Abstract
Molecular motors and machines are essential for all cellular processes that together enable life. Built from proteins with a wide range of properties, functionalities and performance characteristics, biological motors perform complex tasks and can transduce chemical energy into mechanical work more efficiently than human-made combustion engines. Sophisticated studies of biological protein motors have provided many structural and biophysical insights and enabled the development of models for motor function. However, from the study of highly evolved, biological motors, it remains difficult to discern detailed mechanisms, for example, about the relative role of different force generation mechanisms, or how information is communicated across a protein to achieve the necessary coordination. A promising, complementary approach to answering these questions is to build synthetic protein motors from the bottom up. Indeed, much effort has been invested in functional protein design, but so far, the “holy grail” of designing and building a functional synthetic protein motor has not been realized. Here, we review the progress made to date, and we put forward a roadmap for achieving the aim of constructing the first artificial, autonomously running protein motor. Specifically, we propose to break down the task into (i) enzymatic control of track binding, (ii) the engineering of asymmetry and (iii) the engineering of allosteric control for internal communication. We also propose specific approaches for solving each of these challenges.
Keywords: Motor protein, Synthetic biology, Energy transduction, Allostery, Processivity, Thermal fluctuations
Introduction
Molecular motors and machines transduce chemical energy (typically by ATP hydrolysis) to perform mechanical work (Kull and Endow 2013) and are part of essentially all cellular processes that together enable life (Alberts 1998). Examples include linear motors such as myosin (Houdusse and Sweeney 2016), kinesin (Wang et al. 2015) and dynein (Schmidt and Carter 2016) and rotary motors such as the flagellar motors (Albers and Jarrell 2018; Minamino and Imada 2015), V1-ATPase (Ueno et al. 2018) and FoF1-ATPase (Okuno et al. 2011). The archetypal linear molecular motors are bipedal dyneins, kinesins and myosins, which can “walk” along one-dimensional tracks such as microtubules (dynein and kinesin) and actin filaments (myosin) (Fig. 1a–c, respectively).
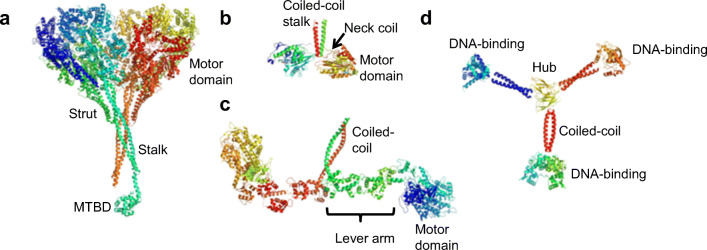
Fig. 1.
The structures of linear protein motors drawn to scale and oriented so that the track would be at the bottom of the figure. a Crystal structure of Dictyostelium discoideum dynein dimer determined at a 3.8-Å resolution (PDB 3VKH) with the motor domain (top) distal from the microtubule-binding domain (MTBD). Only one of the two MTBDs was resolved in the crystal structure (Kon et al. 2012). b The crystal structure of the kinesin dimer (PDB 3KIN) from rat brain determined at a 3.1-Å resolution (Kozielski et al. 1997). The arrow shows the neck coil linker (red line) leading from the right-hand motor domain to the C-terminal coiled-coil helix (red spiral). The green line near the arrowhead is the N-terminus of the right-hand motor domain. The motor domain binds directly to the microtubule track. c The structure of chicken smooth muscle heavy meromyosin dimer (PDB 3J04) determined by electron crystallography at a 20-Å resolution (Baumann et al. 2012). The two myosin heads splay out from the C-terminal coiled-coil. One lever arm and motor domain are labelled. The motor domains bind directly to the actin track. d A model for the tumbleweed artificial clocked walker protein (Bromley et al. 2009). The tumbleweed has three ligand-gated DNA-binding domains that are joined to a central hub via coiled-coil arms
The performance of these nanometre-scale machines is awe-inspiring from an engineering perspective: in spite of their small size, their energy efficiency exceeds that of human-made engines powered by chemical fuel, such as combustion engines (Bustamante et al. 2004). Additionally, molecular motors transform energy to work in an entirely different way: compared with combustion engines, protein motors appear to transduce chemical energy directly into work, without using heat as an intermediary energy form.
Evolution has selected proteins as the optimal material from which to construct these complex molecules with a wide range of properties, functionalities and performance characteristics. Today, we understand that the “magic” by which proteins achieve motor function boils down to a few key properties: (i) proteins can fold into a unique, solid-like, three-dimensional structure; (ii) determined by its sequence and hence its 3D structure, a protein can bind strongly and specifically via matching surfaces, making multiple weak interactions; (iii) a protein can transmit information between remote binding sites as a fundamental control mechanism, which we call allostery; and (iv) proteins operate in aqueous environments at ambient temperatures where thermal fluctuations dominate molecular motion—diffusive stepping (a Brownian ratchet–like mechanism) is thought to be part of the function of most, if not all, molecular machines. Along with achieving motor function, these four principles also underlie enzymatic function performed by proteins.
Principles for molecular motor function
The properties required to build a macromolecular motor are dictated by the physics of molecules operating in aqueous buffer in a biologically relevant temperature range. These requirements can be distilled into the following list: (1) an energy source; (2) far-from-equilibrium conditions; (3) thermal fluctuations; (4) asymmetry; (5) processivity; (6) timing; and (7) communication. Each of these elements is required for building an autonomous protein motor that travels along a molecular track. In the following, we discuss each of these seven requirements in order:
The energy source for almost all known linear biological motors is the hydrolysis of either ATP or GTP. Energy transduction is indirect, as the products of hydrolysis (ADP + Pi or GDP + Pi, respectively, where Pi is inorganic phosphate) are not immediately released into the medium. Instead, the protein orchestrates product release so that it is coupled to the structural state of the motor system in the transduction cycle. This has led to the concept of “free energy transduction” (Hill 1983).
Molecular motors and the surrounding medium must maintain far-from-equilibrium conditions in order to maintain the energy flow from chemical to mechanical (Brown and Sivak 2020). One way to achieve this is to maintain a high, non-equilibrium concentration of a fuel molecule such as ATP. Once the system reaches equilibrium, all energy transduction will cease and the motor will, at best, carry out diffusive motion driven randomly by thermal fluctuations.
Molecular motors, biological macromolecules and cells operate in a low Reynolds number regime, where friction and thermal fluctuations dominate all motion (Purcell 1977). Under these conditions, there is effectively no inertial motion and protein mean free paths are sub-nanometre. Instead of battling thermal fluctuations, molecular motors exploit them for stepping. In the absence of protein geometric constraints, thermally driven motion is randomly directed, with proteins capable of rectifying this random motion, for example, via a Brownian ratchet mechanism (Vale and Oosawa 1990).
Molecular motors require some form of asymmetry in order to gain directionality. Without it, they will just step back and forth in a diffusive manner. The asymmetry can be structural, in either the motor or the track, or it can involve a catalytic cycle or equivalent process controlling motor function.
Processivity is a linear motor’s ability to stay attached to its track for many consecutive steps. In the low Reynolds number regime, processivity is essential for individual motor molecules: if the motor detaches from the track for any significant period of time, it will diffuse away under the action of thermal fluctuations and will be essentially lost from the track system. To ensure processivity, many biological motors are either bipedal or they are arranged in linear arrays of motor modules to prevent migration away from the track.
Biological motors have a chemical cycle (enzymatic cycle) and a mechanical cycle. The transitions and intermediate steps in each of these cycles must be timed and coordinated (Kasper and Sivak 2020). For example, if a motor must make a diffusive step followed by track binding, the timescale for these events must be fast compared with a chemical clock controlling the state of the system; otherwise, the chemical clock will move on before the structural transition (diffusion followed by binding) has occurred.
Finally, internal communication is essential for an efficient molecular motor. For the individual motor domain, communication may involve coupling between the track-binding site and the enzymatic active site so as to coordinate the energetic cycle with the mechanical cycle. For bipedal motors, the state of one motor domain must be synchronized with the other motor domain. For example, if both motor domains release from the track simultaneously, then the motor is lost from the system. Biochemists use the word allostery to describe this molecular communication, particularly within proteins and protein complexes (Monod et al. 1963). The physical basis for this communication between distal sites within a protein is still poorly understood (Hilser et al. 2012; Motlagh et al. 2014; Thirumalai et al. 2019).
Based on our current understanding, integration of all seven of these elements is required to achieve autonomous operation of a molecular motor, that is, the ability to step along a track, powered by a fuel molecule in solution, and without external control.
