Abstract
Our knowledge in the field of cardiac muscle and associated cardiomyopathies has been evolving incrementally over the past 60 years and all was possible due to the parallel progress in techniques and methods allowing to take a fresh glimpse at an old problem. Here, we describe an exciting tool used to examine the various states of the human cardiac myosin at the single molecule level. By imaging single Alexa647-ATP binding to permeabilised cardiomyocytes using total internal reflection fluorescence (TIRF) microscopy, we are able to acquire large populations of events in a short timeframe (~ 5000 sites in ~ 10 min) and measure each binding event with high spatio-temporal resolution. The applied algorithm decomposes the point pattern of single molecule binding events into individually distinct binding sites that enables us to recover kinetic parameters, such as bound or free time per site. This single molecule binding approach is a useful tool used to examine muscle contractility. Of particular importance is its application to probing the dynamic lifetimes and proportion of myosins in the super-relaxed state in human cardiomyopathies.
Keywords: SMLM, TIRF microscopy, Human heart cardiomyocytes, ATP-binding kinetics, ATP analogues, Single molecule imaging of ATP
Introduction
Our understanding of how striated muscles works, particularly cardiac muscle, has undergone a series of incremental developments. Initially, we learned that when the muscle is in the relaxed state, the myosin forms cross-bridges with actin and hydrolyse ATP, but its energy cannot be converted into contraction because the relaxing proteins of the thin filament, troponins TnI, TnC and TnT, and tropomyosin (Ebashi 1963) sterically block the myosin-binding sites on the actin filaments leaving the cross-bridges in a disordered state. In contraction, Ca2+ released from the sarcoplasmic reticulum removes this steric inhibition allowing the activated cross-bridges to bind. Myosin heads projecting from the thick filament stabilise the thin filament through the formation of cross-bridges which generates contraction. The force and velocity of the contraction largely depends on the number of cross-bridges formed that is dictated by the overlap of thick and thin filaments (McGrath and dos Remedios 1974). At low ATP levels (≥ 500 μm) and in the absence of Ca2+, myosin heads form a stable “rigor state” bond with actin and develop a small isometric rigor tension that can be relieved by allowing the fibres to shorten by 2.5–6% (White 1970). The above statements are completely consistent with the sliding filament hypothesis proposed simultaneously by Huxley and Niedergerke (1954) at Cambridge, and by Huxley and Hanson (1954) at King’s College in London. Together they showed that the thick and thin myofilaments do not change length but instead undergo a sliding action that shortens the sarcomere as a whole and somehow produces contractile force. A more detailed history of these developments can be found in Perry (2008).
Is there more than one state of cross-bridges in the state of rigor?
In 1981, Roger Cooke addressed this question by rigidly attaching paramagnetic probes to myosin cross-bridges. The data showed that the orientation of the probes was highly uniform, and it was not altered by applying forces up to 0.2 N mm−2. But what if a small concentration of ATP were introduced, would it weaken the rigor cross-bridges? Later, in an elegant low-angle X-ray diffraction paper, Mike Ferenczi and his colleagues (Tsaturyan et al. 1999) examined frog semitendinosus single muscle fibres initially in the rigor state. They devised a way releasing free ATP next to the cross-bridges and examined the resulting structural changes. To do this, they applied a photoactivatable (“caged”) ATP (P3[1(3,5 dimethoxyphenyl) 2 phenyl 2 oxo]ethyl ester of ATP) that could be activated at a precise location between the myofilaments with a laser. This faster rate of ATP release compared to than un-caged ATP combined with synchrotron X-rays measurements enabled them to obtain a time resolution of 1 ms using only1–2 permeabilized muscle fibres. Contractile force and fibre stiffness were also monitored while they recorded changes in the equatorial 1,1 and 1,0 reflections that reported the state of cross-bridge attachment. The prospect of ADP rebinding to the cross-bridges was prevented using apyrase, and sarcomere damage during rigor formation was minimised using the selective myosin ATPase inhibitor 2,3-butanedione monoxime. Under these technically challenging conditions, they followed the effect of ATP release within their fibres in the absence of free Ca2+ (pCa 8). Fibres were held under slight stretch to generate a small rigor force which decreased when either 1.1 mM (fully relaxed) or 0.3 mM (partially relaxed) ATP was added. At 0.3 mM ATP, decay in rigor force was best described by a double exponential with rate constants of 169 ± 98 and 9.5 ± 17 s−1 (mean ± SEM of the decay estimate obtained from the Marquardt Levenberg algorithm), and the decay in stiffness was best described by a single exponential process of 30 ± 0.8 s−1. It remains to be seen how these data should be interpreted now we know about the formation of DRX and SRX (Stewart et al. 2010). Both the X-ray diffraction and the EPR experiments deliver mean measurements of cross-bridge angle and their distributions with respect to the fibre direction. What if we could develop a technique that measured the time ATP was bound to single cross-bridges?
Here, we describe the application of TIRF microscopy to image the binding of 5 nM concentrations of Alexa647ATP molecules when they are immobilised at single myosin heads in the state of rigor where unlabelled ATP is absent. We describe the quantification of binding kinetics on a per site (local) level, and provide details of the sample preparation, TIRF imaging, and image processing that allow the extraction of the molecular binding events, and finally using the event spatio-temporal maps to extract the local clusters of events and from them the binding statistics. We believe this approach will help uncover more mysteries in the structure and function of sarcomeric myosin.
