Abstract
Cell-to-cell variation of protein expression in genetically homogeneous populations is a common biological trait often neglected during analysis of high throughput (HT) screens, and is rarely used as a metric to characterize chemicals. We have captured single cell distributions of androgen receptor (AR) nuclear levels after perturbations as a means to evaluate assay reproducibility and to characterize a small subset of chemicals. AR, a member of the nuclear receptor family of transcription factors, is the central regulator of male reproduction and is involved in many pathophysiological processes. AR protein levels and nuclear localization often increase following ligand binding, with dihydrotestosterone (DHT) being the natural agonist. HT AR immunofluorescence imaging was used in multiple cell lines to define single cell nuclear values extracted from thousands of cells per condition treated with DHT or DMSO (control). Analysis of numerous biological replicates led to a quality control metric that takes into account the distribution of single cell data, and how it changes upon treatments. Dose-response experiments across several cell lines showed a large range of sensitivity to DHT, prompting us to treat selected cell lines with 45 EPA-provided chemicals that include many endocrine disrupting chemicals (EDCs); data from six of the compounds were then integrated with orthogonal assays. Our comprehensive results indicate that quantitative single cell distribution analysis of AR protein levels is a valid method to detect potential androgenic and anti-androgenic action of environmentally relevant chemicals in a sensitive and reproducible manner.
Keywords: High throughput microscopy, endocrine disruptor chemicals, androgen receptor
Introduction
The need for high throughput screening (HTS) assays in environmental toxicology has spurred worldwide efforts geared towards identification and quantification of potential toxicant effects of thousands of chemicals and, more recently, complex mixtures, in a variety of systems and pathways. One of the largest efforts to date has been the US Environmental Protection Agency’s (EPA) Endocrine Disruptor Screening Program (EDSP), which aimed to combine HTS with computational analyses to characterize chemicals that interfere with endocrine hormones, focusing on three nuclear receptors pathways: estrogen (ER), androgen (AR) and thyroid (TR) receptors1. Through the extensive efforts of the Tox21/ToxCast initiatives, via testing hundreds of chemicals in over a hundred HT assays, the NIEHS and EPA curated sets of reference chemicals for the ER, AR and TR pathways 2-4 that are relevant to in vivo animal studies, that have included development of computational models for quantifying activity on ER and AR3,5,6.
Many studies, including several from our group 7-11, have described the development of High Throughput (HT), High Content Assays (HCA) using the AR as a model system (reviewed in 12). However, the vast majority of efforts have been concentrated on non-native and/or cell-free assays that measured several characteristics of the AR pathway, including: ligand binding (NVS_NR_hAR in ToxCast for example), coregulator recruitment (via protein complementation assay, OT_AR_ARSRC1), reporter gene activation (Tox21_AR_Luc_MDAKB2 etc.), and nuclear translocation (GFP-AR, 8,9,13). More uniquely, we recently showed the novelty and usefulness of measuring endogenous AR levels across several 2D models, using HT microscopy (HTM) to characterize effects of EDCs, and identified BPAP as a cell-line specific AR down-regulator 11. In this study, we continued investigating the use of endogenous AR nuclear levels to analyze effects of chemicals with potential endocrine disruptor activity. When observing AR immunofluorescence images, the marked cell-to-cell variation in nuclear levels is obvious, especially following hormone treatment, a metric that was not considered in previous studies. Here, we use single cell analysis metrics for quality control to describe the distribution of AR nuclear levels, and to measure changes in such distribution by modulating time, dose and chemicals across several cell models.
Materials and Methods
Cell Culture and Treatments.
Cell lines (MCF-7, T47D, BT474, MDA-MB-453, UMUC3, A549) were obtained from BCM Cell Culture Core, which routinely validates their identity by genotyping, or directly acquired from ATCC. All cell lines were constantly tested for the absence of mycoplasma using DNA (DAPI) staining. Cell lines were routinely maintained in their standard media, as recommended by ATCC, except in phenol red-free conditions. Three days prior to experiments, cells were plated in media containing 5% charcoal-dextran stripped and dialyzed FBS-containing media. For all experiments, cells were plated at 3000 cells/well in Aurora 384 well Microplates. Treatments were performed by adding a 2x solution of each chemical in media with final DMSO concentration <0.5%.
Chemicals.
The EPA 45 reference plate 2 was kindly provided by Dr Keith Houck (NIEHS).
Immunofluorescence.
Immunofluorescence experiments were completed as previously described 11. Briefly, cells were fixed in 4% formaldehyde in PBS, and permeabilized with 0.5% Triton X-100 for 30 min. Cells were incubated at room temperature in blotto for 1 hour, and then primary antibody (mouse monoclonal AR-441, 1:500, obtained from the Protein and Monoclonal Antibody Production Core, Baylor College of Medicine; or AR (N-20), 1:500, from Santa Cruz Biotechnologies) was added overnight at 4C prior to 1h of secondary antibody (AlexaFluor conjugates; Molecular Probes) and DAPI staining. The AR N-20 antibody was used to create all the figures, while AR-441 was used in a third of the experiments, in Figure 1 and 2, to determine if results obtained were antibody specific, which was not the case.