Force generation: power strokes and Brownian ratchets
Some biological motors appear to operate as Brownian ratchets (Gu and Rice 2010; Guo and Sousa 2006; Xie 2011), while others, such as conventional linear motors, may implement a “power stroke” (Belyy and Yildiz 2014; Howard 2006) to induce long-range steps (Bier 2007). The physical basis for such power strokes is an active area of research (Hwang and Karplus 2019).
A Brownian ratchet is a device that rectifies thermal noise to do work (Astumian 1997). It operates by trapping positive motion due to a thermal fluctuation and preventing backward motion. This process of detecting a random fluctuation in the correct direction, and acting on this information to trap this motion, necessarily requires the processing of information, which incurs a thermodynamic cost in the form of free energy. Thus, although the work done by the ratchet is provided by thermal fluctuations, they still consume free energy, because the energy cost associated with deleting the acquired information (reset) is necessarily at least as large as the work done by the motor, in full agreement with the second law of thermodynamics (Jun et al. 2014; Landauer 1961). This concept is central to many natural and artificial molecular motors.
Synthetic biology approach
Traditionally, motor function has been studied by determining near-atomic resolution structures, measuring biochemical and biophysical properties and altering the protein using the tools of molecular biology. These approaches have been very successful in understanding the operation of biological linear motors such as the actin-based motors of the myosin family and microtubule-based motors such as the kinesin and dynein families. We now have atomic resolution structures of each of these motor systems in numerous functionally important states (Houdusse and Sweeney 2016; Kull and Endow 2013; Schmidt and Carter 2016). More recently, the structures of less stable or transient states have been determined at atomic resolution by cryo-electron microscopy (Liu et al. 2017; Locke et al. 2017; Mentes et al. 2018; Nishida et al. 2020; Peña et al. 2020; von der Ecken et al. 2016; Zhang et al. 2017). Site-directed mutagenesis guided by the structural information has probed the role of individual residues in effecting various aspects of motor function (Endow 2000). Single-molecule approaches have allowed biophysical processes to be observed and measured on a nanometre scale at sub-millisecond time resolution (Neuman and Nagy 2008). Although rich in detail, these studies still leave questions regarding the mechanisms by which motor proteins achieve their physical properties.
This extensive analysis of natural motor proteins has produced a very detailed picture of the workings of each of these linear motors, and the same is true for well-studied rotary motor proteins. Despite this excellent research, fundamental questions remain as to exactly how these molecular machines transduce energy. There is still considerable debate over mechanistic features, with apparently contradictory data (Houdusse and Sweeney 2016). Finally, in highly evolved motors, it is difficult to discern the presence and/or prevalence of different force-generating mechanisms, such as Brownian ratchet and directed movement (power strokes), as the extant motor seamlessly incorporates all aspects into one action.
An alternative approach to understanding the principles by which biological motors function is to build an autonomous protein motor from scratch. This is an ambitious goal and progress to achieving autonomous motor function has been modest to date, with regard to protein molecules.
Artificial molecular motors from small molecules and DNA
The importance of molecular-scale machines for future science and technology was recognized by the awarding of the 2016 Nobel Prize in Chemistry to Sauvage, Stoddart and Feringa for “the design and synthesis of molecular machines” (Service RF 2016; Van Noorden and Castelvecchi 2016). These scientists laid the foundations for functional molecular machines using a combination of modelling, organic synthesis and assembly and by dissecting the underlying physical mechanisms by which machines operate (Browne and Feringa 2006). For example, small molecules have been synthesized that will walk down a track (Leigh et al. 2014; von Delius et al. 2010) and that can perform a rotary motion. Although these achievements are spectacular, the synthesis of these molecular machines based on the synthesis of small molecules tends to be complex and bespoke.
On a somewhat larger scale, several groups have used DNA and its ability to base pair in a sequence-specific and highly predictable manner to generate molecular walkers and machines (Bath and Turberfield 2007; Cha et al. 2014), including free-running (autonomous) motors with spectacular properties (Bath and Turberfield 2007; Bazrafshan et al. 2020; Wickham et al. 2012; Wickham et al. 2011). However, these machines are limited by their size and the simple structure of DNA duplexes. Alternatively, proteins have been incorporated as design elements in nanoscale machines, such as polyvalent burnt-bridge ratchets (Kovacic et al. 2015). Proteins are superior to nucleic acid materials (DNA and RNA) in that they can form larger, more solid-like structures with much greater geometrical and chemical diversity. These clear advantages, reinforced by the fact that evolution has selected proteins as its material of choice, make a strong case for the value of learning how to design and build molecular motors based on proteins.
Functional protein design
Custom-made protein design with its promise of wide-range applications requires a detailed understanding of how proteins fold and function. Such design has been a long-standing goal in biochemistry and has seen much progress in recent years. Protein structures have been built from scratch with high precision and high stability (Brunette et al. 2015; Huang et al. 2016b). Protein interfaces have been designed to make proteins self-assemble into large cage-like nanomaterials (Bale et al. 2016; King et al. 2014; King et al. 2012), to undergo calcium-responsive large-scale conformational changes (Wei et al. 2020) or to bind therapeutic targets (Chevalier et al. 2017; Fleishman et al. 2011). In many of these cases, computational design has been integrated with mutagenesis and high-throughput screening techniques to improve initial hits. This strategy of combining the power of directed evolution with computational design had already been successfully applied in some of the first enzyme and small-molecule binding designs. Success, in particular in terms of turnover rates or binding affinities, could be significantly improved (Blomberg et al. 2013; Giger et al. 2013; Khersonsky et al. 2012; Lechner et al. 2018; Tinberg et al. 2013). While this shows that protein design “has come of age” (Huang et al. 2016a), functional protein design remains difficult in particular with respect to enzyme function. The integration of dynamics and allostery has been achieved in isolated cases (Davey et al. 2017; Joh et al. 2014) but remains the real challenge in the design of molecular motors.
Radical engineering of natural motors
Whereas building a protein motor from scratch remains an aspirational goal, considerable advances have been made by radically re-engineering functional domains of existing motor proteins. These advances are the subject of excellent reviews (DelRosso and Derr 2017; Furuta and Furuta and Furuta 2018; Iino et al. 2020). Given this, we will in this section only briefly review the advances that have been made by radically re-engineering domains of natural protein motors.
Kinesin chimeras and motor directionality
The kinesin family contains motors (Fig. 1b) that move to either the plus end or the minus end of the microtubule (Kull and Endow 2013), even though the crystal structures of the motor domains from plus-end-directed conventional kinesin (Kull et al. 1996) and the minus-end-directed NCD (Sablin et al. 1996) are almost identical. Thus, a region outside the motor domain determines motor directionality. Chimeras made from plus-end- and minus-end-directed kinesins were constructed by fusing motor domains from one class to neck plus stalk domains from the other (Fig. 1b). These experiments resulted in polarity switching where the polarity is not determined by the motor domain, but rather residues in the neck region (Case et al. 1997; Endow and Waligora 1998; Henningsen and Schliwa 1997). The discovery of naturally bidirectional kinesins, kinesin-5s and a kinesin-14 that switch direction in a context-dependent manner has shown that directionality can easily be reversed by factors outside the motor domain (Popchock et al. 2017; Roostalu et al. 2011).
Reversible control of myosin motor function via lever arm engineering
The concept of a “lever arm” (Uyeda et al. 1996) that determines the size of the working stroke of the actin-myosin interaction is based on the crystal structure of myosin S1 (Rayment et al. 1993). The lever arm is essentially the quasi-rigid rod formed by the C-terminus of the myosin S1 heavy chain that is supported by the myosin light chains (Fig. 1c). The lever arm hypothesis predicts that altering the length of the lever arm will alter both the step size and the velocity generated by the actin-myosin interaction. These predictions were borne out by both truncation and extension mutants of Dictyostelium discoideum myosins (Uyeda et al. 1996).
Replacing the light chain-binding domain of myosin, a central component of the lever arm, with one or two α-actinin repeats (a module of similar length and rigidity), leaves the function of Dictyostelium myosin essentially unaltered; however, the motility is dependent on the length of the lever arm (Anson et al. 1996). Single-molecule tracking showed that inserting either one or two α-actinin repeats results in a step size that is linearly proportional to the length of the lever arm (Ruff et al. 2001). The structure of the construct containing two α-actinin repeats was determined by x-ray crystallography (Kliche et al. 2001).
The direction of myosin motion relative to actin polarity (plus end versus minus end) is determined by the orientation of the lever arm. Thus, motor directionality can be altered by engineering the lever arm structure. Synthetic constructs were generated where an artificial lever arm comprising two α-actinin repeats was attached to either myosin II or myosin I motor domains with the lever arm pointing towards the plus or minus end of actin, respectively (Tsiavaliaris et al. 2004). These motors were shown to travel in opposite directions on actin filaments (F-actin).
