Abstract
The emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) late December 2019 in Wuhan, China, marked the third introduction of a highly pathogenic coronavirus into the human population in the twenty-first century. The constant spillover of coronaviruses from natural hosts to humans has been linked to human activities and other factors. The seriousness of this infection and the lack of effective, licensed countermeasures clearly underscore the need of more detailed and comprehensive understanding of coronavirus molecular biology. Coronaviruses are large, enveloped viruses with a positive sense single-stranded RNA genome. Currently, coronaviruses are recognized as one of the most rapidly evolving viruses due to their high genomic nucleotide substitution rates and recombination. At the molecular level, the coronaviruses employ complex strategies to successfully accomplish genome expression, virus particle assembly and virion progeny release. As the health threats from coronaviruses are constant and long-term, understanding the molecular biology of coronaviruses and controlling their spread has significant implications for global health and economic stability. This review is intended to provide an overview of our current basic knowledge of the molecular biology of coronaviruses, which is important as basic knowledge for the development of coronavirus countermeasures.
Keywords: Cell biology, Microbiology, Genetics, Biochemistry, Molecular biology, Virology, Coronaviruses, MERS-CoV, SARS-CoV, SARS-CoV-2, Covid-19
Cell biology; Microbiology; Genetics; Biochemistry; Molecular Biology; Virology; coronaviruses; MERS-CoV; SARS-CoV; SARS-Cov-2; Covid-19
1. Introduction
1.1. Emergence of coronavirus diseases
1.1.1. Driving factors for coronavirus emergence
The unpredictable emergence of new infectious diseases can be seen as a threat to human health and global stability, despite extraordinary progress in development of countermeasures such as diagnostics, vaccines, and treatments. Diseases caused by coronaviruses are a few of many examples of emerging infectious diseases in the modern world (Morens and Fauci, 2013). Coronaviruses (CoVs) are emerging and re-emerging pathogens and several of them have caused serious problems in humans and animals (Lau and Chan, 2015). These include varying symptoms ranging from mild respiratory illness to severe infections causing death. Apart from the respiratory tract, coronaviruses can also affect other organs in the body, such as the gastrointestinal tract, liver, kidney, and brain of both humans and animals. The pandemic of severe acute respiratory syndrome (SARS) in 2002–2003, the emergence of Middle East respiratory syndrome (MERS) in 2012 and the emergence of a new coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causal agent of the coronavirus disease 2019 (COVID-19) pandemic, are all examples of human infections leading to significant fatality caused by coronaviruses (Anindita et al., 2015; Guarner, 2020; WHO, 2020).
Notably, the key features of the SARS-CoV, MERS-CoV, and SARS-CoV-2 are all similar in that they exhibit dominance of hospital-acquired infection, and pathogenesis driven by a combination of viral replication in the lower respiratory tract and an aberrant host immune response (de Wit et al., 2016; Wang et al., 2020a). In the laboratory, general recommended precautions for handling the highly pathogenic human coronaviruses include biosafety level 2 (BSL2) facilities for diagnosis and biosafety level 3 (BSL3) facilities for propagation (Artika and Ma'roef, 2017). However, in the situations when limited information is available on the newly emerged highly pathogenic coronaviruses, it is prudent to implement additional safeguards until more data are available for laboratory risk assessment (WHO, 2004; BMBL, 2009). In addition, it is important to note that although coronaviruses are enveloped viruses, this does not mean that they are necessarily fragile or quickly inactivated. Coronavirus particles are relatively robust compared to HIV-1. SARS-CoV particles for example, remain infectious for 1–4 days on the relatively harsh environment of hard surfaces. MERS-CoV virions are slightly more fragile than SARS-CoV, with half lives of approximately one hour on hard surfaces and a maximum survival time of 2–3 days. However, MERS-CoV virions are much more robust than the pandemic influenza A virus under the same conditions. The evidence of persistent infectivity of coronaviruses outside the body suggests that direct contact with contaminated surfaces and respiratory droplets is a likely route of MERS-CoV spread (Neuman and Buchmeier, 2016).
Population shift from rural areas to urban areas, and the increasingly frequent mixing of different animal species in densely populated areas, have been thought to facilitate the emergence and re-emergence of some coronaviruses (Lau and Chan, 2015). Increased contact with wild life in developing regions, greater levels of international travel and trade, and different land use have also been found as contributing factors for the rapid emergence of pathogenic viruses (Rosenberg et al., 2013). The nature of viral genetic material has also been suggested to influence the propensity for emergence. About 85% of emerging viruses have single-stranded RNA (ssRNA) genomes, which are prone to uncorrected errors during replication (Rosenberg et al., 2013). In general, the rate of error during RNA replication (about 10−4) is greater than that of DNA (about10−5). In contrast to DNA polymerase, the RNA polymerase which catalyzes the replication of RNA molecule does not have the proofreading capabilities nor post-replication mismatch repair mechanisms. Consequently, the potential for mutation per replication cycle of an RNA genome is high (Rosenberg, 2015). Coronaviruses possess genomic material in the form of single-stranded RNA and have been found to have high mutation and recombination rates, which might allow them to cross species barriers and adapt to new hosts (Lau and Chan, 2015). Today, coronaviruses are known as one of the most rapidly evolving viruses due to their high genomic nucleotide substitution and recombination rates (Lim et al., 2016). SARS viruses, for example, have the capacity to be directly transmitted from animals to humans (Rosenberg, 2015). The evolution of coronaviruses is also a result of their interaction with their hosts. For example, it was reported that the host shift of SARS related coronaviruses (SARSr-CoVs) mostly occurred in different species under the same genus Rhinolophus, indicating that genetic distance between hosts also determines both the host shift and the cross-species transmission of the viruses (Yu et al., 2019).
Some Asian regions are considered as hot spots of viral disease emergence especially the areas of rapid social and environmental change (Horby et al., 2013). For example, the SARS-CoV emerged in Guangdong, China, and then spread to many countries in South East Asia, North America, Europe, and South Africa. Transmission from person to person occurred through droplets, personal contact, or by touching contaminated surfaces. Health professionals, in particular, were reported to be at a high risk of acquiring the disease, as transmission also occurred when isolation precautions were inadequate. The last case of SARS-CoV occurred in September 2003, after having infected over 8,096 persons in 11 countries and causing 774 deaths with a case fatality rate of 9.5% (Luk et al., 2019; Guarner, 2020). The SARS-CoV-2, the etiological agent of COVID-19, emerged in Wuhan, China, at the end of 2019. As of 12 August 2020, the virus has affected more than 200 countries around the world with human cases of more than 20,162,000 and of more than 737,000 deaths (WHO, 2020).
China, in particular, has been predicted by scientists as a region of high potential for pathogenic coronaviruses emergence. This prediction was made based on the association between coronavirus species, bat species, and geographical location in China which potentially lead to cross-species transmission of coronaviruses (Fan et al., 2019). Bats are now regarded as important reservoir hosts of coronaviruses. Prior to the emergence of the SARS-CoV-2 in Wuhan in late 2019, two highly pathogenic coronaviruses of bat origin, the SARS-CoV and the swine-acute-diarrhea-syndrome coronavirus (SADS-CoV) have emerged in China over the past two decades. They caused large-scale disease outbreaks in humans and pigs, respectively. Apart from being the most populous nation in the world, China is the third largest territory with great biodiversity including bats and bat-borne viruses. The majority of the currently identified coronaviruses can be found in China. Moreover, most of the bat hosts of these coronaviruses live in close proximity to humans. According to Chinese food culture, freshly slaughtered animals are more nutritious. This may increase the potential of coronavirus transmission to humans. In particular, the bat SARS related coronaviruses capable of using the human angiotensin-converting enzyme 2 (ACE2) as a receptor are considered to pose a direct threat to humans. Astonishingly, all of the SARS related coronaviruses which are capable of using human ACE2 are found in China. Therefore, it is generally believed that bat-borne coronaviruses will re-emerge to cause future disease outbreaks and China is a likely hotspot (Fan et al., 2019).
The Southeast Asian region is also considered to be susceptible to coronavirus emergence. For instance, from 1 March 2003 to 11 May 2003, a SARS outbreak occurred in Singapore and a total of 206 probable SARS cases were diagnosed. The outbreak was the most severe infectious disease to challenge the public health system of Singapore (Tan, 2006). MERS-CoV infections linked to travel in the Middle East were reported to occur in Malaysia and the Philippines. In addition, a MERS-CoV infection associated to visiting Thailand was also detected in an Omani citizen (Setianingsih et al., 2019). In Indonesia, infection by human coronavirus 229E was detected in samples from 1 out of 13 hospitalized patients suspected of MERS-CoV infection who were admitted to an infectious disease hospital in Jakarta from July 2015 to December 2016 (Setianingsih et al., 2019). Infections of human coronaviruses NL63 and 229E have also been reported in Malaysia. The SARS-CoV-2, which emerged in Wuhan, has also been identified in many Southeast Asian countries including Indonesia, Malaysia, Philippines, Thailand, Viet Nam, Brunei Darussalam, Cambodia, and Timor-Leste (WHO, 2020). In addition, bats harboring coronaviruses have been discovered in the Philippines, Thailand and Indonesia (Anindita et al., 2015). Avian coronavirus, the main representative of the genus Gammacoronavirus, has recently been isolated from the Eclectus parrot (Eclectus roratus) in Indonesia (Suryaman et al., 2019).
1.1.2. Cross-species transmission of coronaviruses
Cross-species transmission has been known to play an important role in the emergence of viral diseases. For example, viruses from wildlife hosts have caused high-impact diseases such as severe acute respiratory syndrome (SARS), Ebola fever, and influenza in humans. The emergence of many human diseases has occurred when established animal viruses switch hosts into humans and then are transmitted within human populations (Parrish et al., 2008). In general, there are at least four major criteria which determine the successful cross-species transmission of a particular virus: the availability of susceptible host cells which have the specific receptor required for viral entry; permissiveness of these host cells to permit the virus to replicate and complete their replication cycle; accessibility of susceptible and permissive cells in the host; and the inability of the host cells’ innate immune response to restrict the viral replication (Hulswit et al., 2016). Most of the emerging viruses are zoonotic, in that, they can be transmitted from animals to humans (Morens and Fauci, 2013). Biological, ecological and epidemiological factors have been suggested to determine the successful cross-species transmission. The high frequency with which RNA viruses jump species boundaries in part reflects their ability to rapidly generate important adaptive variation. As RNA viruses, coronaviruses seem to exhibit a strong zoonotic potential (Leopardi et al., 2018). Host switching has been shown to contribute to coronavirus evolution and the diversity of coronaviruses may be associated with the potential risk of zoonotic emergence (Anthony et al., 2017).
Although the majority of individual virus species seems to be restricted to a narrow host range of a single animal species, genome sequencing and phylogenetic analyses indicate that coronaviruses have often crossed the host-species barrier. Bats harbor great coronavirus genetic diversity. The majority, if not all of coronaviruses which infect humans are believed to originate from bat coronaviruses which are transmitted to humans directly or indirectly through an intermediate host (Hu et al., 2015; Hulswit et al., 2016). The emergence of SARS-CoV, MERS-CoV, and SARS-CoV-2 underpin the threat of cross-species transmission events resulting in outbreaks in humans (Menachery et al., 2015; Lu et al., 2020). Prior to the outbreak of SARS-CoV in 2002–2003, only two human coronaviruses, the HCoV-OC43 and HCoV-229E, were known. They were identified in the 1960s. The emergence of SARS-CoVs sparked the search for novel coronaviruses and led to the identification of HCoV-NL63 in 2004 and HCoV-HKU1 in 2005. The common human CoVs are generally not considered to be highly pathogenic and are associated with relatively mild clinical symptoms in immunocompetent individuals and cause a self-limiting upper respiratory tract disease. In some cases, they may also cause a more severe infection in the lower respiratory tract. It is reported that young, elderly, and immunocompromised individuals are the most susceptible to the coronavirus infections (McBride and Fielding, 2012; Enjuanes et al., 2016). A list of important coronaviruses pathogenic to humans is presented in Table 1 (Lim et al., 2016; Cui et al., 2019; Chen et al., 2020; WHO, 2020; Yee et al., 2020).
Table 1.
Human pathogenic coronaviruses.
| Virus | Genus | Natural Host | Year of discovery | Symptoms |
|---|---|---|---|---|
| HCoV-229E | α-coronavirus | Bats | 1966 | Mild respiratory tract infections |
| HCoV-NL63 | α-coronavirus | Bats | 2004 | Mild respiratory tract infections |
| HCoV-OC43 | β-coronavirus | Rodents | 1967 | Mild respiratory tract infections |
| HCoV-HKU1 | β-coronavirus | Rodents | 2005 | Pneumonia |
| SARS-CoV | β-coronavirus | Bats | 2003 | Severe acute respiratory syndrome, 10% fatality rate |
| MERS-CoV | β-coronavirus | Bats | 2012 | Severe acute respiratory syndrome, 37% fatality rate |
| SARS-CoV-2 | β-coronavirus | Bats? | 2019 | Severe acute respiratory syndrome, 3.7% fatality rate |
The SARS-CoV, MERS-CoV and the SARS-CoV-2 are three highly transmissible pathogens that emerged in human over the past 2 decades (Cui et al., 2019; Andersen et al., 2020). In the case of SARS-CoV, it is most likely that the virus originated from bats through sequential recombination of bat (SARSr-CoVs) and that masked palm civets (Paguma larvata) were intermediate hosts. It is thought that recombination occurred in bats before SARS-CoV was introduced into Guangdong Province through infected civets or other infected mammals from Yunnan (Cui et al., 2019). Epidemiological studies indicated that civets from live animal markets in Guangdong Province, China, played an important role for human exposure to SARS-CoV. However, most of the masked palm civets from the wild, or from farms, were negative for SARS-CoV, indicating that those palm civets were not a reservoir, but intermediate hosts for the SARS-CoV (Su et al., 2016). Subsequent investigations have found that wild horseshoe bats (Rhinolophidae family), which are also present in live animal markets in China, have detectable levels of antibodies against SARS-CoV and also a SARS-CoV-like virus, suggesting that SARS-CoV originated in bats. An evolutionary hypothesis was then proposed that the ancestor for SARS-CoV first spread to bats of the Hipposideridae family, then bats of the Rhinolophidae family, then to masked palm civets and eventually humans (Su et al., 2016). Following studies suggested that Chinese horseshoe bats are the natural reservoirs of SARS-CoV and intermediate hosts might not be needed for direct human infection (Su et al., 2016). Similarly, recent molecular epidemiological studies involving 339 SARS-CoV and SARSr-CoV genome sequences including 274 from human and 18 from civets (collected in 2003/2004) and 47 from bats (continuously isolated for the past 13 years after the SARS epidemic) concluded that the human SARS-CoV was a result of multiple recombination events from a number of SARSr-CoV ancestors in different horseshoe bats species (Luk et al., 2019).
Similarly, MERS-CoV is also believed to have originated in bats. While palm civets have been linked to the emergence of SARS, dromedary camels were suggested to play roles as intermediate host for the emergence of MERS-CoV. The majority of the MERS index cases were reported to have contact with camels. Moreover, MERS-CoV strains isolated from camels were almost identical to those isolated from humans (Cui et al., 2019). As some confirmed cases lacked a contact history with camels, it has been suggested that there has been direct human-to-human MERS-CoV transmission, or through contact with a yet-to-be-identified animal species which maintained as a reservoir of MERS-CoV. Furthermore, studies on HKU4, a coronavirus of bat origin and the most phylogenically closely related to MERS-CoV, showed that HKU4 has the ability to utilize the dipeptidyl peptidase 4 (DPP4) receptor for virus entry. As DPP4 is a known receptor for MERS-CoV, the similarity in receptor specificity of these two CoVs supports the hypothesis that MERS-CoVs is of bat-origin. However, live MERS-CoV has yet to be isolated from wild bats (Su et al., 2016).
In the case of SARS-CoV-2, a number of studies have been carried out in order to investigate the original host of the virus. Again, bats have been suggested as likely reservoir hosts (Lu et al., 2020; Zhou et al., 2020) and pangolins have been suggested as a possible hosts in the emergence of the SARS-CoV-2 (Lam et al., 2020). Although bats are the likely reservoir hosts for this virus, their general ecological separation from humans implies that other mammalian species may act as ‘‘intermediate’’ or ‘‘amplifying’’ hosts (Zhang and Holmes, 2020). In addition, the possibility that the virus originated from a laboratory has also been critically analyzed (Andersen et al., 2020). As the virus is newly discovered, currently, the spectrum of available diagnostic tools is tight. More studies are needed to elucidate its origin, tropism, and pathogenesis (Phan, 2020). Further discussion on molecular characteristics of SARS-CoV-2 is presented in Section 4.
One of the important factors linked to the ability of viruses to cross the species barrier is the accumulation of mutations in their genomes (Djikeng and Spiro, 2009). Cross-species transmission may also be facilitated by homologous recombination events which radically alter or cause deletions in viral RNA genomes (Rowe et al., 1997; Ji et al., 2020). For the SARS-CoVs, comparison of genome sequences of the viruses from market civets and humans revealed that they are almost identical. However, two genes, the S and ORF8, were found to show major variation. Two amino acid residues (479 and 487) in the receptor binding domain of the S gene were found to be important for ACE2 receptor-mediated infection of the SARS-CoV and for the virus transmission from civets to humans (Cui et al., 2019; Yu et al., 2019). In addition, the ORF8 protein was indicated to be important for interspecies transmission, as most human SARS-CoV epidemic strains harbor a signature 29-nucleotide deletion in ORF8 compared to civet SARSr-CoVs. The deletion leads to generation of two different open reading frames, ORF 8a and 8b (Fan et al., 2019). Comparison of full-length genomic sequences of MERS-CoVs isolated from humans and camels also showed that the two genomes are almost identical. Variations were found in S, ORF4b, and ORF3 genes. Notably, although several amino acid substitutions were observed in the S protein, none of them was located in the receptor binding domain (Cui et al., 2019).
As constant cross-species transmission of coronaviruses from animal hosts to human occurs, and this is mainly due to human activities, such as modern agricultural practices, frequent interactions of wild animals with humans and urbanization, it is therefore of great importance to maintain the barrier between natural reservoirs and human society in order to effectively prevent viral zoonosis (Cui et al., 2019; Phan, 2020). In addition, comprehensive studies of bat-borne coronaviruses are critical for mitigating, predicting, and preventing future zoonotic coronavirus outbreaks (Hu et al., 2015). Although it has become increasingly clear that bats are important reservoirs of coronaviruses, currently only 6% of all coronavirus sequences in GenBank are from bats. The rest 94% pimarily consist of known pathogens of public health or agricultural significance, which indicates that current studies are heavily biased towards describing known diseases rather than the ‘pre-emergent’ potential pool in bats (Anthony et al., 2017).
2. Molecular characteristics of coronaviruses
2.1. Virion and ribonucleoprotein
Coronaviruses are members of family Coronaviridae, order Nidovirales. These enveloped viruses possess genomes in the form of single-stranded RNA molecules of positive sense, that is, the same sense as the messenger RNA (mRNA). At present, four genera are known: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, Deltacoronavirus. Members of the genera Alphacoronavirus and Betacoronavirus are identified to cause human disease, whereas those of the genera Gammacoronavirus and Deltacoronavirus are causative agents of animal disease (Masters, 2006; Anindita et al., 2015).
