Abstract
Background and Purpose
Despite a growing awareness, annual losses of honeybee colonies worldwide continue to reach threatening levels for food safety and global biodiversity. Among the biotic and abiotic stresses probably responsible for these losses, pesticides, including those targeting ionotropic GABA receptors, are one of the major drivers. Most insect genomes include the ionotropic GABA receptor subunit gene, Rdl, and two GABA‐like receptor subunit genes, Lcch3 and Grd. Most studies have focused on Rdl which forms homomeric GABA‐gated chloride channels, and a complete analysis of all possible molecular combinations of GABA receptors is still lacking.
Experimental Approach
We cloned the Rdl, Grd, and Lcch3 genes of Apis mellifera and systematically characterized the resulting GABA receptors expressed in Xenopus oocytes, using electrophysiological assays, fluorescence microscopy and co‐immunoprecipitation techniques.
Key Results
The cloned subunits interacted with each other, forming GABA‐gated heteromeric channels with particular properties. Strikingly, these heteromers were always more sensitive than AmRDL homomer to all the pharmacological agents tested. In particular, when expressed together, Grd and Lcch3 form a non‐selective cationic channel that opens at low concentrations of GABA and with sensitivity to insecticides similar to that of homomeric Rdl channels.
Conclusion and Implications
For off‐target species like the honeybee, chronic sublethal exposure to insecticides constitutes a major threat. At these concentration ranges, homomeric RDL receptors may not be the most pertinent target to study and other ionotropic GABA receptor subtypes should be considered in order to understand more fully the molecular mechanisms of sublethal toxicity to insecticides.
Abbreviations
- AmGRD
Apis mellifera GRD
- AmLCCH3
Apis mellifera LCCH3
- AmRDL
Apis mellifera RDL
- DmGRD
Drosophila melanogaster GRD
- DmLCCH3
Drosophila melanogaster LCCH3
- DmRDL
Drosophila melanogaster RDL
- GABAA
vertebrate ionotropic GABA receptor
- PCCP−
pentacyanocyclopentdienyl
- TM
transmembrane domain
What is already known
So far, only homomeric RDL GABA receptors have been studied for insecticide toxicity.
The honeybee genome includes two additional genes coding for GABA‐like receptor subunits, Lcch3 and Grd.
What this study adds
Heterologously expressed RDL, LCCH3, and GRD can form functional, heteromeric, ionotropic GABA receptors.
These heteromeric and homomeric GABA receptors display different pharmacological properties.
What is the clinical significance
To understand mechanism(s) of insecticide action, both heteromeric and homomeric GABA receptors should be considered.
1. INTRODUCTION
GABA is a widespread neurotransmitter in the CNS of vertebrates and invertebrates which primarily mediates fast inhibitory neurotransmission but can also act as an excitatory mediator during critical developmental stages of neuronal wiring or under pathological conditions (Ben‐Ari, 2002; Cossart, 2014; Gou, Wang, & Wang, 2012; Ozoe, 2013; Sigel & Steinmann, 2012). The ionotropic GABA receptors (GABAA receptors in mammals) are ligand‐gated ion channels with either homopentameric or heteropentameric structure (Alexander et al., 2019). All the known GABAA receptor subunits display a similar structural scheme, with a large N‐terminal extracellular domain involved in the formation of an agonist binding pocket, and a pore domain made of four transmembrane α‐helices, TM1–TM4. Residues within the TM2 segment line the edge of this pore and are responsible for the ion channel selectivity (Charnet et al., 1990; Menard, Horvitz, & Cannon, 2005; Thompson & Lummis, 2003; Wotring, Miller, & Weiss, 2003). A “Cys‐loop” motif of 13 amino acids located between the N‐terminal and the transmembrane domains is involved in the coupling between agonist binding and channel opening (Grutter et al., 2005; Kash, Jenkins, Kelley, Trudell, & Harrison, 2003; Keramidas & Harrison, 2010; Lema & Auerbach, 2006). The intracellular loop between the TM3 and TM4 helices makes a modulatory platform for regulating channel functions and trafficking by protein–protein interactions or post‐translational modifications (Houston, Lee, Hosie, Moss, & Smart, 2007; Lee, Song, Jee, Vanoaica, & Ahnn, 2005).
While the architecture of vertebrate and insect GABA receptors share these common traits, the number of genes coding for ionotropic GABA receptor subunits is much larger in vertebrates (Buckingham, 2005; Casida & Durkin, 2013, 2015; Ffrench‐Constant, Williamson, Davies, & Bass, 2016). Pentameric combinations of the 19 mammalian subunits produce GABA receptor subtypes with specific pharmacological and biophysical properties. The predominant GABAA receptor type in the mammalian brain is made of α1, β2,3, and γ2, but all other plausible combinations of subunits have been studied (Ben‐Ari, Gaiarsa, Tyzio, & Khazipov, 2007; Olsen & Sieghart, 2009; Sieghart, Ramerstorfer, Sarto‐Jackson, Varagic, & Ernst, 2012). In insects, the Rdl gene has been shown to encode a ionotropic GABA receptor subunit. When expressed in Xenopus oocytes, the RDL subunit is able to form homomeric GABA receptors responding to GABA application with properties summarizing the main characteristics of native current induced by GABA (Ffrench‐Constant, Rocheleau, Steichen, & Chalmers, 1993). Rdl is also widely expressed in the insect CNS (Enell, Hamasaka, Kolodziejczyk, & Nässel, 2007; Ganeshina & Menzel, 2001; Schäfer & Bicker, 1986) and is implicated in numerous functions like olfactory learning (Liu, Krause, & Davis, 2007). The homomeric RDL GABA receptor is thus believed to be responsible for most of the inhibitory effects of GABA on synaptic transmission in insects, and the majority of the efforts to characterize GABA responses biophysically and pharmacologically have focused on this receptor.
However, native currents induced by GABA recorded from different cell types or developmental stages are diverse and present subtle differences which suggest the existence of several subtypes of GABA receptors (Hosie, Aronstein, Sattelle, & Ffrench‐Constant, 1997; Rauh, Lummis, & Sattelle, 1990; Sattelle, Pinnock, Wafford, & David, 1988). Moreover, as seen in Drosophila, the pharmacological profiles and biophysical properties of native GABA receptors and homopentameric RDL receptors do not match perfectly (sensitivity to benzodiazepine, bicuculline, or picrotoxin, for example) (Buckingham, 2005; Es‐Salah, 2008; Ffrench‐Constant et al., 1993; Hosie, Buckingham, Presnail, & Sattelle, 2001; Hosie & Sattelle, 1996; Jones et al., 2009; Zhang, Lee, Rocheleau, Ffrench‐Constant, & Jackson, 1995). This has been attributed to RNA splicing and editing of the Rdl gene but also to the possible formation of heteromeric receptors with other subunits (Dupuis et al., 2010). Indeed, three other genes, Grd, Lcch3, and Lcch‐14A, have been shown to encode GABA receptor‐like subunits (Gisselmann, Plonka, Pusch, & Hatt, 2004; Harvey et al., 1994; Henderson, Knipple, & Soderlund, 1994; Knipple & Soderlund, 2010; Witte, Kreienkamp, Gewecke, & Roeder, 2002). While the role of Lcch‐14A is still unknown, GRD and LCCH3 subunits can produce functional heteropentamers in heterogeneous systems, when expressed together or in combination with RDL subunits, and thus may also participate in GABAergic neurotransmission in the insect brain. Studies on these other GABA receptor‐like subunits, however, are quite sparse, limited to Drosophila, and, until now, have never demonstrated expression of native heteromeric GABA receptors (Knipple, Henderson, & Soderlund, 1995; Zhang et al., 1995). Co‐expression of LCCH3 with RDL leads to the formation of GABA receptors with specific gating properties, enhanced bicuculline sensitivity, and reduced picrotoxin sensitivity (Zhang et al., 1995). Co‐expression of GRD subunits with RDL has never been tested, and co‐expression of GRD with LCCH3 produces functional GABA responding receptors but with cationic conductance, thus giving an excitatory response (Gisselmann et al., 2004; Zhang et al., 1995). Moreover, studies in Xenopus oocytes have also demonstrated that the pharmacology of the RDL receptor subunits can be modified by co‐assembling them with other receptor subunits such as glutamate‐activated chloride channels (Eguchi et al., 2006; Ludmerer et al., 2002). The exact molecular nature of the receptors responsible for GABA response in the insect brain is therefore still a matter of debate, and heteromeric GABA receptors could be more relevant than expected.
Insect GABA receptors are targets for the insecticides used in agriculture and in veterinary medicine, but these pesticides have also been associated with a marked decrease in the numbers of pollinators and, in particular, with increased bee mortality (Holder, Jones, Tyler, & Cresswell, 2018). During acute intoxication, structure–function studies and resistant insect strains have demonstrated that RDL is the main target (Ffrench‐Constant et al., 1993; Hosie, Baylis, Buckingham, & Sattelle, 1995). However, in the case of off‐target species, the mechanism underlying death from chronic sublethal exposure has been less investigated (Aliouane et al., 2009; Charreton et al., 2015; Decourtye et al., 2005; El Hassani, Dacher, Gauthier, & Armengaud, 2005; Jinguji, Ohtsu, Ueda, & Goka, 2018; Pisa et al., 2015). In particular, there is a lack of the comparative analysis of the different GABA receptor properties needed to identify all the molecular determinants that could be involved.
In this work, we have cloned the Rdl, Grd, and Lcch3 GABA receptor subunits from honeybee brain tissue and carried out a systematic biophysical and pharmacological characterization of the different possible combinations of these three subunits in Xenopus oocytes. Studies at the whole‐cell and single‐channel levels revealed specific pharmacological and biophysical profiles for some combinations and suggest that in vivo the response to GABA receptor antagonists used in pest control is more complex than previously estimated.
2. METHODS
2.1. Cloning of AmGABA receptor subunits
RNAs were isolated from the brain, legs, abdomen, and antennae of adult honeybees (pooled from 2 to 20 bees) while 7d and 18d total RNAs were isolated from whole nymphs developed after 7 or 18 days post‐laying. RNA extracts and first‐strand cDNAs were obtained as previously described (Cens et al., 2013). The cDNAs encoding AmRDL (GenBank accession number KJ485710), AmGRD (KJ485711), and AmLCCH3 (KJ485712) were obtained in two steps combining conventional PCR methodologies and RACE‐PCR (First Choice RLM‐RACE kit, Ambion). Specific primers have been designed based on the sequences deposited in BeeBase: amrdl‐001S CATGCTGAATGACGTCAACATCTCCG (PCR) amrdl‐002AS CTCTTCGCTTATTTCGCCTCCTCG (PCR) amrdl‐003AS GTTCGTCGACGACATTAGGGTGGTC (RACE‐PCR for 3′‐end region amplification) amgrd‐001S GAATTGATGAATGAATCAGGACGTATTG (PCR) amgrd‐002AS CGACCAGTCTCACGTGCCATTTG (PCR) amgrd‐004AS CCAGAAGGAAACCCAAGACAGGAC (RACE‐PCR for 3′‐end region amplification) amlcch3‐001S CATGCATCACAGGATGTGGTTGCAG (PCR) amlcch3‐002AS CTCAGAGGACATAAAATATCCAATAGATGGC (PCR) amlcch3‐003AS GGTAATGGAGACATCCTGAGATCTTG (RACE‐PCR for 3′‐end region amplification)
Amplified fragments were cloned into pBluescript (Stratagene) or pcDNA3.1 (−) (Life Technologies), and each full‐length DNA sequence was sequenced on both strands. in vitro RNA synthesis using the mMESSAGE mMACHINE transcription kits (Ambion) was made from linearized plasmids following the instructions from the manufacturer. The cRNA concentration was adjusted to 1 μg·μl−1 for oocyte injection.
The AmRDL subunit was cloned in frame with GFP (pEGFP‐N1, Clontech) and mCherry (pmCherry‐N1, Clontech) to obtain RDL‐GFP and RDL‐mCherry, respectively. The AmGRD subunit was cloned in frame with mCherry (pmCherry‐N1, Clontech) to obtain GRD‐mCherry. The AmLCCH3 was cloned in frame with CFP (pECFP‐N1, Clontech) to obtain LCCH3‐CFP.