Total internal reflection fluorescence microscopy of ATP-Alexa-647 binding to cardiomyocytes
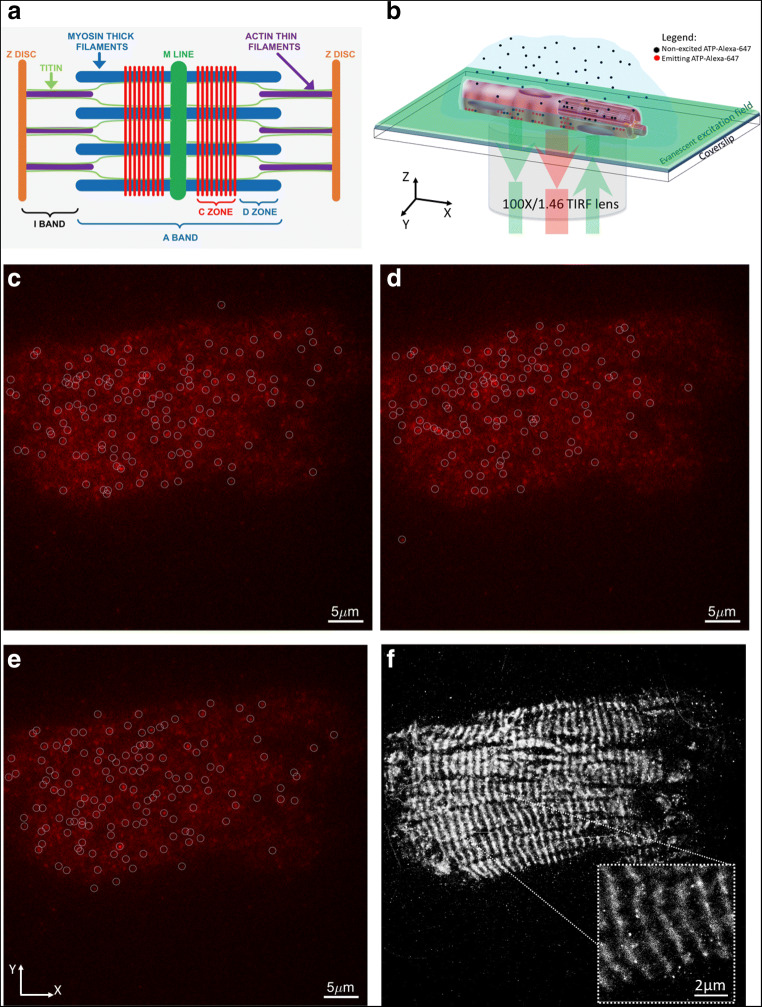
Samples of isolated human cardiomyocytes were imaged (Fig. 1b) on Elyra (Zeiss) TIRF microscope using the × 100 Alpha-Plan APO oil immersion objective lens (NA 1.46). ATP-Alexa-647 were excited using 638 nm laser and emitted light was collected through long pass 640 nm filter. A 512 × 512 pixel imaging area was collected over 10,000–20,000 frames with a 30-ms exposure time on electron-multiplying charge-coupled device (EMCCD) camera (iXon DU-897D by Andor). Opto-var lens of × 1.6 was inserted before the camera, making the pixel size 0.1 μm. The samples were imaged 30 min post mounting on the stage, to ensure that fibrils are stabilised, by sedimentation. Definite focus was used to ensure that the focal plane is stable during ~ 10 min of acquisition. Prior to starting the time series acquisition, laser power was increased to ~ 50% to pre-bleach the bound ATP-Alexa-647 and then reduced to ~ 5% for the imaging. This step ensured that the sample reaches a steady state number of bound visible ATP-Alexa-647. The single ATP detection, per frame, was done in Zen (Zeiss) prior to further analysis of kinetics performed in Matlab (Natick, MA). Each intensity profile of ATP-Alexa-647 (circles in Fig. 1 c–e) in each frame was fitted with a 2D Gaussian function and properties such as molecular position, amplitude (number of photons) and width of the intensity profile, and the data was stored in a table. Molecular observation with signal-to-noise ratio of 6 and above were kept, while low signal (below S/N < 6) were rejected in further analysis. Data was corrected for the linear lateral (xy-plane) drift using Zen model-based de-drifting function. Otherwise, for significant drifts that cannot be corrected for, data sets were not used for further processing. Pre-processed data table was exported into a txt file for further analysis in Matlab. This acquisition approach produced data comparable to those obtained by single molecule localisation microscopy (SMLM) approaches (Fig. 1).
Fig. 1.
