Abstract
Biogenesis of mammalian mitochondrial ribosomes (mitoribosomes) involves several conserved small GTPases. Here, we report that the Obg family protein GTPBP5 or MTG2 is a mitochondrial protein whose absence in a TALEN-induced HEK293T knockout (KO) cell line leads to severely decreased levels of the 55S monosome and attenuated mitochondrial protein synthesis. We show that a fraction of GTPBP5 co-sediments with the large mitoribosome subunit (mtLSU), and crosslinks specifically with the 16S rRNA, and several mtLSU proteins and assembly factors. Notably, the latter group includes MTERF4, involved in monosome assembly, and MRM2, the methyltransferase that catalyzes the modification of the 16S mt-rRNA A-loop U1369 residue. The GTPBP5 interaction with MRM2 was also detected using the proximity-dependent biotinylation (BioID) assay. In GTPBP5-KO mitochondria, the mtLSU lacks bL36m, accumulates an excess of the assembly factors MTG1, GTPBP10, MALSU1 and MTERF4, and contains hypomethylated 16S rRNA. We propose that GTPBP5 primarily fuels proper mtLSU maturation by securing efficient methylation of two 16S rRNA residues, and ultimately serves to coordinate subunit joining through the release of late-stage mtLSU assembly factors. In this way, GTPBP5 provides an ultimate quality control checkpoint function during mtLSU assembly that minimizes premature subunit joining to ensure the assembly of the mature 55S monosome.
INTRODUCTION
Ribosomes are macromolecular RNA-protein assemblies that universally catalyze protein synthesis. Within mitochondria, specialized ribosomes (mitoribosomes) drive the synthesis of a small set of proteins encoded in the mitochondrial DNA (mtDNA). In mammals, these are 13 proteins, all of which are essential components of the oxidative phosphorylation (OXPHOS) enzymatic complexes. The mammalian mitoribosome is a 55S RNA–protein complex, formed by a 39S large subunit (mtLSU) composed of 52 mitoribosome proteins (MRPs), a 16S rRNA and a structural tRNA (Val in human cells), and a 28S small subunit (mtSSU) formed by 30 MRPs and a 12S rRNA (1,2). The intricacies of mitoribosome biogenesis, factors involved and quality control checkpoints during the process remain to be fully understood.
All MRPs are encoded in the nuclear genome, synthesized on cytoplasmic ribosomes and imported into the mitochondrial matrix where they assemble with the subunit-specific RNAs, which are encoded in the mtDNA. The dual genetic origin of the mitoribosome components adds complexity to a biogenetic process that requires the assistance of a growing number of non-ribosomal proteins. These proteins include RNA modification enzymes, guanosine triphosphatases (GTPases), DEAD-box RNA helicases and kinases (3,4). They act as assembly factors to guide the processing and modification of mitoribosomal components and their temporal association to form pre-ribosomal particles during the assembly of individual subunits, and formation of the monosome. Mitoribosome biogenesis follows a maturation pathway that is just starting to emerge, and involves the cooperative assembly of protein sets forming structural clusters and preassembled modules (5,6). For each subunit, the protein components are synthesized in excess and imported into mitochondria, where their stoichiometric accumulation is regulated by degradation of the non-assembled free protein fractions (6). The biogenesis of the two mitoribosome subunits is coordinated. It starts co-transcriptionally with mtLSU proteins forming a subcomplex on an unprocessed RNA containing the 16S rRNA, whose formation is required for precursor RNA processing and liberation of the 12S rRNA as a condition for mtSSU protein incorporation (7). Recent investigations have revealed quality control mechanisms that are in place to ensure that only the mature mtSSU and mtLSU are assembled into functional monosomes (4,8–10).
To gain insight into the mitoribosome assembly process, our group and others have focused on the characterization of the mitoribosome accessory proteome to identify relevant assembly factors (4,11–13). These studies have disclosed the potential involvement in mitoribosome subunit assembly of a set of GTPases belonging to several conserved subfamilies. In mammalian mitochondria, two conserved GTPases participate in mtSSU biogenesis; NOA1 (C4orf14, orthologous to bacterial YqeH) (14) and ERAL1 (bacterial Era1) (15). Additionally, at least two other GTPases are required for mtLSU assembly; MTG1 (bacterial RbgA) (9) and GTPBP10 (bacterial ObgE or CgtA) (4,10,16,17). Several other GTPases that potentially serve as additional mitoribosome assembly factors remain largely uncharacterized (4). Among them, GTPBP5 (also called MTG2 or OBGH1) drew our attention because it is a second homolog of bacterial ObgE, whose precise molecular function in human cells remains unknown. The existence of two Obg proteins in human mitochondria differs from Saccharomyces cerevisiae mitochondria, which contain a single Obg-family protein known as Mtg2 (18)
In Bacillus subtilis, Escherichia coli and other bacteria, Obg proteins bind to several 50S LSU riboproteins (19–21), and their GTPase activity is essential for LSU biogenesis (22). Moreover, Obg proteins from E. coli, Salmonella typhimurium, and mitochondria from S. cerevisiae interact physically or genetically with mtLSU rRNA modification enzymes (18,22,23), further linking Obg proteins to LSU maturation. In mammals, GTPBP10 (OBGH2) is a ribosome biogenesis factor of the mtLSU required for late stages of maturation (4,10) and to coordinate mtSSU and mtLSU accumulation, thus providing a quality control function during mtLSU assembly to prevent premature subunit joining (4). GTPBP5 (OBGH1/MTG2) was shown to associate with the mtLSU specifically, and only its intrinsic GTPase activity was detected (16). However, partial GTPBP5 silencing did not produce any phenotype (16,17), which left its specific role in mitoribosome biogenesis unknown.
In this study, we have used gene-editing approaches to generate GTPBP5-knockout (KO) cell lines, whose characterization demonstrated a substantial increase in unassembled 28S and 39S subunits and a drastic decrease in 55S ribosomes, and therefore the ability to synthesize mtDNA-encoded polypeptides. The description of the composition and modification of the mitoribosome subunits in the absence of GTPBP5, with a combination of molecular and biochemical studies of GTPBP5 in wild-type cells, allowed us to conclude that this GTPase acts at the late stages of mtLSU biogenesis by facilitating 16S mt-rRNA modifications, including Um1369 in the A-loop that is catalyzed by the methyltransferase MRM2. Furthermore, GTPBP5 coordinates the release of late-stage assembly factors required for monosome assembly, such as MTG1, GTPBP10, MALSU1 and particularly MTERF4 from the maturing mtLSU particle. In this way, GTPBP5 provides a final quality control checkpoint function during mtLSU assembly that minimizes premature subunit joining to ensure the assembly of the mature 55S monosome.
MATERIALS AND METHODS
Human cell lines and culture conditions
HEK293T embryonic kidney cells (CRL‐3216), and 143B osteosarcoma cells (CRL‐8303) were obtained from ATCC. HEK293 Flp-In T-REx 293 cells were obtained from Invitrogen. The 143B.TK− rho0 derivative (143B206), obtained from Dr Carlos Moraes, was generated by Dr M.P. King (24). The details of the cell lines are listed in Supplementary Table S4. HEK293T cells KO for MTG1, GTPBP10, or DDX28 were previously generated and reported by our group (4,9,11). All cell lines were cultured at 37°C under 5% CO2 in high-glucose, sodium pyruvate, l-glutamine DMEM medium supplemented with 10% FBS and 100 μg/ml uridine (complete DMEM medium). Analysis of mycoplasma contamination was routinely performed. The chemicals and reagents used in this study are listed in the Supplementary Table S8.
Generation of GTPBP5-edited cell lines and plasmid transfection
To create a stable human GTPBP5-knockout (KO) line in HEK293T cells, we used TALEN constructs obtained from Life technologies. The left and right TALENs for the pair were designed to bind the sequences shown in Supplementary Figure S1B. The details of the sequences of the TALEN pairs are listed in Supplementary Table S2. HEK293T cells grown on a six‐well plate at 30% confluency were transfected with 2 μg of each right and left TALEN plasmid as a pair using 5 μl of Lipofectamine 2000 pre-incubated in 300 μl of Opti-MEM (ThermoFisher). After 8 h of incubation, the media was changed to complete DMEM medium. After five times of repeated transfections every three days, cells were collected and sorted by FACS and seeded as single cells in multiple 96-well plates. Upon growth, each clone was tested by immunoblotting to assess the steady-state levels of GTPBP5 as well as the mtDNA-encoded COX2 protein as a surrogate of mitochondrial protein synthesis capacity. Clones that had attenuated GTPBP5 and COX2 levels were further analyzed by genotyping.
To establish stable lines expressing FLAG-tagged or untagged versions of GTPBP5, the constructs were cloned into a mammalian vector with hygromycin as the selection marker, and 2 μg of empty vector, or the construct containing FLAG-tagged GTPBP5 were transfected to HEK293T cells using a standard Lipofectamine 2000 protocol. Two days after transfection, the medium was supplemented with 200 μg/ml hygromycin (Invitrogen) for 3 weeks. Two vectors were used, pCMV6, in which GTPBP5 expression was placed under the control of a standard human cytomegalovirus (CMV) intermediate early enhancer/promoter, and a modified vector with an attenuated CMV promoter (Δ5), generated by a deletion that eliminates most transcription factor binding sites (25). Supplementary Table S5 lists all recombinant DNAs used in this study.
siRNA oligoribonucleotides and transfection
The sources of the Non‐targeting siRNA and siRNAs targeting MTERF4, MALSU1, bL19m, bL36m, mL45 or mS40 are listed in the Supplementary Table S6.
HEK293T cells grown in a six‐well plate at 30% confluency were transfected with the indicated mRNA-targeting siRNA or non‐targeting (NT) control siRNA at a final concentration of 20 nM using 5 μl of Lipofectamine RNAiMAX (ThermoFisher Scientific) according to the manufacturer's specifications.
Isolation of mitochondria
Mitochondria from wild-type (WT) HEK293T, GTPBP5-KO, and each cell line stably expressing GTPBP5-FLAG were isolated as described previously (26). Briefly, cells were harvested by centrifugation at 1000 rpm for 5 min. The pellet was washed twice with pre-cooled 1× PBS. Following the weighing of the pellet, it was resuspended in pre-cooled buffer (0.15 g/ml; 10 mM Tris–HCl, pH 7.0, 10 mM KCl, 0.15 mM MgCl2), and incubated for 5 min on ice. The cells were lysed with 10 strokes with a glass homogenizer (Kimble/Kontes, Vineland, NJ, USA), and the cell homogenate was brought to a final concentration of 0.25 M sucrose by adding the buffer (1 M sucrose, 10 mM Tris–HCl, pH 7.0). To separate the cell debris and nuclei from mitochondria, the cell homogenate was centrifuged at 1500 g for 3 min, and the supernatant was collected and further centrifuged at 8000 g for 10 min. The crude mitochondrial pellet was gently resuspended in buffer containing 0.6 M sorbitol, 10 mM HEPES, pH 7.4, snap-frozen in liquid nitrogen and stored at -80°C.
