Abstract
PURPOSE
Germline testing (GT) is a central feature of prostate cancer (PCA) treatment, management, and hereditary cancer assessment. Critical needs include optimized multigene testing strategies that incorporate evolving genetic data, consistency in GT indications and management, and alternate genetic evaluation models that address the rising demand for genetic services.
METHODS
A multidisciplinary consensus conference that included experts, stakeholders, and national organization leaders was convened in response to current practice challenges and to develop a genetic implementation framework. Evidence review informed questions using the modified Delphi model. The final framework included criteria with strong (> 75%) agreement (Recommend) or moderate (50% to 74%) agreement (Consider).
RESULTS
Large germline panels and somatic testing were recommended for metastatic PCA. Reflex testing—initial testing of priority genes followed by expanded testing—was suggested for multiple scenarios. Metastatic disease or family history suggestive of hereditary PCA was recommended for GT. Additional family history and pathologic criteria garnered moderate consensus. Priority genes to test for metastatic disease treatment included BRCA2, BRCA1, and mismatch repair genes, with broader testing, such as ATM, for clinical trial eligibility. BRCA2 was recommended for active surveillance discussions. Screening starting at age 40 years or 10 years before the youngest PCA diagnosis in a family was recommended for BRCA2 carriers, with consideration in HOXB13, BRCA1, ATM, and mismatch repair carriers. Collaborative (point-of-care) evaluation models between health care and genetic providers was endorsed to address the genetic counseling shortage. The genetic evaluation framework included optimal pretest informed consent, post-test discussion, cascade testing, and technology-based approaches.
CONCLUSION
This multidisciplinary, consensus-driven PCA genetic implementation framework provides novel guidance to clinicians and patients tailored to the precision era. Multiple research, education, and policy needs remain of importance.
INTRODUCTION
The role of germline testing (GT) for prostate cancer (PCA) has increased, with growing precision treatment implications and expanded testing options.1,2 A primary driver for GT is now precision therapy for metastatic disease where genetic results inform options and strategies for targeted treatment, therapeutic planning, and clinical trials.1-4 Approximately 12% to 17% of men with metastatic PCA harbor germline mutations, primarily in DNA repair genes, such as BRCA2, CHEK2, BRCA1, ATM, PALB2, and the DNA mismatch repair (MMR) genes,5 which are increasingly informing options for poly (ADP-ribose) polymerase (PARP) inhibitors, immune checkpoint inhibitors, platinum chemotherapy, and clinical trials.1-4,6 In early-stage disease, emerging data suggest that men with germline BRCA2 mutations, and possibly ATM mutations, have higher rates of upgrading of prostate biopsies while on active surveillance (AS).7 GT results are considered increasingly in PCA early detection discussions, particularly for men with BRCA2 mutations for which data support higher rates of PCA detection, younger age at diagnosis, and more clinically significant disease.8-10 Many of the genes that are important for PCA therapy, management, and early detection are associated with hereditary cancer syndromes.11 Pathogenic variants in BRCA1 and BRCA2 are associated with hereditary breast and ovarian cancer (HBOC). DNA MMR genes—MLH1, MSH2, PMS2, MSH6, and EPCAM—are associated with Lynch syndrome.11-16 These and other hereditary cancer syndromes confer risks for multiple cancers that must be addressed for men and their kindred.8,16
As PCA GT has increased, new practice and implementation challenges have emerged in three major areas: expanded options for multigene panels, with a resultant lack of clarity regarding optimized panel use and priority genes to test; variability in guidelines regarding GT indications and genetically based management that incorporates emerging data; and a shortage of genetic services.1,17-21 Testing options have expanded rapidly, which include focused, guideline-based, comprehensive, and reflex panels.17,18 Panels include genes with strong, limited, and unknown risk for PCA and that yet confer risks for multiple cancers.18 There is a need for clarity on panel choice and priority genes to test in men with metastatic PCA, nonmetastatic PCA, and men at high risk for PCA that balances the benefits of expanded testing (eg, identifying actionable mutations) with considerations (eg, higher rates of variants of uncertain significance [VUS]).3,8,10
Uniform guidance is also needed regarding GT indications and genetically based PCA management that incorporates rapidly emerging, sometimes conflicting, data. Current National Comprehensive Cancer Network (NCCN) guidelines have variability regarding GT on the basis of pathologic—stage and Gleason/Grade Group—and family history (FH) criteria.3,8,9 Management guidance is also needed in multiple areas with consideration of gene-specific outcomes, such as treatment of metastatic disease with variable responses by DNA repair mutations1-4,6; AS discussions that consider strong data for BRCA2, but limited data for BRCA1 and ATM7; and broader consideration of genes for PCA early detection.1,2,11 In particular, strategies for PCA early detection need clarification regarding age to begin screening on the basis of genetic status.8,9
Furthermore, the rising need for PCA GT has created a critical shortage of genetic counseling (GC) services.1,19 Health care providers, such as oncologists and urologists, increasingly are ordering PCA GT to expedite testing for management.20,21 Concerns include limited guidance on optimal pretest informed consent, optimal panel testing strategies for comprehensive genetic evaluation, inclusion of personal history and FH, and balancing timely GT with appropriate referral to GC to address patient and family needs.1,20,21 As referral of all men to GC for PCA GT is not sustainable, health care and genetic providers need implementation strategies that incorporate alternate genetic evaluation models for the timely and responsible delivery of PCA GT for men and their families.1,19
The 2019 Philadelphia Prostate Cancer Consensus Conference was convened to address challenges in PCA germline evaluation and implementation with attention to evolving genetic and precision medicine data. This meeting was a follow-up to the 2017 Philadelphia Consensus Conference, which focused on the role of GT for inherited PCA risk.18 The 2019 conference had the following 3 goals: to define optimal GT strategies that incorporate expansion of panel testing options and evolving genetic data, to propose consistent PCA GT indications and management, and to propose alternate genetic evaluation models to address the GC shortage. An expert, consensus-driven genetic implementation framework was developed for health care and genetic providers to streamline GT for PCA in the precision medicine era.
METHODS
Overarching Questions Addressing Implementation Gaps
The following questions were primary drivers of the conceptual framework:
Which men should be considered for germline PCA genetic testing?
Which panels should be considered and which genes should be prioritized for testing?
What PCA-specific recommendations should be considered on the basis of genetic results?
What is optimal informed consent for PCA GT?
What collaborative strategies may facilitate PCA genetic evaluation between health care and genetic providers?
What post-test disclosure strategies are most appropriate on the basis of genetic results?
What barriers must be addressed to enhance PCA GT?
Consensus Conference Participants
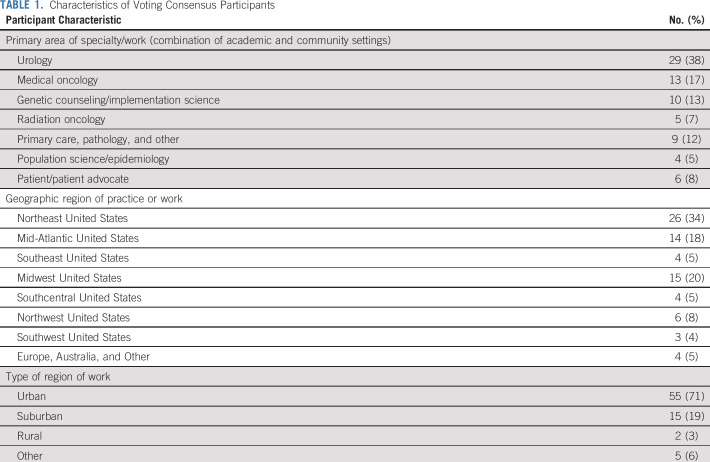
The Consensus Conference included 97 participants spanning the fields of urology, medical oncology, radiation oncology, clinical genetics, genetic counseling, primary care, pathology, implementation science, population science, epidemiology, and basic science. Patient stakeholders and advocates were active participants. Members of several national organizations, which included NCCN representatives, also participated. Academic and community practices were represented, and panelists were from multiple regions of the United States, as well as Europe and Australia. The final voting panel included 76 participants (Table 1).
TABLE 1.
Characteristics of Voting Consensus Participants
Consensus Process
The modified Delphi model was followed that incorporated elements of the Delphi process as previously published.18,22,23 Literature was provided to panel members before the meeting. Multiple expert presentations summarizing evidence relevant to genetic implementation were delivered. Evidence review is summarized in the Data Supplement.
Evidence Review
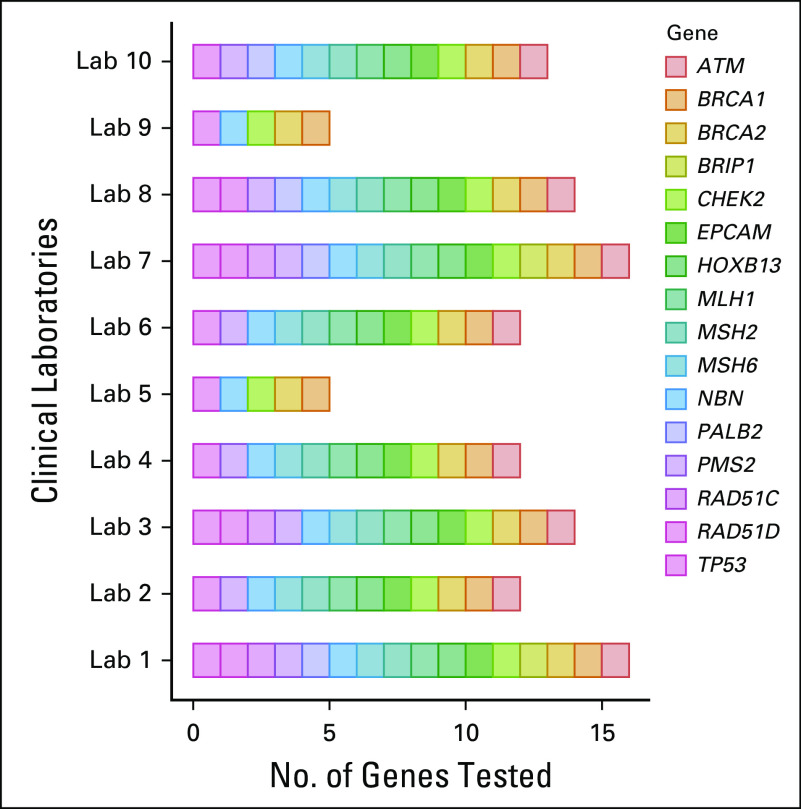
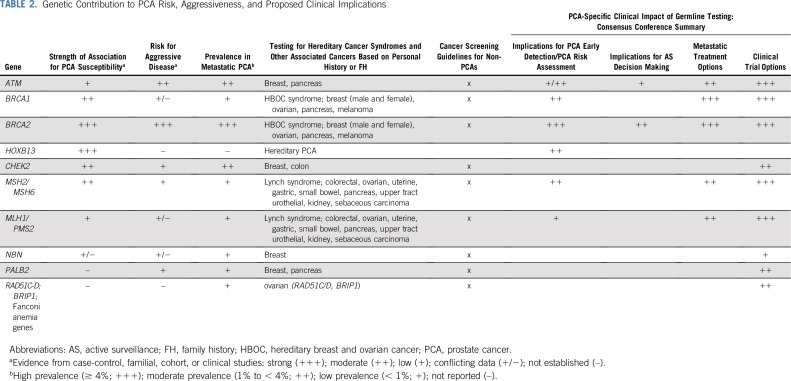
Thematic topics included: genetic contribution to PCA risk/aggressiveness24-54; germline mutations by PCA clinical and molecular characteristics5,55-66; PCA clinical multigene testing data60,61,67; germline mutations in diverse populations5,24,30,49,61,68-74; PCA genetic testing capabilities and considerations17,75-81 (Fig 1); implementation of GC1,3,8,9,17,76,82-93; NCCN PCA genetic testing guidelines and current variability3,8,9; GT for PCA precision medicine in the metastatic setting2,4,6,56,58,94-99; germline implications for AS of early-stage PCA7,35,99,100,101; and germline implications for PCA early detection.8-10,102 Table 2 provides a summary of genetic data for PCA risk and aggressiveness. Full evidence summary is provided in the Data Supplement.