Example of ATP-Alexa-647 binding to fibril time series acquired using TIRF microscope. a A diagrammatic illustration of the cardiac sarcomere showing the Z discs (orange), the M line (dark green), the thin filaments (purple), myosin filaments (blue) and titin (light green). The nine red bars represent the location of cMyBP-C in the C-zone where it makes contact with the LMM portion of myosin and titin. The A band is central to the sarcomere but the I band, defined by the region containing only thin filament proteins, spans both sides of the Z disc and extends into the next sarcomere. b Schematic of TIRF setup with ATP-Alexa-647 binding and unbinding to the cardiomyocytes, while evanescent field excites only those within ~ 100–200 nm from the surface of the coverslip. Panels c, d, and e show 3 different frames from a 10 000 frames time series. At each frame, molecules (white circles) were detected above threshold signal-to-noise ratio of 6. f A single molecule localisation map obtained after combining centroids of all molecules detected in 10,000 frames
Extracting ATP-Alexa-647-binding kinetics parameters from point pattern data
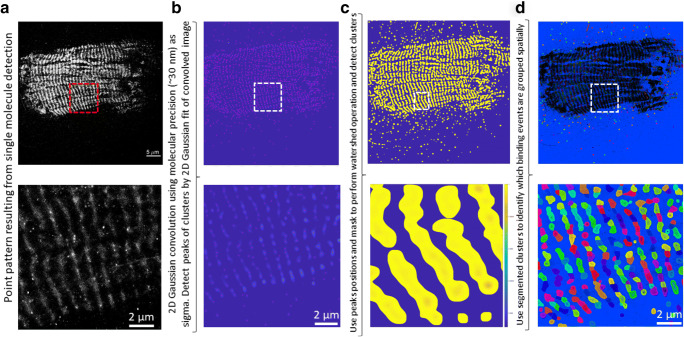
The table containing single molecule localisations was loaded into Matlab (Natick, MA) for further analysis. Single molecule localisations as such are not very useful other than to show super-resolved localisation image of binding sites (Fig. 1f). Nevertheless, grouping spatially the point pattern data into an organised clustered map would allow us to extract the temporal properties, like binding kinetics, on a per cluster basis. Given the spatial complexity of point pattern and hierarchical organisation in cluster sizes generated in this work, it was not trivial to apply existing clustering approaches such as density-based spatial clustering of application with noise (DBSCAN). Instead, we proceeded to convert the point patterns (Fig. 2a) into an image where high intensity pixels reflect higher molecular density (Fig. 2b). We used that image, which is equivalent of super-resolution reconstruction in SMLM data, to detect local peaks which are effectively clusters of SMLM data and break them into separate regions (Fig. 2d and zoom in). In more details, the positions (x,y) of detections from all of the 10,000 frames were grouped into a 2D histogram, using histcounts2 Matlab function, and the resulting image was smoothed using a Laplacian of Gaussian (LoG) filter with 30 nm sigma. The choice of this sigma comes from the average molecular localisation precision, which is defined as microscope’s point spread function sigma divided by square root of the number of photons detected by molecules. Precision is an important number as it specifies the confidence interval of each molecular localisation. The peaks (red dots in Fig. 2b) in convolved 2D histogram were detected using the pkfnd (Dufresne and Blair 2005) function. Then, the inverse image of the 2D histogram was computed and thresholded, while the pixels at the locations of peaks were set to the value of -Infinity. This generated an image (Fig. 2c) where peaks were converted to valleys and actual peak centre pixels became the absolute minima (-Infinity). The watershed function from Matlab was applied to this image which resulted in breaking the image into segregated clusters as shown by variable coloured areas in Fig. 2d. There are some localisations outside of clearly defined bright bands of fibrils, which did not get grouped into a defined cluster (black dots outside of defined coloured areas of Figs. 2d and 3a). These are likely noise and unspecific binding of ATP-Alexa-647 to the fibril structure and are not included in the kinetics analysis as they do not belong to a defined cluster.
Fig. 2.
Workflow for converting the SMLM into a cluster map, defining binding sites for ATP-Alexa-647. Bottom row show the insets defined by dashed lines rectangles area of the images in top row. a SMLM point pattern obtained by TIRF imaging of ATP-Alexa-647 binding. b 2D histogram of molecular localisations, convolved with LoG filter with 30 nm sigma. Peaks are shown as higher amplitudes values (bright features). Red dots represent the locations of peaks detected and representing local clusters central point. c Inverted and mean thresholded (areas outside bright bands of fibrils set to zero) image from b ready to be used in watershed algorithm for clusters segmentation. d Segmented clusters are coloured with different colours and superimposed as black dots is SMLM point pattern. Scale bar in a is equal to 5 μm
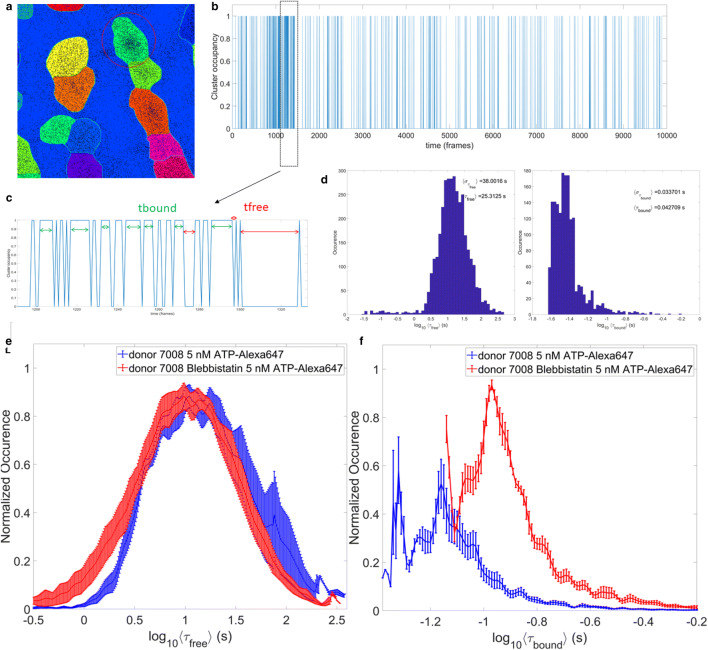
Fig. 3.
