Abstract
Glioma shows progression presenting as malignant transformation or leptomeningeal metastasis (LM). However, longitudinal biopsy of brain parenchyma is difficult due to its critical location, whereas cerebrospinal fluid (CSF) can be obtained serially with a little invasiveness of puncture. Thus, if we could find a biomarker for glioma progression, we could predict such event and determine therapeutic interventions as early as possible. In this study, we examined whether cerebrospinal fluid (CSF) metabolome profiles can reflect glioma grade, difference with non-glial tumor, and LM status. We selected 32 CSF samples from glioma patients, and compared them with 10 non-tumor control and seven non-glial brain tumor (medulloblastoma) samples. A total of 10,408 low-mass ions (LMIs) were detected as a candidate of metabolites using mass spectrometry, and representative LMIs were identified via the Human Metabolome Database. Grade IV gliomas showed eight LMIs, including acetic acid, of higher levels (summed sensitivity and specificity > 180%) than grade III gliomas. Grade IV gliomas demonstrated more abundant 30 LMIs, including glycerophosphate, compared with medulloblastoma, but none was mutually exclusive. Phospholipid derivatives were significantly more abundant in LM (−) than LM (+) gliomas regardless of glioma grade. LMIs representative of LM (+) gliomas were derivatives of glycolysis. We also verified discriminative LMIs based on mean expression level of each LMI (Student t test, p < 0.05) and evaluated the differences of the above analyses. Over 90% of metabolite pathways indicated from two analytical models were common to each other. Non-targeted mass spectrometry of CSF metabolites revealed significantly different profiles across gliomas that possibly permitted differentiation between glioma grades, LM, and non-glial brain tumors.
Electronic supplementary material
The online version of this article (10.1007/s13167-020-00211-4) contains supplementary material, which is available to authorized users.
Keywords: Predictive preventive personalized medicine, Cerebrospinal fluid, Glioma, Grade, Leptomeningeal metastasis, Metabolome
Introduction
Gliomas are incurable tumors that invade critical structures of the brain, preventing curative surgical resection. Although systemic chemotherapy has been attempted, it has met with limited success [1, 2]. Moreover, these gliomas underwent malignant transformation to higher tumor grades during or after the treatment, resulting in aggressive clinical behavior and changed genetic or epigenetic signatures [3]. For diagnosis and treatment decisions, tumor biopsy is required, but serial samples of gliomas are difficult to obtain because the brain is shielded by the cranium, and gliomas are often located in the eloquent area. Thus, diagnosis and treatment decisions largely depend upon neuroimaging such as magnetic resonance imaging (MRI) in case of difficult biopsy, but there are no standardized MRI criteria discriminating grade IV glioma from grade III [4]. Gliomas can also metastasize into the central nervous system (CNS), occasionally resulting in spread to the cerebrospinal fluid (CSF), but gadolinium-enhanced MRI sensitivity for leptomeningeal metastasis (LM) is around 60% [4].
Because CSF is in contact with the brain parenchyma and circulates throughout the neuroaxis, transporting neuro-metabolites, extracellular vesicles, and proteins, CSF has been studied for the evidence of CNS disease and to monitor treatment response as a so-called “liquid biopsy” [5, 6]. In particular, recent advances in metabolomics using high-resolution mass spectrometry (MS) have revealed features of the cancer environment that are expected to permit early diagnosis, monitoring of disease progression, and treatment response in cancer[7]. Metabolic biomarkers inferred from metabolomics could contribute to an advance of predictive, preventive, and personalized medicine (PPPM) in neuro-oncology [8–10]. Nevertheless, questions remain, such as whether the detection of a diluted metabolite, secreted from a relatively small tumor into a large amount of biofluids, reflect the unique characteristics of these tumors unless it is a pathognomonic byproduct of cancer cell metabolism. Previously, we described discriminative metabolomic profiles in 196 patients with different CNS tumors, including LM from solid primary cancers [11]. Using low-mass-ion discriminant equation (LOME) to select the best LMI profile, we could differentiate LM from the high-risk patients with brain metastasis. Other researchers have differentiated the CSF metabolomic profiles of malignant gliomas from those of controls including brain metastasis and non-cancerous patients using targeted MS [12, 13]. However, there is a need to explore CSF metabolomic profiles in gliomas to differentiate between WHO grade, leptomeningeal metastasis, and different invasiveness between glioma and non-glial primary brain tumors.
Here, we performed non-target MS of 32 glioma CSF samples and compared them to seven medulloblastomas and 10 non-tumorous controls (unruptured aneurysm). We compared the LMI profiles between (1) gliomas and controls, (2) gliomas of different WHO grades (III vs. IV), (3) primary brain tumors of the same WHO grade (IV) but different cellular origins and clinical behaviors (glioblastoma vs. medulloblastoma), (4) gliomas with or without LM, and (5) gliomas with different isocitrate dehydrogenase 1 (IDH1) mutation status. The discriminative LMIs were obtained experimentally using two different parameters of summed sensitivity/specificity and the difference of mean peak area of individual LMI between two groups of comparison. Furthermore, we examined the most prevalent metabolomic pathway designated by these discriminative LMIs using MetaboAnalyst (https://www.metaboanalyst.ca, ver. 4.0) to identify possible pathway dysregulation in particular glioma types and to compare the similarity between two analytical methods.
Materials and Methods
CSF collection
All CSF samples were obtained after Institutional Review Board (NCC-150002) approval in May 2015; written informed consent was obtained from each participant before the collection. The ethics approval was given; healthy controls submitted informed consent on their permission for operation of unruptured aneurysm, and in case of minors, their legal guardian submitted the informed consent. CSF samples were centrifuged (2000×g, 20 min) within 1 h of collection for cell down. A 50-μl sample of each supernatant was used for MS analysis.
CSF sample collection has been done under permission of NCC-150002 prospectively. However, this study design came out as a retrospective analysis of gliomas later using CSF archives and electronic medical record. CSF sampling was done from ventricle via Ommaya/shunt valve reservoir/extraventricular drainage catheter for controlling hydrocephalus or intraventricular/intrathecal chemotherapy or via lumbar puncture for the diagnosis of LM (Table 1). Ten samples of non-tumorous control (unruptured aneurysm) CSF were obtained during craniotomy from cisternal space, and seven samples of medulloblastoma were obtained via lumbar puncture for CSF cytology.
Table 1.
