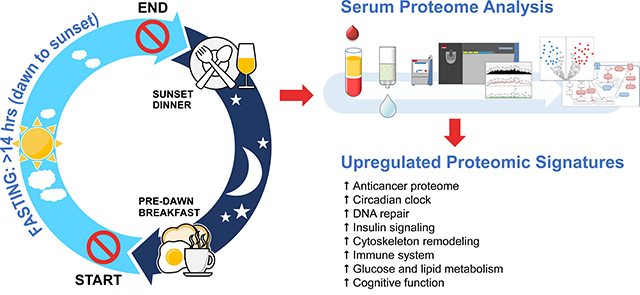
Abstract
Murine studies showed that disruption of circadian clock rhythmicity could lead to cancer and metabolic syndrome. Time-restricted feeding can reset the disrupted clock rhythm, protect against cancer and metabolic syndrome. Based on these observations, we hypothesized that intermittent fasting for several consecutive days without calorie restriction in humans would induce an anticarcinogenic proteome and the key regulatory proteins of glucose and lipid metabolism. Fourteen healthy subjects fasted from dawn to sunset for more than 14 hours daily. Fasting duration was 30 consecutive days. Serum samples were collected before 30-day intermittent fasting, at the end of 4th week during 30-day intermittent fasting, and one week after 30-day intermittent fasting. An untargeted serum proteomic profiling was performed using ultra high-performance liquid chromatography/tandem mass spectrometry. Our results showed that 30-day intermittent fasting was associated with an anticancer serum proteomic signature, upregulated key regulatory proteins of glucose and lipid metabolism, circadian clock, DNA repair, cytoskeleton remodeling, immune system, and cognitive function, and resulted in a serum proteome protective against cancer, metabolic syndrome, inflammation, Alzheimer’s disease, and several neuropsychiatric disorders. These findings suggest that fasting from dawn to sunset for 30 consecutive days can be preventive and adjunct therapy in cancer, metabolic syndrome, and several cognitive and neuropsychiatric diseases.
Graphical Abstract
INTRODUCTION
The disruption of circadian rhythm has been associated with alterations in glucose and lipid metabolism and immune system responses, and carcinogenesis.[1, 2] Resetting the disrupted rhythm of the circadian clock could be a key strategy in the prevention of metabolic syndrome, immune system dysfunction, and cancer.[3, 4] There are two primary mechanisms to reset the circadian clock. The first mechanism functions through the master clock located in the suprachiasmatic nucleus of the anterior hypothalamus that is entrained by dark-light cycles of the day.[5–11] All peripheral clocks are then synchronized by the master clock via neuronal and humoral signals[5–12], and this appears to be the dominant mechanism resetting all peripheral clocks, including hepatic clock during ad libitum food consumption. The second mechanism to reset the circadian clock works in response to mealtime during rhythmic, consecutive, time-restricted feeding-fasting cycles.[5, 7, 10] Rhythmic consecutive time-restricted feeding-fasting cycles have shown to release peripheral clocks, including the hepatic clock, from the control of the master clock and entrain them independent of the master clock.[5, 7, 10] Uncoupling of the peripheral clocks from the master clock shifts and resets the phase of the peripheral clocks.[5, 7, 10] As such, mealtime and duration between meals are critical in resetting and maintaining the circadian rhythmicity of the peripheral clocks.[13]
Murine studies showed that time-restricted access or no access to food during night time/dark phase resets the phase of the hepatic clock, optimizes the amplitude of hepatic clock oscillations, and results in the upregulation of mRNA and various protein synthetic pathways, including enzymes that play a crucial role in carbohydrate and lipid metabolism.[3, 5, 7, 9, 10, 14] Mice are nocturnal feeders; most food consumption and activity occur at night.[5, 7, 9, 10, 14, 15] In contrast, in humans, most activity and meal intake occur during the daytime. Therefore, to reproduce similar optimization in key metabolic regulatory proteins in humans, fasting should occur during the daytime activity for several consecutive days. Preserving daytime activity and timing major food consumption at transition zones of the day with a predawn breakfast and dinner at sunset may be as important as caloric content and composition of the food in the prevention of metabolic syndrome and its complications and cancer.[13]
Since time-restricted access or no access to food during active phase (night time/dark phase for mice) resets the phase of hepatic circadian rhythm and optimizes the functioning of critical regulatory proteins of metabolism in mice [3, 5, 7, 9, 10, 14], we formulated and tested the hypothesis that consecutive rhythmic intermittent fasting during active hours (from dawn to sunset for humans) could produce similar optimization in key regulatory proteins protective against cancer, inflammation, metabolic syndrome, and its complications.
METHODS
Study Subjects
This study was approved by the Institutional Review Board of the Baylor College of Medicine Biomedical Research and Assurance Information Network (BRAIN) under protocol number H-31612 and written informed consent was obtained from all subjects. Inclusion criteria were as follows: 1) Subjects who are 18 years old or older; 2) Subjects who plan to fast during the month of Ramadan[13]; 3) Subjects who are in excellent general health and do not take daily medication for any condition and report no acute illnesses or symptoms at the time of enrollment. Subjects were excluded if they had any of the following: 1) Body mass index equal to or greater than 30 kg/m2; 2) History of acute, sub-acute, or chronic disease; 3) Use of any daily medications (occasional use of over-the-counter medications to relieve pain, such as acetaminophen or ibuprofen, in minimal to moderate amounts, was permitted); 4) Use of alcohol or recreational substances.
Study Procedures
A 1-hour screening visit was scheduled within 12 weeks of initiation of 30-day intermittent fasting at Baylor College of Medicine in the Texas Medical Center Digestive Diseases Center Clinical Research Core E Laboratory. During this visit, the subjects’ eligibility was assessed based on inclusion and exclusion criteria, and written informed consent was taken. Medical history and physical examination were performed. Urine pregnancy for the female subject at childbearing age was performed.
Daily fasting began at dawn after a predawn breakfast and ended at sunset with a dinner for 30 consecutive days. Fasting occurred without eating or drinking between predawn breakfast and dinner at sunset (lunch, liquids, water, snacks were skipped). There was no calorie restriction, otherwise. Participants were instructed to continue with their usual diet during their non-fasting hours.
Blood specimens for biomarkers and proteomic analysis were collected within four weeks before the initiation of 30-day intermittent fasting after an overnight fast, at the end of 4th week during 30-day intermittent fasting after at least 8 hours of fast, and one week after completion of 30-day intermittent fasting after an overnight fast.
To confirm future compliance during the study (a daytime rise in enrichment indicates non-compliance to fasting), a baseline 13C-isotopic breath enrichment test as described by Opekun et al.[16] was performed within four weeks before 30-day intermittent fasting (baseline). Thereafter, 13C-isotopic breath enrichment samples were collected throughout the study until the end of 30-day intermittent fasting to assure protocol compliance during the study.
Serum Proteomics
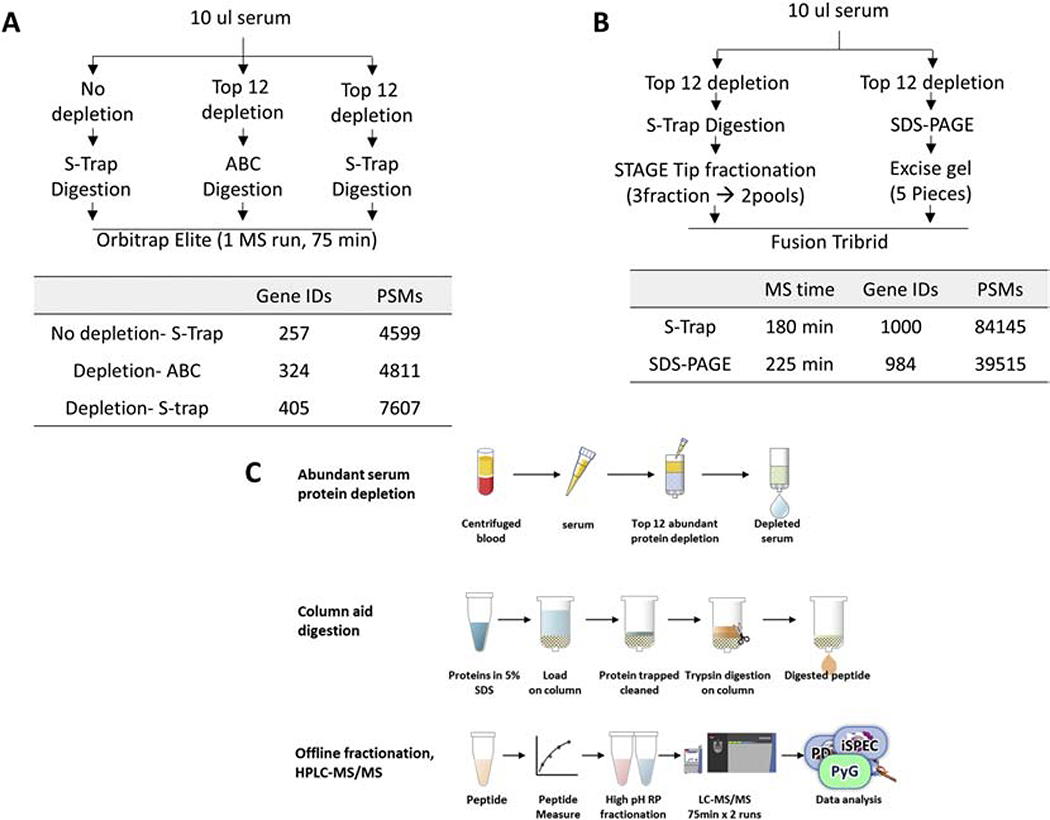
Several methods of serum sample preparation were tested, as shown in Figure 1 to establish a workflow to get the best peptide and proteome coverage. Each of these methods utilized 10μl volume of serum. For the non-depleted serum samples, 10μl serum was subjected directly for S-Trap aided digestion method. The peptides were resuspended in ultra high-performance liquid chromatography (HPLC) loading buffer (5% MeOH, 0.1% FA) and subjected to nLC1000 coupled Orbitrap Elite. For the top 12 abundant protein depletion, 10μl of serum was incubated with the resin slurry of the depletion kit (Thermo Scientific Pierce, Cat# 85164) for an hour at room temperature followed by digestion on S-Trap column (ProtiFi, NY). The S-trap column aid digestion was carried out using the trypsin enzyme for 1 hour at 470C. The digested peptide was extracted by 0.2% formic acid, followed by 50% acetonitrile containing 0.2% formic acid and dried using speed vac. The peptide concentration was measured using Pierce™ Quantitative Colorimetric Peptide Assay (Thermo Scientific 23275). For fractionation, high pH STAGE fractionation of peptide or SDS-PAGE of protein was used. For SDS-PAGE, after top 12 protein depletion, the serum was boiled with 2X SDS-PAGE sample loading buffer (Invitrogen Cat# NP0007), then resolved on 10% Bis-Tris gel (Invitrogen Cat #NP0315BOX). The gel was stained by Coomassie blue and dissected into five slices for in gel digestion, as previously described.[17] For STAGE Tip fractionation, 10μg of dried peptides were dissolved in 100μl of pH 10 buffer (10 mM ammonium bicarbonate, pH 10, adjusted by NH4OH) and subjected to a micro-pipette tip C18 column made from a 200-μl pipette tip by layering 2 mg of C18 matrix (Reprosil-Pur Basic C18, 3 μm, Dr. Maisch GmbH, Germany) on top of the C18 disk (3M, Empore™ C18) plug. The peptides were eluted with step gradient of 100μl of 9%, 21%, and 35% acetonitrile (in pH 10 buffer) and pooled into 2 pools (9% with 35% eluent, and 21% eluent) and vacuum-dried for nano ultra high-performance liquid chromatography/tandem mass spectrometry (nano-HPLC-MS/MS). Dried peptides were dissolved in 20μl of loading solution (5% methanol containing 0.1% formic acid) and subjected to nano-HPLC-MS/MS assay as described previously.[18]
Figure 1.
