radius, Polysciences, Inc) are added to the precursor solution to enable rheological characterization using MPT. Sodium hydroxide (15 mM, Fisher Scientific) is added to the solution to adjust the pH to 7. Finally, hMSCs are added to the precursor solution prior to gelation and results in a final concentration of 2 × 105 cells mL−1.
After mixing, the polymer precursor solution is added to a cylindrical polydimethylsiloxane (PDMS) chamber in the Petri dish described below. Upon exposure to UV light (365 nm, 10 mW cm−2, UVP, LLC) for 3 mins radical-mediated photopolymerization is initiated and the hydrogel is formed. Sample chambers are then filled with 4 mL of growth medium (without FGF) and are incubated overnight at 37°C and 5% CO2 (Eppendorf, Inc.). MPT data are taken 3–6 days after cell encapsulation. This allows hMSCs to relax, spread and migrate in the scaffold. For all thiol:ene ratios, experiments are repeated three times and in each biological replicate two hydrogels are made.
In this work, our goal is to characterize the cellular response due to changes in the stiffness of their microenvironment. We change the stiffness of our material by changing the concentration of the cross-linker. By adding more cross-linker, due to the increase in physical barrier, we hypothesize that hMSCs will change their degradation strategies during migration. Furthermore, since the cross-link density is directly related to the hydrogel modulus, adding more cross-linker will result in changes in the hydrogel modulus. Also, increasing the concentration of the cross-linker will not affect the critical relaxation exponent, n, which defines the critical transition point of our hydrogel. This will be discussed in detail in the Results and Discussion.
3D Cell Encapsulation
hMSCs are cultured in a 150 cm2 tissue culture treated Petri dish in growth medium. Cell media is changed every 3–4 days. hMSCs are passaged after achieving 90 % confluency. For 3D cell encapsulation, hMSCs are suspended in PBS and added to the hydrogel precursor solution prior to gelation at a final concentration of 2 × 105 cells mL−1. This low cell concentration is used to limit cell-cell interactions after encapsulation. The precursor solution is mixed and added to the PDMS tube in the glass-bottomed Petri dish. The solution is exposed to the UV light for 3 mins.
In all experiments, we use growth medium without any chemical cues to ensure that cells are not differentiating to different phenotypes where they would then deposit ECM into the system.17,59,60
Device Fabrication
A glass-bottomed Petri dish (D = 35 mm, no. 1.5 glass coverslip, MatTek Corporation) with a cylindrical PDMS chamber is used to fabricate and measure cell-laden hydrogels. The cylindrical chamber in the Petri dish is made from PDMS (Dow Corning) sheets. This chamber is added to the dish to hold the hydrogel precursor solution and reduce particle drift in the hydrogel after cell-mediated degradation. PDMS is made by mixing silicone elastomer base and curing agent at a 10:1 ratio. This mixture is degassed and cured in an oven at 65°C overnight. 6 mm and 10 mm biopsy punches (Acuderm Inc.) are used to cut the cured PDMS sheets and create tube shape chambers with an inner diameter of 6 mm and an outer diameter of 10 mm. These chambers are attached to the glass bottom of the Petri dishes using uncured PDMS and cured overnight at 65°C.
Petri dishes are sterilized with 70% ethanol and 17 μL of hydrogel precursor solution is added to the PDMS chambers. This volume is used to allow the hydrogel to fully swell after media is added. Hydrogel scaffolds are formed by exposing the precursor solution to UV light for 3 mins. After hydrogel formation, Petri dishes are immediately filled with 4 mL of growth medium (without FGF) and incubated at 37°C and 5% CO2.
Bulk Rheology
Hydrogel unswollen and swollen moduli are measured using bulk rheology. For bulk rheology experiments, hydrogels are made in PDMS tubes (inner diameter of 8 mm and outer diameter of 10 mm) in the absence of cells. This hydrogel diameter is required to ensure that samples are the same size as the geometry being used to measure the scaffold. 100 μL of hydrogel precursor solution is added to the PDMS chambers and photopolymerized. To measure the moduli of unswollen hydrogels, after polymerization hydrogels are loaded onto the rheometer (Ares G2, TA Instruments). These scaffolds are measured with an 8 mm sandblasted parallel plate to minimize slip. The elastic, G′, and viscous, G″, moduli are measured for all different thiol:ene ratios using frequency sweep between 0.1 and 40 Hz at 1% strain.
To measure swollen hydrogel moduli, after hydrogel polymerization PDMS tubes are removed and Petri dishes are filled with 3 mL of growth medium (without FGF) to allow the hydrogels to swell in all directions. Hydrogels are incubated at 37°C overnight. After incubation, media is removed and an 8 mm biopsy punch (Acuderm Inc.) is used to cut the hydrogel to ensure they fit under the geometry. Hydrogel swollen moduli are measured using a bulk rheometer following the same procedure as described above.
For all conditions, 3 hydrogels are tested and scaffolds are measured in the linear viscoelastic regime. All the bulk rheology experiments are done in the absence of cells.
Multiple Particle Tracking Microrheology
Multiple particle tracking microrheology (MPT) is a passive microrheological techniques that is used to measure spatio-temporal cellular re-engineering in the pericellular region. In MPT, fluorescently labeled probe particles are embedded in the hydrogel and their Brownian motion is tracked to measure material properties.44,47,49,51,61,62 Data are collected using an inverted microscope (Zeiss Observer Z1, Carl Zeiss AG) with a 63× objective (water immersion, N.A. 1.3, 1× optovar, Carl Zeiss AG). Probe movement is recorded at a frame rate of 30 frame s−1 for 800 frames using a high speed camera (1024 × 1024 pixels, Phantom Miro M120, Vision Research Inc.) with an exposure time of 1000 μs. These values are chosen to minimize static and dynamic particle tracking errors.62
Particle tracking algorithms developed by Crocker and Grier are used to find particle positions in each frame by identifying their brightness-weighted centroid.44,63 Using a probability distribution function that accounts for Brownian motion, positions of a particle in all frames are linked together into a trajectory that tracks particle movement in the video.44,63 Finally, the ensemble-averaged mean-squared displacement (MSD), ⟨Δr2 (τ)⟩ = ⟨Δx2 (τ)⟩ + ⟨Δy2 (τ)⟩, is calculated from particle trajectories.48,53,54,63-65 τ is the lag time, which is the separation time between frames. The ensemble-averaged mean-squared displacement is related to rheological properties of the hydrogel using the Generalized Stokes-Einstein Relation (GSER). The GSER directly relates the MSD to the rheological properties of the hydrogel by
| (1) |
where a is the particle radius, kBT is the thermal energy and J(t) is the creep compliance.48,50,53,64,66-68 The logarithmic slope of the MSD, , quantitatively identifies the state of the material.48,51-54,68,69 This will be discussed in detail in the Results and Discussion.
Data are collected 3–6 days after cell encapsulation which gives time for cells to spread in and degrade the network. Data are collected in a microscope incubation chamber which maintains samples at 5% CO2 at 37°C to provide a suitable environment for cells to remain viable. Brightfield microscopy is used to locate the position of a cell immediately before collecting MPT data at each time point. After locating a cell, data acquisition is begun and data are collected every 3–8 mins for approximately 15–60 mins around each cell. Time of data collection around each cell changes due to cell migration in the z direction and photobleaching of probe particles. Brightfield images are used to find the cell center, (xi, yi), and cell speed, vcell, using ImageJ (NIH Image). Cell speed is calculated using the cell center at the initial and final time, , where t is time, 0 is the initial and f is the final cell position.
Results and Discussion
In this work, we measure cell-mediated degradation around an hMSC encapsulated in different hydrogel stiffnesses to determine the role of this physical cue in cellular degradation strategies and migration. Hydrogel elastic moduli are varied by changing the cross-linker concentration. We use multiple particle tracking microrheology to measure the change in the degradation profile in the pericellular region in different hydrogel stiffnesses over 6 days. We also measure cell motility on different days. We then use the measured degradation profiles and cell motilities to determine a relationship between cell-mediated pericellular degradation and migration. These measurements give insight into how cellular behavior can be modified and potentially manipulated by the physical microenvironment.
