Abstract
Angiotensin-(1–9) is a peptide from the noncanonical renin-angiotensin system with anti-hypertrophic effects in cardiomyocytes via an unknown mechanism. In the present study we aimed to elucidate it, basing us initially on previous work from our group and colleagues who proved a relationship between disturbances in mitochondrial morphology and calcium handling, associated with the setting of cardiac hypertrophy. Our first finding was that angiotensin-(1–9) can induce mitochondrial fusion through DRP1 phosphorylation. Secondly, angiotensin-(1–9) blocked mitochondrial fission and intracellular calcium dysregulation in a model of norepinephrine-induced cardiomyocyte hypertrophy, preventing the activation of the calcineurin/NFAT signaling pathway. To further investigate angiotensin-(1–9) anti-hypertrophic mechanism, we performed RNA-seq studies, identifying the upregulation of miR-129 under angiotensin-(1–9) treatment. miR-129 decreased the transcript levels of the protein kinase A inhibitor (PKIA), resulting in the activation of the protein kinase A (PKA) signaling pathway. Finally, we showed that PKA activity is necessary for the effects of angiotensin-(1–9) over mitochondrial dynamics, calcium handling and its anti-hypertrophic effects.
Subject terms: Cell biology, Mechanism of action, Cardiovascular diseases
Introduction
Angiotensin-(1–9) is a nine-amino acid peptide produced by hydrolysis of the carboxy terminal leucine from angiotensin I (or angiotensin-(1–10)), by the angiotensin-converting enzyme type 2 (ACE 2) [1]. ACE2 also produces angiotensin-(1–7) through hydrolysis of angiotensin II, thereby reducing available levels of pro-hypertrophic angiotensin II. It was initially thought that angiotensin-(1–9) functioned merely as an intermediate step in the conversion of angiotensin I to angiotensin II; [1] however, new evidence showed that the ACE2/angiotensin-(1–9) axis acts as an antagonist of classical RAS signaling [2]. Angiotensin-(1–9) has significant cardiovascular bioactivity [3–6] through its interaction with the angiotensin receptor type 2 (AT2R) [5–7]. Both in vitro and in vivo studies have shown that angiotensin-(1–9) prevents cardiac hypertrophic growth in response to either angiotensin II or norepinephrine (NE) [3, 6]. A more recent study by Fattah et al. also demonstrated that angiotensin-(1–9) gene therapy, delivered via an adeno-associated viral vector, preserved left ventricular systolic function after myocardial infarction [7].
At the cellular level, cardiac hypertrophy occurs in association with metabolic changes directly related to altered mitochondrial function and morphology. Mitochondrial remodeling helps to meet elevated energy demands and supports modulation of intracellular Ca2+ signaling, leading to changes in gene and protein expression as well as other processes [8, 9]. In the human heart, about 70% of ATP production occurs in the mitochondrial matrix, underscoring the importance of this organelle for maintaining normal myocardial function [10].
Morphological changes alter not only mitochondrial shape and distribution but also metabolic state. Loss of mitochondrial fusion proteins Mitofusin 1/2 (MFN1/2) or Optic Atrophy-1 (OPA1) has been linked to a reduction in cellular growth rate and respiration as well as a dissipation of the mitochondrial membrane potential [11]. However, silencing of the fission protein Dynamin Related Protein-1 (DRP1) can also decrease cellular respiration and intracellular ATP content [12]. Thus, although there is not always a direct relationship between specific mitochondrial morphology and metabolic capacity, current evidence supports the concept that mitochondrial dynamics play a critical role in the metabolic function of this organelle.
We have proposed previously a relationship between the development of pathological cardiac hypertrophy and mitochondrial network fragmentation [13]. In addition, we have shown that treating cultured cardiomyocytes with NE promotes migration of DRP1 to the mitochondria, leading to increased numbers of individual mitochondria and a decreased mean mitochondrial volume [13]. These morphological changes are accompanied by a loss of mitochondrial function. Interestingly, reducing MFN2 levels (using an antisense adenovirus) to decrease fusion was sufficient to promote a hypertrophic response, whereas the use of an adenovirus encoding a dominant-negative form of the fission protein DRP1 elicited a fused phenotype and prevented NE-induced hypertrophic growth [13]. In aggregate, these data point to a new role for mitochondrial dynamics in the onset and regulation of cardiac hypertrophy.
As mentioned above, increased ventricular wall stress in the hypertrophied heart requires increased cytoplasmic Ca2+ levels to maintain cardiac output [14]. This rise in intracellular Ca2+ levels activates a variety of signaling pathways, including calcineurin-NFAT signaling, to mediate gene expression changes involved in the development of pathological cardiac hypertrophy [14]. Finally, our recent work suggests that loss of physical and functional communication between the endoplasmic reticulum (ER) and mitochondria is a key structural event in the development of pathological cardiac hypertrophy [15, 16]. Decreased ER-mitochondria coupling reduces Ca2+ transfer between these organelles and impairs mitochondrial capacity to buffer the increases in intracellular Ca2+ that may be triggered by chronic stimulation with neurohumoral and hypertrophic agents such as NE [15].
MicroRNAs (miRNAs) are short single-stranded RNAs that regulate post-transcriptional mRNA expression by binding to complementary mRNA sequences, resulting in translational repression and gene silencing [17]. These non-coding RNAs play critical roles in several physiological processes such as cellular proliferation and differentiation as well as tissue development and repair [18, 19]. Since their discovery in humans [20], an explosion of knowledge has given rise to the idea that miRNAs may offer new therapeutic alternatives for previously-untreatable and chronic diseases [21–24]. The field of cardiovascular diseases has been the object of intense investigation regarding potential applications for this knowledge. miRNAs have been implicated in the development of several cardiovascular conditions such as cardiomyocyte hypertrophy, heart failure, and ischemic cardiac disease [25–27]. Various deregulated miRNAs have been found to be elevated in cardiac tissue in mice subjected to transverse aortic constriction and/or calcineurin pathway over-activation, including miR-21, miR-23, miR-24, miR-125b, miR-195, miR-199a, and miR-214. Conversely, miR-29c, miR-93, miR-150, and miR-181b levels were reduced in these mice [25]. In parallel, miR-24, miR-125b, miR-195, miR-199a and miR-214 expression has been shown to be elevated in human hearts with terminal heart failure [25], establishing a common pattern of response among miRNAs to hypertrophy and heart failure. However, although multiple miRNAs have been characterized and studied in the setting of cardiac hypertrophy and heart failure, differential miRNA expression after stimulation with physiological anti-hypertrophic agents has yet to be investigated exhaustively.
Here, we show for the first time that the anti-hypertrophic action of angiotensin-(1–9) is linked to prevention of mitochondrial fragmentation and a reduction in the ER-mitochondrial coupling produced by NE-induced hypertrophy in neonatal rat cardiomyocytes, via stimulation of the miR-129-3p/PKIA signaling pathway. To our knowledge, this is the first study showing a relationship between this novel anti-hypertrophic noncanonical RAS peptide and the regulation of mitochondrial dynamics and mitochondrial Ca2+ uptake in cardiomyocytes.
Materials and methods
Bioethics
All studies conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication, 8th Edition, 2011) and were approved by our Institutional Ethics Review Committee.
Reagents
Antibodies against OPA1, MFN2, and phospho- and total CREB were from Abcam. Anti-phospho-Ser616-DRP1 and anti-phospho-Ser637-DRP1 were from Cell Signaling. MitoTracker Green FM, Rhod-FF AM, Fluo3 AM, Fura-2, Alexa fluorescent secondary antibodies, FBS, PKI [14–22]-myr, Opti-MEM and Lipofectamine RNAiMAX were from Thermo Fisher Scientific. Anti-DRP1 antibody was from Becton-Dickinson, and anti-Fis1, anti-mtHsp70 and anti-KDEL antibodies were from ENZO Life Sciences. H89 was purchased from Calbiochem. Anti-β-tubulin and anti-β-actin antibodies, carbonyl cyanide m-chlorophenylhydrazone (CCCP), DMEM, M199 medium, norepinephrine, PD123,319 ditrifluoroacetate, and other reagents were from Sigma-Aldrich Corp. A-779 was from Santa Cruz Biotechnology. Angiotensin-(1–9) was purchased from GL Biochem Ltd. Anti-miR-129-3p (AM10076), Anti-miR negative control #1 (AM17010) and pre-miR-129-3p (PM10076) were from Ambion. All inorganic compounds, salts, and solvents were from Merck. Protein assay reagents were from Bio-Rad.
Cardiomyocyte culture
Cardiomyocytes were isolated from hearts of neonatal Sprague-Dawley rats as described previously [13]. Rats were bred at the University of Chile Animal Breeding Facility. Primary cell cultures were incubated with or without angiotensin-(1–9) (100 μM) and/or NE (10 μΜ) for 0–24 h in DMEM/M199 (4:1) medium, in the presence or absence of the various inhibitors and other genetic reagents. Only spontaneous beating cardiomyocytes were used for experiments.
Mitochondrial dynamics analysis
Cells were incubated for 30 min with MitoTracker Green FM (400 nM) and maintained in Krebs solution. Confocal image stacks were captured with a Zeiss LSM-5, Pascal 5 Axiovert 200 microscope, using LSM 5 3.2 image capture and analysis software and a Plan-Apochromat 63×/1.4 Oil DIC objective, as previously described [28, 29]. Images were deconvolved with ImageJ, and then Z-stacks of thresholded images were volume-reconstituted. The numbers of and individual volumes in voxels (pixels3) of each object (mitochondria) were quantified using the ImageJ-3D Object counter plugin. Each experiment was performed at least four times, and 16–25 cells per condition were quantified. The criterion for mitochondria fusion was increased mitochondrial volume accompanied by decreased numbers of mitochondria, whereas decreased mitochondrial volume and increased numbers of mitochondria were considered to represent mitochondrial fission [29, 30]
Evaluation of cardiomyocyte hypertrophy
Sarcomerization was observed with confocal microscopy (Carl Zeiss LSM 5, Pascal 5 Axiovert 200 microscope) of methanol-permeabilized cells stained with rhodamine-phalloidin (1:400; F-actin staining) as previously described [13, 31]. For the cellular perimeter and mitochondrial area analysis, one focal plane of at least 50 cells from randomly selected fields was analyzed using ImageJ software (NIH).