Clinical characteristics of 39 samples from 32 patients
| Case | Gender/age | Diagnosis | WHO grade | LM status | LM examination (MRI/cytology) | IDH 1 status |
|---|---|---|---|---|---|---|
| 1 | M/56 | Glioblastoma | IV | (−) | (−)/(−) | wt |
| 2 | F/33 | DMG | IV | (−) | (−)/(−) | wt |
| 3 | F/47 | Glioblastoma | IV | (−) | (−)/Atypical | wt |
| 4 | F/18 | Glioblastoma | IV | (−) | (−)/ Atypical | n.d. |
| 5 | M/68 | Glioblastoma | IV | (−) | (−)/(+) | wt |
| 6 | F/58 | Glioblastoma | IV | (+) | (+)/(+) | n.d. |
| 7 | F/29 | Glioblastoma | IV | (−) | (−)/(−) | n.d. |
| 8 | M/51 | Gliosarcoma | IV | (+) | (+)/(−) | wt |
| IV | (+) | (+)/(+) | wt | |||
| IV | (+) | (+)/(−) | wt | |||
| IV | (+) | (+)/(−) | wt | |||
| IV | (+) | (+)/(−) | wt | |||
| 9 | F/48 | Glioblastoma | IV | (−) | (−)/n.d. | wt |
| 10 | F/29 | Glioblastoma | IV | (+) | (+)/(−) | wt |
| 11 | M/56 | Glioblastoma | IV | (−) | (−)/(−) | wt |
| 12 | F/35 | Glioblastoma | IV | (−) | (−)/n.d. | wt |
| 13 | F/57 | Glioblastoma | IV | (−) | (−)/n.d. | wt |
| 14 | M/23 | DMG | IV | (−) | (−)/n.d. | wt |
| 15 | M/74 | Glioblastoma | IV | (− | (−)/(−) | wt |
| 16 | M/5 | Brainstem glioma | III | (−) | (+)/Atypical | n.d. |
| Brainstem glioma | III | (−) | (+)/(−) | n.d. | ||
| 17 | F/2.4 | AA | III | (−) | (−)/(−) | n.d. |
| 18 | M/58 | AA | III | (+) | (+)/Atypical | wt |
| 19 | F/71 | AA | III | (+) | (+)/Atypical | n.d. |
| 20 | M/22 | Brainstem glioma | III | (−) | (−)/(−) | n.d. |
| 21 | F/35 | AA | III | (+) | (+)/Atypical | wt |
| 22 | F/47 | Anaplastic ODG | III | (+) | (+)/Atypical | mt |
| Anaplastic ODG | III | (+) | (+)/n.d. | mt | ||
| Anaplastic ODG | III | (+) | (+)/Suggestive | mt | ||
| 23 | M/72 | AA | III | (−) | (−)/n.d. | wt |
| 24 | F/41 | ODG | II | (−) | (−)/n.d. | mt |
| 25 | M/32 | Gliomatosis | II | (+) | (+)/Suggestive | n.d. |
| 26 | F/5 | Medulloblastoma | IV | (−) | (−)/(−) | n.a. |
| 27 | F/2.8 | Medulloblastoma | IV | (−) | (−)/(−) | n.a. |
| 28 | F/7 | Medulloblastoma | IV | (+) | (+)/Atypical | n.a. |
| 29 | M/7 | Medulloblastoma | IV | (−) | (−)/(−) | n.a. |
| 30 | F/10 | Medulloblastoma | IV | (−) | (−)/Atypical | n.a. |
| 31 | M/11 | Medulloblastoma | IV | (−) | (−)/Atypical | n.a. |
| 32 | M/7 | Medulloblastoma | IV | (+) | (+)/(−) | n.a. |
AA, anaplastic astrocytoma; DMG, diffuse midline glioma; IDH 1, isocitrate dehydrogenase 1; ODG, oligodendroglioma; LM, leptomeningeal metastasis; mt, mutated; n.a., not applicable; n.d., not done; wt, wild-type
Diagnosis of LM
CSF cytology findings positive or suggestive for metastasis were considered LM-positive. However, in the event of atypical cytology, MRI findings and previous CSF cytology were used as references for positive categorization as follows: (1) MRI was definite (leptomeningeal enhancement of cerebral hemisphere gyri, cerebellar folia, subependymal wall, spinal cord, and cauda equina) or suggestive (ventricular seeding or dural enhancement) of LM [14], and (2) previous CSF cytology was positive, and this sample was obtained after treatment for LM.
CSF extraction for metabolite profiling
Metabolites present in the CSF were extracted using a modified Bligh and Dyer method. In brief, 50 μl CSF were added to 1 ml deionized water. After vortexing, 2 ml methanol and 0.9 ml dichloromethane were added. After vortexing and incubating on ice for 30 min, 1 ml water and 0.9 ml dichloromethane were added, then the mixture was centrifuged (1000×g, 10 min, room temperature). Nitrogen gas was used to dry the supernatant for MS analysis. The extracted metabolites were analyzed by TripleTOF 5600+ system (Sciex, Framingham, MA) coupled with a Nexera X2 UHPLC system (Shimadzu, Kyoto, Japan). The metabolites were loaded onto an Atlantis T3 sentry guard cartridge (3 μm, 2.1 mm × 10 mm; Waters, Milford, MA), and separated by an Atlantis T3 column (3 μm, 2.1 mm × 100 mm; Waters) in a two-step linear gradient (solvent A, 0.1% FA in water; solvent B, 100% ACN; with 1% solvent B for 2 min, 1 to 30% solvent B for 6 min, 30 to 90% solvent B for 8 min, 90% solvent B for 4 min, 90 to 1% solvent B for 1 min, and 9 min in 1% solvent B). The MS system was set to perform one full scan (50 to 1200 m/z range) followed by tandem mass spectrometry (MS/MS) of the 10 most abundant parent ions (mass tolerance, 50 mDa; collision energy, 35%). Monoisotopic mass peak was used for the next analysis and the ratio of signal to noise (S/N) was set at 10:1.
Identification of discriminative LMIs
LMIs with outstanding discriminative ability (e.g., distinguishing the control from glioma groups) were identified using logarithmic peak areas [15]. Briefly, individual LMIs were assessed as follows. (1) For each LMI, a discrimination threshold value was determined where the sum of the sensitivity and specificity was highest, at increments of 0.01. When a range of adjacent threshold values showed the same highest discrimination performance, the highest and lowest threshold values (and their difference) were tabulated for later comparison. Each LMI was investigated in two modes. First, it was assumed that logarithmic peak areas of one group were mostly higher than those of the other group. Then a sample whose peak area was more/less than the discrimination threshold in the former group was assigned as a true-positive/false-negative case. A sample whose peak area was more/less than the discrimination threshold in the latter group was assigned as a false-positive/true-negative case. Next, it was assumed that logarithmic peak areas of the former group were mostly lower than those of the latter group. Then, a sample whose peak area was less/more than the discrimination threshold in the former group was assigned as a true-positive/false-negative case. A sample whose peak area was less/more than the discrimination threshold in the latter group was assigned as a false-positive/true-negative case. Finally, the sum of the sensitivity and specificity was calculated for the two modes. Then, the mode with the higher sum of the sensitivity and specificity was assigned as a discrimination mode of the LMI. The sum of the sensitivity and specificity at the discrimination mode of the LMI was considered in subsequence comparison. (2) A few discriminative LMIs were designated as possible candidates based on a “summed sensitivity/specificity” based on at least 80% sensitivity and specificity and a summed sensitivity/specificity greater than 180%.
Identification of metabolite ions
The MS and MS/MS spectra were submitted to the Formula Finder computational tool (SCIEX) that proposes probable elemental compositions within a specified mass tolerance of a given m/z ratio using the PeakView software (SCIEX). Using metabolite databases from the Human Metabolome Database (HMDB, www.hmdb.ca), putative specific compounds were found for the given m/z and listed in rank-order based on the MS and MS/MS data. The compounds list found from the HMDB was used for further analysis of metabolomic pathways in MetaboAnlayst (https://www.metaboanalyst.ca, ver. 4.0).
Statistical analyses
Categorical data were analyzed using chi square, Fisher’s exact, or Mann-Whitney U tests as appropriate. Quantitative data were analyzed using one-way analysis of variance with post hoc comparisons (Scheffe’s test). The Student t test and the false discovery rate (FDR) correction (Benjamini and Hochberg method) were examined to access the difference of mean peak area between groups. All statistical analyses were performed using STATA 10.0 (version 10.0, StataCorp LP, College Station, TX, USA) or R (version 3.6.0, The R Foundation).
Results
Clinical characteristics of patients
Clinical characteristics of 39 samples from 32 primary brain tumor patients are presented in Table 1. Three glioma patients provided nine samples (5 from case 8, 2 from case 16, and 3 from case 22), which were not replicates but different dates of sampling for pre-/post-procedure or treatment. Patients were 43.7% male and 56.3% female, and the median age was 34 years (range 2.4–74). Because the non-glioma group (n = 7) consisted of medulloblastoma only, the median age of these patients was 7.0 years, significantly younger than the median 47 years of the glioma group (n = 25, chi square test, p < 0.001). The control group (n = 10) showed the oldest median age, 61.5 years (range 50–72).