First, bulk rheology is used to characterize the moduli of hydrogels with different thiol:ene ratios, R. For clarity only the elastic moduli, G′, is shown in Figure 1 for swollen gels in the absence of cells. The elastic and viscous moduli for swollen and unswollen hydrogels are reported in the Supplemental Information, Figure S1 and S2, respectively. Cross-link density, ρ, is directly related to the elastic modulus, G′, using the equation G′ ~ αkBT where T is the temperature and kB is the Boltzmann constant. Therefore, changing the amount of cross-linker in the system will affect cross-link density and elastic modulus. As the amount of cross-links is changed, cells will experience different physical microenvironments which will affect the strategy used to degrade the scaffold, the amount of degradation in the pericellular region and motility. Changes in the modulus, cross-linking efficiency and the Flory-Stockmayer critical fraction of PEG reaction sites needed to form a gel for each thiol:ene hydrogel is included in Supplemental Information, Table S2. By increasing the thiol:ene ratio, more cross-links are formed and G′ increases. Our hydrogel swollen elastic moduli vary from 80 ± 83 Pa in a soft gel to 2400 ± 230 Pa in a stiff gel. The thiol:ene ratio R = 0.55 is the minimum ratio that forms a network using these materials. After swelling, the hydrogel is very soft; therefore, there is a large error in the elastic moduli at this stiffness. These elasticities mimic a wide range of elastic microenvironments in the body, such as brain, lung, adipose and endothelial tissue.35 After bulk characterization is complete, hydrogels are made with 3D encapsulated hMSCs and cell-mediated degradation in the pericellular region is characterized using multiple particle tracking microrheology.
Figure 1:
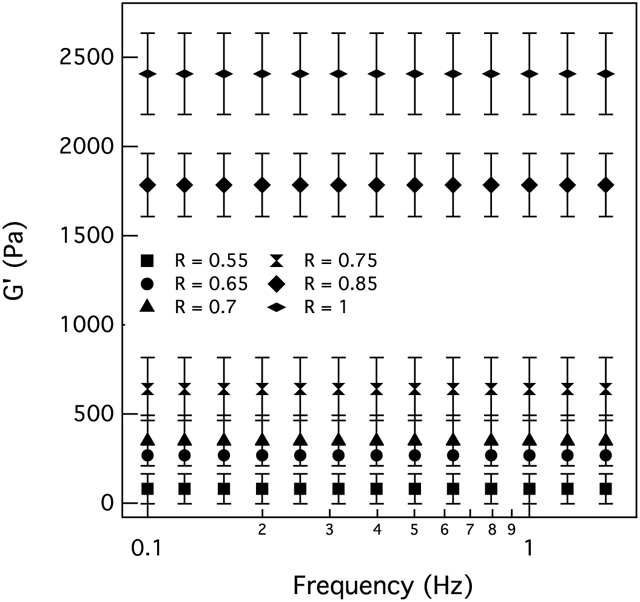
Swollen elastic moduli for hydrogels with different thiol:ene ratios. The thiol:ene ratio, , is varied to make hydrogels with different stiffnesses to mimic elasticity of different tissues in the body.
MPT is used to measure rheological properties in the pericellular region as hMSCs degrade the scaffold. Using MPT, we track probe particle movement and calculate an ensemble-averaged mean-squared displacement (MSD). The logarithmic slope of the MSD, , quantitatively identifies the state of the material. 48,51,52,52,64,70,71 α = 1 measures Brownian motion of particles in a liquid. α → 0 indicates probes are completely arrested in a gel network. 0 < α < 1 indicates that probes are in a viscoelastic liquid or gel state. To determine the state of the material, i.e. when the last sample-spanning network cluster breaks and the material transitions from a gel to a sol, α is compared to the critical relaxation exponent, n. If n < α the material is a viscoelastic liquid and if n > α the material is a viscoelastic solid.
n is determined using time-cure superposition.45,48,64,72-74 n is a material property and only changes when the underlying structure of the material (e.g. size of the polymer, number of arms on the PEG molecules and cross-linking chemistry) is changed. Previous work has shown that changing the cross-link density does not change the value of n.53,75-77 n also indicates the structure of the material. 0.1 < n < 0.5 indicates the material has a tightly cross-linked network that stores energy. 0.5 < n < 1 indicates that the material has a loosely cross-linked network which dissipates energy.48-54,56,64,69,70,72,73,78 For our hydrogel, we previously measured n by degrading a thiol:ene=0.65 hydrogel homogeneously using collagenase, a mixture of enzymes that are mostly MMPs that degrade the peptide cross-linker. 41 Schultz et al. also calculated n for this hydrogel using thiol:ene = 0.55.54 The values from these two different studies are within error of each other. These experiments show that changing the concentration of the cross-linker does not change n. The value of n for this hydrogel is n = 0.25 ± 0.05.41,54 We define the gel-sol phase transition region as 0.2 < α ≤ 0.3. In this region, the material is transitioning from a gel to a sol.41,54 This enables the quantitative identification of the state of the material in the pericellular region around encapsulated hMSCs.
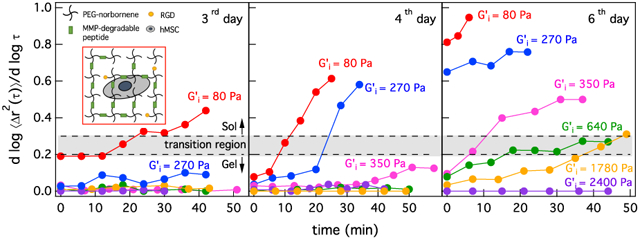
hMSCs are encapsulated in hydrogels and incubated at 37°C and 5% CO2 before and during data acquisition. MPT data are collected 3–6 days after cell encapsulation. Data are organized by the post-encapsulation day. Within this post-encapsulation day, all experiments define t = 0 mins as the time that cells are located using brightfield microscopy and MPT data acquisition begins. MPT measurements are collected every 3–8 mins around each hMSC for 15–60 mins. Degradation and remodeling of the pericellular region is measured around 23–30 cells in each post-encapsulation day at each scaffold stiffness. Figure 2a-c shows the logarithmic slope of the mean-squared displacement, α, over time for hMSC-mediated degradation in R = 0.55, 0.65, 0.7, 0.75, 0.85 and 1 on different post-encapsulation days. Each line represents changes in the state of the material around a single hMSCs. These data are representative of all measured cells. Different symbols in Figure 2a-c, represent data around single hMSCs encapsulated in scaffolds with different values of R. The gray box in each figure indicates the phase transition region, 0.2 ≤ α ≤ 0.3.
Figure 2:

Changes in the logarithmic slope of the mean-squared displacement, , over time for hMSCs encapsulated in , 0.65, 0.7, 0.75, 0.85 and 1 (a) 3, (b) 4 and (c) 6 days post-encapsulation. The gray box in each figure indicates the phase transition region, 0.2 ≤ α ≤ 0.3, which quantitatively identifies the transition from a gel to a sol when hMSC-secreted MMPs degrade the last sample-spanning network cluster in the hydrogel.
Figure 2a shows changes in the state of the material around hMSCs 3 days post-encapsulation. For hMSCs encapsulated in the lowest cross-link density gel, R = 0.55, we measure degradation in the pericellular region with the α value increasing over time. At the time the cell is located, t = 0 mins, the material is in the viscoelastic gel phase, α = 0.19, and is about to enter the transition region. Over time, MMPs degrade the peptide cross-linkers and α increases until it passes through the gel-sol transition region at t = 30 mins. MMPs continue to degrade the network and the material is in the viscoelastic sol phase, α = 0.44 at 40 mins. For hMSCs encapsulated in an R = 0.65 scaffold, after 30 mins the cell initiates degradation of the surrounding material but α does not reach the gel-sol transition region. As the scaffolds get stiffer (R ≥ 0.7) we measure no material degradation in the pericellular region of encapsulated hMSCs on the probe particle measurement length scale (1 μm). For these stiffnesses, hMSCs must degrade more peptide cross-linkers and this requires longer times. To characterize cell-mediated degradation in the pericellular region for higher hydrogel stiffnesses, we measure cellular degradation 4 and 6 days post-encapsulation.
Figure 2b shows changes in the state of the material around hMSCs 4 days post-encapsulation. On the fourth day, hMSCs encapsulated in both low cross-link density gels, R = 0.55 and R = 0.65, degrade the scaffold from the gel phase (α → 0) into the the transition region (0.2 ≤ α ≤ 0.3) and finally to the sol phase (α = 0.61 and α = 0.58 for R = 0.55 and R = 0.65, respectively). For hMSCs encapsulated in R = 0.7 scaffolds 4 days post-encapsulation, we measure the same trend of degradation as hMSCs encapsulated in R = 0.65 gels at 3 days. The cell has started to degrade the scaffold and α is about to enter the gel-sol transition region by the end of the measurement window. We still measure no scaffold degradation around hMSCs encapsulated in R = 0.75, 0.85 and 1. At these higher stiffnesses, cells still require more time to degrade and re-engineer their local microenvironments.
