Abstract
In the Elongator-dependent modification pathway, chemical modifications are introduced at the wobble uridines at position 34 in transfer RNAs (tRNAs), which serve to optimize codon translation rates. Here, we show that this three-step modification pathway exists in Dictyostelium discoideum, model of the evolutionary superfamily Amoebozoa. Not only are previously established modifications observable by mass spectrometry in strains with the most conserved genes of each step deleted, but also additional modifications are detected, indicating a certain plasticity of the pathway in the amoeba. Unlike described for yeast, D. discoideum allows for an unconditional deletion of the single tQCUG gene, as long as the Elongator-dependent modification pathway is intact. In gene deletion strains of the modification pathway, protein amounts are significantly reduced as shown by flow cytometry and Western blotting, using strains expressing different glutamine leader constructs fused to GFP. Most dramatic are these effects, when the tQCUG gene is deleted, or Elp3, the catalytic component of the Elongator complex is missing. In addition, Elp3 is the most strongly conserved protein of the modification pathway, as our phylogenetic analysis reveals. The implications of this observation are discussed with respect to the evolutionary age of the components acting in the Elongator-dependent modification pathway.
INTRODUCTION
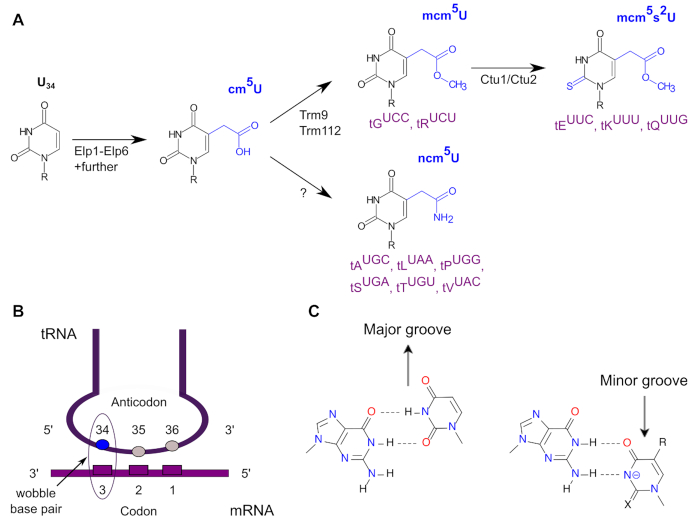
Chemical modifications in nucleic acids are widespread and this holds in particular true for transfer RNA (tRNA) molecules, for which more than 100 types of modifications are known (1–3). The wobble uridines at position 34 (U34) in tRNAs are almost always modified in any organism (3). The first step in the modification pathway of U34 (Figure 1A, and see below) is catalyzed by the eukaryotic Elongator complex (together with Kti11-14) (4,5), which has been reported for mammals, plants, yeast and nematodes (5–8). The Elongator complex introduces a carboxymethyl side chain at C5 (cm5) at U34, which is converted either into a 5-carbamoylmethyl (ncm5) group by an unknown enzymatic activity, or into 5-methoxycarbonylmethyl (mcm5) by the Trm9 and Trm112 proteins (9,10). These two modifications are found at U34 in a total of eight mature tRNAs (Figure 1A). In wild type fission yeast, mcm5U34 is a required modification to allow for subsequent thiolation at C2, resulting in 5-methoxycarbonymethyl-2-thiouridine (mcm5s2U) (11). This modification is found in three tRNAs and introduced by the Ctu1/Ctu2 complex (Figure 1A). The majority of yeast tRNAs with a uridine at the wobble position (Figure 1B) feature either of these Elongator-dependent modifications (3).
Figure 1.
The Elongator-dependent tRNA modification pathway. (A) The anticodon uridine at position 34 (U34) is modified initially to cm5 by the Elongator complex and further modified either by an unknown enzyme to ncm5, which is found in the indicated six tRNAs (purple), or by Trm9/Trm112 to mcm5 that is found in the two tRNAs shown. Ctu1/Ctu2 finally are thiolating the C2 of the pyrimidine ring in mcm5 resulting in mcm5s2U, which is found in the three tRNAs displayed. An alternative order for these steps has also been described (4). (B) Schematic drawing of U34’s position in tRNAs during decoding of an mRNA with codon and anticodon indicated. (C) Formation of a G•U wobble base pair during decoding depends on the absence (left) or presence (right) of chemical modifications (X, R) on the nucleobase. Panel (C) was modified from (72).
The Elongator complex consists of six proteins and was originally identified as a part of the yeast RNA polymerase II holoenzyme (12–15). Structurally, the six subunits reside in two sub-complexes (12,13,15), the Elp123 complex and the Elp456 complex. The latter forms a hexameric ring of alternating Elp4, Elp5 and Elp6 subunits (16,17). Recently, this ring was shown to be asymmetrically attached to a symmetric Elp123 dimer (18–20). In the resulting holocomplex, the tRNA substrate is thought to be associated with the Elp456 complex via the anticodon stem loop, which positions U34 in the vicinity of the catalytic Elp3 subunit (21) that harbors the functionally important radical S-adenosyl methionine (SAM) and histone acetyltransferase (HAT) domains (22,23).
Even though the Elongator complex is highly conserved in various kingdoms of life (5–8), deletion of one or more Elongator components in different organisms leads to an array of diverse macroscopic phenotypes. These include alterations in diverse pathways such as transcriptional elongation or silencing, nucleosome assembly, exocytosis, acetylation of histones and other proteins, cell proliferation, embryogenesis or zygotic paternal DNA demethylation (22,24–32). That bewildering variety of at first glance rather unrelated pathways might actually have a common cause in the Elongator complex-mediated modification of tRNAs (Figure 1), as discussed recently (33). This notion is based on the observation that in yeast, all the stress-induced phenotypes observed in Elongator mutants can be rescued, at least in part, by elevated levels of hypomodified tRNA tKUUU and tQUUG that normally carry mcm5s2U34 (34–37). Ribosome profiling (38) revealed that translation in yeast Elongator mutants is slowed at CAA and AAA triplets, which are normally decoded by the mcm5s2U34-containing tKUUU and tQUUG (39–41). Thus, the presence of the mcm5s2U34 modification in these two tRNAs is required for proper and efficient translation in yeast, and similar data were also obtained for Caenorhabditis elegans (41). Furthermore, strong evidence was provided that the presence of the U34 modification is required for the maintenance of proteome integrity, as Elongator mutant strains displayed a high propensity to aggregate endogenous proteins, which again could be rescued by overexpression of hypomodified tRNAs (41). Thus, as a consequence of lacking U34 modifications and distorted translation speed, loss of function due to inappropriate protein amounts can be envisaged, and equally well misfolded proteins, as discussed recently (33). Recently, seminal structural work has provided a mechanistic basis for the effects that chemical modifications of U34 exert on translation (42): only if modified can U34 properly base pair with both, A and G ending codons. In the resulting G•U wobble base pair, the modified U is displaced into the minor groove (Figure 1C) and thus, the same position as a Watson–Crick base pair is occupied in the ribosome. This is not the case in the absence of these modifications, when the U is displaced in the major groove (42). Thus, both these studies provided direct, mechanistic links between tRNA modifications at wobble uridines and proteome integrity (41,42).
Already earlier, the necessity for wobble uridine modifications for proper decoding of G-ending codons in yeast was recognized (43). In that study, Byström and co-workers revealed that the modifications introduced by the Elongator pathway (Figure 1A) actually promote a functionally redundant decoding system. For example, the single copy gene of tQCUG, which is essential in the wild type, can be deleted if the isoacceptor tQUUG is overexpressed. This conditional knockout, however, could not be obtained in strains being inactivated in the Elongator pathway (Figure 1A). In contrast, several prokaryotes do not contain tQCUG genes (44), which suggests that their CAG codons can be read by tQUUG. S. cerevisiae mRNAs feature about 30% CAG and 70% CAA codons, and for decoding, its genome contains a single tQCUG locus and 13 copies of the tRNA gene tQUUG (see also below). The fraction of CAG codons in yeast is possibly too high to allow for an unconditional deletion of the tQCUG gene. Following this notion, we set out in this study to investigate wobble uridine modifications in an organism with a particularly high AT content in its genome.
Our choice was D. discoideum (45), the model organism of the evolutionary supergroup of Amoebozoa (46). D. discoideum is an established model to study mechanisms governing cell motility, autophagy, development, or social evolution (reviewed in (47)), and as such, a broad range of experimental tools have been established (48). The amoeba features one of the lowest GC contents amongst eukaryotic genomes, with overall 23% GC, and 27% in protein coding sequences (45). Further, the function of the Elongator complex has not been addressed in this organism, nor in any other member of the Amoebozoa supergroup. These features make D. discoideum an ideal model to study the role of the Elongator-dependent tRNA modifications in a particularly AT-rich genome. We show that the Elongator pathway of U34 is largely conserved in the amoeba, but that its disruption affects protein expression. This phenotype is particularly evident in a strain that, uniquely amongst eukaryotes known so far, is unconditionally viable without tQCUG.
MATERIALS AND METHODS
Codon usage
D. discoideum codon usage analysis was performed on D. discoideum primary CDS (downloaded from www.dictybase.org/), from where also tRNA information was obtained. The codon compilation for Saccharomyces cerevisiae and Schizosaccharomyces pombe is based on www.kazusa.or.jp/codon/ and for yeast tRNAs from http://gtrnadb.ucsc.edu/. Sequence analysis was performed with Python 3.7. Data was visualized with matplotlib and excel.
Sequence homology
Protein sequences were retrieved from UniProtKB (49). The relevant protein sequences for D. discoideum that were not available on UniProtKB have been downloaded from Dictybase.org. Protein sequences in fasta format were aligned with Clustal Omega (clustalo, version 1.2.4, (50)) resulting in a multiple sequence alignment and phylogenetic tree for each protein. The phylogenetic trees were then visualised using the R/Bioconductor package ggtree (51).
