Abstract
Recent methyl adiabatic relaxation dispersion experiments provide examination of conformational dynamics across a very wide timescale (102 – 105 sec−1) and, particularly, provide insight into the hydrophobic core of proteins and allosteric effects associated with modulators. The experiments require efficient decoupling of 1H and 13C spin interactions, and some artifacts have been discovered, which are associated with the design of the proton decoupling scheme. The experimental data suggest that the original design is valid; however, pulse sequences with either no proton decoupling or proton decoupling with imperfect pulses can potentially exhibit complications in the experiments. Here, we demonstrate that pulse imperfections in the proton decoupling scheme can be dramatically alleviated by using a single composite π pulse and provide pure single-exponential relaxation data. It allows the opportunity to access high-quality methyl adiabatic relaxation dispersion data by removing the cross-correlation between dipole-dipole interaction and chemical shift anisotropy. The resulting high-quality data is illustrated with the binding of an allosteric modulator (G2BR) to the ubiquitin conjugating enzyme Ube2g2.
Keywords: methyl TROSY, adiabatic relaxation dispersion, composite decoupling, cross-correlation between DD and CSA, methyl relaxation, conformational dynamics
Summary:
Recent methyl adiabatic relaxation dispersion experiments provide examination of conformational dynamics across a very wide timescale (102 – 105 sec−1) and, particularly, provide insight into the hydrophobic core of proteins and allosteric effects associated with modulators. The experiments require efficient decoupling of 1H and 13C spin interactions. Here, we demonstrate that pulse imperfections in the proton decoupling scheme can be dramatically alleviated by using a single composite π pulse and provide pure single-exponential relaxation data. The resulting high-quality data is illustrated with the binding of an allosteric modulator (G2BR) to the ubiquitin conjugating enzyme Ube2g2.
Relaxation dispersion experiments in solution-state NMR spectroscopy are one of the most important tools in studying biologically relevant conformational dynamics of macromolecules1–2. These experiments have provided critical insights in many biological systems3–7. The well-established experiments8–11, as well as their analytic solutions12–14, provide a powerful way to reveal and quantitate minor states that have been demonstrated to play functional roles in many macromolecules. Recently, a new type of relaxation dispersion technique15–16, adiabatic relaxation dispersion, has been developed, which provides an alternative way to reveal the information for conformational exchange and access a broader timescale16–17. Studies applied to methyl groups indicated that revamped proton decoupling schemes are needed to improve the experiments17. Both theoretical and experimental examination of the decoupling design suggested that pulse imperfection could be a factor that impacted some relaxation components in the methyl group due to spin interactions.
Pulse imperfections have long been a culprit for many issues in NMR spectroscopy, such as systematic measurement errors in Car-Purcell-Meiboom-Gill (CPMG) experiments18–19 and decreased sensitivities in triple resonance experiments20–21. The imperfections are reflective of finite instrumental limitations and consist of off-resonance effects and radiofrequency field (RF) inhomogeneity. RF inhomogeneity is a function of probe coil design, which is now well optimized, and off-resonance effects reflect the finite power limitations and pulsewidths. The off-resonance effects correlate with the strength of the static magnetic field and will become much more significant as the available magnetic fields increase. In order to compensate the pulse imperfections, several alternatives have been proposed, such as composite pulses22–24 and shaped pulses25–27 (e.g. adiabatic pulses28–29). The selection of compensated pulses for an application often represents a compromise between excitation performance and timing constraints within a specific pulse sequence. In the following demonstrations, we will show the improvements of methyl adiabatic relaxation experiments17 with composite pulse decoupling and its application to revealing the potential allosteric signaling in ubiquitin ligase proteins that are central to the ubiquitin proteasome system (UPS).
Recently, it has been reported that pulse imperfections can yield artifacts in the methyl spectra when using a simple Heteronuclear Multiple Quantum Coherence (HMQC) sequence with gradients30. These direct spin coupling artifacts are similar to those observed in J-resolved spectra31 and can be removed through well-known composite pulses24, 90x-180y-90x or 90x-240y-90x. Similarly, dipole-dipole spin coupling from remote protons can be removed though the use of fully deuterated macromolecules in deuterated buffer. Under these conditions, the 1H carrier may be placed in the center of the methyl proton region and off-resonance effects are minimized, thus providing the ideal conditions of biological samples for methyl-TROSY spectroscopy. Unfortunately, these ideal conditions may not be satisfied in more sensitive/unstable macromolecules, where perdeuteration may not be possible or may be incomplete and the protein (or conditions) may require the system to be in H2O. Under these conditions, the pulse sequence is most efficient when the 1H carrier is on-resonance for H2O and accommodates water suppression schemes, e.g. Watergate. In order to eliminate the artifacts resulting from the off-resonance effect when the proton carrier frequency is placed on water, we found that an alternative composite pulse23 of 90x-270y-90x, which had been proposed to minimize the off-resonance effect on the spin inversion, may be used to generate high-quality, artifact-free HMQC spectra (Supporting Fig. 1). Additionally, spin simulations confirm that the 90x-270y-90x composite pulse is better at compensating the off-resonance effects than the 90x-180y-90x composite pulse and the regular π pulse (Supporting Fig. 2).