Test of different sample preparation methods. A. Comparison of the effect of Top 12 abundant protein depletion, direct in-solution digestion and S-Trap aid in column digestion method. B. Comparison of the fractionation method between high pH STAGE tip method and SDS-PAGE. Recovered protein (shown in Gene Protein Product, GPs) and Peptide Spectrum Matches (PSMs) numbers are representative results from triple repeat. C. Schematic illustration of established workflow in the serum profiling process. S-Trap aided trypsin digestion, high pH STAGE tip method for preparation of serum samples was used.
For Orbitrap Elite MS analysis, digested peptides were analyzed by a nano-HPLC 1000 system (Thermo Scientific) coupled to an Orbitrap Elite Hybrid mass spectrometer. An in-housed trap column packed with 1.9 μm Reprosil-Pur Basic C18 beads (2 cm X 100 μm) and an in-housed 5 cm X 150 μm capillary column packed with 1.9 μm Reprosil-Pur Basic C18 beads were used. A 75-min discontinuous gradient of 4–26% acetonitrile, 0.1% formic acid at a flow rate of 800 nl/min was applied to column then electro-sprayed into the mass spectrometer. The instrument was operated under the control of Xcalibur software version 2.2 (Thermo Fisher Scientific) in data-dependent mode, acquiring fragmentation spectra of the top 25 strongest ions. Parent mass spectrum was acquired in the Orbitrap with full MS range of 375–1300 m/z at the resolution of 240,000. Collison-induced dissociation (CID) fragmented MS/MS spectrum was acquired in ion-trap with rapid scan mode.
For Fusion Lumos MS analysis, digested peptides were analyzed by a nano-HPLC 1200 system (Thermo Scientific) coupled to an Orbitrap Fusion™ Lumos™ Tribrid™ (Fusion Lumos, Thermo Scientific) mass spectrometer. An in-housed trap column packed with 3 μm Reprosil-Pur Basic C18 beads (2 cm × 100 μm) and an in-housed 5 cm × 150 μm capillary column packed with 1.9 μm Reprosil-Pur Basic C18 beads were used. A 90-min discontinuous gradient of 2– 24% acetonitrile, 0.1% formic acid at a flow rate of 800 nl/min was applied to column then electro-sprayed into the mass spectrometer. The instrument was operated under the control of Xcalibur software version 4.1 (Thermo Fisher Scientific) in data-dependent mode, acquiring fragmentation spectra of the top 50 strongest ions. Parent MS spectrum was acquired in the Orbitrap with full MS range of 300–1400 m/z in the resolution of 120,000. Higher-energy collisional dissociation (HCD) fragmented MS/MS spectrum was acquired in ion-trap with rapid scan mode. The MS/MS spectra were searched against target-decoy Human RefSeq database (release 2015.06, containing 73,637 entries) in Proteome Discoverer 2.1 interface (Thermo Fisher) with Mascot algorithm (Mascot 2.4, Matrix Science). The precursor mass tolerance of 20 ppm and fragment mass tolerance of 0.5 Dalton was allowed. Two maximum missed cleavage, and dynamic modifications of acetylation of N-term and oxidation of methionine were allowed. Assigned peptides were filtered with a 1% false discovery rate (FDR) using Percolator validation based on q-value. The Peptide Spectrum Matches (PSMs) output from PD2.1 was used to group peptides onto gene level using ‘gpGrouper’ algorithm.[19] An in-housed program, gpGrouper, uses a universal peptide grouping logic to accurately allocate and provide MS1 based quantification across multiple gene products. Gene-protein products (GPs) quantification was performed using the label-free, intensity-based absolute quantification (iBAQ) approach and then normalized to FOT (a fraction of the total protein iBAQ amount per experiment). FOT was defined as an individual protein’s iBAQ divided by the total iBAQ of all identified proteins within one experiment.
Statistical Analysis
Statistical analysis of proteomics was done using Microsoft Excel Program (Microsoft, Redmond, WA). Paired two-tailed Student’s t-test using log converted iFOT was used to determine statistically significantly regulated proteins from samples collected before 30-day intermittent fasting, at the end of 4th week during 30-day intermittent fasting and one week after 30-day intermittent fasting. Proteins that showed P < 0.05 and equal to or greater than 4-fold change were considered as significant. Volcano plot analysis was performed to display the GP levels that had an equal to or greater than 4-fold significant change at the end of 4th week during 30-day intermittent fasting and one week after 30-day intermittent fasting compared with the GP levels before 30-day intermittent fasting.
Conventional Metabolic Parameters and Serum Biomarkers
Several metabolic parameters including weight, body mass index, temperature, systolic and diastolic blood pressures, mean arterial pressure, lipid profile (total cholesterol, triglyceride, high-density lipoprotein, low-density lipoprotein), insulin, glucose, homeostatic model assessment-insulin resistance (HOMA-IR)[20], homocysteine, C-reactive protein, interleukin 1b, interleukin 6, interleukin 8, tumor necrosis factor-alpha, leptin, and adiponectin were measured within four weeks before 30-day intermittent fasting, at the end of 4th week during 30-day intermittent fasting, and one week after completion of 30-day intermittent fasting.
Statistical Analysis
Statistical analysis of clinical metabolic parameters and serum biomarkers were performed using SAS Version 9.4 TS Level 1M5 X64_10PRO platform (SAS, Cary, NC).[21] Student’s paired t-test was used to compare the levels of continuous variables measured at the end of 4th week during 30-day intermittent fasting, and one week after completion of 30-day intermittent fasting with the levels measured before 30-day intermittent fasting. A two-tailed P value of < 0.05 was considered statistically significant.
Fecal Microbiota
The fecal microbiota of 70 fecal samples (14 subjects × 5 samples collected from each subject) was profiled for the analysis of samples obtained before 30-day intermittent fasting (before), at the 2nd and 4th week during 30-day intermittent fasting (during), and one week after 30-day intermittent fasting (after). The 16S rRNA gene sequencing methods were adapted from the methods developed for the Earth Microbiome Project [22,23] and NIH-Human Microbiome Project [24,25]. Fecal microbiota analysis is described in Supplementary Materials.
RESULTS
Subjects
Fourteen healthy subjects (13 males:1 female) with a mean age of 32 were enrolled in the study. All subjects fasted for more than 14 hours daily for 30 consecutive days beginning from May 16, 2018, until June 14, 2018, except for one subject who fasted for 26 days. The minimum required duration of daily fasting was 14 hours, 23 minutes for the shortest day (May 16, 2018), and 14 hours, 48 minutes for the longest day (June 14, 2018). All subjects tolerated intermittent fasting (no food or drink) well without any complications. The first blood collection occurred before the initiation of 30-day intermittent fasting. The second blood collection occurred on an average of 28 days after the initiation of 30-day intermittent fasting (at the end of the 4th week during 30-day intermittent fasting). The third blood collection occurred on an average of 8.5 days after the completion of 30-day intermittent fasting (one week after 30-day intermittent fasting).
Serum Proteomics
Several sample preparation methods were tested to find the best way for more in-depth proteome coverage. As shown in Figure 1A, the top 12 abundant protein depletion provides around 58% proteome increase (257 vs. 405 GPs). Suspension traps (S-Trap) were recently reported as a sensitive and time-saving way of proteome profiling sample preparation.[26] Compared to direct in-solution digest, S-Trap provides a 25% better recovery in proteome coverage (324 vs. 405 GPs). Also, we tested two different ways of pre-sample fractionation before UPLC-MS/MS. Comparing high pH STAGE tip and SDS-PAGE, the STAGE tip provides deeper proteome coverage within shorter MS analysis time (Figure 1B). So, we decided to use S-Trap aided trypsin digestion, high pH STAGE tip method for the preparation of fasting serum samples. The established workflow is shown in Figure 1C.
