Abstract
Importance:
All patients who develop multiple myeloma have a preceding asymptomatic expansion of clonal plasma cells, clinically recognized as monoclonal gammopathy of undetermined significance (MGUS) or smoldering multiple myeloma (SMM). During the past decade, significant progresses have been made in the smoldering multiple myeloma (SMM) classification and risk stratification.
Observations:
Here, we summarize and discuss current clinical challenges and available models for risk stratification in the setting of SMM. Thanks to several novel more effective and less toxic drugs, these aspects are increasingly becoming important to identify patients eligible for early treatment. However, all proposed criteria were built around indirect markers of disease burden, and therefore they are generally able to identify a fraction of SMM patients where both the transformation into multiple myeloma and the genomic subclonal diversification are already happening. In contrast, next generation sequencing approaches have the potential to identify myeloma precursor disease that will progress even before the major clonal expansion and progression, providing a potential base for a more effective and better precision regarding the optimal timing of treatment initiation.
Conclusions and Relevance:
Thanks to modern technologies, in the near coming future, prognostic models derived from genomic signatures independent from the disease burden will allow to better identify the optimal timing to treat a plasma cell clonal disorder at the very early stages, when the chances of eradication are higher.
Introduction
Many cancers develop through a Darwinian evolution model along preferred evolutionary trajectories where different driver events accumulate over time and confer a proliferation/survival advantage to a given subclone, progressively shaping the genomic profile and the clinical behavior of that cancer1. Gammopathies has been one of the fields where the concept of pre-malignant clonal evolution was described for the first time. In fact, virtually all patients who develop multiple myeloma have a preceding asymptomatic expansion of clonal plasma cells, clinically recognized as monoclonal gammopathy of undetermined significance (MGUS) or smoldering myeloma (SMM).2–6. For more than 20 years the definition and differentiation between these two clonal entities have been based on indirect measurements of the clonal burden (Table 1).3,4,7,8 These precursor conditions (i.e. MGUS and SMM) provide an extraordinary window of opportunity to study the multi-step evolution of cancer and to design preventive and early treatment strategies. In the last 10 years, the introduction of new drugs has significantly changed and improved the treatment landscape of multiple myeloma (eFigure 1 in the Supplement).9 Many of these new drugs are more effective and less toxic than traditional chemotherapy, providing for the first time attractive agents for early treatment strategies. Combining these novel regimens with modern technologies, such as next generation sequencing, we have the opportunity to identify patients with myeloma precursors that might benefit from early intervention.
Table 1.
Definition of multiple myeloma based on 2014 IMWG criteria.
| Criteria for Multiple Myeloma: |
|---|
| Clonal bone marrow plasma cells ≥10% or biopsy-proven bony or extramedullary plasmacytoma and any one or more of the following myeloma defining events: |
| 1) Evidence of end organ damage that can be attributed to the underlying plasma cell proliferative disorder, specifically: |
| • Hypercalcaemia: serum calcium >0·25 mmol/L (>1 mg/dL) higher than the upper limit of normal or >275 mmol/L (>11 mg/dL) |
| • Renal insufficiency: creatinine clearance <40 mL per min† or serum creatinine>177 μmol/L (>2 mg/dL) |
| • Anaemia: haemoglobin value of >20 g/L below the lower limit of normal, or a haemoglobin value <100 g/L |
| • Bone lesions: one or more osteolytic lesions on skeletal radiography, CT, or PET-CT |
| 2) Any one or more of the following biomarkers of malignancy |
| • Clonal bone marrow plasma cell percentage≥60% |
| • Involved:uninvolved serum free light chain ratio≥100 |
| • >1 focal lesions on MRI study |
| Definition of smoldering multiple myeloma: |
| 1) Serum monoclonal protein (IgG or IgA) ≥30 g/L or urinary monoclonal protein ≥500 mg per 24 h and/or clonal bone marrow plasma cells 10–60% |
| 2) Absence of myeloma defining events or amyloidosis |
Here, we will review the current criteria for risk-stratification in SMM and highlight new research and clinical perspectives in light of novel discoveries in the era of genomics.
