Abstract
Generative adversarial networks (GANs), first published in 2014, are among the most important concepts in modern artificial intelligence (AI). Bridging deep learning and game theory, GANs are used to generate or “imagine” new objects with desired properties. Since 2016, multiple GANs with reinforcement learning (RL) have been successfully applied in pharmacology for de novo molecular design. Those techniques aim at a more efficient use of the data and a better exploration of the chemical space. We review recent advances for the generation of novel molecules with desired properties with a focus on the applications of GANs, RL, and related techniques. We also discuss the current limitations and challenges in the new growing field of generative chemistry.
Keywords: Generative adversarial networks, Reinforcement learning, De novo molecular design, Artificial intelligence, Drug discovery
Introduction
Since the 1960s, computational approaches for drug design and discovery are continuously, including machine learning (ML), have been developed and applied in many forms to the design of compounds. Following the hypothesis that the physiological response of a compound is merely a function of its chemical constitution, these methods were initially designed to predict the properties of compounds without the need to synthesize these compounds. Going a step further, current methods for de novo design aim at creating novel (previously unknown) chemical entities with desired properties (e.g., pharmacological activity) from scratch. This design concept comprises molecule generation, molecule scoring and molecule optimization. For drug discovery and medicinal chemistry specifically, this involves tasks in drug target and lead compound identification, drug design optimization against multiple property profiles of interest, and finally identifying synthetic routes to realize the composition of matter. Today, de novo design tools have the capabilities to provide molecular structures which are often readily synthesizable within a few reaction steps.
Standard de novo design methods often rely on explicit chemical knowledge accumulation in the form of synthesis rules or basic physical models. For this reason, they are limited by our incomplete understanding of how molecules interact because scientists cannot tell conventional software how to find insights in data when they do not themselves know what elements of the data are most important and how they relate to one another. This explains in part why, despite the rapid advances in recent decades in high-throughput screening technologies and the inventiveness of medicinal and synthetic chemists, only a small fraction of the druglike space has been investigated in the search for new therapeutic compounds. The situation is made more difficult by the fact that our fundamental understanding of human disease and its high degree of complexity, which can only be comprehended by combining the information from multiple data types, is still far from complete, and this makes it difficult to decide how to best intervene therapeutically. These factors render decision making in medicinal chemistry and drug design exceptionally demanding.
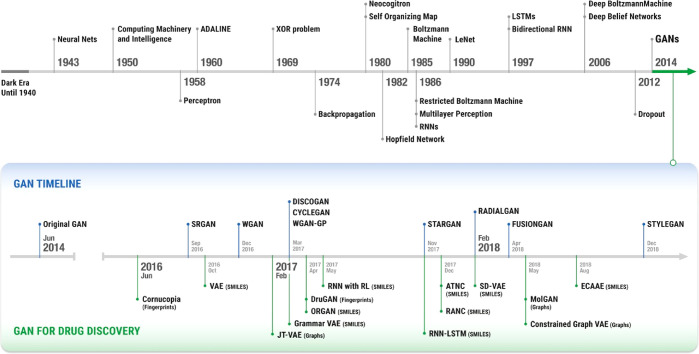
Researchers started to use deep learning (DL) to develop methods for de novo drug design with the goal to overcome our lack of understanding of the disease mechanisms and to more efficiently explore the largely unexplored chemical space. DL started to attract attention recently after DL algorithms encountered major successes when applied in image and speech recognition. DL methods now outperform humans for those tasks, and the capabilities of DL quickly started to be investigated in the field of biomedical sciences1 for addressing problems including biomarkers and new target identification, improvement of a patient’s prognosis by reducing error rate, and selection of the most appropriate treatment by predicting treatment outcome.2 DL-based algorithms often referred to as AI methods in the literature, have many variants, are versatile and flexible, and can treat information from the scientific literature and databases as well as patient-level data. What distinguishes DL from other ML methods and makes it so attractive is its ability to identify relevant patterns within complex, nonlinear data in an automatic fashion without the need for manual feature engineering. Among the DL architectures proposed recently, the generative adversarial networks (GANs) constitute a major breakthrough. GAN, suggested in 2014,3,4 takes its origin at the intersection of DL and game theory. GANs are able to generate new objects with desired properties, and for that reason they are referred to as a form of AI imagination. During the last 5 years, GANs and variants using reinforcement learning (RL) have been deployed in chemistry and pharmacology (Supporting Information Table 1) and the design and use of GAN and RL models for the generation of novel molecules with specific desired properties (Figure 1) has been successful with major milestones recently achieved.
Figure 1.
Timeline summarizing the development of ML, DL, and the learning concepts including GAN and RL. Those technologies were critical for the emergence of generative chemistry.
Recently, case studies using standard automated de novo drug design methods for which designed compounds had their activities verified by synthesis and assay were reviewed.5 These novel potent compounds were obtained with de novo design methods integrated into a larger multidisciplinary pipeline. These de novo design methods showed promising results, although generated compounds were sometimes a long way from a candidate drug compound and additional optimization steps were still required. In some cases, generated compounds elicited high toxicity which prevented further development. While the authors mainly focused on case studies using standard de novo design computational methods, the present work proposes to focus on the DL-based methods for de novo design. This field is relatively new, and the performance of the first models was evaluated by assessing at what extent the basic structural features of the generated molecules matched the average initial distribution of the training sets. Over time, more precise metrics were included to better assess the properties and druggability of the generated compounds. Models were developed to generate molecules eliciting activity against a specific target. The research to build DL-based models for de novo drug design is also concerned with finding the best approach to encode the molecular structures. Recently, several DL-based models for de novo design whose results had undergone in vitro and/or in vivo validations were published. In what follows, we recall the technical caveats of computer-aided drug design. When presenting the progress made in the development of DL-based methods for the design of therapeutic compounds, we summarize the evolution of the main classes of DL models for de novo design and provide examples of application.