Accession number of the proteins used to produce the alignment of the pore forming TM2 segment is as follows: Apis mellifera glycine receptor (AmGlyR): XP_016768424.1; Apis mellifera glutamate‐gated chloride channel (AmGluClR): XP_006560232.1; Apis mellifera nicotinic receptor (AmAchRα8): NP_001011575.1; Homo sapiens serotonin receptor (Hs5HT3α): AAH04453.2; and Caenorhabditis elegans Exp‐1 receptor (CeExp1): NP_495229.3.
2.2. Retrieval, preparation, and two‐electrode voltage clamping of Xenopus oocytes
All animal care and experimental procedures were carried out in strict accordance with the recommendations and relevant guidelines of the CNRS Animal Experimentation Ethics Board which are in compliance with the European convention for the protection of vertebrate animals used for experimental and other scientific purposes ETS no. 123. The experimental protocols, which are in accordance with the ARRIVE guidelines, were approved by the “Direction Départementale des Services Vétérinaires” (authorization no. C34.16). Efforts were made to minimize the number of animals used and animal suffering. Animal studies are reported in compliance with the ARRIVE guidelines (Kilkenny, Browne, Cuthill, Emerson, & Altman, 2010) and with the recommendations made by the British Journal of Pharmacology.
Xenopus laevis were obtained from the CRB Xenope, University of Rennes, France. The CRB Xenope is an accredited French national platform dedicated to Xenopus breeding for experimental research. Frogs were housed at the CRBM Animal facility (authorization no. B 34‐172‐39) in a water‐filled plastic cages in a temperature‐controlled (20°C) and humidity‐controlled (50%) colony room on a 12‐h light/dark cycle. Frogs were fed three times a week with dedicated pellets (Aquatic III). Surgery was performed to female X. laevis under anaesthesia with MS222 (2 g·L−1 added to the water). Parts of one ovary were removed via a mini‐laparotomy (0.5 to 1 cm of abdominal incision), the incision sutured, and the animal allowed to recover for at least 6 months. Animals were not killed for these experimental procedures.
Xenopus oocytes were prepared by collagenase (Sigma) digestion at a concentration of 1 mg·ml−1 in calcium‐free solution for 2 h at room temperature, then incubated at 19°C under gentle agitation in ND96 solution (in mM): NaCI 96, KCl 2, MgCl2 2, CaCl2 1.8, HEPES 5, pH 7.4 with NaOH, and supplemented with 50 mg·ml−1 gentamicin. Stage V and stage VI oocytes were injected with 20–40 nl of an aqueous solution of in vitro transcribed cRNAs of AmGABA receptor subunits, at 1 μg·μl−1, with a stoichiometry 1:1 as previously described (Ménard et al., 2018). Macroscopic currents were recorded 1 to 3 days after injection with the two‐electrode voltage‐clamp technique, in a so‐called ND‐Herg recording solution containing (in mM) NaCl (96), KCl (3), CaCl2 (0.5), MgCl2 (1), and HEPES (5), with pH adjusted to 7.4 with NaOH. This solution was continuously perfused in the recording chamber at a constant rate of 3–5 ml·min−1 by a gravity‐fed system. Microelectrodes were pulled from borosilicate glass (GC150T‐10, Harvard Apparatus) using a horizontal puller (P97, Sutter Company) with a resistance in the range of 0.5 to 2 MΩ after manual backfilling with 3‐M KCl. Oocytes were clamped at −60 mV, and currents recorded at 500 Hz and filtered at 20 Hz using a GeneClamp 500 amplifier (Molecular Devices) controlled by the Clampex software (Molecular Devices) through a Digidata 1200 digitizer (Molecular Devices). A multichannel semi‐automatic perfusion system, fitted with a series of low‐voltage electrovalves (homemade) and controlled by Clampex, allowed rapid changes in the flowing solution, with a negligible dead volume and a constant flowing rate (application of agonists and/or drugs). GABA and muscimol concentration response curves were generated by sequential 3‐s puffs of increasing concentrations with 1‐min intervals (control ND‐Herg solution flowing) between applications. Inhibition curves for the tested insecticides were generated by a 30‐s pre‐incubation period of increasing insecticide concentrations followed by a 3‐s puff of 100‐μM GABA with the corresponding concentration of insecticides. Stock solutions of fipronil, dieldrin, ivermectin, picrotoxin, and pentacyanocyclopentdienyl TEA (TEA‐PCCP) were prepared in DMSO and then diluted in ND‐Herg solution. Final concentration of 0.1% DMSO did not affect electrophysiological recordings. Bicuculline was dissolved at 50 μM in ND‐Herg. Oocytes used for pharmacological tests were selected if stable after three consecutive challenges with 100‐μM GABA at 1‐min interval.
2.3. Single‐channel recording
For single‐channel recordings, the oocyte vitelline membrane was removed using forceps after immersion in a hypertonic solution (200‐mM NaCl, 10‐mM HEPES, pH 7.2 adjusted with NaOH; osmolarity ~500 mOsm), and the oocyte was then allowed to swell in the recording chamber filled with a depolarizing solution (100‐mM KCl, 5‐mM HEPES, 10‐mM EGTA, pH 7.2 adjusted with KOH; osmolarity ~250 mOsm). Firepolished and coated (Sylgard®) patch pipettes had a resistance of 8–12 MΩ when filled with the pipette solution containing 100‐mM KCl, 5‐mM HEPES, and 10‐mM EGTA (pH 7.2 was adjusted with NaOH; ~290 mOsm). Cell‐attached patch‐clamp currents were recorded with an Axopatch 200B amplifier (Molecular Devices), low‐pass filtered at 2 kHz and digitized at 10 kHz using a Digidata 1200 interface, and stored on a computer using the Clampex software. The liquid junction potential was 1–3 mV and was thus neglected. Currents were analysed with the Clampfit software (Molecular Devices). Well‐resolved channel openings were detected by a threshold analysis set at 50% of the elementary current. Channel conductance was calculated from Gaussian fits of amplitude histograms, obtained at different voltages.
2.4. Total internal reflection fluorescence microscopy
Xenopus oocytes co‐injected with in vitro transcribed cRNAs of AmRDL‐GFP and AmGRD‐mChe which produce GABA‐induced current were selected using two‐electrode voltage clamp. The vitelline membranes were manually removed just after the recording, and the oocytes were placed on a 35‐mm imaging dish with a high refractive index glass bottom (n = 1.523; Ibidi) filled with ND96 solution. These oocytes were imaged using a 100X Plan TIRF apochromatic 1.49NA oil immersion objective (Nikon) at room temperature.
2.5. Co‐immunoprecipitation of AmGABA receptor subunits
HEK293 cells were maintained in DMEM supplemented with 10% FBS in a 5% CO2 air incubator at 37°C. One day after plating, HEK cells were co‐transfected using Lipofectamine 2000 (ThermoFisher) by two tagged AmGABA receptor subunits with a stoichiometry of 1:1: GRD‐mCherry with RDL‐GFP, GRD‐mCherry with LCCH3‐CFP, or RDL‐mCherry with LCCH3‐CFP. Each plasmid was concentrated at 1 μg·μl−1. Three days after transfection, cells were harvested in RIPA Lysis Buffer (Santa Cruz) with 1‐mM PMSF and 1‐mM EDTA (pH 8) and Complete Protease Inhibitor (Roche Applied Sci). After mechanical trituration and sonication (3″, 30%), lysates were centrifuged at 17949 x g for 15 min at 4°C. The resulting supernatants were incubated with a prewashed slurry of 20‐μl anti‐GFP‐Trap®_A beads (Chomotek) at 4°C for 2 h under agitation. Beads were washed three times in dilution buffer: 10‐mM Tris/HCl pH 7.5, 150‐mM NaCl, and 0.5‐mM EDTA. Proteins were eluted in 0.2‐M glycine pH 2.5. Cell lysates and immunoprecipitates were analysed separately by western blots using anti‐GFP (1:5,000; MBL), anti‐mCherry (1:5,000, ThermoFisher), and HRP‐conjugated goat anti‐rabbit (1:5,000, Sigma‐Aldrich) antibodies. Detection was performed with the Western Lightning Plus (PerkinElmer) reagents. The antibody‐based procedures used in this study comply with the recommendations made by the British Journal of Pharmacology.
2.6. Data and statistical analyses
Off‐line data analyses were performed with Clampfit 10.5 (Molecular Devices) and Origin 6.0 (OriginLab Corporation). Concentration response data for each oocyte were normalized to the maximum current for that same oocyte. Each experiments correspond to at least five recordings made on different oocytes (n), from at least three different frogs, and data were presented as mean ± SEM (n corresponding to the number of recordings for each receptor type).
The EC50, IC50, and the n H have been calculated by non‐linear regression analysis using the equation for a sigmoid concentration–response curve (Origin 6.0). The data and statistical analysis comply with the recommendations of the British Journal of Pharmacology on experimental design and analysis in pharmacology (Curtis et al., 2018). Statistical significance was determined in comparison with AmRDL data and was performed using SigmaStat software. Data from each group are tested for normality using Shapiro–Wilk test (reject threshold fixed at P < 0.05). If the normality test fails, then a non‐parametric method, that is, ANOVA on rank, is performed. If the normality test succeeds, then equality of variances is tested (reject threshold fixed at P < 0.05). If this test fails, then a non‐parametric method, that is, ANOVA on rank, is performed. Conversely, if this test succeeds, then classical ANOVA was performed. When the differences among the different groups are greater than chance (P < 0.05), post hoc test for a multiple comparison is performed in order to identify which group differ from the others, using either the Holm–Sidak method in case of a parametric test or Dunn's method in case of a non‐parametric test. Differences with a P‐value < 0.05 using Holm–Sidak method or Dunn's method were considered significant.
2.7. Materials
Bicuculline, dieldrin, fipronil, GABA, ivermectin, muscimol and picrotoxin were supplied by Sigma‐Aldrich (St. Louis, MO, USA), TEA‐PCCP was a kind gift from Professor Erwin Sigel (University of Bern, Switzerland) and Professor Dirk Trauner (University of Munich, Germany).
2.8. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20 (Alexander et al., 2019).
3. RESULTS
3.1. Cloning and sequence analysis of A. mellifera RDL, GRD, and LCCH3 genes
The full coding region of A. mellifera GABA receptor subunits was cloned by RT‐PCR from total RNA extract of whole‐brain tissue from A. mellifera. The predicted polypeptide sequence obtained for AmRDL (acc. nb. KJ485710) has 476 amino acids with an estimated MW of 53 kDa and is related to the DmRDL splice variant bd_var1 (Ffrench‐Constant & Rocheleau, 1993; Jones et al., 2009; Taylor‐Wells, Hawkins, Colombo, Bermudez, & Jones, 2017). This sequence is identical to the AmRDL_var1 sequence published recently by Taylor‐Wells et al. The predicted sequences for AmLCCH3 (acc. nb. NM_001077812) and AmGRD (acc. nb. NM_001305884) display 489 amino acids and 514 amino acids, with estimated MWs of 56 and 59 kDa, respectively (Figure 1a). The MW of AmGRD is noticeably smaller than DmGRD which is predicted to be 68 kDa. Multiple small inserts specific to the dipter order are absent in the AmGRD sequence. In particular, a 75‐aa insertion in the extracellular part of the Drosophila GRD channel subunit, which is equivalent to exon 6 of the vertebrate GABAA and glycine receptors, is not present in AmGRD (Harvey et al., 1994).
FIGURE 1.
