Abstract
Background
Invasiveness is a very challenging clinical problem in nonfunctional pituitary adenomas (NFPAs), and currently, there are no effective invasiveness-related molecular biomarkers. The post-neurosurgery treatment is much different as for invasive and noninvasive NFPAs. The aim of this study was to integrate phosphoproteomics and transcriptomics data to reveal phosphorylation-mediated molecular events for invasive characteristics of NFPAs to achieve a potential tool for patient stratification, and prognostic/predictive assessment to discriminate invasive from noninvasive NFPAs for personalized attitude.
Methods
The 6-plex tandem mass tag (TMT) labeling reagents coupled with TiO2 enrichment of phosphopeptides and liquid chromatography-tandem mass spectrometry (LC-MS/MS) were used to identify and quantify each phosphoprotein and phosphosite in NFPAs and controls. Differentially expressed genes (DEGs) between invasive NFPA and control tissues were obtained from the Gene Expression Omnibus (GEO) database. The overlapping analysis was performed between phosphoprotiens and invasive DEGs. Gene Ontology (GO) enrichment, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, and protein–protein interaction (PPI) analyses were used to analyze these overlapped molecules.
Results
In total, 1035 phosphoproteins with 2982 phosphorylation sites were identified in NFPAs vs. controls, and 2751 DEGs were identified in invasive NFPAs vs. controls. Overlapping analysis of these phosphoproteins and DEGs exposed 130 overlapped molecules (phosphoproteins; invasive DEGs). GO enrichment and KEGG pathway analyses of 130 overlapped molecules revealed multiple biological processes and signaling pathway network alterations, including cell–cell adhesion, platelet activation, GTPase signaling pathway, protein kinase signaling, calcium signaling pathway, estrogen signaling pathway, glucagon signaling pathway, cGMP–PKG signaling pathway, GnRH signaling pathway, inflammatory mediator regulation of TRP channels, vascular smooth muscle contraction, and Fc gamma R-mediated phagocytosis, which were obviously associated with tumor invasive characteristics. For 130 overlapped molecules, PPI network-based molecular complex detection (MCODE) identified 10 hub molecules, namely SLC2A4, TSC2, AKT1, SCG3, ALB, APOL1, ACACA, SPARCL1, CHGB, and IGFBP5. These hub molecules are involved in multiple signaling pathways and represent potential predictive/prognostic markers in NFPA patients as well as they represent potential therapeutic targets.
Conclusions
This study provided the first large-scale phosphoprotein profiling and phosphorylation-related signaling pathway network alterations in human NFPA tissues. Further, overlapping analysis of phosphoproteins and invasive DEGs revealed the phosphorylation-mediated signaling pathway network changes in invasive NFPAs. These findings are the precious resource for in-depth insight into the molecular mechanisms of NFPAs, as well as for the discovery of effective phosphoprotein biomarkers and therapeutic targets for invasive NFPAs.
Electronic supplementary material
The online version of this article (10.1007/s13167-020-00215-0) contains supplementary material, which is available to authorized users.
Keywords: Nonfunctional pituitary adenomas, Invasiveness, Tandem mass tag (TMT) labeling, TiO2 enrichment, Quantitative phosphoproteomics, Transcriptomics, Differentially expressed genes, Overlapped molecule (phosphoprotein, invasive DEG), Signaling pathway, Patient stratification, Prognostic/predictive assessment, Personalized treatment
Introduction
Pituitary adenoma is a common intracranial tumor that occurs in the anterior lobe of the pituitary gland, which seriously impacts the human endocrine systems [1–4]. Pituitary adenomas are classified into functional pituitary adenomas (FPAs) and nonfunctional pituitary adenomas (NFPAs) [5]. FPAs have the clinically increased levels of the corresponding blood hormones, whereas NFPAs do not have any clinically increased levels of blood hormones to cause the difficulty in its early diagnosis. NFPA is often diagnosed when it grows up to compress its surrounding tissues and organs. Further, NFPAs are classified into invasive and noninvasive NFPAs [6–8]. Noninvasive NFPA is easily treated with neurosurgery. However, the treatment of invasive NFPAs is a big challenge because its invasive behavior injures or damages tumor-surrounding structures, which cannot be completely removed with neurosurgery and causes a risk of recurrence after neurosurgery. Thus, patients with invasive NFPAs are often treated with radiation therapy after neurosurgery. The clinical diagnosis of invasive NFPAs is mainly derived from nuclear magnetic resonance (NMR) image changes and tumor morphological changes observed by neurosurgery, which actually is not fully correct because it is not easy to determine its invasiveness when this tumor is at its early stage with small size [9, 10]. Currently, there are no effective invasiveness-related biomarkers used in clinical practice. Characterizing any invasiveness-related molecular events in NFPAs may benefit the patient stratification and prognostic assessment to discriminate invasive from noninvasive NFPAs for personalized medical procedures. There is an urgent need to discover the changed molecular events for invasive NFPAs.
It is well-known that invasive NFPA is a multicause, multiprocess, and multiconsequence disease, with a series of molecular changes at the levels of genome, transcriptome, proteome, and metabolome, and those molecules interact mutually in a molecular network system [4, 11, 12]. It is driving one to shift the previous single factor model to multiparameter systematic model. Multiomics is an effective approach to realize this multiparameter systematic model shift [13–15]. The proteome and transcriptome are the functional performer of genome, and the proteome and transcriptome are regulated by extensive post-translational modifications (PTMs) [16, 17].
Among those PTMs, phosphorylation is an important and extensively studied PTM with the addition of phosphorus group (–HPO3 to –OH or –H3PO4 to –NH2) to residues such as serine (Ser, S), threonine (Thr, T), and tyrosine (Tyr, Y) in a protein, which plays crucial roles in signaling pathways and many pathophysiological processes [18]. Phosphorylation and dephosphorylation are reversibly dynamic reactions that are catalyzed by kinases and phosphatases, respectively, which regulate the basic biological functions [19, 20]. Phosphorylation in a protein promotes the conformational changes through interacting with other hydrophilic and hydrophobic residues [19]. Human genome sequencing identifies 107 human phosphatase genes and 518 human protein kinase genes including 90 known tyrosine kinases that include 58 receptor tyrosine kinases. These kinases and phosphatases are the potential targets of anticancer drugs, and tyrosine kinases accounting for 0.3% of genome contribute to 30% of 100 known dominant oncogenes [18, 21]. In eukaryotic cells, protein phosphorylation is a low abundance event, and serine phosphorylation accounts for ~ 90%, threonine phosphorylation for ~ 10%, and tyrosine phosphorylation for ~ 0.05% [18]. Identification and characterization of the altered phosphorylation and functional activities of phosphoproteins in different types of cancers have directly assisted in the discovery of protein kinase inhibitors to treat a tumor [22, 23]. Therefore, it emphasizes the scientific merits of investigating phosphoproteins in pituitary adenomas.
Tandem mass tag (TMT) labeling/TiO2 phosphopeptide enrichment-based quantitative phosphoproteomics [24, 25] is an effective method to identify phosphoprotein amino acid sequence and phosphorylation sites, and quantify the level of phosphorylation in cancers compared to controls. Briefly, the proteins from cancer and control tissues, respectively, are digested with trypsin, followed by TiO2 enrichment of phosphopeptides, and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. The MS/MS data are used to determine the amino acid sequence and phosphorylation sites, and the TMT reporter ions were used to quantify the level of phosphorylation. The studies on phosphorylation in single molecules have been extensively carried out in pituitary adenomas with documented 267 publications when the key words “phosphorylation and pituitary adenoma” were used to search the PubMed database, and those studies are mainly involved in phosphorylation-involved signaling pathways. However, only five publications were found in the PubMed database that are about phosphoproteome or phosphoproteomics in pituitaries or pituitary adenomas [26–30]. Of these publications, Carretero et al. studied the phosphorylation of ERK (extracellular signal–regulated kinase) and RSK (ribosomal S6 kinase) for thyrotropin-releasing hormone (TRH)-induced inhibition of rat ether-à-go-go-related (r-ERG) channel potassium currents in rat pituitary growth hormone 3 (GH3) cells [26]. Zhao et al. used proteomics to reveal abnormally phosphorylated AMPK (AMP-activated protein kinase) and ATF2 (activating transcription factor 2) involved in glucose metabolism and tumorigenesis of growth hormone (GH)-secreting pituitary adenomas [27]. Delcout et al. found the role of phosphorylation in pituitary adenylyl cyclase activating polypeptide (PACAP) type I receptor transactivation for insulin growth factor-1 (IGF-1) receptor signaling and antiapoptotic activity in neurons [28]. Beranova-Giorgianni et al. performed phosphoproteomics analysis of the human pituitary and identified 28 phosphoproteins [29]. Zhan et al. used PTMScan experiment that combined immunoaffinity enrichment and LC-MS/MS to analyze a total of 1006 unique phosphorylation sites within 409 proteins in more than 19 signaling pathways in human NFPAs relative to control pituitary tissues [30], and found that lots of hub molecules in many signaling pathways such as mTOR (mammalian target of rapamycin), PI3K/Akt (phosphatidylinositol 3 kinase/protein kinase B), NFκB (nuclear factor kappa-B), Wnt, p38, ERK/MAPK (extracelluar signal-regulated kinase/mitogen-activated protein kinase), and JNK signaling pathways in NFPAs were phosphorylated in NFPAs, and that mTOR, PI3K/AKT, NFκB, Wnt, p38, ERK/MAPK, and JNK signaling pathways were excessively activated in NFPAs [30]. Those publications clearly demonstrated the important roles of protein phosphorylations in pituitary adenoma pathophysiological processes. However, the large-scale global phosphoproteomics analysis has not been carried out in human NFPA tissues. The large-scale profiling of phosphoproteome in human NFPA tissues has important scientific value to understand in-depth the molecular mechanism of NFPAs and discover effective phosphoprotein biomarkers in NFPA patients.
Transcriptome is another level of functional performer of genome. In-depth investigation of transcriptome alterations in invasive NFPAs will directly benefit for the discovery of invasiveness-related molecular events. The public Gene Expression Omnibus (GEO) dataset includes the transcriptomics data between invasive NFPAs and controls, which can be directly extracted to identify differentially expressed genes (DEGs), followed by integration with phosphoproteomics data for comprehensive consideration of invasiveness-related molecular events in invasive NFPAs, from the view point of the multiparameter systematic model.
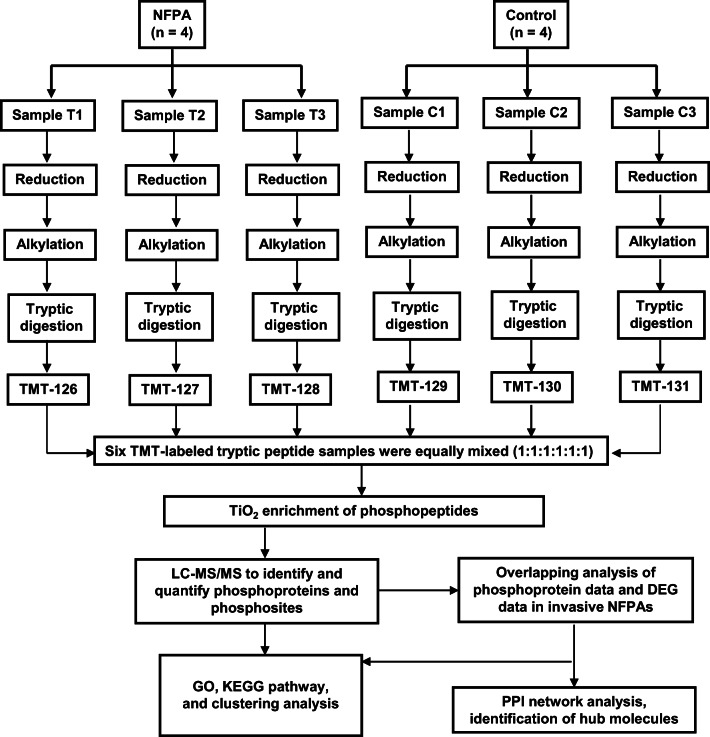
This study used quantitative phosphoproteomics based on TMT labeling in combination with TiO2 enrichment of phosphopeptides to identify the large-scale phosphoprotein profile in human NFPA relative to control pituitary tissues, followed by bioinformatics analysis to determine the functional characteristics of phosphoproteins in NFPAs. Further, these identified phosphoproteins and DEG data identified between invasive NFPAs and controls were integrated to determine the overlapped molecules (phosphoproteins; invasive DEGs). Gene ontology (GO) enrichment and the Kyoto Encyclopedia Gene and Genome (KEGG) pathway analysis were performed to determine the functional roles of invasiveness-related phosphoproteins (the overlapped molecules) in NFPAs. These overlapped molecules (phosphoproteins; invasive DEGs) were the precious resource to understand the molecular mechanisms of invasive NFPAs and discover invasiveness-related phosphoprotein biomarkers for potential prognostic/predictive assessment, and patient stratification to discriminate invasive from noninvasive NFPAs, for personalized attitude in medical services of invasive NFPAs. Figure 1 shows the experimental flowchart to identify phosphoproteins in NFPAs and the integrative analysis of phosphoproteomics and transcriptomics data of invasive NFPAs.
Fig. 1.
Experimental flowchart to identify phosphoproteins in NFPAs. Thanks to the 6-plex TMT labeling, it is possible to run six samples at one MS analysis. T1, T2, and T3 were three equal amounts of NFPA protein samples. C1, C2, and C3 were three equal amounts of control pituitary protein samples. The amount of T1, T2, and T3 was equal to that of C1, C2, and C3. Each protein sample was reduced with dithiothreitol (DTT), alkylated with iodoacetamide, and digested with trypsin. NFPA, nonfunctional pituitary adenoma; TMT, tandem mass tag; LC, liquid chromatography; MS/MS, tandem mass spectrometry; GO, gene ontology; KEGG, Kyoto Encyclopedia Gene and Genome; DEG, differentially expressed gene
Methods
Pituitary adenoma and control tissue samples
Control post-mortem pituitary tissues (n = 4; tissues from 3 white and 1 black patients) were from the Memphis Regional Medical Center (n = 4), which were approved by the University of Tennessee Health Science Center Internal Review Board. Pituitary adenoma biopsy tissues (n = 4; tissues from 4 Chinese patients) were from the Department of Neurosurgery of Xiangya Hospital, China, and were approved by the Xiangya Hospital Medical Ethics Committee of Central South University (Table 1). Written informed consent was obtained from each patient for pituitary adenoma biopsy tissues or the family of each control post-mortem pituitary subject after full explanation of the purpose and nature of all experimental procedures. Quantitative phosphoproteomics was performed with the four mixed NFPA samples and the four mixed control samples.
Table 1.
Clinical information of NFPA and control tissue samples
| Group | Sex | Age | Clinical information | Immunohistochemistry | Experiments |
|---|---|---|---|---|---|
| NFPA | Male | 58 | Chinese, NFPA in sellar region. Sellar floor bone destruction, enriched blood supply in tumor, and tumor size 4.5 × 3 × 3 cm3 | ACTH (−), hGH (−), PRL (−), FSH (−), LH (−), TSH (−) | Proteomicst |
| Male | 53 | Chinese, NFPA in sellar region. Sellar floor bone thinning and tumor size 3 × 3 × 2.5 cm3 | ACTH (−), hGH (−), PRL (−), FSH (−), LH (−), TSH (−) | Proteomics | |
| Female | 43 | Chinese, NFPA in sellar region. Sellar floor bone thinning, enriched blood supply in tumor, and tumor size 4 × 3 × 3 cm3 | ACTH (−), hGH (−), PRL (−), FSH (+), LH (−), TSH (−) | Proteomics | |
| Female | 43 | Chinese, NFPA in sellar region. Adhesion of surrounding tissues, and tumor size 4.5 × 4 × 6 cm3 | ACTH (−), hGH (−), PRL (−), FSH (+), LH (−), TSH (−) | Proteomics | |
| Control | Male | 36 | White American, multiple toxic materials. Blood alcohol = 0.5 g/L. Blood: HepB (+), HepC (−), HIV (−) | DNT | Proteomics |
| Female | White American, 15 h gunshot wound to head. No drugs or alcohol. Blood: HepB (−), HepC (−), HIV (−) | DNT | Proteomics | ||
| Female | 34 | Black African American, gunshot wound to chest. Blood alcohol = 0.3 g/L; no drugs. Blood: HepB (+), HepC (−), HIV (−) | DNT | Proteomics | |
| Female | 40 | White American, multiple toxic compounds. Blood: HepB (+), HepC (+), HIV(−) | DNT | Proteomics |
DNT, did not test, which means hormone expressions were not tested in each control pituitary tissue with immunohistochemistry; ACTH, adrenocorticotropic hormone; hGH, human growth hormone; PRL, prolactin; FSH, follicle-stimulating hormone; LH, luteinizing hormone; TSH, thyrotropin-stimulating hormone or thyrotropin; −, negative; +, positive
Protein extraction and quantification
A volume (1 mL) of urea pyrolysis solution [9 M urea, 2.5 mM sodium pyrophosphate, 20 mM 2-hydroxyethyl (HEPES), 1 mM β-glycerophosphate, and 1 mM sodium orthovanadate, and pH 8.0] was added to each tissue sample (100 mg), which was treated with an ultrasonic ice bath (100 W, 10 s; interval 10 s; 10 times), and then followed by centrifugation (18,000×g, 10 min, 4 °). The supernatant was the sample of extracted proteins, whose protein content was measured with a Bradford protein quantification kit (YEASEN, Cat# 20202ES76).
Protein digestion with trypsin
Four control protein samples were equally mixed as control protein sample (1.5 mg/sample × 4 = 6 mg), and four NFPA protein samples were equally mixed as NFPA protein sample (1.5 mg/sample × 4 = 6 mg). An amount (300 μg) of each mixed sample (control; NFPA) was mixed with a volume of 1 M dithiothreitol (DTT) (the final DTT concentration was 10 mM) (control: n = 3; tumor: n = 3), followed by incubation (37 °C; 2.5 h) and cooling to room temperature, and then mixed with a volume of 1 M iodoacetamide (the final iodoacetamide concentration was 50 mM), followed by incubation (in the dark; 30 min). The urea concentration was diluted to 1.5 M with five volumes of water. Finally, the mixture was mixed with trypsin (2 μg/μL) at 1:50 (v:v) and then was digested (37 °C; 18 h), followed by desaltation and lyophilization using an SPE C18 column (Waters WAT051910).
TMT labeling of tryptic peptides and enrichment of phosphopeptides
For each sample (control and NFPA, respectively), a total of 300 μg tryptic peptides was equally divided into three parts (100 μg/part) (control: n = 3 parts; NFPA: n = 3 parts). The 6-plex TMT sixplexTM isobaric label reagent set (Thermo Scientific) was used to label those six parts of tryptic peptides, respectively. The six labeled tryptic peptide samples were equally mixed and lyophilized in vacuum. The lyophilized TMT-labeled peptide was resuspended in 1× DHB buffer that was mixed (1:4) with one volume of 5× DHB [3% DHB, 80% acetonitrile (ACN), and 0.1% trifluoroacetic acid (TFA)] and four volumes of water. The TiO2 beads were added to the resuspended TMT-labeled peptide mixture and shaken for 40 min, followed by centrifugation (5000×g, 1 min) to remove the supernatant. The TiO2 beads with phosphopeptides were washed with 50 μL of washing buffer I (30% ACN and 3% TFA) (3×) and then with 50 μL of washing buffer II (80% ACN and 0.3% TFA) (3×) to remove the remaining nonbinding peptides. The phosphopeptides were eluted with 50 μL of elution buffer (40% ACN and 15% NH3·H2O) (3×) and then lyophilized. The dried TiO2-enriched phosphopeptides were redissolved in a volume (30 μL) of 0.1% TFA for LC-MS/MS analysis.
LC-MS/MS of enriched phosphopeptides
A volume (20 μL) of TiO2-enriched phosphopeptides was analyzed with LC-MS/MS. First, phosphopeptides were separated with a high-performance liquid chromatography (HPLC) system EASY-nLC1000 at nanoliter flow rate. The enriched phosphopeptide sample was automatically loaded onto Thermo Scientific EASY column (2 cm × 100 μm; 5 μm C18) that was balanced in 95% of liquid A (0.1% formic acid aqueous solution), then the enriched phosphopeptides were separated by an analytical column (75 μm × 250 mm 3 μm C18 at a flow rate of 250 nL/min) with a linear gradient of solution B (0.1% formic acid in 84% ACN aqueous solution): from 0 to 55% during 0–220 min, 55 to 100% during 220–228 min, and maintaining at 100% during 228–240 min. The LC-separated peptides were online input into a Q-Exactive mass spectrometer (Thermo Finnigan) for MS/MS analysis. The MS parameters were set as follows: positive-ion scan mode, precursor ion scan range 350–1800 m/z with a resolution 70,000 at m/z 200, automatic gain control (AGC) target 3e6, maximum inject time 20 ms, number of scan ranges 1, and dynamic exclusion 30.0 s. For each MS scan, the 10 most abundant precursor ions were selected for MS/MS analysis. The MS/MS parameters were set as follows: high-energy collision dissociation (HCD) ion fragmentation, isolation window 2 m/z, resolution 17,500 at m/z 200, maximum injection time 60 ms, normalized collision energy 29 eV, and underfill ratio 0.1%. The MS/MS spectra were used to search the protein database (Uniprot_human_154578_20160815.fasta; 154,578 human sequences; downloaded on 15 August 2016) with MASCOT engine (Matrix Science, London, UK; version 2.2) embedded into Proteome Discoverer 1.4 (Thermo Scientific).
Database searching and functional characteristics of phosphoproteins
Phosphoproteins and phosphosites were identified with MS/MS data using the MASCOT software. The searching parameters were set as follows: MS/MS ion search, trypsin, 2 max missed cleavages, fixed modifications (carbamidomethyl at residue C, TMT 6 plex at the N-terminal, TMT 6 plex at residue K), variable modifications (oxidation at residue M, phosphorylation at residues S, T, and Y), ± 20 ppm for peptide mass tolerance, ± 0.1 Da for fragment mass tolerance, ESI-TRAP (electrospray ionization-ion trap) for instrument type, unrestricted protein mass, true for decoy database pattern, decoy for database pattern, and peptide FDR (false discovery rate) < 0.01. The MS/MS data were used to determine phosphoprotein amino acid sequences and phosphosites. The intensities of TMT-reporter ions were used to determine the phosphorylation level in NFPAs compared to controls. GO enrichments, including cellular components (CCs), molecular functions (MFs), and biological processes (BPs), were analyzed with Cytoscape ClueGO to reveal the functional characteristics of identified phosphoproteins. KEGG pathway enrichment analysis was used to obtain the statistically significant signaling pathways found on the basis of identified phosphoproteins. P value for GO enrichment analysis was obtained by two-sided hypergeometric test and corrected by Benjamini–Hochberg. P value for pathway enrichment analysis was obtained by two-sided hypergeometric test and corrected by Q value. The level of statistical significance was set as P < 0.05.
Transcriptomics data of invasive NFPAs relative to control pituitaries
The microarray gene data GSE51618 datasets of human pituitary adenomas were obtained from the public GEO database (http://www.ncbi.nlm.nih.gov/geo/) at the National Center for Biotechnology Information (NCBI). It contained 4 invasive NFPA tissue samples and 3 control pituitary tissue samples, which were analyzed with a gene chip human genome platform (Agilent-014850 Whole Human Genome Microarray 4x44K G4112F) in another laboratory. The R-software (The R Foundation for Statistical Computing, https://www.r-project.org/) was used to analyze DEGs between invasive NFPAs and controls. FDR < 0.05 and fold changes (FC) ≥ 2 were used to determine each DEG.
Overlapping analysis of phosphoprotein data and invasive DEG data
The gene name of each phosphoprotein was obtained from the UniProt human database. The overlapping analysis was carried out between the gene names of phosphoproteins in NFPAs and DEG data between invasive NFPAs and controls to obtain the overlapped molecules (invasive DEGs; phosphoproteins). DAVID GO and KEGG pathway enrichments were used to analyze those overlapped molecules, with statistically significant parameters (P < 0.05 and gene count > 3). Each P value was corrected with FDR for multiple testing.
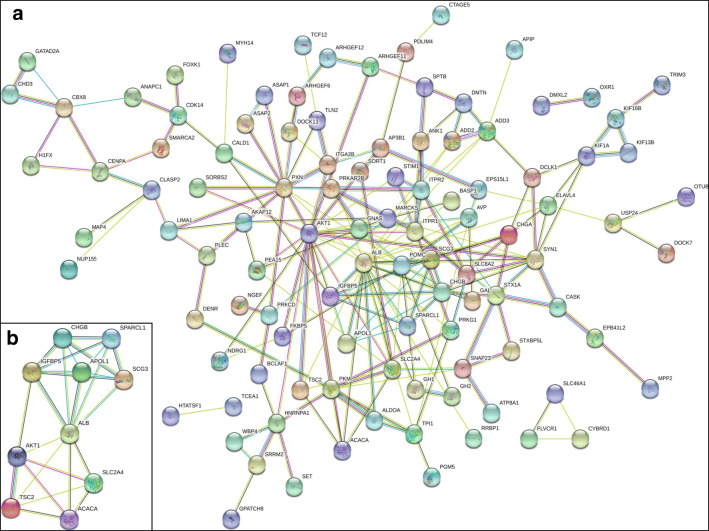
Protein–protein interaction and hub molecules with molecular complex detection based on overlapped molecules (invasive DEGs; phosphoproteins)
To investigate their interactive associations, all overlapped molecules between phosphoprotein data and invasive DEG data were mapped to the STRING database. The protein–protein interactions were analyzed by Cytoscape software (version 3.2.1; National Resource for Network Biology) to obtain the protein–protein interaction (PPI) network. The criteria of hub molecule searching were set as a molecular complex detection (MCODE) score > 5 and a statistically significant difference (P < 0.05).
Results
Phosphorylation profiling in NFPAs
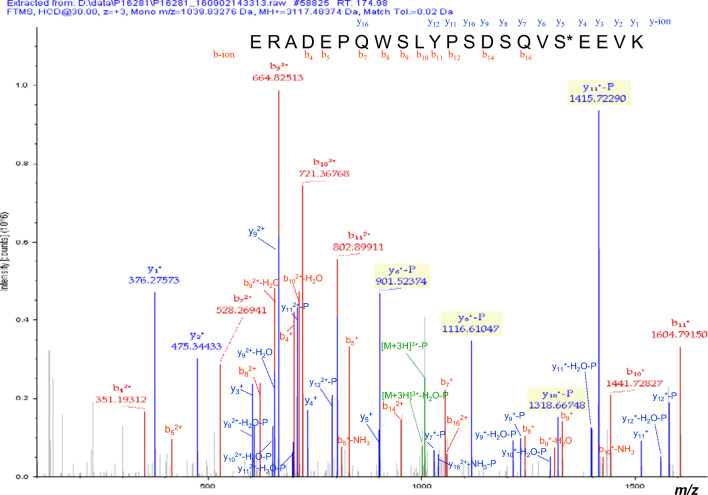
Totally, 2982 phosphorylation sites within 2076 phosphopeptides derived from 1035 phosphoproteins were identified with TMT–TiO2 enrichment–LC-MS/MS in NFPAs and controls, including 1207 (1207/2076 = 58%) quantified phosphopeptides with 1660 phosphorylation sites and 869 (869/2076 = 42%) qualified phosphopeptides with 1322 phosphorylation sites (Supplemental Table 1). A representative MS/MS spectrum was from the phosphorylated peptide 132ERADEPQWSLYPSDSQVS*EEVK153 ([M+3H]3+, m/z = 1039.83, S* = phosphorylated serine residue) derived from secretogranin-1 (Swiss-Prot No. P05060) (Fig. 2), with a high-quality MS/MS spectrum, excellent signal-to-noise (S/N) ratio, and extensive product-ion b-ion and y-ion series (b4, b5, b6, b7, b8, b9, b10, b11, y1, y2, y3, y4, y5, y6, y7, y8, y9, y10, y11, y12, and y16). The phosphorylation site was localized to amino acid residue Ser149, and the phosphorylation level was significantly decreased in NFPAs compared to controls (ratio of T/N = 0.29; P = 2.80E-05) (Supplemental Table 1). With the same method, each phosphopeptide and phosphorylation site was identified with MS/MS data (Supplemental Table 1). Among 2076 identified phosphopeptides, 1362 (1362/2076 = 66%) phosphopeptides only contained one single phosphorylation site, and 714 (714/2076 = 34%) phosphopeptides contained at least two phosphorylation sites. Among 2982 identified phosphorylation sites, 2591 (2591/2982 = 86.89%) phosphorylation sites occurred on residue Ser, 357 (357/2982 = 11.97%) phosphorylation sites on residue Thr, and 34 (34/2982 = 1.14%) phosphorylation sites on residue Tyr. Among 1035 phosphoproteins, 486 phosphoproteins were identified with only one phosphorylation site, 242 phosphoproteins with 2 phosphorylation sites, 133 phosphoproteins with 3 phosphorylation sites, 61 phosphoproteins with 4 phosphorylation sites, 39 phosphoproteins with 5 phosphorylation sites, and 74 phosphoproteins with over 5 phosphorylation sites (Supplemental Table 1), including CIC (n = 6 sites), C2CD2L (n = 6), ARHGEF11 (n = 6), EPB41L2 (n = 6), PHF2 (n = 6), POMC (n = 6), VIM (n = 6), NEFH (n = 6), BRAF (n = 6), HTT (n = 6), MECP2 (n = 6), MSH6 (n = 6), SRSF2 (n = 6), TJP1 (n = 6), EPB49 (n = 6), TCOF1 (n = 6), SAFB (n = 6), SMN1 (n = 6), CTR9 (n = 6), KIF21A (n = 6), ZC3H18 (n = 6), RSF1 (n = 6), IRF2BPL (n = 6), THRAP3 (n = 6), CTAGE5 (n = 7), EIF5B (n = 7), MAP4 (n = 7), SUB1 (n = 7), MFAP1 (n = 7), HNRNPUL2 (n = 7), AAK1 (n = 7), ZC3H13 (n = 7), TNKS1BP1 (n = 7), HNRNPC (n = 8), PRPF4B (n = 8), SMARCC2 (n = 8), LMNA (n = 8), PML (n = 8), ADD1 (n = 8), ADD2 (n = 8), PSIP1 (n = 9), ANK1 (n = 9), ANK2 (n = 9), TOP2B (n = 9), TRIM2 (n = 9), TGOLN2 (n = 10), AHNAK (n = 10), SPARCL1 (n = 10), MARCKS (n = 11), HIRIP3 (n = 11), SCG2 (n = 12), LEO1 (n = 12), ACIN1 (n = 12), SPTBN1 (n = 13), FAM169A (n = 13), ATRX (n = 14), AKAP12 (n = 14), EPB41L3 (n = 14), MAPT (n = 15), NEFM (n = 15), MAP2 (n = 15), BCLAF1 (n = 15), HTATSF1 (n = 17), CHGA (n = 19), NUCKS1 (n = 22), IWS1 (n = 23), MAP1A (n = 24), TP53BP1 (n = 26), FGA (n = 35), SRRM1 (n = 42), MAP1B (n = 42), CHGB (n = 50), and SRRM2 (n = 76). These highly phosphorylated proteins might play important roles in the NFPA pathogenesis.