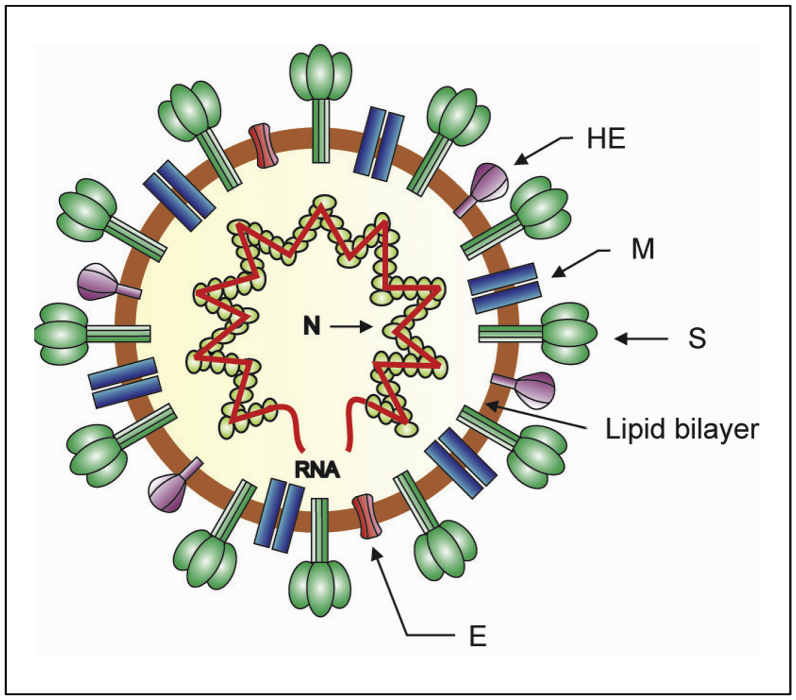
Coronaviruses have a typical characteristic in negative-stained electron microscopy showing a fringe on their surface structure like a spike. This fringe resembles the solar corona, from which the name coronavirus was derived (Masters, 2006). These viruses are roughly spherical with average diameter of 80–120 nm. The surface spikes of the coronaviruses projects about 17–20 nm from the surface of the virus particle and have been described as club-like, pear-shaped, or petal-shaped, having a thin base which swells to a width of approximately 10 nm at the distal extremity (Masters, 2006). A schematic visualization of the coronavirus virion is presented in Figure 1. In infection, the coronavirus particle serves three important functions for the genome: first, it provides the means to deliver the viral genome across the plasma membrane of a host cell; second, it serves as a means of escape for the newly synthesized genome; third, the viral particle functions as a durable vessel which protects the genome integrity on its journey between cells (Neuman and Buchmeier, 2016).
Figure 1.
Schematic diagram of the coronavirus virion. Together with membrane (M) and envelope (E) transmembrane proteins, the spike (S) glycoprotein projects from a host cell-derived lipid bilayer, giving the virion a distinctive appearance. The haemagglutinin esterase (HE) forms small spikes which appear under the tall S protein spikes. The positive-sense viral genomic RNA is associated with the nucleocapsid phosphoprotein (N) forming the ribonucleoprotein with a helical structure (Masters, 2006; de Wit et al., 2016).
Investigation of the internal component of the coronavirus conducted using virions which have burst spontaneously and expelled their content, or using virions which have been treated using detergents, showed that the viruses possess helically symmetric nucleocapsids. Of note, such nucleocapsid symmetry is generally formed by viruses having negative-strand RNA. To the contrary, almost all animal viruses with positive-strand RNA have icosahedral ribonucleoprotein capsids. Although it is generally accepted that coronaviruses have helical nucleocapsids of 14–16 nm in diameter, other studies employing different virus species and methods of preparation, have reported different results such as filamentous structures of 9–11 nm or 11–13 nm in diameter, or a linear strand of 6–7 μm long which may represent unwound helices, etc (Masters, 2006). More recent studies using cryo-electron microscopy to investigate the structural organization of SARS-CoV showed that the ribonucleoprotein particles form a coiled shape, packaged in spherical form with no indication of icosahedral symmetry (Chang et al., 2014). Electron microscopic studies of ribonucleoprotein of mouse hepatitis virus (MHV), also a betacoronavirus, showed that the ribonucleoproteins are in either a loose filamentous structure or in a compact flower-like assembly (Gui et al., 2017).
The genome of the coronaviruses codes four main structural proteins: the spike (S) protein, the nucleocapsid (N) protein, the membrane (M) protein and the envelope (E) protein, each of which play primary roles in the structure of the virus particle as well as in other aspects of the viral replication cycle. Generally, all of these proteins are needed to form a structurally complete virion. Some coronaviruses, however, do not require the full assemblage of the structural proteins to produce a complete, infectious viral particle. This indicates that some structural proteins are likely dispensable, or that those viruses may encode additional proteins with compensatory roles (Schoeman and Fielding, 2019). The envelope of coronaviruses contains three or four viral proteins. The major proteins of the viral envelope are the S and the M proteins. In some, but not all coronaviruses, a third major envelope protein, the hemagglutinin esterase (HE) is found. Lastly, the small E protein constitutes a minor however critical structural component of the viral envelope (de Haan et al., 1999). Many of the coronavirus proteins are modified by post-translational modifications which change the protein structure by proteolytic cleavage and disulfide bond formation or extend the chemical repertoire of the 20 standard amino acids by introducing new functional groups. Functional groups are commonly added through phosphorylation, glycosylation and lipidation (such as palmitoylation and myristoylation). The post-translational modifications play critical roles in regulating folding, stability, enzymatic activity, subcellular localization and interaction of the viral protein with other proteins (Fung and Liu, 2018).
In contrast to the other main structural proteins, the N protein is the only protein which mainly plays roles to bind to the viral RNA genome to form the nucleoprotein. However, apart from its primarily function in packaging and stabilizing the viral genome, the N protein also plays roles in other aspects of the coronavirus replication cycle and in the modulation of host cellular response to viral infection such as regulating the host cell cycle, affecting cell stress response, influencing the immune system, etc. Although the N protein is not required for the viral envelope formation, it may be required for the whole virion formation as transient expression of gene encoding the N protein significantly increases the production of virus like particles in some coronaviruses (Schoeman and Fielding, 2019). The coronavirus has a large sized genome, while the overall size of the viral particle is similar to that of other RNA viruses. It seems therefore that the space inside the coronavirus envelope would not be adequate to encapsulate loosely packed ribonucleoproteins. Surprisingly, the way the coronaviruses package their large genome is similar to that of the eukaryotic cells, that is in the form of a supercoiled dense structure (Gui et al., 2017). The incorporation of the coronavirus genomic RNA into a virion is dependent on the N proteins. Recent studies using mouse hepatitis virus (MHV)-infected cells showed that the cytoplasmic N proteins constitutively form oligomers through a process which does not need binding to genomic RNA. It was hypothesized that constitutive N protein oligomerization allows the optimal loading of the genomic viral RNA into a ribonucleoprotein complex through the presentation of multiple viral RNA binding motifs (Cong et al., 2017).
2.2. Spike (S) protein
The coronavirus spike (S) protein is a large glycosylated transmembrane protein ranging from about 1162 to 1452 amino acid residues. Monomers of the S protein, prior to glycosylation, are 128–160 kDa, but molecular masses of the glycosylated forms of the full-length monomer are 150–200 kDa. Following translation, the proteins fold into a metastable prefusion form and assemble into a homotrimer forming the coronavirus distinctive surface spike of crown-like appearance. The S protein is the most outward envelope protein of the coronaviruses. The S glycoprotein plays critical roles in mediating virus attachment to the host cell receptors and facilitating fusion between viral and host cell membranes. In addition, it is the primary determinant of the coronavirus tropism. Changes in the S protein especially in the regions involved in the interactions with entry receptors, may result in altered host, tissue, or cellular tropism of the coronaviruses (Masters, 2006; Hulswit et al., 2016). The S protein is the main antigen present at the surface of the coronaviruses functioning as a major inducer of host immune responses. During infection, the S protein is the target of the neutralizing antibodies. Therefore, it has been a focus in vaccine design (Li, 2016; Tortorici et al., 2019).
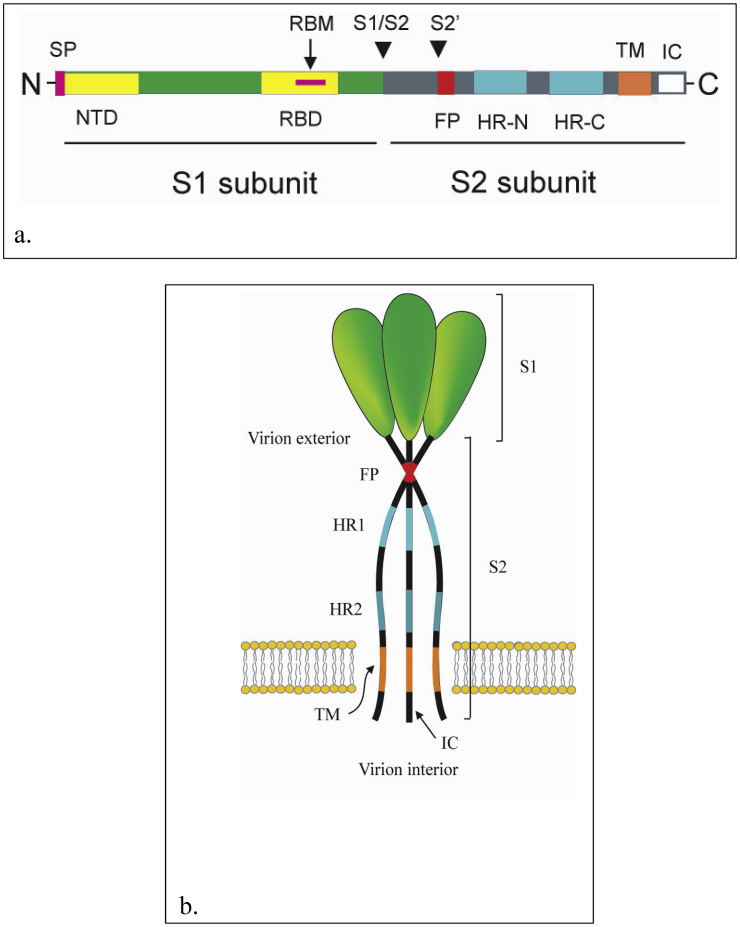
The S protein is inserted into the endoplasmic reticulum through a cleaved, amino-terminal signal peptide. The domain that extends into the outside space of the virus (virion exterior), termed ectodomain, makes up most of the molecule, with only a small N-terminal segment (of 71 residues or fewer) constituting the transmembrane domain and endodomain. The endodomain also called intracellular tail (IC) is located in the inside space of the virus (virion interior) (Masters, 2006; Li, 2016). The multifunctional S protein can be divided into two functionally distinct subunits: the S1 and S2 subunits (Figure 2a). The global S1 subunit is critical for receptor recognition, while S2 subunit is important for membrane fusion and for anchoring the S protein into the viral membrane (Hulswit et al., 2016; Tortorici et al., 2019). The S1 subunit consists of two major domains which fold independently, the N-terminal domain (S1-NTD) and the C-terminal domain (S1-CTD). Depending on the virus, one or both of these domains may bind to receptors and function as a receptor-binding domain (RBD). While the RBD of the mouse hepatitis virus (MHV) is at the S1-NTD, the majority of other coronaviruses, including SARS-CoV and MERS-CoV have the RBDs at the S1-CTDs. The S1-NTDs are responsible for binding sugar receptor molecules except for the betacoronavirus MHV, the S1-NTD of which binds a protein receptor CEACAM1. The roles of the S1-CTDs are to bind to protein receptors ACE2, APN, and DPP4 (Li, 2016; Ou et al., 2020). Structurally, the S1 subunit of the betacoronavirus S protein is divided into 4 distinct β-rich domains, A, B, C, and D. Domains A and B are suggested to serve as RBD. The core structure of domain A shows a galectin-like β-sandwich fold. The domain B contains a core subdomain of antiparallel β-sheets decorated with an extended loop on the viral membrane-distal side. The domains A and B are linked by a linker region. The domain A is located within the functionally S1-NTD, whereas the domains B, C, and D are located within the S1-CTD. The domains C and D form β-sheet-rich structures adjacent to the S2 subunit (Hulswit et al., 2016). Structural studies of SARS-CoV RBD revealed that the RBD contains a core and a motif termed receptor-binding motif (RBM) which is critical for forming contact with receptor (Shang et al., 2020).
Figure 2.
Map and membrane topology model of coronavirus spike (S) protein. a). Map of coronavirus spike (S) protein. The S protein can be divided into two functionally distinct subunits: the S1 and S2 subunits. The S1 subunit consists of two major domains the N-terminal domain (S1-NTD) and the C-terminal domain (S1-CTD). The S1 subunit contains a receptor-binding domain (RBD). The RBD contain a receptor binding motif (RBM). The arrow-heads mark the site of cleavage for the S protein by cellular protease(s). The signal peptide (SP), N-terminal domain (NTD) and regions of RBD and RBM are shown in S1. The S2 subunit contains the heptad repeat regions (HR1 and HR2), fusion peptide (FP), transmembrane domain (TM) and intracellular tail (IC) are shown. b). Model for coronavirus spike (S) trimer and its membrane topology. The S protein is a transmembrane protein which assembles into a homotrimer. The S1 subunits constitute the bulb portion of the spike, in the virion exterior. The S2 subunits anchor the S proteins into the viral membrane. The S2 subunits contain segments which include the fusion peptides (FP), HR1, HR2 and the highly conserved transmembrane domains. The HR2 regions locate close to the C-terminal end of the S ectodomain in the virion exterior. The intracellular tails (ICs) and the C-terminal ends of the S proteins are located in the virion interior (Masters, 2006; Li, 2016).
The S2 subunit of coronaviruses is highly conserved and contains segments which have critical roles to facilitate virus-cell fusion. These segments include the fusion peptide (FP), two heptad repeat regions, the heptad repeat region 1 (HR1 or HR-N), heptad repeat region 2 (HR2 or HR-C) and the highly conserved transmembrane domain (Figure 2b). The HR-2 region locates close to the C-terminal end of the S ectodomain. In the prefusion conformation of the MHV and HKU1 S proteins, the S2 subunit consists of segments of multiple α-helices and a three-stranded antiparallel β-sheet at the viral membrane-proximal end. The fusion peptide forms a short helix of which the conserved hydrophobic residues are buried in an interface with other elements of S2. The conserved fusion peptide is not directly upstream of the HR1 but located about 65 residues upstream HR1. Another fusion peptide (termed FP2) has also been suggested to be exist immediately upstream of the HR1 region. The metastable prefusion structure of the S2 subunit has been suggested to be locked by a cap formed by intertwined S1 protomers (Hulswit et al., 2016; Li, 2016).
Following its synthesis, the coronavirus S protein undergoes post-translational modifications which include glycosylation, disulfide bond formation and palmitoylation. The virion exterior (luminal) ectodomain of the S protein is highly glycosylated and this modification is exclusively N-linked. The S protein ectodomains have from 19 to 39 potential consensus glycosylation sites. For the transmissible gastroenteritis virus (TGEV) S protein, it has been shown that the initial steps of glycosylation occur cotranslationally, but that terminal glycosylation is preceded by trimerization, which may be rate limiting in the S protein maturation. The N-linked glycosylation has been indicated to contribute significantly to the conformation of coronavirus S protein, and therefore undoubtedly affects the receptor binding and antigenicity of S protein. The glycosylation of TGEV S protein, for instance, was suggested to assist monomer folding, given that tunicamycin, a N-glycosylation inhibitor, was found to block trimerization. Notably, not all of the putative glycosylation sites are functional. For example, among the 23 putative glycosylation sites in the SARS-CoV S protein, only 12 sites were really glycosylated (Masters, 2006; Fung and Liu, 2018). In addition, the S protein ectodomain has between 30 and 50 cysteine residues, and within each coronavirus group the position of cysteine residues is well conserved. It has been reported that disulfide bond formation occurs in S proteins of MHV, suggesting that the disulfide bonds are essential for the correct folding, trafficking and trimerization of the S proteins (Masters, 2006; Fung and Liu, 2018). The conserved cysteine residues in the endodomain tail of the S proteins are modified by palmitoylation which in some coronaviruses have been suggested to be important for the S protein trafficking and folding, virion assembly and infectivity, as well as for the interaction between S and M proteins (Fung and Liu, 2018).
In most coronaviruses, the S protein is cleaved by a trypsin-like host protease into two polypeptides, S1 and S2, of approximately the same size which are still covalently bound in the prefusion conformation. Even for uncleaved proteins, that is, such as the SARS-CoV, the designation of S1 and S2 is used for the N-terminal and C-terminal halves of the S protein respectively. Peptide sequencing has demonstrated that cleavage takes place following the last residue in a highly basic motif of the S protein: RRFRR in infectious bronchitis coronavirus (IBV), RRAHR in MHV strain A59, and KRRSRR in bovine coronavirus (BCoV). Similar cleavage sites are predicted from some others S protein, except that of SARS-CoV. During viral entry, the S2 subunit is further proteolytically cleaved at the S2’ site, upstream of the fusion protein (Masters, 2006; Tortorici et al., 2019). The S1 subunit is the most divergent region of the S protein, both across and within the three coronavirus groups. Even among strains and isolates of a single coronavirus species, the sequence of S1 can vary considerably. To the contrary, the most conserved part of the molecule across the three coronavirus groups is the region that encompasses S2 portion of the ectodomain, plus the start of the transmembrane domain (Masters, 2006). It has been hypothesized that the S1 domains of the S protein oligomer constitute the bulb portion of the spike. The stalk portion of the spike, on the other hand, was envisioned to be a coiled-coil-structure, formed by association of heptad repeat regions of the S2 domains of monomers (Masters, 2006).
2.3. Membrane (M) protein
The membrane (M) glycoprotein is the most abundant envelope protein of coronaviruses playing critical roles in the virion assembly through M-M, M-spike (S), and M-nucleocapsid (N) protein interactions (Arndt et al., 2010). Generally, its length is 217–230 amino acids. It is a triple-spanning membrane protein with a short amino-terminal domain located on the exodomain of the virus (in the virion exterior, equivalent to the lumen of intracellular organelles) and a long carboxy-terminal domain in the endodomain of the virion (in the virion interior, equivalent to the cytoplasmic space of intracellular membranes) (de Haan et al., 1999; Masters, 2006; Perrier et al., 2019). The nascent polypeptides, in the preglycosylated forms, are of 25–30 kDa (221–262 amino acids) and the detected glycosylated forms are of higher molecular weights (Masters, 2006). The C-terminal domains of the MERS-CoV and IBV M proteins have been shown to contain signals for the trans-Golgi network and the endoplasmic reticulum-Golgi intermediate compartment (ERGIC)/cis-Golgi localization, of host cells respectively (Perrier et al., 2019).
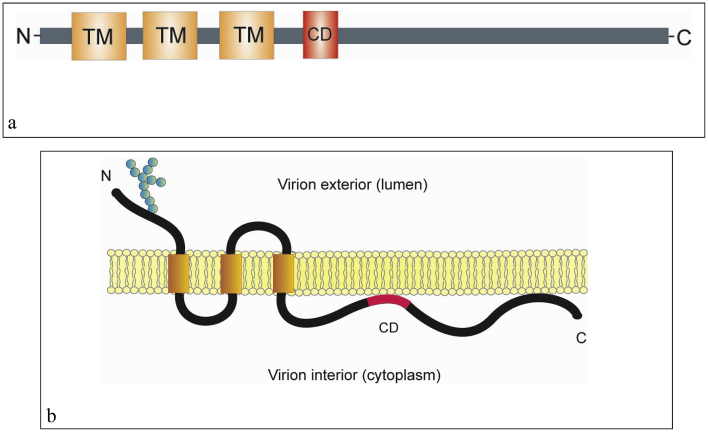
The M proteins from different coronaviruses show the same overall basic structure although their amino acid contents vary. The proteins have three transmembrane (TM) domains flanked by the amino terminal glycosylated domain and the carboxy-terminal domain. Multiple M domains and residues have been indicated to be essential for coronavirus assembly. After the third TM domain, the long intravirion (cytoplasmic) tail of M protein harbors an amphipathic domain and a short hydrophilic region at the carboxyl end of the tail. The amphipathic domain is suggested to be closely associated with the membrane. At the amino terminus of the amphipathic domain, there is a highly conserved 12-amino-acid domain with amino acid sequence SMWSFNPETNIL in the SARS-CoV M protein. This conserved domain (CD) has been suggested to be functionally important for M protein to participate in virus assembly (Arndt et al., 2010). The schematic domain and membrane topology of the M protein is shown in Figure 3.
Figure 3.
The schematic domain and membrane topology of the coronavirus membrane (M) protein. a). The coronavirus M protein has three transmembrane (TM) domains flanked by the amino terminal domain and the carboxy-terminal domain. The carboxy-terminal endodomain contains a conserved domain (CD) following the third transmembrane (TM) domain. b). The transmembrane topology of the coronavirus M protein. The M protein spans the viral membrane three times. The three transmembrane (TM) domains are flanked by the amino-terminal glycosylated domain (in the virion exterior) and the carboxy-terminal endodomain (in the virion interior). The conserved domain (CD) in the long carboxy-terminal endodomain is indicated (Arndt et al., 2010; Perrier et al., 2019).
