Abstract
PURPOSE
The main aim of this study was to evaluate the significance of HLA-DPB1 expression in acute graft-versus-host disease (GVHD) after hematopoietic cell transplantation (HCT) from HLA-A, -B, -C, -DRB1, -DQB1–matched and –mismatched unrelated donors.
PATIENTS AND METHODS
Between January 1, 2017, and January 10, 2019, we assessed 19,136 patients who received HCT from an HLA-A, -B, -C, -DRB1, -DQB1–matched or –mismatched unrelated donor performed in Australia, the European Union, Japan, North America, and the United Kingdom between 1988 and 2016. Among transplant recipients with one HLA-DPB1 mismatch, the patient’s mismatched HLA-DPB1 allotype was defined as low or high expression. Multivariable regression models were used to assess risks of GVHD associated with high expression relative to low expression HLA-DPB1 mismatches. The effect of increasing numbers of HLA-DPB1 mismatches on clinical outcome was assessed in HLA-mismatched transplant recipients.
RESULTS
In HLA-A, -B, -C, -DRB1,-DQB1–matched transplant recipients, donor mismatching against one high-expression patient HLA-DPB1 increased moderate (odds ratio [OR], 1.36; P = .001) and severe acute GVHD (OR, 1.32; P = .0016) relative to low-expression patient mismatches, regardless of the expression level of the donor’s mismatched HLA-DPB1. Among transplant recipients with one HLA-A, -B, -C, -DRB1, or -DQB1 mismatch, the odds of acute GVHD increased with increasing numbers of HLA-DPB1 mismatches (OR, 1.23 for one; OR, 1.40 for two mismatches relative to zero mismatches for moderate GVHD; OR, 1.19 for one; OR, 1.40 for two mismatches relative to zero for severe GVHD), but not with the level of expression of the patient’s mismatched HLA-DPB1 allotype.
CONCLUSION
The level of expression of patient HLA-DPB1 mismatches informs the risk of GVHD after HLA-A, -B, -C, -DRB1, -DQB1–matched unrelated HCT, and the total number of HLA-DPB1 mismatches informs the risk of GVHD after HLA-mismatched unrelated HCT. Prospective consideration of HLA-DPB1 may help to lower GVHD risks after transplantation.
INTRODUCTION
HLA-A, -B, -C, -DRB1, and -DQB1 matching of unrelated donors for hematopoietic cell transplantation (HCT) is undertaken to lower the risks of graft failure, graft-versus-host disease (GVHD), and mortality in patients.1,2 The study of HLA-DPB1 in clinical transplantation has relied heavily on retrospective assessment of patients and donors who were originally selected on the basis of compatibility for HLA-A, -B, -C, -DRB1, and -DQB1. Analysis of HLA-DPB1 in these populations uncovered high rates of HLA-DPB1 mismatching that stymied early research efforts.3 The growth of unrelated-donor HCT worldwide has provided a large transplant experience with which to study HLA-DPB1, and recent data confirm that HLA-DPB1 is a classic transplantation antigen capable of provoking graft-versus-host (GVH) and host-versus-graft (HVG) allorecognition.4-12 The development of robust methods for large-scale HLA typing,13-15 together with upfront typing of HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 in unrelated donors at the time of recruitment, have significantly enhanced the feasibility of 6-locus (HLA-12/12) matching of patients and donors.
CONTEXT
Key Objectives
This article answers a key question about the role of HLA-DPB1 expression in unrelated donor hematopoietic cell transplantation.
Knowledge Generated
In HLA-10/10–matched transplantation, donor mismatching against a high-expression patient HLA-DPB1 allotype is associated with higher GVHD compared with mismatching against a low-expression patient allotype. Any HLA-DPB1 mismatch increases GVHD in HLA-9/10 transplantation.
Relevance
HLA-10/10–matched unrelated donor transplantation may be optimized through the avoidance of donor mismatching against high-expression patient HLA-DPB1 allotypes.