Assessment of intracellular and mitochondrial Ca2+
Cytosolic Ca2+ levels were determined in cardiomyocytes preloaded with Fluo3-AM (5.4 μM, 30 min) or Fura2 (5 μM, 30 min), as described previously [32, 33]. To evaluate mitochondrial Ca2+ levels, images were obtained from cultured cardiomyocytes preloaded with Rhod-FF (5.4 μM, 30 min) [32–34]. At the end of each measurement, 10 µM of CCCP was used as control [15]. Both measurements were performed in an inverted confocal microscope (Carl Zeiss LSM 5, Pascal 5 Axiovert 200 microscope).
Immunofluorescence studies and colocalization analysis
Cells were fixed, permeabilized, blocked, and incubated with primary antibodies (anti-DRP1 or anti-KDEL and anti-FIS1). Secondary antibodies were anti-mouse Alexa 456 or 488 and anti-rabbit 488 or 456 for FIS1. For the colocalization analysis, only one focal plane was analyzed. Images obtained were deconvolved, and the background was subtracted using ImageJ software. Colocalization between proteins was quantified using the Mander’s algorithm, as previously described [15, 35, 36].
Transmission electron microscopy
Cells were fixed in 2.5% glutaraldehyde in sodium cacodylate buffer, embedded in 2% agarose, post-fixed in buffered 1% osmium tetroxide and stained in 2% uranyl acetate, dehydrated with an ethanol graded series, and embedded in EMbed-812 resin. Thin sections were cut on an ultramicrotome and stained with 2% uranyl acetate and lead citrate. Images were acquired on an FEI Tecnai G2 Spirit electron microscope equipped with a LaB6 source and operating at 120 kV. Measurements of mitochondrial area, perimeter, circularity, and mitochondrial density were performed using the Multi Measure ROI tool in the ImageJ (NIH) software package [29].
Western blotting
Cell and tissue total protein extracts were analyzed as described previously [28, 29]. Protein contents were normalized to β-tubulin or β-actin.
DRP1 immunoprecipitation
Immunoprecipitation of DRP1 was performed overnight using 2 μg of anti-DRP1 antibody (Becton-Dickinson) on 400 μg of total protein. DRP1 was precipitated with Sepharose beads conjugated to protein G (Santa Cruz Biotech), resolved by SDS-PAGE. Phosphorylation was then assessed with anti-phospho-DRP1 antibody (Cell Signaling) [37].
RNA extraction
Total RNA was isolated using TRIzol (Thermo Fisher Scientific) according to the manufacturer’s instructions, including the addition of 20 μg of glycogen (Roche) prior to isopropyl precipitation.
RT-qPCR
Real-time PCR was performed with SYBR green (Applied Biosystems) as previously described [29]. RCAN1.4 transcripts were normalized to 18S rRNA, whereas data for PKIA transcripts were normalized to mean β-actin and Hprt1 mRNA as an internal control with the 2-ΔΔCt method. The qPCRs for each of the biological quadruplicates were performed in triplicate. Primers used were as follows: RCAN1.4 rat forward 5′-CCCGTGAAAAAGCAGAATGC-3′; RCAN 1.4 rat reverse 5′-TCCTTGTCATATGTTCTGAAGAGGG-3′; PKIA rat forward 5′-CTACCATCAGAGACTGCCCC-3′; PKIA rat reverse 5′-TTGCTGTTGCCACTTGCAGA-3′.
miQPCR
For miQPCR 10 ng of total RNA were reverse transcribed as described [38] and 0.36 ng of cDNA were used for individual assays. cDNAs were amplified by using 2x Brilliant II SYBR (Agilent) and run on Mic qPCR cycler (Bio Molecular Systems) (95 °C for 10 min, followed by 50 cycles of 95 °C for 10 s, 60 °C for 35 s; all reactions were run in triplicates) including melting curve of the amplification products. The threshold cycle was automatically determined by the cycler manager software (Bio Molecular Sciences), averaged across replicates, and Ct values of each condition of three independent transfections are summarized in Supplementary Table 1. Primers used were as follows: rno-miR-129-2-3p specific primer 5′-CCCTTACCCCAAAAAGCATG-3′; RNU6 specific primer 5′-GCAAGGATGACAGCAAATT-3′, and universal reverse primer Ump2A 5′-CCCAGTTATGGCCGTTTA-3′.
Small RNA cloning (TruSeq System)
Total RNA (3 µg) with an RNA integrity number above 9.5 (Fig. S6) was ligated to a 3′ adenylated adapter with a truncated RNA-ligase (New England Biolabs) and to a 5′-adenylated adapter with T4 RNA-ligase (#M0204S, New England Biolabs) as previously described [39] cDNA synthesis was performed using a PCR amplification and SuperScript III first-strand cDNA Super Mix for 50 min at 50 °C and then 5 min at 85 °C. 10 µL of the cDNA obtained in the previous steps was amplified with index sequences and specific-Illumina sequences (desired TruSeq Index-primer, different for each sample), with a PCR program as described: 1 min at 98 °C, 12 cycles (10 s at 98 °C, 10 s at 60 °C, 30 s at 72 °C), 10 min at 72 °C and ∞ at 4 °C. We used 6% PAA SequaGel electrophoresis to select the successfully cloned and amplified sRNAsand ethidium bromide staining to visualize the results (Fig. S7). The appropriate bands (ca. 140 bp) were cut out shredded, and eluted, and then filtered and precipitated. The resulting dscDNA quality and concentration was measured in each sample. Finally, each pellet was dissolved in water. Pool libraries were normalized and stored at −80 °C, and deep sequencing was run on a HiSeq 1000 platform (Illumina).
Small RNA reconstruction, annotation, and differential expression analysis
Raw reads were trimmed with the clipping of the first ten bases in each side using Trimmomatic version 0.36 [40] if average quality fell below the threshold of Q < 30. A five-base sliding-window quality filtering was also performed, eliminating reads where the quality average of a window fell below Q < 30. To reconstruct small RNA transcripts, transcripts were annotated and expression levels estimated using a similar approach to that described by Orell et al. [41] and Matamala et al. [42]. Reads shorter than 15 nt after trimming were discarded, and the resulting clean data were mapped to the rat genome Rnor_6.0 [43] using Bowtie version 1.0.0 [44], allowing one mismatch and suppressing reads with more than 10 alignments. SAM files generated on the mapping process were converted into BED format and used for coordinate-base assembly with BEDTools [45]. Reads belonging to each assembled small RNA transcript were recovered and used to estimate expression levels. Transcriptional fragments with less than 50 reads in total were eliminated from the analysis. The functional annotation of each small RNA class was recovered based on genomic coordinate overlap using microRNAs from miRBase version 21 [46] and the ncRNAs from Ensembl version 92 [47] as a reference, retaining transcripts with an overlap of at least 50%. The final expression matrix generated from the assembly was used for differential expression analysis with DESeq2 version 1.28.0 [48]. Transcripts were considered to show significant differential expression for p value ≤ 0.05.
Conservation analysis between rat, mouse and human miRs
Mature/primary sequences of mouse and human microRNAs were recovered from miRBase and mapped against the rat genome using Bowtie version 1.0.0 [44]. Genomic coordinates of the mapped miRNAs were then cross-referenced with the small RNA transcripts generated in our RNA-seq assay, retaining regions with an overlap of at least 50%.
Quantification of miRNA expression by northern blotting
Northern blotting was performed as previously described [49]. Briefly, 30 µg of total RNA was separated in a 12% SequaGel at 350 V for ~1 h. The gel was then stained with ethidium bromide and transferred at 20 V for 30 min in a semi-dry chamber to an Amersham Hybond‐N membrane (GE Healthcare), where the RNA was crosslinked to the membrane at 50 °C using an EDC-solution [50]. Next, the membrane was incubated overnight at 50 °C with a hybridization solution containing the miR-129-3p antisense sequence (ATGCTTTTTGGGGTAAGGGCTT) labeled with γ‐32P‐ATP. After incubation, the membrane was washed, exposed, scanned, and analyzed with ImageQuant software (Quantity One 4.6.6).
Bioinformatics tools for microRNA target prediction
Two different software tools were used to assess gene target prediction. Using different approaches and algorithms, both tools retrieved predicted regulatory targets for our microRNAs. The first, TargetScan 7.2 uses an improved method to predict targeting efficacy [51, 52]. The second, miRWalk 2.0, reports miRNA-binding sites within the complete sequence of a gene. It also combines this information with a comparison of binding sites resulting from 12 existing miRNA-target prediction programs to build a novel comparative platform of binding sites for the promoter, cds, and 5′- and 3′-UTR regions [53].
Transient miRNA transfection
Cells were transfected either with anti-miR negative control #1 (Ambion, AM17010; 100 nM), anti-miR-129-3p (Ambion, AM10076; 100 nM), or pre-miR-129-3p (Ambion, PM10076; 50 nM), 24 h before the end of the experiment, using OptiMEM and Lipofectamine RNAiMAX, according to the manufacturer’s specifications. The final effects of anti-miR negative control (anti-miR-Control), anti-miR-129-3p and pre-miR-129-3p on miR-129-3p levels in cardiomyocytes are summarized in Supplementary Table 1.
Statistical analysis
Sample size was determined using mitochondrial dynamic parameters. Assuming a onefold change in mitochondrial volume or mitochondrial number, with an alpha error of 0.05%, two-tailed, the required sample size is three experiments per group. Data shown are mean ± SEM of the number of independent experiments indicated (n) and represent experiments performed on at least three separate occasions with similar outcomes. Data were analyzed by one-way or two-way ANOVA accordingly, and comparisons between groups were performed using a protected Tukey’s test using GraphPad Prism 6 software. Statistical significance was defined as p < 0.05.