In the previous implementation of adiabatic relaxation dispersion experiments on methyl groups (methyl-geoHARD)17, we proposed to use deuterated and ILV-labeled proteins; however it was found that the proton decoupling scheme is important and very sensitive to pulse imperfections. To remove the cross-correlation between the dipole-dipole interaction and chemical shift anisotropy, it is necessary to incorporate a proton decoupling scheme that avoids perturbing the proton-proton spin-flip mechanism and yields clean/mono-exponential intrinsic relaxation decays (and adiabatic relaxation dispersion profiles). It can be shown that decoupling is achieved by uniformly inverting all proton spins within the protein (all methyl protons, residual protonation, and exchangeable protons when the solvent is H2O) at the center of the adiabatic spin-lock blocks (Fig. 1). Spin simulations found that uniform spin inversion causes little or no perturbation on the adiabatic relaxation dispersion profiles, and complete spin inversion effectively removes the adverse effects of cross-correlation between the 1H-13C dipole-dipole interaction and 13C chemical shift anisotropy (Fig. 2). The cross-correlation relaxation effects can become substantial, since, although the methyl 13C CSA is small, the experiment measures the reduced R2 rate from the inner two components of the manifolds, where the R2 rate is reduced by the cross-correlation between 1H-13C dipolar interactions. The cross-correlation effects can become more severe in large protein complexes with anisotropic tumbling. Experimentally, non-ideal inversion with a simple square 180x pulse leads to considerable artifacts (non-exponential decays of longitudinal magnetizations)17, which complicates data analyses. Hence, it is important to remove such effects and improve the quality of relaxation dispersion data. The artifacts will worsen at higher magnetic fields, where off-resonance effects increase, unless the pulse width of the 180x pulse decreases with the field. Therefore, it is crucial to use an efficient composite pulse that can uniformly invert all proton spins within the protein. The observations are first: the effects of the cross-correlation between dipole-dipole interaction and chemical shift anisotropy can be significantly suppressed by efficient composite pulse decoupling, which completely removes the potential bi-exponential decay of transverse magnetization for the methyl groups (Supporting Fig. 3). The deleterious effects of bi-exponential decay of transverse magnetization are exacerbated when the biomolecule has an asymmetric globular shape and experiences anisotropic tumbling. Secondly, our experimental data shows that a single 90x-270y-90x composite pulse has little or no effect on the relaxation component due to the proton-proton spin-flip mechanism between methyl protons and external protons, as desired (Supporting Fig. 4). In this work and our previous report on methyl geoHARD17, we examined four schemes relating to proton decoupling (Supporting Fig. 5). The use of repeated 180x pulses leads to considerable non mono-exponential behavior (Supporting Fig. 5c and Fig. S617). The use of a single π-pulse during the relaxation period (Supporting Figs. 5d,e, 6) significantly reduces undesired relaxation behavior yielding mono-exponential decays, and the best performance is obtained with the single 90x-270y-90x composite pulse. The final pulse sequence is shown in Fig. 1.
Figure 1.