Assessment of sequence polymorphisms that define the peptide-binding region of HLA molecules is the mainstay of donor matching because of the sentinel role of HLA in antigen presentation.16 When HLA-DPB1–matched donors are not available, research efforts have focused on identification of HLA-DPB1 mismatches that are better tolerated than others. One of the earliest models is based on T-cell recognition of epitopes of the peptide-binding region (T-cell epitopes [TCEs]) and sharing of TCE by subsets of alleles that distinguish mismatches that are associated with a lower risk of adverse outcomes (TCE permissive) from those associated with higher risk (TCE nonpermissive).6,12,17-20
More recently, HLA expression has been shown to influence health and disease.21-23 In HCT, levels of HLA-C and HLA-DP expression affect the risk of acute GVHD and is hypothesized to result from donor recognition of a high-expression mismatched patient allotype.24,25 In the case of HLA-DPB1, exon 2 sequence polymorphisms that define the peptide-binding region are in strong positive linkage disequilibrium with variation in exon 3 and the 3′-untranslated region, including the rs9277534 expression marker.13 rs9277534A-positive HLA-DPB1 alleles have lower surface expression than rs9277534G-positive alleles.21,25 In HCT from HLA-A, -B, -C, -DRB1, -DQB1–matched unrelated donors, donor mismatching against one high-expression patient HLA-DPB1 allotype is associated with a higher risk of acute GVHD compared with mismatching against a low-expression patient allotype.25 The expression and TCE models identify similar, but not identical, combinations of low- and high-risk HLA-DPB1 mismatches,19 and the extent to which the two models are complementary remains to be elucidated.
The major goal of the current analysis was to confirm the significance of HLA-DPB1 expression for the risk of acute GVHD in a large, independent cohort of HLA-A, -B, -C, -DRB1, -DQB1–matched patients and to evaluate the role of HLA-DPB1 matching in transplant recipients mismatched for one HLA-A, -B, -C, -DRB1, or -DQB1 allotype. Information on permissive HLA-DPB1 mismatches will enhance donor selection and help to improve outcomes for patients who are in need of HCT as curative therapy.
PATIENTS AND METHODS
Study Design and Population
We assessed outcomes in 19,136 patients who received an unrelated donor transplantation whose data were contributed to the International Histocompatibility Working Group (IHWG) in HCT by transplant centers and transplant registries in Australia, the European Union, Japan, North America, and the United Kingdom (Data Supplement). As the goal of the current study was to validate the role of HLA-DPB1 expression in an independent cohort of HLA-matched transplant recipients, HLA-10/10–matched transplant recipients who were previously studied for HLA-DPB1 expression were excluded.25 Of 19,136 transplant recipients, 11,318 were matched at HLA-A, -B, -C, -DRB1, and -DQB1 (HLA-10/10), of whom 2,047 had no HLA-DPB1 mismatch (HLA-12/12), 5,880 had one HLA-DPB1 mismatch (HLA-11/12), and 3,391 had two HLA-DPB1 mismatches. Among 7,818 transplant recipients with one mismatch at either HLA-A, -B, -C, -DRB1, or -DQB1 (HLA-9/10), 1,112 had zero, 4,357 had one, and 2,349 had two HLA-DPB1 mismatches.
HLA
High-resolution typing of HLA-DPB1 alleles was performed as described, and ambiguity and G-group allele strings were defined by the parent allele.25 The expression level of each high-resolution allele was inferred from known exon 2, exon 3, and rs9277534 variation, where rs9277534A-linked alleles were considered low expression and rs9277534G-linked alleles were considered high expression.13,25 GVH and HVG vectors of mismatching were determined for each of the single HLA-DPB1 mismatches among the 5,880 HLA-10/10 and 4,357 HLA-9/10 cohorts.25 Each single HLA-DPB1 mismatch was defined according to the expression level of the patient and donor mismatched allotypes, and as TCE permissive or TCE nonpermissive according to version 2.0 of the algorithm.6,20,25-27
Protocols were approved by the institutional review boards of the National Institutes of Health Office for Human Research Protections and each participating IHWG center. Funding agencies had no role in study design, data collection, data analysis, data interpretation, writing of the manuscript, or the decision to submit the manuscript for publication. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Statistical Analysis
Primary clinical end points were acute grades II to IV and III to IV GVHD.25 Secondary end points were relapse, death not preceded by relapse, disease-free survival, and overall mortality. For each end point, Cox proportional hazards or logistic—for acute GVHD, as occurrences within 100 days are considered—regression models were fit to compare the hazards or odds of failure between appropriate groups. Regression models were adjusted for patient age, donor age, source of cells, disease status, T-cell depletion, transplantation type, use of total body irradiation, patient sex, donor sex, cytomegalovirus serologic status, patient race, donor race, year of transplantation, patient HLA-B leader genotype, and mean HLA-C and mean HLA-A expressions. Potential interactions between patient expression level (low v high) and various factors were examined by including appropriate terms in the relevant regression model. All regression models were based on patients with complete data; however, we conducted sensitivity analyses by including separate categories for missing data. These approaches yielded the same qualitative conclusions and the complete data results are therefore presented. Two-sided P values from regression models were obtained by using the Wald test, and no adjustments were made for multiple comparisons.
