Abstract
Targeting of genomic quadruplexes is an approach to treating complex human cancers. We describe a series of tetra-substituted naphthalene diimide (ND) derivatives with a phenyl substituent directly attached to the ND core. The lead compound (SOP1812) has 10 times superior cellular and in vivo activity compared with previous ND compounds and nanomolar binding to human quadruplexes. The pharmacological properties of SOP1812 indicate good bioavailability, which is consistent with the in vivo activity in xenograft and genetic models for pancreatic cancer. Transcriptome analysis shows that it down-regulates several cancer gene pathways, including Wnt/β-catenin signaling.
Keywords: quadruplex, naphthalene diimide derivative, pancreatic cancer, xenograft, transcriptome
Nucleic acid sequences containing repetitive guanine (G)-rich sequences can fold into higher order quadruplex arrangements (G4s), stabilized by the π–π stacking of successive G-quartets.1,2 G4s can occur in genomic DNA and RNA and are widely distributed in human and other genomes and in all eukaryotic telomeric sequences. They are nonrandomly distributed in the human genome,3,4 with over-representation in the promoter regions of proliferation genes, notably cancer-related genes,5−7 and in 5′ and 3′ untranslated regions.8 They play a role in the genetic instability of cancer cells,9 can affect transcription, translation, or replication, and may regulate these processes.9−13 Estimates of the number of G4 sequences in the human genome from bioinformatics analyses have varied from ca. 250 0003,4 to ca. 700 000. An experimental study14 has revealed that in the chromatin environment of an immortalized cell, there are ca. 10 000 G4 structures. By contrast, in a noncancer cell, ca. 1500 were observed, supporting the concept that G4s are cancer-selective targets. G4s in cells can be visualized using G4-specific antibodies.15,16
The stabilization of G4 structures in cancer cells by a small molecule can selectively inhibit cellular processes, suggesting a therapeutic strategy for cancer therapy,17−19 with the G4-binding compound generating anticancer activity. Most studies with such compounds have targeted promoter G4s in cancer-associated genes17,20 to down-regulate transcription, for example, a G4 in the promoter sequence of the telomerase complex catalytic subunit hTERT.21
The naphthalene diimide (ND) core has been extensively explored for devising G4-binding compounds.22−26 X-ray crystallography has revealed that the ND core stacks on the exterior G-quartet of G4s,23,24 in common with other categories of G4-binding compounds. Substituted NDs bind with each side chain situated in a G4 groove. Cationic charges on at least some of the side-chain termini are important contributors to G4 affinity and biological activity.
We have previously reported23−26 on a number of ND compounds, including26 the trisubstituted derivative 2,7-bis(3-morpholinopropyl)-4-((2-(pyrrolidin-1-yl)ethyl)amino) benzo[lmn][3,8] phenanthroline-1,3,6,8(2H,7H)-tetraone (CM03). This binds strongly to G4s in vitro, has potent antiproliferative activity in a panel of human pancreatic ductal adenocarcinoma (PDAC) cell lines, and shows antitumor activity in in vivo models for this clinically challenging disease. Transcriptome analysis showed that >600 genes were significantly down-regulated in CM03-treated PDAC cells. 350 out of 600 (∼60%) genes contain G4 sequences in promoter regions, of which >40 out of 600 (∼7%) are involved in PDAC-related pathways. By contrast, many genes down-regulated following PDAC cell treatment with a current standard-of-care drug gemcitabine do not possess G4 elements. We suggested that the effectiveness and potency of CM03 is a consequence of its ability to down-regulate multiple dysregulated gene targets in PDAC cells27 containing G4 elements in regulatory regions.26
The hypothesis of the present study is that enhancing the in vitro G4 affinity of CM03 may result in enhanced biological potency, which is further evidence of G4 targeting. We have recently reported28 quantitative G4 binding data for CM03 and a new ND compound, SOP1812, with ca. 10 times greater cellular and in vivo potency than CM03. We report here on the further characterization of SOP1812.
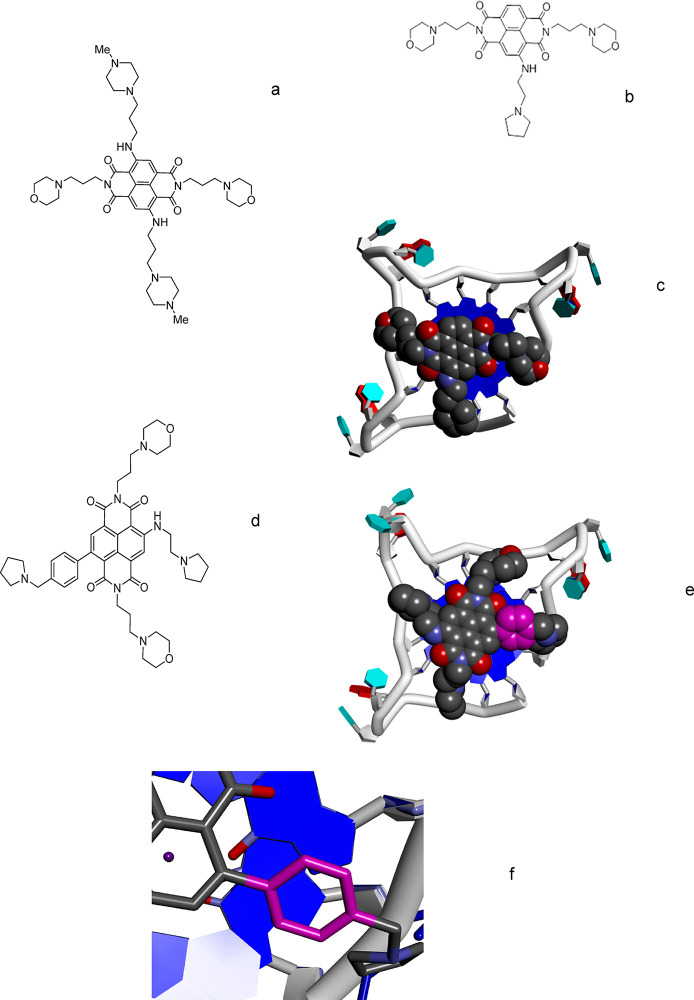
Co-crystal structures of human telomeric G4s with tetra-substituted NDs,23,24 such as MM41 (Figure 1a), and the modeled structure26 of a complex with CM03 (Figure 1b,c) were used to design SOP1812. Ligand binding sites in these structures were modeled onto the hTERT parallel-stranded G4 structure (NMR structure).29 Tetra-substituted ND compounds, typified by MM41, have each cationic acyclic side chain residing in a G4 groove. The three side chains of CM03 were each modeled to interact in a groove;26 the lack of a fourth substituent is compensated by the tight fit of the three. We hypothesized that a fourth substituent group, a pyrrolidino-substituted phenyl ring directly attached to the ND core, could contribute enhanced affinity. The energy-minimized aminopyrrolidino phenyl derivative of CM03 (Figure 1c) has a 27° dihedral angle between the phenyl and the planar ND core. The phenyl thus stacks with the terminal G-quartet (Figure 1d–f), adopting a ruffled arrangement with the G-quartet being slightly noncoplanar, as observed in several G4 cocrystal structures23,24 and theoretical models.30 Modeling also suggests that a cationic tail of the phenyl could reside in the fourth G4 groove (Figure 1d).