Cell growth and transformation
All D. discoideum strains were grown axenically in HL5+ medium (Formedia) containing antibiotics against prokaryotic growth (10 μg/ml Penicillin, 10 μg/μl Streptomycin, 250 ng/ml Amphotericin, 50 μg/ml Ampicillin) and supplemented with Blasticidine S and/or Geneticin at concentrations of 10 μg/ml when required. Transformation of the axenic strain Ax2-214 (DBS0235535 at www.dictybase.org) or derivatives was done by electroporation as described previously (52). When integrating knockout constructs were transformed, cells were subcloned in order to isolate single colonies. After transformation with extrachromosomal vectors, cell populations were used for further analysis.
Generation of null mutants
Left arm and right arm were amplified with primers specific to the region flanking the gene of interest (Supplementary Table S1). The Stargate Transfer Reagent Set (IBA) was used to shuttled the arms in the pKOSG-IBA-dicty1 destination vector (IBA) where they flanked a BsR-cassette, which itself is surrounded by loxP sites in the pKOSG vector system (53). Gene disruption fragments were cut out from the vector backbone with SpeI and transformed into Ax2 wild type cells or other strains. Upon homologous recombination, blasticidin resistant clones could be selected and the gene deletion verified by PCR (Supplementary Table S1). Making use of the loxP sites, the BsR cassette of different strains was removed by transient expression of Cre-recombinase (54).
RNA isolation and modification analysis
Chemicals
All chemicals and reagents were obtained at the highest purity available and were used without further purification unless stated. Benzonase, bacterial alkaline phosphatase, butylated hydroxytoluene, Pentostation (Deoxycoformycin), theophylline, acetonitrile and buffer salts were purchased from Sigma-Aldrich (Steinheim, Germany). Snake venom phosphodiesterase I was purchased from VWR (Darmstadt, Germany). Tetrahydrouridine was purchased from Merck (Darmstadt, Germany). Water purified through a Milli-Q system was used throughout our studies.
RNA fractionation
Total RNA was loaded on a size-exclusion column (Agilent Bio SEC-3, 3μm, 300Å, 7.8 × 300 mm, Agilent, Waldbronn, Gemany) and RNA fractions eluted with 1 ml/min 100 mM ammonium acetate at pH 7 as the mobile phase at 60°C (55). tRNA was separated from rRNA and small RNAs, vacuum concentrated and precipitated with ammonium acetate and ethanol (Supplementary Figure S4). tRNA was reconstituted in pure water and stored at –20°C until further use.
Nucleoside preparation
∼400 ng of purified tRNA was digested using a mixture of benzonase (2.5 U), bacterial alkaline phosphatase (10 U) and phosphodiesterase I (0.1 U) in a final reaction volume of 20 μl. The reaction mixture was supplemented with MgCl2 to a final concentration of 1 mM and Tris–HCl (pH 8.0) to a final concentration of 0.1 M. Nucleobase deaminase inhibitor coformycin and tetrahydrouridine were added at a concentration of 10 and 50 μg/ml, respectively, and butylated hydroxytoluene (an antioxidant) was added at a concentration of 0.5 mM, for further detail see (56). The digestion was allowed to proceed for 2 h at 37°C and was stopped by filtering through a 10 kDa MWCO filter (AcroPrepTM Advance, 350 ml, OmegaTM 10K MWCO, Pall, Dreieich, Germany) at 3000 × g for 30 min. After addition of 10 μl pure water for salt dilution purposes, 18 μl of filtrate was mixed with 2 μl 15N-dA (50 nM 15N-dA mixed with 10 mM theophylline as external standard) to a final concentration of 5 nM as an internal standard to account for mass spectrometric detection fluctuations. 10 μL of each sample was injected to LC–MS/MS analysis (corresponding to around 150 ng tRNA digest). Note that hypermodified uridine nucleosides are unstable after digestion (57).
Preparation of calibration solutions
For calibration, nucleosides were mixed at the given concentration and serially diluted. cm5U, s2U (both TRC, Toronto, Canada), ncm5s2U, mcm5U and mcm5s2U (generous gift from Mark Helm's lab, Mainz, Germany) were mixed to a 10 μM stock solution in water. This stock solution was mixed with 10 mM adenosine (VWR, Darmstadt, Germany) to a final concentration of 0.55 mM adenosine and 1.11 μM of each modified nucleoside. This calibration stock solution was serially diluted 1:10 until a final concentration of 0.055 μM adenosine and 0.11 nM of each modified nucleoside was reached. 18 μl of each calibration dilution was mixed with 2 μl 15N5-dA (50 nM 15N5-dA mixed with 10 mM theophylline as external standard) to a final concentration of 5 nM as an internal standard to account for mass spectrometric detection fluctuations. 10 μl of each calibration solution was injected and analyzed (Supplementary Figures S5 and S6).
LC–MS/MS analysis of RNA nucleosides
Ribonucleosides were separated using a Synergy Fusion RP, 2.5 μm particle size, 100 Å pore size, 100 mm length, 2 mm inner diameter from Phenomenex (Torrance, CA, USA), on an Agilent 1290 series HPLC system equipped with a diode array detector. Mobile phase A was 5 mM ammonium acetate adjusted to pH 5.3 with glacial acetic acid and mobile phase B was pure acetonitrile. Gradient elution started with 100% A for one minute, increase to 10% B after 5 min, 40% B after 7 min which was maintained for an additional minute. From minute 8 to 8.5 starting conditions are re-established and equilibrated for two additional minutes. The flow rate was 0.35 ml/min and the column temperature 35°C. The effluent from the column was directed through the DAD before entering the Agilent 6490 Triple Quadrupole mass spectrometer in dynamic multiple reaction monitoring (MRM) mode. The MS was operated in positive ion mode with the following parameters: electro-spray ionization (ESI-MS, Agilent Jetstream), Fragmentor Voltage (set in tunefile to) 250 V, Cell Accelerator Voltage 2 V, N2-gas temperature 150°C, N2-gas flow 15 l/min, Nebulizer 30 psi, sheath gas (N2) temperature 275°C, sheath gas flow 11 l/min, capillary 2500 V and nozzle voltage 500 V. The mass transitions for each modified nucleoside are found in Supplementary Table S3.
Data analysis: response factor determination from calibration measurements
Using Agilent's Qualitative Data Analysis software, the UV peak area of adenosine at 260 nm and the mass transition peak areas of each modified nucleoside and the internal standard 15N-dA were integrated. The peak area of adenosine was plotted over the amount injected (in pmol) and the slope represents the response factor of adenosine. Each signal area of the modified nucleosides was divided by the signal of the internal standard. The resulting value was plotted over the amount of injected nucleoside (in pmol) and the slope is then used as the response factor for sample quantification. The calibration curves from one exemplary calibration measurement is shown in Supplementary Figure S6. The lower limit of quantification and detection (LOQ and LOD) are given in Supplementary Table S4.
Quantification of tRNA samples
Using Agilent's Qualitative Data Analysis software, the UV peak area of adenosine at 260 nm was integrated and the amount of tRNA injected calculated by using the calibration curve in Supplementary Figure S6 and assuming 15 adenosine per average tRNA. Peaks corresponding to detection of MRM transitions for the modified nucleosides were normalized to the peak area of the internal standard (15N-deoxyadenosine) to account for inter-sample detection fluctuations and the absolute quantity of modified nucleoside was determined using the calibration curves from Supplementary Figure S6 The amount of each injected modified nucleoside was then normalized to the amount of injected tRNA and plotted in Graphpad Prism ©.
Generation of ectopic expression strains
The elp3 cDNA (accession number: DDB_G0290103; dictybase.org) was acquired by oligo-dT primed reverse transcription and amplification by PCR with specific primers (Supplementary Table S1). The amplicons were cloned into the shuttling vector pJet1.2. Upon sequence verification, the elp3 gene was cloned via the restriction sites BglII or SpeI into the designated Plasmid the pDM317 N-term and pDM323 C-term, respectively (58). The plasmids were transformed into the elp3−rox clone for further analysis.
For generation of point mutated Elp3 versions, the elp3 cDNA was amplified in two different PCR reactions using two primer pairs, with one primer containing the mutated site (Supplementary Table S1). The PCR amplicons contain specific recognition sites that were added to the primers. The Stargate Transfer Reagent Set (IBA) was used to shuttle the two fragments into the pENTRY vector. The resulting point mutated elp3 cDNAs was cloned into the pDM317 via SpeI and BglII restriction sites that were included by PCR amplification.
The Q15-GFP expression plasmid was generated by annealing two oligonucleotides with specific overhang sequences and subsequent cloning into a SpeI and BglII linearized pDM323. Upon transformation in selected D. discoideum strains, GFP expression was analyzed by western blotting.
The tRNA genes tQUUG (DDB_G0295309) and tQCUG (DDB_G0294975) were each amplified ±250 bp from genomic DNA with specific primers and cloned into the pDM vectors pDM323 Q15-GFP and pDM353 Q103-GFP (59) (kind gift of Dr L. Malinovska) via XhoI outside the expression cassette (Supplementary Table S1).
Western blot analysis
Western blot analysis to verify expression of fusion proteins or endogenous proteins was performed as described earlier (60).
Analysis of RNA by northern blot
RNA isolation was performed as described previously (60). In brief, 5 μg total RNA were separated on a 11% PAA gel (7 M Urea in 1× TBE) and run at 25 mA (per gel) constant for 4 h. The APM polyacrylamide gel setups and the electrophoresis runs were performed as described (61). APM was a kind gift by Dr Peter Sarin. The gel run was performed at 50–65 mA for 3 h. The size separated RNA molecules were transferred via semi-dry electroblotting to an uncharged nylon membrane (Amersham Hybond TM-NX). Transfer was carried out at 20 V for 30 min at room temperature. Northern Blot analysis of tRNAs was performed as described for small RNAs previously (60). The hybridizing temperature was set to 45°C. Blots were probed with 5′ 32P labeled DNA oligonucleotides that are listed in Supplementary Table S1.