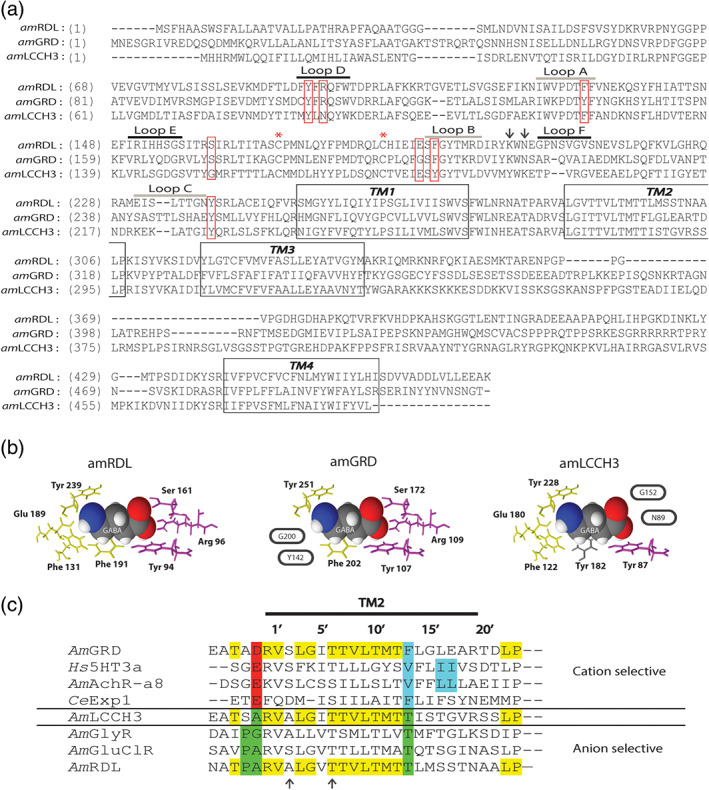
AmGABA receptor sequence analysis. (a) Alignment of honeybee RDL, GRD, and LCCH3 subunit polypeptidic sequences. The four transmembrane segments are marked by black boxes. The two asterisks correspond to the two cysteines involved in the stereotypical motif “Cys‐loop.” The pentameric structure and organization of insect GABA receptors are assumed from mammalian GABA receptors. On this basis, the agonist pocket would be located at the interface between two successive subunits, the principal and the complementary subunits. The six extracellular loops structuring the agonist pocket are underlined above the sequence. Loops A to C (grey line) are on the principal subunit, while loops D to F (black line) are on the complementary subunit. The seven amino acids assumed to directly interact with the GABA molecule are indicated by red boxes. The two arrows locate the charged lysine residues implicated in first events of GABA binding. (b) Compacted representation of a GABA molecule surrounded by its seven interacting residues in the GABA binding pocket of each honeybee GABA receptor subunit (inspired from Comitani, Limongelli, & Molteni, 2016). The GABA binding site is located at the interface of two GABA receptor subunits that both participate in shaping a GABA binding site. The first part of this binding site is made of four amino acids of the principal subunit (in yellow), and the second part is made of three amino acids of the complementary subunit (in purple). The GABA binding site is fully conserved in AmRDL and allows homomeric RDL assembly to exhibit functional GABA binding sites. Two amino acids of AmGRD as principal subunit and two amino acids of AmLCCH3 as complementary subunit are non‐conserved (circled), which probably alters the functionality of these half binding sites and produce homomeric GRD or LCCH3 receptors unable to make functional GABA binding sites. The phenylalanine of loop B in the typical GABA binding site is also not conserved in AmLCCH3, but the functionality of this phenylalanine could be preserved by the presence of a tyrosine residue (Y182) at the same position (coloured in grey). (c) Alignment of the pore forming TM2 segment of Apis mellifera RDL, LCCH3, GRD, glycine receptor (GlyR), nicotinic receptor (AchRα8), glutamate‐gated chloride channel (GluClR), Homo sapiens 5‐HT receptor (5HT3α) and Caenorhabditis elegans Exp‐1 receptor (from Gisselmann et al., 2004). The conserved amino acids in the three sequences AmRDL, AmGRD, and AmLCCH3 are in yellow. AmRDL possesses the key elements of anionic conductance coloured in green, with proline in position −2′ and threonine in position 13′. AmGRD possesses a negatively charged amino acid in position −1′, an aspartic acid, which is a hallmark of cationic conductance (in red). Amino acids crucial for the calcium permeability of cationic receptor are in blue. A mutation of one of the two successive leucine residues in human AchRα7 abolishes calcium permeability. Arrows highlight amino acids important for dieldrin sensitivity. The threonine in position 6′ is conserved in all AmGABA receptor subunits, but the alanine in position 2′ is absent in AmGRD
All these polypeptide subunits present the general tertiary structure and the main features of the ligand‐gated ion channels, including (i) the four transmembrane segments, (ii) the two cysteines of the “Cys‐loop,” and (iii) the loops A, B, and C, on the principal subunit and the loops D, E, and F on the complementary one, which are involved in the agonist binding pocket located at the interface between extracellular domains of two subunits (Lummis, 2009) (Figure 1a). Seven amino acids are described in Drosophila RDL as specifically responsible for the GABA binding in this pocket (Ashby et al., 2012; Comitani et al., 2014; Comitani et al., 2016). These amino acids are all present on the A. mellifera RDL. Interestingly, the four amino acids of the principal subunit are only conserved on the LCCH3 polypeptide, except for the phenylalanine located at position 206 in the DmRDL subunit. However, at the equivalent position in AmLCCH3, the tyrosine Y182 is also able to go into cation–π interaction. Conversely, all of the three amino acids of the complementary subunit are only conserved on the GRD polypeptide (Figure 1b). So, a priori, (i) LCCH3 and GRD homomers could not be functional because their GABA binding sites are incomplete, but (ii) GRD and LCCH3 heteromers could be functional because the two parts of the GABA binding site are complementary. (iii) Association of AmLCCH3 or AmGRD subunits with the AmRDL subunit in heteropentamers will produce receptors with a number of putative binding sites (1–4) depending on their precise stoichiometry but always smaller than the five putative binding sites of the RDL homomers.
The first half of the second transmembrane segment (TM2), which includes most of the amino acids lining the channel pore (in particular the TTVLTMT motif), is conserved in these three GABA receptor subunits (Harvey et al., 1994) (Figure 1c). This implies that the global structure of the pore should be very similar between all these possible channels. The selectivity filter is located within the TM2 but also in the flanking region between positions −4′ and −1 of the TM2 segments (Corringer et al., 1999; Galzi et al., 1992; Keramidas, Moorhouse, Pierce, Schofield, & Barry, 2002; Keramidas, Moorhouse, Schofield, & Barry, 2004; Menard et al., 2005; Wotring et al., 2003). As DmRDL (Gisselmann et al., 2004) and the other honeybee anion ligand‐gated ion channels (glycine receptors or glutamate receptors), AmRDL possesses all the amino acids (P‐2′; A‐1′; T13′) required to be an anion‐selective channel. Inversely, AmGRD displays no proline in position −2′, no threonine in position 13′, but an aspartate in position −1′ (D‐1′), which is a hallmark of cation selective ligand‐gated ion channels like AmnAChRα8. The anion/cation signature for AmLCCH3 is intermediate as this polypeptide has no P‐2′ and no D‐1′ but does haveT13′ (Figure 1c).
All together these data indicate that these three subunits could associate with each other to form a functional GABA responding receptor but with unpredictable selectivity and agonist affinities.
3.2. Expression and assembly of the AmGABA subunits
The three subunits are expressed in larva (J7), in pupa (J18), and in the brain of adult workers (Figure 2a). The broad expression of AmRDL suggests that finding a tissue with an exclusive expression of the AmGRD/LCCH3 heteromers could be a difficult task, which make the recording of this combination in native conditions challenging. On the other hand, some honeybee organs, such as the brain, express concomitantly all of the three Rdl, Grd, and Lcch3 genes. In consequence, we have tested in vitro the possibility of forming heteromeric AmGABA receptors at the biochemical and functional levels. Immunoprecipitations of AmRDL‐GFP or AmLCCH3‐CFP co‐overexpressed in HEK293 cells with GRD‐mCherry or RDL‐mCherry indicate that, under these experimental conditions, AmRDL interacts with AmGRD or AmLCCH3, and AmGRD interacts with AmLCCH3 (Figure 2b). Moreover, total internal reflection fluorescence microscopy (TIRFM) experiments made in oocytes co‐expressing AmRDL‐GFP and AmGRD‐mCherry demonstrate at the scale of single receptor that the RDL GABA receptor subunit is able to co‐assemble with GABA receptor‐like subunits at the plasma membrane (Figure 2c).
FIGURE 2.
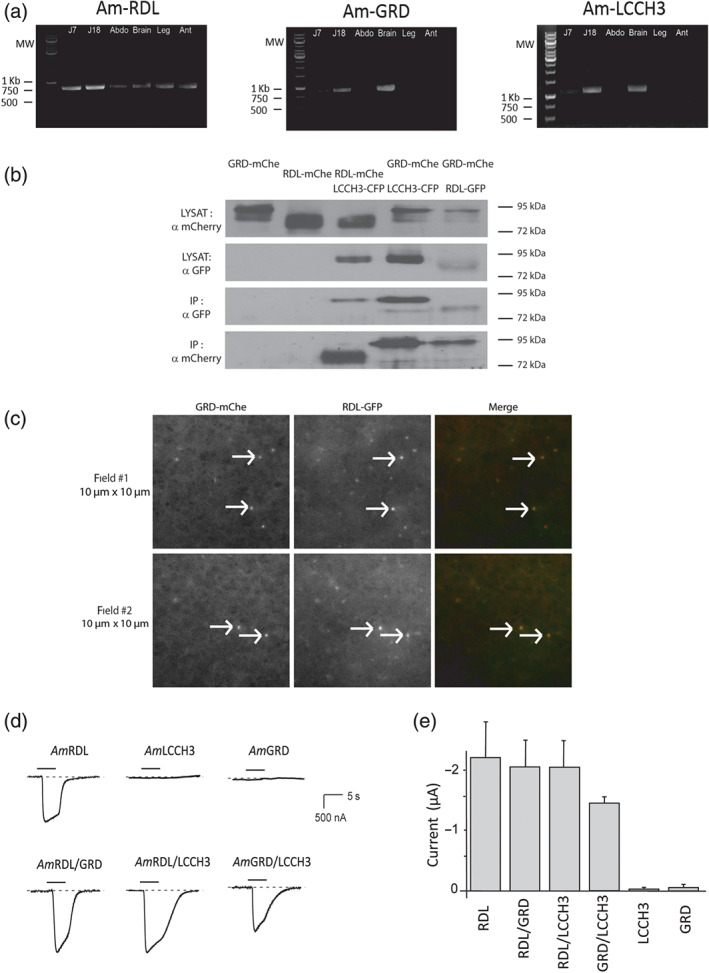
Expression and interaction of AmGABA subunits. (a) RT‐PCR of AmGABA receptor subunits from larvae (J7), nymph (J18), and different parts of adult bodies. RT‐PCR were carried out with the following paired primers: AmRDL: amrdl‐001S and amrdl‐003AS (799pb), AmGRD: amgrd‐001S and amgrd‐004AS (867pb), and AmLCCH3: amlcch3‐001S and amlcch3‐003AS (1143pb). AmRDL transcript is found expressed at all developmental stages and, in the adult honeybee, in all body parts. AmGRD and AmLCCH3 expression are low in larvae, rather high in nymphs, and mostly found in the brain at the adult stage. (Abbreviation: Abdo for abdomen, Ant for antenna). (b) Co‐immunoprecipitation experiments made from HEK293 cells co‐transfected with fluorescently tagged versions of AmGABA receptor subunits. The GFP‐trap system allows the immunoprecipitation of LCCH3‐CFP or RDL‐GFP. Western blot from lysates (input) shows the co‐expression of both proteins in the cells, while anti‐GFP western blot with immunoprecipitation controls both the efficiency and the specificity of the immunoprecipitation. AmLCCH3‐CFP can physically interact with AmGRD‐mChe and AmRDL‐mChe, and AmRDL‐GFP interacts with AmGRD‐mChe. The expected MWs of these tagged proteins are AmGRD‐mChe 85 kDa, AmLCCH3‐CFP 82 kDa, and AmRDL 80 kDa. (c) TIRFM images of oocyte co‐expressing AmGRD‐mCherry and AmRDL‐GFP. Most of the individual protein complex observed on Xenopus oocyte membrane (white arrows) display colocalized spots of mCherry and GFP demonstrating that AmGRD‐mChe and AmRDL‐GFP are able to co‐assemble as a single GABA receptor. (d) Representative current traces obtained by applying 100‐μM GABA to Xenopus oocytes with different AmGABA receptor subunits expressed alone or in combination. Only AmRDL is able to form a functional homopentamer sensitive to GABA application. In contrast, all possible heteromer combinations produce a GABA‐sensitive channel. (e) Averaged values of AmGABA receptor current amplitudes evoked by 100‐μM GABA application (n = 10). The amplitudes of the currents elicited by the stimulation of AmGRD/LCCH3 are slightly smaller than those elicited by the activation of AmRDL‐containing heteromers
A functional test has been performed in Xenopus oocytes using a two‐electrode voltage‐clamp technique. mRNA of honeybee GABA receptor subunits was synthesized in vitro from the linearized plasmids and microinjected individually or in combination with a 1:1 stoichiometry. After 1–4 days, oocytes were placed in the recording chamber, and GABA was perfused at 100 μM. Under these conditions, GABA‐elicited currents could be recorded only upon AmRDL single subunit expression, thus suggesting that solely AmRDL is able to form functional homopentameric GABA receptors. On the other hand, all the tested heteromeric combinations (AmRDL/LCCH3, AmRDL/GRD, and AmGRD/LCCH3) could produce GABA‐sensitive channels, with AmGRD/LCCH3 giving the smallest amplitude currents on average, even if this difference is not statistically significant (P > 0.05 using an ANOVA on rank) (Figure 2d,e).
