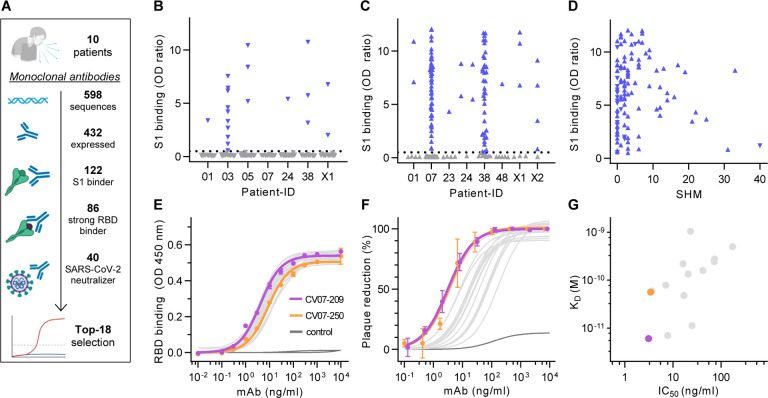
Fig. 1 |. Identification and characterization of potent SARS-CoV-2 neutralizing mAbs.
(A) Diagram depicting the strategy for isolation of 18 potently neutralizing mAbs (Top-18). (B) Normalized binding to S1 of SARS-CoV-2 for mAbs isolated from antibody secreting cells (▼; blue = S1-binding, grey = not S1-binding). (OD=optical density in ELISA) (C) Normalized binding to S1 of SARS-CoV-2 for mAbs isolated from S1-stained memory B cells (▲; colors like in (B)) (D) S1-binding plotted against the number of somatic hypermutations (SHM) for all S1-reactive mAbs. (E) Concentration-dependent binding of Top-18 SARS-CoV-2 mAbs to the RBD of S1 (mean±SD from two wells of one experiment). (F) Concentration-dependent neutralization of authentic SARS-CoV-2 plaque formation by Top-18 mAbs (mean±SD from two independent measurements). (G) Affinity of mAbs to RBDs (KD determined by surface plasmon resonance) plotted against IC50 of authentic SARS-CoV-2 neutralization.