FIG 1.
Variability in prostate cancer–specific multigene panels. Genetic testing registry: As of August 2019. Available at: https://www.ncbi.nlm.gov/gtr/. Courtesy of Saud AlDubayan, MD.
TABLE 2.
Genetic Contribution to PCA Risk, Aggressiveness, and Proposed Clinical Implications
Strength of Consensus
Votes were cast anonymously using a Web-based polling platform. Strength of consensus was ≥ 75% agreement for strong consensus, 50% to 74% agreement for moderate consensus, and < 50% agreement for lack of consensus.22,23
Development of PCA Genetic Evaluation and Management Framework
A conceptual framework for PCA genetic evaluation and management was developed (Fig 2). Criteria that achieved strong consensus were designated as “Recommend” and those with moderate consensus were designated as “Consider” in the final framework.
FIG 2.
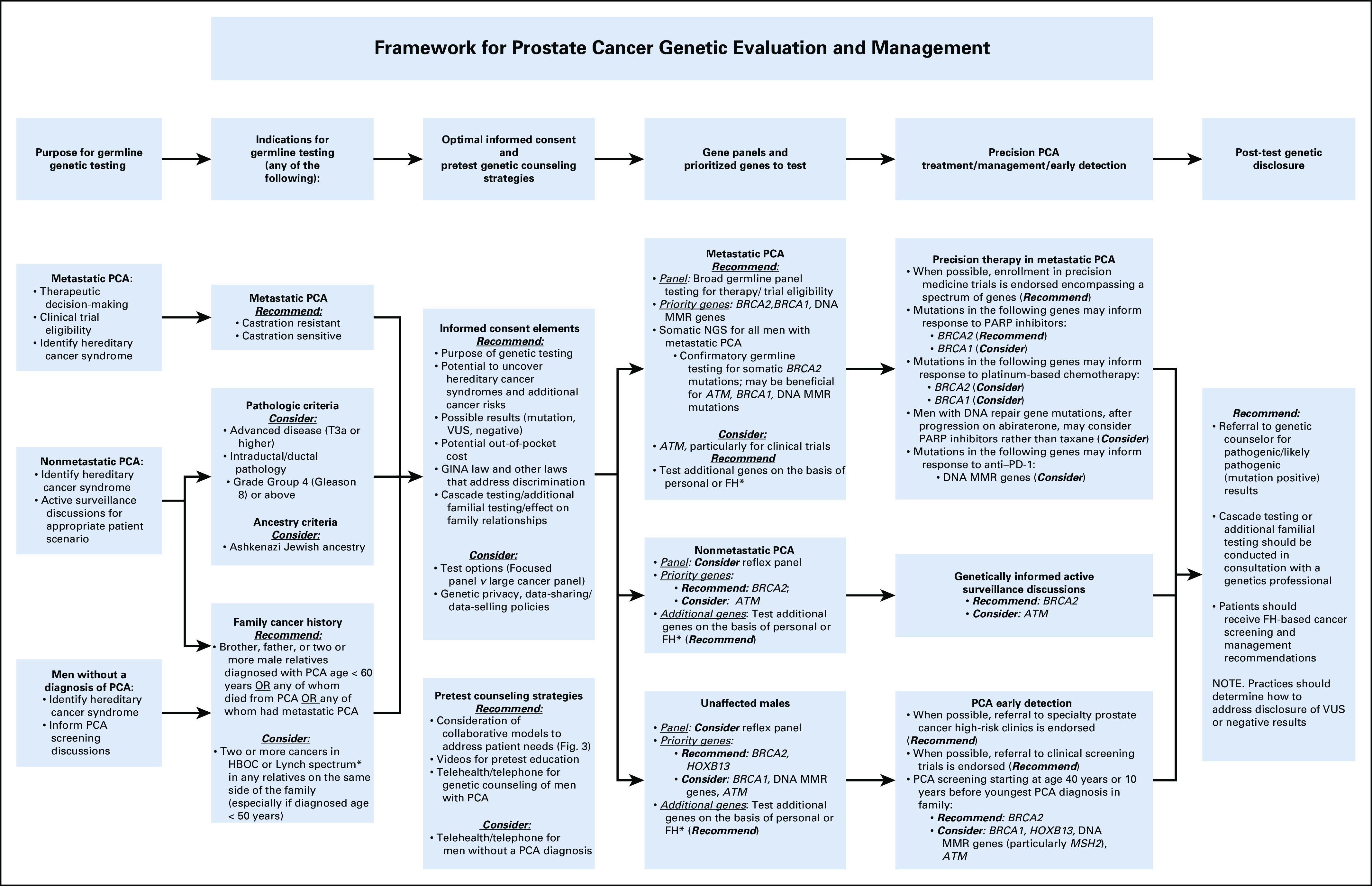
Framework for prostate cancer (PCA) genetic evaluation and management. (*) See Table 2 for personal history or family history (FH) of cancers indicating genes to test. GINA, Genetic Information Nondiscrimination Act; HBOC, hereditary breast and ovarian cancer; MMR, mismatch repair; NGS, next-generation sequencing; PARP, poly (ADP-ribose) polymerase; PD-1, programmed death 1; VUS, variant of uncertain significance.
RESULTS
Key premises
The following are guiding principles for clinical genetic evaluation:
Premises based on prior literature and Consensus Conference expert guidance:
Premises based on consensus voting:
Men should engage in informed decision making for genetic testing (Recommend).
Building collaborations between health care and genetics providers is important for optimal genetic evaluation (Recommend).
1. Which Men Should Be Considered for Germline PCA Genetic Testing?
Gaps addressed.
NCCN guidelines (NCCN Prostate Version 4.2019 and NCCN Breast/Ovary Version 3.2019) at the time of the 2019 Consensus meeting had varying indications for PCA GT.3,8 Data regarding clinical, pathologic, and FH features were summarized (Data Supplement).
Criteria for testing.
Any one of the following criteria may prompt GT:
Men with metastatic PCA (castration resistant or castration sensitive; Recommend).
-
Men with nonmetastatic PCA—one of the following:
○ Ashkenazi Jewish ancestry (Consider).
○ Advanced disease (T3a or higher; Consider).
○ Intraductal/ductal pathology (Consider).
○ Grade Group 4 (Gleason sum 8) or above (Consider).
-
FH criteria:
-
○ PCA FH criteria:
• Men with one brother or father or two or more male relatives with one of the following:
•Diagnosed with PCA at age < 60 years (Recommend).
• Any of whom died of PCA (Recommend).
•Any of whom had metastatic PCA (Recommend).
○ FH of other cancers:
• Two or more cancers in HBOC or Lynch spectrum in any relatives on the same side of the family (especially if diagnosed at age < 50 years; Consider).
-
Additional considerations.
FH consistent with hereditary PCA achieved a strong recommendation for GT. Additional FH criteria were expanded to consider 2 or more cancers in the HBOC or Lynch spectrum to account for limitations in self-reported FH. Genes corresponding to specific cancers are listed in Table 2. Of note, an unremarkable FH does not necessarily negate consideration of GT, particularly for treatment decisions in the metastatic setting.
All pathologic criteria achieved moderate agreement. Universal screening for Lynch syndrome in PCA is not current practice; however, if immunohistochemistry is performed on a prostate specimen revealing loss of the DNA MMR genes, and particularly MSH2, the recommendation is to proceed with GT to determine if the patient has Lynch syndrome given the significant cancer risks and potential treatment implications. Panelists noted that many centers do not report intraductal/ductal pathology or immunohistochemistry for Lynch syndrome markers, which must be addressed with pathologists.
Although multiple unique questions were posed specifically regarding GT for African American men, none met consensus agreement as a result of limited data. Until additional research is completed, testing guidelines as described herein should be applied in under-represented populations.
2. Which Panels Should Be Considered and Which Genes Should Be Prioritized for Testing?
Gaps addressed.
Guidance on the use of various gene panels adapted to clinical scenarios is needed given the rapid expansion of panel options and the inclusion of genes with limited association to PCA risk or PCA treatment implications (Fig 1). Furthermore, NCCN guidelines vary regarding genes to test,3,8 necessitating consensus prioritization of genes for testing (Data Supplement).
Panels considered.
Focused—guidelines-based—panels (approximately 5 to 6 genes), PCA-specific panels (approximately 10 to 15 genes), comprehensive cancer panels (approximately 80 genes), and reflex panels (initial set of genes tested followed by broad gene testing) were considered. Benefits and limitations of various panels were also considered (Data Supplement).
Genes considered.
BRCA1, BRCA2, HOXB13, CHEK2, ATM, NBN, MSH2, MSH6, MLH1, PMS2, PALB2, BRIP1, TP53, and Fanconi anemia genes were considered.
Panels and genes prioritized for testing:
-
Metastatic PCA:
○ Comprehensive (large) panel testing for therapy/clinical trial eligibility (Recommend).
-
○ Priority germline testing:
• BRCA2/BRCA1 (Recommend).
• DNA MMR genes (Recommend).
• ATM (Consider).
• Test additional genes on the basis of personal or FH (Recommend).
-
○ Somatic testing:
• Somatic next-generation sequencing for all men with metastatic PCA (Recommend).
• Confirmatory germline testing for somatic mutations:
• BRCA2 (Recommend).
• BRCA1, DNA MMR genes, ATM (Consider).
• Test additional genes on the basis of personal or FH (Table 2; Recommend).
-
Nonmetastatic PCA:
○ Reflex testing may be optimal (Consider).
-
○ Priority genes particularly to inform AS:
• BRCA2 (Recommend).
• ATM (Consider).
• Test additional genes on the basis of personal or FH (Table 2; Recommend).
-
Men without a diagnosis of PCA meeting FH testing criteria:
○ Reflex testing may be optimal (Consider).
-
○ Priority genes for risk assessment:
• BRCA2 (Recommend).
• HOXB13 (Recommend).
• BRCA1, ATM, DNA MMR genes (Consider).
• Test additional genes on the basis of personal or FH (Table 2; Recommend).