It is proposed that lateral interactions between the coronavirus membrane proteins are important in mediating the formation of the virion envelope. This was based on the observation that when expressed alone, M protein accumulates in the Golgi complex of host cell in the form of homomultimeric complexes. However, when it is expressed in combination with the E protein, virus-like particles (VLPs) more or less of the authentic virion size and shape are assembled. This showed that the M and E proteins are the minimal requirements for envelope biogenesis. Furthermore, by employing the VLP assembly system it was suggested that all domains of the M protein are critical for virion assembly, and the interactions between membrane proteins (M-M interactions) play roles in promoting coronavirus envelope assembly (de Haan et al., 1999; Neuman et al., 2011).
The M protein is also important for the assembly of the S protein in the viral envelope. Heterotypic interactions between M protein and S protein have been indicated to be required for directing the incorporation of the S protein into the viral envelope in spite of the fact that the S protein is not essential for assembly of coronavirus particle. The S protein is incorporated into virions when present. When coronaviruses are grown in the presence of the N-glycosylation inhibitor, tunicamycin, virions are generated although without any spike (de Haan et al., 1999). The S protein is N-glycosylated and therefore is sensitive to tunicamycin (Mounir and Tablot, 1992). Glycosylation is believed to be important for ability of the virus to replicate in the host cells (Oostra et al., 2006). The interactions between M and S proteins have been demonstrated experimentally (de Haan et al., 1999).
The coronavirus M proteins also interact with each other. The M-M interactions constitute the overall scaffold for the viral envelope. In the mature virion, the S protein and a few of E molecules are interspersed in the M protein lattice (Arndt et al., 2010). The M protein was also shown to interact with the HE protein. The interaction was shown by cells infected with the bovine coronavirus expressing an HE protein, which generate complexes of the M, S, and HE proteins detected by co-immunoprecipitation assays. In addition, the M protein also interacts with the nucleocapsid during virus assembly (de Haan et al., 1999). The M protein is anchored by its three transmembrane domains to the viral envelope and by its carboxy-terminal tail interaction to the nucleocapsid (McBride et al., 2014). Recent studies indicated that the M protein of the HCoV-NL63 also plays roles during the early stages of infection by facilitating the viral attachment to the heparan sulfate proteoglycans used by the HCoV-NL63 as initial attachment factors (Naskalska et al., 2019).
2.4. Envelope (E) protein
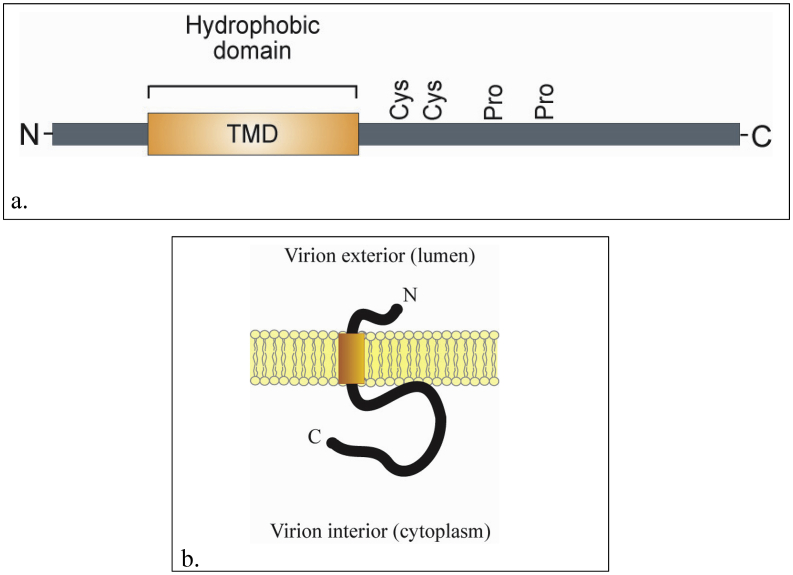
The envelope (E) protein is a small integral membrane polypeptide, ranging from 76 to 109 amino acid residues with molecular weight of 8.4–12 kDa. The E protein plays important roles in a number of aspects of the coronavirus replication cycle, such as assembly, budding, envelope formation, and pathogenesis. Interestingly, although the protein is highly expressed inside the infected cells, only a small portion of the protein is incorporated into the viral envelope. Consequently, the protein is only a small constituent of the virus particle. Due to its small size and limited quantity, the E protein was identified much later compared to the other coronavirus structural proteins. Its primary and secondary structure indicates that the E protein has a short hydrophobic N terminus of 7–12 amino acid residues, followed by a transmembrane domain (TMD) of 25 amino acids, and ends with a long hydrophilic carboxy terminus (Masters, 2006; Schoeman and Fielding, 2019). The E protein harbors conserved cysteine residues in the hydrophilic region that are targets for palmitoylation. In addition, it contains conserved proline residues in the C-terminal tail (Figure 4) (Ruch and Machamer, 2012).
Figure 4.
The schematic domain and membrane topology of coronavirus envelope (E) protein. a). The schematic domain of the coronavirus E protein. The protein has a hydrophobic domain predicted to span the viral membrane. The conserved cysteine and proline residues are indicated. b). Membrane topology of coronavirus E protein. The protein spans the viral membrane once with the N terminal end at the virion exterior and the C terminal end at the virion interior. The transmembrane domain is indicated by bar (Ruch and Machamer, 2012).
The hydrophobic region of the TMD is predicted to contain at least one α-helix which plays roles in the protein E oligomerization to form a membrane ion conductive pore termed viroporin. The amino acid sequence of the SARS-CoV E protein shows that a large portion of the TMD consists of the two non-polar amino acids, valine and leucine, which give the protein strong hydrophobicity. The overall net charge of the molecule is zero as the uncharged middle region is flanked by the negatively charged amino terminus and the variably charged carboxy terminus. The long C-terminus also shows some hydrophobicity due to the presence of a cluster of positively charged residues, however the hydrophobicity level is not as strong as that of the TMD. Interestingly, the C-terminus of the Beta- and Gamma-coronaviruses, has a conserved proline residue in the center of a β-coil-β motif. The motif has been suggested to serve as a Golgi-complex targeting signal, as mutation of the proline residue abolished the localization of the E protein in the host cells Golgi complex, and instead the mutant E protein then targeted the plasma membrane (Schoeman and Fielding, 2019).
One of the unique features of coronaviruses is the source of their membrane envelope. Differ from the other well-known enveloped viruses, coronaviruses bud into the endoplasmic reticulum-Golgi intermediate compartment (ERGIC), from where they obtain their membrane envelope. Therefore, it is not surprising to find that most of the E protein is localized to the ERGIC and Golgi complex where the E protein plays roles in the assembly, budding and trafficking of the nascent virus particle (Schoeman and Fielding, 2019). Similar to the E protein, the S and M proteins are known to co-localize to the ERGIC. However, live-cell imaging studies of MHV E protein using confocal microscopy showed that, in contrast to the S and M proteins which are also localized in the plasma membrane, the E protein does not traffic to the surface of the cells, but remains at the site of viral assembly in the ERGIC. Furthermore, in the Golgi complex, the E protein is mainly concentrated in the cis and medial regions of this organelle. It should be noted that information regarding the precise cellular localization of the coronavirus E protein is critical in order to understand its roles in viral infection as to whether it is involved in morphogenesis or pathogenesis (Venkatagopalan et al., 2015).
Studies of different coronaviruses have been conducted to determine the membrane topology of the E proteins and a variety of different E protein topologies have described and proposed (Schoeman and Fielding, 2019). Studies of the MHV E protein showed that the N-terminus of the protein is located in the lumen of the Golgi complex and the C-terminus is in the host cells cytoplasm (corresponds to the interior of the virus) (Venkatagopalan et al., 2015). Studies of SARS coronavirus E protein also suggested a topological conformation in which the E protein N-terminus is oriented towards the lumen of the intracellular membranes and the C-terminus faces the host cell's cytoplasm (Nieto-Torres et al., 2011). Similarly, experiments of IBV E protein showed that the N-terminus is located in the lumen of the Golgi complex and the C-terminus in the cytoplasm. On the contrary, the TGEV E protein shows a topology of a luminal C-terminus and a cytoplasmic N-terminus. FLAG-tagged SARS-CoV was reported to have an N- and C- terminus cytoplasmic topology. Prediction software has also been employed and resulted in conflicting predictions between both the software and the experimental data. A rationale for different membrane topologies has been proposed, in that, between the different coronavirus species, the E protein may not show a uniform topology depending on the level of protein expression and oligomerization. In addition, the membrane topology of the E protein might be dictated by its function, whether it is required to form viroporin or it is involved in the viral envelope during viral assembly (Schoeman and Fielding, 2019).
In several coronaviruses such as IBV, SARS-CoV, and MHV, the E protein is palmitoylated, i.e. it is modified by the addition of palmitic acid. The target amino acids for palmitoylation are the cysteine residues adjacent to the transmembrane domain. Palmitoylation has been suggested to plays roles in the subcellular protein trafficking as well as modulation of protein-protein interaction. Palmitoylation increases protein hydrophobicity which may facilitate protein association and anchoring to the viral membrane. This interaction might lead to a more stable association of the protein with the membrane. Double or triple alanine substitution for cysteine residues in MHV E protein was reported to significantly reduce virus-like particle (VLP) formation. In addition, the triple-mutated E proteins were found to be unstable, prone to degradation, and significantly reduced in terms of virus production. This indicates that palmitoylation of E protein is of paramount importance for viral assembly. Notably, although palmitoylation was found to be important for correct localization of some viral proteins, in the case of MHV E proteins, the addition of palmitic acid has no influence on the protein localization (Lopez et al., 2008; Schoeman and Fielding, 2019).
The fact that only a small portion of the E protein is incorporated into the viral envelope suggests that the protein has additional functions around the host cell's endoplasmic reticulum and Golgi region. The coronavirus E protein has a unique ability to form homotypic interaction leading to oligomerization and generation of viroporins (Schoeman and Fielding, 2019). Viroporins are integral hydrophobic viral proteins that form pores on host cell membranes, and affect the vesicle system of host cells, affect glycoprotein trafficking, and increase cellular membrane permeability, leading to the promotion of progeny virus particle release (Liao et al., 2006). Viroporins have also been suggested to play roles in pathogenesis. Although viroporins are not required for viral replication, their absence weakens or attenuates the virus and reduces its pathogenicity. The pores of the viroporins are hydrophilic. Generally, in forming a viroporin the hydrophobic residues of the protein line outside the pore oriented toward the phospholipid, while the inside of the pore is formed by the hydrophilic amino acids. The majority of the viroporins have an amphipathic α-helix in the hydrophobic domain and the pore is anchored to membrane by a cluster of positively charged amino acids through electrostatic interaction with the negatively charged phospholipids. Conformational changes in the structure regulate the ion flow by opening and closing of the pore (Schoeman and Fielding, 2019). Viroporins seem to selectively transport positively charged ions such as hydrogen (H+), potassium (K+), sodium (Na+) and calcium (Ca2+). The coronavirus E protein viroporins have been demonstrated to be selectively channeling monovalent cations, Na+ and K+ (Schoeman and Fielding, 2019). It is noteworthy that a deeper analysis of viroporin structure and function may provide novel strategies for development of antiviral therapeutics by blocking viroporin channel activity (Torres et al., 2015). Furthermore, due to the involvement of coronavirus E proteins in multiple critical aspects of the virus replication cycle, virus particles devoid of E protein may be a promising vaccine candidate (Schoeman and Fielding, 2019). The gene encoding the E protein has been targeted for coronavirus molecular detection (Setianingsih et al., 2019).
2.5. Nucleocapsid (N) protein
The coronavirus nucleocapsid (N) protein is a structural phosphoprotein of 43–46 kDa, a component of the helical nucleocapsid. The main function of the N protein is to package the viral genome into a ribonucleoprotein (RNP) particle in order to protect the genomic RNA and for its incorporation into a viable virion. The N protein is thought to bind the genomic RNA in a beads-on-a-string fashion. In addition, it also interacts with the viral membrane protein during virion assembly and plays a critical role in improving the efficiency of virus transcription and assembly. The N protein undergoes rapid phosphorylation following its synthesis. In mouse hepatitis virus (MHV), phosphorylation occurs exclusively on serine residues. In infectious bronchitis virus (IBV), however, phosphorylation also takes place on threonine residues. The role of the phosphorylation is unclear but it has been hypothesized to have a regulatory significance. The 46 kDa N protein of the SARS-CoV shares 20%–30% identity with other coronavirus N proteins. It forms a dimer which constitutes the basic building block of the nucleocapsid through its C-terminus (Masters, 2006; Chang et al., 2014; McBride et al., 2014). The N protein is dynamically associated with the replication-transcription complexes (Verheije et al., 2010).
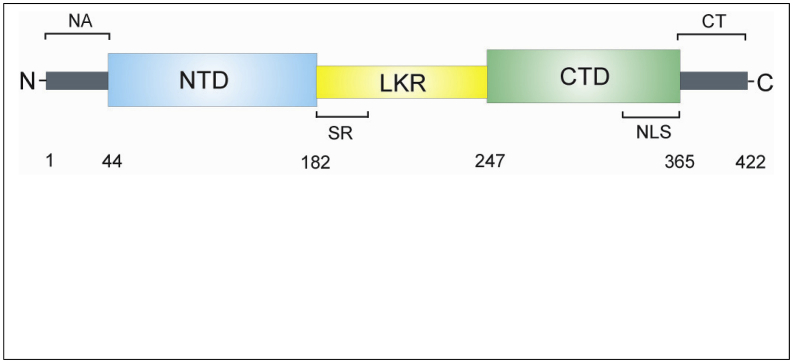
Based on amino acid sequence comparisons it has been shown that the coronavirus N proteins have three distinct and highly conserved domains, namely the N terminal domain (NTD), the linker region (LKR) and the C-terminal domain (CTD). The NTD is separated from the CTD by the LKR, also termed an intrinsically disordered middle region (Figure 5). All of the three domains have been demonstrated to bind with viral RNA. The LKR includes a Ser/Arg-rich region (SR-motif) which contains a number of putative phosphorylation sites. The flexible LKR has the capability of direct interaction with RNA under in vitro conditions. The phosphorylation sites within the LKR are believed to play a role in binding M protein, heterogeneous nuclear ribonucleoprotein A1 (hnRNP-A1) and RNA to the N protein with high binding affinity (McBride et al., 2014; Chang et al., 2014).
Figure 5.
The schematic domain of coronavirus nucleocapsid (N) protein. The coronavirus N protein is a phosphoprotein of 422 amino acid residues (in SARS-CoV). The protein has three distinct and highly conserved domains, the N terminal domain (NTD), the linker region (LKR) and the C-terminal domain (CTD). The NTD is separated from the CTD by the LKR. All of the three domains have been shown to bind with viral RNA. The LKR contains a Ser/Arg-rich region (SR) which contains a number of putative phosphorylation sites. The nuclear localization signal (NLS) motifs are shown. The N-terminal arm (NA) and the C-terminal tail (CT) are shown (McBride et al., 2014; Chang et al., 2014).
The CTD, which is a hydrophobic helix-rich terminal, spans the amino acid residues 248 to 365 in SARS-N protein and amino acid residues 219 to 349 in IBV-N protein and is also called the dimerization domain because it contains residues responsible for self-association to form homodimers. The CTD also facilitates the formation of homo-oligomers through a domain-swapping mechanism. Oligomerization of the N protein is essential in order to generate a stable conformation. In its monomeric form, the CTD is unstable because it folds into an extended conformation with a large cavity in its center. Sequence comparison indicates that the dimerization domain of the N protein is conserved at least among the alpha, beta and gamma groups of coronaviruses, suggesting a common structural and functional role for this domain. The CTD contains the nuclear localization signal (NLS). Crystal structure analysis of the CTD of the SARS-CoV N protein covering residues 248–365, showed that the N protein dimer has the shape of a rectangular slab in which the four-stranded β-sheet forms one face of the slab and the α-helices form the opposite face (McBride et al., 2014).
Self-association of the N protein has been observed in many viruses, and is needed to form the viral capsid which provides protection to the viral genome from extracellular agents. In addition, the viral capsid is important for RNA-binding ability. The N protein fragment of SARS-CoV containing the dimerization domain has been demonstrated to be able to bind a putative packing signal (PS) within the viral RNA, with the most likely RNA binding site located within its basic region between residues 248–280. It was then revealed that the CTD, which spans residues 248–365, harbors eight positively charged lysine and arginine residues, forming a positively charged groove, one of the most positively charged regions of the N protein. The strong electrostatic nature of residues 248–280 suggests that oligonucleotide binding is based on interactions between the positively charged protein and the negatively charged backbone of the RNA molecule. The position of RNA-binding domains near the CTD is important for the formation of a large helical nucleocapsid core, and the association of the N protein dimers is necessary for further assembly of the core. In vitro studies showed that the full-length dimeric N protein has a tendency to form tetramers and higher molecular weight oligomers (McBride et al., 2014). The gene coding for the N protein is among the target genes for coronavirus molecular detection (Artika et al., 2020; Corman et al., 2020).
2.6. Accessory proteins
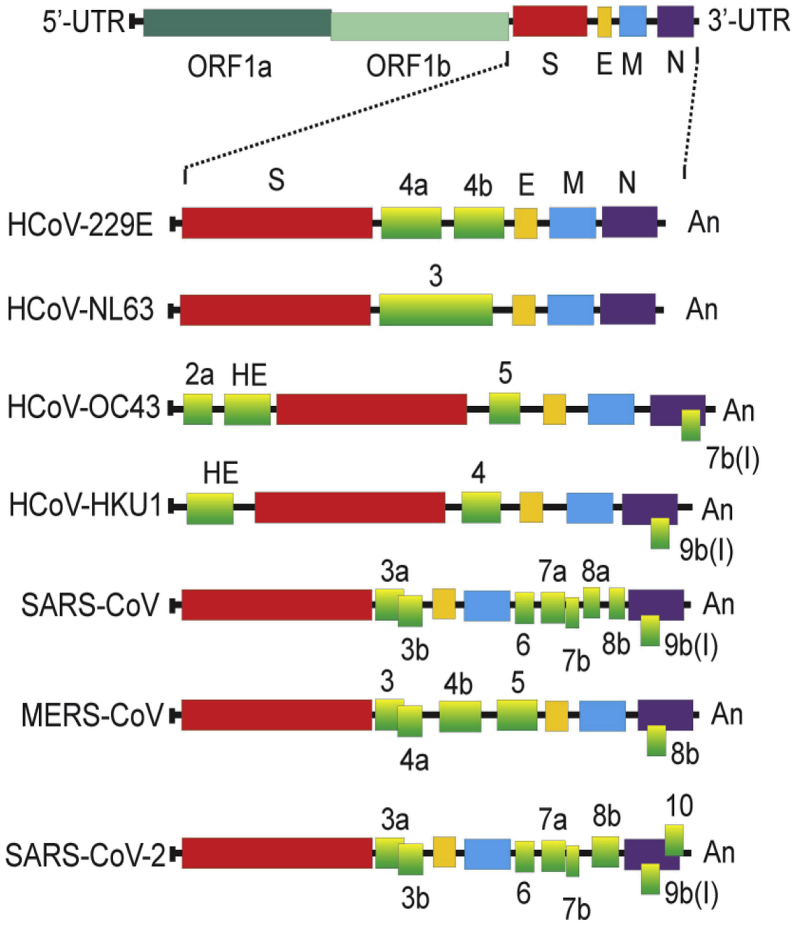
All coronavirus genomes contain accessory genes interspersed among the canonical genes, replicase, S, E, M, N which vary from as few as one (HCoV-NL63) to as many as eight genes (SARS-CoV). These accessory proteins are dispensable for coronavirus replication, however, they may confer biological advantages for the coronaviruses in the environment of the infected host cells. Some accessory proteins have been shown to exhibit roles in virus-host interaction and seem to have functions in viral pathogenesis. For SARS-CoV, some of the accessory proteins have been shown to be able to influence the interferon signaling pathways and the generation of pro-inflammatory cytokines (Masters, 2006; McBride and Fielding, 2012; Liu et al., 2014). The accessory proteins encoded by the coronaviruses that infect humans are listed in Table 2 (Masters, 2006; Wang et al., 2020b; Wu et al., 2020).