All gliomas and medulloblastomas were histologically confirmed except two cases of brainstem glioma (case 16 and 20). Among 25 patients with gliomas, glioblastoma (GBM) was the most common (n = 12, 48%). All GBM was primary except one case of gliosarcoma (case 8) being secondary GBM from pleomorphic xanthoastrocytoma. Two brainstem gliomas without biopsy-proven pathology (n = 2) were considered to be grade III. Of the 17 tumors of 23 CSF samples were examined by immunohistochemistry, four samples of oligodendroglioma (ODG) lineage showed an IDH1 mutation, and the remainder possessed wild-type IDH1. Eighteen samples were collected from 15 patients with LM. Among these, 13 samples were accompanied with CSF cytology, which was positive in 4 patients, atypical in 4 patients, and negative in 5 patients.
Among 32 glioma samples, recent therapeutic interventions were none in 12 samples (8 samples were obtained on operation day, 3 samples were postoperative period, and one received chemotherapy 5 years ago), systemic chemotherapy in 16 samples, intrathecal chemotherapy in 3 samples, and radiation in one sample.
Overview of CSF metabolomic profiling
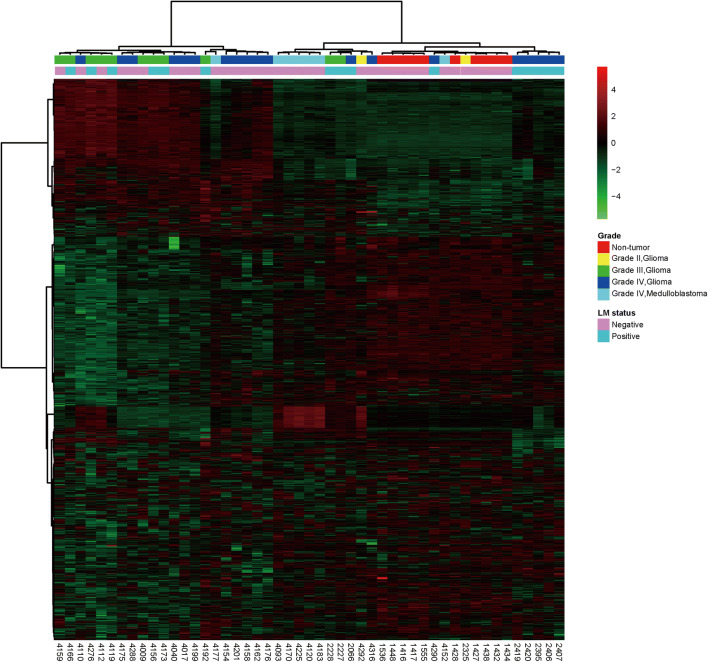
A total of 10,193 LMIs in a peak table using Marker View (version 1.2.1, Sciex, Tokyo, Japan) were evaluated by MS of 49 CSF samples (32 gliomas, 7 medulloblastomas, and 10 controls, Supporting Information Table 1). Each LMI was identified by m/z and retention time (min), and relative expression level was denoted by differences in mean normalized logarithmic peak areas between groups of comparison. To evaluate the possibility of discriminative metabolomic profiles, supervised clustering heatmap was generated using MetaboAnlayst 4.0 (Fig. 1a). In the dendrogram, LMIs were classified into three clusters. The cluster 1 had higher expression in glioma group; the cluster 2, highest in the control group; the cluster 3, mixed profiles. In unsupervised clustering heatmap, all tumor samples separately clustered from non-tumor controls except for three tumor samples (WHO grades II, III, and IV, each, Fig. 1b). Furthermore, among 19 grade IV glioma samples, 11 out of 12 LM (−) samples clustered separately from the 7 LM (+) samples. These finding possibly suggests that CSF LMIs might enable differentiation between primary brain tumors and non-tumorous conditions as well as between gliomas with and without leptomeningeal metastasis.
Fig. 1.
Hierarchical clustering profiles of CSF LMIs. (a) Supervised clustering heatmap of CSF metabolomes was generated using the Pearson correlation–based distance measure and Ward’s linkage method provided from Metaboanalyst 4.0. (b) Unsupervised hierarchical clustering heatmap of metabolomes was generated using the Pearson correlation–based distance measure and Ward’s linkage method provided from Metaboanalyst 4.0. Color key indicates log10 peak area value
Candidate LMIs discriminating gliomas from non-tumorous control
To select discriminative LMIs between groups of comparison, we performed two different methods; (1) the summed sensitivity/specificity (LMI-SSS) at a designated threshold level, and (2) the difference of mean peak areas (LMI-MPA) of each LMI. We identified 99 LMIs that were expressed significantly more abundant in glioma samples than in controls by LMI-SSS, and all of which showed complete discrimination (both 100% sensitivity and specificity). Additionally, 148 LMIs demonstrated abundance in gliomas at 97% sensitivity (one false-negative sample) (Supporting Information Table 1a). Based on the difference of LMI-MPA, we found 2674 discriminating LMIs (Student t test, p < 0.05 and |fold change| > 2). Among these, 2021 of which were more abundant in glioma samples than in controls, and 653 of which showed abundance in the non-tumorous control compared with glioma samples (Supporting Information Fig. 1a). However, the heatmap of 2021 LMIs abundant in gliomas failed to show distinctive profile according to glioma grade with this mean expression level selection (Supporting Information Fig. 1b).
Candidate LMIs discriminating gliomas of different WHO grade (III vs. IV)
Although there were hundreds of discriminative LMIs between grades II and III gliomas (Supporting Information Table 1b), we devoid of analyzing as the number of grade II gliomas was too small (only two cases). From LMI-SSS (> 180%), we found 61 LMIs exhibiting more abundance in grade III gliomas and eight LMIs with abundance in grade IV gliomas (Fig. 2a, scatter plot). Based on the LMI-MPA, 330 LMIs were significantly more abundant in grade III gliomas compared with that in grade IV, whereas only 21 LMIs showed significantly greater abundance in grade IV gliomas than that in grade III (Fig. 2a, volcano plot).
Fig. 2.
Discriminative LMI of CSF metabolites. Expression level and distribution of representative LMIs, which was overly expressed. (a) In grade III (scatter plot, left) and grade IV gliomas (scatter plot, right). (b) In grade IV non-gliomas (scatter plot, left) and glioma grade IV (scatter plot, right). (c) LM (−) (scatter plot, left) and LM (+) (scatter plot, right) grade III gliomas. (d) In LM (−) (scatter plot, left) and LM (+) grade IV gliomas (scatter plot, right). (Volcano plot of LMIs in each comparison) Metabolites which having more than 2 (a, b, and d) or 1.5 (c) fold change (FC) and Student t test p value < 0.05 (FDR-corrected) between two groups were colored in magenta
We identified the representative candidate metabolites from the LMI-SSS in the HMDB (Table 2). Sphingolipid metabolites, such as sphingosine and ceramide, which are involved in lipid signaling and apoptosis in cancer cells [16], were elevated in grade III relative to grade IV gliomas. LMIs more abundant in grade IV gliomas were glycol aldehyde, glyceric acid, and acetic acid, which are known to be related to dysregulated glycolysis and ketoacidosis.
Table 2.