Figure 1.
Structures of naphthalenediimide derivatives (a) MM41, (b) CM03, and (d) SOP1812. (c) Plausible low-energy position of CM03 bound to an external G-quartet of the human telomeric G4. (e) Plausible low-energy position of SOP1812 bound to an external G-quartet of the human telomeric G4, based on panel c, with the substituent phenyl ring colored magenta. (f) Magnified view, from panel e, of the phenyl ring of SOP1812 partially stacked onto a G-quartet.
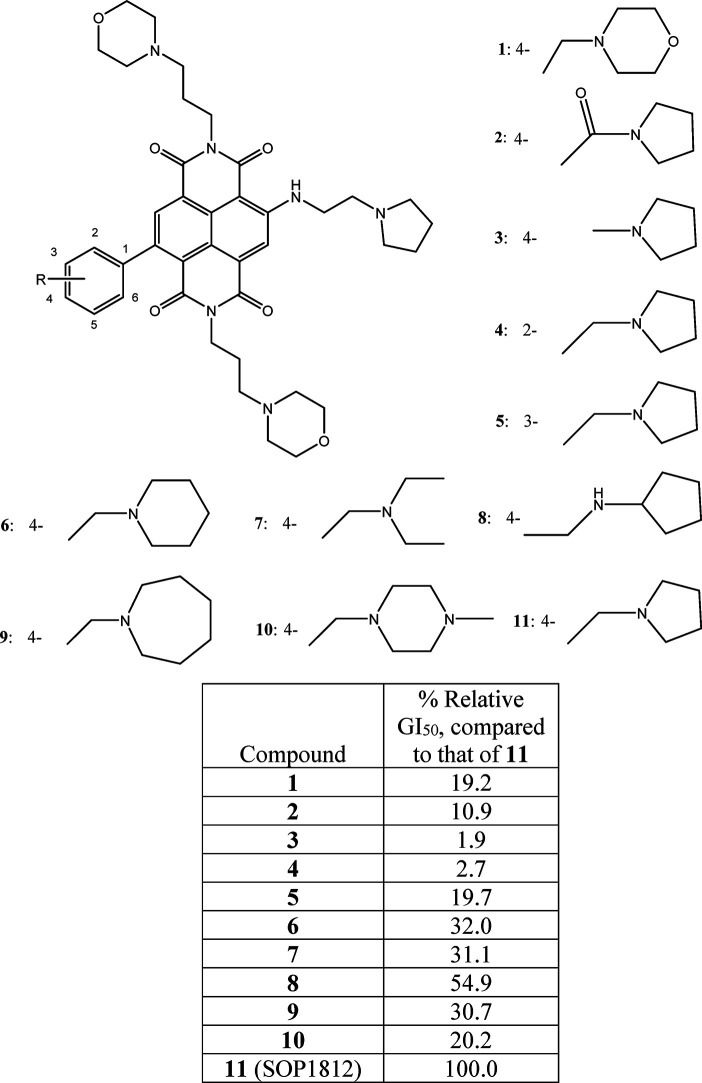
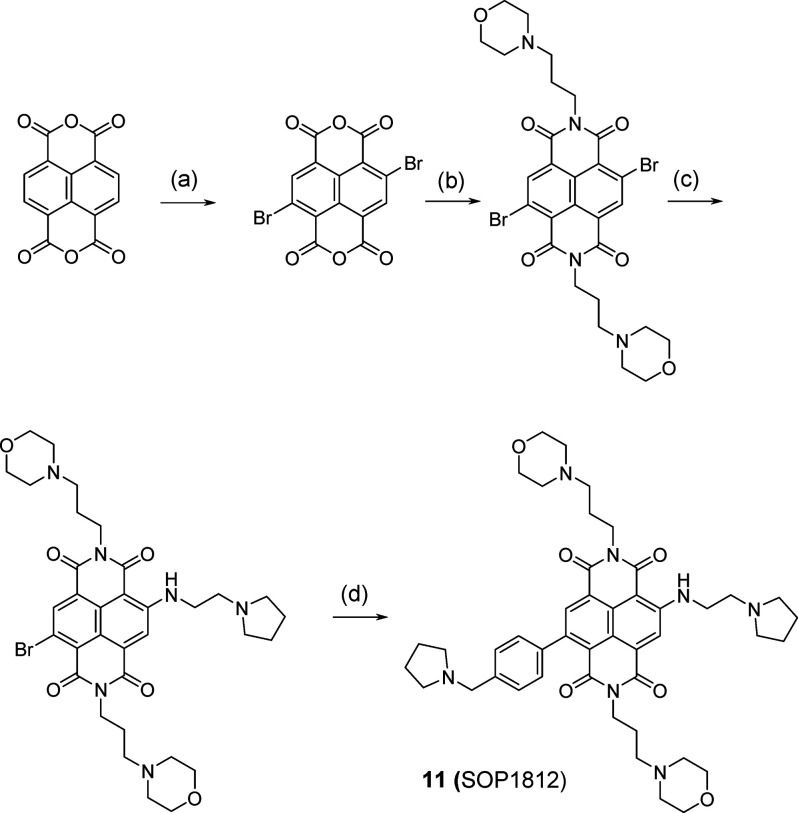
A small library of 11 compounds (Figure 2) based on this concept was synthesized, with a range of substituents on the phenyl ring. The synthetic procedure is a modification of the established route24−26 to other ND derivatives. (See Scheme 1 and the experimental details in the SI.) Final compounds were purified to 95–98% purity, often at a higher yield than hitherto, by using silica and reverse-phase column chromatography rather than HPLC. Good yields and purity may be the consequence of obtaining key intermediates in higher purity than achieved previously, enabling cleaner reaction profiles for the final products.
Figure 2.
Structures of compounds 1–11 and relative GI50 values measured in MIA PaCa-2 cells, scaled to those for SOP1812.
Scheme 1. Scheme for Synthesis of Compound SOP1812,
Syntheses of other compounds in the 11-compound library used appropriate intermediates at step d, as described in detail in the SI.
Conditions: (a) 1,3-dibromo-5,5-dimethylimidazolone-2,4-dione, H2SO4, 80 °C, 0.5 h (82%); (b) 3-morpholinopropan-1-amine (2 equiv), AcOH, 80 °C, 1 h (56%); (c) 2-(pyrrolidin-1-yl)ethanamine (3 equiv), toluene, 120 °C, 1 h (40%); (d) 1-(4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)benzyl-pyrrolidine, S-Phos Pd G3, THF, 70 °C, 3 h (40%).