Overview of Computer-Aided Drug Design
In the last 40 years, the discipline of chemoinformatics has created many computational drug design tools, ranging from classical quantitative structure–activity relationship (QSAR) techniques6 to more recent advances in matched molecular pairs7 and free-energy perturbation.8 QSARs and their relations were proposed by Hansch et al. in 1962,9 and since this time they have been an active area of research. QSAR is widely used in the drug design process to analyze the correlations between molecular structure and biological activity. A QSAR method is defined as an ML application and/or statistical method to the problem of finding empirical relationships where the biological activity (or any property of interest) of molecules is expressed in terms of function of calculated molecular descriptors of compounds. The step of descriptor generation in QSAR modeling is a nondifferentiable transformation which does not allow a straightforward inverse mapping from the descriptor space back to molecules, although a few approaches for such mapping (inverse QSAR) have been proposed.10De novo design aims to generate biologically active molecules from scratch. In that case, the chemotypes obtained from the process represent new chemical entities (NCEs) that not only provide new insight into the atomic scale of molecular–receptor interactions but are also patentable.11 Structure-based de novo drug design (SBDND) and ligand-based de novo drug design (LBDND) are the two most commonly used computational approaches.12 SBDND requires an X-ray structure or reasonably valid homology models of the desired targets, and ligands are generated by joining atoms or fragments so that the resulting product fits into the desired pocket.13 Many approaches are used to generate novel ligand structures.14 Atom-based methods add atoms one by one based on the receptor site, whereas fragment-based approaches search fragment libraries and use the fragments to build new molecules through growth and linkage. Pharmacophore-based methods, another widely used model, generate molecules that are structurally similar or complementary to the pharmacophore retrieved from defined pockets. The fitness of the generated molecules is then evaluated by physicochemical filters or by a scoring function based on molecular docking and molecular dynamics.15−17 When there are no high-resolution X-ray structures available, the ligand-based design can be used as an alternative approach. LBDND aims at creating NCEs by identifying fragments with pharmacophoric features similar to the template scaffolds and then replacing the template with new fragments. This fragment-based molecular design follows the concept of scaffold-hopping. According to this concept, the newly designed molecules should possess property profiles similar to or superior to that of the template but should contain different scaffolds.18 NCEs are assembled by either commercially available building blocks or by building blocks dissected by the pseudo-retrosynthesis of known drugs.19,20 Some successful applications of compound optimization for desired selectivity and pharmacological profiles by coupling evolutionary algorithms with proper QSAR models or similarity indices as fitness functions have been reported.21−24
Deep-Learning for Drug Discovery: Searching for Unexplored Chemical Space
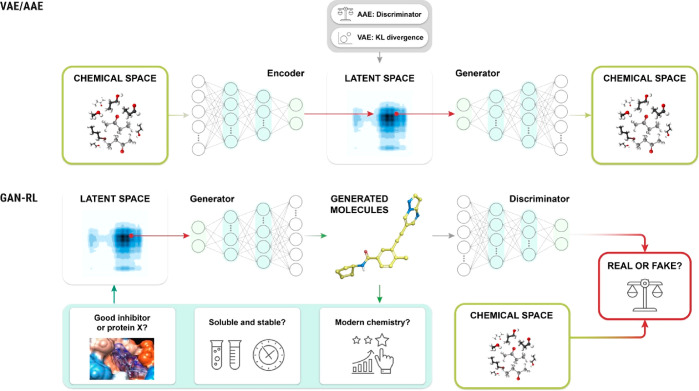
The attractive feature of DL approaches is the implicit chemical knowledge generation by pattern recognition in structural molecular data. This is an important difference with standard de novo design methods which rely on the accumulation of explicit chemical knowledge in the form of synthesis rules or basic physical models. Imbued with the ability to derive their own insights into which data elements matter, DL-based programs can extract better predictions for a wider range of variables. Considering the almost unlimited number of chemical structures that can be generated de novo, conventional computational drug design approaches tend to include limited numbers of fragments and/or to employ sophisticated search strategies to sample hit compounds from a predefined area of the chemical space. It has been estimated that 1060 druglike molecules could be synthetically accessible.25 Chemists have to select and examine molecules from this large space to find molecules that are active toward a biological target. Recent advances in ML and AI techniques can help scientists to more efficiently explore the whole druglike chemical space.26−29 Generative chemistry based on DL models learns the nonlinear probability distribution between molecular chemical structures and their biological and pharmacological properties from massive data sets and then perform in silico design of de novo molecules with desired properties.30−32 Architectures such as recurrent neural networks (RNNs)33 and autoencoders including variational autoencoders (VAEs),34 adversarial encoders (AAEs),35 and GANs3 (Figure 2) can serve as powerful tools for de novo molecular design. Detailed technical reviews of each DL model are available elsewhere.36
Figure 2.
Schematic representation of architectures used in generative chemistry. The VAE/AAE model (top) and the GAN-RL model (bottom) have been successful in generating molecular structures of compounds with desired sets of properties.
Recurrent Neural Networks
RNNs are neural networks with an internal memory that are suitable ML algorithms for sequential data. Segler et al. trained an RNN based on the long short-term memory (LSTM) architecture to generate molecular structures.37 The model was first trained on a large set of molecules to learn the grammar of the simplified molecular input line-entry system (SMILES) and then fine-tuned on a smaller set of molecules active against desired targets where it adopted transfer learning concepts. It was shown that the model could create novel molecules with predicted activities targeting 5-HT2A, the malaria parasite, as well as Staphylococcus aureus. The results demonstrated that LSTM-generative models could be coupled with a docking program to iteratively generate active molecules without the need for a set of known ligands.38 Other LSTM neural networks trained on druglike molecules were able to generate novel molecules occupying the same area of chemical space as the known bioactive molecules with potential activity against various targets.39,40 Merk et al. trained an RNN-LSTM model on the SMILES-encoded ChEMBL compound database. The model was fine-tuned with active compounds against retinoid X receptors (RXRs) and peroxisome proliferator-activated receptors (PPARs) for the de novo design of molecules for these receptors.41 After prioritizing the generated molecules according to their similarity in shapes and charges to known ligands and using predictive binding results from machine-learning models, the authors synthesized and tested five compounds and found that four possessed low-micromolar levels of activity and suitable selectivity profiles.