Results
Angiotensin-(1–9) induces mitochondrial fusion through Drp1 phosphorylation
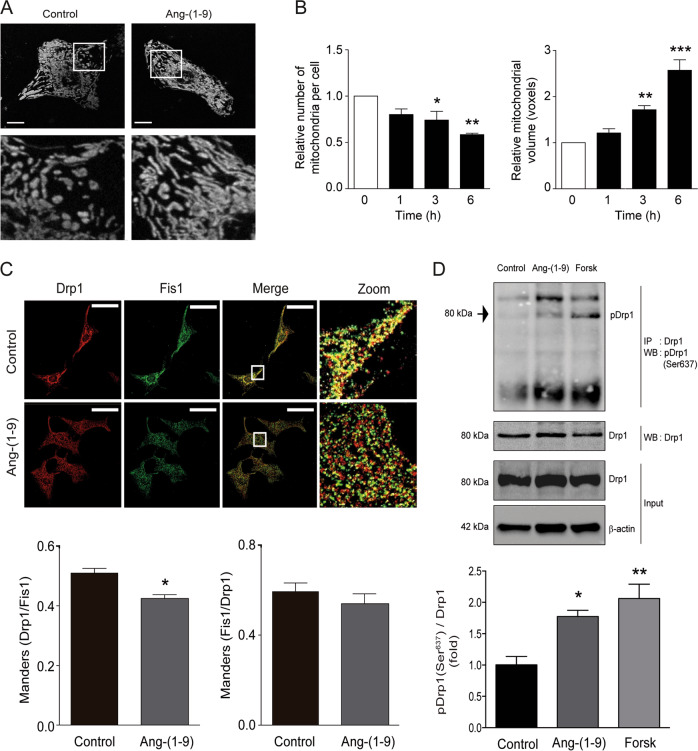
To evaluate whether angiotensin-(1–9) affects mitochondrial morphology, cultured neonatal rat cardiomyocytes were treated with angiotensin-(1–9) (100 μM, 0–6 h) and stained with the mitochondrial-specific probe MitoTracker Green (400 nM, 30 min). Treatment with angiotensin-(1–9) stimulated the appearance of large interconnected mitochondria in cardiomyocytes, mainly in the perinuclear region (Figs. 1a and S1A, C). Mitochondrial network integrity was then assessed according to number and volume of individual mitochondria through 3D reconstitution of confocal stacks [13, 29]. Angiotensin-(1–9) led to a gradual time-dependent decrease in number of mitochondria per cell, to 75% (±9%) and 59% (±3%) of control levels after 3 and 6 h, respectively (Fig. 1b, left panel). Angiotensin-(1–9) concomitantly increased mitochondrial volume by 72% and 158% after 3 and 6 h, respectively (Fig. 1b, right panel). Because 6 h of angiotensin-(1–9) treatment achieved greater statistical significance and more robust results in terms of morphological effects than 3 h of treatment, this time was used for all subsequent experiments. To investigate whether the observed changes in the morphological structure of the mitochondrial network involved alterations in mitochondrial biogenesis or mitochondrial degradation, mitochondrial mass was evaluated by quantifying the abundance of the constitutive mitochondrial protein mtHsp70. As shown in S3A, B, angiotensin-(1–9) did not alter total mtHsp70 levels, suggesting that a change in mitochondrial turnover did not accompany the morphological changes.
Fig. 1. Angiotensin-(1–9) promotes mitochondrial fusion through Drp1 phosphorylation.
a Mitochondrial morphology of cardiomyocytes stimulated with 100 μM angiotensin-(1–9) for 0–6 h and then loaded with MitoTracker Green (MTG). Z-stack images were obtained with confocal microscopy. Scale bar: 2 μm. Lower panels represent a ×15 magnification. b Quantification of the relative number of mitochondria per cell (left) and the relative mean mitochondrial volume (right) as mitochondrial dynamic parameters in cells treated with 100 μM angiotensin-(1–9) for the indicated times (n = 4), *p < 0.05; **p < 0.01 and ***p < 0.001 vs. 0 h. c Sub-cellular localization of the mitochondrial fission protein Drp1 visualized using immunofluorescence. Cardiomyocytes were stimulated with angiotensin-(1–9) for 6 h, incubated with anti-Drp1 and anti-Fis1 antibodies, and then visualized with confocal microscopy. Scale bar: 10 μm. Right panels represent a ×45 magnification. The Manders’ coefficients of the Drp1 and Fis1 signals were used as indicators of Drp1 localization in the mitochondria (n = 3) *p < 0.05 vs. control. d Drp1 phosphorylation (pDrp1; Ser637) was assessed by western blotting after immunoprecipitation of Drp1 protein. Cells were treated with angiotensin-(1–9) for 6 h or a pulse of 100 nM forskolin for 30 min (positive control for pDrp1). A representative images is shown for three independent experiments with similar outcomes.
Recent work from our group and others has shown that the anti-hypertrophic effects of angiotensin-(1–9) are mediated by AT2R activation [6, 54]. Consistent with this evidence, the effects of angiotensin-(1–9) on mitochondrial morphology were completely prevented by the AT2R antagonist PD123,319 (Fig. S1A, B). Angiotensin-(1–7), which can be produced from angiotensin II by ACE2, has also been shown to have cardioprotective effects, acting through the MAS-related G protein-coupled receptors [55]. To rule out the possibility that the changes in mitochondrial morphology were caused by cleavage of angiotensin-(1–9) to angiotensin-(1–7) form, we used the pharmacological MAS receptor antagonist A779. Fig. S1C, D shows that A779 did not prevent the effects of angiotensin-(1–9) on cardiomyocyte mitochondrial morphology. Taken together, these data suggest that angiotensin-(1–9) activates AT2R to either induce mitochondrial fusion or decrease mitochondrial fission in cardiomyocytes
To study the molecular mechanisms by which angiotensin-(1–9) regulates mitochondrial dynamics, levels of mitochondrial fusion (Opa1 and Mfn2) and fission (Drp1 and Fis1) proteins were assessed by western blotting. Neither Opa1/Mfn2 nor Drp1/Fis1 protein levels were altered by angiotensin-(1–9) treatment in cardiomyocytes (Fig S2A–F). Because previous studies have established that DRP1 migration from the cytosol to FIS1-containing fission points on the mitochondrial surface is an initial step in mitochondrial fragmentation [13, 56], we then evaluated whether angiotensin-(1–9)-triggered mitochondrial fusion was associated with changes in Drp1 distribution, especially movement away from the mitochondrial surface. Immunofluorescence studies showed that the punctate distribution pattern of Drp1 was disrupted by 6 h of angiotensin-(1–9) treatment (Fig. 1c). Moreover, angiotensin-(1–9) decreased the effective colocalization of Drp1 with Fis1 (p < 0.05), but not the effective colocalization of Fis1 with Drp1 (Fig. 1c, lower panel), as indicated by the Manders’ coefficient. This result suggests that Drp1 shows a reduced mitochondrial distribution after angiotensin-(1–9) stimulation, whereas Fis1 remains associated with the mitochondria under both conditions. Pursuing this line of research, we assessed Drp1 phosphorylation status at Ser637, a site that can inactivate the protein [37, 57]. Total Drp1 was immunoprecipitated and then probed with a Ser637-specific antibody. As shown in Fig. 1d, angiotensin-(1–9) increased Drp1 phosphorylation to levels only slightly lower than those obtained using forskolin, a known inhibitor of the Drp1-fission pathway [58, 59]. Furthermore, angiotensin-(1–9) also decreased Drp1 phosphorylation on Ser616 (Fig. S2G, H), a fission-promoting modification [60]. In conclusion, these results strongly suggest that angiotensin-(1–9) decreases mitochondrial fission, rather than increasing mitochondrial fusion, to bring about the observed changes in cardiomyocyte mitochondrial morphology.
Angiotensin-(1–9) prevents norepinephrine-elicited mitochondrial fission
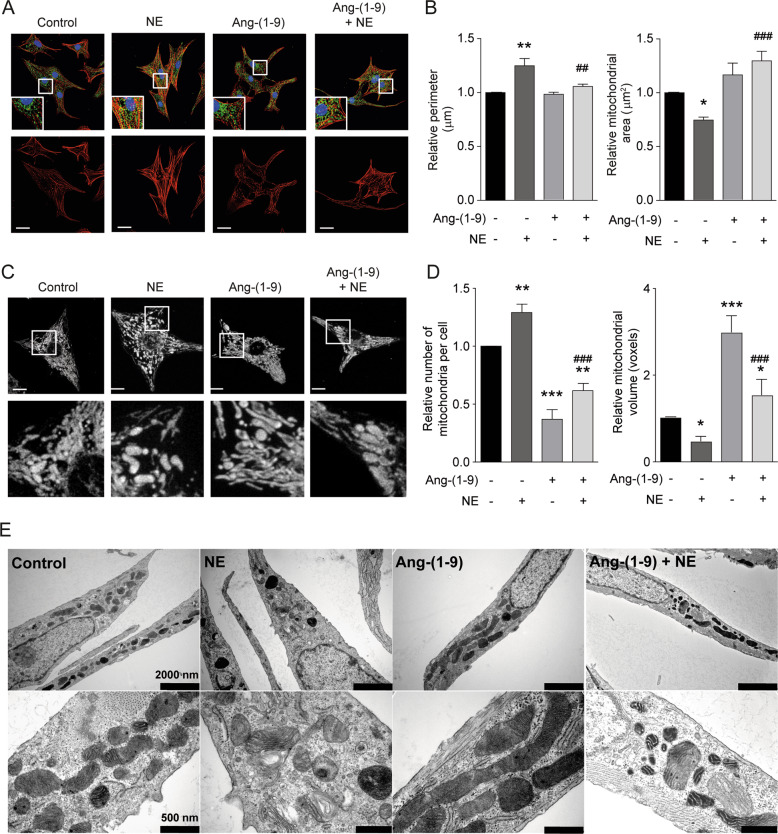
We recently reported that NE triggers mitochondrial fission in a calcineurin- and DRP1-dependent manner [13]. Furthermore, adenovirus-mediated expression of the dominant-negative form of DRP1 prevented mitochondrial fission and hypertrophic cardiomyocyte growth in response to NE. Given the result shown in Fig. 1, we tested whether the use of angiotensin-(1–9) was sufficient to prevent NE-elicited mitochondrial fission. First, we verified that angiotensin-(1–9) prevented NE-induced hypertrophic growth in cardiomyocytes. As seen in Fig. 2a, double-staining revealed that NE-treated cardiomyocytes exhibited a hypertrophic phenotype, as indicated by increased sarcomeric organization (Fig. 2a), larger cell perimeter (Fig. 2b, left panel), and decreased relative mitochondrial area as compared with controls (Fig. 2b, right panel). As expected, angiotensin-(1–9) prevented these NE-dependent changes in sarcomeric structure and abundance (Fig. 2a) as well as relative cell size (Fig. 2b, left panel). Angiotensin-(1–9) also prevented the NE-dependent decrease in the mean size of individual mitochondria (Fig. 2b right panel).