Diagnosis and assessment
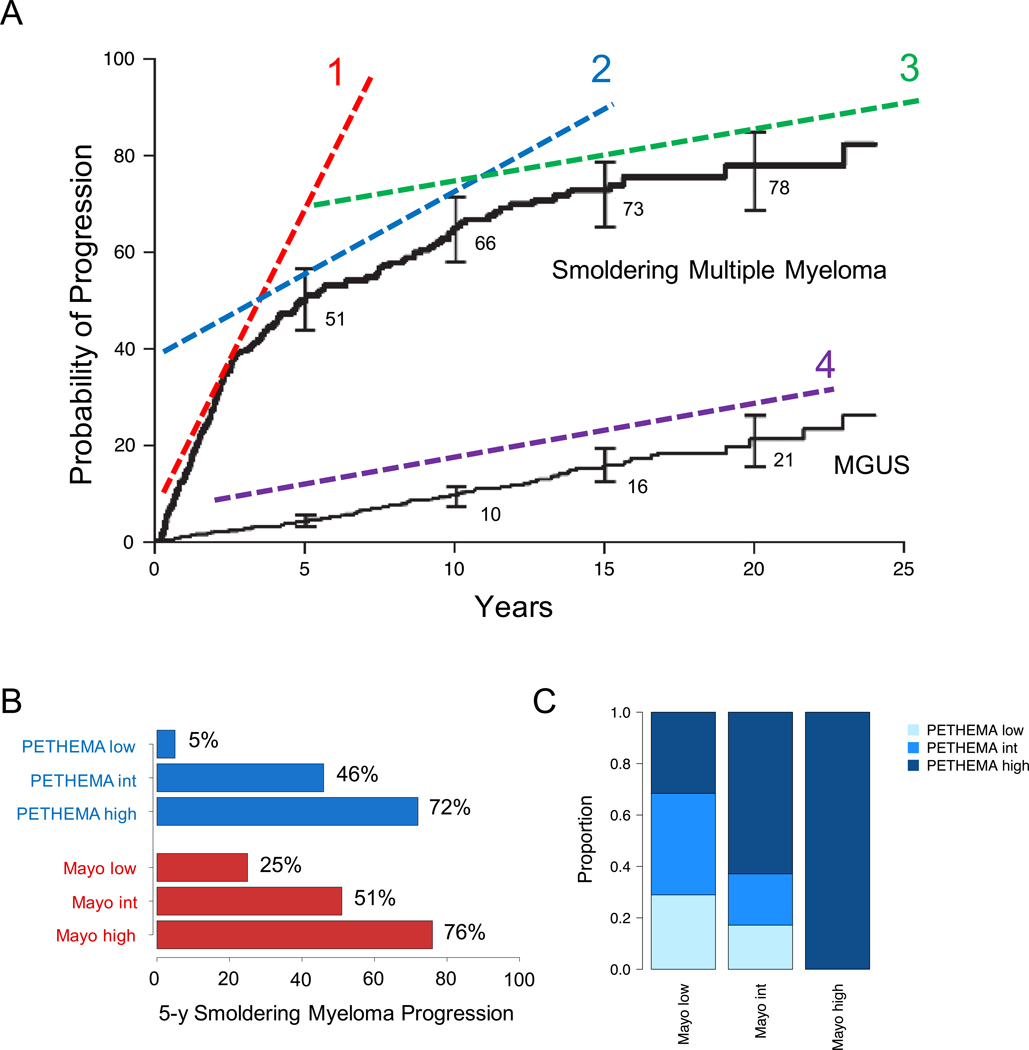
Investigating a large retrospective cohort of 276 SMM patients, Kyle et al reported an overall risk of progression of 10% per year for the first 5 years, approximately 3% per year for the next 5 years, and 1% per year for the last 10 years (Figure 1A-B).10 For the first time, different serum markers were associated with the risk of SMM progression into multiple myeloma. The ability to predict the risk of progression was a turning point in the study of SMM, stimulating many different groups to develop translational studies and clinical trials.
Figure 1.
Clinical outcome of MM precursos. A) Three evolution trajectories for MGUS/SMM progression (adapted from Kyle et al NEJM 200710), potentially reflecting three different biological and genomic profiles. The first trajectories (1) reflect an ongoing transformation/progression. The second (2) reflect patients and clones with high predisposition to acquire new drivers and progress over the time. The third (3) is characterized by low predisposition to acquire new drivers and progress over the time, with a clinical outcome similar to MGUS (4). B) C) Risk of SMM/MGUS progression according to either Mayo or PETHEMA prognostic model. D) Comparison between Mayo and PETHEMA risk models for the identification of patients with high-risk SMM. Adapted from Cherry et al.17
After these preliminary observations, different markers were subsequently reported to be associated with an increased risk of SMM progression and two major models for SMM risk stratification were developed based on combinations of these markers (Figure 1C-D) 11–15: i) the Mayo Clinic model, mostly focused on serum protein abnormalities as surrogate measures of tumor burden;10 and ii) the PETHEMA model, using multi-parametric flow cytometry to phenotype bone marrow plasma cells.16 Discouragingly, a head-to-head comparison of the Mayo Clinic and PETHEMA models in a prospective cohort of 77 patients with SMM showed a significant discordance on the overall risk stratification, with only 28.6% patients overlapping on specific risk categories: low-risk vs high-high (p<0.01), low-risk vs non-low-risk (p<0.01) and high-risk vs non-high-risk (p<0.01) (Figure 1D).17 Furthermore, among the Swedish Myeloma Registry, only 30% of SMM that progressed met the Mayo Clinic criteria for high-risk disease.18 Overall, this highlights the need for more robust strategies to identify individuals with myeloma precursor disease who are at high risk of progression.
Thanks to technological advances, several different approaches have been tested to allow an earlier and more accurate identification of high risk SMM patients. The application of imaging approaches, such as MRI of the spine and pelvis and whole-body PET-CT scans, showed a strong ability to predict the clinical course of SMM.19–21 Specifically, patients with focal lesions and/or bone marrow abnormalities were characterized by a higher risk of progression.22 However, these approaches still reflect the SMM disease burden and end-organ damage.
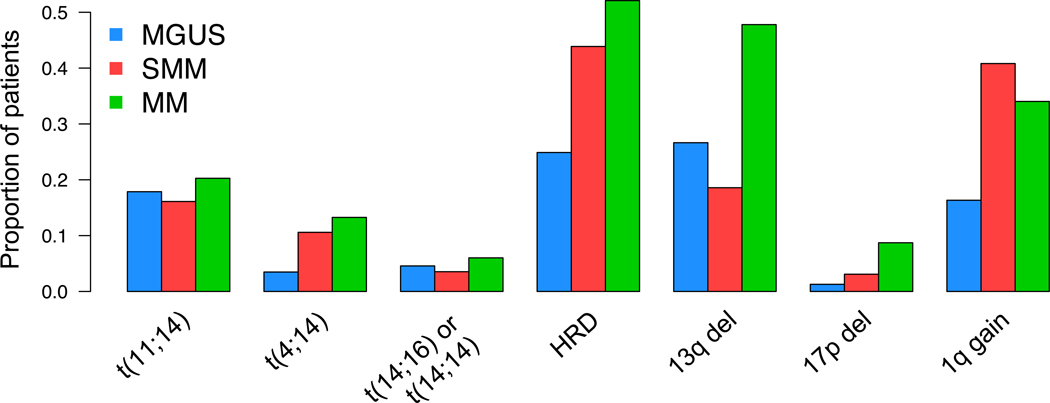
Importantly, alternative efforts have been attempted to identify markers of SMM progression, which are not measures of the disease burden but rather more intrinsically linked to the underlying biology of the disease. In the last 20 years, several studies reported on recurrent translocations and copy number abnormalities (CNAs) in multiple myeloma using karyotype and FISH, some of them linked to prognosis. (Figure 2).23,24 Following on these efforts, recurrent CNAs and translocations were investigated by FISH across several different clinical trials and retrospective cohorts showing that the presence of del17p13 (TP53), t(4;14)(MMSET;IGH) and/or 1q21 gain were associated with shortened time to progression in SMM and MGUS.25,26 Following a different approach, Dhodapkar and colleagues identified a 70-gene expression signature in SMM (GEP70) independently associated with the risk of progression to multiple myeloma.27 However, similarly to many prognostic models based on gene expression, their application into daily clinical practice has always been limited by low standardization and low reproducibility.