3.3. Pharmacological analysis of AmGABA receptors
The GABA concentration–response curves indicate that the potencies (EC50) of GABA for heteromeric GABA receptors containing RDL are similar to homomeric AmRDL but significantly higher for the AmGRD/LCCH3 receptors. Furthermore, the n H of AmGRD/LCCH3 is significantly lower than the n Hs of others combinations. Similarly, while the EC50 of muscimol for homomeric AmRDL and heteromeric AmRDL/GRD and AmRDL/LCCH3 GABA receptors are approximately 20 μM, which is 10 times smaller for the AmGRD/LCCH3 with the lowest n H. Finally, muscimol is a full agonist as potent as GABA for any molecular combination of AmGABA receptors (Figure 3).
FIGURE 3.
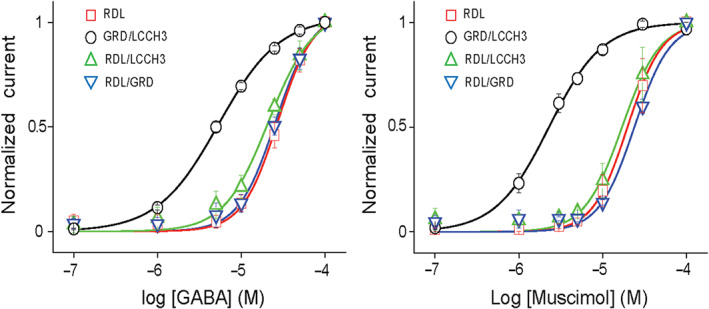
Agonist‐sensitive AmGABA receptors. Agonist concentration‐dependent curves obtained in Xenopus oocytes expressing AmGABA receptors with increasing concentrations of GABA (n = 16) or muscimol (n = 9)
Five GABA receptor antagonists were tested for their capacity to block any of these subunit combinations. Bicuculline, an antagonist of GABAA receptor in mammals, has no effect in any combination containing the AmRDL subunit, and a small inhibitory effect on AmGRD/AmLCCH3 receptors, blocking 26 ± 13% of the current at 50 μM (Figure 4a). Conversely, all AmGABA receptor currents were blocked by fipronil, dieldrin, picrotoxin, and ivermectin in a concentration‐dependent manner (Figure 4b–e). The dose–response curves obtained in each case allowed the estimation of the IC50 and n Hs (Table 1). These parameters for the fipronil and dieldrin concentration–response were approximately similar for all these combinations, albeit subtle statistically significant differences in comparison to AmRDL exist. In particular, AmRDL/LCCH3 and AmGRD/LCCH3 are slightly more sensitive than AmRDL to fipronil and dieldrin, respectively. By contrast, there are marked differences between each combination, in the case of picrotoxin and ivermectin concentration–responses. The AmRDL/LCCH3 heteromer appears to be the most sensitive receptor to picrotoxin while the presence of AmGRD in the other heteromers increased the EC50 significantly. For ivermectin, the n Hs and IC50 are significantly lower for heteromeric AmGABA receptors than for homomeric AmRDL receptors, suggesting that the binding properties of ivermectin on the homomeric AmRDL and on heteromeric AmGABA receptors are different. Interestingly, 5 min pre‐incubation in 0.5‐μM ivermectin induced a full block of AmRDL receptors but only a 50% inhibition for AmGRD/LCCH3 receptors (Figure 4f).
FIGURE 4.
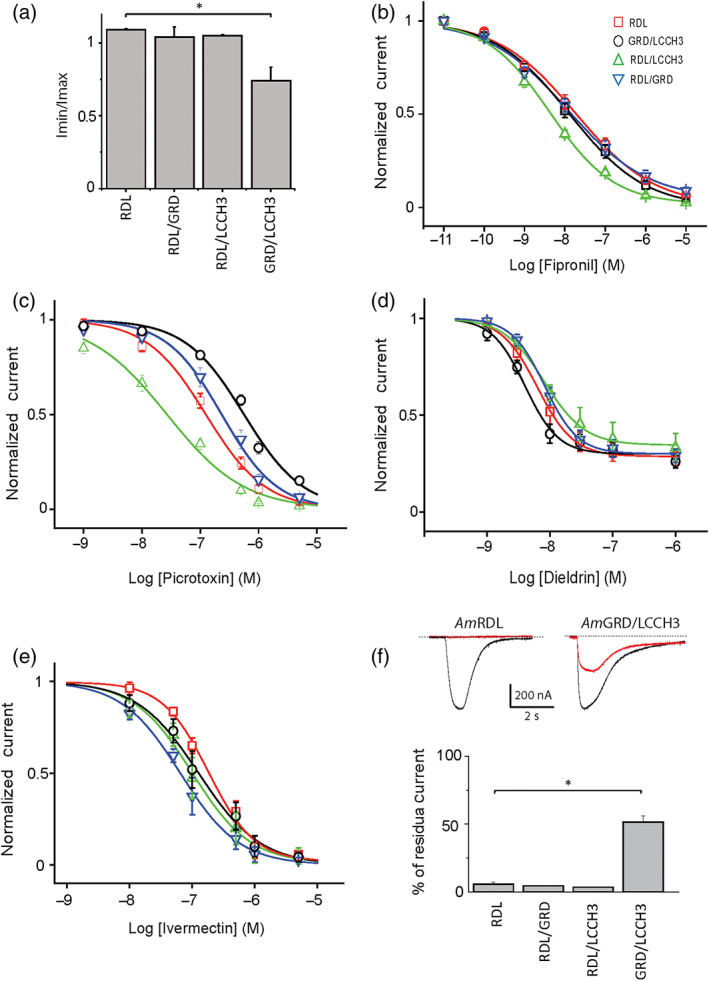
Pharmacological characterization of AmGABA receptors. (a) Effect of 50‐μM bicuculline on 100‐μM GABA‐evoked current from oocyte expressing AmRDL, AmRDL/LCCH3, AmRDL/GRD, or AmLCCH3/GRD (n = 5). All RDL‐containing receptors are insensitive, and AmGRD/LCCH3 is slightly sensitive to bicuculline at 50 μM. (b–e) Concentration‐dependent inhibitory curves obtained from oocytes expressing AmRDL, AmRDL/LCCH3, AmRDL/GRD, or AmLCCH3/GRD; 100‐μM GABA‐evoked current are blocked by successive applications of increasing concentrations of fipronil (n = 11) (b), picrotoxin (n = 12) (c), dieldrin (n = 7) (d), and ivermectin (n = 9) (e). (f) Representative examples of two successive 100‐μM GABA‐evoked currents, spaced by 5 min in the presence of 500‐nM ivermectin, recorded in oocytes expressing AmRDL or AmLCCH3/GRD. The black traces represent the initial GABA‐evoked current, and the red traces show the second one (up). Mean values (n = 7) of the pre‐incubation block with ivermectin on AmRDL, AmRDL/LCCH3, AmRDL/GRD, or AmLCCH3/GRD (bottom). *P < .05, significantly different as indicated; Holm–Sidak method
TABLE 1.
Parameters calculated from agonist and antagonist concentration–response curves for AmGABA receptors obtained in Xenopus oocytes
| GABA (n = 16) | Muscimol (n = 11) | Dieldrin (n = 7) | Fipronil (n = 11) | Ivermectin (n = 9) | Picrotoxin (n = 12) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EC50 (μM) | n H | EC50 (μM) | n H | IC50 (nM) | n H | IC50 (nM) | n H | IC50 (nM) | n H | IC50 (nM) | n H | |
| AmRDL | 24.29 ± 0.02 | 2.01 | 19.74 ± 0.01 | 2.11 | 6.88 ± 0.07 | 1.58 | 12.99 ± 0.04 | 0.46 | 181.18 ± 0.11 | 1.17 | 131.6 ± 0.08 | 0.88 |
| AmRDL/LCCH3 | 29.47 ± 0.01 | 2.17 | 23.88 ± 0.03 | 2.04 | 8.59 ± 0.04 | 1.82 | 4.27 ± 0.04* | 0.55 | 99.29 ± 0.09 # | 0.93* | 26.05 ± 0.11* | 0.67 |
| AmRDL/GRD | 27.59 ± 0.01 | 2.09 | 19.44 ± 0.00 | 2.15 | 8.91 ± 0.03 | 1.43 | 11.85 ± 0.05 | 0.43 | 64.62 ± 0.05 # | 0.91* | 219.5 ± 0.10* | 1.01 |
| AmGRD/LCCH3 | 5.34 ± 0.01* | 1.18* | 2.28 ± 0.01* | 1.36* | 4.18 ± 0.04* | 1.77 | 18.61 ± 0.05 | 0.44 | 126.84 ± 0.10 | 0.88* | 568.4 ± 0.06* | 0.89 |
Note: GRD/LCCH3 receptors display a higher potency for GABA and muscimol, and a n H lower than that observed with RDL homomers and heteromers suggesting a unique GABA binding site.
P < .05, significantly different from corresponding value for AmRDL homomers; Holm–Sidak method.
P < .05, significantly different from corresponding value for AmRDL homomers; Dunn's method.
3.4. Biophysical properties of AmGABA receptors
The differences in the TM2 region between the different subunits suggested that differences in ion selectivity/permeability may exists among the different subunit combinations. This was tested for each subunit combination (i) by analysing the permeability profile of the receptors using different permeant ions and (ii) by measuring the single‐channel conductance in cell‐attached patches.
We first measured the reversal potential of the GABA receptor currents on oocytes expressing different subunit combinations. We applied voltage ramps (−50 to +50 mV) during continuous 100‐μM GABA application in the basic recording solution (NaCl 100 mM, HEPES 10 mM). Under these conditions, currents through AmRDL, AmRDL/LCCH3, and AmRDL/GRD receptors reversed at −20.73 ± 1.35 mV, −16.32 ± 1.02 mV, and −20.80 ± 1.00 mV, respectively, values close to −29 mV, the estimated chloride equilibrium potential for immature oocytes calculated with the known intracellular chloride concentration of 33 mM (Barish, 1983) (Figure 5a). The reversal potential for AmLCCH3/GRD was significantly different (−6.87 ± 0.47 mV, Figure 5a). This experimental reversal potential is far from any known ionic equilibrium potential for immature oocytes (+66 mV for Na+, −91 mV for K+) (Barish, 1983).
FIGURE 5.