Fig. 2.
A representative MS/MS spectrum of phosphopeptide derived from NFPAs. The identified phosphopeptide 132ERADEPQWSLYPSDSQVS*EEVK153 ([M+3H]3+, m/z = 1039.83, S* = phosphorylated serine residue) was derived from secretogranin-1 (Swiss-Prot No. P05060)
Functional characteristics of phosphoproteins
Those identified 1035 phosphoproteins between NFPAs and controls were analyzed with GO enrichment analysis, including CCs (Supplemental Table 2), MFs (Supplemental Table 3), and BPs (Supplemental Table 4).
CC analysis revealed those phosphoproteins were mainly distributed in the nucleoplasm, cytosol, cell–cell adherens junction, cytoplasm, nuclear speck, nucleus, membrane, focal adhesion, cytoskeleton, protein complex, nuclear matrix, spliceosomal complex, perinuclear region of cytoplasm, Golgi apparatus, chromatin, intracellular ribonucleoprotein complex, Z disc, extracellular exosome, actomyosin, secretory granule, postsynaptic density, costamere, spectrin, microtubule, cell cortex, nuclear heterochromatin, spindle, synaptic vesicle, nuclear membrane, microtubule-associated complex, T-tubule, chromosome, transport vesicle, nuclear envelope, transcriptional repressor complex, PcG protein complex, npBAF complex, Sin3 complex, cAMP-dependent protein kinase complex, soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex, and centrosome (Supplemental Table 2).
MF analysis revealed that the identified phosphoproteins were mainly involved in RNA binding, protein binding, cadherin binding involved in cell–cell adhesion, nucleotide binding, actin filament binding, structural constituent of cytoskeleton, chromatin binding, calmodulin binding, structural molecule activity, microtubule binding, mRNA binding, actin binding, protein kinase binding, 14-3-3 protein binding, spectrin binding, vinculin binding, nucleosomal DNA binding, protein domain–specific binding, translation initiation factor activity, helicase activity, protein binding bridging, protein serine/threonine kinase activity, ion channel binding, histone deacetylase binding, kinase activity, intramolecular transferase activity phosphotransferases, and GTPase activator activity (Supplemental Table 3).
BP analysis revealed that those identified phosphoproteins were mainly involved in multiple biological processes, including cell–cell adhesion, mRNA splicing via spliceosome, RNA splicing, mRNA processing, viral process, RNA processing, mRNA export from nucleus, RNA export from nucleus, microtubule cytoskeleton organization, mRNA 3′-end processing, regulation of alternative mRNA splicing via spliceosome, termination of RNA polymerase II transcription, regulation of cellular response to heat, negative regulation of mRNA splicing via spliceosome, ER to Golgi vesicle-mediated transport, protein sumoylation, membrane fusion, ATP-dependent chromatin remodeling, regulation of mRNA stability, chromatin remodeling, platelet aggregation, regulation of translational initiation, positive regulation of axon extension, negative regulation of transcription DNA-templated, protein phosphorylation, IRE1-mediated unfolded protein response, covalent chromatin modification, platelet activation, and actomyosin structure organization (Supplemental Table 4).
KEGG pathway network analysis identified 31 statistically significant signaling pathways involved in phosphoproteins (Table 2), including spliceosome, platelet activation, RNA transport, endocytosis, mTOR signaling pathway, vascular smooth muscle contraction, SNARE interactions in vesicular transport, proteoglycans in cancer, insulin signaling pathway, glucagon signaling pathway, cGMP–PKG signaling pathway, focal adhesion, estrogen signaling pathway, progesterone-mediated oocyte maturation, protein processing in endoplasmic reticulum, gap junction, gonadotropin-releasing hormone (GnRH) signaling pathway, mitogen-activated protein kinase (MAPK) signaling pathway, and mRNA surveillance pathway.
Table 2.
Statistically significant KEGG signaling pathways identified from 1035 phosphoproteins in NFPAs. Count means the number of genes enriched in this pathway
| Category | Pathway name | Count | % | Fold enrichment | P value | Benjamini | Gene name of phosphoprotein |
|---|---|---|---|---|---|---|---|
| KEGG_PATHWAY | hsa03040:Spliceosome | 32 | 3.41 | 4.57 | 8.32E-13 | 1.83E-10 | SRSF1, NCBP1, CHERP, SRSF10, TRA2A, WBP11, CTNNBL1, SF3B1, DDX23, RBM8A, PCBP1, USP39, ACIN1, HNRNPC, PRPF40B, RBM25, PRPF40A, DDX42, SNW1, PRPF3, HNRNPA1, SF3A1, HNRNPU, SRSF2, SRSF7, SRSF6, SRSF9, SNRNP200, SLU7, PRPF38B, THOC1, RBM17 |
| KEGG_PATHWAY | hsa04611:Platelet activation | 22 | 2.35 | 3.22 | 3.32E-06 | 3.65E-04 | TLN1, LYN, TLN2, STIM1, ARHGAP35, MYL12B, PRKG1, ARHGEF12, ITPR1, ITPR2, AKT1, MAPK1, FGA, FGB, GP1BB, MAPK3, PPP1R12A, GP1BA, GNAS, PRKACB, SNAP23, ITGA2B |
| KEGG_PATHWAY | hsa03013:RNA transport | 24 | 2.56 | 2.65 | 2.89E-05 | 2.12E-03 | CLNS1A, NCBP1, NUP98, EIF5B, RNPS1, NUP155, SMN1, PNN, PABPC4L, EIF4B, EIF4G1, EIF3CL, EIF4G2, EIF4G3, NUP214, EIF3B, EIF3G, RAE1, RBM8A, SRRM1, ACIN1, RANBP2, THOC1, EIF2B5 |
| KEGG_PATHWAY | hsa04144:Endocytosis | 28 | 2.99 | 2.21 | 1.40E-04 | 7.70E-03 | CHMP2A, USP8, ASAP2, PML, ASAP1, EPS15L1, ARFGEF2, CHMP2B, GBF1, VPS4B, DNAJC6, VPS4A, VPS35, EHD2, IQSEC1, GIT1, PARD6A, DNM3, FAM21A, EPS15, RABEP1, ARRB1, IGF2R, ACAP2, SH3KBP1, SNX12, BIN1, DNM1 |
| KEGG_PATHWAY | hsa04150:mTOR signaling pathway | 12 | 1.28 | 3.93 | 1.75E-04 | 7.67E-03 | PRKCA, EIF4B, AKT1, MAPK1, RPS6KA3, AKT1S1, BRAF, TSC2, MAPK3, PRKAA1, RRAGD, RPTOR |
| KEGG_PATHWAY | hsa04270:Vascular smooth muscle contraction | 16 | 1.71 | 2.60 | 1.11E-03 | 3.98E-02 | PRKCA, BRAF, CALD1, MRVI1, PRKCE, ARHGEF12, PRKG1, PRKCD, ITPR1, ITPR2, ARHGEF11, MAPK1, MAPK3, PPP1R12A, GNAS, PRKACB |
| KEGG_PATHWAY | hsa04130:SNARE interactions in vesicular transport | 8 | 0.85 | 4.47 | 1.62E-03 | 4.98E-02 | STX1A, STX4, SEC22B, VAMP4, BET1L, SNAP23, VAMP2, STX1B |
| KEGG_PATHWAY | hsa05205:Proteoglycans in cancer | 22 | 2.35 | 2.09 | 1.77E-03 | 4.75E-02 | PRKCA, BRAF, ARHGEF12, PDCD4, FLNA, PXN, ITPR1, ITPR2, AKT1, EIF4B, MAPK1, CTTN, ANK1, ANK2, ANK3, SOS1, MAPK3, PPP1R12A, CAMK2D, PRKACB, SLC9A1, FN1 |
| KEGG_PATHWAY | hsa04910:Insulin signaling pathway | 17 | 1.81 | 2.34 | 2.24E-03 | 5.34E-02 | PHKA2, IRS2, BRAF, PHKB, ACACA, RPTOR, AKT1, PRKAR2B, MAPK1, PRKAR2A, SLC2A4, SOS1, PRKAR1A, MAPK3, TSC2, PRKAA1, PRKACB |
| KEGG_PATHWAY | hsa04666:Fc gamma R-mediated phagocytosis | 12 | 1.28 | 2.71 | 4.21E-03 | 8.86E-02 | PRKCA, AKT1, MAPK1, LYN, MARCKSL1, MAPK3, ASAP2, ASAP1, MARCKS, PRKCE, BIN1, PRKCD |
| KEGG_PATHWAY | hsa04114:Oocyte meiosis | 14 | 1.49 | 2.40 | 5.18E-03 | 9.86E-02 | ANAPC1, YWHAZ, CDC23, YWHAE, SMC3, ITPR1, ITPR2, MAPK1, RPS6KA3, SLK, MAPK3, CAMK2D, PPP3CB, PRKACB |
| KEGG_PATHWAY | hsa04922:Glucagon signaling pathway | 13 | 1.39 | 2.50 | 5.39E-03 | 9.44E-02 | PHKA2, PHKB, PGAM1, ACACA, ITPR1, ITPR2, AKT1, SLC2A1, PPP3CB, CAMK2D, GNAS, PRKAA1, PRKACB |
| KEGG_PATHWAY | hsa04720:Long-term potentiation | 10 | 1.07 | 2.88 | 7.12E-03 | 1.14E-01 | PRKCA, MAPK1, RPS6KA3, BRAF, MAPK3, PPP3CB, CAMK2D, PRKACB, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04962:Vasopressin-regulated water reabsorption | 8 | 0.85 | 3.46 | 7.37E-03 | 1.10E-01 | DYNC1LI1, STX4, DYNC1LI2, AVP, GNAS, PRKACB, VAMP2, DYNC1I2 |
| KEGG_PATHWAY | hsa04022:cGMP–PKG signaling pathway | 17 | 1.81 | 2.04 | 8.48E-03 | 1.17E-01 | MEF2C, IRS2, SLC8A2, MRVI1, PRKCE, VDAC2, PRKG1, ITPR1, ITPR2, VDAC1, ATP2B1, AKT1, MAPK1, ATP2B4, MAPK3, PPP1R12A, PPP3CB |
| KEGG_PATHWAY | hsa04510:Focal adhesion | 20 | 2.13 | 1.84 | 1.15E-02 | 1.47E-01 | PRKCA, TLN1, BRAF, TLN2, TNC, ARHGAP35, MYL12B, PXN, FLNA, VCL, AKT1, MAPK1, ARHGAP5, PAK2, SOS1, MAPK3, PPP1R12A, SPP1, FN1, ITGA2B |
| KEGG_PATHWAY | hsa04730:Long-term depression | 9 | 0.96 | 2.85 | 1.25E-02 | 1.50E-01 | PRKCA, MAPK1, BRAF, LYN, MAPK3, GNAS, PRKG1, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04970:Salivary secretion | 11 | 1.17 | 2.43 | 1.41E-02 | 1.59E-01 | PRKCA, ATP2B1, ATP2B4, SLC12A2, GNAS, PRKACB, VAMP2, PRKG1, ITPR1, SLC9A1, ITPR2 |
| KEGG_PATHWAY | hsa04915:Estrogen signaling pathway | 12 | 1.28 | 2.30 | 1.42E-02 | 1.53E-01 | HSP90AB1, AKT1, MAPK1, HSP90AA1, FKBP5, SOS1, MAPK3, GNAS, PRKACB, PRKCD, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04530:Tight junction | 11 | 1.17 | 2.40 | 1.52E-02 | 1.55E-01 | PARD6A, TJP1, CTTN, MYH11, MYL12B, MYH14, PRKCE, MYH9, TJP2, TJAP1, MYH10 |
| KEGG_PATHWAY | hsa04914:Progesterone-mediated oocyte maturation | 11 | 1.17 | 2.40 | 1.52E-02 | 1.55E-01 | HSP90AB1, ANAPC1, AKT1, MAPK1, CCNB3, RPS6KA3, HSP90AA1, BRAF, MAPK3, CDC23, PRKACB |
| KEGG_PATHWAY | hsa04141:Protein processing in endoplasmic reticulum | 17 | 1.81 | 1.91 | 1.56E-02 | 1.52E-01 | HSP90AB1, SEC31A, HSP90AA1, WFS1, RRBP1, NSFL1C, PDIA6, SEC62, CANX, STUB1, STT3B, HYOU1, DNAJB11, SIL1, DNAJC1, UBE2E2, SSR3 |
| KEGG_PATHWAY | hsa04540:Gap junction | 11 | 1.17 | 2.38 | 1.64E-02 | 1.52E-01 | PRKCA, MAPK1, TJP1, SOS1, MAPK3, GNAS, PRKACB, PRKG1, TUBA1B, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04919:Thyroid hormone signaling pathway | 13 | 1.39 | 2.15 | 1.70E-02 | 1.52E-01 | PRKCA, AKT1, MAPK1, NCOA2, HDAC2, HDAC1, TSC2, MAPK3, SLC2A1, RCAN1, PRKACB, NCOR1, SLC9A1 |
| KEGG_PATHWAY | hsa05100:Bacterial invasion of epithelial cells | 10 | 1.07 | 2.44 | 2.03E-02 | 1.71E-01 | DNM3, CTTN, SEPT2, ARHGEF26, CTNNA1, DNM1, PXN, CTNNA2, FN1, VCL |
| KEGG_PATHWAY | hsa04912:GnRH signaling pathway | 11 | 1.17 | 2.30 | 2.03E-02 | 1.65E-01 | PRKCA, MAPK1, MAP3K3, SOS1, MAPK3, CAMK2D, GNAS, PRKACB, PRKCD, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa05132:Salmonella infection | 10 | 1.07 | 2.29 | 2.91E-02 | 2.21E-01 | MAPK1, DYNC1LI1, TJP1, DYNC1LI2, MAPK3, PKN2, KLC4, KLC2, FLNA, DYNC1I2 |
| KEGG_PATHWAY | hsa04010:MAPK signaling pathway | 21 | 2.24 | 1.58 | 4.26E-02 | 2.99E-01 | PRKCA, MEF2C, BRAF, TAOK2, TAOK1, GNG12, FLNA, AKT1, MAPK1, RPS6KA3, PAK2, MAP3K3, ARRB1, SOS1, MAPT, MAPK3, PPP3CB, HSPB1, MAPK8IP3, STMN1, PRKACB |
| KEGG_PATHWAY | hsa04068:FoxO signaling pathway | 13 | 1.39 | 1.84 | 4.82E-02 | 3.22E-01 | USP7, IRS2, SGK3, BRAF, RBL2, AKT1, MAPK1, CCNB3, SLC2A4, HOMER3, SOS1, MAPK3, PRKAA1 |
| KEGG_PATHWAY | hsa03015:mRNA surveillance pathway | 10 | 1.07 | 2.09 | 4.84E-02 | 3.14E-01 | PABPC4L, NCBP1, FIP1L1, CSTF3, RBM8A, SRRM1, RNPS1, ACIN1, CPSF2, PNN |
| KEGG_PATHWAY | hsa04810:Regulation of actin cytoskeleton | 18 | 1.92 | 1.63 | 4.90E-02 | 3.08E-01 | GIT1, BRAF, ARHGEF6, ARHGAP35, MYL12B, GNG12, ARHGEF12, PXN, VCL, MAPK1, PAK2, SOS1, MAPK3, PIKFYVE, PPP1R12A, SLC9A1, FN1, ITGA2B |
Further, functional clustering analysis based on CCs, MFs, BPs, and KEGG pathways grouped those identified phosphoproteins into 8 functional clusters (Table 3). Cluster 1 mainly functions in cell–cell adherens junction, cadherin binding involved in cell–cell adhesion, and cell–cell adhesion. Cluster 2 mainly functions in SNARE interactions in vesicular transport, SNAP (soluble N-ethylmaleimide-sensitive fusion attachment proteins) receptor activity, and SNARE complex. Cluster 3 mainly functions in histone H2B ubiquitination, histone monoubiquitination, endodermal cell fate commitment, and Cdc73/Paf1 complex. Cluster 4 mainly functions in myosin II complex and myosin II filament. Cluster 5 mainly functions in oxygen transporter activity, oxygen transport, haptoglobin binding, haptoglobin–hemoglobin complex, and hemoglobin complex. Cluster 6 functions in cAMP-dependent protein kinase complex, protein kinase A catalytic subunit binding, and cAMP-dependent protein kinase regulator activity. Cluster 7 functions in ESCRT (endosomal sorting complex required for transport) III complex disassembly and positive regulation of viral release from host cell. Cluster 8 functions in eukaryotic 43S preinitiation complex. Those findings clearly demonstrate that protein phosphorylations are extensively involved in multiple biological processes and molecular functions in NFPAs.
Table 3.
Functional characteristics of 1035 phosphoproteins in NFPAs, clustered with GO and KEGG pathway enrichments. Count means the number of genes enriched in each item
| Category | Function characteristics | Count | % | Fold enrichment | P value | Gene name of phosphoprotein |
|---|---|---|---|---|---|---|
| Annotation cluster 1 | ||||||
| GOTERM_CC_DIRECT | GO:0005913~cell–cell adherens junction | 71 | 7.57 | 4.44 | 2.23E-26 | CAST, HSP90AB1, LIMA1, TLN1, SEPT2, ZC3HAV1, VAPB, H1FX, EIF2A, EPS15L1, VCL, LARP1, BZW1, CTTN, PAK2, SLK, PCBP1, ARHGAP1, FAM129B, AHNAK, GOLGA3, DAB2IP, BSG, LYN, PKN2, KTN1, MYH9, CTNNA1, FLNA, CTNNA2, RSL1D1, EIF4G1, EIF4G2, EPB41L1, PGM5, DHX29, SERBP1, USO1, EEF1D, CD226, DBN1, ADD1, ALDOA, HDLBP, YWHAZ, USP8, PPME1, CALD1, ASAP1, CTNND1, KLC2, ESYT2, TAGLN2, SCRIB, CHMP2B, MACF1, NDRG1, PLEC, ZC3H15, YWHAE, MPRIP, TNKS1BP1, EPS15, TJP1, LASP1, TMOD3, SPTBN1, NOP56, TMPO, TJP2, SPTAN1 |
| GOTERM_MF_DIRECT | GO:0098641~cadherin binding involved in cell–cell adhesion | 68 | 7.25 | 4.49 | 9.96E-26 | CAST, HSP90AB1, LIMA1, TLN1, SEPT2, ZC3HAV1, VAPB, H1FX, EIF2A, EPS15L1, VCL, LARP1, BZW1, CTTN, PAK2, SLK, PCBP1, ARHGAP1, FAM129B, AHNAK, GOLGA3, DAB2IP, BSG, PKN2, KTN1, MYH9, CTNNA1, FLNA, CTNNA2, RSL1D1, EIF4G1, EIF4G2, EPB41L1, DHX29, SERBP1, USO1, EEF1D, DBN1, ADD1, ALDOA, YWHAZ, HDLBP, USP8, PPME1, CALD1, ASAP1, CTNND1, KLC2, ESYT2, TAGLN2, SCRIB, CHMP2B, MACF1, NDRG1, PLEC, ZC3H15, YWHAE, MPRIP, EPS15, TNKS1BP1, TJP1, LASP1, TMOD3, SPTBN1, NOP56, TMPO, TJP2, SPTAN1 |
| GOTERM_BP_DIRECT | GO:0098609~cell–cell adhesion | 60 | 6.40 | 4.33 | 6.80E-22 | CAST, HSP90AB1, LIMA1, SEPT2, ZC3HAV1, VAPB, H1FX, EPS15L1, EIF2A, LARP1, BZW1, CTTN, SLK, PAK2, PCBP1, ARHGAP1, FAM129B, AHNAK, GOLGA3, DAB2IP, BSG, PKN2, KTN1, RSL1D1, EIF4G1, EIF4G2, EPB41L1, DHX29, SERBP1, USO1, EEF1D, DBN1, ADD1, ALDOA, YWHAZ, HDLBP, USP8, PPME1, CALD1, ASAP1, KLC2, TAGLN2, ESYT2, CHMP2B, MACF1, NDRG1, PLEC, ZC3H15, YWHAE, MPRIP, EPS15, TNKS1BP1, TJP1, LASP1, TMOD3, SPTBN1, TMPO, NOP56, TJP2, SPTAN1 |
| Annotation cluster 2 | ||||||
| KEGG_PATHWAY | hsa04130:SNARE interactions in vesicular transport | 8 | 0.85 | 4.47 | 1.62E-03 | STX1A, STX4, SEC22B, VAMP4, BET1L, SNAP23, VAMP2, STX1B |
| GOTERM_MF_DIRECT | GO:0005484~SNAP receptor activity | 8 | 0.85 | 3.93 | 3.66E-03 | STX1A, STX4, SEC22B, VAMP4, BET1L, SNAP23, VAMP2, STX1B |
| GOTERM_CC_DIRECT | GO:0031201~SNARE complex | 9 | 0.96 | 3.43 | 4.28E-03 | STX1A, STX4, SEC22B, VAMP4, BET1L, SNAP23, VAMP2, STX1B, STXBP5L |
| Annotation cluster 3 | ||||||
| GOTERM_BP_DIRECT | GO:0033523~histone H2B ubiquitination | 4 | 0.43 | 9.79 | 6.11E-03 | LEO1, PAF1, RNF20, CTR9 |
| GOTERM_BP_DIRECT | GO:0010390~histone monoubiquitination | 4 | 0.43 | 7.12 | 1.61E-02 | LEO1, PAF1, RNF20, CTR9 |
| GOTERM_BP_DIRECT | GO:0001711~endodermal cell fate commitment | 3 | 0.32 | 9.79 | 3.40E-02 | LEO1, PAF1, CTR9 |
| GOTERM_CC_DIRECT | GO:0016593~Cdc73/Paf1 complex | 3 | 0.32 | 8.65 | 4.35E-02 | LEO1, PAF1, CTR9 |
| Annotation cluster 4 | ||||||
| GOTERM_CC_DIRECT | GO:0016460~myosin II complex | 4 | 0.43 | 11.53 | 3.64E-03 | MYL12B, MYH14, MYH9, MYH10 |
| GOTERM_CC_DIRECT | GO:0097513~myosin II filament | 3 | 0.32 | 20.18 | 7.10E-03 | MYH14, MYH9, MYH10 |
| Annotation cluster 5 | ||||||
| GOTERM_MF_DIRECT | GO:0005344~oxygen transporter activity | 5 | 0.53 | 6.84 | 4.84E-03 | IPCEF1, HBA2, HBA1, HBB, HBD |
| GOTERM_BP_DIRECT | GO:0015671~oxygen transport | 5 | 0.53 | 6.52 | 5.85E-03 | IPCEF1, HBA2, HBA1, HBB, HBD |
| GOTERM_MF_DIRECT | GO:0031720~haptoglobin binding | 3 | 0.32 | 19.14 | 7.88E-03 | HBA2, HBA1, HBB |
| GOTERM_CC_DIRECT | GO:0031838~haptoglobin–hemoglobin complex | 3 | 0.32 | 15.14 | 1.37E-02 | HBA2, HBA1, HBB |
| GOTERM_CC_DIRECT | GO:0005833~hemoglobin complex | 4 | 0.43 | 6.73 | 1.90E-02 | HBA2, HBA1, HBB, HBD |
| Annotation cluster 6 | ||||||
| GOTERM_CC_DIRECT | GO:0005952~cAMP-dependent protein kinase complex | 4 | 0.43 | 11.53 | 3.64E-03 | PRKAR2B, PRKAR2A, PRKAR1A, PRKACB |
| GOTERM_MF_DIRECT | GO:0034236~protein kinase A catalytic subunit binding | 4 | 0.43 | 5.47 | 3.35E-02 | PRKAR2B, PRKAR2A, GSK3A, PRKAR1A |
| GOTERM_MF_DIRECT | GO:0008603~cAMP-dependent protein kinase regulator activity | 3 | 0.32 | 8.20 | 4.80E-02 | PRKAR2B, PRKAR2A, PRKAR1A |
| Annotation cluster 7 | ||||||
| GOTERM_BP_DIRECT | GO:1904903~ESCRT III complex disassembly | 4 | 0.43 | 7.83 | 1.21E-02 | CHMP2A, VPS4B, VPS4A, CHMP2B |
| GOTERM_BP_DIRECT | GO:1902188~positive regulation of viral release from host cell | 4 | 0.43 | 7.12 | 1.61E-02 | CHMP2A, VPS4B, VPS4A, CHMP2B |
| Annotation cluster 8 | ||||||
| GOTERM_CC_DIRECT | GO:0016282~eukaryotic 43S preinitiation complex | 4 | 0.43 | 5.38 | 3.52E-02 | EIF3CL, EIF3B, EIF3G, DHX29 |
Integration of phosphoproteomics data and transcriptomics data in invasive NFPAs relative to controls
Microarray transcriptomic data between invasive NFPAs and controls were obtained from the GEO database, which identified 2751 DEGs, consisting of 1477 (53.69%) downregulated and 1274 (46.31%) upregulated DEGs (Supplemental Table 5). An overlapping analysis was performed between 1035 phosphoproteins identified in NFPAs relative to controls and 2751 DEGs identified in invasive NFPAs relative to controls, which identified 130 overlapped molecules (invasive DEGs; phosphoproteins) (Table 4). Those 130 invasiveness-related molecules (DEGs, phosphoproteins) were the precious resource to identify phosphorylation-mediated invasive characteristics in NFPAs for potential prognostic/predictive assessment, patient stratification, and personalized treatment of NFPA patients. Further, the functional characteristics of those 130 overlapped molecules (phosphoproteins; invasive DEGs) were revealed with GO and clustering enrichment analyses.
Table 4.