Additional considerations.
For men with metastatic PCA, broader panel testing may be appropriate, particularly if considering treatment or clinical trial options (Table 2, Fig 2, and Data Supplement). Reflex testing may be considered for all patients, but especially for men with nonmetastatic disease considering AS or men without PCA for early detection, which allows for initial testing of genes that inform management (Data Supplement). Reflex testing also allows for testing of additional genes to account for personal cancer or FH at a later time for comprehensive genetic evaluation and may also be more amenable to collaborative genetic evaluation models (see below).
Among MMR genes, MSH2 has the strongest association to PCA; however, it is recognized that MLH1, PMS2, MSH6, and EPCAM also need to be tested to establish the diagnosis of Lynch syndrome. Full MMR testing also may be important for treatment consideration or clinical trials in the metastatic setting; therefore, full Lynch syndrome testing is recommended as indicated.
In addition, confirmatory GT is recommended for men with somatic BRCA2 mutations and may be beneficial for somatic mutations in BRCA1, MMR genes, and ATM to identify hereditary cancer predisposition. Additional GT beyond these genes may also be recommended on the basis of personal and FH. Consultation with a genetics professional is advised.
3. What PCA-Specific Recommendations Should Be Considered on the Basis of Genetic Results?
Gaps addressed.
There is a need for consensus agreement on genetically informed PCA treatment, management, and early detection1,2 (Data Supplement). An additional challenge is inconsistency in NCCN genetically based PCA early detection recommendations regarding which genes to consider and the age at which to begin screening8,9 (Data Supplement).
Genetically based recommendations.
Genes considered included BRCA1, BRCA2, HOXB13, CHEK2, ATM, NBN, MSH2, MSH6, MLH1, PMS2, PALB2, BRIP1, TP53, and Fanconi anemia genes.
-
Metastatic PCA: GT to inform precision therapy:
○ Enrollment of men with PCA in precision medicine trials is endorsed (Recommend).
-
○ Mutations in the following genes may inform response to PARP inhibitors:
• BRCA2 (Recommend).
• BRCA1 (Consider).
-
○ Mutations in the following genes may inform response to platinum-based chemotherapy:
• BRCA2 (Consider).
• BRCA1 (Consider).
○ Men with DNA repair gene mutations, after progression on abiraterone, may proceed with PARP inhibitor rather than taxane (Consider).
-
○ Germline mutations in the following genes may inform response to anti–programmed death 1 (PD-1) therapy:
• DNA MMR genes (Consider).
• NOTE. The US Food and Drug Administration has granted accelerated approval for anti–PD-1 therapy for microsatellite instability-high/MMR-deficient tumors.
-
Nonmetastatic PCA: to inform AS discussions:
○ BRCA2 (Recommend).
○ ATM (Consider).
-
Men without a PCA diagnosis to inform PCA early detection:
○ Referral to specialty PCA high-risk clinics and/or early detection trials was endorsed (Recommend).
-
○ PCA early detection starting at age 40 years or 10 years before the youngest PCA diagnosis in family:
• BRCA2 (Recommend).
• BRCA1, HOXB13, ATM, and DNA MMR genes (particularly MSH2; Consider).
Additional considerations.
In the metastatic setting, a broad spectrum of genes may be important in determining clinical trial eligibility, and emerging data should continue to refine recommendations. ATM garnered consideration for testing, primarily for clinical trial eligibility; however, the panel did not feel that there was sufficient data to endorse ATM for informing therapy to PARP inhibitors off study because of the limited independent association to PARP inhibitor response at this time (Data Supplement). ATM also garnered moderate consensus for informing AS, but there are limited data at this time (Data Supplement).
For anti–PD-1 therapy, the US Food and Drug Administration has granted accelerated approval for tumors that are microsatellite instability-high or MMR deficient. The panel had moderate consensus regarding a definitive recommendation for anti–PD-1 therapy off study for men with germline MMR mutations, with stronger consideration for clinical trials.
Regarding AS discussions, clinicopathologic criteria, age, and overall health must be considered. BRCA1 did not achieve consensus for inclusion in AS as a result of limited data for PCA aggressiveness (Data Supplement). Polygenic risk score data were reviewed77-81 and did not achieve consensus.
4. What Is Optimal Informed Consent for PCA GT?
Gaps addressed.
Current practice guidelines do not provide guidance to health care providers regarding optimal informed consent for PCA GT.
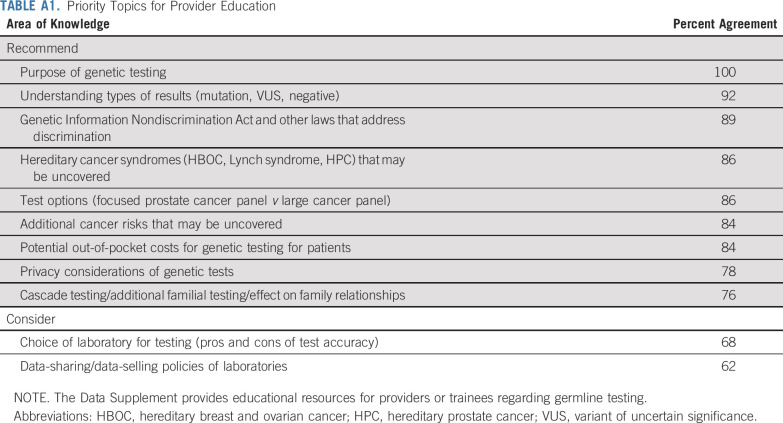
Optimal pretest informed consent elements.
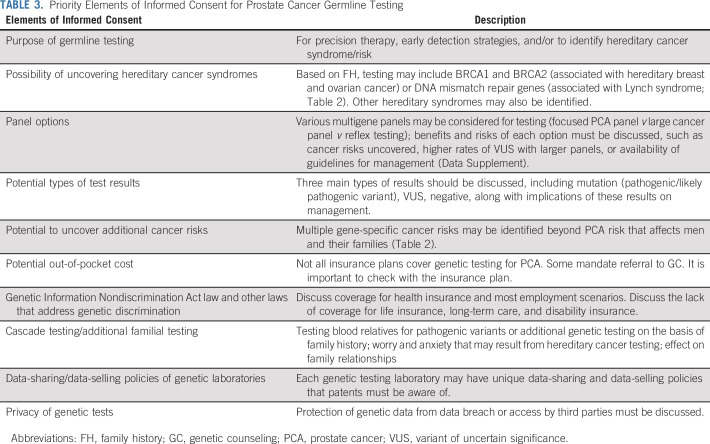
Ethical considerations of GC were reviewed (Data Supplement). The following elements garnered strong or moderate consensus to discuss with men before GT (Fig 2 and Table 3):
TABLE 3.
Priority Elements of Informed Consent for Prostate Cancer Germline Testing
Recommend discussing: (1) the purpose of GT; (2) the possibility of uncovering hereditary cancer syndromes; (3) potential types of test results; (4) the potential to uncover additional cancer risks; (5) potential out-of-pocket cost; (6) Genetic Information Nondiscrimination Act law and other laws that address genetic discrimination; and (7) cascade testing/additional familial testing.
Consider discussing: (1) multigene panel options; (2) data sharing/data selling policies of genetic laboratories; and (3) the privacy of genetic tests.
Additional considerations.
These elements of pretest informed consent apply to all men who are considering PCA GT76,82-84 (Fig 2). Such GC aids as handouts or videos may be useful to deliver this information. However, informed consent is a process during which patients have opportunities to ask questions76,82-84; therefore, a question-and-answer process must be available before testing. Clinicians without specific training/expertise in GC/GT are urged to refer patients to GC before ordering GT. Furthermore, it is important to remain current on the ethics/informed consent process for GT because of the rapidly evolving nature of precision medicine.
5. What Collaborative Strategies May Facilitate PCA Genetic Evaluation Between Health Care and Genetic Providers?
Gaps addressed.
Multidisciplinary guidance on the implementation of collaborative models between health care providers and GC is currently lacking.103 There is a need to address alternate GC models for timely GT with attention to appropriate pretest informed consent and comprehensive evaluation.
Alternate genetic evaluation delivery strategies.
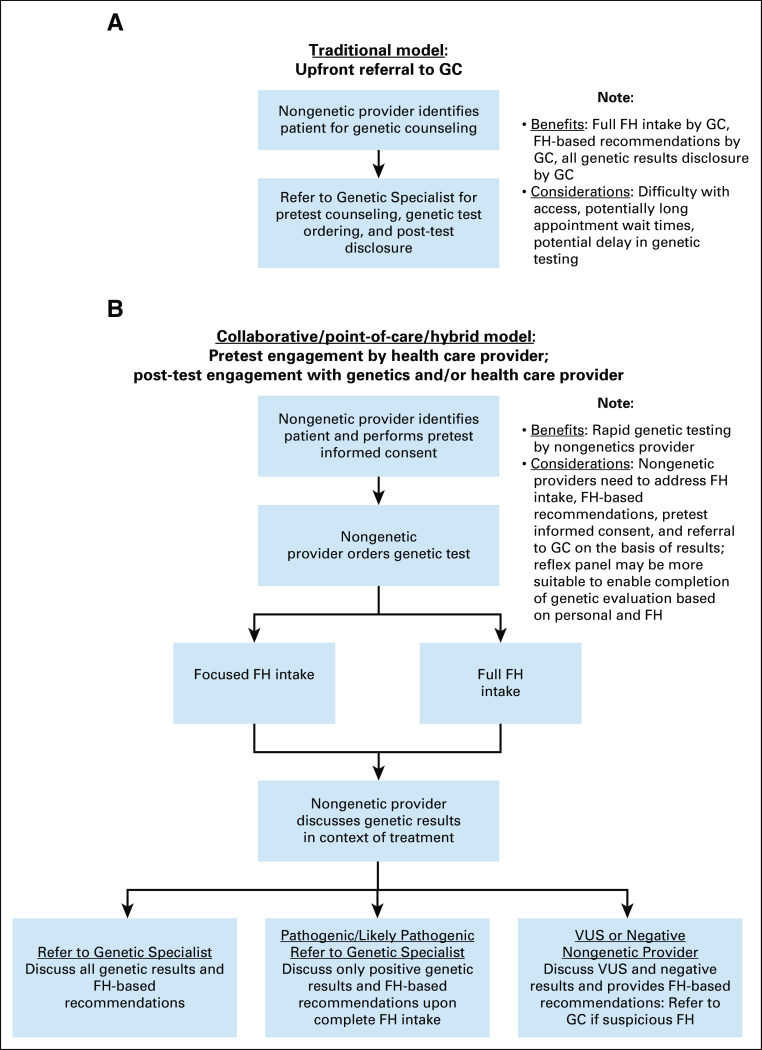
The following strategies were endorsed (Data Supplement and Fig 3):
FIG 3.
Models of collaboration between genetics and health care practices for prostate cancer genetic evaluation. FH, family history; GC, genetic counseling.
Practices should consider multiple models to address patients’ needs (Fig 3), including point-of-care models with limited or full pretest FH collection as well as traditional model with upfront referral to GC (Recommend).
