Abstract
MicroRNAs (miRNAs) are ubiquitous small RNAs guiding post-transcriptional gene repression in countless biological processes. However, the miRNA pathway in mouse oocytes appears inactive and dispensable for development. We propose that marginalization of the miRNA pathway activity stems from the constraints and adaptations of RNA metabolism elicited by the diluting effects of oocyte growth. We report that miRNAs do not accumulate like mRNAs during the oocyte growth because miRNA turnover has not adapted to it. The most abundant miRNAs total tens of thousands of molecules in growing (∅ 40 μm) and fully grown (∅ 80 μm) oocytes, a number similar to that observed in much smaller fibroblasts. The lack of miRNA accumulation results in a 100-fold lower miRNA concentration in fully grown oocytes than in somatic cells. This brings a knock-down-like effect, where diluted miRNAs engage targets but are not abundant enough for significant repression. Low-miRNA concentrations were observed in rat, hamster, porcine and bovine oocytes, arguing that miRNA inactivity is not mouse-specific but a common mammalian oocyte feature. Injection of 250,000 miRNA molecules was sufficient to restore reporter repression in mouse and porcine oocytes, suggesting that miRNA inactivity comes from low-miRNA abundance and not from some suppressor of the pathway.
INTRODUCTION
MicroRNAs (miRNAs, reviewed in detail in (1)) are genome-encoded small RNAs, which guide post-transcriptional repression of gene expression. Their biogenesis starts with nuclear processing of long primary transcripts into pre-miRNAs, small hairpin miRNA precursors. Pre-miRNAs are transported to the cytoplasm and cleaved by RNase III Dicer into 21–23 nucleotide (nt) RNA duplexes. One strand of the duplex is then loaded onto an Argonaute (AGO) protein, the key component of miRNA-induced silencing complex (miRISC), the effector complex repressing cognate mRNAs (reviewed in (2)). Functional miRNAs in somatic cells have a relatively high copy number (3–5). A HeLa cell contains ∼50,000 let-7 miRNA molecules (3), which is just an order of magnitude below ∼580,000 mRNAs estimated to be present in a HeLa cell (6).
There are two distinct modes of miRNA-mediated target repression (Figure 1A). The RNA interference (RNAi)-like mode requires a miRNA loaded on AGO2 (holo-RISC) and a perfect or nearly perfect miRNA:mRNA duplex. In this case, exemplified by mammalian mir-196 and HoxB8 mRNA (7), AGO2 cleaves mRNA in the middle of the miRNA:mRNA duplex. However, mammalian miRNAs loaded on one of the four mammalian AGO proteins (AGO1-4) typically bind their cognate mRNAs with imperfect complementarity. Functional miRNA:mRNA interaction may involve little beyond the ‘seed’ region comprising nucleotides 2–8 of the miRNA (8–11). This results in miRISC-mediated translational repression (12,13) coupled with substantial mRNA degradation (14), which is a common mammalian miRNA mode of target repression.
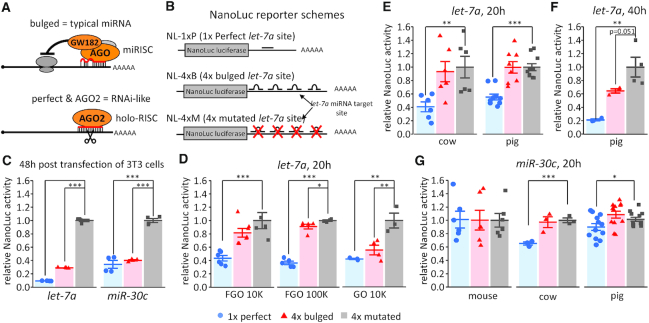
Figure 1.
Analysis of miRNA activity in mammalian oocytes with NanoLuc reporters. (A) Schematic difference between ‘bulged’ and ‘perfect’ miRNA binding sites. Bulged miRNA binding sites, which are typical of animal miRNAs, have imperfect complementarity and lead to translational repression followed by deadenylation and mRNA degradation (1). Perfect complementarity of miRNAs loaded on AGO2 results in RNAi-like endonucleolytic cleavage. (B) Schematic depiction of let-7a nanoluciferase reporter constructs used in the study. The miRNA sites were cloned in the 3′UTR either as a 1×-perfectly complementary site, a 4× site producing a bulged conformation or a 4× mutated site. An analogous set of reporters was constructed for miR-30c. (C) let-7a and miR-30c nanoluciferase reporter activities in 3T3 cells. Data represent an average of three independent transfections performed as described previously (21). (D) Luciferase assay of let-7 activity in meiotically incompetent growing (GO) and fully grown (FGO) mouse oocytes injected with 10,000 or 100,000 molecules of let-7 NanoLuc luciferase reporters; reporter expression was assayed after 20 h of culture in a medium preventing resumption of meiosis. (E) Luciferase assay of let-7 activity in fully grown bovine and porcine oocytes. A total of 10,000 molecules of NanoLuc luciferase let-7a reporters were microinjected; reporter expression was assayed after 20 h of culture. (F) Luciferase assay of let-7 activity in fully grown porcine oocytes. A total of 10,000 molecules of NanoLuc luciferase let-7a reporters were microinjected; reporter expression was assayed after 40 h of culture. (G) Luciferase assay of miR-30 activity in fully grown mouse, bovine, and porcine oocytes. A total of 10,000 molecules of NanoLuc luciferase reporters carrying 1×-perfect, 4×-bulged or 4×-mutant miR-30c binding sites were microinjected; reporter expression was assayed after 20 h of culture in a medium preventing resumption of meiosis. All luciferase data are a ratio of the NanoLuc luciferase reporter activity over a co-injected control firefly luciferase reporter activity; the relative 4×-mutant reporter was set to one. All error bars represent standard deviation (SD). Asterisks indicate statistical significance (P-value) of one-tailed t-test (* < 0.05, ** < 0.01, *** < 0.001).
miRNA-mediated gene regulation is extensive. Thousands of miRNAs have been annotated in mammals (15). Experimental data suggest that miRNAs directly or indirectly regulate thousands of genes in one cell type (14,16–18). A single miRNA might directly suppress hundreds to over a thousand of genes (15,16). Mammalian miRNAs have been implicated in the majority of cellular and developmental processes, and changes in their expression are observed in various diseases (1). One of the notable exceptions are mouse oocytes, where miRNAs are present (19,20), but do not repress their targets and are dispensable for early development (21,22).
The molecular mechanism underlying miRNA inactivity in oocytes is unclear. It was proposed that it could stem from inefficient formation of the miRISC (23), or could be linked to an alternative oocyte-specific AGO2 protein isoform and reduced expression of the full-length functional AGO2 (24). In addition, mouse oocytes express a truncated Dicer isoform that produces two classes of small RNAs: (i) microRNAs from pre-miRNAs, and (ii) endogenous small interfering RNAs (siRNAs) from long dsRNAs, which function in the endogenous RNAi pathway (25). Endogenous RNAi has evolved in the Muridae family and is highly active and essential in mouse oocytes (22,25–27). Endogenous siRNAs make a significant portion of 21–23 nt RNAs in mouse oocytes (19,20) and are loaded on AGO similarly as miRNAs (28), hence they could also restrict miRNA pathway activity.
It is apparent from kinetic studies that miRNA and targeted mRNA concentrations are critical factors for efficient repression (10,11). This is noteworthy when considering oocytes, which are uniquely large cells. While a mammalian somatic cell has a diameter of 10–20 μm, a meiotically incompetent mouse oocyte grows during a ∼2.5 week-long growth phase to 80 μm. Consequently, the cytoplasmic volume of a fully grown germinal vesicle-intact oocyte (∼260 pl) is almost two orders of magnitude larger than that of a somatic cell such as NIH/3T3 fibroblast (denoted 3T3 hereafter), which is ∼3 pl (Table 1). During the growth phase, a fully grown mouse oocyte accumulates up to ∼27 million mRNA molecules, while an average somatic cell contains 100,000–600,000 mRNAs (6,29–33). Maternal mRNA accumulation during the growth phase is facilitated by extended average mRNA half-life (34–36). Control of mRNA stability is essential for gene expression changes during oocyte-to-zygote transition because transcription ceases at the end of the growth phase and appears again only during the zygotic genome activation (reviewed in (37)). Importantly, because mRNAs accumulate while the oocyte grows in volume, the cytoplasmic mRNA concentration in fully grown oocytes remains comparable to that of somatic cells (100–300 nM, Table 1). Because of the size of the maternal transcriptome, we hypothesized that growth-associated changes in miRNA:mRNA stoichiometry could play a role in inefficient miRNA-mediated repression in the oocyte.
Table 1.