The ability of the compound library to inhibit cell proliferation in MIA PaCa-2 PDAC cells was estimated by a 96 h high-throughput MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assay. Compound 11 (SOP1812:2,7-bis(3-morpholinopropyl)-4-((2-(pyrrolidin-1-yl)ethyl)amino)-9-(4-(pyrrolidin-1-ylmethyl) phenyl) benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetraone) was the most potent in the series. For consistency with previous ND data, cell proliferation studies with a panel of PDAC cell lines were subsequently performed using a quantitative 96 h SRB (Sulforhodamine B) assay24,25 (Table 1).
Table 1. Cell Proliferation Data in Four Pancreatic Cancer Cell Lines as GI50 Values (in nM) from 96 h SRB Assaysa.
| compound 11 (SOP1812) | compound CM03 | |
|---|---|---|
| MIA PaCa-2 | 1.3 | 9.0 |
| PANC-1 | 1.4 | 15.6 |
| Capan-1 | 5.9 | 26.5 |
| BxPC-3 | 2.6 | 15.5 |
Standard deviations from the average of >3 independent determinations ±0.2 nM.
The weak relative activity of 2 and 3 (Figure 2) indicates that the substituent attached to the phenyl requires a cationic charge, which these lack and is confirmed by the weakly basic morpholino group, 1. Maximal activity is shown by the para-substituted pyrrolidino ring, 11 (SOP1812). Activity is lost when this substituent is moved around the phenyl (4 and 5). The substituent size is also a factor, with six-membered (6 and 10) and seven-membered rings (9) being deleterious. Extending the substituent length (8) is deleterious, but less so than other changes represented in this small library. The data suggest that optimal activity is narrowly defined: SOP1812 is the outstanding member of the series. A detailed structure–activity analysis is given in the SI.
The SRB cell proliferation assay with a panel of PDAC cell lines (Table 1) shows significantly greater activity for SOP1812 than CM03. In MIA PaCa-2 and PANC-1 cells, the increase in potency is 10-fold, and it is 5-fold for Capan-1, the least sensitive in the panel. Consistent with these differences in cellular activity, SOP1812 has two to three times greater affinity for the two exemplar G4s (a human telomeric and an hTERT promoter G428) than CM03 using SPR (Table 2). Both compounds have enhanced (>5-fold) affinity for hTERT G4.28
Table 2. Dissociation Equilibrium Binding Constants, KD (in nM), for Compounds with Two Quadruplex Sequences Measured by Surface Plasmon Resonance, from Reference (28).
| compound SOP1812 | compound CM03 | |
|---|---|---|
| hTERT G4 | 4.9 ± 0.2 | 9.0 ± 0.4 |
| HuTel21 G4 | 28.4 ± 1.1 | 81.8 ± 5.3 |
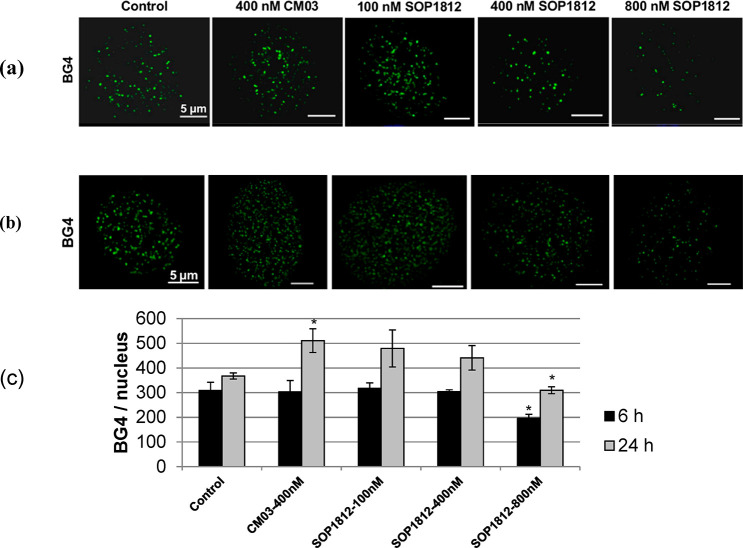
The visualization of CM03 and SOP1812 binding to G4s in cells used the BG4 G4-specific antibody.15 At 400 nM CM03 in PANC-1 cells (Figure 3), the number of BG4 foci is significantly increased following 24 h of exposure compared with the control (P < 0.05). SOP1812 at 100 nM gives the same result, with BG4 foci numbers increasing after 24 h. At 400 and 800 nM SOP1812, BG4 foci numbers are decreased in a dose-dependent manner. Decreased foci are still apparent at 6 h, but only at the highest concentration, and this effect is statistically significant (P < 0.05).
Figure 3.
Effects of CM03 and varying concentrations of SOP1812 on the numbers of G4-specific antibody BG4 foci in PANC-1 cells. (a,b) Confocal Z-stack images after 6 and 24 h of exposure. (c) Quantitation of the numbers of BG4 foci. Images were deconvolved and BG4 and nuclei numbers were obtained with the Imaris image analysis software package (https://imaris.oxinst.com/). Data shown represent the mean ± SEM of three independent experiments, *P < 0.05, using the Student’s t test.
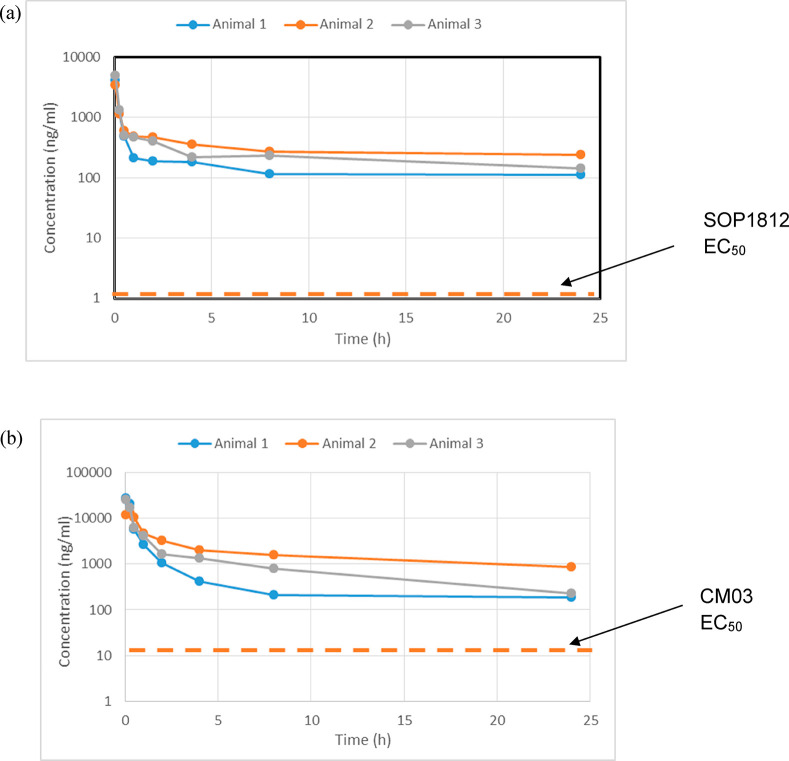
The pharmacokinetic behavior of CM03 and SOP1812 was evaluated in vivo in rodents following IV administration. Their half life in blood is >20 h. SOP1812 in tumor-free mice (Table S1) has a longer (>3 times) half life than CM03, although these differences are not apparent in tumor-bearing mice (Table 3) or tumor-free rats (Table S1). SOP1812 has a half life of ca. 36 h in both models. Figure 4a,b shows that the cellular concentrations of the two compounds remain low for >24 h, and both have windows above the EC50 values (especially SOP1812), so that sufficient compound is available in vivo in blood at these concentrations for therapeutic exposure, which were then used for subsequent in vivo therapeutic studies.