Videos may be useful to deliver pretest informed consent (Recommend).
In point-of-care models, reflex genetic testing may be optimal to enable additional testing on the basis of personal/FH (Consider).
Telehealth/telephone delivery of GC is a suitable alternative to in-person GC (Recommend for men with PCA; Consider for unaffected males).
Additional considerations.
If limited pretest FH is collected, practices must proactively address the collection of FH in the post-test setting. Reflex testing enables future testing to account for personal/FH. Telehealth/telephone GC was endorsed to address geographic barriers to GC, although patient outcomes data in males are lacking. Key process questions for practices to consider when implementing point-of-care versus traditional GC models were discussed (Data Supplement).
6. What Post-Test Disclosure Strategies Are Most Appropriate Based on Genetic Results?
Gaps addressed.
Joint guidance from oncologists, urologists, and genetic counselors for referral to GC is currently lacking.
Optimal post-test disclosure strategies:
Referral to a GC for pathogenic/likely pathogenic results (Recommend).
Patients should receive FH-based recommendations, either in health care or genetic practices (Recommend).
Cascade/additional familial testing should be conducted in consultation with a genetic professional (Recommend).
Additional considerations.
There was no consensus regarding referral of men with VUS or negative results; therefore, providers will need to determine their ability to discuss VUS results and FH-based recommendations. VUS reclassification to “pathogenic/likely pathogenic” and subsequent management are critical for ordering providers to consider and may support the referral of select men with suspicious VUS to GC. Men with FH of cancers may also warrant referral to GC.
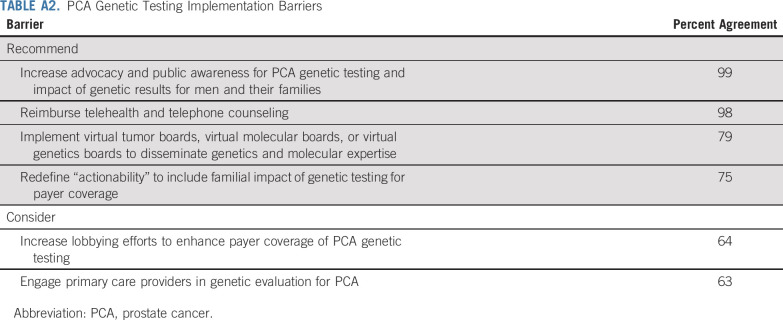
7. What Barriers Must Be Addressed to Enhance PCA GT?
Gaps addressed.
Multiple practice, research, and policy gaps pose barriers to PCA GT.
Areas in need of additional attention.
The following areas achieved strong or moderate consensus to address:
DISCUSSION
As GT for PCA has rapidly increased, responsible implementation of testing and management are of primary concern.1,2,19,23 Current practice challenges that pose barriers to operationalizing PCA GT include the variability in testing indications and genetically based management, the need for guidance on panels and priority genes to test, and guidance regarding alternate evaluation models to address GC demand. The 2019 Philadelphia Prostate Cancer Consensus Conference was a focused attempt to address these critical challenges and practice gaps by developing a first-in-field working framework for PCA genetic evaluation, management, and implementation informed by best evidence and expert guidance.
The strength of the consensus framework is the creation of a unified approach regarding GT indications, genetically informed management and treatment, and the integration of GC. Multiple aspects of the framework had strong evidence and strong expert agreement to deem a definitive action of “Recommend”. The strongest recommendations encompassed testing all men with metastatic PCA or men with FH suggestive of hereditary PCA. Priority genes for testing included BRCA2, BRCA1, and the DNA MMR genes in metastatic disease to inform treatment or clinical trials; BRCA2 for AS discussions; and BRCA2 and HOXB13 for PCA early detection discussions. This was the first formal, multidisciplinary endorsement for broad panel testing among men with metastatic PCA, recognizing that genetic information may enable men to enroll in clinical trials. Consensus emerged regarding strategies for PCA early detection on the basis of genetic status. For male carriers of BRCA2, a recommendation was made to begin PSA screening at age 40 years or 10 years before the youngest PCA diagnosis in a family and is modeled after colorectal cancer guidelines.16
An important aspect to the genetic evaluation framework was the integration of care processes and GC to account for the increasing need for GC. Strong recommendations were made for optimal pretest informed consent. Recommended strategies to deliver GC included collaborative GC models, videos, and telehealth to facilitate GT through health care practices and to collaborate with GC. Reflex testing garnered moderate consensus and may be considered, particularly when using collaborative counseling models to enable upfront testing by health care providers, followed by testing additional genes using GC for comprehensive genetic evaluation. In the post-test setting, strong recommendations were made to refer all men with pathogenic mutations to GC, to conduct cascade testing of relatives under the care of genetics professionals, and to determine the delivery of FH-based recommendations.
The panel dealt with many uncertainties in recommendations which garnered moderate consensus. Whereas many genes have a lower level of evidence for PCA risk, aggressiveness, or treatment response, several clinically available multigene panels include lower evidence genes. To indicate these nuances in limited data or moderate consensus, many criteria were designated as “Consider” in the framework. Pathologic criteria for testing, such as disease stage, intraductal/ductal histology, or Grade Group ≥ 4, garnered moderate consensus and therefore are included as suggestive criteria for testing.63,65,66 Ashkenazi Jewish ancestry as a standalone criterion achieved moderate consensus, but may be a stronger consideration for testing for men with higher Gleason score per current NCCN guidelines.8 Whereas PCA has been linked with HBOC and Lynch syndrome, a working definition of familial features that increase the likelihood of detecting germline mutations is needed. As such, having two or more relatives with cancers in the HBOC or Lynch syndrome spectrum garnered moderate consensus as standalone criteria and may be considered for GT on the basis of patient preference and insurance coverage.
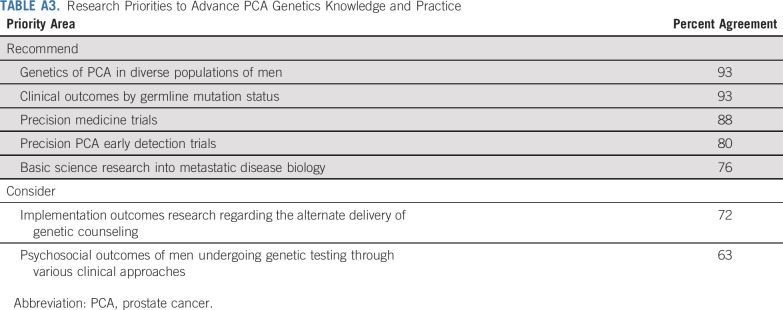
Priority genes to test also presented challenges, particularly regarding ATM, DNA MMR genes, and HOXB13. Initial data have reported that men with ATM mutations experienced clinical response to PARP inhibitors94; however, follow-up studies have reported a limited independent effect of ATM.99 Similarly, studies in AS had limited association of ATM mutations alone with upgrading of biopsies.7 Until additional data are available, ATM was given a designation of “Consider” for testing, recognizing the potential for clinical trial options for ATM carriers. Additional uncertainties were encountered regarding prioritizing MMR genes for GT. Among MMR genes, MSH2 has the highest reported association to PCA.41 Although other MMR genes have lower or limited association to PCA, the potential to uncover Lynch syndrome and clinical trial eligibility drove the suggestion to consider full Lynch syndrome testing. MSH2 status may be more informative for PCA early detection discussions.41 HOXB13 has strong association to PCA risk and early-onset disease, though screening outcomes data are limited. Therefore, the consensus panel recommended testing for HOXB13 and to consider the results in early detection discussions. Overall, BRCA1, HOXB13, and MMR genes were designated as “Consider” for beginning screening at age 40 years or 10 years before the youngest PCA diagnosis in the family because of the currently limited screening data.9 Data from screening studies, such as IMPACT and the National Cancer Institute (ClinicalTrials.gov identifier: NCT03805919), will be important to reconsider strengthening these recommendations.10 However, this is the first time that screening strategies based on a larger genetic spectrum have been proposed. Additional research in African American males is vitally needed. Future consideration of circulating tumor and cell-free DNA is also warranted.
In conclusion, the 2019 Consensus Conference created the first multidisciplinary PCA genetic implementation framework tailored to the precision medicine era. The framework, which importantly had input from NCCN panel leaders, provides guidance to a spectrum of providers to facilitate timely and responsible PCA GT for the benefit of men and their families.
ACKNOWLEDGMENT
The authors are grateful to patients and patient advocates who participated in the Consensus Conference: Buehler J., Hegedus A., Kaye P., Martin S., and Waxman S.P.
Appendix
TABLE A1.
Priority Topics for Provider Education
TABLE A2.
PCA Genetic Testing Implementation Barriers
TABLE A3.
Research Priorities to Advance PCA Genetics Knowledge and Practice
SUPPORT
Supported by National Institutes of Health Cancer Center Support Grant 5P30CA056036-19, Foundation Medicine, Myriad, Bayer, Clovis Oncology, BioReference Laboratories, Ferring Pharmaceuticals, Philadelphia Father’s Day Run, Invitae, AstraZeneca, Janssen Oncology, Roche, UroSeq, Color, OncLive, Physician Education Resource, and MDx Health.
The contents of this manuscript are not to be construed as official or reflecting the views of the Department of Defense, the Uniformed Services University of the Health Sciences, or any other agency of the US Government. This manuscript does not constitute endorsement or implied endorsement on the part of the Department of Defense or any component agency.