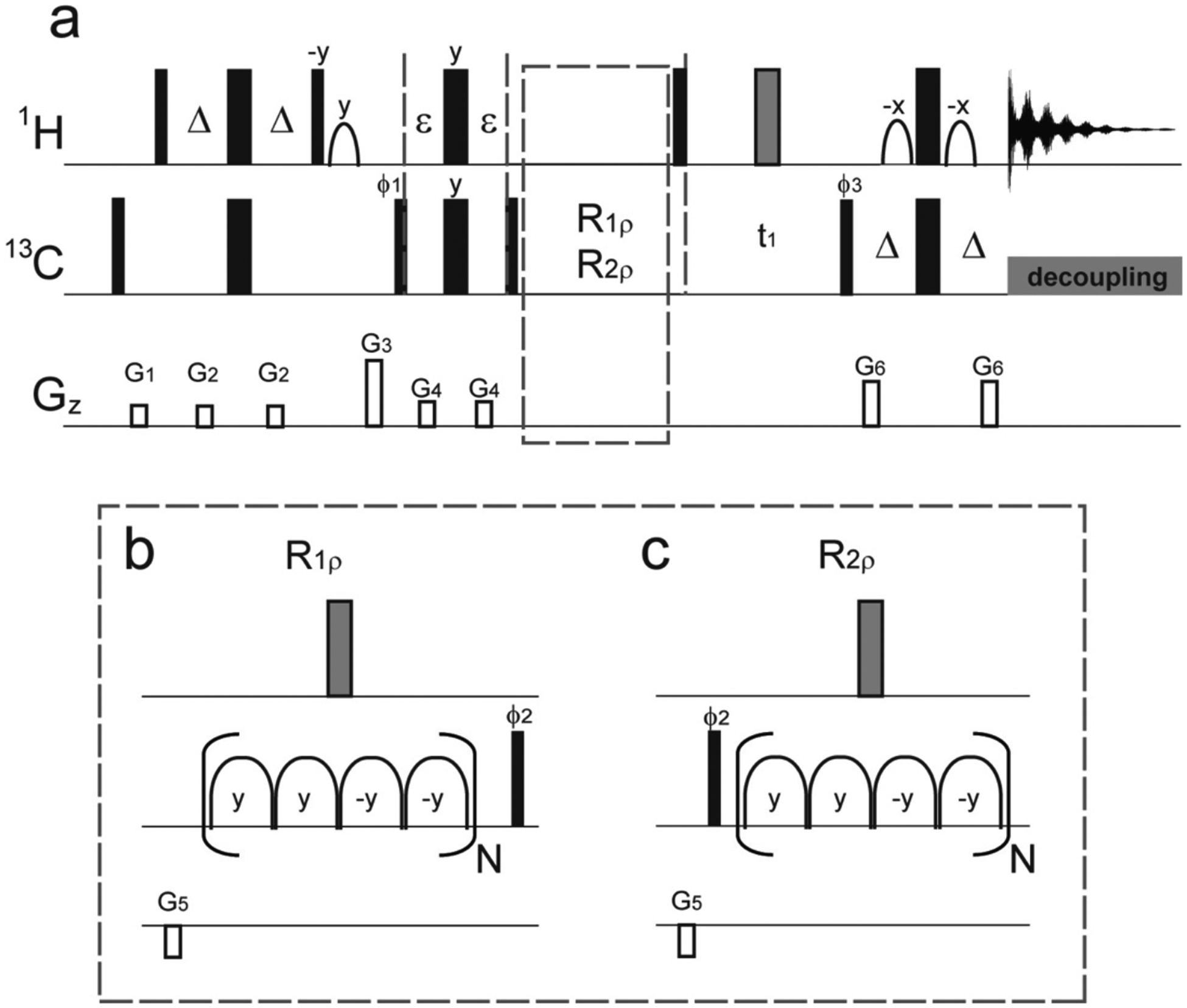
(a) Pulse sequences for the improved version of methyl adiabatic relaxation dispersion experiments. As described previously, adiabatic hyperbolic secant pulses (HSn) are used for the 13C spin-lock periods. R1ρ (or R1) relaxation delays (b) and R2ρ (or R2) relaxation delays (c) are followed after the filter element with ε = 0.6666 ms. All 90° (180°) pulses are indicated by narrow (wide) rectangles and are applied along the x axis, unless indicated otherwise. The shape pulses on the proton channel are for water flip-back and Watergate, and the gray wide rectangles are composite pulses, 90x-270y-90x. Four adiabatic pulses (b and c) are concatenated in a MLEV-4 fashion (y, y, −y, −y), and each spin-lock unit is represented by a composite adiabatic pulse (16 ms) in parentheses for both R1ρ and R2ρ experiments. Δ is set to 1.8 ms. Phases are ϕ1 = x, −x; ϕ2 = x, x, −x, −x; ϕ3 = x; and ϕrec = x, −x, −x, x. Quadrature detection in the t1 dimension is achieved by shifting the phase of ϕ3 by 90° in the states-TPPI manner. Gradient magnitudes for G1–G6 in units of (ms, G/cm) are (1, 10), (0.5, 6), (1, 50), (0.2, 10), (1, −20), and (0.5, 25), respectively.
Figure 2.