Table 3. Blood Pharmacokinetic Parameters in Female Athymic MIA PaCa-2 Tumor-Bearing Nude Mice Following a Single IV Administrationa.
| CM03 (9.1 mg/kg) | CM03 (14 mg/kg) | SOP1812 (0.96 mg/kg) | |
|---|---|---|---|
| T1/2 (hr) | 33(21) | 16(7) | 37(8.0) |
| Clobs (mL/min/kg) | 10.9(2.7) | 8.1(4.0) | 2.2(1.0) |
| Vss (L/kg) | 5.2(0.9) | 2.2(0.4) | 1.2(0.4) |
| Vz (L/kg) | 27.8(10.7) | 11.8(10.4) | 6.9(3.0) |
| Cmax (ng/mL) | 4519(3240) | 22088(7407) | 4178(707) |
| AUCall (ng·h/mL) | 11113(2248) | 30628(13924) | 5863(1868) |
| AUCINF_obs (ng·h/mL) | 14484(3769) | 34356(17974) | 8271(3633) |
Three mice were in each group, and the data shown for each parameter are the mean values. Standard deviations are in parentheses.
Figure 4.
Pharmacokinetics in tumor (MIA PaCa-2)-bearing mice for (a) SOP1812 at 0.96 mg/kg IV and (b) CM03 at 14 mg/kg IV.
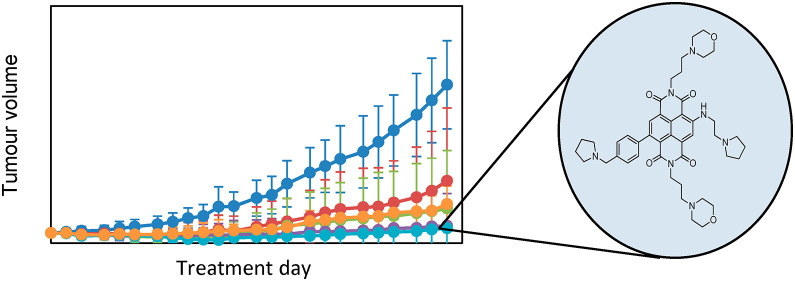
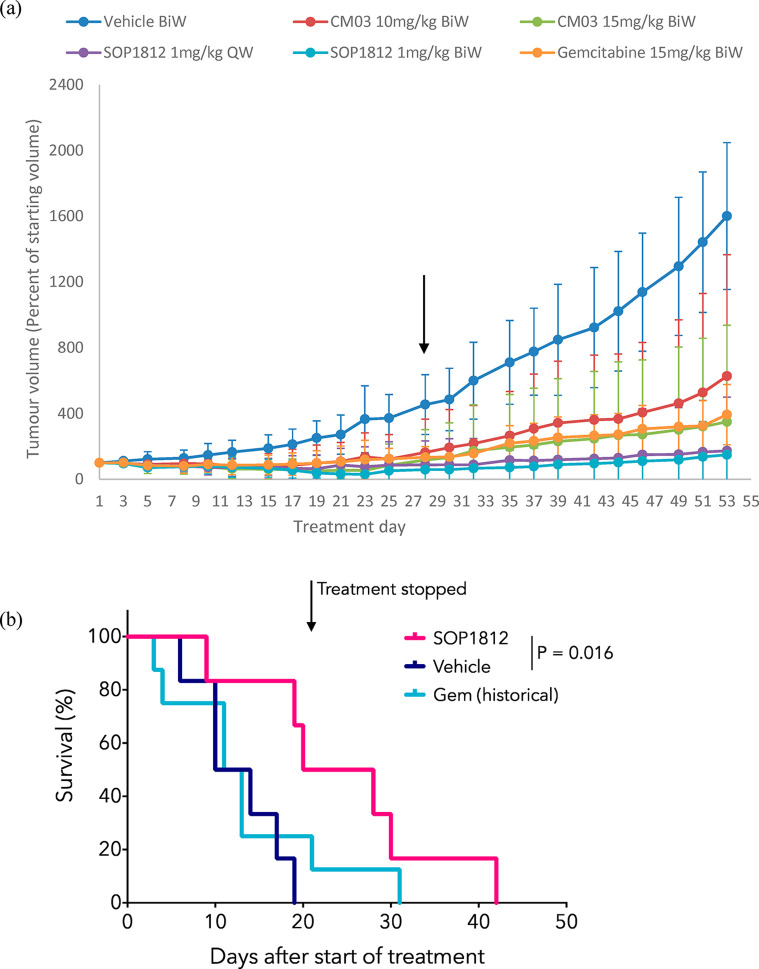
The antitumor activity of SOP1812 and CM03 has been evaluated in PDAC models and compared with a current standard-of-care PDAC drug, gemcitabine.31 Studies with the MIA PaCa-2 xenograft model (Figure 5a) examined each compound versus gemcitabine, with twice weekly dosing regimens over a 28-day period, followed by a period of no-dose observation. One SOP1812 group used once-weekly dosing. Several animals in each group were withdrawn from the study following day 28 and were sacrificed for further analysis. The study was concluded at day 53, when tumors in the vehicle control arm grew beyond acceptable regulatory levels.
Figure 5.
(a) Antitumor data for the MIA PaCa-2 xenograft model, after 28 days of IV administration, followed by 28 days without treatment. Data are mean ± SD; n = 8 mice to day 28 and n = 4 mice to day 53. The black arrow shows 28 days. QW and BiW represent once-weekly and twice-weekly dosing, respectively. (b) Kaplan–Meier survival curves showing the survival of KPC mice treated with 1 mg/kg IV SOP1812 (n = 6 mice) or control (n = 6 mice) 1× weekly for 3 weeks (4 doses), where possible. Data for mice treated 2× weekly with 100 mg/kg I.P. gemcitabine (n = 8 mice) is also shown. SOP1812 versus vehicle, P = 0.016.