A total of 130 overlapped molecules between 1035 phosphoproteins and 2751 invasive DEGs
| DEG name | logFC | AveExpr | t | P value | Adjusted P value | B | Protein accession ID | Protein name | Phosphopeptide | Phosphorylated amino acid residue | Phosphorylated position | Phosphorylated probabilities | PEP | Score | Phosphorylated level (N) | Phosphorylated level (T) | Ratio (T/N) | P value (t test) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABCA2 | 1.9524 | 9.4927 | 8.1782 | 1.28E-05 | 1.26E-03 | 3.7214 | Q9BZC7 | ATP-binding cassette subfamily A member 2 | VSEEDQS*LENS*EADVK | S; S | 1327; 1331 | 1; 1 | 3.62E-06 | 92 | 10,518 | 32,589 | 3.10 | 1.60E-04 |
| ACACA | 1.1822 | 6.8754 | 3.3664 | 7.62E-03 | 4.22E-02 | − 2.8421 | Q13085 | Acetyl-CoA carboxylase 1 | FIIGSVSEDNS*EDEISNLVK | S | 29 | 1 | 3.35E-32 | 154 | 2551 | 7604 | 2.98 | 6.08E-05 |
| FIIGSVS*EDNS*EDEISNLVK | S; S | 25; 29 | 0.82; 0.892 | 7.89E-07 | 54 | |||||||||||||
| ADD2 | 1.6533 | 6.9033 | 5.2921 | 4.08E-04 | 7.59E-03 | 0.1686 | P35612 | Beta-adducin | EAETKS*PLVS*PSK | S | 613; 617 | 0.885; 0.981 | 1.79E-04 | 88 | 11,989 | 36,075 | 3.01 | 1.28E-05 |
| EAETKS*PLVS*PSK | S | 613; 617 | 0.877; 1 | 1.79E-04 | 88 | 11,989 | 36,075 | 3.01 | 1.28E-05 | |||||||||
| KLELDGEKETAPEEPGS*PAK | S | 592 | 1 | 8.36E-06 | 84 | 14,454 | 20,046 | 1.39 | 9.74E-04 | |||||||||
| S*APAS*PVQSPAK | S; S | 596; 600 | 1; 1 | 8.36E-06 | 70 | 1895 | 3649 | 1.93 | 1.08E-03 | |||||||||
| EAET*KSPLVS*PSK | T | 611 | 0.998 | 1.24E-02 | 52 | |||||||||||||
| SAPAS*PVQS*PAK | S; S | 599; 604 | 0.998; 1 | 4.07E-03 | 94 | |||||||||||||
| ADD3 | 1.0662 | 11.6287 | 4.4111 | 1.46E-03 | 1.57E-02 | − 1.1500 | Q9UEY8 | Gamma-adducin | IEEVLSPEGS*PSKS*PS*K | S; S; S | 677; 681; 683 | 0.993; 0.498; 0.498 | 4.80E-10 | 105 | 17,730 | 41,658 | 2.35 | 1.10E-04 |
| IEEVLSPEGS*PS*KSPSK | S; S | 677; 679 | 0.906; 0.548 | 6.72E-04 | 69 | 10,380 | 17,649 | 1.70 | 1.34E-04 | |||||||||
| IEEVLS*PEGS*PSK | S; S | 673; 677 | 1; 0.986 | 3.87E-04 | 72 | |||||||||||||
| IEEVLSPEGS*PSKS*PS*K | S; S; S | 677; 681; 683 | 0.993; 0.498; 0.498 | 4.80E-10 | 105 | |||||||||||||
| IEEVLSPEGS*PSKS*PSK | S | 677; 681 | 0.522; 0.761 | 4.80E-10 | 105 | |||||||||||||
| AKAP12 | − 4.1353 | 10.1654 | − 4.9159 | 6.93E-04 | 1.03E-02 | − 0.3808 | Q02952 | A-kinase anchor protein 12 | GLAEVQQDGEAEEGAT*SDGEK | T | 597 | 0.932 | 7.78E-31 | 146 | 4946 | 11,387 | 2.30 | 9.59E-05 |
| SPPS*PVER | S | 1331 | 0.999 | 4.18E-03 | 91 | 6418 | 4875 | 0.76 | 2.79E-04 | |||||||||
| ETCVSGEDPTQGADLS*PDEK | S | 483 | 1 | 1.91E-22 | 135 | 25,062 | 14,371 | 0.57 | 4.98E-04 | |||||||||
| GLAEVQQDGEAEEGATS*DGEK | S | 598 | 0.847 | 7.54E-39 | 156 | 15,714 | 22,991 | 1.46 | 1.77E-03 | |||||||||
| ADS*QDAGQETEK | S | 1587 | 1 | 1.69E-145 | 133 | 4482 | 8736 | 1.95 | 2.36E-03 | |||||||||
| RGS*S*S*DEEGGPK | S; S; S | 696; 697; 698 | 1; 1; 1 | 1.42E-03 | 87 | 52,712 | 37,364 | 0.71 | 3.64E-03 | |||||||||
| EDEKGDDVDDPENQNSALADT*DASGGLTKESPDTNGPK | T; S | 1717; 1720 | 0.221; 0.221 | 7.03E-11 | 68 | |||||||||||||
| EVSSLEGS*PPPCLGQEEAVCTK | S | 1395 | 0.927 | 1.21E-12 | 58 | |||||||||||||
| SAESPT*S*PVTSETGSTFK | T; S | 285; 286 | 0.749; 0.749 | 3.61E-04 | 50 | |||||||||||||
| VVGQTT*PESFEK | T | 1116 | 0.956 | 1.48E-02 | 84 | |||||||||||||
| AKT1 | 1.1801 | 6.8509 | 5.4900 | 3.11E-04 | 6.55E-03 | 0.4493 | P31749 | RAC-alpha serine/threonine-protein kinase | S*GSPSDNSGAEEMEVSLAKPK | S; S | 122; 124 | 0.457; 0.457 | 1.16E-07 | 92 | ||||
| ALB | 1.9536 | 6.1289 | 4.5055 | 1.27E-03 | 1.45E-02 | − 1.0035 | P02768-1 | Serum albumin | TCVADES*AENCDK | S | 82 | 1 | 1.24E-47 | 197 | 114,813 | 84,746 | 0.74 | 3.24E-04 |
| ALDOA | 1.2343 | 14.3796 | 5.4707 | 3.19E-04 | 6.64E-03 | 0.4222 | P04075 | Fructose-bisphosphate aldolase A | GILAADES*TGSIAK | S | 36 | 0.97 | 2.14E-30 | 166 | ||||
| GILAADEST*GSIAK | T | 37 | 0.555 | 7.28E-05 | 95 | |||||||||||||
| GILAADESTGS*IAK | S | 39 | 0.983 | 1.65E-30 | 168 | |||||||||||||
| LQS*IGTENTEENR | S | 46 | 0.994 | 1.17E-03 | 66 | |||||||||||||
| ANAPC1 | 1.1050 | 8.8290 | 4.9940 | 6.20E-04 | 9.67E-03 | − 0.2650 | Q9H1A4 | Anaphase-promoting complex subunit 1 | NFDFEGSLS*PVIAPK | S | 688 | 0.839 | 1.86E-02 | 40 | ||||
| ANK1 | 2.2500 | 8.5296 | 3.7930 | 3.82E-03 | 2.75E-02 | − 2.1377 | P16157 | Ankyrin-1 | LGYIS*VTDVLK | S | 781 | 1 | 4.25E-15 | 146 | 47,985 | 171,943 | 3.58 | 4.13E-06 |
| ELQFS*VEDINR | S | 1428 | 1 | 1.62E-03 | 107 | 7814 | 13,457 | 1.72 | 1.74E-05 | |||||||||
| LST*PPPLAEEEGLASR | T | 961 | 0.979 | 1.29E-08 | 106 | 18,331 | 29,532 | 1.61 | 4.31E-04 | |||||||||
| LS*T*PPPLAEEEGLASR | S; T | 960; 961 | 0.5; 0.5 | 1.29E-08 | 105 | 10,530 | 17,926 | 1.70 | 5.29E-04 | |||||||||
| LEGALS*EEPR | S | 1607 | 1 | 1.17E-03 | 105 | 10,600 | 8027 | 0.76 | 3.19E-03 | |||||||||
| IT*HSPTVSQVTER | T | 1684 | 0.745 | 2.33E-02 | 51 | |||||||||||||
| ITHS*PTVSQVTER | S | 1686 | 0.962 | 8.25E-04 | 76 | |||||||||||||
| ITHS*PT*VSQVTER | S; T | 1686; 1688 | 0.327; 0.327 | 2.14E-02 | 47 | |||||||||||||
| AP3B1 | − 1.2102 | 11.2042 | − 4.7116 | 9.33E-04 | 1.22E-02 | − 0.6877 | O00203 | AP-3 complex subunit beta-1 | NFYES*DDDQKEK | S | 276 | 1 | 1.03E-45 | 117 | 22,634 | 68,527 | 3.03 | 2.37E-05 |
| APIP | 1.1934 | 8.6558 | 4.5636 | 1.16E-03 | 1.38E-02 | − 0.9140 | Q96GX9 | Probable methylthioribulose-1-phosphate dehydratase | DISGPS*PSK | S | 87 | 0.962 | 4.65E-03 | 81 | ||||
| APOL1 | − 1.1936 | 9.8189 | − 3.2480 | 9.26E-03 | 4.79E-02 | − 3.0397 | A5PL32 | Apolipoprotein L1 | VTEPIS*AES*GEQVER | S; S | 352; 355 | 1; 1 | 2.53E-03 | 59 | 6305 | 4500 | 0.71 | 5.33E-03 |
| ARHGEF11 | 1.5982 | 6.3788 | 7.6133 | 2.33E-05 | 1.74E-03 | 3.1111 | O15085 | Rho guanine nucleotide exchange factor 11 | S*LENPT*PPFTPK | S; T | 663; 668 | 1; 1 | 1.93E-10 | 106 | 12,686 | 29,402 | 2.32 | 4.24E-04 |
| NS*GIWESPELDR | S | 1295 | 1 | 6.04E-03 | 52 | 7700 | 11,195 | 1.45 | 1.11E-03 | |||||||||
| HQVLLEDPEQEGS*AEEEELGVLPCPSTSLDGENR | S | 1155 | 1 | 3.31E-47 | 146 | 1696 | 3062 | 1.81 | 1.02E-02 | |||||||||
| WTDGS*LS*PPAKEPLASDSR | S; S | 1478; 1480 | 0.48; 0.48 | 2.20E-03 | 38 | |||||||||||||
| ARHGEF12 | 1.0544 | 7.6127 | 5.9598 | 1.67E-04 | 4.70E-03 | 1.0924 | Q9NZN5 | Rho guanine nucleotide exchange factor 12 | TDCSSGDASRPSSDNADS*PK | S | 309 | 1 | 1.82E-10 | 109 | 8015 | 23,551 | 2.94 | 9.07E-04 |
| ARHGEF6 | 1.5519 | 6.6394 | 4.1475 | 2.19E-03 | 1.99E-02 | − 1.5656 | Q15052 | Rho guanine nucleotide exchange factor 6 | KDS*IPQVLLPEEEK | S | 684 | 1 | 1.39E-10 | 126 | ||||
| ASAP1 | 1.4735 | 6.5041 | 5.7914 | 2.08E-04 | 5.30E-03 | 0.8655 | Q9ULH1 | Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 1 | QEEIDES*DDDLDDKPSPIK | S | 717 | 1 | 1.20E-107 | 116 | 4217 | 8616 | 2.04 | 1.35E-04 |
| ASAP2 | − 2.6231 | 8.6901 | − 4.5167 | 1.25E-03 | 1.43E-02 | − 0.9862 | O43150 | Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 2 | LLHEDLDES*DDDMDEK | S | 701 | 1 | 2.71E-52 | 185 | 11,289 | 17,685 | 1.57 | 2.02E-03 |
| ATP8A1 | 1.5729 | 8.3920 | 5.5795 | 2.75E-04 | 6.14E-03 | 0.5743 | Q9Y2Q0 | Probable phospholipid-transporting ATPase IA | TDDVS*EKT*S*LADQEEVR | S; T; S | 25; 28; 29 | 0.966; 0.502; 0.502 | 5.82E-03 | 57 | 5114 | 8707 | 1.70 | 3.09E-05 |
| TDDVS*EKT*SLADQEEVR | S; T | 25; 28 | 0.764; 0.563 | 2.73E-03 | 62 | 5114 | 8707 | 1.70 | 3.09E-05 | |||||||||
| TS*LADQEEVR | S | 29 | 0.767 | 2.73E-03 | 63 | 5114 | 8707 | 1.70 | 3.09E-05 | |||||||||
| AVP | − 1.0570 | 7.0007 | − 3.3755 | 7.51E-03 | 4.18E-02 | − 2.8269 | P01185 | Vasopressin-neurophysin 2-copeptin | CQEENYLPS*PCQSGQK | S | 83 | 0.996 | 1.57E-03 | 69 | 6536 | 4234 | 0.65 | 8.03E-03 |
| BAIAP3 | − 7.0454 | 10.8238 | − 22.2592 | 1.46E-09 | 5.00E-06 | 11.8821 | O94812 | BAI1-associated protein 3 | S*GSPAPPEPVDPSLGLR | S | 113 | 0.528 | 4.57E-03 | 58 | 10,732 | 21,332 | 1.99 | 6.38E-04 |
| SGS*PAPPEPVDPSLGLR | S | 115 | 0.981 | 1.52E-16 | 133 | 1866 | 3885 | 2.08 | 3.13E-03 | |||||||||
| BASP1 | − 2.4499 | 11.6391 | − 3.2822 | 8.75E-03 | 4.61E-02 | − 2.9825 | P80723 | Brain acid–soluble protein 1 | S*DGAPASDSKPGSSEAAPSSK | S | 164 | 0.996 | 8.79E-03 | 42 | 7903 | 10,069 | 1.27 | 1.46E-02 |
| AEGAATEEEGT*PK | T | 36 | 1 | 4.87E-24 | 90 | |||||||||||||
| ETPAATEAPS*S*T*PK | S; S; T | 194; 195; 196 | 0.333; 0.333; 0.333 | 1.91E-03 | 61 | |||||||||||||
| ETPAATEAPSST*PK | T | 196 | 0.683 | 3.52E-06 | 63 | |||||||||||||
| BCLAF1 | 1.2108 | 6.8562 | 6.4198 | 9.33E-05 | 3.40E-03 | 1.6913 | Q9NYF8 | Bcl-2-associated transcription factor 1 | KAEGEPQEES*PLK | S | 177 | 1 | 8.49E-31 | 158 | 84,318 | 208,327 | 2.47 | 2.02E-07 |
| AEGEWEDQEALDYFS*DKESGK | S | 385 | 1 | 5.55E-93 | 218 | 31,132 | 135,860 | 4.36 | 4.16E-07 | |||||||||
| DLFDYS*PPLHK | S | 512 | 1 | 8.04E-16 | 107 | 9441 | 20,570 | 2.18 | 3.11E-06 | |||||||||
| ETQS*PEQVK | S | 496 | 1 | 1.35E-19 | 173 | 70,549 | 155,420 | 2.20 | 2.09E-05 | |||||||||
| FNDS*EGDDT*EETEDYR | S | 397; 402 | 1; 1 | 3.19E-129 | 256 | 44,945 | 69,960 | 1.56 | 2.82E-05 | |||||||||
| KET*QSPEQVK | T | 494 | 0.567 | 2.69E-02 | 43 | 4370 | 8650 | 1.98 | 1.10E-04 | |||||||||
| QKS*PEIHR | S | 648 | 1 | 9.96E-03 | 65 | 797 | 2439 | 3.06 | 2.65E-04 | |||||||||
| YS*PSQNSPIHHIPSR | S | 285 | 0.907 | 2.15E-31 | 167 | 23,981 | 39,296 | 1.64 | 3.62E-04 | |||||||||
| YSPS*QNS*PIHHIPSR | S; S | 287; 290 | 0.908; 0.999 | 2.15E-31 | 167 | 20,077 | 36,892 | 1.84 | 4.90E-04 | |||||||||
| IDIS*PSTLR | S | 658 | 0.995 | 2.86E-13 | 162 | 36,901 | 49,609 | 1.34 | 8.48E-04 | |||||||||
| SSATSGDIWPGLSAYDNS*PR | S | 222 | 0.998 | 9.35E-121 | 119 | 16,902 | 13,214 | 0.78 | 7.70E-03 | |||||||||
| DTFEHDPSES*IDEFNK | S | 198 | 0.98 | 2.82E-35 | 94 | |||||||||||||
| SSATSGDIWPGLS*AYDNSPR | S | 217 | 0.528 | 3.48E-29 | 76 | |||||||||||||
| CALD1 | 1.1936 | 4.5492 | 3.8770 | 3.34E-03 | 2.54E-02 | − 2.0009 | Q05682 | Caldesmon | RGS*IGENQIKDEK | S | 207 | 1 | 1.37E-06 | 119 | 98,098 | 323,267 | 3.30 | 2.31E-06 |
| CASC4 | − 1.1339 | 10.5848 | − 4.4287 | 1.42E-03 | 1.54E-02 | − 1.1227 | Q6P4E1 | Protein CASC4 | FFDENES*PVDPQHGSK | S | 366 | 1 | 1.16E-03 | 73 | ||||
| CASK | 2.8830 | 11.3140 | 7.5385 | 2.53E-05 | 1.82E-03 | 3.0275 | B7ZKY2 | Calcium/calmodulin-dependent serine protein kinase (MAGUK family) | TQSS*S*CEDLPSTTQPK | S; S | 570; 571 | 0.78; 0.739 | 5.24E-05 | 88 | 3606 | 7644 | 2.12 | 3.28E-04 |
| TQS*SSCEDLPSTTQPK | S | 569 | 0.379 | 8.01E-16 | 79 | |||||||||||||
| CBX8 | 1.0985 | 6.6357 | 3.9595 | 2.93E-03 | 2.35E-02 | − 1.8672 | Q9HC52 | Chromobox protein homolog 8 | VDDKPS*SPGDSSK | S | 190 | 0.513 | 2.77E-04 | 83 | ||||
| CCDC48 | − 3.3166 | 8.0575 | − 9.1213 | 5.02E-06 | 7.68E-04 | 4.6616 | Q9HA90 | Coiled-coil domain-containing protein 48 | TLGTS*EEEAELQQK | S | 480 | 0.841 | 1.19E-21 | 150 | 3504 | 2952 | 0.84 | 1.04E-01 |
| T*LGTSEEEAELQQK | T | 476 | 0.504 | 2.94E-11 | 136 | |||||||||||||
| CCDC86 | − 2.5059 | 12.1490 | − 4.2785 | 1.79E-03 | 1.76E-02 | − 1.3580 | Q9H6F5 | Coiled-coil domain-containing protein 86 | LQQGAGLESPQGQPEPGAAS*PQR | S | 91 | 1 | 8.60E-16 | 91 | 11,730 | 18,273 | 1.56 | 5.06E-03 |
| LGGLRPES*PESLTSVSR | S | 18 | 0.998 | 1.72E-03 | 61 | 9461 | 12,076 | 1.28 | 1.46E-02 | |||||||||
| ALVEFESNPEETREPGS*PPSVQR | S | 47 | 0.988 | 8.54E-17 | 121 | |||||||||||||
| CDC42EP4 | 2.2410 | 9.6096 | 3.4168 | 7.02E-03 | 4.01E-02 | − 2.7581 | Q9H3Q1 | Cdc42 effector protein 4 | AGPDLPSLPSHALEDEGWAAAAPS*PGSAR | S | 292 | 0.789 | 6.03E-10 | 41 | ||||
| CDK14 | 1.1778 | 11.3632 | 3.5667 | 5.50E-03 | 3.45E-02 | − 2.5096 | O94921 | Cyclin-dependent kinase 14 | VHS*ENNACINFK | S | 95 | 1 | 1.18E-02 | 75 | 2416 | 3471 | 1.44 | 5.63E-03 |
| CENPA | 1.5999 | 4.3832 | 4.5900 | 1.12E-03 | 1.35E-02 | − 0.8733 | P49450 | Histone H3-like centromeric protein A | RRS*PS*PTPTPGPSR | S; S | 17; 19 | 0.998; 0.984 | 7.74E-04 | 68 | 2107 | 18,118 | 8.60 | 3.01E-06 |
| CHD3 | 1.1576 | 8.0852 | 4.7990 | 8.21E-04 | 1.13E-02 | − 0.5558 | Q12873 | Chromodomain-helicase-DNA-binding protein 3 | ELQGDGPPS*SPTNDPTVK | S; S | 712; 713 | 0.481; 0.481 | 1.47E-02 | 50 | ||||
| CHGA | − 3.8263 | 14.4379 | − 4.6354 | 1.04E-03 | 1.30E-02 | − 0.8038 | P10645 | Chromogranin-A | S*GELEQEEERLS*KEWEDSK | S; S | 322; 333 | 1; 1 | 1.06E-26 | 183 | 296,230 | 566,530 | 1.91 | 4.87E-05 |
| S*GELEQEEERLS*K | S; S | 322; 333 | 1; 1 | 1.06E-26 | 147 | 207,873 | 320,807 | 1.54 | 2.39E-04 | |||||||||
| HSGFEDELSEVLENQS*S*QAELK | S; S | 112; 113 | 1; 1 | 0.00E+00 | 287 | 320,767 | 437,363 | 1.36 | 8.38E-04 | |||||||||
| YPGPQAEGDSEGLS*QGLVDR | S | 207 | 1 | 3.51E-32 | 154 | 7755 | 9573 | 1.23 | 1.97E-03 | |||||||||
| GEQEHS*QQKEEEEEMAVVPQGLFR | S | 300 | 1 | 1.45E-131 | 233 | 280,203 | 169,087 | 0.60 | 2.61E-03 | |||||||||
| S*GEATDGARPQALPEPMQESK | S | 142 | 1 | 2.90E-55 | 180 | 437,597 | 568,253 | 1.30 | 3.19E-03 | |||||||||
| EAVEEPS*SKDVMEK | S | 125 | 0.567 | 2.10E-05 | 93 | 18,890 | 23,102 | 1.22 | 7.15E-03 | |||||||||
| EAVEEPSS*KDVMEK | S | 126 | 0.873 | 1.02E-07 | 120 | 20,108 | 27,926 | 1.39 | 1.99E-02 | |||||||||
| REDS*KEAEK | S | 136 | 1 | 7.15E-06 | 108 | 126,907 | 149,643 | 1.18 | 3.08E-02 | |||||||||
| YPGPQAEGDS*EGLSQGLVDR | S | 203 | 1 | 3.97E-102 | 225 | 63,186 | 74,743 | 1.18 | 4.94E-02 | |||||||||
| GWRPS*S*REDS*LEAGLPLQVR | S; S; S | 397; 398; 402 | 0.543; 0.543; 0.914 | 8.64E-27 | 145 | 38,199 | 34,450 | 0.90 | 6.23E-02 | |||||||||
| LEGQEEEEDNRDS*SMK | S | 370 | 0.989 | 3.96E-61 | 200 | 119,240 | 151,717 | 1.27 | 6.51E-02 | |||||||||
| EDS*LEAGLPLQVR | S | 402 | 1 | 1.44E-38 | 185 | 123,567 | 116,303 | 0.94 | 7.89E-02 | |||||||||
| GLS*AEPGWQAK | S | 218 | 1 | 2.58E-25 | 180 | 37,874 | 34,815 | 0.92 | 1.12E-01 | |||||||||
| HSGFEDELS*EVLENQS*SQAELK | S; S | 105; 112 | 0.8261 | 3.88E-90 | 99 | 2083 | 2441 | 1.17 | 1.58E-01 | |||||||||
| RPEDQELESLS*AIEAELEK | S | 438 | 0.98 | 3.04E-120 | 119 | 11,305 | 10,366 | 0.92 | 2.55E-01 | |||||||||
| GWRPS*SREDS*LEAGLPLQVR | S; S | 396; 402 | 0.731; 0.99 | 8.64E-27 | 145 | 67,782 | 66,289 | 0.98 | 4.30E-01 | |||||||||
| KHS*GFEDELSEVLENQS*SQAELK | S | 98; 112 | 0.686; 0.843 | 3.26E-34 | 67 | |||||||||||||
| RPEDQELES*LSAIEAELEK | S | 436 | 0.997 | 1.77E-167 | 127 | |||||||||||||
| CHGB | − 1.3288 | 14.5253 | − 3.2997 | 8.51E-03 | 4.53E-02 | − 2.9533 | P05060 | Secretogranin-1 | SETHAAGHS*QEK | S | 225 | 1 | 4.87E-30 | 175 | 194,330 | 26,293 | 0.14 | 6.87E-07 |
| GRGS*EEYR | S | 367 | 1 | 1.70E-07 | 146 | 85,298 | 10,118 | 0.12 | 7.46E-07 | |||||||||
| AS*EEEPEYGEEIK | S | 335 | 1 | 5.81E-71 | 223 | 553,550 | 73,696 | 0.13 | 2.05E-06 | |||||||||
| EKSS*QES*GEETGS*QENHPQESK | S; S; S | 236; 239; 245 | 0.531; 1; 0.965 | 4.20E-73 | 205 | 248,397 | 14,007 | 0.06 | 3.67E-06 | |||||||||
| ADEPQWSLYPSDS*QVS*EEVK | S; S | 146; 149 | 0.999; 0.999 | 1.30E-150 | 236 | 321,833 | 101,210 | 0.31 | 5.28E-06 | |||||||||
| SSQESGEET*GSQENHPQESK | T | 243 | 0.978 | 1.47E-45 | 161 | 143,047 | 2755 | 0.02 | 5.36E-06 | |||||||||
| GERGEDS*S*EEK | S; S | 182; 183 | 1; 1 | 1.80E-03 | 100 | 80,795 | 13,041 | 0.16 | 7.03E-06 | |||||||||
| AS*EEEPEY*GEEIK | S; Y | 335; 341 | 1; 1 | 2.35E-06 | 98 | 220,190 | 39,598 | 0.18 | 8.29E-06 | |||||||||
| SSQES*GEETGS*QENHPQESK | S; S | 239; 245 | 0.992; 1 | 4.20E-73 | 205 | 407,313 | 23,721 | 0.06 | 1.01E-05 | |||||||||
| S*QREDEEEEEGENYQK | S | 160 | 1 | 1.77E-17 | 140 | 50,520 | 12,595 | 0.25 | 1.17E-05 | |||||||||
| SAEFPDFYDS*EEPVSTHQEAENEKDR | S | 626 | 1 | 1.85E-99 | 214 | 339,777 | 89,045 | 0.26 | 1.18E-05 | |||||||||
| S*QEES*EEGEEDATS*EVDK | S | 259; 263; 272 | 1; 1; 0.857 | 1.51E-53 | 157 | 260,217 | 35,935 | 0.14 | 1.23E-05 | |||||||||
| SSQGGS*LPSEEK | S | 298 | 0.981 | 3.28E-14 | 142 | 88,621 | 25,139 | 0.28 | 1.31E-05 | |||||||||
| DPADASEAHES*SSR | S | 98 | 0.966 | 1.32E-30 | 169 | 16,941 | 1615 | 0.10 | 1.41E-05 | |||||||||
| NYPS*LELDK | S | 391 | 1 | 1.30E-04 | 112 | 209,063 | 29,849 | 0.14 | 1.44E-05 | |||||||||
| ADEPQWSLYPS*DS*QVSEEVK | S; S | 144; 146 | 0.941; 0.751 | 3.58E-98 | 136 | 19,345 | 10,016 | 0.52 | 1.53E-05 | |||||||||
| EKSS*QES*GEET*GSQENHPQESK | S; S; T | 237; 239; 243 | 0.819; 0.935; 0.598 | 1.47E-45 | 161 | 227,790 | 11,221 | 0.05 | 1.53E-05 | |||||||||
| SSQGGSLPS*EEK | S | 301 | 0.999 | 5.51E-20 | 128 | 85,508 | 21,593 | 0.25 | 1.88E-05 | |||||||||
| S*AEFPDFYDSEEPVSTHQEAENEKDR | S | 617 | 1 | 8.30E-152 | 246 | 312,090 | 68,957 | 0.22 | 2.13E-05 | |||||||||
| S*AEFPDFY*DSEEPVSTHQEAENEKDR | Y | 624 | 0.995 | 8.86E-43 | 152 | 68,357 | 15,724 | 0.23 | 2.23E-05 | |||||||||
| SQEESEEGEEDAT*SEVDK | T | 271 | 0.973 | 1.38E-88 | 219 | 322,700 | 69,163 | 0.21 | 2.23E-05 | |||||||||
| M#AHGY*GEES*EEER | Y; S | 401; 405 | 1; 1 | 2.41E-02 | 51 | 7666 | 1759 | 0.23 | 2.34E-05 | |||||||||
| ERADEPQWSLYPSDSQVS*EEVK | S | 149 | 1 | 2.83E-147 | 264 | 584,753 | 166,957 | 0.29 | 2.80E-05 | |||||||||
| PQS*EES*WDEEDKR | S; S | 377; 380 | 1; 1 | 4.73E-42 | 181 | 945,707 | 144,227 | 0.15 | 3.27E-05 | |||||||||
| SAEFPDFYDS*EEPVS*THQEAENEK | S; S | 626; 631 | 0.996; 0.863 | 3.45E-35 | 140 | 84,147 | 25,399 | 0.30 | 3.33E-05 | |||||||||
| SS*QGGSLPSEEK | S | 294 | 0.807 | 3.74E-09 | 133 | 23,609 | 5551 | 0.24 | 3.77E-05 | |||||||||
| S*SQGGSLPSEEK | S | 293 | 0.992 | 1.97E-30 | 180 | 104,456 | 26,549 | 0.25 | 4.48E-05 | |||||||||
| GHPQEES*EESNVSMASLGEK | S | 311 | 1 | 6.58E-209 | 232 | 930,043 | 206,513 | 0.22 | 5.91E-05 | |||||||||
| SAEFPDFYDSEEPVS*T*HQEAENEK | S; T | 631; 632 | 0.591; 0.586 | 3.45E-35 | 124 | 22,832 | 7174 | 0.31 | 5.94E-05 | |||||||||
| EKS*S*QES*GEET*GS*QENHPQESK | S; S; S; T; S | 235; 236; 239; 243; 245 | 0.643; 0.589; 0.589; 0.589; 0.589 | 1.47E-45 | 161 | 146,517 | 7750 | 0.05 | 7.50E-05 | |||||||||
| WQQQGDLQDT*KENR | T | 492 | 1 | 6.83E-05 | 85 | 92,934 | 34,420 | 0.37 | 9.99E-05 | |||||||||
| GHPQEES*EESNVS*M#ASLGEK | S | 317 | 0.988 | 1.45E-59 | 184 | 150,487 | 43,938 | 0.29 | 1.70E-04 | |||||||||
