Abstract
In synthetic circuits, CRISPR-Cas systems have been used effectively for endpoint changes from an initial state to a final state, such as in logic gates. Here, we use deactivated Cas9 (dCas9) and deactivated Cas12a (dCas12a) to construct dynamic RNA ring oscillators that cycle continuously between states over time in bacterial cells. While our dCas9 circuits using 103-nt guide RNAs showed irregular fluctuations with a wide distribution of peak-to-peak period lengths averaging approximately nine generations, a dCas12a oscillator design with 40-nt CRISPR RNAs performed much better, having a strongly repressed off-state, distinct autocorrelation function peaks, and an average peak-to-peak period length of ∼7.5 generations. Along with free-running oscillator circuits, we measure repression response times in open-loop systems with inducible RNA steps to compare with oscillator period times. We track thousands of cells for 24+ h at the single-cell level using a microfluidic device. In creating a circuit with nearly translationally independent behavior, as the RNAs control each others’ transcription, we present the possibility for a synthetic oscillator generalizable across many organisms and readily linkable for transcriptional control.
INTRODUCTION
Oscillators are essential in nature—circadian rhythm, cell cycles and division, heartbeats, breathing. Human-made oscillators also have a history of great importance, from pendulums in clocks, to crystal oscillators in electronics, to pacemaker and ventilator biomedical devices that interface with the human body. Developing a new biological oscillator may be powerful in many applications, as well as for learning fundamental principles to design synthetic biological circuits with controllable, predictable performance. For example, a synthetic oscillator could be coupled to added functions in enhanced gut bacteria, such as small-molecule drug production (1), lysis for cargo delivery (2), or timekeeping (3). Protein-based synthetic oscillators have been powerful demonstrations (4–9), but building oscillators using nucleic acids has been far less explored (10). Here, we use CRISPR-Cas systems to develop new circuits whose performance does not depend on activity of varying translated proteins but rather on a varying transcriptional pool of RNAs.
CRISPR-Cas technologies have been used extensively for genome editing and manipulation (11). For CRISPR-Cas9, the 160 kD Cas9 enzyme forms a complex with a targeting guide RNA, able to bind to specific regions of DNA with base complementarity and cleave. While natural systems typically use two RNAs—a DNA-targeting guide strand and a Cas9 handle strand—these can be combined into a single guide RNA (sgRNA) with both functions. Deactivated Cas9 (dCas9) has the enzymatic active site for DNA cleavage mutated, allowing for gene repression instead of cutting (12–14). Along with using this nuclease-deficient form as a standalone transcriptional repressor, dCas9 has also been fused to transcription factor domains (e.g. VP64 or KRAB) to allow for gene activation or further repression (15). This strategy of dCas9-controlled transcriptional repression through specific sgRNAs has been used to create logic gates, memory data storage and other synthetic genetic circuits (16–21). In these cases, the desired readout was an endpoint, a changed expression level after several hours to a final state.
Here, we build on those endpoint circuits to create free-running ring oscillators, where sgRNAs repress each other's transcription through dCas9 binding of promoters driving sgRNA production. Similarly, we use a deactivated Cas12a (dCas12a) protein along with its targeting CRISPR RNAs in analogous oscillator designs. CRISPR-Cas12a is another bacterial defense system, with the added enzymatic ability of Cas12a to cleave a CRISPR RNA array in addition to its DNA targeting ability (22). Circuits based on endpoint behavior, such as sensing and logic gates, have also been made with dCas12a (23–25).
We use a microfluidics device that allows individual tracking of thousands of cells over many generations: Escherichia coli cells are confined in trenches with media flowed through, for diffusive feeding that supports exponential growth and washout of newly divided cells (26). In addition to oscillators, we make single-cell measurements of dCas9–sgRNA binding repression times, which have mostly been measured at a population level (12,27–29). We compare these response times in inducible RNA cascades with the observed period lengths of the oscillator designs.
Our free-running dCas12a RNA oscillator fluctuates far more regularly than our dCas9-based designs and shows oscillatory behavior, with significant autocorrelation function (ACF) peaks for the population average. Along with another recent effort building dynamic CRISPR-based circuits (30), our circuits represent foundational work toward development of an even more regular oscillator using CRISPR-Cas parts and has the potential to be generalizable in a variety of host organisms across kingdoms.
MATERIALS AND METHODS
Plasmid and strain construction
CRISPR DNA fragments were polymerase chain reaction (PCR) amplified from existing plasmids: dCas9 system from pdCas9-bacteria, pAN-AND and pAN-OR (Addgene # 62308, 62307, 44249) (12,16). For dCas12a, Cas12a pieces were PCR amplified with added D917A mutation from pY001 (pFnCpf1_full; Addgene #69973) (22). Other DNA pieces not obtained by PCR were synthesized as IDT gBlock gene fragments (Integrated DNA Technologies). Plasmids were made by Gibson assembly (31) or restriction enzyme digestion and ligation with T4 ligase in E. coli DH5a. Either a high (ColE1), medium (p15A) or low (SC101) copy number origin of replication was used. Genomic integration into the attB site of E. coli was performed with the pOSIP plasmid system (32), with clonetegration plasmid cloning done in TransforMax EC100D pir+ electrocompetent E. coli (Lucigen).
All mother machine strains were in E. coli MG1655 with ΔmotA, a motility knockout for mother machine growth and an integrated constitutive red fluorescent protein reporter for use as a segmentation marker (strain NDL162) (33). Plasmids and strains are listed in Supplementary Tables S1–3. DNA sequences of parts are in Supplementary Table S4, and select primer sequences are in Supplementary Table S5.
Mother machine and microscopy
Mother machine microfluidic chips made of polydimethylsiloxane (PDMS) were cast from a patterned silicon wafer and bonded to a No. 1.5 glass coverslip (Fisher Scientific), as previously described (5,33). Most runs used chips with trench dimensions of 1.5 μm wide and high, with different lengths (10+ μm) used throughout. EZ Rich Defined Medium (EZ-RDM, TekNova) was used with 0.85 g/l Pluronic F108 (Sigma-Aldrich) to prevent cell adhesion to PDMS. Runs did not have supplemented antibiotic. For induction, typical concentrations were 1.88 ng/ml anhydrotetracycline (aTc) and 50 μM 2,4-diacetylphloroglucinol (DAPG). For most runs, media was flown at 20 μl/min through Tygon microfluidic tubing (US Plastic Corp 56515) with NE-300 syringe pumps (New Era Pump Systems). Each strain presented was run in at least two separate mother machine experiments, with a representative run shown, with the exception of dCas12a two-plasmid oscillator (JK504+506), which was only run once.
Cells were imaged on modified Nikon Ti2-E inverted microscopes with Plan Apo 40× air objective (NA 0.95, Nikon), Perfect Focus System (PFS, Nikon), Nikon Ei-S-ER motorized stage with encoders, and high-resolution CMOS camera (Andor Zyla 4.2 PLUS), all controlled with NIS-Elements software (Nikon). We used a 40× air objective as a balance between resolution/signal to accurately characterize individual cells and the larger field-of-view for rapidly imaging thousands of cells every several minutes, as used in other oscillator efforts (33). All runs were kept at 37°C with a temperature-controlled cage incubator (Okolab). Exposure times varied per experimental run, but RFP was imaged at 100–200 ms and YFP for 10–50 ms using an LED excitation system (SPECTRA X light engine, Lumencor). Images were acquired at frame rates of 6–10 min, as noted throughout.