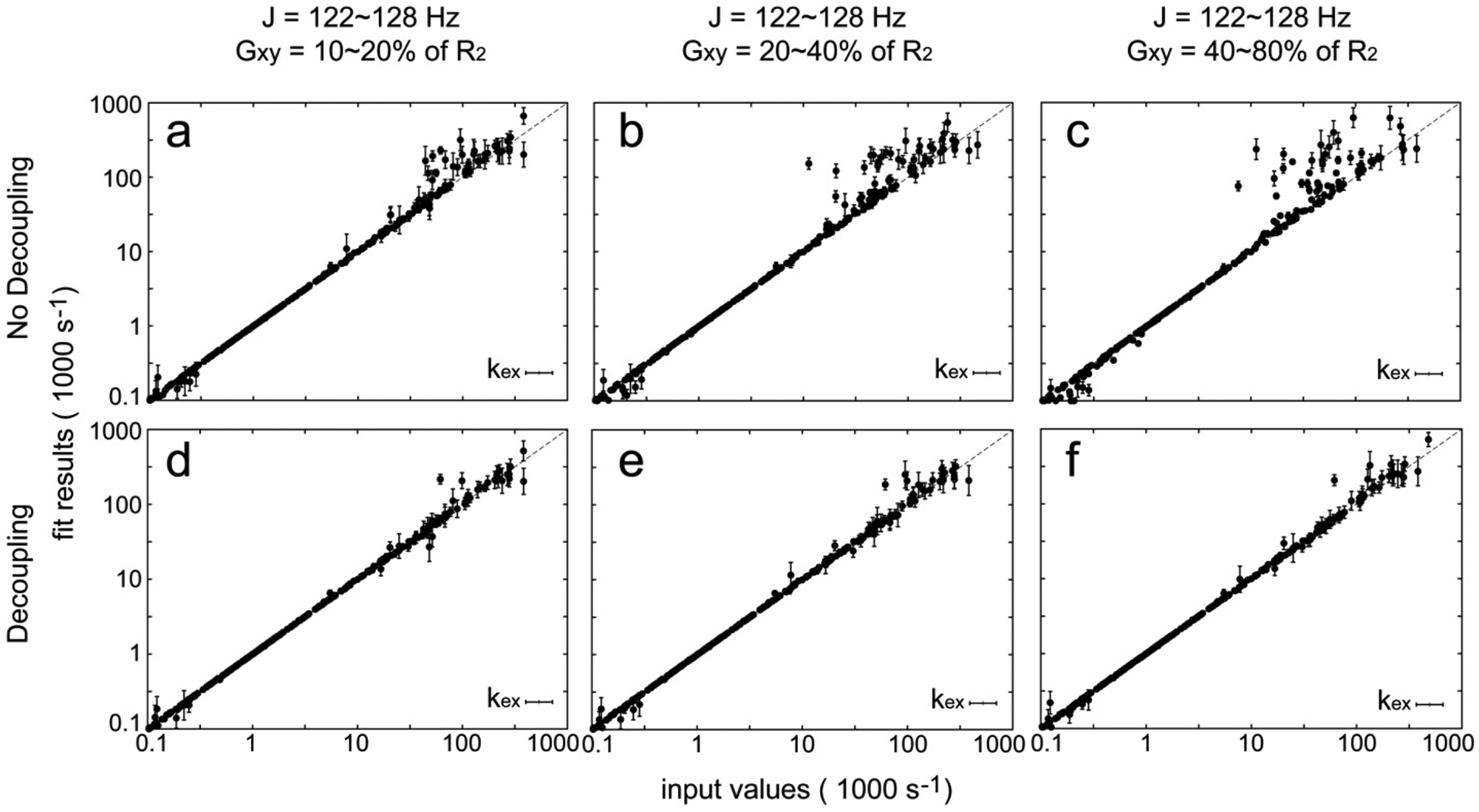
Simulated improvement due to the implementation of the proton decoupling scheme using a π pulse with perfect spin inversion. Data of adiabatic R1ρ, adiabatic R2ρ, and R1 at two (14.1 and 18.8 T) magnetic fields are first simulated and then analyzed using a geometric approximation to output the dynamic parameters. Adiabatic relaxation dispersion data sets (400) are all simulated using the 12 × 12 density matrix with random input dynamic parameters as described previously. No errors are introduced in this data analysis. J coupling values (J) and transverse cross-correlation rates (Gxy) are varied as indicated above panels (a, d), (b, e), and (c, f). In (a–c), the relaxation dispersion data were simulated without a proton spin inversion during the spin lock periods; in (d–f), the relaxation dispersion data were simulated with a proton spin inversion during the spin lock periods. The output results are plotted against the input values, and those with large standard deviations (SD of kex > 100.2) after Monte Carlo sampling are omitted.
In order to demonstrate the broader application of this new sequence, Ube2g2, an ubiquitin-conjugating enzyme E2 with known anisotropic tumbling, will be used to test the new experiments. Ube2g2 functions with the ubiquitin ligase E3, gp78, as part of the endoplasmic reticulum associated degradation (ERAD) pathway32–33, in a concerted mechanism to conjugate ubiquitin to substrates. In particular, different cytosolic domains of gp78 bind to Ube2g2 to induce allosteric effects. Through crystallography and solution-state NMR, structures and interfaces of Ube2g2 with various domains of gp78 bound to Ube2g2 reveal very subtle structural differences. Current studies of conformational dynamics have been focused on the backbone dynamics of Ube2g2 and suggest a role for dynamics driving the allostery34. However, what is unclear is how the allostery is transmitted through Ube2g2 upon gp78 binding. Due to the limited exchange rate window of the conventional CPMG experiments and the lower sensitivity of the backbone amide experiments, it is beneficial to probe the conformational dynamics of Ube2g2 in the presence of other binding partners using side-chain methyl-geoHARD.