Figure 2.
MM precursor cytogenetic. The prevalence of the main cytogenetic aberration evaluated by either FISH or SNP array combining together different published cohort of MGUS, SMM and newly diagnosed MMs.28–32
Prognosis
Overall, looking at progression curves of SMM, 3 main patterns can be identified based on the rate of transformation, likely reflecting differences in underlying biology10 (Figure 1): 1) a group characterized by ongoing transformation already at the time of SMM diagnosis who will meet transformation criteria within few months/years. These patients have likely already acquired the key drivers of progression, and likely have a genomic and microenvironment landscape extremely similar to the one of multiple myeloma; 2) A second group will progress after several years of indolent and asymptomatic clinical course. This group of SMM likely has a genomic background more prone to acquire both new lesions and microenvironment changes with the potential to drive an aggressive evolutionary trajectory and consequently shift the clinical behavior. Differently from the first group, the key drivers of progression to multiple myeloma have not yet been acquired in this intermediate group. 3) The third group is characterized by a clinical course compatible to that of MGUS, despite meeting clinical criteria for SMM diagnosis (i.e. they have a recognizable burden of disease in terms of plasma cell bone marrow infiltration and serum M-component). In this last group of patients, the genomic predisposition to acquire new drivers of progression is very low, the clone is stable, and the clinical course is indolent in the long term. While these patterns can be identified by careful scrutiny of progression curves, we do not have any direct evidence of the biological events causally linked to disease progression and it is therefore not possible at present to assign a patient to a group at the time of SMM diagnosis. However, the identification of biological markers to predict MGUS/SMM progression before the onset of end-organ damage has always been one of the main goals in multiple myeloma research. Thanks to the wide availability of next generation sequencing (NGS) platforms we now have the means to tackle this critical task.
In general, MGUS and SMM shared some recurrent genomic aberrations with multiple myeloma (Figure 2); however others, such as del13q and 17p13 deletions, were observed at lower prevalence compared to multiple myeloma.26,28–34 Furthermore, all but t(11;14)(CCND1;IGH) recurrent cytogenetic and structural aberrations have been reported at lower frequency among MGUS cases compared to multiple myeloma and SMM cases (Figure 2).This confirms the existence of a chronological order of acquisition of cytogenetic aberrations potentially involved in the clone progression from MGUS to SMM and from SMM to multiple myeloma.35
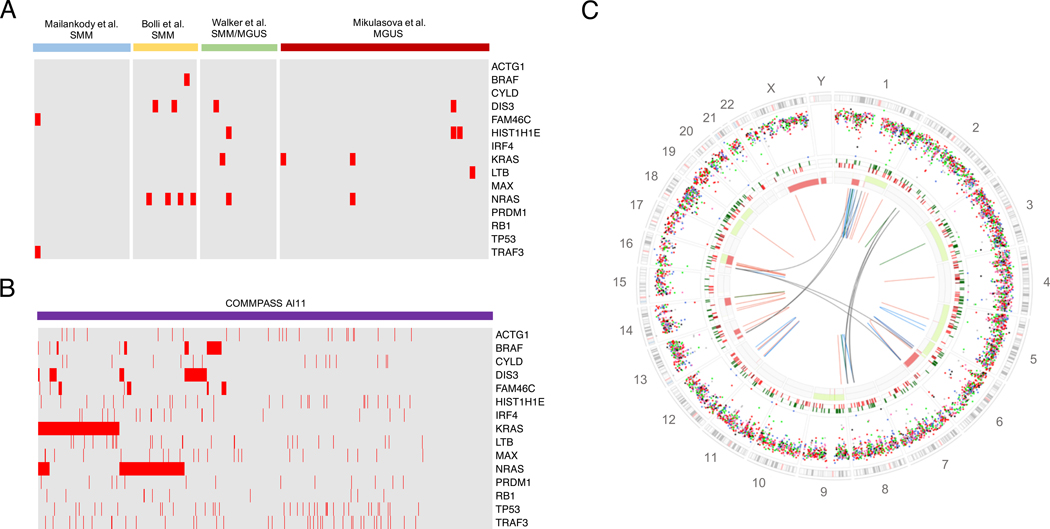
Using whole genome sequencing (WGS) and whole exome sequencing (WES) data, different groups have recently shown that multiple myeloma has a complex and heterogeneous genomic profile.36–41 Significant efforts have been performed to characterize the landscape of driver gene mutations in multiple myeloma.36,37,39,42,43 However, several reports suggested that gene mutations are secondary events, not directly involved in the cancer initiation and early progression.39,42,43 As confirmation of this, initial WES and WGS sequencing studies have shown a lower prevalence of these driver mutations in MGUS/SMM compared to what has been reported among newly diagnosed multiple myeloma (Figure 3A-B).31,44–46 Overall, these observations suggested that the MGUS/SMM genomic landscape is less complex than multiple myeloma, and the identification of secondary events involved in progression could represent an important early and accurate prognostic marker. However, this is not an easy task in multiple myeloma. Some hematological malignancies, such as acute myeloid leukemia, are usually characterized by a low genomic complexity, and the characterization of their genomic landscape based on targeted and/or exome-based approaches has been relatively straightforward.47 Similar approaches have been attempted in multiple myeloma,40,43,48,49 however its genomic landscape is far more complex and heterogeneous (Figure 3C-D).42 Recently, the first WGS characterization of 10 non-high risk SMM10 cases that progressed to multiple myeloma highlighted the great genomic complexity and heterogeneity of this pre-malignant entity.44 Two different progression models were reported, based on gene mutations, translocations, CNAs and mutational signatures in paired SMM and multiple myeloma samples (eFigure 2 in the Supplement): 1) the “static model”, where the same subclonal architecture was retained as the disease progressed to multiple myeloma, and 2) the “spontaneous evolution model”, where the subclonal composition of SMM changed without any external selective pressure. While patients included in the first group generally progressed in less than one year, reflecting an already ongoing transformation process, the second showed a longer time to progression. While the identification of robust novel prognostic markers of SMM progression was beyond the scope of the study, additional studies confirmed that several genomic lesions only explorable by WGS might be involved in SMM progression.42,44,46,50–52 Aside from mutations, CNAs and translocations, analysis of mutational signatures can also provide clues to disease progression. In particular, aberrant activity of the APOBEC family of DNA deaminases, a mutational process particularly active in aggressive multiple myeloma,41,50 was mostly present in late SMM subclones, suggesting a role in progression to multiple myeloma