Quantitative aspects of mRNAs and miRNAs in somatic cells and oocytes
| NIH/3T3 | Growing oocyte | Fully grown oocyte | |
|---|---|---|---|
| Diameter [μm] | 18a | 40 | 80 |
| Total volume [pl] | 3 | 33.5 | 270 |
| Cytoplasmic vol. [pl] | 2.95 | 23.5 | 260 |
| Number of molecules | cytoplasmic molar concentration [nM] | ||
| 25,000 | 14 | 1.7 | 0.15 |
| 50,000 | 28 | 3.4 | 0.3 |
| 100,000 | 56 | 6.8 | 0.6 |
| 250,000 | 140 | 17 | 1.5 |
| 500,000 | 280 | 34 | 3.0 |
| 5,000,000 | 2800 | 340 | 30 |
| 25,000,000 | 14,000 | 1700 | 150 |
aCountess™ NIH/3T3 cell data sheet/B10NUMB3R5, ID #108905.
We report that miRNAs do not accumulate during the oocyte growth, resulting in a two orders of magnitude shift in the miRNA:mRNA ratio, which is likely underlying the apparent inactivity of maternal miRNAs. This lack of accumulation seems to be a consequence of unchanged turnover of miRNAs that, in contrast to mRNA turnover, did not adapt to the diluting effect of the oocyte growth. Maternal miRNAs seem to engage their targets but are not abundant enough to produce a significant silencing impact. The same dilution phenomenon has been observed in oocytes of four other mammalian species. Finally, miRNA activity can be restored in mouse oocytes by injecting as few as 100,000 miRNA molecules. Altogether, our data support a model where the miRNA inactivity primarily stems from a low miRNA concentration caused by the lack of miRNA accumulation during the oocyte growth.
MATERIALS AND METHODS
Oocyte collection
Animal experiments were approved by the Institutional Animal Use and Care Committees (approval no. 34/2014) and were carried out in accordance with the European Union regulations. C57BI/6NCrl mice were obtained from Charles Rivers. Meiotically incompetent oocytes were collected from 12 days old females, whereas fully grown germinal vesicle-intact oocytes (referred to as fully grown oocytes hereafter) were isolated from 7 to 9 weeks old females. Golden hamsters were obtained from Charles Rivers. Oocytes were collected from ovaries of freshly sacrificed 12 months old animals. Porcine and bovine oocytes were obtained from the slaughterhouse material as described previously (38,39).
Meiotically incompetent oocytes were isolated by incubating the ovaries in M2 medium (Sigma) with 1 mg/ml collagenase (Sigma) at 37°C and then collected in M2 medium. Fully grown oocytes and hamster oocytes were collected by puncturing antral follicles with a syringe needle followed by collection in M2 medium containing 0.2 mM isobutylmethylxanthine (IBMX; Sigma) to prevent resumption of meiosis.
cDNA synthesis and qPCR
The oocytes were washed with M2 media to remove any residual IBMX and collected in a minimal amount of M2 media containing 1 μl of Ribolock and incubated at 85°C for 5 min to release RNA. 3T3 cells were counted and then lysed using a RealTime ready Cell Lysis Kit (Roche). cDNA synthesis for miRNA was done with the miRCURY LNA RT kit (Qiagen) according to the manufacturer's protocol. Quantitative polymerase chain reaction (qPCR) was set up using a single oocyte cDNA equivalent per qPCR reaction in all the experiments except for Figure 5B and D, where 0.5 oocyte equivalent was used per reaction. The miRCURY LNA SYBR green kit (Qiagen) as per manufacturer's protocol and Roche LightCycler 480 were used for qPCR. miRCURY LNA miRNA-specific primers were used and are listed in the resource table. The standard curve reactions were carried out using mmu-miR-221 and let-7a oligonucleotides obtained from Integrated DNA Technologies (see Supplementary Table S2 for oligonucleotide sequences).
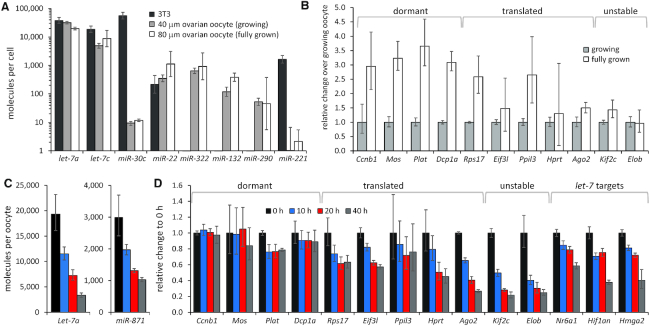
Figure 5.
miRNAs do not accumulate during the oocyte growth and have higher turnover than mRNAs. (A) PCR-based quantification of miRNAs in 3T3 fibroblasts (∅ 18 μm), mouse growing oocyte (∅ 40 μm) and fully grown oocyte (∅ 80 μm). miRNA copy numbers per cell were estimated in three independent experiments using the standard curve in Supplementary Figure S1C. Error bars = standard deviation (SD). Logarithmic scale is used for y-axis. 3T3 cell and fully grown oocyte values are the same as in Figure 2A. (B) mRNA abundance in fully grown oocyte (∅ 80 μm) relative to mouse growing oocyte (∅ 40 μm). The mRNA abundance in the growing oocyte is set to one. mRNA abundance was estimated in three independent experiments per oocyte cDNA equivalent. Error bars = standard deviation (SD). (C) qPCR-based quantification of miRNA per oocyte equivalent during extended culture of fully grown oocytes in the presence of IBMX. miRNA abundance was estimated in two independent experiments. Error bars = standard deviation (SD). (D) mRNA abundance in fully grown oocytes during extended culture in the presence of IBMX relative to oocytes collected at 0 h, which is set to one. mRNA abundance was estimated per oocyte equivalent in two independent experiments. Error bars = standard deviation (SD).
A standard curve was first plotted using let-7a RNA oligonucleotide. The oligonucleotide was either first serially diluted and then cDNA was prepared for each dilution (Supplementary Figure S1A), or cDNA was prepared first and then serially diluted (Supplementary Figure S1B) to rule out the variability in the efficiency of cDNA synthesis. The standard curve was also standardized using miR-221 oligonucleotide as it was not detected in fully grown mouse oocytes (Figure 1A). It could thus be added to fully grown mouse oocyte lysate and then used to set cDNA to avoid any discrepancies that might occur as a consequence of additional oocyte-specific factors (Supplementary Figure S1C).
Northern blot
The sequence for the let-7a standard used for northern blotting was the same as mentioned above but with 5′P. Northern blot probes were labeled by incubating RNA with 20 μCi of γ- 32P-ATP (Hartmann Analytic) and 0.5 U/μl T4 Polynucleotide Kinase (PNK) in 1x PNK buffer A (Thermo Scientific) at 37°C for 30–60 min. The reaction was stopped by adding ethylenediaminetetraacetic acid, pH 8.0, and labeled oligonucleotides were purified with a G-25 column (GE Healthcare). Oocytes were collected in 1× PBS with Ribolock, RNA was isolated with Trizol and separated in 1× TBE in 12% polyacrylamide urea gel. The RNA was then blotted for 30 min at 20 V onto Amersham Hybond-N membrane (GE Healthcare) and crosslinked to the membrane for 1 h at 50°C using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide solution. The membrane was incubated overnight at 50°C in hybridization solution (5× SSC, 7% sodium dodecyl sulphate (SDS), 20 mM sodium phosphate buffer pH 7.2, 1× Denhardt's solution) with a 32P-labeled oligonucleotide antisense to the small RNA. The membrane was washed twice with 5× SSC, 1% SDS, once with 1× SSC, 1% SDS and wrapped in Saran. Signals were detected by exposure to a storage phosphor screen and scanning with Personal Molecular Imager (Bio-Rad). For further details, refer to reference (40).
Western Blot
Injected or non-injected oocytes were cultured in M2 media with IBMX for 20 h. For cycloheximide treatment, they were cultured for 8 h in M2 with IBMX and 20 μM cycloheximide (Sigma). They were washed and collected in 1× PBS with protease inhibitor cocktail and then lysed with RIPA and loaded with SDS dye. Fifty oocytes were used in each lane for HIF1AN (ThermoFisher, MA5-27619) and 25 oocytes for NR6A1 (Bioss, bs-11558R). The blots were developed for the let-7a target proteins first and then washed with sodium azide twice for 5 min each followed by reblotting for tubulin (Sigma #T6074).
Plasmid reporters
Nanoluciferase plasmid pNL1.1 was obtained from Promega. The Nluc gene was cleaved out and ligated in the phRL-SV40 plasmid downstream of the T7 promoter. miRNA binding sites were ordered as oligonucleotides from Sigma. Oligonucleotides were annealed, phosphorylated and then cloned downstream of the Nluc gene in the XbaI site. The firefly control plasmid was made in house with a T7 promoter (see Supplementary Table S2 for oligonucleotide sequences—restriction overhangs underlined).