All NMR experiments for methyl-geoHARD were performed on {U-[15N, 2H]; Ileδ1-[13CH3]; Leu, Val-[13CH3, 12CD3]}-labeled samples of Ube2g2 (~300 μM) at 15 °C in 50 mM Tris buffer (pH 7.4) containing 0.5 mM TCEP and 10% D2O. Data were acquired using Bruker Avance III spectrometers equipped with a helium-temperature TCI cryoprobe at 800 MHz and a nitrogen-temperature TCI Prodigy probe at 600 MHz. As described previously17, the adiabatic R1ρ and adiabatic R2ρ experiments monitor the decay of magnetization for each composite shape (HSnc) beginning at 0 ms and extended by multiples of a 16 ms block of composite adiabatic pulses to spin lock the magnetization (0, 16, 32, 48 ms… for N = 0, 1, 2, 3… in Fig. 1b,c). The total experiments consisted of adiabatic R1ρ with 0~128 ms of the spin lock, adiabatic R2ρ with 0~80 ms of the spin lock, R1 with 0~400 ms of the delay, and R2 with 0~128 ms of the spin echo, which took 3 days of acquisition in each static magnetic field. All relaxation experiments were carried out with a 2 s recycle delay, 16 scans, 128 complex points in the 13C dimension (19 ppm), and 2048 complex points in the 1H dimension (14 ppm). Data are processed with linear prediction for another 128 complex points in the 13C dimension with NMRPipe35 and subsequently analyzed with Sparky36.
Among 55 isotope-labeled methyl groups from Ile, Leu, and Val in Ube2g2 (~18 kDa; PDB: 2CYX), 47 methyl resonances are selected for data analysis, based on resolution and resonance overlap at the two magnetic field strengths. All the selected methyl resonances have mono-exponential decay of transverse magnetizations using the new pulse sequences at the two magnetic fields. For those methyl groups with relaxation dispersion, analyses assumed a two-site exchange model. The chemical exchange rates (kex) of 13 methyl groups can be reported for Ube2g2 (Fig. 3a and Supporting Fig. 7). (Only the kex rates with small standard deviations are reported.) Comparatively, the carbon single-quantum CPMG experiment37 only shows 6 methyl groups with measurable dispersions (Supporting Fig. 8). Our experimental data reveal two clusters of methyl groups in Ube2g2 (Supporting Fig. 7b) with well-defined exchange processes. One group (L9_CD2, L16_CD1, V116_CG1, V116_CG2), with a wider range of chemical exchange rates from 2000 s−1 to 9000 s−1, is located at the junction between helix α1 and helix α2. The second group (I24_CD1, I41_CD1, V124_CG2), with chemical exchange rates around 4000 s−1, is located at the junction between helix α2 and the β turns, which form the binding site for the G2BR peptide38. In previous reports, the allostery crucial for the biological function is observed when G2BR peptide binds to Ube2g2 and greatly enhances (approximately 50-fold) the RING binding affinity to Ube2g233. Interestingly, the binding site for RING domain is located at the N-terminal end of helix α1. In order to reveal the role of conformational dynamics in communicating the allostery as compared to a simple shift in a single conformation, we carried out a second set of experiments when Ube2g2 is saturated with the non-deuterated G2BR peptide (~5 kDa) (PDB:3H8K). Our second set of experimental data reveals detectable kex rates of only 7 methyl groups throughout Ube2g2 (Supporting Fig. 9), and no cluster of methyl groups as observed in free Ube2g2 (Fig. 3b). The decreased number of methyl groups with detectable kex values could result from a significantly decreased population of the minor states, which supports the previous hypothesis that G2BR can lock Ube2g2 into a closed state priming RING binding33–34. The CPMG experiment37 for the Ube2g2:G2BR complex shows all methyl groups with little or no relaxation dispersion (Supporting Fig. 10), similar to previous NH CPMG data34. However, the new version of methyl geoHARD detects conformational exchange for 7 methyl groups in Ube2g2:G2BR. The trimmed average of the observed kex rates for these methyl groups decreases from ~6000 s−1 to ~3000 s−1 upon the binding of G2BR. The reduction implies that the energy barrier between the two exchanging conformations becomes higher when G2BR binds to Ube2g2. This is in agreement with our previous interpretation of exchange between the closed (bound) state of Ube2g2 and the other state (most probably the open state), based on MD trajectories34.
Figure 3.