Profiles of discriminative low-mass-ion candidates between G III and G IV
| Status | Selected LMI (m/z*) | HMDB identifier | Candidate metabolite | Chemical formula | Class |
|---|---|---|---|---|---|
| G III > G IV |
577.5154 577.5187 577.5221 |
HMDB0007300 | Diacylglycerol | C36H64O5 | Fatty acyl |
|
578.5191 578.5225 |
HMDB0010399 | Lysophosphatidylcholine | C30H60NO7P | Lysophospholipid | |
| 650.6416 | HMDB0000831 | N-Lignoceroylsphingosine | C42H83NO3 | Sphingolipid | |
| HMDB0004956 | Ceramide | C42H83NO3 | Sphingolipid | ||
| 651.6494 | HMDB0000918 | 1-Oleoyl-cholesterol | C45H78O2 | Steroid derivative | |
| HMDB0056112 | Diacylglycerol | C41H78O5 | Glycerolipid | ||
| 684.1997 | HMDB0059644 | 6β-Hydroxy-1,4,5,6-tetrahydronicotinamide-adenine dinucleotide | C21H31N7O15P2 | Purine nucleotide | |
| 859.5273 | HMDB0009850 | Phosphatidylinositol | C45H79O13P | Glyserophopholipid | |
| HMDB0116547 | Phosphatidylglycerophosphate | C42H84O13P2 | Glyserophopholipid | ||
| 859.5355 | HMDB0009893 | Phosphatidylinositol | C45H79O13P | Glyserophopholipid | |
| HMDB0116547 | Phosphatidylglycerophosphate | C42H84O13P2 | Glyserophopholipid | ||
| HMDB0059616 | All-trans-decaprenyl diphosphate | C50H84O7P2 | Prenol lipid | ||
| G IV > G III | 60.0458 | HMDB0031645 | Acetamide | C2H5NO | Carboximidic acid |
| HMDB0001842 | Guanidine | CH5N3 | Organonitrogen | ||
| HMDB0000906 | Trimethylamine | C3H9N | Organonitrogen | ||
|
61.0303 61.0304 |
HMDB0000042 | Acetic acid | C2H4O2 | Carboxylic acid | |
| HMDB0003344 | Glycolaldehyde | C2H4O2 | Organooxygen | ||
| HMDB0062508 | N-Nitrosomethanamine | CH4N2O | Organonitrogen | ||
| HMDB0000294 | Urea | CH4N2O | Organic carbonic acid | ||
| HMDB0000820 | 1-Propanol | C3H8O | Organooxygen | ||
| 73.9453 | HMDB0034106 | Methyl isothiocyanic acid | C2H3NS | Isothiocyanate | |
| 107.0857 | HMDB0006372 | l-Glyceric acid | C3H8O4 | Organic oxygen | |
| 123.0913 | HMDB0041570 | Isopropylpyrazine | C7H10N2 | Diazine | |
| HMDB0041571 | Propylpyrazine | C7H10N2 | Diazine | ||
| HMDB0059873 | 3-Ethylphenol | C8H10O | Organic oxygen | ||
| HMDB0029306 | 4-Ethylphenol | C8H10O | Organic oxygen |
Candidate LMIs discriminating between WHO grade IV glioblastoma and medulloblastoma
Both glioblastoma and medulloblastoma are highly malignant primary brain tumors of WHO grade IV, but their cellular origins and clinical behaviors are different [17]. We found no mutually exclusive LMI by the LMI-SSS (Supporting Information Table 1c). However, at the summed sensitivity/specificity of > 180%, 30 LMIs showed higher expression levels in grade IV gliomas and 15 LMIs showed more abundance in the medulloblastoma group (Fig. 2b, scatter plot). Based on the LMI-MPA, 51 LMIs showed higher levels in medulloblastoma and eight LMIs were significantly more abundant in grade IV gliomas (Fig. 2b, volcano plot). Metabolites for representative LMIs discriminating grade IV gliomas and medulloblastomas by the LMI-SSS are identified and listed (Supporting Information Table 2). In glioblastoma, metabolites were carnitine and glycerophosphate (related to mitochondrial energy production) and phosphatidylinositol and phosphatidylserine (cell membrane phospholipid derivatives). Conversely, the representative LMIs of medulloblastoma were carboxylic acid or pteridine, which were not well-identified as products of biological pathway but seem to be intermediate organic compounds or chemical compounds.
Candidate LMIs discriminating gliomas with or without LM according to the same WHO grade
Among grade III gliomas, eight samples corresponded to LM (+) patients and three to LM (−). Based on the LMI-SSS, 94 LMIs were more abundant in LM (+) than LM (−) and 40 LMIs revealed greater peak areas in LM (−) than LM (+) at complete discrimination in grade III gliomas (Fig. 2c, scatter plot, and Supporting Information Table 1d). By the LMI-MPA, 48 LMIs were significantly more abundant in LM (+) relative to LM (−), and 97 LMIs showed significantly greater abundance in LM (−) than LM (+) grade III gliomas (Fig. 2c, volcano plot). In grade IV gliomas, 61 and 32 LMIs showed complete discrimination (Fig. 2d, scatter plot), with peak areas higher and lower, respectively, in LM (+) than in LM (−) by the LMI-SSS (Supporting Information Table 1e). Volcano plot from the LMI-MPA revealed 115 LMIs were significantly more abundant in G IV gliomas with LM (+) compared with LM (−); meanwhile, 548 LMIs showed significantly higher level in LM (−) than LM (+) grade IV gliomas (Fig. 2d, volcano plot).
Representative LMIs discriminating gliomas with or without LM by the LMI-SSS were identified at HMDB and listed in Table 3. In Table 3 comparing LM (+) and LM (-) gliomas, phospholipid derivatives, such as diacylglycerol and phosphatidylserine, major components of the cell membrane, were more abundant in LM (−) gliomas relative to LM (+) regardless of glioma grade. However, certain LMIs were more abundant in LM (+) than that in LM (−) gliomas according to glioma grade. In LM (+) grade IV gliomas, derivatives of abnormal glycolysis, including acetic acid, glycolaldehyde, and pyruvate aldehyde, predominated as representative LMIs. Meanwhile, the representative LMIs of LM (+) grade III gliomas were varied and included ketone derivatives (ethanone and lactone), catecholamines (norepinephrine and hydroxydopamine), pyruvaldehyde, and cysteine.
Table 3.
Profiles of discriminative low-mass-ion between leptomeningeal metastasis (+) and (−)
| Status | Selected LMI (m/z*) | HMDB identifier | Candidate metabolite | Chemical formula | Class |
|---|---|---|---|---|---|
|
G III LM(+) > LM(−) |
60.0466 60.0459 60.0447 |
HMDB0001842 HMDB0031645 HMDB0003656 |
Guanidine Acetamide Acetaldehyde oxime |
CH5N3 C2H5NO C2H5NO |
Organooxygen Organooxygen Organooxygen |
| 61.0304 |
HMDB0003344 HMDB0000042 |
Glycolaldehyde Acetic acid |
C2H4O2 C2H4O2 |
Organooxygen Carboxylic acids |
|
| 73.0662 | HMDB0001167 | Pyruvaldehyde | C3H4O2 | Organic oxygen | |
| 122.0955 | HMDB0002017 | 1-Phenylethylamine | C8H11N | Organonitrogen | |