Flow cytometry
Different D. discoideum strains were grown in sterile filtrated HL5 medium on Petri dishes. At a cell density of 80% confluent cover of the Petri dish, cells were harvested by washing them off the plate. 200–400 μl of the cell suspension was mixed with 1600 μl Soerensen buffer (2 mM Na2HPO4, 15 mM KH2PO4 with H3PO4 pH 6.0) and injected into the flow cytometer. The injection speed was set to 1 μl/s. The measurement was completed when 30 000 events (corresponding to live D. discoideum cells) were detected. A strain, which does not express any fluorescent protein, served as a negative control. Fluorescence intensity detection was performed with a logarithmic scale. The Data analysis was performed with the FlowJo software.
RESULTS
Codon bias in D. discoideum
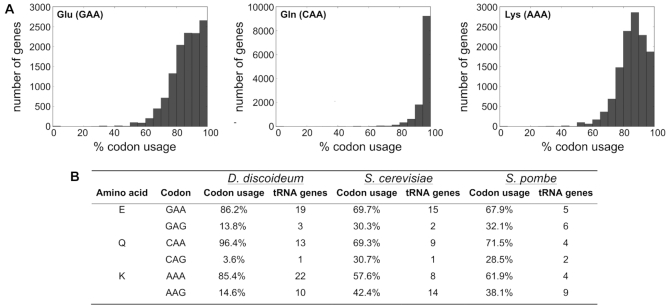
D. discoideum encodes ∼12 500 proteins in its haploid genome (45). In view of its highly AT-rich genome, it was expected that A-ending codons such as GAA, CAA and AAA, which are decoded by tRNAs featuring the mcm5s2 modification at the wobble uridine U34, are preferred in the amoeba over the alternative codons GAG, CAG and AAG. The extent of codon bias, however, was surprising, particularly for glutamine codons, where the great majority of mRNAs contains only CAAs (Figure 2A). To put this into perspective, we determined the per-gene codon usage in D. discoideum and compared it to those in the yeasts S. cerevisiae and S. pombe, together with the number of tRNA genes encoded in either microorganism and the respective codon usage averaged over all genes (Figure 2). Indeed, compared to the yeasts, the A-ending codons for E (GAA), Q (CAA) and K (AAA) appear to be strongly overrepresented. Furthermore, also the gene number of the three tRNAs decoding these codons appears to be significantly expanded in the amoeba, as compared to the afore-mentioned yeast species (Figure 2B). This striking difference encouraged us to investigate the function of the D. discoideum Elongator-dependent modification pathway yielding mcm5s2 in wobble uridines.
Figure 2.
Codon usage in D. discoideum. (A) Relative usage of codons that are decoded by tRNAs, which are fully modified in the Elongator-dependent pathway: GAA for glutamate (left), CAA for glutamine (center) and AAA for lysine (right), in all D. discoideum protein coding genes. (B) Comparison of the relative, proteome-wide codon usage for E, Q and K and decoding tRNA gene number in D. discoideum (Dd), S. cerevisiae (Sc) and S. pombe (Sp). Data for Dd derived from www.dictybase.org; Codon compilation for Sc and Sp based on www.kazusa.or.jp/codon/ and for yeast tRNAs from http://gtrnadb.ucsc.edu/.
Phylogenetic comparison of the D. discoideum proteins of the Elongator-dependent tRNA modification pathway
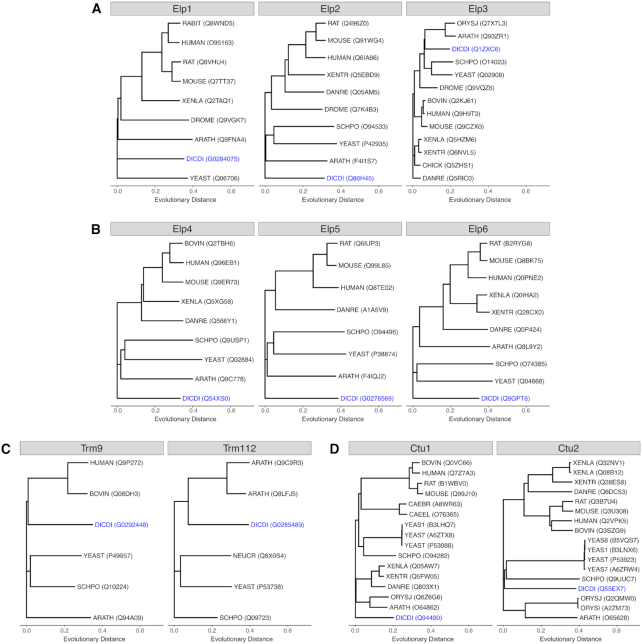
To understand their conservation, we initially compared the Elongator proteins from D. discoideum to those from S. cerevisiae, which revealed that within the Elongator complex, only Elp3, harboring the catalytically important HAT and SAM domains (22,23), displayed a highly significant sequence identity of 70% between the two species (Supplementary Table S2). In contrast, the degree of conservation of the Elp1 and Elp2 proteins was <30% and those of the Elp456 complex 20% or less. This variable conservation amongst Elongator constituents prompted us to compare the protein sequences of the Elongator complex of D. discoideum also to homologues from other species. Unambiguously identified protein sequences available from UniprotKB (49) were used and revealed a similar scenario: Elp3 is by far the most conserved protein of the Elongator complex, followed by Elp1, Elp2 and the proteins of the Elp456 sub-complex (Figure 3A, B). When we carried out similar analyses for the further proteins of the Elongator-dependent modification pathway, we observed a conservation of the Trm9 and Trm112 proteins that is similar to that observed for Elp1 and Elp2 (Figure 3C). In contrast, within the Ctu1/Ctu2 complex, the Ctu1 proteins show a considerably stronger sequence similarity compared to their Ctu2 interaction partners (Figure 3D). Using this information, we next set out to disrupt the genes of the most conserved proteins of the Elongator-dependent modification pathway.
Figure 3.
Phylogenetic trees of proteins acting in the Elongator-dependent tRNA modification pathway. Shown are the trees for proteins identified as homologues of the proteins Elp1-Elp3 (A), Elp4-Elp6 (B), Trm9 and Trm112 (C) and Ctu1 and Ctu2 (D). Protein sequences of the following species were compared (where available): ARATH: Arabidopsis thaliana; BOVIN: Bos taurus; CAEBR: Caenorhabditis briggsae; CAEEL: Caenorhabditis elegans; CHICK: Gallus gallus; CHLRE: Chlamydomonas reinhardtii; DANRE: Danio rerio; DICDI: Dictyostelium discoideum; DROME: Drosophila melanogaster; HUMAN: Homo sapiens; MAIZE: Zea mays; MOUSE: Mus musculus; NEUCR: Neurospora crassa (strain ATCC 24698/74-OR23-1A/CBS 708.71/DSM 1257/FGSC 987); ORYSI: Oryza sativa subsp. indica; ORYSJ: Oryza sativa subsp. japonica; RABIT: Oryctolagus cuniculus; RAT: Rattus norvegicus; SCHPO: Schizosaccharomyces pombe (strain 972/ATCC 24843); XENLA: Xenopus laevis; XENTR: Xenopus tropicalis; YEAS1: Saccharomyces cerevisiae (strain RM11-1a); YEAS6: Saccharomyces cerevisiae (strain AWRI1631); YEAS7: Saccharomyces cerevisiae (strain YJM789); YEAST: Saccharomyces cerevisiae (strain ATCC 204508/S288c). Their relative evolutionary distances (unitless) are plotted using the same scale for all proteins and their UniprotKB or Dictybase identifiers are given in brackets. D. discoideum homologues are shown in blue.
Generation of gene deletion strains of the Elongator-dependent modification pathway in D. discoideum
As Elp3, the catalytic entity of the complex (22,23), is the only highly conserved protein of the Elongator complex in D. discoideum (Supplementary Table S2; Figure 3A, B), we deleted the elp3 gene by homologous recombination (Supplementary Figure S1). To disrupt the Elongator-dependent modification pathway at all major steps (Figure 1A), we also targeted the D. discoideum trm9 and ctu1 genes, encoding the more conserved proteins of the respective complexes (Figure 3C, D). Gene deletions were confirmed for elp3, trm9 and ctu1 by PCR analyses and the Bs(R) cassettes were removed from the mutant alleles, resulting in the respective single deletion strains (Supplementary Figures S1–S3). Additionally, an elp3−/ctu1− double mutant was obtained by transforming the ctu1-targeting construct in the elp3− background (Supplementary Figure S3). The viability of the generated mutant strains indicates that the investigated genes are not essential in D. discoideum, and under standard lab conditions, these strains displayed no reproducible growth defects compared to the Ax2 wild type.
Chemical modifications of nucleosides in the generated mutant strains of the Elongator-dependent modification pathway
To dissect the U34 modification pathway (Figure 1) in D. discoideum, we investigated the presence of Elongator-dependent modified nucleosides in tRNA in the generated mutant strains. For this purpose, total RNA was extracted and total tRNA purified by size exclusion chromatography (SEC) (Supplementary Figure S4). The nucleosides released through enzymatic digestion were separated by liquid chromatography and detected by either UV absorption (λ 260 nm) or tandem mass spectrometry (LC–UV–MS/MS, MS parameters are given in Supplementary Table S3). An exemplary merged chromatogram is given in Supplementary Figure S5. For absolute quantification, calibration solutions were prepared with synthetically available nucleosides and 15N5-deoxyadenosine (15N5-dA) was used as an internal standard (57).
