Abstract
Introduction
A number of MRI methods have been proposed to be useful, quantitative biomarkers of neurodegeneration in ageing. The Calgary Normative Study (CNS) is an ongoing single-centre, prospective, longitudinal study that seeks to develop, test and assess quantitative magnetic resonance (MR) methods as potential biomarkers of neurodegeneration. The CNS has three objectives: first and foremost, to evaluate and characterise the dependence of the selected quantitative neuroimaging biomarkers on age over the adult lifespan; second, to evaluate the precision, variability and repeatability of quantitative neuroimaging biomarkers as part of biomarker validation providing proof-of-concept and proof-of-principle; and third, provide a shared repository of normative data for comparison to various disease cohorts.
Methods and analysis
Quantitative MR mapping of the brain including longitudinal relaxation time (T1), transverse relaxation time (T2), T2*, magnetic susceptibility (QSM), diffusion and perfusion measurements, as well as morphological assessments are performed. The Montreal Cognitive Assessment (MoCA) and a brief, self-report medical history will be collected. Mixed regression models will be used to characterise changes in quantitative MR biomarker measures over the adult lifespan. In this report, we describe the study design, strategies to recruit and perform changes to the acquisition protocol from inception to 31 December 2018, planned statistical approach and data sharing procedures for the study.
Ethics and dissemination
Participants provide signed informed consent. Changes in quantitative MR biomarkers measured over the adult lifespan as well as estimates of measurement variance and repeatability will be disseminated through peer-reviewed scientific publication.
Keywords: dementia, neuroradiology, magnetic resonance imaging
Strengths and limitations of this study.
Both cross-sectional and longitudinal quantitative magnetic resonance data are being acquired in a large sample normal ageing population to characterise changes in these potential neuroimaging biomarkers over the adult lifespan.
Clinical and multiple quantitative imaging data are being collected and shared with the research community.
Measures of repeatability and a process to update the imaging protocol are included in the study design.
Associated self-reported medical history and cognitive assessments are limited.
Introduction
Quantitative imaging methods can be defined as ‘extraction and use of numerical or statistical features from medical images’.1 There are numerous examples where quantitative measures have improved detection of changes or monitoring of the brain.2–4 Specifically, quantitative neuroimaging using magnetic resonance (MR) is thought to be suitable for identifying potential biomarkers of cognitive impairment risk because it may (1) be more sensitive to early detection of pathological changes within otherwise normal appearing tissue5–9; (2) be more directly related to underlying physiological processes of interest; (3) have reduced variability across time and/or equipment platforms when compared with traditional, more qualitative, neuroimaging methods; (4) can provide less subjective interpretation; and (5) allows for a low risk, repeatable measurement.1 10
Numerous quantitative MRI biomarkers have been proposed to assess risk, progression and treatment of age-related neurodegenerative diseases and disorders including cerebral small vessel disease,10 Alzheimer’s disease,3 Parkinson’s disease,11 multiple sclerosis12 13 and Huntington’s disease.14 These quantitative markers typically include measures of brain atrophy, white matter integrity, iron accumulation and cerebral blood flow. Often, these measures can be determined using standard, vendor-provided MR sequences and freely available image processing packages. Less commonly, longitudinal (T1) and transverse (T2, T2*) relaxometry, vascular permeability and quantitative susceptibility mapping using more customised approaches have been used.
Development of quantitative imaging biomarkers requires appropriate validation and qualification as outlined by the Quantitative Imaging Biomarkers Alliance organised by the Radiological Society of North America.15 16 An advantage of quantitative imaging is that the measurements obtained should be, in principle, independent of the specifics of acquisition. While brain atrophy, white matter hyperintensity volume, white matter integrity as measured by diffusion imaging metrics, number of cerebral microbleeds, number and volume of infarcts (including ischaemic, haemorrhagic and lacunar), relative increase in tissue iron have all been associated with one or more neurodegenerative processes, detailed validation of these quantitative biomarkers is relatively lacking.16 Furthermore, quantitative methods like MR relaxometry, for example, T1, T2 and T2* mapping, quantitative susceptibility mapping and arterial spin labelling measurement of cerebral perfusion also require comprehensive evaluation and validation as potential quantitative neuroimaging biomarkers of neurodegenerative disease or pathology.
The Calgary Normative Study (CNS) is a single centre, prospective, longitudinal study with three objectives: (1) to evaluate and characterise the dependence of the selected quantitative neuroimaging biomarkers on age over the adult lifespan; (2) to evaluate the precision, variability and repeatability of quantitative neuroimaging biomarkers as part of biomarker validation providing both proof-of-concept and proof-of-principle16; and (3) provide a shared repository of normative data for comparison to various disease cohorts. In developing this study, we developed a plan to incorporate the ability to revise our data acquisition protocol so that we can refine and accommodate emerging quantitative techniques. We also aimed to complete all imaging procedures and other study evaluations within 2 hours. Our primary research interest is in cerebral small vessel disease and this guided our selection of potential quantitative neuroimaging biomarkers, though in both theory and practice they have proven to be of broad application in other neuroimaging studies. Here we describe the study design and methods used in the CNS and report on recruitment and data sharing. Reporting the specific short-repeatability and long-repeatability or changes in quantitative metrics over the adult lifespan are beyond the scope of this report and will be submitted as separate submissions for publication subsequently.