RESULTS
Previous studies have demonstrated the importance of the vector of HLA compatibility in GVHD risk in which a mismatched patient HLA allotype not present in the donor—GVH vector—is a risk factor for GVHD, whereas a mismatched donor HLA allotype not present in the patient—HVG vector—does not increase GVHD risk.4,25,28 We examined the importance of the vector of HLA-DPB1 mismatching for GVHD risk (Data Supplement). Median follow up among surviving patients was 1,835 days (interquartile range, 927.8 to 2,979.2 days). Relative to HLA-12/12 transplant recipients, one unidirectional GVH and one bidirectional HLA-DPB1 mismatch each increased the risk of grades II to IV and III to IV acute GVHD, as did two HLA-DPB1 mismatches. A single unidirectional HVG mismatch was associated with GVHD risk similar to that of HLA-12/12 transplant recipients. These data demonstrate that, similar to mismatching at other HLA loci, HLA-DPB1 mismatches that involve any GVH vector mismatch increase the risk of acute GVHD, but not mismatches that lack a GVH vector of incompatibility. As such, single HLA-DPB1 mismatches were defined as either unidirectional GVH or bidirectional for analyses with GVHD as the outcome, which is consistent with prior definitions of mismatching.4,25
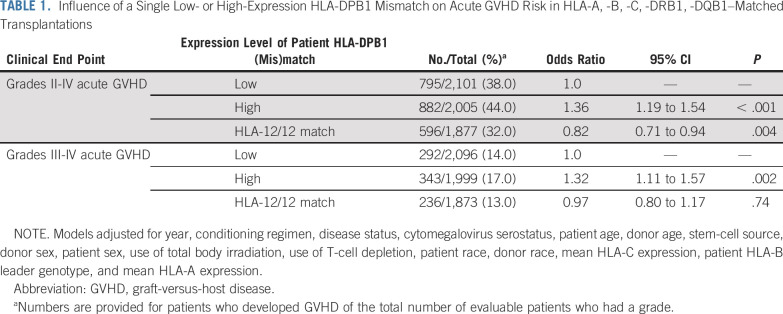
We previously observed a higher risk of acute GVHD after HLA-A, -B, -C, -DRB1, -DQB1–matched unrelated donor HCT when the donor is mismatched against one high-expression patient HLA-DPB1 allotype.25 To confirm these results, we examined the risk of GVHD in an independent cohort of HLA-11/12 transplant recipients according to the expression level of the patient’s mismatched HLA-DPB1 allotype. A total of 3,335 patients had low expression HLA-DPB1 mismatch and 2,545 had high expression HLA-DPB1 mismatch on the basis of known phase with rs9277534A (low) or rs9277534G (high).13,25 Compared with low-expression patient HLA-DPB1 mismatches, high-expression patient HLA-DPB1 mismatches were associated with an increased risk of grades II to IV and III to IV acute GVHD (Table 1). Grades II to IV acute GVHD in HLA-12/12 transplant recipients was lower or similar to that among transplant recipients with one low-expression patient HLA-DPB1 mismatch (Table 1); however, relapse was higher in HLA-12/12 transplant recipients than in HLA-DPB1–mismatched transplant recipients (Data Supplement). These results confirm and extend previous observations of HLA-DPB1 expression on GVHD outcomes after HLA-A, -B, -C, -DRB1, -DQB1–matched HCT.25 There was little suggestion that the impact of a high-expression patient HLA-DPB1 mismatch on the risk of acute GVHD varied according to the expression level of the mismatched allele in the donor, as observed previously (interaction P = .30 and P = .71 for grades II to IV and grades III to IV, respectively). HLA-DP expression did not show any apparent associations with secondary outcomes (mortality, nonrelapse mortality, disease-free survival; Data Supplement).
TABLE 1.
