Abstract
The elevated expression of poly(ADP-ribose) polymerase-1 (PARP1) and increased PARP1 activity, namely, poly(ADP-ribosyl)ation (PARylation), have been observed in cardiac remodeling, leading to extreme energy consumption and myocardial damage. However, the mechanisms underlying the regulation of PARP1 require further study. WWP2, a HECT-type E3 ubiquitin ligase, is highly expressed in the heart, but its function there is largely unknown. Here, we clarified the role of WWP2 in the regulation of PARP1 and the impact of this regulatory process on cardiac remodeling. We determined that the knockout of WWP2 specifically in myocardium decreased the level of PARP1 ubiquitination and increased the effects of isoproterenol (ISO)-induced PARP1 and PARylation, in turn aggravating ISO-induced myocardial hypertrophy, heart failure, and myocardial fibrosis. Similar findings were obtained in a model of ISO-induced H9c2 cells with WWP2 knockdown, while the reexpression of WWP2 significantly increased PARP1 ubiquitination and decreased PAPR1 and PARylation levels. Mechanistically, coimmunoprecipitation results identified that WWP2 is a novel interacting protein of PARP1 and mainly interacts with its BRCT domain, thus mediating the degradation of PARP1 through the ubiquitin–proteasome system. In addition, lysine 418 (K418) and lysine 249 (K249) were shown to be of critical importance in regulating PARP1 ubiquitination and degradation by WWP2. These findings reveal a novel WWP2–PARP1 signal transduction pathway involved in controlling cardiac remodeling and may provide a basis for exploring new strategies for treating heart disorders related to cardiac remodeling.
Subject terms: Epigenetics, Cardiovascular diseases
Introduction
Heart failure is one of the deadliest diseases worldwide. Despite important advances in its diagnosis and treatment, the mortality rate of heart failure has not significantly improved. Cardiac remodeling, including myocardial hypertrophy and fibrosis, is the foundation of the occurrence and development of heart failure [1, 2]. Poly(ADP-ribose) polymerase-1 (PARP1) represents a critical injury factor in cardiovascular diseases, especially in cardiac remodeling caused by various factors [3–7]. The elevated expression of PARP1 and increased poly(ADP-ribosyl)ation (PARylation; PARP1 activity) have been observed in cardiac remodeling, leading to extreme energy consumption by myocardial cells, thereby triggering injury of these cells [3–8]. However, the mechanisms underlying the regulation of PARP1 in cardiac remodeling require further study.
An imbalance of protein homeostasis is a key factor in the disruption of cell homeostasis, which significantly accelerates the progression of cardiac remodeling [9, 10]. The ubiquitin–proteasome system is an important biological machinery for controlling protein homeostasis, and intracellular ubiquitination allows the coordinated removal of nonfunctional, misfolded, or potentially toxic proteins to maintain protein homeostasis. The interaction between E3 ubiquitin ligase and the target protein is a core step in ubiquitin–proteasome system-mediated protein degradation [11]. Hence, exploring the role and molecular mechanism of protein ubiquitination factors, especially E3 ubiquitin ligase, in cardiac remodeling may be key to preventing and treating this condition.
WWP2 is a HECT-type E3 ubiquitin ligase and is one of the major members of the NEDD4 family [12, 13]. WWP2 was initially identified as a cancer-promoting factor [13, 14]. However, recent studies have shown that it actually participates in various biological processes, such as membrane protein transportation, signal transduction, transcription, and the cell cycle, by regulating multiple substrates [15, 16]. Although WWP2 is expressed at a high level in the heart, its function in this organ remains largely unclear.
In the present study, it was found that WWP2 participates in isoproterenol (ISO)-induced cardiac remodeling. The knockout of WWP2 specifically in myocardium resulted in decreased levels of PARP1 ubiquitination and increased effects of ISO-induced PARP1 expression and higher PARylation, leading to the aggravation of ISO-induced heart failure, myocardial hypertrophy, and fibrosis in mice. Similar findings were obtained in a model of H9c2 cells with ISO-induced WWP2 knockdown, while the reexpression of WWP2 significantly increased PARP1 ubiquitination and decreased PARP1 and PARylation levels, revealing that WWP2 is a protective factor that regulates ISO-induced cardiac remodeling. Mechanistically, PARP1 was identified as a new physiological substrate of WWP2, with enhanced interaction between them induced by ISO. Notably, WWP2 mainly interacted with PARP1 at the BRCT domain, and ubiquitination occurred at the K249 and K418 sites of PARP1. This process mediated the degradation of PARP1 through the ubiquitin–proteasome system, which protected against ISO-induced cardiac remodeling.
Materials and methods
MycCre+;WWP2Fl/Fl and MycCre−;WWP2Fl/Fl mouse experiments
Mice with conditional myocardium-specific genotypes of MycCre+;WWP2Fl/Fl and MycCre−;WWP2Fl/Fl were established by Shanghai Biomodel Organism Science & Technology Development. Efficient myocardium-specific silencing of WWP2 was confirmed (Fig. 5a–d). Specific-pathogen-free 8–10-week-old male mice were assessed. In the ISO infusion model, MycCre+; WWP2Fl/Fl and MycCre−;WWP2Fl/Fl mice (a total of 24 mice; n = 6 in each group) were randomized prior to anesthesia via the inhalation of isoflurane/oxygen (2%, ~1500 ml/min; sufficient depth of anesthesia was confirmed by monitoring the lack of paw withdrawal reflex) and then incised in the middle scapular region, followed by osmotic minipump (Alzet) implantation subcutaneously, as directed by the manufacturer. The animals were administered ISO (60 mg/kg/day) by infusion for 14 days with ALZET (Alzet, model 2002; 0.5 µL/h), which involves a widely recognized method of inducing cardiac remodeling in mice [17–19]. Blood pressure measurement was carried out daily by the tail-cuff method. Cervical dislocation after isoflurane inhalation anesthesia was used for mouse euthanasia. The efficiency of WWP2 knockdown in the animals was evaluated by immunofluorescence and immunoblotting at the endpoint. The animals were handled in accordance with animal welfare regulations of China Medical University. All experiments involving animals were approved by the Animal Subjects Committee of China Medical University. The work also conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1985).
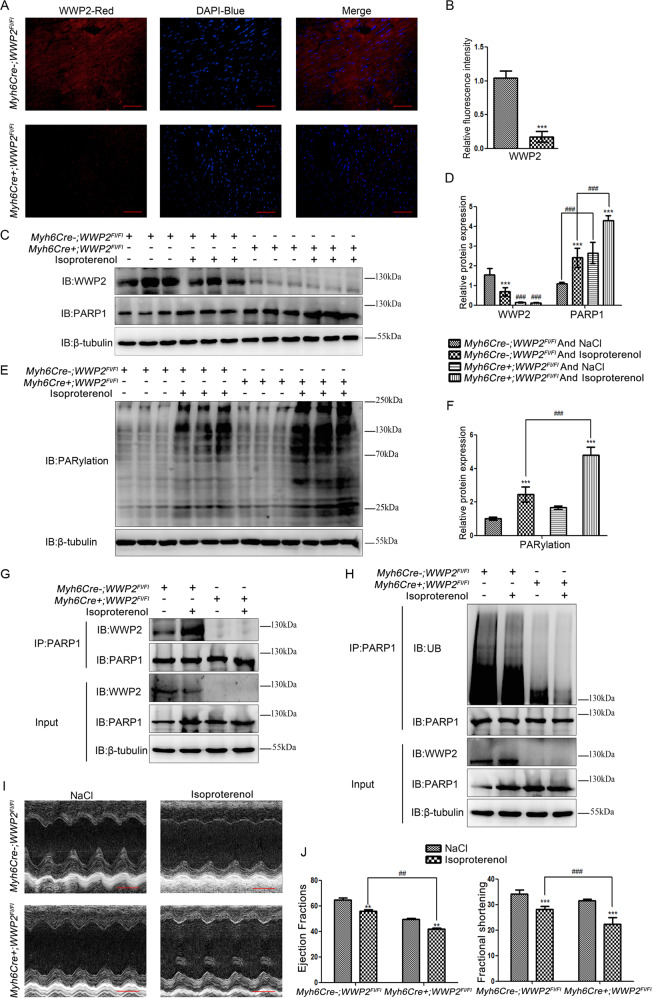
Fig. 5. Myocardium-specific WWP2 knockout significantly aggravated ISO-induced heart failure by decreasing PARP1 ubiquitination and increasing PARP1 and PARylation levels in mice.