Cell lines and cell culture
3T3 mouse fibroblast cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% foetal bovine serum and transfected with miRNA-targeted reporters as described previously (21).
In vitro transcription
Linearized pNL-Let7a-perfect, bulged, mutant and Firefly plasmids were in vitro transcribed, capped and then polyadenylated by Poly(A) tailing kit (ThermoFisher).
Microinjection
RNA for injection was diluted in pure water such that a given number of molecules would be present in five picoliters (pl). For a typical microinjection, an injection mixture consisted of in vitro transcribed firefly and nanoluciferase (NanoLuc) RNA in the ratio of 100,000:10,000 or in a ratio of 100,000:100,000 molecules with or without let-7a or miR-30c mimic (Sigma, HMI0001-5NMOL and HMI0458-5NMOL, respectively). Microinjections were done with a FemtoJet microinjector (Eppendorf). Femtojet injection pressure was set to maintain injection volume of 5 pl for all microinjections. Reliability of the estimated amount of microinjected molecules was examined experimentally (Supplementary Figure S2A).
Injected mouse oocytes were cultured in M16 media (Merck) supplemented with IBMX in 5% CO2 at 37°C for 20 h. Bovine oocytes were cultured in MPM media (prepared in house (39)) containing 0.1 mM milrinone (Sigma) without a paraffin overlay in a humidified atmosphere at 39°C with 5% CO2 for 20 h. Pig oocytes were cultured in M-199 MEDIUM (Gibco) supplemented with 1 mM dbcAMP, 0.91 mM sodium pyruvate, 0.57 mM cysteine, 5.5 mM Hepes, antibiotics and 5% foetal calf serum (Sigma). Injected oocytes were incubated at 38.5°C in a humidified atmosphere of 5% CO2 for 20 h.
Luciferase assay
Five oocytes were collected per aliquot and Nano-Glo Dual-Luciferase reporter assay system was used to measure the samples as per the manufacturer's protocol (Promega).
Mathematical modeling
The mathematical model for miRNA degradation introduces the following simplifying assumptions:
i) miRNA degradation is modeled as a one step process (similar to RNAi pathway), but with lower catalytic efficiency than the RNAi pathway.
ii) The AGO2 concentration is assumed to not be rate limiting, i.e., all miRNAs are assumed to be loaded on AGO2 at any tested concentration.
iii) The concentration of AGO2 + miRNA complex is assumed to be constant, i.e., there is neither synthesis nor degradation of miRNA.
iv) No new target mRNAs are synthesised.
v) All assumed miRNA binding events are to be those of pure seed pairing.
vi) Only uncleaved free mRNA is considered in the simulations (mRNA bound in complex with AGO2 + miRNA is not considered).
These assumptions allow us to describe the system as a set of differential equations, identical to the ones used in Michaelis–Menten kinetics:
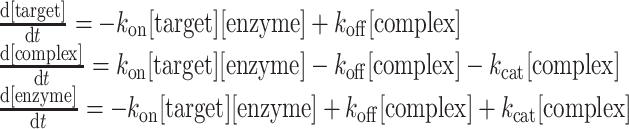 |
where, kon, koff and kcat are the reaction rates in the system and [target], [complex] and [enzyme] are the concentrations of free mRNA molecules with target sites for the miRNA in question, mRNA + miRNA + AGO2 complex and miRNA + AGO2 complex, respectively.
The simulation was written in the R language (https://www.R-project.org/) using packages deSolve (41) and visualised via ggplot2 (42). More details about the simulation, including full source code, are given in the Supplementary Material.
RESULTS
Canonical miRNA-mediated repression is negligible in mammalian oocytes
Previous analysis of miRNA activity in mouse oocytes used firefly and Renilla luciferase reporter assays (21). These reporters followed a common design (13,43) for monitoring two distinct modes of miRNA-mediated repression (Figure 1A): (i) RNAi-like endonucleolytic cleavage of perfectly complementary binding sites by AGO2, which was demonstrated by mapping cleavage sites (17) and (ii) translational repression mediated by partial complementarity between a small RNA and its target (typical of animal miRNAs). Notably, reporter activity could also be partially influenced by related miRNAs with similar sequences, particularly the seed, which is considered the minimum requirement for binding to a cognate RNA (10,11).
The luciferase reporter approach used previously for studying miRNA activity in oocytes required ∼100,000 reporter molecules injected per oocyte (21), which is in the range of the most abundant maternal mRNAs (44). Our concern was that using such a high reporter amount could limit sensitivity of detection of miRNA-mediated repression. For example, 5000 repressed reporter molecules would represent 5% of the injected reporter—such reduction could not be reliably detected with this reporter system. To rectify this issue, we employed NanoLuc luciferase, a new type of luciferase reporter system that should allow reducing the reporter amount significantly (45). We aimed at establishing a reporter system employing 10,000 molecules per oocyte, which could reliably detect suppression of several thousand reporter mRNA molecules. This amount would be in the range of medium-abundant mRNAs such as Plat, which was efficiently knocked down by 10,000 long double-stranded RNA molecules per oocyte (46). A luciferase reporter of this sensitivity would also be useful for other studies of gene expression in oocytes.
NanoLuc luciferase reporters carried either a single perfectly complementary site or four bulged sites to let-7a (Figure 1B). An analogous set of miR-30c-targeted reporters was also produced. Repression of reporters with intact miRNA binding sites was first validated in cultured cells (Figure 1C). When fully grown oocytes were injected with 100,000 and 10,000 molecules of NanoLuc reporters per 5 pl of microinjection mixture, let-7a perfect reporter was repressed substantially (64 and 43%, respectively) as expected for dominating multiple turnover RNAi-like cleavage (Figure 1D). Repression of Let-7a bulged reporter was negligible (9 and 16%, respectively, Figure 1D). Consistent with previous results (21), injection of 10,000 let-7a bulged reporter molecules showed efficient repression in meiotically incompetent oocytes from 12-days-old females (Figure 1D). Altogether, these results showed that the lack of bulged reporter repression in fully grown oocytes in the previous study was not caused by an excessive amount of the reporter.
Let-7a-mediated reporter repression was also examined in porcine and bovine oocytes; the bulged reporters did not show any repression, while the reporter carrying a binding site perfectly complementary to let-7a was repressed by 45 and 60%, respectively (Figure 1E). On one occasion, culture of an extra batch of porcine oocytes was extended to 40 h, which exceeds the time window when oocytes maintain their developmental competence (47). In this case, the bulged reporter was repressed by 36% although the difference was not considered statistically significant (P-value = 0.0506, Figure 1F).
MiR-30c-targeted reporters did not show any repression in mouse oocytes (Figure 1G). This differed from the perfect reporter repression observed in an earlier study (21) but agreed with low miR-30c abundance estimated by qPCR in C57Bl/6 oocytes (Figure 2A). In any case, miR-30c perfect but not bulged reporters exhibited significant repression in bovine and porcine oocytes (Figure 1G).
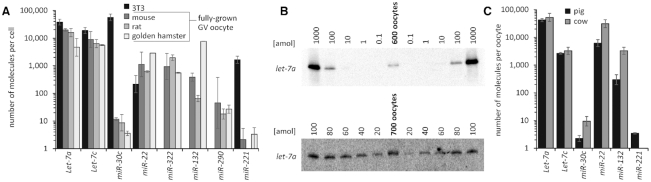
Figure 2.
miRNA quantification in oocytes and 3T3 cells. (A) qPCR-based quantification of miRNAs in 3T3 cells (∅ 18 μm) and rodent fully grown oocytes (∅ 80 μm). miRNA copy numbers per cell were estimated in three independent experiments using the standard curve in Supplementary Figure S1C. Error bars = standard deviation (SD). Logarithmic scale is used for y-axis. miR-322, miR-132 and miR-290 were not detected in 3T3 cells. (B) Northern blots for quantifying let-7a miRNA counts in mouse fully grown oocytes. Serially diluted let-7a synthetic oligonucleotide was used for standard calibration. (C) qPCR-based miRNA quantification in porcine oocytes (∅ 105 μm) and bovine oocytes (∅ 120 μm). miRNA copy numbers per cell were estimated in three independent experiments using the standard curve shown in Supplementary Figure S1C, except for hamster oocyte data, which came from a single oocyte collection. Error bars = SD. Logarithmic scale is used for y-axis.
Taken together, NanoLuc reporter results showed that miRNAs in mammalian oocytes do repress their targets; their activity can be detected with perfect reporters but is not high enough to efficiently repress targets with typical miRNA binding sites. Furthermore, our results demonstrate the suitability of microinjected NanoLuc reporters for studying post-transcriptional control of gene expression in oocytes.