Influence of a Single Low- or High-Expression HLA-DPB1 Mismatch on Acute GVHD Risk in HLA-A, -B, -C, -DRB1, -DQB1–Matched Transplantations
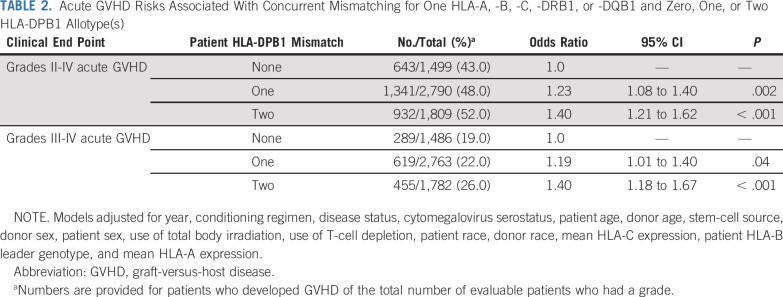
Previous studies suggest that HLA-DPB1 matching may provide information on risks after HLA-A, -B, -C, -DRB1, -DQB1–mismatched unrelated HCT.6 We first examined risks associated with the total number of HLA-DPB1 mismatches among all HLA-9/10 transplant recipients, regardless of which locus was mismatched. Compared with HLA-DPB1–matched transplant recipients, risks of acute GVHD increased stepwise with one and two HLA-DPB1 mismatches (Table 2). The impact of HLA-DPB1 mismatching on GVHD did not depend on the location of the specific HLA mismatch (HLA-A, -B, -C, -DRB1, or -DQB1; interaction P = .21). Results for secondary clinical end points are shown in the Data Supplement. To ascertain the role of HLA-DPB1 expression on acute GVHD risk in HLA-9/10 transplantation, we examined GVHD risks associated with one high-expression compared with one low-expression HLA-DPB1 mismatch in the patient, but did not observe a demonstrable increase in risk (OR, 1.08; 95% CI, 0.94 to 1.25; P = .29 for grades II to IV; OR, 1.08; 95% CI, 0.91 to 1.28; P = .39 for grades III to IV). These results suggest that knowledge of HLA-DPB1 mismatching, regardless of expression level, is an important determinant of acute GVHD in HLA-9/10 transplantation.
TABLE 2.
Acute GVHD Risks Associated With Concurrent Mismatching for One HLA-A, -B, -C, -DRB1, or -DQB1 and Zero, One, or Two HLA-DPB1 Allotype(s)
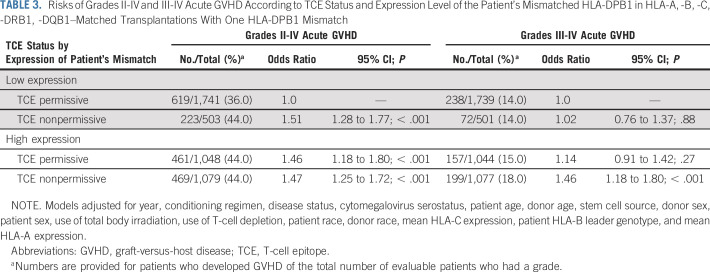
A second model for HLA-DPB1 alloreactivity is based on TCE.6,17,18,20 In the current cohort of HLA-10/10–matched transplant recipients, TCE-nonpermissive HLA-DPB1 mismatches had higher rates of GVHD compared with TCE-permissive mismatches (Data Supplement). However, HLA-DPB1 mismatches as defined by TCE group and expression levels are not mutually exclusive,19,29 and the overlap between the two models gives rise to low-expression TCE-permissive, low-expression TCE-nonpermissive, high-expression TCE-permissive, and high-expression TCE-nonpermissive mismatches (1,804; 521; 1,089; and 1,129 pairs, respectively, in the current study population). In other words, 62% (1,804 of 2,893) of TCE-permissive mismatches are low expression and 68% (1,129 of 1,650) of TCE-nonpermissive mismatches are high expression. It was therefore of interest to examine whether the deleterious impact of a high-expression mismatch is similar for TCE-permissive versus TCE-nonpermissive mismatches. Likewise, we examined whether the deleterious effect of a TCE-nonpermissive mismatch is similar for low-expression versus high-expression mismatches—that is, a statistical interaction between TCE and expression. Odds of grades II to IV acute GVHD among TCE-nonpermissive mismatches relative to TCE-permissive mismatches is larger when the mismatches are low expression (OR, 1.51; Table 3) than when the mismatches are high expression (OR, 1.01 derived from OR, 1.47/OR, 1.46; Table 3; interaction P = .04). In contrast to grades II to IV acute GVHD, the impact of TCE-nonpermissive mismatches compared with TCE-permissive mismatches on grades III to IV GVHD was higher in high-expression mismatches (OR, 1.28 derived from OR, 1.46/OR, 1.14; Table 3) than low-expression mismatches (OR, 1.02), although a statistical test of interaction yields P = .23. Finally, we previously observed that consideration of the expression level of an HLA-DPB1 mismatch added statistically significant information to a model containing only TCE in HLA-10/10–matched transplant recipients.25 In addition, a previous study by the Japan Marrow Donor Program (JMDP) demonstrated a role for both expression and TCE.12 We therefore examined whether there is evidence of an additive effect of TCE to expression and vice versa in the current study population and did so separately for JMDP versus non-JMDP patients. Among non-JMDP transplant recipients, we found that the addition of expression level (low v high) to a model containing only TCE (permissive v nonpermissive) led to a statistically significant improvement of the TCE-only model (likelihood ratio test [LRT] P = .005 for grades II to IV and P = .003 for grades III to IV). In this same subset, the addition of TCE, however, did