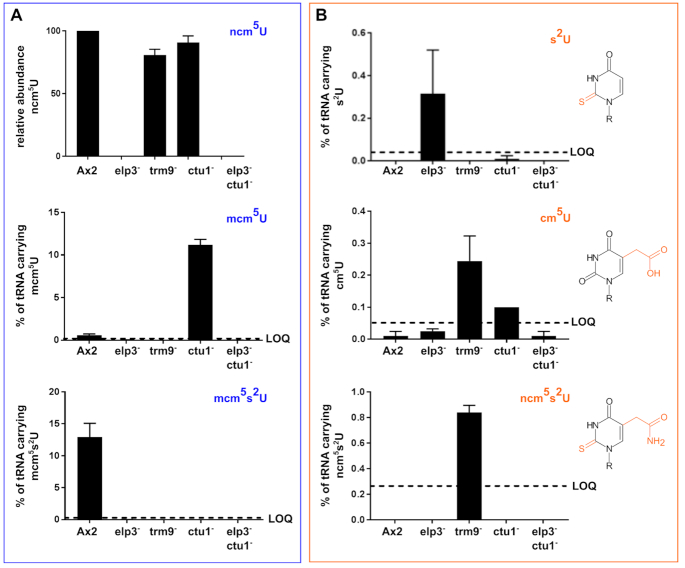
In the wild type Ax2 strain, we observed the Elongator-dependent modifications mcm5s2U, mcm5U and ncm5U (Figure 4A; note: due to the unavailability of ncm5U synthetic standard, the absolute quantities could not be determined for this modified nucleoside). In the elp3− and subsequently the elp3−/ctu1− strain, none of these modified nucleosides is detectable, which is in accordance with the function of the elp3 gene product (Figure 1). As a consequence of elp3 deletion, the abundance of thiolated uridine (s2U) increases in the elp3− strain (Figure 4B), demonstrating that the mcm5U modification is no absolute requirement for uridine thiolation. In the absence of Trm9, we find an elevated abundance of cm5U in accordance with the methyltransferase activity of Trm9, which facilitates the cm5U to mcm5U reaction. Interestingly, the thiolated version of ncm5U, namely ncm5s2U, is increased above the lower limit of quantification in the trm9− strain, while its precursor ncm5U is slightly reduced compared to Ax2 derived tRNAs. Upon loss of the thiotransferase subunit Ctu1, the conversion of mcm5U to mcm5sU is not observable and mcm5U accumulates in tRNAs derived from the ctu1− strain. cm5s2U was not detectable in any samples due to its high LLOQ (lower limit of quantification, Supplementary Table S4). Taken together, these data indicate that the Elongator-dependent modification pathway of wobble uridines (Figure 1) is conserved in D. discoideum (Figure 4), but that the sequential order when individual modification steps are introduced at uridines might vary.
Figure 4.
Chemical modifications of uridines in D. discoideum Ax2 wild type and mutant strains of the Elongator-dependent modification pathway. LC–UV–MS/MS analysis of Elongator-dependent modifications of uridines in purified D. discoideum tRNAs. (A) Chemical modifications (blue) of the Elongator-dependent pathway. Shown are the relative abundance of ncm5U (no chemical standard available, top) and the absolute abundance of tRNAs carrying the mcm5U (center) and mcm5s2U (bottom) modifications in the Ax2 wild type and the indicated mutant strains of the Elongator-dependent pathway. The lower limit of quantification (LOQ) is indicated with a dashed line where appropriate. (B) Additional chemical modifications of uridines observed in mutant strains of the Elongator pathway. Shown are the absolute abundance of tRNAs carrying s2U (top), cm5U (center) and ncm5s2U (bottom) modifications in the Ax2 wild type and the indicated mutant strains of the Elongator-dependent pathway. The LOQ for these modifications is indicated. These modifications (orange) are not observed in the Ax2 wild type and their structures are shown to the right. Please note the different scaling of the y-axes in panels (A) and (B).
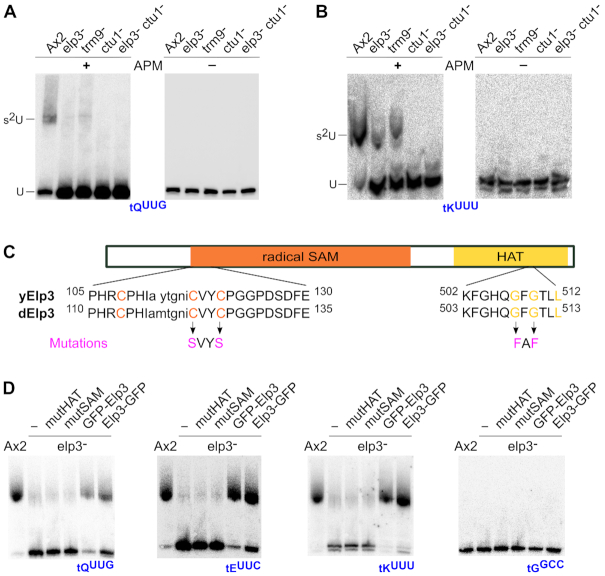
APM Northern blots allow to assess thiolation in specific tRNAs in D. discoideum
In order to specifically address thiolation of the tRNAs tKUUU, tEUUC and tQUUG, p-(N-acrylamino)-phenyl]mercuric chloride (APM) northern blot analysis was performed next. APM is an organic mercury compound that can be embedded into the matrix of normal polyacrylamide gels (PAA) (61,62). Molecules carrying sulfur atoms will be held back in the gel matrix, resulting in a shift that does not correspond to the migration distance of their molecular size. In combination with northern blot analyses, thiolation levels of specific RNA molecules thus can be determined by use of APM (11). This is exemplified by APM northern blots for tQUUG (Figure 5A) and tKUUU (Figure 5B), which display shifted signals for RNA isolated from the Ax2 wild type and, at a lower signal strength, for RNA isolated from the elp3− and trm9− strains. In accordance with our model, no shifted band is observed for strains with disrupted ctu1 gene. This is in line with the global analysis of chemical modifications in nucleosides (Figure 4), and it proves the thiolation of specific tRNA molecules. Taken together, the data shown in Figure 4 and 5 demonstrate that the thiolation of the U34 is conditionally dependent on the mcm5 modification: a small fraction of unmodified or ncm5 modified Us can be thiolated, but only the full conversion to mcm5U leads to a high thiolation level.
Figure 5.
Thiolation of tRNAs in mutant strains of the Elongator-dependent modification pathway. APM Northern blots against tQUUG (A) and tKUUU (B) in the Ax2 wild type and indicated mutant strains. (A, B) In the APM-containing gels (+), thiolated RNA interacts with the embedded mercury, resulting in an electrophoretic shift (indicated by s2U) that does not correspond to the size of the molecule (U). Left: Control Northern blots from gels lacking APM (−). (C) Scheme of the D. discoideum Elp3 protein with its catalytic domains, the radical S-adenosyl methionine (SAM, orange) and histone acetyl transferase (HAT, yellow) domains. Underneath are amino acid sequences of the catalytic center of the respective domain shown from the S. cerevisiae homologue (yElp3) vs. the D. discoideum Elp3 (dElp3). Capital letters indicate strict conservation between Elp3 proteins. Orange and yellow colored amino acids are essential for the catalytic function of the corresponding domain. Mutations (magenta) indicate amino acid changes in these positions, resulting in the mutSAM (CVYC→ SVYS) and mutHAT (GFG→ FAF) D. discoideum Elp3 variants. (D) APM Northern blots (left to right) against tQUUG, tEUUC, tKUUU and tGCCG (control), employing RNA isolated from the Ax2 wild type strain, compared to the elp3— strain, without or with overexpressed Elp3 variants (mutHAT, mutSAM, and N- and C-terminal GFP fusions of the Elp3 wild type sequence).
Ectopic expression of Elp3 GFP fusion proteins complements the molecular phenotype of the elp3− strain
To analyze if the phenotype of the elp3− strain can be rescued by ectopic overexpression, we next set out to generate N- and C-terminal GFP-fusions of Elp3. To this end, the elp3 cDNA was amplified by PCR and cloned into suitable vectors of the pDM series (58). In yeast, the two catalytic domains HAT and radical SAM of Elp3 were shown to be indispensable for the tRNA modification at U34 (63). Therefore, two point mutated GFP-Elp3 constructs were also generated (Figure 5C) to address whether Elp3 is the catalytic subunit of the Elongator complex in D. discoideum and if both catalytic domains are needed also in the amoeba for the modification of U34 in tRNAs. In brief, similar to a recent study (32), two cysteines of the cysteine tetrad in the catalytic core of the radical SAM domain were changed to serines (GFP-mutSAM-Elp3), and in the HAT domain, two glycines in the catalytic core were replaced by phenylalanines (GFP-mutHATElp3; Figure 5C). Whether the different GFP-Elp3 variants can rescue the thiolation phenotype of the elp3− strain was investigated by APM Northern blotting. For this, we employed RNA from elp3− strain expressing Elp3-GFP variants, and for comparison from untransformed elp3− and the Ax2 wild type strains. As seen before (Figure 5A), the upper signal corresponds to the thiolated fraction of the investigated tRNAs and the lower to the unthiolated fraction (Figure 5D). A strong thiolation signal is detectable in the Ax2 strain, while the elp3− strain displays only a faint signal. The tRNAs from the point mutants display a low thiolation signal similar to the elp3− strain, indicating that both mutations result in proteins that cannot rescue the phenotype to the wild type situation. The ectopically overexpressed N- and C-terminal GFP Elp3 fusion proteins, however, are capable of rescuing this phenotype, resulting in the wild type situation. This was observed for the three tRNAs tKUUU, tEUUC and tQUUG, which undergo the complete Elongator-dependent modification pathway (Figure 1A), but not for the non-thiolated tGGGC that served as negative control. Taken together, this indicates that the GFP Elp3 fusion proteins are functional and that both catalytic domains of the D. discoideum Elp3 appear to be indispensable for the Elongator-dependent U34 modifications.