Materials and methods
Participant eligibility and characterisation
This study was approved by the local research ethics board (REB). Study recruitment is ongoing; volunteers over 18 years of age are being recruited from the community, primarily through local poster advertisement and word-of-mouth. Interested individuals provided written informed consent and were screened for eligibility based on self-reported absence of significant neurological disease, psychiatric disorders or contraindications for MRI at 3 T. Participants with uncertain contraindications (poorly characterised implant, and so on) were not imaged and excluded from the study. Participants provided a brief medical history, completed a Montreal Cognitive Assessment (MoCA),17 completed our institutional MR safety screening and underwent MRI.
Demographic and simplified medical information collected includes age, sex, ethnicity, handedness, years of education, smoking history, weight, hypertension or taking medication for the treatment of hypertension, dyslipidaemia or taking medication for the treatment of high cholesterol and presence of diabetes mellitus, and family history of stroke, dementia, Alzheimer’s disease or cardiovascular diseases. Participants were also asked if they are willing to return for a follow-up visit and if they are willing to be contacted in the future for possible participation in other research studies. All data were deidentified and labelled with CNS-specific identification numbers.
The MoCA is a cognitive screening test designed to assist the detection of mild cognitive impairment, which was administered and scored by trained research personnel. If participants were not fluent in English, then alternate language versions of the MoCA (Mandarin and Spanish) were offered and were administered with the aid of a translator. Participants who scored <26 on the MoCA were considered screen failures for normal cognition.17 However, these individuals still completed the study procedures, though we anticipate that their data may be excluded from some subsequent analyses. Participants were not informed of their MoCA scores because the MoCA alone is not sufficient to diagnose a clinical cognitive disorder such as mild cognitive impairment(MCI), and there will be false positive low scores. If concerns about a clinical cognitive disorder arose during the testing the participant was advised to consult their primary care practitioner.
We sought to enrol an approximately balanced distribution of men and women in each of six age categories: 18–29, 30–39, 40–49, 50–59, 60–69 and 70+ years. We anticipated that recruitment of eligible normal older participants would be more difficult due to the increased prevalence of stroke and/or other significant neurological disease with age. For this reason, we decided to group individuals 70 years of age and older into a single age category, anticipating that we would have difficulty recruiting a sufficient number of individuals over 80 years of age (figure 1).
Figure 1.
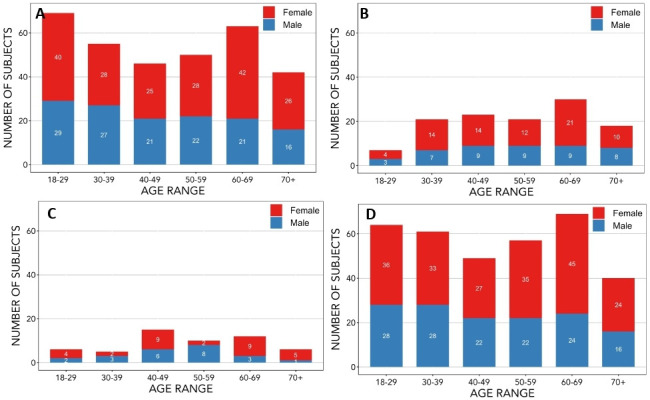
Distribution of age at study entry and sex of participants recruited up to December 2018. The age and sex distribution of participants has been provided for (A) all baseline MR scans completed, (B) follow-up MR scans completed, (C) participants whose scans were completed with phase 1 MR acquisition protocol and (D) participants whose scans were completed with phase 2 MR acquisition protocol. Phase 1 MR acquisition protocol was used only for baseline scans; phase 2 MR acquisition protocol was used for some baseline and all follow-up scans. The exact number of participants for each sex in a given age category are indicated in the appropriate portion of the bar representing that category. MR, magnetic resonance
A nominal sample size of 20 men and 20 women was initially targeted for each age category, for a nominal CNS recruitment of 240 participants. This sample size estimate was based on estimates of variance from previous work using voxel-based analysis of T2 changes in temporal lobe epilepsy.2 The study was initially powered (α=0.05, 1−β=0.8) to detect T2 changes of 3.4 ms or greater. Larger or smaller sample sizes may be required depending on the quantitative measure, region and analysis method chosen. To accommodate differences in power requirements, protocol revisions and secondary analyses (eg, sex differences), recruitment will continue after the nominal sample size has been achieved. Establishing better variance estimates for each quantitative imaging method is one of the objectives of the CNS.