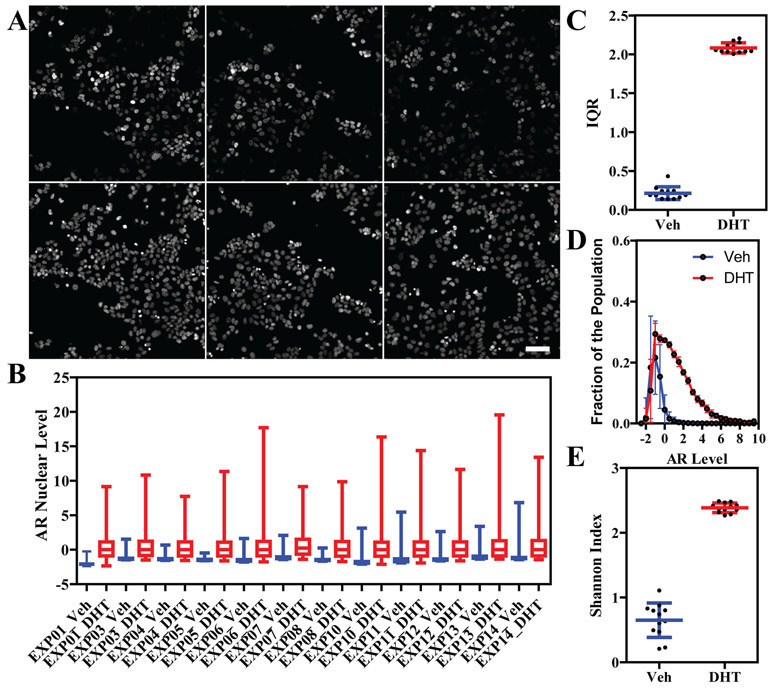
Figure 1. Variation in AR nuclear levels in MCF-7 cells.
A) Random fields (20x/0.75) showing AR immunofluorescence (top) and DAPI staining (bottom) in MCF-7 cells treated with DHT 100nM. Scale bar: 100μm. B) Box plot representing single cell nuclear AR levels (median and MAD normalized) in vehicle (veh) or DHT-treated MCF-7 cells across multiple independent biological replicates. C) Interquartile range variability across the experiments in panel B. D) AR nuclear level distribution in vehicle and DHT-treated cells represented as mean + standard deviation per each bin. E) Shannon index variability across the experiments in panel B.
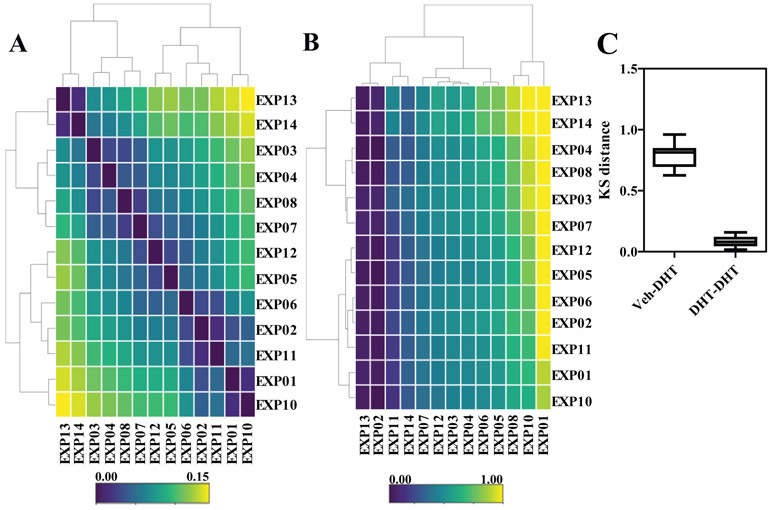
Figure 2. KS test as a metric of quality control for nuclear AR distributions across independent experiments.
A) Pairwise Kolmogorov-Smirnov distance between DHT-treated samples across the indicated experiments. B) Pairwise Kolmogorov-Smirnov distance between vehicle and DHT-treated samples across the indicated experiments. C) Summary of the distances in panels A and B shown as box-plots.
High Throughput Microscopy.
Imaging (four fields per well) was performed on a Vala Sciences IC-200 (Vala Sciences, San Diego, CA) automated HT image cytometer with a 20x/0.75 objective and a standard filter set (DAPI/HTC/TRITC/Cy5). 10μm z-stack was acquired at 2μm z-steps and max projected. The IC-200 is equipped with solid state light engine and a sCMOS camera. Excitation light intensity and exposure times were set in each experiment to fill ~70% of the dynamic range of the camera and were set based on the DHT-treated control wells, which are expected to be the brightest in the plate.
Image Analysis.
16-bit greyscale TIFFs were imported into our custom, PipelinePilot-based software mIA14 for automated image analysis. Briefly, background subtraction was performed using a rolling ball algorithm, nuclei segmented off the DAPI channel and filtered based on size and intensity to eliminate mitotic and dead cells, and cellular debris. Mean pixel intensity values for the AR channel were then extracted and used for subsequent analysis (labeled as “Nuclear AR levels” in the figures).
Data Analysis and Statistics.
Single cell data were first median and MAD (median absolute deviation) normalized based on DHT 100nM samples (selected as positive control treatment), as the AR distribution is non-normal. For histogram and calculation of the Shannon Index27, the data was divided into 25 bins, based on the square root of the smallest number of experimental observations. To measure the distance between experimental distributions across replicates, a two sample Kolmogorov-Smirnov test was applied. Heatmaps were generated in Python and Orange15. Graphs and curve fitting were created in GraphPad Prism. ToxPi16 was used to calculate ToxPi scores, after range normalization, and to generate pie charts. We performed a minimum of two biological replicates (i.e., for the EPA 45 compounds screen) with at least four technical replicates per treatment.
