Abstract
Background
People with human immunodeficiency virus (PWH) demonstrate increased atherosclerotic cardiovascular disease (ASCVD). Statins are being studied to prevent ASCVD in human immunodeficiency virus (HIV), but little is known regarding the effects of statins on a broad range of inflammatory and cardiovascular proteins in this population.
Methods
We used a highly specific discovery proteomic approach (Protein Extension Assay), to determine statin effects on over 350 plasma proteins in relevant ASCVD pathways among HIV and non-HIV groups. Responses to pitavastatin calcium were assessed in 89 PWH in the INTREPID trial and 46 non-HIV participants with features of central adiposity and insulin resistance. History of cardiovascular disease was exclusionary for both studies.
Results
Among participants with HIV, PCOLCE (enzymatic cleavage of type I procollagen) significantly increased after pitavastatin therapy and PLA2G7 (systemic marker of arterial inflammation) decreased. Among participants without HIV, integrin subunit alpha M (integrin adhesive function) and defensin alpha-1 (neutrophil function) increased after pitavastatin therapy and PLA2G7 decreased. At baseline, comparing participants with and without HIV, differentially expressed proteins included proteins involved in platelet and endothelial function and immune activation.
Conclusions
Pitavastatin affected proteins important to platelet and endothelial function and immune activation, and effects differed to a degree within PWH and participants without HIV.
Keywords: atherosclerosis, cardiovascular disease, HIV, inflammation
We assessed the effects of pitavastatin on a broad array of proteins using a highly specific proximity extension assay (PEA) in HIV and metabolic syndrome non-HIV patients. Our results highlight effects on key inflammatory, collagen, neutrophil, and integrin functions.
Atherosclerotic cardiovascular disease (ASCVD) rates are increased in human immunodeficiency virus (HIV) [1]. Although increased rates are thought to relate in part to increased inflammation and immune activation, the specific mechanisms are poorly understood. Moreover, as prevention strategies are considered, assessing the underlying biological pathways contributing to increased ASCVD, and statin effects on these pathways in HIV, remains a critical goal for the field.
Statins are now being studied as a potential primary prevention strategy for ASCVD in people with HIV (PWH) due to their potential pleotropic effects on multiple inflammatory, metabolic, and immune pathways in addition to their known effects on low-density lipoprotein cholesterol (LDL-C). However, no prior study has compared the effects of statins on a broad range of proteins in PWH versus higher risk asymptomatic non-HIV groups, to provide insight into the unique effects of statins in PWH compared with the general population across cardiometabolic, cardiovascular, and inflammatory pathways.
Recent innovations to traditional enzyme-linked immunosorbent assay (ELISA)-based protein assays provide analytical sensitivity and specificity to efficiently detect hundreds of low-abundance proteins in “targeted discovery” with precision to accurately quantify longitudinal changes [2]. Utilizing these techniques, we compared baseline protein patterns as well as longitudinal effects of statin therapy in PWH and among non-HIV-infected participants, with metabolic abnormalities common in PWH, assessing effects on more than 350 proteins with potential roles in cardiovascular, cardiometabolic, and inflammatory disease.
MATERIAL AND METHODS
Participants With Human Immunodeficiency Virus
People with HIV were enrolled in the INTREPID (HIV-infected patieNts and TREatment with Pitavastatin vs pravastatin for Dyslipidemia) Trial (ClinicalTrials.gov: NCT 01301066) [3], which compared the effects of moderate-dose statins—pitavastatin 4 mg daily versus pravastatin 40 mg daily on LDL-C—among dyslipidemic PWH over 52 weeks. Participants with coronary heart disease (CHD) or CHD risk equivalent were excluded. Lipid eligibility was based on an LDL-C ≥130 and <220 mg/dL and triglycerides <400 mg/dL. Institutional review board (IRB) approval was obtained at each participating site, and participants provided written informed consent [3]. Comparison of effects on a limited set of proteins was previously performed in this trial cohort [4].
Participants Without Human Immunodeficiency Virus
Participants without HIV were enrolled in a separate, previously described, randomized, double-blind, placebo-controlled clinical trial (ClinicalTrials.gov: NCT 02290106) [5]. Participants were eligible based on LDL-C ≥100 and <190 mg/dL or 10-year ASCVD risk ≥5% as well as the presence of central obesity and insulin resistance, features prevalent among PWH as well [5]. Participants with known ASCVD, triglycerides ≥500 mg/dL, or diabetes were excluded. Participants were randomized to pitavastatin 4 mg versus placebo once daily for 6 months. This study was approved by the Partners IRB, and participants provided written informed consent.
Study Design
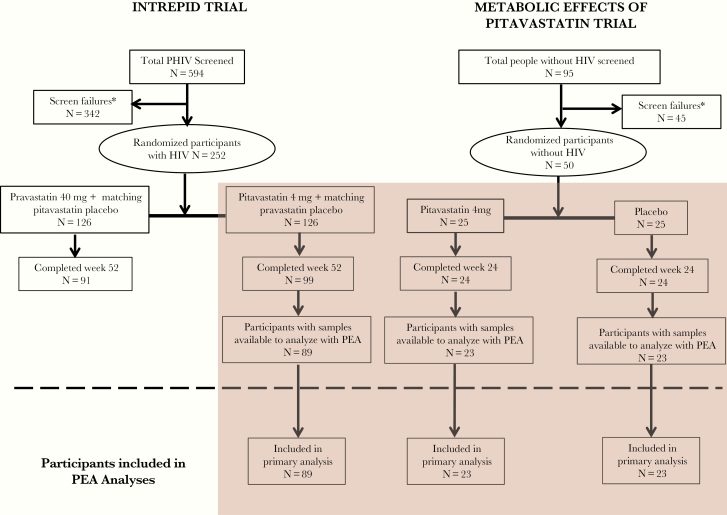
First, we compared the longitudinal effects of pitavastatin 4 mg/day among PWH (N = 89) and participants without HIV (N = 23) (Figure 1). The 89 participants were the group that completed the study to Week 52 and had samples to analyze by Proximity Extension Assay (PEA). In comparison, this represented a subset of participants included in a prior publication [4] for whom paired data were available (N = 111) and which included several participants that did not complete the study to Week 52. Pitavastatin was studied because it has no effect on glucose, effectively decreases LDL-C in patients with and without HIV [3, 5], has minimal interactions with antiretroviral therapy (ART) [6], and is metabolized via glucuronidation not cytochrome P450. Moreover, prior work has shown that effects of pitavastatin on key indices of inflammation are greater with pitavastatin than pravastatin in PWH [4, 7]. Second, we compared baseline differences in these same proteins before statin therapy in the HIV and non-HIV groups. Longitudinal data were assessed in placebo-treated participants without HIV (N = 23) as a natural history control group to assess stability in assay assessment over time. We did not assess protein responses to pravastatin in the INTREPID participants to standardize results and allow comparison between groups receiving pitavastatin. Atherosclerotic cardiovascular disease risk score was calculated identically for both studies based on the guidelines in place at the time of INTREPID (updated ATP III).