Pulse labeling of mitochondrial translation products
Mitochondrial protein synthesis was determined by pulse‐labeling of 60-70% confluent cultures in medium without methionine and in the presence of 100 μg/ml emetine (for pulse) and anisomycin (for chase) to inhibit cytoplasmic protein synthesis as described (27,28). Cells were labeled for 30 min at 37°C with 100 μCi of 35S-Methionine (PerkinElmer Life Sciences, Boston, MA, USA). After incubation, the medium with 35S-Methionine was removed, and the cells were incubated in complete DMEM medium for 5 min at 37°C (For Pulse). For Chase, the cells were washed twice with 1× PBS (to remove the inhibitor) and incubated in complete DMEM medium for the indicated time frame at 37°C. After the brief incubation (for pulse) and the desired time frame (for chase), the cells were washed once with 1× PBS (phosphate-buffered saline), collected by trypsinization, and whole‐cell extracts were prepared by solubilization in RIPA buffer ((1% NP-40, 0.1% SDS, 0.5% Na-deoxycholate, 150 mM NaCl, 2 mM EDTA (ethylenediaminetetraacetic acid) and 50 mM Tris-HCl, pH 7.4)) supplemented with 1 mM PMSF (phenylmethylsulfonyl fluoride) and 1× EDTA-free mammalian protease inhibitor cocktail (Roche). Fifty to hundred micrograms of each sample were separated by SDS (sodium dodecyl sulfate)-PAGE on a 17.5% polyacrylamide gel, transferred to a nitrocellulose membrane, and exposed to Kodak X-OMAT X-ray film. The membranes were then probed with primary antibodies against ß-ACTIN or mitochondrial protein TOM20 as loading controls.
SDS-PAGE and immunoblotting analyses
To estimate steady-state levels of individual proteins, whole-cell protein extracts (WCE) or isolated mitochondria were analyzed by denaturing SDS-PAGE and immunoblotting. Total cellular proteins were extracted in RIPA buffer ((1% NP-40, 0.1% SDS, 0.5% Na-deoxycholate, 150 mM NaCl, 2 mM EDTA and 50 mM Tris-HCl, pH 7.4)) with freshly added 1 mM PMSF and 1× EDTA-free protease inhibitor cocktail (Roche). Mitochondria were isolated as explained earlier. Protein concentration in WCE and mitochondrial preparations was measured with the Folin phenol reagent (29). In general, 30-40 μg of mitochondrial proteins or WCE were separated by SDS-PAGE in the Laemmli buffer system (30). After transfer to a nitrocellulose membrane, the membranes with immobilized proteins were blocked with 5% skim milk and then incubated with antibodies against the indicated proteins followed by a second reaction with anti-mouse or anti-rabbit IgG conjugated to horseradish peroxidase. All the antibodies used in this study are listed in Supplementary Table S3. Signals were detected by chemiluminescence incubation and X-ray film exposure. Upon trials of anti-GTPBP5 antibodies from several companies, the Prestige polyclonal antibody raised in rabbit and provided by Sigma (Cat # HPA047379) was considered the best. However, differences in performance were found between two lots, one of which detected a very prominent cross-reacting band.
Sucrose gradient analysis
The sedimentation properties of GTPBP5 and mitoribosomal proteins were analyzed by sucrose gradient sedimentation. From 1.5 to 2 mg of mitochondria isolated from WT or GTPBP5-KO cells were solubilized in lysis buffer containing 260 mM sucrose, 100 mM KCl, 20 mM MgCl2, 10 mM Tris-HCl, pH 7.5, 1% Triton X100, 1× EDTA-free protease inhibitor cocktail, 0.08 U/μl RNAsin and incubated on ice for 20 min. The lysate was centrifuged at 9200 g for 45 min at 4°C, and the supernatant was loaded onto a 12.5 ml linear 10-30% sucrose gradient containing 10 mM Tris-HCl, pH 7.5, 100 mM KCl, 20 mM MgCl2 1× EDTA-free protease inhibitor cocktail, and centrifuged at 24 000 rpm for 15 h at 4°C in SW 41Ti (Beckman Coulter). The gradients were collected from the top into 15 or 16 fractions, TCA (trichloroacetic acid)-precipitated and the pellets resuspended in 1× Laemmli buffer and analyzed by immunoblotting. The continuous RNA profile (absorbance at 254 nm) obtained during gradient collection using a Brandel fractionation system and Brandel PeakChart software version 2.08.
For mass spectrometry analysis, fractions containing monosome, mtLSU, and mtSSU, confirmed by immunoblotting, were pooled and precipitated with methanol/chloroform and analyzed at the Keck Biotechnology Resource Laboratory (Yale University School of Medicine, New Haven, CT, USA).
RNA analysis by northern blot and qPCR
Total RNA was prepared from whole cells by using Trizol (Ambion) as described previously (31). Briefly, for northern blot analyses, 2-3 μg of RNA was mixed with 10 μl of RNA sample loading buffer (Sigma), and separated on a denaturing 1.2% agarose gel containing 5.49 M formaldehyde. The quality of the RNA was assessed under UV using the BioRad ChemiDoc MP Imaging System prior to transferring the RNA onto a nylon membrane (Nytran®, SuPerCharge, GE Healthcare Life Science Whatman). The RNA bound to the nylon membrane was cross-linked with UV light at 254 nm and pre-hybridized in ultrahybridization ultrasensitive hybridization buffer (Invitrogen) at 42°C for 1 h. Subsequently, 100 ng of the specific DNA probes were added to the membrane and allowed to hybridize overnight at 42°C. The probes were created by PCR on genomic DNA, column-purified (Promega) and labeled with [α-32P] dATP by random priming. For qRT-PCR analysis of the steady-state levels of mitochondrial RNAs, total RNA was extracted from whole cells by using Trizol, followed by DNAse digestion (Invitrogen) and reverse-transcribed to cDNA (Applied Biosystems) and analyzed with specific primer pairs for each gene following standard procedures. Primers used in this study are listed in the Supplementary Table S7.
RNA methylation analysis
Methylation of 16S rRNA at U1369 and G1370 was analyzed by reverse transcription primer extension. Briefly, 2 μg of total cellular RNA from control and GTPBP5-KO cells were extracted as described above and annealed to [32P] 5′-end labeled primer specific to the 5′-end of the 16S rRNA (p5LSU: 5′- CTGGTAGTAAGGTGGAGTG-3′) or to regions near the 16S methylated sites of interest p1369/70, pAL: 5′-GATCACGTAGGACTTTAATC-3′). The primers were extended with SuperScript™ III Reverse Transcriptase (Invitrogen) according to the manufacturer's protocol. Primer extension products were mixed with RNA loading buffer from Sigma; separated on 12% polyacrylamide/7M urea sequencing gel (SequaGel Sequencing System, National Diagnostics); and visualized by exposition to an X-ray film. The same experimental conditions were used with two different dNTPs concentrations (0.04 and 1 mM) to identify the methylation sites on the 16S rRNA. RNA from HEK293T cells treated with siRNA to knockdown MRM2 expression was used as a positive control.
To precisely quantify the 16S rRNA 2′-O-methylations by RiboMethSeq, we used Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA (32). Briefly, total RNA isolated from human cells (150 ng) was subjected to alkaline hydrolysis in 50 mM bicarbonate buffer pH 9.2 for 14 min at 95°C. The reaction was stopped by ethanol precipitation using 3 M Na-OAc, pH 5.2 and glycoblue as a carrier in liquid nitrogen. After centrifugation, the pellet was washed with 80% ethanol and resuspended in nuclease-free water. The sizes of generated RNA fragments were assessed by capillary electrophoresis using a PicoRNA chip on Bioanalyzer 2100 (Agilent, USA) and were ranging from 30 to 200 nt. RNA fragments were directly 3′-end dephosphorylated using 5 U of Antarctic Phosphatase (NEB, UK) for 30 min at 37°C. After phosphatase inactivation for 5 min at 70°C, RNA fragments were phosphorylated at the 5′-end using T4 PNK and 1 mM ATP for 1 h at 37°C. End-repaired RNA fragments were then purified using RNeasy MinElute Cleanup kit (QIAGEN, Germany) according to the manufacturer's recommendations except that 675 μl of 96% ethanol were used for RNA binding. Elution was performed in 10 μl of nuclease-free water. RNA fragments were converted to library using NEBNext® Small RNA Library kit (NEB ref E7330S, UK) following the manufacturer's specifications. DNA library quality was assessed using a High Sensitivity DNA chip on a Bioanalyzer 2100. Library quantification was done using a fluorometer (Qubit 2.0 fluorometer, Invitrogen, USA). Libraries were multiplexed and subjected for high-throughput sequencing using an Illumina HiSeq 1000 instrument with a 50 bp single-end read mode. The bioinformatics analyses involved initial trimming of adapter sequence using Trimmomatic-0.32, alignment to the reference rRNA sequence by Bowtie2 (ver 2.2.4) in End-to-End mode, and k = 1. Simultaneous 5′- and 3′- ends counting was done by bedtools v2.25.0 after conversion to *.bed file. Quantification of RNA methylation was performed by calculation of MethScore (ScoreC) in ±2 nt region (33). Um1369 was also analyzed in synthetic oligos carrying or not the modified nucleotide. However, Um1369 was more ‘protected’ in siMRM2 samples than for the synthetic oligo (most probably due to incomplete modification at Gm1370 during chemical synthesis). For this reason, the MethScore for Um1369 was converted to real methylation level by using siMRM2 RNA as a reference of totally unmodified RNA.
Immunoprecipitation assays
For native interactome analysis, one mg of mitochondria isolated from HEK293T cells or GTPBP5-KO cells stably expressing GTPBP5-FLAG were solubilized in extraction buffer (50 mM Tris-HCl, pH 7.5, 100 mM KCl, 20 mM MgCl2, 1% digitonin, 10% glycerol, 1 mM PMSF, 1× EDTA-free protease inhibitor cocktail) to a final concentration of one mg/ml and incubated for 30 min at 4°C. The lysates were cleared by centrifugation at 16 000 g for 10 min at 4°C. The lysate was incubated with 50 μl of washed recombinant ProteinA agarose beads (control) or Anti-Flag M2 affinity gel beads at 4°C for 1-2 h with gentle rotation. Flag Tag (D6W5B) Rabbit mAb-conjugated sepharose beads were specifically used for pulldown assays to analyze the interaction of GTPBP5-FLAG with MRM2. The supernatant containing unbound material was subsequently collected, and the beads were washed three to four times with wash buffer (50 mM Tris-HCl, pH 7.5, 100 mM KCl, 20 mM MgCl2, 10% glycerol, 1 mM PMSF, 1× EDTA-protease inhibitor cocktail). The bound proteins were eluted using 1× Laemmli buffer containing 300 mM DTT and incubated at 95°C for 5 min.