MRI acquisition protocol
MRI is being completed at a single centre on a 3 T MR scanner (MR750, General Electric Healthcare, Waukesha, Wisconsin, USA) using the vendor-supplied, 12-channel head, neck and spine coil. Standard procedure at our centre is to perform daily quality control scans and to follow the vendor-recommended service guidelines.
Prior to setting up the initial MR protocol, we informally surveyed five local research teams with respect to their MRI protocols and typical outcome measures. This survey guided some of our protocol decisions and ensured that aspects of the MR acquisition parameters were similar to other studies in our centre. This similarity in some protocol acquisitions makes it feasible to share and leverage the data appropriately for comparison with disease cohorts.
The MRI protocol includes conventional structural 3D-T1-weighted (3D-T1), T2-weighted fluid attenuated inversion recovery (FLAIR), as well as diffusion, pseudo-continuous arterial spin labelling (pcASL), resting state functional MRI (rs-fMRI) based on blood oxygen level dependent contrast, quantitative susceptibility mapping (QSM) and both T1 (qT1) and T2 (qT2) relaxometry sequences. Susceptibility-weighted imaging results were derived from the acquired QSM image data. The quantitative neuroimaging metrics of interest generated from each sequence are summarised in table 1. Details of the acquisition protocol are summarised in table 2. The field of view was 240 mm for all sequences except 3D-T1 and QSM, which have a field of view of 256 mm.
Table 1.
Acquired MR sequences, potential quantitative neuroimaging measurement outcomes and associated physiological changes
| Outcome | Associated physiological change(s) | |
| 3D-T1 | Structural brain volumes; infarct detection | Atrophy |
| FLAIR | White matter hyperintensity volume, detection of lesions presumed vascular in origin | Inflammation, demyelination, gliosis, axonal loss |
| rs-fMRI | Functional connectivity | Network changes in brain regions with synchronous activity |
| DWI | FA, MD, RD, PSMD, structural connectivity | Microstructural changes in white matter |
| T1 mapping | Regional T1 relaxation values | Change in local biochemical environment for example, lipid, haemosiderin |
| T2 mapping | Regional T2 relaxation values | Diffuse inflammation, demyelination, gliosis, axonal loss |
| QSM | Regional relative magnetic susceptibility values; regional R2* values; detection of cerebral microbleeds | Iron concentration, bleeding |
| pcASL | Cerebral blood perfusion (CBF) | Tissue perfusion |
BOLD, blood oxygen level dependent; CBF, cerebral blood flow; 3D-T1, 3D T1-weighted; DWI, diffusion-weighted imaging; FA, fractional anisotropy; FLAIR, fluid attenuated inversion recovery; MD, mean diffusivity; MR, magnetic resonance; NEX, number of excitations; pcASL, pseudo-continuous arterial spin labelling; PSMD, peak skeletonised mean diffusivity; QSM, quantitative susceptibility mapping; RD, radial diffusivity; rs-fMRI, resting state functional MRI.
Table 2.
A: MR acquisition parameters for Phase 1 (n=54)
| Sequence type | Slice orientation | TR (ms) | TE (ms) | Flip angle (°) | Acquisition matrix: |
Reconstructed resolution (mm) | Other parameters | Acquisition duration (min:s) |
| A: MR acquisition parameters for phase 1 (n=54) | ||||||||
| 3D-T1 | Coronal | ~7 | ~2.5 | 8 | 256×256 | 0.94×0.94×1.0 | TI=650 ms | 5:03 |
| FLAIR | Axial | 9000 | 148 | 90 | 256×256 | 0.94×0.94×3.0 | TI=2250 ms | 4:50 |
| rs-fMRI | Axial | 2000 | 30 | 70 | 64×64 | 3.75×3.75×3.8 | 200 Volumes | 5:10 |
| DWI | Axial | 9000 | ~80 | 90 | 80×80 | 0.94×0.94×3.0 | b=1000 s/mm2; 31 directions | 6:01 |
| qT118 | Axial | 15 000 | ~24 | 40 | 160×160 | 1.0×1.0×4.0 | Eight echoes; B1 map included | 8:09 |
| qT2 | Coronal | 3000 | ~8 to 128 | 90 | 256×128 | 0.94×0.94×4.0 | 16 Echoes; echo spacing=8 ms | 9:48 |
| QSM19 | Axial | 30 | ~3 to 28 | 20 | 192×192 | 1.0×1.0×1.0 | Eight echoes; echo spacing=3.4 ms | 4:13 |
| pcASL | Axial | 4899 | ~11 | 111 | 512×10 | 1.88×1.88×5.0 | Post label delay=2025 ms; NEX=1 | 6:55 |
| B: MR acquisition parameters for Phase 2 (n=271) | ||||||||
| 3D-T1 | Sagittal | ~7 | ~2.5 | 8 | 256×256 | 0.94×0.94×1.0 | TI=650 ms | 5:44 |
| FLAIR | Axial | 9000 | 148 | 90 | 256×256 | 0.94×0.94×3.0 | TI=2250 ms | 4:50 |
| rs-fMRI | Axial | 2000 | 30 | 70 | 64×64 | 3.75×3.75×3.8 | 200 Volumes | 5:10 |
| DWI | Axial | 9000 | ~80 | 90 | 80×80 | 0.94×0.94×3.0 | b=1000 s/mm2; 31 directions | 6:01 |
| qT120 | Axial | 15 000 | ~24 | 40 | 160×160 | 1.0×1.0×4.0 | Eight echoes; variable TI | 5:35 |
| qT2 | Axial | 3000 | ~8 to 128 | 90 | 256×128 | 0.94×0.94×4.0 | 16 Echoes; echo spacing=8 ms | 9:48 |
| QSM19 | Axial | 30 | ~3 to 28 | 20 | 192×192 | 1.0×1.0×1.0 | Eight echoes; echo spacing=3.4 ms | 3:58 |
| pcASL | Axial | 4899 | ~11 | 111 | 512×8 | 1.79×1.79×3.5 | Post label delay=2025 ms; NEX=3 | 4:45 |
T1, spin–lattice relaxation time constant.