An unconditional knockout of tQCUG
The extreme codon usage for glutamine in D. discoideum (Figure 2) raised the question whether or not the single tQCUG gene might be dispensable. The prokaryotic tQUUG is able to read the CAG codon, while the eukaryotic counterpart may not be able to do so efficiently (64). Attempts to remove the single copy tQCUG gene in S. cerevisiae were only successful, when concomitantly the isoacceptor tQUUG was ectopically expressed. This, however, required a functional Elongator-dependent modification pathway (43). To address if the single tQCUG gene is also essential in D. discoideum, a knockout construct targeting the genomic surrounding of the tQCUG gene (Figure 6A) was generated and used for transformation of the Ax2 wild type and elp3− strains. For Ax2 wild type, PCR on gDNA from a transformed and an untransformed strain revealed two distinct PCR amplicons (Figure 6B) with sizes corresponding to those expected for wild type (∼650 bp) and a tQCUG knockout (1850 bp). This indicated the successful deletion of the targeted genomic area in the Ax2 background. A Northern blot against tQCUG confirmed the successful deletion of the tQCUG gene in the Ax2 background (Figure 6C), despite a weak signal that presumably can be attributed to cross-hybridization of the tQCUG probe to tQUUG. Contrary to this, attempts to delete the tQCUG gene in the elp3− background failed repeatedly. Since a viable, tQCUG-deficient strain was obtained in the Ax2 wild type background, it appears likely that tQUUG is able to read the CAG codon to some extent. For this, however, the functionality of the Elongator-dependent modification pathway appears indispensable, which would explain why a tQCUG-deficient strain could not be generated in the elp3− background.
Figure 6.
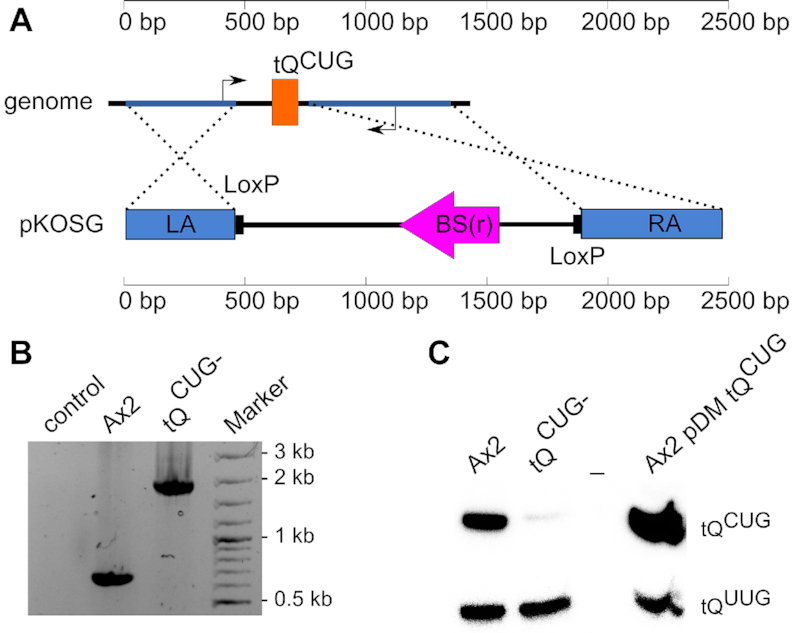
A viable eukaryote without a tQCUG gene. (A) Deletion of the single tQCUG gene in D. discoideum. Shown is on top a sketch of the tQCUG gene (orange box) in its genomic context. Left (LA) and right arm (RA) for homologous recombination (dotted lines) are indicated in blue. They are contained in the pKOSG construct, where they surround a blasticidin S resistance (BS(r)) cassette (magenta) next to LoxP sites. The thin black arrows represent the binding sites for PCR primers. In the Ax2 wild type strain, PCR results in an amplicon of 650 bp and upon homologous recombination of the pKOSG construct, an amplicon of 1850 bp is expected. Except for primer binding sites, the figure is drawn to scale. (B) PCR on genomic DNA of the tQCUG− and the Ax2 wild type strains. The size difference corresponds to expectation and indicates pKOSG integration. Marker = GeneRuler 100 bp DNA Ladder Plus. PCR products were separated electrophoretically on agarose gels and visualized by Ethidium bromide staining. (C) Expression analysis of tQCUG. Shown is a Northern blot, in which 5 μg total RNA from the Ax2 wild type, the tQCUG− strain and the tQCUG overexpressor (in the Ax2 background) were probed against tQCUG. The signal for tQUUG served as loading and transfer control.
To study the effect on translation of GFP reporter constructs (see below), we also addressed, if functional tRNA genes can be overexpressed in D. discoideum. For this, we amplified tRNA genes ± about 250 bp genomic surrounding sequences that were cloned into a high (40-60) copy number pDM-Vector backbone (58). Northern blot analysis of an Ax2 wild type transformed with that construct showed a stronger tQCUG expression compared to parental strain, while the signal of tQUUG remained unchanged (Figure 6C). As the probe thus is specific, this indicates that the tRNA gene is transcribed from the pDM plasmid, and that the transcript is processed into a product of the size of the mature tQCUG.
The effect of the loss of anticodon U34 modification on glutamine codon translation
D. discoideum possesses the most prion-like proteome known to date (59,65), as a significant portion of proteins feature long polyQ stretches within their amino acid sequences. In their translation, D. discoideum genes use almost exclusively one glutamine codon, which leads to an overall glutamine codon usage of 96% CAA to 4% CAG (Figure 2). In an attempt to investigate if there are expression differences between the generated mutant strains and the Ax2 wild type, we made use of a GFP construct with the extended human huntingtin exon 1 that features 103 consecutive Qs in a 1:1 ratio of CAA:CAG as leader sequence (construct kindly provided by Dr Liliana Malinovska (59)). As a control, we generated a synthetic construct with 15 Qs, using only the CAG codon, upstream of the GFP sequence. The constructs were cloned under the control of an actin15 promoter, and tRNA overexpression in leader construct strains was achieved by cloning the tRNA genes into the same plasmid (Supplementary Figure S7). Upon transformation in the generated mutant strains, GFP expression was measured via flow cytometry, and Western blotting using an antibody against GFP.
Q103-GFP expression in the elp3− background was significantly reduced compared to the Ax2 wild type, and this did not change by the concomitant overexpression of tQCUG, while the overexpression of tQUUG reverted GFP expression to wild type levels (Figure 7A). A minor reduction in GFP expression was seen in the trm9− background, and this remained unchanged by the presence of either tRNA (Figure 7B). No significant change in Q103-GFP fluorescence was observed in the ctu1− strain in either set-up (Figure 7C). The elp3−/ctu1− strain displayed for all three conditions fluorescence signals similar to that observed in the elp3− background (Figure 7A), however, with a narrower fluorescence peak width compared to all other mutants (Figure 7D). Finally, the strongest reduction of Q103-GFP expression compared to the wild type was observed in the tQCUG− strain (Figure 7E). The fluorescence signal is, however, still several fold above the signal of the negative control, supporting the notion that tQUUG can decode the CAG codon, but does so at a reduced efficiency as compared to tQCUG. In the tQCUG− strain, the concomitant overexpression of tQCUG increased the Q103-GFP expression to ∼70% of the wild type level. The differences in Q103-GFP expression in the different strains were independently confirmed by Western blotting (Figure 7F).
Figure 7.
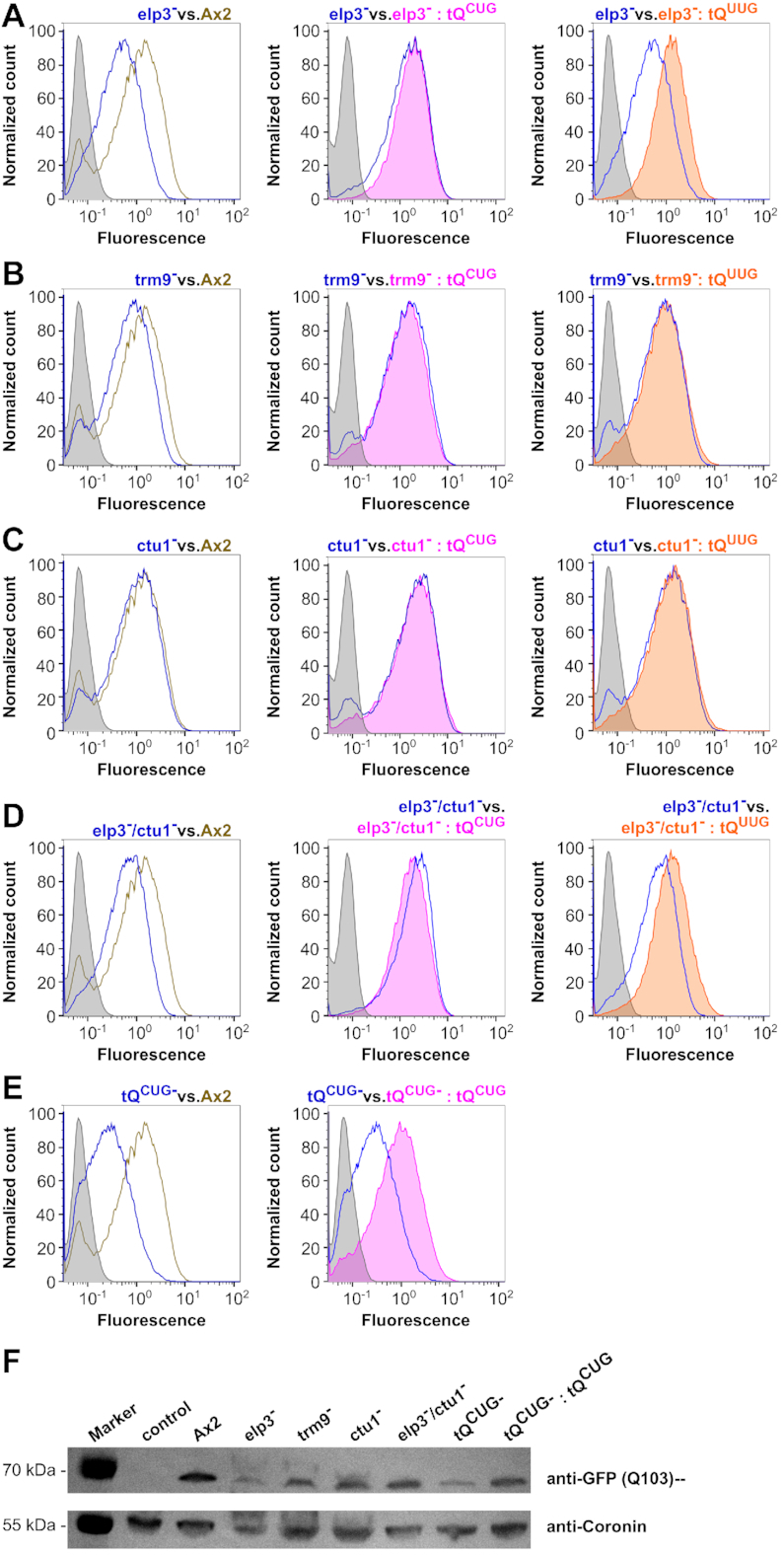
Expression of GFP with the Q103 leader sequence. Representative flow cytometry detected fluorescence profiles of Q103-GFP in Ax2 wild type and elp3− (A), trm9− (B), ctu1− (C), elp3−/ctu1− (D) and tQCUG− (E) mutant strains. Shown are expression profiles from the mutant strain (blue) versus Ax2 (olive, left), versus mutant strain +tQCUG overexpression (magenta, middle) and versus mutant strain +tQUUG overexpression (orange, right). Gray areas indicate the background signal from untransformed strains. Fluorescence is shown in arbitrary units. (F) Western blots against GFP with the Q103-leader expressed in the indicated mutant and recue strains, compared to Ax2 wild type. Coronin antibodies served as loading and transfer controls. Control: untransformed Ax2 wild type; M: PageRuler Plus Prestained Protein Ladder as size marker with relevant sizes shown to the left.