Workflow for extracting a cluster’s ATP-Alexa-647 binding kinetics. a Segmented cluster example zooms in from Fig. 2d, showing each cluster colour coded differently. The black dots are showing the single molecule localisations. b Red circle shows the cluster for which the example of molecular occupancy is shown. When cluster occupancy is equal to 1, it reflects that a molecule is bound to this cluster and 0 signifies that cluster was not occupied for these frames. c The zoom in to the frames 1200 to 1300 of occupancy diagram shown in b). The episodes for which cluster was occupied (green arrows) are defined as bound time, tbound, and frames for which cluster was not occupied (red arrows) are grouped in free time, tfree. d Examples of histograms of average bound and free times extracted for one field of view (FOV) imaged. For each cluster, we obtain one value of average bound and free time. Since there are several thousand of clusters per FOV, histograms like the ones shown are obtained per FOV. e Comparison of average free and f bound time between a donor heart fibril before (blue) and after (red) blebbistatin treatment. Error bars represent standard error of the mean value for 7–10 FOVs recorded
For each cluster identified in last step (Fig. 3a), single molecule localisations were extracted from the table and knowing at what frame, out of total 10,000, given detection occurred, we can define the time when cluster was occupied or not (Fig. 3b). With this information, we can define exactly the frame sequence length for which cluster was bound (τbound) or unoccupied (τfree) as shown in Fig. 3c. Practically, identifying the boundary of a cluster from a binary mask obtained in Fig. 2d was achieved using Matlab function mask2poly. This cluster boundary was then used to define which points inside point pattern belong to it, using function inpolygon. The custom-made function was used to identify frame length of occupied (bound) or free (unbound) segments for each cluster. Sequences of free or bound events that lasted only one frame were filtered out and not considered for further comparisons, given that they could be result of unspecific binding events or Alexa dye spontaneous blinking. Once this decomposition of cluster’s occupancy is done, we can plot histograms of bound and free time, τbound and τfree, respectively. The bound time, τbound, measures the time length for which Alexa-647-ATP was bound to a site, while the free time, τfree, records the time length of episodes for which the site vs. unoccupied or unbound.
What does this all mean?
The approach presented allows for the imaging of functional myosin states within regions of interests of permeabilised human cardiomyocytes. Using TIRF microscopy, we captured fluorescent ATP binding to myosin at a single molecule level. Single molecule point pattern image processing provided a means of separating the binding site to access the kinetics of ATP-Alexa-647 binding for each site individually (Figs. 2d and 3a). For a typical image data series of 512 × 512 × 10,000 frames as shown in example in Fig. 1, we can extract around 4000–5000 binding sites. This allows us to map the spatial heterogeneity of the binding kinetics and furthermore use it to compare kinetics parameters distribution like average free and bound times per site (Fig. 3d–f) between donor control data and various cardiomyopathies. When comparing a healthy donor data pre- and post 40 mM blebbistatin, we find that the recovered distributions (Fig. 3e and f) of bound and unbound (free) times follow the expected trends. Indeed, post-blebbistatin bound time distribution shifted to higher values, with ˂ τbound ˃ ± στbound = 0.13 ± 0.02 s, compared to pre-blebbistatin bound time distribution which were smaller, ˂ τbound ˃ ± στbound = 0.083 ± 0.014 s. Similarly, the average free (unoccupied) state per site shifted slightly to lower values post-blebbistatin, with ˂ τfree ˃ ± στfree = 19.0 ± 3.5 s, compared to the pre-blebbistatin, with ˂ τfree ˃ ± στfree = 25.2 ± 13.4 s. These lifetimes are consistent with active cross-bridges in cardiac muscle where turnover is approximately ~ 25 ATPs/sec. Due to the very low ATP concentration, each cross-bridge with bound ATP is surrounded by rigor cross-bridges. The rigor cross-bridges bind to the thin filament and activate it. It was shown that a relaxed fibre becomes active at low ATP concentration.
Furthermore, it is unsurprising that ATP-bound myosin lifetimes in essence doubled in the presence of blebbistatin (Xu et al. 2009; Wilson et al. 2014). At the same time, the average free (unbound) time has dropped after blebbistatin, which would make sense if overall sites were more bound, as reported by ˂ τbound ˃. It is noteworthy that values recorded here are on the order of <1 s, while most reported accepted results are in 10–100 of seconds. It is likely that our current methodology lacks proper calibration for the blinking of Alexa-647 and could be a potential improvement in the next iteration of this tool. Indeed, taking a fixed tissue post labelling with ATP-Alexa-647, and imaging it in order to retrieve the kinetics of Alexa-647 dye in situ, would be ideal calibration approach. This information would allow us to currently group the binding events as shown in Fig. 4b and c, and effectively combine several bound events into a large one, spanning several seconds. Despite these last missing bits, the currently presented approach promises to be useful and applicable for future studies in structure and function of sarcomeric proteins, and potentially beyond.
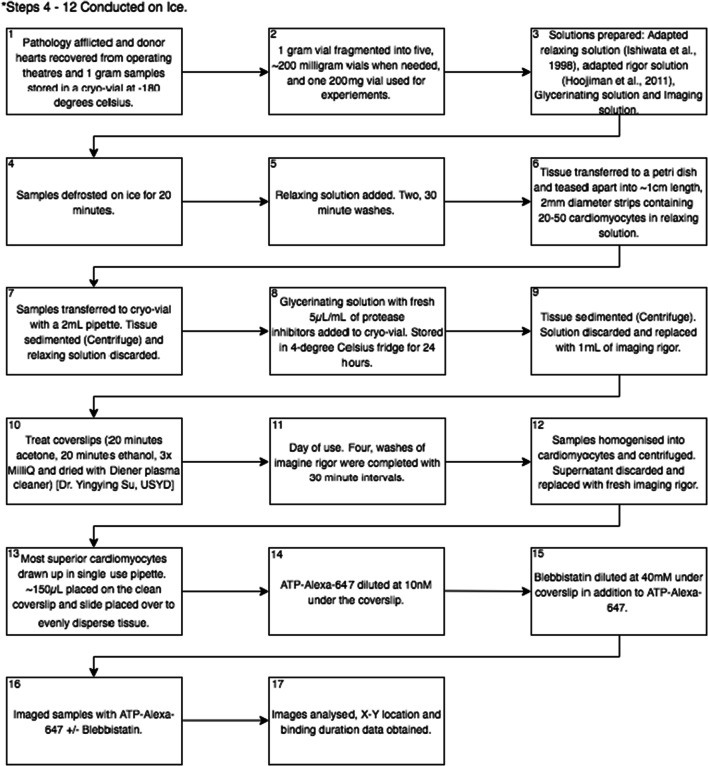
Fig. 4.