AUTHOR CONTRIBUTIONS
Conception and design: Veda N. Giri, Karen E. Knudsen, William K. Kelly, William Dahut, Howard R. Soule, Adam P. Dicker, Amanda E. Toland, Mary B. Daly, Peter R. Carroll, Amie Blanco, Ashley Woodson, Mary-Ellen Taplin, Jacqueline Powers, Richard Wender, Anthony Costello, Anne Calvaresi, Thenappan Chandrasekar, James Eastham, Costas Lallas, Ana Maria Lopez, Mark Mann, Martin M. Miner, Lorelei Mucci, Ronald E. Myers, Brock O’Neil, Peter Pinto, Timothy R. Rebbeck, Charles Ryan, E. Michael D. Scott, Leonard G. Gomella
Administrative support: Leonard G. Gomella
Provision of study materials or patients: Veda N. Giri, Leonard G. Gomella
Collection and assembly of data: Veda N. Giri, William K. Kelly, Heather H. Cheng, Kathleen A. Cooney, Scott Weissman, Adam P. Dicker, Saud AlDubayan, Amanda E. Toland, Colin C. Pritchard, Curtis A. Pettaway, Mary B. Daly, James L. Mohler, Peter R. Carroll, Ashley Woodson, Alanna Rahm, Mary-Ellen Taplin, Thomas J. Polascik, Brian T. Helfand, Colette Hyatt, Alicia K. Morgans, Felix Feng, Raoul Concepcion, Daniel W. Lin, Richard Wender, James Ryan Mark, William B. Isaacs, Jianfeng Xu, Jeffrey Weitzel, Lindsey Byrne, Anne Calvaresi, Thenappan Chandrasekar, Patrick T. Gomella, Nathan Handley, Joseph Izes, R. Jeffrey Karnes, Ana Maria Lopez, S. Bruce Malkowicz, Mark Mann, Patrick Mille, Sarah M. Nielsen, Brock O’Neil, Peter Pinto, Wendy Poage, Timothy R. Rebbeck, Howard Sandler, E. Michael D. Scott, Brittany Szymaniak, Neha Vapiwala, Charnita Zeigler-Johnson, Leonard G. Gomella
Data Analysis and interpretation: Veda N. Giri, William K. Kelly, Heather H. Cheng, Kathleen A. Cooney, Michael S. Cookson, William Dahut, Scott Weissman, Daniel P. Petrylak, Colin C. Pritchard, Curtis A. Pettaway, James L. Mohler, J. Kellogg Parsons, Peter R. Carroll, Robert Pilarski, Ashley Woodson, Alanna Rahm, Mary-Ellen Taplin, Thomas J. Polascik, Brian T. Helfand, Alicia K. Morgans, Felix Feng, Michael Mullane, Richard Wender, Arthur L. Burnett, Oliver Sartor, Jeffrey Weitzel, Gerald L. Andriole, Himisha Beltran, Alberto Briganti, David Y. T. Chen, Robert B. Den, Albert Dobi, E. David Crawford, James Eastham, Scott Eggener, Matthew L. Freedman, Marc Garnick, Mark D. Hurwitz, Joseph Izes, R. Jeffrey Karnes, Lucia Languino, Stacy Loeb, Ana Maria Lopez, Kevin R. Loughlin, Grace Lu-Yao, S. Bruce Malkowicz, Mark Mann, Patrick Mille, Martin M. Miner, Todd Morgan, Jose Moreno, Wayne Pinover, Peter Pinto, Ganesh V. Raj, Matthew Schiewer, William Tester, Edouard J. Trabulsi, Neha Vapiwala, Evan Y. Yu, Leonard G. Gomella
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Implementation of Germline Testing for Prostate Cancer: Philadelphia Prostate Cancer Consensus Conference 2019
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Veda N. Giri
Stock and Other Ownership Interests: Novopyxis (I)
Karen E. Knudsen
Stock and Other Ownership Interests: Pfizer, Genomic Health
Honoraria: CellCentric, Sanofi
Consulting or Advisory Role: CellCentric, Sanofi, Atrin Pharmaceuticals, Context Therapeutics
Research Funding: Celgene
Travel, Accommodations, Expenses: Sanofi, Genentech
William K. Kelly
Honoraria: Janssen Oncology, Bayer
Consulting or Advisory Role: Merck Sharp & Dohme
Research Funding: Sanofi (Inst), Novartis (Inst), Janssen Oncology (Inst), Bayer (Inst), Exelixis (Inst), Seattle Genetics (Inst), Endocyte (Inst), Amgen (Inst), BioClin Therapeutics (Inst), Sarah Cannon Research Institute (Inst), F Hoffman-La Roche (Inst)
Travel, Accommodations, Expenses: Janssen Oncology, Merck Sharp & Dohme
Heather H. Cheng
Research Funding: Inovio Pharmaceuticals (Inst), Sanofi (Inst), Astellas Medivation (Inst), Janssen Oncology (Inst), Clovis Oncology (Inst), Color Foundation (Inst)
Kathleen A. Cooney
Patents, Royalties, Other Intellectual Property: Patent awarded for discovery of HOXB13 as prostate cancer susceptibility gene (Inst)
Travel, Accommodations, Expenses: Boston Scientific (I)
Michael S. Cookson
Honoraria: Merck, Janssen Biotech, Bayer, Astellas Pharma, Myovant Sciences
Consulting or Advisory Role: Merck, Janssen Biotech, MDxHealth, Bayer, Astellas Pharma, Myovant Sciences, TesoRx Pharma, Genomic Health, Ferring Pharmaceuticals, Precision Biopsy
Scott Weissman
Employment: Genome Medical
Stock and Other Ownership Interests: Genome Medical
Howard R. Soule
Leadership: WindMIL
Consulting or Advisory Role: Compugen, WindMIL
Travel, Accommodations, Expenses: Compugen, Sanofi, WindMIL
Daniel P. Petrylak
Stock and Other Ownership Interests: Bellicum Pharmaceuticals, TYME
Consulting or Advisory Role: Bayer, Exelixis, Pfizer, Roche, Astellas Pharma, AstraZeneca, Eli Lilly, Amgen, Boehringer Ingelheim, Bristol Myers Squibb, Clovis Oncology, Incyte, Janssen Oncology, Pharmacyclics, Seattle Genetics, Urogen Pharma, Advanced Accelerator Applications, Ipsen
Research Funding: Progenics (Inst), Sanofi (Inst), Endocyte (Inst), Genentech (Inst), Merck (Inst), Astellas Medivation (Inst), Novartis (Inst), AstraZeneca (Inst), Bayer (Inst), Eli Lilly (Inst), Innocrin Pharma (Inst), MedImmune (Inst), Pfizer (Inst), Roche (Inst), Seattle Genetics (Inst), Clovis Oncology (Inst), Bristol Myers Squibb (Inst), Advanced Accelerator Applications (Inst)
Expert Testimony: Celgene, Sanofi
Adam P. Dicker
Leadership: Department of Defense-Prostate Cancer Research Program, NRG Oncology, American Society for Radiation Oncology
Stock and Other Ownership Interests: Oncohost, Self Care Catalyst
Consulting or Advisory Role: EMD Serono, Janssen Oncology, Self Care Catalyst, Celldex, Johnson & Johnson, Roche, Apex, Cybrexa Therapeutics, Oncohost, Thirdbridge, Accordant
Research Funding: Prostate Cancer Foundation
Patents, Royalties, Other Intellectual Property: Recently filed patent “Doped BEO Compounds for Optically Stimulated Luminescence (OSL) and Thermoluminescence (TL) Radiation Dosimetry”
Expert Testimony: Wilson, Socini
Travel, Accommodations, Expenses: Merck, Ferring Pharmaceuticals, Self Care Catalyst, EMD Serono, Oncohost
Other Relationship: Dreamit Ventures
Uncompensated Relationships: Google
Colin C. Pritchard
Consulting or Advisory Role: Promega
Curtis A. Pettaway
Consulting or Advisory Role: Wolters-Kluwer
Research Funding: Beckmann-Coulter, MDxHealth
James L. Mohler
Patents, Royalties, Other Intellectual Property: Mohler JL, Fiandalo M, Watt D, Sviripa V: Compounds and methods to impair androgen receptor (AR) activation, impair dimerization, and/or impair AR transregulation. US provisional patent application 62/839,676, filed 4/27/2019, by Health Research & University of Kentucky Research Foundation (Inst); Mohler JL, Fiandalo M, Watt D, Sviripa V: Inhibitors of androgen receptor activation and methods of making and using same. US provisional patent application 62/890,292, filed 8/22/2019, by Health Research & University of Kentucky Research Foundation (Inst); Mohler JL, Fiandalo M, Watt D, Sviripa V: Spirocyclic dihydrotestosterone as ligand for proteolysis chimeras for AR degradation, imaging agents, and screening tools for the treatment of prostate cancer. US provisional patent application 62/844,062, filed 5/6/2019, by Health Research & University of Kentucky Research Foundation (revised; Inst)
J. Kellogg Parsons
Stock and Other Ownership Interests: Urigen, Pfizer, Johnson & Johnson, Omega Healthcare Investors
Honoraria: Sophiris Bio
Travel, Accommodations, Expenses: Sophiris Bio
Other Relationship: MDxHealth
Peter R. Carroll
Honoraria: Intuitive Surgical
Consulting or Advisory Role: Nutcracker Therapeutics
Amie Blanco
Employment: Biomarin (I)
Stock and Other Ownership Interests: Biomarin (I)
Ashley Woodson
Employment: Genome Medical
Travel, Accommodations, Expenses: Genome Medical
Mary-Ellen Taplin
Honoraria: Janssen-Ortho, Clovis Oncology, Astellas Pharma, Incyte, UpToDate, Research to Practice, Pfizer, Bayer, Amgen, AstraZeneca, Progenics, Guidepoint Global, Celegen, Merck
Consulting or Advisory Role: Janssen-Ortho, Bayer, Guidepoint Global, Best Doctors, UpToDate, Clovis Oncology, Research to Practice, Myovant Sciences, Incyte, Pfizer, AstraZeneca
Research Funding: Janssen-Ortho (Inst), Medivation (Inst), Bayer (Inst), Pfizer (Inst)
Travel, Accommodations, Expenses: Medivation, Janssen Oncology, Tokai Pharmaceuticals, Astellas Pharma, Incyte, Pfizer, Clovis Oncology, Bayer
Thomas J. Polascik
Honoraria: Endocare
Brian T. Helfand
Speakers' Bureau: Exact Sciences, Ambry Genetics
Colette Hyatt
Employment: GenomeSmart
Stock and Other Ownership Interests: GenomeSmart
Consulting or Advisory Role: GenomeSmart
Alicia K. Morgans
Honoraria: Genentech, Janssen Oncology, Sanofi, AstraZeneca, Astellas Scientific and Medical Affairs, Astellas Colombia, Janssen Oncology, Bayer
Consulting or Advisory Role: Genentech, AstraZeneca, Sanofi, Bayer, Astellas Pharma, Janssen Oncology
Research Funding: Bayer, Seattle Genetics, Astellas Pharma, Genentech, AstraZeneca
Travel, Accommodations, Expenses: Sanofi