Table 2.
Accessory proteins of human coronaviruses∗.
| Virus | Accessory genes (Proteins) |
|---|---|
| HCoV-229E | [rep]-[S]-4a,4b-[E]-[M]-[N] |
| HCoV-NL63 | [rep]-[S]-3-[E]-[M]-[N] |
| HCoV-HKU1 | [rep]-2(HE)-[S]-4-[E]-[M]-[N], 7b(I) |
| HCoV-OC43 | [rep]-2a-2b (HE)-[S]-5 (12.9k)-[E]-[M]-[N], 7b(I) |
| SARS-CoV | [rep]-[S]-3a,3b-[E]-[M]-6-7a,7b-8a,8b-[N], 9b(I) |
| MERS-CoV | rep]-[S]-3-4a,4b-5-[E]-[M]-8b-[N] |
| SARS-CoV-2 | [rep]-[S]-3a,3b [E]-[M]-6-7a,7b-8b-[N],9b,10 |
Rep denotes replicase; the canonical coronavirus genes are indicated in brackets; the protein is indicated in parentheses in cases where it has a different designation to the gene.
The eight SARS-CoV ORFs encoding for accessory protein are 3a, 3b, 6, 7a, 7b, 8a and 9b. Interestingly, these accessory proteins are found to be specific for SARS-CoV and have no significant homology to accessory proteins from other coronaviruses. The protein 3a is the largest accessory protein and is thought to play a role as a structural component of the SARS-CoV. It has been demonstrated to be incorporated into the virus-like particles (VLPs) although it is not essential for the VLP formation. In addition, the 3a protein has been shown to interact with the SARS-CoV structural proteins M, S, E, and the accessory protein 7a and may facilitate the SARS-CoV assembly. The 3a protein may also play roles in evading the host immune system. Moreover, it has been proposed that it functions as an ion channel through the use of its transmembrane domains (Liu et al., 2014). The protein 3b has been indicated to have the ability to induce necrosis and apoptosis and is also able to inhibit the host antiviral response by repressing type-I interferon production. The 3b protein is regarded as an interferon antagonist. The protein 6 is incorporated in VLPs when co-expressed with the SARS-CoV structural proteins S, M, E. The physical interaction of protein 6 with these structural proteins is hypothesized to be critical for its assembly into the VLP. The p6 protein has been identified as a β-interferon antagonist (Liu et al., 2014). The protein 7a is a minor structural protein which may facilitate viral assembly. It has been suggested that the 7a protein also important in SARS-CoV pathogenesis by inducing inflammatory responses (McBride and Fielding, 2012; Liu et al., 2014). More detailed molecular characterization is required for 7b, 8a, and 8b proteins. The 7b protein may act as an attenuation factor. The 8a protein has been indicated to have an ability to induce caspase-dependent apoptosis, while the 8b protein has been suggested to have an ability to induce DNA synthesis (Liu et al., 2014). The 9b protein may be a structural component of the SARS-CoV particle. It has been shown to be incorporated into the mature virion and packaged into VLPs when co-expressed with E and M proteins. The 9b protein may play a role during SARS-CoV assembly. In addition, it has been indicated to have an ability to induce caspase-dependent apoptosis (Liu et al., 2014).
Most of the characterized coronavirus accessory proteins have been indicated to have a role in antagonizing the host response. The MERS-CoV accessory proteins have also been shown to be important for the virus pathogenesis. Deletion of four ORFs (ORF3, ORF4a, ORF4b, and ORF5) causes major impacts on viral replication and pathogenesis (Menachery et al., 2017). The ORF4a protein of the human coronavirus 229E has been shown to form homo-oligomers have ion channel activity and is suggested to function as a viroporin which is critical for regulating the viral reproduction. Functionally, it is analogous to the SARS-CoV 3a protein, which also plays a role as a viroporin that regulates virus production (Zhang et al., 2014). Similarly, the ns12.9 accessory protein of the human coronavirus OC43 has been shown to act as a viroporin involved in virion morphogenesis and pathogenesis (Zhang et al., 2015). The human coronavirus NL63 has one ORF encoding an accessory protein 3 (ORF3). The hCoV-NL63 ORF3 protein has been demonstrated to colocalize extensively with the E and M proteins within the ERGIC. It is incorporated into virions and therefore it functions as an additional structural protein (Müller et al., 2010).
One of the coronavirus accessory proteins which has been extensively studied is the haemagglutinin esterase (HE). The HE gene is found in the genome of Betacoronaviruses of A lineage, between orf1b and the S gene, and the encoded HE protein constitutes the fourth protein component of the viral membrane. The HE forms small spikes which appear below the tall S protein spikes. The HE monomer has an N-exo, and C-endo transmembrane topology. The mature protein forms a homodimer stabilized by disulfide bonds (Masters, 2006; Liu et al., 2014). The HE has haemagglutinating and acetylesterase activities. The protein facilitates viral-reversible attachment to O-acetylated sialic acids by acting both as lectin and as receptor-destroying enzyme (RDE). The HCoV-OC43, for example, uses 9-O-acetylated sialic acids as a receptor and possesses sialate-9-O-acetylesterases as its RDE. The HE also functions as a cofactor for S protein, facilitating viral attachment to the host cells (Masters, 2006; Zeng et al., 2008).
2.7. Genome
The genome of coronaviruses is a nonsegmented, single-stranded RNA molecule with positive sense (+ssRNA), which is, of the same sense as the mRNA. Structurally it is similar to most eukaryotic mRNAs, in having 5'caps and 3′ poly-adenine tails. One of the distinctive features of the coronavirus genome is its remarkably large size ranging from 26 to 32 kb. For comparison, this is approximately three times the size of alphavirus or flavivirus genomes and four times the size of picornavirus genomes. Indeed, the size of the coronavirus genomes is among the largest known viral genomic RNAs. The genomes contain multiple ORFs, encoding a fixed array of structural and nonstructural proteins, as well as a variety of accessory proteins which differ in number and sequence among the coronaviruses (Masters, 2006; Chen et al., 2020).
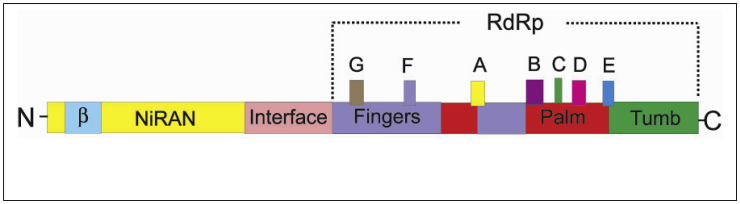
About two-thirds of the 5′-most end of the genome is occupied by two large overlapping open reading frames, ORF1a and ORF1b. There is a -1 frameshift between ORF1a and ORF1b, leading to the synthesis of two polypeptides, pp1a and pp1ab, which are further processed by the viral proteases into 16 nonstructural proteins (nsps) which form the coronavirus replicase-transcriptase complex. This complex is an assembly of viral and hosts cellular proteins, which facilitate the synthesis of the genome and subgenome-sized mRNAs in the infected cell. The replicase-transcriptase complex plays an important role to amplify the genomic RNA and synthesize subgenomic mRNAs. Amplification of the genomic RNA involves full-length negative-strand templates, while the synthesis of subgenomic mRNA involves subgenome length negative-strand templates. The 16 nsps consist of nsp1– nsp11 encoded in ORF1a and nsp12–16 encoded in ORF1b. Studies in MHV-A59 have suggested that these proteins have multiple enzymatic functions, including papain-like proteases (nsp3), adenosine diphosphate-ribose 1,9-phosphatase (nsp3), 3C-like cysteine proteinase (nsp5), RNA-dependent RNA polymerase (nsp12), superfamily 1 helicase (nsp13), exonuclease (nsp14), endoribonuclease (nsp15), and S-adenosylmethionine-dependent 29-O-methyl transferase (nsp16) (Sawicki et al., 2005). The ORF1a and ORF1b have been targeted for molecular detection of coronaviruses (Setianingsih et al., 2019).
The remaining about one-third of the genome clustered at the 3′ end is transcribed into a nested set of subgenomic RNAs which contain ORFs for the structural proteins: spike (S), envelope (E), membrane (M) and nucleoprotein (N) as well as a variable number of accessory proteins depending on the viruses. The genes of accessory proteins are interspersed among the structural protein genes. Interestingly, there is a conserved gene order in all members of the coronavirus family, 5′-replicase-S-E-M-N-3’. However, genetic engineering experiments suggested that this evolutionary native order is not essential for functionality (Masters, 2006; Forni et al., 2017; Chen et al., 2020). Additionally, the genome has a 5′ UTR (untranslated region), ranging from 210 to 530 nucleotides, and 3′ UTR, ranging from 270 to 500 nucleotides (Masters, 2006). The 5′ 350 nucleotides folds into a set of RNA secondary structures which are well conserved, and in the Betacoronaviruses, have been suggested to play a critical role in the discontinuous synthesis of subgenomic RNAs. These functionally important cis-acting elements extend the 3′ of the 5′UTR into ORF1a. All of the 3′UTRs have a 3′-terminal poly(A) tail. The 3′UTR is similarly conserved and harbors all of the cis-acting sequences necessary for viral replication. All of the mRNAs carry identical 70–90 nucleotide leader sequences at their 5′ ends (Yang and Leibowitz, 2015). The organization of human-infecting coronavirus genomes is shown in Figure 6.
Figure 6.
The schematic diagram of structure of the human-infecting coronavirus genomes. Each bar represents the genomic organization of each coronavirus. The genomic regions or open-reading frames (ORFs) are compared. The structural proteins, including spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins, as well as non-structural proteins translated from ORF 1a and ORF 1b and accessory proteins are indicated. The tags indicate the name of the ORFs. 5′UTR = 5′ untranslated region, 3′UTR = 3′ untranslated region, An = poly(A) tail (Masters, 2006; Chen et al., 2020; Wang et al., 2020b).
The coronavirus genome plays multiple functions during viral infection. It acts initially as an mRNA which is translated into two large replicase polyproteins. In fact, these polyproteins are the only translational products derived directly from the genome. All of the downstream ORFs are expressed from subgenomic RNAs. The genome then serves as a template for replication and transcription. Finally the genome plays a role in assembly, as progeny genomes are incorporated into progeny coronavirus particles (Masters, 2006). The expanded genome size of coronaviruses compared to other RNA viruses has been linked to the improved replication fidelity by acquiring genes for RNA processing enzymes. These include the RNA 3′-to-5′ exonuclease, and possibly an endonuclease. In addition, genome expansion is also considered to facilitate the acquisition of genes encoding for accessory proteins which are beneficial for coronaviruses to adapt to a specific host. These features are thought to underlie the propensity of coronaviruses to jump across species barriers to new hosts (Forni et al., 2017; Fan et al., 2019).
3. Life cycle of coronaviruses
3.1. Viral entry and membrane fusion
The infection of coronaviruses is initiated by the binding of the virus particles to the cellular receptors which leads to viral entry followed by fusion of the viral and host cellular membranes (Figure 7). The membrane fusion event allows the release of the viral genome into the host cells cytoplasm, a process known as uncoating, which makes the viral genome available for translation. Coronavirus entry is facilitated by the trimeric transmembrane spike (S) glycoprotein, which mediates receptor binding and fusion of the viral and host membranes. The interaction between the S protein and the cellular receptor is a main determinant of host species range and tissue tropism (Masters, 2006; Burkard et al., 2014). The S1 subunit (domain) of the coronavirus S proteins plays an important role in mediating the S protein binding to the host receptor. This S1 subunit shows the most diversity among coronaviruses and partly accounts for the wide host range of this virus family (Walls et al., 2017). Coronaviruses show complex patterns regarding receptor recognition and the diversity of receptor usage is one of the most profound features of coronaviruses (Li, 2016). The human cellular receptor for the coronaviruses is listed in Table 3.
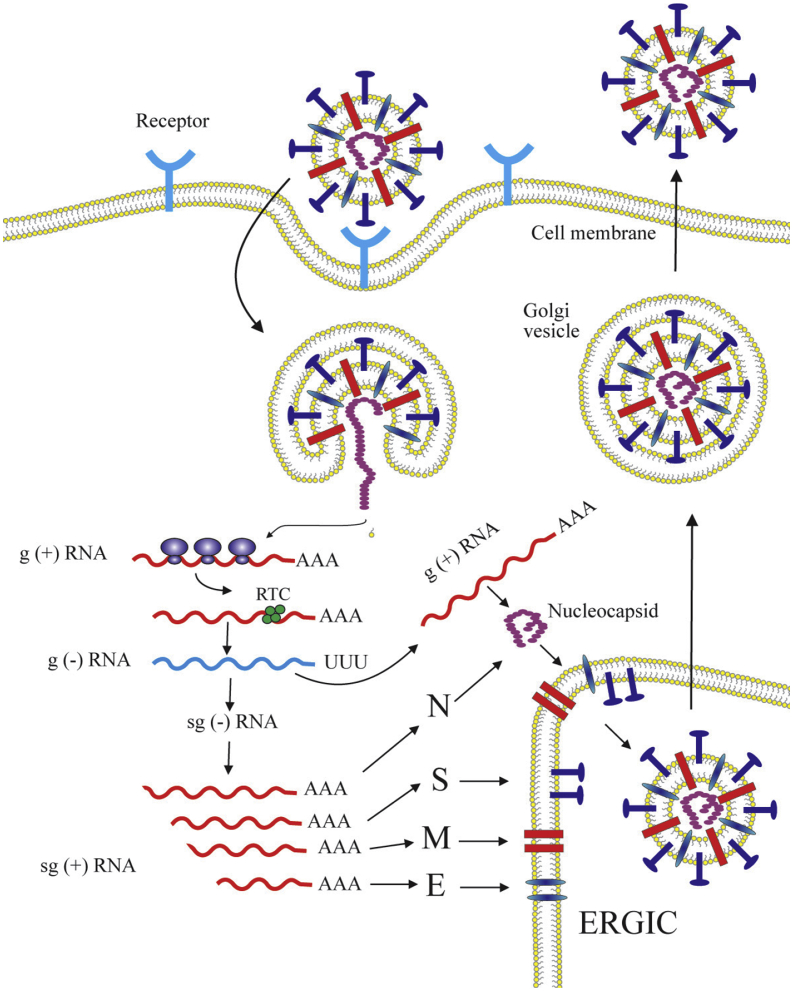
Figure 7.
The schematic diagram of coronavirus life cycle. The coronavirus infection is initiated by the binding of the virus particles to the cellular receptors leading to viral entry followed by the viral and host cellular membrane fusion. After the membrane fusion event, the viral RNA is uncoated in the host cells cytoplasm. The ORF1a and ORF1ab are translated to produce pp1a and pp1ab, which are subsequently processed by the proteases encoded by ORF1a to produce 16 non-structural proteins (nsps) which form the RNA replicase–transcriptase complex (RTC). This complex localizes to modified intracellular membranes which are derived from the rough endoplasmic reticulum (ER) in the perinuclear region, and it drives the generation of negative-sense RNAs ((–)RNAs) through both replication and transcription. During replication, the full-length (–)RNA copies of the genome are synthezied and used as templates for the production of full-length (+)RNA genomes. During transcription, a subset of 7–9 subgenomic RNAs, including those encoding all structural proteins, is produced through discontinuous transcription. In this process, subgenomic (–)RNAs are synthesized by combining varying lengths of the 3′end of the genome with the 5′ leader sequence necessary for translation. These subgenomic (–)RNAs are then transcribed into subgenomic (+)mRNAs. The subgenomic mRNAs are then translated. The generated structural proteins are assembled into the ribonucleocapsid and viral envelope at the ER–Golgi intermediate compartment (ERGIC), followed by release of the newly produced coronavirus particle from the infected cell (Masters, 2006; de Wit et al., 2016).
Table 3.
Receptor of human pathogenic coronaviruses.
| Virus | Receptor | Reference |
|---|---|---|
| CoV-229E | Human aminopeptidase N (CD13) | Yeager et al. (1992) |
| CoV-NL63 | Heparan sulfate proteoglycan | Milewska et al. (2014) |
| CoV-HKU1 | 9-O-acetylated sialicacid (9-O-Ac-Sia) | Huang et al. (2015) |
| CoV-OC43 | 9-O-Acetylated sialic acid (9-O-Ac-Sia) | Vlasak et al. (1988) |
| SARS-CoV | Angiotensin-converting enzyme 2 (ACE2) | Li et al. (2003) |
| MERS-CoV | Dipeptidyl peptidase 4 (DPP4; CD26) | Raj et al. (2013) |
| SARS-CoV-2 | Angiotensin-converting enzyme 2 (ACE2) | Zhou et al. (2020) |
The human CoV-229E employs human aminopeptidase N as a receptor. The human aminopeptidase N is a cell-surface metalloprotease on intestinal, lung, and kidney epithelial cells and is identical to CD13, a glycoprotein identified on granulocytes, monocytes, and their bone marrow progenitors (Yeager et al., 1992). In contrast to the human alphacoronavirus CoV-229E, the human alphacoronavirus NL63 utilizes heparan sulfate proteoglycans for its attachment to target cells. The human CoV-NL63 requires the ACE2 protein for entry but ACE2 is not the primary binding site on the cell surface. On the contrary, heparan sulfate proteoglycans was found to function as adhesion molecules, increasing the virus density on the surface of the cells and likely facilitating the interaction between human CoV-NL63 and its receptor. The heparan sulfate S proteoglycans therefore constitute the human CoV-NL63 adhesion receptors (Milewska et al., 2014). The human CoV-HKU1 (Huang et al., 2015) and human CoV-OC43 (Vlasak et al., 1988) use 9-O-acetylated sialic acid (9-O-Ac-Sia) as their receptor. Sialic acid is an ubiquitous residue of glycoconjugate terminally linked to oligosaccharide decorating glycoprotein and ganglioside at the surface of the host cells. It occurs in a wide variety of forms as a result of modifications of the core N-acetyl neuraminic acid molecule and of variations in glycosidic linkages. Cryo-electron microscopic structural data of human CoV-OC43 revealed that the sialic acid receptor binds to the groove located at the surface of domain A of the S1 subunit of the S glycoprotein (Tortorici et al., 2019). It should be noted that the human CoV-HKU1 (Huang et al., 2015) and human CoV-OC43 (Desforges et al., 2013) possess another viral surface protein, the hemagglutinin-esterase (HE), which is also a type I transmembrane glycoprotein. The HE protein plays a role as receptor-destroying enzyme, through sialate-O-acetyl-esterase activity, to promote release of viral progeny from infected cells and escape from attachment to resistant host cells (Tortorici et al., 2019).
The highly pathogenic human SARS-CoV (Li et al., 2003) and SARS-CoV-2 (Zhou et al., 2020) recognize the same receptor, the human angiotensin-converting enzyme 2 (ACE2). ACE2 is a type I membrane protein found in lung, heart, kidneys, and intestine. It is a zinc-binding carboxypeptidase which plays a critical role in the maturation of angiotensin, a peptide hormone which regulates vasoconstriction and blood pressure. Additionally, ACE2 also functions as a chaperone for membrane trafficking of the amino acid transporter B0AT1 which facilitates uptake of neutral amino acids into intestine cells (Masters, 2006; Yan et al., 2020). The ACE2 protein has an N terminal peptidase domain (PD) and a C-terminal collectrin-like domain (CLD) that ends with a single transmembrane helix and an intracellular segment of more or less 40 amino acid residues. High resolution structural data of SARS-CoV-2 show that two S protein trimers can simultaneously bind to an ACE2 homodimer. In this interaction, each ACE2 PD accommodates one receptor binding domain (RBD) of S protein. The dimerization of ACE2 is mainly mediated by the neck domain of the protein involving an extensive network of polar interactions which stabilize dimer formation. Furthermore, structural information suggests that the overall SARS-CoV-2 and SARS-CoV interfaces with ACE2 are similar, in spite that a number of sequence variations and conformational deviations are observed in their respective interfaces with ACE2 (Yan et al., 2020). Structural information at the atomic level also revealed that the overall structures of RBD and receptor binding motif (RBM) of the SARS-CoV-2 and SARS-CoV are similar, supporting the nearly identical mode of their interaction with the ACE2 receptor (Lan et al., 2020). The overall structural similarity of SARS-CoV-2 and SARS-CoV binding to ACE2 receptor supports a close evolutionary relationship between the two viruses (Shang et al., 2020).