| 165.0549 | HMDB0060267 | l-Fuculose | C6H12O5 | Fatty acyls | |
| 215.0158 |
HMDB0001213 HMDB0001031 HMDB0001351 |
1-Deoxy-d-xylulose 5-phosphate Deoxyribose 5-phosphate Deoxyribose 1-phosphate |
C5H11O7P C5H11O7P C5H11O7P |
Organooxygen Organooxygen Organooxygen |
|
| 319.2242 | HMDB0013633 | 12-Ketoeicosatetraenoic acid | C20H30O3 | Fatty acyls | |
| 332.0947 | HMDB0000905 | Deoxyadenosine monophosphate | C10H14N5O6P | Purine nucleotide | |
| 351.2016 |
HMDB0000268 HMDB0001425 |
Tetrahydrocorticosterone Estrone sulfate |
C21H34O4 C18H22O5S |
Steroids Steroids |
|
| 399.0329 | HMDB0011606 | 4-alpha-Methyl-5-alpha-cholest-7-en-3-one | C28H46O | Steroids | |
| 536.1656 | HMDB0059635 | Lipoyl-AMP | C18H26N5O8PS2 | Purine nucleotide | |
| 603.5273 | HMDB0007503 | Diacylglycerol (35:4) | C38H66O5 | Glycerolipids | |
|
G III LM(−) > LM(+) |
59.0516 |
HMDB0001659 HMDB0003366 HMDB0031652 |
Acetone Propanal Allyl alcohol |
C3H6O C3H6O C3H6O |
Organooxygen Organooxygen Alcohols |
| 111.1168 | HMDB0002434 | Hydroquinone | C6H6O2 | Phenols | |
| 125.0905 | HMDB0004181 | Methylimidazole acetaldehyde | C6H8N2O | azoles | |
| 149.0232 |
HMDB0000227 HMDB0002050 HMDB0000606 HMDB0000567 |
Mevalonic acid Dihydroxyfumaric acid d-2-Hydroxyglutaric acid Cinnamic acid |
C6H12O4 C4H4O6 C5H8O5 C9H8O2 |
Fatty acyls Keto acids Hydroxy acids Cinnamic acids |
|
| 629.5409 | HMDB0007090 | Diacylglycerol (37:5) | C40H68O5 | Glycerolipids | |
| 709.5068 | HMDB0116692 | Phosphatidylglycerol (31:0) | C37H73O10P | Glycerophospholipids | |
|
887.5629 887.5587 |
HMDB0009900 HMDB0009888 |
Phosphatidylinositol (38:4) | C47H83O13P | Glycerophospholipids | |
|
905.5981 905.5874 |
HMDB0116497 | Phosphatidylglycerophosphate (40:5) | C46H82O13P2 | Glycerophospholipids | |
| 910.5443 | HMDB0061595 | Phosphatidylserine (9D5/13m5) | C49H84NO12P | Glycerophospholipids | |
|
G IV LM(+) > LM(−) |
56.0509 56.0516 |
HMDB0061870 | N-Methylene-ethenamine | C3H5N | Organonitrogen |
| 61.0293 | HMDB0000042 | Acetic acid | C2H4O2 | Carboxylic acid | |
| HMDB0003344 | Glycolaldehyde | C2H4O2 | Organooxygen | ||
| 60.0460 | HMDB0031645 | Acetamide | C2H5NO | Carboximidic acid | |
| HMDB0001842 | Guanidine | CH5N3 | Organonitrogen | ||
| 61.0304 | HMDB0003344 | Glycolaldehyde | C2H4O2 | Organooxygen | |
| HMDB0000042 | Acetic acid | C2H4O2 | Carboxylic acid | ||
|
73.0657 73.0668 |
HMDB0001167 | Pyruvaldehyde | C3H4O2 | Organooxygen | |
| 98.9608 | HMDB0000973 | Hydrogen phosphate | H3O4P | Non-metal phosphate | |
| 102.0914 | HMDB0062600 | N-Butylformamide | C5H11NO | Carboxylic acid | |
| HMDB0001252 | Betaine aldehyde | C5H12NO | Organonitrogen | ||
| HMDB0006509 | Nervonyl carnitine | C6H16N | Organonitrogen | ||
|
G IV LM(−) > LM(+) |
377.1543 | HMDB0000244 | Riboflavin | C17H20N4O6 | Pteridine |
| 384.2955 | HMDB0013332 | 3-Hydroxy-5, 8-tetradecadiencarnitine | C21H37NO5 | Carboxylic acid | |
| 403.0912 | HMDB0001117 | 4′-Phosphopantothenoyl cysteine | C12H23N2O9PS | Peptidomimetic | |
| 433.2061 | HMDB0114743 | Lysophosphatidic acid | C21H37O7P | Glyserophopholipid | |
| 722.5060 | HMDB0009215 | Phophatidylethanolamine | C41H72NO7P | Glyserophopholipid | |
| 790.5873 | HMDB0011321 | Phosphatidylcholine | C46H80NO7P | Glyserophopholipid | |
| 796.5393 | HMDB0112816 | Phosphatidylserine (37:5) | C43H74NO10P | Glyserophopholipid | |
| HMDB0113780 | Phosphatidyl-N-methylethanolamine | C43H74NO10P | Glyserophopholipid | ||
| 810.5534 | HMDB0116758 | Phosphatidylserine (38:5) | C44H76NO10P | Glyserophopholipid | |
| HMDB0113782 | Phosphatidyl-N-methylethanolamine | C44H76NO10P | Glyserophopholipid | ||
| 812.5632 | HMDB0112575 | Phosphatidylserine (38:4) | C44H78NO10P | Glyserophopholipid | |
| 898.6161 | HMDB0112923 | Phosphatidylserine (44:3) | C50H92NO10P | Glyserophopholipid |
Analysis for gliomas according to different IDH1 mutation status
Despite insufficient sample numbers (21 IDH1 wild-type samples from 15 patients vs. 4 IDH1 mutant samples from 2 patients), individual LMIs showed significantly different peak areas according to IDH1 mutational status (Supporting Information Fig. 2). Two LMIs showed complete discrimination as the peak areas of these LMIs were higher in IDH1 wild-type compared with IDH1 mutant, but no LMI showed exclusively higher peak areas in IDH1 mutants (Supporting Information Table 1f). However, a total of four and 32 LMIs showed discrimination at the LMI-SSS > 180% in IDH1 wild-type and mutant samples, respectively. Representative LMIs including 2-hydroxyglutarate (2-HG) discriminating between IDH1 mutation status are identified and listed (Supporting Information Table 3). The mean expression level of 2-HG, a well-known product of mutant IDH1 from alpha-ketoglutarate, was significantly higher in IDH1 mutant samples at a threshold level gap of 0.05 in log10 scale compared with IDH1 wild-type samples (Fig. 3).
Fig. 3.

The mean normalized peak area value of a candidate metabolite of 2-hydroxyglutarate (2-HG). Low-mass-ions 149.0229 m/z at a retention time of 18.04 min. which were identified as a candidate metabolite of 2-HG. The mean expression level of 2-HG in IDH1 mutant gliomas is elevated at a threshold gap of 0.05 in log10 scale compared with IDH1 wild-type gliomas
Metabolic pathway analysis using discriminative LMIs
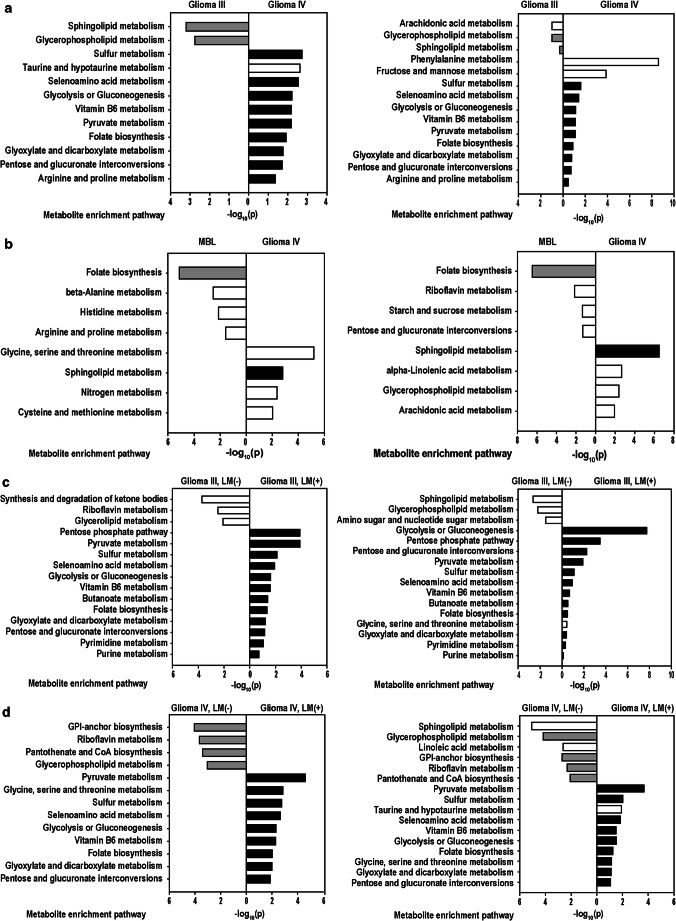
In comparing grade III and grade IV gliomas (Fig. 4a), the LMI-SSS of grade III gliomas denoted two metabolic pathways, the most highly involved of which was sphingolipid metabolism, and the LMI-MPA were greatest the glycerophospholipid metabolism pathways, which were shared across these two different selection methods. The LMI-SSS of grade IV compared with grade III gliomas indicated 10 pathways, and the LMI-MPA indicated 11 pathways overrepresented in grade IV. Among these, nine pathways involving glycolysis or gluconeogenesis; interconversion of pentose and glucoronate; biosynthesis of folate biosynthesis; and metabolism of pyruvate, selenoamino acid, sulfur, taurine/hypotaurine, and vitamin B6 were common in both methods.