Fig. 2. Angiotensin-(1–9) prevents mitochondrial fission elicited by norepinephrine.
a Representative confocal images of cardiomyocytes treated with 100 μM angiotensin-(1–9) for 6 h and then stimulated with 10 μM of norepinephrine (NE) for 24 h. Cardiomyocytes were stained with rhodamine phalloidin to detect sarcomeric structures and immunolabeled for the mtHsp70 protein to identify the mitochondrial network. Scale bar: 25 μm. b Quantitative analysis (n = 4) of relative cellular perimeter and mitochondrial area of cardiomyocytes. c Mitochondrial morphology of cardiomyocytes pretreated as in a. Scale bar: 2 μm. Bottom panels represent a ×13.5 magnification. d Relative number of mitochondria per cell (left) and mean mitochondrial volume (right) were quantified as mitochondrial dynamics parameters (n = 4). *p < 0.05; **p < 0.01 and ***p < 0.001 vs. control; ##p < 0.01 and ###p < 0.001 vs. NE. e Representative transmission electron microscopy images from angiotensin-(1–9)- and NE-treated cells showing two different magnifications of the same cells.
We next assayed mitochondrial morphology in live cardiomyocytes using the mitochondrial-specific probe MitoTracker Green (400 nM, 30 min) and 3D imaging reconstruction. Angiotensin-(1–9) inhibited mitochondrial fission in NE-treated cardiomyocytes (Fig. 2c), as indicated by both relative number of mitochondria per cell and relative mitochondrial volume (Fig. 2d). To corroborate these results, we performed electronic microscopy studies on untreated cardiomyocytes and cardiomyocytes treated with angiotensin-(1–9) and/or NE (Fig. 2e). The results for mitochondrial area, perimeter, circularity, and density corroborate our findings described above (Fig. S3C–F).
Angiotensin-(1–9) prevents Ca2+ dysregulation induced by norepinephrine treatment
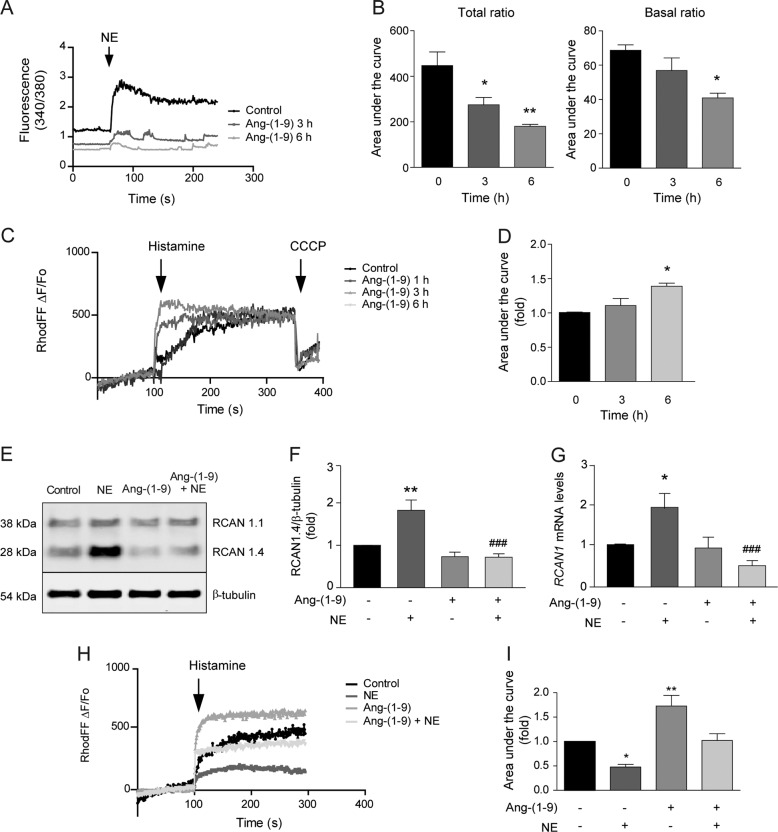
It has been shown that a more connected mitochondrial network, with a higher membrane potential, endows the mitochondria with greater Ca2+ buffering efficiency [61]. Moreover, inhibition of fusion or induction of fission weakens mitochondrial metabolic reserve, producing deregulated Ca2+ oscillations in skeletal muscle cells [62]. Therefore, we asked whether angiotensin-(1–9) might exert its action by modulating mitochondrial or cytosolic Ca2+ levels. Using a cytoplasmic Ca2+ dye, Fura-2, we monitored NE-triggered cytosolic Ca2+ levels [13] and found that this signal was reduced in cells pretreated with angiotensin-(1–9) (p < 0.05 and <0.01 at 3 and 6 h, respectively) (Fig. 3a, b). To determine whether angiotensin-(1–9) increases mitochondrial Ca2+ uptake capacity, Rhod-FF-preincubated cardiomyocytes were treated with histamine, which evokes a PtdIns-(3,4,5)-P3-dependent release of Ca2+ (Fig. S4A–D) and changes in mitochondrial Ca2+ uptake were assessed. Histamine-stimulated mitochondrial Ca2+ uptake was significantly higher in cardiomyocytes pretreated with angiotensin-(1–9) (p < 0.01; 3 and 6 h) than in vehicle-treated control cells (Fig. 3c, d). Similar to its impact on mitochondrial morphology, PD123,319 (AT2R antagonist) abolished this effect, whereas A779 (MAS antagonist) did not (Fig. S4E–G).
Fig. 3. Angiotensin-(1–9) regulates intracellular calcium preventing NFAT pathway activation.
a Cytosolic calcium levels in cardiomyocytes treated with angiotensin-(1–9) and then challenged with NE. Cells were loaded with Fura-2AM and then visualized in a confocal microscope. Fura-2AM fluorescence ratio (340 nm/380 nm) was quantified. An NE pulse (10 µM) was added to elicit an increase in cytosolic Ca2+. b Total (left) and basal (right) area under the curve were quantified (n = 3). *p < 0.05 and **p < 0.01 vs. control. c Mitochondrial Ca2+ levels in cardiomyocytes treated with 100 μM angiotensin-(1–9) for 6 h and then loaded with Rhod-FF. Rhod-FF fluorescence kinetics were captured using confocal microscopy. A histamine pulse (100 μM) was added to elicit Ca2+ release from the endoplasmic reticulum, while CCCP (50 μM) was used to inhibit mitochondrial Ca2 uptake. Neither angiotensin-(1–9) nor NE were able to directly increase mitochondrial Ca2+ levels as histamine did (Fig. S4A–D). d The area under the curve of the Rhod-FF signal was determined from the curves obtained in a (n = 4). e Western blotting for RCAN1 isoforms in cardiomyocytes treated with angiotensin-(1–9) and NE. Cells were stimulated and then subjected to western blotting using RCAN1 antibody. β-tubulin was used as a loading control. f Quantification of RCAN1.4 protein and g mRNA levels (n = 4). *p < 0.05 and **p < 0.01 vs. control; ###p < 0.001 vs. NE. h Mitochondrial Ca2+ levels were measured in Rhod-FF-loaded cardiomyocytes. Cells were pre-incubated with 100 μM angiotensin-(1–9) for 6 h and then stimulated for 24 h with NE. Rhod-FF fluorescence kinetics were visualized with confocal microscopy. A histamine pulse (100 mM) was added to trigger Ca2+ release from the ER. i The area under the curve of Rhod-FF signals are shown (n = 3). *p < 0.05 and **p < 0.01 vs. control.
NE activates the Ca2+/calcineurin-dependent pathway in cardiomyocytes [13, 63] with its molecular target, the nuclear factor of activated T cells (NFAT), mediating the hypertrophic transcriptional response. Genes responsive to calcineurin/NFAT include the exon 4 isoform of the Rcan1 gene, which encodes an endogenous feedback inhibitor of calcineurin activity [13, 37]. An increase in Rcan1.4 expression or RCAN1.4 protein levels is often used as a molecular marker of calcineurin pathway activation. Interestingly, angiotensin-(1–9) blocked NE-induced activation of calcineurin signaling, as demonstrated by its effects on RCAN1.4 protein (Fig. 3e, f) and transcript levels (Fig. 3g). These findings are consistent with a model in which angiotensin-(1–9) stimulation prevents downstream activation of calcineurin by NE, thereby preventing DRP1 dephosphorylation at Ser637 and subsequent mitochondrial fission and as readout of intracellular calcium regulation
Our group previously reported that NE-stimulated cardiomyocytes lose a significant number of ER-mitochondria contact sites, resulting in a functional decline in mitochondrial Ca2+ uptake capacity [15]. Sites of physical contact between the ER and mitochondria are crucial for Ca2+ transfer between these organelles [64]. To evaluate whether ER-mitochondria distance was altered in the angiotensin-(1–9)- and/or NE-treated cardiomyocytes, the colocalization of ER and mitochondria was measured using immunofluorescence and indirect labeling of specific proteins [15, 35, 65]. Fig. S5A shows representative images of control and angiotensin-(1–9)- and/or NE-treated cardiomyocytes labeled with Fis1 (red) and KDEL (green), to detect mitochondria and ER, respectively. The colocalization of the two organelles was assessed in the whole cell by calculating the Manders’ coefficient [35, 36]. The Manders’ coefficient for Fis1/KDEL denotes the fraction of mitochondria that colocalize with ER, whereas the Manders’ coefficient for KDEL/Fis1 denotes the fraction of ER that colocalize with mitochondria. Fis1/KDEL was significantly reduced in NE-treated cardiomyocytes as compared with controls, indicating movement of mitochondria away from sites of contact with the ER. This change was absent in the cells pretreated with angiotensin-(1–9) (Fig. S5B, C). Mitochondrial Ca+2 uptake is a functional readout of communication between the ER and mitochondria. Our data showed that mitochondrial Ca2+ uptake was reduced and less efficient in NE-treated cells and that angiotensin-(1–9) prevented these effects (Fig. 3h, i). Together with the colocalization results, these data suggest that angiotensin-(1–9) can prevent NE-dependent loss of physical and functional communication between the ER and mitochondria; furthermore, these changes were temporally correlated with changes in mitochondrial morphology.