not lead to a statistically significant improvement to the model containing only expression (LRT P = .26 for grades II to IV and P = .44 for grades III to IV). Among JMDP patients, the addition of expression level to a model containing only TCE led to a statistically significant improvement of the TCE-only model for grades II to IV acute GVHD (LRT P = .003), but not for grades III to IV (LRT P = .91). In the JMDP subset, adding TCE to the model containing only expression did not lead to a statistical improvement for either grades II to IV (LRT P = .20) or grades III to IV (LRT P = .13). Forty-one percent of patients developed grades II to IV acute GVHD and 16% developed grades III to IV acute GVHD in the entire population. Reasons for the differences in incidence by grade are not entirely known.
TABLE 3.
Risks of Grades II-IV and III-IV Acute GVHD According to TCE Status and Expression Level of the Patient’s Mismatched HLA-DPB1 in HLA-A, -B, -C, -DRB1, -DQB1–Matched Transplantations With One HLA-DPB1 Mismatch
Given that the above results demonstrate a potential difference between JMDP and non-JMDP groups in the improvement to the grade III to IV model after adding expression to TCE, we separately examined the four expression/TCE groups listed in Table 3 for JMDP and non-JMDP patients (Data Supplement). Although these results suggest numerically a differential effect of expression/TCE between JMDP and non-JMDP transplant recipients for grades III to IV acute GVHD, a statistical interaction test yields P = .18 (interaction test for grades II to IV, P = .64). Whether population differences in the HLA-DPB1 region lead to differential effects of expression and TCE on grades III to IV acute GVHD in JMDP and non-JMDP patients remains an important question for future studies.
DISCUSSION
HLA-DPB1 is a classic transplantation determinant because mismatching at this locus is associated with GVH alloreactivity, resulting in an increased risk of GVHD after unrelated HCT. We previously observed higher acute GVHD with mismatching between high-expression patient and low-expression donor HLA-DPB1 allotypes, but not between high-expression patient and high-expression donor mismatches.25 In the current study, GVHD was higher with any high-expression patient HLA-DPB1 mismatch, regardless of the expression level of the donor’s mismatched allotype. These findings align with the underlying mechanism of GVH donor recognition that is enhanced when the mismatched patient allotype is expressed at high levels and depends less on the expression level of the donor’s mismatched allotype. Validation of HLA-DPB1 expression in HLA-11/12 unrelated donor transplantations in the current study and in other recent series strongly supports the judicious use of HLA-DPB1 expression levels in the prospective selection of HLA-DPB1–mismatched unrelated donors for transplantation.12,30
The second goal of the current study was to better understand the role for HLA-DPB1 when a known mismatch at either HLA-A, -B, -C, -DRB1, or -DQB1 is present. We found that risks increase with increasing numbers of HLA-DPB1 mismatches; however, we did not observe a demonstrable increase in GVHD risk between high- and low-expression mismatches in these HLA-9/10 transplant recipients. We surmise that a higher overall GVHD rate among HLA-9/10 transplantation might obscure an effect of expression. These data suggest that when an HLA-9/10 donor is considered, additional matching for HLA-DPB1 may help to lower risks of GVHD in the patient.
The TCE model is founded on alloreactive T-cell cross-reactivity against different HLA-DP allotypes that can be clustered into TCE groups according to epitopes encoded by exon 2.17,18 The expression model is founded on strong positive linkage disequilibrium across exons 2 and 3 and the expression-associated rs9277534 variant in the 3′-untranslated region.13,15,21,25 Considering that exon 2 is a feature that is common to both models, we asked whether knowledge of expression could uncover GVHD risks in addition to what the TCE status informs, and vice versa. We found that whereas low-expression mismatches and TCE-permissive mismatches each provide information on low-risk mismatch combinations, consideration of both expression and TCE provides more information on the risk of acute GVHD than does either alone.