Felix Feng
Leadership: PFS Genomics
Stock and Other Ownership Interests: PFS Genomics, Nutcracker Therapeutics, SerImmune
Honoraria: Genentech
Consulting or Advisory Role: Bayer, Blue Earth Diagnostics, Celgene, Medivation, Astellas Pharma, Sanofi, Genzyme, EMD Serono, Janssen Biotech
Research Funding: Zenith Epigenetics
Patents, Royalties, Other Intellectual Property: Develop a molecular signature to predict radiation resistance in breast cancer, and this signature was patented by the University of Michigan; in the process of being licensed to PFS Genomics, a company that the author helped found (Inst)
Jacqueline Powers
Employment: Carevive Systems
Honoraria: CureConnect, Myriad Genetics
Consulting or Advisory Role: Carevive Systems
Travel, Accommodations, Expenses: Hospital of the University of Pennsylvania
Raoul Concepcion
Honoraria: Clovis Oncology, InVitae
Consulting or Advisory Role: IntegraConnect
Speakers' Bureau: Astellas Medivation, Janssen Oncology, Dendreon, Amgen
Daniel W. Lin
Consulting or Advisory Role: Astellas Pharma, Clovis Oncology, Dendreon
Research Funding: Genomic Health (Inst), GenomeDx (Inst), MDxHealth (Inst), Magforce
Anthony Costello
Honoraria: Sandoz
Arthur L. Burnett
Honoraria: Myriad Genetics, Novartis Pharmaceuticals, Futura Medical, Astellas Pharma, Boston Scientific (Inst)
Consulting or Advisory Role: Myriad Genetics, Novartis Pharmaceuticals, Futura Medical, Astellas Pharma
Patents, Royalties, Other Intellectual Property: Patents for the development of devices that may be used in pelvic surgeries, such as penile prosthesis implantation
Uncompensated Relationships: Comphya, Reflexonic
Oliver Sartor
Stock and Other Ownership Interests: Eli Lilly, GlaxoSmithKline, AbbVie, Cardinal Health, United Health Group, Varian Medical Systems, PSMA Therapeutics
Consulting or Advisory Role: Bayer, Johnson & Johnson, Sanofi, AstraZeneca, Dendreon, Endocyte, Constellation Pharmaceuticals, Advanced Accelerator Applications, Pfizer, Bristol Myers Squibb, Bavarian Nordic, EMD Serono, Astellas Pharma, Progenics, Noxo, Blue Earth Diagnostics, Myovant, Myriad Genetics, Novartis, Clovis Oncology, Novartis
Research Funding: Bayer (Inst), Johnson & Johnson (Inst), Sanofi (Inst), Endocyte (Inst), Innocrin Pharma (Inst), Merck (Inst), InVitae (Inst), Constellation Pharmaceuticals (Inst), Advanced Accelerator Applications (Inst), AstraZeneca (Inst), Dendreon (Inst), SOTIO
Expert Testimony: Sanofi
Travel, Accommodations, Expenses: Bayer, Johnson & Johnson, Sanofi, AstraZeneca, Progenics
William B. Isaacs
Honoraria: AstraZeneca
Travel, Accommodations, Expenses: AstraZeneca
Jianfeng Xu
Patents, Royalties, Other Intellectual Property: US9534256 B2: Methods and compositions for correlating genetic markers with risk of aggressive prostate cancer; US9534256 B2: Methods and compositions for correlating genetic markers with risk of aggressive prostate cancer; US9732389 B2: Methods and compositions for correlating genetic markers with prostate cancer risk; informal title: 33 SNPs for PCa risk
Jeffrey Weitzel
Speakers' Bureau: AstraZeneca
Himisha Beltran
Consulting or Advisory Role: Janssen Oncology, Genzyme, GlaxoSmithKline, AbbVie, Astellas Pharma, AstraZeneca, Pfizer
Research Funding: Janssen Oncology (Inst), AbbVie (Inst), Stemcentrx (Inst), Eli Lilly (Inst)
Travel, Accommodations, Expenses: Janssen Oncology
Alberto Briganti
Consulting or Advisory Role: Astellas Pharma, Janssen-Cilag, OPKO Health, MDxHealth, Ferring Pharmaceuticals
Speakers' Bureau: Astellas Pharma
Research Funding: Sandoz-Novartis, Merck Sharp & Dohme
David Y. T. Chen
Stock and Other Ownership Interests: Pfizer, Pfizer (I)
Robert B. Den
Employment: Alpha TAU
Albert Dobi
Patents, Royalties, Other Intellectual Property: Inventor of the ERG monoclonal antibody 9FY, licensed by the Biocare Medical; inventor of a urine biomarker panel, licensed by Exosome Diagnostics
E. David Crawford
Speakers' Bureau: Bayer, Ferring Pharmaceuticals
James Eastham
Stock and Other Ownership Interests: 3D Biopsy
Scott Eggener
Consulting or Advisory Role: Sophiris Bio, Francis Medical, InSightec, Profound Medical
Speakers' Bureau: Janssen Pharmaceuticals
Travel, Accommodations, Expenses: Janssen Biotech, InSightec, Sophiris Bio
Uncompensated Relationships: Steba Biotech
Marc Garnick
Stock and Other Ownership Interests: Immunogen, Exelixis, Dr. Consulta (Sao Paulo Brazil), Myovant
Consulting or Advisory Role: Dr. Consulta (Sao Paulo Brazil), Eli Lilly, Amag, Steba Biotech, Agile Therapeutics, Janssen Oncology, Karyop
Expert Testimony: Fitzpatrick Cella Harper and Scinto, US Department of Justice, Meyers ad Flowers
Nathan Handley
Research Funding: Nektar Therapeutics (Inst)
Mark D. Hurwitz
Honoraria: Pyrexar
Consulting or Advisory Role: Neotherma
Speakers' Bureau: Pyrexar
Patents, Royalties, Other Intellectual Property: Provision patent holder for hyperthermia delivery system
R. Jeffrey Karnes
Patents, Royalties, Other Intellectual Property: GenomeDx
Travel, Accommodations, Expenses: GenomeDx
Lucia Languino
Stock and Other Ownership Interests: Johnson & Johnson
Stacy Loeb
Stock and Other Ownership Interests: Gilead Sciences (I)
Consulting or Advisory Role: Bayer, Lumenis
Travel, Accommodations, Expenses: Sanofi
Grace Lu-Yao
Employment: Sun Pharma Advanced Research Company (I)
Leadership: Sun Pharma Advanced Research Company (I)
Stock and Other Ownership Interests: Merck (I)
Todd Morgan
Consulting or Advisory Role: Myriad Genetics, TerumoBCT
Research Funding: Myriad Genetics (Inst), MDxHealth (Inst), GenomeDx (Inst)
Jose Moreno
Stock and Other Ownership Interests: Illumina, Invitae, ThermoFisher Scientific, Exact Sciences, Guardant Health, Bio-Techne
Research Funding: Janssen Pharmaceuticals, Pfizer, Pillar Biosciences
Lorelei Mucci
Research Funding: Sanofi (Inst), Astellas Pharma (Inst), Bayer (Inst), Janssen Pharmaceuticals (Inst)
Ronald E. Myers
Consulting or Advisory Role: Exact Sciences
Sarah M. Nielsen
Employment: Invitae
Stock and Other Ownership Interests: Invitae
Consulting or Advisory Role: AstraZeneca, Merck, Myriad Genetics
Speakers' Bureau: AstraZeneca
Travel, Accommodations, Expenses: Myriad Genetics, AstraZeneca, Invitae
Peter Pinto
Patents, Royalties, Other Intellectual Property: Royalties from US Patent No. 8948845: “System, methods, and instrumentation for image guided prostate treatment”, with inventors/authors Brad Wood and Peter Pinto; the National Institutes of Health (NIH) and Philips (InVivo) have a licensing agreement. NIH does not endorse or recommend any commercial products, processes, or services. The views and personal opinions of authors expressed herein do not necessarily state or reflect those of the US Government, nor any official recommendation or opinion of the NIH or National Cancer Institute.
Wendy Poage
Employment: Servier
Stock and Other Ownership Interests: 3D Biopsy
Honoraria: Janssen Oncology, Myriad Genetics
Travel, Accommodations, Expenses: Pfizer, Dendreon, Janssen Oncology, Myriad Genetics
Ganesh V. Raj
Stock and Other Ownership Interests: EtiraRx,C-Diagnostics
Honoraria: Medivation, Janssen Biotech, Sanofi, Astellas Pharma
Consulting or Advisory Role: Pfizer, Bayer
Speakers' Bureau: Astellas Pharma
Research Funding: Janssen Biotech, Bayer
Patents, Royalties, Other Intellectual Property: Licensing
Timothy R. Rebbeck
Honoraria: AstraZeneca (I)
Consulting or Advisory Role: AstraZeneca (I)
Charles Ryan
Honoraria: Janssen Oncology, Bayer
Consulting or Advisory Role: Bayer, Dendreon, AAA
Research Funding: Clovis Oncology (Inst), Sanofi (Inst), Genzyme (Inst)
Howard Sandler
Stock and Other Ownership Interests: Radiogel
Consulting or Advisory Role: Janssen Pharmaceuticals
Other Relationship: Caribou Publishing
E. Michael D. Scott
Employment: Johnson & Johnson (I), Ex Archa, Calcium USA
Consulting or Advisory Role: Vavotar Life Sciences
Travel, Accommodations, Expenses: Vavotar Life Sciences
Other Relationship: International Myeloma Foundation, Prostate Cancer International
William Tester
Honoraria: DAVA Pharmaceuticals
Consulting or Advisory Role: Janssen Oncology
Edouard J. Trabulsi
Consulting or Advisory Role: GenomeDx
Speakers' Bureau: Johnson & Johnson, Janssen Oncology, Astellas Medivation, Pfizer
Neha Vapiwala
Consulting or Advisory Role: Magellan HealthRx
Evan Y. Yu
Consulting or Advisory Role: Janssen Oncology, Bayer, Merck, AstraZeneca, Amgen, QED, Dendreon, Seattle Genetics, Pharmacyclics, Clovis Oncology, Advanced Accelerator Applications, Sanofi, AbbVie, Myovant Sciences
Research Funding: Dendreon (Inst), Merck (Inst), Seattle Genetics (Inst), Daiichi Sankyo (Inst), Taiho Pharmaceutical (Inst), Pharmacyclics (Inst)
Leonard G. Gomella
Consulting or Advisory Role: Janssen Oncology, Astellas Pharma, Pfizer, Clovis Oncology, Bayer
Patents, Royalties, Other Intellectual Property: Patents held by Thomas Jefferson University
No other potential conflicts of interest were reported.