Flow diagram of steps taken between obtaining a heart sample and image analysis
Indeed, this approach opens the possibilities to assessing the in situ binding kinetics, while still being able to change ligand concentration in bulk while imaging very large number (several thousands) of binding sites per field of view, in just 5–10 min of experimental recording time. These properties make it an ideal tool for bridging a high spatio-temporal resolution of small scales to the macroscopic scale problems such as cardiomyopathies, and potentially opens the door to use it as diagnostic tool one day, given that very small quantities of tissue (< 10 mg) are required to perform the experiments. We believe that this modality of imaging and image processing is capable of deciphering myosin lifetimes and proportion of bound myosins dynamically, such as measuring the super-relaxed myosin state.
Role of SRX in sarcomere function
Most muscle biophysicists probably thought there would probably be no more major discoveries in the structure and function of the sarcomeric proteins, but they were wrong. Roger Cooke and colleagues were thinking about thermogenesis in striated muscles where body temperature is regulated by myosin ATPase activity. In particular, he was aware of the earlier work of Mike Ferenczi and his colleagues (Ferenczi et al. 1978) who reported a substantial disparity between the low myosin ATPase activity measured in vivo in relaxed frog skeletal muscle fibres and the fast ATPase rate for the same amount of purified frog myosin subfragment-1 in solution. A similar but smaller difference was obtained for mammalian (rabbit) skeletal muscle was reported by Myberg et al. (1995) where ATP turned over rates for isolated myofibrils at 42 °C (rabbit body temperature) was 0.06 ± 0.005 s−1, while the rate for purified myosin was 0.16 ± 0.005 s−1. Later, Cooke and his colleagues (Stewart et al. 2010; Hooijman et al. 2011) introduced what turned out to be a major discovery in the way striated muscles work. They concluded that, in the presence of excess MgATP (e.g. < 500 μm) and at low free Ca (pCa ≤ 8), the majority of relaxed myosin cross-bridges were in a highly disordered (DRX) relaxed state (where one ATP is hydrolyzed every 30 s). However, some cross-bridges had much lower ATP turnover and were in a new state; they called the super-relaxed (SRX) state where hydrolysis was slower. They argued that SRX cross-bridges may play an important cardioprotective role under normal conditions by slowing myosin ATPase activity, and maintaining myocardial contractile energetics, and homeostasis (Hooijman et al. 2011). Subsequently, Alamo et al. (2017) showed the SRX state is due to the formation of an “interacting head motif” (IHM) conformation. Here, the trailing head strongly inhibits the ATPase activity of the leading head in a cross-bridge. These SRX cross-bridges are aligned along the thick filament, unable to participate in force generation.
The role of MyBP-C
The protein, myosin-binding protein C (MyBP-C), was first discovered in 1971 by Gerald Offer and Roger Starr as a contaminant of rabbit skeletal muscle myosin preparation (Offer and Starr 1971). To put this discovery into perspective, 50 years ago sodium dodecyl sulphate (SDS) polyacrylamide gel electrophoresis (PAGE) had only recently been discovered (Weber and Osborn 1969), and gels had to be prepared using in hand-made equipment. Gerald was curious to see how clean his myosin preparations were, so when he heavily overloaded his gels with myosin he was somewhat surprised to find several other proteins were present besides the expected myosin heavy chain (200 kDa) he labelled “A”. They were sensibly named the smaller molecular weight bands B, C, D, E etc… and he modestly suggested that C protein as he called it, might be physiologically important, especially as his colleague, Carl Moos, suggested protein C produced much more uniform myosin thick filaments (Offer et al. 1973). That turned out to be something of an understatement. The arrangement of the three myofilaments is schematically shown in Fig. 1 where MyBP-C (red stripes) is located in the C-zone of the A band.
In 1993, Carrier and her colleagues (Carrier et al. 1993) were the first to identify MYBPC3 as a gene responsible for hypertrophic cardiomyopathy (the other one being cardiac myosin heavy chain, MYH7). Subsequently, truncations in its encoded protein, cardiac MyBP-C (cMyBP-C) were identified but were associated with a range of phenotypes (Weber et al. 1993). The roles cMyBP-C were reviewed in 2015 (Carrier et al. 2015; McNamara and Li 2015), but since then several key reports have focussed our thinking on the way it regulates myosin cross-bridge activity, ATP consumption, and in particular the fraction of cross-bridges in the SRX state (Nelson et al. 2020).
The C-terminus of this 135 kDa protein is integrated into the thick filaments (Fig. 1a) where it binds both the LMM portion of myosin heavy chain to titin. Its N-terminus binds to actin and can sensitise relaxing proteins, troponin and tropomyosin to Ca2+ and therefore enhancing the probability of myosin-binding and cross-bridge formation (Lin et al. 2018). However, at saturating Ca2+ and ATP concentrations, cMyBP-C acts as a viscous load on the cross-bridges impeding maximal sliding velocity (Walcott et al. 2015). Intriguingly, these cross-bridge modulatory properties seem to be unique to the cardiac isoform of MyBP-C. Its closely related skeletal counterparts (skMyBP-C) modulate contractility at either low or high calcium concentrations but not both (Li et al. 2019). Thus, in the cardiac sarcomere, cMyBP-C (1) can activate and inhibit thin filament sliding in a calcium dependent manner through the action of its N-terminal extension (Lin et al. 2018), (2) increases the rate of detachment of myosin cross-bridge (Tonino et al. 2019), and (3) can influence the interactive heads motif (IHM) (Alamo et al. 2017) and therefore modify the number of SRX cross-bridges aligned along the surface of the thick filaments.