Angiotensin-(1–9) activates the PKA signaling pathway in cardiomyocytes by upregulating miR-129-3p and inhibiting PKIA
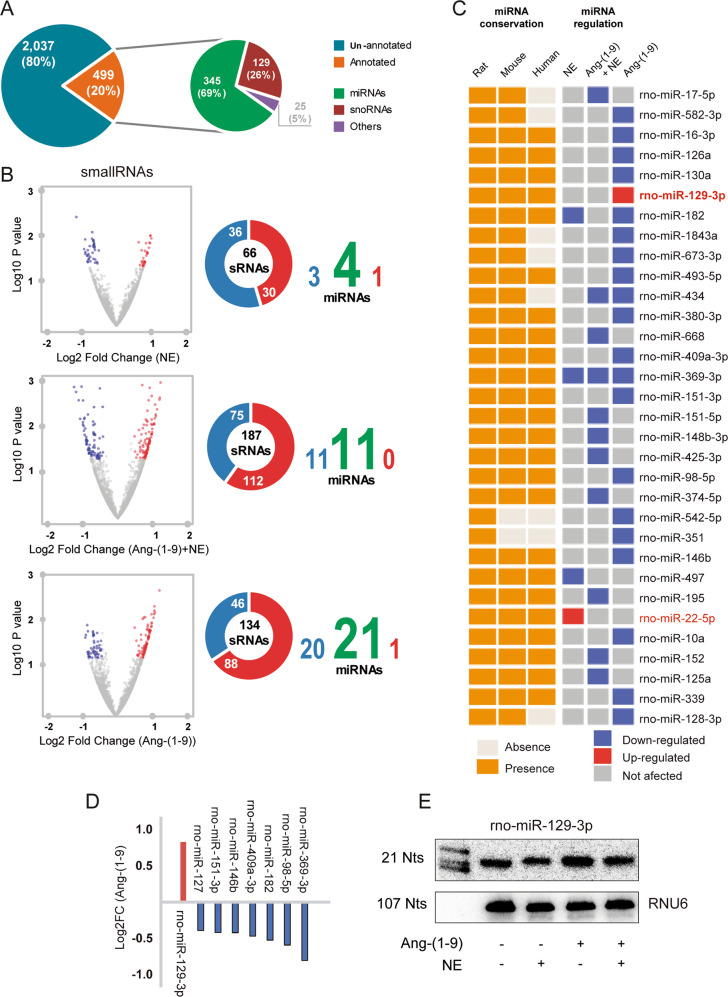
To study the effect of angiotensin-(1–9) on miRNA expression and signaling, cultured neonatal rat cardiomyocytes were treated with angiotensin-(1–9) and/or NE. Afterward, RNA was isolated, purified, and subjected to small RNA expression profile analysis using an RNA-seq approach. The RNA-seq data for the three conditions (NE, angiotensin-(1–9), and angiotensin-(1–9)/NE) identified a total of 2536 transcripts (Fig. 4a), each of which was later compared with the small RNA classes reported in GFF format in the NCBI database as well as miRNAs recovered from miRBase for the Rnor_6.0 genome assembly [46]. The analysis showed that 20% (499) of those transcripts shared annotation with known sRNA classes. We identified 345 transcripts (69% of annotated features) as miRNAs (Fig. 4a), which were later used to perform a differential expression analysis using DESeq [48]. We found 4, 11, and 21 miRNAs that were differentially expressed after NE, combined NE/angiotensin-(1–9) and angiotensin-(1–9) alone treatment, respectively (Fig. 4b). Some miRNAs were differentially expressed in more than one condition; a total of 32 miRNAs showed distinct expression patterns compared with the control condition. Only two of these 32 miRNAs were differentially upregulated, one after NE and the other after angiotensin-(1–9) treatment (Fig. 4b). Interestingly, a conservation analysis indicated that 24 of these differentially expressed neonatal rat cardiomyocyte miRNAs were present in both human and mouse, 6 were present in mouse but not human, and 2 were exclusive to the rat genome (Fig. 4c). We selected the 24 conserved human/mouse miRNAs to generate a new filter, using the base mean values provided by DESeq2. Using a p value ≤ 0.05 as the cutoff, the only upregulated miRNA that was fully conserved among these mammals was miR-129-3p. Given the novelty of this finding in the context of cardiac hypertrophy, we evaluated this miRNA as a candidate for regulating the effects of angiotensin-(1–9) in cardiomyocytes (Fig. 4d). Northern blotting was then performed to confirm the change in miR-129–3p expression following angiotensin-(1–9) treatment identified by our miRNA expression profiling (Fig. 4e). Further, the pretreatment with the AT2R antagonist, PD 123,319 1 μM, prevented the increase of the miR-129-3p triggered by angiotensin-(1–9) (Fig. S8).
Fig. 4. Angiotensin-(1–9) increases miR-129–39 levels in cardiomyocytes.
a miRNA identification from the RNA-seq data of cardiomyocytes treated with norepinephrine (NE), angiotensin-(1–9)/NE and angiotensin-(1–9) versus control. Each transcript was compared with the small RNA classes reported in the GFF format in the NCBI database and the miRNAs recovered from miRBase for the Rnor_6.0 genome assembly. b Differential expression analysis using DESeq2. NE treatment triggered the differential expression of 4 miRNAs, whereas angiotensin-(1–9)/NE and angiotensin-(1–9) increased expression of 11 and 21 miRNAs, respectively. c Conservation analysis against human, mouse, and rat of the differentially expressed neonatal rat cardiomyocytes-miRNAs from the conditions in b. 24 of these miRNAs were present in human and mouse, 6 in mouse, and 2 of them presented no conservation. d Relative expression using the 24 differentially expressed miRNAs that were conserved in mouse, rat, and human, a new filter was generated using their base mean values provided by DESeq2. Finally, the highest LFC led us to focus on the only differentially upregulated miRNA, miR-129-3p in the angiotensin-(1–9) treatment. e To validate the bioinformatic analysis, we compared miR-129-3p expression levels in control MEFs cells and cells treated with NE and/or angiotensin-(1–9) by northern blot.
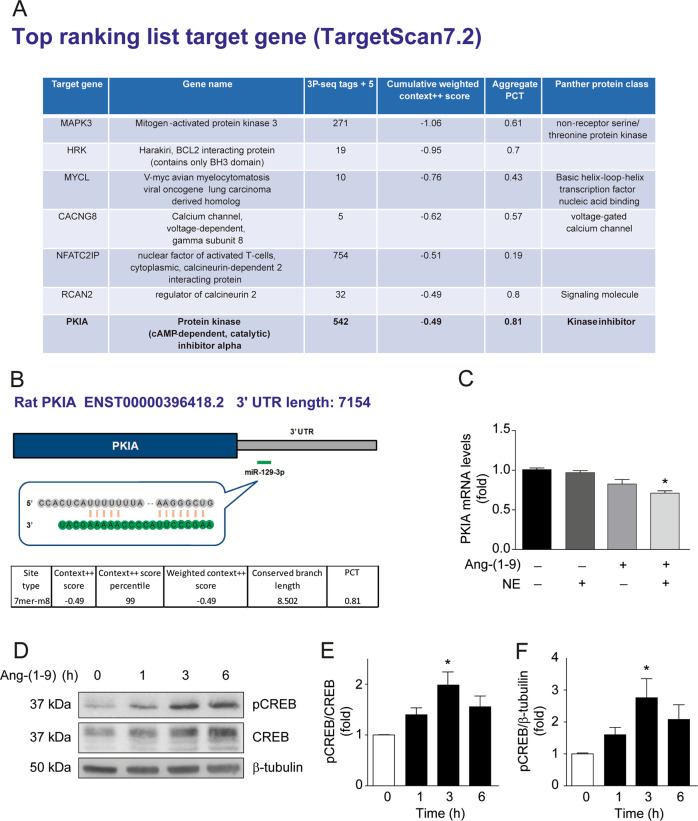
In silico analysis of specific target genes for miR-129-3p, using two independent prediction tools, TargetScan [51] and miRWalk [66], identified the cAMP-dependent protein kinase inhibitor alpha (PKIA) as an important target of miR-129-3p (Fig 5a, b). This protein has been shown to interact with PKA and to inhibit the activities of both C alpha and C beta catalytic subunits [67], which notably had already been implicated in the regulation of mitochondrial dynamics through DRP1 phosphorylation [57, 65]. The target site prediction revealed a 7mer-m8 seed region for miR-129-3p in position 588-594 of the 3′ untranslated region (3′ UTR) of the PKIA mRNA (Fig. 5b). This prediction was functionally validated by qRT-PCR of the PKIA mRNA in cardiomyocytes treated with angiotensin-(1–9) and/or NE, which showed a decrease in PKIA mRNA after angiotensin-(1–9) plus NE administration (Fig. 5c). Consistently, angiotensin-(1–9) treatment of cardiomyocytes resulted in CREB phosphorylation, as a readout of PKA activation in these cells (Fig. 5d–f).
Fig. 5. Angiotensin-(1–9) activates PKA signaling via miR-129-3p-dependent inhibition of PKIA.
a Top ranking list of target genes obtained from TargetScan 7.2, filtered using http://pantherdb.org (reference proteome dataset), by biological process and biological regulation. b Schematic representation of the miR-129-3p and its seed region targeting the 3’UTR PKIA gene, displaying different prediction parameters in terms of conservation, mismatching, and predicted efficacy based on TargetScan 7.2. c Quantification of PKIA mRNA levels using RT-qPCR (n = 5), *p < 0.05. d pCREB, CREB, and β-tubulin levels were assessed by western blotting of cells treated with 100 μM angiotensin-(1–9) for the indicated times (n = 3). e, f Densitometric quantification of the western blots depicted in d. *p < 0.05 vs. 0 h.