Myosin VI naturally travels in the reverse direction (towards the minus end of the actin filament) to most myosin (plus end) molecules along the actin filament. The structural basis for this minus-end-directed myosin VI is an insertion between the converter (the last domain of the motor) and the lever arm. The net result of this insertion is that it redirects the orientation of the lever arm (Menetrey et al. 2005). Additionally, myosin VI has an unusually large power stroke or step size which results from a unique pre-power stroke state (Menetrey et al. 2007).
A series of truncated myosin VI constructs probed the function of the converter and lever arm (Bryant et al. 2007). Myosin VI truncated near the converter domain became a plus-end motor, while longer constructs move towards the minus end. Motility assays showed that the longer construct produced higher velocity motion, while direct measurement of stroke sizes showed that they were longer for the longer constructs (Bryant et al. 2007). The resulting model for motor action is consistent with the crystal structures (Menetrey et al. 2005; Menetrey et al. 2007).
Given the success in altering motor properties such as velocity, step size and directionality by changing the lever arm, new studies included a switchable conformational change in modified lever arms of various myosins (including the plus-end-directed myosins V and XI and the minus-end-directed myosin VI). Myosin VI was rendered Ca2+ sensitive by incorporating an IQ domain from the lever arm of myosin V. The IQ domain binds calmodulin in the presence of Ca2+, and it can be used to determine the directionality of the chimeric motor (Chen et al. 2012).
Photo-switchable myosin VI and XI chimeras were generated by incorporating a LOV domain sandwiched between two α-actinin domains in the lever arm (Nakamura et al. 2014). Depending on the construct, the synthetic motors could either change direction or change speed in a reversible, light-dependent manner (Nakamura et al. 2014).
Finally, hybrid protein-RNA molecules have been generated incorporating structured RNA into the lever arm of myosin VI (Omabegho et al. 2018). The structure of the RNA-based lever arm can be altered by specific oligonucleotide signals, switching the direction of the hybrid motor (Omabegho et al. 2018).
Controlling dynein directionality via engineering the length of its stalk
The dynein motor domain is separated from the microtubule-binding domain (MTBD) by an elongated coiled-coil stalk domain (Fig. 1a) (Carter et al. 2011; Kon et al. 2012), which is reminiscent of the myosin lever arm (Fig. 1c). The length of the coiled-coil stalk is fully conserved in all species; however, it can be changed by adding or subtracting heptads from its coiled-coil structure (Fig. 1a). Although varying the length of the stalk by simply removing or adding several heptads did not change the direction of dynein movement with respect to microtubule polarity (Carter et al. 2008), it did alter the helical direction by which Saccharomyces cerevisiae dynein spirals around the microtubule (Can et al. 2019). More importantly, two conserved proline residues at the base of the MTBD along the coiled-coil of the stalk appear to determine the polarity of dynein motion. All natural dyneins move towards that minus end of the microtubule. However, shifting these two conserved prolines by two residues reversed the direction of dynein so that it moves towards the plus end of the microtubule, implying that the power stroke contributes significantly to the directional movement of dynein (Can et al. 2019).
Engineering for increased processivity
Processivity is a key property of molecular motors. If a motor releases completely from the track, it will diffuse away, thereby ceasing directed motion. The two-headed structure of nature’s linear motors (most myosins, kinesins and dyneins) is likely an evolutionary adaptation to ensure processivity.
The neck coiled-coil region of kinesin was shown to influence conventional kinesin processivity (Thorn et al. 2000). In particular, adding positive charge to the neck coiled-coil region increased the already high processivity of kinesin, while adding negative charge decreased the run length. The charge is thought to maintain the engineered kinesin in the proximity of the microtubule through an electrostatic tethering with the C-terminus of tubulin, thus enhancing processivity (Thorn et al. 2000).
Using a more radical synthetic approach, hybrid DNA-kinesin motors were generated, where a DNA duplex joined two kinesin motor domains (Miyazono et al. 2010). The DNA linker could be used to vary the separation of the heads in a systematic manner. The results show that strain communicated between the two motor domains via a 13-residue neck linker at the C-terminus of the head was critical for maintaining processivity (Fig. 1b) (Miyazono et al. 2010).
Synthetic biology has been used to explore the processivity of myosin (Schindler et al. 2014). These studies found that strategies that enhanced the effectiveness of random, uncoordinated stepping improved processivity. Specifically, processivity was enhanced by increasing the number of heads on both myosin VI and myosin XI to either three or four. Processivity was also increased by the insertion of flexible elements between heads (Schindler et al. 2014).
Re-engineering the dynein track-binding domain
One normally assumes that track binding and motor domain activity need to be tightly coupled in order to create a new motor. However, experiments on dynein show that this need not be the case (Furuta et al. 2017). Dynein differs from myosin and kinesin, in that the motor domain is distant from the track (Fig. 1a), with the microtubule-binding domain (MTBD) separated from the motor by an elongated coiled-coil stalk domain, supported by a strut/buttress (Carter et al. 2011; Kon et al. 2012). It is unclear as to how or if any information is transmitted from the motor domain to the track-binding domain, where microtubules form the dynein track.
By replacing the microtubule-binding domain with various actin-binding domains inserted into the tip of the dynein stalk, hybrid dynein motors were observed to step along actin filaments (Furuta et al. 2017). The motor activity of this hybrid structure did not critically depend on the nature of the dynein-actin-binding domain fusion, with several circular permutations of the actin-binding domain producing comparable motor activity (Furuta et al. 2017). The authors conclude that these hybrid motors do not have tight coupling between ATPase activity of the motor domain and the track-binding domain. Instead, motor activity results from a Brownian ratchet mechanism (Vale and Oosawa 1990), with directionality achieved by asymmetric unbinding from the actin track (Cleary et al. 2014). Dynein might be a mechanical ratchet that rectifies fluctuations by utilizing the asymmetric nature of motor-track interfaces (Ezber et al. 2020).
Lessons learnt from radical engineering and other studies on natural protein motors
From the above work and other studies, one sees a variety of different modes of motor operation. For some motors, such as myosin, there appears to be a tight coupling between track binding/unbinding, enzymatic cycle and work done by the motor. In contrast, the radical engineering of the dynein track-binding domain indicates that this tight coupling is unnecessary for successful motor action (Furuta et al. 2017). Lever arm engineering on a variety of myosins indicates that the motor domain generates a small structural change that is amplified by the lever arm, which is consistent with the concept of an active power stroke (Howard 2006). In contrast, kinesins appear more akin to Brownian ratchets in their force-generating mechanism. Kinesins also rely on signalling between the two motor domains to maintain high processivity; however, there is also a contribution from electrostatics, keeping the motor in proximity of the track. All this indicates that evolution has discovered numerous ways to affect protein motor function.
In addition to gaining an understanding of natural linear motor proteins, these engineering studies reveal the power of using foreign protein domains as elements to alter motor properties. Modules used in these studies include the tandem α-actinin domains as a rigid element, coiled-coils for dimerization, LOV domains for light activation and structured nucleic acid domains—DNA and RNA. The modularity of many aspects of protein motor function lends itself to this synthetic hybrid approach. This modularity simplifies the task of ab initio protein motor design (Fig. 2a).
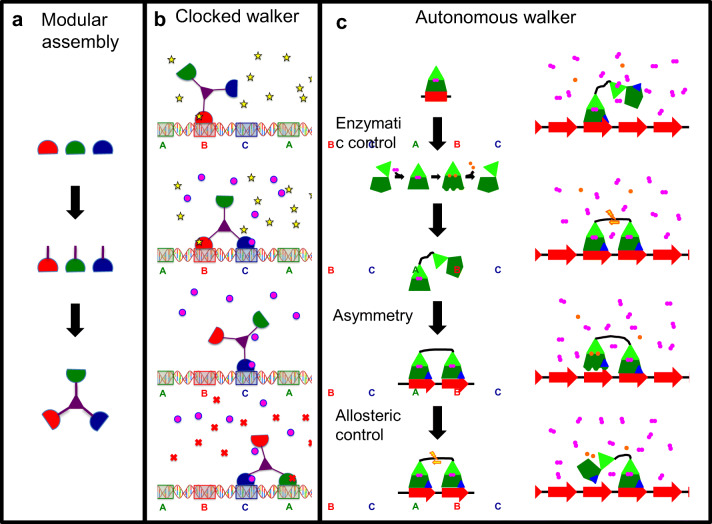
Fig. 2.