In Myh6Cr+;WWP2Fl/Fl and Myh6Cr−;WWP2Fl/Fl mice following 2 weeks of ISO or NaCl infusion in vivo, (a) immunofluorescence (WWP2 red, DAPI blue) was used to assess the expression of WWP2 (scale bar 50 µm). b Data are shown as mean ± SD (each group of mice, n = 6; ***P < 0.001, unpaired Student’s t test). c, e Western blotting was used to assess the expression of WWP2 and PARP1, and PARylation. d, f Data are shown as mean ± SD (each group of mice, n = 6; ***P < 0.001, unpaired Student’s t test; ###P < 0.001, two-way ANOVA with Bonferroni post hoc testing). g Coimmunoprecipitation was performed to determine the interaction between PARP1 and WWP2. h As did the ubiquitination level of PARP1. i Echocardiography (scale bar: 2 mm), (j) ejection fraction, and fractional shortening were used to assess heart failure. Data are shown as mean ± SD (each group of mice, n = 6; **P < 0.01, ***P < 0.001, unpaired Student’s t test; ##P < 0.01, ###P < 0.001, two-way ANOVA with Bonferroni post hoc testing).
Histopathological analysis
The mouse myocardial tissues underwent fixation with 4% formalin (4 h), paraffin embedding, and sectioning at a thickness of 5 µm. After dewaxing with xylene and rehydration through graded ethanol, the specimens underwent staining with hematoxylin and eosin (H&E) and Masson’s Trichrome Stain Kit (G1340; Solarbio, China).
Echocardiography and LVEF determination
Cardiac function in the MycCre+;WWP2Fl/Fl and MycCre−;WWP2Fl/Fl groups was evaluated on a Visual Sonics Vevo 2100 real-time, high-resolution in vivo micro-imaging system (Visualsonic, Canada). A total of 24 mice underwent anesthesia using 1.5% isoflurane, after which cardiac function was determined with a 40 MHz transducer, with oxygen continuously supplied. Two-dimensional M-mode recording was employed to assess left ventricular function (LVEF). Cardiac function was determined by measuring interventricular septal dimension, left ventricular posterior wall dimension, LV internal dimension at systole, LV internal dimension at diastole, and LV mass. In addition, the left ventricular ejection fraction (EF%) and fractional shortening (FS%) were calculated.
WWP2 knockdown by shRNA and siRNA
Normal Control and WWP2 shRNAs produced by GeneChem (China) were used on H9c2 cells. WWP2 silencing was performed with lentivirus. For WWP2 targeting, three sequences were employed to rule out off-target effects:
WWP2 shRNA-1: CACCTCACCTACTTCCGCTTT
WWP2 shRNA-2: TGGCAGAAGAACGCCATCTAT
WWP2 shRNA-3: TAGGAGCCATACATGCTTCAA
Control and WWP2 siRNAs produced by RIBOBIO (China) were used on 293T cells. WWP2 silencing was performed with jetPRIME transfection reagent (PolyPlus, France). For WWP2 targeting, three sequences were employed to rule out off-target effects:
WWP2 siRNA-1: GATCTGGGAAATGTGCCTA
WWP2 siRNA-2: GGTGCTTCAGCCAGAACAA
WWP2 siRNA-3: CGGACGTGTCTATTATGTT
WWP2 knockdown efficiency was determined by immunoblotting.
Plasmid construction and transfection
Plasmids harboring full-length human WWP2 (GeneChem) were cloned into Flag GV366 and full-length rat WWP2 (GeneChem) was cloned into the HA vector. Ubiquitin was cloned into the HA vector (GeneChem). Full-length human PARP1 and lysine-site-mutated human PARP1 were cloned into the Myc GV219 vector (GeneChem). Four truncated human PARP1 plasmids containing different domains were constructed as follows: Zinc 1, Zinc 2, BRCT, and PARP A-helical domains with a Myc-tag; Zinc 1, Zinc 2, and BRCT domains with a Myc-tag; Zinc 1 and Zinc 2 domains with a Myc-tag; and BRCT, PARP A-helical, and PARP catalytic domains with a Myc-tag. Next, six mutant PARP1 plasmids were constructed as follows: K131R, K249R, K418R, K548R, K579R, and K633R. Plasmid transfection was performed with Lipofectamine 3000 (L3000015; Invitrogen, USA), as directed by the manufacturer.
Antibodies and reagents
Polyclonal rabbit anti-WWP2 (1:1000; ab103527, Abcam, USA; 1:500; 12197-1-AP, Proteintech), monoclonal rabbit anti-HA (1:1000; 3724S, Cell Signaling, USA), monoclonal mouse anti-Myc (1:1000; 2276S, Cell Signaling, USA), polyclonal rabbit anti-Flag (1:1000; 14793S, Cell Signaling, USA), polyclonal rabbit anti-caspase3 (1:1000; 9662S, Cell Signaling, USA; 19677-1-AP, Proteintech, USA), monoclonal rabbit anti-PARP1 (1:1000; 9532S, Cell Signaling, USA), monoclonal mouse anti-PARylation (1:500; 4335-MC-100-AC, Trevigen, USA), monoclonal mouse anti-ANP (1:200; sc-515701, Santa, USA), polyclonal rabbit anti-BNP (1:1000; ab19645, Abcam, USA), monoclonal mouse anti-Ub antibodies (1:1000; 3933S, Cell Signaling, USA), monoclonal mouse anti-β-tubulin (1:2000; 66240-1-Ig, Proteintech, USA), and monoclonal mouse anti-GAPDH (1:2000; 10494-1-AP, Proteintech, USA) were obtained commercially, as were polyclonal rabbit anti-collagen-I (1:1000; 14695-1-AP, Proteintech, USA) and polyclonal rabbit anti-α-SMA (1:1000; 55135-1-AP, Proteintech, USA). MG132 (A2585; 20 µM) and cycloheximide (CHX) (A8244; 100 µM) were obtained from Apexbio (USA) and reconstituted in DMSO. ISO was obtained from Sigma (I5627, USA) and used at a concentration of 10 µM in DMSO. H9c2 cells were provided, identified, and tested for mycoplasma contamination by ATCC and cultured in Dulbecco’s modified Eagle’s medium (HyClone, Logan, UT, USA) with 10% fetal bovine serum (HyClone, Logan, UT, USA) in a humidified atmosphere of 5% CO2 at 37 °C.
Coimmunoprecipitation and immunoblotting
Myocardial tissue of mice and cells underwent lysis with flag lysis buffer (50 mM Tris, 137 mM NaCl, 1 mM EDTA, 10 mM NaF, 0.1 mM Na3VO4, 1% NP-40, 1 mM DTT, 10% glycerol, pH 7.8) with protease inhibitors (Roche). The lysates were incubated with 30 µl of anti-Flag/Myc Affinity Gel (B23102/B26302; Biotool, USA) for 12 h at 4 °C or followed by treatment with antibodies (antibody/cell lysate = 1 µg/mg) for 2–3 h and incubation with 30 µl of Protein A/G immunoprecipitate (B23202; Biotool) for 12 h at 4 °C. The binding complexes were resolved by SDS–PAGE followed by electrotransfer onto PVDF membranes, which were blocked with 5% bovine serum albumin for 1 h at ambient temperature. Then, the samples underwent successive incubations with primary (4 °C, overnight) and secondary (ambient conditions, 1 h) antibodies. GAPDH and β-tubulin were used for normalization, and Image J v1.46 (National Institutes of Health, USA) was employed to quantify the immunoreactive bands.