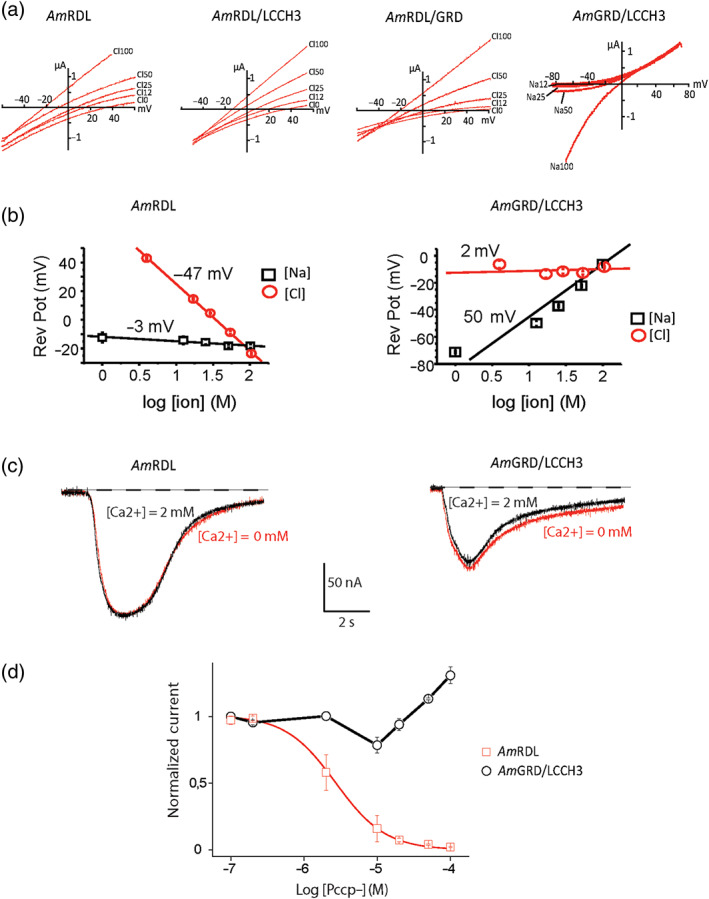
Ion permeability of AmGABA receptors. (a) Reversal potential analysis of current–voltage curves obtained by applying −50 mV/+50 mV voltage ramps in the presence of 100‐μM GABA. Reversal potentials of AmRDL‐containing GABA receptors are sensitive to increased Cl− concentrations but insensitive to external Na+ concentration changes. The opposite results are obtained with AmGRD/LCCH3, for which the reversal potential is only sensitive to increasing sodium concentrations. (b) Semi‐log representation of reversal potential values. Mean values of reversal potentials obtained are plotted versus the logarithm of the Na+ or Cl− concentrations. This allows the calculation of the slope of the regression line for each current studied. The absolute value of the slope for a monovalent permeant ion should be close to 58 mV according to the Nernst equation. For AmRDL receptors, the calculated slopes for Na+ and Cl− are −47 and −3 mV, respectively, meaning that these receptors are mainly permeable to Cl−. On the contrary, these values are +2 and +50 mV respectively for AmGRD/LCCH3, suggesting that these receptors are mainly permeable to Na+, exhibiting a substantial permeability to K+. (c) AmRDL and AmGRD/LCCH3 currents recorded in the presence or in the absence of external Ca2+. (d) Concentration‐dependant curves obtained in Xenopus oocytes expressing AmRDL or AmGRD/LCCH3 with increasing concentrations of the anionic open channel blocker PCCP− (n = 4). PCCP− blocked only the 100‐μM GABA‐evoked currents elicited by AmRDL activation and therefore is a pharmacological tool useful in discriminating between the anion or cation permeability of insect GABA receptors
All the reversal potentials obtained for the RDL‐containing GABA receptors were highly dependent on changes in external chloride concentrations, but insensitive to changes in external Na+ concentrations. The reverse situation was observed for the AmGRD/LCCH3 receptors. The relationships between the current reversal potential and the different concentrations of external Cl− or Na+ are shown in Figure 5a. The regression line for chloride ions yielded a slope of −47 mV (per concentration decade) for AmRDL confirming that Cl− ions are the main permeant species through these channels. The AmGRD/LCCH3 slope is 2 mV suggesting that this channel does not carry Cl−. However, the slope for Na+ is close to 50 mV meaning that under these conditions AmGRD/LCCH3 channels carry Na+ ions (Figure 5b). The permeability behaviours of AmRDL/LCCH3 and AmRDL/GRD are similar to that of AmRDL, as both have a −38 mV slope indicating that they also carry Cl− ions (data not shown).
The reversal potential obtained with AmGRD/LCCH3 in a 100‐mM KCl bathing solution was 6.11 ± 0.49 mV (data not shown) which gave a calculated permeability ratio between Na+ and K+ of 1.6. Furthermore, the presence or absence of Ca2+ did not modify the current amplitude elicited by AmGRD/LCCH3 stimulation, implying that Ca2+ is not able to permeate through this channel (Figure 5c). AmGRD/LCCH3 is therefore a non‐selective, monovalent cation channel.
Recently, pentacyanocyclopentdienyl (PCCP−), a new GABAA anionic open channel blocker, was designed by Carta et al. (2014). We tested it on the anionic RDL and cationic GRD/LCCH3 GABA receptors. PCCP− also inhibited AmRDL receptors activated by 100‐μM GABA application with an IC50 = 2.51 ± 0.02 μM. As expected, PCCP− was clearly not an inhibitor of AmGRD/LCCH3. It even displayed a potentiation effect at high doses (>100 μM) (Figure 5d). PCCP− is therefore a pharmacological tool able to discriminate between anionic and cationic AmGABA receptors.
Finally, cell‐attached patch‐clamp recordings were performed on these four combinations of honeybee GABA receptors using an internal pipette solution comprising KCl (100 mM), HEPES (10 mM), and GABA (100 μM) (Figure 6a). The single‐channel conductance measured from least‐square fit of the current–voltage curves in the negative voltage range yielded single‐channel conductances ranging from 28 pS for AmRDL to 52 pS for AmRDL/LCCH3 (Figure 6b). Fitting the distribution of the dwell time for opening displayed by the different subunit combinations always indicated a bi‐exponential decay (see, e.g., AmRDL/LCCH3, Figure 6c) but with specific time constants for each combinations. In comparison with AmRDL time constants, the presence of LCCH3 in the heteromeric combination significantly prolonged the tau fast (τfast (AmRDL/LCCH3)/τfast (AmRDL) = 2.48; τfast (AmLCCH3/GRD)/τfast (AmRDL) = 1.73) while the presence of GRD reduced the tau slow (τslow (AmRDL/GRD)/τslow (AmRDL) = 0.20; τslow (AmLCCH3/GRD)/τslow (AmRDL) = 0.46).
FIGURE 6.
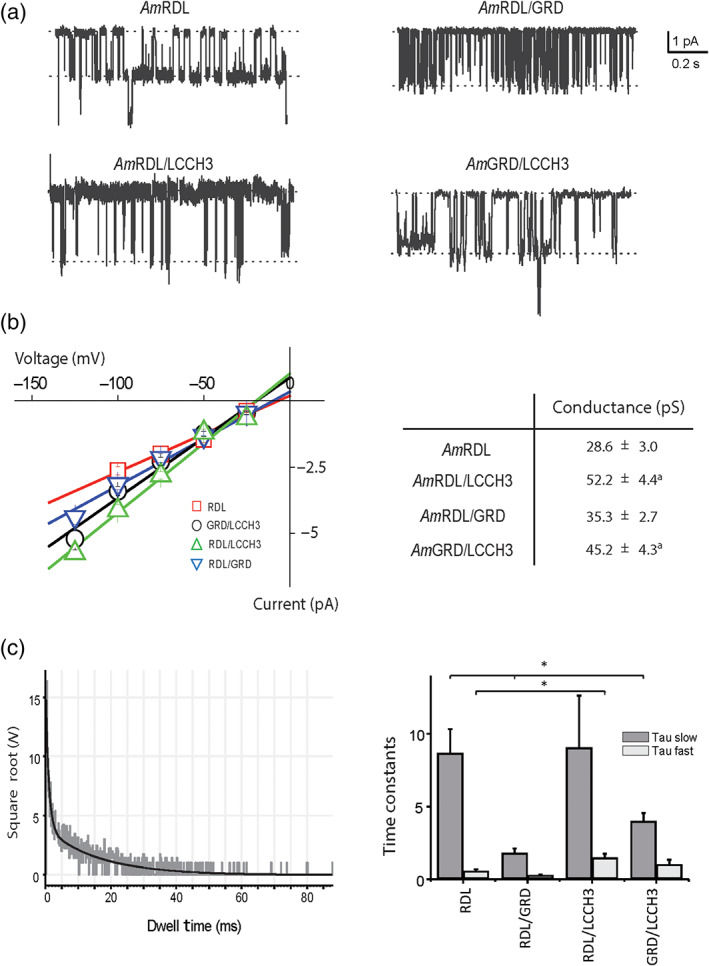
Single‐channel recording of AmGABA receptors. (a) Representative single‐channel current traces recorded from Xenopus oocyte expressing AmRDL, AmRDL/LCCH3, AmRDL/GRD, and AmLCCH3/GRD under symmetrical concentrations of KCl (100 mM). (b) Current–voltage curves (left) for AmRDL, AmRDL/LCCH3, AmRDL/GRD, and AmLCCH3/GRD, and corresponding deduced conductances (right). (c) Open dwell time distribution for AmGABA receptors. On the left, a representative example illustrating AmRDL/LCCH3 receptors open dwell times. On this graph, the black curve plots the distribution which follows bi‐exponential decay. On the right are shown histograms depicting tau fast and tau slow obtained from the fitting of the open dwell time for AmRDL (n = 8), AmRDL/LCCH3 (n = 5), AmRDL/GRD (n = 5), and AmLCCH3/GRD (n = 11). *P < .05, significantly different as indicated; Holm–Sidak method
4. DISCUSSION
This work is the first comparative analysis of biophysical and pharmacological properties of insect homomeric and heteromeric GABA receptors encoded by the Rdl, Grd, or Lcch3 genes.
While all tested heteromeric combinations produced a functional GABA receptor, only AmRDL can be expressed heterologously as a homomer, a situation that has already been described for other insects including Drosophila melanogaster (Harvey et al., 1994; Zhang et al., 1995). Sequence analysis indicates that only the AmRDL subunit presents a functional GABA binding pocket, fully conserved with respect to the stereotypical GABA binding pocket of DmRDL. AmGRD and AmLCCH3 polypeptides lack two key amino acids able to produce an ionic interaction with the zwiterrion GABA within the GABA binding pocket. AmLCCH3 possesses an uncharged asparagine (N89) on the complementary subunit in place of an arginine of DmRDL (R111), and AmGRD presents a glycine (G200) on the principal subunit while DmRDL possesses a glutamic acid at the equivalent place (E204). As mutations R111N and E204G have been shown to generate a non‐functional DmRDL (Ashby et al., 2012), it is not surprising that homomeric forms of AmGRD and AmLCCH3 also do not respond to GABA application.
Additionally, loop C, which plays the role of a gate for the GABA binding pocket (Comitani et al., 2016), is also quite variable between these three subunits. Besides the GABA binding pocket, an arginine at position 218 of DmRDL also seems essential for the first event of GABA binding. At the equivalent position, AmLCCH3 also bears a charged amino acid (K194), but AmRDL (N203) and AmGRD (N214) display uncharged residues. However, two positions upstream, a lysine present on both AmRDL (K201) and AmGRD (K212) sequences could play this role (Figure 1a arrows). Therefore, as a GABA binding pocket is formed by the interface of two successive GABA subunits, each GABA binding pocket has subtle structural disparities depending on the type of subunit involved. This should have some consequences on the affinity of agonists at any type of heteromeric GABA receptor. Actually, the calculation of EC50 from recording of native GABA‐induced currents led to a broad range of values from 12 to 285 μM in adult Kenyon cells and pupa antennal lobe neurons, respectively (Déglise, Grünewald, & Gauthier, 2002; Dupuis et al., 2010; Grünewald & Wersing, 2008). Interestingly, the potencies of GABA or muscimol at the heteromeric AmLCCH3/RDL or AmGRD/RDL receptors are similar to the homomeric RDL ones (Huang et al., 2019; Taylor‐Wells et al., 2017). Only the heteromeric AmGRD/LCCH3 receptors exhibited potencies for GABA and muscimol fourfold and eightfold higher respectively, than those of the homomeric RDL receptors. According to Flybase, Grd and Lcch3 seem to be transcribed at very low levels in Drosophila, and the strongest expression is observed during the larval and pupal stages (Jia et al., 2019; Knipple & Soderlund, 2010). Furthermore, LCCH3 is expressed in the nervous system of Drosophila larvae earlier than RDL (Aronstein, Auld, & Ffrench‐Constant, 1996). One possibility, now under study, is that the GRD/LCCH3 heteromer would be expressed at precise steps during development and would be activated by very low GABA concentrations.