Pathway to generating synthetic motor proteins. a Modular assembly of protein motors using a tool kit of discrete protein modules each with well-characterized molecular functions. Here, three track-binding domains (red, green and blue semicircles) are linked to spacer domains (purple rods) and then assembled into a three-legged structure, such as the tumbleweed (Bromley et al. 2009). b The principle of action for the tumbleweed clocked walker Brownian ratchet (Bromley et al. 2009). The track comprises double-stranded DNA with three cognate sites (A, B and C) for each of the DNA-binding domains in the tumbleweed. Top panel: the red domain binds to its cognate DNA site (labelled B) in the presence of the activating ligand (yellow star). Second panel: the addition of the activating ligand (magenta circle) for the blue domain traps it bound to its cognate ligand once it encounters this DNA site (labelled C) via Brownian motion. Third panel: the removal of the red domain ligand (yellow star) allows it to detach from the DNA track. Bottom panel: the introduction of the cognate ligand for the green domain (red cross) traps it once it collides with its cognate site on DNA (labelled A). c Design principles for an autonomous walker. Left side: the track-binding domain (green) is controlled by enzymatic activity. This is shown as a cycle, where the two-domain enzyme opens a cleft so that a substrate (pink dumbbell) enters the active site and is cleaved (two yellow circles); these products are released when the site reopens. Some form of asymmetry is needed in the design (shown as the polarity in the track (red arrows)) and polarity of the track binder (blue triangle on the bottom right). Finally, allosteric control is represented by communication between two track-binding domains (yellow arrow and lightning). The right-hand side of the panel puts these principles into action. Top panel: one binder attaches to the track with substrate (pink dumbbell) bound. Second panel: the second binder attaches to the same track upstream from the first. As it does so, it sends a signal to the original bound domain, activating the enzymatic activity of this latter domain. Third panel: the originally bound domain completes its enzymatic cycle, detaches from the track and releases the products of catalysis. Bottom panel: the system is now in the same state as the top panel with only one module bound to the track; however, the walker has progressed by one site along the track
Building an artificial protein motor from scratch
Designing and building an artificial motor from scratch is a tall order. Given the variety of options used by natural protein motors, the protein engineer can make different choices when selecting energy source, asymmetry, force-generating mechanism, timing and communication. Some options are easier to engineer than others. Here, we review key approaches approximately in order of increasing complexity.
Clocked walker Brownian ratchets
The first successful artificial molecular motors (small-molecule or DNA-based) were non-autonomous clocked walkers that operated as Brownian ratchets. A “clocked walker” is a motor that requires external, cyclical signals (often changes in the bathing solution or light pulses) to both coordinate motor action and supply energy to the motor, keeping the system far from equilibrium (Bath and Turberfield 2007). This massively simplifies motor design, since one does not need to engineer a sophisticated (enzymatic) energy supply nor does one have to engineer allosteric control and communication systems into the motor.
A Brownian ratchet mechanism is also much simpler to engineer than a power stroke, particularly since the physics of the latter is still poorly understood. For a ratchet, one effectively needs to control binding and unbinding events but can leave all motion to diffusion or thermal fluctuations. The energetic cost for a Brownian ratchet comes from the binding/unbinding reactions, and this lends itself to the clocked walker design, where the changes in chemical potential (e.g. ligands that trigger binding are supplied at high chemical potential and are removed at low potential) or the application of controlling light pulses supply energy to the motor.
Finally, ab initio protein design is still in its infancy (see above); hence, simpler approaches circumventing ab initio design provide expediency to an already difficult task. One such approach is to use known protein modules with well-characterized molecular function as building blocks in the design process (Fig. 2a). This approach has been used widely in the radical engineering of natural protein motors (see above).
The first designs for protein-based artificial motors are essentially clocked walkers operating as Brownian ratchets: they are the three-legged tumbleweed (Fig. 1d) (Bromley et al. 2009), the two-legged SKIP (Zuckermann et al. 2015) and the light-activated bipedal bar-hinge motor (Small et al. 2019). These proposed clocked walkers use double-stranded DNA as a track (Kovacic et al. 2012), ligand-gated DNA-binding proteins as “motor domains” and coiled-coils as lever arms (Bromley et al. 2009; Small et al. 2019; Zuckermann et al. 2015). The tumbleweed is a three-legged structure with three different motor domains, where each binds to a specific DNA motif in the presence of a specific ligand (Bromley et al. 2009). The three-legged geometry provides the structural asymmetry conferring directionality (Fig. 2b).
The two-legged SKIP uses two coiled-coil lever arms each attached to a pair of DNA-binding domains, which are specific for each lever arm (Zuckermann et al. 2015). Its mode of operation is similar to an earlier bipedal nanomotor proposal (Wang 2007). Structural asymmetry is lacking in SKIP, and so directional movement is achieved by docking SKIP in an appropriate orientation at the start of the track (or at any randomly selected binding site). SKIP has the unusual advantage of acting as a shuttle, capable of running processively from one end of the track to the other and back (Zuckermann et al. 2015). Both SKIP and tumbleweed are proposed to use a microfluidic system to supply appropriate sequences of ligand pulses, which both control and power the motor (Niman et al. 2013). The bar-hinge motor differs from the other designs in that it incorporates a light-sensing azobenzene molecule that controls the angle between the two lever arm coiled-coils in a light-dependent manner (Small et al. 2019). Whereas detailed, numerical models exist for each of these designs for clocked walker Brownian ratchets (Bromley et al. 2009; Small et al. 2019; Zuckermann et al. 2015), to date, there are no reports on their experimental realizations.
Towards autonomous artificial motor proteins
Three key principles that are thought to be essential to natural protein motors have to be incorporated into the design of an autonomous artificial protein motor. These principles are enzymatic control of track binding, structural asymmetry to get unidirectional motion and allosteric regulation of the enzymatic activity (Fig. 2c). Attaining these goals will be a major advance beyond clocked walker Brownian ratchets (Table 1).
Table 1.
Strategy for design and construction of an autonomous bipedal motor
| Objective | Strategy |
|---|---|
| Enzymatic control of track binding |
• Engineer existing small-molecule ligand (called a co-repressor)a binding pocket to acquire enzymatic activity • Couple to an enzyme to acquire catalytic activity then alter binding pocket to accommodate co-repressor mimetic • Use directed evolution to refine enzymatic activity and DNA binding |
| Structural asymmetry |
• Break symmetry of the repressor by altering one DNA-binding motif per repressor dimer • Make single polypeptide versions of dimeric repressors to engineer asymmetry • Couple asymmetric monomers to engineer asymmetric dimers |
| Allosteric control of enzymatic activity |
• Split catalytic site so catalysis only occurs when 2nd foot binds adjacent to 1st foot • Use strain generated when 2nd foot binds to alter active site of original bound foot |
aAssumes that a ligand-gated repressor, activated by a co-repressor (small-molecule ligand), is the starting protein for a DNA track-binding module
Engineering enzymatic control of track binding
The designs that we have explored in our groups use ligand-dependent DNA-binding proteins (called repressors, where the activating small-molecule ligand is called a co-repressor) as the basis for our clocked walker Brownian ratchets. An obvious enhancement that could be the first step towards autonomy would be to convert these repressor proteins into enzymes, where track binding will now be controlled autonomously by the phase of the engineered enzymatic cycle.
One ligand-dependent DNA-binding protein that is currently being used in the tumbleweed design is the Trp repressor (TrpR): it binds to the trp operator DNA site in the presence of the ligand (or co-repressor) tryptophan which forms part of the repressor-DNA interface (Phillips and Stockley 1996). TrpR has already been engineered to accommodate a series of tryptophan-related ligands (Stiel et al. 2020). The challenge will be to select an appropriate ligand and use an engineering approach to convert TrpR into an enzyme (Lechner et al. 2018), thereby autonomously controlling track binding (see below).
Engineering allosteric control by exploiting dimerization interfaces
Engineering allosteric control is an even more difficult protein engineering task. In general, the structural basis of allostery, where binding of a ligand to one site on a protein influences the affinity or activity of the protein for a distal site, is not well understood. However, there are some simpler allosteric mechanisms that may be amenable to current protein engineering. One possibility is to place the active site on a protein-protein or domain-domain interface, so that the enzyme is only active if the interface has been correctly formed. Such a split active site is used in many examples in biology where dimerization is essential for enzymatic activity (e.g. dimeric ATPases like MinD (Lutkenhaus and Sundaramoorthy 2003) and nitrogenase iron protein and small GTPases which require a GTPase accelerating protein, GAP, to complete the active site (Rittinger et al. 1997)).