Results
Analysis of AR non-genetic heterogeneity by HT imaging.
Based on our previous study11, AR immunofluorescence of MCF-7 cells revealed dihydrotestosterone (DHT) treatment for 24 hours resulted in a marked increase of the mean nuclear levels of AR; moreover, the cell-to-cell variation was very high (up to two logs), representing a good example of non-genetic or phenotypic heterogeneity in the steroid hormone field (Figure 1A shows random images of DHT-treated MCF-7 cells). A key question from these results was whether or not the distribution of single cell data from vehicle vs. DHT-treated cells was reproducible and, if so, could a quality control metric be generated for a new experiment and/or to assess quantitative changes exerted by known/unknown chemicals. With this objective in mind, we repeated the same experiment 12 independent times, collecting over 600,000 single cell values, represented as a box plot in Figure 1B. Remarkably, the DHT response appeared to be quite constant, measured, for example, as an interquartile range (Figure 1C) and when visualized as a continuous distribution where every bin represents mean +/− standard deviation of all 12 experiments (Figure 1D). Not surprisingly, the highest variations were evident in the vehicle treated samples, as AR expression in MCF-7 is very low, with minimal signal-to-noise ratio, making it quite sensitive to experimental conditions. As an alternative measure of phenotypic heterogeneity, we used the Shannon Index (Figure 1E), a common diversity index, that also showed a large difference between vehicle and DHT-treated cells (3.6-fold), but only a small variation between experiments, especially in DHT-treated cells (standard deviation 0.07 vs. 0.27 in vehicle treated cells).
To directly compare and to quantify the distance between distributions of DHT-treated AR cells across multiple experiments (thirteen independent biological replicates, each with a minimum of four technical replicates per treatment), we employed a pairwise two-sample Kolmogorov-Smirnov (KS) test (Figure 2A). When the KS statistic (“distance”) was represented as a heatmap, clustering analysis identified two groups of experiments of almost equal size; however, the absolute distance between the two groups was small (Figure 2C), indicating that the DHT treatments were overall comparable across experiments. This was also evident when we used the KS test to quantify the pairwise differences between vehicle and DHT-treated samples (Figure 2B) that indicated the difference between distributions is also quite stable during repetitive assays. As expected, the KS distance between vehicle and DHT distributions was much larger compared to the distance between DHT treatments across multiple experiments (Figure 2C).
The KS test metric thus can then be used as a quality control method to classify whether or not experiments/wells/treatments are good or bad; for example, when the KS value exceeds three times the standard deviation of the mean value from good experiments. Alternative QC methods could also be employed, as shown by17-19, and by other novel pipelines in development (Stossi et al., manuscript in preparation).
AR single cell distribution across multiple cell models.