To address how efficiently tQUUG can decode CAG, we challenged the system by performing expression analyses on the mutant strains, which we transformed with the Q15 (CAG only)-GFP fusion construct, representing a highly unnatural situation for D. discoideum (Figure 2). A small reduction in GFP expression was seen for the Q15-leader construct in the elp3− background, but not for the ctu− and trm9− strains, when compared to the Ax2 wild type (Figure 8A). In the tQCUG− strain, however, GFP expression was greatly reduced with almost no Q15-GFP signal above the negative control. This was confirmed independently in a western blot (Figure 8B). This indicates that if challenged by a for the amoeba unnatural stretch of 15 CAGs, the decoding of these triplets by tQUUG reaches its limits. Taken together, the data gained with the artificial constructs (Figures 7 and 8) support the notion that the mcm5s2U modification has a positive effect on the translation of glutamine codons and further, that tQUUG is able to read the CAG codon, albeit with a significantly reduced efficiency if challenged by an unnaturally high sequence of CAG triplets in the model system.
Figure 8.
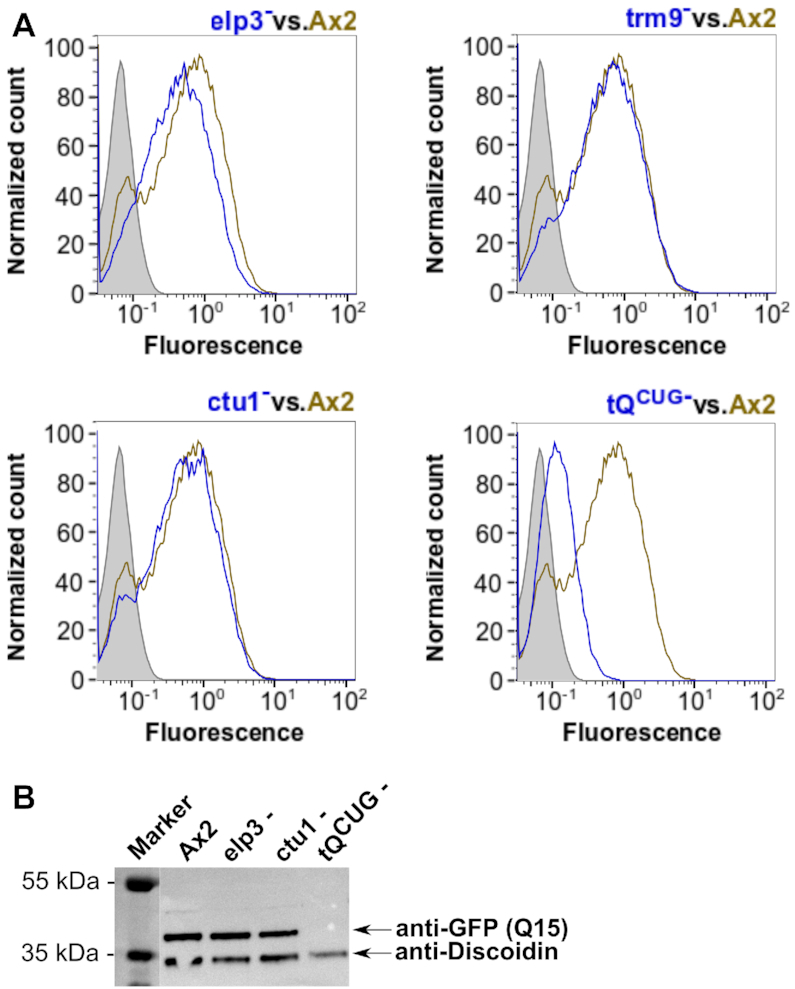
Expression of GFP with the Q15 leader sequence. (A) Representative flow cytometry detected fluorescence profiles of Q15-GFP in Ax2 wild type compared to elp3−, trm9−, ctu1− and tQCUG− mutant strains. Shown are expression profiles from the mutant strain (blue) vs. Ax2 (olive). Grey areas indicate the background signal from untransformed strains. Fluorescence is shown in arbitrary units. (B) Western blots against GFP with the Q15-leader expressed in the indicated mutant strains, compared to Ax2 wild type. Discoidin antibodies served as loading and transfer controls. M: PageRuler Plus Prestained Protein Ladder as size marker with relevant sizes shown to the left.
DISCUSSION
There are over 100 different modifications known in mature tRNA molecules, which makes it arguably the most heavily modified class of RNA molecules known (2). On average, 17% of tRNA residues are modified (3). The modification of wobble uridines at position 34 (U34) in tRNAs has been established for plants and various organisms of the evolutionary supergroup of the Opisthokonta, encompassing as variable organisms as yeasts, mammals or nematodes (3). We have shown here that the modifications that are introduced by the Elongator-dependent pathway (Figure 1A) are largely conserved also in D. discoideum, the model organism of the evolutionary supergroup of Amoebozoa (46). Deleting the genes (Supplementary Figures S1-S3) that encode the most conserved proteins (Figure 3) of every step in the pathway was possible in the amoeba, and also a double mutant of the elp3 and ctu1 genes was achieved in D. discoideum, unlike in budding yeast (35,66). The obtained deletion strains allowed us to analyse the chemical modifications that are present at wobble uridines in the amoeba (Figure 4 and Supplementary Figures S4–S6, Tables S3–S4). These analyses revealed not only the general conservation of the pathway (Figure 1A), but also a certain plasticity in the sequence, in which the modifications are introduced. For example, modifications like s2U, cm5U and ncm5s2U, which are not part of the linear pathway (Figure 1A) were observed to accumulate in the generated mutant strains, albeit at low levels (Figure 4B). That is unlike the situation seen in fission yeast (11), and it indicates that the thiolating activity of the Ctu1/Ctu2 dimer in D. discoideum does not pre-require the presence of the cm5 modification by the Elongator complex, and that also ncm5-modified and unmodified uridines can be thiolated. Furthermore, these results support and expand previous analyses from budding yeast by Byström et al. (4), who discussed alternative models for the modification pathway shown in Figure 1A. Overall, the chemical U34 modification profiles present in the different deletion strains are perfectly matched by our data generated from APM Northern blots (Figure 5). They confirm the tRNA-specific thiolation, and furthermore show that expression of Elp3 GFP fusion proteins in D. discoideum allows to complement the molecular phenotype of the elp3 gene deletion strain.
In yeast, the modifications introduced by the Elongator-dependent modification pathway (Figure 1A) actually promote a functionally redundant decoding system (43). This was concluded from the observation that the single copy gene tQCUG, which is essential in the wild type, can be deleted if the isoacceptor tQUUG is overexpressed. This conditional knockout, however, could not be obtained in strains with a distorted Elongator-dependent modification pathway (Figure 1A). In stark contrast, the single copy gene of tQCUG can be deleted in D. discoideum (Figure 6) without further genetic manipulations. To our knowledge, this makes D. discoideum the only (first?) eukaryotic organism that is unconditionally viable without tQCUG. Presumably, this is only possible due to the highly unusual codon usage for glutamine in the amoeba, where <4% are encoded by the CAG triplet, compared to ∼30% in S. cerevisiae and S. pombe (Figure 2B). Our attempts to knockout the tQCUG gene in the elp3– background, however, failed repeatedly. While we cannot formally rule out alternative explanations and given the use of the same construct for gene deletion in both strains, we take this observation as an indication to surmise that the mcm5s2U modification, which is missing in the elp3– background, might be essential for tQUUG to read over the CAG codon. Thus, we consider the tQCUG gene in D. discoideum essential for viability upon loss of the mcm5s2U modification.