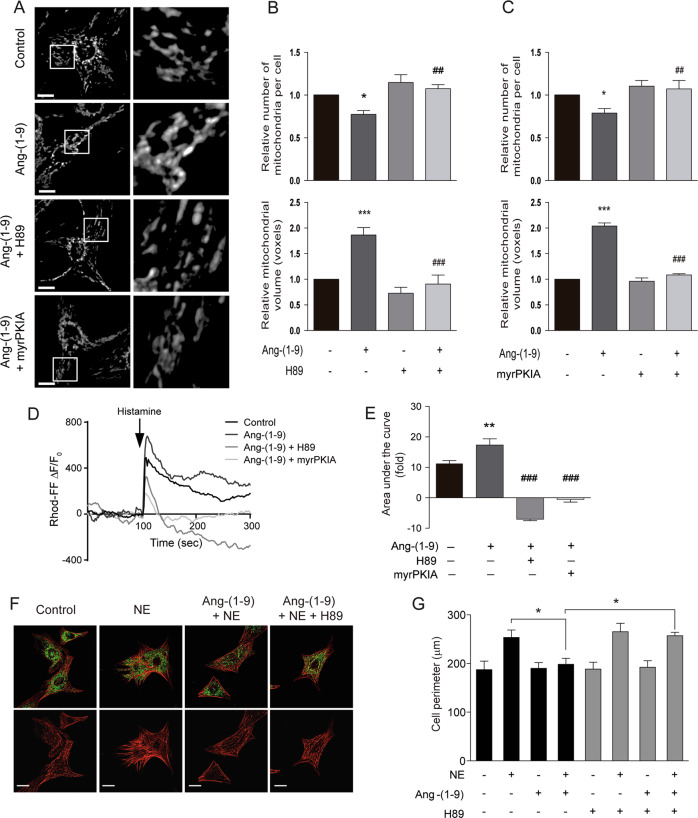
To study the participation of the miR-129-3p/PKIA/PKA signaling pathway on angiotensin-(1–9) action, we used several approaches. First, the pre-incubation of cardiomyocytes with H89, a chemical inhibitor of PKA, or a cell-permeable myristoylated form of PKIA prevented all the effects of angiotensin-(1–9) on mitochondrial fusion (Fig. 6a–c) and mitochondrial calcium uptake (Fig. 6d, e), suggesting that angiotensin-(1–9) triggers PKA signaling activation through miR-129-3p-dependent inhibition of PKIA. Moreover, pre-incubation of cardiomyocytes with H89 also abrogated the anti-hypertrophic effect of angiotensin-(1–9) in NE-treated cells (Fig. 6f, g). Finally, the transient transfection of cardiomyocytes with antimiR-129-3p also prevented the increase in mitochondrial fusion (Fig. 7a, b) and mitochondrial calcium uptake (Fig. 7c, d) observed with angiotensin-(1–9). Moreover, the solely transfection of cardiomyocytes with an artificial precursor of miR-129-3p mimics the effects of angiotensin-(1–9) on mitochondrial morphology and calcium uptake (Fig. 7a–d), reinforcing the idea that angiotensin-(1–9) could exert these effects through this signaling pathway.
Fig. 6. Inhibition of PKA signaling blunts the effects of angiotensin-(1–9) on mitochondrial dynamics and mitochondrial Ca2+ uptake in cardiomyocytes.
a Confocal MitoTracker Green microscopy images of cardiomyocytes treated with angiotensin-(1–9) (100 μM, 6 h) and the PKA inhibitors H89 (10 μM) or myrPKIA (10 μM). Scale bar: 2 μm. Right panels represent a ×15 magnification. Mitochondrial number and mean mitochondrial volume for each condition were quantified (n = 7 for b and n = 5 for c). d Mitochondrial Ca2+ levels in cells stimulated with angiotensin-(1–9) after pre-treatment with the PKA inhibitors H89 (10 μM) or myrPKIA (10 μM). e Quantification of the area under the curve for Rhod-FF fluorescence (n = 6). f Representative confocal images of cardiomyocytes treated with angiotensin-(1–9) 100 μM for 6 h and/or H89 10 μM and then stimulated with norepinephrine (NE) 10 μM for 24 h. The cells were stained with phalloidine rhodamine to detect sarcomeric structures, and immunolabeled for the mtHsp70 protein to identify mitochondrial network. Scale bar: 25 μm. g Quantitative analysis (n = 3) of cellular perimeter of cells in f. *p < 0.05; **p < 0.01 and ***p < 0.001 vs. control; #p < 0.05; ##p < 0.01 and ###p < 0.001 vs. angiotensin-(1–9).
Fig. 7. Antagonizing the miR-129-39 signaling avoids angiotensin-(1–9) effects on mitochondrial dynamics and Ca2+ uptake in cardiomyocytes.
a Confocal MitoTracker Green microscopy images of cardiomyocytes treated with anti-miR-Control (100 nM), anti-miR-129-3p (100 nM) or pre-miR-129-3p (50 nM) for 24 h and then stimulated with angiotensin-(1–9) (100 μM, 6 h). Scale bar: 2 μm. Bottom panels represent a ×15 magnification. b Mitochondrial number and mean mitochondrial volume for each condition were quantified (n = 4). c Mitochondrial Ca2+ levels in cells stimulated as in a. d Quantification of the area under de curve for Rhodd-FF fluorescence (n = 3) *p < 0.05, **p < 0.01 and ##p < 0.05 vs Anti-miR-Control/Ang-(1–9). e Working model: Via miR-129-3p and PKA activation, angiotensin-(1–9) induces a more fused mitochondrial network through DRP1 phosphorylation and prevents NE-induced fission. These changes are associated with diminished cytoplasmic Ca2+ levels, increased ER-mitochondrial Ca2+ transport efficiency, and prevention of ER-mitochondria coupling loss.
Discussion
Angiotensin-(1–9) is a noncanonical RAS peptide with anti-hypertrophic effects both in vitro and in vivo. These effects are mediated through AT2R activation; [5, 6, 54] however, the downstream signaling pathways and intracellular effects of this peptide remain unknown. Alterations in mitochondrial dynamics and Ca2+ handling have been associated with the onset and progression of pathological cardiac hypertrophy [68]. The data presented here show for the first time that angiotensin-(1–9) regulates both mitochondrial dynamics and Ca2+ handling in cultured rat cardiomyocytes, via upregulation of miR-129-3p and inhibition of PKIA. Angiotensin-(1–9) also prevents the mitochondrial fission triggered by NE. These changes are associated with diminished cytoplasmic Ca2+ levels, increased ER-mitochondrial Ca2+ transport efficiency, and preservation of ER-mitochondria communication (Fig. 7e).
Several studies have shown that mitochondrial dynamics proteins play a key role in maintaining normal cardiomyocyte function. Deletion of genes encoding pro-fusion proteins, such as Mfn1/2 [69] or Opa1 [70], or genes encoding pro-fission proteins, such as Drp1 [71], disrupts myocardial function, underlining the importance of mitochondrial dynamics in cardiac health. Manipulation of mitochondrial dynamics, mainly through promotion of a fused mitochondrial network, has been proposed as a novel and promising cardioprotective strategy [68]. Angiotensin-(1–9) promoted a fused mitochondrial network in cardiomyocytes without altering the levels of proteins involved in mitochondrial dynamics, suggesting a post-translational regulation of their activities. The pro-fission activity of Drp1 has been associated with the development of cardiac diseases in mouse models. In a mouse model of ischemia/reperfusion injury, for instance, Mdivi-1, a pharmacological DRP1 inhibitor, prevented mitochondrial fragmentation and cell death [72]. In an in vivo study, Mdivi-1 prevented left ventricular dysfunction in response to pressure overload [73]. Our previous work also showed that NE induces cardiomyocyte hypertrophy and mitochondrial fission by decreasing Drp1 phosphorylation at Ser637 [13], a substrate of calcineurin [74]. Adenoviral-mediated expression of a human dominant-negative form of DRP1 prevented mitochondrial fragmentation and hypertrophy, supporting the idea that DRP1 plays a central role in pathological remodeling of the myocardium. Here we show that angiotensin-(1–9) increases inhibitory phosphorylation of Drp1 (Ser637) and decreases its activating phosphorylation (Ser616), together with its migration to the mitochondria. Moreover, angiotensin-(1–9) prevented NE-triggered mitochondrial fission, a key step underlying its pro-hypertrophic effects.
NE increases cytoplasmic Ca2+ levels, thereby promoting calcineurin-mediated dephosphorylation of DRP1 to increase mitochondrial fission [13]. Here, we show that angiotensin-(1–9) prevents NE-triggered calcineurin pathway activation, as indicated by changes in Rcan1.4. mRNA and protein levels. In the absence of calcineurin activation, DRP1 would remain phosphorylated at Ser637 and the mitochondrial network intact. Angiotensin-(1–9) decreased both baseline and NE-stimulated cytoplasmic Ca2+ levels, and we propose that the impact of angiotensin-(1–9) on Ca2+ signaling is fundamental to its anti-hypertrophic effects. Mitochondria function as buffers for Ca2+, particularly Ca2+ released from the ER. The efficiency of Ca2+ transfer from the ER to mitochondria is inversely dependent on the distance between these organelles [75]. NE causes a loss of ER-mitochondria contact sites, decreasing the efficiency of Ca2+ transport between these organelles [15]. Angiotensin-(1–9) treatment maintained Ca2+ transfer efficiency and prevented the loss of ER-mitochondria contact sites (Figs. 3, S5). Maintenance of ER-mitochondrial communication could indeed underlie the observed blunting of the cytoplasmic Ca2+ response induced by NE. However, angiotensin-(1–9) treatment did not increase the extent of ER-mitochondria contact at baseline. Therefore, physical changes in ER/mitochondrial coupling cannot explain the increase in Ca2+ transfer efficiency observed in the angiotensin-(1–9)-treated cells at baseline (Fig. 3c–e). Another possible explanation for this change is the increased mitochondrial fusion observed in the angiotensin-(1–9)-treated cardiomyocytes (Fig. 1). Mitochondria that are elongated or have a higher membrane potential show more efficient Ca2+ uptake [61].
The work of Fattah et al. suggests another potential mechanism for the observed increase in ER-mitochondrial Ca2+ transfer. These authors reported positive inotropic effects of angiotensin-(1–9) in isolated cardiomyocytes attributable to increased ER Ca2+ stores, likely through a protein kinase A (PKA)-dependent mechanism [7]. Certainly, their findings agree with our results, and increased ER Ca2+ stores may well contribute to the enhanced interorganellar Ca2+ transfer documented here. However, increased ER stores alone would not be sufficient to explain the suppression of cytoplasmic Ca2+ in response to NE. We propose that remodeling of the mitochondrial network toward a more fused state represents a key aspect of the ability of angiotensin-(1–9) to buffer ER Ca2+ release and underlies its anti-hypertrophic effects. It is relevant to note that DRP1 Ser637 can be phosphorylated by PKA, thereby acting in opposition to calcineurin to promote fusion. Thus, DRP1 phosphorylation may be an important target of action for PKA in response to angiotensin-(1–9), in addition to the increased ER Ca2+ stores documented by Fattah et al. Notably, our miRNAs expression pattern studies indicated that miR-129–3p was the only miRNA upregulated after angiotensin-(1–9) treatment. This miRNA, which has been implicated in angiotensin II signaling in HEK293 cells [76], would in this case act downstream of the AT2R involved in the inhibition of PKIA translation. Decreased PKIA signaling would directly impact PKA activation, potentially inducing DRP1 phosphorylation and increasing ER Ca2+ stores. Further studies are required to determine the relative contributions and possible interdependence of these two potential mechanisms of action.