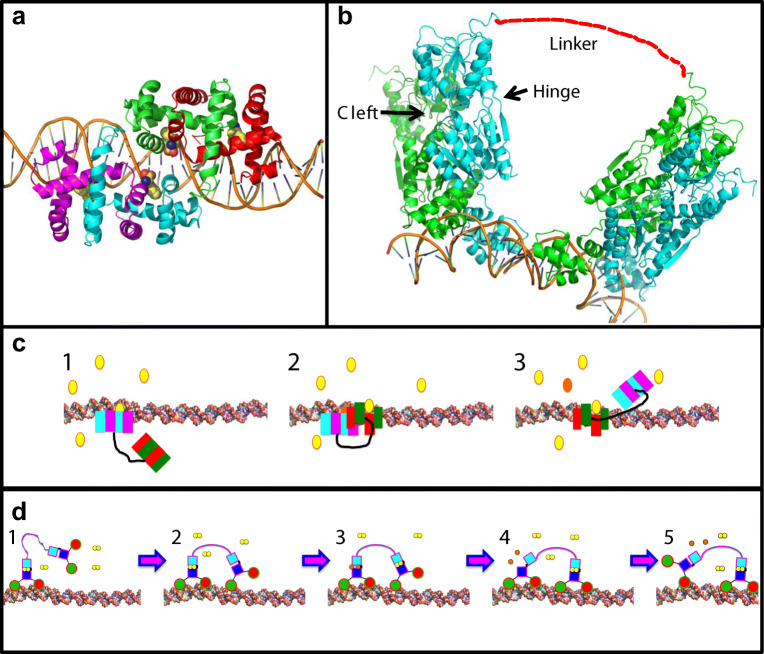
TrpR, which we use in our artificial motor designs as a DNA-binding domain, might be a suitable engineering target. The crystal structure of full-length TrpR shows binding of two TrpR dimers to two adjacent trp operator sites with the tryptophan ligands (co-repressors) sitting at the interface between the two adjacent TrpR proteins (Fig. 3a) (Lawson and Carey 1993). It may be possible to create an enzymatic site using residues from the two adjacent TrpR dimers with the substrate at the interface so that the catalytic site is only complete when the two “feet” (each containing a TrpR dimer) are bound to adjacent sites.
Fig. 3.
Designing allosteric communication to create an autonomous walker protein. a Allostery by putting the active site on an interface. Crystal structure of two adjacent TrpR dimers bound to tandem trp DNA sites (Lawson and Carey 1993). One TrpR dimer is shown in cyan and magenta, the other in red and green ribbon representation. The tryptophan molecules (sphere representation coloured: carbon, yellow; nitrogen, blue; and oxygen, red) lie near the interface between the two dimers. b Allosteric communication via strain. Two dimeric PurR repressors (green-cyan) bound to neighbouring sites on DNA. PurR is engineered to be a heterodimer with only the cyan monomer actively controlling DNA binding. Strain induced by the right-hand PurR biases the cleft in the left-hand PurR to the open position, releasing products and hence detaching from the track. For the right-hand PurR, the same strain keeps the active site closed, preventing premature product release from the active site. c Concept for a TrpR-based autonomous bipedal walker where allosteric communication is achieved by having the active site at an interface. Two asymmetric TrpR feet are tethered together where each asymmetric foot has only one partial catalytic site. The full active site requires both TrpR feet to bind to adjacent sites in the DNA major groove in a forward direction (due to the asymmetry of the engineered TrpR). The walker cycle proceeds as follows: Step 1: One foot (cyan/magenta) is bound to the track with substrate (yellow ellipse) bound to the partial active site. Step 2: 2nd foot (red/green) binds adjacent to the 1st foot completing the catalytic site, generating products (orange). Step 3: After catalysis, the original foot (cyan/magenta) is released from the track. d PurR-based autonomous walker where allostery is communicated by strain. Two PurR chimeras with asymmetric feet are linked by a tether. The tether length is such that when one foot is bound to the DNA track, tight binding of the second foot will be impeded unless it generates tension in the linker. This tension will serve to open the active site cleft in the rear, previously bound foot and hence induce product release and unbinding from the track. The cycle proceeds as follows: Step 1: One leg binds to DNA track in the presence of ligand (yellow dumbbell). 2nd foot is free to diffuse. Step 2: Second leg discovers a cognate site on DNA in a forward direction and commences to bind this site. Step 3: Forward leg binds activating ligand (yellow dumbbell) while the initially bound rear foot hydrolyses the ligand (two orange circles). Ligand binding to front and hydrolysis in rear foot need not be concomitant. Step 4: The front foot now binds DNA strongly. It induces strain in the previously bound rear foot, opening the active site and facilitating release of products (orange circles). Step 5: The rear foot has no ligand; hence, it releases from the DNA track
Engineering asymmetry
All repressor molecules (such as TrpR) are homodimers, and all repressor binding sites (such as the trp operator DNA site) are palindromic DNA sequences. As such, any motor built of repressor proteins will not be directional without additional constraints.
Many repressors bind to DNA via helix-turn-helix (HTH) reading head structures that recognize specific DNA sequences via interactions in the major groove. Thus, an engineered heterodimer can be generated by altering the DNA sequence specificity of only one monomer by grafting a HTH recognition motif from a different repressor. To ensure that the engineered molecule is a heterodimer, the two different repressors can be linked into a single chain. Since only the DNA-interacting residues would be altered, the impact on protein structure and stability would be minimal.
For TrpR, it has been shown that a TrpR dimer can be expressed as a single polypeptide chain with a fluorescent protein (FP) as “linker” (Kaper et al. 2007). Using such a single-chain construct, it would be possible to replace the HTH DNA recognition motif from one of the two copies of TrpR with a HTH from a different DNA-binding protein, thus generating a non-symmetric protein that binds to a non-palindromic DNA site.
Two such identical engineered asymmetric TrpR heterodimers could then be coupled to generate an autonomous bipedal walker (Fig. 3c). The autonomous walker cycle could be implemented where a substrate only binds to one side of an asymmetric TrpR heterodimer (again, this would require engineering), biasing it to the DNA-bound state (Fig. 3c, step 1). When a second asymmetric TrpR heterodimer binds to the adjacent downstream trp operator site on DNA (Fig. 3c step 2), it completes an engineered enzymatic active site, turning over the substrate and releasing the original TrpR from the DNA track (Fig. 3c, step 3). This cycle will continue in a unidirectional manner (Fig. 3c). Here, directionality of the bipedal walker ensues from the asymmetry within each heterodimer (directing the polarity of each foot binding to the track) and the coupling of the enzymatic cycle to the already-bound substrate in the trailing foot on the track.
Allosteric control via strain generated by a bipedal walker
Many enzymes contain active sites at the interface of two domains, where the apo form has an open active-site cleft which closes on substrate binding via domain rotation (e.g. the GroEL ATPase (Xu et al. 1997)). For this type of enzyme, mechanical strain could be used to bias the structural equilibrium to favour either the open or the closed state of the protein, thereby allowing the strain to control the enzyme (Bustamante et al. 2004; Choi et al. 2005b).
Mechanical strain is used to control many enzymes and motor proteins. Many kinases are found in an inactive form when regulator protein domains (e.g. SH2 and SH3) bind to linear protein segments that anchor the upper kinase domain (Sicheri and Kuriyan 1997). When these regulator proteins bind, they induce strain in the connecting linker and thus prevent closure of the kinase active site. Under these conditions, the kinase is rendered inactive. Similarly (and more relevant), the molecular motor myosin V has a large step, with the two motor domains separated by multiple actin monomers (Tyska and Mooseker 2003). It is thought that product release of the rear motor domain is activated by strain transmitted through the linker as the forward motor domain binds to the actin filament (Craig and Linke 2009). Thus, one should be able to utilize mechanical strain to communicate binding information, i.e. allostery.
A suitable protein family for engineering allostery in such a way are the LacI/PurR repressor family proteins, whose binding to DNA is controlled by a two-domain co-repressor binding domain (CBD; Fig. 3b) (Lewis 2005), which is a member of the substrate-binding protein (SBP) structural family (Berntsson et al. 2010). Co-repressor (small-molecule ligand) binding controls the open-closed state of the CBD, controlling DNA binding. Structurally related SBPs have already been converted into enzymes (Clifton et al. 2018). Thus, it should be possible to convert a member of the LacI/PurR family into an enzyme where the enzymatic state will control DNA binding.