GTPBP5 interactome analysis
For specific interacting partners analysis, one mg of mitochondria isolated from HEK293T cells after stably expressing GTPBP5-FLAG were incubated for 2 h at 4°C in the presence of 1 mM cleavable crosslinker DSP (dithiobis[succinimidylpropionate]) (Thermo Scientific) or the vehicle DMSO as a negative control, and then the reactions were quenched by adding 50 mM Tris (pH 7.5) and incubated for 15 min at 4°C. DSP has NHS-ester groups, which crosslink amino groups with a 12 Å spacer arm. The mitochondria was pelleted by centrifugation at 8000 g for 10 min at 4°C and was solubilized in 600 μl of extraction buffer ((1× PBS, 1% lauryl-β-d-maltoside (LM) and 1× EDTA-free protease inhibitor cocktail)) or 150 μl of extraction buffer (1× PBS, 1% SDS and 1× EDTA-free protease inhibitor cocktail) and incubated for 10 min at 4°C. The extract with 1% SDS was diluted to 0.1% SDS. The lysates were cleared by centrifugation at 15 000 rpm (maximum speed on benchtop centrifuge) for 30 min at 4°C. The lysate was incubated with 50 μl of washed Flag Tag (D6W5B) Rabbit mAb-conjugated sepharose beads for 2 h at 4°C with gentle rotation. The supernatant containing unbound material was subsequently collected, and the beads were washed two to three times with wash buffer (1× PBS, 0.1% LM or 0.05% SDS and 1X EDTA-protease inhibitor cocktail). The bound proteins were eluted using 1× Laemmli buffer containing 8% ßME and incubated at 65°C for 1 h. The flowthrough or unbound fraction and the input fraction were treated with 1× Laemmli buffer containing 8% ßME and incubated at 37°C for 1 h.
BioID proximity labeling
The BirA*-FLAG construct was generated using Gateway cloning into the pDEST5-BirA*-FLAG-C-ter vector with GTPBP5 lacking stop codon in pENTR223 (HsCD00288276, DNASU). Flp-In T-REx 293 cells were seeded at 250 000 cells per well in a six-well plate in 2 ml DMEM supplemented with 10% FBS and 1% Pen/Strep (100 U/ml). The next day, cells were transfected with 200 ng pDEST5-ProteinX-BirA*-FLAG, and 2 μg of pOG44 in 250 μl of 1× Opti-MEM (Invitrogen) mixed with 5 μl of Lipofectamine 2000 reagent in 250 μl of 1X Opti-MEM. The medium was changed 4 h after transfection. On the following day, transfected cells were passaged into 10 cm2 plates, and selection medium containing hygromycin (Wisent) at a final concentration of 200 μg/ml was added. The selection media was changed every 2–3 days until clear visible colonies were present. Six colonies were picked and expanded, and the localization and the expression level of the construct were assessed by immunofluorescence using an anti-FLAG antibody. A single clone was selected and scaled up to six 15 cm2 plates for treatment and harvesting. Cells were grown to 70% confluency before induction of protein expression using 1 μg/ml tetracycline (Sigma), and media supplementation with 50 μM biotin for protein labeling. Cells were harvested 24 h later as follows: cell medium was decanted; cells were washed twice with 5 ml PBS per 15 cm2 plate and then harvested by scraping in 5 ml of PBS. Cells from 3 × 15 cm2 plates were pelleted at 800 rpm for 3 min, PBS aspirated, and pellets transferred to a -80°C freezer. Purification of biotinylated proteins followed by their identification by mass spectrometry, was performed in biological duplicates (as defined by two separate harvests) described previously (34), and data were compared to negative controls expressing the BirA* tag fused to GFP, and un-transfected cells using SAINTexpress with default parameters (35). Forty eight independent negative controls were performed; to increase the stringency in scoring, the 24 maximal spectral counts across the 48 controls were used for each prey protein, generating 24 ‘virtual controls’ against which the GTPBP5 results were scored. The dataset was jointly analyzed within a larger group of 100 mitochondrial baits. ((Antonicka et al., published on bioRxiv 2020.04.01.020479; (https://doi.org/10.1101/2020.04.01.020479)). All preys detected with GTPBP5 which passed a Bayesian False Discovery Rare (BFDR) cutoff of ≤0.01 are listed in Supplementary Table S1. The prey specificity module of the ProHits-viz software (prohits-viz.lunenfeld.ca) was used to score specific prey enrichment with GTPBP5 against all other mitochondrial baits profiled, using spectral counts as a proxy for relative abundance. GO term analysis was performed using PANTHER Overrepresentation Test (pantherdb.org; Released 20190711).
GTPBP5-RNA interaction analysis
The protocol was modified from the PAR-CLIP assay (36). Cells stably expressing Flag-tagged GTPBP5 proteins were grown in a 15 cm plate in medium supplemented with 100 μM 4-thiouridine (SU) for 15-17 h and subjected to UV-mediated protein-RNA crosslinking once with 150 mJ/cm2 at 365 nm using a Stratalinker UV crosslinker (Stratagene). The cells were collected by scraping off the plate and washed once with 1× PBS before disrupting them with 500 μl of 1% NP40 lysis buffer (1× PBS, 1% NP40, 0.1% SDS, 0.5% sodium deoxycholate) supplemented with 1× EDTA-free protease inhibitor cocktail (Roche), and 10 mM ribonucleoside vanadyl complexes (NEB) for 10 min at 4°C. 4SU-labeled non-irradiated cells were used as control. After incubation for 10 min at 4°C the lysate was centrifuged at 30 000 rpm for 20 min at 4°C in a TLA 100 rotor, and the eluted protein-RNA complexes were applied for immunoprecipitation with anti-FLAG M2 beads or agarose beads for 4-5 h at 4°C. Bound protein-RNA complexes were washed twice with high salt buffer (50 mM Tris–HCl pH 7.4, 1 M NaCl, 1 mM EDTA, 1% NP40, 0.1% SDS, 0.5% sodium deoxycholate) and three times with wash buffer (PBS, 0.02% Tween 20, 1× EDTA-free protease inhibitor cocktail and 10 mM ribonucleoside vanadyl complexes). The eluate was further treated with DNase I digestion for 30 min at 37°C, and RNA was extracted from the beads by proteinase K treatment and Trizol extraction (Ambion). RNA extracts were converted to cDNA and analyzed by quantitative PCR analysis using SYBR Green (Biorad) and specific primer pairs for each gene following standard procedures. Primers used in this study are listed in the Supplementary Table S7.
Mitochondrial respiratory chain (MRC) enzyme activities
The activity of the MRC complex CIV was determined spectrophotometrically in 3× frozen-thawed cells, as described previously (Barrientos et al., 2009). The rate of complex CIV activity was normalized by the amount of total protein in the assay.
GTPBP5 localization experiments
Two hundred μg of mitochondria from HEK293T were briefly sonicated, and then soluble and insoluble fractions were obtained by centrifugation at 35 000 rpm for 15 min at 4°C. The membrane pellet was then resuspended in 200 mM Na2CO3, pH 11.5. After 30 min on ice, the sample was centrifuged at 35 000 rpm for 15 min at 4°C to separate the soluble extrinsic from the insoluble intrinsic membrane proteins. Two hundred μg of purified mitochondria were also resuspended in buffer (10 mM HEPES pH 7.4) with or without 0.6 M sorbitol to allow mitochondrial swelling and conversion to mitoplasts. Where indicated, 0.5 μl of 0.63 mg/ml proteinase K for 60 min on ice. 0.2 M PMSF was added to stop the reaction of Proteinase K and mitochondria and mitoplasts were recovered by centrifugation at 20 000 rpm for 15 min at 4°C and analyzed by immunoblotting. Cytoplasmic (C) and nuclear (N) fractions extraction were performed following manufacturer's protocol using the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit from Thermo Scientific.
Immunocytochemistry
Wild-type and cells overexpressing Flag-tagged GTPBP5 HEK293T cells were plated on poly-l-lysine (Sigma) coated coverslips and grown overnight at 37°C. Cells were stained for 30 min with 50 nM Mitotracker red (Molecular Probes, Invitrogen). The cells were washed once with 1× PBS and then fixed with 4% paraformaldehyde for 10 min at room temperature. After rapid washing three times with 1× PBS, cells were treated with 0.1 M glycine, followed by rapid washing two times with 1× PBS and then treated with 0.1% Triton X-100 for 10 min at room temperature. The cells were further washed rapidly two times with 1× PBS and blocked with 2% bovine serum albumin (BSA) and 2% normal goat serum for 45 min at room temperature. Next, the cells were incubated with an anti-FLAG primary monoclonal antibody (dilution 1:50) in 0.2% BSA and 1% normal goat serum for 45 min at room temperature. Following primary antibody incubation, the cells were washed twice with 1× PBS each for 5 min at room temperature. The secondary antibody Alexa Fluor™ Plus 488, green (Invitrogen), was added at a dilution of 1:200 and incubated for 2 h at room temperature. The cells were washed three times with 1× PBS each for 5 min, and then the slides were mounted in Vectashield mounting medium with DAPI (Vector Laboratories Inc) that were used for immunofluorescence detection. The images were captured using Zeiss 710 confocal microscope at 63X magnification, 1.3 ZOOM.
Statistical analysis
All of the experiments were done in triplicate or otherwise indicated. Data in X-ray films were digitalized and analyzed using the ImageJ software or the Adobe Photoshop histogram option. Statistical analyses were performed using the Prism-5/6 software and Microsoft Excel. The details of the software used in this study are listed in Supplementary Table S9. The data are presented as the means ± S.D. or mean ± S.E.M. of absolute values or percentages of control. The values obtained for WT and the GTPBP5-KO strains for the different parameters studied were compared using a Student's two-tailed unpaired t-test for comparison of two groups. For comparison of multiple groups, we performed two-way analysis of variance (ANOVA) followed by a Dunnett's multiple comparisons test: (*P < 0.05; **P < 0.01; ***P < 0.001).
Key reagents
Tables presenting the list of sequences of TALEN pairs, cell lines, antibodies, recombinant DNAs, siRNA oligonucleotides, oligoribonucleotides for qPCR, chemicals, and reagents and software used in this study are included in the Supplementary Tables S2–S9.