Image processing and analysis
Microscopy images were processed using ImageJ and MATLAB to segment mother cells using code originally developed by Somenath Bakshi. Further analysis was performed in MATLAB with code from S. Bakshi, J. Kuo and R. Yuan. ACF and power spectrum of the windowed ACF were calculated in MATLAB and as described previously (5). Interpeak distances and peak heights were determined using MATLAB findpeaks function to find local maxima on a Savitzky–Golay filter smoothed signal. After minimal smoothing by fitting a polynomial order 5 with an 11-frame window using MATLAB function sgolayfilt, the peak distances were found using MATLAB function findpeaks. The minimum peak prominence value was adjusted if the fluctuation heights were not captured based on visual inspection. Inducible cascade measurements plotted as average plus shaded region standard deviation used MATLAB function confplot from Michele Giugliano (Brain Mind Institute, EPFL, Lausanne, Switzerland).
Bulk strain analysis: plate reader and agarose pads
Strains were evaluated in 200 μl test cultures in 96-well flat-bottom black with clear bottom fluorescence plates (Corning 3631) with low evaporation lids (Corning 3931). Plates were grown in a BioTek Synergy H1 plate reader with continuous shaking at 37°C, including a higher gradient temperature above to reduce condensation. Seed cultures were grown overnight in a separate 96-well plate. Agarose pads to qualitatively look at cultures were prepared with 1% agarose with microscope slides (34).
RESULTS
Design overview: dCas9 RNA oscillator
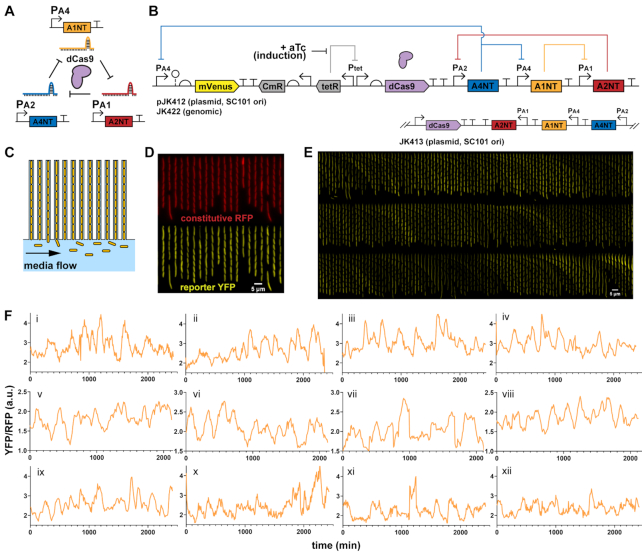
We designed a ring oscillator using dCas9 and sgRNAs based on the ‘repressilator’ synthetic circuit (35), a three-component ring oscillator with protein repressor parts. At that time, the repressilator was built using well-characterized protein transcription factors (lacI, tetR, phage cI), more recently greatly improved after reducing plasmid noise and removing protein degradation tags (5). Similarly, we chose from a characterized set of orthogonal sgRNAs and targeted promoters used in making logic gates with dCas9 repression (16) to assemble a ring oscillator (Figure 1A). The sgRNAs are expressed from strong bacterial σ70 promoters with variations for sgRNA binding between the conserved −35 and −10 regions (Supplementary Figure S1), with reported repression fold-changes of 56–250 (16). The protospacer adjacent motif (PAM) sites for dCas9 binding are also between the −35 and −10. Each sgRNA consists of a 20 nucleotide (nt) targeting region and an 83 nt Cas9 handle plus terminator. Additional strong synthetic terminators used previously were also inserted. To preserve any supercoiling effects (36), terminators were set close to the promoter of the downstream sgRNA, separated by several bases from a restriction site. Streptococcus pyogenes dCas9 (with deactivating mutations D10A and H840A) was placed under control of an anhydrotetracycline (aTc) inducible TetR promoter (Figure 1B), as in another study (12).
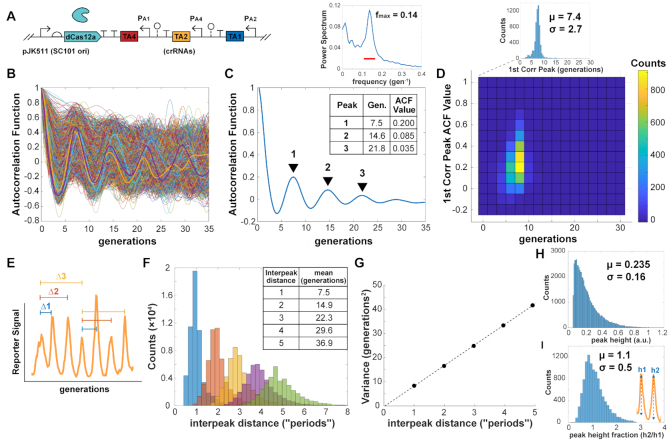
Figure 1.
dCas9 RNA oscillator imaged in microfluidic device. (A) The ring oscillator has guide RNAs targeting each other through deactivated Cas9 (dCas9), which forms a complex with a single guide RNA (sgRNA) to create an active transcriptional repressor. Each guide RNA targets the sigma factor binding region of another promoter, which produces its own sgRNA. (B) A design of the dCas9 RNA oscillator with YFP fluorescent reporter mVenus, Tet-inducible dCas9 and the sgRNA ring. The circuit is combined in a single low-copy plasmid (pSC101 origin of replication), genome-integrated or split across two plasmids. Another form has the sgRNAs transcriptionally reversed. (C) The ‘mother machine’ microfluidic device allows imaging of thousands of cells. Cells are confined in trenches, where ‘mother’ cells are perpetually trapped to allow for single-cell tracking over many generations. (D) Escherichia coli MG1655 cells with the designed dCas9 RNA oscillator plasmid (pJK412) are in a mother machine device, with constitutive RFP expressed genomically as a segmentation marker and YFP reporter expressed from the circuit. A field-of-view at one timepoint (6 h post- aTc induction) shows many cells. (E) In a kymograph looking at a single trench over time, YFP reporter fluorescence appears to fluctuate. Each frame is 8 min, with one generation ∼32 min. (F) Example representative traces of mother cell reporter YFP fluorescence normalized by segmentation RFP signal show fluctuations over time for (i–iv) 1-plasmid circuit (pJK412), (v–viii) genome-integrated circuit (JK422) and (ix–xii) 1-plasmid circuit with flipped sgRNA direction (pJK413).
A yellow fluorescent protein (YFP) mVenus reporter was chosen for its fast maturation rate (37), allowing dynamics to be captured, and placed on the same low-copy plasmid as dCas9. A reporter was made for each of the three different sgRNA promoters, but promoter PA4 outperformed the others in brightness and dynamics, with the lowest reported fold change (16). Different copy number forms of the circuit were created—a two-plasmid system, a one-plasmid version, and a genome-integrated version—with the idea of reducing potential noise from copy number fluctuations.
Free-running dCas9 RNA oscillator
The dCas9 circuits were transformed into E. coli MG1655 with genomic red fluorescent protein (RFP) constitutively expressed as a segmentation marker. Cells were imaged in real time in a ‘mother machine’ microfluidic device that traps cells in trenches while fresh media is flown through, for diffusive feeding that supports exponential growth and washout of newly divided cells (Figure 1C and D). The mVenus YFP reporter showed fluctuations only with dCas9 induction, where all cells remained bright without added aTc inducer and these fluctuations occurred continuously for 24+ h (Figure 1E and Supplementary Video S1). The RFP segmentation marker did not fluctuate significantly compared to YFP reporter and was used for reporter signal normalization of gene expression noise (Supplementary Figure S2). Tracking single mother cells, irregular fluctuations resembling oscillations were observed, with some cells appearing to have some level of regularity (Figure 1F). A genome-integrated form of the circuit and a form where sgRNAs were transcribed convergent to dCas9 behaved similarly. A two-plasmid system also showed similar fluctuations (Supplementary Figure S3), and using a different reporter promoter had much lower signal and dynamic range (Supplementary Figure S4). We also tried a different set of orthogonal sgRNAs, which had similar behavior but repressed too strongly for very low reporter signal and range (Supplementary Figure S5).