CM03 dosing at 10 mg/kg produced a significant antitumor effect at 28 days (P = 0.035), but by day 53, tumor regrowth had occurred, so that the overall antitumor effect was modest and not statistically significant (Table 4 and Figure 5a). CM03 dosing at 15 mg/kg gave significant decreases in tumor volumes (P = 0.043), although regrowth after day 28 was observed (Table 4 and Figure 5a). SOP1812-dosed groups showed statistically significant tumor volume reductions at day 28 and at day 53 (P < 0.05 and P < 0.005; Table 4). Several animals showed complete tumor regression, no significant tumor regrowth after day 28, and no statistically significant difference between these two groups. Gemcitabine produced a significant antitumor effect (P = 0.004) and modest tumor regrowth after day 28. SOP1812 dosing schedules were well-tolerated, with no sign of adverse cardiac or neurological effects.
Table 4. Changes in Tumor Volume (TV) (cm3) Relative to Controls for the MIA PaCa-2 Xenograft Experiment (MTV: Mean Tumor Volume (cm3))a.
| drug | MTV ± SEM (day 28) | % TV change (day 28) | P value | MTV ± SEM (day 53) | % TV change (day 53) | P value |
|---|---|---|---|---|---|---|
| CM03, 10 mg/kg, BiW | 0.16 ± 0.20 | –64.4 | 0.035* | 0.63 ± 0.74 | –60.6 | 0.574 |
| CM03, 15 mg/kg, BiW | 0.12 ± 0.19 | –73.3 | 0.126 | 0.35 ± 0.59 | –78.1 | 0.043* |
| SOP1812, 1 mg/kg, QW | 0.08 ± 0.14 | –82.2 | 0.020* | 0.17 ± 0.33 | –89.3 | 0.004*** |
| SOP1812, 1 mg/kg, BiW | 0.06 ± 0.08 | –86.6 | 0.018* | 0.15 ± 0.24 | –90.6 | 0.008** |
| gemcitabine, 15 mg/kg, BiW | 0.13 ± 0.07 | –71.1 | 0.016* | 0.39 ± 0.18 | –75.6 | 0.004*** |
| control | 0.45 ± 0.18 | 1.60 ± 0.44 |
Statistical analyses of average tumor volumes of treated versus control mice cohorts at the given time points used. Student’s t test: * P < 0.05, ** P < 0.01, *** P < 0.005.
Antitumor activity for SOP1812 was observed in a genetic model of PDAC. KPC (Pdx1-Cre; LSL-KrasG12D/+; LSL-Trp53R172H/+) mice express constitutively active KRASG12D and mutant p53R172H specifically in the pancreas and develop PDAC with many of the same features as humans32 and resistance to most chemotherapies. KPC mice were aged until palpable tumors were confirmed by ultrasound and then enrolled in treatment. SOP1812 was administered IV, 1 mg/kg 1× weekly, with six mice in each group (Figure 5b) for 3 weeks (four doses) and monitored until symptoms progressed, when animals were culled. Survival was significantly extended in mice treated with SOP1812, with a median survival in those mice of 24 days compared with 12 days in both the control group and the gemcitabine-treated mice (P = 0.016). One animal survived in the control group to day 18, whereas 3/6 of the SOP1812-treated group survived for 28 days or more, and 1 survived to 43 days. By contrast, in the gemcitabine group,26,33 only one animal survived >20 days.
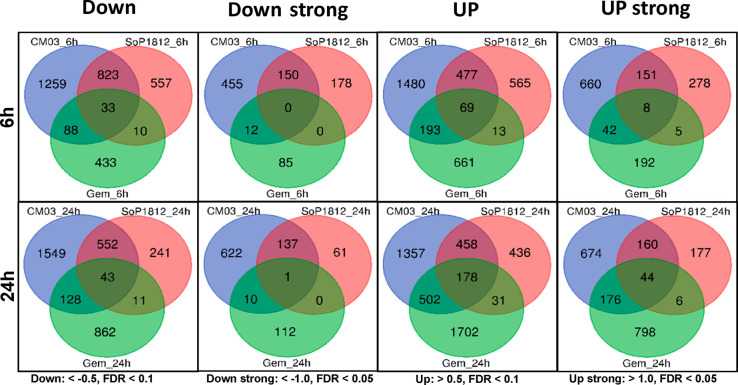
RNA-seq technology was used to assess transcriptional changes following dosing with SOP1812. The protocol employed was close to that used in the CM03 study.26 MIA PaCa-2 cells were exposed for 6 and 24 h to the compound. After 6 h of SOP1812 treatment, many genes were solely affected by SOP1812 but not by CM03 or gemcitabine; 178 genes were strongly down-regulated (<−1.0 log2 FC (fold change) values), and 557 had some change (log2 FC < −0.5) (Figure 6). These are two times fewer than the number of CM03-affected genes. With both, the number of affected genes is fewer at 24 h. There are few SOP1812-altered genes in common with those down- (or up-) regulated by gemcitabine (Figure 6) at 6 and 24 h. We conclude that SOP1812 is more potent than CM03 in cells and more selective of the number of genes that it down-regulates. Few of the genes down-regulated by SOP1812 are affected by gemcitabine.
Figure 6.
Venn plots showing numbers of down- and up-regulated genes common to CM03-,26 SOP1812-, and gemcitabine26-treated MIA PaCa-2 pancreatic cancer cells from RNA-seq analysis.
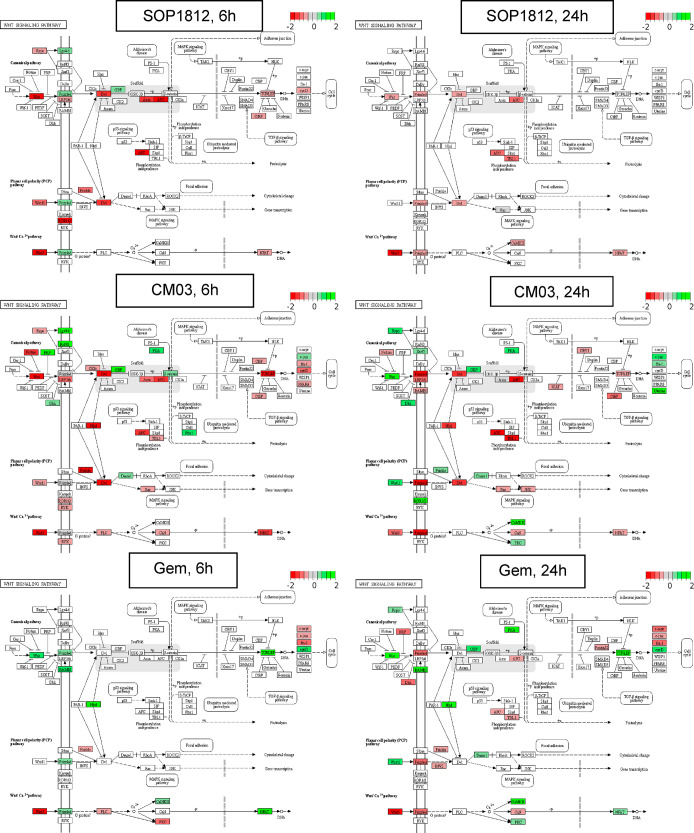
Table 5 highlights the pathways most affected by expression changes. Those in common for CM03 and SOP1812 treatment include Wnt/β-catenin (Figure 7), axon guidance, Hippo, MAPK, and Rap1 (Figure S1a–d). Several genes in the Wnt/β-catenin pathway were affected by SOP1812 (Figure 7): WNT5B, DVL1, AXIN1, and APC2 (Table 6). Their expression changes (and that of hTERT) are consistently greater at 6 h. Changes in several other genes are also noted in Table 6, with GLI1, MAPK11, and BCL-2 all increasing at 24 h.