Analysis of PARP1 ubiquitination
Myocardial tissue of mice and cells underwent lysis with 1% SDS buffer (Tris pH 7.5, 0.5 mM EDTA, 1 mM DTT) and boiling (10 min). HA-ubiquitin, blank, full-length human WWP2, full-length human PARP1, and mutant PARP1 plasmids were transfected into cells for 48 h, followed by lysis as described above. All cell lysates were incubated with anti-PARP1 antibody (antibody/cell lysate = 1 µg/mg) and 30 µl of protein A/G immunoprecipitation magnetic beads or 30 µl of anti-Myc (B26302; Biotool) magnetic beads for 12 h at 4 °C. PARP1 ubiquitination was analyzed by immunoblotting with anti-HA antibodies or anti-Ub antibodies.
Statistical analysis
Data are presented as mean ± standard deviation (SD) and F-test (two groups) or Brown–Forsythe test (three or more groups) was used to assess the homogeneity of variance. Shapiro–Wilk test was used to assess the normality of data. Student’s t test or Welch’s t test was used for equal variance or unequal variance, respectively (two groups). One-way ANOVA and two-way ANOVA were employed to determine the significance of differences between more than two groups for one factor involved and two factors involved, respectively, followed by Bonferroni post hoc testing. The P values were adjusted for multiple comparisons where appropriate, and SPSS 22.0 (SPSS, USA) was used for statistical analyses, with P < 0.05 indicating statistical significance.
Results
WWP2 interacts with PARP1 and is mainly positioned at the PARP1 BRCT region, mediating proteasome-associated PARP1 degradation
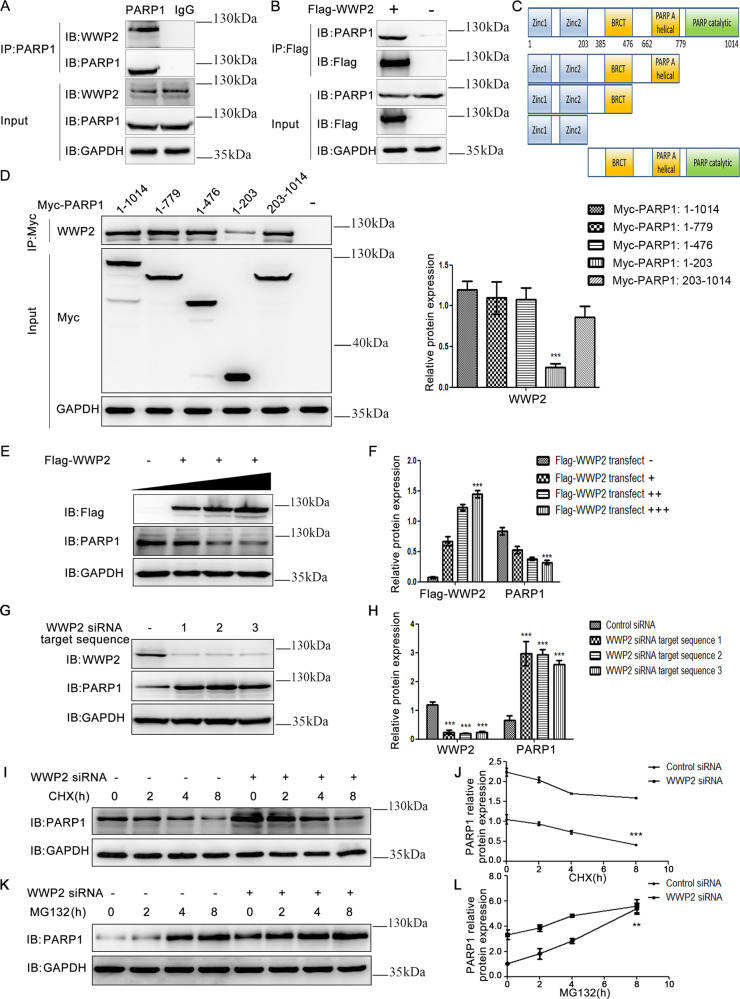
PARP1 is known to be an important inducer of cardiac remodeling induced by various factors by leading to extreme energy consumption by myocardial cells, thereby triggering injury of these cells [3–7]. Based on the limited understanding and potential biological importance of PARP1 regulation, we explored the proteins interacting with PARP1 and the mechanisms underlying its regulation in 293T cells. Coimmunoprecipitation results showed that endogenous and exogenous WWP2 interacted with PARP1 (Fig. 1a, b). Furthermore, to clarify on which domain of PARP1 WWP2 acts, coimmunoprecipitation was performed to assess the interaction between WWP2 and full-length PARP1 or various truncated mutants of it. The results demonstrated that WWP2 mainly interacted with the BRCT domain of PARP1 (Fig. 1c, d).
Fig. 1. WWP2 interacts with PARP1 and is mainly positioned at the PARP1 BRCT region, where it participates in proteasome-associated PARP1 degradation.
a Coimmunoprecipitation was performed to determine the interaction between endogenous WWP2 and PARP1 in 293T cells. b As did exogenous WWP2 and PARP1. c Schematic representation of functional domains of PARP1. d Coimmunoprecipitation was performed to determine the interaction between endogenous WWP2 and functional domains of PARP1 including amino acids 1–1014, 1–779, 1–476, 1–203, and 203–1014 in 293T cells. And data are shown as mean ± SD (***P < 0.001, one-way ANOVA with Bonferroni post hoc testing). e Western blotting was carried out to assess PARP1 expression levels with the overexpression of WWP2 gradually in 293T cells. f Data are shown as mean ± SD (***P < 0.001, one-way ANOVA with Bonferroni post hoc testing). g Three target sequences of siRNA-WWP2 were used to determine the expression of PARP1 and efficiency of WWP2 knockdown in 293T cells. h Data are shown as mean ± SD (***P < 0.001, unpaired Student’s t test). i Expression of PARP1 was assessed at different durations of CHX administration with or without WWP2 knockdown in 293T cells. j Data are shown as mean ± SD (***P < 0.001, two-way ANOVA with Bonferroni post hoc testing). k Expression of PARP1 was assessed at different durations of MG132 administration with or without WWP2 knockdown in 293T cells. l Data are shown as mean ± SD (**P < 0.01, two-way ANOVA with Bonferroni post hoc testing).
WWP2 is an E3 ubiquitin ligase, which exerts biological effects by regulating particular substrates [12–16]. Therefore, WWP2’s function in regulating PARP1 protein stability was next assessed in 293T cells. The results showed that gradated overexpression of WWP2 resulted in a gradual decrease in the expression of PARP1 (Fig. 1e, f). In contrast, WWP2 silencing significantly increased the expression of PARP1 (Fig. 1g, h).
To determine whether WWP2 decreases the expression of PARP1 by inhibiting PARP1 transcription or promoting PARP1 proteasome degradation, the protein synthesis inhibitor CHX and the proteasome inhibitor MG132 were used to determine the effects of WWP2 on PARP1 in 293T cells. The results showed that, compared with the WWP2 siRNA group, the control siRNA group displayed markedly reduced expression of PARP1 in a CHX-time-dependent manner (Fig. 1i, j), suggesting that WWP2 did not affect PARP1 protein synthesis.
Moreover, the expression of PARP1 gradually increased with the prolongation of MG132 stimulation time in both the control siRNA group and the WWP2 siRNA group. However, the increased speed and proportion of the expression of PARP1 in the control siRNA group were greater than in the WWP2 siRNA group (Fig. 1k, l), suggesting that WWP2 decreases the expression of PARP1 by promoting proteasome-mediated PARP1 degradation.