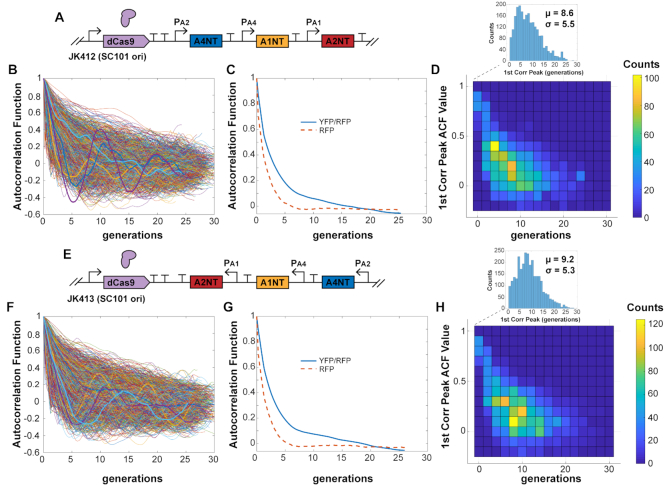
With the wide range of fluctuations across cells, the population-averaged ACF did not show correlation peaks characteristic of oscillators, but the slow decays of multiple generations showed a history-dependence of the reporter signal (Figure 2). The population-averaged ACF needed over 15 generations to decay to zero, compared to the constitutive RFP signal averaged ACF that decayed to zero after approximately five generations (Figure 2C and G). For individual traces that did show a first correlation peak, the average was approximately nine generations. These traces had many different characteristic periods estimated from first correlation peaks (Figure 2D and H), which were averaged out when looking over the whole population (Figure 2C and G).
Figure 2.
dCas9 RNA oscillator ACF. (A) The 1-plasmid dCas9 RNA oscillator circuit (JK412) with (B) ACF calculated for individual cell traces. Selected representative traces are bolded. (C) The population-averaged ACF shows a slower decay, with the reporter signal YFP/RFP ACF decaying much slower than the ACF for segmentation marker RFP. (D) Some individual traces showed first correlation peaks across a wide range, with an average of 8.6 generations. (E–H) The same properties were seen for a version with sgRNAs transcriptionally flipped. Cells measured: (B) JK412, 2358; (F) JK413, 3037.
The dCas9–sgRNA complex is fairly stable with nanomolar dissociation constants, predominantly unbinding only during DNA replication (27,28,38). This helps explain strong multi-generational repression, as a single dCas9–sgRNA complex could bind to a DNA target for the duration of growth between divisions. Many of the cells did not appear to oscillate obviously, perhaps due to these long binding lifetimes: binding of a lower-population, ‘wrong’ sgRNA could throw off the ring cycle, since the dCas9–sgRNA could effectively bind for an entire generation. This is contrasted with protein repressors such as LacI that individually bind for minutes at a time (39). As with the protein-based repressilator, fluctuations were asynchronous across a population. An effort to synchronize the system by initially producing a high level of one sgRNA from another plasmid failed to entrain the population (Supplementary Figure S6).
Measuring response times for individual dCas9–sgRNA repression steps
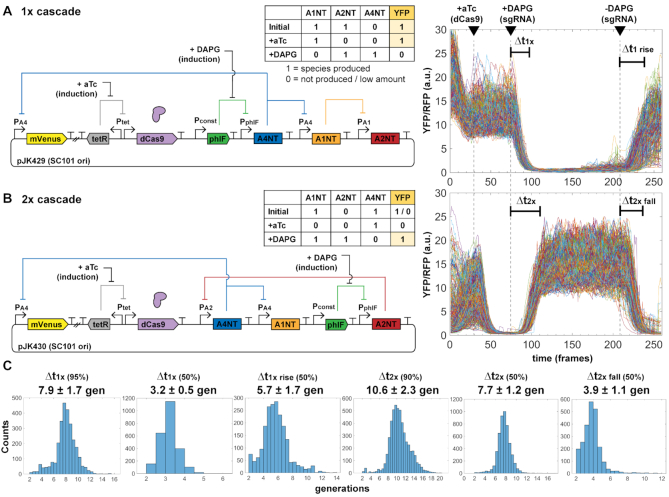
To compare with our observed oscillator periods, we created inducible guide RNA circuits using PhlF protein repressor, with an inducible promoter by small molecule 2,4-diacetylphloroglucinol (DAPG). A PhlF promoter replaced an sgRNA promoter in the free-running oscillator for an open-loop version with chemically inducible control of an sgRNA species (Figure 3). We could control production of both dCas9 (with aTc) and sgRNA (with DAPG) separately, which we used to characterize response times of individual cells in mother machine experiments for a global single-cell look at dCas9 kinetics.
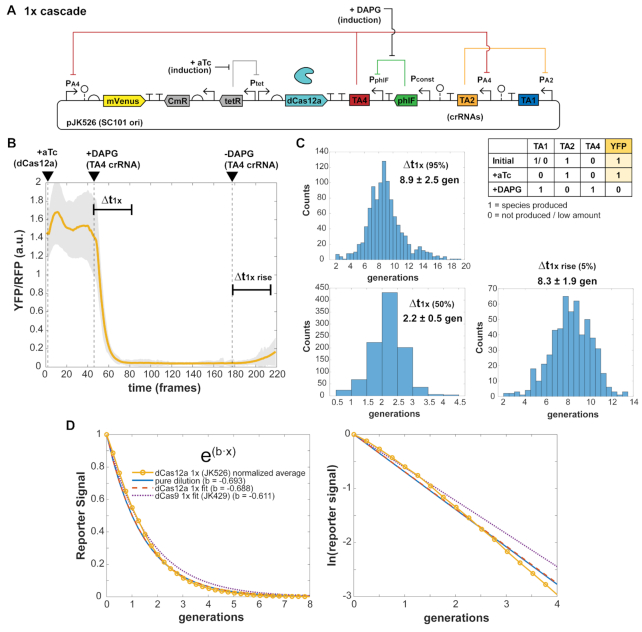
Figure 3.
Inducible sgRNA response times are consistent with the free-running oscillator. A single constitutive promoter (PAx) of the free-running system was replaced with a PhlF repressor-based promoter system, allowing inducible control with small molecule DAPG. Measurement of cells in the mother machine allows explicit tracking of individual cell responses to added inducers to determine characteristic times Δt of sgRNA repression cascade steps. Escherichia coli MG1655 cells with plasmids were grown in EZ-RDM. (A) One-step sgRNA repression is measured. Expected behavior is YFP bright to dim upon DAPG induction (A4NT sgRNA). Cells are re-entering exponential growth and equilibrating prior to aTc induction. Plot shows 599 cell reporter traces of plasmid version (pJK429). 1 frame = 6 min. (B) Two-step sgRNA repression is measured. Expected behavior is YFP dim to bright upon DAPG induction (A2NT sgRNA). (The initial 1/0 YFP accounts for repression from leaky dCas9 expression.) Plot shows 515 cell traces of plasmid version (pJK430). (C) Histograms of measured characteristic time steps indicated in (A) and (B). Percentages are percent repression or de-repression. Distributions were measured from (A) 2819 cells for pJK429 and (B) 3136 cells for pJK430.