Variational Autoencoders
Gomez-Bombarelli et al. proposed a VAE generative model that was trained on SMILES representations of publicly available chemical structures.42 The model encoded molecules into low-dimensional vectors in latent space as continuous and smooth probability distributions, and the decoder of VAEs converted these continuous vectors back to discrete molecular representations. The continuous representations of molecules allow sampling of the chemical space stepwise, leading to the successful optimization of molecules with desired druglike properties. The model was trained on the ZINC and QM9 databases, and the authors compared the water–octanol partition coefficient (log P), the synthetic accessibility score (SAS), and the quantitative estimation of drug-likeness (QED) of the generated molecules with those in the training sets. The VAE-generated molecules showed chemical properties similar to the original data set but with diverse chemical scaffolds. Moreover, when applied to optimize the QED and SAS scores of molecules, the model outperformed the genetic algorithm, which required manual specification of mutation rates and rules. Conditional variational autoencoder (CVAE) incorporated molecular properties directly into both the encoder and decoder that allowed one to handle the structures and the properties independently, which is particularly useful in optimizing a given molecule toward a certain property by marginal structure modification.43 The author adopted CVAE to generate druglike molecules and to concurrently optimize molecules toward a desired molecular weight, log P, number of hydrogen bond donors (HBDs), number of hydrogen acceptors (HBAs), and topological polar surface area (TPSA). The model was tested to generate molecules with similar properties to those of aspirin and Tamiflu. The property values of the generated molecules were found to be within an error range of 10% compared to the properties of the two molecules used as references. Junction tree variational autoencoder (JT-VAE), another VAE-based generative chemistry model, views molecules as graphs and generates new molecules by assembling building blocks derived from subgraphs of the molecules.44 It has been shown that JT-VAE could successfully generate valid molecules with the desired log P and optimize the log P value for the template molecules.
Generative Adversarial Networks
Insilico Medicine was the first to publish a deep adversarial model, AAE, trained on publicly available biological and chemical data including 6252 compounds profiled on the MCF-7 cell line for new compound generation.45 The system takes a vector of binary fingerprints and the molecule’s cell inhibition concentration in log scale as inputs, and it outputs an inhibition concentration and a vector consisting of probabilities assigned to each bit of the fingerprint. The generated vectors were screened against 72 million compounds from PubChem, and the maximum likelihood function was used to select top hits for each of the vectors. Sixty nine compounds were identified as belonging to various chemical classes with profiled anticancer activities. This work was followed by a second AAE model, the drug generative adversarial network (druGAN).46 Compared to the first model, druGAN improved the performance of the discriminator by introducing an additional hyperparameter to improve the training. DruGAN also uses fingerprints to represent molecules and adopts Tanimoto similarity to measure the similarity between the generated molecules and the original data. Overall, druGAN exhibited higher adjustability in generating molecular fingerprints and had a better capacity to process very large data sets of molecules. The study included a comparison between druGAN and a VAE model. Both the AAE and VAE models were shown to perform very well depending on the kind of task to be solved. Consequently, both VAE and AAE can be considered valuable tools for use with fingerprints and with other molecular structure representations. Another study constructed various types of generative adversarial autoencoder models and applied them to inverse QSAR to generate novel chemical structures.47 By implementing Bayesian optimization to search for molecules in the latent space of the models with desired properties predicted by a QSAR model, novel structures with predicted activities were revealed, indicating the usefulness of the generative models in tackling drug discovery problems. Recently, Polykovskiy et al. proposed the entangled conditional adversarial autoencoder (ECAAE), which was conditioned on generating selective molecules for JAK3.48 The model demonstrated higher performance in the generation of novel chemical structures given complex conditions, such as activity against a specific protein, solubility, and ease of synthesis. The 300 000 conditioned compounds generated by this model were screened with a series of filters, including medicinal chemistry filters, log P, SAS, and docking as well as MD simulation, to identify 100 of the most promising hits. The most promising molecule was chosen by experienced medicinal chemists and was synthesized and tested in vitro against JAK3, JAK2, B-Raf, and c-Raf. The molecule was shown to have low-micromolar activity against JAK3 (IC50 = 6.73 μM) but was inactive against JAK2 (IC50 = 17.58 mM), B-Raf (IC50 = 85.55 μM), and c-Raf (IC50 = 64.86 μM). Another model called generative tensorial reinforcement learning (GENTRL) was used to generate in vivo active DDR1 and DDR2 inhibitors.49 DDR1 and DDR2 inhibitors with different property and selectivity profiles were assayed in vitro, followed by in vivo mouse experiments that validated the pharmacokinetics of the inhibitors. This study illustrates the effectiveness of this generative approach by showing that the molecules can be generated in a time and cost efficient manner and that they can be synthesized, are active in vitro, are metabolically stable, and show in vivo activity in disease-relevant models. However, it was pointed out that the GENTRL-generated molecule was similar to ponatinib and that the molecules described still required optimization.50 However, considering that the design/make/test/evaluate workflow in drug discovery can only afford a limited number of cycles because each step is resource- and time-consuming, this case study represents an important milestone for generative chemistry because it showed how AI methods can be integrated within the drug design cycle and contribute to its optimization.