In a next step, this enzymatically controlled repressor has to be engineered to be sensitive to strain in a way that maintains processivity—i.e. the rear DNA-bound foot cannot complete its enzymatic reaction and thus will not leave the DNA track until the other foot binds in front of it (Fig. 3b, d). Another member of the SBP family, maltose-binding protein, has successfully been engineered so as it is allosterically controlled by mechanical strain (Choi et al. 2005a). Thus, strain-dependent allosteric control should be attainable for an engineered LacI/PurR protein.
Searching for power stroke generating modules
All of the above autonomous designs are still Brownian ratchets: their stepping is driven purely by random thermal motion and then is locked in by binding to the substrate in the presence of the appropriate ligand. It remains an open challenge to discover a non-motor protein module that can be engineered in such a fashion that it generates a power stroke akin to members of the myosin family.
Conclusion: design and engineering challenges
The design and construction of artificial protein motors as described above include a number of design and engineering challenges. Synthetic biologists and protein designers have successfully constructed proteins never before seen in nature. Successes include the de novo design of pre-defined protein topologies, the generation of new protein-protein interfaces and, in some cases, the installation of enzymatic functions in previously non-catalytic protein scaffolds. However, the design of proteins with pre-defined function still remains a major difficulty. Moreover, the integration of dynamics and allostery is an even bigger challenge. As such, it is not surprising that the design of a functional motor protein capable of autonomously transducing chemical energy into mechanical work is regarded as an exciting but outstanding challenge to the field.
To succeed in this challenge, we have developed a possible roadmap for the design and construction of an autonomous bipedal motor (Table 1). To achieve this goal, multiple engineering milestones have to be met simultaneously: mutual tuning of allosteric control, track binding and coupling of both so that autonomous stepping is accomplished. A bottom-up approach must confront these challenges in understanding and design. We envision that modularity provides an excellent path to the targeting of more complex functions and that protein motors can be designed bottom-up from existing protein domains that have no motor function. At the same time, the full advantage of a bottom-up approach is maintained, namely, the ability to study the emergence of function from the synergistic interplay of its parts.
Authors’ contributions
All authors contributed to the conception of this work, the generation of ideas, the analysis, the review of the literature and the writing of the manuscript.
Funding information
NRF is funded by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC; 2020-04680). BH is funded by an ERC Consolidator Grant from the European Research Council (647548 “Protein Lego”). PMGC is funded by a Discovery Grant from the Australian Research Council (DP170103153). KF is funded by a Grant-in-Aid for Scientific Research on Innovative Areas “Molecular Engine” (JP18H05420). HL is funded by NanoLund, by the European Union (732482 Bio4Comp), the Swedish Research Council and the Knut and Alice Wallenberg Foundation (2016.0089).
Data availability
Not applicable.
Code availability
Not applicable.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Albers SV, Jarrell KF. The Archaellum: an update on the unique archaeal motility structure. Trends Microbiol. 2018;26:351–362. doi: 10.1016/j.tim.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- Anson M, Geeves MA, Kurzawa SE, Manstein DJ. Myosin motors with artificial lever arms. EMBO J. 1996;15:6069–6074. doi: 10.1002/j.1460-2075.1996.tb00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astumian RD. Thermodynamics and kinetics of a Brownian motor. Science. 1997;276:917–922. doi: 10.1126/science.276.5314.917. [DOI] [PubMed] [Google Scholar]
- Bale JB, et al. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science. 2016;353:389–394. doi: 10.1126/science.aaf8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath J, Turberfield AJ. DNA nanomachines. Nat Nanotechnol. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- Baumann BA, Taylor DW, Huang Z, Tama F, Fagnant PM, Trybus KM, Taylor KA. Phosphorylated smooth muscle heavy meromyosin shows an open conformation linked to activation. J Mol Biol. 2012;415:274–287. doi: 10.1016/j.jmb.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazrafshan A, et al. Tunable DNA origami motors translocate ballistically over mum distances at nm/s speeds. Angew Chem. 2020;59:9514–9521. doi: 10.1002/anie.201916281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyy V, Yildiz A. Processive cytoskeletal motors studied with single-molecule fluorescence techniques. FEBS Lett. 2014;588:3520–3525. doi: 10.1016/j.febslet.2014.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berntsson RP, Smits SH, Schmitt L, Slotboom DJ, Poolman B. A structural classification of substrate-binding proteins. FEBS Lett. 2010;584:2606–2617. doi: 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- Bier M. The stepping motor protein as a feedback control ratchet. Bio Systems. 2007;88:301–307. doi: 10.1016/j.biosystems.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Blomberg R, et al. Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature. 2013;503:418–421. doi: 10.1038/nature12623. [DOI] [PubMed] [Google Scholar]
- Bromley EHC, et al. The tumbleweed: towards a synthetic protein motor. Hfsp J. 2009;3:204–212. doi: 10.2976/1.3111282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AI, Sivak DA. Theory of nonequilibrium free energy transduction by molecular machines. Chem Rev. 2020;120:434–459. doi: 10.1021/acs.chemrev.9b00254. [DOI] [PubMed] [Google Scholar]
- Browne WR, Feringa BL. Making molecular machines work. Nat Nanotechnol. 2006;1:25–35. doi: 10.1038/nnano.2006.45. [DOI] [PubMed] [Google Scholar]
- Brunette TJ, et al. Exploring the repeat protein universe through computational protein design. Nature. 2015;528:580–584. doi: 10.1038/nature16162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant Z, Altman D, Spudich JA. The power stroke of myosin VI and the basis of reverse directionality. Proc Natl Acad Sci U S A. 2007;104:772–777. doi: 10.1073/pnas.0610144104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu Rev Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- Can S, Lacey S, Gur M, Carter AP, Yildiz A. Directionality of dynein is controlled by the angle and length of its stalk. Nature. 2019;566:407–410. doi: 10.1038/s41586-019-0914-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, et al. Structure and functional role of dynein’s microtubule-binding domain. Science. 2008;322:1691–1695. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case RB, Pierce DW, Hom-Booher N, Hart CL, Vale RD. The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell. 1997;90:959–966. doi: 10.1016/s0092-8674(00)80360-8. [DOI] [PubMed] [Google Scholar]
- Cha TG, Pan J, Chen H, Salgado J, Li X, Mao C, Choi JH. A synthetic DNA motor that transports nanoparticles along carbon nanotubes. Nat Nanotechnol. 2014;9:39–43. doi: 10.1038/nnano.2013.257. [DOI] [PubMed] [Google Scholar]
- Chen L, Nakamura M, Schindler TD, Parker D, Bryant Z. Engineering controllable bidirectional molecular motors based on myosin. Nat Nanotechnol. 2012;7:252–256. doi: 10.1038/nnano.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier A, et al. Massively parallel de novo protein design for targeted therapeutics. Nature. 2017;550:74–79. doi: 10.1038/nature23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B, Zocchi G, Canale S, Wu Y, Chan S, Perry LJ. Artificial allosteric control of maltose binding protein. Phys Rev Lett. 2005;94:038103. doi: 10.1103/PhysRevLett.94.038103. [DOI] [PubMed] [Google Scholar]
- Choi B, Zocchi G, Wu Y, Chan S, Jeanne Perry L. Allosteric control through mechanical tension. Phys Rev Lett. 2005;95:078102. doi: 10.1103/PhysRevLett.95.078102. [DOI] [PubMed] [Google Scholar]
- Cleary FB, et al. Tension on the linker gates the ATP-dependent release of dynein from microtubules. Nat Commun. 2014;5:4587. doi: 10.1038/ncomms5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton BE, Kaczmarski JA, Carr PD, Gerth ML, Tokuriki N, Jackson CJ. Evolution of cyclohexadienyl dehydratase from an ancestral solute-binding protein. Nat Chem Biol. 2018;14:542–547. doi: 10.1038/s41589-018-0043-2. [DOI] [PubMed] [Google Scholar]