RESULTS
GTPBP5 is a mitochondrial soluble/peripheral inner membrane protein that co-sediments with the mtLSU
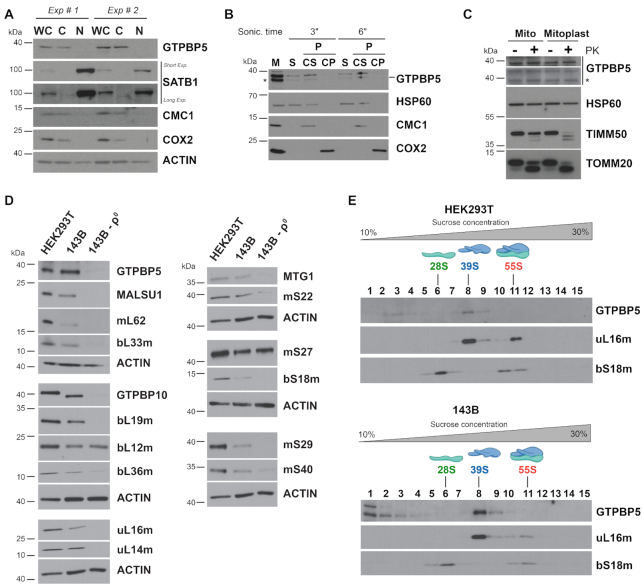
Human GTPBP5 was previously localized to mitochondria in HeLa cells (16). We used several approaches to confirm and extend these observations in HEK293T cells. Fluorescence immunocytochemistry showed co-localization of FLAG-tagged GTPBP5 with Mito-tracker Red that stains mitochondria in live cells (Supplementary Figure S1A). The cytoplasmic localization of GTPBP5 was confirmed by a cell fractionation assay showing that GTPBP5 follows a pattern similar to other mitochondrial proteins such as CMC1 and COX2 and not like the nuclear protein SATB1 (Figure 1A). To determine whether GTPBP5 is a mitochondrial soluble, peripheral or integral membrane protein, we used isolated mitochondria, which, upon brief sonication, allows the separation of soluble and insoluble fractions by centrifugation, followed by alkaline carbonate (pH 11.5) extraction of the pellet to further separate peripheral and integral membrane proteins. GTPBP5 was detected in the fraction of extrinsic proteins present in the supernatant after alkaline carbonate extraction (CS in Figure 1B), which indicates that GTPBP5 is loosely associated with mitochondrial membranes. Finally, to unveil the submitochondrial localization and topology of GTPBP5, we performed a proteinase K protection assay (Figure 1C) that showed that GTPBP5 is protected against degradation in both mitochondria and mitoplasts generated by hypotonic swelling of mitochondria, similar to the mitochondrial matrix protein HSP60. These data (Figure 1A-C) lead us to conclude that GTPBP5 is a mitochondrial matrix protein loosely associated with the inner membrane, the location, and behavior expected of a ribosome assembly factor.
Figure 1.
GTPBP5 is a mitochondrial protein that sediments with the mitoribosome large subunit (mtLSU). (A) Immunoblot analyses of GTPBP5 levels in HEK293T whole-cell lysate (WCL), cytoplasmic (C) and nuclear (N) fractions. Antibodies against mitochondrial proteins CMC1 and COX2 and nuclear protein SATB1 were used as controls. (B) Mitochondria (M) isolated from HEK293T cells were first fractionated into soluble (S) and membrane-bound (P) proteins by brief sonication and centrifugation. The pellet was submitted to alkaline carbonate extraction (pH: 11.5) to allow the separation of the extrinsic proteins present in the supernatant (CS) from the intrinsic proteins in the pellet (CP). Equivalent volumes of each fraction were analyzed by immunoblotting using antibodies against GTPBP5, the matrix-soluble protein HSP60, the extrinsic membrane-associated protein CMC1, and the inner membrane intrinsic protein COX2. In the anti-GTPBP5 immunoblot, the asterisks mark the GTPBP5 protein. (C) Proteinase K protection assay in mitochondria (Mt) and mitoplasts (Mp) prepared by hypotonic swelling of mitochondria. The samples were analyzed by immunoblotting using antibodies against GTPBP5, the matrix protein HSP60, the inner membrane protein TIM50, and the outer membrane protein TOM20. In the anti-GTPBP5 immunoblot, the asterisk marks the GTPBP5 protein. (D) Immunoblot analyses of the steady-state levels of mitoribosome LSU, SSU proteins and assembly factors in whole-cell extracts from HEK293T, 143B and 143B-206 rho0 (ρ0) cells. ACTIN was used as a loading control. (E) Sucrose gradient sedimentation analyses of GTPBP5 and mitoribosomal proteins in mitochondrial extracts from wild-type HEK293T or 143B mitochondria prepared in the presence of the 20 mM MgCl2. The fractions were analyzed by immunoblotting using antibodies against the indicated proteins.
To test whether GTPBP5 stability depends on the presence of mitochondrial ribosomes, we estimated GTPBP5 steady-state levels in a derivative of an osteosarcoma 143B cell line devoid of mtDNA (rho° cells) and therefore of all mtRNAs including rRNAs. Several ribosome proteins tested were not detected in rho° cells (uL16m, bL19m, bS18m); others had decreased levels (bL36m, mS22) and some accumulated as in wild-type cells, despite being rRNA-interacting proteins (bL12m and mS27) (Figure 1D). Only traces of GTPBP5 were detected. Similar behavior was observed for established mitoribosome assembly factors, including GTPBP10, MALSU1, or MTG1 (Figure 1D). These data could link GTPBP5 to mitoribosome biogenesis, as for its bacterial homologs, but in principle, also to mtDNA or mtRNA transactions.
To explore a possible association of GTPBP5 with the mitoribosome, we used sucrose gradient sedimentation analysis of mitochondrial extracts prepared in the presence of 20 mM Mg2+ to preserve ribosome subunit interactions within monosomes (Figure 1E). The GTPBP5 sedimentation pattern was similar in mitochondrial extracts prepared from either HEK293T or 143B WT cells. In both samples a portion of GTPBP5 (∼60% of total) co-sedimented with mtLSU markers in the mtLSU-containing fractions, whereas the rest accumulated in lighter fractions and did not co-sediment with the mtSSU or the monosome, suggesting a role for GTPBP5 in mtLSU biogenesis.
GTPBP5 is required for efficient mitochondrial translation and OXPHOS function in HEK293T cells
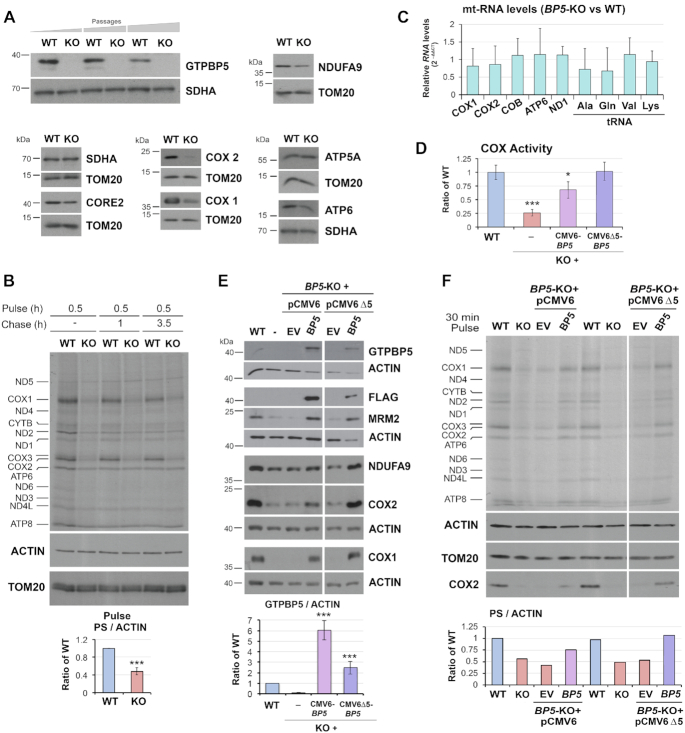
To gain insight into the possible role of GTPBP5 in mitochondrial translation and mitoribosome assembly, we generated a GTPBP5 knockout (KO) in HEK293T cells using a transcription activator-like effector nuclease (TALEN) - mediated gene-editing approach (Supplementary Figure S1B). By screening nearly 100 TALEN transformants by immunoblot against GTPBP5 and subsequent genotyping, we were able to identify one homozygous KO (Figure 2A and Supplementary Figure S1C). The KO clone grows reasonably well in glucose-rich medium, which becomes acidified only slightly faster than for WT cell, thus suggesting the retention of some residual respiratory capacity.
Figure 2.
GTPBP5 is required for efficient mitochondrial translation and OXPHOS function in HEK293T cells. (A) Immunoblot analysis of the steady‐state levels of GTPBP5 and OXPHOS complex subunits in HEK293T (WT), and GTPBP5-knockout (KO) cell lines. For GTPBP5, KO cells collected at three different passages are presented. NDUFA9 is a subunit of complex I, CORE2 of complex III, COX1 and COX2 of complex IV, ATP5α and ATP6 of the F1Fo‐ATP synthase of complex V. Immunoblotting for SDHA and TOM20 are used as loading controls. (B) Metabolic labeling with 35S-methionine of newly synthesized mitochondrial translation products in whole cells from the indicated lines during a 30-min pulse in the presence of emetine to inhibit cytoplasmic protein synthesis. Immunoblotting for ACTIN and TOM20 were used as loading controls. Newly synthesized polypeptides are identified on the left. Please see below in panel 2F for the description of the quantification. (C) Quantitative PCR (qPCR) analyses of the steady-state levels of several mtDNA-encoded mRNAs (COX1, COX2, COB, ATP6, and ND1) and tRNAs (Ala, Gln, Val, and Lys) in WT and the GTPBP5-KO cells. Data represent the mean ± SEM from four WT and GTPBP5-KO samples. t-test: * P <0.05, **P <0.01; ***P <0.001. (D) Enzymatic activity of CIV or cytochrome c oxidase (COX) normalized by whole-cell protein concentration and expressed as a fraction of WT. GTPBP5-KO (KO) cells transfected with an empty vector (EV) or a construct expressing FLAG-tagged GTPBP5. GTPBP5-FLAG was expressed under the control of either standard human cytomegalovirus (CMV) intermediate early enhancer/promoter (plasmid pCMV6) or an attenuated CMV promoter (pCMV6Δ5), generated by a deletion that eliminates a large proportion (4/5) of the transcription factor binding sites (25). Data represent the mean ± SD of three independent repetitions; t-test: * P < 0.05, **P < 0.01; ***P < 0.001. (E) Immunoblot analysis of the steady‐state levels of the indicated proteins in HEK293T wild-type (WT) and GTPBP5-KO (KO) cells transfected with an empty vector (EV) or a construct expressing FLAG-tagged GTPBP5 (pCMV6 or pCMV6Δ5). Actin was used as a loading control. Data represent the mean ± SD of four independent repetitions; t-test: * P < 0.05, **P < 0.01; ***P < 0.001. (F) Metabolic labeling as in panel (B) using GTPBP5-KO (KO) cells reconstituted with a construct expressing FLAG-tagged GTPBP5 under the control of either a standard CMV promoter (pCMV6) or a truncated promoter (pCMV6Δ5). In panels (B) and (F), the overall 35S signal of the pulses was quantified by densitometric integration of the lines of the total signal of the mitochondrial protein synthesis (PS) of each sample using the histogram panel of Adobe Photoshop, normalized by the signal of ACTIN immunobloting and plotted as the ratio of the WT in the bottom graphs. For the graph in panel (B), data represent the mean ± SD of only the pulse phase of mitochondrial protein synthesis (PS) of three independent repetitions; t-test: * P < 0.05, **P < 0.01; ***P < 0.001.
GTPBP5-KO HEK293T cells exhibited decreased steady-state levels of nDNA and mtDNA-encoded OXPHOS components, including NDUFA9 (a CI subunit, nDNA encoded), COX1 and COX2 (two CIV subunits), and ATP6 (a CV subunit) as analyzed by denaturing immunoblotting (Figure 2A). This deficiency stems from a ∼60% decrease in the capacity of GTPBP5-KO cells to undergo mitochondrial protein synthesis (Figure 2B), which is not due to a general decrease in mitochondrial mRNA and tRNA levels (Figure 2C). These data suggest a role for GTPBP5 in the biogenesis and function of the mitochondrial translation machinery, and consequently, for the biogenesis and function of mitochondrial OXPHOS complexes. As a marker, the activity of MRC complex IV (cytochrome c oxidase) was significantly decreased in the absence of GTPBP5 (Figure 2D).