In our system, we see repression times of several generations dependent on dCas9 expression (Figure 3). As with the oscillator designs, signal changes were from YFP reporter while constitutive RFP segmentation marker was mostly stable (Supplementary Figure S7). Times measured were similar in genome-integrated forms, with lower signal levels (Supplementary Figure S8) and in a two-plasmid cascade (Supplementary Figure S9). The times are consistent with those measured by others in different systems at a population level (12). Interestingly, the time difference between the 1× and 2× cascades should be roughly the time for one sgRNA step, as the 1× time includes de-repression of phlF promoter. This time difference is about 2.7 generations (10.6–7.9 generations). For the three steps of the free running oscillator, this corresponds to 2.7 × 3 = 8.1 generations, similar to the ∼8.6 generation average period seen for the oscillator plasmid design JK412 (Figure 2D). Of course, the large variations for the measured oscillator periods make this comparison difficult but suggestive nonetheless.
Exploring dCas9 degradation
With tight dCas9 binding and off rates longer than E. coli division times, we thought dCas9 protein degradation might help remove dCas9–sgRNA complexes to more accurately reflect a dynamically changing pool of sgRNAs. We tried adding ssrA (small stable RNA A) degradation tags of varying strengths (40), but did not see improved oscillatory behavior (Supplementary Figure S10). Because of this, our main designs use an untagged dCas9. However, roughly setting dCas9 expression level with concentration of aTc inducer was important (Supplementary Figures S11 and 12).
dCas12a RNA oscillator
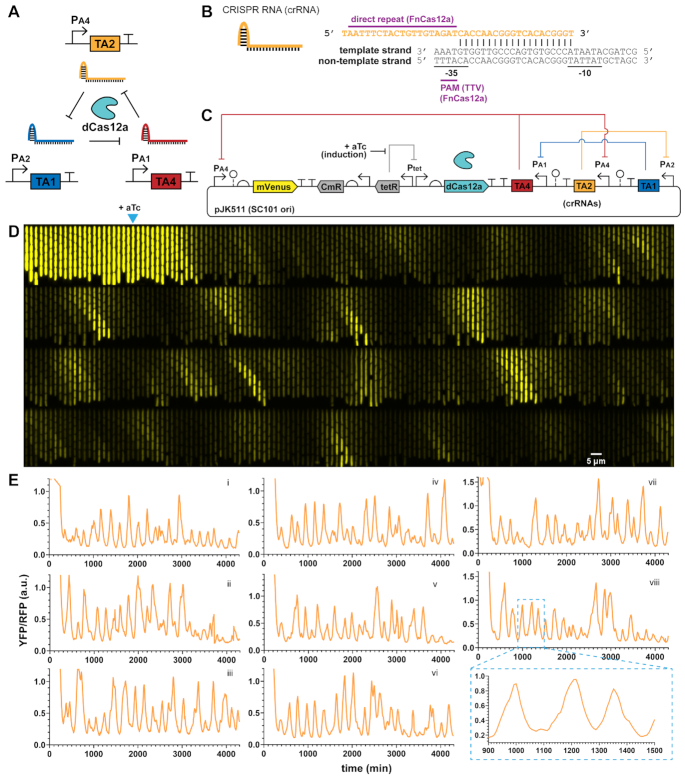
CRISPR-Cas12a systems also use a main effector enzyme that acts on DNA by forming a complex with targeting CRISPR RNAs (crRNA) and binding to DNA. We used the ∼150 kD Cas12a protein from Francisella novicida, with a nuclease-deactivating D917A mutation (22,41) to create another ring oscillator circuit. The single mutation preserves crRNA array processing ability. Here we use a 19-nt direct repeat as a Cas12a handle along with a 20-nt DNA targeting region (Figure 4B). The F. novicida Cas12a uses a ‘TTV’ or ‘TTTV’ PAM (22,42), which targets the −35 box of the same σ70 promoters in the dCas9 designs. We made our Cas12a crRNAs target these promoter regions on the template strand, with the idea that the dCas9 orthogonality with sgRNAs would carry over to the dCas12a system.
Figure 4.
dCas12a RNA oscillator shows regularity. (A) An analogous RNA ring oscillator design replaces dCas9 with deactivated Cas12a (dCas12a). (B) Cas12a CRISPR RNA (crRNA) architecture. Designs use a 19-nt direct repeat dCas12a handle plus 20-nt targeting region. The PAM is ‘TTV’ for Francisella novicida Cas12a (FnCas12a). (C) Plasmid design of the dCas12a RNA oscillator circuit has a PA4-mVenus reporter, Tet-inducible dCas12a and crRNAs in a transcriptionally convergent direction. (D and E) Escherichia coli MG1655 cells with the circuit were grown in mother machine chips with EZ-RDM. (D) A kymograph of a single trench over time shows YFP reporter fluctuation. Each frame is 8 min, with one generation ∼25 min. Arrow marks induction for dCas12a production at frame 23 (184 min). (E) Example reporter traces show fluctuations resembling oscillations and a lower signal repressed state. Average generation ∼25 min. Pullout for trace viii zooms in on the peak shapes.
As before, we made one- and two- plasmid designs with the same architecture as the dCas9 RNA oscillator (Figure 4C). To accommodate the shorter 40-nt targeting RNAs for Cas12a, instead of 103-nt for dCas9, we added ∼80-nt ribozyme spacer sequences (43) between crRNAs to reduce transcriptional readthrough effects. Notably, strains fluctuated with a much lower baseline, meaning cells looked darker for most of the run with pulses of bright (Figure 4D and E; Supplementary Video S2). Overall, the fluctuations were much more regular, with some cells appearing quite oscillatory (Figure 4E). Individual cell ACF traces showed multiple correlation peaks, and the averaged ACF for the population also showed multiple distinct peaks (Figure 5). We saw a first correlation peak at 7.5 generations and second peak at 14.6 generations. The amplitude of the ACF peaks decays rapidly at first to 0.200, and then drops in smaller increments (∼2.4-fold), similarly to an improved protein repressilator with fluorescent reporter integrated in the main plasmid and an interfering degradation-tagged reporter removed (5). The period length of ∼7.5 generations was similar to the dCas9 oscillators, but slightly faster. Unlike the dCas9 forms, individual cell traces had a much narrower distribution (Figure 5D), showing the more homogenous behavior across cells. Measuring interpeak distances of apparently oscillating cells, the estimated period length of mean ∼7.5 generations was nearly identical to that determined by ACF (Figure 5E and F). Peak distances changed as regular multiples of this first, with the variance increasing linearly with increasing interpeak distances by periods (Figure 5G). This indicated that the period lengths were independent, where the circuit did not show memory effects between periods. For interpeak distance of one period, the standard deviation was 2.9 generations, for an estimated coefficient of variation (CV, standard deviation over mean) of 0.39. The estimated peak heights were seemingly exponentially distributed, with a CV of ∼0.68 (Figure 5H). Peak heights were also seen to be independent, where consecutive peaks did not show memory/carryover effects in amplitude (Figure 5I).
Figure 5.
dCas12a RNA oscillator ACF and interpeak distance. (A) The 1-plasmid dCas12a oscillator circuit with (B) ACF calculated for 6335 individual cell traces. Selected representative traces are bolded. (C) The population-averaged ACF showed clear correlation peaks, with the first correlation peak of ∼7.5 generations. The power spectrum of the ACF shows a max frequency around 0.14, or 7.1 generations. The bar indicates the width of the window function. (D) Individual cell trace first correlation peaks were mostly clustered around the same period length, with some individual trace ACF peaks showing higher values. (E) Interpeak distance was calculated for peaks of oscillatory traces, where ACF first minimum and first correlation peaks exceeded values ±0.05. (F) Calculated interpeak distances showed distinct distributions following multiples of the first. These are labeled as ‘periods’, multiples of the 1-interpeak distance. (G) Variances of the interpeak distances increase linearly with period distance. (H) Distribution of estimated peak heights for peaks of oscillatory traces, where ACF first minimum and correlation peaks exceeded ±0.05. (I) Peak height fractions for consecutive peaks, comparing one peak with its preceding one.