A combined model can identify high-expression TCE-nonpermissive combinations that confer high risk of grades III to IV acute GVHD in non-JMDP patients and should be avoided. Data from the current study therefore suggest that both qualitative (TCE) and quantitative (expression) measures of HLA-DPB1 mismatching contribute to the success of HLA-10/10–matched unrelated donor transplantation. When an HLA-A, -B, -C, -DRB1, or -DQB1–mismatched donor (HLA-9/10) is considered, then matching the patient and donor for HLA-DPB1 may help to lower nonrelapse mortality.
In clinical practice, efforts are made to limit the total number of donor HLA mismatches, but criteria based on function are limited. That HLA-DPB1 allotypes can be described as low or high expression and as TCE permissive or TCE nonpermissive contributes to the ease of translating HLA-DP expression to donor selection. Increasing use of new sequencing platforms for routine HLA typing in support of unrelated donor evaluation provides laboratories with upfront information on rs9277534, obviating the need to perform any specialized testing.13-15 As new HLA-DPB1 alleles are discovered, their haplotype association to expression markers will remain an important area of research.13 Together with the increasing ease of upfront HLA-DPB1 typing of patients and unrelated donors,13-15 the use of HLA-DPB1 expression in day-to-day clinical practice may enhance donor selection and lower GVHD risks for patients.
ACKNOWLEDGMENT
The authors thank Dawn Miller and Mark Gatterman for outstanding support.
SUPPORT
Supported by National Institutes of Health Grants No. AI069197 (E.W.P., T.G., M.H., M.M., C.M., S.R.S., P.S.), CA100019 (E.W.P., T.G., M.M., P.S., C.M.), CA18029 (E.W.P., T.G., M.M.), CA72978 (E.W.P.), CA015704 (T.G., P.S.), 5U24CA076518 (M.H., S.R.S.), and HL069294 (M.H., S.R.S.); US Office of Naval Research Grants No. N00014-17-1-2388 and N00014-17-1-2850 (M.H., S.R.S.); and Swiss FNRS N°310030_173237/1 and the Philanthropy Settlement Foundation (J.V.). This project has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research (to M.C.), under Contract No. HHSN261200800001E. This Research was supported in part by the Intramural Research Program of the NIH, Frederick National Lab, Center for Cancer Research.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
The views expressed in this article do not necessarily reflect the official policy or position of the National Institutes of Health, the Department of the Navy, the Department of Defense, or any other agency of the US government.
Written on behalf of the International Histocompatibility Working Group in Hematopoietic-Cell Transplantation.
AUTHOR CONTRIBUTIONS
Conception and design: Effie W. Petersdorf, Katharina Fleischhauer, Mary Horowitz, J. Alejandro Madrigal, Machteld Oudshoorn, Mary Carrington
Financial support: Effie W. Petersdorf, Ted Gooley, Mary Horowitz, Mari Malkki, Caroline McKallor, Stephen R. Spellman, Jean Villard, Phil Stevenson, Mary Carrington
Provision of study materials or patients: Effie W. Petersdorf, Mats Bengtsson, Dianne De Santis, Valerie Dubois, Katharina Fleischhauer, Mary Horowitz, J. Alejandro Madrigal, Mari Malkki, Caroline McKallor, Yasuo Morishima, Machteld Oudshoorn, Stephen R. Spellman, Jean Villard, Mary Carrington
Collection and assembly of data: Effie W. Petersdorf, Mats Bengtsson, Dianne De Santis, Valerie Dubois, Katharina Fleischhauer, Mary Horowitz, J. Alejandro Madrigal, Mari Malkki, Caroline McKallor, Yasuo Morishima, Machteld Oudshoorn, Stephen R. Spellman, Jean Villard, Mary Carrington
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Role of HLA-DP Expression in Graft-Versus-Host Disease After Unrelated Donor Transplantation
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Dianne De Santis
Travel, Accommodations, Expenses: OneLambda
Katharina Fleischhauer
Research Funding: Gennome Diagnostics (Inst)
Patents, Royalties, Other Intellectual Property: Royalties for a commonly developed commercial product from Genome Diagnostics, royalties are paid to my institution and used for research funding purposes (Inst)
Ted Gooley
Consulting or Advisory Role: Kiadis Pharma, Pharmacyclics, Regimmune
Mary Horowitz
Consulting or Advisory Role: Magenta (Inst)
Research Funding: Biovitrum (Inst), Telomere Diagnostics (Inst), Gamida Cell (Inst), Jazz Pharmaceuticals (Inst), Magenta (Inst)
Caroline McKallor
Stock and Other Ownership Interests: Pfizer (I)
Jean Villard
Honoraria: Astellas Pharma
Consulting or Advisory Role: Astellas Pharma
Mary Carrington
Travel, Accommodations, Expenses: Gilead Sciences
No other potential conflicts of interest were reported.