PARP1 is degraded by WWP2 via the ubiquitin–proteasome pathway, depending on the PARP1 K249 and K418 sites
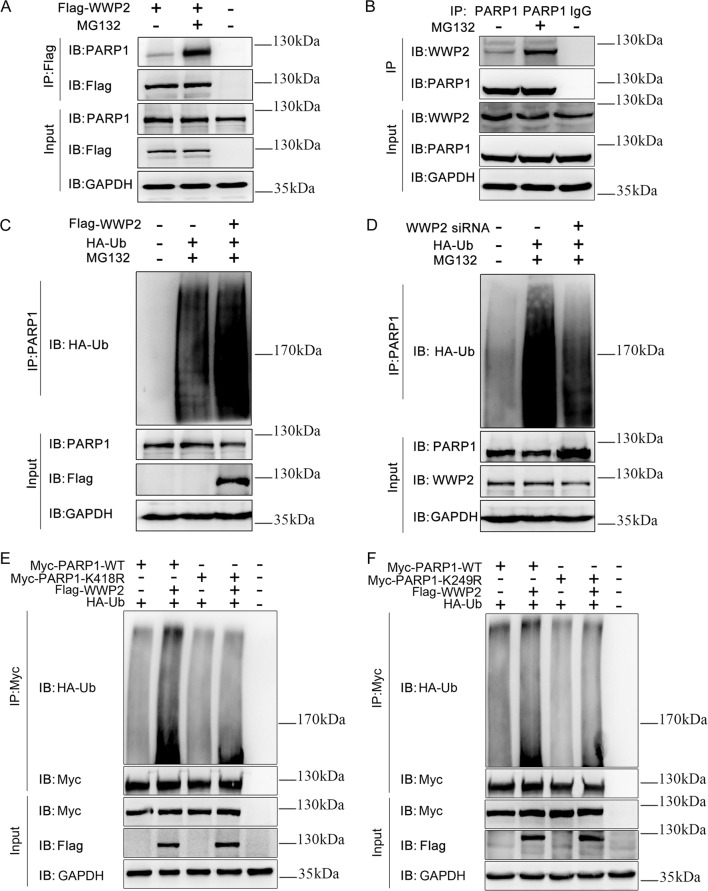
Next, to determine whether WWP2 induces PARP1 degradation via PARP1 polyubiquitination, coimmunoprecipitation was performed to determine the ubiquitination effect of WWP2 on PARP1 in 293T cells. The results showed that the interaction between exogenous WWP2 and PARP1 was significantly enhanced in the presence of MG132 (Fig. 2a) as well as the interaction between endogenous WWP2 and PARP1 (Fig. 2b). Consistent with these results, overexpression of WWP2 significantly increased the level of PARP1 polyubiquitination in the presence of MG132 (Fig. 2c). By contrast, the knockdown of WWP2 resulted in decreased levels of PARP1 polyubiquitination in the presence of MG132 (Fig. 2d). These findings indicated that WWP2 promoted the polyubiquitination of PARP1, thus mediating PARP1 degradation via the ubiquitin–proteasome pathway.
Fig. 2. PARP1 is degraded by WWP2 via the ubiquitin–proteasome pathway, depending on the PARP1 K249 and K418 sites.
a Coimmunoprecipitation was performed to determine the interaction between endogenous WWP2 and PARP1 with or without MG132 treatment in 293T cells. b As did exogenous WWP2 and PARP1. c Flag-WWP2 and HA-Ub were coexpressed and applied with or without MG132 in 293T cells. PARP1 was isolated by IP, and PARP1 ubiquitination levels were assessed with anti-HA antibodies. d WWP2 siRNA and HA-Ub were coexpressed and applied with or without MG132 in 293T cells. PARP1 was isolated by IP, and PARP1 ubiquitination levels were assessed with anti-HA antibodies. e Flag-WWP2, HA-Ub, Myc-WT-PARP1, and Myc-K418R-PARP1 were coexpressed and Myc-PARP1 was isolated by IP in 293T cells. PARP1 ubiquitination levels were assessed with anti-HA antibodies. f Flag-WWP2, HA-Ub, Myc-WT-PARP1, or Myc-K249R-PARP1 was coexpressed and Myc-PARP1 was isolated by IP in 293T cells. PARP1 ubiquitination levels were assessed with anti-HA antibodies.
In addition, to provide the most direct and important evidence that WWP2 promoted polyubiquitination of PARP1, we further determined the polyubiquitination modification sites of PARP1 regulated by WWP2 in 293T cells. Mass spectrometry analysis was used to screen ubiquitination modification sites of PARP1. Furthermore, coimmunoprecipitation assay results showed that the polyubiquitination function of WWP2 on PARP1 was lost when lysine 249 (K249) of PARP1 was substituted by arginine (K249R) (Fig. 6e), as well as PARP1 K418R (Fig. 6f). In contrast, PARP1 ubiquitination was not altered compared with the level for wild-type PARP1 after the mutation of other residues (PARP1 K131R/K548R/K579R/K633R) (Supplementary Fig. 1).
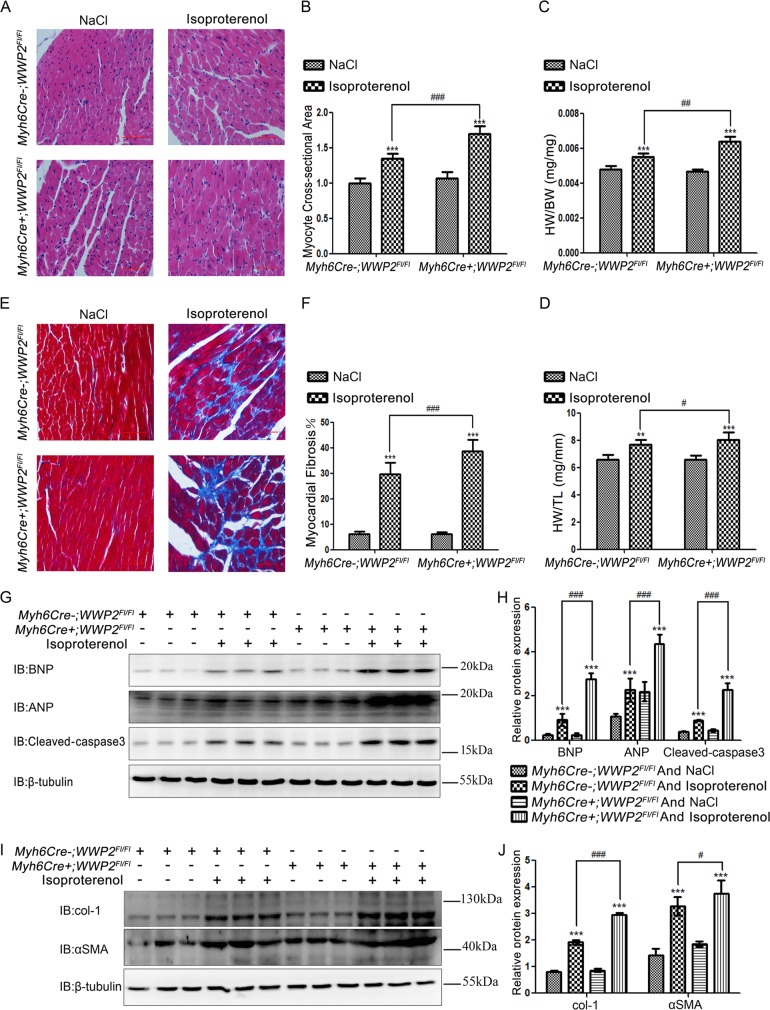
Fig. 6. Myocardium-specific WWP2 knockout markedly aggravated ISO-induced myocardial hypertrophy and fibrosis in mice.