Figure 2c shows changes in the logarithmic slope of mean-squared displacement 6 days post-encapsulation. At the beginning of data acquisition, material around the hMSCs encapsulated in R = 0.55 and R = 0.65 gels is already degraded and is a viscoelastic liquid. 6 days post-encapsulation we measure scaffold degradation from a gel to a sol around hMSCs encapsulated in R = 0.7 gels. On this day, hMSCs encapsulated in R = 0.75 and R = 0.85 scaffolds have degraded their local microenvironments and α has reached the transition region. We still measure no degradation around hMSCs encapsulated in the highest cross-link density hydrogels, R = 1. More examples of MPT measurements of the logarithmic slope of the MSD over time for each stiffness are provided in Figure 3S and 4S in the Supplemental Information.
From these measurements, we quantify the time that it takes an hMSC to degrade the scaffold past the gel-sol transition at each stiffness. We determine that facile degradation occurs in the lowest cross-link density gels, as is expected. We also determine that at the highest cross-link gel (R = 1), hMSCs do not substantially degrade their pericellular region, which we hypothesize will be correlated with minimal changes in the pericellular region and cellular speed. Next, we calculate cell speed to understand the impact of the extent of degradation and cell-mediated re-engineering of the pericellular region on migration.
Average cellular speed, vavg, is measured each day post-encapsulation for hMSCs encapsulated in hydrogels with different thiol:ene ratios, Figure 3a-c. We calculate cell speed for each day separately to determine how the extent of degradation changes motility. Figure 3a shows cellular speed for all measured days. To show there is a significant increase in cellular speed after 6 days, we plot average cellular speed for 3 and 4 days and 6 days post-encapsulation separately, Figure 3b and c, respectively. After 3 days of cell encapsulation, cellular speed is significantly decreased from vavg–day3 = 28 ± 27 μm hr−1 in low cross-link density gels (R = 0.55) to vavg–day3 = 3.6 ± 3.3 μm hr−1 in high cross-link density gels (R = 1), Figure 3b. As the networks become stiffer, hMSCs must degrade more peptide cross-linkers to create microchannels for migration. Therefore, stiffer networks with a higher cross-link density present a physical barrier for cell migration. We measure the same trend for 4 and 6 days post-encapsulation where cellular speed significantly decreases as initial cross-link density increases. 4 days post-encapsulation, vavg decreases an order of magnitude from vavg–day4 = 46±21 μm hr−1 in R = 0.55 to vavg–day4 = 3±2.5 μm hr−1 in R = 1, Figure 3b. 6 days post-encapsulation, vavg decreases 10× from vavg–day6 = 330 ± 200 μm hr−1 in R = 0.55 to vavg–day6 = 3 ± 2.4 μm hr−1 in R = 1, Figure 3c.
Figure 3:
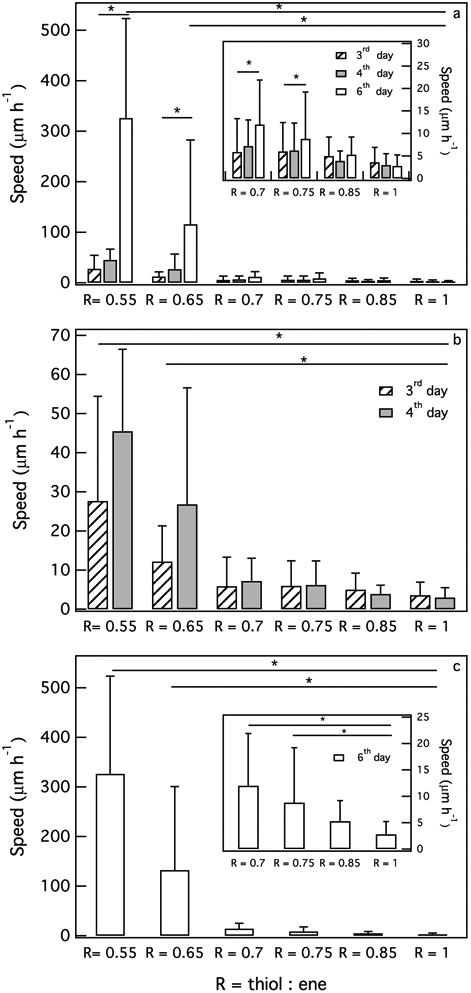
Effect of hydrogel stiffness on average migration speed of hMSCs (a) 3–6 days, (b) 3 and 4 days and (c) 6 days post-encapsulation. Cellular speed decreases significantly as or cross-link density increases. Cells encapsulated in low cross-link density hydrogels have a significant increase in speed due to the lower physical barrier for cell migration (*p < 0.05).
In the lowest cross-link density gel, R = 0.55, the material is fully degraded in the pericellular region 6 days post-encapsulation. Due to the minimal material barrier for migration, cell speed, vavg–day6 = 330±200 μm hr−1, is significantly higher than cell speed 3 and 4 days post-encapsulation, Figure 3c. As we discussed earlier, after 6 days hMSCs have more time to degrade the hydrogel and create microchannels, which enable facile migration. 6 days post-encapsulation, vavg for hMSCs encapsulated in all thiol:ene ratios except R = 0.85 and R = 1 is significantly increased, Figure 3c. Due to the significant increase in cellular speed for R = 0.55 and R = 0.65 on this day, the inset in Figure 3c is plotted to better illustrate cellular speed for R = 0.7, 0.75, 0.85 and 1. This shows that increased hMSC-mediated degradation results in increased hMSC motility. Spatial rheological properties in the pericellular region also affect cellular migration in the hydrogel. Next, we use MPT to measure hydrogel spatial evolution in the pericellular region to understand its role in cellular migration.
Prior to characterizing the degradation profile cells create in the pericellular region at each stiffness, we first quantify the percentage of cells that have remodeled their microenvironment 3, 4 and 6 days post-encapsulation. We define cell-mediated remodeling as an increase in the α value in the pericellular region over time. Figure 4 is the percentage of cells that have remodeled their surrounding microenvironment at each hydrogel stiffness. is the initial hydrogel swollen moduli without encapsulated cells. As hydrogel stiffness increases, 3 days post-encapsulation the percentage of cells that have remodeled their pericellular region decreases from 86% in R = 0.55 to 0% in R = 1. We measure the same decreasing trend in the percentage of cells remodeling the material 4 and 6 days post-encapsulation. As post-encapsulation time increases, the percentage of cells that have remodeled their microenvironment encapsulated in the same stiffness increases. As time passes, cells have more time to secrete MMPs and degrade their microenvironment. This increased remodeling is also directly correlated to the increase in hMSC motility. To better understand how re-engineering and remodeling in the pericellular region changes with hydrogel stiffness, we measure spatial changes in the α value in the pericellular region to characterize the profile of degradation at each hydrogel stiffness.
Figure 4:
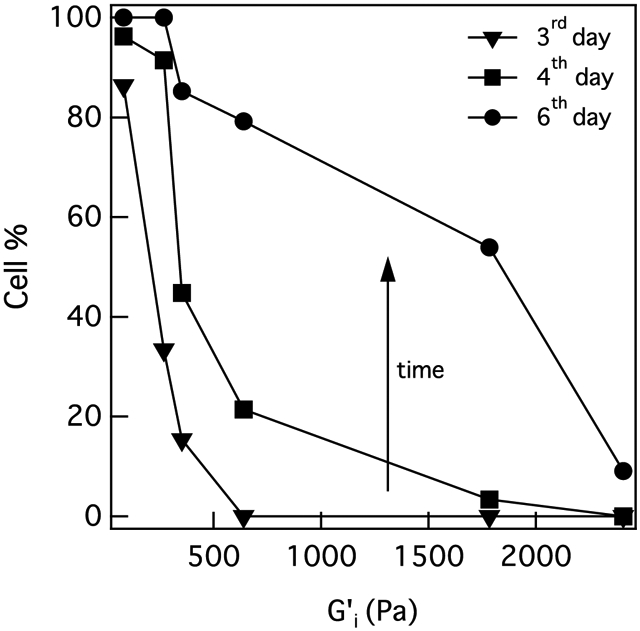
The percentage of cells that have remodeled the pericellular region at each stiffness. The percentage of cells that have remodeled their pericellular region increases with time when they are encapsulated in the same stiffness. This percentage also decreases as hydrogel stiffness increases.