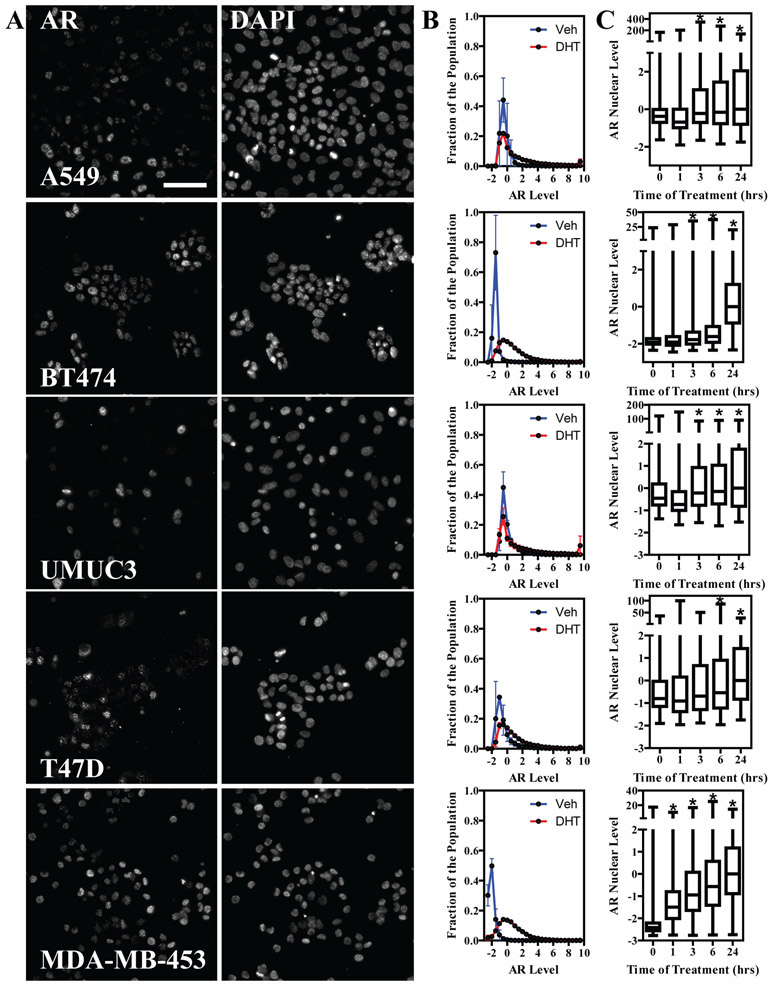
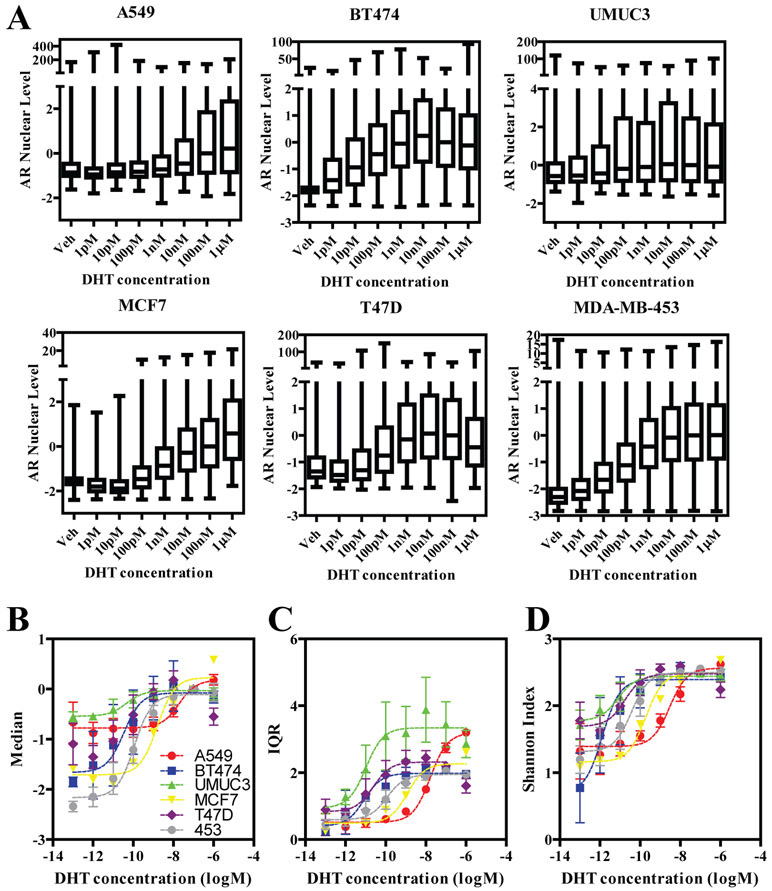
As AR is expressed in many models from different tissues of origin, we performed AR immunofluorescence in 384 well plates on 5 additional cell lines: three from breast cancer subtypes (T47D, BT474 and MDA-MB-453), one from lung cancer (A549), and one from bladder cancer (UMUC3). Images from 24h DHT-treated cells AR immunofluorescence are shown in Figure 3A, along with DAPI staining for reference, and distribution of AR levels across the population of vehicle vs. DHT-treated cells (100nM) is shown in Figure 3B. Apart from MCF-7 cells (Figure 1D), BT474 and MDA-MB-453 showed the largest response to DHT, while the others had a lesser shift in the population. To ensure that time of treatment was not a main factor in the observed responses, we performed a time-course analysis of 100nM DHT treatment (Figure 3C). All cell lines showed a time-dependent increase in AR nuclear levels, typically starting around three to six hours; with MDA-MB-453 being a notable exception, as it showed a response at only one-hour of agonist. Next, we wanted to determine the sensitivity of the different cell lines to DHT. We treated cells with seven doses of DHT, ranging from 1pM to 1μM, for 24 hours. Figure 4A shows box-plots of the dose-response analysis after median and MAD normalization using DHT 100nM as the normalizing sample. We compared three metrics in repeated biological replicates to calculate EC50 values: 1) AR median level (Figure 4B); 2) interquartile range (IQR, Figure 4C); and, 3) Shannon Index (Figure 4D). The calculated logEC50 values are shown in Supplementary Table 1. Interestingly, sensitivity to DHT covered almost three logs across the different cell lines, ranging from a low pM to a low nM range, indicating that investigating multiple models is important in characterizing chemical responses. Overall, Shannon Index, a phenotypic heterogeneity measure, when examined across all cell models, had a tendency of being slightly more sensitive (logEC50: −10.4+/−1.2) as compared to IQR (logEC50: −9.9+/−1.6) and median AR (logEC50: −9.5+/−1.1), indicating that measuring single cell indexes can be relevant and provide additional information, however, several more observations will be needed to determine significance of this analysis.
Figure 3. Single cell AR distribution in other cell models.
A) Representative images of AR immunofluorescence (left column) and DAPI (right column) in the indicated cell lines, after 24h of treatment with DHT 100nM. Scale bar: 100μm. B) AR distribution curves in vehicle vs DHT 100nM treatments in the indicated cell lines. For each bin, mean + stdev from a minimum of three biological replicates is represented. C) Time course analysis of AR nuclear level increase in all cell lines, represented as box plots. Kruskal-Wallis test with Dunn’s post test was performed for statistical significance (* p<0.05)
Figure 4. Various cell lines sensitivity to DHT as measured by single cell AR levels.
Seven-point DHT dose-response was performed in all cell lines at the 24h time point. Data is shown as box plots (A), median AR level (B), IQR (C), and Shannon index (D).