Furthermore, cMyBP-C is only fully functional when it is phosphorylated, and hypo-phosphorylation produces reduced of SRX cross-bridges (McNamara and Li 2015). More recently, David Warshaw and colleagues were the first to assess the single molecule myosin ATPase lifetimes and the effects of MyBP-C in the skeletal muscle (Nelson et al. 2020). They showed under relaxed conditions that soleus muscle exhibits two myosin hydrolysis rates with a five-fold difference consistent with the DRX and SRX rates previously reported by Roger Cooke. However, using single molecule microscopy, they were able to functionally segregate the myosin ATPase lifetimes in the presence (C-zone) and absence (D-zone) of MyBP-C. They also showed that a predominance of the SRX lifetime is localised to the C-zone. This presents exciting opportunities to investigate whether this holds true in the cardiac muscle and may present itself to be a valuable therapeutic target. A drug, now in clinical trials, mavacamten, shows promise as a treatment for hypertrophic cardiomyopathy patients’ because it can shift hyperactive cross-bridges from the DRX state to the SRX state (and are being considered as potential treatment for hypertrophic cardiomyopathy) (Anderson et al. 2018). Thus, cMyBP-C is effectively an integrator of multiple signalling pathways in the sarcomere that determines downstream physiological function (Ponnam et al. 2019).
Is there a better way to measure the SRX state?
Until recently, all previous report of SRX involved the use of small bundles of permeabilized cardiomyocytes where a washout step is needed to replace mant-ATP with unlabelled ATP, a step that can take several seconds. Furthermore, the emission wavelength of mant-ATP epifluorescence is close to the muscle autofluorescence, particularly in human cardiomyocytes. This prompted us to consider alternatives that might improve the signal-to-noise to the point where single molecular imaging might be possible. That would enable us to simultaneously quantify individual ATP molecules in real time as they are rapidly hydrolyzed and released in DRX and more slowly released from the SRX cross-bridges. While total internal reflection of fluorescence (TIRF) microscopy cannot image molecules deep within cardiomyocytes attached to a glass coverslip (Axelrod 2001), it is well-suited to examine the behaviour of bright, photobleaching-resistant fluorescent probes. The advantages TIRF microscopy are summarised by Fish (2009) in TIRF microscopy: (1) There is a significant reduction in the background resulting in an excellent signal-to-noise ratio; (2) in inverted microscopes, the exponentially decaying evanescent field extends for ~ 100 nm above the coverslip surface leaving everything else focus; and (3) cells are exposed to much less damage by the exciting laser. Moreover, imaging on modern TIRF microscopes like the Zeiss Elyra, equipped with high efficiency electron-multiplying charge-coupled device (EMCCD) cameras allow us to capture 100 × 100 μm field of view at very fast frame rates (> 100 fps) resulting in unprecedented spatio-temporal resolution that achieves molecular resolution. We use Alexa-647 labelled ATP because its far-red emission channel avoids cardiomyocyte autofluorescence (Morkel et al. 2019). Furthermore, Alexa-647 is brighter than mant-ATP and is less prone to photobleaching.
Detailed methods and materials
Heart tissue collection
The Sydney Heart Bank (SHB) began in collaboration with the late Victor Chang in 1989 at St Vincent’s Hospital, Sydney. The SHB has since grown its collection with ~ 450 human hearts of both healthy donors and those with end-stage cardiac pathologies such as hypertrophic, dilated and peripartum cardiomyopathy. Samples were dissected and stored from all four chambers of the heart including inter-chamber septum and major vasculatures within 40 min of aortic cross clamping to ensure sample viability. Approximately 1 g of tissue is placed into individual cryo-vials and deposited into liquid nitrogen at – 180 °C. These 1-g vials are then fragmented into several ~ 200 mg cryo-vials when required (HREC No. #2814).
Cardiomyocyte solutions
Several solutions were prepared prior to sample preparation. All chemicals were from Sigma unless otherwise stated.
The relaxing solution is adapted from Ishiwata et al. (1998): 5 mM ATP; 40 mM BES; 1 mM MgCl2; 1 mM dithiothreitol (DTT) (added fresh); 15 μ/mL phosphocreatine kinase type I; 15 mM phosphocreatine; 1% Triton X-100 at pH 6.8).
The rigor solutions is adapted from Hooijman et al. (2011): 2.5 mM K2HPO4; 2.5 mM KH2PO4; 120 mM KOAc; 10 mM EGTA; 5 mM Mg(OAc); 50 mM MOPS, at pH 6.8. This rigor solution differs slightly from Rassier (2008) as although the pH values were similar (pH 6.8 and pH 7.4); there were significant differences in the buffers used.
Glycerinating solution was made from rigor solution with v/v 50% glycerol. The solution was thoroughly homogenised through vigorous inversion by hand.
Imaging solution (made fresh on the day of imaging) is rigor with the following additions 1% NaN3 [26628-22-8]; 17.99 mM creatine phosphate; 0.012 mM creatine phosphokinase; 2 mM dithiothreitol (DTT); 5.33 mM glucose; 0.6uM glucose oxidase [9001-37-0]; 5 μL/mL protease inhibitor cocktail (P8340) when 25 mL of solution were made.
Tissue preparation
All steps were conducted on ice unless otherwise specified. Samples were removed from liquid nitrogen and defrosted on ice for 20 min. The relaxing solution was added to the cryo-vial for two 30-min washes. The tissue was transferred into a petri dish (50 mm diameters) with approximately 1 mL of relaxing solution. A 5 μL/mL of protease inhibitor was added to the solution at every step. Forceps were used to tease apart the tissue to form strips containing 20–50 cardiomyocytes approximately 5–10 mm long and 2 mm in width. These strips were then pipetted into a cryo-vial. A DLAB D10008 was spun at 2000 RPM at room temperature for 15 s. The supernatant was discarded and replaced by ~ 1 mL of glycerinating solution with protease inhibitors. Samples were stored at 4° overnight.