Mammalian oocytes have low miRNA concentration
To assess how much miRNA abundance in oocytes could contribute to minimal detectable miRNA activity, we quantified selected maternal miRNAs by qPCR. To select candidate miRNAs, we analyzed small RNA sequencing (RNA-seq) data (19,48–49) and chose eight miRNAs either because of their high abundance in mammalian oocytes (let-7a, c (21,27,49–51)), association with pluripotency (miR-290 (52)), or because they were unique so that there were no additional family members that could interfere with their analysis. Next, we quantified the number of molecules per cell of these miRNAs in 3T3 fibroblasts and rodent (mouse, rat and hamster) fully grown oocytes (Figure 2A). Particular care was taken of potential biases in producing calibration curves and primer specificity (Supplementary Figure S1).
Remarkably, qPCR results revealed that the most abundant miRNAs were present in the order of 104 copies both in 3T3 cells and in rodent oocytes (Figure 2A and Supplementary Table S1). We estimated by qPCR that let-7a, the most abundant of our studied maternal miRNAs, has ∼20,000 copies/oocyte. To validate the qPCR data, we analyzed maternal let-7a directly by quantitative northern blotting (Figure 2B), which does not include reverse transcription and geometric amplification of the signal in contrast to qPCR. Northern blot analysis estimation yielded ∼ 66,000 let-7a molecules per oocyte (Figure 2B). Northern blot also showed that maternal pre-let-7a processing is intact, as was indicated by the absence of a detectable miRNA precursor signal. While the northern blot-based estimate is higher than the qPCR-based estimate, we cannot rule out that unlike qPCR, which specifically quantified let-7a (Supplementary Figure S1D), other let-7 family members contributed to the higher northern blot-based estimation. In any case, both experimental strategies suggest that the miRNA abundance in fully grown oocytes is in the subnanomolar range (Table 1).
To examine whether the presence of highly active RNAi in rodent oocytes could be associated with the reduced miRNA levels, we quantified maternal miRNAs in bovine and porcine oocytes, where a truncated Dicer isoform does not exist according to RNA-seq data (53). This analysis showed miRNA tallies similar to those in rodent oocytes (Figure 2C and Table 1). While slightly smaller rodent oocytes (Figure 2A) had lower miRNA counts than porcine and bovine oocytes, the most abundant quantified miRNA in bovine and porcine oocytes was let-7a, which had around 50,000 molecules per bovine oocyte (Figure 2C and Supplementary Table S1). In available RNA-seq data from bovine (51) and porcine (50) oocytes, let-7a is among the top five most abundant maternal miRNAs. These data suggest that the subnanomolar miRNA abundance is a common feature of fully grown mammalian oocytes and is independent of Dicer and AGO2 isoforms that emerged during the rodent evolution.
Mathematical modeling supports the impact of subnanomolar miRNA concentration
To further examine the plausibility of the stoichiometric explanation, we performed computational simulations of target repression using kinetic parameters of target binding and repression derived from experimental analyses of miRNAs (10,11) (Figure 3A). The model simulated miRNA-mediated repression at different concentrations of targets and miRNAs. We assumed no transcription, which is indeed absent in transcriptionally quiescent fully grown oocytes. The binding of a miRNA to a target was modeled as a one step process using KD 26 pM, which applies to the seed-only mammalian miRNA:mRNA interaction (11). Typical miRNA-mediated repression is a multistep process with unknown rate constant(s). Therefore, we replaced typical miRNA-mediated repression with RNAi-like cleavage for which the kcat was measured (11). The model was then adjusted by a variable coefficient representing the strength of miRNA repression relative to RNAi-like cleavage.
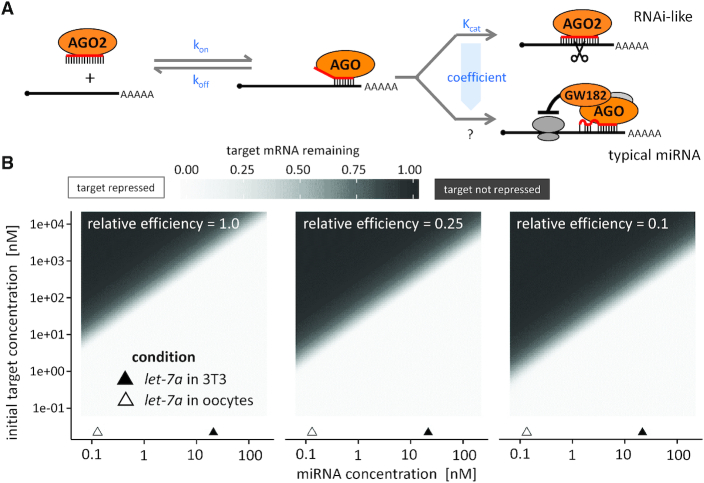
Figure 3.
Mathematical simulation of mRNA repression in mouse oocytes. (A) A scheme of the miRNA pathway and parameters used for the mathematical simulation. (B) Mathematical simulation showing the proportion of mRNA target repression after 20 hours. Three different simulation results are shown for different values of efficiency of typical miRNA-mediated repression relative to RNAi-like cleavage. Left: assuming miRNA repression is as effective as RNAi-like cleavage (relative efficiency = 1). Centre and right: assuming four and ten times less effective typical miRNA-mediated repression than RNAi-like cleavage (relative efficiency = 0.25 and 0.1, respectively).
For a large set of parameters, the model predicted fast mRNA degradation when the miRNA concentration was similar to the measured concentration of let-7a in 3T3 cells, while minimal degradation was predicted for miRNA concentrations of let-7a in oocytes (Figure 3B, see also Table 1 for concentration calculations). The model predicted that subnanomolar miRNA concentrations observed in fully grown oocytes could be inefficient for repression of targets above one nM. The entire maternal transcriptome is ∼150 nM; should a miRNA target 5% of the transcriptome through the seed interaction (let-7 has 1074 predicted mRNA targets with 15,028 binding sites (54)), its target pool concentration could be just below 10 nM. Taken together, mathematical modeling supports the importance of the two orders of magnitude difference in the concentration of miRNAs in somatic cells and fully grown oocytes relative to the target concentration even when using a variable coefficient to address the fact that the miRNA-mediated repression is a multistep process with unknown rate constant(s).
Increased miRNA levels are sufficient to restore miRNA-mediated repression
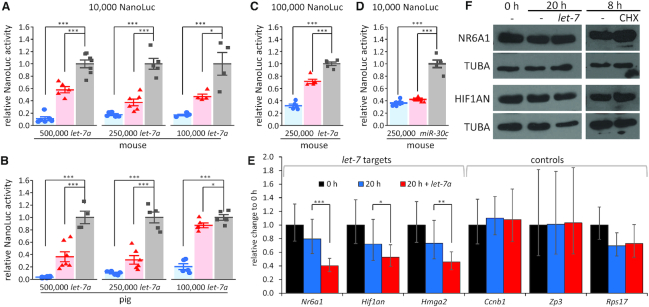
Since unfavourable miRNA stoichiometry was observed in all examined mammalian oocytes, we tested whether the increased miRNA abundance could restore the repressive potential of the pathway. This was directly tested by injecting let-7a mimics into mouse and porcine oocytes. We observed that 500,000 and 250,000 miRNA mimic molecules per 5 pl of injected mixture were sufficient to restore the miRNA pathway activity in oocytes of both species, as evidenced by robust repression of 10,000 molecules of let-7 bulged reporters (Figure 4A,B). In mouse oocytes, which are smaller than porcine oocytes, we observed ∼50% repression of bulged reporter even with 100,000 microinjected let-7 mimic molecules (Figure 4A). At the same time, when the amount of the bulged reporter was increased to 100,000 molecules, the repression was reduced upon injection of 250,000 mimic molecules (Figure 4C). Given the qPCR estimates and northern blot data (Figure 2), it appears that a 5- to 10-fold increase of the let-7a abundance, which brings its concentration to above ∼1 nM, is sufficient to establish significant miRNA-mediated repression. This result was confirmed using 250,000 miR-30c mimic and miR-30c-targeted set of reporters (Figure 4D). We validated that interactions between mimic and reporter molecules in the microinjection mixture did not influence the outcome of the experiment by performing a dual microinjection where a microinjection of 250,000 miR-30c mimics was followed by another microinjection of reporters (Supplementary Figure S2).
Figure 4.