Even if some neurons have been shown to co‐express RDL and LCCH3 (Dupuis et al., 2010; Okada, Awasaki, & Ito, 2009), expressions of heteromeric RDL/LCCH3 and RDL/GRD receptors have not been clearly demonstrated in vivo, mainly because of the lack of tools able to discriminate them from homomeric RDL receptors. Unfortunately, even if physical interaction of AmRDL with AmLCCH3 or AmGRD has been shown, our characterization in vitro does not deliver a usable biophysical or pharmacological profile of these heteromeric channels to conclusively discriminate them from homomeric RDL receptors. These results are in opposition with those obtained by Zhang et al. (1995) who identified RDL/LCCH3 heteromers as picrotoxin‐insensitive but bicuculline‐sensitive GABA receptors. Until now, recording on A. mellifera neurons have shown that GABA‐induced currents are bicuculline‐insensitive but inhibited by the usual antagonists with the following IC50 ranking order: fipronil (823 ± 19 nM) ≥ dieldrin (7.1 ± 0.3 μM) > picrotoxin (27.1 ± 1.8 μM) (Dupuis et al., 2010). Homomeric RDL receptors present the same profile which led to considering these receptors as the molecular determinants of inhibitory GABA receptors. However, from the present work which clearly demonstrates that this profile is also shared by heteromeric GABA receptors, the role of these heteromeric receptors in honeybee inhibitory GABAergic pathways should now be considered.
One common feature of Drosophila and honeybee is the formation of AmGRD/LCCH3 cationic receptors with a specific pharmacology when expressed in Xenopus oocytes. Notably, the agonist sensitivity is higher than that for the other combinations as discussed previously. However, dieldrin, which has been shown to be ineffective on DmGRD/LCCH3 (Gisselmann et al., 2004), is a potent antagonist of AmGRD/LCCH3. Sensitivity to dieldrin critically relies on an alanine in position 2′ of the TM2 region (Figure 1c), and its mutation into serine in the sequence of DmRDL is sufficient to confer resistance to dieldrin (Ffrench‐Constant et al., 1993; Ffrench‐Constant, Mortlock, Shaffer, MacIntyre, & Roush, 1991). However, a serine is present at this position in the AmGRD sequence implying that other amino acids of GRD or LCCH3 determine the sensitivity to dieldrin in honeybees. In the tobacco budworm Heliothis virescens, the reverse mutation of serine to alanine at position 2′ in the TM2 segment of RDL does not change significantly the insensitivity of these subunits to dieldrin, confirming that other molecular determinants could be involved in dieldrin sensitivity (Wolff & Wingate, 1998).
The cationic conductance, although clearly demonstrated here, is also puzzling. The charge and the spatial orientation of the amino acids lining the TM2 segment determine the selectivity for cations or anions. In A. mellifera, all RDL‐containing combinations are anionic channels selective for Cl− ions while the AmGRD/LCCH3 receptor is a non‐specific cationic channel with the permeability sequence K+ > Na+, but which is not permeable to Ca2+ ions. The D‐1′ of the AmGRD polypeptide seems to be sufficient to confer cationic selectivity to the AmGRD/LCCH3 receptor. Analysis of the pore lining residues shows that the ring of adjacent leucines 16′ and 17′, which is required for Ca2+ selectivity in nicotinic receptors, is absent in GRD and LCCH3 subunits. The fact that AmRDL/LCCH3 and AmRDL/GRD are Cl− ion conducting channels raises the question of the influence of different subunits in the determination of the ion selectivity of the heteromeric receptor. Here, the presence of the D‐1′ and the absence of P‐2′ (A‐2′ instead) on the GRD polypeptide are not sufficient to induce a selectivity switch. It thus appears that the presence of RDL seems to be decisive for the selectivity filter (Jensen et al., 2002) but the reasons for this predominance are unknown.
The RDL‐containing receptors, as anion channels, are probably involved in fast inhibitory transmission. We can speculate that GRD/LCCH3 receptors, as cation channels, form an insect excitatory conductance. Such excitatory GABA receptors have been described, indeed, in many physiological situations as, for example, during immature rodent neuron development and synaptogenesis (Ben‐Ari et al., 2007; Khalilov, Minlebaev, Mukhtarov, & Khazipov, 2015; Valeeva, Tressard, Mukhtarov, Baude, & Khazipov, 2016). Exp‐1 and LGC‐35, which are cation channels gated by GABA binding expressed in C. elegans neurons and muscles, have been implicated in physiological functions like defecation (Beg & Jorgensen, 2003; Branicky & Hekimi, 2005; Jobson et al., 2015). However, one major concern is the demonstration of the functional expression of GRD/LCCH3 receptors in native tissues. A biophysical characterization of GABA‐responding conductance on neurons remains to be done at different developmental stages when expression of these two subunits is suspected. Application of 100‐μM PCCP− or ivermectin could be useful tools to discriminate these from other GABA receptors. Ivermectin is indeed able to induce a full block of RDL‐containing GABA receptors even in the closed state, that is, at resting conditions without GABA application, while this block is only partial for GRD/LCCH3.
Our work is of primary importance when considering the mechanism of action of insecticides. At high doses, acute intoxications block all GABA receptors and produce death by down‐regulating the main inhibitory systems of neuronal excitability supported by RDL‐containing GABA receptors. The origin of the physiological perturbations elicited by chronic exposure to sublethal doses is more complicated to determine. Although pentameric RDL GABA receptors have so far been the only ones considered in assessing the toxicity of insecticides directed against GABA receptors, we find here that GRD/LCCH3 GABA receptors display comparable sensitivity to insecticides. In addition, the arthropod phylum presents a large molecular diversity of GABA receptor subunit genes. For example, Varroa destructor, the honeybee mite, has six genes encoding GABA receptor subunits (four RDL, one GRD, and one LCCH3) (Ménard et al., 2018), while Acyrthosiphon pisum, the pea aphid, has only two genes encoding RDL subunits (Del Villar & Jones, 2018). These molecular and pharmacological diversities must lead to species‐specific physiological perturbations induced by GABA receptor‐targeting insecticides that probably act with different mechanisms and at distinct time windows. In the context of general pollinator decline, further detailed knowledge of the pharmacology of insect GABA receptors is required to fully decipher insecticide toxicities and preserve global biodiversity.
AUTHOR CONTRIBUTIONS
C.H., N.M., and M.R. performed the experiments and analysed the two‐electrode voltage‐clamp data, C.H. and P.C. performed the experiments and analysed the patch‐clamp data, T.C. conducted and performed molecular biology, J.V.D., M.J.F., and J.G. performed biochemical analysis, C.C.S., C.M., and J.R. carried out technical assistance and gave valuable suggestions, J.B.T., M.V., and C.C. participated in data interpretation and revised the manuscript, P.C. and M.R. designed the study and interpreted data, and M.R. drafted the manuscript.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
DECLARATION OF TRANSPARENCY AND SCIENTIFIC RIGOUR
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research as stated in the BJP guidelines for Design & Analysis, Immunoblotting and Immunochemistry, and Animal Experimentation, and as recommended by funding agencies, publishers, and other organizations engaged with supporting research.
ACKNOWLEDGEMENTS
The authors thank Professor Erwin Sigel (University of Bern, Switzerland) and Professor Dirk Trauner (University of Munich, Germany) who kindly provided the tetraethyl ammonium salt of PCCP−. This manuscript benefited from discussions in the ANR “bee channels” (ANR‐13‐BSV7‐0010) plenary meeting in Avignon (2016) and in Montpellier (2017) with Dr JC Sandoz (Gif sur Yvette) and Prof M Chahine (Québec, Ca). The authors thank Professor Valérie Raymond for her critical reading of, and suggestions for, this manuscript. This work was supported by INSERM, CNRS, the Université de Montpellier, Agence Nationale de la Recherche Grant (SuperBeelive, ANR‐16‐IDEX‐0006) (Bee channels, ANR‐13‐BSV7‐0010) and the Fondation Lune de Miel (http://fondation.lunedemiel.fr/).
Henry C, Cens T, Charnet P, et al. Heterogeneous expression of GABA receptor‐like subunits LCCH3 and GRD reveals functional diversity of GABA receptors in the honeybee Apis mellifera . Br J Pharmacol. 2020;177:3924–3940. 10.1111/bph.15135
REFERENCES
- Alexander, S. P. H. , Mathie, A. , Peters, J. A. , Veale, E. L. , Striessnig, J. , Kelly, E. , … CGTP Collaborators . (2019). THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels. British Journal of Pharmacology, 176, S142–S228. 10.1111/bph.14749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliouane, Y. , El Hassani, A. K. , Gary, V. , Armengaud, C. , Lambin, M. , & Gauthier, M. (2009). Subchronic exposure of honeybees to sublethal doses of pesticides: Effects on behavior. Environmental Toxicology and Chemistry, 28(1), 113–122. 10.1897/08-110.1 [DOI] [PubMed] [Google Scholar]
- Aronstein, K. , Auld, V. , & Ffrench‐Constant, R. (1996). Distribution of two GABA receptor‐like subunits in the Drosophila CNS. Invertebrate Neuroscience: IN, 2(2), 115–120. 10.1007/BF02214114 [DOI] [PubMed] [Google Scholar]
- Ashby, J. A. , McGonigle, I. V. , Price, K. L. , Cohen, N. , Comitani, F. , Dougherty, D. A. , … Lummis, S. C. R. (2012). GABA binding to an insect GABA receptor: A molecular dynamics and mutagenesis study. Biophysical Journal, 103(10), 2071–2081. 10.1016/j.bpj.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish, M. E. (1983). A transient calcium‐dependent chloride current in the immature Xenopus oocyte. The Journal of Physiology, 342, 309–325. 10.1113/jphysiol.1983.sp014852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg, A. A. , & Jorgensen, E. M. (2003). EXP‐1 is an excitatory GABA‐gated cation channel. Nature Neuroscience, 6(11), 1145–1152. 10.1038/nn1136 [DOI] [PubMed] [Google Scholar]
- Ben‐Ari, Y. (2002). Excitatory actions of gaba during development: The nature of the nurture. Nature Reviews. Neuroscience, 3(9), 728–739. 10.1038/nrn920 [DOI] [PubMed] [Google Scholar]
- Ben‐Ari, Y. , Gaiarsa, J.‐L. , Tyzio, R. , & Khazipov, R. (2007). GABA: A pioneer transmitter that excites immature neurons and generates primitive oscillations. Physiological Reviews, 87(4), 1215–1284. 10.1152/physrev.00017.2006 [DOI] [PubMed] [Google Scholar]
- Branicky, R. , & Hekimi, S. (2005). Specification of muscle neurotransmitter sensitivity by a Paired‐like homeodomain protein in Caenorhabditis elegans . Development (Cambridge, England), 132(22), 4999–5009. 10.1242/dev.02069 [DOI] [PubMed] [Google Scholar]