Optimization with Deep Generative Models Using Reinforcement Learning
RL51 is used to fine-tune generative neural networks by rewarding or penalizing generative behaviors. Olivecrina et al.52 proposed a method to tune a sequence-based generative model for molecular de novo design that through augmented episodic likelihood can learn to generate structures with certain specified desirable properties. The RNN-based model was pretrained on the ChEMBL database to generate molecular structures similar to that of the drug celecoxib and to generate predicted actives against dopamine receptor type 2 (DRD2). The similarity was evaluated using a variant of the Jaccard index. When trained toward generating predicted actives against DRD2, the model generated structures of which more than 95% were predicted to be active and could recover test set actives even in the case where they were not included in either the activity model or the Prior. When tasked to generate structures similar to that of the drug celecoxib, the model could locate the intended region of chemical space which was not part of the Prior even when all analogues of celecoxib were removed from it. However, none of these predictions were tested through in vitro or in vivo experiments. A comparable deep RL framework integrating generative models trained with a stack-augmented memory network and predictive QSAR models was also proposed.10 The model was able to generate chemical libraries for creating compounds with desired structural complexity or with desired physical properties, such as melting point or hydrophobicity. The model could also be used to generate compounds with predefined inhibitory activity against desired targets. The structural properties and SAS of the generated molecules were evaluated. More than 99.5% of de novo generated molecules had SAS values below 6. SAS is scaled to be between 1 and 10. Molecules with high SAS values, typically above 6, are considered to be difficult to synthesize, whereas molecules with low SAS values are easily synthetically accessible. Therefore, despite their high novelty, most generated compounds were considered to be synthetically accessible. The advantage of this deep RL framework is that it does not rely on predefined chemical descriptors; the models are trained on chemical structures represented by SMILES strings only. Using DNNs directly on SMILES is fully differentiable, and it also enables direct mapping of properties to the SMILES sequence of characters. This distinction differentiates this approach from traditional QSAR methods and makes it simpler to both use and execute. SMILES strings were also used for QSAR model building but in most cases to derive string-based numerical descriptors. The architecture called sequence generative adversarial network (SeqGAN) combines GANs with an RL-based generator to create sequences of discrete tokens.53 Another extension of SeqGAN, called objective-reinforced generative adversarial network (ORGAN), was also proposed.54 This model added an objective-reinforced reward function for particular sequences into the SeqGAN reward loss. Considering the success of the ORGAN model, architectures based on objective functions for molecular design within the ORGAN paradigm were later developed.55 For instance, the model called objective-reinforced generative adversarial network for inverse-design chemistry (ORGANIC) used various criteria as objective filters to train the ORGAN model. The results showed how different objective reward functions made it possible to bias the generation process to generate molecules with desired user-specified properties, such as QED. Other recent architectures based on the GAN and RL paradigms include the RANC model, which used a differentiable neural computer (DNC) as a generator.56 DNC is a category of neural networks that features the addition of an explicit memory bank to increase generation capabilities. The comparisons with ORGANIC showed that RANC performed better in terms of number of unique structures and number of generated molecules that passed the medicinal chemistry filters. The molecules generated by this model did not undergo in vitro and in vivo assessment. The focus was to ensure that the model was capable of generating molecules which matched the distributions of chemical features/descriptors. Another similar model called adversarial threshold neural computer (ATNC) was also published.57 ATNC also used DNC as a generator but included a supplementary computational unit called the adversarial threshold (AT), which acted as a filter between the agent (generator) and the environment (discriminator and objective reward functions). ATNC also included a new objective reward function, called internal diversity clustering (IDC), to improve the diversity of generated molecules. ATNC generated 72% of valid and 77% of unique SMILES. Moreover, the druglikeness properties of the molecules were estimated using chemical descriptors. The performances were compared with that of ORGANIC, and both models were trained on the SMILES string representation of molecules. The results showed that ATNC could generate more valid molecules with better druglike properties than ORGANIC could. In order to perform in vitro validation, de novo molecules generated by the ATNC model trained on 30 000 kinase inhibitors from a public database were screened against in-house small molecule collections by similarity searches. The inhibition potency of the similar compounds found in the collections was tested against a panel of kinases, and some hits were observed, indicating the usefulness of ATNC for generating hit compounds. In ref (58), the authors described a SMILES-based generative model, called Bidirectional Adversarial Autoencoder, which infers molecular structures inducing a predefined change in gene expression. The model separates cellular processes captured in gene expression changes into two feature sets: those related and unrelated to the drug incubation. The model uses the first set to formulate a drug hypothesis. The model was validated using the LINCS L1000 data set and it was shown that the model can generate novel molecular structures which can induce the desired gene expression change or predict a gene expression difference after incubation of a given molecular structure.
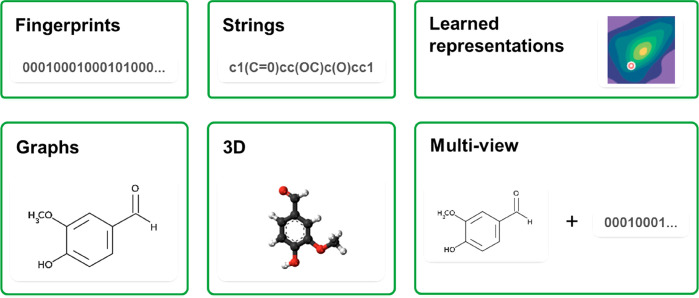
Although the initial AI-based generative chemistry models offered promising possibilities, their performance was limited by the methods used for representing and encoding the molecules. Most of the first generative models are SMILES-based models and cannot properly employ fragment-based objective reward functions because the SMILES format notation does not allow fragments to be found in the SMILES string of a molecule. To circumvent this issue, researchers have proposed models using other molecular representations, (Figure 3), such as graph representations, where each molecule is represented in a unique way. Kearnes et al. proposed a graph convolution architecture to encode small molecules as undirected graphs of atoms connected by bonds for virtual screening.59 The results demonstrated comparable performance to that of neural networks models trained on molecules encoded by fingerprints. JT-VAE, for example, generated molecules by building blocks derived from subgraphs of valid molecules. The key advantage of this technique is that the decoder can use valid components and correct interactions to realize a valid molecule piece-by-piece.44 Another example addressing these problems is a GAN-RL-based generative model called MolGAN that uses graph-structured data.60 MolGAN is an implicit, likelihood-free generative model for small molecular graphs that circumvents the need for expensive graph-matching procedures or node-ordering heuristics. MolGAN includes an RL objective that favors the generation of molecules with specific desired chemical properties. Another example is Mol-CycleGAN.61 This CycleGAN-based model is designed to generate optimized compounds with a chemical scaffold of interest. Given a molecule, this model could generate a structurally similar molecule with an optimized value of a given property, such as log P. A direct 3D representation of a molecule could also be an alternative for molecular representation. A study was recently published to describe the wave transform in which atoms were extended to fill nearby voxels with a transformation. The results demonstrated that this representation reached a better performance in training autoencoders.62
Figure 3.
Different approaches suggested for representing molecular structures. Although the first published DL-based model for de novo drug design used fingerprints to represent the molecules, SMILES is currently the most used format for encoding molecular structures. Other representations such as graphs are also attracting interest. Combining different encoding formats allow building a more detailed description of the molecular structures leading to better performance of de novo design methods.