The receptor recognized by the MERS-CoV was identified to be dipeptidyl peptidase 4 (DPP4) also known as CD26. The DPP4 is a multifunctional type-II transmembrane glycoprotein of 766 amino acid residues. It presents as a dimeric form on the cell surface. It has exopeptidase activity and preferentially cleaves dipeptides from hormones and chemokines at a site following a proline amino acid residue, important for controlling their bioactivity (Raj et al., 2013). This enzyme activity, however, is not required for viral entry. Intriguingly, DPP4 does not share any sequence or structural similarities to the previously identified human coronavirus receptors (Wang et al., 2013). Its abundance on epithelial and endothelial tissues has been thought to be the reason of its use as a receptor for the MERS-CoV (Raj et al., 2013). The crystal structure of RBD of the MERS-CoV spike (S) protein showed that the MERS-CoV RBD binds to the extracellular domain of human DPP4. MERS-CoV RBD is made up of a core and a receptor-binding subdomain. The extracellular domain of DPP4 is comprised by an N terminal eight-bladed β-propeller domain and a C-terminal α/β-hydrolase domain. The receptor-binding subdomain of the MERS-CoV RBD was revealed to interact with the DPP4 β-propeller but not with the intrinsic hydrolase domain. The β-propeller domain is comprised by eight blades, each consists of four antiparallel β-strands. The DPP4 employs the blades 4 and 5 to interact to MERS-CoV RBD. This contact site is located distant from the hydrolase domain (Wang et al., 2013). Structural studies of the MERS-CoV spike (S) trimers using single particle cryo-electron microscopy showed that the S protein has a flexible RBD which can readily be approached by the receptors to bind and guarantee virus entry (Yuan et al., 2017).
Following receptor binding, the fusion between the viral envelope and host cell membranes occurs mediated by the viral transmembrane fusion proteins, termed fusogens. In general, based on their structure, there are four different classes of virus-cell membrane fusion proteins. Class I virus-cell fusion proteins are α-helix-rich prefusion trimers which form central coiled-coil structures which insert hydrophobic fusion peptides (or loops) into membranes and refold into postfusion trimmers of α-helical hairpins. Class II virus-cell fusogens have a structural signature of β-sheet-rich prefusion homo- or hetero-dimers which insert fusion loops into membranes, ending in postfusion trimers. These Class II proteins lack of the central coiled coil. Class III virus-cell fusogens exhibit a combination of α-helical and β-structure identified in classes I and II. They are trimers with both α-helices and β-sheets that dissociate into monomers, insert fusion loops into membranes, and oligomerize into postfusion trimers. Class IV reoviral cell-cell fusogens are the smallest identified viral-encoded fusion proteins with fusion loops which oligomerize to fuse membranes (Podbilewicz, 2014). The coronavirus spike (S) protein belongs to the class I viral fusion protein which has a similar function to the fusion proteins of phylogenetically distant RNA viruses such as influenza virus, HIV, and Ebola virus. It requires protease cleavage for activation of fusion capability (Masters, 2006; Walls et al., 2017; Ou et al., 2020). Betacoronavirus spike (S) proteins are processed into S1 and S2 subunits by host proteases. The proteolytic cleavage of the S proteins is essential to induce dissociation of S1 from S2 as a trigger that directly leads to membrane fusion (Li, 2016; Kirchdoerfer et al., 2016). This proteolytic activation step permits for controlled release of the fusion peptide into target cellular membranes. The host proteases shown to function in cleaving the coronavirus spike proteins include, but are not limited to, furin, trypsin, elastase, transmembrane protease/serine subfamily member 2 (TMPRSS2), lysosomal cathepsin L and cathepsin B. In addition to receptor binding and proteolysis of S proteins, membrane fusion may also be triggered by low pH. Protease cleavage at S2′ (Figure 2a) is thought to follow S1/S2 cleavage and may not occur until host-receptor engagement at the plasma membrane or viral endocytosis (Millet and Whitakker, 2015; Li, 2016; Kirchdoerfer et al., 2016). It should be noted that SARS-CoV S protein is not proteolytically cleaved during biosynthesis and the virus does not require a S1/S2 pre-cleavage event for plasma membrane fusion. As the S1/S2 cleavage event is believed to be essential for conformational changes that further expose the S2’ site for immediate plasma membrane fusion, there may be alternative mechanisms to cause these conformational rearrangements, such as receptor binding. Interestingly, the S protein of the SARS-CoV-2 possesses a potential furin cleavage site in the S1/S2 region, which is unique to SARS-like coronaviruses (Tang et al., 2020).
Membrane fusion is a critical event in the coronavirus life cycle which occurs following receptor binding, when the viral and the host cell membranes are proximal. Depending on protease availability, there are two routes for coronavirus entry and membrane fusion, the early (plasma membrane) pathway and the late (endosome) pathway. If the plasma membrane proteases are available, the virus can fuse through the early pathway at the plasma membrane. For example, the presence of exogeneous and membrane bound proteases, such as trypsin and TMPRSS2, can stimulate the early fusion pathway. In the absence of plasma membrane proteases, the coronavirus will be internalized via clathrin- and non-clathrin-mediated endocytosis and thus achieve membrane fusion via a late pathway at the endosomal membrane. As the virus is transported towards the host cell interior, the pH in the endosome decreases. This increasing acidity can activate cathepsin L to trigger fusion at the endosomal membrane. It has been shown that SARS-CoV, MERS-CoV, and SARS-CoV-2 can enter cells either using an early pathway or a late pathway, depending on protease availability and cell type (Tang et al., 2020).
Of note, the membrane fusion is not a spontaneous process, as it needs high energy to bring the membranes close together. In this process, the viral fusion protein plays a critical role as a catalyst by providing the energy required to drive the reaction (Tang et al., 2020). The coronavirus S glycoprotein exists as a metastable prefusion protein at the viral surface (Walls et al., 2017). Studies of HKU1 S protein revealed that in the prefusion conformation, the receptor-binding subunits (S1) rest above the fusion-mediating subunits (S2), preventing their conformational rearrangement. The membrane fusion needs progressive S protein destabilization through receptor binding and proteolytic cleavage (Kirchdoerfer et al., 2016). As for MERS-CoV, the S protein may have two proteolytic cleavage sites, the S1/S2 and the S2′ cleavage sites. It is suggested that all coronaviruses need the cleavage site on S2’ for membrane fusion to be accomplished (Hulswit et al., 2016). The receptor binding and proteolytic cleavages trigger large-scale conformation changes which initiate the fusion reaction involving insertion of the hydrophobic fusion peptide into the host membrane. This irreversible refolding of the fusion machinery provides the energy needed to bring the viral and host membrane close together leading to membrane fusion through the S2 domain. The postfusion conformation state of the S protein represents its most stable conformation with the lowest energy point (Lim et al., 2016; Walls et al., 2017).
3.2. Replication of coronavirus genome
The replication of the coronavirus genome is viewed as the most fundamental aspect of the coronavirus biology. As the largest group of RNA virus, coronaviruses require an RNA synthesis machinery with the fidelity to faithfully replicate their RNA. Coronavirus replication is achieved by employing complex mechanisms involving various proteins encoded by both viral and host cell genomes. Evolutionary, the virus genome contains relatively constant replicative genes which are indispensable for viral replication. Despite undergoing high mutation rates, RNA viral genomes still encode proteins with arrays of conserved sequence motifs playing roles in facilitating their genome replication and expression. Such proteins include the RNA-dependent RNA polymerase (RdRp), RNA helicase, chymotrypsin-like proteases, papain-like proteases, and metal binding proteins. In coronavirus genomes, all of the genes encoding these proteins are located in the ORF1 strategically located at the 5′-most end of the genome. In addition, viruses also exploit cellular proteins for multiple purposes in their replication cycle, including the attachment and entry into the cells, the initiation and regulation of RNA replication and transcription, protein synthesis, and the assembly of progeny virions. For these purposes, viruses typically subvert the normal components of cellular RNA processing and translational machinery to play both integral and regulatory roles in the replication, transcription, and translation of the viral genomes (Shi and Lai, 2005).
Soon after the accomplishment of receptor binding and membrane fusion events which lead to the release and uncoating of the viral RNA genome, the genomic replication cycle is started. In line with all other positive (+)-stranded RNA viruses, a coronavirus replicates its genome through synthesis of a complementary negative (‒)-strand RNA using the genomic RNA as a template. Firstly, using a continuous transcription process, the genome-size positive (+) stranded RNA is used as a template to make the genome-size negative (‒)-stranded RNA which subsequently serves as a template for the synthesis of the genome-size positive (+) stranded RNA progenies. Astonishingly, a coronavirus also synthesizes a number of shorter negative (‒)-stranded RNA of various sizes through discontinuous transcription process. These subgenome-length negative (‒)-stranded RNA molecules subsequently serve as templates for producing a number of positive (+) stranded RNAs of various sizes, termed subgenomic RNAs. For examples, during replication of MHV-A59, six subgenomic mRNA molecules are produced. The coronavirus genome and subgenomic mRNAs share identical 3′ sequences and form a 3′ nested set of RNA molecules. Interestingly, only the ORF at the 5’ region of each subgenomic mRNA is translated into a unique protein. Notably, the positive strands (genomes and subgenomic mRNA) are produced in relatively large amounts compared to the negative strands of genome- and subgenome-length RNA which serve as templates for genome and subgenomic mRNA synthesis (Sawicki and Sawicki, 2005).
As mentioned earlier, about two-third of the 5′-end of the coronavirus genomic RNA (ORF1a and ORF1b) are translated into two polypeptides, pp1a and pp1ab, which further undergo proteolytic cleavage process by proteases encoded by the ORF1a into 16 nonstructural proteins (nsps). Together with cellular proteins, these 16 nonstructural proteins are thought to form the replicase-transcriptase complex (RTC). The nonstructural proteins generated include the papain-like proteases (PLpro), adenosine diphosphate-ribose 19-phosphatase, 3chymotrypsin-like cysteine proteinase (3CLpro), RdRp, helicase (Hel), exonuclease (ExoN), endoribonuclease, and S-adenosylmethionine-dependent 29-O-methyl transferase, etc. The roles of most nonstructural proteins have been reported. However, the roles of nsp2 and nsp 11 are unknown. The remaining one-third of the genome is transcribed into subgenomic RNAs for production of structural proteins: spike (S), envelope (E), membrane (M), nucleoprotein (N) and a variable number of accessory proteins (Sawicki et al., 2005; de Wit et al., 2016; Chen et al., 2020).
Similar to many other positive (+) sense RNA viruses, coronaviruses use proteolytic processing to control expression of their replicative protein machineries. The critical roles of the pp1a/pp1ab polyprotein processing in genomic replication of coronaviruses are demonstrated by the prevention of RNA biosynthesis by proteinase inhibitors blocking essential proteolytic cleavages. Based on their physiological role, coronavirus proteinases are classified into main proteinases and accessory proteinases. All coronaviruses encode one main proteinase (Ziebuhr, 2005), called 3chymotrypsin-like cysteine proteinase (3CLpro). These are indications of a similarity of cleavage-site specificity with that of picornavirus 3C proteinases (3Cpro), although the structural similarities were found to be limited (Anand et al., 2003). The coronavirus 3CLpro is a cysteine protease that forms a homodimer for its proteolytic activity with one active site per subunit. Dimerization of this enzyme is critical for shaping a substrate-binding pocket at its active site (Muramatsu et al., 2016; Zhang et al., 2020a). The proposed catalytic residues His41 and Cys145 are essential for 3CLpro catalytic activity. Mutations His41 to Ala, and Cys145 to Ser, resulted in a 40-fold reduction in activity (Huang et al., 2004). A number coronavirus 3CLpro crystal structures have been reported. The 3CLpro subunit is made up of N-terminal finger (residues 1–8), catalytic domain (residues 8–184), and a C-terminal domain (residues 201–306). The overall domain structures are the same among all of the reported 3CLpro enzymes (Muramatsu et al., 2016). Recently, the crystal structure of the SARS-CoV-2 3CLpro was elucidated. Its 3D structure was found to be very similar to that of SARS-CoV, consistent with a 96% sequence identity between the two polypeptides. The recognition sequence of the main proteinases at most sites is Leu-Gln↓(Ser, Ala, Gly) (↓ indicates the cleavage site) (Zhang et al., 2020a). The roles of the 3CLpro is to cleave the major part of the polyproteins at 11 conserved sites and release the conserved replicative machinery such as the RdRp, helicase, and three RNA processing domains. The 3CL pro is also responsible for cleaving itself from the polyproteins. The coronavirus 3CLpro shows both cis and trans activity (Chuck et al., 2011). Generally, most of the pp1a/pp1ab cleavages are mediated by the trans activity of the fully processed form of 3CLpro (Ziebuhr, 2005).
Depending on the virus, there may be one or two accessory proteinases produced. The function of the accessory proteinases is to cleave the more divergent N-proximal pp1a/pp1ab regions at two or three sites. The accessory proteinases are papain-like cysteine proteinases (PLpro). The MHV and HCoV-229E encode two PLpros, the PL1pro and the PL2pro. In the case of MHV, the PL1pro cuts the nsp1/nsp2 and nsp2/nsp3 sites, and the PL2pro cleaves the nsp3/nsp4 site. For HCoV-229E, the PL1pro cleaves the nsp1/nsp2 and nsp2/nsp3 sites, however, the PL2pro cleaves the nsp3/nsp4 site and can also act at the nsp2/nsp3 site. The infectious bronchitis virus (IBV) was reported to have only one proteolytically active PLpro, the PL2pro, because the PL1pro domain has lost its proteolytic activity due to accumulation of mutations in its active site during IBV evolution. Similarly, the SARS-CoV encodes only one PLpro corresponding to the PL2pro. It is therefore suggested that the SARS-CoV PL2pro is responsible for processing all of the three sites at the N-proximal pp1a/pp1ab regions (Ziebuhr, 2005). Due to their critical roles in coronavirus replication, especially in processing the polyproteins translated from the viral RNA, the 3CLpro along with the PLpro have been considered as putative antiviral drug targets. By targeting the viral proteases, the production of RdRp and helicase is inhibited, hence, the replication and transcription of the coronavirus genome will be disrupted (Gaurav and Al-Nema, 2019). It worthy to note, as no human proteases have a similar cleavage specificity to 3CLpro, specific inhibitors of 3CLpro are expected to be nontoxic (Zhang et al., 2020a). Three flavonoid compounds, herbacetin, rhoifolin and pectolinarin were reported to efficiently block the proteolytic activity of SARS-CoV 3CLpro (Jo et al., 2020). It is important to note that although the main function of the PLpro and 3CLpro is to proteolytically cleave the viral polyprotein in a coordinated manner, PLpro has an additional role to strip ubiquitin and ubiquitin-like interferon-stimulated gene product 15 (ISG15) from host-cell proteins, in order to help coronaviruses in generally to evade the host innate immune responses. Therefore, targeting PLpro with antiviral drugs has the potential advantage of not only inhibiting viral replication, but also preventing the dysregulation of signaling cascades in infected cells which may lead to cell death in surrounding, healthy cells (Báez-Santos et al., 2015).
The coronavirus RNA helicase represents the second most conserved protein for RNA synthesis and resides in the nsp13 domain. Based on conservation of specific sequence motifs, RNA helicases of positive (+) sense RNA viruses are classified into three large super-families, SF1, SF2 and SF3. The coronavirus RNA helicase belongs to the SF1 super-family (Ziebuhr, 2005). It is a motor protein which functions in an energy-dependent manner responsible for unwinding double-stranded RNA molecule using energy derived from the hydrolysis of nucleoside triphosphates (NTPs). All natural nucleotides and deoxynucleotides are substrates for coronavirus helicases, with ATP, dATP, and GTP being hydrolyzed slightly more efficiently than other nucleotides (Ivanov et al., 2004). Crystal structure analysis of full-length MERS-CoV helicase revealed that the enzyme possesses multiple domains, including an N-terminal Cys/His rich domain (CH) with three zinc atoms, a beta-barrel domain and two helicase core domains, the RecA1 and RecA2 domains. In addition, there is a stalk region connecting the CH domain and the beta-barrel domain. The CH domain has 15 conserved Cys/His residues, twelve of which participate in the coordination of the three zinc ions. In general, organization of the helicase domain is conserved throughout Nidoviruses and the individual domains of MERS-CoV are closely related to the equivalent eukaryotic domains of SF1 Up-frameshift 1 (Upf1) helicases (Hao et al., 2017). Although coronaviruses replicate their genomic RNA in the hosts cytoplasm, and the viral helicase may not localize to the nucleus of the cells (Ziebuhr, 2005), it has been shown that the MERS-CoV helicase possesses both RNA and DNA unwinding activity (Adedeji and Lazarus, 2016). The helicase activity can be enhanced by the RNA-dependent RNA polymerase (RdRp). The two enzymes are central components of the RTC. The coronavirus helicase has been identified as an ideal target for the development of anti-viral drugs because of its sequence conservation and indispensability across all coronavirus species (Hao et al., 2017; Jia et al., 2019).
The majority of viruses spend their entire life cycle in the cytoplasm of the host cells and have no access to the host polymerases. Therefore, viruses have to encode polymerases essential for their own transcription and replication (Gaurav and Al-Nema, 2019). For RNA viruses, the RdRp is the most conserved viral domain and is the most fundamental component of the viral replicase machinery (Shi and Lai, 2005; Ziebuhr, 2005). The RdRp domain of coronaviruses locates in the C-terminal part of nsp12 which catalyzes the replication and transcription of the coronavirus RNA genome. The size of the coronavirus nsp12 is about 930 amino acid residues which is larger than other known viral RdRp's, commonly about 500–600 amino acid residues. The C-terminal part, which represents about two-thirds of nsp12, has been found to align with the common viral RdRp subunit (Gaurav and Al-Nema, 2019). Structure analysis of the SARS-CoV nsp12 polymerase showed that the nsp12 polymerase binds to its essential co-factors, nsp7-nsp8 heterodimer, with a second nsp8 subunit occupying a distinct binding site. The presence of nsp7 and nsp8 co-factors significantly increases the RdRp activity. The polymerase domain consists of a fingers domain, a palm domain and a thumb domain. The SARS-CoV nsp12 also contains a Nidovirus-unique N-terminal extension. Notably, the SARS-CoV nsp12 contains two zinc-binding sites, one in the Nidovirus-unique extension and the other in the fingers domain. Both of these zinc-binding sites are distal to the active sites, suggesting that the ions are structural components of the folded protein and probably not involved in the enzymatic activity. Interestingly, all viral polymerases possess seven conserved motif regions (A, B, C, D, E, F, G) involved in template and nucleotide binding and catalysis (Kirchdoerfer and Ward, 2019). Similarly, cryo-electron microcopy structure of the nsp12 of the SARS-CoV-2 showed that it has a “right hand” RdRp domain and Nidovirus-unique N-terminal extension domain that adopts a Nidovirus RdRp-associated nucleotidyltransferase (NiRAN) architecture. The polymerase domain and NiRAN domain are connected by an interface (Gao et al., 2020). Domain organization of the COVID-19 virus nsp12 is shown in Figure 8.
Figure 8.
Schematic diagram of domain organization of the SARS-CoV-2 nsp12, a typical coronavirus nsp12. The C-terminal of coronavirus nsp12 contains an RNA dependent RNA polymerase (RdRp) domain. The polymerase domain consists of a fingers domain, a palm domain and a thumb domain. All viral polymerases possess seven conserved motif regions (A, B, C, D, E, F, G) involved in template and nucleotide binding and catalysis (Kirchdoerfer and Ward, 2019). The nsp12 of the SARS-CoV-2 has a nidovirus-unique N-terminal extension domain that adopts a nidovirus RdRp-associated nucleotidyltransferase (NiRAN) architecture. The polymerase domain and NiRAN domain are connected by an interface (Gao et al., 2020).