Unlike in the dCas9 oscillators, having the crRNAs be transcriptionally convergent with dCas12a had a noticeable improvement in oscillation performance and increased overall brightness, for greater dynamic range of the reporter fluctuations (Supplementary Figure S13 and Video S3). A two-plasmid version of the circuit also showed oscillatory-like fluctuations but was not as regular as the one-plasmid form at the population level (Supplementary Figure S14 and Video S4). Also unlike the dCas9 case, genome-integrating the circuit led to much poorer performance (Supplementary Figure S15), suggesting the higher copy numbers of the plasmid are needed for timing accuracy. Perhaps the crRNAs are degraded at a faster rate than sgRNAs, so plasmid expression levels are needed to maintain the correct active pool for dCas12a repression.
Measuring response times for individual dCas12a–crRNA repression steps
As with the dCas9 system, we created inducible crRNA circuits using PhlF protein repressor, to compare with our measured periods of the observed fluctuations. Repression times were characterized as with the dCas9 systems, with a one-step repression time of ∼9 generations (Figure 6). (Single traces are in Supplementary Figure S16.) However, the time for 50% repression was rapid at only ∼2 generations, likely accounting for the faster cycling time of the oscillator. As with dCas9, dCas12a–crRNA repression of YFP began soon after crRNA induction and the repression was strong at ∼36-fold (1.5–0.041 a.u. signal change high to low) and maintained in most cells for at least 600 min (∼25 generations).
Figure 6.
dCas12a crRNA repression response times. (A) A single constitutive promoter of the free-running dCas12a oscillator was replaced with a PhlF repressor-based promoter system, allowing inducible control with small molecule DAPG and measurement of characteristic times Δt of crRNA repression cascade steps. Escherichia coli MG1655 cells with plasmids were grown in EZ-RDM in the mother machine. (B) One-step crRNA repression is measured. Expected behavior is YFP bright to dim upon DAPG induction (TA4 crRNA). Averaged trace from 1049 cells of plasmid version (pJK526), with shaded region showing one standard deviation of the averaged values. 1 frame = 6 min. (C) Histograms of measured characteristic time steps indicated in (B) for single cell traces. Percentages are percent repression or de-repression. Distributions measured from 1045 cells. (D) Exponential fitting of the averaged trace (JK526) and dCas9 1× cascade trace. The ‘pure dilution’ curve (solid line) corresponds to that expected for dilution from cell division alone.
Fitting the average trace with a simple exponential (a·exp(b·t)), the exponent coefficient of b = −0.688, was very close to that expected for pure dilution based on cell division alone (b = −ln(2) = −0.693) (Figure 6D). This was also close to that observed from the averaged trace of the dCas9 single step cascade (b = −0.611). These results show that in these inducible step strains, the reporter decay processes are nearly pure dilution from cell division.
A two-step crRNA cascade circuit did not behave as expected (Supplementary Figure S17). Cell reporter signal was repressed upon aTc addition for dCas12a induction as expected, but unexpectedly then steadily increased before crRNA induction. The steady-state signal with crRNA induction was about half that of before dCas12a induction, and removal of crRNA inducer caused only a small drop in signal. Cells grew normally, with average division time of ∼25.8 min. The unexpected output suggested some feedback coupling between the crRNAs, perhaps from bound dCas12a hindrance and/or transcriptional readthrough in spite of the strong terminators and ribozyme spacer sequences. Some level of this crRNA coupling is likely also present in the oscillator, though it is unclear how it affects performance.
Cell growth comparisons
Cell growth rates of dCas12a strains were fairly normal in mother machine runs, with average division times of ∼25.7 min for the dCas12a oscillator (Figure 7A). These rates are close to those without circuit or with repressilator or dual-feedback oscillator circuits of ∼24–25 min (33), suggesting low circuit burden. In contrast, the dCas9 oscillator strains had slowed growth, with increased division rates up to 30+ min. (It was difficult to assess exact growth impacts of individual circuits because other variable factors of media condition and PDMS chip could affect growth rate much more. We had runs where different media and chip preparations increased division rates by a few minutes without changing the strain's circuit performance by generations timescale.) While division times could vary due to conditions, the dCas12a and dCas9 oscillator strains were from the same mother machine run for direct comparison in the results presented. The oscillator circuits appeared to be greater burden than the inducible RNA circuits, whose strains all grew faster on average. The distributions give a look at the heterogeneity of the populations, with a relatively narrow distribution for the dCas12a oscillator and wide distributions for other strains (Figure 7B).
Figure 7.
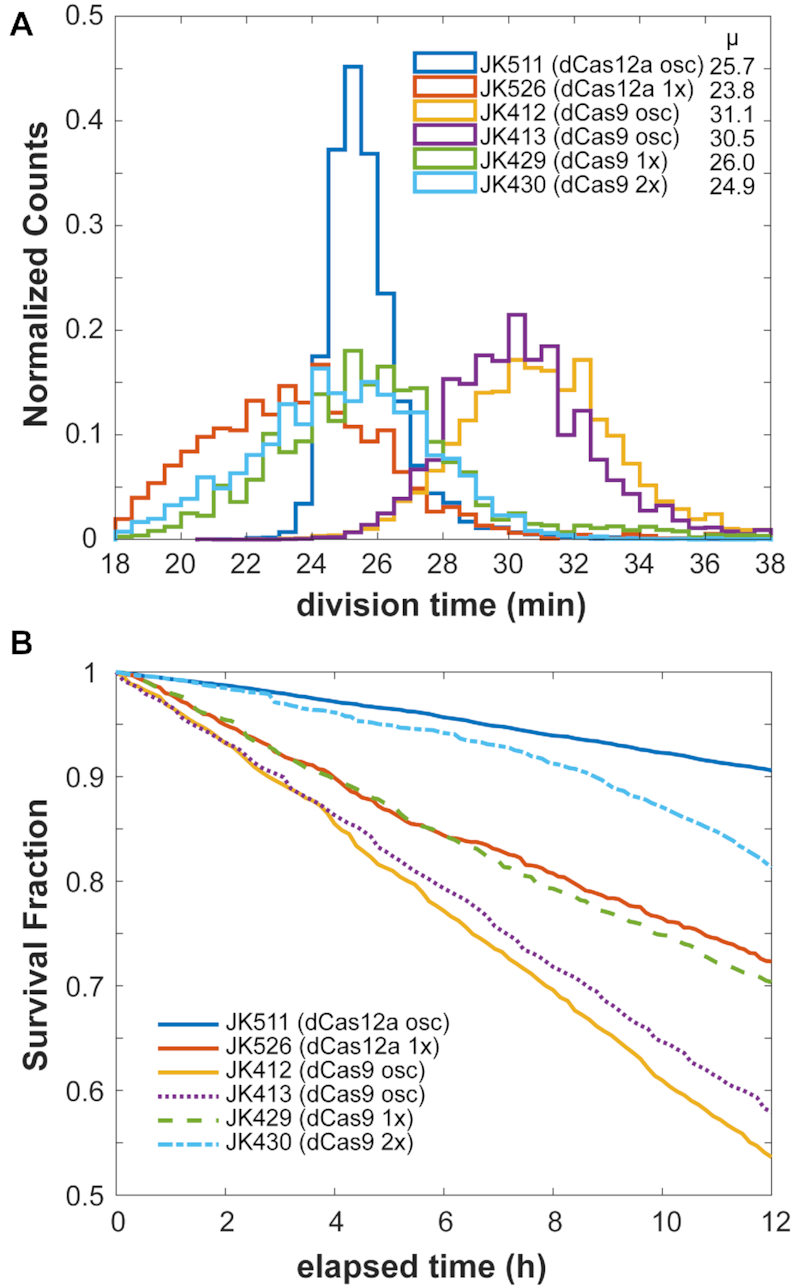
Cell growth comparisons. Estimates on growth efficiency of RNA oscillator and cascade strains. Cell division times depended on media and chip conditions. (A) Average growth division times, as observed in mother machine runs. Each count is a single mother cell's average division time over the course of the run. Cell counts: JK511, 7998; JK526, 2263; JK412, 7158; JK413, 7396; JK429, 5921; JK430, 7779. (B) ‘Survival’ curves of mother cells, estimated in a 12-h window within the first 24 h of imaging. The fraction of mother cell trajectories remaining after the indicated time estimates cell survival.