Table 5. Results of the Transcriptome Study with SOP1812 and MIA PaCa-2 Cells: The Major Signalling Pathways Down-Regulated by CM03, SOP1812, and gemcitabine.
| compound/time | no. of down-regulated genes | top affected signaling pathways |
|---|---|---|
| CM03 6 h | 2203 | Axon guidance, Hippo, Rap1, MAPK, endocytosis, insulin resistance, TNF, Wnt |
| CM03 24 h | 2272 | ErbB, neurotrophin, insulin, Rap1, mTOR, axon guidance, AMPK, TNF, Wnt |
| SOP1812 6 h | 1423 | Hippo, TNF, Rap1, NF-kappa B, Wnt |
| SOP1812 24 h | 847 | Axon guidance, cell cycle, DNA replication, Rap1, Wnt |
| gemcitabine 6 h | 564 | cell cycle, TNF, FoxO, |
| gemcitabine 24 h | 1044 | ribosome biogenesis, spliceosome, RNA transport, Hippo, Wnt |
Figure 7.
Enriched Wnt/β-catenin pathway and gene set after 6 or 24 h of SOP1812, CM03, and gemcitabine treatment in MIA PaCa-2 cells from RNA-seq experiments. The KEGG pathway enrichment analysis used the complete down-regulated gene set (Log2FC < −0.5 and FDR < 0.1). Significant DEGs: red, down-regulated genes (Log2FC < −0.5 and FDR < 0.1); green, up-regulated genes (Log2FC > 0.5 and FDR < 0.1).
Table 6. Reduction in Expression in MIA PaCa-2 Cells for Selected Cancer-Related Genes Following Exposure to SOP1812, CM03, and Gemcitabinea.
| gene | Gem 6 h | Gem 24 h | CM03 6 h | CM03 24 h | SOP1812 6 h | SOP1812 24 h | protein function | no. of PQs |
|---|---|---|---|---|---|---|---|---|
| WNT5B | –0.56 | 0.06 | –1.77 | –0.97 | –2.25 | –0.57 | promotes cancer cell migration, invasion | 8 |
| DVL1 | –0.01 | –0.26 | –1.35 | –1.31 | –1.48 | –0.77 | key regulator of Wnt signaling pathway | 30 |
| AXIN1 | –0.33 | –0.11 | –0.73 | –1.43 | –1.59 | –0.39 | negative regulator of β-catenin in Wnt pathway | 18 |
| APC2 | –0.13 | –0.57 | –1.62 | –1.38 | –1.88 | –1.06 | regulator of Wnt signaling | 38 |
| GLI1 | –0.52 | –0.33 | –0.61 | –0.64 | –0.66 | –1.84 | oncogenic PDAC transcription factor | 15 |
| MAPK11 | 0.10 | 0.61 | –0.53 | –2.56 | –0.71 | –1.72 | essential effector kinase of p38 MAPK pathway | 18 |
| BCL-2 | 1.05 | 0.84 | –1.12 | –1.24 | –0.58 | –1.75 | antiapoptosis gene | 21 |
| hTERT | –0.10 | 0.31 | –1.66 | –2.47 | –2.29 | –1.02 | telomerase catalytic subunit | 15 |
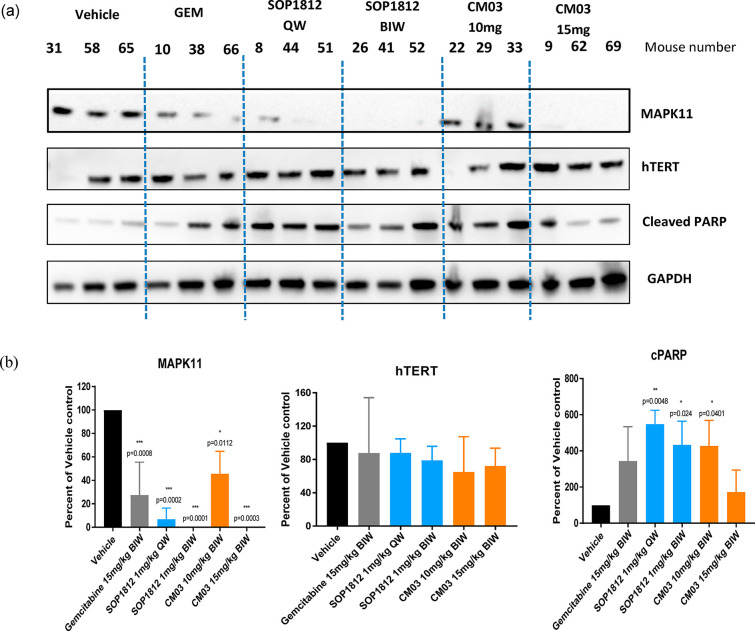
The expression of MAPK11 and hTERT proteins and the levels of cleaved PARP protein (an indicator of apoptosis) were estimated at the termination of xenograft experiment dosing. MAPK11 was reduced to undetectable levels (Figure 8) following SOP1812 (1 mg/kg) and CM03 (15 mg/kg) 2× weekly dosing (P ≤ 0.003) but not with gemcitabine or the lower CM03 dose. No significant changes in hTERT protein levels were found. The level of the cleaved PARP protein34 was the greatest in the SOP1812-dosed tumors (Figure 8a).
Figure 8.
(a) Western blots on a subset of proteins encoded by G4-containing genes, together with the PARP protein, extracted from tumors taken at day 28 in the MIA PaCa-2 xenograft experiments. (b) Densitometric quantitation of MAPK11, hTERT, and PARP proteins from the data in Figure 8a, averaged over tumors in three animals (panel a above). Student’s t test was used.