AR changes upon a set of potential endocrine disruptor chemicals.
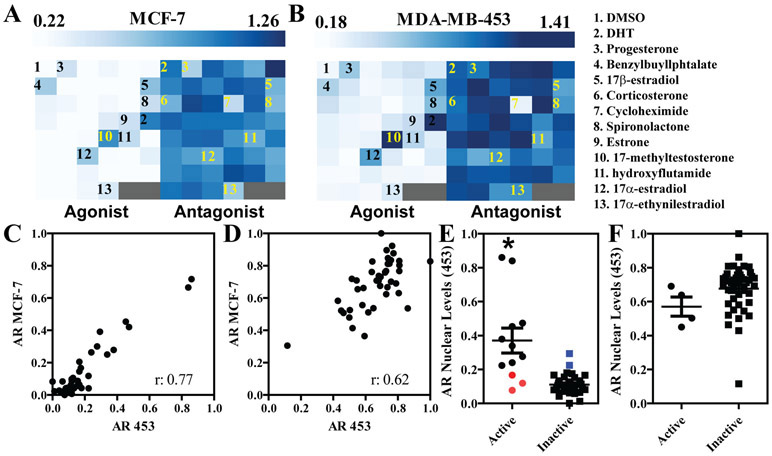
To determine if the described pipeline can be used to identify and quantify the effects of potential endocrine disruptor chemicals on the AR system, we tested a battery of 45 chemicals obtained from the EPA, and previously used as a benchmarking test for the estrogen receptor (Supplementary Table 22). We treated MCF-7 and MDA-MB-453 cells for 24 hours with 10μM of each compound, both individually (“agonist mode”) or in combination with DHT (100nM, “antagonist mode”). Average well data measuring changes in AR levels (where DHT wells are set as 1) are presented as heatmaps (Figure 5A-B), with numbers indicating control treatments (DMSO, DHT), and selected compounds. Some of the hits are known androgenic and anti-androgenic drugs (e.g., 17-methyltestosterone and hydroxyflutamide); others are steroids that lose receptor specificity at high concentration (e.g., progesterone, corticosterone and estrogens), and EDCs (e.g., benzylbutylphtalate). As expected for studies involving endogenous systems, assay interference, a common issue for any high throughput assays, is also present, as exemplified here by cycloheximide, a protein translation inhibitor, that is of course non-specific for AR. Data from MCF-7 and MDA-MB-453 correlated fairly well, especially in the plates treated in agonist mode (Spearman r: 0.77, Figure 5C) as compared to antagonist mode (r: 0.62, Figure 5D). In previous ToxCast results20, one of the assays is an ARE (androgen response element)-based luciferase transcriptional reporter in MDA-MB-453 cells, that was performed in both agonist and antagonist mode; allowing us to compare changes in single cell AR levels with an activity assay (Figure 5E-F). Based on the luciferase assay results, we divided the 45 compounds into active (defined as having an AC50<10μM) or inactive categories, and determined if there was a correlation between AR levels in the nucleus and activity. Although AR levels were not a good indicator of responses to antagonists, the approach was quite accurate with agonists, with only three active compounds in the ARE assay (genistein, daidzein and 17α-ethynilestradiol; red dots) that did not affect AR levels, and two inactive compounds (benzylbutylphtalate and 4-hydroxytamoxifen; blue dots) that only slightly increased AR levels.
Figure 5. Changes in AR levels upon treatment with a panel of reference compounds.
A) MCF-7 cells were treated with 45 compounds at 10μM +/− DHT 100nM. Average well results are shown as heatmap where the left-hand side is compounds alone and the right-hand side is compounds + DHT. DHT is set as 1. B) Same as panel A but in MDA-MB-453 cells. On the right-hand side, a legend links the numbers on the heatmaps to the corresponding compounds. C-D) Correlation analysis (Spearman rho) between AR level changes from the 45 chemicals in MCF-7 and MDA-MB-435. Compounds alone are in panel C, plus DHT 100nM in panel D. E-F) compounds were stratified as active or inactive based on ToxCast ARE luciferase data and plotted according to their AR level changes in MDA-MB-453 cells. Red dots represent compounds that were active in the ARE assay but did not change AR levels; blue dots are compounds with an opposite profile. Compounds alone are in panel E, plus DHT 100nM in panel F. *p<0.0001 using Mann-Whitney test.