However, we did see much greater rates of filamentation and cell death from all circuits. By looking at a 12-h window in the first 24 h of imaging, we could get an estimate of circuit toxicity based on surviving mother cells. Over these 24+ generations, while the dCas12a oscillator (JK511) had over 90% mother cell survival, the dCas9 oscillators (JK412, JK413) had only ∼55–60% survival. In a different mother machine study, E. coli cells constitutively expressing fluorescent protein had survival rates of ∼90% (33). (Note that even 50% survival over 24 generations means an ∼98% survival rate per generation, clearly not inhibiting for an exponentially growing population but maybe deleterious in resource-limited conditions.) Other dCas12a circuits showed similar toxicity (Supplementary Figure S18). As with growth rates, death rates also varied due to chip conditions. The higher rates of cell death, often by filamentation, were likely due to toxicity from dCas9 expression and potentially exacerbated by off-target effects of a guide RNA (44,45), and a similar toxicity from dCas12a. The greater toxicity seen from dCas9 compared to dCas12a is consistent with that seen in other bacteria (46,47). Perhaps toxicity may be reduced through protein engineering efforts, as was done for dCas9 (48). It is worth noting again that even with these toxicity rates, a growing population will still increase substantially: these death rates are in a 12-h window of 24+ new generations of exponentially growing cells.
DISCUSSION
We present construction of new translationally independent synthetic oscillators based on transcription of CRISPR-associated RNAs, a dynamic use of CRISPR-Cas systems over time. The only other example we are aware of is an experimental effort to make a dCas9-based CRISPR interference oscillator (30), where their periods were much longer at 14–17 generations, as well as a modeling-only exploration (49). Most previous CRISPR component circuits, while impressive, used endpoint as the behavior, showing before-and-after changes from incubations of several hours or more. That is excellent for sensors and logic gates, but we show a time-varying system that fluctuates between on/off reporter states in an analog manner. In addition, our characterization method is another example of single-cell analysis of a dCas9 or dCas12a circuit in bacteria (27,50,51). We also use our setup to explicitly measure single-cell dCas9–sgRNA and dCas12a–crRNA repression times in thousands of individual cells.
While we do not see performance as regular as the high-precision improved protein-based repressilator (5), we see clear signs of regularity that are promising for further improvement or immediate applications with less stringent timing requirements. For the dCas12a RNA oscillator, some cells have quite regular timing, and the circuit performs very similarly to an improved version of the protein-based repressilator on a single plasmid, with an even better population-averaged ACF (5). We used parts from a set of 5 orthogonal sgRNA-promoter pairs (16), but in principle parts could be drawn from any orthogonal pool. In this way, the set of modular parts may be enormous to assemble multiple oscillator rings that not only are orthogonal to each other in a cell, but also to the host DNA, minimizing off-target effects. Our proof-of-concept may allow construction of chained RNA ring oscillators, such as two three-component rings or larger 5+ component rings. Logic gate type functional output can be created with two rings, such as AND or NOR gates where each ring partly controls repression of a single promoter output. We point out that many original proof-of-concept oscillator designs (7,35) had performed irregularly at first but were greatly improved in separate major efforts (5,33).
The dCas12a RNA oscillator may also be an interesting model system for further study in itself, as the oscillatory behavior is seen even without direct binding cooperativity in CRISPR systems (16,30). However, the system likely has non-linearity from other mechanisms, such as bound-repressor degradation (52), with DNA-bound dCas12a-crRNA removed by dilution from cell division and possibly protein degradation, or Michaelis–Menten degradation of the RNAs (53). Estimating parameters for these mechanisms may require measurements in different contexts, such as in yeast (54,55) or cell-free in vitro systems (56,57). Addition of proteases, RNases, or interfering RNAs may allow tuning of the parameters without measuring. Further investigation of these alternatives to cooperative binding for nonlinearity may lead to better understanding and new strategies for designing other oscillator circuits.
In the dCas12a design, another crRNA can be added at the end of any existing oscillator RNA to form a crRNA array (22,58,59). These added crRNAs can target other desired sequences for regulation beyond the core oscillator crRNAs, with the CRISPR array being processed by dCas12a. While commonly used protein transcription factors typically target promoter regions, the programmability of CRISPR has expanded targeting to elsewhere along a gene, not just regulatory regions (16,59,60). In this way, our circuits may be readily coupled for transcription regulation controlled by the oscillator, including potential control of unmodified endogenous genes (58,60). Other versions of dCas12a may be useful: Acidaminococcus sp. BV3L6 (AsCas12a) and Lachnospiracaeae bacterium ND2006 (LbCas12a) Cas12a both have perhaps even more specific PAM targeting sites of ‘TTTV’ (22). The targeted promoters in our circuits already have a ‘TTTG’ PAM-motif in the −35 region of the strong σ70 promoters, so these other Cas12as may reduce any off-target effects. Variants of Cas12a can also target different PAM sites, further increasing the versatility to target many sequences (61,62). Also, AsCas12a and LbCas12a have had faster association on-rates measured (63), so these may be ways to tune the period lengths of the oscillator. The FnCas12a on-rate (kon ∼2.3 × 106 M−1·s−1, (63)) was measured as roughly half or a third that of Cas9 (kon ∼6 × 106 M−1·s−1, (64)), maybe a factor in the dCas12a oscillator's better performance than dCas9. Swapping FnCas12a with faster AsCas12a or LbCas12a could show any effects of DNA binding on-rate. Addition of ribozyme sequences between the crRNAs, not added for the sgRNA, may have also helped, regulating RNA amounts by insulating from context effects such as from transcriptional readthrough (43,65).
The other dCas9 RNA oscillator (30), built on two plasmids, is particularly interesting in suggesting an importance of insulating the RNAs. They used csy4 RNase cleavage sequences extensively to insulate their parts, around each sgRNA and by each reporter RBS, and had 200 bp spacer sequences and fluorescent reporter genes further separating their sgRNAs. This sgRNA insulation may have contributed to their greater accuracy in timing for dCas9 and longer 14–17 generation periods, further suggesting transcriptional coupling between our sgRNAs. The success of their circuit suggests better insulation of our sgRNAs may improve performance. Besides spacer sequences, spatially separating the sgRNAs across the plasmid may also help, though this may remove advantages of a single compact cassette. Our dCas12a RNA oscillator may also benefit from greater spatial separation of the crRNAs. Also, their work perhaps shows a generalization of the CRISPR RNA oscillator strategy, demonstrating programmability by using a different set of sgRNAs and promoters.
We have built prototype oscillators that are nearly translationally independent in their output behavior. While transfer to other proteobacteria like Salmonella may be direct, changes are needed for other desired bacteria with different promoter architectures. A promising host is Bacteroides thetaiotaomicron, gut bacteria in which others have built dCas9–sgRNA logic gates (66)—their study included seven orthogonal sgRNA-promoter pairs, much like the pairs we used for our E. coli circuits. With functional prototypes in bacteria, it may be worth exploring construction in eukaryotic organisms like yeast where CRISPR-Cas has been much explored, though a complete circuit reconstruction will be required. Mammalian cells may also be interesting mechanistically, since bound dCas–RNA protein degradation may be the dominant removal process, as opposed to dilution from cell division. Enhancing degradation of bound repressor may be needed, such as with protease or ubiquitin tags (67). Since dCas9 and dCas12a can be functionally expressed in many unique hosts across kingdoms, a generalizable oscillator circuit may be possible.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Somenath Bakshi, Emanuele Leoncini, Luis Gutiérrez-López, Laurent Potvin-Trottier, Scott Luro, Jeffrey Way and other lab members for their help. Some microscopy experiments were performed at The Nikon Imaging Center at Harvard Medical School with help from Jennifer Waters and Anna Payne-Tobin Jost. Thanks also to K.B.