T2, spin–spin relaxation time constant.
TI, inversion time.
DTI, diffusion tensor imaging; DWI, diffusion-weighted imaging; FLAIR, fluid attenuated inversion recovery; IR, inversion recovery; MR, magnetic resonance; NEX, number of excitations; pcASL, pseudo-continuous arterial spin labelling; QSM, quantitative susceptibility mapping; rs-fMRI, resting state functional MRI.
In designing this study, we recognised a need to evolve elements of the acquisition protocol, potentially in response to improved or new quantitative imaging techniques. The study was designed to incorporate distinct phases in order to accommodate protocol revisions. Revisions are proposed to and reviewed by the CNS protocol committee which evaluates the impact of the change, the number of participants completed in the current phase and the number of overall protocol changes. The committee selects and recommends the appropriate timing of any proposed changes. This approach allows for a formal, orderly, well specified, evolution of the study data acquisition over time.
To date, two study phases have been completed. The assigned CNS study identification number is used to help identify into which phase a subject is enrolled. In the first phase of the study (table 2A), 3D-T1 images and qT2 were acquired with coronal slice orientation for consistency with other existing research protocols at our centre. Driven equilibrium single observation of T1 (DESPOT1) method18 was implemented. QSM acquisition used unipolar gradients.19 After 54 participants were recruited and imaged, the data were reviewed and it was decided to modify the protocol. A phase 2 protocol (table 2B) was implemented with changes in slice orientation for the 3D-T1 and qT2, approach for qT1 and resolution of ASL. We found the DESPOT1 approach provided inconsistent T1 maps with values outside those reported in the literature. A T1 mapping method that is independent of B1 (and thus did not require a B1 map) was developed for brain T1 mapping.20 The T2 mapping slice orientation was changed from coronal to axial to harmonise the protocol with newer ongoing studies within our centre. To improve the resolution and reduce through-plane blurring, the acquisition matrix and slice thickness of the ASL sequence were changed. The number of excitations was increased to maintain a similar signal-to-noise ratio.
MRI was completed by trained staff (>15 years MRI research experience) or registered MR technicians. If any possible abnormalities in the images were noted, a neuroradiologist reviewed the images to determine if findings were of potential clinical significance, per the standard policy at our centre. Clinically significant findings as determined by the radiologist were reported to the participant by the radiologist following the procedure as outlined in our REB-approved ethics application.
Image assessment and processing
Images were visually inspected for imaging artefacts like motion or spike noise, prior to any processing steps. The 3D-T1, FLAIR, diffusion-weighted and susceptibility-weighted images were reviewed more thoroughly to identify individuals with possible covert disease pathology associated with cerebral small vessel disease by an expert reader (FS) with >15 years of experience. Presence and number of lacunar stroke, cerebral microbleeds, recent subcortical infarcts and scoring of white matter hyperintensities were recorded using the criteria for cerebral small vessel disease outlined in the STRIVE paper.21 Other incidental findings were noted and recorded. Standard and custom image processing pipelines will be used to generate and analyse the quantitative images and maps. These procedures are briefly summarised in table 3 with example images and maps shown in figure 2.
Table 3.
Image processing pipelines and measurement outcomes
| Sequence type | Processing pipelines | Measurement outcome |
| 3D-T1-weighted | Freesurfer V.6.0 | Whole brain and regional volumes cortical thickness |
| FLAIR | Cerebra-LET (CIPAC) | White matter hyperintensity volumes |
| rs-fMRI | FSL | Default mode network connectivity |
| DWI | FSL, ExploreDTI | FA, MD, RD, PSMD, structural connectivity |
| qT1 | CIPAC custom pipeline | Mean regional T1 values, VBM |
| qT2 | StimFit27 | Mean regional T2 values, VBM |
| QSM | CIPAC custom pipeline | Mean regional susceptibility values, mean regional T2* values, cerebral microbleed detection |
| pcASL | GE scanner generated CBF maps; BASIL (FSL) | Mean regional CBF values |
T1, spin–lattice relaxation time constant; T2, spin–spin relaxation time constant.