Assessment and Validation of Generative Chemistry Approaches
AI-based approaches require rigorous evaluation to determine at what extent they might be applied in real world drug discovery settings. Unfortunately, publications describing AI-based approaches do not always disclose all documentation needed to perform an objective evaluation of their abilities. With the increasing number of approaches proposed in the field, there is a need to develop a unified set of benchmarks to evaluate and compare generative models. This includes the formulation of practices about how large a training data set is required, how long the model should be trained, and which metrics and loss functions are the most appropriate for monitoring the performance and assessing the validity of the model outputs. Initiatives are underway to establish a range of standards in generative chemistry. For instance, the Alliance for Artificial Intelligence in Healthcare (AAIH), cofounded by Insilico Medicine, has proposed the Molecular Sets (MOSES).63 MOSES is a benchmarking platform to support research on DL for drug discovery which contains a set of molecular generative models and metrics for evaluating the novelty and quality of generated molecules. GuacaMol is another similar effort supported by BenevolentAI. Another issue to be considered when assessing the performance of a generative model is whether a simpler approach that does not employ AI could have produced the same molecules. Although it may be difficult to perform a head-to-head comparison, the existence of these alternatives should be taken into account. For standard computational methods, the requirements for the novelty, activity, and breadth of structure–activity relationship are defined by guidelines specially designed for computational papers. This contributes to improve the quality of computational medicinal chemistry papers and to ensure that articles are reviewed in a consistent manner. Similar guidelines should be established by journals publishing AI-based case studies. This will enable a better systematic evaluation of the results presented. Criteria by which molecules generated using DL-based design methods could be assessed were recently suggested.50
As discussed above, the systemic pharmacological effect of a drug is governed by nonlinear relationships between contributing factors whose biological effects are not always well understood. The chemical structure of a drug alone does not necessarily account for the observed pharmacological effect in a simple fashion and most drugs have multiple biological targets and activities, and their relative importance is highly dependent on the individual genetic profile of patients, and many other factors. This poses a particular challenge for de novo drug design because in this context drug design appears to be an ill-posed problem due to the number of often unknown factors. This leads to an unpredictable system behavior which does not always follow the correspondence between structure and function, a prerequisite for systematic optimization. This complexity is not always precisely taken into account when representing molecular structures in terms of molecular graphs for instance, as evidenced by the limitations of graph-based molecular fingerprints, and this makes molecule scoring a complex task. The limited appropriateness of the quantitative scoring functions also makes difficult the fully automated compound fine-tuning and hit-to-lead expansion. Advanced AI methods could be developed to create more adapted scoring functions thanks to their abilities to combine data of different types (genomics, proteomics, metabolomics, lipidomics, etc.) and to use biological information encoded within the data without the need for hand-crafted rules.
What distinguishes healthcare from other fields when it comes to validating AI technologies is that while the advances in high performance computing and data management allow training the systems very quickly, the time needed to test the output is much longer than that in other industries dealing with pictures, videos, or text. The time it takes to perform in vivo validation of molecules produced by AI technologies by far exceeds the time it takes to build and train the models. It also takes effort and money and necessitates collaborations involving experts in AI and biological, chemical, and medical sciences. Synthesis and experimental validation will remain the main gating factor for the transformation of drug discovery using AI over the next few years, and the process will require a significant amount of investment to demonstrate its worth prospectively to commit to make and test the identified compounds. For successful AI-aided compounds, the rate of entry into the market also depends on how the regulatory agencies will consider the potential and safety of the use of AI within the drug discovery and healthcare industries.
Challenges and Outlook
The lack of synthetic tractability has been a weakness of many of earlier computational de novo molecular design approaches. In generative models, the synthetic tractability or accessibility is evaluated using the synthetic accessibility score method64 which relies on the knowledge extracted from known synthetic reactions and adds a penalty for high molecular complexity. Recent case studies (Supporting Information Table 2) showed that DL-based methods were able to generate molecular structures that are synthetically tractable and elicit the desired biological activity. This suggests that DL-based design methods can learn implicit rules about this important aspect from the training structures provided. Nevertheless, there is still significant manual input to the process of de novo design with generated structures requiring manual curation to make them more amenable for synthesis. This happens when certain chemical building blocks are unavailable, are unstable, require lengthy preparation, or lack sufficient reactivity. It is worth emphasizing that the design of molecular structures is only a component of a larger workflow that includes docking and other in silico filtering. In the future, research will focus on the amelioration of the assessment of synthetic accessibility and this might require the definition of more adapted scoring functions. Finally, considering that de novo drug design is primarily dependent on data, in terms of quantity and quality, additional resources should be invested in data curation and integration in order to realize the full impact of AI techniques in the future. We have listed (Supporting Information Table 2) representative examples of AI-based de novo molecular methods. These case studies, published in peer-reviewed journals, provide an overview of the current capabilities of AI.
Disruptive technologies bridging multiple scientific domains are impacting and reshaping the future of chemical biology and drug discovery. AI algorithms can learn to identify subtle information, enabling them to efficiently and precisely analyze the correlations between molecular structure and biological activity as well as build up a reliable QSAR model that can generate de novo compounds with desired properties.21,24,48,65 This could lead to a major disruption of the pharmaceutical industry by AI,66 as this field will be entering into an era where scientists will be able to deliver drugs to patients in a more efficient and more cost-effective manner.1,30 Within the next few years, the integration of AI within drug discovery pipelines is most likely to be incremental and will depend on the ability of AI to provide scientists with reliable solutions to the challenges faced by the industry. Where AI can make a huge difference is having drugs that fail early on, to avoid making all that investment in them. In addition to the AI-based computational design of novel compounds against selected targets, other ambitious challenges would be to identify molecules that are pharmacologically active and orally bioavailable.
The often nonlinear structure–activity relationships of drugs, as well as their complex physicochemical properties, lead to complicated optimization problems when applying automated in silico drug design.5 Although the recent growing interest in applying AI for solving key challenges in drug discovery has revealed the capacities of generative deep networks for molecule design, the reliable and practical models that can predict binding affinities to prioritize thousands of generated molecules for synthesis and testing remain lacking.67 One primary reason for this insufficiency is that the binding of the ligand to its receptor is driven not only by the enthalpy contribution but also the entropy contribution, particularly the influence of the free-energy cost of displacing ordered or partially ordered water.68 Interestingly, a recent study seems to address this unmet need by demonstrating the successful application of virtual screening on ultra large compound libraries.69 The authors constructed libraries of 170 million diverse compounds from the well-characterized chemical reaction products of 70 000 building blocks and performed docking against AmpC β-lactamase and the D4 dopamine receptor. The study carefully designed the docking protocol, avoiding a large chemical space that could overwhelm the true active compounds in order to successfully identify potent binders with new scaffolds against the targets. Though the team encountered problems in designing selective compounds by large-library dockings,70 they demonstrated the practical application of docking a large compound library in de novo drug design, an approach that could be used as a scoring function for prioritizing generated molecules. It could be implemented in RL generative models as a reward function to guide the models to efficiently explore the whole chemical space.37 Moreover, the synthetic accessibility of generated compounds remains a challenge to fully exploiting AI-based generative chemistry tools in drug discovery. Advances in chemical intelligence approaches that utilize automated robotic systems to allow the universal assembly of complex molecules on demand,29,71 as well as DL-based chemical synthesis planning,72 may provide solutions for the synthetic tractability.