REFERENCES
- 1.Petersdorf EW, Hansen JA, Martin PJ, et al. Major-histocompatibility-complex class I alleles and antigens in hematopoietic-cell transplantation. N Engl J Med. 2001;345:1794–1800. doi: 10.1056/NEJMoa011826. [DOI] [PubMed] [Google Scholar]
- 2.Lee SJ, Klein J, Haagenson M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110:4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 3.Petersdorf EW, Smith AG, Mickelson EM, et al. The role of HLA-DPB1 disparity in the development of acute graft-versus-host disease following unrelated donor marrow transplantation. Blood. 1993;81:1923–1932. [PubMed] [Google Scholar]
- 4.Petersdorf EW, Gooley T, Malkki M, et al. The biological significance of HLA-DP gene variation in haematopoietic cell transplantation. Br J Haematol. 2001;112:988–994. doi: 10.1046/j.1365-2141.2001.02655.x. [DOI] [PubMed] [Google Scholar]
- 5.Shaw BE, Mayor NP, Russell NH, et al. Diverging effects of HLA-DPB1 matching status on outcome following unrelated donor transplantation depending on disease stage and the degree of matching for other HLA alleles. Leukemia. 2010;24:58–65. doi: 10.1038/leu.2009.239. [DOI] [PubMed] [Google Scholar]
- 6.Fleischhauer K, Shaw BE, Gooley T, et al. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: A retrospective study. Lancet Oncol. 2012;13:366–374. doi: 10.1016/S1470-2045(12)70004-9. [Erratum: Lancet Oncol 13:e134-135, 2012] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernández-Viña MA, Klein JP, Haagenson M, et al. Multiple mismatches at the low expression HLA loci DP, DQ, and DRB3/4/5 associate with adverse outcomes in hematopoietic stem cell transplantation. Blood. 2013;121:4603–4610. doi: 10.1182/blood-2013-02-481945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fürst D, Müller C, Vucinic V, et al. High-resolution HLA matching in hematopoietic stem cell transplantation: A retrospective collaborative analysis. Blood. 2013;122:3220–3229. doi: 10.1182/blood-2013-02-482547. [Erratum: Blood 123:1768, 2014] [DOI] [PubMed] [Google Scholar]
- 9.Rutten CE, van Luxemburg-Heijs SA, Halkes CJ, et al. Patient HLA-DP-specific CD4+ T cells from HLA-DPB1-mismatched donor lymphocyte infusion can induce graft-versus-leukemia reactivity in the presence or absence of graft-versus-host disease. Biol Blood Marrow Transplant. 2013;19:40–48. doi: 10.1016/j.bbmt.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 10.Pidala J, Lee SJ, Ahn KW, et al. Nonpermissive HLA-DPB1 mismatch increases mortality after myeloablative unrelated allogeneic hematopoietic cell transplantation. Blood. 2014;124:2596–2606. doi: 10.1182/blood-2014-05-576041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gagne K, Loiseau P, Dubois V, et al. Is there any impact of HLA-DPB1 disparity in 10/10 HLA-matched unrelated hematopoietic SCT? Results of a French multicentric retrospective study. Bone Marrow Transplant. 2015;50:232–236. doi: 10.1038/bmt.2014.253. [DOI] [PubMed] [Google Scholar]
- 12.Morishima S, Shiina T, Suzuki S, et al. Evolutionary basis of HLA-DPB1 alleles affects acute GVHD in unrelated donor stem cell transplantation. Blood. 2018;131:808–817. doi: 10.1182/blood-2017-08-801449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schöne B, Bergmann S, Lang K, et al. Predicting an HLA-DPB1 expression marker based on standard DPB1 genotyping: Linkage analysis of over 32,000 samples. Hum Immunol. 2018;79:20–27. doi: 10.1016/j.humimm.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayor NP, Hayhurst JD, Turner TR, et al. Recipients receiving better HLA-matched hematopoietic cell transplantation grafts, uncovered by a novel HLA typing method, have superior survival: A retrospective study. Biol Blood Marrow Transplant. 2019;25:443–450. doi: 10.1016/j.bbmt.2018.12.768. [DOI] [PubMed] [Google Scholar]