In Myh6Cre+;WWP2Fl/Fl and Myh6Cre−;WWP2Fl/Fl mice following 2 weeks of ISO or NaCl infusion in vivo, (a) H&E staining (scale bar: 50 µm), (b) relative myocyte cross-sectional area, (c) heart weight to body weight ratio, and (d) heart weight to tibia length ratio were used to assess myocardial hypertrophy. Data are shown as mean ± SD (each group of mice, n = 6; **P < 0.01, ***P < 0.001, unpaired Student’s t test; #P < 0.05, ##P < 0.01, ###P < 0.001, two-way ANOVA with Bonferroni post hoc testing). e Masson trichrome staining (scale bar: 50 µm) and (f) fibrotic area were used to assess myocardial fibrosis. Data are shown as mean ± SD (each group of mice, n = 6; ***P < 0.001, unpaired Student’s t test; ###P < 0.001, two-way ANOVA with Bonferroni post hoc testing). g Western blotting was carried out to assess the expression of markers of myocardial hypertrophy and heart failure (BNP, ANP, and cleaved caspase3). h Data are shown as mean ± SD (each group of mice, n = 6; ***P < 0.001, unpaired Student’s t test; ###P < 0.001, two-way ANOVA with Bonferroni post hoc testing). i As did myocardial fibrosis markers (col-1 and αSMA). j Data are shown as mean ± SD (each group of mice, n = 6; ***P < 0.001, unpaired Student’s t test; #P < 0.05, ###P < 0.001, two-way ANOVA with Bonferroni post hoc testing).
Taking these findings together, WWP2 interacts with PARP1 and is mainly positioned at the PARP1 BRCT region, promoting PARP1 polyubiquitination modification by regulating PARP1 K249 and K418 sites, leading to the degradation of PARP1 via the ubiquitin–proteasome pathway. Therefore, WWP2 may play a key role in cardiac remodeling by regulating PARP1.
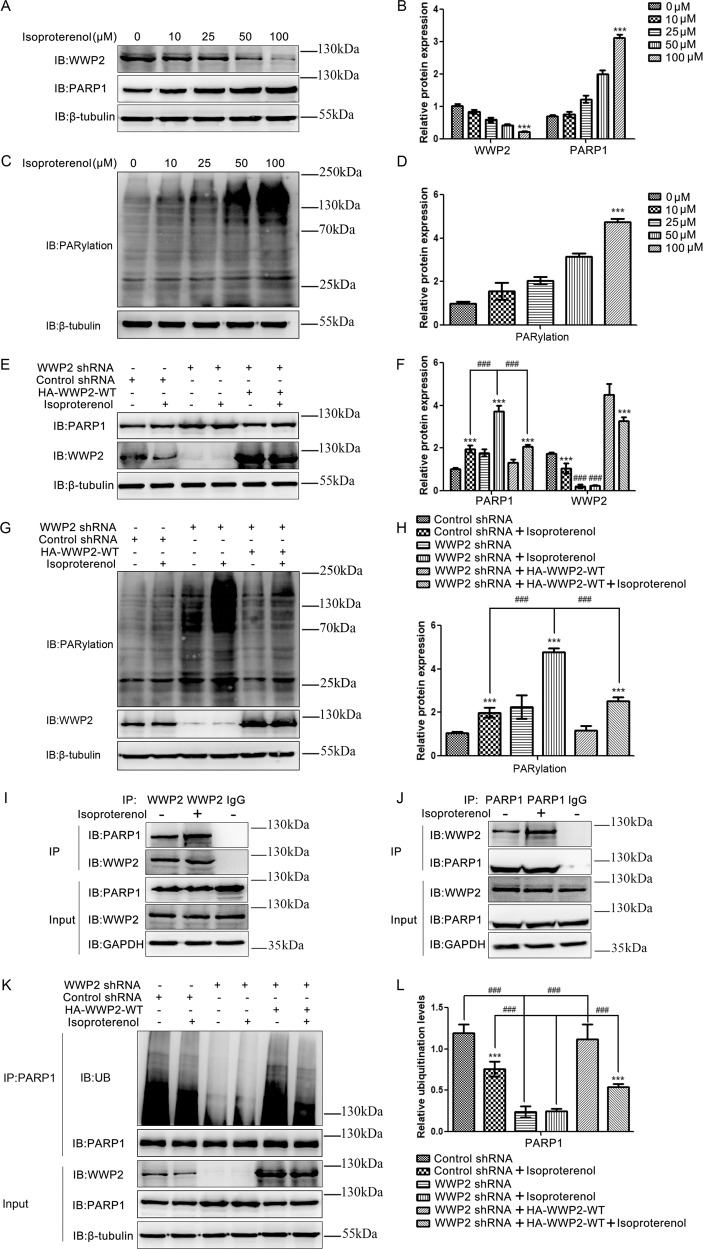
WWP2 significantly increased PARP1 ubiquitination and decreased PARP1 and PARylation levels in ISO-induced H9c2 cell model
As PARP1 and its activity, namely, PARylation, represent key factors related to injury in cardiovascular diseases, especially in ISO-induced cardiac remodeling [3–8], we next determined the role of E3 ubiquitin ligase WWP2 in the regulation of PARP1 and PARylation in the ISO-induced H9c2 cell model. First, different concentrations of ISO were used to establish the H9c2 cell model in vitro; the results showed that, with increasing ISO concentrations, WWP2 level gradually decreased, while PARP1 and PARylation levels significantly increased (Fig. 3a–d). Second, we determined the effects of WWP2 on PARP1 and PARylation upon the administration of ISO in H9c2 cells. The results showed that the expression of PARP1 and PARylation were markedly increased in ISO-induced H9c2 cells with stable WWP2 knockdown, while they were significantly decreased after the expression of exogenous WWP2 in H9c2 cells with stable WWP2 knockdown (Fig. 3e–h). Third, given the physical interaction of WWP2 with PARP1, we assessed whether ISO might affect the interaction between WWP2 and PARP1 in H9c2 cells. As expected, stimulation with ISO resulted in the enhancement of this interaction in H9c2 cells (Fig. 3i, j). Finally, we explored the effects of WWP2 on PARP1 ubiquitination upon the administration of ISO in H9c2 cells. The results showed that the ubiquitination of PARP1 was markedly decreased in ISO-induced H9c2 cells with stable WWP2 knockdown, while it was significantly increased after the expression of exogenous WWP2 in H9c2 cells with stable WWP2 knockdown (Fig. 3k, l). Taking these findings together, the above in vitro experiments confirmed that WWP2 increased PARP1 ubiquitination and decreased PAPR1 and PARylation levels in the ISO-induced H9c2 cell model.
Fig. 3. WWP2 significantly increased PARP1 ubiquitination and decreased PARP1 and PARylation levels in the ISO-induced H9c2 cell model.
a, c Western blotting was performed to examine the expression of WWP2 and PARP1, and PARylation with increasing concentration of ISO in H9c2 cells. b, d Data are shown as mean ± SD (***P < 0.001, one-way ANOVA with Bonferroni post hoc testing). e, g Western blotting was performed to examine the expression of PARP1 and PARylation after WWP2 knockdown and reexpression in ISO-induced H9c2 cells. f, h Data are shown as mean ± SD (***P < 0.001, unpaired Student’s t test; ###P < 0.001, two-way ANOVA with Bonferroni post hoc testing). i, j Coimmunoprecipitation was performed to determine the interaction between endogenous WWP2 and PARP1 with or without ISO treatment in H9c2 cells. k Coimmunoprecipitation was performed to examine the ubiquitination levels of PARP1 after WWP2 knockdown and reexpression in ISO-induced H9c2 cells. l Data are shown as mean ± SD (***P < 0.001, unpaired Student’s t test; ###P < 0.001, two-way ANOVA with Bonferroni post hoc testing).