The attenuated MRC complex IV activity, the steady-state levels of mtDNA-encoded OXPHOS subunits, and the mitochondrial translation-deficient phenotype were restored by expression of wild-type levels of recombinant FLAG-tagged GTPBP5 (Figure 2D-F), thus minimizing the possibility of TALEN-induced off-target effects. However, to fully understand the function of GTPBP5, it is essential to mention that excess of GTPBP5 is deleterious. During the reconstitution assays, we used two kinds of constructs, in which GTPBP5-FLAG expression was under either the control of standard human cytomegalovirus (CMV) intermediate early enhancer-promoter (plasmid pCMV6), or from an attenuated CMV promoter (Δ5) (25). When we used pCMV6-Δ5 promoter, near endogenous levels of GTPBP5-FLAG (∼2.5-fold) were expressed and full complementation of the mitochondrial protein synthesis rate and steady-state of mtDNA-encoded OXPHOS subunits was achieved (Figure 2D-F). However, when using pCMV6, GTPBP5-FLAG levels were ∼6.5-fold of WT endogenous GTPBP5 levels (Figure 2E), the protein synthesis defect was not fully complemented (Figure 2F), and the restoration of MRC complex IV activity (Figure 2D) and OXPHOS enzyme subunit levels were only partial (see levels of COX1 and COX2 in Figure 2E and F), suggesting a dominant negative effect of the overexpressed protein. Consistent with this interpretation, expression of GTPBP5 from a pCMV6 promoter in WT cells also attenuated OXPHOS enzyme subunit levels (Supplementary Figure S2). Therefore, for all the complementation experiments in this study, we used the construct with the CMV6-Δ5 promoter, if not otherwise indicated. The deleterious effect of GTPBP5 overexpression is reminiscent of the effect we and others have reported for its homolog GTPBP10 (4,10), as it will be discussed later. The two human OBG proteins, however, do not seem to play redundant roles because the enhancement of GTPBP10 expression (controlled by either the full or truncated CMV6 promoter) did not complement the deleterious phenotype of the GTPBP5-KO cell line (Supplementary Figure S3A and B).
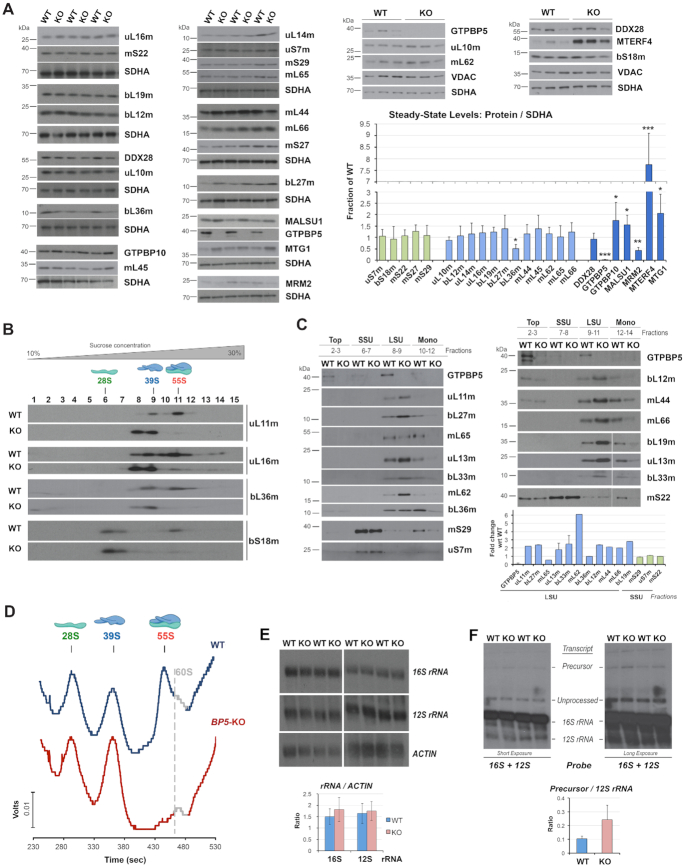
GTPBP5 is essential for the formation of functional mtLSU and monosomes
To explore whether the absence of GTPBP5 affects mitoribosome accumulation, we first measured the steady-state levels of mtLSU and mtSSU subunit markers, as well as known mitoribosome assembly factors (Figure 3A). Immunoblot analyses showed that the late assembly mtLSU protein bL36m was significantly attenuated in GTPBP5-KO mitochondria, whereas all other mtLSU proteins tested were unchanged (Figure 3A). Regarding mtLSU assembly factors, the levels of the 16S rRNA methyltransferase MRM2 were decreased to ∼35% in GTPBP5-KO mitochondria (Figures 2E and 3A), whereas the levels of MALSU1, MTG1 and GTPBP10 were enhanced by 1.5-2- fold of WT, and the levels of MTERF4 were elevated by 7-fold (Figure 3A).
Figure 3.
GTPBP5 is essential for the formation of monosomes. (A) Immunoblot analysis of the steady‐state levels of mitoribosome proteins and assembly factors in mitochondria isolated from HEK293T (WT) and GTPBP5-KO (KO) cells. SDHA or VDAC were used as loading controls. The right panel shows the densitometry values normalized by the signal of SDHA and expressed relative to the WT. Data represent the mean ± SD of at least three WT and GTPBP5-KO samples; t-test: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (B) Sucrose gradient sedimentation analyses of mitoribosome mtSSU (bS18m) and mtLSU (uL11m, uL16m and bL36m) markers in mitochondria prepared from HEK293T (WT) or GTPBP5-KO (KO) cells. (C) Immunoblot analysis of MRP levels in the sucrose gradients fractions presented in panel (B) in which the monosome, mtLSU, mtSSU, and unassembled subunits (top) peak. Equal volumes of fractions corresponding to each cell line were loaded. The lower panel shows the densitometry values of the proteins in either the mtLSU or mtSSU fraction and expressed as fold change relative to the WT. Data represent only one set of each protein and the mean and range of two sets of GTPBP5, uL13m and bL33m. (D) Continuous RNA profile (absorbance at 254 nm) obtained during the collection of the gradient fractions presented in panel (B), using a Brandel fractionation system and Brandel Peak Chart Software. The fractions where the 28S mtSSU, 39S mtLSU, and 55S monosome sediment are indicated. The presence of traces of contaminating 60S cytoplasmic ribosomes is marked in grey. (E, F) Northern blot analyses of the steady-state levels of mitochondrial rRNAs in WT or the GTPBP5-KO cells. Multiple experimental repetitions and X-ray film exposures are presented to display the steady-state levels of 12S-16S precursor transcript, 12S, and 16S unprocessed and fully processed transcripts. The lower panels show the densitometry values normalized by the signal of ACTIN mRNA or the 12S rRNA. Data represent the mean ± SD of three independent repetitions.
Subsequently, we followed the formation of the mtLSU ribonucleoprotein particle in GTPBP5-KO mitochondria by sucrose gradient sedimentation analysis of mitochondrial extracts prepared in the presence of magnesium. The most apparent effect of the absence of GTPBP5 was a profound decrease of 55S monosomes (Figure 3B-D) and the concomitant accumulation of the mtLSU subunit.
Otherwise, the sedimentation pattern of the mtLSU and mtSSU protein markers within the individual subunits was unchanged in GTPBP5-KO cells (Figure 3B). These data suggest that the 39S mtLSU particle can form near to completion when GTPBP5 is absent. To better compare the accumulation of mitoribosome protein markers in WT and GTPBP5-KO cell extracts, we ran samples from the relevant sucrose gradient fractions in parallel in a single gel (Figure 3C). We confirmed that in the GTPBP5-KO fractions where the mtLSU peaks, the levels of most mtLSU proteins were increased 2- to 5-fold compared to WT. Two exceptions for which levels in mtLSU particles were similar to WT were bL36m, consistent with the low overall mitochondrial steady-state levels of this protein, and the late-assembly protein mL65. Together, our data indicate that in the absence of GTPBP5, at least bL36m cannot be efficiently incorporated into the mtLSU assembly line and is presumably removed by proteolytic degradation.
The rRNA profile of the gradients presented in Figure 3B showed that the levels of 12S rRNA in the mtSSU fractions were similar in WT and GTPBP5-KO cells, but the relative levels of 16S rRNA in the mtLSU fractions were enhanced in the KO cells (Figure 3D), likely reflecting the lack of monosomes. Consistently, quantification of total 12S and 16 rRNAs by Northern blot did not identify any significant changes in their steady-state levels (Figure 3E). When the Northern blots were overexposed, we could detect the accumulation of the precursor polycistronic transcripts that contain both the 12S and 16S rRNAs (∼2-fold) in GTPBP5-KO samples, but levels of unprocessed transcripts were similar to wild-type (Figure 3F). In mammalian cells, mtRNA processing is rate-limiting for the assembly of the two mitoribosomal subunits, which has been shown to occur co-transcriptionally (7). The defect in mtLSU assembly in the absence of GTPBP5 only modestly affects precursor transcript processing, and does not affect mtSSU maturation, in contrast to what we have previously reported for GTPBP10 (4). These data suggest that GTPBP5 must act after GTPBP10 in the mtLSU biogenesis pathway.
GTPBP5 is required to facilitate the correct mtLSU maturation after the association of MTG1 and MTERF4-NSUN4
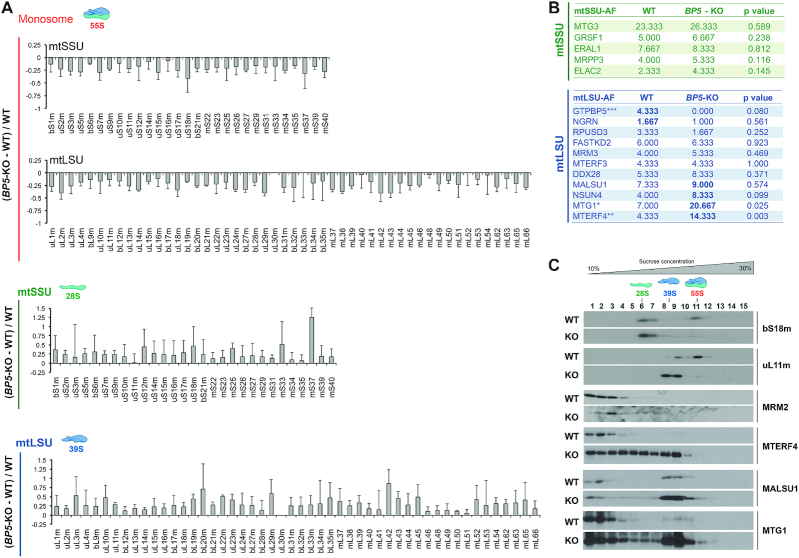
To further analyze the differences in the composition and abundance of assembled mitoribosomal particles, sucrose gradient fractions corresponding to the monosome, mtLSU and mtSSU were methanol/chloroform precipitated and analyzed by mass spectrometry (Figure 4A). Consistent with the data presented in Figure 3B-D, most mtLSU and mtSSU proteins from GTPBP5-KO mitochondria showed a decrease in the monosome fraction and were increased in the disassociated subunits, although bL36m was not detected even in WT samples. Together, our data indicate that, despite the presence of virtually all protein components in the mtLSU ribonucleoprotein particles, there is an inability to finalize their proper assembly and maturation of the monosome in the absence of GTPBP5.