Figure 1.
Consort diagram. Participants from 2 randomized double-blind clinical trials were eligible for this analysis. Participants with human immunodeficiency virus (HIV) from the INTREPID (HIV-infected patieNts and TREatment with Pitavastatin vs Pravastatin for Dyslipidemia) Trial were included in the primary analysis if they completed to Week 52 of the study and had received pitavastatin as part of the intervention for the trial. Participants from the Metabolic Effects of Pitavastatin Trial were included in the primary analysis if they completed to Week 24 and received either pitavastatin or a placebo as part of the intervention for the trial. PEA, proximity extension assay; PHIV, people with HIV.
Study Procedures
Analyses were performed on EDTA plasma using PEA technology (Olink Proteomics, Watertown, MA; https://www.olink.com/resources-support/document-download-center/) [4, 8]. We chose 4 panels: the cardiometabolic, cardiovascular II and III, and the inflammation panels. Using oligonucleotide-labeled antibody probe pairs, PEA permits simultaneous assessment of multiple proteins, maintaining the precision of dual epitope ELISA without the loss of specificity of earlier generation multiplex assays. Signal generation requires both recognition of DNA barcoding from sequence-specific oligonucleotides and dual recognition of correctly matched antibody pairs. The PEA assay reports fold change in log2 (Normalized Protein eXpression [NPX]) units. Using these panels, the levels of 358 unique proteins were measured, with >99% passing quality control requirements (Supplemental Methods).
Statistical Analysis
The primary endpoint was the change in protein markers with pitavastatin therapy among participants with HIV and without HIV. Secondary endpoints included the difference in baseline levels of protein biomarkers by HIV status.
The changes in protein levels before and after statin therapy were determined using repeated measures longitudinal mixed-effects model analysis of variance with fixed treatment and time effects and random subject intercepts. T-statistics were determined for the mixed effects model-based linear contrast of intrasubject longitudinal changes. We controlled for false discovery rate (FDR) using the Benjamini-Hochberg method [9]. False discovery rate P < .05 were considered significant. The detectable standardized statistical differences (ie, Cohen’s d) were calculated at a 5% FDR rate with 80% power for 2-sided paired samples t test under 1%, 5%, and 10%, and 20% assumptions of the truly different proportion among 368 simultaneously evaluated proteins were d = 0.48, 0.43, 0.40, and 0.37 for the analysis of 89 PWH, and d = 0.69, 0.61, 0.57, and 0.52 for the analysis of 46 participants without HIV, respectively. The observed effect sizes were larger than the calculated prestudy target effect sizes.
Baseline characteristics are presented as mean ± standard deviation and compared using an independent samples t test for continues variables and χ 2 test for categorical variables. Baseline differences in mean protein levels between participants with and without HIV were examined using independent sample t tests. T-statistics for differences between groups for each protein were calculated. False discovery rate-adjusted P values were calculated accounting for the number of proteins examined and grouping into panels.
Principal component analysis (PCA) was conducted on the subset of significantly differentially expressed proteins between participants with and without HIV across all 135 baseline samples to determine potential biological clustering. Proteins were clustered by their expression patterns across all participants using the fuzzy C-means algorithm in R by utilizing the fcm function with default parameter setting and desired cluster number. The number of clusters n = 5 was selected based on the visual inspection of the clustering results. Network analyses, performed using the GeneMANIA prediction tool, were used to infer functional connections between proteins and visualize these connections based on coexpression, physical interactions, genetic interactions, colocalization, predicted, shared protein domains, and pathways. This was performed to delineate relationships among the proteins in one of the clusters identified in our clustering analysis and also to delineate potential relationships with other proteins not assessed (see Supplemental Methods).
Analyses examining the effect of pitavastatin on protein levels and comparing baseline protein levels by HIV status were controlled for age, body mass index (BMI), and ASCVD risk scores. Sensitivity analyses of longitudinal and baseline data controlling for glucose and triglyceride levels in addition to age, BMI, and ASCVD risk scores were also performed. We performed tests for interaction to determine whether use of specific ART classes modified baseline protein levels, and the effect of statins on proteins assessed. Sensitivity analyses were performed limiting PWH to lipid and ASCVD criteria, LDL-C ≥100 and <190 mg/dL and 10-year ASCVD risk ≥5%, that were similar to the non-HIV group.
Statistical analyses for longitudinal and cross-sectional analyses were performed using SAS software (version 9.5; SAS Institute) and SAS JMP software (version 12.0; SAS Institute). Principal component and cluster analyses were performed using R software (version 3.5.1).
RESULTS
Baseline Demographics
Baseline demographics, risk parameters, and ART use are shown in Table 1. Relatively large differences were seen in BMI and other factors based on the inclusion criteria of each study. Overall, Framingham risk scores were generally similar between groups, 6.8% (HIV) and 8.4% (non-HIV), respectively. Log viral load was 1.1 ± 0.2 copies/mL and CD4 count was 682 ± 264 cells/mm3.
Table 1.