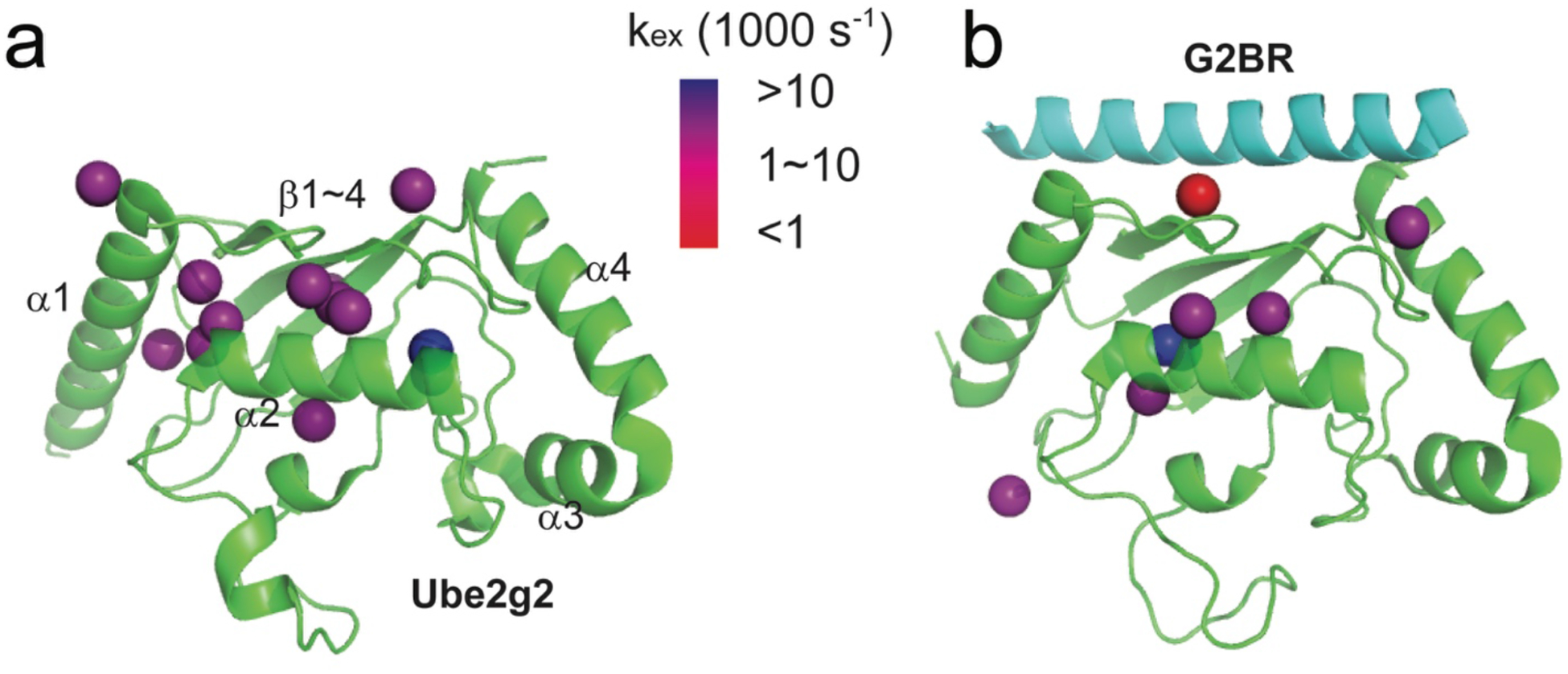
Conformational dynamics of methyl groups in Ube2g2 without (a) or with (b) the binding of G2BR, which are characterized by the new version of methyl-geoHARD. A total of 55 methyl groups are available as reporters throughout Ube2g2. Those with large standard deviations (SD of k ex > 100.2) after Monte Carlo sampling are not reported. (a) The kex rates of 13 methyl groups are reported in Ube2g2 without the binding of G2BR. (PDB: 2CYX) (b) The kex rates of 7 methyl groups are reported for Ube2g2 bound to G2BR. (PDB: 3H8K)
Finally, we conclude that the improved version of methyl-geoHARD experiments with composite pulse decoupling can provide high-quality data to reveal complex conformational dynamics. One may envision other, more complex composite pulses39 to be used for proton decoupling schemes to minimize the effects of pulse imperfections; however, the total length of the proton pulse should be less than ~160 μs (<1% of the spinlock period), to avoid undesired mixing of trajectories and creation of multiple quantum terms during overlap of the proton pulse and the adiabatic pulses. Such pulses must be validated to leave the proton-proton spin-flip mechanism unperturbed, which is especially important for proteins bound with non-deuterated ligands, such as synthetic peptides, nucleotides, or small molecule drugs. The present use of 90x-270y-90x performs very well and will be effective for all accessible magnetic field strengths, including the forthcoming 1.2 GHz systems. Utilizing these new tools, we reveal a potential allosteric path in Ube2g2 that is essential for its biological functions. Subsequent applications in studying conformational dynamics of multi-component complexes, as well as the hydrophobic cores of proteins, will improve our understanding in allosteric effects due to interactions among different components and allosteric signaling transmitted within the cores of proteins.