- 15.Balgansuren G, Regen L, Sprague M, et al. Identification of the rs9277534 HLA-DP expression marker by next generation sequencing for the selection of unrelated donors for hematopoietic cell transplantation. Hum Immunol. 2019;80:828–833. doi: 10.1016/j.humimm.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Morishima Y, Kashiwase K, Matsuo K, et al. Biological significance of HLA locus matching in unrelated donor bone marrow transplantation. Blood. 2015;125:1189–1197. doi: 10.1182/blood-2014-10-604785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zino E, Frumento G, Marktel S, et al. A T-cell epitope encoded by a subset of HLA-DPB1 alleles determines nonpermissive mismatches for hematologic stem cell transplantation. Blood. 2004;103:1417–1424. doi: 10.1182/blood-2003-04-1279. [DOI] [PubMed] [Google Scholar]
- 18.Fleischhauer K, Locatelli F, Zecca M, et al. Graft rejection after unrelated donor hematopoietic stem cell transplantation for thalassemia is associated with nonpermissive HLA-DPB1 disparity in host-versus-graft direction. Blood. 2006;107:2984–2992. doi: 10.1182/blood-2005-08-3374. [DOI] [PubMed] [Google Scholar]
- 19.Fleischhauer K. Immunogenetics of HLA-DP: A new view of permissible mismatches. N Engl J Med. 2015;373:669–672. doi: 10.1056/NEJMe1505539. [DOI] [PubMed] [Google Scholar]
- 20.Crivello P, Heinold A, Rebmann V, et al. Functional distance between recipient and donor HLA-DPB1 determines nonpermissive mismatches in unrelated HCT. Blood. 2016;128:120–129. doi: 10.1182/blood-2015-12-686238. [Erratum: Blood 128:2744, 2016] [DOI] [PubMed] [Google Scholar]
- 21.Thomas R, Thio CL, Apps R, et al. A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. J Virol. 2012;86:6979–6985. doi: 10.1128/JVI.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Apps R, Qi Y, Carlson JM, et al. Influence of HLA-C expression level on HIV control. Science. 2013;340:87–91. doi: 10.1126/science.1232685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramsuran V, Naranbhi V, Horowitz A, et al. Elevated HLA-A expression impairs HIV control through inhibition of nKG2A-expressing cells. Science. 2018;359:86–90. doi: 10.1126/science.aam8825. [Erratum: Science 365:pii:eaay7985, 2019] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersdorf EW, Gooley TA, Malkki M, et al. HLA-C expression levels define permissible mismatches in hematopoietic cell transplantation. Blood. 2014;124:3996–4003. doi: 10.1182/blood-2014-09-599969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersdorf EW, Malkki M, O’hUigin C, et al. High HLA-DP expression and graft-versus-host disease. N Engl J Med. 2015;373:599–609. doi: 10.1056/NEJMoa1500140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.EMBL-EBI IPD-IMGT/HLA: DPB1 T-cell epitope algorithm v2.0 (2016-08) https://www.ebi.ac.uk/ipd/imgt/hla/dpb_v2.html
- 27.Arrieta-Bolaños E, Crivello P, Shaw BE, et al. In silico prediction of nonpermissive HLA-DPB1 mismatches in unrelated HCT by functional distance. Blood Adv. 2018;2:1773–1783. doi: 10.1182/bloodadvances.2018019620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurley CK, Woolfrey A, Wang T, et al. The impact of HLA unidirectional mismatches on the outcome of myeloablative hematopoietic stem cell transplantation with unrelated donors. Blood. 2013;121:4800–4806. doi: 10.1182/blood-2013-01-480343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meurer T, Arrieta-Bolaños E, Metzing M, et al. Dissecting genetic control of HLA-DPB1 expression and its relation to structural mismatch models in hematopoietic stem cell transplantation. Front Immunol. 2018;9:2236. doi: 10.3389/fimmu.2018.02236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorentino F, Sacchi N, Oldani E, et al. Comparative evaluation of biological human leukocyte antigen DPB1 mismatch models for survival and graft-versus-host disease prediction after unrelated donor hematopoietic cell transplantation. Haematologica. 2020;105:e186–e189. doi: 10.3324/haematol.2019.225177. [DOI] [PMC free article] [PubMed] [Google Scholar]