Baseline Demographicsa
| Characteristic | Participants With HIV N = 89 | Participants Without HIV N = 46 |
|---|---|---|
| Sex (%male) | 88 (78/89) | 100 (46/46) |
| Race | ||
| White (%) | 85 (76/89) | 89 (41/46) |
| Black (%) | 14 (12/89) | 7 (3/46) |
| Other (%) | 1 (1/89) | 4 (2/46) |
| BMI (kg/m2) | 27.5 ± 3.9 | 35.9 ± 5.6 |
| LDL-C (mg/dL) | 158 ± 27 | 120 ± 20 |
| Triglycerides (mg/dL) | 176 ± 99 | 143 ± 65 |
| Fasting glucose (mg/dL) | 95 ± 10 | 97 ± 9 |
| Hemoglobin A1c (%) | 5.4 ± 0.4 | 5.7 ± 0.3 |
| Framingham risk score (%) | 6.7 ± 5.1 | 8.4 ± 4.0 |
| Age (years) | 50.1 ± 7.5 | 53.0 ± 6.6 |
| Current smokingb (%) | 21 (18/84) | 15 (7/46) |
| Total cholesterol (mg/dL) | 241 ± 31 | 190 ± 24 |
| HDL-C (mg/dL) | 50 ± 14 | 42 ± 8 |
| Systolic BP (mmHg) | 123 ± 13 | 135 ± 16 |
| Current antihypertensive (%) | 24 (21/89) | 28 (13/46) |
| Log HIV-1 viral load (copies/mL) | 1.1 ± 0.2 | — |
| CD4 count (cells/mm3) | 682 ± 264 | — |
| Protease inhibitor-based therapy (%) | 25/89 (28) | — |
| NNRTI-based therapy (%) | 47/89 (53) | — |
| Integrase inhibitor-based therapy (%) | 3/89 (3) | — |
| Protease inhibitor and NNRTI (%) | 7/89 (8) | — |
| Integrase inhibitor and NNRTI (%) | 6/89 (7) | |
| NRTI only (%) | 1/89 (1) | — |
Abbreviations: BMI, body mass index; BP, blood pressure; CD4, cluster of differentiation 4; HDL-C, high-density lipoprotein cholesterol; HIV, human immunodeficiency virus; LDL-C, low-density lipoprotein cholesterol; NNRTI, nonnucleoside reverse-transcriptase inhibitor; NRTI, nucleoside reverse-transcriptase inhibitor.
aNormally distributed data are reported as mean ± standard deviation.
bSmoking status was not reported for 5 participants with HIV.
Baseline Differences in Protein Levels Among Participants With and Without Human Immunodeficiency Virus
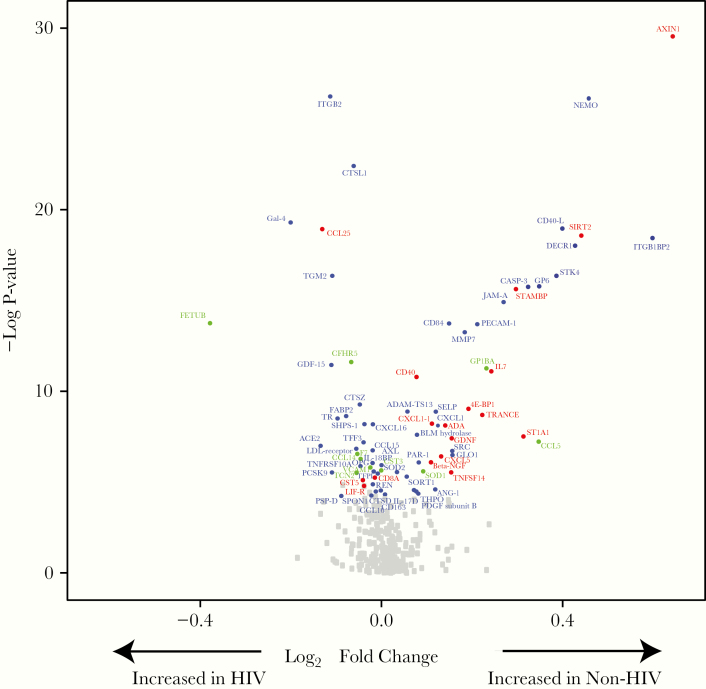
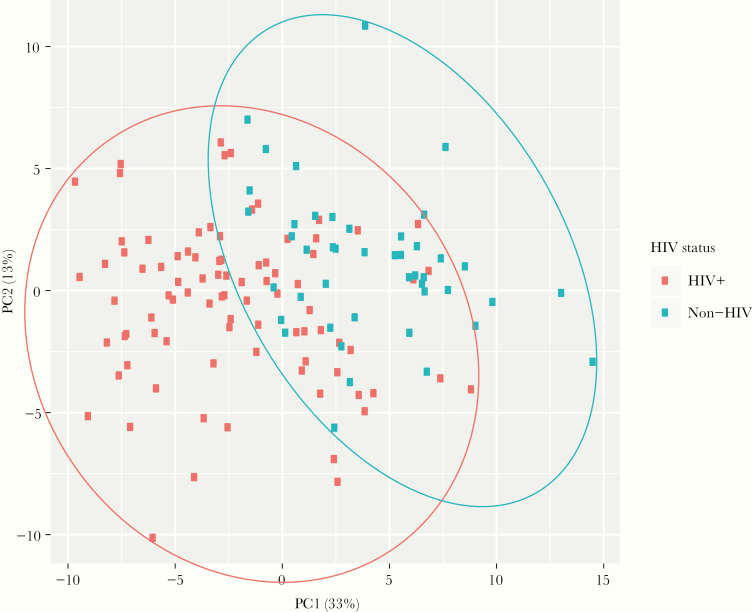
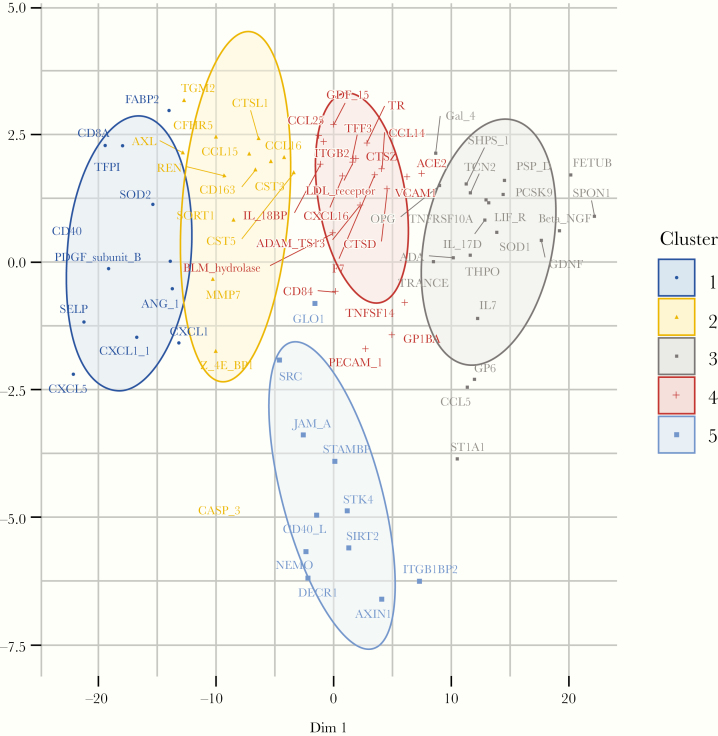
A significant number of differentially expressed proteins are shown in the volcano plot in Figure 2. Fold differences, shown in log2 units, between PWH and non-HIV participants were generally modest, up to 54% (0.625 log2 fold difference) for AXIN1 (Axin-1, negative regulator of WNT signaling pathway). Of note, relatively more proteins were differentially upregulated in the non-HIV group compared with the HIV group, but for some of these proteins, upregulation is consistent with lower risk of CVD, for example, SIRT2 (SIR-2 like protein 2, deacetylase involved in macrophage polarization), and STAMBP (STAM-Binding Protein, zinc metalloproteinase involved in endosomal deubiqitinization). Principal component analysis revealed relatively distinct clustering by HIV status (Figure 3). Fuzzy C-means clustering demonstrated one cluster distinctly separated from the others (Figure 4). Eight of 11 proteins in this distinct cluster also were the proteins that accounted for most of the variance between groups in our PCA: SIRT2, STAMBP, AX1N1, ITGB1BP2 (integrin subunit beta-1 binding protein 2, chaperone protein), NEMO (nuclear factor [NF]-кB essential modulator, activates NF-кB), CD40-L (CD40 ligand, tumor necrosis factor [TNF]-related activation protein, may activate matrix metalloproteinases and coagulation cascade), JAM-A (junctional adhesion molecule A, regulates tight junctions in multiple tissues), and DECR1 (2,4-dienoyl-CoA reductase 1, beta oxidation of unsaturated fatty CoA esters) (see Supplemental Table 1A for further information on protein functions). Additional proteins