Supplementary Material
Acknowledgements:
The authors gratefully acknowledge the use of the Biophysics Resource, Structural Biophysics Laboratory, and the assistance of Dr. Sergey Tarasov and Ms. Marzena Dyba. The research was supported by the Intramural Research Program of the National Cancer Institute, Projects ZIA BC 011131 and ZIA BC 011132. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reference
- 1.Palmer AG 3rd, NMR characterization of the dynamics of biomacromolecules. Chem Rev 2004, 104 (8), 3623–40. [DOI] [PubMed] [Google Scholar]
- 2.Boehr DD; Dyson HJ; Wright PE, An NMR perspective on enzyme dynamics. Chem Rev 2006, 106 (8), 3055–79. [DOI] [PubMed] [Google Scholar]
- 3.Boehr DD; McElheny D; Dyson HJ; Wright PE, The dynamic energy landscape of dihydrofolate reductase catalysis. Science 2006, 313 (5793), 1638–42. [DOI] [PubMed] [Google Scholar]
- 4.Henzler-Wildman KA; Thai V; Lei M; Ott M; Wolf-Watz M; Fenn T; Pozharski E; Wilson MA; Petsko GA; Karplus M; Hubner CG; Kern D, Intrinsic motions along an enzymatic reaction trajectory. Nature 2007, 450 (7171), 838–44. [DOI] [PubMed] [Google Scholar]
- 5.Tzeng SR; Kalodimos CG, Dynamic activation of an allosteric regulatory protein. Nature 2009, 462 (7271), 368–72. [DOI] [PubMed] [Google Scholar]
- 6.Kim J; Ahuja LG; Chao FA; Xia Y; McClendon CL; Kornev AP; Taylor SS; Veglia G, A dynamic hydrophobic core orchestrates allostery in protein kinases. Sci Adv 2017, 3 (4), e1600663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimsey IJ; Szymanski ES; Zahurancik WJ; Shakya A; Xue Y; Chu CC; Sathyamoorthy B; Suo Z; Al-Hashimi HM, Dynamic basis for dG*dT misincorporation via tautomerization and ionization. Nature 2018, 554 (7691), 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loria JP; Rance M; Palmer AG, A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. Journal of the American Chemical Society 1999, 121 (10), 2331–2332. [Google Scholar]
- 9.Korzhnev DM; Skrynnikov NR; Millet O; Torchia DA; Kay LE, An NMR experiment for the accurate measurement of heteronuclear spin-lock relaxation rates. Journal of the American Chemical Society 2002, 124 (36), 10743–10753. [DOI] [PubMed] [Google Scholar]
- 10.Massi F; Johnson E; Wang CY; Rance M; Palmer AG, NMR R-1 rho rotating-frame relaxation with weak radio frequency fields. Journal of the American Chemical Society 2004, 126 (7), 2247–2256. [DOI] [PubMed] [Google Scholar]
- 11.Hansen DF; Vallurupalli P; Kay LE, An improved 15N relaxation dispersion experiment for the measurement of millisecond time-scale dynamics in proteins. J Phys Chem B 2008, 112 (19), 5898–904. [DOI] [PubMed] [Google Scholar]
- 12.Carver JP; Richards RE, General 2-Site Solution for Chemical Exchange Produced Dependence of T2 Upon Carr-Purcell Pulse Separation. J Magn Reson 1972, 6 (1), 89–&. [Google Scholar]
- 13.Trott O; Abergel D; Palmer AG, An average-magnetization analysis of R-1 rho relaxation outside of the fast exchange. Mol Phys 2003, 101 (6), 753–763. [Google Scholar]
- 14.Palmer AG; Koss H, Chemical Exchange. Biological Nmr, Pt B 2019, 615, 177–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mangia S; Traaseth NJ; Veglia G; Garwood M; Michaeli S, Probing slow protein dynamics by adiabatic R(1rho) and R(2rho) NMR experiments. J Am Chem Soc 2010, 132 (29), 9979–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao FA; Byrd RA, Geometric Approximation: A New Computational Approach To Characterize Protein Dynamics from NMR Adiabatic Relaxation Dispersion Experiments. J Am Chem Soc 2016, 138 (23), 7337–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chao FA; Li Y; Zhang Y; Byrd RA, Probing the Broad Time Scale and Heterogeneous Conformational Dynamics in the Catalytic Core of the Arf-GAP ASAP1 via Methyl Adiabatic Relaxation Dispersion. J Am Chem Soc 2019, 141 (30), 11881–11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ross A; Czisch M; King GC, Systematic errors associated with the CPMG pulse sequence and their effect on motional analysis of biomolecules. J Magn Reson 1997, 124 (2), 355–365. [Google Scholar]