We have shown here that addition of a fourth directly attached aminopyrrolidino-phenyl group to the CM03 core results in a compound, SOP1812, with superior G4 affinity and potency. This design concept has two features that increase G4 binding: (i) the additional aryl ring, creating an extended π-system and increasing aromatic interactions of the core with G-quartets, and (ii) an additional basic amine that enhances electrostatic interactions with the phosphate backbone. We suggest that the ability of high levels of SOP1812 to efficiently compete with G4-specific BG4 antibody from nucleic acids in cell nuclei is due to its high G4 affinity, which is comparable to that of BG4 itself,15 and its greater concentration in the staining experiments (Figure 3). This result is supportive of the hypothesis that SOP1812 binds to cellular G4s.
The 10-fold increase in the cellular GI50 value for SOP1812 versus CM03 is consistent with a 10 times lower dose for significant antitumor activity in MIA PaCa-2 xenografts. The pharmacokinetics of CM03 and SOP1812 are broadly comparable to their in vivo responses. Overall, the high t1/2 values together with the cellular GI50 values indicate good bioavailability for both compounds. The SOP1812 activity in the KPC model (P = 0.016) is more significant than that with CM03 (P = 0.217). SOP1812 dosage regimens have not been optimized; our data from the xenograft experiments suggest that four 1 mg/kg doses suffice, but not necessarily in the KPC study. Overall, SOP1812 has a superior antitumor profile to CM03, at least in these two models. The KPC model is more representative of human PDAC, and the negligible responses with gemcitabine monotherapy35 are in accord with clinical experience.31,36 The differences are discussed in more detail in the SI.
The transcriptome data show that SOP1812 has greater selectivity compared with CM03. The effectiveness of SOP1812 in the PDAC models is, we suggest, due to its ability to affect several targets, which is a necessity in the case of advanced PDAC, a cancer with multiple genomic dysfunction.37 The observation that several key genes in the Wnt/β-catenin signaling pathway (Table 6, Figure 7) containing putative G4 elements are significantly down-regulated is in accord with other studies highlighting this pathway for small-molecule therapy in PDAC.38 The pathway genes in Table 6 are also affected by CM03 but not gemcitabine. SOP1812 dosing results in consistently greater changes in mRNA levels for the genes in the Wnt/β-catenin pathway at 6 h than at 24 h (Table 6), indicating that these effects occur early after dosing.
Changes in several other genes are highlighted in Table 6 and Figure 8. SOP1812 has high affinity for the hTERT promoter G4,28 which is consistent with the down-regulated expression in MIA PaCa-2 cells. hTERT G4-targeted down-regulation has been previously reported,20 with the down-regulation of hTERT protein levels in an in vivo prostate cancer xenograft model, in contrast with the unchanged hTERT tumor levels here (Figure 8). This difference may be functionally significant, a consequence of the differences between the tumor types. BCL-2 contains several G4 sites39,40 and is an apoptosis marker: It may also be a direct target for SOP1812 (Table 6). The MAPK11 gene, encoding for p38β MAP kinase in the MAP kinase pathway, is overexpressed in several human cancers. It is efficiently down-regulated in both cells and tumors and may be a potential marker of the SOP1812 response.
Acknowledgments
Work in the S.N. laboratory has been supported by the Medical Research Council, the Wellcome Trust, and the UCL Technology Fund. We thank Tony Brooks and the staff of the UCL Genomics Facility for expert assistance with RNA-seq studies at UCL. We are grateful to Professor Daniel Hochhauser (UCL Cancer Institute) for helpful discussions. S.A.K. and J.P.M. acknowledge the support of CRUK PRECISION-Panc (A25233), the Cancer Research UK Glasgow Centre (A25142), and the BSU facilities at the Cancer Research UK Beatson Institute (CRUK Experimental Medicine Programme Award (A25233 and A25265)). T.V. was supported by the Molecular Basis of Disease (MBD) fellowship from the College of Arts and Sciences, Georgia State University. Work in Atlanta was supported by the U.S. National Institutes of Health (NIH) (grant no. GM111749 to W.D.W. and D. W. Boykin).
Glossary
Abbreviations
- ND
naphthalene diimide
- G4
quadruplex
- PDAC
pancreatic ductal adenocarcinoma
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- SRB
sulforhodamine B
- KPC
Pdx1-Cre; LSL-KrasG12D/+; LSL-Trp53R172H/+
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.0c00317.
Experimental details, supplementary figures, and extended results of the RNA-seq experiments (PDF)
Author Contributions
S.N. conceived the overall project and coordinated the project with R.A. S.N. undertook the modeling, was involved in analyzing the RNA-seq and in vivo data, and oversaw writing the manuscript. A.A.A. undertook the detailed cell biology, the immunofluorescence experiments, and the transcriptome analyses and participated in the writing. M.M., R.A., and S.O. designed the chemistry and developed the synthetic route with the help of N.B., T.G.F., D.E.O., and M.S. J.W. supervised the xenograft and pharmacokinetic studies and their interpretation. S.A.K. and J.P.M. were responsible for the KPC mouse study. T.V. and W.D.W. performed the quantitative quadruplex binding studies. All authors interpreted their own data and contributed to sections of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Burge S.; Parkinson G. N.; Hazel P.; Todd A. K.; Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel J.; Adhikari S.; Balasubramanian S. The structure and function of DNA G-Quadruplexes. Trends in Chem. 2020, 2, 123–136. 10.1016/j.trechm.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert J. L.; Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd A. K.; Johnston M.; Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui-Jain A.; Grand C. L.; Bearss D. J.; Hurley L. H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 11593–11598. 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert J. L.; Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 406–413. 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigo R.; Palumbo M.; Sissi C. G-quadruplexes in human promoters: A challenge for therapeutic applications. Biochim. Biophys. Acta, Gen. Subj. 2017, 1861, 1399–1413. 10.1016/j.bbagen.2016.12.024. [DOI] [PubMed] [Google Scholar]