REFERENCES
- 1.Giri VN, Hyatt C, Gomella LG. GT for men with prostate cancer: Navigating an expanding new world of genetic evaluation for precision therapy and precision management. J Clin Oncol. 2019;37:1455–1459. doi: 10.1200/JCO.18.02181. [DOI] [PubMed] [Google Scholar]
- 2.Cheng HH, Sokolova AO, Schaeffer EM, et al. Germline and somatic mutations in prostate cancer for the clinician. J Natl Compr Canc Netw. 2019;17:515–521. doi: 10.6004/jnccn.2019.7307. [DOI] [PubMed] [Google Scholar]
- 3.National Comprehensive Cancer Network NCCN Clinical Practice Guidelines in Oncology: Prostate cancer (version 4.2019) doi: 10.6004/jnccn.2010.0012. https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf [DOI] [PubMed]
- 4.Morgans AK, Szymaniak BM. Genetically-informed treatment for advanced and metastatic prostate cancer. Can J Urol. 2019;26:54–56. [PubMed] [Google Scholar]
- 5.Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375:443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlo MI, Giri VN, Antonarakis ES, et al. Evolving intersection between inherited cancer genetics and therapeutic clinical trials in prostate cancer: A white paper from the Germline Genetics Working Group of the Prostate Cancer Clinical Trials Consortium. JCO Precis Oncol. doi: 10.1200/PO.18.00060. 10.1200/PO.18.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter HB, Helfand B, Mamawala M, et al. Germline mutations in ATM and BRCA1/2 are associated with grade reclassification in men on active surveillance for prostate cancer. Eur Urol. 2019;75:743–749. doi: 10.1016/j.eururo.2018.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Comprehensive Cancer Network NCCN Clinical Practice Guidelines in Oncology: Genetic/familial high-risk assessment: Breast, ovarian, and pancreatic (version 3.2019) doi: 10.6004/jnccn.2021.0001. https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf [DOI] [PubMed]
- 9.National Comprehensive Cancer Network NCCN Clinical Practice Guidelines in Oncology: Prostate cancer early detection (version 1.2019) doi: 10.6004/jnccn.2010.0016. https://www.nccn.org/professionals/physician_gls/pdf/prostate_detection.pdf [DOI] [PubMed]
- 10.Page EC, Bancroft EK, Brook MN, et al. Interim results from the IMPACT study: Evidence for prostate-specific antigen screening in BRCA2 mutation carriers. Eur Urol. 2019;76:831–842. doi: 10.1016/j.eururo.2019.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Cancer Institute PDQ cancer information summaries: Genetics. https://www.cancer.gov/publications/pdq/information-summaries/genetics
- 12.Mersch J, Jackson MA, Park M, et al. Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer. 2015;121:269–275. doi: 10.1002/cncr.29041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan S, Jenkins MA, Win AK. Risk of prostate cancer in Lynch syndrome: A systematic review and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2014;23:437–449. doi: 10.1158/1055-9965.EPI-13-1165. [DOI] [PubMed] [Google Scholar]
- 14.National Cancer Institute Genetics of breast and gynecologic cancers (PDQ) –Health professional version. https://www.cancer.gov/types/breast/hp/breast-ovarian-genetics-pdq [PubMed]
- 15.National Cancer Institute Genetics of colorectal cancer (PDQ) –Health professional version. https://www.cancer.gov/types/colorectal/hp/colorectal-genetics-pdq [PubMed]
- 16.National Comprehensive Cancer Network NCCN Clinical Practice Guidelines in Oncology: Genetic/familial high-risk assessment: Colorectal (version 2.2019) https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf
- 17.Hall MJ, Forman AD, Pilarski R, et al. Gene panel testing for inherited cancer risk. J Natl Compr Canc Netw. 2014;12:1339–1346. doi: 10.6004/jnccn.2014.0128. [DOI] [PubMed] [Google Scholar]
- 18.Giri VN, Knudsen KE, Kelly WK, et al. Role of genetic testing for inherited prostate cancer risk: Philadelphia Prostate Cancer Consensus Conference 2017. J Clin Oncol. 2018;36:414–424. doi: 10.1200/JCO.2017.74.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abacan M, Alsubaie L, Barlow-Stewart K, et al. The global state of the genetic counseling profession. Eur J Hum Genet. 2019;27:183–197. doi: 10.1038/s41431-018-0252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paller CJ, Antonarakis ES, Beer TM, et al. Germline genetic testing in advanced prostate cancer, practices and barriers: Survey results from the Germline Genetics Working Group of the Prostate Cancer Clinical Trials Consortium. Clin Genitourin Cancer. 2019;17:275.e1–282.e1. doi: 10.1016/j.clgc.2019.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giri VN. Genetic education and practice considerations of non-genetic providers. Can J Urol. 2019;26:44–45. [PubMed] [Google Scholar]
- 22.Goggins M, Overbeek KA, Brand R, et al. Management of patients with increased risk for familial pancreatic cancer: Updated recommendations from the International Cancer of the Pancreas Early detection (CAPS) Consortium. Gut. 2020;69:7–17. doi: 10.1136/gutjnl-2019-319352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomella LG, Knudsen KE, Giri VN. Introduction to the 2019 Philadelphia Prostate Cancer Consensus Program: Implementation of genetic testing for inherited prostate cancer. Can J Urol. 2019;26:1–4. [PubMed] [Google Scholar]
- 24.National Cancer Institute Genetics of prostate cancer (PDQ) –Health professional version. https://www.cancer.gov/types/prostate/hp/prostate-genetics-pdq [PubMed]
- 25.Breast Cancer Linkage Consortium Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst. 1999;91:1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- 26.Thompson D, Easton DF, Breast Cancer Linkage Consortium Cancer incidence in BRCA1 mutation carriers. J Natl Cancer Inst. 2002;94:1358–1365. doi: 10.1093/jnci/94.18.1358. [DOI] [PubMed] [Google Scholar]
- 27.Tryggvadóttir L, Vidarsdóttir L, Thorgeirsson T, et al. Prostate cancer progression and survival in BRCA2 mutation carriers. J Natl Cancer Inst. 2007;99:929–935. doi: 10.1093/jnci/djm005. [DOI] [PubMed] [Google Scholar]
- 28.Nyberg T, Frost D, Barrowdale D, et al. Prostate cancer risks for male BRCA1 and BRCA2 mutation carriers: A prospective cohort study. Eur Urol. 2020;77:24–35. doi: 10.1016/j.eururo.2019.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards SM, Evans DG, Hope Q, et al. Prostate cancer in BRCA2 germline mutation carriers is associated with poorer prognosis. Br J Cancer. 2010;103:918–924. doi: 10.1038/sj.bjc.6605822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallagher DJ, Gaudet MM, Pal P, et al. Germline BRCA mutations denote a clinicopathologic subset of prostate cancer. Clin Cancer Res. 2010;16:2115–2121. doi: 10.1158/1078-0432.CCR-09-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorne H, Willems AJ, Niedermayr E, et al. Decreased prostate cancer-specific survival of men with BRCA2 mutations from multiple breast cancer families. Cancer Prev Res (Phila) 2011;4:1002–1010. doi: 10.1158/1940-6207.CAPR-10-0397. [DOI] [PubMed] [Google Scholar]
- 32.Castro E, Goh C, Olmos D, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol. 2013;31:1748–1757. doi: 10.1200/JCO.2012.43.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akbari MR, Wallis CJ, Toi A, et al. The impact of a BRCA2 mutation on mortality from screen-detected prostate cancer. Br J Cancer. 2014;111:1238–1240. doi: 10.1038/bjc.2014.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castro E, Goh C, Leongamornlert D, et al. Effect of BRCA mutations on metastatic relapse and cause-specific survival after radical treatment for localised prostate cancer. Eur Urol. 2015;68:186–193. doi: 10.1016/j.eururo.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 35.Na R, Zheng SL, Han M, et al. Germline mutations in ATM and BRCA1/2 distinguish risk for lethal and indolent prostate cancer and are associated with early age at death. Eur Urol. 2017;71:740–747. doi: 10.1016/j.eururo.2016.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyle J, Cooney KA. DNA repair genes: Contributions to prostate cancer predisposition and aggressiveness. Can J Urol. 2019;26:10–11. [PubMed] [Google Scholar]
- 37.Ewing CM, Ray AM, Lange EM, et al. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366:141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J, Lange EM, Lu L, et al. HOXB13 is a susceptibility gene for prostate cancer: Results from the International Consortium for Prostate Cancer Genetics (ICPCG) Hum Genet. 2013;132:5–14. doi: 10.1007/s00439-012-1229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salinas CA, Tsodikov A, Ishak-Howard M, et al. Prostate cancer in young men: An important clinical entity. Nat Rev Urol. 2014;11:317–323. doi: 10.1038/nrurol.2014.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bauer CM, Ray AM, Halstead-Nussloch BA, et al. Hereditary prostate cancer as a feature of Lynch syndrome. Fam Cancer. 2011;10:37–42. doi: 10.1007/s10689-010-9388-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dominguez-Valentin M, Sampson JR, Seppälä TT, et al. Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: Findings from the Prospective Lynch Syndrome Database. Genet Med. 2020;22:15–25. doi: 10.1038/s41436-019-0596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Angèle S, Falconer A, Edwards SM, et al. ATM polymorphisms as risk factors for prostate cancer development. Br J Cancer. 2004;91:783–787. doi: 10.1038/sj.bjc.6602007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer A, Wilhelm B, Dörk T, et al. ATM missense variant P1054R predisposes to prostate cancer. Radiother Oncol. 2007;83:283–288. doi: 10.1016/j.radonc.2007.04.029. [DOI] [PubMed] [Google Scholar]
- 44.Schumacher FR, Al Olama AAA, Berndt SI, et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018;50:928–936. doi: 10.1038/s41588-018-0142-8. [Erratum: Nat Genet 51:363, 2019] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cybulski C, Górski B, Debniak T, et al. NBS1 is a prostate cancer susceptibility gene. Cancer Res. 2004;64:1215–1219. doi: 10.1158/0008-5472.can-03-2502. [DOI] [PubMed] [Google Scholar]
- 46.Leongamornlert DA, Saunders EJ, Wakerell S, et al. Germline DNA repair gene mutations in young-onset prostate cancer cases in the UK: Evidence for a more extensive genetic panel. Eur Urol. 2019;76:329–337. doi: 10.1016/j.eururo.2019.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rusak B, Kluźniak W, Wokołorczykv D, et al. Inherited NBN mutations and prostate cancer risk and survival. Cancer Res Treat. 2019;51:1180–1187. doi: 10.4143/crt.2018.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Dai B, Ye D. CHEK2 mutation and risk of prostate cancer: A systematic review and meta-analysis. Int J Clin Exp Med. 2015;8:15708–15715. [PMC free article] [PubMed] [Google Scholar]
- 49.Conti DV, Wang K, Sheng X, et al. Two novel susceptibility loci for prostate cancer in men of African ancestry. J Natl Cancer Inst. 2017;109:djx084. doi: 10.1093/jnci/djx084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Southey MC, Goldgar DE, Winqvist R, et al. PALB2, CHEK2 and ATM rare variants and cancer risk: Data from COGS. J Med Genet. 2016;53:800–811. doi: 10.1136/jmedgenet-2016-103839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pakkanen S, Wahlfors T, Siltanen S, et al. PALB2 variants in hereditary and unselected Finnish prostate cancer cases. J Negat Results Biomed. 2009;8:12. doi: 10.1186/1477-5751-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mai PL, Best AF, Peters JA, et al. Risks of first and subsequent cancers among TP53 mutation carriers in the National Cancer Institute Li-Fraumeni syndrome cohort. Cancer. 2016;122:3673–3681. doi: 10.1002/cncr.30248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amadou A, Achatz MIW, Hainaut P. Revisiting tumor patterns and penetrance in germline TP53 mutation carriers: Temporal phases of Li-Fraumeni syndrome. Curr Opin Oncol. 2018;30:23–29. doi: 10.1097/CCO.0000000000000423. [Erratum: Curr Opin Oncol 31:52, 2019] [DOI] [PubMed] [Google Scholar]