Figure 3.
MM precursors genomic landscape. A-B) Heatmap showing the prevalence of recurrent MM mutations among MGUS/SMM/MMS according to the 4 main next generation sequencing studies and among newly diagnosed MM enrolled within the COMMPASS trial.31,44–46 The CoMMpass data were generated as part of the Multiple Myeloma Research Foundation Personalized Medicine Initiatives (https://research.themmrf.org and www.themmrf.org). C) Example of SMM whole genome landscape. All main genomic events are reported in this genome plot: mutations (external circle), indels (middle circle; dark green and brown lines represent insertion and deletion respectively), copy number variants (red = deletions, green = gain) and rearrangements (blue = inversion, red = deletions, green = ITD, black = translocation). This patients was a non-high risk SMM10 and progressed after 42 months.
The idea that we can identify biological hallmarks of disease aggressiveness among MGUS/SMM has been further confirmed by a recent single cell RNA sequencing study, where authors showed among MGUS the existence of small clonal populations indistinguishable at the molecular level from individuals with multiple myeloma.53
One limitation of genomic studies is that they are focused on the cancer (i.e. myeloma) cells, and thus blind to its complex interaction with microenvironment. The “cancer-naïve” niche is not suited for cancer cell growth. Cancer cells may however acquire the ability to induce phenotypic changes in the microenvironment, promoting the production of cellular and humoral factor that may sustain cancer proliferation, as well as escape from immune tumor-surveillance so to make it permissive to cancer cell seeding.54,55 Overall the microenvironment can influence the myeloma-genesis in different ways: 1) creating the condition for the cancer initiations within the germinal center; 2) inducing a selective pressure limiting the expansion of pre-malignant entities; 3) promoting the pre-malignant clonal expansion and progression into symptomatic disease. The role of the microenvironment in multiple myeloma has been extensively studied, and comparative studies have highlighted changes that may be linked to progression from asymptomatic and symptomatic stages.56,57 A direct, in vivo demonstration of the role of the microenvironment on progression of monoclonal gammopathies recently came from the study of a humanized mouse model, where xenografts from patients with asymptomatic gammopathies showed progressive growth, suggesting that the clinical stability of these conditions may be in part due to extrinsic growth restriction of tumor cells from the host microenvironment.58 Interestingly, progression occurred after a Darwinian competition where a minor subclone in the patient acquired clonal dominance in the xenograft. This is a clear example of how the heterogeneous genomic landscape of plasma cell disorders can dynamically adapt to different microenvironments, in an interplay required for tumor progression. This concept was also recently supported by a study on simultaneous biopsies in in NDMM that revealed an extensive heterogeneity between spatially distinct lesions, possibly reflecting the influence of different microenvironmental contexts on subclonal selection within tumor cells.59
Recently, the first large, population-based prospective screening study from myeloma precursor disease to MM provided novel insight on how serum immune markers – indicators of disease-burden – can change over time, emphasizing the importance of repeated blood tests and re-assessment of risk annually.71 Certainly, a logical extension of the results from the population-based prospective screening study71 is the application of genomic assays on longitudinally collected samples, with the aim to define key evolutionary and clinical trajectories involved in the progression from myeloma precursor disease to multiple myeloma.
The integration of cell-intrinsic and cell-extrinsic analyses will offer the extraordinary opportunity to design new prognostic models that are related to biologic features of the disease rather than to its burden. Harnessing the selective evolutionary pressure from the microenvironment for therapeutic purposes, before a more active subclone takes over and the disease (eventually) becomes symptomatic, will also be a relevant challenge for future.