- Bugaut A.; Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012, 40, 4727–4741. 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney D.; Spiegel J.; Zyner K.; Tannahill D.; Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 10.1038/s41580-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wietmarschen N.; Merzouk S.; Halsema N.; Spierings D. C. J.; Guryev V.; Lansdorp P. M. BLM helicase suppresses recombination at G-quadruplex motifs in transcribed genes. Nat. Commun. 2018, 9, 271. 10.1038/s41467-017-02760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyner K. G; Mulhearn D. S; Adhikari S.; Martinez Cuesta S.; Di Antonio M.; Erard N.; Hannon G. J; Tannahill D.; Balasubramanian S.; et al. Genetic interactions of G-quadruplexes in humans. eLife 2019, 8, e46793 10.7554/eLife.46793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Yang J.; Wild A. T.; Wu W. H.; Shah R.; Danussi C.; Riggins G. J.; Kannan K.; Sulman E. P.; Chan T. A.; et al. G-quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX-deficient malignant glioma. Nat. Commun. 2019, 10, 943. 10.1038/s41467-019-08905-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner L. K.; Sale J. E. Replication of G quadruplex DNA. Genes 2019, 10, 95. 10.3390/genes10020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel-Hertsch R.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- Biffi G.; Tannahill D.; McCafferty J.; Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A.; Wu Y.; Huang Y. C.; Chavez E. A.; Platt J.; Johnson F. B.; Brosh R. M. Jr.; Sen D.; Lansdorp P. M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014, 42, 860–869. 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S.; Hurley L. H.; Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy?. Nat. Rev. Drug Discovery 2011, 10, 261–275. 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle S. Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem. 2016, 59, 5987–6011. 10.1021/acs.jmedchem.5b01835. [DOI] [PubMed] [Google Scholar]
- Asamitsu S.; Bando T.; Sugiyama H. Ligand design to acquire specificity to intended G-quadruplex structures. Chem. - Eur. J. 2019, 25, 417–430. 10.1002/chem.201802691. [DOI] [PubMed] [Google Scholar]
- Calabrese D. R.; Chen X.; Leon E. C.; Gaikwad S. M.; Phyo Z.; Hewitt W. M.; Alden S.; Hilimire T. A.; He F.; Michalowski A. M.; et al. Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex. Nat. Commun. 2018, 9, 4229. 10.1038/s41467-018-06315-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. H.; et al. Small-molecule-targeting hairpin loop of hTERT promoter G-quadruplex induces cancer cell death. Cell Chem. Biol. 2019, 26, 1110–1121. 10.1016/j.chembiol.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirota V.; Nadai M.; Doria F.; Richter S. N. Naphthalene diimides as multimodal G-quadruplex-selective ligands. Molecules 2019, 24, 426. 10.3390/molecules24030426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collie G. W.; Promontorio R.; Hampel S. M.; Micco M.; Neidle S.; Parkinson G. N. Structural basis for telomeric G-quadruplex targeting by naphthalene diimide ligands. J. Am. Chem. Soc. 2012, 134, 2723–2731. 10.1021/ja2102423. [DOI] [PubMed] [Google Scholar]
- Micco M.; Collie G. W.; Dale A. G.; Ohnmacht S. A.; Pazitna I.; Gunaratnam M.; Reszka A. P.; Neidle S. Structure-based design and evaluation of naphthalene diimide G-quadruplex ligands as telomere targeting agents in pancreatic cancer cells. J. Med. Chem. 2013, 56, 2959–2974. 10.1021/jm301899y. [DOI] [PubMed] [Google Scholar]
- Ohnmacht S. A; Marchetti C.; Gunaratnam M.; Besser R. J; Haider S. M; Di Vita G.; Lowe H. L; Mellinas-Gomez M.; Diocou S.; Robson M.; et al. A G-quadruplex-binding compound showing anti-tumour activity in an in vivo model for pancreatic cancer. Sci. Rep. 2015, 5, 11385. 10.1038/srep11385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti C.; et al. Targeting multiple effector pathways in pancreatic ductal adenocarcinoma with a G-quadruplex-binding small molecule. J. Med. Chem. 2018, 61, 2500–2517. 10.1021/acs.jmedchem.7b01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin A. V.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- Vo T.; Oxenford S.; Angell R.; Marchetti C.; Ohnmacht S. A.; Wilson W. D.; Neidle S. Substituted naphthalenediimide compounds bind selectively to two human quadruplex structures with parallel topology. ACS Med. Chem. Lett. 2020, 11, 991–999. 10.1021/acsmedchemlett.0c00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K. W.; Lacroix L.; Yue D. J.; Lim J. K.; Lim J. M.; Phan A. T. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J. Am. Chem. Soc. 2010, 132, 12331–12342. 10.1021/ja101252n. [DOI] [PubMed] [Google Scholar]
- Gkionis K.; Kruse H.; Šponer J. Derivation of reliable geometries in QM calculations of DNA structures: explicit solvent QM/MM and restrained implicit solvent QM optimizations of G-quadruplexes. J. Chem. Theory Comput. 2016, 12, 2000–2016. 10.1021/acs.jctc.5b01025. [DOI] [PubMed] [Google Scholar]
- Taieb J.; Prager G. W.; Melisi D.; Westphalen C. B.; D’Esquermes N.; Ferreras A.; Carrato A.; Macarulla T. First-line and second-line treatment of patients with metastatic pancreatic adenocarcinoma in routine clinical practice across Europe: a retrospective, observational chart review study. ESMO Open 2020, 5, e000587 10.1136/esmoopen-2019-000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani S. R.; et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005, 7, 469–483. 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Steele C. W.; et al. CXCR2 inhibition profoundly suppresses metastases and augments immunotherapy in pancreatic ductal adenocarcinoma. Cancer Cell 2016, 29, 832–845. 10.1016/j.ccell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker P.; Isenberg D.; Muller S. Inhibition of caspase-3-mediated poly(ADP-ribose) polymerase (PARP) apoptotic cleavage by human PARP autoantibodies and effect on cells undergoing apoptosis. J. Biol. Chem. 2000, 275, 9043–9046. 10.1074/jbc.275.12.9043. [DOI] [PubMed] [Google Scholar]
- Deer E. L.; et al. Phenotype and genotype of pancreatic cancer cell lines. Pancreas 2010, 39, 425–35. 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht S.; Feldmann G. Animal models for modeling pancreatic cancer and novel drug discovery. Expert Opin. Drug Discovery 2019, 14, 127–142. 10.1080/17460441.2019.1566319. [DOI] [PubMed] [Google Scholar]
- Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 2017, 32, 185–203. 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javadinia S. A.; et al. Therapeutic potential of targeting the Wnt/β-catenin pathway in the treatment of pancreatic cancer. J. Cell. Biochem. 2019, 120, 6833–6840. 10.1002/jcb.27835. [DOI] [PubMed] [Google Scholar]
- Montoya J. J.; Turnidge M. A.; Wai D. H.; Patel A. R.; Lee D. W.; Gokhale V.; Hurley L. H.; Arceci R. J.; Wetmore C.; Azorsa D. O.; et al. In vitro activity of a G-quadruplex-stabilizing small molecule that synergizes with Navitoclax to induce cytotoxicity in acute myeloid leukemia cells. BMC Cancer 2019, 19, 1251. 10.1186/s12885-019-6464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaratnam M.; Collie G. W.; Reszka A. P.; Todd A. K.; Parkinson G. N.; Neidle S. A naphthalene diimide G-quadruplex ligand inhibits cell growth and down-regulates BCL-2 expression in an imatinib-resistant gastrointestinal cancer cell line. Bioorg. Med. Chem. 2018, 26, 2958–2964. 10.1016/j.bmc.2018.04.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.