- Craig EM, Linke H. Mechanochemical model for myosin V. Proc Natl Acad Sci U S A. 2009;106:18261–18266. doi: 10.1073/pnas.0908192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JA, Damry AM, Goto NK, Chica RA. Rational design of proteins that exchange on functional timescales. Nat Chem Biol. 2017;13:1280–1285. doi: 10.1038/nchembio.2503. [DOI] [PubMed] [Google Scholar]
- DelRosso NV, Derr ND. Exploiting molecular motors as nanomachines: the mechanisms of de novo and re-engineered cytoskeletal motors. Curr Opin Biotechnol. 2017;46:20–26. doi: 10.1016/j.copbio.2016.10.011. [DOI] [PubMed] [Google Scholar]
- Endow SA. Molecular motors--a paradigm for mutant analysis. J Cell Sci. 2000;113(Pt 8):1311–1318. doi: 10.1242/jcs.113.8.1311. [DOI] [PubMed] [Google Scholar]
- Endow SA, Waligora KW. Determinants of kinesin motor polarity. Science. 1998;281:1200–1202. doi: 10.1126/science.281.5380.1200. [DOI] [PubMed] [Google Scholar]
- Ezber Y, Belyy V, Can S, Yildiz A. Dynein harnesses active fluctuations of microtubules for faster movement. Nat Phys. 2020;16:312–316. doi: 10.1038/s41567-019-0757-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishman SJ, et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta K, Furuta A. Re-engineering of protein motors to understand mechanisms biasing random motion and generating collective dynamics. Curr Opin Biotechnol. 2018;51:39–46. doi: 10.1016/j.copbio.2017.11.009. [DOI] [PubMed] [Google Scholar]
- Furuta A, Amino M, Yoshio M, Oiwa K, Kojima H, Furuta K. Creating biomolecular motors based on dynein and actin-binding proteins. Nat Nanotechnol. 2017;12:233–237. doi: 10.1038/nnano.2016.238. [DOI] [PubMed] [Google Scholar]
- Giger L, Caner S, Obexer R, Kast P, Baker D, Ban N, Hilvert D. Evolution of a designed retro-aldolase leads to complete active site remodeling. Nat Chem Biol. 2013;9:494–498. doi: 10.1038/nchembio.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci U S A. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Sousa R. Translocation by T7 RNA polymerase: a sensitively poised Brownian ratchet. J Mol Biol. 2006;358:241–254. doi: 10.1016/j.jmb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Henningsen U, Schliwa M. Reversal in the direction of movement of a molecular motor. Nature. 1997;389:93–96. doi: 10.1038/38022. [DOI] [PubMed] [Google Scholar]
- Hill TL. Some general principles in free energy transduction. Proc Natl Acad Sci U S A. 1983;80:2922–2925. doi: 10.1073/pnas.80.10.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilser VJ, Wrabl JO, Motlagh HN. Structural and energetic basis of allostery. Annu Rev Biophys. 2012;41:585–609. doi: 10.1146/annurev-biophys-050511-102319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houdusse A, Sweeney HL. How myosin generates force on actin filaments. Trends Biochem Sci. 2016;41:989–997. doi: 10.1016/j.tibs.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J. Protein power strokes. Curr Biol. 2006;16:R517–R519. doi: 10.1016/j.cub.2006.06.045. [DOI] [PubMed] [Google Scholar]
- Huang PS, Boyken SE, Baker D. The coming of age of de novo protein design. Nature. 2016;537:320–327. doi: 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]
- Huang PS, Feldmeier K, Parmeggiani F, Velasco DAF, Hocker B, Baker D. De novo design of a four-fold symmetric TIM-barrel protein with atomic-level accuracy. Nat Chem Biol. 2016;12:29–34. doi: 10.1038/nchembio.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W, Karplus M. Structural basis for power stroke vs. Brownian ratchet mechanisms of motor proteins. Proc Natl Acad Sci U S A. 2019;116:19777–19785. doi: 10.1073/pnas.1818589116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino R, Kinbara K, Bryant Z. Introduction: molecular motors. Chem Rev. 2020;120:1–4. doi: 10.1021/acs.chemrev.9b00819. [DOI] [PubMed] [Google Scholar]
- Joh NH, et al. De novo design of a transmembrane Zn(2)(+)-transporting four-helix bundle. Science. 2014;346:1520–1524. doi: 10.1126/science.1261172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun Y, Gavrilov M, Bechhoefer J. High-precision test of Landauer’s principle in a feedback trap. Phys Rev Lett. 2014;113:190601. doi: 10.1103/PhysRevLett.113.190601. [DOI] [PubMed] [Google Scholar]
- Kaper T, Looger LL, Takanaga H, Platten M, Steinman L, Frommer WB. Nanosensor detection of an immunoregulatory tryptophan influx/kynurenine efflux cycle. PLoS Biol. 2007;5:e257. doi: 10.1371/journal.pbio.0050257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper AKS, Sivak DA. Modeling work-speed-accuracy trade-offs in a stochastic rotary machine. Phys Rev E. 2020;101:032110. doi: 10.1103/PhysRevE.101.032110. [DOI] [PubMed] [Google Scholar]
- Khersonsky O, et al. Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59. Proc Natl Acad Sci U S A. 2012;109:10358–10363. doi: 10.1073/pnas.1121063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King NP, et al. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science. 2012;336:1171–1174. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King NP, et al. Accurate design of co-assembling multi-component protein nanomaterials. Nature. 2014;510:103–108. doi: 10.1038/nature13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliche W, Fujita-Becker S, Kollmar M, Manstein DJ, Kull FJ. Structure of a genetically engineered molecular motor. EMBO J. 2001;20:40–46. doi: 10.1093/emboj/20.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, Kurisu G. The 2.8 A crystal structure of the dynein motor domain. Nature. 2012;484:345–350. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- Kovacic S, Samii L, Woolfson DN, Curmi PMG, Linke H, Forde NR, Blab GA (2012) Design and construction of a one-dimensional DNA track for an artificial molecular motor. J Nanomater:Artn 109238. 10.1155/2012/109238
- Kovacic S, Samii L, Curmi PMG, Linke H, Zuckermann MJ, Forde NR. Design and construction of the lawnmower, an artificial burnt-bridges motor Ieee T. Nanobiosci. 2015;14:305–312. doi: 10.1109/Tnb.2015.2393872. [DOI] [PubMed] [Google Scholar]
- Kozielski F, et al. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 1997;91:985–994. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- Kull FJ, Endow SA. Force generation by kinesin and myosin cytoskeletal motor proteins. J Cell Sci. 2013;126:9–19. doi: 10.1242/jcs.103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landauer R. Irreversibility and heat generation in the computing process. IBM J Res Dev. 1961;5:183–191. doi: 10.1147/rd.53.0183. [DOI] [Google Scholar]
- Lawson CL, Carey J. Tandem binding in crystals of a trp repressor/operator half-site complex. Nature. 1993;366:178–182. doi: 10.1038/366178a0. [DOI] [PubMed] [Google Scholar]
- Lechner H, Ferruz N, Hocker B. Strategies for designing non-natural enzymes and binders. Curr Opin Chem Biol. 2018;47:67–76. doi: 10.1016/j.cbpa.2018.07.022. [DOI] [PubMed] [Google Scholar]
- Leigh DA, Lewandowska U, Lewandowski B, Wilson MR. Synthetic molecular walkers. Top Curr Chem. 2014;354:111–138. doi: 10.1007/128_2014_546. [DOI] [PubMed] [Google Scholar]
- Lewis M. The lac repressor. C R Biol. 2005;328:521–548. doi: 10.1016/j.crvi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Liu D, Liu X, Shang Z, Sindelar CV (2017) Structural basis of cooperativity in kinesin revealed by 3D reconstruction of a two-head-bound state on microtubules. Elife 6. 10.7554/eLife.24490 [DOI] [PMC free article] [PubMed]
- Locke J, Joseph AP, Pena A, Mockel MM, Mayer TU, Topf M, Moores CA. Structural basis of human kinesin-8 function and inhibition. Proc Natl Acad Sci U S A. 2017;114:E9539–E9548. doi: 10.1073/pnas.1712169114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J, Sundaramoorthy M. MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation. Mol Microbiol. 2003;48:295–303. doi: 10.1046/j.1365-2958.2003.03427.x. [DOI] [PubMed] [Google Scholar]
- Menetrey J, Bahloul A, Wells AL, Yengo CM, Morris CA, Sweeney HL, Houdusse A. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature. 2005;435:779–785. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetrey J, Llinas P, Mukherjea M, Sweeney HL, Houdusse A. The structural basis for the large powerstroke of myosin VI. Cell. 2007;131:300–308. doi: 10.1016/j.cell.2007.08.027. [DOI] [PubMed] [Google Scholar]