| ADEPQWSLY*PSDSQVS*EEVK | Y; S | 142; 149 | 0.981; 0.96 | 6.42E-108 | 134 | 22,916 | 11,918 | 0.52 | 1.87E-04 | |||||||||
| GHPQEES*EESNVS*MAS*LGEK | S; S; S | 314; 320; 323 | 0.739; 1; 1 | 1.83E-50 | 161 | 57,813 | 21,261 | 0.37 | 4.40E-04 | |||||||||
| WAEGGGHS*R | S | 130 | 1 | 4.86E-03 | 80 | 1039 | 214 | 0.21 | 5.96E-04 | |||||||||
| GHPQEES*EES*NVS*MASLGEK | S; S; S | 314; 317; 320 | 0.959; 0.959; 0.959 | 8.63E-27 | 145 | 41,766 | 12,932 | 0.31 | 6.63E-04 | |||||||||
| CIIEVLSNALS*K | S | 46 | 1 | 3.06E-04 | 94 | 994 | 401 | 0.40 | 1.63E-02 | |||||||||
| DHHS*THYRASEEEPEY*GEEIK | S; T; Y | 329; 330; 341 | 0.355; 0.355; 0.98 | 1.56E-02 | 43 | |||||||||||||
| DPADASEAHESS*S*RGEAGAPGEEDIQGPTK | S; S | 99; 100 | 0.651; 0.651 | 8.57E-07 | 62 | |||||||||||||
| LLRDPADAS*EAHESSSR | S | 93 | 0.989 | 5.88E-05 | 88 | |||||||||||||
| S*ETHAAGHS*QEK | S | 217 | 0.757 | 1.02E-02 | 78 | |||||||||||||
| CHMP2B | − 1.5915 | 9.4237 | − 9.3648 | 3.99E-06 | 6.71E-04 | 4.8895 | Q9UQN3 | Charged multivesicular body protein 2b | ATIS*DEEIER | S | 199 | 1 | 3.83E-06 | 121 | 11,233 | 17,076 | 1.52 | 6.05E-03 |
| CIC | 1.0491 | 11.3820 | 6.0126 | 1.56E-04 | 4.52E-03 | 1.1627 | I3L2J0 | Protein capicua homolog | KNS*TDLDSAPEDPTS*PK | S; S | 2303; 2315 | 0.697; 0.741 | 2.08E-04 | 75 | ||||
| KNST*DLDSAPEDPTS*PK | S | 2304; 2315 | 0.563; 0.831 | 2.08E-04 | 75 | |||||||||||||
| S*S*PPLPPPAEER | S; S | 2152; 2153 | 0.5; 0.5 | 1.43E-02 | 49 | |||||||||||||
| SS*PPLPPPAEER | S | 2153 | 0.795 | 1.43E-02 | 49 | |||||||||||||
| CLASP2 | 1.1382 | 9.5975 | 6.8774 | 5.38E-05 | 2.59E-03 | 2.2577 | O75122 | CLIP-associating protein 2 | VLNT*GSDVEEAVADALK | T | 594 | 0.618 | 2.50E-06 | 99 | 7092 | 16,081 | 2.27 | 2.64E-04 |
| VLNTGS*DVEEAVADALK | S | 596 | 0.848 | 7.26E-05 | 57 | |||||||||||||
| CTAGE5 | − 1.5080 | 8.1531 | − 6.2874 | 1.10E-04 | 3.78E-03 | 1.5219 | O15320 | Cutaneous T cell lymphoma-associated antigen 5 | AFLS*PPTLLEGPLR | S | 536 | 0.954 | 4.01E-03 | 54 | 5494 | 8403 | 1.53 | 1.09E-03 |
| APSDTGS*LS*PPWDQDRR | S; S | 594; 596 | 0.962; 0.997 | 2.71E-03 | 51 | 2316 | 3679 | 1.59 | 2.24E-03 | |||||||||
| S*NS*ELEDEILCLEK | S; S | 137; 139 | 0.5; 0.5 | 8.61E-34 | 89 | 2602 | 3220 | 1.24 | 3.50E-02 | |||||||||
| SNS*ELEDEILCLEK | S | 139 | 0.861 | 8.61E-34 | 89 | 2602 | 3220 | 1.24 | 3.50E-02 | |||||||||
| SFNMPS*LDK | S | 647 | 1 | 1.79E-02 | 59 | |||||||||||||
| CYBRD1 | − 2.4812 | 7.6514 | − 4.6620 | 1.00E-03 | 1.27E-02 | − 0.7633 | Q53TN4 | Cytochrome b reductase 1 | GSMPAY*SGNNMDK | Y | 252 | 0.662 | 1.47E-02 | 49 | ||||
| DCLK1 | − 4.0867 | 9.7240 | − 7.5568 | 2.48E-05 | 1.80E-03 | 3.0480 | O15075 | Serine/threonine-protein kinase DCLK1 | SPSPSPTS*PGSLR | S | 337 | 0.911 | 9.87E-04 | 71 | ||||
| VCSS*MDENDGPGEEVSEEGFQIPATITER | S | 364 | 0.807 | 1.08E-03 | 48 | |||||||||||||
| DENR | 1.0387 | 8.3264 | 3.5168 | 5.96E-03 | 3.62E-02 | − 2.5921 | O43583 | Density-regulated protein | LTVENS*PK | S | 73 | 1 | 4.03E-03 | 61 | ||||
| DMXL2 | 1.2471 | 8.1839 | 7.6581 | 2.22E-05 | 1.70E-03 | 3.1609 | Q8TDJ6 | DmX-like protein 2 | KQS*EVEADLGYPGGK | S | 2640 | 1 | 3.59E-05 | 79 | ||||
| RQS*ENISAPPVLSEDIDK | S | 2399 | 0.839 | 2.63E-04 | 58 | |||||||||||||
| DOCK11 | − 2.9596 | 8.1805 | − 7.3717 | 3.05E-05 | 1.98E-03 | 2.8384 | Q5JSL3 | Dedicator of cytokinesis protein 11 | ETVETAQDDET*SSQGK | T; S; S | 294; 295; 296 | 0.333; 0.333; 0.333 | 1.02E-02 | 45 | ||||
| DOCK7 | − 1.0075 | 9.0073 | − 3.4301 | 6.87E-03 | 3.96E-02 | − 2.7360 | Q96N67 | Dedicator of cytokinesis protein 7 | SLSNS*NPDISGTPT*SPDDEVR | S; T | 900; 909 | 0.9; 0.402 | 2.45E-06 | 47 | 1576 | 2651 | 1.68 | 9.54E-03 |
| ELAVL4 | 2.1977 | 9.9185 | 4.3468 | 1.61E-03 | 1.66E-02 | − 1.2507 | P26378 | ELAV-like protein 4 | NCPS*PMQTGATTDDSK | S | 33 | 0.999 | 4.54E-04 | 80 | 6680 | 11,145 | 1.67 | 9.07E-04 |
| EPB41L2 | 1.9271 | 8.3372 | 6.2524 | 1.15E-04 | 3.86E-03 | 1.4768 | O43491 | Band 4.1-like protein 2 | EIS*PGSGPGEIR | S | 715 | 0.607 | 1.49E-02 | 56 | 4934 | 9864 | 2.00 | 7.50E-05 |
| S*YTLVVAK | S | 87 | 1 | 5.02E-03 | 99 | 9974 | 25,384 | 2.54 | 1.55E-04 | |||||||||
| EISPGS*GPGEIR | S | 718 | 0.971 | 9.87E-03 | 81 | 4771 | 9142 | 1.92 | 4.81E-03 | |||||||||
| EVAENQQNQS*S*DPEEEK | S; S | 38; 39 | 0.5; 0.5 | 1.38E-02 | 50 | |||||||||||||
| EVAENQQNQSS*DPEEEK | S | 39 | 0.88 | 3.99E-81 | 111 | |||||||||||||
| EPB49 | 1.1990 | 8.2266 | 6.2015 | 1.22E-04 | 4.01E-03 | 1.4108 | Q08495 | Dematin | GNS*LPCVLEQK | S | 333 | 1 | 4.66E-15 | 152 | 21,088 | 48,603 | 2.30 | 2.12E-05 |
| RGAEEEEEEEDDDS*GEEMK | S | 226 | 1 | 2.45E-73 | 205 | 66,329 | 117,453 | 1.77 | 1.15E-04 | |||||||||
| STS*PPPS*PEVWADSR | S; S | 92; 96 | 0.734; 0.978 | 2.86E-03 | 56 | 4181 | 9523 | 2.28 | 3.97E-04 | |||||||||
| STS*PPPSPEVWADSR | S | 92 | 0.994 | 4.97E-08 | 103 | 27,533 | 42,218 | 1.53 | 5.58E-04 | |||||||||
| ESVGGS*PQTK | S | 156 | 1 | 6.79E-03 | 92 | 2009 | 4972 | 2.47 | 1.18E-03 | |||||||||
| SSS*LPAYGR | S | 289 | 0.998 | 3.78E-05 | 134 | 8120 | 15,700 | 1.93 | 1.35E-03 | |||||||||
| DSSVPGS*PSSIVAK | S | 26 | 0.927 | 5.87E-04 | 69 | |||||||||||||
| EPS15L1 | 1.0173 | 7.5204 | 5.6410 | 2.54E-04 | 5.89E-03 | 0.6596 | Q9UBC2 | Epidermal growth factor receptor substrate 15-like 1 | TVFPGAVPVLPAS*PPPK | S | 229 | 1 | 1.50E-12 | 79 | 1597 | 2107 | 1.32 | 8.98E-02 |
| STPSHGSVSSLNSTGSLS*PK | S | 255 | 0.759 | 1.29E-04 | 79 | |||||||||||||
| FKBP5 | 2.6927 | 9.7556 | 4.9688 | 6.43E-04 | 9.84E-03 | − 0.3023 | Q13451 | Peptidyl-prolyl cis-trans isomerase FKBP5 | NNEES*PTATVAEQGEDITSK | S | 13 | 0.91 | 9.79E-06 | 43 | ||||
| FLVCR1 | − 1.0780 | 9.4860 | − 5.7358 | 2.24E-04 | 5.54E-03 | 0.7898 | Q9Y5Y0 | Feline leukemia virus subgroup C receptor-related protein 1 | AIPADS*PTDQEPK | S | 536 | 0.965 | 5.83E-04 | 83 | 5787 | 10,532 | 1.82 | 2.55E-04 |
| FOXK1 | − 1.0565 | 7.7540 | − 4.9965 | 6.18E-04 | 9.66E-03 | − 0.2613 | P85037 | Forkhead box protein K1 | EGS*PI*DPEFGSK | S | 445 | 1 | 1.27E-06 | 87 | 3949 | 9911 | 2.51 | 1.37E-03 |
| GAL | − 8.5314 | 9.5656 | − 12.9818 | 2.17E-07 | 1.45E-04 | 7.6993 | P22466 | Galanin peptides | LLDLPAAAS*SEDIERS | S | 116 | 0.991 | 1.09E-82 | 227 | 161,450 | 28,762 | 0.18 | 1.20E-06 |
| LLDLPAAAS*SEDIERS* | S; S; S | 116; 117; 123 | 0.658; 0.658; 0.684 | 3.21E-10 | 98 | 11,171 | 6908 | 0.62 | 4.26E-03 | |||||||||
| LLDLPAAAS*SEDIERS* | S | 116; 123 | 0.878; 0.716 | 2.09E-04 | 74 | 11,171 | 6908 | 0.62 | 4.26E-03 | |||||||||
| GATAD2A | 1.0255 | 8.6559 | 4.3609 | 1.58E-03 | 1.64E-02 | − 1.2285 | Q86YP4 | Transcriptional repressor p66-alpha | RPPS*PDVIVLS*DNEQPS*S*PR | S | 100; 107; 113; 114 | 1; 0.87; 0.581; 0.548 | 3.91E-07 | 52 | ||||
| GH1 | − 12.5607 | 10.1374 | − 20.6323 | 2.98E-09 | 7.99E-06 | 11.3526 | P01241 | Somatotropin | FDTNS*HNDDALLK | S | 176 | 1 | 3.20E-216 | 194 | 580,220 | 43,527 | 0.08 | 1.93E-06 |
| FDT*NSHNDDALLK | T | 174 | 0.578 | 1.80E-03 | 70 | |||||||||||||
| SVFANSLVYGAS*DSNVYDLLK | S; S | 132; 134 | 0.428; 0.428 | 1.85E-10 | 51 | |||||||||||||
| GH2 | − 11.4395 | 7.8750 | − 30.7801 | 6.85E-11 | 5.16E-07 | 13.8468 | P01242 | Growth hormone variant | S*HNDDALLK | S | 176 | 1 | 6.34E-05 | 131 | ||||
| YSFLQNPQT*SLCFSESIPTPSNR | T; S | 76; 77 | 0.481; 0.481 | 1.63E-02 | 37 | |||||||||||||
| GNAS | − 2.6651 | 5.5317 | − 5.7789 | 2.11E-04 | 5.36E-03 | 0.8486 | Q5JWF2 | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas | ISTAS*GDGR | S | 995 | 1 | 1.21E-04 | 115 | 11,879 | 32,931 | 2.77 | 2.32E-05 |
| GPATCH8 | 1.0279 | 9.8805 | 7.4027 | 2.94E-05 | 1.96E-03 | 2.8738 | Q9UKJ3 | G patch domain-containing protein 8 | GSEGDCS*PEDK | S | 1081 | 1 | 1.74E-02 | 60 | 2087 | 4025 | 1.93 | 5.01E-03 |
| H1FX | 1.2641 | 10.5253 | 5.0038 | 6.11E-04 | 9.60E-03 | − 0.2505 | Q92522 | Histone H1x | AGGSAALS*PSK | S | 31 | 0.996 | 1.62E-02 | 80 | 5113 | 13,633 | 2.67 | 4.62E-05 |
| HIST1H1D | 2.2738 | 9.1170 | 3.4293 | 6.87E-03 | 3.96E-02 | − 2.7374 | P16402 | Histone H1.3 | GTGASGS*FK | S | 105 | 0.969 | 7.22E-03 | 74 | 5296 | 22,242 | 4.20 | 8.93E-06 |
| KAS*GPPVSELITK | S | 37 | 1 | 3.45E-10 | 128 | 57,689 | 15,516 | 0.27 | 2.58E-04 | |||||||||
| HNRNPA1 | − 1.0812 | 8.4803 | − 4.0578 | 2.51E-03 | 2.15E-02 | − 1.7091 | P09651 | Heterogeneous nuclear ribonucleoprotein A1 | SES*PKEPEQLR | S | 6 | 1 | 1.83E-13 | 144 | 59,421 | 186,063 | 3.13 | 7.49E-06 |
| HTATSF1 | − 2.5483 | 13.6709 | − 11.7593 | 5.31E-07 | 2.15E-04 | 6.8570 | O43719 | HIV Tat-specific factor 1 | LFEDDDS*NEKLFDEEEDSSEK | S | 702 | 1 | 2.14E-175 | 268 | 288,110 | 160,883 | 0.56 | 1.95E-04 |
| EFDEDS*DEKEEEEDTYEK | S | 624 | 1 | 1.72E-69 | 202 | 204,193 | 125,980 | 0.62 | 2.23E-04 | |||||||||
| ESEDDLNKES*EEEVGPTK | S | 529 | 1 | 1.68E-56 | 192 | 88,307 | 60,457 | 0.68 | 2.99E-04 | |||||||||
| LFDEEEDS*S*EKLFDDSDER | S; S | 713; 714 | 1; 1 | 1.56E-26 | 142 | 44,719 | 23,576 | 0.53 | 4.06E-04 | |||||||||
| LFDDS*DER | S | 721 | 1 | 2.71E-03 | 131 | 34,573 | 22,836 | 0.66 | 1.16E-03 | |||||||||
| ELHENVLDKELEENDS*ENS*EFEDDGS*EK | S; S; S | 597; 600; 607 | 1; 1; 1 | 1.27E-106 | 208 | 20,769 | 11,906 | 0.57 | 2.31E-03 | |||||||||
| VFDDES*DEKEDEEYADEK | S | 642 | 1 | 1.66E-199 | 294 | 239,173 | 191,277 | 0.80 | 5.55E-03 | |||||||||
| DLDEEGS*EKELHENVLDK | S | 579 | 1 | 3.61E-80 | 210 | 202,810 | 176,810 | 0.87 | 3.93E-02 | |||||||||
| LFEES*DDKEDEDADGKEVEDADEK | S | 676 | 1 | 7.74E-152 | 254 | 366,127 | 394,907 | 1.08 | 1.47E-01 | |||||||||
| ESS*PEKEAEEGCPEK | S | 453 | 0.989 | 1.37E-31 | 165 | 93,516 | 101,601 | 1.09 | 2.04E-01 | |||||||||
| TEDGGEFEEGAS*ENNAKES*SPEKEAEEGCPEK | S; S | 445; 452 | 0.704; 0.857 | 8.12E-04 | 56 | 15,981 | 14,532 | 0.91 | 3.02E-01 | |||||||||
| VLDEEGS*ER | S | 616 | 1 | 1.23E-04 | 117 | 11,426 | 11,535 | 1.01 | 9.22E-01 | |||||||||
| ESEEDDSEKES*DEDCSEK | S | 548 | 0.997 | 4.34E-04 | 69 | |||||||||||||
| IGFBP5 | − 2.6828 | 10.6639 | − 4.1430 | 2.20E-03 | 1.99E-02 | − 1.5728 | P24593 | Insulin-like growth factor-binding protein 5 | EHEEPT*TSEMAEETYSPK | T | 123 | 0.346 | 2.19E-04 | 56 | ||||
| IPCEF1 | 1.0260 | 5.0367 | 4.7825 | 8.41E-04 | 1.15E-02 | − 0.5806 | Q8WWN9 | Interactor protein for cytohesin exchange factors 1 | AS*PAPDDTDDTPQELK | S | 411 | 1 | 6.48E-10 | 113 | 2761 | 8305 | 3.01 | 4.03E-05 |
| ITGA2B | 2.0755 | 7.4627 | 7.3744 | 3.04E-05 | 1.98E-03 | 2.8416 | P08514 | Integrin alpha-IIb | AGSEPQALFCS*GVPK | S | 880 | 0.996 | 2.38E-03 | 60 | ||||
| ITPR1 | − 1.2600 | 12.0938 | − 5.0407 | 5.80E-04 | 9.29E-03 | − 0.1962 | Q14643 | Inositol 1,4,5-trisphosphate receptor type 1 | PQKHES*T*SS*Y*NYRVVK | S; T; S; S; Y; Y | 1177; 1178; 1180; 1181; 1183 | 0.843; 0.855; 0.896; 0.55; 0.55 | 2.27E-02 | 37 | ||||
| ITPR2 | 1.8582 | 7.8515 | 10.0154 | 2.22E-06 | 5.18E-04 | 5.4714 | Q14571 | Inositol 1,4,5-trisphosphate receptor type 2 | DS*FVEEGNTLRK | S | 1687 | 0.966 | 8.73E-03 | 44 | 125,980 | 84,783 | 0.67 | 2.20E-03 |
| KBTBD11 | − 3.5584 | 10.5689 | − 4.7908 | 8.31E-04 | 1.14E-02 | − 0.5681 | O94819 | Kelch repeat and BTB domain-containing protein 11 | AGS*RPQS*PSGDADAR | S; S | 310; 314 | 0.989; 0.838 | 1.26E-03 | 57 | 2029 | 5855 | 2.89 | 1.34E-04 |
| KIF13B | − 1.0053 | 12.0356 | − 5.8909 | 1.82E-04 | 4.94E-03 | 1.0001 | Q9NQT8 | Kinesin-like protein KIF13B | SIS*SPNVNR | S | 1381 | 0.98 | 5.62E-03 | 78 | 6201 | 20,094 | 3.24 | 4.31E-05 |
| SISS*PNVNR | S | 1382 | 0.654 | 7.49E-03 | 74 | |||||||||||||
| KIF16B | − 1.3826 | 8.3241 | − 5.2223 | 4.49E-04 | 8.02E-03 | 0.0682 | Q96L93 | Kinesin-like protein KIF16B | S*KT*T*IT*NLK | S; T; T; T | 31; 33; 34; 36 | 0.25; 0.25; 0.25; 0.25 | 2.62E-02 | 47 | ||||
| KIF1A | 1.6590 | 7.9552 | 6.7867 | 5.98E-05 | 2.74E-03 | 2.1477 | Q12756 | Kinesin-like protein KIF1A | SDS*LILDHQWELEK | S | 1370 | 0.98 | 1.89E-33 | 131 | 20,803 | 21,162 | 1.02 | 9.30E-01 |
| LIMA1 | 1.5971 | 11.6365 | 8.0040 | 1.53E-05 | 1.41E-03 | 3.5371 | Q53GG0; Q9UHB6 | LIM domain and actin-binding protein 1 | ET**SPGVEDAPIAK | T | 487 | 0.577 | 5.99E-04 | 75 | 2533 | 3843 | 1.52 | 2.94E-02 |
| LIM domain and actin-binding protein 1 | QQS*PQEPK | S | 698 | 1 | 9.90E-03 | 79 | 4890 | 18,709 | 3.83 | 1.00E-05 | ||||||||
| ET*S*PGVEDAPIAK | S | 490 | 0.983 | 1.43E-10 | 119 | 6800 | 11,621 | 1.71 | 3.83E-05 | |||||||||
| EGHSLEMENENLVENGADS*DEDDNSFLK | S | 686 | 1 | 0.00E+00 | 141 | 3725 | 5941 | 1.59 | 1.17E-02 | |||||||||
| S*QDVELWEGEVVK | S | 726 | 1 | 2.48E-04 | 57 | |||||||||||||
| MAP4 | 1.0684 | 8.1047 | 4.7053 | 9.42E-04 | 1.22E-02 | − 0.6974 | P27816 | Microtubule-associated protein 4 | DVT*PPPETEVVLIK | T | 521 | 1 | 3.18E-40 | 153 | 66,008 | 110,437 | 1.67 | 1.71E-04 |
| VGS*LDNVGHLPAGGAVK | S | 1073 | 1 | 6.49E-06 | 91 | 11,125 | 28,354 | 2.55 | 2.17E-04 | |||||||||
| DM#ES*PTKLDVTLAK | S | 280 | 0.981 | 5.00E-05 | 82 | 30,879 | 33,386 | 1.08 | 1.59E-01 | |||||||||
| DMESPT*KLDVTLAK | T | 282 | 0.605 | 7.94E-06 | 109 | |||||||||||||
| DMS*PLSETEMALGK | S | 507 | 0.999 | 8.64E-07 | 115 | |||||||||||||
| KCS*LPAEEDSVLEK | S | 636 | 1 | 8.51E-06 | 77 | |||||||||||||
| LATNTS*APDLK | S | 928 | 0.975 | 1.89E-03 | 105 | |||||||||||||
| MARCKS | − 1.2368 | 12.4473 | − 4.0541 | 2.53E-03 | 2.16E-02 | − 1.7149 | P29966 | Myristoylated alanine-rich C-kinase substrate | GEAAAERPGEAAVASS*PSK | S | 27 | 0.96 | 1.32E-26 | 143 | 9538 | 82,612 | 8.66 | 3.86E-06 |
| GEAAAERPGEAAVAS*SPSK | S | 26 | 0.878 | 8.86E-22 | 126 | 10,104 | 57,201 | 5.66 | 8.79E-06 | |||||||||
| AEDGATPSPSNET*PK | T | 150 | 0.998 | 2.15E-16 | 140 | 7219 | 27,996 | 3.88 | 1.52E-05 | |||||||||
| AEDGATPS*PSNETPK | S | 145 | 0.959 | 1.58E-04 | 79 | 2120 | 3439 | 1.62 | 8.15E-03 | |||||||||
| AEDGATPSPS*NETPK | S | 147 | 0.936 | 5.11E-04 | 73 | |||||||||||||
| EAPAEGEAAEPGS*PTAAEGEAASAASSTSSPK | S; T | 118; 120 | 0.499; 0.499 | 2.84E-58 | 66 | |||||||||||||
| EAPAEGEAAEPGS*PTAAEGEAASAASSTSSPK | S; T; S | 128; 130; 138 | 0.323; 0.323; 0.323 | 3.44E-05 | 53 | |||||||||||||
| EAPAEGEAAEPGS*PTAAEGEAASAASSTSSPK | S; T | 118; 120 | 0.499; 0.499 | 2.84E-58 | 66 | |||||||||||||
| GEPAAAAAPEAGAS*PVEK | S | 101 | 1 | 7.07E-58 | 115 | |||||||||||||
| MPP2 | 2.5276 | 8.8393 | 8.7985 | 6.85E-06 | 8.95E-04 | 4.3503 | Q14168 | MAGUK p55 subfamily member 2 | TYET*PPPS*PGLDPTFSNQPVPPDAVR | T; S | 141; 145 | 0.833; 0.998 | 1.77E-68 | 80 | 4500 | 5589 | 1.24 | 1.22E-03 |
| T*YETPPPS*PGLDPTFSNQPVPPDAVR | T; S | 138; 145 | 0.537; 0.988 | 6.46E-35 | 63 | |||||||||||||
| MYH14 | − 1.1390 | 8.6919 | − 7.3970 | 2.96E-05 | 1.97E-03 | 2.8674 | Q7Z406 | Myosin-14 | LEEGVAS*DEEAEEAQPGSGPSPEPEGSPPAHPQ | S | 1969 | 1 | 5.84E-67 | 84 | 249 | 413 | 1.66 | 2.86E-01 |
| NBEAL1 | − 1.0097 | 7.8731 | − 4.4773 | 1.32E-03 | 1.47E-02 | − 1.0471 | H7C3C8 | Neurobeachin-like protein 1 (Fragment) | SSDQTNYETEGASIQS*RK | S | 24 | 0.805 | 1.12E-02 | 35 | ||||
| NCOA7 | − 1.4212 | 7.9855 | − 5.1172 | 5.21E-04 | 8.71E-03 | − 0.0842 | H0UI55 | Nuclear receptor coactivator 7 | DALPS*PGEWEDLASEK | S | 295 | 0.99 | 3.21E-03 | 45 | ||||
| VLS*ST*S*EEDEPGVVK | S; T; S | 208; 210; 211 | 0.87; 0.475; 0.475 | 6.29E-06 | 61 | |||||||||||||
| VLSS*TS*EEDEPGVVK | S; S | 209; 211 | 0.77; 0.849 | 1.45E-04 | 79 | |||||||||||||
| NDRG1 | − 3.0949 | 12.3453 | − 3.9310 | 3.07E-03 | 2.42E-02 | − 1.9133 | Q92597 | Protein NDRG1 | TASGSS*VTS*LDGTR | S; S | 333; 336 | 0.728; 0.85 | 1.03E-02 | 50 | 3618 | 7439 | 2.06 | 3.36E-04 |
| NGEF | − 3.0287 | 7.9883 | − 4.7608 | 8.68E-04 | 1.17E-02 | − 0.6133 | Q8N5V2 | Ephexin-1 | SVPIIS*HS*RWLLK | S; S | 485; 487 | 0.914; 0.914 | 1.29E-02 | 44 | 12,102 | 18,559 | 1.53 | 1.83E-03 |
| NPDC1 | 1.0179 | 15.1785 | 5.9750 | 1.63E-04 | 4.64E-03 | 1.1127 | Q5SPY9 | Neural proliferation differentiation and control protein 1 | APGS*PAAPR | S | 307 | 1 | 9.47E-04 | 96 | 4714 | 20,392 | 4.33 | 4.33E-05 |
| IS*PGDQR | S | 314 | 1 | 1.93E-02 | 86 | 4139 | 5242 | 1.27 | 4.50E-02 | |||||||||
| NUP155 | 1.1429 | 8.4281 | 5.2846 | 4.12E-04 | 7.62E-03 | 0.1579 | O75694 | Nuclear pore complex protein Nup155 | AAPQS*PSVPK | S | 992 | 0.997 | 5.58E-06 | 120 | 6885 | 16,363 | 2.38 | 7.92E-05 |
| AAPQSPS*VPK | S | 994 | 0.602 | 5.58E-06 | 120 | 6885 | 16,363 | 2.38 | 7.92E-05 | |||||||||
| OGFR | − 2.1155 | 15.1596 | − 4.0398 | 2.59E-03 | 2.19E-02 | − 1.7379 | Q9NZT2 | Opioid growth factor receptor | SQGDEAGGHGEDRPEPLS*PK | S | 378 | 1 | 7.10E-04 | 61 | ||||
| OTUB1 | 1.4872 | 9.2341 | 8.6065 | 8.28E-06 | 9.95E-04 | 4.1600 | F5GYJ8 | Ubiquitin thioesterase OTUB1 | QEPLGSDS*EGVNCLAYDEAIMAQQDR | S | 18 | 0.48 | 7.04E-03 | 34 | ||||
| OXR1 | 1.1946 | 5.7526 | 3.3704 | 7.57E-03 | 4.20E-02 | − 2.8353 | Q8N573 | Oxidation resistance protein 1 | VVSSTS*EEEEAFTEK | S | 204 | 0.889 | 2.36E-07 | 100 | ||||
| PDLIM4 | − 3.5737 | 9.5064 | − 5.5832 | 2.74E-04 | 6.13E-03 | 0.5795 | P50479 | PDZ and LIM domain protein 4 | IHIDPEIQDGS*PTTSR | S | 112 | 0.923 | 3.16E-04 | 75 | 9266 | 15,989 | 1.73 | 2.03E-03 |
| PEA15 | 1.1346 | 10.2397 | 4.5028 | 1.27E-03 | 1.45E-02 | − 1.0077 | Q15121 | Astrocytic phosphoprotein PEA-15 | QPS*EEEIIK | S | 116 | 1 | 8.87E-06 | 138 | 18,264 | 123,410 | 6.76 | 1.32E-05 |
| PGM5 | − 1.0253 | 5.8422 | − 3.2170 | 9.75E-03 | 4.94E-02 | − 3.0915 | Q15124 | Phosphoglucomutase-like protein 5 | AAGGIILTAS*HCPGGPGGEFGVK | S | 122 | 0.54 | 2.66E-64 | 85 | ||||
| PKM2 | 1.1738 | 12.2601 | 6.1513 | 1.31E-04 | 4.15E-03 | 1.3453 | P14618 | Pyruvate kinase | LDIDS*PPITAR | S | 37 | 0.912 | 1.63E-02 | 47 | ||||
| PLEC | 1.1685 | 10.5703 | 3.6270 | 4.98E-03 | 3.25E-02 | − 2.4100 | Q15149 | Plectin | SDEGQLS*PATR | S | 720 | 0.995 | 1.49E-02 | 82 | 6049 | 13,023 | 2.15 | 3.16E-04 |
| RAS*FAEK | S | 1721 | 1 | 1.18E-02 | 55 | 3016 | 5604 | 1.86 | 9.12E-04 | |||||||||
| TQLAS*WSDPTEETGPVAGILDTETLEK | S | 4406 | 0.909 | 1.61E-15 | 97 | |||||||||||||