A homology model of SARS-CoV RdRp based on crystal structures of other known RNA viruses provided pivotal information about the potential functional roles of the conserved motifs and specific residues in the polymerization reaction. For example, the highly conserved Asp618 and Asp623 in motif A are proposed to function in metal ion chelation and recognition of a rNTP sugar ring, respectively. The Ser682 and Thr687 in motif B are hypothesized to play roles in recognition of template-primer, etc. Furthermore, it was suggested that the catalytic core of the SARS-CoV RdRp is formed by the Asp618 of motif A along with the Asp760 and Asp761 in motif C. These three aspartates are likely involved in binding divalent metal ions required for catalysis (Gaurav and Al-Nema, 2019). RdRp performs essentially the same basic replication and transcription functions as all the other viral polymerases, i.e., to copy the RNA template strand to generate a daughter strand. The process involves transfer of a nucleotidyl moiety of incoming NTP (complementary to template strand) to the 3ʹ-end of a growing RNA daughter strand. The polymerase needs two divalent metal cations, Mg2+/Mn2+ for activity. The polymerase active site has binding sites for the template strand, primer and the incoming NTP. The reaction is started by binding of template-primer and NTP, followed by incorporation of nucleoside monophosphate into the growing daughter strand with the release of pyrophosphate, and then translocation of the template strand and growing daughter strand (Gaurav and Al-Nema, 2019). The RdRp appears to be the primary target for the antiviral drug, Remdesivir (Gao et al., 2020). Remdesivir is a nucleotide analog inhibitor of RdRps. The triphosphate form of Remdesivir (RDV-TP) competes with its natural counterpart ATP. Interestingly, RDV-TP seems to be more efficiently incorporated than ATP. After incorporated, Remdesivir arrests RNA synthesis which most likely leads to delayed RNA chain termination (Gordon et al., 2020). In addition, the RdRp gene, located at the 5’ region of ORF 1b (Figure 6), has been an important target for coronavirus molecular detection by PCR (Artika et al., 2020; Corman et al., 2020).
Apart from virally encoded proteins, coronavirus replication also employs several cellular proteins, such as the heterogeneous nuclear ribonucleoprotein A 1 (hnRNP-A1), polypyrimidine-tract-binding (PTB) protein, poly(A)-binding protein (PABP), and mitochondrial aconitase. As typical for viruses, coronavirus may hijack these cellular proteins from their normal roles to function in the viral replication process. The fact that no coronavirus proteins in an infected cell extract could be cross-linked to the viral RNA in vitro, suggests that viral proteins may interact with viral RNA only indirectly through the mediation of cellular proteins. Of note is that a number of cellular proteins have been shown to bind to the regulatory elements of MHV RNA. In MHV, the RNA-binding cellular hnRNP-A1 has been suggested to play roles in the regulation of coronavirus transcription. It may also function in facilitating mRNA translation. Similarly, the PTB has also been suggested to regulate viral mRNA translation. The PABP has been revealed to interact specifically with poly(A), which is an important cis-acting signal for coronavirus RNA replication. Because coronavirus RNA is capped and polyadenylated similar to the host mRNAs, PABP is also thought to function in the translation of the coronavirus genome upon virus entry into the cells, which is required for efficient coronavirus RNA replication. The mitochondrial aconitase has been shown to bind specifically to the 3′ protein-binding element of the MHV genomic RNA and has been suggested to interact with the MHV replication complexes. In addition, it is hypothesized that the binding of the mitochondrial aconitase to the 3′-UTR of the MHV genomic RNA increases the stability of the viral mRNAs and hence improves the translation of viral proteins. It is believed that additional host cellular proteins may also interact with coronavirus RNA and are essential for viral replication (Shi and Lai, 2005).
Coronavirus genomic replication and transcription occurs in the cytoplasm of the hosts involving coronavirus-induced host membranous rearrangements of varying morphologies that serve as platforms for the viral replication and transcription complexes (RTCs). These organelle-like replicative structures act as a framework for viral genome replication by localizing and concentrating the necessary factors and most likely providing protection from the anti-viral host defense mechanisms of the infected cell. The biogenesis of these replicative structures involves the concerted actions of hijacked host and viral membrane shaping proteins, lipid-modifying enzymes and various exploited cellular pathways. The coronavirus-induced replicative structures are mostly in the form of double-membrane vesicles (DMVs) and convoluted membranes (CMs), interconnected with a reticulovesicular network of modified membranes, which seem to be continuous with the endoplasmic reticulum (ER). The replicase proteins are localized to the DMVs and CMs. These replicative structures together with their localized proteins, are called the replication-transcription complex (RTC). The double-stranded RNA (dsRNA) believed to function as replicative intermediate during viral RNA synthesis was detected in the interior of the DMVs. Additional small double-membrane spherule-like structures associated with “zippered” ER membranes were also observed in infectious bronchitis virus (IBV)-infected cells, but not in SARS-CoV, MHV- or MERS-CoV-infected cells (Angelini et al., 2013: Hagemeijer et al., 2014). Studies of SARS-CoV-infected cells showed that the nonstructural proteins, nsp3, nsp4 and nsp6 play roles in inducing the formation of the double-membrane vesicles. It is worth noting that among the 16 nsps, these three proteins contain multiple hydrophobic, membrane-spanning domains. The luminal loops of the nsp3 and nsp4 are critical for formation of the replicative structures (Angelini et al., 2013; Hagemeijer et al., 2014). Together, nsp3 and nsp4 were shown to be able to pair membranes. The nsp6 alone has the ability to induce the formation small spherical single-membrane vesicles around host cells microtubule-organizing centers. In collaboration, the nsp3, nsp4, and nsp6 have the ability to induce double-membrane vesicles (Angelini et al., 2013). A small chemical compound designated K22 has been identified as a potent inhibitor of double-membrane vesicle formation. Furthermore, the K22 nearly totally prevented viral RNA synthesis. Hence, it has been suggested as a potential anti-coronavirus drug (Lundin et al., 2014).
Being obligate intracellular parasites, coronaviruses also exploit the translational apparatus of the infected cell in order to synthesize their proteins, in a process which may accompanied by inhibition of the hosts cellular protein synthesis. The process of protein synthesis in typical eukaryotic cells, which occurs in cytoplasm, involves an initiation step, in which the mRNA is recognized by the host translational machinery, and a methionyl initiator tRNA (Met-tRNAMet) binds at the ribosomal peptidyl (P) site to read the start codon of the mRNA. This is followed by an elongation step in which aminoacyl tRNAs enter the acceptor (A) site and, if the correct tRNA is bound, the formation of a peptide bond is catalyzed by the ribosome. The following step is the translocation of the tRNAs and mRNA such that the next codon is moved into the A site, and the process is repeated. When a stop codon is encountered, the translation process is terminated, followed by the release of the peptide from the ribosome (Nakagawa et al., 2016). It is important to note that similar to the eukaryotic mRNAs, the 5’ ends of coronavirus mRNAs are capped and methylated. This is believed to help viral RNAs escape from recognition by the host innate immune system (Chen and Guo, 2016). Coronavirus protein synthesis involves a cap-dependent translation mechanism. In addition, the process also employs regulatory mechanisms, such as ribosomal frameshifting. After the virus successfully enters the cell, the viral genome RNA is translated to synthesize the viral proteins which are necessary for subsequent RNA replication and transcription. This process results in polyprotein (pp) 1a and 1ab. As mentioned earlier, the synthesis of pp1ab involves a minus 1 (-1) ribosomal frameshift until the translation stop codon is reached. Therefore, the polyprotein 1ab is encoded by a (functionally) fused ORF produced from the two ORFs 1a and 1b (Nakagawa et al., 2016). For the structural proteins, after synthesis, the S, E, and M proteins are inserted into the rough endoplasmic reticulum. From there, these structural proteins travel along the secretory pathway to the endoplasmic reticulum-Golgi apparatus intermediate compartment (ERGIC) which is the location of the coronavirus particle assembly (Tang et al., 2020).
3.3. Virion assembly and budding
One of the distinctive features of coronaviruses is the location of their virion assembly. For most enveloped viruses, virion assembly takes place at the host cells plasma membrane. For coronaviruses, however, virion budding and assembly occurs at the endoplasmic reticulum-Golgi intermediate compartment (ERGIC). Coronaviruses, therefore, obtain their membrane envelope from ERGIC (Ujike and Taguchi, 2015; Schoeman and Fielding, 2019). For efficient coronavirus virion assembly, the three membrane (enveloped) proteins must be retained near the ERGIC. In fact, the M, E and some S proteins have intracellular trafficking signals which target these structural proteins to the budding site where they accumulate. Therefore, the efficiency of viral proteins incorporation into coronavirus virions is determined by protein trafficking to the ERGIC and protein–protein interactions at the ERGIC (Ujike and Taguchi, 2015). Studies of SARS-CoV have revealed that following translation, four structural proteins, the spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins, enter the secretory pathway in the ERGIC, where they are assembled into virions (Woo et al., 2019). Most of the protein-protein interactions required for coronavirus assembly are mediated by the M proteins. The coronavirus packaging signal (PS), a cis-regulatory element encoded within the viral RNA, functions in packaging the viral genome into the ribonucleocapsid (Woo et al., 2019). The nucleocapsid (N) phosphoprotein plays a fundamental role during viral self-assembly and one of its critical functions is to form the viral genome into a helical ribonucleocapsid (ribonucleoprotein, RNP). Viral N–N self-interactions are thought to be necessary for formation of the ribonucleocapsid and subsequent assembly of the viral particles (Chang et al., 2014).
In the presence of a great excess of subgenomic RNA species, coronaviruses have the ability to select the genomic positive (+) sense single stranded RNA to be packaged into assembled virions. This high degree of selectivity is mediated by the coronaviruses genomic PS, a critical element for genomic RNA packaging, originally identified in MHV (Kuo and Masters, 2013). One of the most characterized PS elements, called psi, is located at the 5′ leader region of the HIV genome. Two viral proteins, the N protein and the M protein, have been suggested to play roles in recognizing the PS. The coronavirus N protein has two highly basic domains, the NTD and CTD, and a mostly acidic carboxy-terminal domain, termed N3 within the C-terminal tail (CT) (Figure 5). The CTD and the N3 domains have been proposed to recognize the PS (Masters, 2019). In vivo studies of SARS-CoV have also indicated that both the N-terminal and C-terminal domains of the N protein are crucial for recognition in the packaging RNA (Woo et al., 2019). Notably, the N protein plays a critical role to wrap the viral genome into a helical nucleoprotein which is bound to the network of M endodomains at the site of virion budding, at the endoplasmic reticulum-Golgi intermediate compartment. The association of the N and M molecules is mediated by their respective carboxy termini. Because the M protein is the most abundant virion structural protein, there should be sufficient M endodomain tails on the internal surface of the virion membrane to interact with the domain N protein in the nucleocapsid. In addition, the M protein (possibly with the assistance of E) has also been hypothesized to recognize and bind to the PS (Masters, 2019).
Once the viral gene expression and genome replication is ongoing, coronavirus progenies can begin to assemble. The coronavirus M protein has been recognized as the central organizer for the virion assembly (Masters, 2006). The M protein has the ability to form virus-like particles (VLPs) in the presence of N protein or E protein, suggesting its pivotal role in the virion assembly (Tseng et al., 2010). The coronavirus assembly is likely mediated by specific interactions of the M protein with S, N, and E proteins. However, the detailed molecular mechanism of N protein packaging inside the virion and the interaction between N and other proteins has yet to be elucidated (Chang et al., 2014). The E protein has been suggested to play roles in inducing membrane curvature which permits coronavirus particles to acquire their spherical shape and morphology (Schoeman and Fielding, 2019). The generation of mature virions involves insertion into the endoplasmic reticulum (ER) of the coronavirus structural proteins, S, E, and M. These proteins travel along the secretory pathway into the ERGIC and are inserted into the membrane of the ERGIC. The ERGIC is also a location where the viral genomes are encapsidated by the N protein. The structural proteins then interact with the encapsidated viral genomes and assemble into mature coronavirus particles by budding (Fehr and Perlman, 2015). Following assembly, the progeny virions accumulate in smooth-walled vesicles are transported to the cell surface, and released into the extracellular space through exocytosis or cell lysis (Orenstein et al., 2008) (Figure 7).
4. Molecular characteristics of SARS-CoV-2
Analysis of full-length genome sequences of five patients from the early outbreak stages of the SARS-CoV-2 (alternatively called 2019-nCoV) showed that these genomes sequences were almost identical to each other, with more than 99.9% sequence identity. The length of the genomes obtained was 29,891 bases which shared 79.6% identity to SARS-CoV. As with other coronaviruses, the SARS-CoV-2 genome harbors six major ORFs and a number of ORFs which encode accessory proteins. Further analysis indicated that some of the SARS-CoV-2 genes have less than 80% nucleotide sequence identity to the corresponding genes of SARS-CoV. However, when amino acid sequences of seven conserved replicase domains in ORF1ab (used for coronavirus species classification) were compared, there was 94.4% sequence identity between SARS-CoV-2 and SARS-CoV. It was concluded that the SARS-CoV-2 and SARS-CoV are of the same species, both being SARS related coronavirus (SARSr-CoV) (Zhou et al., 2020). Interestingly, further studies showed that the full-length genome of SARS-CoV-2 has high similarity to the genome of a bat coronavirus, BatCoVRaTG13, detected in Rhinolophus affinis, with overall sequence identity of 96.2%. Phylogenetic analysis was then carried out for the full-length genome, the spike gene, and the RdRp gene. Results showed that RaTG13 is the closest relative of the SARS-CoV-2 and they form a distinct lineage from the other SARSr-CoVs. The close phylogenetic relationship to RaTG13 suggests that the SARS-CoV-2 may have originated in bats (Zhou et al., 2020). Similarly, other studies showed that genome sequences of SARS-CoV-2 obtained from nine patients among the early outbreak cases are almost identical to each other. The genome sequences also suggested that the SARS-CoV-2 is most closely related to other betacoronaviruses of bat origin, indicating that bats are the most likely reservoir hosts for SARS-CoV-2 (Lu et al., 2020). However, several facts suggested that another animal might function as an intermediate host between bats and humans. In addition to the general ecological separation of bats from humans, the outbreaks occurred when most bats in Wuhan were hibernating and no bats were sold or found in the Huanan seafood market the site which was linked to the emergence of the SARS-CoV-2. Therefore, other mammalian species may act as an intermediate or amplifying host, allowing SARS-CoV-2 to acquire mutations needed for efficient transmission to humans. Notably, in the case of SARS and MERS, civets and camels, played a role as intermediate hosts, respectively (Lu et al., 2020; Zhang and Holmes, 2020).
Exploring the potential intermediate hosts of SARS-CoV-2 is critical for blocking its interspecies transmission. In attempt to discover intermediate hosts of SARS-CoV-2, Zhang et al. (2020b) analyzed published genomic sequence data of SARS-CoV-like coronaviruses. They found that a SARS-CoV-2-like coronavirus, named Pangolin-CoV, isolated from Malayan pangolins shows 91.02% genomic identity to SARS-CoV-2 and 90.55% identity to the BatCoV RaTG13. The high overall sequence identity of the Pangolin-CoV to the SARS-CoV-2 and RaTG13 indicated that Pangolin-CoV could be the second closest relative of SARS-CoV-2 after RaTG13, and Pangolin-CoV is likely the common origin of SARS-CoV-2 and RaTG13 (Zhang et al., 2020b). Interestingly, the S1 protein of Pangolin-CoV is much more closely related to that in SARS-CoV-2 compared to that in RaTG13, with five key amino acids in the RBD being involved in recognition of human ACE2, are completely consistent between Pangolin-CoV and SARS-CoV-2. In contrast, four amino acid changes are found in RaTG13 (Zhang et al., 2020b). Similarly, Lam et al. (2020) identified SARS-CoV-2-related coronaviruses in Malayan pangolins (Manis javanica) with genome sequence similarity of 85.5%–92.4% to genome sequence of SARS-CoV-2. In addition, they also investigated SARS-CoV-2-related coronaviruses from Guangdong pangolins. Interestingly, the RBD domain of the Guangdong pangolin coronaviruses shared 97.4% sequence similarity to the RBD of SARS-CoV-2. The Guangdong pangolin coronaviruses and SARS-CoV-2 possess identical amino acids at the five critical residues of the RBD, while RaTG13 only shares one amino acid with SARSCoV-2. The high similarity of pangolin SARS-CoV-2-related coronaviruses to SARS-CoV-2 suggests that these pangolins should be considered as possible hosts of SARS-CoV-2 and may play critical roles in the emergence of novel coronaviruses generally (Lam et al., 2020). As most of the laboratory-confirmed cases of the initial SARS-CoV-2 outbreaks were linked to the Wuhan seafood market, identification of source or intermediate host of SARS-CoV-2 was focused on animals which also sold in the market such as snakes, birds and other small mammals. However, no specific animal associated with SARS-CoV-2 was conclusively identified. Owing to fact that the genetic sequences of pangolin SARS-CoV-2-related coronaviruses and SARS-CoV-2 show high similarity, the most likely intermediate host candidate is believed to be pangolin (Prompetchara et al., 2020).
The SARS-CoV-2 has been spreading rapidly and globally. The surface-exposed transmembrane spike (S) glycoprotein plays an important role in mediating coronavirus entry into the host cells. Therefore, molecular characterization of SARS-CoV-2 S protein is highly important. It was found that the S gene of SARS-CoV-2 and RaTG13 was longer than those of other SARS related coronaviruses. Other important differences in the S gene of SARS-CoV-2 include three short insertions in the N terminal domain and changes in four out of five key residues in the receptor binding motif (RBM) compared with the S gene sequences of SARS-CoV. The significant of these changes needs further elucidation (Zhou et al., 2020). Furthermore, virus infectivity experiments employing HeLa cell lines which express or do not express human ACE2 proteins were aimed to determine the SARS-CoV-2 receptor usage. Results confirmed that the SARS-CoV-2 uses the human ACE2 as an entry receptor, the same as the SARS-CoV. The experiments also showed that the SARS-CoV-2 does not employ other coronavirus receptors such as the aminopeptidase N (APN) and the dipeptidylpeptidase 4 (DPP4) (Zhou et al., 2020).
In attempt to reveal the structural basis of receptor recognition by SARS-CoV-2, Shang and coworkers (2020) elucidated the crystal structure of the SARS-CoV-2 receptor binding domain (RBD) in a complex with human ACE2. They found that compared to the SARS-CoV RBD, the binding of SARS-CoV-2 RBD to human ACE2 results in a more compact conformation. In addition, some residues changes in SARS-CoV-2 RBD stabilize the virus-binding hotspots at the interface between SARS-CoV-2 RBD and human ACE2. In particular, structural changes were observed in SARS-CoV-2 receptor binding motif (RBM) due to a four-residue motif (residues 482–485: Gly-Val-Glu-Gly) which lead to a tighter contact between the SARS-CoV-2 RBM and the human ACE2. Moreover, the Phe486 of SARS-CoV-2 RBM inserts into a hydrophobic pocket of human ACE2 causing stronger contact compared to a corresponding smaller leucine residue in SARS-CoV RBM. These data have provided important structural and molecular characteristics information of SARS-CoV-2 as a basis for its enhanced binding affinity to its human ACE receptor. Taken together, the SARS-CoV-2 RBD recognizes the human ACE2 receptor better than SARS-CoV RBD does (Shang et al., 2020).