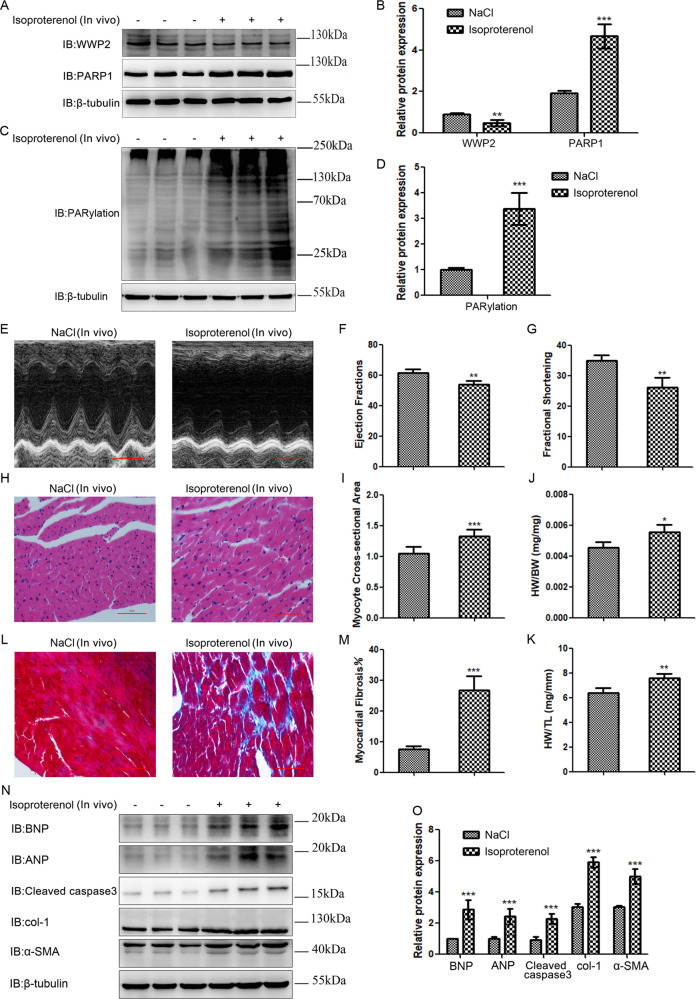
WWP2 level significantly decreased, while PARP1 and PARylation levels markedly increased in ISO-induced cardiac remodeling in mice
The elevated expression of PARP1 and increased PARP1 activity, namely, PARylation, have been observed in cardiac remodeling, leading to extreme energy consumption by myocardial cells, triggering injury of these cells [3–8, 20–23]. Therefore, to clarify the expression of WWP2 in ISO-induced cardiac remodeling in mice, the animals were administered ISO (60 mg/kg/day) or NaCl for 14 days to induce cardiac remodeling. The results showed that ISO significantly decreased the expression of WWP2 and markedly increased the expression of PARP1 and PARylation (Fig. 4a–d), with the occurrence of ISO-induced heart failure (Fig. 4e–g, n, o), cardiac hypertrophy (Fig. 4h–k, n, o), and myocardial fibrosis (Fig. 4l–o) in mice.
Fig. 4. WWP2 level significantly decreased, while PARP1 and PARylation levels markedly increased in ISO-induced cardiac remodeling in mice.
a, c In C57BL/6 mice following 2 weeks of ISO or NaCl infusion in vivo, western blotting was carried out to assess the expression of WWP2 and PARP1, and PARylation. b, d Data are shown as mean ± SD (each group of mice, n = 6; **P < 0.01, ***P < 0.001, unpaired Student’s t test). e Echocardiography (scale bar: 2 mm), (f) ejection fraction, and (g) fractional shortening were used to assess heart failure. h H&E staining (scale bar: 50 µm), (i) relative myocyte cross-sectional area, (j) heart weight to body weight ratio, and (k) heart weight to tibia length ratio were used to assess myocardial hypertrophy. l Masson trichrome staining (scale bar: 50 µm) and (m) fibrotic area were used to assess myocardial fibrosis. Data (e–m) are shown as mean ± SD (each group of mice, n = 6; *P < 0.05, **P < 0.01, ***P < 0.001, unpaired Student’s t test). n Western blotting was carried out to assess the expression of markers of myocardial hypertrophy, heart failure, and myocardial fibrosis (BNP, ANP, cleaved caspase3, col-1, and α-SMA). o Data are shown as mean ± SD (each group of mice, n = 6; ***P < 0.001, unpaired Student’s t test).
Myocardium-specific WWP2 knockout significantly aggravated ISO-induced heart failure by decreasing PARP1 ubiquitination and increasing PARP1 and PARylation levels in mice
Next, to investigate the role of E3 ubiquitin ligase WWP2 in the regulation of PARP1 and PARP1 activity, and the impact of this regulatory process on cardiac remodeling in vivo, a mouse model with the specific deletion of WWP2 in cardiomyocytes was used to test the role of WWP2 in regulating PARP1 and PARylation during ISO-induced cardiac remodeling. Immunofluorescence and western blotting were used to prove the efficient myocardium-specific silencing of WWP2 in MycCre+;WWP2Fl/Fl mice compared with that in the MycCre−;WWP2Fl/Fl group (Fig. 5a–d). Furthermore, myocardium-specific WWP2 knockout (MycCre+;WWP2Fl/Fl) significantly increased ISO-induced PARP1 and PARylation levels in mice (Fig. 5c–f). In addition, coimmunoprecipitation and western blotting results showed that PARP1 interacted with WWP2 in myocardial tissue of MycCre−;WWP2Fl/Fl mice and that this interaction was enhanced upon treatment with ISO, but not in the MycCre+;WWP2Fl/Fl group (Fig. 5g). And myocardium-specific WWP2 knockout (MycCre+;WWP2Fl/Fl) significantly decreased ISO-induced PARP1 ubiquitination in mice (Fig. 5h).
We further investigated the impact of this regulatory process on cardiac remodeling in vivo. The echocardiography results showed that ISO-induced heart failure in MycCre+;WWP2Fl/Fl mice was significantly aggravated compared with that in MycCre−;WWP2Fl/Fl mice (Fig. 5i). Meanwhile, compared with those in MycCre−;WWP2Fl/Fl mice, LVEF (EF%) and FS% of ISO-induced MycCre+;WWP2Fl/Fl mice were significantly decreased (P < 0.01 and P < 0.001, respectively) (Fig. 5j). In addition, the myocardium-specific knockout of WWP2 remarkably exacerbated the elevation of heart rate due to ISO in mice (Supplementary Fig. 2).
Myocardium-specific WWP2 knockout markedly aggravated ISO-induced myocardial hypertrophy and fibrosis in mice
Myocardial hypertrophy and fibrosis are two critical phenotypes for cardiac remodeling induced by ISO-promoted overactivation of PARP1 in mice [3–8, 20–23]. Therefore, we further explored the role of WWP2 in ISO-induced myocardial hypertrophy and fibrosis in mice via the regulation of PARP1.
The results showed that the myocardial cross-sectional area upon ISO administration was significantly increased in MycCre+;WWP2Fl/Fl mice (Fig. 6a, b), with marked elevations of heart weight to body weight ratio and heart weight to tibial length ratio (Fig. 6c, d), compared with those in MycCre−;WWP2Fl/Fl mice. In addition, the ISO-induced myocardial hypertrophy and heart failure markers ANP, BNP, and cleaved caspase3 were markedly increased in MycCre+;WWP2Fl/Fl mice compared with those in MycCre−;WWP2Fl/Fl mice (Fig. 6g, h). Consistent with these results, Masson staining in ISO-induced MycCre+;WWP2Fl/Fl heart tissue showed more intense signals compared with the case in MycCre−;WWP2Fl/Fl heart samples (Fig. 6e, f). In addition, the markers of myocardial fibrosis α-SMA and collagen-I were markedly increased in the ISO-induced MycCre+;WWP2Fl/Fl group compared with those in the MycCre−;WWP2Fl/Fl group (Fig. 6i, j).
Taken together, the above results showed that WWP2 interacts with PARP1, mainly at PARP1’s BRCT region, thus degrading PARP1 through the ubiquitin–proteasome system by regulating its ubiquitination at K418 and K249, leading to ISO-induced heart failure, myocardial hypertrophy, and fibrosis, indicating that WWP2 plays a core role in ISO-induced cardiac remodeling.