The anti-hypertrophic effects of angiotensin-(1–9) are mediated by AT2R activation [5, 6, 54]. Signaling through the G-protein-coupled receptor is not well understood, although it is generally thought of as Giα coupled. Therefore, the mechanism through which it might couple to PKA activity is not immediately obvious. Thus, understanding the signaling mechanisms of angiotensin-(1–9) immediately downstream of AT2R is an important contribution of this work. To our knowledge, this is the first report that associates miR-129–3p with AT2R activation in cardiomyocytes, despite the fact that this miRNA was previously described to be elevated in HEK293 cells after angiotensin II treatment [76]. Notably, in this same work, treating cardiomyocytes with a biased angiotensin II analog significantly downregulated miR-129-3p [76], which is consistent with the known pro-hypertrophic effects exerted by angiotensin II in cardiomyocytes and correlates with our own results with angiotensin-(1–9) and its anti-hypertrophic properties.
Taken together, our studies show that angiotensin-(1–9) acts through AT2R to regulate mitochondrial morphology and Ca2+ uptake capacity via the AT2R/miR-129-3p/PKA signaling pathway. These effects are mediated by suppression of DRP1-dependent mitochondrial fission, increased mitochondrial Ca2+ buffering capacity, and preservation of ER-mitochondria contact sites following NE exposure. To our knowledge, this is the first study that links the angiotensin-(1–9)/AT2R/miR-129-3p/PKA signaling axis with control over mitochondrial dynamics. This also opens the question of whether other members of the canonical and noncanonical RAS family could act through similar mechanisms. Given the newly-identified roles of the noncanonical RAS peptides angiotensin-(1–7) and angiotensin-(1–9) as counter-regulators of the ACE/angiotensin II axis and their recently described ability to regulate blood pressure as well as cardiovascular and renal remodeling, our findings reveal an important new aspect of their modes of action, thereby providing novel approaches for therapeutic manipulation of these peptides and their biological actions in the cardiovascular system.
Supplementary information
Acknowledgements
The authors thank Fidel Albornoz, Gindra Latorre and Sebastian Leiva for their excellent technical assistance.
Funding
This work was supported by grants from the Agencia Nacional de Investigacion y Desarrollo (ANID), Chile: (FONDECYT 1120212 to SL, 1190743 to VP, 3130749 to CP, 11181330 to IP, 1140329 to MC, 11161020 to VM-C and; FONDAP 15130011 to SL, AHC, MC, VP, VM-C, LG, MPO; Anillo ACT192144 to MPO; PAI Insertion Program 79150007 to VP and 79170021 to VM-C; Becas Chile Postdoctoral Fellowship 74120010 to VP), NIH (HD087351 and HD101006 to BAR); American Heart Association (Postdoctoral Fellowship 13POST16520009 to VP and AHA grant 19TPA34920001 to BAR) and ICGEB (CRP-ICGEB Research Grant CRP/CHL18-04 to VP). We are also thankful for the PhD or MSc fellowships from CONICYT Chile to CS-F, PR-M, CV-T, CL-C and PEM.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by M. Piacentini
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Cristian Sotomayor-Flores, Pablo Rivera-Mejías
Contributor Information
Valentina Parra, Email: vparra@ciq.uchile.cl.
Sergio Lavandero, Email: slavander@uchile.cl.
Supplementary information
The online version of this article (10.1038/s41418-020-0522-3) contains supplementary material, which is available to authorized users.
References
- 1.Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, et al. A novel angiotensin-converting enzyme–related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ Res. 2000;87:1–9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 2.Ocaranza P, Godoy I, Jalil JE, Varas M, Collantes P, Pinto M, et al. Enalapril attenuates downregulation of angiotensin-converting enzyme 2 in the late phase of ventricular dysfunction in myocardial infarcted rat. Hypertension. 2006;572–8. [DOI] [PubMed]
- 3.Ocaranza MP, Lavandero S, Jalil JE, Moya J, Pinto M, Novoa U, et al. Angiotensin-(1–9) regulates cardiac hypertrophy in vivo and in vitro. J Hypertens. 2010;28:1054–64. doi: 10.1097/hjh.0b013e328335d291. [DOI] [PubMed] [Google Scholar]
- 4.Ocaranza P, Rivera P, Novoa U, Pinto M, Gonza L, Chiong M, et al. Rho kinase inhibition activates the homologous angiotensin- converting enzyme-angiotensin-(1–9) axis in experimental hypertension. J Hypertens. 2011;29:706–15. doi: 10.1097/HJH.0b013e3283440665. [DOI] [PubMed] [Google Scholar]
- 5.Flores-Munoz M, Work LM, Douglas K, Denby L, Dominiczak AF, Graham D, et al. Angiotensin-(1-9) attenuates cardiac fibrosis in the stroke-prone spontaneously hypertensive rat via the angiotensin type 2 receptor. Hypertension. 2012;59:300–7. doi: 10.1161/HYPERTENSIONAHA.111.177485. [DOI] [PubMed] [Google Scholar]
- 6.Flores-Muñoz M, Smith NJ, Haggerty C, Milligan G, Nicklin SA. Angiotensin1-9 antagonises pro-hypertrophic signalling in cardiomyocytes via the angiotensin type 2 receptor. J Physiol. 2011;4:939–51. doi: 10.1113/jphysiol.2010.203075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fattah C, Nather K, McCarroll CS, Hortigon-Vinagre MP, Zamora V, Flores-Munoz M, et al. gene therapy with angiotensin-(1-9) preserves left ventricular systolic function after myocardial infarction. J Am Coll Cardiol. 2016;68:2652–66. doi: 10.1016/j.jacc.2016.09.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai D-F, Johnson SC, Villarin JJ, Chin MT, Nieves-Cintron M, Chen T, et al. Mitochondrial oxidative stress mediates angiotensin II-induced cardiac hypertrophy and Gαq overexpression-induced heart failure. Circ Res. 2013;108:837–46. doi: 10.1161/CIRCRESAHA.110.232306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H, Zhang Y, Sun J, Wei J, Sun L, Zhang J. Effect of distinct sources of Ca2+ on cardiac hypertrophy in cardiomyocytes. Exp Biol Med. (Maywood) 2012;237:271–8. doi: 10.1258/ebm.2011.011273. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu I, Minamino T. Physiological and pathological cardiac hypertrophy. J Mol Cell Cardiol. 2016;97:245–62. doi: 10.1016/j.yjmcc.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–92. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 12.Benard G, Bellance N, James D, Parrone P, Fernandez H, Letellier T, et al. Mitochondrial bioenergetics and structural network organization. J Cell Sci. 2007;120:838–48. doi: 10.1242/jcs.03381. [DOI] [PubMed] [Google Scholar]
- 13.Pennanen C, Parra V, Lopez-Crisosto C, Morales PE, Del Campo A, Gutierrez T, et al. Mitochondrial fission is required for cardiomyocyte hypertrophy via a Ca2+ -calcineurin signalling pathway. J Cell Sci. 2014;127:1–33. doi: 10.1242/jcs.139394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houser SR, Molkentin JD. Does contractile Ca2+ control calcineurin-NFAT signaling and pathological hypertrophy in cardiac myocytes? Sci Signal. 2008;1:1–7. doi: 10.1126/scisignal.125pe31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutiérrez T, Parra V, Troncoso R, Pennanen C, Contreras-Ferrat A, Vasquez-Trincado C, et al. Alteration in mitochondrial Ca2+ uptake disrupts insulin signaling in hypertrophic cardiomyocytes. Cell Commun Signal. 2014;12:1–14. doi: 10.1186/s12964-014-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.López-Crisosto C, Bravo-Sagua R, Rodriguez-Peña M, Mera C, Castro PF, Quest AFG, et al. ER-to-mitochondria miscommunication and metabolic diseases. Biochim Biophys Acta. 2015;1852:2096–105. doi: 10.1016/j.bbadis.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP, Lee R, Feinbaum R. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 18.Hwang H, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;94:776–80. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau NC, Lim LP, Weinstein EG, Bartel DP, Lau NC, Lim LP, et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 20.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:1998–2001. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 21.Wu X, Piper-Hunter MG, Crawford M, Nuovo GJ, Clay B, Otterson GA, et al. Micrornas in the pathogenesis of lung cancer. J Thorac Oncol. 2009;4:1028–34. doi: 10.1097/JTO.0b013e3181a99c77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang B, Lu Y, Wang Z. MicroRNAs and apoptosis: implications in the molecular therapy of human disease. Clin Exp Pharm Physiol. 2009;36:951–60. doi: 10.1111/j.1440-1681.2009.05245.x. [DOI] [PubMed] [Google Scholar]
- 23.Zorio E, Medina P, Rueda J, Millán JM, Arnau MA, Beneyto M, et al. Insights into the role of microRNAs in cardiac diseases: from biological signalling to therapeutic targets. Cardiovasc Hematol Agents Med Chem. 2009;7:82–90. doi: 10.2174/187152509787047676. [DOI] [PubMed] [Google Scholar]
- 24.Galasso M, Sana ME, Volinia S. Non-coding RNAs: a key to future personalized molecular therapy? Genome Med. 2010;2:1–10. doi: 10.1186/gm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong S, Cheng Y, Yang J, Li J, Liu X, Wang X, et al. MicroRNA expression signature and the role of microRNa-21 in the early phase of acute myocardial infarction. J Biol Chem. 2009;284:29514–25. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y, Zhang Y, Wang N, Pan Z, Gao S, Zhang F, et al. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation. 2010;122:2378–87. doi: 10.1161/CIRCULATIONAHA.110.958967. [DOI] [PubMed] [Google Scholar]
- 28.Parra V, Eisner V, Chiong M, Criollo A, Moraga F, Jaimovich E, et al. Changes in mitochondrial dynamics during ceramide-induced cardiomyocyte early apoptosis. Cardiovasc Res. 2008;77:387–97. doi: 10.1093/cvr/cvm029. [DOI] [PubMed] [Google Scholar]