| PLXNC1 | 1.8664 | 7.1756 | 5.5181 | 2.99E-04 | 6.40E-03 | 0.4886 | O60486 | Plexin-C1 | KQS*QQLELLESELR | S | 978 | 1 | 3.76E-03 | 47 | 2450 | 4156 | 1.70 | 3.71E-03 |
| POMC | − 7.4874 | 13.4887 | − 3.8110 | 3.71E-03 | 2.70E-02 | − 2.1083 | P01189 | Pro-opiomelanocortin | ACKPDLS*AETPMFPGNGDEQPLTENPR | S | 55 | 0.997 | 1.64E-56 | 167 | 499,933 | 41,941 | 0.08 | 7.79E-07 |
| REDVS*AGEDCGPLPEGGPEPR | S | 108 | 1 | 7.15E-166 | 277 | 379,447 | 90,008 | 0.24 | 3.01E-06 | |||||||||
| ACKPDLSAET*PM#FPGNGDEQPLTENPR | T | 58 | 0.759 | 1.26E-45 | 159 | 75,285 | 1928 | 0.03 | 8.83E-06 | |||||||||
| VYPNGAEDES*AEAFPLEFK | S | 168 | 1 | 3.56E-34 | 127 | 17,243 | 8453 | 0.49 | 8.78E-04 | |||||||||
| EGDGPDGPADDGAGAQADLEHS*LLVAAEK | S | 208 | 1 | 1.38E-05 | 58 | |||||||||||||
| S*DGAKPGPR | S | 125 | 1 | 1.04E-02 | 54 | |||||||||||||
| PRKAR2B | − 1.0157 | 9.9746 | − 3.4835 | 6.29E-03 | 3.75E-02 | − 2.6473 | P31323 | cAMP-dependent protein kinase type II-beta regulatory subunit | RAS*VCAEAYNPDEEEDDAESR | S | 114 | 1 | 4.14E-31 | 139 | 37,944 | 26,707 | 0.70 | 6.75E-04 |
| PRKCD | 1.9942 | 9.0704 | 7.0482 | 4.40E-05 | 2.33E-03 | 2.4620 | Q05655 | Protein kinase C delta type | ASTFCGTPDY*IAPEILQGLK | Y | 514 | 0.511 | 4.36E-02 | 33 | ||||
| PRKG1 | − 2.0409 | 5.5427 | − 4.3580 | 1.58E-03 | 1.65E-02 | − 1.2331 | Q13976 | cGMP-dependent protein kinase 1 | T*WTFCGTPEYVAPEIILNK | T | 515 | 0.543 | 4.49E-15 | 123 | ||||
| PXN | 1.0331 | 7.0094 | 3.5188 | 5.94E-03 | 3.61E-02 | − 2.5888 | P49023 | Paxillin | S*SPGGQDEGGFMAQGK | S; S | 302; 303 | 0.5; 0.5 | 1.30E-03 | 55 | 1185 | 1696 | 1.43 | 2.16E-02 |
| TGSSS*PPGGPPKPGSQLDSMLGSLQSDLNK | S | 322 | 0.702 | 5.72E-19 | 89 | 4056 | 7646 | 1.88 | 4.60E-02 | |||||||||
| T*GS*SSPPGGPPKPGSQLDSMLGSLQSDLNK | T; S | 318; 320 | 0.259; 0.259 | 4.20E-06 | 54 | |||||||||||||
| T*GSSSPPGGPPKPGSQLDSMLGSLQSDLNK | T | 318 | 0.291 | 4.31E-19 | 96 | |||||||||||||
| QSOX2 | − 1.2837 | 6.3713 | − 4.1443 | 2.20E-03 | 1.99E-02 | − 1.5708 | Q6ZRP7 | Sulfhydryl oxidase 2 | DNLLDTYSADQGDS*SEGGTLAR | S | 578 | 0.821 | 1.21E-12 | 81 | ||||
| DNLLDTYSADQGDSS*EGGTLAR | S | 579 | 0.556 | 5.21E-33 | 72 | |||||||||||||
| RCAN1 | − 1.0836 | 9.4397 | − 3.4404 | 6.75E-03 | 3.91E-02 | − 2.7189 | P53805 | Calcipressin-1;Calcipressin-3 | QFLIS*PPAS*PPVGWK | S; S | 163; 167 | 1; 1 | 1.21E-25 | 100 | 5957 | 12,269 | 2.06 | 1.21E-03 |
| RRBP1 | − 1.3657 | 10.4681 | − 3.9869 | 2.81E-03 | 2.30E-02 | − 1.8230 | Q9P2E9 | Ribosome-binding protein 1 | NTDVAQS*PEAPK | S | 615 | 0.996 | 3.41E-04 | 55 | ||||
| SCG3 | 1.2843 | 10.4326 | 4.3052 | 1.72E-03 | 1.72E-02 | − 1.3159 | Q8WXD2 | Secretogranin-3 | ELS*AERPLNEQIAEAEEDKIK | S | 37 | 1 | 2.80E-38 | 149 | 54,239 | 25,082 | 0.46 | 1.50E-04 |
| LFPAPS*EKSHEETDSTK | S | 359 | 0.468 | 1.85E-02 | 36 | |||||||||||||
| SET | − 1.7108 | 7.9953 | − 8.2175 | 1.23E-05 | 1.23E-03 | 3.7624 | Q01105 | Protein SET | LNEQAS*EEILK | S | 63 | 1 | 1.18E-16 | 87 | ||||
| SLC2A4 | − 1.3065 | 5.1362 | − 4.4148 | 1.45E-03 | 1.56E-02 | − 1.1444 | P14672 | Solute carrier family 2, facilitated glucose transporter member 4 | T*PSLLEQEVKPSTELEYLGPDEND | T; S | 486; 488 | 0.5; 0.5 | 1.85E-10 | 91 | ||||
| SLC46A1 | − 1.1586 | 7.9882 | − 4.9777 | 6.35E-04 | 9.78E-03 | − 0.2891 | A0A024QZ15 | Proton-coupled folate transporter | AD*LEFQQFPQS*P | S | 458 | 1 | 1.60E-05 | 107 | 19,669 | 26,410 | 1.34 | 4.09E-02 |
| SLC7A14 | − 1.3388 | 10.1699 | − 4.9424 | 6.68E-04 | 1.01E-02 | − 0.3414 | Q8TBB6 | Probable cationic amino acid transporter | EACS*PVS*EGDEFSGPATNTCGAK | S | 465; 468 | 0.998; 0.939 | 1.03E-05 | 67 | ||||
| SLC8A2 | 2.0827 | 10.6502 | 5.7643 | 2.15E-04 | 5.42E-03 | 0.8287 | Q9UPR5 | Sodium/calcium exchanger 2 | GIS*ALLLNQGDGDR | S | 622 | 1 | 7.55E-34 | 97 | 1039 | 1666 | 1.60 | 1.40E-01 |
| SMARCA2 | − 1.0693 | 9.4559 | − 3.2347 | 9.47E-03 | 4.85E-02 | − 3.0619 | P51531 | Probable global transcription activator SNF2L2 | AKPVVS*DFDS*DEEQDER | S | 1568; 1572 | 1; 1 | 3.09E-10 | 108 | 8702 | 21,740 | 2.50 | 1.06E-05 |
| SNAP23 | 1.0040 | 6.7468 | 4.8842 | 7.26E-04 | 1.05E-02 | − 0.4280 | O00161 | Synaptosomal-associated protein; synaptosomal-associated protein 23 | TTWGDGGENS*PCNVVSK | S | 110 | 1 | 3.54E-04 | 83 | 5323 | 11,357 | 2.13 | 6.07E-04 |
| SNX17 | 1.1624 | 8.6272 | 4.3682 | 1.56E-03 | 1.63E-02 | − 1.2171 | Q15036 | Sorting nexin-17 | VTSSVPLPSGSTSS*PGR | S | 336 | 0.658 | 4.46E-05 | 89 | 5947 | 11,510 | 1.94 | 1.05E-03 |
| VTSSVPLPS*GS*T*S*S*PGR | S; S; T; S; S | 331; 333; 334; 335; 336 | 0.2; 0.2; 0.2; 0.2; 0.2 | 1.79E-03 | 67 | |||||||||||||
| SORBS2 | − 2.2906 | 6.9395 | − 3.2557 | 9.15E-03 | 4.75E-02 | − 3.0267 | O94875 | Sorbin and SH3 domain-containing protein 2 | QNAEIWSS*T*EET*VS*PKIK | S; T; T; S | 370; 371; 374; 376 | 0.238; 0.238; 0.238; 0.238 | 1.88E-02 | 34 | ||||
| SORT1 | 1.0872 | 11.4517 | 5.0676 | 5.58E-04 | 9.09E-03 | − 0.1567 | Q99523 | Sortilin | SGYHDDS*DEDLLE | S | 825 | 0.999 | 1.99E-04 | 99 | 4842 | 5538 | 1.14 | 1.42E-01 |
| SPAG9 | 1.1290 | 9.1942 | 3.9292 | 3.08E-03 | 2.42E-02 | − 1.9163 | O60271 | C-Jun-amino-terminal kinase-interacting protein 4 | SAS*QS*SLDKLDQELK | S; S | 730; 732 | 0.834; 0.554 | 2.29E-17 | 145 | 21,879 | 108,870 | 4.98 | 2.23E-07 |
| SAS*QS*S*LDKLDQELK | S | 730; 732; 733 | 0.577; 0.507; 0.91 | 5.14E-44 | 145 | 11,070 | 44,110 | 3.98 | 4.65E-06 | |||||||||
| SPARCL1 | − 3.1871 | 13.6134 | − 4.7627 | 8.66E-04 | 1.17E-02 | − 0.6103 | Q14515 | SPARC-like protein 1 | EES*HEQS*AEQGK | S | 80 | 9.37E-38 | 176 | 44,342 | 10,604 | 0.24 | 1.86E-05 | |
| SSS*QELGLK | S | 92 | 0.999 | 1.61E-28 | 188 | 100,790 | 28,141 | 0.28 | 1.91E-05 | |||||||||
| SKEES*HEQS*AEQGK | S; S | 80; 84 | 0.999; 1 | 9.37E-38 | 176 | 57,751 | 20,002 | 0.35 | 5.38E-05 | |||||||||
| EHANS*KQEEDNTQSDDILEESDQPTQVSK | S | 231 | 1 | 5.61E-70 | 176 | 20,394 | 7396 | 0.36 | 5.25E-04 | |||||||||
| HIQETEWQS*QEGK | S | 295 | 1 | 2.83E-21 | 152 | 24,777 | 14,058 | 0.57 | 1.15E-03 | |||||||||
| DQGNQEQDPNIS*NGEEEEEKEPGEVGTHNDNQER | S | 198 | 0.997 | 1.10E-05 | 53 | 2334 | 2083 | 0.89 | 1.92E-01 | |||||||||
| EHANS*KQEEDNT*QS*DDILEESDQPTQVSK | S; T; S | 231; 238; 240 | 0.331; 0.331; 0.331 | 1.44E-02 | 38 | |||||||||||||
| S*KEES*HEQSAEQGK | S; S | 76; 80 | 0.999; 0.995 | 7.77E-05 | 77 | |||||||||||||
| SPTB | 1.0905 | 6.8402 | 3.9457 | 3.00E-03 | 2.38E-02 | − 1.8895 | P11277 | Spectrin beta chain, erythrocyte | LS*S*S*WESLQPEPSHPY | S | 2123; 2124; 2125 | 0.653; 0.693; 0.653 | 1.43E-23 | 154 | 9038 | 16,844 | 1.86 | 4.11E-04 |
| WDAPDDELDNDNSS*AR | S | 36 | 0.964 | 1.15E-18 | 148 | 6818 | 10,720 | 1.57 | 7.93E-04 | |||||||||
| LSS*S*WESLQPEPSHPY | S | 2124; 2125 | 0.582; 0.983 | 4.77E-17 | 137 | 18,958 | 25,412 | 1.34 | 5.27E-03 | |||||||||
| LSSS*WES*LQPEPSHPY | S; S | 2125; 2128 | 0.86; 1 | 8.08E-10 | 116 | 12,384 | 10,854 | 0.88 | 7.41E-02 | |||||||||
| VLS*PVDSGNK | S | 1226 | 1 | 4.33E-03 | 63 | |||||||||||||
| SRRM2 | 1.1850 | 7.1163 | 6.1599 | 1.29E-04 | 4.14E-03 | 1.3566 | Q9UQ35 | Serine/arginine repetitive matrix protein 2 | TS*PPLLDR | S | 2398 | 0.899 | 1.67E-02 | 70 | 2052 | 6736 | 3.28 | 7.74E-07 |
| EISS*S*PTSK | S | 455; 456 | 0.657; 0.864 | 4.72E-04 | 99 | 8692 | 22,507 | 2.59 | 1.78E-06 | |||||||||
| RPS*PQPS*PR | S | 2702; 2706 | 1; 1 | 1.13E-03 | 78 | 6822 | 21,190 | 3.11 | 4.31E-06 | |||||||||
| SGMS*PEQSR | S | 1132 | 0.767 | 5.10E-03 | 79 | 6423 | 23,060 | 3.59 | 4.83E-06 | |||||||||
| SST*PPRQS*PSR | S; S | 903; 908 | 0.756; 0.93 | 1.17E-02 | 89 | 7024 | 17,287 | 2.46 | 1.19E-05 | |||||||||
| S*S*T*PPRQSPSR | S; S; T | 901; 902; 903 | 0.4476; 0.447; 0.849 | 1.17E-02 | 89 | 7916 | 19,551 | 2.47 | 1.62E-05 | |||||||||
| SS*RSSPELTR | S | 1691 | 0.505 | 1.44E-02 | 60 | 3402 | 9542 | 2.81 | 1.81E-05 | |||||||||
| S*RS*PLAIR | S; S | 2044; 2046 | 1; 1 | 1.07E-02 | 63 | 11,261 | 26,216 | 2.33 | 2.12E-05 | |||||||||
| ENSFGS*PLEFR | S | 1329 | 1 | 3.46E-05 | 113 | 12,581 | 33,168 | 2.64 | 2.86E-05 | |||||||||
| EIS*SS*PTSK | S | 454 | 0.946 | 1.16E-03 | 94 | 9678 | 23,505 | 2.43 | 2.97E-05 | |||||||||
| S*S*RS*SPELTR | S; S; S | 1690; 1691; 1693 | 0.501; 0.501; 0.664 | 1.44E-02 | 60 | 1233 | 3849 | 3.12 | 3.92E-05 | |||||||||
| T*SPPLLDR | T | 2397 | 0.949 | 9.09E-03 | 125 | 1856 | 5793 | 3.12 | 4.14E-05 | |||||||||
| S*RS*PATAK | S; S | 484; 486 | 1; 1 | 8.82E-03 | 66 | 2800 | 13,671 | 4.88 | 5.19E-05 | |||||||||
| SCFES*S*PDPELK | S; S | 875; 876 | 1; 1 | 2.22E-05 | 118 | 15,580 | 35,770 | 2.30 | 5.73E-05 | |||||||||
| SLS*YS*PVER | S; S | 2692; 2694; | 0.997; 0.999 | 2.02E-04 | 103 | 9902 | 30,084 | 3.04 | 7.07E-05 | |||||||||
| VPS*PTPAPK | S | 2581 | 0.964 | 1.55E-02 | 61 | 3557 | 11,007 | 3.09 | 7.34E-05 | |||||||||
| GEFSAS*PMLK | S | 1124 | 0.999 | 1.38E-03 | 104 | 14,424 | 35,744 | 2.48 | 9.98E-05 | |||||||||
| SGS*S*PGLR | S; S | 1443; 1444 | 0.933; 0.899 | 5.07E-03 | 82 | 4248 | 12,065 | 2.84 | 1.49E-04 | |||||||||
| QGSITS*PQANEQSVT*PQRR | S; T | 857; 866 | 0.761; 0.779 | 5.23E-03 | 63 | 1771 | 5578 | 3.15 | 1.74E-04 | |||||||||
| AQT*PPGPSLSGSKS*PCPQEK | S | 1014 | 0.984 | 5.61E-04 | 65 | 2459 | 11,814 | 4.81 | 2.24E-04 | |||||||||
| SATRPS*PS*PER | S; S | 351; 353 | 0.982; 0.994 | 5.54E-03 | 85 | 9186 | 27,782 | 3.02 | 2.36E-04 | |||||||||
| S*RT*PPVTR | S; T | 1925; 1927 | 1; 1 | 1.78E-02 | 72 | 3013 | 9448 | 3.14 | 3.54E-04 | |||||||||
| GDS*RS*PSHK | S; S | 2727; 2729 | 1; 0.999 | 1.24E-02 | 57 | 2764 | 11,511 | 4.16 | 4.64E-04 | |||||||||
| S*PS*PASGR | S; S | 295; 297 | 1; 0.999 | 3.32E-04 | 77 | 8935 | 20,961 | 2.35 | 7.87E-04 | |||||||||
| S*SSPVTELASRS*PIR | S | 1112 | 0.834 | 4.10E-05 | 83 | 3678 | 7472 | 2.03 | 1.16E-03 | |||||||||
| S*VS*PCSNVESR | S; S | 952; 954 | 1; 0.978 | 1.24E-02 | 84 | 6685 | 12,365 | 1.85 | 1.24E-03 | |||||||||
| S*LS*GSSPCPK | S; S | 778; 780 | 1; 0.999 | 1.17E-04 | 112 | 31,177 | 58,376 | 1.87 | 1.42E-03 | |||||||||
| AQT*PPGPSLSGS*KSPCPQEK | T; S | 1003; 1012 | 1; 0.693 | 1.98E-15 | 123 | 1958 | 4374 | 2.23 | 3.06E-03 | |||||||||
| SS*SPVTELASR | S | 1102 | 0.567 | 4.10E-05 | 87 | 10,436 | 15,924 | 1.53 | 5.33E-03 | |||||||||
| S*RS*PSSPELNNK | S; S | 1497; 1499 | 0.93; 0.93 | 5.98E-03 | 66 | 11,049 | 15,474 | 1.40 | 1.06E-02 | |||||||||
| RGEGDAPFSEPGTTSTQRPS*S*PETATK | S | 322; 323 | 0.646; 0.847 | 2.10E-17 | 101 | 1813 | 2549 | 1.41 | 1.28E-02 | |||||||||
| SSS*PVTELASR | S | 1103 | 0.993 | 3.29E-15 | 154 | 8557 | 9123 | 1.07 | 3.32E-01 | |||||||||
| GRS*ECDS*S*PEPK | S; S; S | 1477; 1482; 1483 | 1; 1; 1 | 1.00E-07 | 128 | 10,775 | 11,809 | 1.10 | 3.56E-01 | |||||||||
| AQT*PPGPSLS*GSKSPCPQEK | S | 1010 | 0.996 | 3.50E-03 | 55 | |||||||||||||
| EISSSPT*SK | T | 458 | 0.551 | 1.25E-02 | 75 | |||||||||||||
| ERS*GS*ESSVDQK | S; S | 1517; 1519 | 0.993; 0.995 | 5.19E-03 | 65 | |||||||||||||
| GQSQT*SPDHR | T; S | 1063; 1064 | 0.496; 0.496 | 2.73E-02 | 79 | |||||||||||||
| HGGS*PQPLATTPLSQEPVNPPSEAS*PT*R | S | 377; 398; 400 | 0.991; 0.415; 0.415 | 3.40E-04 | 37 | |||||||||||||
| HGGS*PQPLATTPLSQEPVNPPS*EASPTR | S | 395 | 0.953 | 3.40E-04 | 37 | |||||||||||||
| MALPPQEDAT*ASPPR | T | 1177 | 0.855 | 1.04E-02 | 55 | |||||||||||||
| QSHSES*PSLQSK | S | 1083 | 0.994 | 3.88E-03 | 96 | |||||||||||||
| RGEGDAPFSEPGT*T*S*T*QRPS*S*PETATK | T; T; S; T; S; S | 315; 316; 317; 318; 322; 323 | 0.325; 0.325; 0.325; 0.325; 0.325; 0.325 | 3.79E-03 | 50 | |||||||||||||
| RGEGDAPFSEPGT*T*S*T*QRPS*S*PETATK | T; S; T | 316; 317; 318 | 0.399; 0.399; 0.399 | 1.72E-04 | 57 | |||||||||||||
| S*DTSSPEVR | S | 1069 | 0.307 | 6.58E-03 | 66 | |||||||||||||
| S*GS*S*QELDVKPS*AS*PQER | S; S; S; S; S | 1539; 1541; 1542; 1550; 1552 | 0.334; 0.334; 0.334; 0.499; 0.499 | 2.63E-03 | 54 | |||||||||||||
| S*SS*PVTELASR | S; S | 1101; 1103 | 0.96; 0.955 | 4.10E-05 | 83 | |||||||||||||
| SSBP3 | 1.2570 | 7.2589 | 4.6141 | 1.08E-03 | 1.32E-02 | − 0.8364 | Q9BWW4 | Single-stranded DNA-binding protein 3 | NS*PNNISGISNPPGTPR | S | 347 | 1 | 4.55E-14 | 121 | 5331 | 8383 | 1.57 | 4.80E-03 |
| STIM1 | 1.0481 | 9.8564 | 4.9079 | 7.01E-04 | 1.03E-02 | − 0.3927 | Q13586 | Stromal interaction molecule 1 | AEQS*LHDLQER | S | 257 | 1 | 1.74E-02 | 78 | ||||
| STK39 | 1.4339 | 11.9102 | 6.9767 | 4.79E-05 | 2.42E-03 | 2.3769 | Q9UEW8 | STE20/SPS1-related proline-alanine-rich protein kinase | TEDGDWEWS*DDEMDEK | S | 385 | 1 | 1.13E-219 | 188 | 27,341 | 40,397 | 1.48 | 7.42E-04 |
| STX1A | 1.8823 | 8.2932 | 5.5539 | 2.85E-04 | 6.25E-03 | 0.5386 | Q16623 | Syntaxin-1A | TAKDS*DDDDDVAVTVDR | S | 14 | 1 | 4.88E-57 | 182 | 72,880 | 183,350 | 2.52 | 9.68E-06 |
| STXBP5L | 1.5925 | 9.9115 | 4.8616 | 7.50E-04 | 1.07E-02 | − 0.4617 | Q9Y2K9 | Syntaxin-binding protein 5-like | SS*SISS*IDKDSK | S; S; S | 819; 820; 823 | 0.346; 0.346; 0.831 | 1.68E-02 | 44 | ||||
| SYN1 | − 1.0498 | 11.9626 | − 5.4376 | 3.34E-04 | 6.79E-03 | 0.3755 | P17600 | Synapsin-1 | PVAGGPGAPPAARPPAS*PS*PQR | S; S | 551; 553 | 1; 1 | 1.08E-22 | 130 | 8100 | 33,111 | 4.09 | 3.59E-04 |
| TCEA1 | − 1.0289 | 8.8480 | − 4.2602 | 1.84E-03 | 1.79E-02 | − 1.3868 | P23193 | Transcription elongation factor A protein 1 | EPAITSQNS*PEAR | S | 100 | 1 | 1.20E-47 | 197 | 66,104 | 140,043 | 2.12 | 8.61E-05 |
| TCF12 | − 1.1897 | 9.1467 | − 3.9273 | 3.09E-03 | 2.42E-02 | − 1.9193 | A0A024R5Z0 | Transcription factor 12 | T*S*ST*NEDEDLNPEQK | T; S; T | 581; 582; 584 | 0.256; 0.256; 0.256 | 3.44E-02 | 44 | ||||
| TLN2 | 1.5289 | 7.8000 | 4.3424 | 1.62E-03 | 1.67E-02 | − 1.2575 | Q9Y4G6 | Talin-2 | LDEGT*PPEPK | T | 1843 | 1 | 1.10E-02 | 93 | 7613 | 8825 | 1.16 | 1.17E-02 |
| TNKS1BP1 | 1.0225 | 7.5659 | 4.1437 | 2.20E-03 | 1.99E-02 | − 1.5717 | Q9C0C2 | 182 kDa tankyrase-1-binding protein | AS*PEPPGPESSSR | S | 672 | 1 | 1.03E-03 | 70 | 6115 | 19,567 | 3.20 | 2.19E-05 |
| VPS*S*DEEVVEEPQSR | S; S | 1620; 1621 | 1; 1 | 1.38E-16 | 138 | 7581 | 16,197 | 2.14 | 4.57E-05 | |||||||||
| GWS*QEGPVK | S | 221 | 1 | 1.47E-02 | 62 | 6177 | 15,957 | 2.58 | 6.76E-05 | |||||||||
| LDS*PPPSPITEASEAAEAAEAGNLAVSSR | S | 494 | 0.984 | 1.15E-12 | 88 | |||||||||||||
| LDS*PPPSPITEASEAAEAAEAGNLAVSSR | S | 494; 498 | 0.492; 0.492 | 1.22E-05 | 38 | |||||||||||||
| TPI1 | 1.7746 | 10.1649 | 6.8527 | 5.53E-05 | 2.64E-03 | 2.2279 | P60174 | Triosephosphate isomerase | KQS*LGELIGTLNAAK | S | 58 | 1 | 5.49E-61 | 200 | 57,287 | 111,203 | 1.94 | 2.76E-05 |
| IIYGGS*VTGATCK | S | 249 | 0.992 | 7.14E-37 | 110 | |||||||||||||
| TPMT | 1.0815 | 6.1567 | 4.4016 | 1.48E-03 | 1.58E-02 | − 1.1650 | P51580 | Thiopurine S-methyltransferase | TSLDIEEYS*DTEVQK | S | 14 | 0.806 | 4.23E-05 | 86 | ||||
| TRIM3 | 1.4660 | 5.4120 | 4.1545 | 2.16E-03 | 1.97E-02 | − 1.5544 | O75382 | Tripartite motif-containing protein 3 | REDS*PGPEVQPMDK | S | 7 | 1 | 8.45E-07 | 73 | ||||
| TSC2 | 2.1795 | 10.7866 | 6.3857 | 9.73E-05 | 3.49E-03 | 1.6479 | P49815 | Tuberin | S*SSSPELQTLQDILGDPGDK | S; S; S | 1385; 1386; 1387 | 0.313; 0.313; 0.313 | 1.56E-11 | 74 | ||||
| USP24 | 1.3950 | 11.1223 | 9.0626 | 5.31E-06 | 7.86E-04 | 4.6057 | Q9UPU5 | Ubiquitin carboxyl-terminal hydrolase 24 | VSDQNS*PVLPK | S | 2047 | 1 | 7.77E-03 | 92 | 24,339 | 33,164 | 1.36 | 4.29E-03 |
| WBP4 | 1.0929 | 7.4877 | 4.0214 | 2.66E-03 | 2.23E-02 | − 1.7675 | O75554 | WW domain-binding protein 4 | NSDGGS*DPETQK | S | 262 | 0.996 | 5.26E-03 | 90 | 4151 | 7080 | 1.71 | 4.96E-04 |
DEG, differentially expressed gene; LogFC, log2(fold change); LogFC > = 1, upregulated DEG; LogFC <= − 1, downregulated DEGs; T, NFPA; N, control pituitaries; Ratio (T/N), ratio of phosphorylation level of NFPAs to controls
Gene ontology enrichment analysis
The GO enrichment analysis of those identified 130 overlapped molecules (phosphoproteins; invasive DEGs) was annotated with CCs (Supplemental Table 6), MFs (Supplemental Table 7), and BPs (Supplemental Table 8). CC analysis revealed those 130 overlapped molecules (phosphoproteins; invasive DEGs) were mainly distributed in the cytosol, focal adhesion, secretory granule, actin cytoskeleton, cell–cell adherens junction, cytoskeleton, plasma membrane, cell–cell junction, cell cortex, membrane, stress, protein complex, COP9 signalosome, cytoplasm, Golgi apparatus, nuclear speck, SNARE complex, and kinesin complex (Supplemental Table 6). MF analysis revealed that those overlapped molecules (phosphoproteins; invasive DEGs) were significantly involved in cadherin binding involved in cell–cell adhesion, actin filament binding, protein binding, actin binding, structural constituent of cytoskeleton, guanyl-nucleotide exchange factor activity, Rho guanyl-nucleotide exchange factor activity, calmodulin binding, GTPase activator activity, ankyrin binding, structural molecule activity, calcium ion transmembrane transporter activity, spectrin binding, poly(A) RNA binding, microtubule binding, neuropeptide hormone activity, inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity, phosphatidylinositol binding, hormone activity, phosphatidylinositol-3,4,5-trisphosphate binding, and protein kinase activity (Supplemental Table 7). BP analysis revealed multiple statistically significant biological processes, including cell–cell adhesion, regulation of insulin secretion, striated muscle cell differentiation, positive regulation of GTPase activity, neurotransmitter secretion, glucose homeostasis, cellular response to prostaglandin E stimulus, regulation of small GTPase-mediated signal transduction, regulation of Rho protein signal transduction, positive regulation of apoptotic process, renal water homeostasis, glucose transport, positive regulation of endodeoxyribonuclease activity, peptidyl-threonine phosphorylation, transmembrane transport, positive regulation of cAMP biosynthetic process, cellular response to hydroperoxide, signal transduction, platelet degranulation and activation, cellular iron ion homeostasis, and peripheral nervous system myelin maintenance (Supplemental Table 8).