Discussion
The main findings of this study are that, in a physiological state, WWP2 interacts with PARP1, mainly acting on the PARP1 BRCT domain and ubiquitinating PARP1 K249 and K418 lysine sites to degrade PARP1 through the ubiquitin–proteasome system. In the case of myocardial WWP2 deficiency, WWP2 loses its ability to interact with PARP1, and the PARP1 K249/K418 ubiquitination levels decrease, elevating PARP1 and PARylation, which finally aggravates ISO-induced cardiac remodeling.
The processing and maturation of intracellular proteins require posttranslational modifications (PTMs). PTMs adjust the form of proteins and further affect their activities, ultimately leading to functional changes [24, 25]. Ubiquitination is an extremely important part of PTMs and has a critical function in cardiac remodeling [26, 27]. Specific recognition of substrates by the E3 ubiquitin ligase is the core of ubiquitination modifications. In the present study, we found the HECT-type E3 ubiquitin ligase WWP2 is a novel regulatory factor involved in ISO-induced cardiac remodeling. Both in vivo and in vitro WWP2 expression levels were found to be significantly decreased in ISO-induced cardiac remodeling. Therefore, we generated myocardium-specific WWP2 knockout mice, which showed aggravated ISO-induced heart failure, myocardial hypertrophy, and fibrosis, indicating that WWP2 is a protective factor in ISO-induced cardiac remodeling.
WWP2 was originally described as an oncogenic factor. PTEN mutation at the tyrosine (Tyr) 155 residue causes WWP2 to directly interact with and ubiquitinate PTEN, reducing tumor cell apoptosis via the PI3K/AKT pathway and promoting tumor growth [14]. In addition, WWP2 has been shown to play a crucial role in immune regulation and embryonic differentiation [28–30]. Previous studies demonstrated that WWP2 interacts with Trif and promotes its ubiquitination by binding to K48, which activates the TLR3 pathway. Meanwhile, WWP2 knockdown results in decreased Trif ubiquitination and the enhanced expression of IFN-β [28]. Moreover, WWP2 catalyzes the polyubiquitination of Oct-4 in a dose-dependent manner by binding to lysine 63 of Oct-4 during ECC differentiation [29]. The present study revealed a novel biological function of WWP2, identifying PARP1 as its novel substrate. WWP2 specifically bound to the PARP1 BRCT domain and ubiquitination modified the PARP1 K249 and K418 sites to promote the degradation of PARP1 by the ubiquitin–proteasome system, protecting against ISO-induced cardiac remodeling in mice. This study thus provides a theoretical basis for exploring new targets of cardiac remodeling-related heart disorders.
The elevated expression of PARP1 and increased PARP1 activity, namely, PARylation, have been observed in many cardiovascular diseases [3–8]. PARP1 and PARylation represent critical injury factors and are involved in cardiomyocyte damage and cardiac remodeling caused by various factors [8, 20–23]. PARP1 is activated to generate PARylation via the recognition of damaged DNA fragments [20, 31, 32]. In conditions, such as ISO-induced cardiac remodeling and myocardial ischemia–reperfusion injury, cardiomyocytes produce reactive oxygen and reactive nitrogen species. These reactive substances cause oxidative DNA damage and thus activate PARP1 to generate PARylation, leading to extensive energy loss and finally causing cardiomyocyte damage as well as apoptosis [4, 31, 32]. Mice with knockout of PARP1 show significant resistance to myocardial reperfusion injury and cardiomyocyte apoptosis [22, 23]. In mild heart failure with the retention of mitochondrial function, PARP1 activation generates PARylation, leading to the release of apoptosis-inducing factor from the mitochondria, which, in turn, trigger the apoptosis of cardiomyocytes. In advanced heart failure, excessive PARP1 activation generates PARylation, leading to severe decreases in the levels of myocardial nicotinamide adenine dinucleotide (NAD+) and adenosine triphosphate, triggering the necrosis of myocardial cells [5, 23]. The present study demonstrated that WWP2-mediated PARP1 ubiquitination directly reduced the expression of PARP1 and PARylation, thereby affecting ISO-induced cardiac remodeling. Moreover, as a substrate of WWP2, PARP1, and PARP1 activity further reflected the effects of WWP2 in ISO-induced heart failure, myocardial hypertrophy, and fibrosis in mice.
PARP1, as a key facilitator in ISO-induced cardiac remodeling, is regulated by PTM. Tyr kinase c-Met phosphorylates Tyr at position 907 in PARP1, which enhances the resistance of cells to PARP1 inhibitors. Moreover, the combined application of c-Met and PARP1 inhibitors was shown to effectively inhibit PARP1 activity [33]. In addition, histone acetyltransferase P300 acetylates PARP1 through poly(ADP-ribosyl)ated K498, K521, and K524, affecting PARP1 transactivation [34]. This study found that PARP1 was modified by E3 ligase WWP2-regulated ubiquitination. Notably, we also identified that WWP2 mainly interacts with the BRCT domain of PARP1. PARP1 consists of at least four functional domains, namely, the N-terminal DNA binding (Zinc 1 and Zinc 2), self-modifying (BRCT), PARP A-helical, and C-terminal catalytic (PARP catalytic) domains. The BRCT domain was first described in the breast cancer-associated protein BRCA1 and later found to be associated with the repair of eukaryotic DNA [35]. We also demonstrated that K249 and K418 are important in PARP1 ubiquitination by WWP2, revealing a novel signaling pathway (WWP2/PARP1 axis) that regulates ISO-induced cardiac remodeling. This reflects the association of protein ubiquitination with cardiac remodeling and provides deeper insights into the pathogenetic mechanisms of cardiac remodeling.
This study has some limitations. For example, we have not definitively proven that it is indeed the downregulation of PARP1 (and not the downregulation of any other potential substrate of WWP2) that prevents ISO-induced cardiac remodeling. It is thus important to further investigate whether other potential substrates of WWP2 are involved in cardiac remodeling. In addition, the roles of WWP2 in other cardiovascular diseases and whether WWP2 functions in patients with cardiac remodeling should be further assessed.
Conclusion
WWP2 interacts with PARP1 and degrades it through the ubiquitin–proteasome system by regulating its K418 and K249 ubiquitination, leading to an impact on ISO-induced cardiac remodeling. These findings reveal a novel WWP2–PARP1 signal transduction pathway involved in controlling cardiac remodeling and may provide a basis for exploring new therapeutic strategies of cardiac remodeling-related heart disorders.
Supplementary information
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos 81900355, 81670231, 81900372, and 81970211).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by M. Piacentini
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Liu Cao, Email: lcao@cmu.edu.cn.
Yingxian Sun, Email: yxsun@cmu.edu.cn.
Supplementary information
The online version of this article (10.1038/s41418-020-0523-2) contains supplementary material, which is available to authorized users.