SARS-CoV, MERS-CoV and SARS-CoV-2 are three highly pathogenic coronaviruses which have crossed the species barrier to cause deadly pneumonia in humans (Walls et al., 2020). Similar to SARS-CoV (Artika and Ma'roef, 2017) and MERS-CoV (Xiao et al., 2018), the SARS-CoV-2 was proposed to be an airborne transmitted pathogen (Zhou et al., 2020), and therefore all possible precautions against airborne transmission in indoor scenarios should be taken (Morawska and Cao, 2020). Although the case fatality rate of SARS-CoV-2 (3.7%) (WHO, 2020) is lower compared to those of SARS-CoV (9.14%) and MERS-CoV (34.4%), it is evident that the SARS-CoV-2 is more infectious leading to very different epidemiological dynamics. Elucidation of the molecular characteristics underlying such adaptability and transmissibility in humans is therefore very important (Prompetchara et al., 2020; Zhang and Holmes, 2020). The genomic sequence data obtained from patients at the early stages of the COVID-19 outbreak clearly identified the SARS-CoV-2 belongs to betacoronavirus lineage B (Sarbecovirus), within the same lineage as the SARS-CoV. Notably, the MERS-CoV falls in lineage C (Marbecovirus) (Zhang and Holmes, 2020). One of the notable molecular characteristics of the SARS-CoV-2 is the presence of a polybasic cleavage site (PRRARSV) at the junction of S1 and S2 of the S protein due to four amino acid residue insertion (PRRA) at SARS-CoV-2 S positions 681–684 (Andersen et al., 2020; Walls et al., 2020). This furin recognition motif permits effective cleavage by furin and other protease and determines viral infectivity and host range. The functional consequences of the polybasic cleavage site of the SARS-CoV-2 have yet to be elucidated, but may increase the virus infectivity and have impact on transmissibility and pathogenicity of the virus (Andersen et al., 2020; Zhang et al., 2020b; Walls et al., 2020). Surprisingly, the polybasic cleavage sites are absent in S protein of RaTG13, the bat virus closest relative of SARS-CoV-2. These cleavage sites also have not been identified in lineage B coronaviruses such as SARS-CoV, although it is found in other human betacoronaviruses such as HKU1 (lineage A). The polybasic cleavage site is a feature of the highly pathogenic avian influenza viruses (Karo-karo et al., 2019; Andersen et al., 2020). In these viruses, acquisition of a polybasic cleavage site in hemagglutinin (HA) protein converts the low pathogenic avian influenza viruses into highly pathogenic viruses. The polybasic cleavage site is selected for rapid replication and transmission by avian influenza viruses (Andersen et al., 2020). In addition, there is also a proline insertion in the polybasic cleavage sites of the S protein of SARS-CoV-2. The presence of the proline residue creates a turn which is predicted to result in attachment of O-linked glycans to residues S673, T678 and S686 flanking the cleavage site. The consequences of these sets of glycosylation, which are unique to SARS-CoV-2 S protein are not known, but they may create a mucin-like domain covering epitopes or key residues of SARS-CoV-2 S protein involved in evading the host immune response (Andersen et al., 2020).
Depending on virus strains and the availability of the host cell proteases, the S protein may be processed by one or several proteases such as furin, trypsin, cathepsin, transmembrane protease serine protease-2 (TMPRSS-2), TMPRSS-4, or human airway trypsin-like protease (HAT) to facilitate virus entry. The cleavage by type II membrane serine protease (TMPRSS) can activate fusion potential of the S protein and induce receptor-dependent formation of giant, multinucleated cells, termed syncytia. Using a lentiviral pseudotype system, Ou et al. (2020) showed that SARS-CoV-2 S protein is proteolytically activated by cathepsin L. Similar to SARS-CoV, the SARS-CoV-2 S protein-mediated cell-cell fusion was enhanced by TMPRSS 2, 4, 11A, 11D and 11E. In addition, trypsin was also found to trigger SARS-CoV and SARS-CoV-2 to induce synsytia formation. Interestingly, the S protein of SARS-CoV-2 (but not SARS-CoV) was shown to induce syncytia formation without the presence of trypsin, suggesting that SARS-CoV-2 S protein could be triggered upon receptor binding in the absence of exogenous protease activation (Ou et al., 2020). Similarly, Xia et al. (2020) also reported a typical phenomenon of natural syncytia formation in cells infected by SARS-CoV-2. To the contrary, SARS-CoV S protein lacked the ability to mediate the cell–cell fusion under the same conditions of cell–cell fusion system they used (Xia et al., 2020). Syncytia formation has been proposed as a strategy of coronaviruses to promote cell-cell fusion between infected and neighboring uninfected cells so as to permit direct spreading of the virus between cells, evading virus-neutralizing antibodies (Schoeman and Fielding, 2019).
In general, the SARS-CoV-2 nucleocapsid (N) protein-encoding regions are conserved. However, a few variations such as S194L, K249I, P344S, in the N protein of different SARS-CoV-2 strains were found (Kang et al., 2020). In addition, the codon usage pattern of SARS-CoV-2 has been analyzed by comparing its codon usage with that of other viruses of the subfamily of Orthocoronavirinae. It was found that SARS-CoV-2 has a high AU content which significantly influences its codon usage and may lead to better adaptation to the human host. Studies of evolutionary pressures which dictate codon usage of genes encoding the viral replicase, spike, envelope, membrane and nucleocapsid proteins suggested that different patterns of mutational bias and natural selection affects the codon usage of these genes. The matrix (M) and envelope (E) genes tend to evolve slowly by accumulating nucleotide mutations, while genes encoding nucleocapsid (N), viral replicase and spike proteins (S), tend to evolve relatively faster (Dilucca et al., 2020). Moreover, analysis of codon usage bias, especially in spike, envelope and main protease genes suggested that SARS-CoV-2 has a higher gene expression efficiency compared to SARS-CoV and MERS-CoV. SARS-CoV-2 prefers pyrimidine rich codons to purines. Most of high frequency codons end with A or T (Kandeel et al., 2020). Currently, the coding potential of SARS-CoV-2 is not fully elucidated. The presence ORF9a (internal to N), ORF3h (within ORF3a) and a putative ORF10 have been proposed (Cagliani et al., 2020).
Since the beginning of its emergence, there has been considerable discussion regarding the origin of the virus, including whether the SARS-CoV-2 is a laboratory construct or a purposefully engineered virus (Andersen et al., 2020). Indeed, with the current advanced powerful genetic engineering techniques, a virus can be modified, redesigned, reconstructed or even synthesized. This can potentially bring about a novel virus which is unprecedented in nature (Artika and Ma'roef, 2018). The molecular characteristics of the SARS-CoV-2 indicated that the virus is not a product of purposeful manipulation. For example, although the RBD of the SARS-CoV-2 seems to bind with high affinity to ACE2 from humans and other animals with high receptor homology, the interaction is predicted to be not ideal. In addition, the sequence of the SARS-CoV-2 RBD is different from that of SARS-CoV which has optimal receptor binding. This implies that the high-affinity binding of the SARS-CoV-2 spike protein to human ACE2 is most likely due to natural selection (Andersen et al., 2020). Moreover, if genetic engineering had been carried out, one of the several known genetic engineering systems available for betacoronaviruses would probably have been applied. The genetic data did not indicate that the SARS-CoV-2 is derived from any previously used virus backbone. Therefore it is hypothesized that the SARS-CoV-2 emerged through natural selection either in an animal host or directly in humans. The facts that some pangolin coronavirus RBDs show strong similarity to that of SARS-CoV-2 suggest that the S protein of the SARS-CoV-2 has undergone optimization for binding to human-like ACE2 by natural selection (Andersen et al., 2020).
To date, no vaccines or therapeutics are approved against any human-infecting coronaviruses including SARS-CoV-2. Several options for countermeasures can be developed to control or prevent SARS-CoV-2 infections, including vaccines, monoclonal antibodies, oligonucleotide-based therapies, peptides, interferon therapies and low molecular weight drugs (Li and De Clercq, 2020). Prior to the emergence of SARS-CoV-2, various strategies have been developed for a coronavirus vaccine which can potentially be adopted for SARS-CoV-2 vaccine development. These include viral vector-based vaccines, subunit vaccines, recombinant proteins, and DNA vaccines (Enjuanes et al., 2016; Chen et al., 2020). Molecular biology techniques which have been used for development of viral countermeasures, can also be employed to develop vaccine and antiviral drugs against SARS-CoV-2. For example, the molecular techniques can be applied for rapid development of subunit vaccines against SARS-CoV-2 based on recombinant viral antigenic proteins. This type of vaccine primarily contains specific viral antigenic fragments, without inclusion of any infectious viruses, therefore eliminating the concerns of incomplete inactivation, virulence recovery, or pre-existing immunity. Previous studies showed that recombinant SARS-CoV receptor binding domains (RBDs) which are stably or transiently expressed in Chinese hamster ovary (CHO) cells, bind strongly to RBD-specific monoclonal antibodies, elicit high titer anti-SARS-CoV neutralizing antibodies, and have the ability to protect most or all of the SARS-CoV-challenged mice (Wang et al., 2020b). Molecular biology techniques can also be employed in development and production of antiviral drugs such as recombinant human interferon (Wipf et al., 1994; Artika et al., 2013; Landowski et al., 2016). Recently, Mantlo et al. (2020) reported that SARS-CoV-2 is sensitive to recombinant human interferons α and β (IFNα/β). Treatment with IFN-α or IFN-β was found to significantly reduce viral titers in Vero cells. Importantly, they observed that SARS-CoV-2 is more sensitive than many other human pathogenic viruses, including SARS-CoV, to human type I interferons. These suggested that of human type I interferons have the potential efficacy in suppressing SARS-CoV-2 infection, and may be one of the potential future treatment options for COVID-19 (Mantlo et al., 2020). Recombinant human interferon can efficiently be produced in microbial cells by cloning the interferon genes into an expression vector followed by expressing the cloned genes in a host system. The interferon molecules are then purified. Microbial cells which have been used as host systems include Escherichia coli, Saccharomyces cerevisiae, Bacillus subtilis, Pichia pastoris, Lactococcus lactis, Yarrowia lipolytica, and Trichoderma reesei (Wipf et al., 1994; Artika et al., 2013; Landowski et al., 2016). In general, recombinant proteins for treating diseases are mainly produced using prokaryotic and eukaryotic expression host systems such as bacteria, yeast, insect cells, mammalian cells, and transgenic plants at laboratory scale as well as in large-scale settings (Tripathi and Shrivastava, 2019).
5. Conclusions
Coronaviruses regularly emerge and pose a major threat to both humans and animal health. The rapid global spread and the high death toll being claimed by COVID-19 clearly demonstrates that both the developed and developing nations are unprepared in controlling the latest highly-pathogenic human coronavirus emergence. Better understanding of the molecular biology of coronaviruses is critical to elucidate their emergence, origin, evolution, diversity, pathogenesis and epidemiology. Complete information of molecular characteristics of circulating coronaviruses is important for development of effective diagnostic tools to detect these viruses. Whenever new coronaviruses emerge, it is most important to have the capacity for their rapid detection in order to implement appropriate control measures and limit their spread. Detailed insights into the molecular mechanisms underlying their pathogenesis are also crucial for development of effective and safe vaccines and therapeutics. In addition, it is important to identify, at the molecular level, the biological and environmental factors which may contribute to the distribution and prevention of coronavirus diseases across populations. The highly pathogenic human coronaviruses are believed to be zoonotic. Therefore, understanding the molecular mechanism which drives the cross-species transmission of coronaviruses is critical. Future studies aimed at elucidating how animal viruses cross species barriers and efficiently infect humans will help in the prevention of future zoonotic events. Continuous monitoring and analysis of genome sequences is vital to understand the genetic evolution and rates of genomic nucleotide substitution of the coronaviruses.
Declarations
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We thank Dr. John Acton for his assistance at the manuscript stage.
References
- Adedeji A.O., Lazarus H. Biochemical characterization of Middle East respiratory syndrome coronavirus helicase. mSphere. 2016;1:1–14. doi: 10.1128/mSphere.00235-16. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5014916/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. https://science.sciencemag.org/content/300/5626/1763 [DOI] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020 doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini A.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4:1–10. doi: 10.1128/mBio.00524-13. https://mbio.asm.org/content/4/4/e00524-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anindita P.D., Sasaki M., Setiyono A., Handharyani E., Orba Y., Kobayashi S., Rahmadani I., Taha S., Adiani S., Subangkit M., Nakamura I., Sawa H., Kimura T. Detection of coronavirus genomes in Moluccan naked-backed fruit bats in Indonesia. Arch. Virol. 2015;160:1113–1118. doi: 10.1007/s00705-015-2342-1. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7086880/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony S.J., Johnson C.K., Greig D.J., Kramer S., Che X., Wells H., Hicks A.L., Joly D.O., Nathan D., Wolfe N.D., Daszak P., Karesh W., Lipkin W.I., Morse S.S., By PREDICT Consortium. Mazet J.A.K., Goldstein T. Global patterns in coronavirus diversity. Virus Evol. 2017;3:1–15. doi: 10.1093/ve/vex012. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5467638/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt A.L., Larson B.J., Hogue B.G. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. J. Virol. 2010;84:11418–11428. doi: 10.1128/JVI.01131-10. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2953170/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artika I.M., Budirahardja Y., Ekowati A.L. Molecular cloning and heterologous expression of human interferon alpha2b gene. Am. J. Biochem. Biotechnol. 2013;9:423–429. https://thescipub.com/pdf/10.3844/ajbbsp.2013.423.429.pdf [Google Scholar]
- Artika I.M., Ma’roef C.N. Laboratory biosafety for handling emerging viruses. Asian Pac. J. Trop. Biomed. 2017;7:483–491. doi: 10.1016/j.apjtb.2017.01.020. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7103938/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artika I.M., Ma’roef C.N. Current laboratory biosecurity for handling pathogenic viruses. Am. J. Biochem. Biotechnol. 2018;14:108–116. https://thescipub.com/pdf/10.3844/ajbbsp.2018.108.116.pdf [Google Scholar]
- Artika I.M., Wiyatno A., Ma’roef C.N. Pathogenic viruses: molecular detection and characterization. Infect. Genet. Evol. 2020;81:1–14. doi: 10.1016/j.meegid.2020.104215. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7106233/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez-Santos Y.M., St. John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5896749/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biosafety in Microbiological and Biomedical Laboratories (BMBL) fifth ed. National Institutes of Health; USA: 2009. U.S. Department of Health and Human Services: Public Health Service, Centers for Disease Control and Prevention.https://www.cdc.gov/labs/BMBL.html [Google Scholar]
- Burkard C., Verheije M.H., Wicht O., van Kasteren S., van Kuppeveld F.J., Haagmans B.L., Pelkmans L., Rottier J.M., Bosch B.J., de Haan C.A.M. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10:1–17. doi: 10.1371/journal.ppat.1004502. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4223067/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliani R., Forni D., Clerici M., Sironi M. Coding potential and sequence conservation of SARS-CoV-2 and related animal viruses. Infect. Genet. Evol. 2020;5(83):104353. doi: 10.1016/j.meegid.2020.104353. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7199688/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Hou M.-H., Chang C.-F., Hsiao C.-D., Huang T.-H. The SARS coronavirus nucleocapsid protein – forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7113676/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Guo D. Molecular mechanisms of coronavirus RNA capping and methylation. Virol. Sin. 2016;31:3–11. doi: 10.1007/s12250-016-3726-4. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7091378/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7167049/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck C.-P., Chow H.-F., Wan D.C.-C., Wong K.-B. Profiling of substrate specificities of 3C-like proteases from group 1, 2a, 2b, and 3 coronaviruses. PloS One. 2011;6:1–6. doi: 10.1371/journal.pone.0027228. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0027228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Kriegenburg F., de Haan C.A.M., Reggiori F. Coronavirus nucleocapsid proteins assemble constitutively in high molecular oligomers. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-06062-w. https://www.nature.com/articles/s41598-017-06062-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:1–8. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6988269/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. www.nature.com/articles/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., Smeets M., Vernooij F., Vennema H., Rottier P.J.M. Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein. J. Virol. 1999;73:7441–7452. doi: 10.1128/jvi.73.9.7441-7452.1999. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC104271/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. https://www.nature.com/articles/nrmicro.2016.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desforges M., Desjardins J., Zhang C., Talbot P.J. The acetyl-esterase activity of the hemagglutinin-esterase protein of human coronavirus OC43 strongly enhances the production of infectious virus. J. Virol. 2013;87:3097–3107. doi: 10.1128/JVI.02699-12. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3592170/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A., Spiro D. Advancing full length genome sequencing for human RNA viral pathogens. Future Virol. 2009;4:47–53. doi: 10.2217/17460794.4.1.47. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2638583/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilucca M., Forcelloni S., Georgakilas A.G., Giansanti A., Pavlopoulou A. Codon usage and phenotypic divergences of SARS-CoV-2 genes. Viruses. 2020;12:1–21. doi: 10.3390/v12050498. https://www.mdpi.com/1999-4915/12/5/498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L., Zuñiga S., Castaño-Rodriguez C., Gutierrez-Alvarez J., Canton J., Sola I. Molecular basis of coronavirus virulence and vaccine development. Adv. Virus Res. 2016;96:245–286. doi: 10.1016/bs.aivir.2016.08.003. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7112271/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Zhao K., Shi Z.-L., Zhou P. Bat coronaviruses in China. Viruses. 2019;11:1–14. doi: 10.3390/v11030210. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6466186/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. https://link.springer.com/protocol/10.1007/978-1-4939-2438-7_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7111218/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 2018;13:405–443. doi: 10.2217/fvl-2018-0008. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7080180/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yan Y., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. https://science.sciencemag.org/content/368/6492/779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaurav A., Al-Nema M. Polymerases of coronaviruses: structure, function, and inhibitors. Viral Polymerases. 2019:271–300. [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. The antiviral compound Remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. https://www.jbc.org/content/295/15/4773.full [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner J. Three emerging coronaviruses in two decades: the story of SARS, MERS, and Now COVID-19. Am. J. Clin. Pathol. 2020;153:420–421. doi: 10.1093/ajcp/aqaa029. https://academic.oup.com/ajcp/article/153/4/420/5735509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M., Liu X., Guo D., Zhang Z., Yin C.-C., Chen Y., Xiang Y. Electron microscopy studies of the coronavirus ribonucleoprotein complex. Protein Cell. 2017;8:219–224. doi: 10.1007/s13238-016-0352-8. https://link.springer.com/article/10.1007/s13238-016-0352-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Monastyrska I., Griffith J., van der Sluijs P., Voortman J., van Bergen en Henegouwen P.M., Vonk A.M., Rottier P.J.M., Reggiori F., de Haan C.A.M. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology. 2014;458–459:125–135. doi: 10.1016/j.virol.2014.04.027. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7111329/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W., Wojdyla J.A., Zhao R., Han R., Das R., Zlatev I., Manoharan M., Wang M., Cui S. Crystal structure of Middle East respiratory syndrome coronavirus helicase. PLoS Pathog. 2017:1–19. doi: 10.1371/journal.ppat.1006474. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5501694/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P.W., Pfeiffer D., Oshitani H. Prospects for emerging infections in East and Southeast Asia 10 years after severe acute respiratory syndrome. Emerg. Infect. Dis. 2013;19:853–860. doi: 10.3201/eid1906.121783. https://wwwnc.cdc.gov/eid/article/19/6/12-1783_article [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Ge X., Wang L.-F., Shi Z. Bat origin of human coronaviruses. Virol. J. 2015;2015:1–10. doi: 10.1186/s12985-015-0422-1. https://virologyj.biomedcentral.com/track/pdf/10.1186/s12985-015-0422-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wei P., Fan K., Liu Y., Lai L. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry. 2004;43:4568–4574. doi: 10.1021/bi036022q. https://pubs.acs.org/doi/pdf/10.1021/bi036022q [DOI] [PubMed] [Google Scholar]