Before discussing spatial MPT data in the pericellular region, first we define six different profiles of degradation: (1) reaction-diffusion, (2) reverse reaction-diffusion, (3) no pattern, (4) uniform, (5) not degraded and (6) degraded. All profiles are illustrated in Figure 5. In Figure 5, α is plotted versus the distance from the cell center, which is the x-axis labeled radius. Radius equal to zero is the center of the cell and an increasing radius value moves spatially further from the cell center. The gray box in these graphs indicates the gel-sol phase transition region. Each of these profiles shows that there is spatial heterogeneity in the pericellular region due to cell-mediated re-engineering. This cannot be captured with bulk rheological measurements. Bulk rheology will measure the average stiffness of these heterogeneous cell-engineered microenvironments along with scaffold that has not been degraded.
Figure 5:
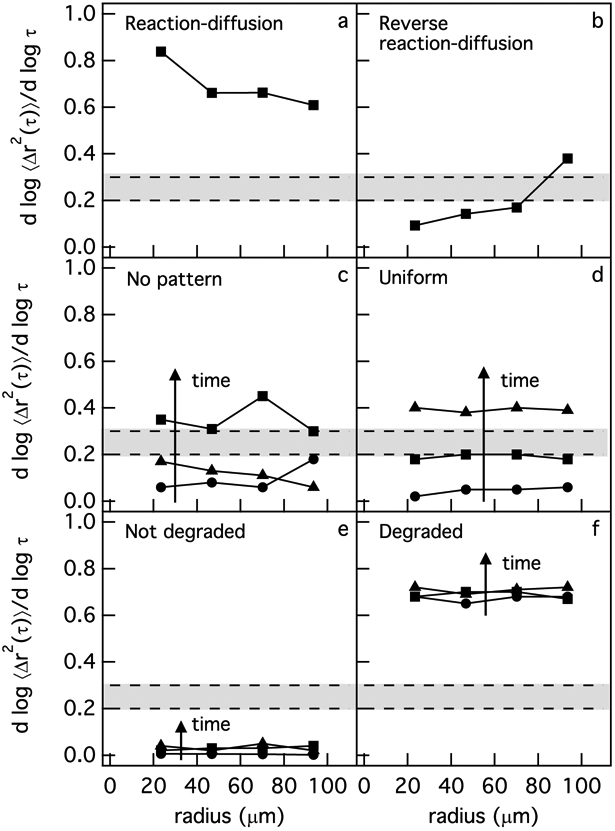
Six different profiles of degradation are defined in the pericellular region from MPT measurements. The types of degradation profiles are (a) reaction-diffusion, (b) reverse reaction-diffusion, (c) no pattern (d) uniform, (e) not degraded and (f) degraded. The data presented in this figure are examples taken from actual measurements.
The reaction-diffusion profile, Figure 5a, is where α is increasing as a function of distance away from the cell center. The reverse reaction-diffusion profile, Figure 5b, is a profile where α is decreasing as a function of distance from the cell center. A no pattern profile is defined as a profile where α does not follow any specific pattern as a function of distance from the cell center or when α at different time points is a mixture of different types of degradation profiles, Figure 5c. Figure 5d is a uniform profile where α at different time points is not changing as a function of distance from the cell center. Not degraded profiles, Figure 5e, are where α ≃ 0 at every distance from the cell center at each time point. Figure 5f is a degraded profile, which is defined as a profile where the pericellular region is in a viscoelastic liquid phase at the time that the cell is located. In this profile, the value of α is measured for the entire pericellular region and is greater than 0.6 at t = 0 mins. This value remains constant over the data acquisition window. Since this profile is calculated for the entire pericellular region, there may be variations as a function of distance from the cell center.
To define the type of degradation, we compare the values of α at each specified region around an encapsulated hMSC. First, a linear regression model is fit to the mean-squared displacement data at each defined region (region 1 is 23 μm < r < 46 μm, region 2 is 46 μm < r < 69 μm and region 3 is 69 μm < r < 92 μm). The slope of the regression line is the α value in these regions (α1, α2 and α3). In order to find whether two α values (α1 and α2, α2 and α3 and α1 and α3) are different, we perform a hypothesis test on the two α values from the regression models at two distance from the cell center with a p-value less than 0.05 to determine whether the two values are significantly different. Once the slopes at each specified region are compared we define a degradation profile.
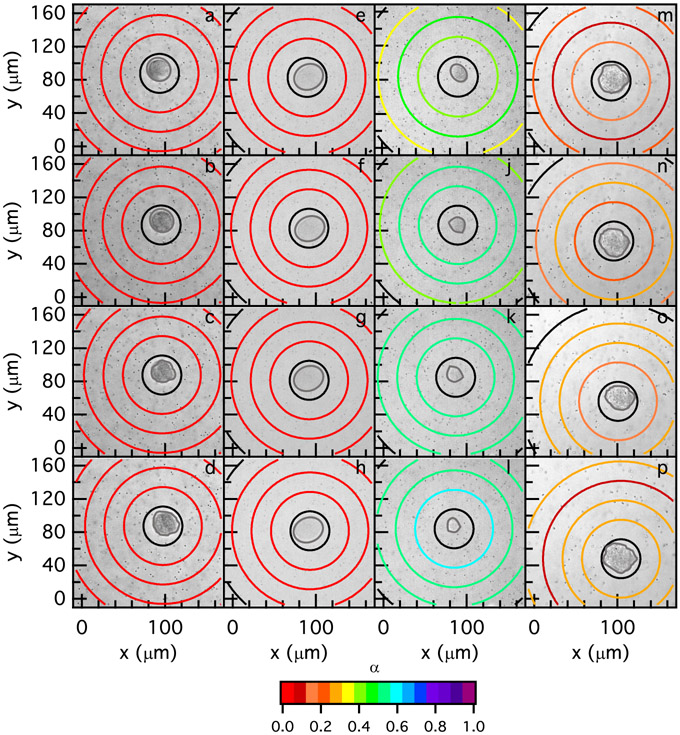
Figure 6 shows profiles of degradation in the pericellular region in R = 0.75 hydrogels 3, 4 and 6 days post-encapsulation. To characterize the degradation profile in the pericellular region, we measure α for different regions around the cell. The radius of the first ring is 23 μm and and the radius of each subsequent ring increases with an increment of 23 μm, (ri+1 = ri + 23 μm). These are the same types of MPT measurements shown in Figure 5 except in Figure 6 they are plotted as a spatial map. The color of each ring corresponds to the value of α for the particles within each specified region. Warm colors indicate the material is a gel and cool colors indicate the material is a liquid. The red-orange to light orange colors are the transition region, where the material undergoes a phase transition from a gel to a liquid, 0.2 ≤ n ≤ 0.3. Black rings indicate that there are not enough statistics or probe particles in that specified area to calculate a statistically significant value of α. A brightfield image of each cell at each time point is behind the MPT data with the cell outlined in gray. t = 0 mins is the time that data acquisition is begun. Degradation profiles in Figure 6, show the types of degradation that the majority of the cells are experiencing on each day in R = 0.75 hydrogels.
Figure 6:
Spatial degradation profiles around hMSCs encapsulated in R = 0.75 hydrogels measured with MPT (a-d) 3 (e-h) 4 and (i-p) 6 days post-encapsulation. Two degradation profiles 6 days post-encapsulation are shown, which are (i-l) a reaction-diffusion and (m-p) a no pattern degradation profile. MPT data are collected after locating the cell 3 days post-encapsulation at (a) 0, (b) 12, (c) 24 and (d) 48 mins, 4 days post-encapsulation at (e) 0, (f) 21, (g) 35 and (h) 49 mins and 6 days post-encapsulation at (i) 0, (j) 14, (k) 28, (l) 42, (m) 0, (n) 18, (o) 42 and (p) 55 mins. The color of each ring represents , which determines the state of the material in the scaffold.