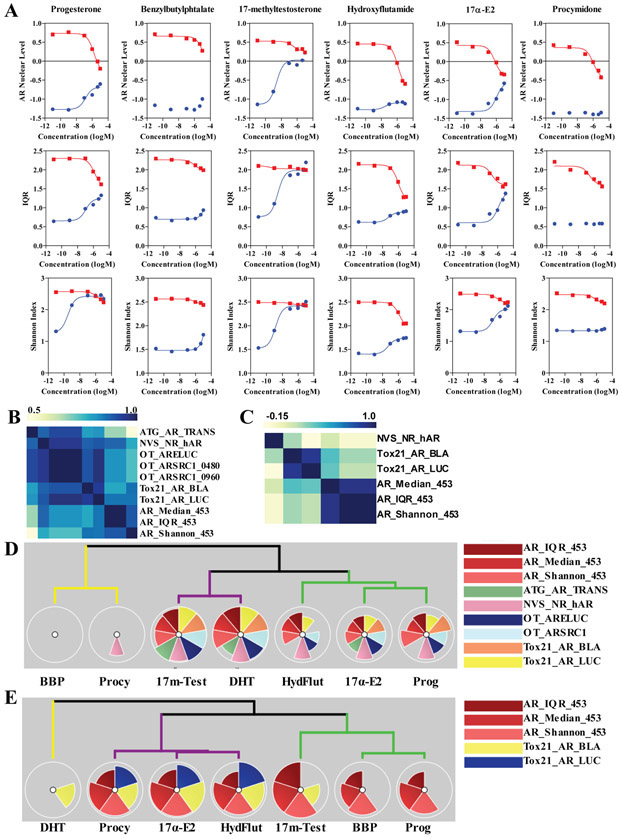
We then selected six of the potential hits (progesterone, 17 methyl testosterone, 17 alpha estradiol, benzylbutylphtalate, procymidone and hydroxyflutamide) out of the 45 compounds for a more detailed dose-response analysis in MDA-MB-453. Figure 6A shows the dose-response curves for the three indexes used above (median AR level, IQR and Shannon Index) for the compounds treated either alone (blue dots) or together with DHT 100nM (red dots) for 24 hours. Four out of the six increased AR nuclear levels: 17-methyl-testosterone reached similar effects as DHT, even when used at nanomolar levels, while other compounds had smaller effects (micromolar range). Perhaps interestingly, all six compounds showed antagonism of 100 nM DHT; some not effectively (e.g., 17β-estradiol and progesterone), and others being more active (e.g., hydroxyflutamide, procymidone). Supplementary Tables 3 and 4 list the log EC50 for all the compounds and the measured indexes. Although the AR single cell analysis assay is not directly measuring compound activity and can be influenced by more complex cellular noise when as compared to pure in vitro assays, we correlated the data obtained for the six compounds plus a DHT control across all the AR ToxCast assays20 in both agonist and antagonist modes. Figure 6B-C show heatmaps of the pairwise Spearman’s r coefficient of correlation between each pair of assays, including the three metrics of the heterogeneity assay. Overall, the correlation between all assays in agonist mode was good (>0.6), with the only exception being ATG_AR_Trans vs. Shannon index which reached 0.497. Also, the correlation in antagonist mode was much worse, being high only in assay-specific readouts (i.e., luciferase assays) indicating that for antagonism there is a stronger need for activity based and orthogonal assays as compared to compound only treatments.
Figure 6. Characterization of six compounds affecting AR levels.
A) Six-point dose response analysis in MDA-MB-435 cells for the indicated compounds, alone (blue line) or with DHT 100nM (red line). Data are shown for median AR level, IQR and Shannon index. B-C) Correlation between activity of the six compounds across the AR assays in ToxCast and the indexes presented in this study. Panel B is for compounds alone, while C is for compounds plus DHT 100nM. D-E) Hierarchical clustering for the six compounds using ToxPi, combining the ToxCast results plus the ones from this study. Panel D is for compounds alone, while E is for compounds plus DHT 100nM.
We then used the ToxPi approach (toxpi.org,16) to represent and cluster effects of the 6 compounds, as determined by the logEC50. ToxPi was developed for data integration and visualization of complex toxicological end points. Here we used it to integrate the data from the ToxCast initiative, which contains the indicated disparate assays, with our single cell data. Figure 6D shows the clustering of the compounds when treated alone, which separates them into three groups: 1) compounds strongly active in all assays (DHT and 17 methyltestosterone); less active compounds across most assays (hydroxyflutamide, 17α E2 and progesterone); and, 3) largely inactive compounds (procymidone and BBP). The same was true in antagonism mode (Figure 6E) where three compounds were antagonistic across the assays (17α-estradiol, hydroxyflutamide, and procymidone) and three compounds (17-methyltestosterone, BBP and progresterone) were uniquely active in the AR heterogeneity assay, which might be due to assay interference (e.g., completely unrelated to AR) and/or specific pathways/conditions that are only present when working in an endogenous setting. This last possibility is likely at least for progesterone, which is known to bind AR at high concentration; while 17 methyltestosterone, being a strong agonist, will compete with DHT at moderate concentrations, having ~50% RBA21 (DHT=100%). Surprisingly, BBP was completely inactive in the ToxCast assays, although it is known as an anti-androgen in vivo, and is active as an antagonist in our assay22. Overall, even sampling a small set of compounds highlights how relevant and informative HT single cell analysis of AR nuclear levels is and how it can augment more classical HT assays.