Contributor Information
James Kuo, Department of Systems Biology, Blavatnik Institute at Harvard Medical School, Boston, MA 02115, USA; Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA.
Ruoshi Yuan, Department of Systems Biology, Blavatnik Institute at Harvard Medical School, Boston, MA 02115, USA.
Carlos Sánchez, Department of Systems Biology, Blavatnik Institute at Harvard Medical School, Boston, MA 02115, USA.
Johan Paulsson, Department of Systems Biology, Blavatnik Institute at Harvard Medical School, Boston, MA 02115, USA.
Pamela A Silver, Department of Systems Biology, Blavatnik Institute at Harvard Medical School, Boston, MA 02115, USA; Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Defense Advanced Research Projects Agency (DARPA) [HR0011-16-2-0049]; National Institute of General Medical Sciences of the National Institutes of Health, Ruth L. Kirschstein National Research Service Award [F32GM125108to J.K.]; National Science Foundation (NSF) [1615487, 1562497, 1517372 to J.P.]. Funding for open access charge: DARPA [HR0011-16-2-0049].
Conflict of interest statement. None declared.
REFERENCES
- 1. Claesen J., Fischbach M.A.. Synthetic microbes as drug delivery systems. ACS Synth. Biol. 2015; 4:358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Din M.O., Danino T., Prindle A., Skalak M., Selimkhanov J., Allen K., Julio E., Atolia E., Tsimring L.S., Bhatia S.N. et al.. Synchronized cycles of bacterial lysis for in vivo delivery. Nature. 2016; 536:81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Riglar D.T., Richmond D.L., Potvin-Trottier L., Verdegaal A.A., Naydich A.D., Bakshi S., Leoncini E., Lyon L.G., Paulsson J., Silver P.A.. Bacterial variability in the mammalian gut captured by a single-cell synthetic oscillator. Nat. Commun. 2019; 10:4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Danino T., Mondragon-Palomino O., Tsimring L., Hasty J.. A synchronized quorum of genetic clocks. Nature. 2010; 463:326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Potvin-Trottier L., Lord N.D., Vinnicombe G., Paulsson J.. Synchronous long-term oscillations in a synthetic gene circuit. Nature. 2016; 538:514–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang Z.B., Wang Q.Y., Ke Y.X., Liu S.Y., Ju J.Q., Lim W.A., Tang C., Wei P.. Design of tunable oscillatory dynamics in a synthetic NF-kappaB signaling circuit. Cell Syst. 2017; 5:460–470. [DOI] [PubMed] [Google Scholar]
- 7. Stricker J., Cookson S., Bennett M.R., Mather W.H., Tsimring L.S., Hasty J.. A fast, robust and tunable synthetic gene oscillator. Nature. 2008; 456:516–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Scott S.R., Din M.O., Bittihn P., Xiong L., Tsimring L.S., Hasty J.. A stabilized microbial ecosystem of self-limiting bacteria using synthetic quorum-regulated lysis. Nat. Microbiol. 2017; 2:17083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Butzin N.C., Hochendoner P., Ogle C.T., Mather W.H.. Entrainment of a bacterial synthetic gene oscillator through proteolytic queueing. ACS Synth. Biol. 2017; 6:455–462. [DOI] [PubMed] [Google Scholar]
- 10. Srinivas N., Parkin J., Seelig G., Winfree E., Soloveichik D.. Enzyme-free nucleic acid dynamical systems. Science. 2017; 358:eaal2052. [DOI] [PubMed] [Google Scholar]
- 11. Dominguez A.A., Lim W.A., Qi L.S.. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016; 17:5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A.. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013; 152:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L.A.. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013; 41:7429–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larson M.H., Gilbert L.A., Wang X., Lim W.A., Weissman J.S., Qi L.S.. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013; 8:2180–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pickar-Oliver A., Gersbach C.A.. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019; 20:490–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nielsen A.A., Voigt C.A.. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol. Syst. Biol. 2014; 10:763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gander M.W., Vrana J.D., Voje W.E., Carothers J.M., Klavins E.. Digital logic circuits in yeast with CRISPR-dCas9 NOR gates. Nat. Commun. 2017; 8:15459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jusiak B., Cleto S., Perez-Pinera P., Lu T.K.. Engineering synthetic gene circuits in living cells with CRISPR technology. Trends Biotechnol. 2016; 34:535–547. [DOI] [PubMed] [Google Scholar]
- 19. Sheth R.U., Yim S.S., Wu F.L., Wang H.H.. Multiplex recording of cellular events over time on CRISPR biological tape. Science. 2017; 358:1457–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nakamura M., Srinivasan P., Chavez M., Carter M.A., Dominguez A.A., La Russa M., Lau M.B., Abbott T.R., Xu X., Zhao D. et al.. Anti-CRISPR-mediated control of gene editing and synthetic circuits in eukaryotic cells. Nat. Commun. 2019; 10:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim H., Bojar D., Fussenegger M.. A CRISPR/Cas9-based central processing unit to program complex logic computation in human cells. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:7214–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. et al.. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015; 163:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kempton H.R., Goudy L.E., Love K.S., Qi L.S.. Multiple input sensing and signal integration using a split Cas12a system. Mol. Cell. 2020; 78:184–191. [DOI] [PubMed] [Google Scholar]
- 24. Nihongaki Y., Otabe T., Ueda Y., Sato M.. A split CRISPR-Cpf1 platform for inducible genome editing and gene activation. Nat. Chem. Biol. 2019; 15:882–888. [DOI] [PubMed] [Google Scholar]
- 25. Oesinghaus L., Simmel F.C.. Switching the activity of Cas12a using guide RNA strand displacement circuits. Nat. Commun. 2019; 10:2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang P., Robert L., Pelletier J., Dang W.L., Taddei F., Wright A., Jun S.. Robust growth of Escherichia coli. Curr. Biol. 2010; 20:1099–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones D.L., Leroy P., Unoson C., Fange D., Curic V., Lawson M.J., Elf J.. Kinetics of dCas9 target search in Escherichia coli. Science. 2017; 357:1420–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E.. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016; 34:339–344. [DOI] [PubMed] [Google Scholar]
- 29. Shibata M., Nishimasu H., Kodera N., Hirano S., Ando T., Uchihashi T., Nureki O.. Real-space and real-time dynamics of CRISPR-Cas9 visualized by high-speed atomic force microscopy. Nat. Commun. 2017; 8:1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Santos-Moreno J., Tasiudi E., Stelling J., Schaerli Y.. Multistable and dynamic CRISPRi-based synthetic circuits. Nat. Commun. 2020; 11:2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 32. St-Pierre F., Cui L., Priest D.G., Endy D., Dodd I.B., Shearwin K.E.. One-step cloning and chromosomal integration of DNA. ACS Synth. Biol. 2013; 2:537–541. [DOI] [PubMed] [Google Scholar]
- 33. Luro S., Potvin-Trottier L., Okumus B., Paulsson J.. Isolating live cells after high-throughput, long-term, time-lapse microscopy. Nat. Methods. 2020; 17:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Skinner S.O., Sepulveda L.A., Xu H., Golding I.. Measuring mRNA copy number in individual Escherichia coli cells using single-molecule fluorescent in situ hybridization. Nat. Protoc. 2013; 8:1100–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Elowitz M.B., Leibler S.. A synthetic oscillatory network of transcriptional regulators. Nature. 2000; 403:335–338. [DOI] [PubMed] [Google Scholar]