Fig. 4.
Metabolite enrichment analysis at KEGG pathway database. Representative LMIs selected based on sensitivity/specificity summation (left), and metabolites representing significantly different mean expression level (right), between (a) grade III and grade IV gliomas, (b) grade IV gliomas and medulloblastoma, (c) grade III gliomas with or without leptomeningeal metastasis (LM), and (d) grade IV gliomas with or without LM
The pathways identified by comparing grade IV gliomas and medulloblastoma were four in both methods (Fig. 4b). Among pathways overpresented in grade IV gliomas, sphingolipid metabolism was shared across these. In selection of pathways involved in medulloblastoma, folate biosynthesis is common among these.
In LM (+) or (−) grade III gliomas (Fig. 4c), the LMI-SSS and the LMI-MPA indicated three significantly involved metabolic pathways overrepresented in LM (−) tumors, respectively. Among those three pathways, glycero(phospho)lipid metabolism was common to both methods. The LMI-SSS and the LMI-MPA indicated 12 and 13 pathways, respectively, overrepresented in LM (+) tumors. Among those, 12 pathways were common to both methods, including glycolysis or gluconeogenesis; biosynthesis of fatty acids or folate; interconversion of pentose and glucoronate; and metabolism of glyoxylate/dicarboxylate, linoleic acid, pyruvate, selenoamino acid, sulfur, taurine/hypotaurine, and vitamin B6.
In LM (+) and (−) grade IV gliomas (Fig. 4d), the LMI-SSS and the LMI-MPA of LM (−) grade IV gliomas showed four and six overrepresented pathways, respectively. Among those, glycerophospholipid metabolism is common to both methods as it was in LM (−) grade III gliomas. In identifying pathways overrepresented in LM (+) grade IV gliomas, the LMI-SSS indicated nine pathways, and the LMI-MPA significantly involved in 10 pathways. Among these, all nine pathways indicated by the LMI-SSS were presented in the pathways of the LMI-MPA. Those were metabolism of pyruvate, “glycine, serine, and threonine,” sulfur, selenoamino acid, vitamin B6, “glyoxylate and diacarboxylate,” folate biosynthesis and “pentose and glucoronate interconversions” pathway.
Discussion
Our study is the first report of CSF metabolomic profiles that discriminate among glioma grade, WHO grade IV primary brain tumors (glioblastoma and medulloblastoma), and development of LM.
Advancement of MS to identify discriminating LMIs
High-performance liquid chromatography/MS (HPLC/MS) coupled with time-of-flight (TOF) MS is widely used to yield accurate qualitative and quantitative analysis in metabolomics. Previous work using matrix-assisted laser desorption ionization (MALDI)-TOF MS in 2005 was capable of identifying only 164 discernible peaks with 76% sensitivity for predicting LM from breast cancer [18]. The current MS technique provides much higher resolution when identifying LMIs in biofluids. Relative to targeted metabolomics, a non-targeted approach may offer global information on whole-individual LMIs in biofluids such as CSF; because the identification of LMI metabolites in non-targeted metabolomics is dependent on readily available HMDB information and tandem MS patterns of standard metabolites, the mass peak area of LMIs may provide information on the relative abundance of selected metabolites. In the present study, a total of 10,193 LMIs were evaluated by non-targeted metabolomics in 32 glioma, 7 medulloblastoma, and 10 non-tumor control CSF samples. This high-resolution triple-TOF MS identified metabolic profiles that can discriminate between detailed characteristics of glioma.
Clinical usefulness of CSF metabolomics profiles for glioma
Recently, studies searching for biomarkers of cancers in biofluids have proliferated to include methods such as circulating tumor cells, cell-free DNA, extracellular microRNA, and exosome sequencing. However, the minute concentration of biomarkers in serum and technical difficulties in identification and quantification of this mutation limits its clinical use. However, given the direct contact of CNS tumors with CSF, CSF should express such biomarkers more prolifically than serum. Although CSF sampling is somewhat invasive, it is less invasive than brain tumor biopsy, and using CSF as a liquid biopsy permits longitudinal sampling, which has special utility in gliomas because they are not a fixed form of disease but instead show progression to malignant transformation or LM. Although targeted MS-based metabolomic analysis of CSF has indeed identified signatures of malignant glioma[12, 19], the clinical usefulness of metabolomics analysis of CSF has not yet been verified through large sample sizes or prospective clinical trials.
However, the use of CSF as a liquid biopsy for glioma needs to be accompanied by the presence of a tumor. In a study of metabolite profiles of serum from patients with lung cancer [20], profiles differed between preoperative and postoperative samples in the same patient, indicating that serum metabolites depend on the existence of tumor tissue. Monitoring of glioma recurrence using CSF microRNA expression level was tried by Teplyuk et al. They tried to correlate the onco-miR expression level and tumor size on MRI [21]. But the evidence level was a case report and they could only suggest the relative change of microRNA expression level. Unfortunately, we could not find any study dealing with metabolite abundance and glioma recurrence yet. Nakamizo et al. performed targeted MS of CSF from 32 patients with histologically proven gliomas, which was collected at preoperative period [19]. They found that citric and isocitric acid levels discriminate glioma grades, and lactic acid levels were significantly higher in IDH1 mutant gliomas compared with IDH1 wild-type. However, tumor location and gadolinium enhancement did not show significant difference of their targeted LMIs. Recently, many clinical trials have been performed to inhibit IDH1 mutant gliomas with a concept of synthetic lethality using direct IDH1 mutant inhibitor, depletion of intermediate with provoking autophagy, and vaccination recognizing IDH1 R132H as a neo-antigen [22]. We expect an encouraging result from this trial of metabolic pathway target as a personalized medicine.
Relevance of identified discriminative LMIs for different gliomas
In malignant gliomas, the activation of EGFR signaling stimulates cellular membrane biogenesis and energy utilization, such as the Warburg effect, leading to increased fatty acid and glucose metabolism [23]. Furthermore, altered metabolic enzymes utilize the tricarboxylic acid cycle and oxidative phosphorylation differently than normal tissue [22]. Thus, the distinctive LMI profile in gliomas is likely due to an increase in the energy substrate or cell membrane components derived from these cells.
Indeed, we found LMIs related to sphingolipid metabolism abundant in grade III relative to grade IV gliomas. Inhibiting sphingosine kinase activity increases ceramide, and sphingosine levels inhibit cell proliferation and promote apoptosis [24]. Conversely, glycolaldehyde and l-glyceric acid were more abundant in grade IV compared with grade III gliomas, which is perhaps unsurprising given their role in energy metabolism [25, 26] and association with tumor aggressiveness [27], respectively. We identified several lipid derivatives differentially observed between glioblastoma and medulloblastoma. Due to the aberration of lipid metabolism not only in glioma but also in medulloblastoma, several studies have tried to identify a lipid marker to evaluate a disease status [27], whereas others describe elevation of choline-containing phospholipids in both glioblastoma and medulloblastoma tissues [28], the relative ratio of lipid components was different between the tumors, as measured by nuclear magnetic resonance, and these differences were manifest in the CSF samples. In our study, relatively higher m/z products (e.g., 887.5594, 888.5598, 905.5939) were found in glioblastoma than medulloblastoma CSF (Supporting Information Table 1). These metabolites were phosphatidylinositol, phosphatidylserine, or galabiosylceramide.
Differences among lipid metabolites in CSF metastasis have been previously reported in medulloblastomas, in which disease progression frequently includes LM. A mouse model of human medulloblastoma features a distinctive lipid signature of tumor tissue according to LM status, including reduced phosphatidic acids (PA), phosphatidylethanolamine (PE), and phosphatidylserine (PS) and increased phosphatidylinositol (PI) in metastasizing medulloblastoma tissues [29]. In our study, LM status was not balanced between grade IV gliomas and medulloblastomas. Thus, the difference of LMIs between these tumors should be cautioned to interpret. Pyruvaldehyde, a detrimental product of glucose, protein, and fatty acid metabolism, was increased in LM (+) relative to LM (−) grade III glioma. Because pyruvaldehyde is rapidly eliminated by the glyoxalase system [30], its accumulation could suggest a disorder in energy metabolism in LM (+) glioma.