Clustering analysis
The clustering analysis of 130 overlapped molecules (phosphoproteins; invasive DEGs) identified 6 significantly functional categories (Table 5). Cluster 1 was mainly associated with cell–cell adherens and regulation, which was involved in the important overlapped molecules (phosphoproteins; invasive DEGs), including ALDOA, TNKS1BP1, LIMA1, CALD1, ASAP1, EPS15L1, H1FX, NDRG1, PLEC, CHMP2B, and PGM5. Cluster 2 was involved in platelet activation, which was involved in the important overlapped molecules (phosphoproteins; invasive DEGs), including AKT1, TLN2, STIM1, GNAS, SNAP23, PRKG1, ARHGEF12, ITPR1, ITPR2, and ITGA2B. Cluster 3 was mainly associated with GTPase signaling and regulation, which was involved in the important overlapped molecules (phosphoproteins; invasive DEGs), including AKT1, ARHGEF11, ARHGEF11, ARHGEF12, ARHGEF6, ASAP1, ASAP2, BCLAF1, CDC42EP4, DOCK11, DOCK7, GAL, GNAS, NGEF, SPTB, STXBP5L, and TSC2. Cluster 4 was associated with the microtubule and its regulation, which was involved in the overlapped molecules (phosphoproteins; invasive DEGs), including CLASP2, KIF13B, KIF16B, KIF1A, MAP4, NDRG1, and STIM1. Cluster 5 was mainly associated with protein kinase activity and peptidyl-threonine phosphorylation, which was involved in the important overlapped molecules (phosphoproteins; invasive DEGs), including AKT1, AVP, CASK, STK39, PRKCD, CDK14, and DCLK1. Cluster 6 was mainly associated with calcium signaling pathway and regulation, which was involved in the overlapped molecules (phosphoproteins; invasive DEGs), including SLC8A2, STIM1, ITPR1, ITPR2, and GNAS. Thereby, those phosphoproteins participated in the corresponding biological functions in NFPAs. It clearly demonstrated the important roles of phosphorylation in invasive NFPA pathogenesis.
Table 5.
Functional characteristics of 130 overlapped molecules between phosphoproteins and invasive DEGs in NFPAs, clustered with GO and KEGG pathway enrichments. Count means the number of genes enriched in each item
| Category | Functional characteristics | Count | % | Fold enrichment | P value | Gene name of overlapped molecule |
|---|---|---|---|---|---|---|
| Annotation cluster 1 | ||||||
| GOTERM_CC_DIRECT | GO:0005913~cell–cell adherens junction | 11 | 8.73 | 5.05 | 6.35E-05 | ALDOA, TNKS1BP1, LIMA1, PGM5, CALD1, ASAP1, EPS15L1, H1FX, NDRG1, PLEC, CHMP2B |
| GOTERM_BP_DIRECT | GO:0098609~cell–cell adhesion | 10 | 7.94 | 5.16 | 1.34E-04 | ALDOA, TNKS1BP1, LIMA1, CALD1, ASAP1, EPS15L1, H1FX, NDRG1, PLEC, CHMP2B |
| GOTERM_MF_DIRECT | GO:0098641~cadherin binding involved in cell–cell adhesion | 10 | 7.94 | 4.73 | 2.58E-04 | ALDOA, TNKS1BP1, LIMA1, CALD1, ASAP1, EPS15L1, H1FX, NDRG1, PLEC, CHMP2B |
| Annotation cluster 2 | ||||||
| KEGG_PATHWAY | hsa04611:Platelet activation | 10 | 7.94 | 10.58 | 2.53E-07 | AKT1, TLN2, STIM1, GNAS, SNAP23, PRKG1, ARHGEF12, ITPR1, ITPR2, ITGA2B |
| Annotation cluster 3 | ||||||
| GOTERM_MF_DIRECT | GO:0005085~guanyl-nucleotide exchange factor activity | 6 | 4.76 | 6.98 | 1.66E-03 | NGEF, ARHGEF6, DOCK7, ARHGEF12, DOCK11, ARHGEF11 |
| GOTERM_BP_DIRECT | GO:0043547~positive regulation of GTPase activity | 12 | 9.52 | 2.97 | 2.25E-03 | NGEF, ARHGEF6, TSC2, ASAP2, ASAP1, GNAS, ARHGEF12, CDC42EP4, DOCK11, ARHGEF11, STXBP5L, SPTB |
| GOTERM_MF_DIRECT | GO:0005089~Rho guanyl-nucleotide exchange factor activity | 5 | 3.97 | 8.91 | 2.34E-03 | NGEF, ARHGEF6, ARHGEF12, DOCK11, ARHGEF11 |
| GOTERM_MF_DIRECT | GO:0005096~GTPase activator activity | 8 | 6.35 | 3.94 | 4.17E-03 | ARHGEF6, TSC2, ASAP2, ASAP1, ARHGEF12, CDC42EP4, ARHGEF11, STXBP5L |
| GOTERM_BP_DIRECT | GO:0051056~regulation of small GTPase mediated signal transduction | 5 | 3.97 | 5.22 | 1.52E-02 | NGEF, ARHGEF6, TSC2, ARHGEF12, ARHGEF11 |
| GOTERM_BP_DIRECT | GO:0035023~regulation of Rho protein signal transduction | 4 | 3.17 | 6.91 | 1.98E-02 | NGEF, ARHGEF6, ARHGEF12, ARHGEF11 |
| GOTERM_BP_DIRECT | GO:0043065~positive regulation of apoptotic process | 7 | 5.56 | 3.27 | 2.00E-02 | AKT1, NGEF, BCLAF1, ARHGEF6, ARHGEF12, GAL, ARHGEF11 |
| Annotation cluster 4 | ||||||
| GOTERM_MF_DIRECT | GO:0008017~microtubule binding | 6 | 4.76 | 3.96 | 1.76E-02 | KIF1A, MAP4, NDRG1, CLASP2, KIF16B, KIF13B |
| GOTERM_CC_DIRECT | GO:0005874~microtubule | 7 | 5.56 | 3.33 | 1.83E-02 | KIF1A, STIM1, MAP4, NDRG1, CLASP2, KIF16B, KIF13B |
| GOTERM_CC_DIRECT | GO:0005871~kinesin complex | 3 | 2.38 | 8.39 | 4.91E-02 | KIF1A, KIF16B, KIF13B |
| Annotation cluster 5 | ||||||
| GOTERM_BP_DIRECT | GO:0018107~peptidyl-threonine phosphorylation | 3 | 2.38 | 11.05 | 2.97E-02 | AKT1, STK39, PRKCD |
| GOTERM_MF_DIRECT | GO:0004672~protein kinase activity | 7 | 5.56 | 2.68 | 4.61E-02 | AKT1, AVP, CASK, STK39, PRKCD, CDK14, DCLK1 |
| Annotation cluster 6 | ||||||
| GOTERM_BP_DIRECT | GO:1903779~regulation of cardiac conduction | 4 | 3.17 | 10.00 | 7.32E-03 | SLC8A2, STIM1, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04020:Calcium signaling pathway | 5 | 3.97 | 3.84 | 3.79E-02 | SLC8A2, STIM1, GNAS, ITPR1, ITPR2 |
| Annotation cluster 7 | ||||||
| KEGG_PATHWAY | hsa04611:Platelet activation | 10 | 7.94 | 10.58 | 2.53E-07 | AKT1, TLN2, STIM1, GNAS, SNAP23, PRKG1, ARHGEF12, ITPR1, ITPR2, ITGA2B |
| KEGG_PATHWAY | hsa04270:Vascular smooth muscle contraction | 8 | 6.35 | 9.41 | 1.64E-05 | CALD1, GNAS, PRKG1, ARHGEF12, PRKCD, ITPR1, ARHGEF11, ITPR2 |
| KEGG_PATHWAY | hsa04915:Estrogen signaling pathway | 6 | 4.76 | 8.34 | 6.44E-04 | AKT1, FKBP5, GNAS, PRKCD, ITPR1, ITPR2 |
| GOTERM_BP_DIRECT | GO:0050796~regulation of insulin secretion | 5 | 3.97 | 10.44 | 1.30E-03 | STX1A, MARCKS, GNAS, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04922:Glucagon signaling pathway | 5 | 3.97 | 6.95 | 5.21E-03 | AKT1, ACACA, GNAS, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04730:Long-term depression | 4 | 3.17 | 9.17 | 8.74E-03 | GNAS, PRKG1, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa05205:Proteoglycans in cancer | 6 | 4.76 | 4.13 | 1.34E-02 | AKT1, ANK1, ARHGEF12, ITPR1, PXN, ITPR2 |
| KEGG_PATHWAY | hsa04970:Salivary secretion | 4 | 3.17 | 6.40 | 2.30E-02 | GNAS, PRKG1, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04540:Gap junction | 4 | 3.17 | 6.25 | 2.44E-02 | GNAS, PRKG1, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04022:cGMP–PKG signaling pathway | 5 | 3.97 | 4.35 | 2.55E-02 | AKT1, SLC8A2, PRKG1, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04912:GnRH signaling pathway | 4 | 3.17 | 6.05 | 2.66E-02 | GNAS, PRKCD, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04750:Inflammatory mediator regulation of TRP channels | 4 | 3.17 | 5.62 | 3.22E-02 | GNAS, PRKCD, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04020:Calcium signaling pathway | 5 | 3.97 | 3.84 | 3.79E-02 | SLC8A2, STIM1, GNAS, ITPR1, ITPR2 |
| GOTERM_BP_DIRECT | GO:0030168~platelet activation | 4 | 3.17 | 4.87 | 4.84E-02 | AKT1, PRKCD, ITPR1, ITPR2 |
Signaling pathways involved in overlapped molecules (phosphoproteins; invasive DEGs)
The KEGG pathway enrichment analysis of 130 overlapped molecules (phosphoproteins; invasive DEGs) identified 14 statistically significant signaling pathways (Table 6; Supplemental Fig. 1) that were related to tumor invasiveness and involved in protein phosphorylations (Table 4), including (i) platelet activation involved in phosphorylation at residues Ser122 and Ser124 in AKT1; Thr1843 (ratio of T/N = 1.16, P = 1.17E-02) in TLN2; Ser257 in STIM1; Ser995 (ratio of T/N = 2.77, P = 2.32E-05) in GNAS; Ser110 (ratio of T/N = 2.13, P = 6.07E-04) in SNAP23; Thr515 in PRKG1; Ser309 (ratio of T/N = 2.94, P = 9.07E-04) in ARHGEF12; Ser1177, Thr1178, Ser1180, and Tyr1181 in ITPR1; Ser1687 (ratio of T/N = 0.67, P = 2.20E-03) in ITPR2; and Ser880 in ITGA2B (Table 6; Supplemental Fig. 1.1); (ii) vascular smooth muscle contraction involved in phosphorylation at residues Ser207 (ratio of T/N = 3.30, P = 2.31E-06) in CALD1; Ser995 (ratio of T/N = 2.77, P = 2.32E-05) in GNAS; Thr515 in PRKG1; Ser309 (ratio of T/N = 2.94, P = 9.07E-04) in ARHGEF12; Tyr514 in PRKCD; Ser1177, Thr1178, Ser1180, and Tyr1181 in ITPR1;, Ser663 (ratio of T/N = 2.32, P = 4.24E-04); Thr668 (ratio of T/N = 2.32, P = 4.24E-04); Ser1295 (ratio of T/N = 1.45, P = 1.11E-03); Ser115 (ratio of T/N = 1.81, P = 1.02E-02); Ser1478 and Ser1480 in ARHGEF11; and Ser1687 (ratio of T/N = 0.67, P = 2.20E-03) in ITPR2 (Table 6; Supplemental Fig. 1.2); (iii) estrogen signaling pathway involved in phosphorylation at residues Ser122 and Ser124 in AKT1; Ser13 in FKBP5; Ser995 (ratio of T/N = 2.77, P = 2.32E-05) in GNAS; Tyr514 in PRKCD; Ser1177, Thr1178, Ser1180, and Tyr1181 in ITPR1; and Ser1687 (ratio of T/N = 0.67, P = 2.20E-03) in ITPR2 (Table 6; Supplemental Fig. 1.3); (iv) Fc gamma R-mediated phagocytosis involved in phosphorylation at residues Ser122 and Ser124 in AKT1; Ser701 (ratio of T/N = 1.57, P = 2.02E-03) in ASAP2; Ser717 (ratio of T/N = 2.04, P = 1.35E-04) in ASAP1; Ser27 (ratio of T/N = 8.66, P = 3.86E-06); Ser26 (ratio of T/N = 5.66, P = 8.79E-06), Thr150 (ratio of T/N = 3.88, P = 1.52E-05), Ser145 (ratio of T/N = 1.62, P = 8.15E-03), Ser147, Ser118, Thr120, Ser128, Thr130, Ser138, and Ser101 in MARCKS; and Tyr514 in PRKCD (Table 6; Supplemental Fig. 1.4); (v) glucagon signaling pathway involved in phosphorylation at residues Ser122 and Ser124 in AKT1; Ser29 (ratio of T/N = 2.98, P = 6.08E-05) in ACACA; Ser995 (Ratio of T/N = 2.77, P = 2.32E-05) in GNAS; Ser1177, Thr1178, Ser1180, and Tyr1181 in ITPR1; and Ser1687 (ratio of T/N = 0.67, P = 2.20E-03) in ITPR2 (Table 6; Supplemental Fig. 1.5); (vi) proteoglycans in cancer involved in phosphorylation at residues Ser122 and Ser124 in AKT1; Ser781 (ratio of T/N = 3.58, P = 4.13E-06), Ser1428 (ratio of T/N = 1.72, P = 1.74E-05), Thr961 (ratio of T/N = 1.61, P = 4.31E-04), Ser960 (ratio of T/N = 1.70, P = 5.29E-04), Thr961 (ratio of T/N = 1.70, P = 5.29E-04), Ser1607 (ratio of T/N = 0.76, P = 3.19E-03), Thr1684, Ser1686, Ser1686, and Thr1688 in ANK1; Ser309 (ratio of T/N = 2.94, P = 9.07E-04) in ARHGEF12; Ser1177, Thr1178, Ser1180, and Tyr1181 in ITPR1; Ser302 (ratio of T/N = 1.43, P = 2.16E-02), Ser303 (ratio of T/N = 1.43, P = 2.16E-02), Ser322 (ratio of T/N = 1.88, P = 4.60E-02), Thr318, and Ser320 in PXN; and Ser1687 (ratio of T/N = 0.67, P = 2.20E-03) in ITPR2 (Table 6; Supplemental Fig. 1.7); (vii) insulin signaling pathway involved in phosphorylation in residues Ser122 and Ser124 in AKT1; Ser114 (ratio of T/N = 0.7, P = 6.75E-04) in PRKAR2B; Thr486 and Ser488 in SLC2A4; Ser1385, Ser1386, and Ser1387 in TSC2; and Ser29 (ratio of T/N = 2.98, P = 6.08E-05) in ACACA (Table 6; Supplemental Fig. 1.8); (viii) gap junction involved in phosphorylation at residues Ser995 (ratio of T/N = 2.77, P = 2.32E-05) in GNAS; Thr515 in PRKG1; Ser1177, Thr1178, Ser1180, and Tyr1181 in ITPR1; and Ser1687 (ratio of T/N = 0.67, P = 2.20E-03) in ITPR2 (Table 6; Supplemental Fig. 1.10); (ix) cGMP–PKG signaling pathway involved in phosphorylation at residues Ser122 and Ser124 in AKT1; Ser622 (ratio of T/N = 1.60, P = 1.40E-01) in SLC8A2; Thr515 in PRKG1; Ser1177, Thr1178, Ser1180, and Tyr1181 in ITPR1; and Ser1687 (ratio of T/N = 0.67, P = 2.20E-03) in ITPR2 (Table 6; Supplemental Fig. 1.11); (x) GnRH signaling pathway involved in phosphorylation at residues Ser995 (ratio of T/N = 2.77, P = 2.32E-05) in GNAS; Tyr514 in PRKCD; Ser1177, Thr1178, Ser1180, and Tyr1181 in ITPR1; and Ser1687 (ratio of T/N = 0.67, P = 2.20E-03) in ITPR2 (Table 6; Supplemental Fig. 1.12); (xi) inflammatory mediator regulation of TRP channels involved in phosphorylation at residues Ser995 (ratio of T/N = 2.77, P = 2.32E-05) in GNAS; Tyr514 in PRKCD; Ser1177, Thr1178, Ser1180, and Tyr1181 in ITPR1; and Ser1687 (ratio of T/N = 0.67, P = 2.20E-03) in ITPR2 (Table 6; Supplemental Fig. 1.13); and (xii) calcium signaling pathway involved in phosphorylation at residues Ser622 (ratio of T/N = 1.60, P = 1.40E-01) in SLC8A2; Ser257 in STIM1; Ser995 (ratio of T/N = 2.77, P = 2.32E-05) in GNAS; Ser1177, Thr1178, Ser1180, and Tyr1181 in ITPR1; and Ser1687 (ratio of T/N = 0.67, P = 2.20E-03) in ITPR2 (Table 6; Supplemental Fig. 1.14). These findings clearly demonstrate that phosphorylation participates in invasiveness-related signaling pathways in invasive NFPAs.
Table 6.
Statistically significant KEGG signaling pathways identified from 130 overlapped molecules between phosphoproteins and invasive DEGs in NFPAs. Count means the number of genes enriched in each pathway
| Category | Pathway name | Count | % | Fold enrichment | P value | Benjamini | Gene name of overlapped molecule |
|---|---|---|---|---|---|---|---|
| KEGG_PATHWAY | hsa04611:Platelet activation | 10 | 7.94 | 10.58 | 2.53E-07 | 3.61E-05 | AKT1, TLN2, STIM1, GNAS, SNAP23, PRKG1, ARHGEF12, ITPR1, ITPR2, ITGA2B |
| KEGG_PATHWAY | hsa04270:Vascular smooth muscle contraction | 8 | 6.35 | 9.41 | 1.64E-05 | 1.17E-03 | CALD1, GNAS, PRKG1, ARHGEF12, PRKCD, ITPR1, ARHGEF11, ITPR2 |
| KEGG_PATHWAY | hsa04915:Estrogen signaling pathway | 6 | 4.76 | 8.34 | 6.44E-04 | 3.02E-02 | AKT1, FKBP5, GNAS, PRKCD, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04666:Fc gamma R-mediated phagocytosis | 5 | 3.97 | 8.19 | 2.89E-03 | 9.82E-02 | AKT1, ASAP2, ASAP1, MARCKS, PRKCD |
| KEGG_PATHWAY | hsa04922:Glucagon signaling pathway | 5 | 3.97 | 6.95 | 5.21E-03 | 1.39E-01 | AKT1, ACACA, GNAS, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04730:Long-term depression | 4 | 3.17 | 9.17 | 8.74E-03 | 1.89E-01 | GNAS, PRKG1, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa05205:Proteoglycans in cancer | 6 | 4.76 | 4.13 | 1.34E-02 | 2.40E-01 | AKT1, ANK1, ARHGEF12, ITPR1, PXN, ITPR2 |
| KEGG_PATHWAY | hsa04910:Insulin signaling pathway | 5 | 3.97 | 4.98 | 1.64E-02 | 2.55E-01 | AKT1, PRKAR2B, SLC2A4, TSC2, ACACA |
| KEGG_PATHWAY | hsa04970:Salivary secretion | 4 | 3.17 | 6.40 | 2.30E-02 | 3.09E-01 | GNAS, PRKG1, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04540:Gap junction | 4 | 3.17 | 6.25 | 2.44E-02 | 2.98E-01 | GNAS, PRKG1, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04022:cGMP-PKG signaling pathway | 5 | 3.97 | 4.35 | 2.55E-02 | 2.85E-01 | AKT1, SLC8A2, PRKG1, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04912:GnRH signaling pathway | 4 | 3.17 | 6.05 | 2.66E-02 | 2.75E-01 | GNAS, PRKCD, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04750:Inflammatory mediator regulation of TRP channels | 4 | 3.17 | 5.62 | 3.22E-02 | 3.02E-01 | GNAS, PRKCD, ITPR1, ITPR2 |
| KEGG_PATHWAY | hsa04020:Calcium signaling pathway | 5 | 3.97 | 3.84 | 3.79E-02 | 3.26E-01 | SLC8A2, STIM1, GNAS, ITPR1, ITPR2 |
Hub molecules identified with protein–protein interaction analysis of 130 overlapped molecules (phosphoproteins; invasive DEGs)
The PPI network was constructed using molecules with combined scores greater than 0.4, based on 130 overlapped molecules (phosphoproteins; invasive DEGs) (Fig. 3a; Supplemental Table 9). Some overlapped molecules (phosphoproteins; invasive DEGs) in PPI networks trend to be co-expressed or co-localized with high combined score, the difference might be related to functional differences between the overlapped molecules, and various co-expression and co-localization might be reflected in the molecular characteristics or structures of two molecules [31], which help to understand the function and biological mechanisms of NFPA invasiveness. Among 130 overlapped molecules (phosphoproteins; invasive DEGs), some between molecules had very high combined score; for example, SNAP23 and STX1A, AKT1 and TSC2, CHD3 and GATAD2A, SPTB and ANK1, ALDOA and TPI1, POMC and AVP, STIM1 and ITPR1, SCG3 and CHGA, DMTN and ADD2, CHGB and SCG3, ALDOA and PKM, STIM1 and ITPR2, PKM and TPI1, POMC and GAL, SYN1 and STX1A, DMTN and ADD3, and HNRNPA1 and SRRM2 had the combined score over 0.95 (Supplemental Table 9).
Fig. 3.
The hub molecules were identified with protein–protein interaction (PPI)-based molecular complex detection (MCODE) using 130 overlapped molecules (phosphoproteins; invasive DEPs) in NFPAs. a The protein–protein interactions (PPIs) with combined scores greater than 0.4 were selected to construct PPI network. b The entire PPI network was analyzed using molecular complex detection (MCODE), and one module was obtained with module score = 5.778. The different colored nodes represent different query proteins and first shell of interactors. The white nodes represent second shell of interactors. The empty nodes represent the proteins with unknown 3D structure. The filled nodes represent the proteins with known or predicted 3D structure
The hub molecules based on the PPI network play important roles in a molecular network system, which also reflects its crucial roles in a biological system such as NFPA invasive behavior system. The entire PPI network was analyzed using MCODE, and one module was obtained with module score = 5.778 (Fig. 3b). Thus, a total of 10 hub molecules were identified in invasive NFPAs (Table 7), including SCG3 (secretogranin-3), TSC2 (tuberin), ALB (serum albumin), AKT1 (RAC-alpha serine/threonine-protein kinase), APOL1 (apolipoprotein L1), ACACA (acetyl-CoA carboxylase 1), SPARCL1 (SPARC-like protein 1), SLC2A4 (solute carrier family 2 facilitated glucose transporter member 4), CHGB (secretogranin-1), and IGFBP5 (insulin-like growth factor-binding protein 5). These hub molecules not only had the differential expression at the mRNA level but also had the changed level of phosphorylation. These hub molecules assisted in improving the understanding of the key molecular mechanisms underlying NFPA invasiveness, and the results may help the further study of the biological mechanism of NFPA invasive behavior. It also clearly demonstrated that phosphorylation of these hub molecules played important roles in NFPA invasive behavior; for example, phosphorylation occurred at residues Ser37 (ratio T/N = 0.46, P = 1.5E-04) in SCG3; Ser1385, Ser1386, and Ser1387 in TSC2; Ser82 (ratio T/N = 0.74, P = 3.24E-04) in ALB; Ser122 and Ser124 in AKT1; Ser352 (ratio T/N = 0.71, P = 5.33E-03) and Ser355 (ratio T/N = 0.71, P = 5.33E-03) in APOL1; Ser29 (ratio of T/N = 2.98, P = 6.08E-05) in ACACA; Ser76, Ser80 (ratio T/N = 0.24, P = 1.86E-05), Ser84 (ratio T/N = 0.35, P = 5.38E-05), Ser92 (ratio T/N = 0.28, P = 1.91E-05), Ser198 (ratio T/N = 0.89, P = 1.92E-01), Ser231, Thr238, Ser240, and Ser295 (ratio T/N = 0.57, P = 1.15E-053) in SPARCL1; Thr486 and Ser488 in SLC2A4; and Thr123 in IGFBP5; and CHGB was phosphorylated at 50 residues (41 Ser, 5 Thr, and 4 Tyr residues) (Table 7). The hub molecules (phosphoproteins; invasive DEGs) SLC2A4 and TSC2 were involved in insulin signaling pathway, ACACA was involved in insulin signaling pathway and glucagon signaling pathway, and AKT1 was involved in insulin signaling pathway, glucagon signaling pathway, cGMP–PKG signaling pathway, proteoglycans in cancer, Fc gamma R-mediated phagocytosis, estrogen signaling pathway, and platelet activation pathway (Table 6). It clearly demonstrated that one signaling pathway included multiple hub molecules, and one hub molecule was involved in multiple signaling pathways, to display a real signaling pathway network system in NFPA invasive behavior. Those hub molecules played the crucial roles in the signaling pathway network system in the invasive behavior of NFPAs, which might be the effective panel biomarker and multitherapeutic targets for NFPA invasiveness.