in this cluster were GLO1 (lactoylglutathione lyase, involved in regulation of TNF-induced transcriptional activating of NF-кB), STK4 (serine/threonine-kinase 4, stress-activated proapoptotic kinase), and SRC (proto-oncogene tyrosine-protein kinase SRC, nonreceptor protein tyrosine kinase). Network analyses revealed a distinct network of relationships, which include GLO1, STK4, and SRC in addition to the other proteins identified in our PCA and cluster analyses: SIRT2, AX1N1, ITGB1BP2, JAM-A, and DECR1. Through our network analyses, we also identified an additional 18 proteins that were not measured by our PEA panels but were found to be related to the proteins assessed (Supplemental Figure 1).
Figure 2.
Differential expression of proteins between participants with and without human immunodeficiency virus (HIV) at baseline. A volcano plot demonstrates differential protein expression in HIV-infected vs non-HIV-infected participants. Fold change refers to the difference in the mean Normalized Protein eXpression ([NPX] log2) units between the HIV (N = 89 participants) and non-HIV (N = 46 participants) groups. Proteins with significantly different expression at baseline between HIV and non-HIV groups are shown in color and named. A false discovery rate P < .05 was considered statistically significant. Proteins are colored based on their respective panel using the following classifications: cardiometabolic panel proteins in green, cardiovascular panels II and III proteins in blue, and inflammation panel proteins in red. Gray squares represent other proteins assessed through proximity extension assay (PEA) technology, which was not significantly different at baseline between participants in the HIV and non-HIV groups. A total of 368 proteins were assessed. A limited number of proteins (n = 10) were included in more than 1 panel. All data are presented and analyzed. Substantive differences between measurements of duplicated proteins were generally not seen at baseline, and no duplicated proteins were significant in the longitudinal analyses. See Supplemental Methods for abbreviations.
Figure 3.
Principal component analysis. Plot demonstrating a principal component analysis, including the 2 principal components (principal component 1 [PC1] on the x-axis and principal component 2 [PC2] on the y-axis), which explain the most variance across the 78 proteins that were differentially expressed in human immunodeficiency virus (HIV)-infected and non-HIV-infected participants. Principal component 1 explained 33% of the variance, whereas PC2 explained 13% of the variance. Ellipses were drawn to contain all points from each disease category to highlight their different distributions in this space. Participants with HIV are demonstrated using red dots and are contained within a red ellipse; participants without HIV are demonstrated using blue dots and are contained within a blue ellipse. Although there is some overlap in the participants with and without HIV, the majority of participants with HIV are not in overlapping regions of the principal component plot.
Figure 4.
Clustering analysis. Fuzzy C-means clustering demonstrating 5 clusters of proteins differentially expressed between participants with and without human immunodeficiency virus (HIV) at baseline. Cluster 5 is located below all of the other clusters and comprises the following proteins: GLO1, SRC, JAM-A, STAMBP, STK4, CD40-L, SIRT2, NEMO, DECR1, ITGB1BP2, and AXIN1. See Supplemental Methods for abbreviations.