Treatment
Classically, asymptomatic stages are managed with a watch and wait approach. The rationale for this comes from unsuccessful early treatment approaches using melphalan-prednisone, thalidomide or bisphosphonates.60–66
Driven by the increased access to novel drugs that are highly effective and generally safe, early high risk SMM treatment is in the spotlight again. Rationale for early treatment of SMM patients consists in better potential of disease control, if not cure, in early stages where the subclonal complexity (and hence drug resistance) could be lower. Furthermore, patients may better tolerate treatment in the absence of end-organ damage. On the other hand, there is a risk of over-treating a fraction of patients that would otherwise be stable for years. Also, treatment could represent an early bottleneck of evolution for cancer cells, thereby promoting earlier expansion of more aggressive and refractory clones as compared to conventional watch and wait approaches. Finally, while the anti-myeloma activity of chemotherapeutical agents is widely accepted, their short- and long-term toxicity would still represent an important concern in the SMM setting.
Recently, the 2014 updated diagnostic criteria for multiple myeloma suggested that ultra-high risk SMM (now defined as multiple myeloma) should be treated even if asymptomatic, owing to a 80% risk of developing end-organ damage in the next 2 years.22 For the SMM cases with a high risk of progression – who do not fit the new criteria for multiple myeloma – several attempts have been made to challenge this practice again since the advent of novel drugs. In 2013, Mateos et al published the first phase III trial where high risk SMM cases were randomized between watch and wait and treatment with lenalidomide-dexamethasone for nine cycles, followed by a fixed 2 year maintenance regimen.67,68 Authors reported a survival advantage in the experimental arm: 94% vs. 80% alive patients at 3 years.
Inspired by these data, in the early 2010s, the combination of Carfilzomib/ Lenalidomide/Dexamethasone was developed as the first 3-drug combination therapy study targeting high-risk SMM patients, and the results showed a promising high rate of MRD negativity (92%).45,69 Despite results from these important trials, follow-up is still short and the paradigm of SMM treatment has not changed in routine clinical practice; however, several clinical trials targeting high-risk SMM patients are currently ongoing and, considering the efficacy of novel agent combinations in newly diagnosed patients, most of these have been designed without standard chemotherapy (eTable1 in the Supplement).
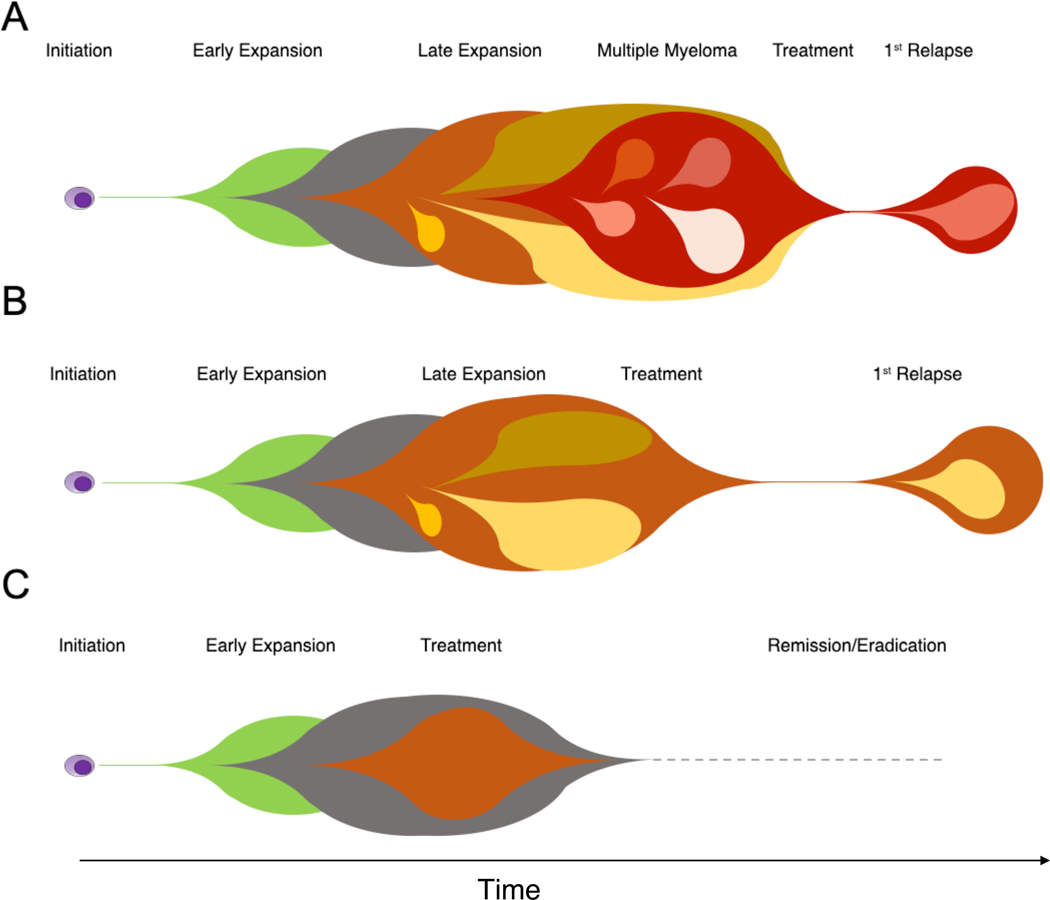
Before treatment of SMM can be accepted as standard practice, several biological and clinical questions must be addressed. Clinical and laboratory markers associated with risk of progression are mainly based on surrogate measures of disease burden, and the chosen cut-offs are generally based on arbitrary values extracted from retrospective series; it is every day’s experience of any clinician that these risk scores are inaccurate for some patients. We believe that through genomic studies we will, in the future, detect aggressive disease cases in asymptomatic phases, also allowing for control of subclonal diversification and long-term toxicities on the hematopoietic stem cells from treatment (Figure 4).
Figure 4.