Restoration of miRNA pathway activity in mammalian oocytes. Luciferase reporter activities upon co-injection of mouse (A) or porcine (B) fully grown oocytes with 5 × 105, 2.5 × 105 and 1 × 105 molecules of let-7a mimic and 10,000 molecules of let-7a reporters. (C) Repression of 100,000 reporter molecules in mouse oocytes co-injected with 2.5 × 105let-7a mimic molecules. (D) Repression of 10,000 mir-30c reporter molecules in mouse oocytes co-injected with 2.5 × 105mir-30c mimic molecules. All luciferase data are a ratio of the NanoLuc luciferase reporter activity over a co-injected control firefly luciferase reporter activity; the relative 4×-mutant reporter was set to one. Data are from two independent microinjection experiments. Error bars = SD. Asterisks indicate statistical significance (P-value) of one-tailed t-test, (* < 0.05, ** < 0.01, *** < 0.001). (E) Repression of endogenous let-7 target mRNAs upon injection of 2.5 × 105let-7a mimic molecules. Oocytes were collected at 0 h non-injected or cultured for 20 h in IBMX non-injected or injected with let-7a mimic. Experiments were performed twice, each time in duplicate, i.e. with four independent cDNA. Data are normalized to 0 h, which is set to 1. Error bars = SD. Asterisks indicate statistical significance (P-value) of one-tailed t-test, (* < 0.05, ** < 0.01, *** < 0.001) (F) Analysis of protein levels of endogenous let-7 targets upon injection of 2.5 × 105let-7a mimic molecules. Fifty oocytes were used for each lane of Hif1an and 25 oocytes for Nr6a1. The blots were treated with sodium azide and reblotted for tubulin. Cycloheximide treatment was carried out for 8 h and the oocytes were cultured in IBMX. The experiment was repeated twice.
Analysis of mutant oocytes lacking miRNA biogenesis factor Dgcr8, the key component of miRNA biogenesis, did not reveal any relevant transcriptome change (22). The notion that miRNAs loaded on AGO2 can suppress artificial targets by RNAi-like cleavage, but do not suppress endogenous partially complementary targets, also seemed supported by other observations such as the absence of processing bodies (P-bodies), lack of co-localization of AGO2 and GW182 miRISC-factors, and dormancy of deadenylation and decapping (23,55–56). It was proposed that repression of these endogenous miRNA targets is compromised by reduced deadenylation and/or some deficiency in full miRISC formation (37). We therefore tested whether endogenous let-7 targets were detectably repressed upon increasing let-7 abundance by injecting 250,000 molecules of the let-7a mimic into fully grown oocytes. Nr6a1, Hmga2 and Hif1an were selected because they are highly scoring predicted conserved let-7a targets in mice (15), are well expressed in mouse oocytes (57) and their paralogs were shown to be targeted by let-7 in human cells (58–60). As non-targeted controls, we analyzed the levels of one dormant mRNA (Ccnb1) and two translated mRNAs (Rps17 and Zp3), which are not predicted let-7 targets. mRNA levels were estimated by qPCR 20 h post-injection. All three examined let-7 targets were significantly reduced (Nr6a1 by 49%, Hif1an by 26% and Hmga2 by 38%) relative to non-injected controls; such reduction was not observed for non-targeted controls (Figure 4E). While Zp3 and Ccnb1 transcripts are highly abundant, abundance of Rps17 mRNA was comparable to that of Nr6a1, as estimated from ERCC-spiked RNA sequencing data (61) (Supplementary Figure S3). We also analyzed the protein levels of NR6A1 and HIF1AN, which could be detected with available antibodies using 25 and 50 oocytes per western blot lane, respectively. In this case, reduced protein levels were not observed (Figure 4F). However, this result is likely due to the high abundance and stability of NR6A1 and HIF1AN proteins in fully grown oocytes (Figure 4F), which would obscure repressive effects.
Taken together, analysis of reporters and endogenous targets showed that nanomolar amounts of let-7a mimic are sufficient to restore repression of endogenous let-7 targets in mammalian oocytes. This result also suggests that miRNA expression and/or turnover could be one of the limiting factors for functional miRNA pathway in mouse oocytes.
miRNAs fail to accumulate during the growth phase
The miRNA pathway in growing oocytes was capable of repressing the bulged reporter ((21) and Figure 1D). Quantification of miRNAs in meiotically incompetent growing oocytes showed that a growing oocyte with a diameter of 40 μm contains approximately the same number of miRNA molecules as a fully grown oocyte with a diameter of 80 μm (Figure 5A). This means that the cytoplasmic concentration of let-7 in a 40 μm oocyte would be above 1 nM and the apparent lack of miRNA accumulation would result in dilution into a subnanomolar level in a fully grown oocyte (Supplementary Table S1).
To formally demonstrate that qPCR-based quantification is capable of detecting accumulation of maternal mRNAs, we selected 10 different maternal mRNAs and compared their abundance per oocyte between meiotically incompetent and fully grown oocytes. These included four dormant mRNAs (Ccnb1, Mos, Plat and Dcpa1), which are untranslated and particularly stable, five translated mRNAs (Rps17, Eif3l, Ppil3, Hprt and Ago2) and two actively destabilized mRNAs (endogenous RNAi targets Kif2c and Elob). This analysis showed that dormant mRNAs accumulated the most (Figure 5B). Translated mRNAs Rps17 and Ppil3 also accumulated well, while Eif3l and Ago2 showed mild ∼1.5-fold accumulation and Hprt even lower. This was similar to accumulation of Kif2c and Elob, which are targeted by endogenous RNAi, hence are expected to have a high turnover (Figure 5B). Number of transcripts per oocyte estimated from ERCC-spiked RNA-sequencing data (61) for each gene is available in Supplementary Figure S3.
A steady-state level of a transcript is determined as a ratio between transcription and degradation. While we cannot rule out that transcriptional control contributes to the observed phenomenon, the transcriptionally quiescent fully grown oocyte offers a unique environment for analysis of the transcript turnover. The fully grown oocyte is still resting in the first meiotic block and can be maintained as such for up to 40 h, which allows investigation of the maternal transcript stability (62). To assess miRNA and mRNA stability in oocytes, we cultured fully grown oocytes in the presence of IBMX for up to 40 h and quantified selected miRNAs and mRNAs at 10-h intervals (Figure 5C and D) This analysis showed that miRNAs are unstable, as evidenced by a gradual decrease of miRNA levels during the experiment (Figure 5C). Dormant mRNAs exhibited high stability while translated mRNAs showed variable stability (Figure 5D). Among less stable ones were let-7 targets as well as Eif3l, Hprt and Ago2 transcripts. In fact, Ago2 mRNA decay was comparable to that of endogenous RNAi targets Kif1c and Elob (Figure 5D). There was a good correlation between transcript stability and its accumulation in oocytes; less stable transcripts did not accumulate during the oocyte growth (Figure 5B)
Taken together, our results indicate that let-7 and other mammalian miRNAs do not significantly accumulate in oocytes and have a half-life of 10–20 h (Figure 5C), while accumulating mRNAs have extended half-lives (Figure 5D). A plausible explanation for our observations is that the miRNA turnover remains unaffected during oogenesis, and thus the amount of maternal miRNA molecules does not proportionally increase during the mouse oocyte growth like that of maternal mRNAs. Consequently, the miRNA steady-state amount would not change during the oocyte growth, but its cytoplasmic concentration would dive two orders of magnitude as the oocyte grows to a diameter of 80 μm. This would drive apart the miRNA concentration from the concentration of their target mRNAs, which remain relatively unchanged.
DISCUSSION
Our quantitative insight into the miRNA pathway in mammalian oocytes implies that the apparent miRNA inactivity in mammalian oocytes is associated with unfavourable miRNA:mRNA stoichiometry, which minimizes the impact and significance of miRNA repression. We propose that dilution caused by the increased cytoplasmic volume during the oocyte growth necessitates adaptations to preserve functionally important molecular mechanisms. Thus, what appears as miRNA pathway suppression in mammalian oocytes could be largely explained by a normal miRNA turnover that did not adapt to the oocyte growth. Because mRNAs have adapted through extended half-life, a fully grown oocyte manifests the miRNA:mRNA stoichiometry skewed to such extent that it would impede efficient miRNA-mediated repression. This model is supported by (i) quantification of miRNAs and mRNAs in growing and fully grown oocytes, (ii) greater instability of miRNAs than mRNAs and (iii) recovery of miRNA-mediated repression upon bringing let-7a or miR-30c into nanomolar concentration.