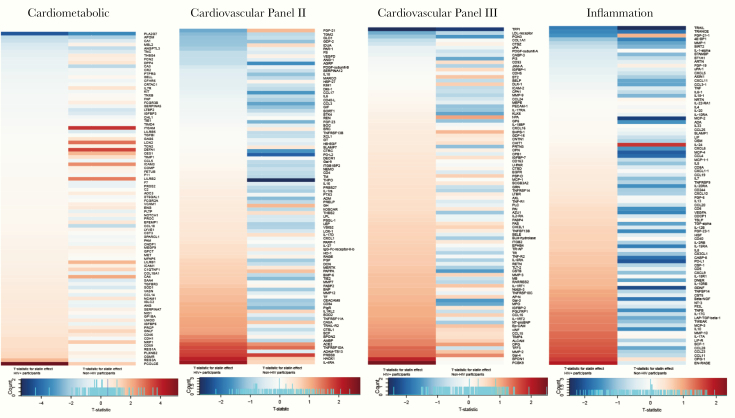
Change in Protein Levels in Response to Pitavastatin Therapy
Differences in the magnitude and directionality of change with statin therapy among participants with and without HIV are demonstrated in panel-specific heat maps (Figure 5). The directionality of change with pitavastatin differed for many of the proteins among the 2 groups, particularly for the inflammation panel. In general, greater effects of pitavastatin to reduce a number of inflammatory and other proteins were seen among the non-HIV group, as seen by the greater relative abundance of blue (decreased) on the heat maps in the non-HIV columns versus red (increased) in the non-HIV columns for each panel. As a caveat, beneficial directionality cannot be universally linked to up- or downregulation for all proteins: see Supplemental Table 1 for implication of directionality of key proteins changing with pitavastatin and also for those differentially expressed at baseline. These results were recapitulated in heat maps using fold change data rather than data from t-statistics (Supplemental Figure 2).
Figure 5.
Differential expression of proteins after pitavastatin therapy among participants with and without human immunodeficiency virus (HIV). Heat maps are shown for each protein biomarker panel (from left to right: Cardiometabolic, Cardiovascular II, Cardiovascular III, and Inflammation). Within each heat map, the t-statistic for statin treatment effect is demonstrated on the left for participants with HIV (N = 89) and on the right for participants without HIV (N = 23). The protein order of each heat map was determined by order of the t-statistic for the participants with HIV from most negative to most positive for each panel. Blue indicates a decrease in the protein level after pitavastatin therapy, and the red indicates an increase in the protein level after pitavastatin therapy. Among the participants with HIV, 2 proteins changed significantly with pitavastatin therapy, PCOLCE and PLA2G7. Among participants without HIV, 3 proteins changed significantly with pitavastatin therapy PLA2G7, ITGAM, and DEFA1. See Supplemental Methods for abbreviations.
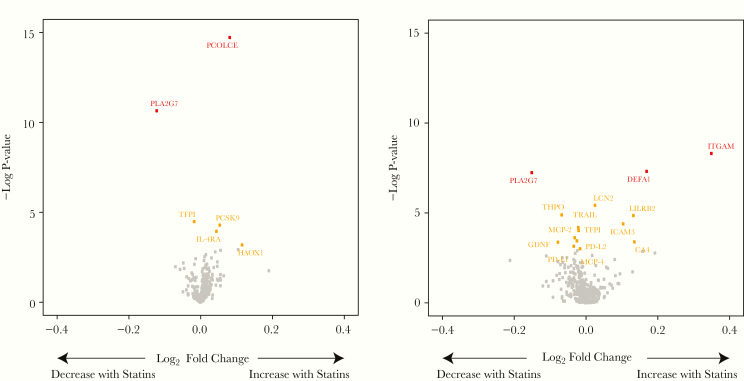
The mean change in LDL-C was similar in each study, −49 mg/dL for HIV and non-HIV. Changes with statin therapy among participants with and without HIV are shown in volcano plots (Figure 6). Among PWH, PCOLCE (procollagen C-endopeptidase enhancer, drives enzymatic cleavage of type 1 procollagen and heightens C-proteinase activity) significantly increased with pitavastatin. PLA2G7 (phospholipase A2 group VII, degrades platelet-activating factor [PAF] and hydrolyzes oxidized LDL-C to release proinflammatory molecules), also known as Lp-PLA2, significantly decreased (see Supplemental Table 1B for protein functions). For several other proteins of interest, including TFPI (tissue factor pathway inhibitor, inhibitor of tissue factor-mediated coagulation), PCSK9 (proprotein convertase subtilisin/kexin type 9, enzyme that plays key role in cholesterol homeostasis), and IL-4RA (interleukin-4 receptor subunit alpha, plays a role in alternative activation of macrophages), the change was significant based on nominal P values.
Figure 6.
Change in proteins after pitavastatin therapy among participants with and without HIV. Volcano plots for the fold change in proteins with pitavastatin for participants with HIV (N = 89) is shown on the left and for participants without HIV (N = 23) on the right. Fold change refers to change in Normalized Protein eXpression ([NPX] (log2) units. Proteins that significantly changed with pitavastatin therapy with a false discovery rate P < .05 are shown in red and with a nominal P < .05 shown in yellow. See Supplemental Methods for abbreviations.
Among participants without HIV, ITGAM (integrin subunit alpha M, plays a role in adhesive interactions of monocytes and macrophages) and DEFA1 (neutrophil defensin 1, plays a role in phagocyte-mediated host defense) significantly increased with pitavastatin, whereas PLA2G7 significantly decreased after pitavastatin therapy. The magnitude of change in PLA2G7, an approximate 15% reduction, was similar in both the HIV and non-HIV groups. Overall, for proteins changing significantly with pitavastatin among the HIV and non-HIV groups, the relative fold change in these proteins was relatively modest, varying from approximately −15% for PLA2G7 to up to approximately 30% for ITGAM. Among participants without HIV randomized to placebo, no proteins changed significantly, demonstrating assay stability in repeated measurements.