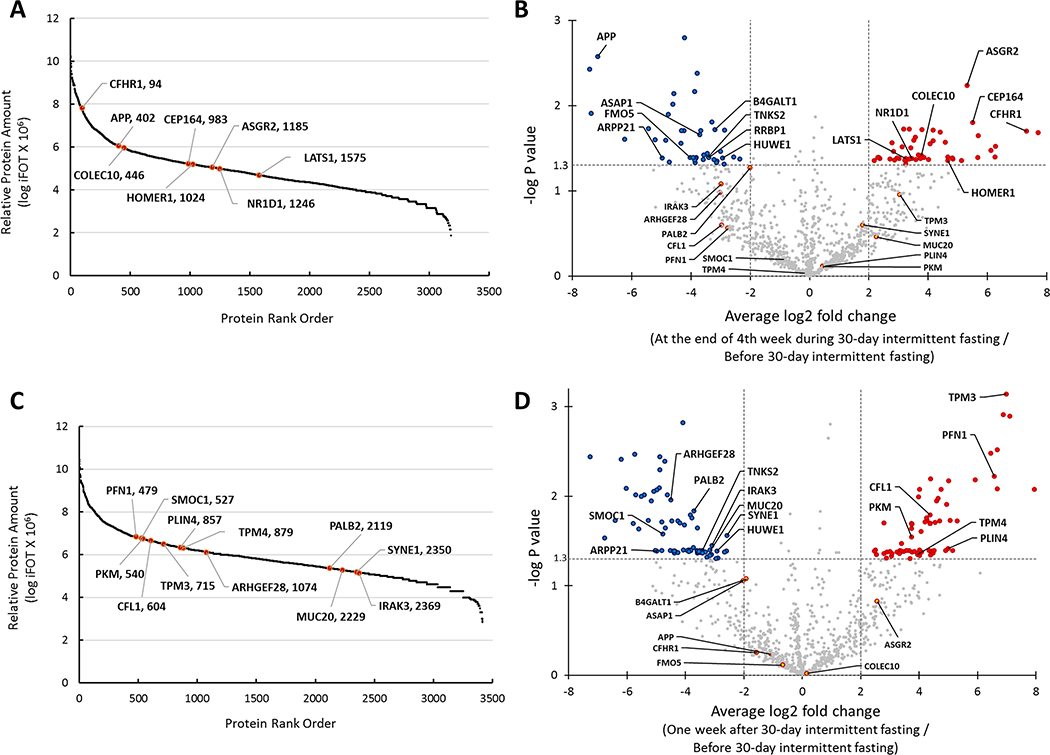
Proteome coverage and its dynamic order of average iFOT values from the end of 4th week during 30-day intermittent fasting samples are shown in Figure 2A. A total of 3181 GPs were recovered with over eight orders of magnitude of dynamic range. There was significant fold change in the levels of multiple GPs at the end of 4th week during 30-day intermittent fasting compared with the levels before 30-day intermittent fasting (Figure 2B, Supplementary Table S1). Figure 2A, Figure 2B, and Table 1A display selected GPs of interest that are associated with immune system regulation, DNA repair, carcinogenesis, tumor suppression, circadian clock, Alzheimer’s disease, and neuropsychiatric disorders. There was an average 40 fold increase in asialoglycoprotein receptor 2 (ASGR2) (log2 fold=5.315, P=0.0058), 45 fold increase in the centrosomal protein 164 (CEP164) (log2 fold=5.499, P=0.0157), 160 fold increase in complement factor H related 1 (CFHR1) (log2 fold=7.320, P=0.0199), 14 fold increase in collectin subfamily member 10 (COLEC10) (log2 fold=3.781, P=0.0383), 9 fold increase in large tumor suppressor kinase 1 (LATS1) (log2=3.243, P=0.0415), 11 fold increase in NR1D1 nuclear receptor subfamily 1 group D member 1 (NR1D1) (log2=3.455, P=0.0417), and 25 fold increase in homer scaffold protein 1 (HOMER1) (log2 fold=4.664, P=0.0443) GP levels at the end of 4th week during 30-day intermittent fasting compared with the levels before 30-day intermittent fasting. The amount of these GPs of interest was relatively high, located in the top 50% rank order (Figure 2A). We found a significant reduction in the amyloid beta precursor protein (APP) (log2 fold=−7.147, P=0.0026), beta-1,4-galactosyltransferase 1 (B4GALT1) (log2 fold=−3.194, P=0.0192), ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 (ASAP1) (log2 fold=−3.715, P=0.0219), tankyrase 2 (TNKS2) (log2 fold=−3.416, P=0.0402), flavin containing dimethylaniline monoxygenase 5 (FMO5) (log2 fold=−4.031, P=0.0406), ribosome binding protein 1 (RRBP1) (log2 fold=−3.403, P=0.0408), cAMP regulated phosphoprotein 21 (ARPP21) (log2 fold=−4.977, P=0.0410) and HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1 (HUWE1) (log2 fold=−2.931, P=0.0411) GP levels at the end of 4th week during 30-day intermittent fasting compared with the levels before 30-day intermittent fasting.
Figure 2.
A. Distribution of normalized relative gene protein product (GP) amount and location of significantly changed selected proteins in serum samples taken at the end of 4th week during 30-day intermittent fasting shown in GP name and rank order. B. Volcano plot shows GPs that had an equal to or greater than 4-fold significant change (blue and red colors represent a significant decrease and increase in the levels of GPs, respectively) at the end of 4th week during 30-day intermittent fasting compared with the levels before 30-day intermittent fasting. C. Distribution of normalized relative GP amount and location of significantly changed selected proteins in serum samples taken one week after 30-day intermittent fasting shown in GP name and rank order. D. Volcano plot shows GPs that had an equal to or greater than 4-fold significant change (blue and red colors represent a significant decrease and increase in the levels of GPs, respectively) one week after 30-day intermittent fasting compared with the levels before 30-day intermittent fasting.
Table 1.
Selected gene protein products (gp)s that are significantly up- or downregulated at the end of 4th week during 30-day intermittent fasting (A) and one week after 30-day intermittent fasting compared with baseline (before 30-day intermittent fasting) in healthy subjects.
| A. Selected Gene Protein Products (GP)s that Are Significantly Up- or Downregulated at the End of 4th Week During 30-Day Intermittent Fasting Compared with Baseline (Before 30-Day Intermittent Fasting) | ||||
| Upregulated Gene Symbol | Upregulated Gene Name | Upregulated Gene ID | Average Log2 Fold Change | Paired P Value |
| ASGR2 | asialoglycoprotein receptor 2 | 433 | 5.315 | 0.0058 |
| CEP164 | centrosomal protein 164 | 22897 | 5.499 | 0.0157 |
| CFHR1 | complement factor H related 1 | 3078 | 7.320 | 0.0199 |
| COLEC10 | collectin subfamily member 10 | 10584 | 3.781 | 0.0383 |
| LATS1 | large tumor suppressor kinase 1 | 9113 | 3.243 | 0.0415 |
| NR1D1 | nuclear receptor subfamily 1 group D member 1 | 9572 | 3.455 | 0.0417 |
| HOMER 1 | homer scaffold protein 1 | 9456 | 4.664 | 0.0443 |
| Downregulated Gene Symbol | Downregulated Gene Name | Downregulated Gene ID | Average Log2 Fold Change | Paired P-Value |
| APP | amyloid beta precursor protein | 351 | −7.147 | 0.0026 |
| B4GALT1 | beta-1,4-galactosyltransferase 1 | 2683 | −3.194 | 0.0192 |
| ASAP1 | ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 | 50807 | −3.715 | 0.0219 |
| TNKS2 | tankyrase 2 | 80351 | −3.416 | 0.0402 |
| FMO5 | flavin containing dimethylaniline monoxygenase 5 | 2330 | −4.031 | 0.0406 |
| RRBP1 | ribosome binding protein 1 | 6238 | −3.403 | 0.0408 |
| ARPP21 | cAMP regulated phosphoprotein 21 | 10777 | −4.977 | 0.0410 |
| HUWE1 | HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1 | 10075 | −2.931 | 0.0411 |
| B. Selected GPs that Are Significantly Up- or Downregulated One Week after 30-Day Intermittent Fasting Compared with Baseline (Before 30-Day Intermittent Fasting) | ||||
| Upregulated Gene Symbol | Upregulated Gene Name | Upregulated Gene ID | Average Log2 Fold Change | Paired P Value |
| TPM3 | tropomyosin 3 | 7170 | 6.988 | 0.0007 |
| PFN1 | profilin 1 | 5216 | 6.566 | 0.0060 |
| CFL1 | cofilin 1 | 1072 | 4.375 | 0.0162 |
| PKM | pyruvate kinase M1/2 | 5315 | 3.743 | 0.0287 |
| PLIN4 | perilipin 4 | 729359 | 4.997 | 0.0383 |
| TPM4 | tropomyosin 4 | 7171 | 3.938 | 0.0446 |
| Downregulated Gene Symbol | Downregulated Gene Name | Downregulated Gene ID | Average Log2 Fold Change | Paired P-Value |
| ARHGEF28 | Rho guanine nucleotide exchange factor 28 | 64283 | −4.510 | 0.0111 |
| PALB2 | partner and localizer of BRCA2 | 79728 | −3.715 | 0.0147 |
| SMOC1 | SPARC related modular calcium binding 1 | 64093 | −4.776 | 0.0264 |
| SYNE1 | spectrin repeat containing nuclear envelope protein 1 | 23345 | −2.576 | 0.0274 |
| IRAK3 | interleukin 1 receptor associated kinase 3 | 11213 | −3.097 | 0.0357 |
| TNKS2 | tankyrase 2 | 80351 | −3.416 | 0.0402 |
| MUC20 | mucin 20, cell surface associated | 200958 | −3.149 | 0.0404 |
| ARPP21 | cAMP regulated phosphoprotein 21 | 10777 | −4.977 | 0.0410 |
| HUWE1 | HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1 | 10075 | −2.931 | 0.0411 |
The proteome coverage and its dynamic order from triplicate of samples collected one week after 30-day intermittent fasting are shown in Figure 2C. A total of 3416 GPs recovered with over seven orders of magnitude of dynamic range. There was a significant fold change in the levels of multiple GPs one week after 30-day intermittent fasting compared with the levels before 30-day intermittent fasting (Figure 2D, Supplementary Table S2). Figure 2C, Figure 2D, and Table 1B display selected GPs of interest that are associated with insulin signaling, cytoskeleton remodeling, glucose and lipid metabolism, carcinogenesis, Alzheimer’s disease and neuropsychiatric disorders. There was an average 127-fold increase in the tropomyosin 3 (TPM3), (log2 fold=6.988, P=0.0007), 95-fold increase in profilin 1 (PFN1) (log2 fold= 6.566, P=0.0060), 21-fold increase in cofilin 1 (CFL1) (log2 fold=4.375, P=0.0162), 13-fold increase in pyruvate kinase M1/2 (PKM) (log2 fold= 3.743, P=0.0287), 32-fold increase in perilipin 4 (PLIN4) (log2 fold=4.997, P=0.0383), and 15-fold increase in tropomyosin 4 (TPM4) (log2 fold=3.938, P=0.0446) GP levels one week after 30-day intermittent fasting compared with the levels before 30-day intermittent fasting (Table 1B). The amount of these GPs was relatively high, rank order between 479–879 out of 3416 GPs (Figure 2C). We found a significant reduction in the Rho guanine nucleotide exchange factor 28 (ARHGEF28) (log2 fold=−4.510, P=0.0111), partner and localizer of BRCA2 (PALB2) (log2 fold=−3.715, P=0.0147), SPARC related modular calcium binding 1 (SMOC1) (log2 fold=−4.776, P=0.0264), spectrin repeat containing nuclear envelope protein 1 (SYNE1) (log2 fold=−2.576, P=0.0274), interleukin 1 receptor associated kinase 3 (IRAK3) (log2 fold=−3.097, P=0.0357), TNKS2 (log2 fold=−3.416, P=0.0402), mucin 20, cell surface associated (MUC20) (log2 fold=−3.149, P=0.0404), ARPP21 (log2 fold=−4.977, P=0.0410) and HUWE1 (log2 fold=−2.931, P=0.0411) GP levels one week after 30-day intermittent fasting compared with the levels before 30-day intermittent fasting.