In 2016, WHO classification of gliomas was revised, and IDH1 enzyme mutation status became a criterion for glioma classification [17]. The pathological role of IDH1 mutation in gliomas has not yet been described, but mutation occurs early in glioma development [31], and accumulation of its byproduct, 2-HG, may provoke malignant progression via hypermethylation [32]. In our study, LMI 149.0229, a candidate LMI of 2-HG, showed higher expression in IDH1 mutant than IDH1 wild-type samples, in keeping with others’ findings [13]. Among 21 IDH1 wild-type samples, four samples showed higher levels of 2-HG than IDH1 mutant samples (19% false negative at a sensitivity 81% and a specificity 100%); conversely, others have reported 15% false-positive rates [13]. All of our high-2-HG IDH1 wild-type samples were from fulminant, large glioblastomas, whereas three out of our four IDH1 mutant samples were from a patient with a small, anaplastic oligodendroglioma (0.5 × 1.5 cm at the largest diameter). Standardization of LMI expression based on tumor size should be considered in future studies.
Limitations and future directions
Limitations of this study were as follows: (1) LMIs are not specific molecules but instead suggest a candidate for targeted MS verification, (2) an absolute threshold level of LMIs in each glioma group was not analyzed, and (3) the relevance of metabolomic profiles to discriminate among types of primary brain tumors was not a proof-of-concept but, rather, suggested specific pathways based on metabolic pathway database analysis, and (4) as CSF sample collection in this study was not for study only but for instance of CSF cytology, or diversion, the number of samples is both limited and uneven to analyze each glioma grade and characteristics. Thus, we had to omit some meaningful analysis here.
In this study, we analyzed potential pathways involved in malignant transformation of gliomas (grade III vs. grade IV), leptomeningeal metastasis (LM (+) vs. LM (−)), and invasiveness (glioblastoma vs. medulloblastoma) using discriminative LMIs. The LMI-SSS might be a more appropriate approach for further investigation with targeted MS than the LMI-MPA in terms of exclusiveness. However, the exclusivity of discriminative LMIs should be re-evaluated with additional samples (or underwent external validation) unless the LMI was a metabolite unique to the glioma mutation, which never existed in the control group. Alternatively, profiling representative LMIs based on p value (the LMI-MPA) could be complemented using an algorithm such as LOME. Fortunately, differences among these two methods seemed not quite different in our study; as the pathways indicated by the LMI-SSS and the LMI-MPA were common to both methods, especially in comparing malignant characteristics (grade IV over grade IV, LM (+) over (−)), where more than 90% of designated pathways were common to each other.
Metabolomic profiles can reflect tumor behavior because they are the end product of tumor cells’ effort to survive and interact with the microenvironment. Relevant metabolomic targets downstream of predicted mutations will likely provide us with valid metabolomic targets as biomarkers [33–35]. As a chemical fingerprint combined with multi-omics data, metabolomic profiles of biofluids can help us understand the biology of glioma development and provide novel therapeutic targets [36–38].
Conclusions and expert recommendations
CSF metabolomic profiles from non-targeted MS analysis revealed significantly different profiles that varied according to glioma characteristics such as glioma grade, LM, and non-glial primary brain tumors of the same tumor grade. These results suggest that the important progression of glioma including malignant transformation and leptomeningeal metastasis could be predicted by CSF metabolomic analysis. Through a future validation trial of targeted MS based on candidate LMIs of this study, we expect that CSF analysis can support or replace the need for brain tumor biopsy and CSF cytology. Although we provide just discriminative metabolomic profiles, candidate molecules identified from HMDB indicate relevant metabolites such as lysophosphatidylcholine of membranous signaling control and acetic acid of ketone acidosis. As evidences of dysregulated metabolism and membrane moiety modulation in cancer cells have been built up, we expect to find a node to switch off glioma progression as a beginning of preventive medicine. CSF analysis as a liquid biopsy could verify genomics and transcriptomic/ proteomic profiles with recently advanced technology. Integrative analysis of CSF multi-omics results is expected to provide not only understanding of pathophysiology at molecular level but also finding druggable molecular targets. Regarding with an advancement of high-throughput drug screening system [39], we expect to find an inhibitor that achieve a personalized medicine in neuro-oncology as early as possible.
Electronic supplementary material
(PNG 3633 kb)
(PNG 1345 kb)
Abbreviations
- CNS
central nervous system
- CSF
cerebrospinal fluid
- HMDB
Human Metabolome Database
- HPLC/MS
high-performance liquid chromatography/MS
- IDH1
isocitrate dehydrogenase 1
- LM
leptomeningeal metastasis
- LMIs
low-mass ions
- LOME
low-mass-ion discriminant equation
- MALDI
matrix-assisted laser desorption ionization
- MS
mass spectroscopy
- ODG
oligodendroglioma
- PA
phosphatidic acids
- PE
phosphatidylethanolamine
- PPPM
predictive preventive personalized medicine
- PS
phosphatidylserine
- TOF
time-of-flight
Authors’ contributions
BC. Y, JH. L, and H-S. G designed and supervised the study, and analyzed data. JW. K, SH. S, H. Y, and K-Y. L contributed to acquisition of data. JH. I, TH. K, and KH. K contributed to the analysis and interpretation of data. JH. K and JB. P designed the study. All authors read and approved the final manuscript.
Funding information
This work was supported by grants from the National Cancer Center, Korea (NCC-1710871-3, 1910090-1 and 1910294-1), the Korea Health Industry Development Institute of Ministry of Health and Social Welfare, Republic of Korea (H1731340-2), and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (NRF-2017R1A2B4007859).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
The study procedure was approved by National Cancer Center and all patients signed consent forms. All CSF samples were obtained after Institutional Review Board (NCC-150002) approval in May 2015; written informed consent was obtained from each participant before the collection. The ethics approval was given in compliance with the Declaration of Helsinki.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ji Hye Im and Byong Chul Yoo contributed equally to this work.