MPT measures no degradation in the pericellular region of hMSCs 3 days post-encapsulation in R = 0.75 hydrogels, Figure 6a-d. On this day, α is approximately 0 across the measured field of view through time or a not degraded profile. Figure 6e-h is the spatial rheological changes around an hMSCs 4 days post-encapsulation. We again measure a not degraded profile in the pericellular region of the hMSC on this day. 6 days post-encapsulation, two types of degradation profiles are measured around the majority of cells. First, Figure 6i-l is a reaction-diffusion profile, which is measured around 25% of cells. At t = 0 mins the material has passed the gel-sol transition region and the greatest degradation is immediately around the cell. In Figure 6i, we measure high α values in the area closest to the cell and α decreases as a function of distance from the cell center. In this profile, MMPs are secreted from the hMSC and are reacting with the peptide cross-linkers while diffusing through the hydrogel network. A reaction-diffusion profile around encapsulated hMSCs can be seen at different time points of data acquisition around this cell, Figure 6j-l.
We also measure a no pattern profile 6 days post-encapsulation around 38% of encapsulated hMSCs in R = 0.75 scaffolds, Figure 6m-p. At t = 0 mins and t = 18 mins we do not measure any specific pattern in the pericellular region. For these time points α has a higher value immediately around the cell, but as the distance from the cell center increases α first decreases and then increases in the farthest region from the cell center. At t = 42 mins, Figure 6o, we measure the greatest degradation furthest from the cell center, which is a reverse reaction-diffusion profile. At t = 55 mins, Figure 6p, the profile of degradation changes to a reaction-diffusion degradation profile. The degradation profile around the hMSC on this day does not following any specific pattern. Most of the measured no pattern degradation profiles, 80%, in all hydrogel stiffnesses happen when α is near and in the gel-sol transition region, which is the case in Figure 6m-p.
We hypothesize that as hydrogel stiffness is increased, cells are changing the molecules that they secrete to regulate scaffold degradation. Cells are sensing the physical microenvironment surrounding them and responding to it. Specifically, we hypothesize that hMSCs are secreting more MMPs and less TIMPs to overcome the physical material barrier presented to them in the scaffold to migrate. This increase in the amount of MMPs secreted by cells changes the degradation profile to a no pattern profile when degrading to the phase transition region and, finally, to a reaction-diffusion profile after the phase transition. Previously, Leight et al. have encapsulated hMSCs in this synthetic hydrogel with an MMP fluorogenic substrate to monitor MMP activity when different physical and chemical cues are presented to cells from the network.79 They measure an increase in MMP activity when hydrogel stiffness is increased.79 As the stiffness increases, cells secrete more MMPs to overcome the physical barrier for migration, which results in faster degradation of the scaffold. For R = 0.75 gels, we measure an increase in pericellular degradation 6 days post-encapsulation and also a significant increase in cell motility, Figure 3c. Examples of the profiles of degradation in the pericellular region for other hydrogel stiffnesses are provided in Figure 5S-9S in the Supplemental Information.
To further quantify cell-mediated pericellular degradation profiles at each stiffness, we categorize degradation profiles to one of the six different degradation profiles defined above. Then, we plot the percentage of cells that create each type of degradation profile at each stiffness in a stacked bar graph, Figure 7. Figure 7a-c shows the percentage of cells in each degradation category at each stiffness 3, 4 and 6 days post-encapsulation, respectively. For hMSCs encapsulated in R = 0.55 gels, all six different types of degradation profiles are measured 3 days post-encapsulation, Figure 7a. At this stiffness the initial cross-link density is low, therefore, cells can easily degrade the hydrogel and become motile. As the material is degraded to the phase transition region, spatial heterogeneity increases and the majority of the cells, 23%, degrade their surrounding environment using a no pattern mechanism of degradation. As hydrogel stiffness is increased to 270 Pa (R = 0.65) the main type of degradation changes to a reverse reaction-diffusion profile. Previously we showed that this profile of degradation is created by an hMSC simultaneously secreting MMPs and TIMPs. These two molecules create MMP-TIMP complexes, which inhibit MMP activity creating an environment immediately around the cell that remains stiff to enable cell relaxation, spreading and attachment to the network prior to migration.20,39,41 By further increasing the thiol:ene ratio we did not measure any degradation in the pericellular region of hMSCs 3 days post-encapsulation, Figure 7a.
Figure 7:
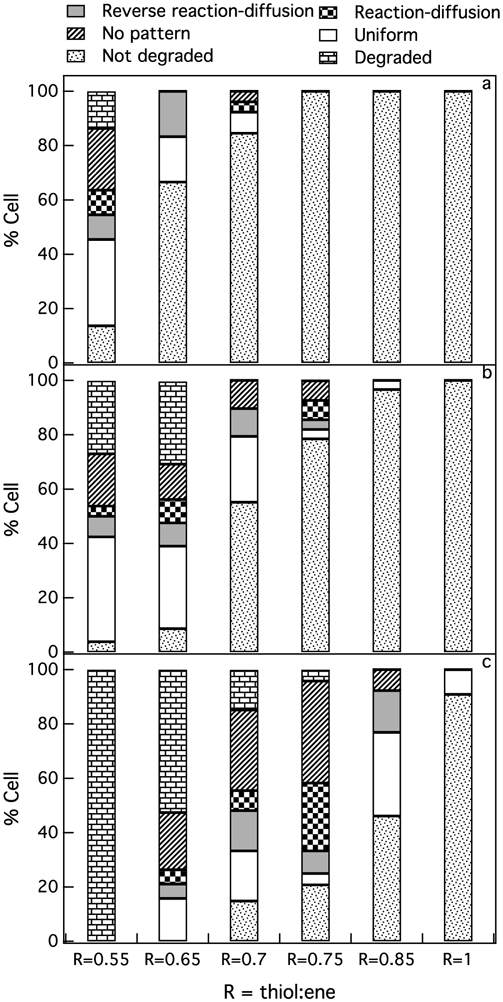
Percent of cells in each degradation category (a) 3, (b) 4 and (c) 6 days after hMSC encapsulation. We measure six different profiles of degradation in the pericellular region: reaction-diffusion, reverse reaction-diffusion, no pattern, uniform, not degraded and degraded. Cells change the amount and types of molecules they secrete to create different degradation profiles in response to microenvironmental stiffness.
Figure 7b shows the profiles of degradation for each stiffness 4 days post-encapsulation. On this day, for both low cross-link density hydrogels, R = 0.55 and R = 0.65, we measure all six types of degradation profiles. Since cells have more time to degrade their microenvironment, the number of cells that have completely degraded their pericellular region is increased from 14% to 27% and 0% to 30% in R = 0.55 and R = 0.65, respectively. For R = 0.7 gels, 10% of the cells create a no pattern profile and another 10% create a reverse reaction-diffusion degradation profile. By increasing the stiffness of the hydrogel, previous studies have shown that cells are secreting more MMPs to degrade the hydrogel and become motile. This increase in MMP activity can be the reason why we measure no pattern profiles around encapsulated hMSCs on this day because we are measuring more hMSCs transitioning their pericellular region from a gel to a sol. By further increasing the thiol:ene ratio to R = 0.75, we measure a low percentage of cells creating reaction-diffusion and no pattern profiles. We again hypothesize that the increase in scaffold stiffness causes an increase in MMP secretion and decrease in TIMP secretion. This will change the profiles to reaction-diffusion and no pattern profiles. We measure no degradation in the pericellular region in our highest cross-link density hydrogels, R = 0.85 and R = 1.
Six days after cell encapsulation, the percentage of the cells that have fully degraded their pericellular region increased to 100% in R = 0.55 and 53% in R = 0.65, Figure 7c. In R = 0.65 gels, much of the scaffold is degraded 6 days post-encapsulation and due to the increase in spatial heterogeneity we measure no pattern profiles in the pericellular region of 11% of the cells. As we discussed earlier, 6 days gives time for cells encapsulated in medium and high cross-link density hydrogel scaffolds to degrade their microenvironments. On this day, we measure an increase in the percentage of the cells that have degraded their microenvironments for R = 0.7, 0.75 and 0.85 hydrogels. For R = 0.7 gels, 30% of the cells are creating a no pattern profile when they degrade the hydrogel scaffold to the gel-sol transition. At this stiffness, MMP secretions are increasing and results in phase transitions in the pericellular region.79 We hypothesize that at this stiffness, TIMP secretion is not high enough to create a reverse reaction-diffusion profile. The second dominant degradation profile at R = 0.7 is the uniform profile. We hypothesize that at this intermediate scaffold stiffness, hMSCs are increasing MMP secretion but are also secreting TIMPs, which are regulating scaffold degradation and enabling uniform degradation in the pericellular region.