BASIL, Bayesian inference for arterial spin labeling; CBL, cerebral blood flow (mL/min/100 g tissue); Cerebra-LET, Cerebra Lesion Extraction Tool; CIPAC, calgary image processing and analysis centre; DWI, diffusion-weighted imaging; FA, fractional anisotropy; MD, mean diffusivity; pcASL, pseudo-continuous arterial spin labelling; PSMD, peak skeletonised mean diffusivity; QSM, quantitative susceptibility mapping; RD, radial diffusivity; rs-fMRI, resting state functional magnetic resonance imaging.
Figure 2.
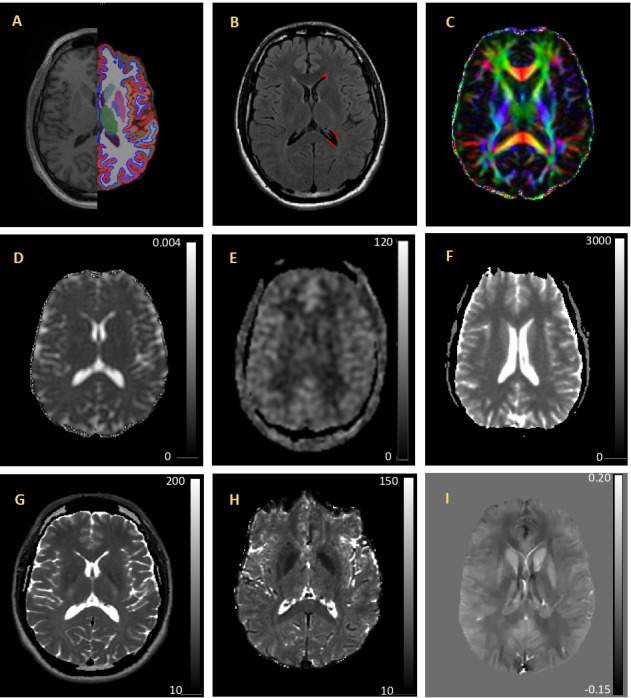
Example quantitative imaging maps and associated processing overlays. (A) 3D-T1 image shown with FreeSurfer segmentation and parcellation results overlay on the left hemisphere. (B) FLAIR with WMH mask (red) on the left hemisphere. (C) FA colour map indicating the primary diffusion direction of white matter tracts (red indicates left/right; green indicates anterior/posterior; and blue indicates inferior/superior). (D) MD map with voxel intensity values are in mm2/s. (E) CBF map. Voxel intensity values are in mm/min/100 g tissue. (F) Quantitative T1 map. Voxel intensity values are in ms. (G) T2 map voxel intensity values are in ms. (H) T2* map. Voxel intensity values are in ms. (I) Quantitative susceptibility map. Voxel intensity values are in PPM. All images are from the same individual and are shown in radiological orientation. CBF, cerebral blood flow; FA, fractional anisotropy; FLAIR, fluid attenuated inversion recovery; PPM, parts per million; WMH, white matter hyperintensity.
Repeatability and measurement validation
A subset of four participants was asked at the time of the first scan to undergo repeated scanning for estimates of precision, variance and repeatability of the proposed quantitative neuroimaging biomarkers. Additionally, these data were used to determine if changes in the MR scanner software and hardware or in the MR acquisition parameters between protocol phases could be pooled or not depending on the sequence and processing pipelines used. Three additional time points were selected to include scanning before and after MR system hardware and software upgrades. Not all sequences were acquired at all time points due to scanner and subject availability. In these cases, a subset of data was acquired, which included 3D-T1, diffusion, T1 mapping, T2 mapping, ASL, QSM, but not FLAIR or rs-fMRI. Figure 3 illustrates the data acquisition timeline with scanner hardware or software changes.
Figure 3.
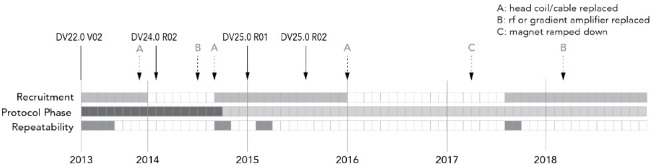
Timeline of MR protocols, recruitment phases and scanner hardware and software upgrades. The filled squares, each representing 1 month, indicate when participants were actively recruited and scanned, when the MR acquisition protocol was revised and when repeatability measurements were completed. The solid arrows indicate version changes to the scanner software with the version number indicated above each arrow. The update between DV24.0 and DV25.0 included installation of some additional hardware. The dashed arrows indicate hardware repair or changes. MR, magnetic resonance.