This study stands largely on analysis of let-7, which is one of the most abundant maternal miRNAs in mouse (21,27,49), bovine (51) and porcine (50) oocytes. While RNA-sequencing is not an optimal tool for estimating miRNA abundance (63) it is useful to identify most abundant miRNAs. Janas et al. reported correlation coefficient of 0.9347 for comparison of small RNA-sequencing and absolute miRNA copy number estimation (64). In addition, qPCR data from mouse oocytes support the high abundance of let-7a and other family members (27). We paid particular attention to uncovering possible artefacts during the construction of calibration curves and specificity of let-7a primers (Supplementary Figure S1). qPCR results were corroborated by quantitative northern blot analysis of let-7a, which detects endogenous miRNAs directly. The northern blot-based estimate is three times higher than that of qPCR, but it could be influenced by cross-hybridization of other let-7 family members, while qPCR specifically detects let-7a (Supplementary Figure S1D). Importantly, both estimates are in a subnanomolar range (Table 1) and represent a two orders of magnitude difference in the concentration of the same number of miRNA molecules in somatic cells: 50,000 miRNA molecules would represent ∼28 nM in 3T3 cells and ∼0.3 nM in fully grown oocytes. Such a drop of miRNA concentration in somatic cells would be equivalent to highly efficient miRNA knock-down. Another line of evidence that our miRNA quantification does not underestimate the number of miRNA molecules in oocytes comes from rescue experiments where 250,000 molecules of let-7 mimic were sufficient to induce significant repression of the injected reporters and endogenous mRNA targets.
The miRNA levels in mouse, rat, and hamster oocytes could also be influenced by the presence of the endogenous RNA interference (RNAi) pathway. All three rodent species express a unique truncated Dicer isoform, which facilitates robust production of siRNAs in the oocyte (25,65–66). Enhanced siRNA production could potentially restrict miRNA accumulation through competition for proteins that otherwise serve the miRNA pathway. However, the miRNA pathway was ineffective in all examined mammalian species regardless of expression of Dicer and Ago2 variants.
It was proposed that the non-functionality of miRNAs in mouse oocytes is associated with an oocyte-specific truncated Ago2 transcript variant, whose production limits expression of the full-length functional Ago2 (24). While this Ago2 regulation could contribute to the lack of miRNA accumulation in mouse oocytes, this alternative isoform is apparently mouse-specific; the alternative terminal exon is not conserved in the rat genome and alternative processing of Ago2 transcript is not observed in bovine oocytes (53) (Supplementary Figure S4). Furthermore, RNA-seq data from Veselovska et al. (67) show nearly identical expression of Ago2 isoforms in growing and fully grown oocytes (Supplementary Figure S5), while miRNAs mediate efficient repression in growing but not fully grown oocytes.
The question of the role of control of AGO levels and the relationship between miRNA abundance and AGO levels in mammalian oocytes remains open. At mRNA level, Ago2 and Ago3 transcripts are the most abundant ones in mouse oocytes (Supplementary Figure S5) while recent proteomic analysis identified AGO1 and AGO2 proteins (68). Our data presented here suggest that one should also consider Ago2 transcript turnover as a factor influencing the abundance of AGO2. The mechanism destabilizing Ago2 mRNA is unknown; small RNA-sequencing data suggests that it is not RNAi (21,27,49). Further research is necessary to determine the number of AGO molecules in mammalian oocytes and the molecular mechanisms controlling their levels.
In this work, we approached the issue from the perspective of miRNAs as it allowed to overcome the limitations of direct AGO analysis in oocytes. Importantly, we have shown that increased miRNA levels were sufficient to induce significant repression of bulged NanoLuc reporters. This suggests that the miRNA machinery has sufficient capacity to accommodate 2.5 × 105 miRNAs on AGO proteins in mouse and porcine oocytes. This observation differs from that of Freimer et al., who did not observe repression of the bulged reporter or endogenous targets upon injection of 2 μM miRNA mimic (24). Given the Renilla reporter properties and previous experimental design (21), it is safe to assume that at least 100,000 reporters were injected in that experiment. This is still within the physiological range since Zp3, one of the most abundant maternal transcripts, was estimated to have 250,000 mRNA copies per oocyte (44). Apart from potential differences in injected miRNA mimics, it is possible that the amount of the reporter was bordering the capacity of the miRNA pathway. When we coinjected 100,000 NanoLuc reporter molecules with 250,000 let-7 mimic molecules, repression of the bulged reporter reached only ∼30% (10,000 reporters showed 60% repression (Figure 4A and C)). The NanoLuc-based system operating at mRNA levels corresponding to medium-abundant transcripts appears more sensitive to miRNA repression. Likewise, microarray analysis of miRNA targets in oocytes microinjected and cultured overnight may not be sensitive enough to detect suppression of endogenous targets since microarray apparently underestimated by 3- fold a 10-fold gene overexpression detected by qPCR (24). This suggests that our results may complement rather than contradict the work by Freimer et al., as we investigated miRNA-mediated repression at concentrations that are less exceptional for maternal transcripts and below those used by Freimer et al.
Notably, the ‘dilution model’ proposed here would be consistent with the disappearance of P-bodies during the oocyte growth (23). P-bodies are cytoplasmic foci forming as liquid–liquid phase separation from proteins involved in the RNA metabolism, miRNA effector complexes and their targets (reviewed in (69)). We originally hypothesized that the P-body disappearance could be caused by inhibition of the miRNA pathway. However, in the light of the current data, the disappearance of P-bodies could be also plausibly explained by dilution caused by the oocyte growth.
It is remarkable that the number of maternal miRNA molecules does not proportionally increase during the mouse oocyte growth like that of maternal mRNAs. Our results are consistent with previous results showing that the relative amounts of let-7 miRNAs observed between P15-16 (growing) and P20-21 (fully grown) oocytes remain similar (27). So far, there is no evidence for active suppression of the miRNA biogenesis and function. Our data imply that the reduced miRNA concentration could occur if the miRNA turnover remained the same during oogenesis. Unlike miRNAs, mRNAs accumulate during the growth phase, so that the mRNA density in the cytoplasm of fully grown oocytes is comparable to that in somatic cells. Therefore, there must be some mechanism through which mRNAs cope with the diluting effect of oocyte growth. It seems to be the extended half-life, which facilitates accumulation of mRNAs. Adaptations extending mRNA half-life include coating of maternal transcripts with MSY2, as well as dormancy of decapping and deadenylation (reviewed in (37)). Consequently, maternal mRNA acquires a half-life that could be as long as 10 days (34–36). Whether mRNA accumulation follows the oocyte growth or accumulation of mRNAs drive the growth is not clear. While it is likely that these processes are interdependent, the mRNA metabolism is clearly adapted to the diluting effect of the growth.
In contrast, it appears that miRNAs did not evolve a specific mechanism facilitating their accumulation through altered turnover. This is possible because the post-transcriptional miRNA turnover is regulated in a number of ways but separately from mRNAs (reviewed in (70)). We examined the turnover of two miRNAs and discovered that it was higher than that of most examined mRNAs. Remarkably, we observed that the half-life of the two analyzed miRNAs was between 10 and 20 h. This is comparable to median miRNA half-lives in mouse embryonic stem cells (11 h) and contact-inhibited MEFs (34 h) (71).
The relatively passive mechanism by which mammalian oocytes would abandon the miRNA pathway implies a lack of positive selection for miRNA function in growing and fully grown oocytes. Consequently, miRNA metabolism would not adapt to the accumulation of maternal mRNAs during the oocyte growth. While we show that this is a common mammalian phenomenon, the lack of miRNA activity was also observed in frog oocytes (24) and miRNA depletion seems to exist in zebrafish oocytes as well (72). Therefore, this phenomenon appears to be a common feature of vertebrate oocytes and we speculate that it could exist in many other taxa. It remains to be investigated how AGO and miRNA levels are inter-related and how AGO expression is controlled in mammalian oocytes, as AGO protein levels may be a significant factor. It should be noted that miR-27a in porcine oocytes (67) and miR-130b in bovine oocytes (68) were proposed to be active and important; however, there is no strong evidence supporting these claims as abundance of these miRNAs and reporter repression by endogenous miRNA levels have not been assessed. Therefore, whether these miRNAs represent unique adaptations that are functional exceptions to the rule remains to be investigated.
In any case, it is surprising that such an omnipresent post-transcriptional regulatory pathway would lose its repressive potential in mammalian oocytes and play no significant role in the mammalian transition from maternal-to-zygotic (MZT) transcriptome control as directly shown for mice (22). There are several hypothetical scenarios to be investigated. First, the pressure to stabilize the maternal gene expression program in non-dividing oocytes through miRNAs may not be strong once they enter meiosis. Second, retaining a functional miRNA pathway may not be compatible or may even be detrimental for the mechanisms needed for MZT transition. This could concern post-transcriptional control of the maternal transcriptome such as storage of deadenylated dormant maternal mRNAs or accumulation of totipotency factors in the presence of let-7 miRNA (reviewed in (73)). Third, the lack of miRNA accumulation could be adopted as a strategy for replacing maternal miRNAs with pluripotent zygotic miRNAs of the miR-290 family, where let-7 could be seen as an anti-pluripotency factor (52). This notion is consistent with the miRNA activity during MZT in zebrafish, where zygotic miR-430 has dominant functional significance in the transition (74). miR-430 is also an excellent example of an adaptation to the large cytoplasmic volume: miR-430 rapidly reaches functionally relevant amounts because it is expressed from multiple tandemly arrayed copies (74).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Anna Tetkova and Ales Petelak for help with establishing microinjection experiments, Tatiana Spitzova and Jovana Sadlova for assistance with hamster oocyte isolation.