- Buckingham, S. D. (2005). Insect GABA receptors: Splicing, editing, and targeting by antiparasitics and insecticides. Molecular Pharmacology, 68(4), 942–951. 10.1124/mol.105.015313 [DOI] [PubMed] [Google Scholar]
- Carta, V. , Pangerl, M. , Baur, R. , Puthenkalam, R. , Ernst, M. , Trauner, D. , & Sigel, E. (2014). A pentasymmetric open channel blocker for cys‐loop receptor channels. PLoS ONE, 9(9), e106688 10.1371/journal.pone.0106688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casida, J. E. , & Durkin, K. A. (2013). Neuroactive insecticides: Targets, selectivity, resistance, and secondary effects. Annual Review of Entomology, 58, 99–117. 10.1146/annurev-ento-120811-153645 [DOI] [PubMed] [Google Scholar]
- Casida, J. E. , & Durkin, K. A. (2015). Novel GABA receptor pesticide targets. Pesticide Biochemistry and Physiology, 121, 22–30. 10.1016/j.pestbp.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Cens, T. , Rousset, M. , Collet, C. , Raymond, V. , Démares, F. , Quintavalle, A. , … Charnet, P. (2013). Characterization of the first honeybee Ca2+ channel subunit reveals two novel species‐ and splicing‐specific modes of regulation of channel inactivation. Pflugers Archiv European Journal of Physiology, 465(7), 985–996. 10.1007/s00424-013-1223-2 [DOI] [PubMed] [Google Scholar]
- Charnet, P. , Labarca, C. , Leonard, R. J. , Vogelaar, N. J. , Czyzyk, L. , Gouin, A. , … Lester, H. A. (1990). An open‐channel blocker interacts with adjacent turns of α‐helices in the nicotinic acetylcholine receptor. Neuron, 4(1), 87–95. 10.1016/0896-6273(90)90445-L [DOI] [PubMed] [Google Scholar]
- Charreton, M. , Decourtye, A. , Henry, M. , Rodet, G. , Sandoz, J.‐C. , Charnet, P. , & Collet, C. (2015). A locomotor deficit induced by sublethal doses of pyrethroid and neonicotinoid insecticides in the honeybee Apis mellifera . PLoS ONE, 10(12), e0144879 10.1371/journal.pone.0144879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comitani, F. , Cohen, N. , Ashby, J. , Botten, D. , Lummis, S. C. R. , & Molteni, C. (2014). Insights into the binding of GABA to the insect RDL receptor from atomistic simulations: A comparison of models. Journal of Computer‐Aided Molecular Design, 28(1), 35–48. 10.1007/s10822-013-9704-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comitani, F. , Limongelli, V. , & Molteni, C. (2016). The free energy landscape of GABA binding to a pentameric ligand‐gated ion channel and its disruption by mutations. Journal of Chemical Theory and Computation, 12(7), 3398–3406. 10.1021/acs.jctc.6b00303 [DOI] [PubMed] [Google Scholar]
- Corringer, P. J. , Bertrand, S. , Galzi, J. L. , Devillers‐Thiéry, A. , Changeux, J. P. , & Bertrand, D. (1999). Mutational analysis of the charge selectivity filter of the α7 nicotinic acetylcholine receptor. Neuron, 22(4), 831–843. Retrieved from. http://www.ncbi.nlm.nih.gov/pubmed/10230802 [DOI] [PubMed] [Google Scholar]
- Cossart, R. (2014). Operational hub cells: A morpho‐physiologically diverse class of GABAergic neurons united by a common function. Current Opinion in Neurobiology, 26, 51–56. 10.1016/j.conb.2013.12.002 [DOI] [PubMed] [Google Scholar]
- Curtis, M. J. , Alexander, S. , Cirino, G. , Docherty, J. R. , George, C. H. , Giembycz, M. A. , … Ahluwalia, A. (2018, April 1). Experimental design and analysis and their reporting II: Updated and simplified guidance for authors and peer reviewers. British Journal of Pharmacology, 175, 987–993. 10.1111/bph.14153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decourtye, A. , Devillers, J. , Genecque, E. , Le Menach, K. , Budzinski, H. , Cluzeau, S. , & Pham‐Delegue, M. H. (2005). Comparative sublethal toxicity of nine pesticides on olfactory learning performances of the honeybee Apis mellifera . Archives of Environmental Contamination and Toxicology, 48(2), 242–250. 10.1007/s00244-003-0262-7 [DOI] [PubMed] [Google Scholar]
- Déglise, P. , Grünewald, B. , & Gauthier, M. (2002). The insecticide imidacloprid is a partial agonist of the nicotinic receptor of honeybee Kenyon cells. Neuroscience Letters, 321(1–2), 13–16. 10.1016/S0304-3940(01)02400-4 [DOI] [PubMed] [Google Scholar]
- Del Villar, S. G. , & Jones, A. K. (2018). Cloning and functional characterisation of the duplicated RDL subunits from the pea aphid, Acyrthosiphon pisum . International Journal of Molecular Sciences, 19(8), 2235 10.3390/ijms19082235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis, J. P. , Bazelot, M. , Barbara, G. S. , Paute, S. , Gauthier, M. , & Raymond‐Delpech, V. (2010). Homomeric RDL and heteromeric RDL/LCCH3 GABA receptors in the honeybee antennal lobes: Two candidates for inhibitory transmission in olfactory processing. Journal of Neurophysiology, 103(1), 458–468. 10.1152/jn.00798.2009 [DOI] [PubMed] [Google Scholar]
- Eguchi, Y. , Ihara, M. , Ochi, E. , Shibata, Y. , Matsuda, K. , Fushiki, S. , … Ozoe, Y. (2006). Functional characterization of Musca glutamate‐ and GABA‐gated chloride channels expressed independently and coexpressed in Xenopus oocytes. Insect Molecular Biology, 15, 773–783. 10.1111/j.1365-2583.2006.00680.x [DOI] [PubMed] [Google Scholar]
- El Hassani, A. K. , Dacher, M. , Gauthier, M. , & Armengaud, C. (2005). Effects of sublethal doses of fipronil on the behavior of the honeybee (Apis mellifera). Pharmacology Biochemistry and Behavior, 82(1), 30–39. 10.1016/j.pbb.2005.07.008 [DOI] [PubMed] [Google Scholar]
- Enell, L. , Hamasaka, Y. , Kolodziejczyk, A. , & Nässel, D. R. (2007). γ‐Aminobutyric acid (GABA) signaling components in Drosophila: Immunocytochemical localization of GABAB receptors in relation to the GABAA receptor subunit RDL and a vesicular GABA transporter. The Journal of Comparative Neurology, 505(1), 18–31. 10.1002/cne.21472 [DOI] [PubMed] [Google Scholar]
- Es‐Salah, Z. (2008). Analyse électrophysiologique, pharmacologique et moléculaire de facteurs modulant les effets d'un insecticide, le fipronil, sur des recepteurs gabaergiques d'insectes (Université d'Angers). Retrieved from https://tel.archives-ouvertes.fr/tel-00439681/
- Ffrench‐Constant, R. H. , Mortlock, D. P. , Shaffer, C. D. , MacIntyre, R. J. , & Roush, R. T. (1991). Molecular cloning and transformation of cyclodiene resistance in Drosophila: An invertebrate γ‐aminobutyric acid subtype A receptor locus. Proceedings of the National Academy of Sciences of the United States of America, 88(16), 7209–7213. 10.1073/pnas.89.16.7849b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ffrench‐Constant, R. H. , & Rocheleau, T. A. (1993). Drosophila γ‐aminobutyric acid receptor gene Rdl shows extensive alternative splicing. Journal of Neurochemistry, 60(6), 2323–2326. 10.1111/j.1471-4159.1993.tb03523.x [DOI] [PubMed] [Google Scholar]
- Ffrench‐Constant, R. H. , Rocheleau, T. A. , Steichen, C. J. , & Chalmers, A. E. (1993). A point mutation in a Drosophila GABA receptor confers insecticide resistance. Nature, 363(3 JUNE 1993),, 449–451. [DOI] [PubMed] [Google Scholar]
- Ffrench‐Constant, R. H. , Williamson, M. S. , Davies, T. G. E. , & Bass, C. (2016). Ion channels as insecticide targets. Journal of Neurogenetics, 7063(November), 1–15. 10.1080/01677063.2016.1229781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galzi, J.‐L. , Devillers‐Thiery, A. , Hussy, N. , Bertrand, S. , Changeux, J.‐P. , & Bertrand, D. (1992). Mutations in the channel domain of a neuronal nicotinic receptor convert ion selectivity from cationic to anionic. Nature, 359(6395), 500–505. 10.1038/359500a0 [DOI] [PubMed] [Google Scholar]
- Ganeshina, O. , & Menzel, R. (2001). GABA‐immunoreactive neurons in the mushroom bodies of the honeybee: An electron microscopic study. The Journal of Comparative Neurology, 437(3), 335–349. Retrieved from. http://www.ncbi.nlm.nih.gov/pubmed/11494260 [DOI] [PubMed] [Google Scholar]
- Gisselmann, G. , Plonka, J. , Pusch, H. , & Hatt, H. (2004). Drosophila melanogaster GRD and LCCH3 subunits form heteromultimeric GABA‐gated cation channels. British Journal of Pharmacology, 142(3), 409–413. 10.1038/sj.bjp.0705818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou, Z.‐H. , Wang, X. , & Wang, W. (2012). Evolution of neurotransmitter gamma‐aminobutyric acid, glutamate and their receptors. Dong Wu Xue Yan Jiu, 33(E5–6), E75–E81. 10.3724/SP.J.1141.2012.E05-06E75 [DOI] [PubMed] [Google Scholar]
- Grünewald, B. , & Wersing, A. (2008). An ionotropic GABA receptor in cultured mushroom body Kenyon cells of the honeybee and its modulation by intracellular calcium. Journal of Comparative Physiology a: Neuroethology, Sensory, Neural, and Behavioral Physiology, 194(4), 329–340. 10.1007/s00359-007-0308-9 [DOI] [PubMed] [Google Scholar]
- Grutter, T. , de Carvalho, L. P. , Dufresne, V. , Taly, A. , Edelstein, S. J. , & Changeux, J.‐P. (2005). Molecular tuning of fast gating in pentameric ligand‐gated ion channels. Proceedings of the National Academy of Sciences of the United States of America, 102(50), 18207–18212. 10.1073/pnas.0509024102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, S. D. , Sharman, J. L. , Faccenda, E. , Southan, C. , Pawson, A. J. , Ireland, S. , … NC‐IUPHAR . (2018). The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: Updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Research, 46, D1091–D1106. 10.1093/nar/gkx1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, R. J. , Schmitt, B. , Hermans‐Borgmeyer, I. , Gundelfinger, E. D. , Betz, H. , & Darlison, M. G. (1994). Sequence of a Drosophila ligand‐gated ion‐channel polypeptide with an unusual amino‐terminal extracellular domain. Journal of Neurochemistry, 62, 2480–2483. 10.1046/j.1471-4159.1994.62062480.x [DOI] [PubMed] [Google Scholar]
- Henderson, J. E. , Knipple, D. C. , & Soderlund, D. M. (1994). PCR‐based homology probing reveals a family of GABA receptor‐like genes in Drosophila melanogaster . Insect Biochemistry and Molecular Biology, 24(4), 363–371. 10.1016/0965-1748(94)90029-9 [DOI] [PubMed] [Google Scholar]
- Holder, P. J. , Jones, A. , Tyler, C. R. , & Cresswell, J. E. (2018). Fipronil pesticide as a suspect in historical mass mortalities of honey bees. Proceedings of the National Academy of Sciences, 115(51), 13033–13038. 10.1073/pnas.1804934115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosie, A. M. , Buckingham, S. D. , Presnail, J. K. , & Sattelle, D. B. (2001). Alternative splicing of a Drosophila GABA receptor subunit gene identifies determinants of agonist potency. Neuroscience, 102(3), 709–714. 10.1016/S0306-4522(00)00483-8 [DOI] [PubMed] [Google Scholar]
- Hosie, A. M. , Baylis, H. A. , Buckingham, S. D. , & Sattelle, D. B. (1995). Actions of the insecticide fipronil, on dieldrin‐sensitive and‐ resistant GABA receptors of Drosophila melanogaster . British Journal of Pharmacology, 115(6), 909–912. 10.1111/j.1476-5381.1995.tb15896.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosie, A. M. , & Sattelle, D. B. (1996). Agonist pharmacology of two Drosophila GABA receptor splice variants. British Journal of Pharmacology, 119, 1577–1585. 10.1111/j.1476-5381.1996.tb16075.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosie, A. M. , Aronstein, K. , Sattelle, D. B. , & Ffrench‐Constant, R. H. (1997). Molecular biology of insect neuronal GABA receptors. Trends in Neurosciences, 20(12), 578–583. 10.1016/S0166-2236(97)01127-2 [DOI] [PubMed] [Google Scholar]