Statistical analyses
From the repeatability data, the within-subject variance, minimum detectable difference and repeatability will be estimated at select regions-of-interest from repeated scans at each time point. Within-subject variance will include the possible impact of MR system changes on these repeated measures across time points. Changes in quantitative neuroimaging outcomes over the adult lifespan will be assessed using linear mixed-effects models. Linear and quadratic functions with age will be considered to determine the most appropriate fitting model based on Akaike information criterion and Bayesian information criterion.
Data storage
Deidentified demographic, medical and cognitive assessment data were entered and maintained in a database (REDCap; Vanderbilt University, Nashville, Tennessee, USA). MRI data were organised in an Osirix database where images can be reviewed clinically, or the database can be queried for specific criteria. Approximately 17 000 DICOM image files are acquired per imaging session requiring 1.3 GB of disk space for storage without compression, 510 MB with lossless compression. The exact number of images for each individual can vary depending on head size and the number of slices required to cover the whole brain and whether sequences needed to be repeated due to motion, for example.
Data sharing, ethics and dissemination
MRI data and demographic information are available on request for academic purposes from qualified researchers. Investigators interested in accessing these data are required to complete a data sharing agreement available on request by emailing CalgaryNormStudy@ucalgary.ca. This brief, two-page form requests a brief description of the study objectives and intended purpose, a list of MR sequences of interest, required medical information and demographic information. The sharing agreement also requires that the study requesting data has approval from the appropriate REB. Once the agreement is approved by the CNS leadership and requesting principal investigators, the selected data are provided through secure electronic transfer. All data are deidentified and labelled with CNS-specific identification numbers. Information regarding MR system upgrades, protocol changes and repeatability data may be provided on request. Groups requesting data are asked to limit use and analysis of the data to those outlined in their data sharing agreement, not sharing with other groups or use the data for other unplanned purposes and to acknowledge the CNS and the CNS funding agencies in presentations or publications. Further or previously unplanned analyses of requested data would require a simple amendment to the data sharing agreement.
Characterisation of the changes in quantitative MR biomarker measures over the adult lifespan as well as estimates of measurement variance and repeatability will be disseminated through peer-reviewed scientific publication.
Patient and public involvement
This research was done without patient or public involvement in the study design and they were not consulted to develop relevant outcomes or interpret the results. Patients were not invited to contribute to the writing or editing of this document for readability or accuracy.
Discussion
This report demonstrates a successful recruitment strategy for a prospective, community-recruited quantitative MR study and describes the flexible design and data collection. The data collected will provide estimates of variability and repeatability of each quantitative method and provide data essential for the validation of potential biomarkers of neurodegeneration associated with ageing. A distinctive aspect of this study is that it has been designed to incorporate both typical clinical MR sequences and multiple, research-based, quantitative biomarkers including MR relaxometry, diffusion imaging, quantitative susceptibility mapping and cerebral perfusion within a 60 min scan time. While there are a number of studies that have evaluated one or two of these methods, there are very few that have incorporated this extensive multimodal approach within an imaging session.10
Recruitment and population characteristics
The prevalence of neuroimaging findings indicative of covert cerebrovascular small vessels disease in participants in the 70+ age category was up to 20.9%. Similar prevalence of covert lacunar infarcts of presumed vascular origin, white matter hyperintensities and cerebral microbleeds in older community-dwelling populations have been reported previously.22 We may need to consider increasing our recruitment numbers in the older age categories to maintain a balanced design of healthy individuals over the adult lifespan. The decision on whether to oversample the older participants is dependent on the definition of what is ‘normal’ ageing. Should individuals with possible evidence of cerebral small vessel disease be excluded from the analyses? This question raises some controversy and is as yet unresolved. Given the limited self-reported medical history collected in this study, it will be difficult to determine a definitive diagnosis of cerebral small vessel disease, or other potential confounds like cardiovascular disease.
MR protocol
Typically, quantitative MR relaxometry, susceptibility, diffusion and perfusion methods are not all incorporated into either clinical or research studies. This may be due to the additional time required for data acquisition or the expertise and time required for data processing to generate quantitative maps. MR fingerprinting (MRF) has been proposed to overcome a number of these limitations, specifically addressing the longer acquisition times. This emerging approach to quantitative imaging simultaneously generates T1, T2, T2* and proton density maps from a single acquisition with an approximate scan duration of as little as 16 s.23 24 MRF uses voxel-wise pattern matching to a dictionary of MR signal evolution to estimate the MR tissue characteristics. While MRF methods overcome the time limitation for acquisition, these methods are still evolving and have limited availability.
The study design has taken into consideration the evolution of quantitative imaging techniques. Ongoing changes in the MR acquisition protocol can be implemented once an adequate number of participants in each age category have been acquired. The number of participants required can be determined for each quantitative imaging technique. An important outcome of this study will be to establish the measurement variability for each of these quantitative metrics. In practice, a limited number of changes are implemented at any given time. One limitation of this approach is the additional time required for repeatability measures prior to implementation of any intended changes to the MR acquisition protocol.