The integration of AI in the activities of scientists will be facilitated with the formation of multidisciplinary teams developing this technology even as they test hypotheses in the laboratory to make the systems better able to learn. Enabling those feedback loops to improve the algorithms through testing their predictions and assumptions contributes to augment the abilities of the scientists and also to improve the understanding of AI capabilities and limitations. As AI-based technologies are integrated, more scientists will be responsible for establishing research directions and produce data. The scientists will have major roles to play in the evaluation of the results from the point of view of accuracy in the sense that scientific data is sometimes contradictory. For instance, some biological facts may be true in animal models but not in humans. Scientists are also important to interpret the result within the right experimental context because in biology context matters with protein interactions taking place in one organ but not in others. AI-based technologies are often not yet sophisticated enough to pick up on such context.
Looking ahead, one can foresee that the increase in computational power and the improvement of the overall performance of generative models will contribute to optimize the early stage of drug discovery with new capabilities to generate higher-quality target-oriented diverse compound libraries with well-designed druglike properties. These new capabilities combined with the steady progress in automated chemical synthesis and in the development of reliable computational simulation techniques for predicting the binding energy of generated compounds73 will allow a major step toward fully automated de novo drug design.74,75
Glossary
Abbreviations
- AI
artificial intelligence
- GAN
generative adversarial network
- RL
reinforcement learning
- ML
machine learning
- DL
deep learning
- QSAR
quantitative structure–activity relationship
- NCEs
new chemical entities
- SBDND
structure-based de novo drug design
- LBDND
ligand-based de novo drug design
- RNN
recurrent neural network
- VAE
variational autoencoder
- AAE
adversarial encoder
- LSTM
long short-term memory
- SMILES
simplified molecular input line-entry system
- RXR
retinoid X receptors
- PPAR
peroxisome proliferator-activated receptors
- SAS
synthetic accessibility score
- QED
quantitative estimation of drug-likeness
- CVAE
conditional variational autoencoder
- TPSA
topological polar surface area
- JT-VAE
junction tree variational autoencoder
- druGAN
drug generative adversarial network
- DRD2
dopamine receptor type 2
- SeqGAN
sequence generative adversarial network
- ORGAN
objective-reinforced generative adversarial network
- ORGANIC
objective-reinforced generative adversarial network for inverse-design chemistry
- DNC
differentiable neural computer
- ATNC
adversarial threshold neural computer
- MOSES
molecular sets
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.0c00088.
The authors declare the following competing financial interest(s): The authors are affiliated with Insilico Medicine, a company developing an AI-based end-to-end integrated pipeline for target identification and drug discovery and engaged in aging and cancer research.
Supplementary Material
References
- Mamoshina P.; et al. Applications of Deep Learning in Biomedicine. Mol. Pharmaceutics 2016, 13 (5), 1445–54. 10.1021/acs.molpharmaceut.5b00982. [DOI] [PubMed] [Google Scholar]
- Zhavoronkov A.; et al. Artificial intelligence for aging and longevity research: Recent advances and perspectives. Ageing Res. Rev. 2019, 49, 49–66. 10.1016/j.arr.2018.11.003. [DOI] [PubMed] [Google Scholar]
- Goodfellow I.NIPS 2016 Tutorial: Generative Adversarial Networks. arXiv (Machine Learning), April 3, 2017, 1701.00160, ver. 4. https://arxiv.org/abs/1701.00160v4.
- Goodfellow I. J.et al. Generative Adversarial Networks. arXiv (Machine Learning), June 10, 2014, 1406.2661, ver. 1. https://arxiv.org/abs/1406.2661.
- Schneider G.; Clark D. E. Automated De Novo Drug Design - “Are we nearly there yet?. Angew. Chem., Int. Ed. 2019, 58 (32), 10792–10803. 10.1002/anie.201814681. [DOI] [PubMed] [Google Scholar]
- Cherkasov A.; et al. QSAR modeling: where have you been? where are you going to? J. J. Med. Chem. 2014, 57 (12), 4977–5010. 10.1021/jm4004285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrchan C.; et al. Matched molecular pair analysis in short: algorithms, applications and limitations. Comput. Struct. Biotechnol. J. 2017, 15, 86–90. 10.1016/j.csbj.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar D.; et al. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J. Chem. Theory Comput. 2010, 6 (5), 1509–1519. 10.1021/ct900587b. [DOI] [PubMed] [Google Scholar]
- Hansch C.; et al. Correlation of biological activity of phenoxyacetic acids with Hammett substituent constants and partition coefficients. Nature 1962, 194, 178. 10.1038/194178b0. [DOI] [Google Scholar]
- Popova M.; et al. Deep reinforcement learning for de novo drug design. Sci. Adv. 2018, 4 (7), eaap7885 10.1126/sciadv.aap7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G. De novo design - hop(p)ing against hope. Drug Discovery Today: Technol. 2013, 10 (4), e453 10.1016/j.ddtec.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Kuhn B.; et al. A Real-World Perspective on Molecular Design. J. Med. Chem. 2016, 59 (9), 4087–102. 10.1021/acs.jmedchem.5b01875. [DOI] [PubMed] [Google Scholar]