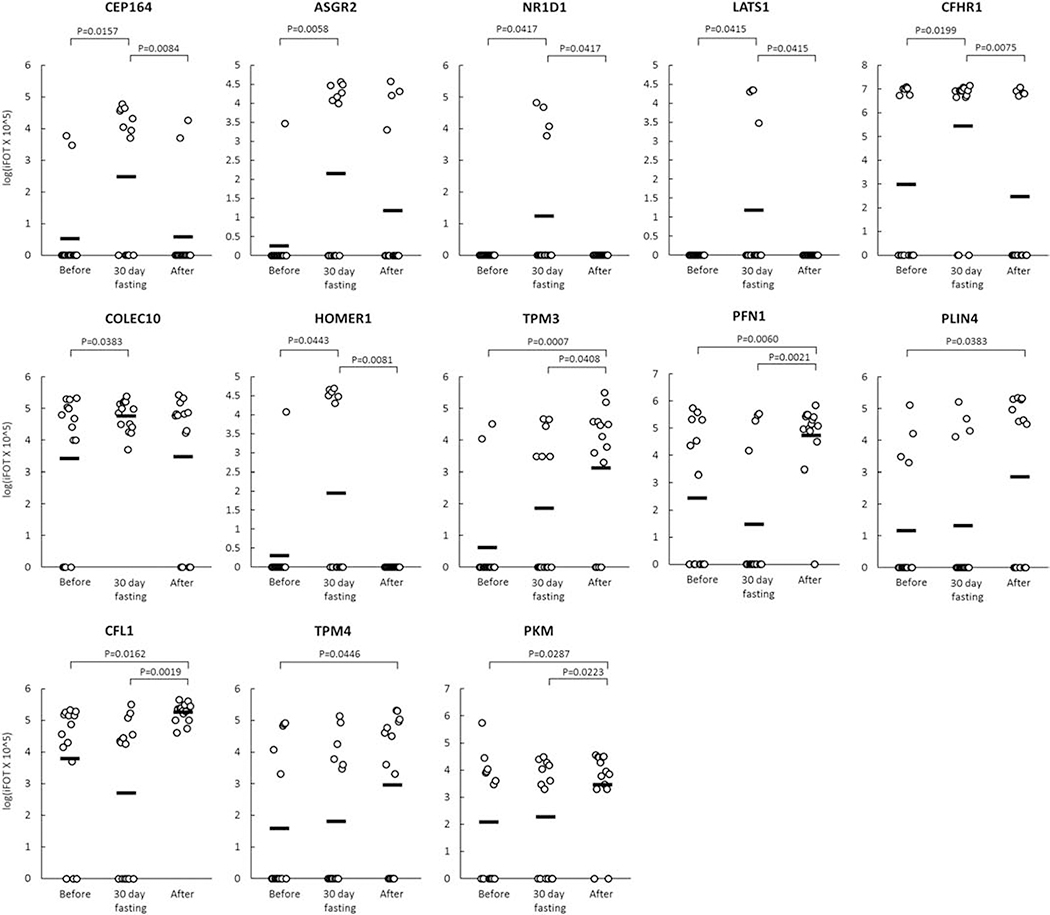
Figure 3 shows the change (logiFOT X 105) in the levels of 13 selected GPs that significantly increased at the end of 4th week during 30-day intermittent fasting or one week after 30-day intermittent fasting compared with the levels before 30-day intermittent fasting.
Figure 3.
Change (logiFOT X 105) in the levels of 13 selected GPs that significantly increased at the end of 4th week during 30-day intermittent fasting (30-day fasting) or one week after 30-day intermittent fasting (After) compared with the levels before 30-day intermittent fasting (Before).
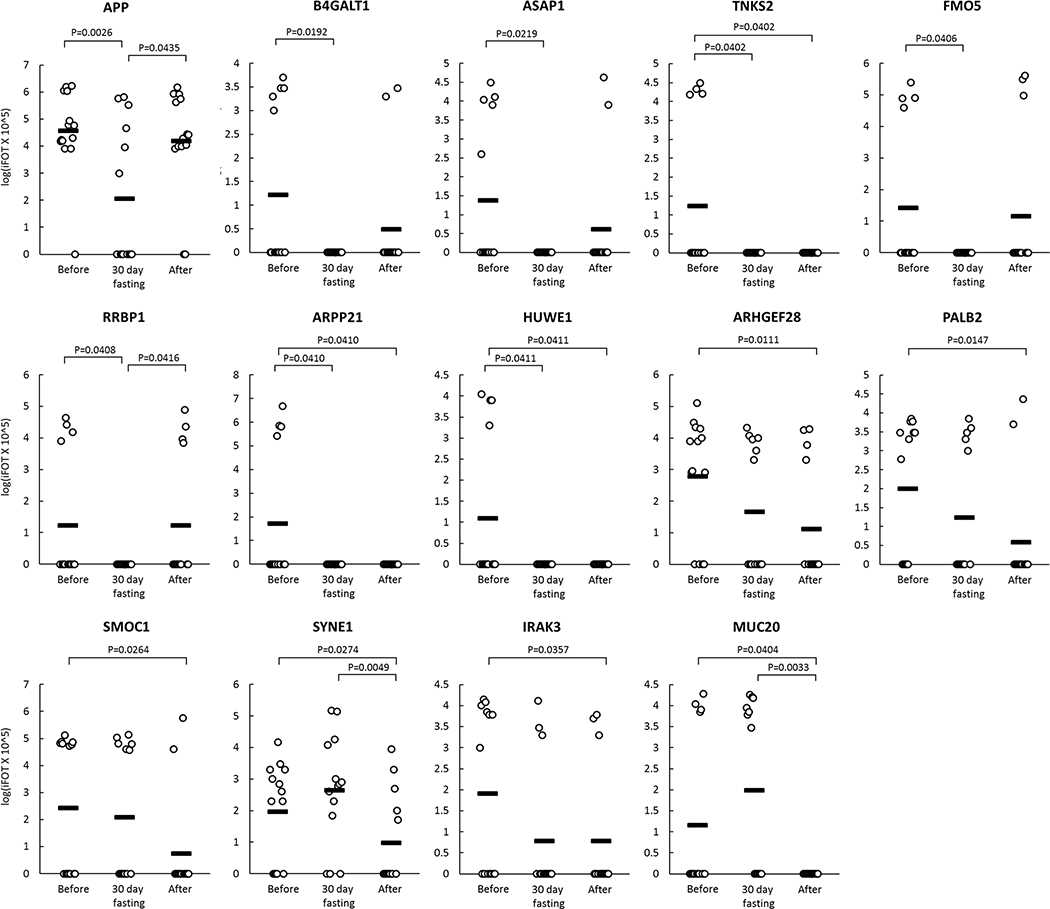
Figure 4 shows the change (logiFOT X 105) in the levels of 14 selected GPs that significantly decreased at the end of 4th week during 30-day intermittent fasting or one week after 30-day intermittent fasting or both at the end of 4th week during 30-day intermittent fasting and one week after 30-day intermittent fasting compared with the levels before 30-day intermittent fasting.
Figure 4.
Change (logiFOT X 105) in the levels of 14 selected GPs that significantly decreased at the end of 4th week during 30-day intermittent fasting (30-day fasting) or one week after 30-day intermittent fasting (After) or both at the end of 4th week during 30-day intermittent fasting and one week after 30-day intermittent fasting compared with the levels before 30-day intermittent fasting (Before).
Conventional Metabolic Parameters and Serum Biomarkers
Overall, there was no significant change in clinical metabolic parameters and serum metabolic biomarkers at the end of 4th week during 30-day intermittent fasting and one week after 30-day intermittent fasting compared with the levels before 30-day intermittent fasting (Supplementary Table S3).
Fecal Microbiota
Although increased alpha diversity richness was present among the subjects, no significant statistical differences were observed in richness and diversity (Shannon and Simpson’s) when comparing the collection periods (Supplementary Figure S4A). Thus, bacterial richness and diversity did not change significantly over the three collection periods. Similarly, the beta-diversity analysis using Weighted and Unweighted UniFrac distance metrics did not reveal differences in the microbial structure between the three periods (Supplementary Figure S4B). Overall, the grouped subjects from the three periods shared two dominant orders among the abundant taxa: Clostridiales (Firmicutes) and Bacteroidales (Bacteroidetes) (Supplementary Figure S4C). Further results of fecal microbiota analysis are included in Supplementary Materials.
DISCUSSION
Herein, we conducted the first human study of serum proteomics of 30-day dawn to sunset intermittent fasting with simultaneous assessment of clinical metabolic parameters, multiple serum biomarkers, and fecal microbiota in 14 healthy subjects. Our study has important clinical implications: Our results showed that intermittent fasting from dawn to sunset for more than 14 hours daily for 30 consecutive days was associated with an anticancer serum proteomic signature and upregulated the key regulatory proteins of glucose and lipid metabolism, insulin signaling, circadian clock, DNA repair, cytoskeleton remodeling, immune system and cognitive function, and resulted in a serum proteome protective against cancer, metabolic syndrome, inflammation, Alzheimer’s disease, and several neuropsychiatric disorders. Importantly, these findings occurred in the absence of any calorie restriction and significant weight loss. We assessed changes in the serum proteome, serum biomarkers, fecal microbiome, and clinical parameters not only at the end of 4th week during 30-day intermittent fasting but also one week after completion of 30-day intermittent fasting. To our knowledge, this is also the first study where compliance with fasting was monitored with objective measures (13C-isotopic breath enrichment test) rather than relying on study participants’ self-reporting.