Figure 4.
The absence of GTPBP5 alters the abundance and composition of the mtLSU and mtSSU proteome. (A) Identity and abundance of mitoribosome proteins and assembly factors that accumulate in mitoribosome particles from WT and GTPBP5-KO HEK293T mitochondrial extracts, following their accumulation in the fractions from sucrose gradient sedimentation studies presented in Figure 3B. The proteins in the fractions in which the mtSSU, mtLSU and monosome peak, were precipitated using methanol-chloroform and identified by mass spectrometry. The bar graphs represent the total unique spectrum count difference between GTPBP5-KO and WT normalized by the WT count. Results represent the average ± SD of three independent repetitions. Mitoribosome proteins are identified at the bottom. (B) Accumulation of mitoribosome SSU and LSU assembly factors in the fractions corresponding to each subunit, analyzed by mass spectrometry in panel (A). Data represent the average ± SD of total unique spectrum counts in WT and GTPBP5-KO samples from three independent repetitions. t-test: *P < 0.05; **P < 0.01, ***P < 0.001 (C) Sucrose gradient sedimentation analyses of mitoribosome LSU assembly factors in mitochondria prepared from HEK293T (WT) or GTPBP5-KO (KO) cells.
Mass spectrometry analysis of the mitoribosome subunits from GTPBP5-KO cells allowed us to evaluate further the place GTPBP5 occupies within the hierarchical incorporation of assembly factors during mtSSU and mtLSU assembly (Figure 4B). The levels of all mtSSU assembly factors detected were similar to WT, in agreement with GTPBP5-independent assembly of the mtSSU. In contrast, levels of late-stage mtLSU assembly factors were elevated, including MALSU1, NSUN4 and particularly MTG1 and MTERF4 (Figure 4B), consistent with the steady-state levels of these proteins in mitochondria (Figure 3A). Sucrose gradient sedimentation analyses confirmed the augmented levels of mtLSU assembly factors that associate with the mtLSU particle in GTPBP5-KO mitochondria (Figure 4C). We conclude that GTPBP5 acts at the very late-stage of mtLSU maturation, once MTG1 and the heterodimer MTERF4-NSUN4 have associated with the maturing mtLSU particle.
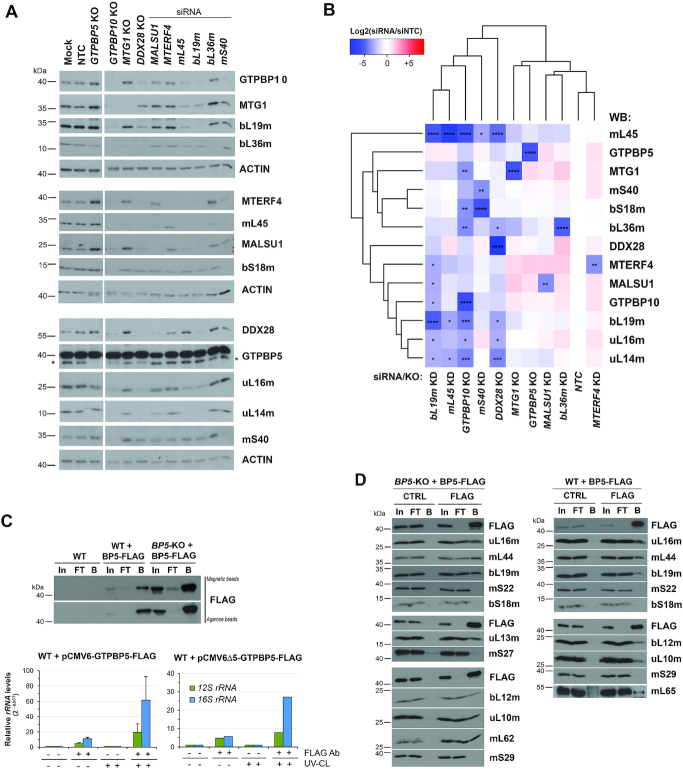
To further examine the role of GTPBP5, we used available knockout cell lines for mtLSU assembly factors or introduced siRNA-induced silencing of selected mitoribosome proteins (Figure 5A) and assembly factors in WT cells. In these cell lines, we assessed the steady-state levels of these proteins by immunoblotting to gauge their hierarchical order of association with the growing mtLSU particle (Figure 5A and B). The selected mtLSU proteins included early-assembled proteins such as bL19m and mL45 (6). As expected, silencing of bL19m or mL45 significantly lowered the steady-state levels of most mtLSU proteins, prevented mtLSU assembly, and attenuated GTPBP10, MTG1, MALSU1 and MTERF4 levels (Figure 5A and B). In contrast, the levels of GTPBP5 were stable in these conditions, and were only attenuated in GTPBP10-KO and DDX28-KO lines (Figure 5A and B), or rho zero cells (Figure 1D), suggesting that GTPBP5 stability may depend on the presence of 16S rRNA. Silencing of the late-assembled bL36m phenocopied the GTPBP5-KO, with increased levels of MTERF4, MALSU1 and MTG1, as we have previously reported (4) but did not affect any other protein, as expected. Our studies allow us to propose a broad hierarchical order on the association of mtLSU assembly factors to the maturing particle (DDX28-GTPBP10 → MTG1-MALSU1 → MTERF4-GTPBP5) that may end with the facilitation of monosome formation by the simultaneous action of MTERF4 and GTPBP5.
Figure 5.
GTPBP5 directly interacts with the 16S rRNA and mtLSU proteins. (A, B) Knockdown (KD) of mitoribosome assembly factors and mitoribosome subunits in HEK293T cells using siRNAs for 3 days, verified by immunoblotting of whole-cell lysates. Lysates from HEK293T cells KO for GTPBP5, GTPBP10, MTG1, or DDX28 were also included in the study. (A) Representative image of immunoblot analysis of the steady-state levels of mitoribosome proteins after the silencing of target proteins. NTC is a non-targeting silencing control, and Mock consisted of transfection reagent only. Antibodies are listed on the right side, and ACTIN was used as a loading control. In the anti-GTPBP5 immunoblot, the asterisk marks the GTPBP5 protein. (B) Following analysis in panel (A), the densitometric data obtained on the abundance of mitoribosome proteins and assembly factors accumulated after silencing or knocking out of each target protein was used for cluster analysis (see Materials and Methods). The heat map, generated with the R studio software, represents the average value of log2 scale of the ratio of the protein levels in knockdown or knockout samples to control (NTC) in three independent repetitions of immunoblotting analyses. 2-way ANOVA was performed followed by a Dunnett's multiple comparisons test: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (C) qPCR analyses of reverse-transcribed control or GTPBP5-FLAG co-immunopurified RNAs after 4-thiouridine treatment and either UV-mediated protein-RNA crosslinking (UV-CL) or not crosslinking. GTPBP5-FLAG was expressed under the control of either a standard CMV6 promoter or an attenuated CMV promoter (pCMV6Δ5) (25). In, input. FT, flow-through or unbound. B is bound. (D) Co-immunoprecipitation analysis of GTPBP5-FLAG and interacting mitoribosome proteins and assembly factors in mitochondrial lysates prepared in the presence of 1% digitonin, by using anti-FLAG agarose beads (FLAG) or plain beads as control (CTRL). In, input. FT, flow-through or unbound. B is bound.
GTPBP5 directly interacts with the 16S ribosomal RNA, the mtLSU and the 16S rRNA methyltransferase MRM2
OBG proteins, including bacterial Obg (37) and human mitochondrial GTPBP10 (4), have mtLSU rRNA binding activity and have been proposed to participate in rRNA refolding or stabilization of rRNA/ribosomal protein structures. To test whether also GTPBP5 directly binds to the 16S rRNA, we used whole-cell extracts after treating with 4-thiouridine prepared in the presence of 1% NP40 detergent from either WT or GTPBP5-KO cells stably expressing functional GTPBP5-FLAG that was efficiently immunoprecipitated (IP) using anti-FLAG agarose beads (Figure 5C). The cells were subjected or not to UV-mediated protein-nucleic acid crosslinking, followed by IP, and isolation of the co-immunopurified RNA. Following reverse transcription and quantitative PCR analysis, although some 12S rRNA was detected, the 16S rRNA was significantly enriched in the FLAG co-IP eluted samples compared to the control IPs (Figure 5C). These data demonstrate the interaction of GTPBP5 with the 16S rRNA.
To validate that GTPBP5 interacts with mitoribosome proteins or assembly factors, we used several approaches. In the assay presented in Figure 5D, mitochondrial extracts from HEK293T cells stably expressing FLAG-tagged GTPBP5 were lysed with 1% digitonin in native conditions, and the GTPBP5-FLAG-interacting proteins were isolated by FLAG-affinity IP and analyzed by immunoblotting. The immunoprecipitate did not contain any of mtSSU proteins, but substantial amounts of an array of eight mtLSU proteins tested (Figure 5D).