Sensitivity Analyses
At baseline, minimal association was seen between ART class and assessed proteins, and there was no interaction in longitudinal analyses between ART and pitavastatin for any proteins using FDR-corrected interaction terms (Supplemental Table 2). In the sensitivity analyses limiting subjects to similar entry criteria used for the non-HIV cohort (Supplemental Table 3), the differences in baseline proteins were similar to the full cohorts; again, the most significant pitavastatin effects were seen on PCOLCE and PLA2G7 (Supplemental Figures 3–4). Results of the longitudinal changes in each group were recapitulated controlling for baseline age, BMI, ASCVD score, glucose, and triglyceride in each group.
Discussion
In this study, we used a novel proteomics targeted discovery approach, based on a highly precise technique, to identify longitudinal effects of statin therapy across a broad range of relevant proteins in participants with and without HIV. We also compared relative expression of these proteins at baseline. The pathways assessed to be different at baseline are critical to atherogenesis, including platelet adhesion, activation and procoagulant activity, endothelial function and permeability, and immune activation. It is notable that, in longitudinal analyses, pitavastatin significantly reduced a key protein involved in platelet activation, arterial inflammation, and LDL oxidation in each group, but it had differential effects on collagen pathways (significant in HIV) and integrin adhesive functions as well as neutrophil function (significant in non-HIV).
Among the proteins differentially regulated between participants with HIV and without HIV, several have notable functions related to atherogenesis. JAM-A was significantly lower among our participants with HIV. JAM-A is a critical regulator of tight junction assembly, endothelial permeability, and leukocyte extravasation [10]. In vitro experiments utilizing HIV-infected monocytes have demonstrated decreased endothelial expression of JAM-A, which was associated with increased endothelial permeability and monocyte transmigration.
CD40-L, a protein that affects platelet-platelet and leukocyte-endothelium interactions [11], was significantly lower among participants with HIV. The interaction of CD40-L on the surface of T cells with CD40 results in activation of the coagulation cascade, release of proinflammatory mediators, and increased activity of matrix metalloproteinases. Complete inhibition of CD40-L signaling results in a decrease in atherosclerosis and promotes the development of stable (versus nonstable) plaque [12–14]. Soluble CD40-L (sCD40-L), in turn, is a marker of platelet activation, and levels are predictive of ASCVD and ASCVD risk factors [15, 16]. In chronic HIV, the accessory protein gp120, detectable among PWH despite ART [17], inhibits the upregulation of CD40-L [18]. Thus, our findings of lower sCD40-L among our participants with HIV may be related to the effects of HIV accessory proteins on the expression of CD40-L. However, the relationship of lower CD40-L to atherogenesis is unclear.
Our longitudinal analyses showed a distinct pattern of protein changes in participants with and without HIV, particularly true for the inflammation panel. These data may reflect differential pitavastatin effects, with potentially greater effects to lower inflammation in non-HIV, perhaps due to resistance in PWH, an altered natural history of changes in inflammatory and other relevant ASCVD-related proteins that may increase more over time in PWH, despite statin therapy, or effects of baseline differences in the study populations. Of note, we specifically chose non-HIV patients with metabolic syndrome characteristics, to make the comparison more meaningful than versus a group of healthy non-HIV patients. Nonetheless, key differences in baseline characteristics might have contributed to our results, but these results were supported by sensitivity analyses, limiting analysis to identical inclusion criteria and controlling for key baseline parameters. Moreover, this distinct pattern of protein changes among the HIV and non-HIV groups occurred in the context of a similar effect of pitavastatin on LDL-C in both cohorts, among participants with and without HIV, suggesting that our results are not due to differences in the relative potency of pitavastatin among the groups.
Using stringent FDR analyses, we demonstrated 5 novel proteins (PCOLCE and PLA2G7 among participants with HIV and PLA2G7, ITGAM, and DEFA1 among participants without HIV) changing with statin therapy that may offer beneficial pleotropic effects via reducing atherogenic pathways. The magnitude of change in these proteins, between 15% and 30%, was generally consistent with the known effects of this moderate dose statin to reduce LDL in this study (−30%). PCOLCE, which significantly increased with pitavastatin among participants with HIV but not among participants without HIV, drives the enzymatic cleavage of type 1 procollagen and promotes C-proteinase activity [19]. C-proteinase is relevant to atherogenesis given that it cleaves type I and III collagen [20]—the predominant forms of collagen in the vascular extracellular matrix. Atorvastatin also significantly increases PCOLCE among participants with HIV, and this increase in PCOLCE related to a decrease in total coronary plaque [8]. Alterations in PCOLCE with statin therapy may have downstream effects on collagen processing that may affect plaque stability. Given that plaque vulnerability features are increased among PWH [21], significant changes in a key protein affecting collagen processing, such as PCOLCE, may have significant implications in plaque vulnerability among PWH.
A marker of systemic arterial inflammation, PLA2G7 (known as Lp-PLA2) [22], significantly decreased with pitavastatin among both participants with and without HIV. PLA2G7 is an enzyme that catalyzes the degradation of PAF [23]—a unique phospholipid that can activate platelets, increase vascular permeability, and hydrolyze oxidized LDL-C resulting in the release of proinflammatory and pro-oxidative molecules. [22] PLA2G7 is predictive of ASCVD outcomes [22]. Decreases in PLA2G7 levels relate to decreases in coronary plaque in PWH [24], a population with increased evidence of arterial inflammation [25].
The results in this study aimed to compare responses in HIV versus non-HIV in contrast to a prior paper, which aimed to compare pitavastatin to pravastatin [4]. This prior paper, which demonstrated relatively larger effects of pitavastatin than pravastatin on multiple inflammatory markers, along with other work [7], informed the focus of the current analysis to compare effects of pitavastatin in HIV and non-HIV among an even broader array of proteins. Although the studies cannot be formally compared due to different sample sizes (n = 111 vs 89), the results in the subset of overlapping patients again showed the largest relative reduction in TFPI, PON3, and the LDL receptor protein, among the proteins in the CVD III panel, providing assurance of consistency between the studies. With respect to our study assessing the effects of atorvastatin among PHIV, our results here overlapped with, but were not entirely complimentary to the prior study, and showed the largest relative effects on TFPI [8]. Therefore, individual statins have differential effects in HIV, but some effects, particularly to reduce TFPI, shown to be increased in PWH [26] are more uniform across multiple statins in PWH. This is relevant because activated monocytes may produce increased tissue factor levels, which may stimulate thrombosis, suggesting that reduced levels in response to statins may be of benefit.