- Huang X., Dong W., Milewska A., Golda A., Qi Y., Zhu Q.K., Marasco W.A., Baric R.S., Sims A.C., Pyrc K., Li W., Sui J. Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J. Virol. 2015;89:7202–7213. doi: 10.1128/JVI.00854-15. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4473545/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulswit R.J.G., de Haan C.A.M., Bosch B.-J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7112277/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. https://jvi.asm.org/content/78/11/5619.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W., Wang W., Zhao X., Zai J., Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 2020;92:433–440. doi: 10.1002/jmv.25682. https://onlinelibrary.wiley.com/doi/full/10.1002/jmv.25682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., Zheng L., Ming Z., Zhang L., Lou Z., Rao Z. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nuc. Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. https://academic.oup.com/nar/article/47/12/6538/5498756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Kim S., Shin D.H., Kim M.-S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6882434/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeel M., Ibrahim A., Fayez M., Al-Nazawi M. From SARS and MERS CoVs to SARS-CoV-2: moving toward more biased codon usage in viral structural and nonstructural genes. J. Med. Virol. 2020;2020:1–7. doi: 10.1002/jmv.25754. https://onlinelibrary.wiley.com/doi/epdf/10.1002/jmv.25754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q., Yan Y., Shang C., Shan H., Chen S. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.04.009. https://pubmed.ncbi.nlm.nih.gov/32363136/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karo-karo D., Bodewes R., Wibawa H., Artika I.M., Pribadi E.S., Diyantoro D., Pratomo W., Sugama A., Hendrayani N., Indasari I., Wibowo M.H., Muljono D.H., Stegeman J.A., Koch G. Reassortments among avian influenza A(H5N1) viruses circulating in Indonesia, 2015-2016. Emerg. Infect. Dis. 2019;25:465–472. doi: 10.3201/eid2503.180167. https://wwwnc.cdc.gov/eid/article/25/3/18-0167_article [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. https://www.nature.com/articles/nature17200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:1–9. doi: 10.1038/s41467-019-10280-3. https://www.nature.com/articles/s41467-019-10280-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. Functional analysis of the murine coronavirus genomic RNA packaging signal. J. Virol. 2013;87:5182–5192. doi: 10.1128/JVI.00100-13. https://jvi.asm.org/content/87/9/5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T.-Y., Shum M.H.-H., Zhu H.-C., Tong Y.-G., Ni X.-B., Liao Y.-S., Wei W., Cheung W.Y.-M., Li W.-J., Li L.-F., Leung G.M., Holmes E.C., Hu Y.-L., Guan Y. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nat. Accel. Artic. Prev. 2020:1–5. doi: 10.1038/s41586-020-2169-0. https://www.nature.com/articles/s41586-020-2169-0_reference.pdf [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. https://www.nature.com/articles/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- Landowski C.P., Mustalahti E., Wahl R., Croute L., Sivasiddarthan D., Westerholm-Parvinen A., Sommer B., Ostermeier C., Helk B., Saarinen J., Saloheimo M. Enabling low cost biopharmaceuticals: high level interferon alpha-2b production in Trichoderma reesei. Microb. Cell Factories. 2016;15:1–15. doi: 10.1186/s12934-016-0508-5. https://microbialcellfactories.biomedcentral.com/articles/10.1186/s12934-016-0508-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Chan J.F.W. Coronaviruses: emerging and re-emerging pathogens in humans and animals. Virol. J. 2015;12:209. doi: 10.1186/s12985-015-0432-z. https://virologyj.biomedcentral.com/articles/10.1186/s12985-015-0432-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopardi S., Holmes E.C., Gastaldelli M., Tassoni L., Priori P., Scaravelli D., Zamperin G., De Benedictis P. Interplay between co-divergence and cross-species transmission in the evolutionary history of bat coronaviruses. Infect. Genet. Evol. 2018;58:279–289. doi: 10.1016/j.meegid.2018.01.012. https://www.sciencedirect.com/science/article/pii/S1567134818300121?via%3Dihub [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. https://www.nature.com/articles/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. https://www.annualreviews.org/doi/full/10.1146/annurev-virology-110615-042301?url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org&rfr_dat=cr_pub++0pubmed [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. https://www.nature.com/articles/d41573-020-00016-0 [DOI] [PubMed] [Google Scholar]
- Liao Y., Tam J.P., Liu D.X. Viroporin activity of SARS-CoV E protein. Adv. Exp. Med. Biol. 2006;581:199–202. doi: 10.1007/978-0-387-33012-9_34. https://link.springer.com/chapter/10.1007%2F978-0-387-33012-9_34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: a review of virus–host interactions. Viruses. 2016;4:2–28. doi: 10.3390/diseases4030026. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5456285/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Fung T.S., Chong K.K.-L., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. https://www.sciencedirect.com/science/article/pii/S0166354214001752?via%3Dihub [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez L.A., Riffle A.J., Pike S.L., Gardner D., Hogue B.G. Importance of conserved cysteine residues in the coronavirus envelope protein. J. Virol. 2008;82:3000–3010. doi: 10.1128/JVI.01914-07. https://jvi.asm.org/content/82/6/3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30251-8/fulltext [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin A., Dijkman R., Bergström T., Kann N., Adamiak B., Hannoun C., Kindler E., Jónsdóttir H.R., Muth D., Kint J., Forlenza M., Müller M.A., Drosten C., Thiel V., Trybala E. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the Middle East respiratory syndrome virus. PLoS Pathog. 2014;10:1–15. doi: 10.1371/journal.ppat.1004166. https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1004166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk H.K.H., Li X., Fung J., Lau S.K.P., Woo P.C.Y. Molecular epidemiology, evolution and phylogeny of SARS coronavirus. Infect. Genet. Evol. 2019;71:21–30. doi: 10.1016/j.meegid.2019.03.001. https://www.sciencedirect.com/science/article/pii/S1567134819300310?via%3Dihub [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantlo E., Bukreyeva N., Maruyama J., Paessler S., Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antivir. Res. 2020;179:1–4. doi: 10.1016/j.antiviral.2020.104811. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7188648/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. https://www.sciencedirect.com/science/article/pii/S0065352706660053?via%3Dihub [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. Coronavirus genomic RNA packaging. Virology. 2019;537:198–207. doi: 10.1016/j.virol.2019.08.031. https://www.sciencedirect.com/science/article/pii/S0042682219302569?via%3Dihub [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., Fielding B.C. The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis. Viruses. 2012;4:2902–2923. doi: 10.3390/v4112902. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3509677/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4147684/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.-Y., Donaldson E.F., Randell S.H., Lanzavecchia A., Marasco W.A., Shi Z.-L., Baric R.S. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. https://www.nature.com/articles/nm.3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Mitchell H.D., Cockrell A.S., Gralinski L.E., Yount B.L., Jr., Graham R.L., McAnarney E.T., Douglas M.G., Scobey T., Beall A.K.D., III, Kocher J.F., Hale A.E., Stratton K.G., Waters K.M., Baric R.S. MERS-CoV accessory ORFs play key role for infection and pathogenesis. mBio. 2017;8:1–14. doi: 10.1128/mBio.00665-17. https://mbio.asm.org/content/8/4/e00665-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewska A., Zarebski M., Nowak P., Stozek K., Potempa J., Pyrc K. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J. Virol. 2014;88:13221–13230. doi: 10.1128/JVI.02078-14. https://jvi.asm.org/content/88/22/13221.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. https://www.sciencedirect.com/science/article/pii/S0168170214004961?via%3Dihub [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska L., Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ. Int. 2020;139:1–3. doi: 10.1016/j.envint.2020.105730. https://www.sciencedirect.com/science/article/pii/S016041202031254X?via%3Dihub [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D.M., Fauci A.S. Emerging infectious diseases: threats to human health and global stability. PLoS Pathog. 2013;9:1–3. doi: 10.1371/journal.ppat.1003467. https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1003467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounir S., Talbot P.J. Sequence analysis of the membrane protein gene of human coronavirus 0C43 and evidence for O-glycosylation. J. Gen. Virol. 1992;73:2731–2736. doi: 10.1099/0022-1317-73-10-2731. https://www.microbiologyresearch.org/content/journal/jgv/10.1099/0022-1317-73-10-2731#tab2 [DOI] [PubMed] [Google Scholar]
- Müller M.A., van der Hoek L., Voss D., Bader O., Lehmann D., Schulz A.R., Kallies S., Suliman T., Fielding B.C., Drosten C., Niedrig M. Human coronavirus NL63 open reading frame 3 encodes a virion-incorporated N-glycosylated membrane protein. Virol. J. 2010;7:1–12. doi: 10.1186/1743-422X-7-6. https://virologyj.biomedcentral.com/articles/10.1186/1743-422X-7-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu T., Takemoto C., Kim Y.-T., Wang H., Nishii W., Terada T., Shirouzu M., Yokoyama S. SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proc. Natl. Acad. Sci. U.S.A. 2016;46:12997–13002. doi: 10.1073/pnas.1601327113. https://www.pnas.org/content/113/46/12997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa K., Lokugamage K.G., Makino S. Viral and cellular mRNA translation in coronavirus infected cells. Adv. Virus Res. 2016;96:165–192. doi: 10.1016/bs.aivir.2016.08.001. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5388242/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naskalska A., Dabrowska A., Szczepanski A., Milewska A., Jasik K.P., Pyrc K. Membrane protein of human coronavirus NL63 is responsible for interaction with the adhesion receptor. J. Virol. 2019;93:1–14. doi: 10.1128/JVI.00355-19. https://jvi.asm.org/content/93/19/e00355-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh F., Connelly S., Droese B., 3 Klaus J.P., Makino S., Sawicki S.G., Siddell S.G., Stamou D.G., Wilson I.A., Kuhn P., Buchmeie M.J. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4486061/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Buchmeier M.J. Supramolecular architecture of the coronavirus particle. Adv. Virus Res. 2016;96:1–27. doi: 10.1016/bs.aivir.2016.08.005. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7112365/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., DeDiego M.L., Álvarez E., Jiménez-Guardeño J.M., Regla-Nava J.A., Llorente M., Kremer L., Shuo S., Enjuanes L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415:69–82. doi: 10.1016/j.virol.2011.03.029. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4726981/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., de Haan C.A.M., de Groot R.J., Rottier P.J.M. Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J. Virol. 2006;80:2326–2336. doi: 10.1128/JVI.80.5.2326-2336.2006. https://jvi.asm.org/content/80/5/2326.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein J.M., Banach B., Baker S.C. Morphogenesis of coronavirus HCoV-NL63 in cell culture: a transmission electron microscopic study. Open Infect. Dis. J. 2008;2:52–58. doi: 10.2174/1874279300802010052. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2763395/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15562-9. https://www.nature.com/articles/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish C.R., Holmes E.C., Morens D.M., Park E., Burke D.D., Calisher C.H., Laughlin C.A., Saif L.J., Peter Daszak P. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2546865/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier A., Bonnin A., Desmarets L., Daneels A., Goffard A., Rouillé Y., Dubuisson J., Belouzard S. The C-terminal domain of the MERS coronavirus M protein contains a trans-Golgi network localization signal. J. Biol. Chem. 2019;294:14406–14421. doi: 10.1074/jbc.RA119.008964. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6768645/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020;81:1–3. doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbilewicz B. Virus and cell fusion mechanisms. Annu. Rev. Cell Dev. Biol. 2014;30:111–139. doi: 10.1146/annurev-cellbio-101512-122422. https://www.annualreviews.org/doi/pdf/10.1146/annurev-cellbio-101512-122422 [DOI] [PubMed] [Google Scholar]
- Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immun. 2020;38:1–9. doi: 10.12932/AP-200220-0772. http://apjai-journal.org/wp-content/uploads/2020/03/AP-200220-0772.pdf [DOI] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M., Thiel V., Drosten C., Rottier P.J.M., Osterhaus A.D.M.E., Bosch B.J., Haagmans B.L. Dipeptidylpeptidase4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. https://www.nature.com/articles/nature12005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg R., Johansson M.A., Powers A.M., Miller B.R. Search strategy has influenced the discovery rate of human viruses. Proc. Natl. Acad. Sci. 2013;110:13961–13964. doi: 10.1073/pnas.1307243110. https://www.pnas.org/content/110/34/13961.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg R. Detecting the emergence of novel, zoonotic viruses pathogenic to humans. Cell. Mol. Life Sci. 2015;72:1115–1125. doi: 10.1007/s00018-014-1785-y. https://link.springer.com/article/10.1007/s00018-014-1785-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe C.L., Fleming J.O., Nathan M.J., Sgro J.-Y., Palmenberg A.C., Baker S.C. Generation of coronavirus spike deletion variants by high-frequency recombination at regions of predicted RNA secondary structure. J. Virol. 1997;71:6183–6190. doi: 10.1128/jvi.71.8.6183-6190.1997. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC191880/pdf/716183.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch T.R., Machamer C.E. The coronavirus E protein: assembly and beyond. Viruses. 2012;4:363–382. doi: 10.3390/v4030363. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3347032/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Younker D., Meyer Y., Thiel V., Stokes H., Siddell S.G. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 2005;1:310–322. doi: 10.1371/journal.ppat.0010039. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1298938/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. Coronavirus transcription: a perspective. Curr. Top. Microbiol. Immunol. 2005;287:31–55. doi: 10.1007/3-540-26765-4_2. https://link.springer.com/chapter/10.1007/3-540-26765-4_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:1–22. doi: 10.1186/s12985-019-1182-0. https://virologyj.biomedcentral.com/articles/10.1186/s12985-019-1182-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setianingsih T.Y., Wiyatno A., Hartono T.S., Hindawati E., Rosamarlina Dewantari A.K., Myint K.S., Lisdawati V., Safari D. Detection of multiple viral sequences in the respiratory tract samples of suspected Middle East respiratory syndrome coronavirus patients in Jakarta, Indonesia 2015-2016. Int. J. Infect. Dis. 2019;86:102–107. doi: 10.1016/j.ijid.2019.06.022. https://www.ijidonline.com/article/S1201-9712(19)30267-X/fulltext [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. https://www.nature.com/articles/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S.T., Lai M.M.C. Viral and cellular proteins involved in coronavirus replication. Curr. Top. Microbiol. Immunol. 2005;287:95–131. doi: 10.1007/3-540-26765-4_4. https://link.springer.com/chapter/10.1007%2F3-540-26765-4_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetics, recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7125511/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryaman G.K., Soejoedono R.D., Setiyono A., Poetri O.N., Handharyani E. Isolation and characterization of avian coronavirus from healthy Eclectus parrots (Eclectus roratus) from Indonesia. Vet. World, EISSN. 2019:916–2231. doi: 10.14202/vetworld.2019.1797-1805. www.veterinaryworld.org/Vol.12/November-2019/16.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.-C. SARS in Singapore – key lessons from an epidemic. Ann. Acad. Med. 2006;35:345–349. http://www.annals.edu.sg/pdf/35VolNo5200606/V35N5p345.pdf [PubMed] [Google Scholar]
- Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Corona virus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020;178:1–16. doi: 10.1016/j.antiviral.2020.104792. https://www.sciencedirect.com/science/article/pii/S0166354220302060?via%3Dihub [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J., Surya W., Li Y., Liu D.X. Protein-protein interactions of viroporins in coronaviruses and Paramyxoviruses: new targets for antivirals? Viruses. 2015;7:2858–2883. doi: 10.3390/v7062750. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4488717/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Walls A.C., Lang Y., Wang C., Li Z., Koerhuis D., Boons G.-J., Bosch B.-J., Rey F.A., de Groot R.J., Veesler D. Structural basis for human coronavirus attachment to sialic acid receptors. Nature Struct. Mol. Biol. 2019;26:481–489. doi: 10.1038/s41594-019-0233-y. https://www.nature.com/articles/s41594-019-0233-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi N.K., Shrivastava A. Recent developments in bioprocessing of recombinant proteins: expression hosts and process development. Front. Bioeng. Biotechnol. 2019;7:1–35. doi: 10.3389/fbioe.2019.00420. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6932962/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y.-T., Wang S.-M., Huang K.-J., Lee A.I.-R., Chiang C.-C., Wang C.-T. Self-assembly of severe acute respiratory syndrome coronavirus membrane protein. J. Biol. Chem. 2010;285:12862–12872. doi: 10.1074/jbc.M109.030270. https://www.jbc.org/content/285/17/12862.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike M., Taguchi F. Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses. 2015;7:1700–1725. doi: 10.3390/v7041700. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4411675/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatagopalan P., Daskalova S.M., Lopez L.A., Dolezal K.A., Hogue B.G. Coronavirus envelope (E) protein remains at the site of assembly. Virology. 2015;478:75–85. doi: 10.1016/j.virol.2015.02.005. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4550588/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheije M.H., Hagemeijer M.C., Ulasli M., Reggiori F., Rottier P.J.M., Masters P.S., de Haan C.A.M. The coronavirus nucleocapsid protein is dynamically associated with the replication-transcription complexes. J. Virol. 2010;84:11575–11579. doi: 10.1128/JVI.00569-10. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2953146/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasak R., Luytjes W., Spaan W., Palese P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. U.S.A. 1988;85:4526–4529. doi: 10.1073/pnas.85.12.4526. https://www.pnas.org/content/85/12/4526.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Snijder J., Xiong X., Bosch B.-J., Rey F.A., Veesler D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. https://www.pnas.org/content/114/42/11157.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., David Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;180:1–12. doi: 10.1016/j.cell.2020.02.058. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7102599/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., Cui Y., Liu X., Arledge K.C., Chen Y.-H., Zhang L., Wang X. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. https://www.nature.com/articles/cr201392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang Y., Chen Y., Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J. Med. Virol. 2020:1–9. doi: 10.1002/jmv.25748. https://onlinelibrary.wiley.com/doi/full/10.1002/jmv.25748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Shang J., Jiang S., Du L. Subunit vaccines against emerging pathogenic human coronaviruses. Front. Microbiol. 2020;11:1–19. doi: 10.3389/fmicb.2020.00298. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7105881/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipf B., Weibel E.K., Vogel S. Computer controlled large scale production of α-interferon by E. coli. Bioprocess Eng. 1994;10:145–153. https://link.springer.com/article/10.1007/BF00387522 [Google Scholar]
- Woo J., Lee E.Y., Lee M., Kim T., Cho Y.-E. An in vivo cell-based assay for investigating the specific interaction between the SARS-CoV N-protein and its viral RNA packaging sequence. Biochem. Biophys. Res. Commun. 2019;520:499–506. doi: 10.1016/j.bbrc.2019.09.115. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7092827/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) Laboratory Biosafety Manual; Geneva, Switzerland: 2004. https://www.who.int/csr/resources/publications/biosafety/Biosafety7.pdf [Google Scholar]
- World Health Organization (WHO) 2020. Coronavirus Disease. (COVID-19) Situation Report – 205.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200812-covid-19-sitrep-205.pdf?sfvrsn=627c9aa8_2 [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7154514/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7104723/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Li Y., Sung M., Wei J., Yang Z. A study of the probable transmission routes of MERS- CoV during the first hospital outbreak in the Republic of Korea. Indoor Air. 2018;28:51–63. doi: 10.1111/ina.12430. https://onlinelibrary.wiley.com/doi/full/10.1111/ina.12430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7164635/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Leibowitz J.L. The structure and functions of coronavirus genomic 3’ and 5’ ends. Virus Res. 2015;206:120–133. doi: 10.1016/j.virusres.2015.02.025. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4476908/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager C.L., Ashmunt R.A., Williams R.K., Cardeillchio C.B., Shapirot L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7095410/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee J., Unger L., Zadravecz F., Cariello P., Seibert A., Johnson M.A., Fuller M.J. Novel coronavirus 2019 (COVID-19): emergence and implications for emergency care. J. Am. College Emerg. Physician Open. 2020:1–7. doi: 10.1002/emp2.12034. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7228264/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Hu B., Shi Z., Cui J. Geographical structure of bat SARS-related coronaviruses. Infect. Genet. Evol. 2019;69:224–229. doi: 10.1016/j.meegid.2019.02.001. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7106260/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y., Zhang X., Gao G.F. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:1–9. doi: 10.1038/ncomms15092. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5394239/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q., Langereis M.A., van Vliet A.L.W., Huizinga E.G., de Groot R.J. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9065–9069. doi: 10.1073/pnas.0800502105. https://www.pnas.org/content/105/26/9065.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang K., Lv W., Yu W., Xie S., Xu K., Schwarz W., Xiong S., Sun B. The ORF4a protein of human coronavirus 229E functions as a viroporin that regulates viral production. Biochim. Biophys. Acta. 2014;1838:1088–1095. doi: 10.1016/j.bbamem.2013.07.025. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7094429/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang K., Ping X., Yu W., Qian Z., Xiong S., Sun B. The ns12.9 Accessory protein of human coronavirus OC43 is a viroporin involved in virion morphogenesis and pathogenesis. J. Virol. 2015;89:11383–11395. doi: 10.1128/JVI.01986-15. https://jvi.asm.org/content/89/22/11383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:1–4. doi: 10.1016/j.cell.2020.03.035. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7194821/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7164518/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30:1346–1351. doi: 10.1016/j.cub.2020.03.022. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7156161/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. https://www.nature.com/articles/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7121973/ [DOI] [PMC free article] [PubMed] [Google Scholar]