- Anderson A. C. The process of structure-based drug design. Chem. Biol. 2003, 10 (9), 787–97. 10.1016/j.chembiol.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Glaab E. Building a virtual ligand screening pipeline using free software: a survey. Briefings Bioinf. 2016, 17 (2), 352–66. 10.1093/bib/bbv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounnas V.; et al. Current progress in Structure-Based Rational Drug Design marks a new mindset in drug discovery. Comput. Struct. Biotechnol. J. 2013, 5, e201302011 10.5936/csbj.201302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okimoto N.; et al. Evaluation of protein-ligand affinity prediction using steered molecular dynamics simulations. J. Biomol. Struct. Dyn. 2017, 35 (15), 3221–3231. 10.1080/07391102.2016.1251851. [DOI] [PubMed] [Google Scholar]
- Bickerton G. R.; et al. Quantifying the chemical beauty of drugs. Nat. Chem. 2012, 4 (2), 90–8. 10.1038/nchem.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider P.; Schneider G. De Novo Design at the Edge of Chaos. J. Med. Chem. 2016, 59 (9), 4077–86. 10.1021/acs.jmedchem.5b01849. [DOI] [PubMed] [Google Scholar]
- Hartenfeller M.; et al. DOGS: reaction-driven de novo design of bioactive compounds. PLoS Comput. Biol. 2012, 8 (2), e1002380 10.1371/journal.pcbi.1002380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth N. C.; et al. MOARF, an Integrated Workflow for Multiobjective Optimization: Implementation, Synthesis, and Biological Evaluation. J. Chem. Inf. Model. 2015, 55 (6), 1169–80. 10.1021/acs.jcim.5b00073. [DOI] [PubMed] [Google Scholar]
- Rodrigues T.; et al. Multidimensional de novo design reveals 5-HT2B receptor-selective ligands. Angew. Chem., Int. Ed. 2015, 54 (5), 1551–5. 10.1002/anie.201410201. [DOI] [PubMed] [Google Scholar]
- Reutlinger M.; et al. Multi-objective molecular de novo design by adaptive fragment prioritization. Angew. Chem., Int. Ed. 2014, 53 (16), 4244–8. 10.1002/anie.201310864. [DOI] [PubMed] [Google Scholar]
- Rodrigues T.; et al. Steering target selectivity and potency by fragment-based de novo drug design. Angew. Chem., Int. Ed. 2013, 52 (38), 10006–9. 10.1002/anie.201304847. [DOI] [PubMed] [Google Scholar]
- Besnard J.; et al. Automated design of ligands to polypharmacological profiles. Nature 2012, 492 (7428), 215–20. 10.1038/nature11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond J.-L.; et al. The enumeration of chemical space. Wiley Interdisc. Rev. Comp. Mol. Sci. 2012, 2, 717–733. 10.1002/wcms.1104. [DOI] [Google Scholar]
- Mullard A. The drug-maker’s guide to the galaxy. Nature 2017, 549 (7673), 445–447. 10.1038/549445a. [DOI] [PubMed] [Google Scholar]
- Sellwood M. A.; et al. Artificial intelligence in drug discovery. Future Med. Chem. 2018, 10 (17), 2025–2028. 10.4155/fmc-2018-0212. [DOI] [PubMed] [Google Scholar]
- Reymond J. L. The chemical space project. Acc. Chem. Res. 2015, 48 (3), 722–30. 10.1021/ar500432k. [DOI] [PubMed] [Google Scholar]
- Gromski P. S.; et al. How to explore chemical space using algorithms and automation. Nature Reviews Chemistry 2019, 3 (2), 119–128. 10.1038/s41570-018-0066-y. [DOI] [Google Scholar]
- Zhavoronkov A. Artificial Intelligence for Drug Discovery, Biomarker Development, and Generation of Novel Chemistry. Mol. Pharmaceutics 2018, 15 (10), 4311–4313. 10.1021/acs.molpharmaceut.8b00930. [DOI] [PubMed] [Google Scholar]
- Sanchez-Lengeling B.; Aspuru-Guzik A. Inverse molecular design using machine learning: Generative models for matter engineering. Science 2018, 361 (6400), 360–365. 10.1126/science.aat2663. [DOI] [PubMed] [Google Scholar]
- Chen H.; et al. The rise of deep learning in drug discovery. Drug Discovery Today 2018, 23 (6), 1241–1250. 10.1016/j.drudis.2018.01.039. [DOI] [PubMed] [Google Scholar]
- Hochreiter S.; Schmidhuber J. Long Short-Term Memory 1997, 9 (8), 1735–1780. 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- Doersch C.Tutorial on Variational Autoencoders. arXiv (Machine Learning), August 13, 2016, 1606.05908, ver. 2. https://arxiv.org/abs/1606.05908.
- Kingma D. P.; Welling M.. Auto-Encoding Variational Bayes. arXiv (Machine Learning), May 1, 2014, 1312.6114, ver. 10. https://arxiv.org/abs/1312.6114.
- Elton D. C.et al. Deep learning for molecular design - a review of the state of the art. arXiv (Machine Learning), May 6, 2019, 1903.04388, ver. 2. https://arxiv.org/abs/1903.04388.
- Segler M. H. S.; et al. Generating Focused Molecule Libraries for Drug Discovery with Recurrent Neural Networks. ACS Cent. Sci. 2018, 4 (1), 120–131. 10.1021/acscentsci.7b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. Generative Recurrent Networks for De Novo Drug Design. Mol. Inf. 2018, 37 (1–2), 1880141. 10.1002/minf.201880141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertl P.et al. In silico generation of novel, drug-like chemical matter using the LSTM neural network. arXiv (Machine Learning), January 8, 2018, 1712.07449, ver. 2. https://arxiv.org/abs/1712.07449.
- Jannik Bjerrum E.; Threlfall R.. Molecular Generation with Recurrent Neural Networks (RNNs). arXiv (Machine Learning), May 17, 2017, 1705.04612, ver. 2. https://arxiv.org/abs/1705.04612.
- Merk D.; et al. De Novo Design of Bioactive Small Molecules by Artificial Intelligence. Mol. Inf. 2018, 37 (1–2), 1700153. 10.1002/minf.201700153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Bombarelli R.; et al. Automatic Chemical Design Using a Data-Driven Continuous Representation of Molecules. ACS Cent. Sci. 2018, 4 (2), 268–276. 10.1021/acscentsci.7b00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J.; et al. Molecular generative model based on conditional variational autoencoder for de novo molecular design. J. Cheminf. 2018, 10 (1), 31. 10.1186/s13321-018-0286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W.et al. Junction Tree Variational Autoencoder for Molecular Graph Generation. arXiv (Machine Learning), March 29, 2019, 1802.04364, ver. 4. https://arxiv.org/abs/1802.04364.