- 54.Rana HQ, Gelman R, LaDuca H, et al. Differences in TP53 mutation carrier phenotypes emerge from panel-based testing. J Natl Cancer Inst. 2018;110:863–870. doi: 10.1093/jnci/djy001. [DOI] [PubMed] [Google Scholar]
- 55.Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;162:454. doi: 10.1016/j.cell.2015.06.053. [DOI] [PubMed] [Google Scholar]
- 56.Annala M, Struss WJ, Warner EW, et al. Treatment outcomes and tumor loss of heterozygosity in germline DNA repair-deficient prostate cancer. Eur Urol. 2017;72:34–42. doi: 10.1016/j.eururo.2017.02.023. [DOI] [PubMed] [Google Scholar]
- 57.Giri VN, Obeid E, Gross L, et al. Inherited mutations in males undergoing multigene panel testing for prostate cancer: Emerging implications for personalized prostate cancer genetic evaluation. JCO Precis Oncol. 2017 doi: 10.1200/PO.16.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Antonarakis ES, Lu C, Luber B, et al. Germline DNA-repair gene mutations and outcomes in men with metastatic castration-resistant prostate cancer receiving first-line abiraterone and enzalutamide. Eur Urol. 2018;74:218–225. doi: 10.1016/j.eururo.2018.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mateo J, Cheng HH, Beltran H, et al. Clinical outcome of prostate cancer patients with germline DNA repair mutations: Retrospective analysis from an international study. Eur Urol. 2018;73:687–693. doi: 10.1016/j.eururo.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giri VN, Hegarty SE, Hyatt C, et al. Germline genetic testing for inherited prostate cancer in practice: Implications for genetic testing, precision therapy, and cascade testing. Prostate. 2019;79:333–339. doi: 10.1002/pros.23739. [DOI] [PubMed] [Google Scholar]
- 61.Nicolosi P, Ledet E, Yang S, et al. Prevalence of germline variants in prostate cancer and implications for current genetic testing guidelines. JAMA Oncol. 2019;5:523–528. doi: 10.1001/jamaoncol.2018.6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lang SH, Swift SL, White H, et al. A systematic review of the prevalence of DNA damage response gene mutations in prostate cancer. Int J Oncol. 2019;55:597–616. doi: 10.3892/ijo.2019.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schweizer MT, Antonarakis ES, Bismar TA, et al. Genomic characterization of prostatic ductal adenocarcinoma identifies a high prevalence of DNA repair gene mutations. JCO Precis Oncol. 2019 doi: 10.1200/PO.18.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Isaacsson Velho P, Silberstein JL, Markowski MC, et al. Intraductal/ductal histology and lymphovascular invasion are associated with germline DNA-repair gene mutations in prostate cancer. Prostate. 2018;78:401–407. doi: 10.1002/pros.23484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pritchard CC. Molecular insights into the germline for prostate cancer initiation, progression, and aggressiveness. Can J Urol. 2019;26:24–26. [PubMed] [Google Scholar]
- 66.Cheng HH. Germline contributions to metastatic prostate cancer. Can J Urol. 2019;26:19–21. [PubMed] [Google Scholar]
- 67.AlDubayan SH. Considerations of multigene test findings among men with prostate cancer: Knowns and unknowns. Can J Urol. 2019;26:14–16. [PubMed] [Google Scholar]
- 68.Giusti RM, Rutter JL, Duray PH, et al. A twofold increase in BRCA mutation related prostate cancer among Ashkenazi Israelis is not associated with distinctive histopathology. J Med Genet. 2003;40:787–792. doi: 10.1136/jmg.40.10.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kirchhoff T, Kauff ND, Mitra N, et al. BRCA mutations and risk of prostate cancer in Ashkenazi Jews. Clin Cancer Res. 2004;10:2918–2921. doi: 10.1158/1078-0432.ccr-03-0604. [DOI] [PubMed] [Google Scholar]
- 70.Agalliu I, Gern R, Leanza S, et al. Associations of high-grade prostate cancer with BRCA1 and BRCA2 founder mutations. Clin Cancer Res. 2009;15:1112–1120. doi: 10.1158/1078-0432.CCR-08-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chandrasekar T, Gross L, Gomella LG, et al. Prevalence of suspected hereditary cancer syndromes and germline mutations among a diverse cohort of probands reporting a family history of prostate cancer: Toward informing cascade testing for men. Eur Urol Oncol. doi: 10.1016/j.euo.2019.06.010. [epub ahead of print on July 2, 2019] [DOI] [PubMed] [Google Scholar]
- 72.Pettaway CA. African American and Asian males: What do we know about germline predisposition to prostate cancer. Can J Urol. 2019;26:27–28. [PubMed] [Google Scholar]
- 73.Kwon DH, Borno HT, Cheng HH, et al. Ethnic disparities among men with prostate cancer undergoing GT. Urol Oncol. 2020;38:80.e1–80.e7. doi: 10.1016/j.urolonc.2019.09.010. [DOI] [PubMed] [Google Scholar]
- 74.Momozawa Y, Iwasaki Y, Hirata M, et al. Germline pathogenic variants in 7,636 Japanese patients with prostate cancer and 12,366 controls. J Natl Cancer Inst. 2019;2019:djz124. doi: 10.1093/jnci/djz124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pilarski R. Current prostate cancer genetic testing capabilities and considerations. Can J Urol. 2019;26:38–39. [PubMed] [Google Scholar]
- 76.Robson ME, Bradbury AR, Arun B, et al. American Society of Clinical Oncology policy statement update: Genetic and genomic testing for cancer susceptibility. J Clin Oncol. 2015;33:3660–3667. doi: 10.1200/JCO.2015.63.0996. [DOI] [PubMed] [Google Scholar]
- 77.Al Olama AA, Kote-Jarai Z, Berndt SI, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet. 2014;46:1103–1109. doi: 10.1038/ng.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benafif S, Kote-Jarai Z, Eeles RA. A review of prostate cancer genome-wide association studies (GWAS) Cancer Epidemiol Biomarkers Prev. 2018;27:845–857. doi: 10.1158/1055-9965.EPI-16-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Toland AE. Polygenic risk scores for prostate cancer: Testing considerations. Can J Urol. 2019;26:17–18. [PubMed] [Google Scholar]
- 80.Seibert TM, Fan CC, Wang Y, et al. Polygenic hazard score to guide screening for aggressive prostate cancer: Development and validation in large scale cohorts. BMJ. 2018;360:j5757. doi: 10.1136/bmj.j5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fritsche LG, Gruber SB, Wu Z, et al. Association of polygenic risk scores for multiple cancers in a phenome-wide study: Results from the Michigan Genomics Initiative. Am J Hum Genet. 2018;102:1048–1061. doi: 10.1016/j.ajhg.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Riley BD, Culver JO, Skrzynia C, et al. Essential elements of genetic cancer risk assessment, counseling, and testing: Updated recommendations of the National Society of Genetic Counselors. J Genet Couns. 2012;21:151–161. doi: 10.1007/s10897-011-9462-x. [DOI] [PubMed] [Google Scholar]
- 83.Hampel H, Bennett R, Buchanan A, et al. A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: Referral indications for cancer predisposition assessment. Genet Med. 2015;17:70–87. doi: 10.1038/gim.2014.147. [DOI] [PubMed] [Google Scholar]
- 84.Institute of Medicine (US) Committee on Assessing Genetic Risks Andrews LB, Fullarton JE, et al.(eds)Assessing Genetic Risks: Implications for Health and Social Policy Washington, DC: National Academies Press; 1994 [PubMed] [Google Scholar]
- 85.Stoll K, Kubendran S, Cohen SA. The past, present and future of service delivery in genetic counseling: Keeping up in the era of precision medicine. Am J Med Genet C Semin Med Genet. 2018;178:24–37. doi: 10.1002/ajmg.c.31602. [DOI] [PubMed] [Google Scholar]
- 86.Patrick-Miller L, Egleston BL, Daly M, et al. Implementation and outcomes of telephone disclosure of clinical BRCA1/2 test results. Patient Educ Couns. 2013;93:413–419. doi: 10.1016/j.pec.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schwartz MD, Valdimarsdottir HB, Peshkin BN, et al. Randomized noninferiority trial of telephone versus in-person genetic counseling for hereditary breast and ovarian cancer. J Clin Oncol. 2014;32:618–626. doi: 10.1200/JCO.2013.51.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gray J, Brain K, Iredale R, et al. A pilot study of telegenetics. J Telemed Telecare. 2000;6:245–247. doi: 10.1258/1357633001935329. [DOI] [PubMed] [Google Scholar]
- 89.Hilgart JS, Hayward JA, Coles B, et al. Telegenetics: A systematic review of telemedicine in genetics services. Genet Med. 2012;14:765–776. doi: 10.1038/gim.2012.40. [DOI] [PubMed] [Google Scholar]
- 90.Coelho JJ, Arnold A, Nayler J, et al. An assessment of the efficacy of cancer genetic counselling using real-time videoconferencing technology (telemedicine) compared to face-to-face consultations. Eur J Cancer. 2005;41:2257–2261. doi: 10.1016/j.ejca.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 91.d’Agincourt-Canning L, McGillivray B, Panabaker K, et al. Evaluation of genetic counseling for hereditary cancer by videoconference in British Columbia. BC Med J. 2008;50:554–559. [Google Scholar]
- 92.Trepanier AM, Allain DC. Models of service delivery for cancer genetic risk assessment and counseling. J Genet Couns. 2014;23:239–253. doi: 10.1007/s10897-013-9655-6. [DOI] [PubMed] [Google Scholar]
- 93.Buchanan AH, Rahm AK, Williams JL. Alternate service delivery models in cancer genetic counseling: A mini-review. Front Oncol. 2016;6:120. doi: 10.3389/fonc.2016.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mateo J, Carreira S, Sandhu S, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pomerantz MM, Spisák S, Jia L, et al. The association between germline BRCA2 variants and sensitivity to platinum-based chemotherapy among men with metastatic prostate cancer. Cancer. 2017;123:3532–3539. doi: 10.1002/cncr.30808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Abida W, Bryce AH, Vogelzang NJ, et al. Preliminary results from TRITON2: A phase 2 study of rucaparib in patients (Pts) with metastatic castration-resistant prostate cancer (mCRPC) associated with homologous recombination repair (HRR) gene alterations. Ann Oncol. 2018;29(suppl 8):viii271–viii302. [Google Scholar]
- 97.Castro E, Romero-Laorden N, Del Pozo A, et al. PROREPAIR-B: A prospective cohort study of the impact of germline DNA repair mutations on the outcomes of patients with metastatic castration-resistant prostate cancer. J Clin Oncol. 2019;37:490–503. doi: 10.1200/JCO.18.00358. [DOI] [PubMed] [Google Scholar]
- 98.European Society for Medical Oncology Olaparib outperforms enzalutamide or abiraterone acetate in men with mCRPC and HRR alterations. https://www.esmo.org/oncology-news/olaparib-outperforms-enzalutamide-or-abiraterone-acetate-in-men-with-mcrpc-and-hrr-alterations
- 99.Marshall CH, Sokolova AO, McNatty AL, et al. Differential response to olaparib treatment among men with metastatic castration-resistant prostate cancer harboring BRCA1 or BRCA2 versus ATM mutations. Eur Urol. 2019;76:452–458. doi: 10.1016/j.eururo.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu Y, Yu H, Zheng SL, et al. A comprehensive evaluation of CHEK2 germline mutations in men with prostate cancer. Prostate. 2018;78:607–615. doi: 10.1002/pros.23505. [DOI] [PubMed] [Google Scholar]
- 101.Helfand BT, Xu J. GT for prostate cancer prognosis: Implications for AS. Can J Urol. 2019;26:48–49. [PubMed] [Google Scholar]
- 102.Polascik TJ, Orabi H. Considerations of GT in prostate cancer early detection. Can J Urol. 2019;26:46–47. [PubMed] [Google Scholar]
- 103.Hyatt C, Russo J, McDougall C. Genetic counseling perspective of engagement with urology and primary care. Can J Urol. 2019;26:52–53. [PubMed] [Google Scholar]