Both, reduced protein amounts and protein misfolding have been suggested to be potential consequences of missing wobble uridine modifications in yeast and other organisms (33). In this context, it is worth noting that the proteome of D. discoideum was shown recently to harbor highly abundant low complexity stretches with extended poly glutamine (polyQ) and poly asparagine (polyN) stretches, which in wild type cells do not lead to protein aggregation (59,65), unlike observed for yeast and other organisms (67,68). In the amoeba, this resilience against protein aggregation can be accounted for by the high activity of chaperones that apparently counteracts amyloid formation (59,65,69). Therefore, we have addressed whether protein misfolding and/or distorted protein expression can be observed in the mutant strains of the Elongator-dependent modification pathway. For this we employed glutamine leader constructs with varying Q codon composition that were fused to GFP. While fluorescence microscopy showed no obvious formation of protein aggregates in either of the D. discoideum strains, Western blotting and flow cytometry showed a significant reduction of protein amounts (Figures 7 and 8). For the leader construct featuring a 1:1 ratio of CAA and CAG this was seen particularly in the elp3– strain and to a lesser extent in the trm9– mutant, but not in the ctu1- strain (Figure 7). This situation is distinct from that in yeast, where lack of s2U thiolation leads to a similar translation phenotype as in Elongator mutants (41). Most dramatic, however, was the reduction in GFP signal for this construct in the strain lacking tQCUG as observed by either technique (Figure 7E, F). The concomitant overexpression of tQUUG in the elp3– strain or of tQCUG in the tQCUG– strain increased the GFP expression strongly, although not fully to that seen in the Ax2 wild type (Figure 7), possibly due to an incomplete chemical modification of the overexpressed tRNA. With the GFP fusion featuring only CAG codons in the leader sequence, in strains featuring the cognate tQCUG only a minor reduction in GFP amounts was observed for the elp3– strain compared to the wild type, but not for the other protein gene deletion strains (Figure 8). Here, however, the use of the construct in the tQCUG– strain resulted, not unexpectedly, in barely any GFP signal. Taken together, these data indicate that lack of the modifications introduced by the Elongator-dependent pathway has predominantly an effect on the protein amounts in D. discoideum. Furthermore, the introduced modifications foster glutamine codon translation, in which tQUUG is able to read the CAG codon, albeit with lower efficiency. Whether this is the case in the presence of tQCUG, cannot be answered with certainty, however, the small reduction of the GFP signal with the CAG only leader observed in the elp3- strain (Figure 8) might point in this direction. Taken together, it stands to surmise that only the low CAG codon usage in D. discoideum (Figure 2) enables life without tQCUG.
Our analysis of the conservation of proteins acting in the Elongator-dependent modification pathway (Figue 1A) revealed a significant variability (Figure 4). By far most conserved within the Elongator complex is Elp3, which harbours its catalytic activity. Not only is the protein sequence highly conserved amongst the investigated species, the obtained phylogenetic tree of Elp3 mirrors also the general evolutionary classification of the diversion of D. discoideum within the eukaryotic branch of the tree of life. In this, the Amoebozoa branched off after the split between plants and Opisthokonta, but before the latter supergroup separated into the yeasts and metazoa (46). As this scenario is seen for Elp3, it is tempting to speculate that the encoding gene was already present at least in the last common ancestor of the investigated species, encompassing organisms of the three evolutionary supergroups Amoebozoa, Archaeplastida and Opisthokonta. In line with this interpretation is the stand-alone activity of Elp3 in archaeal organisms that do not appear to feature either of the other five elongator components (23,70). Albeit indirectly, this scenario is further supported by a comparative genomic analysis that revealed the presence of genes for RNA modifying enzymes already in the last universal common ancestor (LUCA) and thus before the split in the three kingdoms of life (71). Thus, Elp3, and also Trm112 and Ctu2 follow the position of D. discoideum in the evolutionary tree, but the other five components of the Elongator complex do not, and nor does Trm9. This might point towards an early evolutionary situation, in which Elp3, Trm112 and Ctu2 were already present before the aforementioned supergroups have split up. This situation represents the last eukaryotic common ancestor (LECA) (46) and we thus speculate that LECA might already have featured this core set of enzymes required for the modification of wobble uridines.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sina Gronemann and Dr Balachandar A.V. Ammapatti for their initial work on the Dictyostelium Elongator complex. We are most grateful for the generous support by Dr L. Malinovska who provided the plasmid pDM353 Q103-GFP, by Dr Markus Maniak who provided antibodies and by Dr Peter Sarin who provided APM. Also, Dr Mark Helm is acknowledged for his support in the initial phase of the project and the kind gift of synthetic nucleosides. S.K. is grateful to Dr Thomas Carell and his lab for advice and instrument time (LC–UV–MS/MS). We furthermore thank our colleague Dr Monica Hagedorn for discussion and helpful comments on the manuscript.
Notes
Present address: Kim Philipp Jablonski, Department of Biosystems Science and Engineering, ETH Zürich, Basel, Switzerland.
Contributor Information
Manfred A Schäck, Ribogenetics Biochemistry Lab, Department of Life Sciences and Chemistry, Jacobs University Bremen gGmbH, DE 28759 Bremen, Germany.
Kim Philipp Jablonski, Ribogenetics Biochemistry Lab, Department of Life Sciences and Chemistry, Jacobs University Bremen gGmbH, DE 28759 Bremen, Germany.
Stefan Gräf, Department of Medicine, University of Cambridge, Cambridge Biomedical Campus, Cambridge CB2 0QQ, UK.
Roland Klassen, Institut für Biologie, Fachgebiet Mikrobiologie, Universität Kassel, Heinrich-Plett-Str. 40, 34132 Kassel, Germany.
Raffael Schaffrath, Institut für Biologie, Fachgebiet Mikrobiologie, Universität Kassel, Heinrich-Plett-Str. 40, 34132 Kassel, Germany.
Stefanie Kellner, Department of Chemistry and Pharmacy, Ludwig-Maximilians University Munich, Butenandtstr. 5-13, 81377 Munich, Germany.
Christian Hammann, Ribogenetics Biochemistry Lab, Department of Life Sciences and Chemistry, Jacobs University Bremen gGmbH, DE 28759 Bremen, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
DFG Priority Program 1784 Chemical Biology of Native Nucleic Acid Modifications [HA3459/17 to C.H., SCHA750/20 to R.S., KL2937/1 to R.K., KE1943/4 to S.K.]. Funding for open access charge: DFG [HA3459/17].
Conflict of interest statement. None declared.
REFERENCES
- 1. Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crecy-Lagard V., Ross R., Limbach P.A., Kotter A. et al.. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018; 46:D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. El Yacoubi B., Bailly M., de Crecy-Lagard V.. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012; 46:69–95. [DOI] [PubMed] [Google Scholar]
- 3. Helm M., Alfonzo J.D.. Posttranscriptional RNA modifications: playing metabolic games in a cell's chemical Legoland. Chem. Biol. 2014; 21:174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen C., Huang B., Anderson J.T., Bystrom A.S.. Unexpected accumulation of ncm(5)U and ncm(5)S(2) (U) in a trm9 mutant suggests an additional step in the synthesis of mcm(5)U and mcm(5)S(2)U. PLoS One. 2011; 6:e20783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang B., Johansson M.J., Bystrom A.S.. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005; 11:424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen C., Tuck S., Bystrom A.S.. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLos Genet. 2009; 5:e1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin F.J., Shen L., Jang C.W., Falnes P.O., Zhang Y.. Ikbkap/Elp1 deficiency causes male infertility by disrupting meiotic progression. PLos Genet. 2013; 9:e1003516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mehlgarten C., Jablonowski D., Wrackmeyer U., Tschitschmann S., Sondermann D., Jager G., Gong Z., Bystrom A.S., Schaffrath R., Breunig K.D.. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol. Microbiol. 2010; 76:1082–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jablonowski D., Zink S., Mehlgarten C., Daum G., Schaffrath R.. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator's key role for zymocin-induced cell death in yeast. Mol. Microbiol. 2006; 59:677–688. [DOI] [PubMed] [Google Scholar]
- 10. Lu J., Huang B., Esberg A., Johansson M.J., Bystrom A.S.. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA. 2005; 11:1648–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dewez M., Bauer F., Dieu M., Raes M., Vandenhaute J., Hermand D.. The conserved Wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc. Natl Acad. Sci. U.S.A. 2008; 105:5459–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krogan N.J., Greenblatt J.F.. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001; 21:8203–8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Y., Takagi Y., Jiang Y., Tokunaga M., Erdjument-Bromage H., Tempst P., Kornberg R.D.. A multiprotein complex that interacts with RNA polymerase II elongator. J. Biol. Chem. 2001; 276:29628–29631. [DOI] [PubMed] [Google Scholar]
- 14. Otero G., Fellows J., Li Y., de Bizemont T., Dirac A.M., Gustafsson C.M., Erdjument-Bromage H., Tempst P., Svejstrup J.Q.. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell. 1999; 3:109–118. [DOI] [PubMed] [Google Scholar]
- 15. Winkler G.S., Petrakis T.G., Ethelberg S., Tokunaga M., Erdjument-Bromage H., Tempst P., Svejstrup J.Q.. RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J. Biol. Chem. 2001; 276:32743–32749. [DOI] [PubMed] [Google Scholar]
- 16. Glatt S., Letoquart J., Faux C., Taylor N.M., Seraphin B., Muller C.W.. The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat. Struct. Mol. Biol. 2012; 19:314–320. [DOI] [PubMed] [Google Scholar]
- 17. Lin Z., Zhao W., Diao W., Xie X., Wang Z., Zhang J., Shen Y., Long J.. Crystal structure of elongator subcomplex Elp4-6. J. Biol. Chem. 2012; 287:21501–21508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dauden M.I., Kosinski J., Kolaj-Robin O., Desfosses A., Ori A., Faux C., Hoffmann N.A., Onuma O.F., Breunig K.D., Beck M. et al.. Architecture of the yeast Elongator complex. EMBO Rep. 2017; 18:264–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dauden M.I., Jaciuk M., Muller C.W., Glatt S.. Structural asymmetry in the eukaryotic Elongator complex. FEBS Lett. 2017; 592:502–515. [DOI] [PubMed] [Google Scholar]