References
- 1.Stewart LA. Chemotherapy in adult high-grade glioma: a systematic review and meta-analysis of individual patient data from 12 randomised trials. Lancet. 2002;359(9311):1011–1018. doi: 10.1016/s0140-6736(02)08091-1. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Cazzato E, Ladewig E, Frattini V, Rosenbloom DI, Zairis S, et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. doi: 10.1038/ng.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Straathof CS, de Bruin HG, Dippel DW, Vecht CJ. The diagnostic accuracy of magnetic resonance imaging and cerebrospinal fluid cytology in leptomeningeal metastasis. J Neurol. 1999;246(9):810–814. doi: 10.1007/s004150050459. [DOI] [PubMed] [Google Scholar]
- 5.Frankfort SV, Tulner LR, van Campen JP, Verbeek MM, Jansen RW, Beijnen JH. Amyloid beta protein and tau in cerebrospinal fluid and plasma as biomarkers for dementia: a review of recent literature. Curr Clin Pharmacol. 2008;3(2):123–131. doi: 10.2174/157488408784293723. [DOI] [PubMed] [Google Scholar]
- 6.Romeo MJ, Espina V, Lowenthal M, Espina BH, Petricoin EF, 3rd, Liotta LA. CSF proteome: a protein repository for potential biomarker identification. Expert Rev Proteomics. 2005;2(1):57–70. doi: 10.1586/14789450.2.1.57. [DOI] [PubMed] [Google Scholar]
- 7.Lee JH, Kim KH, Park JW, Chang HJ, Kim BC, Kim SY, Kim KG, Lee ES, Kim DY, Oh JH, Yoo BC, Kim IH. Low-mass-ion discriminant equation: a new concept for colorectal cancer screening. Int J Cancer. 2014;134(8):1844–1853. doi: 10.1002/ijc.28517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golubnitschaja O, Baban B, Boniolo G, Wang W, Bubnov R, Kapalla M, et al. Medicine in the early twenty-first century: paradigm and anticipation - EPMA position paper 2016. EPMA J. 2016;7:23. doi: 10.1186/s13167-016-0072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu M, Zhan X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J. 2018;9(1):77–102. doi: 10.1007/s13167-018-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grech G, Zhan X, Yoo BC, Bubnov R, Hagan S, Danesi R, Vittadini G, Desiderio DM. EPMA position paper in cancer: current overview and future perspectives. EPMA J. 2015;6(1):9. doi: 10.1186/s13167-015-0030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoo BC, Lee JH, Kim KH, Lin W, Kim JH, Park JB, et al. Cerebrospinal fluid metabolomic profiles can discriminate patients with leptomeningeal carcinomatosis from patients at high risk for leptomeningeal metastasis. Oncotarget. 2017;8(60):101203–101214. doi: 10.18632/oncotarget.20983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Locasale JW, Melman T, Song S, Yang X, Swanson KD, Cantley LC, et al. Metabolomics of human cerebrospinal fluid identifies signatures of malignant glioma. Mol Cell Proteomics. 2012;(6):11, M111 014688. 10.1074/mcp.M111.014688. [DOI] [PMC free article] [PubMed]
- 13.Ballester LY, Lu G, Zorofchian S, Vantaku V, Putluri V, Yan Y, Arevalo O, Zhu P, Riascos RF, Sreekumar A, Esquenazi Y, Putluri N, Zhu JJ. Analysis of cerebrospinal fluid metabolites in patients with primary or metastatic central nervous system tumors. Acta Neuropathol Commun. 2018;6(1):85. doi: 10.1186/s40478-018-0588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freilich RJ, Krol G, DeAngelis LM. Neuroimaging and cerebrospinal fluid cytology in the diagnosis of leptomeningeal metastasis. Ann Neurol. 1995;38(1):51–57. doi: 10.1002/ana.410380111. [DOI] [PubMed] [Google Scholar]
- 15.Lee JH, Kim YH, Kim KH, Cho JY, Woo SM, Yoo BC, Kim SC. Profiling of serum metabolites using MALDI-TOF and Triple-TOF mass spectrometry to develop a screen for ovarian cancer. Cancer Res Treat. 2018;50(3):883–893. doi: 10.4143/crt.2017.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radin NS. Killing tumours by ceramide-induced apoptosis: a critique of available drugs. Biochem J. 2003;371(Pt 2):243–256. doi: 10.1042/BJ20021878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 18.Dekker LJ, Boogerd W, Stockhammer G, Dalebout JC, Siccama I, Zheng P, Bonfrer JM, Verschuuren JJ, Jenster G, Verbeek MM, Luider TM, Smitt PAS. MALDI-TOF mass spectrometry analysis of cerebrospinal fluid tryptic peptide profiles to diagnose leptomeningeal metastases in patients with breast cancer. Mol Cell Proteomics. 2005;4(9):1341–1349. doi: 10.1074/mcp.M500081-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Nakamizo S, Sasayama T, Shinohara M, Irino Y, Nishiumi S, Nishihara M, Tanaka H, Tanaka K, Mizukawa K, Itoh T, Taniguchi M, Hosoda K, Yoshida M, Kohmura E. GC/MS-based metabolomic analysis of cerebrospinal fluid (CSF) from glioma patients. J Neurooncol. 2013;113(1):65–74. doi: 10.1007/s11060-013-1090-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Ma Z, Min L, Li H, Wang B, Zhong J, Dai L. Biomarker identification and pathway analysis by serum metabolomics of lung cancer. Biomed Res Int. 2015;2015:183624–183629. doi: 10.1155/2015/183624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teplyuk NM, Mollenhauer B, Gabriely G, Giese A, Kim E, Smolsky M, Kim RY, Saria MG, Pastorino S, Kesari S, Krichevsky AM. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol. 2012;14(6):689–700. doi: 10.1093/neuonc/nos074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agnihotri S, Zadeh G. Metabolic reprogramming in glioblastoma: the influence of cancer metabolism on epigenetics and unanswered questions. Neuro Oncol. 2016;18(2):160–172. doi: 10.1093/neuonc/nov125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strickland M, Stoll EA. Metabolic reprogramming in glioma. Front Cell Dev Biol. 2017;5:43. doi: 10.3389/fcell.2017.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mencarelli C, Martinez-Martinez P. Ceramide function in the brain: when a slight tilt is enough. Cell Mol Life Sci. 2013;70(2):181–203. doi: 10.1007/s00018-012-1038-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Enezi KS, Alkhalaf M, Benov LT. Glycolaldehyde induces growth inhibition and oxidative stress in human breast cancer cells. Free Radic Biol Med. 2006;40(7):1144–1151. doi: 10.1016/j.freeradbiomed.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 26.Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, Luthra S, Chandran UR, Benos PV, Smith L, Wang M, Hu B, Cheng SY, Sobol RW, Nakano I. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A. 2013;110(21):8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo D, Bell EH, Chakravarti A. Lipid metabolism emerges as a promising target for malignant glioma therapy. CNS Oncol. 2013;2(3):289–299. doi: 10.2217/cns.13.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srivastava NK, Pradhan S, Gowda GA, Kumar R. In vitro, high-resolution 1H and 31P NMR based analysis of the lipid components in the tissue, serum, and CSF of the patients with primary brain tumors: one possible diagnostic view. NMR Biomed. 2010;23(2):113–122. doi: 10.1002/nbm.1427. [DOI] [PubMed] [Google Scholar]
- 29.Paine MRL, Liu J, Huang D, Ellis SR, Trede D, Kobarg JH, Heeren RMA, Fernández FM, MacDonald TJ. Three-dimensional mass spectrometry imaging identifies lipid markers of medulloblastoma metastasis. Sci Rep. 2019;9(1):2205. doi: 10.1038/s41598-018-38257-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhla B, Luth HJ, Haferburg D, Boeck K, Arendt T, Munch G. Methylglyoxal, glyoxal, and their detoxification in Alzheimer’s disease. Ann N Y Acad Sci. 2005;1043:211–216. doi: 10.1196/annals.1333.026. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174(4):1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, Ye D, Guan KL, Xiong Y. IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clin Cancer Res. 2012;18(20):5562–5571. doi: 10.1158/1078-0432.CCR-12-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Batra S, Adekola KU, Rosen ST, Shanmugam M. Cancer metabolism as a therapeutic target. Oncology (Williston Park). 2013;27(5):460–467. [PubMed] [Google Scholar]
- 34.Kishton RJ, Rathmell JC. Novel therapeutic targets of tumor metabolism. Cancer J. 2015;21(2):62–69. doi: 10.1097/PPO.0000000000000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bajpai R, Shanmugam M. Targeting cancer metabolism through synthetic lethality-based combinatorial treatment strategies. Curr Opin Oncol. 2018;30(5):338–344. doi: 10.1097/CCO.0000000000000467. [DOI] [PubMed] [Google Scholar]
- 36.Janssens JP, Schuster K, Voss A. Preventive, predictive, and personalized medicine for effective and affordable cancer care. EPMA J. 2018;9(2):113–123. doi: 10.1007/s13167-018-0130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng T, Zhan X. Pattern recognition for predictive, preventive, and personalized medicine in cancer. EPMA J. 2017;8(1):51–60. doi: 10.1007/s13167-017-0083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu R, Wang X, Zhan X. Multi-parameter systematic strategies for predictive, preventive and personalised medicine in cancer. EPMA J. 2013;4(1):2. doi: 10.1186/1878-5085-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan Y, Nguyen DT, Akay Y, Xu F, Akay M. Engineering a brain cancer chip for high-throughput drug screening. Sci Rep. 2016;6:25062. doi: 10.1038/srep25062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PNG 3633 kb)
(PNG 1345 kb)