Author Contributions: S.K., V.K. and D.Z. performed the experiments. M.M. did the mathematical modeling. F.H. analyzed RNA-sequencing data. S.K, R.M., J.K., G.M. and P.S. designed the experiments, supervised and wrote the manuscript.
Contributor Information
Shubhangini Kataruka, Institute of Molecular Genetics of the Czech Academy of Sciences, Videnska 1083, 142 20 Prague 4, Czech Republic.
Martin Modrak, Institute of Microbiology of the Czech Academy of Sciences, Videnska 1083, 142 20 Prague 4, Czech Republic.
Veronika Kinterova, Institute of Animal Physiology and Genetics of the Czech Academy of Sciences, Rumburská 89, 277 21 Liběchov, Czech Republic.
Radek Malik, Institute of Molecular Genetics of the Czech Academy of Sciences, Videnska 1083, 142 20 Prague 4, Czech Republic.
Daniela M Zeitler, RNA Biology, Biochemistry Center Regensburg, University of Regensburg, 93053 Regensburg, Germany.
Filip Horvat, Institute of Molecular Genetics of the Czech Academy of Sciences, Videnska 1083, 142 20 Prague 4, Czech Republic; Bioinformatics Group, Division of Molecular Biology, Department of Biology, Faculty of Science, University of Zagreb, Horvatovac 102a, 10000 Zagreb, Croatia.
Jiri Kanka, Institute of Animal Physiology and Genetics of the Czech Academy of Sciences, Rumburská 89, 277 21 Liběchov, Czech Republic.
Gunter Meister, RNA Biology, Biochemistry Center Regensburg, University of Regensburg, 93053 Regensburg, Germany.
Petr Svoboda, Institute of Molecular Genetics of the Czech Academy of Sciences, Videnska 1083, 142 20 Prague 4, Czech Republic.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Main funding H2020 European Research Council (ERC) [647403, DFENS]. Additional funding from Ministry of Education, Youth, and Sports (MEYS) Project [NPU1 LO1419]; Deutsche Forschungsgemeinschaft [SFB960 to D.Z., G.M., in part]; IAPG Institutional Support RVO [67985904 to J.K., V.K., in part]; MEYS, ELIXIR CZ Research Infrastructure [LM2015047, LM2018131 to M.M., in part]; Charles University in Prague, PhD Student Fellowship (S.K.) - this work will be in part used to fulfill the requirements for a PhD degree and hence can be considered “school work”. Funding for open access charge: H2020 ERC.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bartel D.P. Metazoan microRNAs. Cell. 2018; 173:20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dueck A., Meister G.. Assembly and function of small RNA—argonaute protein complexes. Biol. Chem. 2014; 395:611–629. [DOI] [PubMed] [Google Scholar]
- 3. Bosson A.D., Zamudio J.R., Sharp P.A.. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell. 2014; 56:347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Denzler R., Agarwal V., Stefano J., Bartel D.P., Stoffel M.. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell. 2014; 54:766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Denzler R., McGeary S.E., Title A.C., Agarwal V., Bartel D.P., Stoffel M.. Impact of microRNA levels, target-site complementarity, and cooperativity on competing endogenous RNA-regulated gene expression. Mol. Cell. 2016; 64:565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bishop J.O., Morton J.G., Rosbash M., Richardson M.. Three abundance classes in HeLa cell messenger RNA. Nature. 1974; 250:199–204. [DOI] [PubMed] [Google Scholar]
- 7. Yekta S., Shih I.H., Bartel D.P.. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004; 304:594–596. [DOI] [PubMed] [Google Scholar]
- 8. Brennecke J., Stark A., Russell R.B., Cohen S.M.. Principles of microRNA-target recognition. PLoS Biol. 2005; 3:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sontheimer E.J. Assembly and function of RNA silencing complexes. Nat. Rev. Mol. Cell Biol. 2005; 6:127–138. [DOI] [PubMed] [Google Scholar]
- 10. Salomon W.E., Jolly S.M., Moore M.J., Zamore P.D., Serebrov V.. Single-molecule imaging reveals that argonaute reshapes the binding properties of its nucleic acid guides. Cell. 2015; 162:84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wee L.M., Flores-Jasso C.F., Salomon W.E., Zamore P.D.. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012; 151:1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doench J.G., Petersen C.P., Sharp P.A.. siRNAs can function as miRNAs. Genes Dev. 2003; 17:438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hutvagner G., Zamore P.D.. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002; 297:2056–2060. [DOI] [PubMed] [Google Scholar]
- 14. Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M.. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005; 433:769–773. [DOI] [PubMed] [Google Scholar]
- 15. Kozomara A., Birgaoanu M., Griffiths-Jones S.. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019; 47:D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krutzfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M.. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005; 438:685–689. [DOI] [PubMed] [Google Scholar]
- 17. Schmitter D., Filkowski J., Sewer A., Pillai R.S., Oakeley E.J., Zavolan M., Svoboda P., Filipowicz W.. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006; 34:4801–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R.. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007; 39:380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tam O.H., Aravin A.A., Stein P., Girard A., Murchison E.P., Cheloufi S., Hodges E., Anger M., Sachidanandam R., Schultz R.M. et al.. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008; 453:534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Watanabe T., Totoki Y., Toyoda A., Kaneda M., Kuramochi-Miyagawa S., Obata Y., Chiba H., Kohara Y., Kono T., Nakano T. et al.. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008; 453:539–543. [DOI] [PubMed] [Google Scholar]
- 21. Ma J., Flemr M., Stein P., Berninger P., Malik R., Zavolan M., Svoboda P., Schultz R.M.. MicroRNA activity is suppressed in mouse oocytes. Curr. Biol. 2010; 20:265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suh N., Baehner L., Moltzahn F., Melton C., Shenoy A., Chen J., Blelloch R.. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr. Biol. 2010; 20:271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Flemr M., Ma J., Schultz R.M., Svoboda P.. P-body loss is concomitant with formation of a messenger RNA storage domain in mouse oocytes. Biol. Reprod. 2010; 82:1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Freimer J.W., Krishnakumar R., Cook M.S., Blelloch R.. Expression of alternative Ago2 isoform associated with loss of microRNA-driven translational repression in mouse oocytes. Curr. Biol. 2018; 28:296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Flemr M., Malik R., Franke V., Nejepinska J., Sedlacek R., Vlahovicek K., Svoboda P.. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell. 2013; 155:807–816. [DOI] [PubMed] [Google Scholar]
- 26. Murchison E.P., Stein P., Xuan Z., Pan H., Zhang M.Q., Schultz R.M., Hannon G.J.. Critical roles for Dicer in the female germline. Genes Dev. 2007; 21:682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tang F., Kaneda M., O’Carroll D., Hajkova P., Barton S.C., Sun Y.A., Lee C., Tarakhovsky A., Lao K., Surani M.A.. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007; 21:644–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T.. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004; 15:185–197. [DOI] [PubMed] [Google Scholar]
- 29. Hastie N.D., Bishop J.O.. The expression of three abundance classes of messenger RNA in mouse tissues. Cell. 1976; 9:761–774. [DOI] [PubMed] [Google Scholar]
- 30. Piko L., Clegg K.B.. Quantitative changes in total RNA, total poly(A), and ribosomes in early mouse embryos. Dev. Biol. 1982; 89:362–378. [DOI] [PubMed] [Google Scholar]
- 31. Carter M.G., Sharov A.A., VanBuren V., Dudekula D.B., Carmack C.E., Nelson C., Ko M.S.. Transcript copy number estimation using a mouse whole-genome oligonucleotide microarray. Genome Biol. 2005; 6:R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marinov G.K., Williams B.A., McCue K., Schroth G.P., Gertz J., Myers R.M., Wold B.J.. From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res. 2014; 24:496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fan X., Zhang X., Wu X., Guo H., Hu Y., Tang F., Huang Y.. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 2015; 16:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jahn C.L., Baran M.M., Bachvarova R.. Stability of RNA synthesized by the mouse oocyte during its major growth phase. J. Exp. Zool. 1976; 197:161–171. [DOI] [PubMed] [Google Scholar]
- 35. Brower P.T., Gizang E., Boreen S.M., Schultz R.M.. Biochemical studies of mammalian oogenesis: synthesis and stability of various classes of RNA during growth of the mouse oocyte in vitro. Dev. Biol. 1981; 86:373–383. [DOI] [PubMed] [Google Scholar]