Repeatability and measurement validation
These repeatability assessments were coordinated with selected scheduled changes in the MR operating system. These included major software and hardware upgrades and after the MR system main magnetic field was ramped down for renovations to the MR suite. However, we did not capture repeatability measurements after all unscheduled system changes, including repairs to or replacement of the 12-channel head, neck and spine coil. While this may be a limitation of the repeatability measurements’ ability to determine the impact of these changes, our MR system is regularly maintained and met the manufacturer specification and operation tolerances after each of these changes. Regular weekly quality assurance and manufacturer service to maintain the MR system within manufacturer specifications is performed, which should mitigate any systems changes.
Another limitation of the repeatability assessment is the limited number and long-term availability of selected participants for evaluation of repeatability and variability. Two participants have recently moved from the local area and new participants have been recruited for future repeatability measures. For purposes of repeatability assessment, we have generally assumed that no significantly physiological changes have occurred in the brains of these participants over either the short or long term. However, if changes were to occur within a given participant, we should be able to identify them as we have nominally three measurements at each time point. These three independent measurements are usually completed within 3 weeks for each individual.
Data sharing
Including conventional MR sequences in a purportedly healthy adult population, along with research-based sequences, has generated greater interest in data sharing with other local and national studies. A subset of the imaging and other data has been shared with eight investigators at the University of Calgary. Internal sharing of data was and remains the principle anticipated target group of the CNS study; however, we also have had opportunity to share data with three investigative teams external to our institution (University of Alberta, Canada; State University of Campinas, Brazil; and University Medical Centre Utrecht, Netherlands). Data equivalent of over 1400 scans have been shared; data from some participants have been shared more than once. The research area of the investigators accessing the CNS data range from depression, inflammatory bowel disease, machine learning, migraine, epilepsy and stroke. In most cases, only a portion of the acquired data was requested. The most frequently requested imaging data in descending order of demand were 3D-T1, rs-fMRI, FLAIR, pcASL, QSM and qT2. It also allows evaluation of these research-based sequences with the context of clinical findings that may help to better understand changes leading to traditional clinical findings. While this study does not match the size or extent of other larger MR initiatives, such as the UK Biobank,25 we have been able to successfully leverage the data by providing normal control data for other studies.
Summary
Over 55 months from study inception in May 2013, the acquisition protocol has undergone one revision, data for four repeatability time points have been collected, 42 month follow-up data have been collected and the data have been shared effectively. Our study sample includes ethnically diverse people represented in our local community.26 Data collection and analyses remain ongoing. Preparation of detailed reports on repeatability of the quantitative measures, cross-sectional and longitudinal changes over the adult lifespan are underway.
Supplementary Material
Acknowledgments
This ongoing study would not be possible without assistance from a number of individual trainees, technicians and physicians including Brie Crawston-Grant, BSc; Qian (Lucy) Lu, MSc; Paul Romo, MRT; Courtney Seefeldt, Robert J Sevick, MD; and Tamara Trainor, MRT.
Footnotes
Contributors: CRMC helped in conceptualisation, investigation, data collection and curation, formal analyses, writing original manuscript draft, review and editing. MS performed conceptualisation, investigation, data collection and curation, analyses, review and editing manuscript. LBA helped in conceptualisation, data collection and curation, review and editing manuscript. DG and LL helped in methodology, review and editing manuscript. FS helped in investigation, review and editing the manuscript. EES helped in methodology, investigation, review and editing manuscript. RF performed conceptualisation, funding acquisition, review and editing manuscript.
Funding: This work is supported by the Canadian Institutes of Health Research, the Seaman Family MR Research Centre and the Hopewell Professorship (RF).