- 36. Yeung E., Dy A.J., Martin K.B., Ng A.H., Del Vecchio D., Beck J.L., Collins J.J., Murray R.M.. Biophysical constraints arising from compositional context in synthetic gene networks. Cell Syst. 2017; 5:11–24. [DOI] [PubMed] [Google Scholar]
- 37. Balleza E., Kim J.M., Cluzel P.. Systematic characterization of maturation time of fluorescent proteins in living cells. Nat. Methods. 2018; 15:47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Josephs E.A., Kocak D.D., Fitzgibbon C.J., McMenemy J., Gersbach C.A., Marszalek P.E.. Structure and specificity of the RNA-guided endonuclease Cas9 during DNA interrogation, target binding and cleavage. Nucleic Acids Res. 2015; 43:8924–8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hammar P., Wallden M., Fange D., Persson F., Baltekin O., Ullman G., Leroy P., Elf J.. Direct measurement of transcription factor dissociation excludes a simple operator occupancy model for gene regulation. Nat. Genet. 2014; 46:405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Andersen J.B., Sternberg C., Poulsen L.K., Bjorn S.P., Givskov M., Molin S.. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 1998; 64:2240–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miao C., Zhao H., Qian L., Lou C.. Systematically investigating the key features of the DNase deactivated Cpf1 for tunable transcription regulation in prokaryotic cells. Synth. Syst. Biotechnol. 2019; 4:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Specht D.A., Xu Y., Lambert G.. Massively parallel CRISPRi assays reveal concealed thermodynamic determinants of dCas12a binding. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:11274–11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lou C., Stanton B., Chen Y.J., Munsky B., Voigt C.A.. Ribozyme-based insulator parts buffer synthetic circuits from genetic context. Nat. Biotechnol. 2012; 30:1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cho S., Choe D., Lee E., Kim S.C., Palsson B., Cho B.K.. High-level dCas9 expression induces abnormal cell morphology in Escherichia coli. ACS Synth. Biol. 2018; 7:1085–1094. [DOI] [PubMed] [Google Scholar]
- 45. Cui L., Vigouroux A., Rousset F., Varet H., Khanna V., Bikard D.. A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat. Commun. 2018; 9:1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Knoot C.J., Biswas S., Pakrasi H.B.. Tunable repression of key photosynthetic processes using Cas12a CRISPR interference in the fast-growing Cyanobacterium Synechococcus sp. UTEX 2973. ACS Synth. Biol. 2020; 9:132–143. [DOI] [PubMed] [Google Scholar]
- 47. Jiang Y., Qian F., Yang J., Liu Y., Dong F., Xu C., Sun B., Chen B., Xu X., Li Y. et al.. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat. Commun. 2017; 8:15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang S., Voigt C.A.. Engineered dCas9 with reduced toxicity in bacteria: implications for genetic circuit design. Nucleic Acids Res. 2018; 46:11115–11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Clamons S.E., Murray R.M.. Modeling dynamic transcriptional circuits with CRISPRi. 2017; bioRxiv doi:27 November 2017, preprint: not peer reviewed 10.1101/225318. [DOI]
- 50. Camsund D., Lawson M.J., Larsson J., Jones D., Zikrin S., Fange D., Elf J.. Time-resolved imaging-based CRISPRi screening. Nat. Methods. 2020; 17:86–92. [DOI] [PubMed] [Google Scholar]
- 51. Martens K.J.A., van Beljouw S.P.B., van der Els S., Vink J.N.A., Baas S., Vogelaar G.A., Brouns S.J.J., van Baarlen P., Kleerebezem M., Hohlbein J.. Visualisation of dCas9 target search in vivo using an open-microscopy framework. Nat. Commun. 2019; 10:3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Loinger A., Biham O.. Stochastic simulations of the repressilator circuit. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2007; 76:051917. [DOI] [PubMed] [Google Scholar]
- 53. Ananthasubramaniam B., Herzel H.. Positive feedback promotes oscillations in negative feedback loops. PLoS One. 2014; 9:e104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chan L.Y., Mugler C.F., Heinrich S., Vallotton P., Weis K.. Non-invasive measurement of mRNA decay reveals translation initiation as the major determinant of mRNA stability. Elife. 2018; 7:e32536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Baudrimont A., Jaquet V., Wallerich S., Voegeli S., Becskei A.. Contribution of RNA degradation to intrinsic and extrinsic noise in gene expression. Cell Rep. 2019; 26:3752–3761. [DOI] [PubMed] [Google Scholar]
- 56. Strohkendl I., Saifuddin F.A., Rybarski J.R., Finkelstein I.J., Russell R.. Kinetic basis for DNA target specificity of CRISPR-Cas12a. Mol. Cell. 2018; 71:816–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Westbrook A., Tang X., Marshall R., Maxwell C.S., Chappell J., Agrawal D.K., Dunlop M.J., Noireaux V., Beisel C.L., Lucks J. et al.. Distinct timescales of RNA regulators enable the construction of a genetic pulse generator. Biotechnol. Bioeng. 2019; 116:1139–1151. [DOI] [PubMed] [Google Scholar]
- 58. Campa C.C., Weisbach N.R., Santinha A.J., Incarnato D., Platt R.J.. Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts. Nat. Methods. 2019; 16:887–893. [DOI] [PubMed] [Google Scholar]
- 59. Zhang X., Wang J., Cheng Q., Zheng X., Zhao G., Wang J.. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov. 2017; 3:17018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kim S.K., Kim H., Ahn W.C., Park K.H., Woo E.J., Lee D.H., Lee S.G.. Efficient transcriptional gene repression by Type V-A CRISPR-Cpf1 from Eubacterium eligens. ACS Synth. Biol. 2017; 6:1273–1282. [DOI] [PubMed] [Google Scholar]
- 61. Kleinstiver B.P., Sousa A.A., Walton R.T., Tak Y.E., Hsu J.Y., Clement K., Welch M.M., Horng J.E., Malagon-Lopez J., Scarfo I. et al.. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019; 37:276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Toth E., Varga E., Kulcsar P.I., Kocsis-Jutka V., Krausz S.L., Nyeste A., Welker Z., Huszar K., Ligeti Z., Talas A. et al.. Improved LbCas12a variants with altered PAM specificities further broaden the genome targeting range of Cas12a nucleases. Nucleic Acids Res. 2020; 48:3722–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Singh D., Mallon J., Poddar A., Wang Y., Tippana R., Yang O., Bailey S., Ha T.. Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a). Proc. Natl. Acad. Sci. U.S.A. 2018; 115:5444–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Singh D., Sternberg S.H., Fei J., Doudna J.A., Ha T.. Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nat. Commun. 2016; 7:12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nielsen A.A., Der B.S., Shin J., Vaidyanathan P., Paralanov V., Strychalski E.A., Ross D., Densmore D., Voigt C.A.. Genetic circuit design automation. Science. 2016; 352:aac7341. [DOI] [PubMed] [Google Scholar]
- 66. Taketani M., Zhang J., Zhang S., Triassi A.J., Huang Y.J., Griffith L.G., Voigt C.A.. Genetic circuit design automation for the gut resident species Bacteroides thetaiotaomicron. Nat. Biotechnol. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tu Z., Yang W., Yan S., Yin A., Gao J., Liu X., Zheng Y., Zheng J., Li Z., Yang S. et al.. Promoting Cas9 degradation reduces mosaic mutations in non-human primate embryos. Sci. Rep. 2017; 7:42081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.