- Houston, C. M. , Lee, H. H. C. , Hosie, A. M. , Moss, S. J. , & Smart, T. G. (2007). Identification of the sites for CaMK‐II‐dependent phosphorylation of GABAA receptors. Journal of Biological Chemistry, 282(24), 17855–17865. 10.1074/jbc.M611533200 [DOI] [PubMed] [Google Scholar]
- Huang, Q.‐T. , Sheng, C.‐W. , Jiang, J. , Jia, Z.‐Q. , Han, Z.‐J. , Zhao, C.‐Q. , & Liu, G.‐Y. (2019). Functional integrity of honeybee (Apis mellifera L.) resistant to dieldrin γ‐aminobutyric acid receptor channels conjugated with three fluorescent proteins. Insect Molecular Biology, 28(3), 313–320. 10.1111/imb.12552 [DOI] [PubMed] [Google Scholar]
- Jensen, M. L. , Timmermann, D. B. , Johansen, T. H. , Schousboe, A. , Varming, T. , & Ahring, P. K. (2002). The β subunit determines the ion selectivity of the GABAA receptor. Journal of Biological Chemistry, 277(44), 41438–41447. 10.1074/jbc.M205645200 [DOI] [PubMed] [Google Scholar]
- Jia, Z.‐Q. , Sheng, C.‐W. , Tang, T. , Liu, D. , Leviticus, K. , Zhao, C.‐Q. , & Chang, X.‐L. (2019). Identification of the ionotropic GABA receptor‐like subunits from the striped stem borer, Chilo suppressalis Walker (Lepidoptera: Pyralidae). Pesticide Biochemistry and Physiology, 155, 36–44. 10.1016/j.pestbp.2019.01.001 [DOI] [PubMed] [Google Scholar]
- Jinguji, H. , Ohtsu, K. , Ueda, T. , & Goka, K. (2018). Effects of short‐term, sublethal fipronil and its metabolite on dragonfly feeding activity. PLoS ONE, 13(7), e0200299 10.1371/journal.pone.0200299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobson, M. A. , Valdez, C. M. , Gardner, J. , Garcia, L. R. , Jorgensen, E. M. , & Beg, A. A. (2015). Spillover transmission is mediated by the excitatory GABA receptor LGC‐35 in C. elegans . The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 35(6), 2803–2816. 10.1523/JNEUROSCI.4557-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A. K. , Buckingham, S. D. , Papadaki, M. , Yokota, M. , Sattelle, B. M. , Matsuda, K. , & Sattelle, D. B. (2009). Splice‐variant‐ and stage‐specific RNA editing of the Drosophila GABA receptor modulates agonist potency. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 29(13), 4287–4292. 10.1523/JNEUROSCI.5251-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash, T. L. , Jenkins, A. , Kelley, J. C. , Trudell, J. R. , & Harrison, N. L. (2003). Coupling of agonist binding to channel gating in the GABAA receptor. Nature, 421(6920), 272–275. 10.1038/nature01280 [DOI] [PubMed] [Google Scholar]
- Keramidas, A. , & Harrison, N. L. (2010). The activation mechanism of α1β2γ2S and α3β3γ2S GABAA receptors. The Journal of General Physiology, 135(1), 59–75. 10.1085/jgp.200910317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keramidas, A. , Moorhouse, A. J. , Pierce, K. D. , Schofield, P. R. , & Barry, P. H. (2002). Cation‐selective mutations in the M2 domain of the inhibitory glycine receptor channel reveal determinants of ion‐charge selectivity. The Journal of General Physiology, 119(May 2002), 393–410. 10.1085/jgp.20028552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keramidas, A. , Moorhouse, A. J. , Schofield, P. R. , & Barry, P. H. (2004). Ligand‐gated ion channels: Mechanisms underlying ion selectivity. Progress in Biophysics and Molecular Biology, 86(2), 161–204. 10.1016/j.pbiomolbio.2003.09.002 [DOI] [PubMed] [Google Scholar]
- Khalilov, I. , Minlebaev, M. , Mukhtarov, M. , & Khazipov, R. (2015). Dynamic changes from depolarizing to hyperpolarizing GABAergic actions during giant depolarizing potentials in the neonatal rat hippocampus. Journal of Neuroscience, 35(37), 12635–12642. 10.1523/JNEUROSCI.1922-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny, C. , Browne, W. , Cuthill, I. C. , Emerson, M. , & Altman, D. G. (2010). Animal research: Reporting in vivo experiments: The ARRIVE guidelines. British Journal of Pharmacology, 160, 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipple, D. C. , & Soderlund, D. M. (2010). The ligand‐gated chloride channel gene family of Drosophila melanogaster . Pesticide Biochemistry and Physiology, 97(2), 140–148. 10.1016/j.pestbp.2009.09.002 [DOI] [Google Scholar]
- Knipple, D. , Henderson, J. , & Soderlund, D. (1995). Structural and functional characterization of insect genes encoding ligand‐gated chloride‐channel subunits In Molecular action of insecticides on ion channels, ACS Symposium Series (Vol. 591, pp. 205–215). Washington DC: American Chemical Society; https://pubs.acs.org/doi/abs/10.1021/bk-1995-0591.ch013 [Google Scholar]
- Lee, J. , Song, H. O. , Jee, C. , Vanoaica, L. , & Ahnn, J. (2005). Calcineurin regulates enteric muscle contraction through EXP‐1, excitatory GABA‐gated channel, in C. elegans . Journal of Molecular Biology, 352(2), 313–318. 10.1016/j.jmb.2005.07.032 [DOI] [PubMed] [Google Scholar]
- Lema, G. M. C. , & Auerbach, A. (2006). Modes and models of GABAA receptor gating. Journal of Physiology, 572(1), 183–200. 10.1016/j.yhbeh.2006.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Krause, W. C. , & Davis, R. L. (2007). GABAA receptor RDL inhibits Drosophila olfactory associative learning. Neuron, 56(6), 1090–1102. 10.1016/j.neuron.2007.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludmerer, S. W. , Warren, V. A. , Williams, B. S. , Zheng, Y. , Hunt, D. C. , Ayer, M. B. , … McHardy, M. S. (2002). Ivermectin and nodulisporic acid receptors in Drosophila melanogaster contain both γ‐aminobutyric acid‐gated Rdl and glutamate‐gated GluClα chloride channel subunits. Biochemistry, 41, 6548–6560. 10.1021/bi015920o [DOI] [PubMed] [Google Scholar]
- Lummis, S. C. R. (2009). Locating GABA in GABA receptor binding sites. Biochemical Society Transactions, 37(Pt 6), 1343–1346. 10.1042/BST0371343 [DOI] [PubMed] [Google Scholar]
- Ménard, C. , Folacci, M. , Brunello, L. , Charreton, M. , Collet, C. , Mary, R. , … Cens, T. (2018). Multiple combinations of RDL subunits diversify the repertoire of GABA receptors in the honey bee parasite Varroa destructor . Journal of Biological Chemistry, 293(49), 19012–19024. 10.1074/jbc.RA118.005365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard, C. , Horvitz, H. R. , & Cannon, S. (2005). Chimeric mutations in the M2 segment of the 5‐hydroxytryptamine‐gated chloride channel MOD‐1 define a minimal determinant of anion/cation permeability. Journal of Biological Chemistry, 280(30), 27502–27507. 10.1074/jbc.M501624200 [DOI] [PubMed] [Google Scholar]
- Okada, R. , Awasaki, T. , & Ito, K. (2009). Gamma‐aminobutyric acid (GABA)‐mediated neural connections in the Drosophila antennal lobe. Journal of Comparative Neurology, 514(1), 74–91. 10.1002/cne.21971 [DOI] [PubMed] [Google Scholar]
- Olsen, R. W. , & Sieghart, W. (2009). GABAA receptors: Subtypes provide diversity of function and pharmacology. Neuropharmacology, 56(1), 141–148. 10.1038/jid.2014.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozoe, Y. (2013). γ‐Aminobutyrate‐ and glutamate‐gated chloride channels as targets of insecticides. Advances in Insect Physiology, 44(1), 211–286. 10.1016/B978-0-12-394389-7.00004-1 [DOI] [Google Scholar]
- Pisa, L. W. , Amaral‐Rogers, V. , Belzunces, L. P. , Bonmatin, J. M. , Downs, C. A. , Goulson, D. , … Wiemers, M. (2015). Effects of neonicotinoids and fipronil on non‐target invertebrates. Environmental Science and Pollution Research, 22(1), 68–102. 10.1007/s11356-014-3471-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauh, J. J. , Lummis, S. C. R. , & Sattelle, D. B. (1990). Pharmacological and biochemical properties of insect GABA receptors. Trends in Pharmacological Sciences, 11(8), 325–329. 10.1016/0165-6147(90)90236-2 [DOI] [PubMed] [Google Scholar]
- Sattelle, D. B. , Pinnock, R. D. , Wafford, K. A. , & David, J. A. (1988). GABA receptors on the cell‐body membrane of an identified insect motor neuron. Proceedings of the Royal Society of London. Series B, Biological Sciences, 232(1269), 443–456. Retrieved from. http://www.ncbi.nlm.nih.gov/pubmed/2451252 [DOI] [PubMed] [Google Scholar]
- Schäfer, S. , & Bicker, G. (1986). Distribution of GABA‐like immunoreactivity in the brain of the honeybee. Journal of Comparative Neurology, 246(3), 287–300. 10.1002/cne.902460302 [DOI] [PubMed] [Google Scholar]
- Sieghart, W. , Ramerstorfer, J. , Sarto‐Jackson, I. , Varagic, Z. , & Ernst, M. (2012). A novel GABAA receptor pharmacology: Drugs interacting with the a+b− interface. British Journal of Pharmacology, 166(2), 476–485. 10.1111/j.1476-5381.2011.01779.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigel, E. , & Steinmann, M. E. (2012). Structure, function, and modulation of GABAA receptors. The Journal of Biological Chemistry, 287(48), 40224–40231. 10.1074/jbc.R112.386664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor‐Wells, J. , Hawkins, J. , Colombo, C. , Bermudez, I. , & Jones, A. K. (2017). Cloning and functional expression of intracellular loop variants of the honey bee (Apis mellifera) RDL GABA receptor. Neurotoxicology, 60(5), 207–213. 10.1016/j.neuro.2016.06.007 [DOI] [PubMed] [Google Scholar]
- Thompson, A. J. , & Lummis, S. C. R. (2003). A single ring of charged amino acids at one end of the pore can control ion selectivity in the 5‐HT3 receptor. British Journal of Pharmacology, 140(2), 359–365. 10.1038/sj.bjp.0705424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valeeva, G. , Tressard, T. , Mukhtarov, M. , Baude, A. , & Khazipov, R. (2016). An optogenetic approach for investigation of excitatory and inhibitory network GABA actions in mice expressing channelrhodopsin‐2 in GABAergic neurons. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 36(22), 5961–5973. 10.1523/JNEUROSCI.3482-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte, I. , Kreienkamp, H. J. , Gewecke, M. , & Roeder, T. (2002). Putative histamine‐gated chloride channel subunits of the insect visual system and thoracic ganglion. Journal of Neurochemistry, 83(3), 504–514. 10.1046/j.1471-4159.2002.01076.x [DOI] [PubMed] [Google Scholar]
- Wolff, M. A. , & Wingate, V. P. (1998). Characterization and comparative pharmacological studies of a functional γ‐aminobutyric acid (GABA) receptor cloned from the tobacco budworm, Heliothis virescens (Noctuidae: Lepidoptera). Invertebrate Neuroscience: IN, 3(4), 305–315. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10212398 [DOI] [PubMed] [Google Scholar]
- Wotring, V. E. , Miller, T. S. , & Weiss, D. S. (2003). Mutations at the GABA receptor selectivity filter: A possible role for effective charges. The Journal of Physiology, 548(Pt 2), 527–540. 10.1113/jphysiol.2002.032045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. G. , Lee, H. J. , Rocheleau, T. , Ffrench‐Constant, R. H. , & Jackson, M. B. (1995). Subunit composition determines picrotoxin and bicuculline sensitivity of Drosophila γ‐aminobutyric acid receptors. Molecular Pharmacology, 48(5), 835–840. [PubMed] [Google Scholar]