Six days post-encapsulation and further increasing the stiffness to R = 0.75, approximately 38% of cells create a no pattern profile and 25% create a reaction-diffusion profile in their surrounded microenvironment. At this stiffness, we hypothesize that hMSCs change their degradation strategy by increasing MMP secretions and decreasing TIMP secretions, which would change the degradation profile to a reaction-diffusion profile. By further increasing hydrogel stiffness, R = 0.85, we no longer measure a reaction-diffusion profile. At this stiffness, 15% of cells create a reverse reaction-diffusion profile to degrade the scaffold and 31% of the cells are degrading their pericellular region uniformly. Here, we hypothesize that the increase in the number of cross-links is higher than the increased MMP secretion. Therefore, the majority of the encapsulated cells at this stiffness are degrading the scaffold uniformly, Figure 7c. At R = 1, we only measure remodeling by 13% of the encapsulated hMSCs and they have remodeled their surrounded microenvironment uniformly. These results suggest that since cells continuously adapt to their environment, understanding changes in cell behavior in scaffolds that recapitulate different tissue stiffnesses is the first step to better understanding cell-material interactions within materials that are being designed to mimic these environments.
Microenvironmental stiffness changes cellular degradation profiles and hMSC degradation strategies. In soft hydrogels, R = 0.65 (), TIMPs regulate the activity of MMPs and create a reverse reaction-diffusion profile in the pericellular region. This environment gives hMSCs more time to relax and spread in the network prior to migration.41,56 As hydrogel stiffness increases, we hypothesize that cells secrete more MMPs and less TIMPs to degrade and re-engineer the hydrogel faster and become motile.79-81 This change in cell-mediated biomolecule secretion changes the degradation profile to a no pattern profile at R = 0.7 (), which correlates with pericellular regions that are undergoing a phase transition. By further increasing hydrogel stiffness to R = 0.75 (), cells secrete more MMPs resulting in a reaction-diffusion degradation profile. At R = 0.85 (), the increase in MMP secretion measured by Leight et al. is not high enough compared to the increased number of cross-links that need to be degraded to enable motility.79-81 At this stiffness, the degradation profile changes to a uniform profile. For the stiffest gels, R = 1 (), cells are not able to degrade the gel during the data acquisition window and at the length scale of our measurements.
Conclusion
In this work, we use passive microrheological measurements to characterize dynamic re-engineering in the pericellular region around hMSCs encapsulated in well-defined hydrogel scaffolds with different cross-link densities. The stiffness of the hydrogel is varied by changing the concentration of the cross-linker. The initial swollen elastic moduli of our hydrogels vary from 80–2400 Pa. By using MPT, we characterize rheological changes around hMSCs at each stiffness. MPT measurements enable the characterization of the time and extent of degradation of the material on different days. MPT measures that as the elastic modulus of our hydrogels increase, the time for cells to initiate material degradation increases. At higher hydrogel stiffnesses, cells need more time to degrade cross-linkers and become motile. MPT also measures changes in the degradation profiles created by hMSCs as hydrogel stiffness is increased. Changes in the degradation profiles indicate changes in hMSC degradation strategies, which we hypothesize includes changes in cell secretions, i.e. MMPs and TIMPs, at each stiffness. Measurements of the degradation profiles in the pericellular region are related to the initial modulus of the material and show that as the scaffold stiffness increases cells no longer create environments where they spread and attach prior to becoming motile. Instead in stiffer regions we measure uniform or reaction-diffusion degradation profiles, which is related to increased MMP secretion. This change in degradation profile will change cell motility from a two step process of stretching and then becoming motile to a process where cells migrate once the material is soft enough to enable motility.
We measure cell migration speed to understand the effect of stiffness. As the time post-encapsulation increases, we measure an increase in cellular migration. After days of encapsulation, cells have more time to secrete MMPs and degrade the scaffold to create microchannels to migrate through. By increasing cellular stiffness, we measure a decrease in cellular speed due to an increased physical barrier for migration. Understanding cellular speed and extent of degradation for different stiffnesses could inform design of implantable materials that mimic native environments and deliver cells to wounds in different tissues.
Overall, cell-material interactions are complex and the physical environment around a cell plays an important role in regulating cellular function and behavior. Not only are the physical environments of native tissue diverse but ECM elasticity also changes during fibrotic and degenerative diseases and cancer development. Therefore, the use of 3D hydrogels that mimic aspects of the native ECM to study these dynamic and complex interactions will increase our knowledge of how cells interact with their microenvironment within each tissue. Our study characterizes dynamic cell-material interactions at different elasticities which can be used to better understand changes in cellular behavior during various diseases and will lead to better design of biomaterials for cell delivery to wounded areas.
Supplementary Material
Acknowledgement
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R15GM119065.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI:10.1021/acs.biomac.0c00432.
Changes in the wt% of the MMP-degradable peptide, calculated cross-link density (ρ), hydrogel swollen and unswollen moduli, changes in the state of the material over time for each stiffness, profiles of degradation in the pericellular region of encapsulated hMSCs for different hydrogel stiffnesses
Publisher's Disclaimer: “Just Accepted” manuscripts have been peer-reviewed and accepted for publication. They are posted online prior to technical editing, formatting for publication and author proofing. The American Chemical Society provides “Just Accepted” as a service to the research community to expedite the dissemination of scientific material as soon as possible after acceptance. “Just Accepted” manuscripts appear in full in PDF format accompanied by an HTML abstract. “Just Accepted” manuscripts have been fully peer reviewed, but should not be considered the official version of record. They are citable by the Digital Object Identifier (DOI®). “Just Accepted” is an optional service offered to authors. Therefore, the “Just Accepted” Web site may not include all articles that will be published in the journal. After a manuscript is technically edited and formatted, it will be removed from the “Just Accepted” Web site and published as an ASAP article. Note that technical editing may introduce minor changes to the manuscript text and/or graphics which could affect content, and all legal disclaimers and ethical guidelines that apply to the journal pertain. ACS cannot be held responsible for errors or consequences arising from the use of information contained in these “Just Accepted” manuscripts.
References
- (1).Caplan AI Why are MSCs Therapeutic? New Data: New Insight. J. Pathol 2009, 217, 318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Singer AJ; Clark RA Cutaneous Wound Healing. N. Engl. J. Med 1999, 341, 738–746. [DOI] [PubMed] [Google Scholar]
- (3).Bryant SJ; Anseth KS Controlling the Spatial Distribution of ECM Components in Degradable PEG Hydrogels for Tissue Engineering Cartilage. J. Biomed. Mater. Res., Part A 2003, 64A, 70–79. [DOI] [PubMed] [Google Scholar]