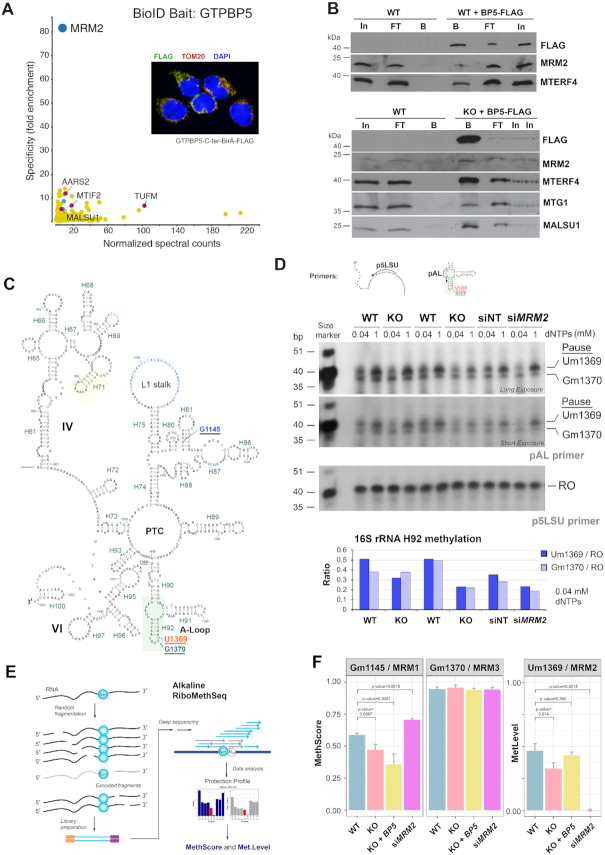
To determine whether we could identify specific proteins that associate with GTPBP5, we first performed a proximity-biotinylation assay (BioID), that identifies proximal protein partners in living cells (38,39). For this purpose, and as part of a more extensive study, we generated HEK293 Flp-In T-REx cell lines expressing the GTPBP5-BirA* fusion protein from a stably integrated inducible construct. The mitochondrial localization of the fusion protein was established by immunofluorescence microscopy (Figure 6A). We performed BioID in biological replicates and analyzed the data by mass spectrometry against a set of negative controls consisting of cells expressing the BirA* fused to GFP, or untransfected cells, ultimately identifying a list of high-confidence proximity partners for GTPBP5 (Supplementary Table S1). GO term analysis shows that these were enriched for mitochondrial matrix proteins involved, in particular, in mitochondrial RNA metabolism, ribosome assembly, and translation. To define whether any of these proteins were specifically enriched when GTPBP5 was used as bait, we took advantage of a larger dataset of ∼100 mitochondrial baits (published on bioRxiv (https://doi.org/10.1101/2020.04.01.020479) (40) and performed specificity enrichment analysis of the preys in the GTPBP5 against the larger mitochondrial BioID dataset. The specificity enrichment analysis calculates the fold enrichment of the proximity interaction (based on the detected spectral counts) for each prey for the bait of interest (GTPBP5) and compares it to all the other baits in the dataset. To identify the most specific proximity partners, we set a specificity enrichment limit to a minimum of 5-fold. The specificity plot of preys detected with GTPBP5-BirA* highlights MRM2 as the most specific interactor of GTPBP5 as this protein was not confidently detected with any of the other baits in the larger dataset (Figure 6A, Supplementary Table S1). MRM2 is an enzyme responsible for 2′-O-methyl modification at position U1369 in the human mitochondrial 16S rRNA. Other enriched interactors involved in mitochondrial translation were MTIF2 (Mitochondrial translation initiation factor 2), MALSU1 (Mitochondrial assembly of ribosomal large subunit protein 1), and TUFM (Mitochondrial elongation factor Tu), AARS2 (Alanyl-tRNA-synthetase 2), as well as two proteins of the mtLSU, uL11m, and mL54. To test whether GTPBP5 could interact with MRM2, we performed co-immunoprecipitation of GTPBP5-FLAG using mitochondria extracted in the presence of 1% of non-ionic detergent digitonin (Figure 6B) or mitochondria treated or not with the chemical crosslinker DSP and extracted in the presence of the more stringent non-ionic detergent 1% lauryl-β-d-maltoside (LM), or 1% denaturing SDS (Supplementary Figure S4A-B) (see Materials and Methods for each extraction condition). In the latter case, the extracts were diluted to 0.1% SDS before proceeding to the IP assay. In all the cases, MRM2 was enriched in the pulldown fraction, indicating a physical interaction with GTPBP5 (Figure 6B and Supplementary Figure S4A-B). The enrichment was detected even in the non-crosslinked SDS-treated extract, suggesting that these proteins undergo a high-affinity interaction that is established as the SDS concentration in the extract was lowered to 0.1%. (Supplementary Figure S4A-B). The digitonin immunoprecipitate was also enriched for MALSU1, MTG1 and MTERF4 (Figure 6B), suggesting a concurrent and coordinated action of these proteins in finalizing mtLSU maturation, preventing premature subunit joining and ultimately promoting monosome formation.
Figure 6.
GTPBP5 interacts physically and functionally with the 16S rRNA methyltransferase MRM2. (A) Prey specificity graph for BioID proximity interactome of GTPBP5 protein, where the prey specificity was determined as the relative enrichment of interaction of individual preys and GTPBP5, compared to their interaction with 100 other mitochondrial baits. The most significant prey is highlighted in dark blue. Enriched mitochondrial ribosome assembly and translation factors are highlighted in red, and enriched ribosomal proteins of the mtLSU are depicted in light blue. The inset represents an immunofluorescence labeling of cells that express the fusion protein using antibodies against FLAG and TOM20 (mitochondrial marker), and DAPI staining of the nucleus. (B) Co-immunoprecipitation analysis of GTPBP5-FLAG and the highly ranked GTPBP5 BioID hits as well as additional native interacting mtLSU assembly factors with Flag Tag (D6W5B) Rabbit mAb-conjugated sepharose beads. Mitochondrial extracts prepared from WT cells not expressing GTPBP5-FLAG were used as control. Two independent experiments are presented. In, input. FT, flow-through or unbound. B is bound. (C) Diagram of the secondary structure of a portion of the human mitochondrial 16S rRNA deduced directly from the reported cryo-EM structure (41). Unbuilt regions are marked in blue lettering, rRNA in Roman numerals, helices and nucleotide numbering in green, modified residues in the A-loop are underlined, and Um1369 is marked in red. (D) Primer extension to measure levels of the A-loop methyl modifications of human mitochondrial 16S rRNA residues at U1369 and G1370, using the primers pAL that detect the modifications, or p5LSU that is used to standardize the loading of 16S rRNA as previously reported (42). Total RNA preparations from HEK293T (WT) or GTPBP5-KO (KO) cells were used in the assay. RNA samples from WT cells treated for 3 days with non-targeting siRNA (siNT) or siMRM2 were used as controls. Radiotracer-labeled primers were added separately in an RT- primer extension reaction using two concentrations of dNTPs as indicated. Reaction products were separated on a 12% denaturing urea-polyacrylamide gel and subjected to autoradiography. Images after two exposure times are presented. The specific pausing sites upon extension of the pAL primer at nucleotide positions Um1369 and Gm1370 are marked. RO is the runoff product resulting from extending the p5LSU primer. In the bottom graphs, Um1369 and Gm1370 signals were quantified by densitometric integration of the lines using the histogram panel of Adobe Photoshop, normalized by the signal of RO signal and plotted. (E) General overview of the Illumina-based RiboMethSeq method for mapping of 2′-O-Me residues in RNA that was implemented to analyze the samples as reported (32). In this approach, alkaline fragmentation of RNA excludes RNA fragments ending with 2′-O-Me and, subsequently, also starting with N + 1 orN + 2 nucleotide. After conversion to the sequencing library these fragments become underrepresented (grey arrows). When sequencing reads are mapped to the reference sequence, 5′-end and 3′-end coverage show a characteristic drop, resulting from protection (see Supplemental Figure S5). These profiles are subsequently merged (with –1 nt or –2 nt backshift for the 5′-end coverage) to obtain a cumulated profile used for calculation of RiboMethSeq scores (43). (F) RiboMethSeq analysis results showing the methylation scores at the sites G1145, U1369, G1370 for WT, GTPBP5-KO, GTPBP5-KO reconstituted with GTPBP5 and silencing of MRM2 (siMRM2). For U1369, we used siMRM2 as unmodified reference to normalize the data and calculate methylation levels. The error bar represents +/- SD values from three replicates of WT, GTPBP5-KO overexpressing GTPBP5, and siMRM2, and four replicates of GTPBP5-KO. The P-values are calculated by t-test (two-tailed/equal variance): *P < 0.05; **P < 0.01, ***P < 0.001.
Methylation of the 16S rRNA at positions U1369 and G1145 is attenuated in the absence of GTPBP5
MRM2 is responsible for 2′-O-methyl modification at position U1369 in the human mitochondrial 16S rRNA (44,42). The 16S rRNA Um1369 modification and the adjacent Gm1370 methylation (Figure 6C) catalyzed by MRM2 and MRM3, respectively, are located in the LSU A-loop, which is an essential component of the peptidyl transferase center (PTC) involved in the interaction of the ribosome with an aminoacyl (A)-site tRNA. Modification of equivalent residues in the A-loop is an evolutionary conserved process among bacterial, cytosolic and organellar ribosomes, and studies in yeast mrm2 deletion mutants or MRM2-silenced human cells show partial attenuation of the rate of mitochondrial translation (42,45). The GTPBP5-MRM2 interaction prompted us to assess whether GTPBP5 is required for the MRM2-catalyzed 16S rRNA Um1369 modification. For this purpose, we used a previously reported primer extension assay (42), using low and high nucleotide concentrations. The silencing of MRM2 resulted in a significant loss of the Um1369 modification, although the Gm1370 was also slightly affected (Figure 6D) as reported (42). When using GTPBP5-KO RNA, the primer extension pattern was similar to that of siMRM2, indicating the necessity of GTPBP5 for MRM2 function. This requirement could be indirect, related to a direct role of GTPBP5 in remodeling the rRNA conformation during PTC maturation, and to the stabilizing effect that GTPBP5 produces on MRM2 (Figures 2E and 3A).
To obtain a highly quantitative and specific assessment of the overall modification state of 2′-O-Me residues in the 16S rRNA we used an Illumina-based RiboMethSeq approach ((32), Figure 6E). The region of interest, containing position 1369/1370 has a read depth of ∼200–300, which is sufficient for precise measurements. The position Um1369 is only partially protected even in WT samples, suggesting only partial methylation, compared with Gm1370, which is fully modified (Supplementary Figure S5A). This has been observed previously for unrelated human 16S rRNA samples. However, variations of protection and methylation at the U1369 position among WT, GTPBP5-KO, and GTPBP5 overexpression samples were confidently measured. From the protection profile, we calculated the MethScore (ScoreC), which represents the degree of protection, with 100% indicating totally modified nucleotide (like Gm1370). For Um1369, methylation levels were then calculated using MRM2 knockdown (siMRM2) samples as unmodified reference (see Materials and Methods). Whereas Gm1370 was unchanged in all cell lines, the U1369 methylation levels were significantly decreased (15–20%, P = 0.014) in the absence of GTPBP5 compared to the wild-type (Figure 6F). Importantly, the U1369 methylation levels were recovered to wild-type levels (P = 0.366) in GTPBP5-KO cells overexpressing GTPBP5. The data indicate that the methylation of U1369 is not abolished in the absence of GTPBP5, but significantly attenuated, most probably as a consequence of the decreased MRM2 steady-state levels measured in the GTPBP5-KO cell line (Figures 2E and 3A).
The RiboMethSeq approach allowed us to additionally analyze the third 2′-O-Me residue in the 16S rRNA, Gm1145, catalyzed by MRM1 (Supplementary Figure S5B). Unexpectedly, the G1145 methylation levels were also significantly decreased in the absence of GTPBP5 (Figure 6F). In addition, Gm1145 levels were decreased even further by overexpression of GTPBP5 in the KO cell line (Figure 6F), providing some evidence for the dominant negative effect of GTPBP5 overexpression.
The action of GTPBP5 precedes MRM2-mediated Um1369 formation in the 16S rRNA and the subsequent recruitment of bL36m
To further clarify whether GTPBP5 acts primarily on bL36m incorporation in the late assembly of the mtLSU particle or 16S rRNA methylation, and to explain the interdependence between these two events, we performed two experiments.