The current study has several strengths but some limitations. We used a broad but highly precise proteomics approach to permit serial assessment of change in multiple proteins over time within HIV and non-HIV groups as the primary focus of the study. Our studies compared participants with and without HIV with generally similar estimated ASCVD risk but recruited for different studies, of long but differing durations. Prior studies have shown that changes in key inflammatory proteins due to statin therapy generally occur within 1 month and remain steady over 2 years on statin therapy [27], well within the time frame of each study (6–12 months). Moreover, each study was sufficiently long enough to result in similar changes in LDL-C levels and changes in key proteins in each group, eg, PLA2G7. Non-HIV participants were specifically chosen from a study enrolling participants based on parameters of metabolic dysregulation common in HIV patients. We controlled for key metabolic and anthropometric variables in our comparison of HIV and non-HIV subjects and recapitulated our results in sensitivity analyses limited to participants meeting similar entry criteria. More important, in both studies, participants with known ASCVD were excluded. Although we compared the same statin, pitavastatin, other statins may have differential effects and should be tested. Additional studies with more women are needed. These studies were performed in North America, and inflammatory pathways may relate to regional and genetic differences. Finally, care must be used not to equate directionality of protein changes seen in this study with biological effects, because unique proteins may regulate inflammation and cardiovascular disease differently, such that up- or downregulation may be beneficial. Careful understanding of the biology of the proteins changing in this study can provide this information.
Conclusions
This study provides novel information and serves as a road map for future investigation of the unique biology and pleotropic, but differing, effects of statin therapy in participants with and without HIV. The study demonstrates some key effects of pitavastatin that are potentially beneficial. The study also highlights many proteins that do not change significantly with pitavastatin treatment. Validation and further exploration of these pathways and statin effects in events driven trials, such as REPRIEVE, will be important.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the participants in INTREPID Trial and the Metabolic Effects of Pitavastatin Trial. We also thank the study staff and the nursing staff for both studies.
Author contributions. C. D. and M. T. contributed to the study design, data collection, data analysis, data interpretation, figure and table preparation, reference search, and writing and editing the manuscript; K. V. F. contributed to data collection and editing the manuscript; L. P. W. and R. S. contributed to data analysis, data interpretation, figure preparation, and writing and editing the manuscript; I. G. contributed to data collection, data analysis, data interpretation, and editing the manuscript; M. V. Z. contributed to data interpretation and writing and editing the manuscript; J. L. contributed to data interpretation and editing the manuscript; C. A. S. contributed to study design, writing, and editing the manuscript; E. S. contributed to data collection, data analysis, data interpretation, figure preparation, and writing and editing the manuscript; N. R. contributed to data collection, data analysis, data interpretation, and editing the manuscript; M. A. T. contributed to parent study design and served as an investigator and was involved in data interpretation and writing and editing the manuscript; D. C. contributed to data collection, data analysis, figure and table preparation, and editing the manuscript; J. A. A. contributed to parent study design and served as an investigator and was involved in data interpretation, writing, and editing the manuscript; L. R. B. contributed to parent study design, data collection, data analysis, and editing the manuscript; T. L. S. contributed to parent study design, data collection, data analysis, and writing and editing the manuscript; H. L. contributed to data analysis, data interpretation, reference search, figure and table preparation, and writing and editing the manuscript; S. K. G. contributed to study design, data collection, data analysis, data interpretation, reference search, figure and table preparation, and writing and editing the manuscript.
Disclaimer. The funding organizations had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Financial support. Performance of the Olink proteomic analyses was supported through funding to C. D. from the Inova Heart and Vascular Institute. This research was also funded by the National Institutes of Health (P30 DK040561; Nutrition Obesity Research Center at Harvard and M01RR01066, 1UL1RR025758, and 8UL1TR000170 to the Harvard Clinical and Translational Science Center). M. T. was supported by the National Institutes of Health (5KL2TR001100-05; Harvard Catalyst KL2 Grant).
Potential conflicts of interest. I. G., E. S., and N. R. are employees of Olink Proteomics. K. V. F. has been supported by an educational grant from Gilead Sciences, Inc., unrelated to this project. M. V. Z. has received investigator-initiated research grant support from Gilead Sciences to her institution (Massachusetts General Hospital), unrelated to this project. J. L. has served as a consultant to Viiv Healthcare and Gilead Sciences, unrelated to this project. C. A. S. is an employee of Kowa Pharmaceuticals. M. A. T., through AIDS Research Consortium of Atlanta, received research support from Kowa Pharmaceuticals for the INTREPID trial and for clinical trials from Bristol Myers Squibb, Cepheid, Inc., Cytodyn, Inc., Glaxo Smith Kline, Gilead Sciences, Merck Sharp Dohme, and ViiV Healthcare, unrelated to this project. J. A. A. receives research support from Frontier Technology, Gilead Sciences, Glaxo-Smith-Kline, and Viiv Healthcare, serves on advisory boards for Gilead Sciences, Glaxo-Smith-Kline Janssen, Merck, Theratechnologies, and Viiv Healthcare, and has served as a consultant for Medicure. All unrelated to this project. T. L. S. has received investigator-initiated grant support from Novo Nordisk., Inc., unrelated to this project. S. K. G. has received grant support from Gilead, Viiv Healthcare, and KOWA Pharmaceuticals and served as a consultant to Theratechnologies and Viiv Healthcare, unrelated to this project. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Freiberg MS, Chang CC, Kuller LH, et al. HIV infection and the risk of acute myocardial infarction. JAMA Intern Med 2013; 173:614–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lam MP, Ping P, Murphy E. Proteomics research in cardiovascular medicine and biomarker discovery. J Am Coll Cardiol 2016; 68:2819–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aberg JA, Sponseller CA, Ward DJ, Kryzhanovski VA, Campbell SE, Thompson MA. Pitavastatin versus pravastatin in adults with HIV-1 infection and dyslipidaemia (INTREPID): 12 week and 52 week results of a phase 4, multicentre, randomised, double-blind, superiority trial. Lancet HIV 2017; 4:e284–94. [DOI] [PubMed] [Google Scholar]
- 4. Toribio M, Fitch KV, Stone L, et al. Assessing statin effects on cardiovascular pathways in HIV using a novel proteomics approach: analysis of data from INTREPID, a randomized controlled trial. EBioMedicine 2018; 35:58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braun LR, Feldpausch MN, Czerwonka N, et al. Effects of pitavastatin on insulin sensitivity and liver fat: a randomized clinical trial. J Clin Endocrinol Metab 2018; 103:4176–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chauvin B, Drouot S, Barrail-Tran A, Taburet AM. Drug-drug interactions between HMG-CoA reductase inhibitors (statins) and antiviral protease inhibitors. Clin Pharmacokinet 2013; 52:815–31. [DOI] [PubMed] [Google Scholar]
- 7. Toribio M, Fitch KV, Sanchez L, et al. Effects of pitavastatin and pravastatin on markers of immune activation and arterial inflammation in HIV. AIDS 2017; 31:797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. deFilippi C, Lo J, Christenson R, et al. Novel mediators of statin effects on plaque in HIV: a proteomics approach. AIDS 2018; 32:867–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B 1995; 57:289–300 [Google Scholar]
- 10. Weber C, Fraemohs L, Dejana E. The role of junctional adhesion molecules in vascular inflammation. Nat Rev Immunol 2007; 7:467–77. [DOI] [PubMed] [Google Scholar]
- 11. Lievens D, Zernecke A, Seijkens T, et al. Platelet CD40L mediates thrombotic and inflammatory processes in atherosclerosis. Blood 2010; 116:4317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lutgens E, Cleutjens KB, Heeneman S, Koteliansky VE, Burkly LC, Daemen MJ. Both early and delayed anti-CD40L antibody treatment induces a stable plaque phenotype. Proc Natl Acad Sci U S A 2000; 97:7464–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lutgens E, Gorelik L, Daemen MJ, et al. Requirement for CD154 in the progression of atherosclerosis. Nat Med 1999; 5:1313–6. [DOI] [PubMed] [Google Scholar]
- 14. Lutgens E, Lievens D, Beckers L, Donners M, Daemen M. CD40 and its ligand in atherosclerosis. Trends Cardiovasc Med 2007; 17:118–23. [DOI] [PubMed] [Google Scholar]
- 15. Varo N, Vicent D, Libby P, et al. Elevated plasma levels of the atherogenic mediator soluble CD40 ligand in diabetic patients: a novel target of thiazolidinediones. Circulation 2003; 107:2664–9. [DOI] [PubMed] [Google Scholar]
- 16. de Lemos JA, Zirlik A, Schönbeck U, et al. Associations between soluble CD40 ligand, atherosclerosis risk factors, and subclinical atherosclerosis: results from the Dallas Heart Study. Arterioscler Thromb Vasc Biol 2005; 25:2192–6. [DOI] [PubMed] [Google Scholar]
- 17. Santosuosso M, Righi E, Lindstrom V, Leblanc PR, Poznansky MC. HIV-1 envelope protein gp120 is present at high concentrations in secondary lymphoid organs of individuals with chronic HIV-1 infection. J Infect Dis 2009; 200:1050–3. [DOI] [PubMed] [Google Scholar]
- 18. Zhang R, Fichtenbaum CJ, Hildeman DA, Lifson JD, Chougnet C. CD40 ligand dysregulation in HIV infection: HIV glycoprotein 120 inhibits signaling cascades upstream of CD40 ligand transcription. J Immunol 2004; 172:2678–86. [DOI] [PubMed] [Google Scholar]
- 19. Scott IC, Clark TG, Takahara K, Hoffman GG, Greenspan DS. Structural organization and expression patterns of the human and mouse genes for the type I procollagen COOH-terminal proteinase enhancer protein. Genomics 1999; 55:229–34. [DOI] [PubMed] [Google Scholar]
- 20. Prockop DJ, Sieron AL, Li SW. Procollagen N-proteinase and procollagen C-proteinase. Two unusual metalloproteinases that are essential for procollagen processing probably have important roles in development and cell signaling. Matrix Biol 1998; 16:399–408. [DOI] [PubMed] [Google Scholar]
- 21. Zanni MV, Abbara S, Lo J, et al. Increased coronary atherosclerotic plaque vulnerability by coronary computed tomography angiography in HIV-infected men. AIDS 2013; 27:1263–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Toth PP, McCullough PA, Wegner MS, Colley KJ. Lipoprotein-associated phospholipase A2: role in atherosclerosis and utility as a cardiovascular biomarker. Expert Rev Cardiovasc Ther 2010; 8:425–38. [DOI] [PubMed] [Google Scholar]
- 23. Cao Y, Stafforini DM, Zimmerman GA, McIntyre TM, Prescott SM. Expression of plasma platelet-activating factor acetylhydrolase is transcriptionally regulated by mediators of inflammation. J Biol Chem 1998; 273:4012–20. [DOI] [PubMed] [Google Scholar]
- 24. Nou E, Lu MT, Looby SE, et al. Serum oxidized low-density lipoprotein decreases in response to statin therapy and relates independently to reductions in coronary plaque in patients with HIV. AIDS 2016; 30:583–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Subramanian S, Tawakol A, Burdo TH, et al. Arterial inflammation in patients with HIV. JAMA 2012; 308:379–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barska K, Kwiatkowska W, Knysz B, Arczyńska K, Karczewski M, Witkiewicz W. The role of the tissue factor and its inhibitor in the development of subclinical atherosclerosis in people living with HIV. PLoS One 2017; 12:e0181533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ridker PM, Cannon CP, Morrow D, et al. C-reactive protein levels and outcomes after statin therapy. N Engl J Med 2005; 352:20–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.