Intermittent Fasting from Dawn to Sunset is Associated with Anticancer Proteomic Signature
We found a significant increase in the levels of specific proteins that are downregulated in several cancers, and therefore associated with metastasis and poor prognosis, at the end of 4th week during 30-day intermittent fasting. LATS1 is a large tumor suppressor kinase 1 that was shown to suppress proliferation, progression, and invasion of several tumors[27], e.g. hepatocellular carcinoma[28], cervical cancer[29], and non-small cell lung cancer[30]. A genomic mapping using data collected from the Catalogue of Somatic Mutations in Cancer (COSMIC) and cBioPortal databases showed mutations in LATS1 and LATS2 in numerous cancers.[31] We found an average 9 fold increase in the LATS1 GP level at the end of 4th week during 30-day intermittent fasting compared with the level before 30-day intermittent fasting.
CFHR1 (also known as CFHL1) and COLECT10 are abundantly expressed in the liver.[27] CFHR1 and COLEC10 were shown to be downregulated in hepatocellular carcinoma and associated with poor prognosis.[32, 33] We found an average 160 and 14 fold increase in the CFHR1 and COLEC10 GP levels, respectively, at the end of 4th week during 30-day intermittent fasting compared with the levels before 30-day intermittent fasting.
We also found a significant reduction in the levels of specific proteins that are overexpressed in several cancers, and therefore associated with metastasis and poor prognosis at the end of 4th week during 30-day intermittent fasting or one week after 30-day intermittent fasting.
B4GALT1 is overexpressed in several tumors, including hepatocellular carcinoma[34], lung cancer[35], and breast cancer[36]. Inhibition of B4GALT1 was shown to remove multidrug resistance in leukemia, which can potentially increase sensitivity to chemotherapy. [37] We found a significant reduction in the B4GALT1 GP level at the end of 4th week during 30-day intermittent fasting compared with the level before 30-day intermittent fasting.
ASAP1 overexpression was reported in laryngeal squamous cell carcinoma [38] and epithelial ovarian cancer [39]. We found a significant reduction in the ASAP1 GP level at the end of 4th week during 30-day intermittent fasting compared with the level before 30-day intermittent fasting.
FMO5 is a flavin-containing dimethylaniline monooxygenase that has a biased expression in the liver.[27] Overexpression of FMO5 was shown to be a poor prognostic indicator in colorectal cancer.[40] We found a significant reduction in the FMO5 GP level at the end of 4th week during 30-day intermittent fasting compared with the level before 30-day intermittent fasting.
RRBP1 was shown to be overexpressed in colorectal cancer [41] and endometrial endometrioid adenocarcinoma [42]. We found a significant reduction in the RRBP1 GP level at the end of 4th week of intermittent fasting compared with the level before 30-day intermittent fasting.
TNKS2 (also known as TANK2 and TNKL), which is a TTAGGG repeat binding factor 1 (TRF1) [43]-associated poly(ADP-ribose) polymerase was shown to induce rapid necrotic cell death.[44] A potential association between TNKS2 and cancer was reported.[45] Overexpression of TNKS2 was found in breast cancer.[46] We found a significant reduction in the TNKS2 GP level at the end of 4th week during and one week after 30-day intermittent fasting compared with the level before 30-day intermittent fasting.
HUWE1, an E3 ubiquitin protein ligase, ubiquitinates the tumor suppressor p53 leading to its degradation. [27] The inactivation or deletion of HUWE1 upregulates p53 and thereby inhibits the development and proliferation of non-small lung cancer.[47] HUWE1 overexpression was reported in several cancers, including breast, colon, lung, prostate, larynx, stomach and uterus.[48] We found a significant reduction in HUWE1 GP level at the end of 4-week during and one week after 30-day intermittent fasting compared with the level before 30-day intermittent fasting.
ARHGEF28, also known as RGNEF, was shown to play a key role in tumor progression and invasion in colon cancer via focal adhesion kinase (FAK).[49] We observed a significant reduction in the ARHGEF28 GP level one week after 30-day intermittent fasting compared with the level before 30-day intermittent fasting.
Pathogenic variants of the PALB2 gene was implicated as a risk factor in the development of bilateral breast cancer.[50] We found a significant reduction in the PALB2 GP level one week after 30-day intermittent fasting compared with the level before 30-day intermittent fasting.
SMOC1 that has a biased expression in the brain [27] was found to be overexpressed in brain tumors including oligodendrogliomas, glioblastomas and astrocytomas compared with control brain tissues.[51] We found a significant reduction in SMOC1 GP level one week after 30-day intermittent fasting compared with the level before 4-week intermittent fasting.
The role of interleukin 1 receptor-associated kinase (IRAK) signaling in tumor development and progression is well-defined. [52] IRAK3 (also known as IRKM) deficient mice were shown to be protected against tumor development. [53] We found a significant reduction in IRAK3 GP level one week after 30-day intermittent fasting compared with the level before 30-day intermittent fasting.
Overexpression of MUC20 was reported in colorectal, endometrial and ovarian cancers as a predictor of tumor progression and aggressiveness. [54] [55] [56] We found a significant reduction in the MUC20 GP level one week after 30-day intermittent fasting compared with the level before 30-day intermittent fasting.
Altogether, our findings showed that 30-day dawn to sunset intermittent fasting resulted in an anticancer serum proteome in healthy subjects. Our results suggest that 30-day intermittent fasting can be a preventive and adjunct treatment in several cancers and increase sensitivity to chemotherapy.
Intermittent Fasting from Dawn to Sunset Upregulates Expression of CEP164, A Key DNA Repair Protein
CEP164 gene plays a primary role in the development and function of the primary cilium.[27] It is also a mediator protein in the ultraviolet and ionizing radiation-induced DNA damage repair signaling pathway[57] and recruited to DNA sites damaged by ultraviolet.[58] We found an average 45 fold increase in the CEP164 GP level at the end of 4th week during 30-day intermittent fasting compared with the level before 30-day intermittent fasting. These findings suggest that dawn to sunset 30-day intermittent fasting induces the repair of DNA damaged by ultraviolet and ionizing radiation.
Intermittent Fasting from Dawn to Sunset Upregulates Expression of NR1D1, a Circadian Clock Protein
NR1D1 (also known as REV-ERBα), a component of the circadian clock, regulates the expression of genes involved in metabolism and inflammation.[27] A murine study demonstrated that NR1D1 plays a critical role in the prevention of metabolic syndrome; the deletion of REV-ERBα (NR1D1) resulted in lipoprotein lipase overexpression in peripheral tissues, increased fat storage in the liver and adipose tissues, and susceptibility to obesity induced by a high-fat diet.[59] Pharmacologic NR1D1 activation was shown to reduce the severity of acute peritonitis and prevent fulminant hepatitis by inhibiting the NLRP3 inflammasome pathway. [60] We observed an average 11 fold increase in the NR1D1 GP level at the end of 4th week during 30-day intermittent fasting compared with the level before 30-day intermittent fasting. These findings suggest that dawn to sunset 30-day intermittent fasting can reduce fat accumulation in the liver and adipose tissues, and attenuates NLRP3-driven inflammation, therefore it can be an adjunct treatment of patients with metabolic syndrome and nonalcoholic fatty liver disease.
Intermittent Fasting from Dawn to Sunset Upregulates Expression of ASGR2, a Subunit of Asialogylcoprotein Receptor which is a Key Hepatic Immunoregulatory Protein
ASGR2 gene encodes for one of the subunits of the asialoglycoprotein receptor and abundantly expressed in the liver.[27] The asialoglycoprotein receptor plays a vital role in the clearance of apoptotic cell debris and immune regulation in the liver.[61, 62] Apoptosis, which is programmed cell death, is a fundamental mechanism to prevent inflammation, fibrosis, and liver cancer.[61] In several chronic liver diseases, cirrhosis, and hepatocellular carcinoma, there is a reduction in the surface distribution of ASGPRs and ASGPR mRNA expression[63, 64], and this can result in dysregulation of apoptosis, and inefficient clearance of apoptotic cell debris leading to chronic inflammation, and breakdown of self-tolerance.[61–64] We observed an average 40 fold increase in the ASGR2 GP level at the end of 4th week of intermittent fasting compared with the level before 30-day intermittent fasting. Altogether, these findings suggest that 30-day intermittent fasting from dawn to sunset can enhance the hepatic clearance of apoptotic cell debris, reduce inflammation, and optimize immune function.
Intermittent Fasting from Dawn to Sunset Results in a Serum Proteome Protective Against Cognitive Dysfunction, Alzheimer’s Disease and Several Neuropsychiatric Diseases
HOMER1 gene is abundantly expressed in the brain [27]. Alterations in HOMER1 levels in the hippocampus and cingulate gyrus were reported in schizophrenia, major depression, and bipolar disorder.[65] HOMER1 variants were reported in various neurological disorders, including Alzheimer’s disease, schizophrenia, chronic pain, traumatic brain injury, and drug-induced addiction.[66] HOMER1 knockout mice were shown to have neurochemical and behavioral abnormalities similar to the abnormalities observed in schizophrenia. [67] Amyloid precursor protein and presenilin 1 transgenic mice as an Alzheimer’s disease model was shown to have reduced HOMER1 mRNA expression.[68] In addition to these reports, HOMER1 expression was found to be increased in the hippocampus post-synaptic densities of Long Evans aged rats with unimpaired memory compared with those with impaired memory.[69] We found an average 25 fold increase in the HOMER1 GP level at the end of 4th week during 30-intermittent fasting compared with the level before 30-day intermittent fasting.
APP, the precursor of amyloid β, appears to play a significant role in the development of Alzheimer’s disease.[27] APP was proposed to trigger atherothrombosis after the accumulation of amyloid β peptides in the cerebral vessels in Alzheimer’s disease.[70] We observed a significant reduction in the APP GP level at the end of 4th week during 30-day intermittent fasting compared with the level before 30-day intermittent fasting.
ARPP-21 that is abundantly expressed in the brain, is another gene that plays a significant role in the development of Alzheimer’s disease.[27] A genome-wide study conducted among subjects with Alzheimer’s disease showed a single-nucleotide polymorphism (SNP) located proximal to the ARPP-21 gene. [71] We found a significant reduction in the ARPP21 GP level at the end of 4th week during and one week after 30-day intermittent fasting compared with the level before 30-day intermittent fasting.