- 20. Setiaputra D.T., Cheng D.T., Lu S., Hansen J.M., Dalwadi U., Lam C.H., To J.L., Dong M.Q., Yip C.K.. Molecular architecture of the yeast Elongator complex reveals an unexpected asymmetric subunit arrangement. EMBO Rep. 2017; 18:280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Glatt S., Zabel R., Kolaj-Robin O., Onuma O.F., Baudin F., Graziadei A., Taverniti V., Lin T.Y., Baymann F., Seraphin B. et al.. Structural basis for tRNA modification by Elp3 from Dehalococcoides mccartyi. Nat. Struct. Mol. Biol. 2016; 23:794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wittschieben B.O., Otero G., de Bizemont T., Fellows J., Erdjument-Bromage H., Ohba R., Li Y., Allis C.D., Tempst P., Svejstrup J.Q.. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell. 1999; 4:123–128. [DOI] [PubMed] [Google Scholar]
- 23. Paraskevopoulou C., Fairhurst S.A., Lowe D.J., Brick P., Onesti S.. The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol. Microbiol. 2006; 59:795–806. [DOI] [PubMed] [Google Scholar]
- 24. Li Q., Fazly A.M., Zhou H., Huang S., Zhang Z., Stillman B.. The elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. PLoS Genet. 2009; 5:e1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rahl P.B., Chen C.Z., Collins R.N.. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol. Cell. 2005; 17:841–853. [DOI] [PubMed] [Google Scholar]
- 26. Winkler G.S., Kristjuhan A., Erdjument-Bromage H., Tempst P., Svejstrup J.Q.. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl Acad. Sci. U.S.A. 2002; 99:3517–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Creppe C., Malinouskaya L., Volvert M.L., Gillard M., Close P., Malaise O., Laguesse S., Cornez I., Rahmouni S., Ormenese S. et al.. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 2009; 136:551–564. [DOI] [PubMed] [Google Scholar]
- 28. Solinger J.A., Paolinelli R., Kloss H., Scorza F.B., Marchesi S., Sauder U., Mitsushima D., Capuani F., Sturzenbaum S.R., Cassata G.. The Caenorhabditis elegans Elongator complex regulates neuronal alpha-tubulin acetylation. PLos Genet. 2010; 6:e1000820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miskiewicz K., Jose L.E., Bento-Abreu A., Fislage M., Taes I., Kasprowicz J., Swerts J., Sigrist S., Versees W., Robberecht W. et al.. ELP3 controls active zone morphology by acetylating the ELKS family member Bruchpilot. Neuron. 2011; 72:776–788. [DOI] [PubMed] [Google Scholar]
- 30. Nelissen H., Fleury D., Bruno L., Robles P., De Veylder L., Traas J., Micol J.L., Van Montagu M., Inze D., Van Lijsebettens M.. The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc. Natl Acad. Sci. U.S.A. 2005; 102:7754–7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen Y.T., Hims M.M., Shetty R.S., Mull J., Liu L., Leyne M., Slaugenhaupt S.A.. Loss of mouse Ikbkap, a subunit of elongator, leads to transcriptional deficits and embryonic lethality that can be rescued by human IKBKAP. Mol. Cell. Biol. 2009; 29:736–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Okada Y., Yamagata K., Hong K., Wakayama T., Zhang Y.. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010; 463:554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schaffrath R., Leidel S.A.. Wobble uridine modifications-a reason to live, a reason to die?. RNA Biol. 2017; 14:1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bauer F., Matsuyama A., Candiracci J., Dieu M., Scheliga J., Wolf D.A., Yoshida M., Hermand D.. Translational control of cell division by Elongator. Cell Rep. 2012; 1:424–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bjork G.R., Huang B., Persson O.P., Bystrom A.S.. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA. 2007; 13:1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Esberg A., Huang B., Johansson M.J., Bystrom A.S.. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell. 2006; 24:139–148. [DOI] [PubMed] [Google Scholar]
- 37. Leidel S., Pedrioli P.G., Bucher T., Brost R., Costanzo M., Schmidt A., Aebersold R., Boone C., Hofmann K., Peter M.. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009; 458:228–232. [DOI] [PubMed] [Google Scholar]
- 38. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zinshteyn B., Gilbert W.V.. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 2013; 9:e1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chou H.J., Donnard E., Gustafsson H.T., Garber M., Rando O.J.. Transcriptome-wide analysis of roles for tRNA modifications in translational regulation. Mol. Cell. 2017; 68:978–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nedialkova D.D., Leidel S.A.. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015; 161:1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rozov A., Demeshkina N., Khusainov I., Westhof E., Yusupov M., Yusupova G.. Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat. Commun. 2016; 7:10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Johansson M.J., Esberg A., Huang B., Bjork G.R., Bystrom A.S.. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell. Biol. 2008; 28:3301–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tran T.T., Belahbib H., Bonnefoy V., Talla E.. A comprehensive tRNA genomic survey unravels the evolutionary history of tRNA arrays in prokaryotes. Genome Biol Evol. 2015; 8:282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eichinger L., Pachebat J.A., Glöckner G., Rajandream M.A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q. et al.. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005; 435:43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Adl S.M., Simpson A.G., Lane C.E., Lukes J., Bass D., Bowser S.S., Brown M.W., Burki F., Dunthorn M., Hampl V. et al.. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012; 59:429–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bozzaro S. The model organism Dictyostelium discoideum. Methods Mol. Biol. 2013; 983:17–37. [DOI] [PubMed] [Google Scholar]
- 48. Fey P., Kowal A.S., Gaudet P., Pilcher K.E., Chisholm R.L.. Protocols for growth and development of Dictyostelium discoideum. Nat. Protoc. 2007; 2:1307–1316. [DOI] [PubMed] [Google Scholar]
- 49. UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D. et al.. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019; 47:W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yu G., Lam T.T., Zhu H., Guan Y.. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol. Biol. Evol. 2018; 35:3041–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gaudet P., Pilcher K.E., Fey P., Chisholm R.L.. Transformation of Dictyostelium discoideum with plasmid DNA. Nat. Protoc. 2007; 2:1317–1324. [DOI] [PubMed] [Google Scholar]
- 53. Wiegand S., Kruse J., Gronemann S., Hammann C.. Efficient generation of gene knockout plasmids for Dictyostelium discoideum using one-step cloning. Genomics. 2011; 97:321–325. [DOI] [PubMed] [Google Scholar]
- 54. Faix J., Kreppel L., Shaulsky G., Schleicher M., Kimmel A.R.. A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucleic Acids Res. 2004; 32:e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chionh Y.H., Ho C.H., Pruksakorn D., Ramesh Babu I., Ng C.S., Hia F., McBee M.E., Su D., Pang Y.L., Gu C. et al.. A multidimensional platform for the purification of non-coding RNA species. Nucleic Acids Res. 2013; 41:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cai W.M., Chionh Y.H., Hia F., Gu C., Kellner S., McBee M.E., Ng C.S., Pang Y.L., Prestwich E.G., Lim K.S. et al.. A platform for discovery and quantification of modified Ribonucleosides in RNA: application to stress-induced reprogramming of tRNA modifications. Methods Enzymol. 2015; 560:29–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Borland K., Diesend J., Ito-Kureha T., Heissmeyer V., Hammann C., Buck A.H., Michalakis S., Kellner S.. Production and application of stable Isotope-Labeled internal standards for RNA modification analysis. Genes (Basel). 2019; 10:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Veltman D.M., Akar G., Bosgraaf L., Van Haastert P.J.. A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid. 2009; 61:110–118. [DOI] [PubMed] [Google Scholar]
- 59. Malinovska L., Palm S., Gibson K., Verbavatz J.M., Alberti S.. Dictyostelium discoideum has a highly Q/N-rich proteome and shows an unusual resilience to protein aggregation. Proc. Natl Acad. Sci. U.S.A. 2015; 112:E2620–E2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Boesler B., Meier D., Förstner K.U., Friedrich M., Hammann C., Sharma C.M., Nellen W.. Argonaute proteins affect siRNA levels and accumulation of a novel extrachromosomal DNA from the Dictyostelium retrotransposon DIRS-1. J. Biol. Chem. 2014; 289:35124–35138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Igloi G.L. Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry. 1988; 27:3842–3849. [DOI] [PubMed] [Google Scholar]
- 62. Lemau de Talance V., Bauer F., Hermand D., Vincent S.P.. A simple synthesis of APM ([p-(N-acrylamino)-phenyl]mercuric chloride), a useful tool for the analysis of thiolated biomolecules. Bioorg. Med. Chem. Lett. 2011; 21:7265–7267. [DOI] [PubMed] [Google Scholar]
- 63. Chen C., Huang B., Eliasson M., Ryden P., Bystrom A.S.. Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet. 2011; 7:e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bloom-Ackermann Z., Navon S., Gingold H., Towers R., Pilpel Y., Dahan O.. A comprehensive tRNA deletion library unravels the genetic architecture of the tRNA pool. PLoS Genet. 2014; 10:e1004084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Santarriaga S., Petersen A., Ndukwe K., Brandt A., Gerges N., Bruns Scaglione J., Scaglione K.M.. The social amoeba Dictyostelium discoideum is highly resistant to Polyglutamine aggregation. J. Biol. Chem. 2015; 290:25571–25578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xu F., Bystrom A.S., Johansson M.J.O.. SSD1 suppresses phenotypes induced by the lack of Elongator-dependent tRNA modifications. PLos Genet. 2019; 15:e1008117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Balch W.E., Morimoto R.I., Dillin A., Kelly J.W.. Adapting proteostasis for disease intervention. Science. 2008; 319:916–919. [DOI] [PubMed] [Google Scholar]
- 68. Halfmann R., Alberti S., Lindquist S.. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 2010; 20:125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Santarriaga S., Haver H.N., Kanack A.J., Fikejs A.S., Sison S.L., Egner J.M., Bostrom J.R., Seminary E.R., Hill R.B., Link B.A. et al.. SRCP1 conveys resistance to polyglutamine aggregation. Mol. Cell. 2018; 71:216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Selvadurai K., Wang P., Seimetz J., Huang R.H.. Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat. Chem. Biol. 2014; 10:810–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Anantharaman V., Koonin E.V., Aravind L.. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002; 30:1427–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Westhof E., Yusupov M., Yusupova G.. The multiple flavors of GoU pairs in RNA. J. Mol. Recognit. 2019; 32:e2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.