References
- 1.Jessup M, Brozena S. Heart failure. N Engl J Med. 2003;348:2007–18. doi: 10.1056/NEJMra021498. [DOI] [PubMed] [Google Scholar]
- 2.Aimo A, Gaggin HK, Barison A, Emdin M, Januzzi JL., Jr. Imaging, biomarker, and clinical predictors of cardiac remodeling in heart failure with reduced ejection fraction. JACC Heart Fail. 2019;7:782–94. doi: 10.1016/j.jchf.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Henning RJ, Bourgeois M, Harbison RD. Poly(ADP-ribose) polymerase (PARP) and PARP inhibitors: mechanisms of action and role in cardiovascular disorders. Cardiovasc Toxicol. 2018;18:493–506. doi: 10.1007/s12012-018-9462-2. [DOI] [PubMed] [Google Scholar]
- 4.Wang C, Xu W, Zhang Y, Zhang F, Huang K. PARP1 promote autophagy in cardiomyocytes via modulating FoxO3a transcription. Cell Death Dis. 2018;9:1047. doi: 10.1038/s41419-018-1108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wen JJ, Yin YW, Garg NJ. PARP1 depletion improves mitochondrial and heart function in Chagas disease: effects on POLG dependent mtDNA maintenance. PLoS Pathog. 2018;14:e1007065. doi: 10.1371/journal.ppat.1007065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pillai JB, Russell HM, Raman J, Jeevanandam V, Gupta MP. Increased expression of poly(ADP-ribose) polymerase-1 contributes to caspase-independent myocyte cell death during heart failure. Am J Physiol Heart Circ Physiol. 2005;288:H486–96. doi: 10.1152/ajpheart.00437.2004. [DOI] [PubMed] [Google Scholar]
- 7.Feng GS, Zhu CG, Li ZM, Wang PX, Huang Y, Liu M, et al. Synthesis of the novel PARP-1 inhibitor AG-690/11026014 and its protective effects on angiotensin II-induced mouse cardiac remodeling. Acta Pharm Sin. 2017;38:638–50. doi: 10.1038/aps.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J, Zhang R, Hong H, Yang Z, Sun D, Sun S, et al. The poly(ADP-ribosyl)ation of FoxO3 mediated by PARP1 participates in isoproterenol-induced cardiac hypertrophy. Biochim Biophys Acta. 2016;1863:3027–39. doi: 10.1016/j.bbamcr.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W, Song M, Qu J, Liu GH. Epigenetic modifications in cardiovascular aging and diseases. Circ Res. 2018;123:773–86. doi: 10.1161/CIRCRESAHA.118.312497. [DOI] [PubMed] [Google Scholar]
- 10.Nishida K, Yamaguchi O, Otsu K. Degradation systems in heart failure. J Mol Cell Cardiol. 2015;84:212–22. doi: 10.1016/j.yjmcc.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Morreale FE, Walden H. Types of ubiquitin ligases. Cell. 2016;165:248. doi: 10.1016/j.cell.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Zou W, Chen X, Shim JH, Huang Z, Brady N, Hu D, et al. The E3 ubiquitin ligase Wwp2 regulates craniofacial development through mono-ubiquitylation of goosecoid. Nat Cell Biol. 2011;13:59–65. doi: 10.1038/ncb2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, Jiang H, Xu W, Li X, Dempsey DR, Zhang X, et al. A tunable brake for HECT ubiquitin ligases. Mol Cell. 2017;66:345–57. doi: 10.1016/j.molcel.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maddika S, Kavela S, Rani N, Palicharla VR, Pokorny JL, Sarkaria JN, et al. WWP2 is an E3 ubiquitin ligase for PTEN. Nat Cell Biol. 2011;13:728–33. doi: 10.1038/ncb2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen W, Jiang X, Luo Z. WWP2: a multifunctional ubiquitin ligase gene. Pathol Oncol Res. 2014;20:799–803. doi: 10.1007/s12253-014-9838-y. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Zhang Z, Wang B, Zhang J, Zhao Y, Jin Y. Wwp2-mediated ubiquitination of the RNA polymerase II large subunit in mouse embryonic pluripotent stem cells. Mol Cell Biol. 2007;27:5296–305. doi: 10.1128/MCB.01667-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishizue N, Niwano S, Niwano H, Oikawa J, Nakamura H, Hashikata T, et al. Linagliptin suppresses electrical and structural remodeling in the isoproterenol induced myocardial injury model. Int Heart J. 2019;60:411–8. doi: 10.1536/ihj.18-226. [DOI] [PubMed] [Google Scholar]
- 18.Hu H, Jiang M, Cao Y, Zhang Z, Jiang B, Tian F, et al. HuR regulates phospholamban expression in isoproterenol-induced cardiac remodeling. Cardiovasc Res. 2019;cvz205. 10.1093/cvr/cvz205. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 19.Goonasekera SA, Hammer K, Auger-Messier M, Bodi I, Chen X, Zhang H, et al. Decreased cardiac L-type Ca2+ channel activity induces hypertrophy and heart failure in mice. J Clin Invest. 2012;122:280–90. doi: 10.1172/JCI58227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pacher P, Szabó C. Role of poly(ADP-ribose) polymerase 1 (PARP-1) in cardiovascular diseases: the therapeutic potential of PARP inhibitors. Cardiovasc Drug Rev. 2007;25:235–60. doi: 10.1111/j.1527-3466.2007.00018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jagtap P, Szabó C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Disco. 2005;4:421–40. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 22.Yamazaki K, Tanaka S, Sakata R, Miwa S, Oriyanhan W, Takaba K, et al. Protective effect of cardioplegia with poly (ADP-ribose) polymerase-1 inhibitor against myocardial ischemia-reperfusion injury: in vitro study of isolated rat heart model. J Enzym Inhib Med Chem. 2013;28:143–7. doi: 10.3109/14756366.2011.642373. [DOI] [PubMed] [Google Scholar]
- 23.Zingarelli B, Hake PW, O’Connor M, Denenberg A, Wong HR, Kong S, et al. Differential regulation of activator protein-1 and heat shock factor-1 in myocardial ischemia and reperfusion injury: role of poly(ADP-ribose) polymerase-1. Am J Physiol Heart Circ Physiol. 2004;286:H1408–15. doi: 10.1152/ajpheart.00953.2003. [DOI] [PubMed] [Google Scholar]
- 24.Zee BM, Levin RS, DiMaggio PA, Garcia BA. Global turnover of histone post-translational modifications and variants in human cells. Epigenetics Chromatin. 2010;3:22. doi: 10.1186/1756-8935-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liddy KA, White MY, Cordwell SJ. Functional decorations: post-translational modifications and heart disease delineated by targeted proteomics. Genome Med. 2013;5:20. doi: 10.1186/gm424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fessart D, Martin-Negrier ML, Claverol S, Thiolat ML, Crevel H, Toussaint C, et al. Proteomic remodeling of proteasome in right heart failure. J Mol Cell Cardiol. 2014;66:41–52. doi: 10.1016/j.yjmcc.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 27.Barac YD, Emrich F, Krutzwakd-Josefson E, Schrepfer S, Sampaio LC, Willerson JT, et al. The ubiquitin-proteasome system: a potential therapeutic target for heart failure. J Heart Lung Transpl. 2017;36:708–14. doi: 10.1016/j.healun.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Liao B, Wang S, Yan B, Jin Y, Shu HB, et al. E3 ligase WWP2 negatively regulates TLR3-mediated innate immune response by targeting TRIF for ubiquitination and degradation. Proc Natl Acad Sci USA. 2013;110:5115–20. doi: 10.1073/pnas.1220271110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao B, Jin Y. Wwp2 mediates Oct4 ubiquitination and its own auto-ubiquitination in a dosage-dependent manner. Cell Res. 2010;20:332–44. doi: 10.1038/cr.2009.136. [DOI] [PubMed] [Google Scholar]
- 30.Aki D, Li H, Zhang W, Zheng M, Elly C, Lee JH, et al. The E3 ligases Itch and WWP2 cooperate to limit TH2 differentiation by enhancing signaling through the TCR. Nat Immunol. 2018;19:766–75. doi: 10.1038/s41590-018-0137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci USA. 2006;103:18308–13. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Virág L, Robaszkiewicz A, Rodriguez-Vargas JM, Oliver FJ. Poly(ADP-ribose) signaling in cell death. Mol Asp Med. 2013;34:1153–67. doi: 10.1016/j.mam.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 33.Du Y, Yamaguchi H, Wei Y, Hsu JL, Wang HL, Hsu YH, et al. Blocking c-Met-mediated PARP1 phosphorylation enhances anti-tumor effects of PARP inhibitors. Nat Med. 2016;22:194–201. doi: 10.1038/nm.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, et al. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J Biol Chem. 2005;280:40450–64. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- 35.Szabo C, Pacher P, Swanson RA. Novel modulators of poly(ADP-ribose) polymerase. Trends Pharm Sci. 2006;27:626–30. doi: 10.1016/j.tips.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.