- 19.Korzhnev DM; Tischenko EV; Arseniev AS, Off-resonance effects in N-15 T-2 CPMG measurements. J Biomol Nmr 2000, 17 (3), 231–237. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen NC; Bildsoe H; Jakobsen HJ; Sorensen OW, Composite Refocusing Sequences and Their Application for Sensitivity Enhancement and Multiplicity Filtration in Inept and 2d Correlation Spectroscopy. J Magn Reson 1989, 85 (2), 359–380. [Google Scholar]
- 21.Xia YL; Rossi P; Subrahmanian MV; Huang CD; Saleh T; Olivieri C; Kalodimos CG; Veglia G, Enhancing the sensitivity of multidimensional NMR experiments by using triply-compensated pi pulses. J Biomol Nmr 2017, 69 (4), 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levitt MH; Freeman R, Nmr Population-Inversion Using a Composite Pulse. J Magn Reson 1979, 33 (2), 473–476. [DOI] [PubMed] [Google Scholar]
- 23.Levitt MH; Freeman R, Compensation for Pulse Imperfections in Nmr Spin-Echo Experiments. J Magn Reson 1981, 43 (1), 65–80. [Google Scholar]
- 24.Freeman R; Kempsell SP; Levitt MH, Radiofrequency Pulse Sequences Which Compensate Their Own Imperfections. J Magn Reson 1980, 38 (3), 453–479. [DOI] [PubMed] [Google Scholar]
- 25.Baum J; Tycko R; Pines A, Broadband and adiabatic inversion of a two-level system by phase-modulated pulses. Phys Rev A Gen Phys 1985, 32 (6), 3435–3447. [DOI] [PubMed] [Google Scholar]
- 26.Warren WS; Silver MS, The Art of Pulse Crafting: Applications to Magnetic Resonance and Laser Spectroscopy In Advances in Magnetic and Optical Resonance, Waugh JS, Ed. Academic Press: 1988; Vol. 12, pp 247–384. [Google Scholar]
- 27.Emsley L; Bodenhausen G, Self-Refocusing Effect of 270-Degrees Gaussian Pulses - Applications to Selective Two-Dimensional Exchange Spectroscopy. J Magn Reson 1989, 82 (1), 211–221. [Google Scholar]
- 28.Tannus A; Garwood M, Adiabatic pulses. NMR Biomed 1997, 10 (8), 423–34. [DOI] [PubMed] [Google Scholar]
- 29.Garwood M; DelaBarre L, The return of the frequency sweep: designing adiabatic pulses for contemporary NMR. J Magn Reson 2001, 153 (2), 155–77. [DOI] [PubMed] [Google Scholar]
- 30.Kay LE, Artifacts can emerge in spectra recorded with even the simplest of pulse schemes: an HMQC case study. J Biomol Nmr 2019, 73 (8–9), 423–427. [DOI] [PubMed] [Google Scholar]
- 31.Freeman R; Keeler J, Suppression of Artifacts in Two-Dimensional J-Spectra. J Magn Reson 1981, 43 (3), 484–487. [Google Scholar]
- 32.Metzger MB; Liang YH; Das R; Mariano J; Li S; Li J; Kostova Z; Byrd RA; Ji X; Weissman AM, A structurally unique E2-binding domain activates ubiquitination by the ERAD E2, Ubc7p, through multiple mechanisms. Mol Cell 2013, 50 (4), 516–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das R; Liang YH; Mariano J; Li J; Huang T; King A; Tarasov SG; Weissman AM; Ji X; Byrd RA, Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine. EMBO J 2013, 32 (18), 2504–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakrabarti KS; Li J; Das R; Byrd RA, Conformational Dynamics and Allostery in E2:E3 Interactions Drive Ubiquitination: gp78 and Ube2g2. Structure 2017, 25 (5), 794–805 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delaglio F; Grzesiek S; Vuister GW; Zhu G; Pfeifer J; Bax A, NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol Nmr 1995, 6 (3), 277–93. [DOI] [PubMed] [Google Scholar]
- 36.Lee W; Tonelli M; Markley JL, NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31 (8), 1325–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lundstrom P; Vallurupalli P; Religa TL; Dahlquist FW; Kay LE, A single-quantum methyl C-13-relaxation dispersion experiment with improved sensitivity. J Biomol Nmr 2007, 38 (1), 79–88. [DOI] [PubMed] [Google Scholar]
- 38.Das R; Mariano J; Tsai YC; Kalathur RC; Kostova Z; Li J; Tarasov SG; McFeeters RL; Altieri AS; Ji X; Byrd RA; Weissman AM, Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol Cell 2009, 34 (6), 674–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manu VS; Veglia G, Genetic algorithm optimized triply compensated pulses in NMR spectroscopy. J Magn Reson 2015, 260, 136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