The rational of early treatment in SMM/MGUS patients. MM pathogenesis is characterized by a typical Darwinian evolution model, characterized by the accumulation over the time of different drivers along preferred evolutionary trajectories. When we treat MM (A), most of our patients achieve a good response and durable remission. However, a significant fraction still progresses, generally after a clonal/subclonal competition and selection induced by our treatment. This clonal heterogeneity and genomic complexity represent one of the main reasons why it is so hard to completely eradicate this cancer. B) A significant fraction of high risk SMM already have the key drivers and have already expanded their clonal and subclonal architecture, reducing our chance to eradicate the disease. C) Conversely, at the very early stage of MGUS/SMM clonal expansion, the genomic and clonal complexity is likely lower, potentially increasing the pre-malignant clone eradication chances.
Conclusions:
In the next years we need to better understand the biological determinants and the dynamics of evolution of myeloma precursor disease to identify more robust genomic prognostic markers to be combined within current risk models. This is finally achievable in the current genomic era and will potentially allow clinicians to better identify the optimal timing to treat a plasma cell clonal disorder at the very early stages, when the chances of eradication are higher.
Supplementary Material
Key points:
Question:
All criteria for smoldering multiple myeloma (SMM) risk stratification are built around indirect markers of disease burden, and therefore they are able to identify a fraction of SMM patient where both the transformation and the genomic subclonal diversification are already happening.
Findings:
Next generation sequencing approaches have the potentiality to identify SMMs that will progress even before the major clonal expansion and progression, providing a potential base for a more effective early treatment in asymptomatic MM.
Meaning:
A better understanding of the biological determinants of SMM will allow clinicians to better identify the optimal timing to treat a plasma cell clonal disorder at the very early stages, when the chances of eradication are higher.
Tweet:
Smoldering myeloma: we need to move from cancer burden to cancer genomics to predict progression vs non-progression
Acknowledgements:
F.M. and O.L. received funding support from the Memorial Sloan Kettering Core Grant (P30 CA008748) and the Perelman Family Foundation in collaboration with the Multiple Myeloma Research Foundation (MMRF). N.B. has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No. 817997).
Disclosure of Conflicts of Interest:
Dr. Landgren’s disclosures:
• Grant support: NIH, FDA, MMRF, IMF, LLS, Perelman Family Foundation, Rising Tides Foundation, Amgen, Celgene, Janssen, Takeda, Glenmark, Seattle Genetics, Karyopharm
• Honoraria/ad boards: Adaptive, Amgen, Binding Site, BMS, Celgene, Cellectis, Glenmark, Janssen, Juno, Pfizer
• Chairman for “Medscape myeloma”: 2014–2019
• Independent Data Monitoring Committee (IDMC): Takeda, Merck, Janssen, Theradex
Prof Bolli’s disclosures:
• Honoraria/ad boards: Celgene, Janssen, Novartis
Footnotes
Other authors: No conflict of interests to declare.
References
- 1. Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Landgren O, Weiss BM. Patterns of monoclonal gammopathy of undetermined significance and multiple myeloma in various ethnic/racial groups: support for genetic factors in pathogenesis. Leukemia. 2009;23(10):1691–1697. [DOI] [PubMed] [Google Scholar]
- 3. Kyle RA, Therneau TM, Rajkumar SV, et al. Prevalence of monoclonal gammopathy of undetermined significance. N Engl J Med. 2006;354(13):1362–1369. [DOI] [PubMed] [Google Scholar]
- 4. Kyle RA, Greipp PR. Smoldering multiple myeloma. N Engl J Med. 1980;302(24):1347–1349. [DOI] [PubMed] [Google Scholar]
- 5. Fidler IJ. Tumor heterogeneity and the biology of cancer invasion and metastasis. Cancer Res. 1978;38(9):2651–2660. [PubMed] [Google Scholar]
- 6. Dexter DL, Kowalski HM, Blazar BA, Fligiel Z, Vogel R, Heppner GH. Heterogeneity of tumor cells from a single mouse mammary tumor. Cancer Res. 1978;38(10):3174–3181. [PubMed] [Google Scholar]
- 7. Rajkumar SV, Landgren O, Mateos MV. Smoldering multiple myeloma. Blood. 2015;125(20):3069–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Landgren O, Kyle RA, Pfeiffer RM, et al. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood. 2009;113(22):5412–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gay F, Jackson G, Rosinol L, et al. Maintenance Treatment and Survival in Patients With Myeloma: A Systematic Review and Network Meta-analysis. JAMA Oncol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kyle RA, Remstein ED, Therneau TM, et al. Clinical course and prognosis of smoldering (asymptomatic) multiple myeloma. N Engl J Med. 2007;356(25):2582–2590. [DOI] [PubMed] [Google Scholar]
- 11. Agarwal A, Ghobrial IM. Monoclonal gammopathy of undetermined significance and smoldering multiple myeloma: a review of the current understanding of epidemiology, biology, risk stratification, and management of myeloma precursor disease. Clin Cancer Res. 2013;19(5):985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dispenzieri A, Stewart AK, Chanan-Khan A, et al. Smoldering multiple myeloma requiring treatment: time for a new definition? Blood. 2013;122(26):4172–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Landgren O. Monoclonal gammopathy of undetermined significance and smoldering multiple myeloma: biological insights and early treatment strategies. Hematology Am Soc Hematol Educ Program. 2013;2013:478–487. [DOI] [PubMed] [Google Scholar]