- (4).Mackenzie TC; Flake AW Human Mesenchymal Stem Cells Persist, Demonstrate Site-Specific Multipotential Differentiation, and are Present in sites of Wound Healing and Tissue Regeneration after Transplantation into Fetal Sheep. Blood Cells, Mol., Dis 2001, 27, 601–604. [DOI] [PubMed] [Google Scholar]
- (5).Ries C; Egea V; Karow M; Kolb H; Jochum M; Neth P MMP-2, MT1-MMP, and TIMP-2 are Essential for the Invasive Capacity of Human Mesenchymal Stem Cells: Differential Regulation by Inflammatory Cytokines. Blood 2007, 109, 4055–4063. [DOI] [PubMed] [Google Scholar]
- (6).Kawada H; Fujita J; Kinjo K; Matsuzaki Y; Tsuma M; Miyatake H; Muguruma Y; Tsuboi K; Itabashi Y; Ikeda Y; Ogawa S; Okano H; Hotta T; Ando K; Fukuda K Nonhematopoietic Mesenchymal Stem Cells can be Mobilized and Differentiate into Cardiomyocytes after Myocardial Infarction. Blood 2004, 104, 3581–3587. [DOI] [PubMed] [Google Scholar]
- (7).Maxson S; Lopez EA; Yoo D; Danilkovitch-Miagkova A; LeRoux MA Concise Review: Role of Mesenchymal Stem Cells in Wound Repair. Stem Cells Transl. Med 2012, 4, 142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Badiavas E; Abedi M; Butmarc J; Falanga V; Quesenberry P Participation of Bone Marrow Derived Cells in Cutaneous Wound Healing. J. Cell. Physiol 2003, 196, 245–250. [DOI] [PubMed] [Google Scholar]
- (9).Lee DE; Ayoub N; Agrawal DK Mesenchymal Stem Cells and Cutaneous Wound Healing: Novel Methods to Increase Cell Delivery and Therapeutic Efficacy. Stem Cell Res. Ther 2016, 7, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Schlosser S; Dennler C; Schweizer R; Eberli D; Stein JV; Enzmann V; Giovanoli P; Erni D; Plock JA Paracrine Effects of Mesenchymal Stem Cells Enhance Vascular Regeneration in Ischemic Murine Skin. Microvasc. Res 2012, 83, 267–275. [DOI] [PubMed] [Google Scholar]
- (11).Otero-Viñas M; Falanga V Mesenchymal Stem Cells in Chronic Wounds: The Spectrum from Basic to Advanced Therapy. Adv. Wound Care 2016, 149–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).West JL; Hubbell JA Polymeric Biomaterials with Degradation Sites for Proteases Involved in Cell Migration. Macromolecules 1999, 32, 241–244. [Google Scholar]
- (13).Lutolf MP; Lauer-Fields JL; Schoekel HG; Metters AT; Weber FE; Fields GB; Hubbell JA Synthetic Matrix Metalloproteinase-Sensitive Hydrogels for the Conduction of Tissue Regeneration: Engineering Cell-Invasion Characteristics. PNAS 2003, 100, 5413–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kyburz KA; Anseth KS Three-Dimensional hMSC Motility Within Peptide-Functionalized PEG-Based Hydrogels of Varying Adhesivity and Crosslinking Density. Acta Biomater. 2013, 9, 6381–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ferreira LS; Gerecht S; Fuller J; Shieh HF; Vunjak-Novakovic G; Langer R Bioactive Hydrogel Scaffolds for Controllable Vascular Differentiation of Human Embryonic Stem Cells Bioactive Hydrogel Scaffolds for Controllable Vascular Differentiation of Human Embryonic Stem Cells. Biomaterials 2007, 28, 2706–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Peppas NA; Hilt JZ; Khademhosseini A; Langer R Hydrogels in Biology and Medicine: From Molecular Principles to Bionanotechnology. Adv. Mater 2006, 18, 1345–1360. [Google Scholar]
- (17).Anderson SB; Lin C-C; Kuntzler DV; Anseth KS The erformance of Human Mesenchymal Stem Cells Encapsulated in Cell-Degradable Polymer-Peptide Hydrogels. Biomaterials 2011, 32, 3564–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Guvendiren M; Burdick JA Engineering Synthetic Hydrogel Microenvironments to Instruct Stem Cells. Curr. Opin. in Biotech 2013, 24, 841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Benton JA; Fairbanks BD; Anseth KS Characterization of Valvular Interstitial Cell Function in Three Dimensional Matrix Metalloproteinase Degradable PEG Hydrogels. Biomaterials 2009, 30, 6593–6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Schultz KM; Kyburz KA; Anseth KS Measuring Dynamic Cell-Material Interactions and Remodeling During 3D Human Mesenchymal Stem Cell Migration in Hydrogels. PNAS 2015, 112, E3757–E3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Metzger S; Blache U; Lienemann PS; Karlsson M; Weber FE; Weber W; Ehrbar M Cell-Mediated Proteolytic Release of Growth Factors from Poly(Ethylene Glycol) Matrices. Macromol. Biosci 2016, 16, 1703–1713. [DOI] [PubMed] [Google Scholar]
- (22).Wolf K; Lindert MT; Krause M; Alexander S; Riet JT; Willis AL; Hoffman RM; Figdor CG; Weiss SJ; Friedl P Physical Limits of Cell Migration: Control by ECM Space and Nuclear Deformation and Tuning by proteolysis and Traction Force. J. Cell Biol 2013, 7, 1069–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Vu TH; Werb Z Matrix Metalloproteinases: Effectors of Development and Normal Physiology. Genes Dev. 2000, 14, 2123–2133. [DOI] [PubMed] [Google Scholar]
- (24).Engler AJ; Sen S; Lee Sweeney H; Discher DE Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126, 677–689. [DOI] [PubMed] [Google Scholar]
- (25).Schwartz MP; Fairbanks BD; Rogers RE; Rangarajan R; Zaman MH; Anseth KS A synthetic Strategy for Mimicking the Extracellular Matrix Provides Insight about Tumor Cell Migration. Integr. Biol 2010, 2, 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Aimetti AA; Machen AJ; Anseth KS Poly(ethylene glycol) Hydrogels Formed by thiol-ene Photopolymerization for Enzyme-Responsive Protein Delivery. Biomaterials 2009, 30, 6048–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Tirrell DA Biomaterials: Important Areas for Future Investment. National Science Foundation 2012, December 2019, http://nsfbiomatworkshop2012.caltech.edu/report/index.html. [Google Scholar]
- (28).Fairbanks BD; Schwartz MP; Halevi AE; Nuttelman CR; Bowman CN; Anseth KS A Versatile Synthetic Extracellular Matrix Mimic via Thiol-Norbornene Photopolymerization. Adv. Mater 2009, 10, 3114–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Raeber GP; Lutolf MP; Hubbell JA Mechanisms of 3D Migration and Matrix Remodeling of Fibroblasts within Artificial ECMs. Acta Biomater. 2007, 3, 615–629. [DOI] [PubMed] [Google Scholar]
- (30).Raeber GP; Lutolf MP; Hubbell JA Molecularly Engineered PEG Hydrogels: a Novel Model System for Proteolytically Mediated Cell Migration. Biophys. J 2005, 89, 1374–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Lutolf MP; Weber FE; Schmoekel HG; Schense JC; Kohler T; Muller R; Hubbell JA Repair of Bone Defects Using Synthetic Mimetics of Collagenous Extracellular Matrices. Nat. Biotech 2003, 21, 513–518. [DOI] [PubMed] [Google Scholar]
- (32).Ponte AL; Marais E; Gallay N; Langonné A; Delorme B; Hérault O; Charbord P; Domenech J The in Vitro Migration Capacity of Human Bone Marrow Mesenchymal Stem Cells: Comparison of Chemokine and Growth Factor Chemotactic Activities. Stem Cells 2007, 25, 1737–1745. [DOI] [PubMed] [Google Scholar]
- (33).Cox TR; Erler JT Remodeling and Homeostasis of the Extracellular Matrix: Implications for Fibrotic Diseases and Cancer. Dis. Models Mech 2009, 4, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Buxboim A; Ivanovska IL; Discher DE Matrix Elasticity, Cytoskeletal Forces and Physics of the Nucleus: How Deeply do Cells ‘Feel’ Outside and In? J. Cell Sci 2010, 123, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Barnes JM; Przybyla L; Weaver VM Tissue Mechanics Regulate Brain Development, Homeostasis and Disease. J Cell Sci 2017, 130, 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Kloxin AM; Kloxin CJ; Bowman CN; Anseth KS Mechanical Properties of Cellularly Responsive Hydrogels and their Experimental Determination. Biomaterials 2010, 22, 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Peyton SR; Raub CB; Keschrumrus VP; Putnam AJ The Use of Poly(ethylene glycol) Hydrogels to Investigate the Impact of ECM Chemistry and Mechanics on Smooth Muscle Cells. Biomaterials 2006, 27, 4881–4893. [DOI] [PubMed] [Google Scholar]
- (38).Grim JC; Marozas IA; Anseth KS Thiol-ene and Photo-cleavage Chemistry for Controlled Presentation of Biomolecules in Hydrogels. J. Controlled Release 2015, 219, 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Daviran M; Caram HS; Schultz KM Role of Cell-Mediated Enzymatic Degradation and Cytoskeletal Tension on Dynamic Changes in the Rheology of the Pericellular Region Prior to Human Mesenchymal Stem Cell Motility. ACS Biomater. Sci. Eng 2018, 4, 468–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Patterson J; Hubbell J Enhanced Proteolytic Degradation of Molecularly Engineered PEG Hydrogels in Response to MMP-1 and MMP-2. Biomaterials 2010, 31, 7836–7845. [DOI] [PubMed] [Google Scholar]