- 29.Parra V, Verdejo HE, Iglewski M, Del Campo A, Troncoso R, Jones D, et al. Insulin stimulates mitochondrial fusion and function in cardiomyocytes via the Akt-mTOR-NFkB-Opa-1 signaling pathway. Diabetes. 2014;63:75–88. doi: 10.2337/db13-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu T, Sheu S-S, Robotham JL, Yoon Y. Mitochondrial fission mediates high glucose-induced cell death through elevated production of reactive oxygen species. Cardiovasc Res. 2008;79:341–51. doi: 10.1093/cvr/cvn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munoz JP, Chiong M, García L, Troncoso R, Toro B, Pedrozo Z, et al. Iron induces protection and necrosis in cultured cardiomyocytes: role of reactive oxygen species and nitric oxide. Free Radic Biol Med. 2010;48:526–34. doi: 10.1016/j.freeradbiomed.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 32.Contreras-Ferrat A, Toro B, Bravo-Sagua R, Parra V, Vasquez-Trincado C, Ibarra C, et al. An Inositol 1,4,5-triphosphate (IP3)-IP3 receptor pathway is required for insulin-stimulated glucose transporter 4 translocation and glucose uptake in cardiomyocytes. Endocrinology. 2010;151:4665–77. doi: 10.1210/en.2010-0116. [DOI] [PubMed] [Google Scholar]
- 33.Chiong M, Parra V, Eisner V, Ibarra C, Maldonado C, Criollo A, et al. Parallel activation of Ca2+-induced survival and death pathways in cardiomyocytes by sorbitol-induced hyperosmotic stress. Apoptosis. 2010;15:887–903. doi: 10.1007/s10495-010-0505-9. [DOI] [PubMed] [Google Scholar]
- 34.Rojas-Rivera D, Díaz-Elizondo J, Parra V, Salas D, Contreras A, Toro B, et al. Regulatory volume decrease in cardiomyocytes is modulated by calcium influx and reactive oxygen species. FEBS Lett. 2009;583:3485–92. doi: 10.1016/j.febslet.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Bravo-Sagua R, López-Crisosto C, Parra V, Rodriguez M, Rothermel BA, Quest AFG, et al. mTORC1 inhibitor rapamycin and ER stressor tunicamycin induce differential patterns of ER- mitochondria coupling. Sci Rep. 2016;6:1–12. doi: 10.1038/srep36394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manders EMM, Verbeek FJ, Aten JA. Measurement of co-localization of objects in dual-colour confocal images. J Microsc. 1993;169:375–82. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 37.Parra V, Altamirano F, Hernandez-Fuentes CP, Tong D, Kyrychenko V, Rotter D, et al. Down syndrome critical region 1 gene, rcan1, helps maintain a more fused mitochondrial network. Circ Res. 2018;122:1–26. doi: 10.1161/CIRCRESAHA.117.311522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benes V, Collier P, Kordes C, Stolte J, Rausch T, Muckentaler MU, et al. Identification of cytokine- induced modulation of microRNA expression and secretion as measured by a novel microRNA specific qPCR assay. Sci Rep. 2015;5:1–14. doi: 10.1038/srep11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hasler D, Lehmann G, Murakawa Y, Rajewsky N, Landthaler M, Meister G, et al. The lupus autoantigen La prevents Mis-channeling of t-RNA fragments into the human microRNA pathway. Mol Cell. 2016;63:110–24. doi: 10.1016/j.molcel.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 40.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orell A, Tripp V, Aliaga-Tobar V, Albers S, Maracaja-Coutinho V, Randau L. A regulatory RNA is involved in RNA duplex formation and biofilm regulation in Sulfolobus acidocaldarius. Nucleic Acids Res. 2018;46:4794–806. doi: 10.1093/nar/gky144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matamala JM, Arias-Carrasco R, Sanchez C, Uhrig M, Matus S, Maracaja-Coutinho V, et al. Genome-wide circulating microRNA expression profiling reveals potential biomarkers for amyotrophic lateral sclerosis. Neurobiol Aging. 2018;64:123–38. doi: 10.1016/j.neurobiolaging.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 43.Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinforma. 2010;11:1–24. doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinlan AR. BEDTools: the Swiss-army tool for genome feature analysis. Curr Protoc Bioinforma. 2015;47:12.1–12.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2018;47:155–62. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, et al. Ensembl 2019. Nucleic Acids Res. 2019;47:745–51. doi: 10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dueck A, Ziegler C, Eichner A, Berezikov E, Meister G. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 2012;40:9850–62. doi: 10.1093/nar/gks705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3:1077–84. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 51.Agarwal V, Bell GW, Nam J, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4(e05005):1–38. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nam J, Rissland OS, Koppstein D, Abreu-Goodger C, Jan CH, Agarwal V, et al. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol Cell. 2015;53:1031–43. doi: 10.1016/j.molcel.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 54.Ocaranza MP, Moya J, Barrientos V, Alzamora R, Hevia D, Morales C, et al. Angiotensin-(1-9) reverses experimental hypertension and cardiovascular damage by inhibition of the angiotensin converting enzyme/Ang II axis. J Hypertens. 2014;32:771–83. doi: 10.1097/HJH.0000000000000094. [DOI] [PubMed] [Google Scholar]
- 55.Fyhrquist F, Saijonmaa O. Renin-angiotensin system revisited. J Intern Med. 2008;264:224–34. doi: 10.1111/j.1365-2796.2008.01981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoon Y, Krueger EW, Oswald BJ, Mcniven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–20. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulatesmitochondrial fission and cell death. EMBO Rep. 2007;8:939–44. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang C-R, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of drp1 regulates its gtpase activity and mitochondrial morphology. J Biol Chem. 2007;282:21583–7. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- 59.Santel A, Frank S. Shaping mitochondria: the complex posttranslational regulation of the mitochondrial fission protein DRP1. IUBMB Life. 2008;60:448–55. doi: 10.1002/iub.71. [DOI] [PubMed] [Google Scholar]
- 60.Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–9. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 61.Eisner V, Parra V, Lavandero S, Hidalgo C, Jaimovich E. Mitochondria fine-tune the slow Ca2+ transients induced by electrical stimulation of skeletal myotubes. Cell Calcium. 2010;48:358–70. doi: 10.1016/j.ceca.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 62.Eisner V, Lenaers G, Hajnóczky G. Mitochondrial fusion is frequent in skeletal muscle and supports excitation–contraction coupling. J Cell Biol. 2014;205:179–95. doi: 10.1083/jcb.201312066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rothermel BA, Vega RB, Williams RS. The role of modulatory calcineurin-interacting proteins in calcineurin signaling. Trends Cardiovasc Med. 2003;13:15–21. doi: 10.1016/s1050-1738(02)00188-3. [DOI] [PubMed] [Google Scholar]
- 64.Giorgi C, De Stefani D, Bononi A, Rizzuto R, Pinton P. Structural and functional link between the mitochondrial network and the endoplasmic reticulum. Int J Biochem Cell Biol. 2010;41:1817–27. doi: 10.1016/j.biocel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bravo-Sagua R, Parra V, Ortiz-Sandoval C, Navarro-Marquez M, Rodriguez AE, Díaz-Valdivia N, et al. Caveolin-1 impairs PKA-DRP1-mediated remodelling of ER–mitochondria communication during the early phase of ER stress. Cell Death Differ. 2019;1:1195–212. doi: 10.1038/s41418-018-0197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ding J, Li X, Hu H. TarPmiR: a new approach for microRNA target site prediction. Bioinformatics. 2016;32:2768–75. doi: 10.1093/bioinformatics/btw318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olsen SR, Uhler MD. Inhibition of protein kinase-A by overexpression of the cloned human protein kinase inhibitor. Mol Endocrinol. 1991;5:1246–56. doi: 10.1210/mend-5-9-1246. [DOI] [PubMed] [Google Scholar]
- 68.Vásquez-Trincado C, García-Carvajal I, Pennanen C, Parra V, Hill JA, Rothermel BA, et al. Mitochondrial dynamics, mitophagy and cardiovascular disease. J Physiol. 2016;3:509–25. doi: 10.1113/JP271301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Y, Liu Y, Dorn GW., II Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ Res. 2011;109:1327–31. doi: 10.1161/CIRCRESAHA.111.258723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen L, Liu T, Tran A, Lu X, Tomilov AA, Davies V, et al. OPA1 mutation and late-onset cardiomyopathy: mitochondrial dysfunction and mtDNA instability. J Am Heart Assoc. 2012;1:1–12. doi: 10.1161/JAHA.112.003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ashrafian H, Docherty L, Leo V, Towlson C, Neilan M, Lygate CA, et al. A Mutation in the mitochondrial fission gene dnm1l leads to cardiomyopathy. Plos Genet. 2010;6:1–18. doi: 10.1371/journal.pgen.1001000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ong S, Subrayan S, Lim SY, Yellon DM, Davidson SM, Hausenloy DJ. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion Injury. Circulation. 2010;121:2012–22. doi: 10.1161/CIRCULATIONAHA.109.906610. [DOI] [PubMed] [Google Scholar]
- 73.Givvimani S, Munjal C, Tyagi N, Sen U, Metreveli N, Tyagi SC. Mitochondrial division/mitophagy inhibitor (mdivi) ameliorates pressure overload induced heart failure. PLoS ONE. 2012;7:e32388. doi: 10.1371/journal.pone.0032388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cereghetti GM, Stangherlin A, Martins de Brito O, Chang CR, Blackstone C, Bernardi P, et al. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci USA. 2008;105:15803–8. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rizzuto R, De Stefani D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol. 2012;13:566–78. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- 76.Jeppesen PL, Christensen GL, Mikael S, Yaël Nossent A, Jensen Bronnum H, Andersen DC, et al. Angiotensin II type 1 receptor signalling regulates microRNA differentially in cardiac fibroblasts and myocytes. Br J Pharm. 2011;164:394–404. doi: 10.1111/j.1476-5381.2011.01375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.