- 14. Larsen JT, Kumar SK, Dispenzieri A, Kyle RA, Katzmann JA, Rajkumar SV. Serum free light chain ratio as a biomarker for high-risk smoldering multiple myeloma. Leukemia. 2013;27(4):941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rajkumar SV, Larson D, Kyle RA. Diagnosis of smoldering multiple myeloma. N Engl J Med. 2011;365(5):474–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perez-Persona E, Vidriales MB, Mateo G, et al. New criteria to identify risk of progression in monoclonal gammopathy of uncertain significance and smoldering multiple myeloma based on multiparameter flow cytometry analysis of bone marrow plasma cells. Blood. 2007;110(7):2586–2592. [DOI] [PubMed] [Google Scholar]
- 17. Cherry BM, Korde N, Kwok M, et al. Modeling progression risk for smoldering multiple myeloma: results from a prospective clinical study. Leuk Lymphoma. 2013;54(10):2215–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kristinsson SY, Holmberg E, Blimark C. Treatment for high-risk smoldering myeloma. N Engl J Med. 2013;369(18):1762–1763. [DOI] [PubMed] [Google Scholar]
- 19. Hillengass J, Fechtner K, Weber MA, et al. Prognostic significance of focal lesions in whole-body magnetic resonance imaging in patients with asymptomatic multiple myeloma. J Clin Oncol. 2010;28(9):1606–1610. [DOI] [PubMed] [Google Scholar]
- 20. Kastritis E, Moulopoulos LA, Terpos E, Koutoulidis V, Dimopoulos MA. The prognostic importance of the presence of more than one focal lesion in spine MRI of patients with asymptomatic (smoldering) multiple myeloma. Leukemia. 2014;28(12):2402–2403. [DOI] [PubMed] [Google Scholar]
- 21. Zamagni E, Nanni C, Gay F, et al. 18F-FDG PET/CT focal, but not osteolytic, lesions predict the progression of smoldering myeloma to active disease. Leukemia. 2016;30(2):417–422. [DOI] [PubMed] [Google Scholar]
- 22. Rajkumar SV, Dimopoulos MA, Palumbo A, et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014;15(12):e538–548. [DOI] [PubMed] [Google Scholar]
- 23. Manier S, Salem KZ, Park J, Landau DA, Getz G, Ghobrial IM. Genomic complexity of multiple myeloma and its clinical implications. Nat Rev Clin Oncol. 2017;14(2):100–113. [DOI] [PubMed] [Google Scholar]
- 24. Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12(5):335–348. [DOI] [PubMed] [Google Scholar]
- 25. Neben K, Jauch A, Hielscher T, et al. Progression in smoldering myeloma is independently determined by the chromosomal abnormalities del(17p), t(4;14), gain 1q, hyperdiploidy, and tumor load. J Clin Oncol. 2013;31(34):4325–4332. [DOI] [PubMed] [Google Scholar]
- 26. Rajkumar SV, Gupta V, Fonseca R, et al. Impact of primary molecular cytogenetic abnormalities and risk of progression in smoldering multiple myeloma. Leukemia. 2013;27(8):1738–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dhodapkar MV, Sexton R, Waheed S, et al. Clinical, genomic, and imaging predictors of myeloma progression from asymptomatic monoclonal gammopathies (SWOG S0120). Blood. 2014;123(1):78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lakshman A, Rajkumar SV, Buadi FK, et al. Risk stratification of smoldering multiple myeloma incorporating revised IMWG diagnostic criteria. Blood Cancer J. 2018;8(6):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lopez-Corral L, Gutierrez NC, Vidriales MB, et al. The progression from MGUS to smoldering myeloma and eventually to multiple myeloma involves a clonal expansion of genetically abnormal plasma cells. Clin Cancer Res. 2011;17(7):1692–1700. [DOI] [PubMed] [Google Scholar]
- 30. Mikulasova A, Smetana J, Wayhelova M, et al. Genomewide profiling of copy-number alteration in monoclonal gammopathy of undetermined significance. Eur J Haematol. 2016;97(6):568–575. [DOI] [PubMed] [Google Scholar]
- 31. Mikulasova A, Wardell CP, Murison A, et al. The spectrum of somatic mutations in monoclonal gammopathy of undetermined significance indicates a less complex genomic landscape than that in multiple myeloma. Haematologica. 2017;102(9):1617–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmidt-Hieber M, Gutierrez ML, Perez-Andres M, et al. Cytogenetic profiles in multiple myeloma and monoclonal gammopathy of undetermined significance: a study in highly purified aberrant plasma cells. Haematologica. 2013;98(2):279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brousseau M, Leleu X, Gerard J, et al. Hyperdiploidy is a common finding in monoclonal gammopathy of undetermined significance and monosomy 13 is restricted to these hyperdiploid patients. Clin Cancer Res. 2007;13(20):6026–6031. [DOI] [PubMed] [Google Scholar]
- 34. Chng WJ, Van Wier SA, Ahmann GJ, et al. A validated FISH trisomy index demonstrates the hyperdiploid and nonhyperdiploid dichotomy in MGUS. Blood. 2005;106(6):2156–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Nieuwenhuijzen N, Spaan I, Raymakers R, Peperzak V. From MGUS to Multiple Myeloma, a Paradigm for Clonal Evolution of Premalignant Cells. Cancer Res. 2018;78(10):2449–2456. [DOI] [PubMed] [Google Scholar]
- 36. Bolli N, Avet-Loiseau H, Wedge DC, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chapman MA, Lawrence MS, Keats JJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Keats JJ, Chesi M, Egan JB, et al. Clonal competition with alternating dominance in multiple myeloma. Blood. 2012;120(5):1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lohr JG, Stojanov P, Carter SL, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25(1):91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walker BA, Boyle EM, Wardell CP, et al. Mutational Spectrum, Copy Number Changes, and Outcome: Results of a Sequencing Study of Patients With Newly Diagnosed Myeloma. J Clin Oncol 2015;33(33):3911–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walker BA, Wardell CP, Murison A, et al. APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nat Commun. 2015;6:6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maura F, Bolli N, Angelopoulos N, et al. Genomic landscape and chronological reconstruction of driver events in multiple myeloma. bioRxiv. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Walker BA, Mavrommatis K, Wardell CP, et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood. 2018;132(6):587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bolli N, Maura F, Minvielle S, et al. Genomic patterns of progression in smoldering multiple myeloma. Nat Commun. 2018;9(1):3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mailankody S, Kazandjian D, Korde N, et al. Baseline mutational patterns and sustained MRD negativity in patients with high-risk smoldering myeloma. Blood Adv. 2017;1(22):1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Walker BA, Wardell CP, Melchor L, et al. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia. 2014;28(2):384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374(23):2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bolli N, Biancon G, Moarii M, et al. Analysis of the genomic landscape of multiple myeloma highlights novel prognostic markers and disease subgroups. Leukemia. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Walker BA, Mavrommatis K, Wardell CP, et al. A high-risk, Double-Hit, group of newly diagnosed myeloma identified by genomic analysis. Leukemia. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Maura F, Petljak M, Lionetti M, et al. Biological and prognostic impact of APOBEC-induced mutations in the spectrum of plasma cell dyscrasias and multiple myeloma cell lines. Leukemia. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao S, Choi M, Heuck C, et al. Serial exome analysis of disease progression in premalignant gammopathies. Leukemia. 2014;28(7):1548–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dutta AK, Fink JL, Grady JP, et al. Subclonal evolution in disease progression from MGUS/SMM to multiple myeloma is characterised by clonal stability. Leukemia. 2019;33(2):457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ledergor G, Weiner A, Zada M, et al. Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat Med. 2018;24(12):1867–1876. [DOI] [PubMed] [Google Scholar]
- 54. Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–458. [DOI] [PubMed] [Google Scholar]
- 56. Manier S, Sacco A, Leleu X, Ghobrial IM, Roccaro AM. Bone marrow microenvironment in multiple myeloma progression. J Biomed Biotechnol. 2012;2012:157496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bianchi G, Munshi NC. Pathogenesis beyond the cancer clone(s) in multiple myeloma. Blood. 2015;125(20):3049–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Das R, Strowig T, Verma R, et al. Microenvironment-dependent growth of preneoplastic and malignant plasma cells in humanized mice. Nat Med. 2016;22(11):1351–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rasche L, Chavan SS, Stephens OW, et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat Commun. 2017;8(1):268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. D’Arena G, Gobbi PG, Broglia C, et al. Pamidronate versus observation in asymptomatic myeloma: final results with long-term follow-up of a randomized study. Leuk Lymphoma. 2011;52(5):771–775. [DOI] [PubMed] [Google Scholar]
- 61. Hjorth M, Hellquist L, Holmberg E, Magnusson B, Rodjer S, Westin J. Initial versus deferred melphalan-prednisone therapy for asymptomatic multiple myeloma stage I--a randomized study. Myeloma Group of Western Sweden. Eur J Haematol. 1993;50(2):95–102. [DOI] [PubMed] [Google Scholar]
- 62. Martin A, Garcia-Sanz R, Hernandez J, et al. Pamidronate induces bone formation in patients with smouldering or indolent myeloma, with no significant anti-tumour effect. Br J Haematol. 2002;118(1):239–242. [DOI] [PubMed] [Google Scholar]
- 63. Musto P, Petrucci MT, Bringhen S, et al. A multicenter, randomized clinical trial comparing zoledronic acid versus observation in patients with asymptomatic myeloma. Cancer. 2008;113(7):1588–1595. [DOI] [PubMed] [Google Scholar]
- 64. Rajkumar SV, Gertz MA, Lacy MQ, et al. Thalidomide as initial therapy for early-stage myeloma. Leukemia. 2003;17(4):775–779. [DOI] [PubMed] [Google Scholar]
- 65. Riccardi A, Mora O, Tinelli C, et al. Long-term survival of stage I multiple myeloma given chemotherapy just after diagnosis or at progression of the disease: a multicentre randomized study. Cooperative Group of Study and Treatment of Multiple Myeloma. Br J Cancer. 2000;82(7):1254–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Weber D, Rankin K, Gavino M, Delasalle K, Alexanian R. Thalidomide alone or with dexamethasone for previously untreated multiple myeloma. J Clin Oncol. 2003;21(1):16–19. [DOI] [PubMed] [Google Scholar]
- 67. Mateos MV, Hernandez MT, Giraldo P, et al. Lenalidomide plus dexamethasone versus observation in patients with high-risk smouldering multiple myeloma (QuiRedex): long-term follow-up of a randomised, controlled, phase 3 trial. Lancet Oncol. 2016;17(8):1127–1136. [DOI] [PubMed] [Google Scholar]
- 68. Mateos MV, Hernandez MT, Giraldo P, et al. Lenalidomide plus dexamethasone for high-risk smoldering multiple myeloma. N Engl J Med. 2013;369(5):438–447. [DOI] [PubMed] [Google Scholar]
- 69. Korde N, Roschewski M, Zingone A, et al. Treatment With Carfilzomib-Lenalidomide-Dexamethasone With Lenalidomide Extension in Patients With Smoldering or Newly Diagnosed Multiple Myeloma. JAMA Oncol. 2015;1(6):746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kumar SK, Rajkumar SV, Dispenzieri A, et al. Improved survival in multiple myeloma and the impact of novel therapies. Blood. 2008;111(5):2516–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Landgren O, Hofmann JN, McShane CM, et al. Association of Immune marker Changes With Progression of Monoclonal Gammopathy of Undetermined Significance to Multiple Myeloma. JAMA Oncol 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.