Discussion
Endocrine disrupting chemicals (EDCs) are a subclass of toxicants present in the environment that interfere with the action of endogenous hormones affecting central physiological pathways that can cause defects in, for example, reproduction and metabolism. While many efforts are in progress to define and classify potential EDCs, both computationally and experimentally, the universe of chemicals, their metabolites and mixtures remain a vast and untested black box. The ToxCast and Tox21 efforts, for example, spearheaded by the Environmental Protection Agency (EPA) and the National Toxicology Program (NTP), were designed to develop and qualify a large number of HT assays to test chemicals that cover a large set of biological pathways1,23-26. One focus in particular has been to study three hormone nuclear receptors, estrogen (ER), androgen (AR) and thyroid (TR) receptors, which participate as transcription regulators in many essential physiological and pathological roles. Recently, computational frameworks obtained from the ToxCast datasets have been built for all three receptors and the in vitro to in vivo data revealed, at least for ER, that a set of in vitro assays can substitute more expensive and time-taking in vivo assays2,4,20. Perhaps one of the drawbacks of most current efforts is that the HT assays are largely out-of-context, meaning they are performed in test tubes (i.e., ligand binding assays), or using engineered cell models (i.e., luciferase assays or GFP-tagged proteins) that, inarguably, do not fully recapitulate the inherent complexity of endogenous systems. One such complexity, often ignored, is the intrinsic cell-to-cell variation in genetically-homogeneous populations (phenotypic heterogeneity), a metric that is rarely used in HT assays17-19, and, to our knowledge, has never been used to assess EDC effects on nuclear receptors. In this study, we used the AR as a test case, following our previous observation that mean levels of AR per well were useful in identifying active agonists and antagonists;11 here, however, we elected to extract and analyze single cell data. First, we focused on MCF-7 breast cancer cells, were DHT treatment causes a large response in terms of increased nuclear AR levels, to determine whether or not single cell distributions were reproducible across multiple experiments to the degree that we could use them as a metric when monitoring the effects of perturbagens. Indeed, using KS statistics, we showed that the distribution of AR levels in a cell population is quite reproducible across many biological replicates and distribution metrics can be described using the interquartile range (IQR) and Shannon index. We then expanded our HT single cell analysis to five additional cell lines of different origin, and evaluated their DHT time- and dose-dependent changes in AR levels within the population. Perhaps not surprisingly, we found that the magnitude and sensitivity of response to DHT was cell line specific, spanning two orders of magnitude, highlighting how model choice and testing multiple models might prove beneficial to characterize perturbagens by single cell analyses. We further tested this notion by treating two cell lines with a small library of 45 EPA-curated perturbagens that have been used in ToxCast analysis to build nuclear receptor models, which included several validated chemicals that are agonists and antagonists to many NRs2. We performed experiments in both agonist (compounds alone) and antagonist (compounds plus DHT) modes and identified several known androgens and anti-androgens. Because one of the cell lines we used (MDA-MB-453) was also part of the ToxCast effort20,13, where it was engineered to contain an Androgen Response Element (ARE) driven luciferase reporter gene, we could compare AR levels with an activity metric (e.g., gene expression). Overall, we found AR levels are quite well-correlated with agonist responses, although they are not informative in terms of antagonism. Another important point, as in all HT screens, assay interference is a problem that is likely exacerbated in endogenous systems where more confounding events can, and do, experimentally occur – i.e., modulating receptor levels due to stimulation of unrelated pathways (e.g., protein translation, gene transcription, protein degradation, intracellular pathways, etc). A clear example in this study is cycloheximide, a universal inhibitor of protein translation. While this can be seen as a drawback, it is also a more integrated way to identify effects of the target protein, and assay interference hits can be easily filtered out by combining endogenous with orthogonal assays, including those in ToxCast. In fact, we have shown here how the integration of single cell metrics with orthogonal assays can be used to improve clustering of AR perturbagens. While this approach has been specific to an AR model, we anticipate that the quality control metrics of this approach have wide applicability to other single cell-oriented HT high content assays designed to in measure the effects of many types of perturbagens (ligands, small molecule inhibitors, RNAi, etc.).
Supplementary Material
Supplementary Table 1. LogEC50 values across cell lines and single cell indexes for DHT.
Supplementary Table 2. List of the 45 EPA compounds used in HTM
Supplementary Table 3. LogEC50 values across indexes for the compounds selected in Figure 6 when treated alone
Supplementary Table 4. LogEC50 values across indexes for the compounds selected in Figure 6 when treated in combination with DHT.
Acknowledgments
M.A.M., F.S., M.G.M. and A.T.S. are funded by Project 4 as part of a NIEHS Superfund Research Program (P42ES027704; PI, Rusyn). M.A.M, F.S., H.J, R.M.M, M.G.M. and P.S. are supported via the CPRIT-funded GCC Center for Advanced Microscopy and Image Informatics (RP170719; PI, Mancini). Imaging is also supported by the Integrated Microscopy Core at Baylor College of Medicine with funding from NIH (DK56338, and CA125123), CPRIT (RP150578), the Dan L. Duncan Comprehensive Cancer Center, and the John S. Dunn Gulf Coast Consortium for Chemical Genomics.