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting or dissemination plans of this research.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Abramson RG, Burton KR, Yu J-PJ, et al. Methods and challenges in quantitative imaging biomarker development. Acad Radiol 2015;22:25–32. 10.1016/j.acra.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kosior RK, Lauzon ML, Federico P, et al. Algebraic T2 estimation improves detection of right temporal lobe epilepsy by Mr T2 relaxometry. Neuroimage 2011;58:189–97. 10.1016/j.neuroimage.2011.06.002 [DOI] [PubMed] [Google Scholar]
- 3.Filippi M, Agosta F, Frisoni GB, et al. Magnetic resonance imaging in Alzheimer's disease: from diagnosis to monitoring treatment effect. Curr Alzheimer Res 2012;9:1198–209. 10.2174/156720512804142949 [DOI] [PubMed] [Google Scholar]
- 4.Bonnier G, Maréchal B, Fartaria MJ, et al. The combined quantification and interpretation of multiple quantitative magnetic resonance imaging metrics enlightens longitudinal changes compatible with brain repair in relapsing-remitting multiple sclerosis patients. Front Neurol 2017;8:506. 10.3389/fneur.2017.00506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacKay AL, Laule C. Magnetic Resonance of Myelin Water: An in vivo Marker for Myelin. Brain Plast 2016;2:71–91. 10.3233/BPL-160033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyers SM, Kolind SH, Laule C, et al. Measuring water content using T2 relaxation at 3T: phantom validations and simulations. Magn Reson Imaging 2016;34:246–51. 10.1016/j.mri.2015.11.008 [DOI] [PubMed] [Google Scholar]
- 7.Vavasour IM, Meyers SM, Mädler B, et al. Multicenter Measurements of T1 Relaxation and Diffusion Tensor Imaging: Intra and Intersite Reproducibility. J Neuroimaging 2019;29:42–51. 10.1111/jon.12559 [DOI] [PubMed] [Google Scholar]
- 8.Langkammer C, Krebs N, Goessler W, et al. Quantitative MR imaging of brain iron: a postmortem validation study. Radiology 2010;257:455–62. 10.1148/radiol.10100495 [DOI] [PubMed] [Google Scholar]
- 9.Laule C, Leung E, Lis DKB, et al. Myelin water imaging in multiple sclerosis: quantitative correlations with histopathology. Mult Scler 2006;12:747–53. 10.1177/1352458506070928 [DOI] [PubMed] [Google Scholar]
- 10.De Guio F, Jouvent E, Biessels GJ, et al. Reproducibility and variability of quantitative magnetic resonance imaging markers in cerebral small vessel disease. J Cereb Blood Flow Metab 2016;36:1319–37. 10.1177/0271678X16647396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langkammer C, Pirpamer L, Seiler S, et al. Quantitative susceptibility mapping in Parkinson's disease. PLoS One 2016;11:e0162460. 10.1371/journal.pone.0162460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta A, Al-Dasuqi K, Xia F, et al. The use of noncontrast quantitative MRI to detect gadolinium-enhancing multiple sclerosis brain lesions: a systematic review and meta-analysis. AJNR Am J Neuroradiol 2017;38:1317–22. 10.3174/ajnr.A5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodini B, Louapre C, Stankoff B. Advanced imaging tools to investigate multiple sclerosis pathology. Presse Med 2015;44:e159–67. 10.1016/j.lpm.2015.02.011 [DOI] [PubMed] [Google Scholar]
- 14.Eskreis-Winkler S, Zhang Y, Zhang J, et al. The clinical utility of QSM: disease diagnosis, medical management, and surgical planning. NMR Biomed 2017;30:nbm.3668. 10.1002/nbm.3668 [DOI] [PubMed] [Google Scholar]
- 15.Raunig DL, McShane LM, Pennello G, et al. Quantitative imaging biomarkers: a review of statistical methods for technical performance assessment. Stat Methods Med Res 2015;24:27–67. 10.1177/0962280214537344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith EE, Biessels GJ, De Guio F, et al. Harmonizing brain magnetic resonance imaging methods for vascular contributions to neurodegeneration. Alzheimers Dement 2019;11:191–204. 10.1016/j.dadm.2019.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nasreddine ZS, Phillips NA, Bédirian V, et al. The Montreal cognitive assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc 2005;53:695–9. 10.1111/j.1532-5415.2005.53221.x [DOI] [PubMed] [Google Scholar]
- 18.Deoni SCL, Peters TM, Rutt BK. High-Resolution T1 and T2 mapping of the brain in a clinically acceptable time with DESPOT1 and DESPOT2. Magn Reson Med 2005;53:237–41. 10.1002/mrm.20314 [DOI] [PubMed] [Google Scholar]
- 19.Lauzon ML, McCreary CR, McLean DA, et al. Quantitative susceptibility mapping at 3 T: comparison of acquisition methodologies. NMR Biomed 2017;30:nbm.3492. 10.1002/nbm.3492 [DOI] [PubMed] [Google Scholar]
- 20.Lauzon ML, McCreary CR, Frayne R. Multislice T1 -prepared 2D single-shot epi: analysis of a clinical T1 mapping method unbiased by B0 or B1 inhomogeneity. NMR Biomed 2016;29:1056–69. 10.1002/nbm.3566 [DOI] [PubMed] [Google Scholar]
- 21.Wardlaw JM, Smith EE, Biessels GJ, et al. Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol 2013;12:822–38. 10.1016/S1474-4422(13)70124-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das AS, Regenhardt RW, Vernooij MW, et al. Asymptomatic cerebral small vessel disease: insights from population-based studies. J Stroke 2019;21:121–38. 10.5853/jos.2018.03608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature 2013;495:187–92. 10.1038/nature11971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panda A, Mehta BB, Coppo S, et al. Magnetic resonance Fingerprinting-An overview. Curr Opin Biomed Eng 2017;3:56–66. 10.1016/j.cobme.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller KL, Alfaro-Almagro F, Bangerter NK, et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat Neurosci 2016;19:1523–36. 10.1038/nn.4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canada Population Canada population 2019, 2019. Available: https://canadapopulation.org/calgary-population/#Ethnicity_in_Calgary
- 27.Lebel RM, Wilman AH. Transverse relaxometry with stimulated echo compensation. Magn Reson Med 2010;64:1005–14. 10.1002/mrm.22487 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