Coverslip preparation
Cover slips were prepared prior with a 20-min soak in acetone, followed by a 20-min soak in absolute ethanol. This was followed by three MilliQ washes. The coverslips were then dried with a Diener plasma cleaner to maximise the amount of tissue bound to the coverslip. This coverslip technique was created and performed by Dr. Yingying Su of the Australian Centre for Microscopy and Microanalysis at the University of Sydney.
Sample and imaging preparation
On the day of use, the sample was removed from the fridge and spun down with a centrifuge (DLAB D10008) spun at 2000 RPM at room temperature for 15 s and supernatant discarded. This solution was replaced with 1 mL of imaging rigor. The tissue was washed four times with Imaging Rigor with 30-min intervals. A KemaKeur Model Pro 200 (Pro Scientific Inc Oxford CT, USA) homogeniser with a 5-mm external and 4-mm internal diameter head was used to break down the tissue into individual cardiomyocytes. This was done through ~ 5 × 5–10-s half maximum speed bursts with the head slightly above the base of the cryo-vial. The cardiomyocytes were centrifuged with a DLAB D10008 spun at 2000RPM at room temperature for 15 s in the cryo-vial and supernatant was replaced with fresh Imaging Rigor solution. Cardiomyocytes were drawn up from the floating fluid above the clumped tissue in the cryo-vial into a single use pipette and three even drops equating to approximately 150 μL were placed onto a clean coverslip. A clean glass slide was placed on top of the coverslip, dispersing the tissue evenly. ATP-Alexa-647 (Alexa Fluor 647) (Thermofisher–A22362), a far-red fluorescent ATP analogue with a Quantum Yield of 0.33 was used due to its Excitation Peak of 651 nm and an Emission Peak of 665, ideal for TIRF microscopy. We found 10 nM ATP-Alexa-647 in imaging rigor to be the optimal concentration to view binding events under TIRF. A 40 mM blebbistatin (856925-71-8) selective ATPase inhibitor was added concurrently with the ATP-Alexa-647 in some treatments to evaluate its effects on myosin head populations. The samples were then imaged with TIRF microscopy and ATP-Alexa-647 binding observed with and without blebbistatin to yield X-Y coordinates and length of bind data.
Technical notes on sample preparation
Sodium azide from aspergillus niger (NaN3) was used to prevent bacterial contamination of tissue. Creatine phosphate and creatine phosphatase were used in combination to regenerate any hydrolyzed ATP analogue. A glucose oxidase and glucose combination was used to mitigate the effects of oxygen free radicals created by the TIRF laser.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Elvis Pandzic, Email: e.pandzic@unsw.edu.au.
Cristobal G. dos Remedios, Email: c.dosremedios@victorchang.edu.au
References
- Alamo L, Ware JS, Pinto A, et al. Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes. eLife. 2017;6:e24634. doi: 10.7554/eLife.24634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RL, Trivedi DV, Sarkar SS, et al. Deciphering the super relaxed state of human beta-cardiac myosin and the mode of action of mavacamten from myosin molecules to muscle fibres. Proc Natl Acad Sci U S A. 2018;115:E8143–E8152. doi: 10.1073/pnas.1809540115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Traffic. 2001;2:764–774. doi: 10.1034/j.1600-0854.2001.21104.x. [DOI] [PubMed] [Google Scholar]
- Carrier L, Hengstenberg C, Beckmann JS et al (1993) Mapping of a novel gene for familial hypertrophic cardiomyopathy to chromosome 11. Nature Genetics. 4:311–313 [DOI] [PubMed]
- Carrier L, Mearini G, Stathopoulou K, Cuello F. Cardiac myosin-binding protein C (MYBPC3) in cardiac pathophysiology. Gene. 2015;573(2):188–197. doi: 10.1016/j.gene.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke R. Stress does not alter the conformation of a domain of the myosin cross-bridge in rigor muscle fibres. Nature. 1981;294:570–571. doi: 10.1038/294570a0. [DOI] [PubMed] [Google Scholar]
- Dufresne E and Blair D (2005) The Matlab particle tracking code repository, http://site.physics.georgetown.edu/matlab/
- Ebashi S. Third component participating in the superprecipitation of ‘Natural actomyosin’. Nature. 1963;200:2010. doi: 10.1038/2001010a0. [DOI] [PubMed] [Google Scholar]
- Ferenczi MA, Homsher E, Simmons RM, et al. Reaction mechanism of the magnesium ion-dependent adenosine triphosphatase of frog muscle myosin and subfragment 1. Biochem J. 1978;171:165–175. doi: 10.1042/bj1710165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish KN (2009) Total internal reflection fluorescence (TIRF) microscopy. Curr Protoc Cytom. Chapter 12 [DOI] [PMC free article] [PubMed]
- Hooijman P, Stewart MA, Cooke R. A new state of cardiac myosin with very slow ATP turnover: a potential cardioprotective mechanism in the heart. Biophys J. 2011;100:1969–1976. doi: 10.1016/j.bpj.2011.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley HE, Hanson J. Changes in the cross-striations of muscle during contractions and stretch and their structural interpretation. Nature. 1954;173:973–976. doi: 10.1038/173973a0. [DOI] [PubMed] [Google Scholar]
- Huxley AF, Niedergerke R. Structural changes in muscle during contraction. Nature. 1954;173:971–973. doi: 10.1038/173971a0. [DOI] [PubMed] [Google Scholar]