References
- 1.Richard AM; Judson RS; Houck KA; et al. ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chem. Res. Toxicol 2016, 29, 1225–1251. [DOI] [PubMed] [Google Scholar]
- 2.Judson RS; Magpantay FM; Chickarmane V; et al. Integrated Model of Chemical Perturbations of a Biological Pathway Using 18 in Vitro High-Throughput Screening Assays for the Estrogen Receptor. Toxicol. Sci 2015, 148, 137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleinstreuer NC; Browne P; Chang X; et al. Evaluation of Androgen Assay Results Using a Curated Hershberger Database. Reprod. Toxicol 2018, 81, 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paul-Friedman K; Martin M; Crofton KM; et al. Limited Chemical Structural Diversity Found to Modulate Thyroid Hormone Receptor in the Tox21 Chemical Library. Environ. Health Perspect 2019, 127, 097009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mansouri K; Abdelaziz A; Rybacka A; et al. CERAPP: Collaborative Estrogen Receptor Activity Prediction Project. Environ. Health Perspect 2016, 124, 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Judson RS; Houck KA; Watt ED; et al. On Selecting a Minimal Set of in Vitro Assays to Reliably Determine Estrogen Agonist Activity. Regul. Toxicol. Pharmacol 2017, 91, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartig SM; Newberg JY; Bolt MJ; et al. Automated Microscopy and Image Analysis for Androgen Receptor Function. Methods Mol. Biol 2011, 776, 313–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcelli M; Stenoien DL; Szafran AT; et al. Quantifying Effects of Ligands on Androgen Receptor Nuclear Translocation, Intranuclear Dynamics, and Solubility. J. Cell. Biochem 2006, 98, 770–788. [DOI] [PubMed] [Google Scholar]
- 9.Szafran AT; Szwarc M; Marcelli M; et al. Androgen Receptor Functional Analyses by High Throughput Imaging: Determination of Ligand, Cell Cycle, and Mutation-Specific Effects. PLoS One 2008, 3, e3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szafran AT; Hartig S; Sun H; et al. Androgen Receptor Mutations Associated with Androgen Insensitivity Syndrome: A High Content Analysis Approach Leading to Personalized Medicine. PLoS One 2009, 4, e8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stossi F; Dandekar RD; Bolt MJ; et al. High Throughput Microscopy Identifies Bisphenol AP, a Bisphenol A Analog, as a Novel AR down-Regulator. Oncotarget 2016, 7, 16962–16974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campana C; Pezzi V; Rainey WE Cell-Based Assays for Screening Androgen Receptor Ligands. Semin. Reprod. Med 2015, 33, 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch C; Sakamuru S; Huang R; et al. Identifying Environmental Chemicals as Agonists of the Androgen Receptor by Using a Quantitative High-Throughput Screening Platform. Toxicology 2017, 385, 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szafran AT; Mancini MA The MylmageAnalysis Project: A Web-Based Application for High-Content Screening. Assay Drug Dev Technol 2014, 12, 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demsar J, Curk T, Erjavec A, et al. Orange: Data Mining Toolbox in Python. J. Mach. Learn. Res 2013, 14, 2349–2353. [Google Scholar]
- 16.Marvel SW; To K; Grimm FA; et al. ToxPi Graphical User Interface 2.0: Dynamic Exploration, Visualization, and Sharing of Integrated Data Models. BMC Bioinformatics 2018, 19, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gough A; Shun TY; Lansing Taylor, et al. A Metric and Workflow for Quality Control in the Analysis of Heterogeneity in Phenotypic Profiles and Screens. Methods 2016, 96, 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gough AH; Chen N; Shun TY; et al. Identifying and Quantifying Heterogeneity in High Content Analysis: Application of Heterogeneity Indices to Drug Discovery. PLoS One 2014, 9, e102678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gough A; Stern AM; Maier J; et al. Biologically Relevant Heterogeneity: Metrics and Practical Insights. SLAS Discovery. 2017. 22 213–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleinstreuer NC; Ceger P; Watt ED; et al. Development and Validation of a Computational Model for Androgen Receptor Activity. Chem. Res. Toxicol 2017, 30, 946–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiita B; Artis A; Ackerman DM; et al. Binding of 17-Alpha-Methyltestosterone in Vitro to Human Sex Hormone Binding Globulin and Rat Ventral Prostate Androgen Receptors. Ther. Drug Monit. 1995, 17, 377–380. [DOI] [PubMed] [Google Scholar]
- 22.Hotchkiss AK; Parks-Saldutti LG; Ostby JS; et al. A Mixture of the “Antiandrogens” Linuron and Butyl Benzyl Phthalate Alters Sexual Differentiation of the Male Rat in a Cumulative Fashion. Biol. Reprod 2004, 71, 1852–1861. [DOI] [PubMed] [Google Scholar]
- 23.Janesick AS; Dimastrogiovanni G; Vanek L; et al. On the Utility of ToxCast™ and ToxPi as Methods for Identifying New Obesogens. Environ. Health Perspect 2016, 124, 1214–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kavlock R; Chandler K; Houck K; et al. Update on EPA’s ToxCast Program: Providing High Throughput Decision Support Tools for Chemical Risk Management. Chem. Res. Toxicol 2012, 25, 1287–1302. [DOI] [PubMed] [Google Scholar]
- 25.Dix DJ; Houck KA; Martin MT; et al. The ToxCast Program for Prioritizing Toxicity Testing of Environmental Chemicals. Toxicol. Sci 2007, 95, 5–12. [DOI] [PubMed] [Google Scholar]
- 26.Huang R; Sakamuru S; Martin MT; et al. Profiling of the Tox21 10K Compound Library for Agonists and Antagonists of the Estrogen Receptor Alpha Signaling Pathway. Sci. Rep 2014, 4, 5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon CE A Mathematical Theory of Communication. The Bell System Tech. Journal 1948, 27, 379–423. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. LogEC50 values across cell lines and single cell indexes for DHT.
Supplementary Table 2. List of the 45 EPA compounds used in HTM
Supplementary Table 3. LogEC50 values across indexes for the compounds selected in Figure 6 when treated alone
Supplementary Table 4. LogEC50 values across indexes for the compounds selected in Figure 6 when treated in combination with DHT.