According to a genome-wide study conducted among 1527 patients with bipolar disorders (1579 controls) and 1159 patients with recurrent unipolar depression (2592 controls), a SNP located in the SYNE1 gene was significantly associated with the risk of bipolar depression and recurrent major depression. [72] We found a significant reduction in the SYNE1 GP level one week after 30-day intermittent fasting compared with the level before 30-day intermittent fasting.
Altogether, these findings suggest that 30-day dawn to sunset intermittent fasting can have implications in the improvement of cognitive function, prevention, and treatment of Alzheimer’s disease and several neuropsychiatric disorders, including major depression, bipolar disorder, and schizophrenia.
Intermittent Fasting from Dawn to Sunset Upregulates Expression of Key Regulatory Proteins of Glucose and Lipid Metabolism, Insulin Signaling, Actin Cytoskeleton Remodeling
We found significant upregulation of several signature genes that play a key role in cytoskeleton remodeling, glucose, and lipid metabolism, and blood pressure regulation one week after completion of 30-day intermittent fasting. There was an average 127-fold increase in the TPM3 and an average 15-fold increase in the TPM4 GP levels one week after 30-day intermittent fasting compared with the levels before 30-day intermittent fasting. Tropomyosins are essential proteins that are well-known for their role in skeletal and cardiac muscle contraction.[73, 74] They are also components of the actin cytoskeleton in non-muscle cells and play a key role in stabilization, regulation, and remodeling of the actin cytoskeleton. [73–75] The actin cytoskeleton is thought to orchestrate cell division, proliferation, support, movement, intra- and intercellular communications, and specifically, organelle trafficking.[76] Dysfunction of the actin cytoskeleton has been shown to result in impaired exocytosis of glucose transporter (GLUT4) and thereby in insulin resistance.[77–79] TPM3 gene encodes for tropomyosin 3.1 that plays a crucial role in remodeling insulin-induced actin cytoskeleton, thereby increasing insulin sensitivity, and protecting against high-fat diet.[77] Similar to TPM3, the TPM4 gene encodes for a protein that binds cytoskeletal actin [27] that plays a key role in insulin responsiveness.
Tropomyosins play a vital role not only in glucose regulation but also in blood pressure regulation by stabilizing and remodeling the actin cytoskeleton.[80] Polymorphisms in TPM3 gene expression in erythrocytes and leukocytes may result in dysfunctional TPMN protein, and thereby, it can lead to essential hypertension.[80] Earlier reports of dawn to sunset fasting showed that there was a significant reduction in systolic blood pressure and pulse-pressure at the end of fourth week of dawn-to-sunset fasting compared with pressures measured before fasting. [81, 82]
In addition to the upregulation of TPM3 and TPM4, there was an average 95 fold increase in the PFN1 GP level one week after 30-day intermittent fasting compared with the level before fasting. PFN1 encodes for cytoskeleton proteins that are involved in actin dynamics.[27] It mediates communication between the cytoskeleton and cell membrane [83] and regulates migration, invasion, and morphogenesis of endothelial cells.[84]
Our results also showed that 30-day intermittent fasting upregulated PLIN4 GP. PLIN4 (also known as S3–12) has a biased expression in fat tissues.[27] A study conducted in Zucker rats showed that S3–12 (PLIN4) was downregulated in obese rats compared with lean ones, the peroxisome proliferator-activated receptor-γ (PPAR- γ) was the key regulator of PLIN4 expression in adipocytes, and activation of PPAR-γ resulted in upregulation of PLIN4.[85] PPAR- γ activation improves insulin resistance and lipid metabolism.[86] Upregulation of PLIN4 expression one week after 30-day intermittent fasting suggests that 30-day intermittent fasting from dawn to sunset mimics PPAR-γ activators (e.g., pioglitazone), and this may be one of the mechanisms of intermittent fasting to improve insulin resistance and protect against adipose tissue dysfunction.
Another GP that was upregulated one week after 30-day intermittent fasting was CFL1. It has been shown that CFL1 plays a critical role in insulin-induced GLUT4 translocation, thereby glucose uptake.[87] We found an average 21 fold increase in the CFL1 GP level one week after 30-day intermittent fasting compared with the level before 30-day intermittent fasting, suggestive of another evidence toward improvement in insulin resistance with dawn to sunset intermittent fasting.
There was an average 13 fold increase in the PKM GP level one week after 30-day intermittent fasting compared to the level before 30-day intermittent fasting. PKM encodes pyruvate kinase enzyme that is one of the key enzymes of glycolysis.[27] The upregulation of PKM2 in diabetes was shown to protect against the progression of diabetic nephropathy and mitochondrial dysfunction by reducing the production of toxic glucose metabolites.[88]
In contrast to serum proteome, we found no significant change in the levels of conventional clinical metabolic parameters and serum metabolic biomarkers. This could be related to the fact that our study subjects were healthy, and therefore a significant change in the levels of clinical metabolic parameters and serum metabolic biomarkers that would be otherwise expected in non-healthy subjects did not occur at the end of 30-day intermittent fasting compared with the levels before 30-day intermittent fasting.
Upregulation of TPM3, TPM4, PLIN4, CFL1, and PKM GP expression suggests that 30-day intermittent fasting can play a significant role in the prevention and treatment of metabolic syndrome. Metabolic syndrome is associated with insulin resistance, lipotoxicity, and inflammation.[89] It is also a significant risk factor for atherosclerotic cardiovascular disease and nonalcoholic fatty liver disease that can result in cirrhosis and hepatocellular carcinoma.[89, 90] As such, the prevention of metabolic syndrome is a major public health concern, and better understandings are needed to guide effective interventions. The current daily dietary habit of many people is to eat periodically throughout the day into the evening, such that their bodies predominantly remain in the fed state. It is well known that nutrient utilization and body store utilization is influenced by macronutrient intake and meal timing.[91, 92] It appears that intermittent fasting from dawn to sunset could offer a new therapeutic approach in metabolic syndrome and its complications.
Our proteomics study of intermittent fasting has several distinct features in terms of design and findings compared with the proteomics study conducted in healthy subjects by Harney et al. [93]. First and foremost, the study conducted by Harney et al. [93] did not assess the effect of strict intermittent fasting but rather the effect of a diet allowing the subjects to eat energy-free foods and low-energy broth and drink water, coffee, and tea during their fasting period that was scheduled for three nonconsecutive days of the week. In contrast, our study subjects fasted from dawn to sunset without eating or drinking for more than 14 hours daily for 30 consecutive days. This type of rhythmic consecutive intermittent fasting from dawn to sunset is compliant with circadian rhythm because daily fast starts at dawn (the first transition zone of the day) and ends at sunset (the second transition zone of the day) without any calorie restriction. Second, our subjects were not allowed to drink water during fasting; this ensured a complete lack of stimulus to the digestive system minimizing metabolic activities. Third, our study subjects did not have calorie restriction. Fourth, our results were distinct, showing an anticancer proteomic signature and significant fold changes in multiple key proteins that play a key role in glucose and lipid metabolism, insulin signaling, DNA repair, immune system regulation, cytoskeletal remodeling and cognitive function associated with 30-day intermittent fasting.
Our study had limitations. Although we tested the effect of 30-day intermittent fasting, our study did not assess the impact of shorter duration of intermittent fasting (e.g., one or two weeks) or include a parallel control group of healthy subjects who did not fast for 30 days. A future study of 30-day intermittent fasting needs to be conducted in healthy subjects for external validation of the selected GPs discovered in our study.
Conclusions
In summary, our results suggest that 30-day intermittent fasting from dawn to sunset can be a preventive and therapeutic approach in cancer as well as in several metabolic, inflammatory and immune diseases, Alzheimer’s disease and neuropsychiatric disorders by resulting in a proteome protective against carcinogenesis, obesity, diabetes, metabolic syndrome, inflammation, cognitive dysfunction, and mental health. Further studies are needed to test the effect of dawn to sunset intermittent fasting in larger cohorts with consideration given to shorter durations of fasting and longer longitudinal follow-up after completion of intermittent fasting.
Supplementary Material
SIGNIFICANCE.
Our study has important clinical implications. Our results showed that intermittent fasting from dawn to sunset for more than 14 hours daily for 30 consecutive days was associated with an anticancer serum proteomic signature and upregulated key regulatory proteins of glucose and lipid metabolism, insulin signaling, circadian clock, DNA repair, cytoskeleton remodeling, immune system, and cognitive function, and resulted in a serum proteome protective against cancer, obesity, diabetes, metabolic syndrome, inflammation, Alzheimer’s disease, and several neuropsychiatric disorders. Importantly, these findings occurred in the absence of any calorie restriction and significant weight loss. These findings suggest that intermittent fasting from dawn to sunset can be a preventive and adjunct therapy in cancer, metabolic syndrome and Alzheimer’s disease and several neuropsychiatric diseases.
HIGHLIGHTS.
First human serum proteomics study of 30-day intermittent fasting from dawn to sunset in healthy subjects
The 30-day intermittent fasting from dawn to sunset is associated with a serum proteome protective against cancer
Intermittent fasting from dawn to sunset for 30 days upregulates proteins protective against obesity, diabetes, and metabolic syndrome
Intermittent fasting from dawn to sunset for 30 days induces key regulatory proteins of DNA repair and immune system
Intermittent fasting from dawn to sunset for 30 days upregulates proteins protective against Alzheimer’s disease and neuropsychiatric disorders
Acknowledgments
Funding
This project was supported in part by NIH Public Health Service grant P30DK056338, which funds the Texas Medical Center Digestive Diseases Center and P30CA125123, which funds the Baylor College of Medicine (BCM) Proteomics Core and its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of Diabetes and Digestive and Kidney Diseases, National Cancer Institute or the NIH.
This project was also supported in part by the Cancer Prevention and Research Institute of Texas (CPRIT) Core Facility Award (RP170005) and by the Alkek Center for Metagenomics and Microbiome Research, Baylor College of Medicine.
Antone R. Opekun, M.S., P.A.-C, DFAAPA was partially supported by an unrestricted institutional grant from DR and GP Laws.
Prasun K. Jalal, M.D. was partially supported by a grant from Dora Roberts Foundation Grant.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflict of interest.