- Mentes A, et al. High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing. Proc Natl Acad Sci U S A. 2018;115:1292–1297. doi: 10.1073/pnas.1718316115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Imada K. The bacterial flagellar motor and its structural diversity. Trends Microbiol. 2015;23:267–274. doi: 10.1016/j.tim.2014.12.011. [DOI] [PubMed] [Google Scholar]
- Miyazono Y, Hayashi M, Karagiannis P, Harada Y, Tadakuma H. Strain through the neck linker ensures processive runs: a DNA-kinesin hybrid nanomachine study. EMBO J. 2010;29:93–106. doi: 10.1038/emboj.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J, Changeux JP, Jacob F. Allosteric proteins and cellular control systems. J Mol Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Chen L, Howes SC, Schindler TD, Nogales E, Bryant Z. Remote control of myosin and kinesin motors using light-activated gearshifting. Nat Nanotechnol. 2014;9:693–697. doi: 10.1038/nnano.2014.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niman CS, Beech JP, Tegenfeldt JO, Curmi PMG, Woolfson DN, Forde NR, Linke H. Controlled microfluidic switching in arbitrary time-sequences with low drag. Lab Chip. 2013;13:2389–2396. doi: 10.1039/C3lc50194a. [DOI] [PubMed] [Google Scholar]
- Nishida N, et al. Structural basis for two-way communication between dynein and microtubules. Nat Commun. 2020;11:1038. doi: 10.1038/s41467-020-14842-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno D, Iino R, Noji H. Rotation and structure of FoF1-ATP synthase. J Biochem. 2011;149:655–664. doi: 10.1093/jb/mvr049. [DOI] [PubMed] [Google Scholar]
- Omabegho T, et al. Controllable molecular motors engineered from myosin and RNA. Nat Nanotechnol. 2018;13:34–40. doi: 10.1038/s41565-017-0005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña A, Sweeney A, Cook AD, Locke J, Topf M, Moores CA. Structure of microtubule-trapped human kinesin-5 and its mechanism of inhibition revealed using cryoelectron microscopy. Structure. 2020;28:450–457.e455. doi: 10.1016/j.str.2020.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips SE, Stockley PG. Structure and function of Escherichia coli met repressor: similarities and contrasts with trp repressor. Philos Trans R Soc Lond Ser B Biol Sci. 1996;351:527–535. doi: 10.1098/rstb.1996.0051. [DOI] [PubMed] [Google Scholar]
- Popchock AR, Tseng KF, Wang P, Karplus PA, Xiang X, Qiu W. The mitotic kinesin-14 KlpA contains a context-dependent directionality switch. Nat Commun. 2017;8:13999. doi: 10.1038/ncomms13999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell EM. Life at low Reynolds number. Am J Phys. 1977;45:3–11. doi: 10.1119/1.10903. [DOI] [Google Scholar]
- Rayment I, et al. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Rittinger K, Walker PA, Eccleston JF, Smerdon SJ, Gamblin SJ. Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997;389:758–762. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- Roostalu J, Hentrich C, Bieling P, Telley IA, Schiebel E, Surrey T. Directional switching of the kinesin Cin8 through motor coupling. Science. 2011;332:94–99. doi: 10.1126/science.1199945. [DOI] [PubMed] [Google Scholar]
- Ruff C, Furch M, Brenner B, Manstein DJ, Meyhofer E. Single-molecule tracking of myosins with genetically engineered amplifier domains. Nat Struct Biol. 2001;8:226–229. doi: 10.1038/84962. [DOI] [PubMed] [Google Scholar]
- Sablin EP, Kull FJ, Cooke R, Vale RD, Fletterick RJ. Crystal structure of the motor domain of the kinesin-related motor ncd. Nature. 1996;380:555–559. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- Schindler TD, Chen L, Lebel P, Nakamura M, Bryant Z. Engineering myosins for long-range transport on actin filaments. Nat Nanotechnol. 2014;9:33–38. doi: 10.1038/nnano.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Carter AP. Review: Structure and mechanism of the dynein motor. ATPase Biopolymers. 2016;105:557–567. doi: 10.1002/bip.22856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service RF Chemistry Nobel heralds age of molecular machines. Science. 2016;354:158–159. doi: 10.1126/science.354.6309.158. [DOI] [PubMed] [Google Scholar]
- Sicheri F, Kuriyan J. Structures of Src-family tyrosine kinases. Curr Opin Struct Biol. 1997;7:777–785. doi: 10.1016/S0959-440X(97)80146-7. [DOI] [PubMed] [Google Scholar]
- Small LSR, Zuckermann MJ, Sessions RB, Curmi PMG, Linke H, Forde NR, Bromley EHC. The bar-hinge motor: a synthetic protein design exploiting conformational switching to achieve directional motility. New J Phys. 2019;21:ARTN 013002. doi: 10.1088/1367-2630/aaf3ca. [DOI] [Google Scholar]
- Stiel AC, Shanmugaratnam S, Herud-Sikimic O, Jürgens G, Höcker B (2020) Ligand promiscuity in the tryptophan repressor – from structural understanding towards rational design bioRxiv
- Thirumalai D, Hyeon C, Zhuravlev PI, Lorimer GH. Symmetry, rigidity, and allosteric signaling: from monomeric proteins to molecular machines. Chem Rev. 2019;119:6788–6821. doi: 10.1021/acs.chemrev.8b00760. [DOI] [PubMed] [Google Scholar]
- Thorn KS, Ubersax JA, Vale RD. Engineering the processive run length of the kinesin motor. J Cell Biol. 2000;151:1093–1100. doi: 10.1083/jcb.151.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinberg CE, et al. Computational design of ligand-binding proteins with high affinity and selectivity. Nature. 2013;501:212–216. doi: 10.1038/nature12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiavaliaris G, Fujita-Becker S, Manstein DJ. Molecular engineering of a backwards-moving myosin motor. Nature. 2004;427:558–561. doi: 10.1038/nature02303. [DOI] [PubMed] [Google Scholar]
- Tyska MJ, Mooseker MS. Myosin-V motility: these levers were made for walking. Trends Cell Biol. 2003;13:447–451. doi: 10.1016/S0962-8924(03)00172-7. [DOI] [PubMed] [Google Scholar]
- Ueno H, Suzuki K, Murata T. Structure and dynamics of rotary V1 motor. Cell Mol Life Sci. 2018;75:1789–1802. doi: 10.1007/s00018-018-2758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda TQ, Abramson PD, Spudich JA. The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc Natl Acad Sci U S A. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Oosawa F. Protein motors and Maxwell’s demons: does mechanochemical transduction involve a thermal ratchet? Adv Biophys. 1990;26:97–134. doi: 10.1016/0065-227X(90)90009-I. [DOI] [PubMed] [Google Scholar]
- Van Noorden R, Castelvecchi D. World’s tiniest machines win chemistry Nobel. Nature. 2016;538:152–153. doi: 10.1038/nature.2016.20734. [DOI] [PubMed] [Google Scholar]
- von Delius M, Geertsema EM, Leigh DA. A synthetic small molecule that can walk down a track. Nat Chem. 2010;2:96–101. doi: 10.1038/nchem.481. [DOI] [PubMed] [Google Scholar]
- von der Ecken J, Heissler SM, Pathan-Chhatbar S, Manstein DJ, Raunser S. Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature. 2016;534:724–728. doi: 10.1038/nature18295. [DOI] [PubMed] [Google Scholar]
- Wang Z. Synergic mechanism and fabrication target for bipedal nanomotors. Proc Natl Acad Sci U S A. 2007;104:17921–17926. doi: 10.1073/pnas.0703639104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Cao L, Wang C, Gigant B, Knossow M. Kinesin, 30 years later: recent insights from structural studies. Protein Sci. 2015;24:1047–1056. doi: 10.1002/pro.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei KY, et al. Computational design of closely related proteins that adopt two well-defined but structurally divergent folds. Proc Natl Acad Sci U S A. 2020;117:7208–7215. doi: 10.1073/pnas.1914808117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham SF, Endo M, Katsuda Y, Hidaka K, Bath J, Sugiyama H, Turberfield AJ. Direct observation of stepwise movement of a synthetic molecular transporter. Nat Nanotechnol. 2011;6:166–169. doi: 10.1038/nnano.2010.284. [DOI] [PubMed] [Google Scholar]
- Wickham SF, Bath J, Katsuda Y, Endo M, Hidaka K, Sugiyama H, Turberfield AJ. A DNA-based molecular motor that can navigate a network of tracks. Nat Nanotechnol. 2012;7:169–173. doi: 10.1038/nnano.2011.253. [DOI] [PubMed] [Google Scholar]
- Xie P. A nucleotide binding rectification Brownian ratchet model for translocation of Y-family DNA polymerases. Theor Biol Med Modell. 2011;8:22. doi: 10.1186/1742-4682-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- Zhang K, Foster HE, Rondelet A, Lacey SE, Bahi-Buisson N, Bird AW, Carter AP. Cryo-EM reveals how human cytoplasmic dynein is auto-inhibited and activated. Cell. 2017;169:1303–1314 e1318. doi: 10.1016/j.cell.2017.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckermann MJ, et al. Motor properties from persistence: a linear molecular walker lacking spatial and temporal asymmetry. New J Phys. 2015;17:Artn 055017. doi: 10.1088/1367-2630/17/5/055017. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.