- (41).Daviran M; Longwill SM; Casella JF; Schultz KM Rheological Characterization of Dynamic Remodeling of the Pericellular Region by Human Mesenchymal Stem Cell-Secreted Enzymes in Well-Defined Synthetic Hydrogel Scaffolds. Soft Matter 2018, 14, 3078–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Bao M; Xie J; Huck WT Recent Advances in Engineering the Stem Cell Microniche in 3D. Adv. Sci 2018, 5, 1800448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Dikovsky D; Bianco-Peled H; Seliktar D Defining the Role of Matrix Compliance and Proteolysis in Three-Dimensional Cell Spreading and Remodeling. Biophys. J 2008, 94, 2914–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Crocker JC; Grier DG Methods of Digital Video Microscopy for Colloidal Studies. J. Colloid Interface Sci 1996, 179, 298–310. [Google Scholar]
- (45).Winter HH Can the Gel Point of a Cross-linking Polymer be Detected by the G’-G” Crossover? Polym. Eng. Sci 1987, 27, 1698–1702. [Google Scholar]
- (46).Mason TG; Ganesan K; van Zanten JH; Wirtz D; Kuo SC Particle Tracking Microrheology of Complex Fluids. Phys. Rev. Lett 1997, 79, 3282–3285. [Google Scholar]
- (47).Waigh TA Microrheology of Complex Fluids. Rep. Prog. Phys 2005, 68, 685–742. [DOI] [PubMed] [Google Scholar]
- (48).Larsen TH; Furst EM Microrheology of the Liquid-Solid Transition During Gelation. Phys. Rev. Lett 2008, 100, 146001–4. [DOI] [PubMed] [Google Scholar]
- (49).Furst EM; Squires TM Microrheology, 1st ed.; Oxford University Press: New York, NY, 2017. [Google Scholar]
- (50).Schultz KM; Furst EM Microrheology of Biomaterial Hydrogelators. Soft Matter 2012, 8, 6198–6205. [Google Scholar]
- (51).Wehrman MD; Lindberg S; Schultz KM Quantifying the Dynamic Transition of Hydrogenated Castor Oil Gels Measured via Multiple Particle Tracking Microrheology. Soft Matter 2016, 12, 6463–6472. [DOI] [PubMed] [Google Scholar]
- (52).Escobar F; Anseth KS; Schultz KM Dynamic Changes in Material Properties and Degradation of Poly(ethylene glycol)-Hydrazone Gels as a Function of pH. Macromolecules 2017, 50, 7351–7360. [Google Scholar]
- (53).Schultz KM; Baldwin AD; Kiick KL; Furst EM Measuring the Modulus and Reverse Percolation Transition of a Degrading Hydrogel. ACS Macro Lett. 2012, 1, 706–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Schultz KM; Anseth KS Monitoring Degradation of Matrix Metalloproteinases Cleavable PEG Hydrogels via Multiple Particle Tracking Microrheology. Soft Matter 2013, 9, 1570–1579. [Google Scholar]
- (55).Lozito TP; Jackson WM; Nesti LJ; Tuan RS Human Mesenchymal Stem Cells Generate a Distinct Pericellular Zone of MMP Activities via Binding of MMPs and Secretion of High Levels of TIMPs. Matrix Biol. 2014, 34, 132–143. [DOI] [PubMed] [Google Scholar]
- (56).Daviran M; Schultz KM Characterizing the Dynamic Rheology in the Pericellular Region by Human Mesenchymal Stem Cell Re-engineering in PEG-Peptide Hydrogel Scaffolds. Rheol. Acta 2019, 58, 421–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Miller JS; Shen CJ; Legant WR; Baranski JD; Blakely BL; Chen CS Bioactive Hydrogels Made from Step-Growth Derived PEG-Peptide Macromers. Biomaterials 2010, 31, 3736–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Fairbanks BD; Schwartz MP; Bowman CN; Anseth KS Photoinitiated Polymerization of PEG-Diacrylate with Lithium Phenyl-2,4,6-trimethylbenzoylphosphinate: Polymerization Rate and Cytocompatibility. Biomaterials 2009, 30, 6702–6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Hafner J; Grijalva D; Ludwig-Husemann A; Bertels S; Bensinger L; Raic A; Gebauer J; Oelschlaeger C; Bastmeyer M; Bieback K; Lee-Thedieck C; Willenbacher N Monitoring matrix remodeling in the cellular microenvironment using microrheology for complex cellular systems. Acta Biomater. 2020, 5, 54. [DOI] [PubMed] [Google Scholar]
- (60).Loebel C; Kwon MY; Wang C; Han L; Mauck RL; Burdick JA Metabolic Labeling to Probe the Spatiotemporal Accumulation of Matrix at the Chondrocyte–Hydrogel Interface. Adv. Funct. Mater 2020, 1909802–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Mason TG; Weitz DA Optical Measurements of Frequency-Dependent Linear Viscoelastic Moduli of Complex Fluids. Phys. Rev. Lett 1995, 74, 1250–1253. [DOI] [PubMed] [Google Scholar]
- (62).Savin T; Doyle PS Static and Dynamic Errors in Particle Tracking Microrheology. Biophys. J 2005, 88, 623–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Crocker JC; Weeks ER Particle Tracking Using IDL. 2011, December 2019, http://www.physics.emory.edu/~weeks/idl/index.html. [Google Scholar]
- (64).Larsen T; Schultz K; Furst EM Hydrogel Microrheology Near the Liquid-Solid Transition. Korea-Aust Rheol J 2008, 20, 165–173. [Google Scholar]
- (65).Squires TM; Mason TG Fluid Mechanics of Microrheology. Annu. Rev. Fluid Mech. 2010, 42, 413–438. [Google Scholar]
- (66).Ferry JD Viscoelastic Properties of Polymers; John Wiley & Sons, Inc.: New York, NY, 1980. [Google Scholar]
- (67).Palmer A; Xu J; Wirtz D High-Frequency Viscoelasticity of Crosslinked Actin Filament Networks Measured by Diffusing Wave Spectroscop. Rheol. Acta. 1998, 33, 97–106. [Google Scholar]
- (68).Schultz KM; Bayles AV; Baldwin AD; Kiick KL; Furst EM Rapid, High Resolution Screening of Biomaterial Hydrogelators by μ2rheology. Biomacromolecules 2011, 12, 4178–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Wu N; Schultz KM Microrheological Characterization of Covalent Adaptable Hydrogels for Applications in Oral Delivery. Soft Matter 2019, 15, 5921–5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Corrigan AM; Donald AM Passive Microrheology of Solvent-Induced Fibrillar Protein Networks. Langmuir 2009, 25, 8599–8605. [DOI] [PubMed] [Google Scholar]
- (71).Wehrman MD; Lindberg S; Schultz KM Multiple Particle Tracking Microrheology Measured Ssing Bi-Disperse Probe Diameters. Soft Matter 2018, 14, 5811–5820. [DOI] [PubMed] [Google Scholar]
- (72).Muthukumar M; Winter HH Fractal Dimension of a Crosslinking Polymer at the Gel Point. Macromolecules 1986, 19, 1284–1285. [Google Scholar]
- (73).Chambon F; Winter HH Linear Viscoelasticity at the Gel Point of a Crosslinking PDMS with Imbalanced Stoichiometry. J. Rheol 1987, 31, 683–697. [Google Scholar]
- (74).Adolf D; Martin JE Time-Cure Superposition During Crosslinking. Macromolecules 1990, 23, 3700–3704. [Google Scholar]
- (75).Winter HH; Izuka A; DeRosa ME Experimental Observation of the Molecular Weight Dependence of the Critical Exponents for the Rheology Near the Gel Point. Polym. Gels Networks 1994, 2, 239–245. [Google Scholar]
- (76).Zhang H; Wehrman MD; Schultz KM Structural Changes in Polymeric Gel Scaffolds around the Overlap Concentration. Front. Chem 2019, 7, 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Schultz KM; Baldwin AD; Kiick KL; Furst EM Gelation of Covalently Cross-Linked PEG-Heparin Hydrogels. Macromolecules 2009, 42, 5310–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Scanlan JC; Winter HH Composition Dependence of the Viscoelasticity of End-Linked Poly(dimethylsiloxane) at the Gel Point. Macromolecules 1991, 24, 47–54. [Google Scholar]
- (79).Leight JL; Alge DL; Maier AJ; Anseth KS Direct Measurement of Matrix Metalloproteinase Activity in 3D Cellular Microenvironments Using a Fluorogenic Peptide Substrate. Biomaterials 2013, 34, 7344–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Fakhouri AS; Weist JL; Tomusko AR; Leight JL High-Throughput Three-Dimensional Hydrogel Cell Encapsulation Assay for Measuring Matrix Metalloproteinase Activity. Assay Drug Dev. Techn 2019, 17, 100–115. [DOI] [PubMed] [Google Scholar]
- (81).Leight JL; Tokuda EY; Jonesa CE; Lin AJ; Anseth KS Multifunctional Bioscaffolds for 3D Culture of Melanoma Cells Reveal Increased MMP Activity and Migration with BRAF Kinase Inhibition. PNAS 2015, 112, 5366–5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.