- Kadurin A.; et al. The cornucopia of meaningful leads: Applying deep adversarial autoencoders for new molecule development in oncology. Oncotarget 2017, 8 (7), 10883–10890. 10.18632/oncotarget.14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurin A.; et al. druGAN: An Advanced Generative Adversarial Autoencoder Model for de Novo Generation of New Molecules with Desired Molecular Properties in Silico. Mol. Pharmaceutics 2017, 14 (9), 3098–3104. 10.1021/acs.molpharmaceut.7b00346. [DOI] [PubMed] [Google Scholar]
- Blaschke T. Application of Generative Autoencoder in De Novo Molecular Design. Mol. Inf. 2018, 37 (1–2), 1700123. 10.1002/minf.201700123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polykovskiy D.; et al. Entangled Conditional Adversarial Autoencoder for de Novo Drug Discovery. Mol. Pharmaceutics 2018, 15 (10), 4398–4405. 10.1021/acs.molpharmaceut.8b00839. [DOI] [PubMed] [Google Scholar]
- Zhavoronkov A.; et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 2019, 37, 1038–1040. 10.1038/s41587-019-0224-x. [DOI] [PubMed] [Google Scholar]
- Walters W. P.; Murcko M. Assessing the impact of generative AI on medicinal chemistry. Nat. Biotechnol. 2020, 38 (2), 143–145. 10.1038/s41587-020-0418-2. [DOI] [PubMed] [Google Scholar]
- Arulkumaran K.et al. A Brief Survey of Deep Reinforcement Learning. arXiv (Machine Learning), September 28, 2017, 1708.05866, ver. 2. https://arxiv.org/abs/1708.05866.
- Olivecrona M.; et al. Molecular de-novo design through deep reinforcement learning. J. Cheminf. 2017, 9 (1), 48. 10.1186/s13321-017-0235-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L.et al. SeqGAN: Sequence Generative Adversarial Nets with Policy Gradient. arXiv (Machine Learning), August 25, 2017, 1609.05473, ver. 6. https://arxiv.org/abs/1609.05473.
- Lima Guimaraes G.et al. (2017) Objective-Reinforced Generative Adversarial Networks (ORGAN) for Sequence Generation Models. arXiv (Machine Learning), February 7, 2018, 1705.10843, ver. 3. https://arxiv.org/abs/1705.10843.
- Sanchez-Lengeling B.et al. Optimizing distributions over molecular space. An Objective-Reinforced Generative Adversarial Network for Inverse-design Chemistry (ORGANIC). ChemRxiv, August 16, 2017, ver. 3. 10.26434/chemrxiv.5309668.v3. [DOI]
- Putin E.; et al. Reinforced Adversarial Neural Computer for de Novo Molecular Design. J. Chem. Inf. Model. 2018, 58 (6), 1194–1204. 10.1021/acs.jcim.7b00690. [DOI] [PubMed] [Google Scholar]
- Putin E.; et al. Adversarial Threshold Neural Computer for Molecular de Novo Design. Mol. Pharmaceutics 2018, 15 (10), 4386–4397. 10.1021/acs.molpharmaceut.7b01137. [DOI] [PubMed] [Google Scholar]
- Shayakhmetov R.; et al. Molecular Generation for Desired Transcriptome Changes With Adversarial Autoencoders. Front. Pharmacol. 2020, 11, 269. 10.3389/fphar.2020.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearnes S.; et al. Molecular graph convolutions: moving beyond fingerprints. J. Comput.-Aided Mol. Des. 2016, 30 (8), 595–608. 10.1007/s10822-016-9938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cao N.; Kipf T.. MolGAN: An implicit generative model for small molecular graphs. arXiv (Machine Learning), May 30, 2018, 1805.11973, ver. 1. https://arxiv.org/abs/1805.11973.
- Maziarka Ł.et al. Mol-CycleGAN - a generative model for molecular optimization. arXiv (Machine Learning), February 6, 2019, 1902.02119, ver. 1. https://arxiv.org/abs/1902.02119. [DOI] [PMC free article] [PubMed]
- Kuzminykh D.; et al. 3D Molecular Representations Based on the Wave Transform for Convolutional Neural Networks. Mol. Pharmaceutics 2018, 15 (10), 4378–4385. 10.1021/acs.molpharmaceut.7b01134. [DOI] [PubMed] [Google Scholar]
- Polykovskiy D.et al. Molecular Sets (MOSES): A Benchmarking Platform for Molecular Generation Models. arXiv (Machine Learning), October 15, 2019, 1811.12823, ver. 3. https://arxiv.org/abs/1811.12823. [DOI] [PMC free article] [PubMed]
- Ertl P.; Schuffenhauer A.; et al. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J. Cheminf. 2009, 1, 8. 10.1186/1758-2946-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. C.; et al. Multidimensional Design of Anticancer Peptides. Angew. Chem., Int. Ed. 2015, 54 (35), 10370–4. 10.1002/anie.201504018. [DOI] [PubMed] [Google Scholar]
- Wainberg M.; et al. Deep learning in biomedicine. Nat. Biotechnol. 2018, 36 (9), 829–838. 10.1038/nbt.4233. [DOI] [PubMed] [Google Scholar]
- Rocklin G. J.; et al. Blind prediction of charged ligand binding affinities in a model binding site. J. Mol. Biol. 2013, 425 (22), 4569–83. 10.1016/j.jmb.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiten B.; et al. Water networks contribute to enthalpy/entropy compensation in protein-ligand binding. J. Am. Chem. Soc. 2013, 135 (41), 15579–84. 10.1021/ja4075776. [DOI] [PubMed] [Google Scholar]
- Lyu J.; et al. Ultra-large library docking for discovering new chemotypes. Nature 2019, 566 (7743), 224–229. 10.1038/s41586-019-0917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D. R.; et al. Selectivity Challenges in Docking Screens for GPCR Targets and Antitargets. J. Med. Chem. 2018, 61 (15), 6830–6845. 10.1021/acs.jmedchem.8b00718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner S.; et al. Science 2019, 363 (6423), eaav2211 10.1126/science.aav2211. [DOI] [PubMed] [Google Scholar]
- Segler M. H. S.; et al. Planning chemical syntheses with deep neural networks and symbolic AI. Nature 2018, 555, 604. 10.1038/nature25978. [DOI] [PubMed] [Google Scholar]
- Chodera J. D.; Mobley D. L. Entropy-enthalpy compensation: role and ramifications in biomolecular ligand recognition and design. Annu. Rev. Biophys. 2013, 42, 121–42. 10.1146/annurev-biophys-083012-130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.; et al. Deep learning for molecular generation. Future Med. Chem. 2019, 11 (6), 567–597. 10.4155/fmc-2018-0358. [DOI] [PubMed] [Google Scholar]
- Feng F.; et al. Computational Chemical Synthesis Analysis and Pathway Design. Front. Chem. 2018, 6, 199. 10.3389/fchem.2018.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.