Table 7.
A total of 10 hub molecules in invasive NFPAs identified with PPI analysis of 130 overlapped molecules (invasive DEG; phosphoproteins)
| Protein accession ID | DEG name | DEG level | Protein name | Phosphorylation level | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| logFC | AveExpr | t | P value | Phosphopeptide | Phosphorylated site | Phosphorylated position | Phosphorylated probabilities | PEP | Score | Ratio (T/N) | P value (t test) | |||
| Q8WXD2 | SCG3 | 1.2843 | 10.4326 | 4.3052 | 1.72E-03 | Secretogranin-3 | ELS*AERPLNEQIAEAEEDKIK | S | 37 | 1 | 2.80E-38 | 149 | 0.46 | 1.50E-04 |
| LFPAPS*EKSHEETDSTK | S | 359 | 0.468 | 1.85E-02 | 36 | |||||||||
| P49815 | TSC2 | 2.1795 | 10.7866 | 6.3857 | 9.73E-05 | Tuberin | S*SSSPELQTLQDILGDPGDK | S; S; S | 1385; 1386; 1387 | 0.313; 0.313; 0.313 | 1.56E-11 | 74 | ||
| P02768-1 | ALB | 1.9536 | 6.1289 | 4.5055 | 1.27E-03 | Serum albumin | TCVADES*AENCDK | S | 82 | 1 | 1.24E-47 | 197 | 0.74 | 3.24E-04 |
| P31749 | AKT1 | 1.1801 | 6.8509 | 5.4900 | 3.11E-04 | RAC-alpha serine/threonine-protein kinase | S*GSPSDNSGAEEMEVSLAKPK | S; S | 122; 124 | 0.457; 0.457 | 1.16E-07 | 92 | ||
| A5PL32 | APOL1 | − 1.1936 | 9.8189 | − 3.2480 | 9.26E-03 | Apolipoprotein L1 | VTEPIS*AES*GEQVER | S; S | 352; 355 | 1; 1 | 2.53E-03 | 59 | 0.71 | 5.33E-03 |
| Q13085 | ACACA | 1.1822 | 6.8754 | 3.3664 | 7.62E-03 | Acetyl-CoA carboxylase 1 | FIIGSVSEDNS*EDEISNLVK | S | 29 | 1 | 3.35E-32 | 154 | 2.98 | 6.08E-05 |
| Q14515 | SPARCL1 | − 3.1871 | 13.6134 | − 4.7627 | 8.66E-04 | SPARC-like protein 1 | EES*HEQS*AEQGK | S | 80 | 9.37E-38 | 176 | 0.24 | 1.86E-05 | |
| SSS*QELGLK | S | 92 | 0.999 | 1.61E-28 | 188 | 0.28 | 1.91E-05 | |||||||
| SKEES*HEQS*AEQGK | S; S | 80; 84 | 0.999; 1 | 9.37E-38 | 176 | 0.35 | 5.38E-05 | |||||||
| EHANS*KQEEDNTQSDDILEESDQPTQVSK | S | 231 | 1 | 5.61E-70 | 176 | 0.36 | 5.25E-04 | |||||||
| HIQETEWQS*QEGK | S | 295 | 1 | 2.83E-21 | 152 | 0.57 | 1.15E-03 | |||||||
| DQGNQEQDPNIS*NGEEEEEKEPGEVGTHNDNQER | S | 198 | 0.997 | 1.10E-05 | 53 | 0.89 | 1.92E-01 | |||||||
| EHANS*KQEEDNT*QS*DDILEESDQPTQVSK | S; T; S | 231; 238; 240 | 0.331; 0.331; 0.331 | 1.44E-02 | 38 | |||||||||
| S*KEES*HEQSAEQGK | S; S | 76; 80 | 0.999; 0.995 | 7.77E-05 | 77 | |||||||||
| P14672 | SLC2A4 | − 1.3065 | 5.1362 | − 4.4148 | 1.45E-03 | Solute carrier family 2, facilitated glucose transporter member 4 | T*PSLLEQEVKPSTELEYLGPDEND | T; S | 486; 488 | 0.5; 0.5 | 1.85E-10 | 91 | ||
| P05060 | CHGB | − 1.3288 | 14.5253 | − 3.2997 | 8.51E-03 | Secretogranin-1 | SETHAAGHS*QEK | S | 225 | 1 | 4.87E-30 | 175 | 0.14 | 6.87E-07 |
| GRGS*EEYR | S | 367 | 1 | 1.70E-07 | 146 | 0.12 | 7.46E-07 | |||||||
| AS*EEEPEYGEEIK | S | 335 | 1 | 5.81E-71 | 223 | 0.13 | 2.05E-06 | |||||||
| EKSS*QES*GEETGS*QENHPQESK | S; S; S | 236; 239; 245 | 0.531; 1; 0.965 | 4.20E-73 | 205 | 0.06 | 3.67E-06 | |||||||
| ADEPQWSLYPSDS*QVS*EEVK | S; S | 146; 149 | 0.999; 0.999 | 1.30E-150 | 236 | 0.31 | 5.28E-06 | |||||||
| SSQESGEET*GSQENHPQESK | T | 243 | 0.978 | 1.47E-45 | 161 | 0.02 | 5.36E-06 | |||||||
| GERGEDS*S*EEK | S; S | 182; 183 | 1; 1 | 1.80E-03 | 100 | 0.16 | 7.03E-06 | |||||||
| AS*EEEPEY*GEEIK | S; Y | 335; 341 | 1; 1 | 2.35E-06 | 98 | 0.18 | 8.29E-06 | |||||||
| SSQES*GEETGS*QENHPQESK | S; S | 239; 245 | 0.992; 1 | 4.20E-73 | 205 | 0.06 | 1.01E-05 | |||||||
| S*QREDEEEEEGENYQK | S | 160 | 1 | 1.77E-17 | 140 | 0.25 | 1.17E-05 | |||||||
| SAEFPDFYDS*EEPVSTHQEAENEKDR | S | 626 | 1 | 1.85E-99 | 214 | 0.26 | 1.18E-05 | |||||||
| S*QEES*EEGEEDATS*EVDK | S | 259; 263; 272 | 1; 1; 0.857 | 1.51E-53 | 157 | 0.14 | 1.23E-05 | |||||||
| SSQGGS*LPSEEK | S | 298 | 0.981 | 3.28E-14 | 142 | 0.28 | 1.31E-05 | |||||||
| DPADASEAHES*SSR | S | 98 | 0.966 | 1.32E-30 | 169 | 0.10 | 1.41E-05 | |||||||
| NYPS*LELDK | S | 391 | 1 | 1.30E-04 | 112 | 0.14 | 1.44E-05 | |||||||
| ADEPQWSLYPS*DS*QVSEEVK | S; S | 144; 146 | 0.941; 0.751 | 3.58E-98 | 136 | 0.52 | 1.53E-05 | |||||||
| EKSS*QES*GEET*GSQENHPQESK | S; S; T | 237; 239; 243 | 0.819; 0.935; 0.598 | 1.47E-45 | 161 | 0.05 | 1.53E-05 | |||||||
| SSQGGSLPS*EEK | S | 301 | 0.999 | 5.51E-20 | 128 | 0.25 | 1.88E-05 | |||||||
| S*AEFPDFYDSEEPVSTHQEAENEKDR | S | 617 | 1 | 8.30E-152 | 246 | 0.22 | 2.13E-05 | |||||||
| S*AEFPDFY*DSEEPVSTHQEAENEKDR | Y | 624 | 0.995 | 8.86E-43 | 152 | 0.23 | 2.23E-05 | |||||||
| SQEESEEGEEDAT*SEVDK | T | 271 | 0.973 | 1.38E-88 | 219 | 0.21 | 2.23E-05 | |||||||
| M#AHGY*GEES*EEER | Y; S | 401; 405 | 1; 1 | 2.41E-02 | 51 | 0.23 | 2.34E-05 | |||||||
| ERADEPQWSLYPSDSQVS*EEVK | S | 149 | 1 | 2.83E-147 | 264 | 0.29 | 2.80E-05 | |||||||
| PQS*EES*WDEEDKR | S; S | 377; 380 | 1; 1 | 4.73E-42 | 181 | 0.15 | 3.27E-05 | |||||||
| SAEFPDFYDS*EEPVS*THQEAENEK | S; S | 626; 631 | 0.996; 0.863 | 3.45E-35 | 140 | 0.30 | 3.33E-05 | |||||||
| SS*QGGSLPSEEK | S | 294 | 0.807 | 3.74E-09 | 133 | 0.24 | 3.77E-05 | |||||||
| S*SQGGSLPSEEK | S | 293 | 0.992 | 1.97E-30 | 180 | 0.25 | 4.48E-05 | |||||||
| GHPQEES*EESNVSMASLGEK | S | 311 | 1 | 6.58E-209 | 232 | 0.22 | 5.91E-05 | |||||||
| SAEFPDFYDSEEPVS*T*HQEAENEK | S; T | 631; 632 | 0.591; 0.586 | 3.45E-35 | 124 | 0.31 | 5.94E-05 | |||||||
| EKS*S*QES*GEET*GS*QENHPQESK | S; S; S; T; S | 235; 236; 239; 243; 245 | 0.643; 0.589; 0.589; 0.589; 0.589 | 1.47E-45 | 161 | 0.05 | 7.50E-05 | |||||||
| WQQQGDLQDT*KENR | T | 492 | 1 | 6.83E-05 | 85 | 0.37 | 9.99E-05 | |||||||
| GHPQEES*EESNVS*M#ASLGEK | S | 317 | 0.988 | 1.45E-59 | 184 | 0.29 | 1.70E-04 | |||||||
| ADEPQWSLY*PSDSQVS*EEVK | Y; S | 142; 149 | 0.981; 0.96 | 6.42E-108 | 134 | 0.52 | 1.87E-04 | |||||||
| GHPQEES*EESNVS*MAS*LGEK | S; S; S | 314; 320; 323 | 0.739; 1; 1 | 1.83E-50 | 161 | 0.37 | 4.40E-04 | |||||||
| WAEGGGHS*R | S | 130 | 1 | 4.86E-03 | 80 | 0.21 | 5.96E-04 | |||||||
| GHPQEES*EES*NVS*MASLGEK | S; S; S | 314; 317; 320 | 0.959; 0.959; 0.959 | 8.63E-27 | 145 | 0.31 | 6.63E-04 | |||||||
| CIIEVLSNALS*K | S | 46 | 1 | 3.06E-04 | 94 | 0.40 | 1.63E-02 | |||||||
| DHHS*THYRASEEEPEY*GEEIK | S; T; Y | 329; 330; 341 | 0.355; 0.355; 0.98 | 1.56E-02 | 43 | |||||||||
| DPADASEAHESS*S*RGEAGAPGEEDIQGPTK | S; S | 99; 100 | 0.651; 0.651 | 8.57E-07 | 62 | |||||||||
| LLRDPADAS*EAHESSSR | S | 93 | 0.989 | 5.88E-05 | 88 | |||||||||
| S*ETHAAGHS*QEK | S | 217 | 0.757 | 1.02E-02 | 78 | |||||||||
| P24593 | IGFBP5 | − 2.6828 | 10.6639 | − 4.1430 | 2.20E-03 | Insulin-like growth factor-binding protein 5 | EHEEPT*TSEMAEETYSPK | T | 123 | 0.346 | 2.19E-04 | 56 | ||
DEG, differentially expressed gene; LogFC, log2(fold change). LogFC > = 1, upregulated DEG; LogFC <= − 1, downregulated DEGs; Ratio (T/N), the ratio of phosphorylation level of NFPAs to controls
Discussion
Phosphorylation is one of the important protein PTMs (ranging from 400 to 600 PTMs in humans [16]) that cause the diversity of proteins, namely proteoforms or protein species [17]. Proteoform or protein species is defined as a primary amino acid sequence + PTMs + spatial conformation + binding partners + cofactors + localization + a defined function [17]. The number of proteoforms (at least 1,000,000 proteoforms in humans) is much more than the number of transcripts (at least 100,000 transcripts in humans) and genes (about 20,300 genes in humans) [32, 33]. Proteoform is the basic unit in a proteome and is the final performer of gene functions. In-depth investigation of proteoforms in human proteome will directly benefit the discovery of reliable biomarkers for accurate molecular mechanisms, therapeutic targets, personalized medicine, or precision medicine in human pituitary adenomas [32]. Therefore, proteoforms have important scientific merit in the entire medical science and life science [34, 35]. The studies on PTMs directly benefit the characterization of proteoforms. This study focused on phosphoproteomic profiling in human NFPA tissues, which belongs to our long-term research program in proteomics, PTMs, and proteoforms in human pituitary adenomas. TMT labeling–TiO2 enrichment–LC-MS/MS was an effective quantitative proteomics approach [36] and identified a total of 2982 phosphorylation sites within 1035 phosphoproteins, with quantitative information for 1660 (1660/2982 = 55.67%) phosphorylation sites and qualitative information for 1322 (1322/2982 = 44.33%) phosphorylation sites, in NFPA tissues relative to controls. It is the first large-scale phosphoproteomic profiling of NFPAs with quantitative information, and those identified phosphoproteins participated in multiple biological process and signaling pathways, including cell–cell adherens junction, cadherin binding involved in cell–cell adhesion, cell–cell adhesion, SNARE interactions in vesicular transport, SNAP receptor activity, SNARE complex, histone H2B ubiquitination, histone monoubiquitination, endodermal cell fate commitment, Cdc73/Paf1 complex, myosin II complex and filament, oxygen transporter activity, oxygen transport, haptoglobin binding, haptoglobin–hemoglobin complex, hemoglobin complex, cAMP-dependent protein kinase complex, protein kinase A catalytic subunit binding, cAMP-dependent protein kinase regulator activity, ESCRT III complex disassembly, positive regulation of viral release from host cell, and eukaryotic 43S preinitiation complex. Also, 31 statistically significant signaling pathways were mined with KEGG pathway analysis from 1035 phosphoproteins in NFPAs, including platelet activation, mRNA surveillance pathway, RNA transport, spliceosome, endocytosis, proteoglycans in cancer, mTOR signaling pathway, insulin signaling pathway, MAPK signaling pathway, SNARE interactions in vesicular transport, gap junction, focal adhesion, glucagon signaling pathway, estrogen signaling endoplasmic reticulum, and GnRH signaling pathway. Those data provided the largest phosphorylation profiling and phosphorylation-mediated signaling pathway systems, which are the precious resource to deeply investigate biological functions of protein phosphorylations in NFPAs, and identify the reliable and effective new biomarkers for patient stratification, prognostic/predictive assessment, and personalized treatment of NFPAs.
Tumor invasiveness is a very challenging clinical problem in NFPAs, which causes the different therapeutic strategies after neurosurgery [37–39]. Investigation of tumor invasiveness in NFPAs is always very interesting and meaningful. Some researchers have focused on several invasiveness-related molecules including pituitary tumor-transforming gene (PTTG), vascular endothelial growth factor (VEGF), hypoxia inducible factor-1a (HIF-1a), fibroblast growth factor-2 (FGF-2), and matrix metalloproteinases (MMPs) such as MMP-2 and MMP-9, and their interacted complex molecular network in human pituitary adenoma [4, 38, 40]. However, the world of molecules of invasive NFPAs is very complex; these molecules [4, 38, 40] are only the partial molecules, which cannot represent the entire molecular world of NFPA invasive behavior. Multiomics-based molecular pathway networks [4, 15, 30] are an effective strategy and approach to study NFPA invasiveness, and annotate the interactome in invasive NFPA, and also are the real way to resolve its clinical invasiveness challenge, for in-depth clarification of its molecular mechanism of NFPA invasiveness and discovery of reliable invasiveness-related biomarkers for diagnosis, prognostic/predictive assessment, and therapeutic targets for personalized therapy [6, 7, 16, 41, 42]. This study used transcriptomics data between invasive NFPAs and control pituitaries, from the public GEO database, which was integrated with the identified phosphoprotein data. An overlapped analysis between 1035 phosphoproteins and 2751 DEGs in invasive NFPAs relative to controls revealed 130 overlapped molecules (phosphoproteins; invasive DEGs). Those 130 overlapped molecules were involved in multiple biological processes, including cell–cell adherens and regulation, platelet activation, GTPase signaling and regulation, microtubule and its regulation, protein kinase activity and peptidyl-threonine phosphorylation, and calcium signaling pathway and regulation. Thereby, those phosphoproteins participated in the corresponding biological functions in NFPA invasive behaviors. We have clearly demonstrated the important roles of phosphorylation in invasive NFPA pathogenesis.
Furthermore, based on 130 overlapped molecules (phosphoproteins; invasive DEGs), 12 statistically significant signaling pathways (Table 6; Supplemental Fig. 1) were identified to associate with tumor invasiveness, and the identified phosphoproteins were involved in these signaling pathways. For example, the platelet activation pathway included phosphoproteins (invasive DEGs) AKT1, TLN2, STIM1, GNAS, SNAP23, PRKG1, ARHGEF12, ITPR1, ITPR2, and ITGA2B; the vascular smooth muscle contraction pathway included phosphoproteins (invasive DEGs) CALD1, GNAS, PRKG1, ARHGEF12, PRKCD, ITPR1, ARHGEF11, and ITPR2; the estrogen signaling pathway included phosphoproteins (invasive DEGs) AKT1, FKBP5, GNAS, PRKCD, ITPR1, and ITPR2; Fc gamma R-mediated phagocytosis included phosphoproteins (invasive DEGs) AKT1, ASAP2, ASAP1, MARCKS, and PRKCD; the glucagon signaling pathway included phosphoproteins (invasive DEGs) AKT1, ACACA, GNAS, ITPR1, and ITPR2; proteoglycans in cancer included phosphoproteins (invasive DEGs) AKT1, ANK1, ARHGEF12, ITPR1, PXN, and ITPR2; the insulin signaling pathway included phosphoproteins (invasive DEGs) AKT1, PRKAR2B, SLC2A4, TSC2, and ACACA; the gap junction pathway included phosphoproteins (invasive DEGs) GNAS, PRKG1, ITPR1, and ITPR2; the cGMP–PKG signaling pathway included phosphoproteins (invasive DEGs) AKT1, SLC8A2, PRKG1, ITPR1, and ITPR2; the GnRH signaling pathway included phosphoproteins (invasive DEGs) GNAS, PRKCD, ITPR1, and ITPR2; inflammatory mediator regulation of TRP channels included phosphoproteins (invasive DEGs) GNAS, PRKCD, ITPR1, and ITPR2; and the calcium signaling pathway included phosphoproteins (invasive DEGs) SLC8A2, STIM1, GNAS, ITPR1, and ITPR2. These KEGG signaling pathways formed the signaling pathway profiling of NFPA invasive behavior and mutually interacted through hub molecules. Interestingly, PPI network-based MCODE analysis identified 10 hub molecules (phosphoproteins; DEGs) in invasive NFPAs (Table 7; Fig. 3b), namely SLC2A4, TSC2, AKT1, SCG3, ALB, APOL1, ACACA, SPARCL1, CHGB, and IGFBP5. Comparative analysis of these hub molecules with the nodes of signaling pathways (Table 6; Supplemental Fig. 1) found that one signaling pathway might include multiple hub molecules, and one hub molecule might be involved in multiple signaling pathways [43]; for example, the insulin signaling pathway included hub molecules SLC2A4, TSC2, and AKT1; and AKT1 was involved in insulin signaling pathway, cGMP–PKG signaling pathway, glucagon signaling pathway, estrogen signaling pathway, platelet activation pathway, Fc gamma R-mediated phagocytosis, and proteoglycans in cancer. These signaling pathways and hub molecules formed the real signaling pathway network system for NFPA invasive behavior, which is offering great promise to clarify the molecular mechanisms of NFPA invasive behavior; determine the effective panel of biomarkers [11] for patient stratification, prognostic/predictive assessment, and personalized treatment of invasive NFPAs; and discover multitherapeutic targets for personalized therapy for patients suffering of invasive NFPAs [44–46].
Currently, the diagnosis of NFPA invasive behaviors mainly relies on tumor morphological changes observed by neurosurgery and nuclear magnetic resonance (NMR) image changes at its middle or late stages, and it is very difficult to determine its invasiveness at its early stage [9, 10]. Even at its middle or late stages, NMR image and morphological change-based diagnosis of invasive behavior of NFPAs are also not fully accurate. In this study, the identified invasiveness-related, phosphorylation-mediated signaling pathways and pathway network-based hub molecules (phosphoproteins; invasive DEGs) have the potential for establishing panel of biomarkers allowing early prediction of invasive behavior and focusing on therapeutic targets to prevent its progression and accurate prognostic/predictive assessment of invasive behavior after neurosurgery, in order to personalize the treatment of invasive NFPA patients. Here, 10 pathway–network-based hub molecules (phosphoproteins; invasive DEGs) were taken for further discussion in the context of PPPM in invasive NFPAs. AKT1 is a crucial component in the PI3K–Akt signaling pathway that is required for VEGF-A mRNA expression in GH3 cells induced by 17alpha-estradiol [47], and the anticancer drug precursor methylseleninic acid (MSeA) can decrease AKT phosphorylation to inhibit the growth and survival of human umbilical vein endothelial cells [48], and AKT1-mutant estrogen receptor (ER)-positive metastatic breast cancer has a longer survival duration on mTOR inhibitor therapy [49]. Thus, AKT is an important therapeutic target potentially capable of improving the treatment results of solid tumors [50]. ACACA is acetyl-CoA carboxylase 1 that catalyzes the ATP-dependent carboxylation of acetyl-CoA, a rate-limiting step in fatty acid biosynthesis [51]. A study found that ACACA is a potent therapeutic target for anticancer therapy [52]. Also, phospho-ACACA protein is an independent prognostic/predictive factor for human gastric cancer without lymph node metastasis [53]. TSC2, tuberin, is a well-known suppressor of the mTOR pathway [54]. SPARCL1 can suppress cancer metastasis and recruit macrophases by activation of canonical WNT/β-catenin signaling [55], or it can suppress cancer cell proliferation and migration via the MEK/ERK signaling [56]. SLC2A4, namely solute carrier family 2 facilitated glucose transporter member 4, encodes glucose transporter 4 protein (GLUT4), which has been identified as a promising therapeutic target for cancer, because the putative antimicrobial peptides (AMPs) can serve as a therapeutic drug in treating cancer by inhibiting SLC2A4 that is responsible for the production of energy for cancer cells during angiogenesis [57]. IGFBP5 is the insulin-like growth factor binding protein 5, which acts as a tumor suppressor, and is often dysregulated in human cancers to associate with tumorigenicity and metastasis; thus, IGFBP5 might be a novel therapeutic target for human melanoma [58, 59]. Furthermore, clinical findings demonstrate the potential of IGFBP5 as an effective biomarker predicting the response to therapy and clinical outcome of cancer patients [60]. SCG3 is secretogranin-3 [61, 62], CHGB is secretogranin-1 [63, 64], ALB is serum albumin [65, 66], and APOL1 is apolipoprotein L1 [67–69]; these molecules were found to be phosphorylated in this study, and they can be secreted into body fluid thus having the potential for the early prediction and prognostic assessment of invasive behavior of NFPAs from the molecular view of point.
Thereby, PTMs are the main factor to result in proteoforms—the final functional forms of a gene/protein. In the context of proteoforms, we focused on an important PTM—phosphorylation that is involved in many biological processes and cell signaling transductions, in NFPA pathogenesis. Further, we integrated the phosphoproteomics and transcriptomics data in invasive NFPAs to obtain phosphorylation-mediated molecular events in invasive NFPAs, which is the precious resource to (i) benefit the understanding of molecular mechanisms of NFPA invasive behavior, (ii) discover effective therapeutic targets to prevent the occurrence and progression of invasive NFPAs, and (iii) develop effective phosphorylation-related biomarkers for prognostic/predictive assessment to stratify NFPA patients into invasive and noninvasive groups who will undergo personalized therapeutic approaches after neurosurgery. This study may serve as an example of effective contribution to the paradigm shift from experimental medicine to PPPM in pituitary adenomas.
Strength and limitation
This study provided the first large-scale phosphoproteomic profiling with 2982 phosphorylation sites within 1035 phosphoproteins in NFPAs and the corresponding phosphorylation-mediated functional characteristics and signaling pathway networks. Moreover, 1035 phosphoproteins were integrated with transcriptomics data (2751 DEGs) in invasive NFPAs relative to controls, which obtained 130 overlapped molecules (phosphoproteins; invasive DEGs), followed by pathway network analysis to obtain 12 statistically significant signaling pathways and 10 hub molecules associated with NFPA invasive behaviors. These findings offer an increasing promise for insights into the molecular mechanisms, determination of effective therapeutic targets, and discovery of effective pathway network-based panel of biomarkers for patient stratification, prognostic/predictive assessment, and personalized treatment of NFPA patients. However, one must realize that those phosphoproteomic data (4 NFPAs vs. 4 controls) and transcriptomics (4 invasive NFPAs vs. 3 controls from GEO database) were derived from a limited sample size. In order to transit those findings into routine practice, a significantly expanded sample size will be needed to further validate and study in detail the molecular mechanisms, functional roles, and potentially therapeutic targets of these identified phosphorylation-involved signaling pathways and hub molecules in NFPA patients. One should also try their best to address the signaling pathway alterations at the proteoform level for real evidence-based PPPM in invasive NFPAs.
Moreover, one should note the potential bias resource of phosphoproteomics analysis due to the different ethnic origin of the samples. Because the post-mortem control pituitary tissue samples were very difficult to be obtained, this phosphoproteomics study used control pituitary tissues from three subjects from the USA of Caucasian ethnic origin and one subject of African-American ethnic origin, and NFPA tissues from 4 Chinese patients (Table 1). It is also important to emphasize yet another limitation as a source of potential differences in phosphorylation—it is the fact that all control samples were taken post-mortem, while NFPA tissues were taken as biopsies from the living patients. For future thorough investigation of phosphorylation level of each phosphoprotein in NFPAs and control pituitary tissues, we would recommend to investigate the variations of phosphorylation levels among different ethnic origin of the samples (American Caucasian, African-American, and Chinese tissue samples). However, for transcriptomics data (DEG data) between 4 invasive NFPAs and 3 control pituitary tissues in the GEO database, these NFPAs and control pituitary tissue samples were from the same ethnic origin. After the overlapped analysis of phosphoproteins and DEG data, the bias of phosphoproteomics derived from ethnic origin of samples would be adjusted. Also, our previous study investigated the heterogeneity of human control pituitary proteome and identified differentially expressed proteins (DEPs) in the group of different genders (male vs. female), in the group of different ages (30-, 40-, and 50-year-old groups), and in the group of different ethnic origin (white vs. black), of control pituitary tissues [70]. The heterogeneity of human control pituitary proteomes did not significantly affect the differences (DEPs) between NFPAs and control pituitary tissues [71, 72]. It clearly indicated that the pituitary adenoma disease-induced differences might be much larger than, or different from, the gender-, age-, and ethnic origin-induced differences, or the potential differences arising from the type of the tissue origin (post-mortem vs. biopsies), in pituitary tissue proteome, which might help to assure our phosphoproteomics results.