Meeting Materials
1. Mindikoglu AL, Abdulsada MM, Jain A, Jung SY, Jalal PK, Opekun AR. Dawn to Sunset Fasting for 30 Days Induces Tropomyosin 1, 3 and 4 Genes in Healthy Volunteers: Its Clinical Implications in Metabolic Syndrome and Non-Alcoholic Fatty Liver Disease. Gastroenterology 2019, Vol. 156, Issue 6, S-1509–S-1510. Late-Breaking abstract was selected for a lecture presentation on May 21, 2019 at Digestive Disease Week (DDW) 2019, San Diego, CA. Control ID: 3194352.
2. Jain A, Jung SY, Abdulsada M, Opekun A, Malovannaya A, Jalal P, Mindikoglu AL. Dawn to Sunset Fasting for Four Weeks Has A Unique Proteomic Signature in Healthy Subjects. Abstract was selected for poster presentation on June 4, 2019 at ASMS Conference on Mass Spectrometry and Allied Topics, Atlanta, Georgia. Abstract ID number: 297726.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Greene MW, Circadian rhythms and tumor growth. Cancer letters, 2012. 318(2): p. 115–123. [DOI] [PubMed] [Google Scholar]
- 2.Shetty A, et al. , Role of the Circadian Clock in the Metabolic Syndrome and Nonalcoholic Fatty Liver Disease. Digestive diseases and sciences, 2018. 63(12): p. 3187–3206. [DOI] [PubMed] [Google Scholar]
- 3.Hatori M, et al. , Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab, 2012. 15(6): p. 848–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X-M, et al. , Cancer inhibition through circadian reprogramming of tumor transcriptome with meal timing. Cancer research, 2010. 70(8): p. 3351–3360. [DOI] [PubMed] [Google Scholar]
- 5.Damiola F, et al. , Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev, 2000. 14(23): p. 2950–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans JA, Collective timekeeping among cells of the master circadian clock. J Endocrinol, 2016. 230(1): p. R27–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hara R, et al. , Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells, 2001. 6(3): p. 269–78. [DOI] [PubMed] [Google Scholar]
- 8.Reppert SM and Weaver DR, Coordination of circadian timing in mammals. Nature, 2002. 418(6901): p. 935–41. [DOI] [PubMed] [Google Scholar]
- 9.Satoh Y, et al. , Time-restricted feeding entrains daily rhythms of energy metabolism in mice. Am J Physiol Regul Integr Comp Physiol, 2006. 290(5): p. R1276–83. [DOI] [PubMed] [Google Scholar]
- 10.Stokkan KA, et al. , Entrainment of the circadian clock in the liver by feeding. Science, 2001. 291(5503): p. 490–3. [DOI] [PubMed] [Google Scholar]
- 11.Yamazaki S, et al. , Resetting central and peripheral circadian oscillators in transgenic rats. Science, 2000. 288(5466): p. 682–5. [DOI] [PubMed] [Google Scholar]
- 12.Dibner C, Schibler U, and Albrecht U, The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol, 2010. 72: p. 517–49. [DOI] [PubMed] [Google Scholar]
- 13.Mindikoglu AL, et al. , Impact of Time-Restricted Feeding and Dawn-to-Sunset Fasting on Circadian Rhythm, Obesity, Metabolic Syndrome, and Nonalcoholic Fatty Liver Disease. Gastroenterol Res Pract, 2017. 2017: p. 3932491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherman H, et al. , Long-term restricted feeding alters circadian expression and reduces the level of inflammatory and disease markers. J Cell Mol Med, 2011. 15(12): p. 2745–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherman H, et al. , Timed high-fat diet resets circadian metabolism and prevents obesity. FASEB J, 2012. 26(8): p. 3493–502. [DOI] [PubMed] [Google Scholar]
- 16.Opekun AR, Balesh AM, and Shelby HT, Use of the Biphasic (13)C-Sucrose/Glucose Breath Test to Assess Sucrose Maldigestion in Adults with Functional Bowel Disorders. Biomed Res Int, 2016. 2016: p. 7952891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung SY, et al. , Proteomic analysis of steady-state nuclear hormone receptor coactivator complexes. Mol Endocrinol, 2005. 19(10): p. 2451–65. [DOI] [PubMed] [Google Scholar]
- 18.Jung SY, et al. , An Anatomically Resolved Mouse Brain Proteome Reveals Parkinson Disease-relevant Pathways. Mol Cell Proteomics, 2017. 16(4): p. 581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saltzman AB, et al. , gpGrouper: A Peptide Grouping Algorithm for Gene-Centric Inference and Quantitation of Bottom-Up Proteomics Data. Mol Cell Proteomics, 2018. 17(11): p. 2270–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthews DR, et al. , Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia, 1985. 28(7): p. 412–419. [DOI] [PubMed] [Google Scholar]
- 21.SAS software. Http://www.Sas.Com/ The data analysis for this paper was generated using SAS software, Version 9.4 of the SAS System for Windows. Copyright © 2016 SAS Institute Inc. SAS and all other SAS Institute Inc. product or service names are registered trademarks or trademarks of SAS Institute Inc., Cary, NC, USA. [Google Scholar]
- 22.Caporaso JG, et al. , Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The Isme Journal, 2012. 6: p. 1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso JG, et al. , Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A, 2011. 108 Suppl 1: p. 4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The Human Microbiome Project, C., et al. , Structure, function and diversity of the healthy human microbiome. Nature, 2012. 486: p. 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The Human Microbiome Project, C., et al. , A framework for human microbiome research. Nature, 2012. 486: p. 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig KR, Schroll MM, and Hummon AB, Comparison of In-Solution, FASP, and S-Trap Based Digestion Methods for Bottom-Up Proteomic Studies. J Proteome Res, 2018. 17(7): p. 2480–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gene [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; 2004. – [cited 2019 Nov 17]. Available from: https://www.ncbi.nlm.nih.gov/gene/. [Google Scholar]
- 28.Cheng Y, et al. , LMO3 promotes hepatocellular carcinoma invasion, metastasis and anoikis inhibition by directly interacting with LATS1 and suppressing Hippo signaling. Journal of Experimental & Clinical Cancer Research, 2018. 37(1): p. 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng J, et al. , LATS1 suppresses proliferation and invasion of cervical cancer. Mol Med Rep, 2017. 15(4): p. 1654–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin XY, et al. , Expression of LATS1 contributes to good prognosis and can negatively regulate YAP oncoprotein in non-small-cell lung cancer. Tumour Biol, 2014. 35(7): p. 6435–43. [DOI] [PubMed] [Google Scholar]
- 31.Yu T, Bachman J, and Lai ZC, Mutation analysis of large tumor suppressor genes LATS1 and LATS2 supports a tumor suppressor role in human cancer. Protein Cell, 2015. 6(1): p. 6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng H, et al. , Downregulated expression of CFHL1 is associated with unfavorable prognosis in postoperative patients with hepatocellular carcinoma. Exp Ther Med, 2019. 17(5): p. 4073–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang B and Wu H, Decreased expression of COLEC10 predicts poor overall survival in patients with hepatocellular carcinoma. Cancer Manag Res, 2018. 10: p. 2369–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei Y, et al. , Identification of β−1,4-galactosyltransferase I as a target gene of HBx-induced cell cycle progression of hepatoma cell. Journal of Hepatology, 2008. 49(6): p. 1029–1037. [DOI] [PubMed] [Google Scholar]
- 35.Zhu X, et al. , Elevated β1,4-Galactosyltransferase I in Highly Metastatic Human Lung Cancer Cells: IDENTIFICATION OF E1AF AS IMPORTANT TRANSCRIPTION ACTIVATOR. Journal of Biological Chemistry, 2005. 280(13): p. 12503–12516. [DOI] [PubMed] [Google Scholar]
- 36.Choi H-J, et al. , Estrogen induced β−1,4-galactosyltransferase 1 expression regulates proliferation of human breast cancer MCF-7 cells. Biochemical and Biophysical Research Communications, 2012. 426(4): p. 620–625. [DOI] [PubMed] [Google Scholar]
- 37.Zhou H, et al. , B4GALT1 gene knockdown inhibits the hedgehog pathway and reverses multidrug resistance in the human leukemia K562/adriamycin-resistant cell line. IUBMB Life, 2012. 64(11): p. 889–900. [DOI] [PubMed] [Google Scholar]
- 38.Li M, et al. , ASAP1 mediates the invasive phenotype of human laryngeal squamous cell carcinoma to affect survival prognosis. Oncol Rep, 2014. 31(6): p. 2676–82. [DOI] [PubMed] [Google Scholar]
- 39.Hou T, et al. , Overexpression of ASAP1 is associated with poor prognosis in epithelial ovarian cancer. International journal of clinical and experimental pathology, 2013. 7(1): p. 280–287. [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang T, et al. , Overexpression of flavin-containing monooxygenase 5 predicts poor prognosis in patients with colorectal cancer. Oncol Lett, 2018. 15(3): p. 3923–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan Y, et al. , Endoplasmic reticulum ribosome-binding protein 1, RRBP1, promotes progression of colorectal cancer and predicts an unfavourable prognosis. British journal of cancer, 2015. 113(5): p. 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu S, et al. , RRBP1 overexpression is associated with progression and prognosis in endometrial endometrioid adenocarcinoma. Diagnostic Pathology, 2019. 14(1): p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Broccoli D, et al. , Human telomeres contain two distinct Myb–related proteins, TRF1 and TRF2. Nature Genetics, 1997. 17(2): p. 231–235. [DOI] [PubMed] [Google Scholar]
- 44.Kaminker PG, et al. , TANK2, a New TRF1-associated Poly(ADP-ribose) Polymerase, Causes Rapid Induction of Cell Death upon Overexpression. Journal of Biological Chemistry, 2001. 276(38): p. 35891–35899. [DOI] [PubMed] [Google Scholar]
- 45.Kuimov AN, et al. , Cloning and characterization of TNKL, a member of tankyrase gene family. Genes and immunity, 2001. 2(1): p. 52–55. [DOI] [PubMed] [Google Scholar]
- 46.Sidorova N, et al. , Immunohistochemical detection of tankyrase 2 in human breast tumors and normal renal tissue. Cell and tissue research, 2006. 323(1): p. 137–145. [DOI] [PubMed] [Google Scholar]