- 36. De Leon V., Johnson A., Bachvarova R.. Half-lives and relative amounts of stored and polysomal ribosomes and poly(A) + RNA in mouse oocytes. Dev. Biol. 1983; 98:400–408. [DOI] [PubMed] [Google Scholar]
- 37. Svoboda P., Franke V., Schultz R.M.. Sculpting the transcriptome during the oocyte-to-embryo transition in mouse. Curr. Top. Dev. Biol. 2015; 113:305–349. [DOI] [PubMed] [Google Scholar]
- 38. Blaha M., Nemcova L., Kepkova K.V., Vodicka P., Prochazka R.. Gene expression analysis of pig cumulus-oocyte complexes stimulated in vitro with follicle stimulating hormone or epidermal growth factor-like peptides. Reprod. Biol. Endocrinol. 2015; 13:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kinterova V., Kanka J., Petruskova V., Toralova T.. Inhibition of Skp1-Cullin-F-box complexes during bovine oocyte maturation and preimplantation development leads to delayed development of embryos. Biol. Reprod. 2019; 100:896–906. [DOI] [PubMed] [Google Scholar]
- 40. Hasler D., Lehmann G., Murakawa Y., Klironomos F., Jakob L., Grasser F.A., Rajewsky N., Landthaler M., Meister G.. The Lupus Autoantigen La Prevents Mis-channeling of tRNA Fragments into the Human MicroRNA Pathway. Mol. Cell. 2016; 63:110–124. [DOI] [PubMed] [Google Scholar]
- 41. Soetaert K., Petzoldt T., Setzer R.W.. Solving differential equations in R: package deSolve. J. Stat. Softw. 2010; 33:1–25.20808728 [Google Scholar]
- 42. Wickham H. ggplot2: Elegant Graphics for Data Analysis. 2009; NY: Springer. [Google Scholar]
- 43. Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W.. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005; 309:1573–1576. [DOI] [PubMed] [Google Scholar]
- 44. Roller R.J., Kinloch R.A., Hiraoka B.Y., Li S.S., Wassarman P.M.. Gene expression during mammalian oogenesis and early embryogenesis: quantification of three messenger RNAs abundant in fully grown mouse oocytes. Development. 1989; 106:251–261. [DOI] [PubMed] [Google Scholar]
- 45. England C.G., Ehlerding E.B., Cai W.. NanoLuc: a small luciferase is brightening up the field of bioluminescence. Bioconjug. Chem. 2016; 27:1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Svoboda P., Stein P., Hayashi H., Schultz R.M.. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development. 2000; 127:4147–4156. [DOI] [PubMed] [Google Scholar]
- 47. Grupen C.G., Fung M., Armstrong D.T.. Effects of milrinone and butyrolactone-I on porcine oocyte meiotic progression and developmental competence. Reprod. Fertil. Dev. 2006; 18:309–317. [DOI] [PubMed] [Google Scholar]
- 48. Garcia-Lopez J., Hourcade Jde D., Alonso L., Cardenas D.B., del Mazo J.. Global characterization and target identification of piRNAs and endo-siRNAs in mouse gametes and zygotes. Biochim. Biophys. Acta. 2014; 1839:463–475. [DOI] [PubMed] [Google Scholar]
- 49. Yang Q., Lin J., Liu M., Li R., Tian B., Zhang X., Xu B., Liu M., Zhang X., Li Y. et al.. Highly sensitive sequencing reveals dynamic modifications and activities of small RNAs in mouse oocytes and early embryos. Sci. Adv. 2016; 2:e1501482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gad A., Nemcova L., Murin M., Kanka J., Laurincik J., Benc M., Pendovski L., Prochazka R.. microRNA expression profile in porcine oocytes with different developmental competence derived from large or small follicles. Mol. Reprod. Dev. 2019; 86:426–439. [DOI] [PubMed] [Google Scholar]
- 51. Roovers E.F., Rosenkranz D., Mahdipour M., Han C.T., He N., Chuva de Sousa Lopes S.M., van der Westerlaken L.A., Zischler H., Butter F., Roelen B.A. et al.. Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Rep. 2015; 10:2069–2082. [DOI] [PubMed] [Google Scholar]
- 52. Svoboda P., Flemr M.. The role of miRNAs and endogenous siRNAs in maternal-to-zygotic reprogramming and the establishment of pluripotency. EMBO Rep. 2010; 11:590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Graf A., Krebs S., Zakhartchenko V., Schwalb B., Blum H., Wolf E.. Fine mapping of genome activation in bovine embryos by RNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:4139–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Agarwal V., Bell G.W., Nam J.W., Bartel D.P.. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015; 4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ma J., Flemr M., Strnad H., Svoboda P., Schultz R.M.. Maternally recruited DCP1A and DCP2 contribute to messenger RNA degradation during oocyte maturation and genome activation in mouse. Biol. Reprod. 2013; 88:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ma J., Fukuda Y., Schultz R.M.. Mobilization of dormant Cnot7 mRNA promotes deadenylation of maternal transcripts during mouse oocyte maturation. Biol. Reprod. 2015; 93:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Karlic R., Ganesh S., Franke V., Svobodova E., Urbanova J., Suzuki Y., Aoki F., Vlahovicek K., Svoboda P.. Long non-coding RNA exchange during the oocyte-to-embryo transition in mice. DNA Res. 2017; 24:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gurtan A.M., Ravi A., Rahl P.B., Bosson A.D., JnBaptiste C.K., Bhutkar A., Whittaker C.A., Young R.A., Sharp P.A.. Let-7 represses Nr6a1 and a mid-gestation developmental program in adult fibroblasts. Genes Dev. 2013; 27:941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lee Y.S., Dutta A.. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007; 21:1025–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen S., Xue Y., Wu X., Le C., Bhutkar A., Bell E.L., Zhang F., Langer R., Sharp P.A.. Global microRNA depletion suppresses tumor angiogenesis. Genes Dev. 2014; 28:1054–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu D., Dean J.. EXOSC10 sculpts the transcriptome during the growth-to-maturation transition in mouse oocytes. Nucleic Acids Res. 2020; 48:5349–5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Puschendorf M., Stein P., Oakeley E.J., Schultz R.M., Peters A.H., Svoboda P.. Abundant transcripts from retrotransposons are unstable in fully grown mouse oocytes. Biochem. Biophys. Res. Commun. 2006; 347:36–43. [DOI] [PubMed] [Google Scholar]
- 63. Linsen S.E., de Wit E., Janssens G., Heater S., Chapman L., Parkin R.K., Fritz B., Wyman S.K., de Bruijn E., Voest E.E. et al.. Limitations and possibilities of small RNA digital gene expression profiling. Nat. Methods. 2009; 6:474–476. [DOI] [PubMed] [Google Scholar]
- 64. Janas M.M., Wang B., Harris A.S., Aguiar M., Shaffer J.M., Subrahmanyam Y.V., Behlke M.A., Wucherpfennig K.W., Gygi S.P., Gagnon E. et al.. Alternative RISC assembly: binding and repression of microRNA-mRNA duplexes by human Ago proteins. RNA. 2012; 18:2041–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Franke V., Ganesh S., Karlic R., Malik R., Pasulka J., Horvat F., Kuzman M., Fulka H., Cernohorska M., Urbanova J. et al.. Long terminal repeats power evolution of genes and gene expression programs in mammalian oocytes and zygotes. Genome Res. 2017; 27:1384–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stein P., Rozhkov N.V., Li F., Cardenas F.L., Davydenko O., Vandivier L.E., Gregory B.D., Hannon G.J., Schultz R.M.. Essential Role for endogenous siRNAs during meiosis in mouse oocytes. PLoS Genet. 2015; 11:e1005013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Veselovska L., Smallwood S.A., Saadeh H., Stewart K.R., Krueger F., Maupetit-Mehouas S., Arnaud P., Tomizawa S., Andrews S., Kelsey G.. Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biol. 2015; 16:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Israel S., Ernst M., Psathaki O.E., Drexler H.C.A., Casser E., Suzuki Y., Makalowski W., Boiani M., Fuellen G., Taher L.. An integrated genome-wide multi-omics analysis of gene expression dynamics in the preimplantation mouse embryo. Sci. Rep. 2019; 9:13356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Luo Y., Na Z., Slavoff S.A.. P-bodies: composition, properties, and functions. Biochemistry. 2018; 57:2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Krol J., Loedige I., Filipowicz W.. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010; 11:597–610. [DOI] [PubMed] [Google Scholar]
- 71. Kingston E.R., Bartel D.P.. Global analyses of the dynamics of mammalian microRNA metabolism. Genome Res. 2019; 29:1777–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen P.Y., Manninga H., Slanchev K., Chien M., Russo J.J., Ju J., Sheridan R., John B., Marks D.S., Gaidatzis D. et al.. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005; 19:1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Svoboda P. Why mouse oocytes and early embryos ignore miRNAs. RNA Biol. 2010; 7:559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F.. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006; 312:75–79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.