- Ishiwata S, Funatsu T, Fujita H. Contractile properties of thin (actin) filament-reconstituted muscle fibres. In: Pollack GH, editor. Sugi H. Springer Science & Business Media: Mechanisms of work production and work absorption in muscle; 1998. pp. 319–329. [DOI] [PubMed] [Google Scholar]
- Li A, Nelson SR, Rahmanseresht S, Braet S, Cornachione AS, Previs SB, O’Leary TS, McNamara JW, Rassier DE, Sadayappan S, Previs MJ, Warshaw DM. Skeletal MyBP-C isoforms tune the molecular contractility of divergent skeletal muscle systems. Proc Natl Acad Sci. 2019;116(43):21882–21892. doi: 10.1073/pnas.1910549116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BL, Li A, Mun JY, Previs MJ, Previs SB, Campbell SG, dos Remedios CG, Tombe PdP, Craig R, Warshaw DM, Sadayappan S. (2018) Skeletal myosin binding protein-C isoforms regulate thin filament activity in a Ca2+-dependent manner. Scientific Reports. 8:2604 [DOI] [PMC free article] [PubMed]
- Mamidi R, Li J, Doh CY, et al. Impact of the myosin modulator mavacamten on force generation and cross-bridge behavior in a murine model of hypercontractility. J Am Heart Assoc. 2018;7(17):e009627. doi: 10.1161/JAHA.118.009627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PA, dos Remedios CG. The dependence of rigor tension on sarcomere length in vertebrate muscle. Experientia. 1974;30:1036–1038. doi: 10.1007/BF01938995. [DOI] [PubMed] [Google Scholar]
- McNamara JW, Li A. The role of super-relaxed myosin in skeletal and cardiac muscle. Biophys Rev. 2015;7:5–15. doi: 10.1007/s12551-014-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morkel C, Pandzic E, Su Y-Y et al (2019) Direct observation of myosin cross-bridge heads while they hydrolyze ATP in cardiomyocytes from healthy donors and end-stage human heart failure. 43rd Ann. Conf Proc Aust Soc Biophys
- Myberg KH, Franks-Skiba K, Cooke R. Nucleotide turnover rate measured in fully relaxed rabbit skeletal muscle myofibrils. J Gen Physiol. 1995;106:957–973. doi: 10.1085/jgp.106.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SR, Li A, Beck-Previs S, Kennedy GG, Warshaw DM (2020) Imaging ATP consumption in resting skeletal muscle: one molecule at a time. bioRxiv 2020.05.27.119065. 10.1101/2020.05.27.119065 [DOI] [PMC free article] [PubMed]
- Offer G, Starr R. Polypeptide chains of intermediate molecular weight in myosin preparations. FEBS Lett. 1971;15:40–44. doi: 10.1016/0014-5793(71)80075-3. [DOI] [PubMed] [Google Scholar]
- Offer G, Moos C, Starr R. A new protein of the thick filaments of vertebrate skeletal myofibrils: extraction, purification and characterization. J Mol Biol. 1973;74:653–662. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- Perry SV (2008) Background to the discovery of troponin and Setsuro Ebashi’s contribution to our knowledge of the mechanism of relaxation in striated muscle. Biochem Biophys Res Commun 369:43–48. 10.1016/j.bbrc.2007.11.185 [DOI] [PubMed]
- Ponnam S, Sevrieva I, Sun YB, Irving M, Kampourakis T (2019) Site-specific phosphorylation of myosin binding protein-C coordinates thin and thick filament activation in cardiac muscle. PNAS 116(31):15485–15494 [DOI] [PMC free article] [PubMed]
- Rassier DE. Pre-power stroke cross bridge contribute to force during stretch of skeletal muscle myofibrils. Proc R Soc B. 2008;275:2577–2586. doi: 10.1098/rspb.2008.0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MA, Franks-Skiba K, Chen S, et al. Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers. Proc Natl Acad Sci U S A. 2010;107:430–435. doi: 10.1073/pnas.0909468107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonino P, Kiss B, Gohlke J, Smith JE, Granzier H (2019) Fine mapping titin's C-zone: Matching cardiac myosin-binding protein C stripes with titin's super-repeats. J Mol Cell Cardiol 133:47–56 [DOI] [PMC free article] [PubMed]
- Tsaturyan AK, Bershitsky SY, Burns R, He Z-H, Ferenczi MA. J Physiol. 1999;520(3):681–696. doi: 10.1111/j.1469-7793.1999.00681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walcott S, Docken S, Harris SP. Effects of cardiac myosin binding protein-C on actin motility are explained with a drag-activation-competition model. Biophys J. 2015;108(1):10–13. doi: 10.1016/j.bpj.2014.11.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K, Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969;244:4406–4412. [PubMed] [Google Scholar]
- Weber FE, Vaughan KT, Reinach FC, Fischman (1993) Complete Sequence of Human Fast-Type and Slow-Type Muscle Myosin-Binding-Protein C (MyBP-C). Differential Expression, Conserved Domain Structure and Chromosome Assignment. Eur J Biochem. 216(2):661–669 [DOI] [PubMed]
- White DCS. Rigor contraction and the effect of various phosphate compounds on glycerinated insect flight and vertebrate muscle. J Physiol. 1970;208:583–605. doi: 10.1113/jphysiol.1970.sp009138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, Naber N, Pate E, Cooke R. The myosin inhibitor blebbistatin stabilizes the super-relaxed state in skeletal muscle. Biophys J. 2014;107:1637–1646. doi: 10.1016/j.bpj.2014.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, White HD, Yu LC. Stabilization of helical order in the thick filaments by blebbistatin: further evidence of coexisting multiple conformations of myosin. Biophys J. 2009;96:3673–3681. doi: 10.1016/j.bpj.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]