- 47.Yang D, et al. , HUWE1 controls the development of non-small cell lung cancer through down-regulation of p53. Theranostics, 2018. 8(13): p. 3517–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Confalonieri S, et al. , Alterations of ubiquitin ligases in human cancer and their association with the natural history of the tumor. Oncogene, 2009. 28(33): p. 2959–2968. [DOI] [PubMed] [Google Scholar]
- 49.Masià-Balagué M, et al. , Gastrin-stimulated Gα13 Activation of Rgnef Protein (ArhGEF28) in DLD-1 Colon Carcinoma Cells. The Journal of biological chemistry, 2015. 290(24): p. 15197–15209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castéra L, et al. , Landscape of pathogenic variations in a panel of 34 genes and cancer risk estimation from 5131 HBOC families. Genetics in medicine : official journal of the American College of Medical Genetics, 2018. 20(12): p. 1677–1686. [DOI] [PubMed] [Google Scholar]
- 51.Brellier F, et al. , SMOC1 is a tenascin-C interacting protein over-expressed in brain tumors. Matrix Biology, 2011. 30(3): p. 225–233. [DOI] [PubMed] [Google Scholar]
- 52.Rhyasen GW and Starczynowski DT, IRAK signalling in cancer. British Journal of Cancer, 2015. 112(2): p. 232–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie Q, et al. , Loss of the innate immunity negative regulator IRAK-M leads to enhanced host immune defense against tumor growth. Molecular Immunology, 2007. 44(14): p. 3453–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao X, et al. , Role of MUC20 overexpression as a predictor of recurrence and poor outcome in colorectal cancer. Journal of translational medicine, 2013. 11: p. 151–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen C-H, et al. , MUC20 overexpression predicts poor prognosis and enhances EGF-induced malignant phenotypes via activation of the EGFR-STAT3 pathway in endometrial cancer. Gynecologic oncology, 2013. 128(3): p. 560–567. [DOI] [PubMed] [Google Scholar]
- 56.Chen C-H, et al. , MUC20 promotes aggressive phenotypes of epithelial ovarian cancer cells via activation of the integrin β1 pathway. Gynecologic oncology, 2016. 140(1): p. 131–137. [DOI] [PubMed] [Google Scholar]
- 57.Sivasubramaniam S, et al. , Cep164 is a mediator protein required for the maintenance of genomic stability through modulation of MDC1, RPA, and CHK1. Genes Dev, 2008. 22(5): p. 587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan Y-R and Lee EYHP, UV-dependent interaction between Cep164 and XPA mediates localization of Cep164 at sites of DNA damage and UV sensitivity. Cell Cycle, 2009. 8(4): p. 655–664. [DOI] [PubMed] [Google Scholar]
- 59.Delezie J, et al. , The nuclear receptor REV-ERBα is required for the daily balance of carbohydrate and lipid metabolism. FASEB journal : official publication of the Federation of American Societies for Experimental Biology, 2012. 26(8): p. 3321–3335. [DOI] [PubMed] [Google Scholar]
- 60.Pourcet B, et al. , Nuclear Receptor Subfamily 1 Group D Member 1 Regulates Circadian Activity of NLRP3 Inflammasome to Reduce the Severity of Fulminant Hepatitis in Mice. Gastroenterology, 2018. 154(5): p. 1449–1464 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dini L, et al. , The clearance of apoptotic cells in the liver is mediated by the asialoglycoprotein receptor. FEBS Lett, 1992. 296(2): p. 174–8. [DOI] [PubMed] [Google Scholar]
- 62.Guy CS, Rankin SL, and Michalak TI, Hepatocyte cytotoxicity is facilitated by asialoglycoprotein receptor. Hepatology, 2011. 54(3): p. 1043–50. [DOI] [PubMed] [Google Scholar]
- 63.Burgess JB, Baenziger JU, and Brown WR, Abnormal surface distribution of the human asialoglycoprotein receptor in cirrhosis. Hepatology, 1992. 15(4): p. 702–6. [DOI] [PubMed] [Google Scholar]
- 64.Dalton SR, et al. , Carbon tetrachloride-induced liver damage in asialoglycoprotein receptor-deficient mice. Biochem Pharmacol, 2009. 77(7): p. 1283–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leber SL, et al. , Homer1a protein expression in schizophrenia, bipolar disorder, and major depression. J Neural Transm (Vienna), 2017. 124(10): p. 1261–1273. [DOI] [PubMed] [Google Scholar]
- 66.Luo P, et al. , Scaffold protein Homer 1: implications for neurological diseases. Neurochem Int, 2012. 61(5): p. 731–8. [DOI] [PubMed] [Google Scholar]
- 67.Szumlinski KK, et al. , Behavioral and neurochemical phenotyping of Homer1 mutant mice: possible relevance to schizophrenia. Genes Brain Behav, 2005. 4(5): p. 273–88. [DOI] [PubMed] [Google Scholar]
- 68.Dickey CA, et al. , Selectively reduced expression of synaptic plasticity-related genes in amyloid precursor protein + presenilin-1 transgenic mice. The Journal of neuroscience : the official journal of the Society for Neuroscience, 2003. 23(12): p. 5219–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ménard C and Quirion R, Successful Cognitive Aging in Rats: A Role for mGluR5 Glutamate Receptors, Homer 1 Proteins and Downstream Signaling Pathways. PLOS ONE, 2012. 7(1): p. e28666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Visconte C, et al. , Amyloid precursor protein is required for in vitro platelet adhesion to amyloid peptides and potentiation of thrombus formation. Cellular Signalling, 2018. 52: p. 95–102. [DOI] [PubMed] [Google Scholar]
- 71.Furney SJ, et al. , Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer’s disease. Molecular psychiatry, 2011. 16(11): p. 1130–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Green EK, et al. , Association at SYNE1 in both bipolar disorder and recurrent major depression. Molecular Psychiatry, 2013. 18(5): p. 614–617. [DOI] [PubMed] [Google Scholar]
- 73.Geeves MA, Hitchcock-DeGregori SE, and Gunning PW, A systematic nomenclature for mammalian tropomyosin isoforms. J Muscle Res Cell Motil, 2015. 36(2): p. 147–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vlahovich N, et al. , Tropomyosin 4 defines novel filaments in skeletal muscle associated with muscle remodelling/regeneration in normal and diseased muscle. Cell Motil Cytoskeleton, 2008. 65(1): p. 73–85. [DOI] [PubMed] [Google Scholar]
- 75.Gunning PW, et al. , Tropomyosin - master regulator of actin filament function in the cytoskeleton. J Cell Sci, 2015. 128(16): p. 2965–74. [DOI] [PubMed] [Google Scholar]
- 76.Blanchoin L, et al. , Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev, 2014. 94(1): p. 235–63. [DOI] [PubMed] [Google Scholar]
- 77.Kee AJ, et al. , An actin filament population defined by the tropomyosin Tpm3.1 regulates glucose uptake. Traffic, 2015. 16(7): p. 691–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lim CY, et al. , Tropomodulin3 is a novel Akt2 effector regulating insulin-stimulated GLUT4 exocytosis through cortical actin remodeling. Nat Commun, 2015. 6: p. 5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu Z, et al. , The role of cytoskeleton in glucose regulation. Biochemistry (Mosc), 2006. 71(5): p. 476–80. [DOI] [PubMed] [Google Scholar]
- 80.Dunn SA, et al. , Altered tropomyosin expression in essential hypertension. Hypertension, 2003. 41(2): p. 347–54. [DOI] [PubMed] [Google Scholar]
- 81.Nematy M, et al. , Effects of Ramadan fasting on cardiovascular risk factors: a prospective observational study. Nutr J, 2012. 11: p. 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Al-Shafei AI, Ramadan fasting ameliorates arterial pulse pressure and lipid profile, and alleviates oxidative stress in hypertensive patients. Blood Press, 2014. 23(3): p. 160–7. [DOI] [PubMed] [Google Scholar]
- 83.Machesky LM and Poland TD, Profilin as a potential mediator of membrane-cytoskeleton communication. Trends Cell Biol, 1993. 3(11): p. 381–5. [DOI] [PubMed] [Google Scholar]
- 84.Ding Z, et al. , Both actin and polyproline interactions of profilin-1 are required for migration, invasion and capillary morphogenesis of vascular endothelial cells. Exp Cell Res, 2009. 315(17): p. 2963–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dalen KT, et al. , Adipose tissue expression of the lipid droplet-associating proteins S3–12 and perilipin is controlled by peroxisome proliferator-activated receptor-gamma. Diabetes, 2004. 53(5): p. 1243–52. [DOI] [PubMed] [Google Scholar]
- 86.Sharma AM and Staels B, Peroxisome Proliferator-Activated Receptor γ and Adipose Tissue—Understanding Obesity-Related Changes in Regulation of Lipid and Glucose Metabolism. The Journal of Clinical Endocrinology & Metabolism, 2007. 92(2): p. 386–395. [DOI] [PubMed] [Google Scholar]
- 87.Chiu TT, et al. , Arp2/3- and cofilin-coordinated actin dynamics is required for insulin-mediated GLUT4 translocation to the surface of muscle cells. Mol Biol Cell, 2010. 21(20): p. 3529–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qi W, et al. , Pyruvate kinase M2 activation may protect against the progression of diabetic glomerular pathology and mitochondrial dysfunction. Nat Med, 2017. 23(6): p. 753–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grundy SM, et al. , Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation, 2005. 112(17): p. 2735–52. [DOI] [PubMed] [Google Scholar]
- 90.Marchesini G and Marzocchi R, Metabolic syndrome and NASH. Clin Liver Dis, 2007. 11(1): p. 105–17, ix. [DOI] [PubMed] [Google Scholar]
- 91.Labayen I, et al. , Basal and postprandial substrate oxidation rates in obese women receiving two test meals with different protein content. Clin Nutr, 2004. 23(4): p. 571–8. [DOI] [PubMed] [Google Scholar]
- 92.Motil KJ, et al. , Leucine oxidation changes rapidly after dietary protein intake is altered in adult women but lysine flux is unchanged as is lysine incorporation into VLDL-apolipoprotein B-100. J Nutr, 1994. 124(1): p. 41–51. [DOI] [PubMed] [Google Scholar]
- 93.Harney DJ, et al. , Proteomic Analysis of Human Plasma during Intermittent Fasting. Journal of Proteome Research, 2019. 18(5): p. 2228–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.