Conclusions and expert recommendation
Protein phosphorylation is an important molecular event in the pathological process of NFPA. This study is the first report to provide the large-scale phosphorylation site profiling with quantitative information and the potential biological functions of protein phosphorylation in human NFPA tissues relative to controls, with TMT-labeled TiO2 enrichment–LC-MS/MS method, bioinformatics, and pathway network analysis. A total of 2982 phosphorylation sites within 1035 phosphoproteins and their involved functional characteristics and signaling pathways were identified in NFPAs. Further, the identified phosphoproteins in NFPAs were integrated with DEG data in invasive NFPAs relative to controls, which revealed a set of overlapped molecules (phosphoproteins; invasive DEGs), followed by analysis of functional characteristics and signaling pathway alterations of those overlapped molecules to reveal the important protein phosphorylation in the pathological process of invasive NFPAs. A total of 130 overlapped molecules (phosphoproteins; invasive DEGs) and their functional characteristics were revealed, and 12 statistically significant signaling pathways were identified to associate the invasive characteristics of invasive NFPAs. Ten hub molecules (phosphoproteins; DEGs) were identified with PPI network-based MCODE analysis in invasive NFPAs, including SLC2A4, TSC2, AKT1, SCG3, ALB, APOL1, ACACA, SPARCL1, CHGB, and IGFBP5. Those findings are the precious resource for new phosphoprotein biomarkers to take deep insight into the molecular mechanisms of NFPAs, prognostic/predictive assessment, patient stratification, and personalized treatment of invasive NFPAs, which might contribute to the development of PPPM in pituitary adenomas [45, 46, 73, 74].
We recommend to strengthen the studies of the large-scale phosphoproteins and their involved signaling pathways in NFPA pathogenesis, and to integrate phosphoproteomics data and transcriptomics data [15, 74] for invasive NFPAs to discover reliable and effective phosphoprotein biomarkers for molecular mechanism clarification of NFPA invasive behavior. These invasiveness-related phosphoprotein biomarkers might be used to discriminate invasive NFPAs from noninvasive NFPAs for patient stratification. The stratified patients (invasive vs. noninvasive NFPAs) will accept the corresponding prognostic assessment and personalized treatment, which will directly contribute to the predictive medicine and personalized medicine in NFPAs. Further, the invasiveness-related biomarkers might benefit the early-stage diagnostics of invasive NFPAs to let invasive NFPA patients undergo an early-stage medical treatment and prevent its progression, or the invasiveness-specific biomarkers might be developed into therapeutic targets for personalized drug treatment to prevent the occurrence and progression of NFPA invasive behavior. These will directly contribute to the predictive, preventive, and personalized medicine applied in NFPA patients.
Electronic supplementary material
(PPT 1739 kb)
(PDF 802 kb)
(XLS 68 kb)
(XLS 55 kb)
(XLS 72 kb)
(XLS 436 kb)
(XLS 28 kb)
(XLS 26 kb)
(XLS 27 kb)
(XLS 66 kb)
Acknowledgments
The authors also acknowledge Professor Xuejun Li from Xiangya Hospital and Professor Dominic M. Desiderio from University of Tennessee Health Science Center to assist in obtaining human tissues.
Abbreviations
- ACACA
acetyl-CoA carboxylase 1
- ACN
acetonitrile
- AGC
automatic gain control
- AKT
protein kinase B
- AKT1
RAC-alpha serine/threonine-protein kinase
- ALB
serum albumin
- AMPK
AMP-activated protein kinase
- APOL1
apolipoprotein L1
- ATF2
activating transcription factor 2
- BP
biological process
- CC
cellular component
- cGMP
cyclic nucleotide cGMP
- CHGB
secretogranin-1
- DEG
differentially expressed gene
- DEP
differentially expressed protein
- DTT
dithiothreitol
- r-ERG channel
rat ether-à-go-go-related (ERG) channel
- ERK
extracellular signal–regulated kinase
- ESCRT
endosomal sorting complex required for transport
- ESI-TRAP
electrospray ionization-ion trap
- FC
fold change
- FDR
false discovery rate
- FGF-2
fibroblast growth factor-2
- FPA
functional pituitary adenoma
- GEO
Gene Expression Omnibus
- GH
growth hormone
- GH3
pituitary growth hormone 3
- GnRH
gonadotropin-releasing hormone
- GO
Gene Ontology
- HCD
high-energy collision dissociation
- HIF-1a
hypoxia-inducible factor-1a
- HPLC
high-performance liquid chromatography
- IGF-1
insulin growth factor-1
- IGFBP5
insulin-like growth factor-binding protein 5
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LC
liquid chromatography
- MAPK
mitogen-activated protein kinase
- MCODE
molecular complex detection
- MF
molecular function
- MMPs
matrix metalloproteinases
- MS/MS
tandem mass spectrometry
- mTOR
mammalian target of rapamycin
- NCBI
National Center for Biotechnology Information
- NFκB
nuclear factor kappa-B
- NFPA
nonfunctional pituitary adenoma
- NMR
nuclear magnetic resonance
- PACAP
pituitary adenylyl cyclase activating polypeptide
- PI3K
phosphatidylinositol 3 kinase
- PKG
protein kinase G
- PPI
protein–protein interaction
- PTM
post-translational modification
- PTTG
pituitary tumor-transforming gene
- RSK
ribosomal S6 kinase
- SCG3
secretogranin-3
- Ser or S
serine
- SLC2A4
solute carrier family 2 facilitated glucose transporter member 4
- S/N
signal-to-noise
- SNAP
soluble N-ethylmaleimide-sensitive fusion attachment protein
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- SPARCL1
SPARC-like protein 1
- TFA
trifluoroacetic acid
- Thr or T
threonine
- TMT
tandem mass tag
- TSC2
Tuberin
- TRH
thyrotropin-releasing hormone
- Tyr or Y
tyrosine
- VEGF
vascular endothelial growth factor
Author contributions
D.L. analyzed data, prepared figures and tables, and wrote the manuscript draft. J.J., N.L., M.L., and S.W. participated in partial data analysis and bioinformatics. X.Z. conceived the concept, designed and instructed the experiments, analyzed the data, obtained the phosphoproteomic data, supervised the results, coordinated, wrote and critically revised the manuscript, and was responsible for its financial supports and the corresponding works. All authors approved the final manuscript.
Funding information
This work was supported by the Shandong First Medical University Talent Introduction Funds (to X.Z.), the Hunan Provincial Hundred Talent Plan (to X.Z.), the SCIBP supported project (No. SCIBP2019090006), and China “863” Plan Project (Grant No. 2014AA020610-1 to XZ).
Compliance with ethical standards
Competing interests
The authors declare that they have no conflict of interest.
Ethical approval
Four pituitary adenoma tissue samples, obtained from the Department of Neurosurgery of Xiangya Hospital, Central South University, were approved by the Xiangya Hospital Medical Ethics Committee of Central South University. Post-mortem control pituitary tissue samples, obtained from the Memphis Regional Medical Center (n = 5), were approved by the University of Tennessee Health Science Center Internal Review Board.
Footnotes
Abbreviations for all particular genes and proteins can be found in the Supplemental Table 1 and the UniProtKB database at the following link: https://www.expasy.org/.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Melmed S. Mechanisms for pituitary tumorigenesis: the plastic pituitary. J Clin Invest. 2003;112:1603–1618. doi: 10.1172/JCI20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melmed S. Pathogenesis of pituitary tumors. Nat Rev Endocrinol. 2011;7:257–266. doi: 10.1038/nrendo.2011.40. [DOI] [PubMed] [Google Scholar]
- 3.Melmed S. Pituitary tumors. Endocrinol Metab Clin N Am. 2015;44:1–9. doi: 10.1016/j.ecl.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhan X, Desiderio DM. Editorial: Molecular network study of pituitary adenomas. Front Endocrinol. 2020;11:26. doi: 10.3389/fendo.2020.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng T, Wang Y, Lu M, Zhan X, Zhou T, Li B, Zhan X. Quantitative analysis of proteome in non-functional pituitary adenomas: clinical relevance and potential benefits for the patient. Front Endocrinol. 2019;10:854. doi: 10.3389/fendo.2019.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Cheng T, Lu M, Mu Y, Li B, Li X, Zhan X. TMT-based quantitative proteomics revealed follicle-stimulating hormone (FSH)-related molecular characterizations for potentially prognostic assessment and personalized treatment of FSH-positive non-functional pituitary adenomas. EPMA J. 2019;10:395–414. doi: 10.1007/s13167-019-00187-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhan X, Desiderio DM, Wang X, Zhan X, Guo T, Li M, Peng F, Chen X, Yang H, Zhang P, Li X, Chen Z. Identification of the proteomic variations of invasive relative to noninvasive nonfunctional pituitary adenomas. Electrophoresis. 2014;35(15):2184–2194. doi: 10.1002/elps.201300590. [DOI] [PubMed] [Google Scholar]
- 8.Losa M, Mortini P, Barzaghi R, Ribotto P, Terreni MR, Marzoli SB, Pieralli S, Giovanelli M. Early results of surgery in patients with nonfunctioning pituitary adenoma and analysis of the risk of tumor recurrence. J Neurosurg. 2008;108(3):525–532. doi: 10.3171/JNS/2008/108/3/0525. [DOI] [PubMed] [Google Scholar]
- 9.Meij BP, Lopes MB, Ellegala DB, Alden TD, Laws ER., Jr The long-term significance of microscopic dural invasion in 354 patients with pituitary adenomas treated with transsphenoidal surgery. J Neurosurg. 2002;96(2):195–208. doi: 10.3171/jns.2002.96.2.0195. [DOI] [PubMed] [Google Scholar]
- 10.Selman WR, Laws ER, Jr, Scheithauer BW, Carpenter SM. The occurrence of dural invasion in pituitary adenomas. J Neurosurg. 1986;64(3):402–407. doi: 10.3171/jns.1986.64.3.0402. [DOI] [PubMed] [Google Scholar]
- 11.Cheng T, Zhan X. Pattern recognition for predictive, preventive, and personalized medicine in cancer. EPMA J. 2017;8:51–60. doi: 10.1007/s13167-017-0083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grech G, Zhan X, Yoo BC, Bubnov R, Hagan S, Danesi R, Vittadini G, Desiderio D. EPMA position paper in cancer: current overview and future perspectives. EPMA J. 2015;6:9. doi: 10.1186/s13167-015-0030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhan X, Desiderio DM. The use of variations in proteomes to predict, prevent, personalize treatment for clinically non-functional pituitary adenomas. EPMA J. 2010;1:439–459. doi: 10.1007/s13167-010-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu R, Wang X, Zhan X. Multi-parameter systematic strategy for predictive, preventive, and personalized medicine in cancer. EPMA J. 2013;4:2. doi: 10.1186/1878-5085-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu M, Zhan X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J. 2018;9(1):77–102. doi: 10.1007/s13167-018-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhan X, Long Y, Lu M. Exploration of variations in proteome and metabolome for predictive diagnostics and personalised treatment algorithms: innovative approach and examples for potential clinical application. J Proteome. 2018;188:30–40. doi: 10.1016/j.jprot.2017.08.020. [DOI] [PubMed] [Google Scholar]
- 17.Zhan X, Li B, Zhan X, Schlüter H, Jungblut PR, Coorssen JR. Innovating the concept and practice of two-dimensional gel electrophoresis in the analysis of proteomes at the proteoform level. Proteomes. 2019;7(4):36. doi: 10.3390/proteomes704003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo T, Wang X, Li M, Yang H, Li L, Peng F, Zhan X. Identification of glioblastoma phosphotyrosine-containing proteins with two-dimensional Western blotting and tandem mass spectrometry. Biomed Res Int. 2015;2015:134050. doi: 10.1155/2015/134050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh V, Ram M, Kumar R, Prasad R, Roy BK, Singh KK. Phosphorylation: implications in cancer. Protein J. 2017;36:1–6. doi: 10.1007/s10930-017-9696-z. [DOI] [PubMed] [Google Scholar]
- 20.Golden RJ, Chen B, Li T, Braun J, Manjunath H, Chen X, Wu J, Schmid V, Chang TC, Kopp F, Ramirez-Martinez A, Tagliabracci VS, Chen ZJ, Xie Y, Mendell JT. An argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature. 2017;542:197–202. doi: 10.1038/nature21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai CF, Wang YT, Yen HY, Tsou CC, Ku WC, Lin PY, Chen HY, Nesvizhskii AI, Ishihama Y, Chen YJ. Large-scale determination of absolute phosphorylation stoichiometries in human cells by motif-targeting quantitative proteomics. Nat Commun. 2015;6:6622. doi: 10.1038/ncomms7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, Moasser MM. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445(7126):437–441. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah KN, Bhatt R, Rotow J, Rohrberg J, Olivas V, Wang VE, Hemmati G, Martins MM, Maynard A, Kuhn J, Galeas J, Donnella HJ, Kaushik S, Ku A, Dumont S, Krings G, Haringsma HJ, Robillard L, Simmons AD, Harding TC, McCormick F, Goga A, Blakely CM, Bivona TG, Bandyopadhyay S. Aurora kinase A drives the evolution of resistance to third-generation EGFR inhibitors in lung cancer. Nat Med. 2019;25(1):111–118. doi: 10.1038/s41591-018-0264-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreuzer J, Edwards A, Haas W. Multiplexed quantitative phosphoproteomics of cell line and tissue samples. Methods Enzymol. 2019;626:41–65. doi: 10.1016/bs.mie.2019.07.027. [DOI] [PubMed] [Google Scholar]
- 25.Li Z, Li M, Li X, Xin J, Wang Y, Shen QW, Zhang D. Quantitative phosphoproteomic analysis among muscles of different color stability using tandem mass tag labeling. Food Chem. 2018;249:8–15. doi: 10.1016/j.foodchem.2017.12.047. [DOI] [PubMed] [Google Scholar]
- 26.Carretero L, Llavona P, López-Hernández A, Casado P, Cutillas PR, de la Peña P, Barros F, Domínguez P. ERK and RSK are necessary for TRH-induced inhibition of r-ERG potassium currents in rat pituitary GH3 cells. Cell Signal. 2015;27(9):1720–1730. doi: 10.1016/j.cellsig.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 27.Zhao S, Feng J, Li C, Gao H, Lv P, Li J, Liu Q, He Y, Wang H, Gong L, Li D, Zhang Y. Phosphoproteome profiling revealed abnormally phosphorylated AMPK and ATF2 involved in glucose metabolism and tumorigenesis of GH-PAs. J Endocrinol Investig. 2019;42(2):137–148. doi: 10.1007/s40618-018-0890-4. [DOI] [PubMed] [Google Scholar]
- 28.Delcourt N, Thouvenot E, Chanrion B, Galéotti N, Jouin P, Bockaert J, Marin P. PACAP type I receptor transactivation is essential for IGF-1 receptor signalling and antiapoptotic activity in neurons. EMBO J. 2007;26(6):1542–1551. doi: 10.1038/sj.emboj.7601608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beranova-Giorgianni S, Zhao Y, Desiderio DM, Giorgianni F. Phosphoproteomic analysis of the human pituitary. Pituitary. 2006;9(2):109–120. doi: 10.1007/s11102-006-8916-x. [DOI] [PubMed] [Google Scholar]
- 30.Long Y, Lu M, Cheng T, Zhan X, Zhan X. Multiomics-based signaling pathway network alterations in human non-functional pituitary adenomas. Front Endocrinol. 2019;10:835. doi: 10.3389/fendo.2019.00835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ota M, Gonja H, Koike R, Fukuchi S. Multiple-localization and hub proteins. PLoS One. 2016;11:e0156455. doi: 10.1371/journal.pone.0156455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhan X, Li N, Zhan X, Qian S. Revival of 2DE-LC/MS in proteomics and its potential for large-scale study of human proteoforms. Med One. 2018;3:e180008. doi: 10.20900/mo.20180008. [DOI] [Google Scholar]
- 33.Zhan X, Yang H, Peng F, Li J, Mu Y, Long Y, Cheng T, Huang Y, Li Z, Lu M, Li N, Li M, Liu J, Jungblut PR. How many proteins can be identified in a 2-DE gel spot within an analysis of a complex human cancer tissue proteome? Electrophoresis. 2018;39:965–980. doi: 10.1002/elps.201700330. [DOI] [PubMed] [Google Scholar]
- 34.Aebersold R, Agar JN, Amster IJ, Baker MS, Bertozzi CR, Boja ES, Costello CE, Cravatt BF, Fenselau C, Garcia BA, Ge Y, Gunawardena J, Hendrickson RC, Hergenrother PJ, Huber CG, Ivanov AR, Jensen ON, Jewett MC, Kelleher NL, Kiessling LL, Krogan NJ, Larsen MR, Loo JA, Ogorzalek Loo RR, Lundberg E, MacCoss MJ, Mallick P, Mootha VK, Mrksich M, Muir TW, Patrie SM, Pesavento JJ, Pitteri SJ, Rodriguez H, Saghatelian A, Sandoval W, Schlüter H, Sechi S, Slavoff SA, Smith LM, Snyder MP, Thomas PM, Uhlén M, Van Eyk JE, Vidal M, Walt DR, White FM, Williams ER, Wohlschlager T, Wysocki VH, Yates NA, Young NL, Zhang B. How many human proteoforms are there? Nat Chem Biol. 2018;14(3):206–214. doi: 10.1038/nchembio.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith LM, Kelleher NL. Proteoforms as the next proteomics currency. Science. 2018;359(6380):1106–1107. doi: 10.1126/science.aat1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broncel M, Treeck M. Label-based mass spectrometry approaches for robust quantification of the phosphoproteome and total proteome in Toxoplasma gondii. Methods Mol Biol. 2020;2071:453–468. doi: 10.1007/978-1-4939-9857-9_23. [DOI] [PubMed] [Google Scholar]
- 37.Serioli S, Doglietto F, Fiorindi A, Biroli A, Mattavelli D, Buffoli B, et al. Pituitary adenomas and invasiveness from anatomo-surgical, radiological, and histological perspectives: a systematic literature review. Cancers (Basel). 2019;11(12). 10.3390/cancers11121936. [DOI] [PMC free article] [PubMed]
- 38.Zheng X, Li S, Zhang W, Zang Z, Hu J, Yang H. Current biomarkers of invasive sporadic pituitary adenomas. Ann Endocrinol (Paris) 2016;77(6):658–667. doi: 10.1016/j.ando.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Øystese KA, Evang JA, Bollerslev J. Non-functioning pituitary adenomas: growth and aggressiveness. Endocrine. 2016;53(1):28–34. doi: 10.1007/s12020-016-0940-7. [DOI] [PubMed] [Google Scholar]
- 40.Yang Q, Li X. Molecular network basis of invasive pituitary adenoma: a review. Front Endocrinol. 2019;10:7. doi: 10.3389/fendo.2019.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhan X, Desiderio DM. Editorial: Systems biological aspects of pituitary tumors. Front Endocrinol. 2016;7:86. doi: 10.3389/fendo.2016.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhan X, Long Y. Exploration of molecular network variations in different subtypes of human nonfunctional pituitary adenomas. Front Endocrinol. 2016;7:13. doi: 10.3389/fendo.2016.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhan X, Long Y, Zhan X, Mu Y. Consideration of statistical vs. biological significances for omics data-based pathway network analysis. Med One. 2017;1:e170002. doi: 10.20900/mo.20170002. [DOI] [Google Scholar]
- 44.Seifirad S, Haghpanah V. Inappropriate modeling of chronic and complex disorders: how to reconsider the approach in the context of predictive, preventive and personalized medicine, and translational medicine. EPMA J. 2019;10(3):195–209. doi: 10.1007/s13167-019-00176-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janssens JP, Schuster K, Voss A. Preventive, predictive, and personalized medicine for effective and affordable cancer care. EPMA J. 2018;9(2):113–123. doi: 10.1007/s13167-018-0130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhan X, Desiderio DM, editors. Molecular network study of pituitary adenomas. Lausanne: Frontiers Media SA; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banerjee S, Saxena N, Sengupta K, Banerjee SK. 17alpha-Estradiol-induced VEGF-A expression in rat pituitary tumor cells is mediated through ER independent but PI3K-Akt dependent signaling pathway. Biochem Biophys Res Commun. 2003;300(1):209–215. doi: 10.1016/s0006-291x(02)02830-9. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z, Jiang C, Ganther H, Lü J. Antimitogenic and proapoptotic activities of methylseleninic acid in vascular endothelial cells and associated effects on PI3K-AKT, ERK, JNK and p38 MAPK signaling. Cancer Res. 2001;61(19):7171–7178. [PubMed] [Google Scholar]
- 49.Smyth LM, Zhou Q, Nguyen B, Yu C, Lepisto EM, Arnedos M, Hasset MJ, Lenoue-Newton ML, Blauvelt N, Dogan S, Micheel CM, Wathoo C, Horlings H, Hudecek J, Gross BE, Kundra R, Sweeney SM, Gao J, Schultz N, Zarski A, Gardos SM, Lee J, Sheffler-Collins S, Park BH, Sawyers CL, André F, Levy M, Meric-Bernstam F, Bedard PL, Iasonos A, Schrag D, Hyman DM, AACR Project GENIE Consortium Characteristics and outcome of AKT1 E17K-mutant breast cancer defined through AACR Project GENIE, a clinicogenomic registry. Cancer Discov. 2020;10(4):526–535. doi: 10.1158/2159-8290.CD-19-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iida M, Harari PM, Wheeler DL, Toulany M. Targeting AKT/PKB to improve treatment outcomes for solid tumors. Mutat Res. 2020;819-820:111690. doi: 10.1016/j.mrfmmm.2020.111690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hunkeler M, Hagmann A, Stuttfeld E, Chami M, Guri Y, Stahlberg H, Maier T. Structural basis for regulation of human acetyl-CoA carboxylase. Nature. 2018;558(7710):470–474. doi: 10.1038/s41586-018-0201-4. [DOI] [PubMed] [Google Scholar]
- 52.Stoiber K, Nagło O, Pernpeintner C, Zhang S, Koeberle A, Ulrich M, Werz O, Müller R, Zahler S, Lohmüller T, Feldmann J, Braig S. Targeting de novo lipogenesis as a novel approach in anti-cancer therapy. Br J Cancer. 2018;118(1):43–51. doi: 10.1038/bjc.2017.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang W, Cui H, Yu D, Chen Y, Wang J, Yu G. Increased expression of phospho-acetyl-CoA carboxylase protein is an independent prognostic factor for human gastric cancer without lymph node metastasis. Med Oncol. 2014;31(7):15. doi: 10.1007/s12032-014-0015-7. [DOI] [PubMed] [Google Scholar]
- 54.Alkharusi A, Lesma E, Ancona S, Chiaramonte E, Nyström T, Gorio A, Norstedt G. Role of prolactin receptors in lymphangioleiomyomatosis. PLoS One. 2016;11(1):e0146653. doi: 10.1371/journal.pone.0146653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao SJ, Jiang YQ, Xu NW, Li Q, Zhang Q, Wang SY, Li J, Wang YH, Zhang YL, Jiang SH, Wang YJ, Huang YJ, Zhang XX, Tian GA, Zhang CC, Lv YY, Dai M, Liu F, Zhang R, Zhou D, Zhang ZG. SPARCL1 suppresses osteosarcoma metastasis and recruits macrophages by activation of canonical WNT/β-catenin signaling through stabilization of the WNT-receptor complex. Oncogene. 2018;37(8):1049–1061. doi: 10.1038/onc.2017.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma Y, Xu Y, Li L. SPARCL1 suppresses the proliferation and migration of human ovarian cancer cells via the MEK/ERK signaling. Exp Ther Med. 2018;16(4):3195–3201. doi: 10.3892/etm.2018.6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aruleba RT, Adekiya TA, Oyinloye BE, Kappo AP. Structural studies of predicted ligand binding sites and molecular docking analysis of Slc2a4 as a therapeutic target for the treatment of cancer. Int J Mol Sci. 2018;19(2):386. doi: 10.3390/ijms19020386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Ding N, Li Y, Cheng H, Wang D, Yang Q, Deng Y, Yang Y, Li Y, Ruan X, Xie F, Zhao H, Fang X. Insulin-like growth factor binding protein 5 (IGFBP5) functions as a tumor suppressor in human melanoma cells. Oncotarget. 2015;6(24):20636–20649. doi: 10.18632/oncotarget.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duan C, Allard JB. Insulin-like growth factor binding protein-5 in physiology and disease. Front Endocrinol. 2020;11:100. doi: 10.3389/fendo.2020.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Güllü G, Karabulut S, Akkiprik M. Functional roles and clinical values of insulin-like growth factor-binding protein-5 in different types of cancers. Chin J Cancer. 2012;31(6):266–280. doi: 10.5732/cjc.011.10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lloyd RV, Jin L. Analysis of chromogranin/secretogranin messenger RNAs in human pituitary adenomas. Diagn Mol Pathol. 1994;3(1):38–45. doi: 10.1097/00019606-199403010-00007. [DOI] [PubMed] [Google Scholar]
- 62.Lloyd RV, Jin L, Kulig E, Fields K. Molecular approaches for the analysis of chromogranins and secretogranins. Diagn Mol Pathol. 1992;1(1):2–15. doi: 10.1097/00019606-199203000-00002. [DOI] [PubMed] [Google Scholar]
- 63.Jin L, Chandler WF, Smart JB, England BG, Lloyd RV. Differentiation of human pituitary adenomas determines the pattern of chromogranin/secretogranin messenger ribonucleic acid expression. J Clin Endocrinol Metab. 1993;76(3):728–735. doi: 10.1210/jcem.76.3.7680355. [DOI] [PubMed] [Google Scholar]
- 64.d'Herbomez M, Do Cao C, Vezzosi D, Borzon-Chasot F, Baudin E, groupe des tumeurs endocrines (GTE France) Chromogranin A assay in clinical practice. Ann Endocrinol (Paris) 2010;71(4):274–280. doi: 10.1016/j.ando.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 65.Cruz-Topete D, Christensen B, Sackmann-Sala L, Okada S, Jorgensen JO, Kopchick JJ. Serum proteome changes in acromegalic patients following transsphenoidal surgery: novel biomarkers of disease activity. Eur J Endocrinol. 2011;164(2):157–167. doi: 10.1530/EJE-10-0754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang KT, Yang HJ, Choo KB, Lin HD, Fang SL, Braverman LE. A point mutation in the albumin gene in a Chinese patient with familial dysalbuminemic hyperthyroxinemia. Eur J Endocrinol. 1999;141(4):374–378. doi: 10.1530/eje.0.1410374. [DOI] [PubMed] [Google Scholar]
- 67.Liu X, Zheng W, Wang W, Shen H, Liu L, Lou W, Wang X, Yang P. A new panel of pancreatic cancer biomarkers discovered using a mass spectrometry-based pipeline. Br J Cancer. 2017;117(12):1846–1854. doi: 10.1038/bjc.2017.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin X, Hong S, Huang J, Chen Y, Chen Y, Wu Z. Plasma apolipoprotein A1 levels at diagnosis are independent prognostic factors in invasive ductal breast cancer. Discov Med. 2017;23(127):247–258. [PubMed] [Google Scholar]
- 69.Hu CA, Klopfer EI, Ray PE. Human apolipoprotein L1 (ApoL1) in cancer and chronic kidney disease. FEBS Lett. 2012;586(7):947–955. doi: 10.1016/j.febslet.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhan X, Desiderio DM. Heterogeneity analysis of the human pituitary proteome. Clin Chem. 2003;49(10):1740–1751. doi: 10.1373/49.10.1740. [DOI] [PubMed] [Google Scholar]
- 71.Moreno CS, Evans CO, Zhan X, Okor M, Desiderio DM, Oyesiku NM. Novel molecular signaling and classification of human clinically nonfunctional pituitary adenomas identified by gene expression profiling and proteomic analyses. Cancer Res. 2005;65(22):10214–10222. doi: 10.1158/0008-5472.CAN-05-0884. [DOI] [PubMed] [Google Scholar]
- 72.Zhan X, Wang X, Long Y, Desiderio DM. Heterogeneity analysis of the proteomes in clinically nonfunctional pituitary adenomas. BMC Med Genet. 2014;7:69. doi: 10.1186/s12920-014-0069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Golubnitschaja O, Costigliola V, EPMA General report & recommendations in predictive, preventive and personalised medicine 2012: white paper of the European Association for Predictive, Preventive and Personalised Medicine. EPMA J. 2012;3(1):14. doi: 10.1186/1878-5085-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu R, Wang X, Zhan X. Multi-parameter systematic strategies for predictive, preventive and personalised medicine in cancer. EPMA J. 2013;4(1):2. doi: 10.1186/1878-5085-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PPT 1739 kb)
(PDF 802 kb)
(XLS 68 kb)
(XLS 55 kb)
(XLS 72 kb)
(XLS 436 kb)
(XLS 28 kb)
(XLS 26 kb)
(XLS 27 kb)
(XLS 66 kb)