First, we assessed mitochondrial translation in whole cells upon knockdown of MRM2 for 3 and 8 days. After 8 days of MRM2 silencing, we observed a protein translation defect comparable to the GTPBP5-KO (Supplementary Figure S6A), as reported (42).
Second, we compared the pattern of steady-state levels of key mitoribosome LSU proteins and assembly factors in siMRM2 and GTPBP5-KO cell lines. Additional knockdown (KD) or KO cell lines of these factors were used as controls. Data obtained from three independent repetitions were used to generate a heat map of steady-state protein levels (Supplementary Figure S6B-C). Whereas the absence of GTPBP5 attenuates the steady-state levels of MRM2 and bL36m, silencing MRM2 increases the levels of GTPBP5 and has no significant effect on bL36m, and silencing bL36m does not significantly affect GTPBP5 or MRM2 levels (Supplementary Figure S6B-C). We thus conclude that GTPBP5 acts before MRM2-mediated 16S rRNA Um1369 formation, and before the incorporation of bL36m.
The heat map also revealed that late mtLSU assembly factors MALSU1, MTERF4, and the GTPases MTG1 and GTPBP10 accumulate in the absence of GTPBP5, MRM2 or bL36m, indicating that these assembly factors remain bound to the mtLSU particle until maturation is finalized, probably to coordinate final quality control steps.
DISCUSSION
A substantial amount of experimental evidence links the function of OBG GTPases to ribosome biogenesis and maturation in both bacteria and eukaryotic organelles with bacterial ancestry (4,10,16,46,47). Although the S. cerevisiae OBG protein Mtg2 was linked to mitoribosome assembly 15 years ago, the roles of the two human OBG proteins, GTPBP10 (OBGH2) and GTPBP5 (OBGH1 or MTG2), are just beginning to emerge. Recent characterization of GTPBP10 from our group and others has shown that this protein physically interacts with the 16S rRNA and crosslinks with several mtLSU proteins. GTPBP10 primarily ensures proper mtLSU maturation and ultimately serves to coordinate the accumulation of mtSSU and mtLSU, thus establishing a checkpoint function during mtLSU assembly to ensure balanced levels of ribosomal subunits. The role of GTPBP5 in mitoribosome biogenesis have has remained relatively unexplored.
Genome engineering has become a widespread approach to generate cell lines for analyzing the phenotypic consequences of a specific genetic modification. Here, we used a TALEN-mediated approach to produce a GTPBP5-KO cell line whose characterization allowed us to gain mechanistic insight into the function of GTPBP5. Collectively, the data presented in this study leads to a model for mtLSU biogenesis in which GTPBP5 join the mtLSU assembly line after GTPBP10 and several other assembly factors, including MALSU1, MTG1, and MTERF4 to finalize mtLSU maturation and its assembly into the monosome. We have demonstrated that GTPBP5 binds to the 16S rRNA and connects 2′-O-methyl modification at conserved position U1369 with the incorporation of the protein bL36m to finalize mtLSU maturation, prior to monosome formation, thus coordinating late-stage mtLSU assembly with mtSSU joining (Figure 7C).
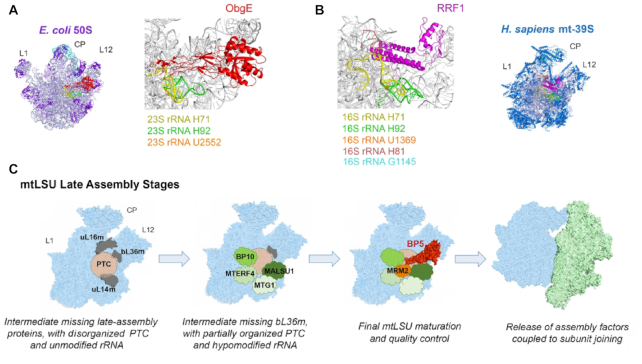
Figure 7.
Model of GTPBP5 action during mtLSU biogenesis. (A) Interaction of E. coli ObgE with the 50S LSU (PDB 4CSU) (46). (B) Interaction of the human 39S mtLSU (PDB 6NU2) with mitochondrial ribosome recycling factor RRF1 (48). (C) Model depicting the mtLSU late-stages of assembly and the roles broadly performed by GTPBP5 and additional assembly factors in mtLSU maturation (see text for extended explanation). CP, central protuberance; L1 and L12 mark the position of the corresponding stalks. The surface representation of the structural profile of the human mitoribosome (PDB 3J9M) (1) and the structure of the ObgE homolog of GTPBP5 (PDB 4CSU) (46) were used. Figures in panels A and B were prepared using PYMOL software.
A primary, and evolutionarily conserved, role for Obg proteins in LSU maturation was first proposed based on their physical or genetic interaction with the LSU and LSU rRNA modification enzymes. The Obg protein from S. typhimurium was shown to interact physically with the 23S rRNA pseudouridine synthetase RluD that modifies several residues of helix 69 (22). In E. coli, increased levels of ObgE suppress the ribosome assembly defect in strains deleted for the methyltransferase RrmJ that catalyzes 2′-O-methylation of uridine at position 2552 (Um2552) of the 23S rRNA A-loop (Helix 92) (23). Notably, Saccharomyces cerevisiae Mtg2 was initially identified as a suppressor of mutant mtLSU 21S rRNA methyltransferase mrm2 (E. coli RrmJ) (18), and the data presented here demonstrate physical and functional interactions of human GTPBP5 with MRM2, suggesting a conserved role for bacterial and mitochondrial OBG proteins.
GTPBP5-KO mitochondria have increased levels of mitoribosome subunits and only residual monosome amounts. In the presence of the high magnesium concentrations (20 mM MgCl2) used in our experiments, the GTPBP5-KO mtLSU sediments as the wild-type particle and contains all subunits, although bL36m is largely underrepresented. E. coli deletion mutants of either obgE or rrmJ, have a similar phenotype, consisting of a significant buildup of ribosomal subunits at the expense of functional 70S ribosomes, and accumulation of a 45S particle that undergoes Mg2+- dependent alteration in density (37,49). This 45S particle is an on-pathway assembly intermediate that lacks L36 (50). Structural probing of 23S rRNA and high-salt stripping of 45S components has shown that RrmJ-mediated methylation promotes 23S rRNA interdomain interactions via the association between helices 92 and 71, stabilized by the 2′-O-methylation of Um2552 (the equivalent of mitochondrial Um1369), in concert with the incorporation of L36, triggering late steps of 50S subunit assembly (50). Two native late-stage assembly intermediates of the human mtLSU were recently solved by cryo-EM (51). In a subclass, 16S rRNA helices H34-H35, H65, H67-H71 and H89-H93 that form the PTC in the mature mitoribosome were not detected, indicating unfolded interfacial rRNA. In this late-stage assembly intermediate, only the protein bL36m was absent. The recruitment of bL36m and the folding of 16S rRNA has been proposed to be interdependent (51), because in the LSU purified from a ΔL36 strain of E. coli, H71 and H89-H93 showed increased susceptibility to chemical probes (52). Our data suggest that correct folding of this region in the mtLSU might rely on the OBG protein GTPBP5 and the MRM2-catalyzed 2′-O-methyl modification at position U1369. Furthermore, our RiboMethSeq data have revealed decreased methylation of G1145, catalyzed by MRM1. Gm1145 locates at the 16S rRNA H81, near the potential area of influence of GTPBP5, as explained below (Figure 7A, B).
In Figure 7C, we propose a model for the final stages of mtLSU maturation in which several assembly factors with rRNA binding activity join the mtLSU assembly line in a sequential manner (GTPBP10 → MTG1-MALSU1 → MTERF4-GTPBP5) to promote the folding of the 16S rRNA catalytic site and the incorporation of late-assembly proteins. In the absence of GTPBP10, mtLSU proteins such as uL16m, bL33m, bL34m or bL36m are particularly underrepresented in assembly intermediates (4). MTG1 interacts with bL19m and catalyzes a step that is required for the efficient incorporation of at least bL36m and bL35m (9). MALSU1 interacts with uL14m (53) and was found bound to the native intermediates lacking bL36m, as part of a module that also contained two new assembly factors L0R8F8 or mitochondrial elongation factor 1 microprotein (MIEF1-MP) (54) and mt-ACP (acyl carrier protein) (51). MTERF4 (mitochondrial transcription termination factor 4) forms a complex with the 12S rRNA methyltransferase NSUN4 that has been proposed to coordinate monosome formation (8). We propose that GTPBP5 facilitates the correct folding of several 16S rRNA helices, including H81 and H92, G1145 and U1369 methylations, and the subsequent recruitment of bL36m.
The GTPase GTPBP5 was previously shown to specifically associate with the mtLSU in a GTP-dependent manner (16), similar to bacterial ObgE (46). However, although the intrinsic GTPase activity of GTPBP5 was detectable, it was not found to be stimulated by 55S mitoribosomes in vitro (16). Similarly, in vitro dissociation of bacterial 70S ribosomes by ObgE did not require energy input from GTP-hydrolysis (46). Recent cryo-EM studies of ObgE-bound ribosomes have provided insights into the mode of action of this GTPase (46). The GTP-binding domain of ObgE in the 50S subunit is placed interacting directly with the bL12 stalk base and the sarcin-rich loop (SRL) of the 23S rRNA, which is different from the translational GTPases. This position may explain the moderate activation of ObgE GTPase activity by 50S particles, suggesting that GTP hydrolysis alone may not be sufficient to promote the release of OBG proteins from the mtLSU. The Obg fold (N-terminal domain) of ObgE binds to the intersubunit face of the 50S subunit protruding into the PTC at a position commonly used for the docking of translational GTPases (46) (Figure 7A). The ObgE binding site is precisely next to several rRNA modification sites, including RrmJ in the A-loop. These data further support a role for OBG proteins in monitoring and facilitating the correct folding of the PTC helices and the methylation status of the A-loop, thus establishing a quality control mechanism to prevent premature subunit joining. The effect of GTPBP5 on the methylation of both 16S rRNA U1369 and G1145 supports this conserved function.
It has been noted that OBG interacts with the acceptor arm of the A-site tRNA using the same tRNA mimicry strategy than translation factors and the ribosome release factor 2 (RF2) (46). Despite the differences between bacterial and mitochondrial ribosomes, a comparison of the binding sites for Obg/GTPBP5 and the mitoribosome recycling factor (RRFmt) (48) indicates that the two factors interact with the PTC in a similar manner (Figure 7A and B). Therefore, GTPBP5 is envisioned to play a subunit anti-association role to prevent premature monosome formation. In support of this possibility, excess of ObgE has been shown to promote the dissociation of 70S ribosomes in vitro by disrupting several essential intersubunit bridges (46), and our data in cultured human cells show that excess of GTPBP5 is deleterious by, at least, interfering with 16S rRNA G1145 methylation. In the process of human mitoribosome assembly, several mtLSU maturation factors have been proposed to prevent subunit association until they are released from the individual subunits. GTPBP10 coordinates mtSSU and mtLSU accumulation (Maiti et al., 2018) as mentioned earlier; MTG1 couples late-stage mtLSU assembly with intersubunit bridge formation (9); MALSU1 belongs to the family of ribosome release factors (55) and release of the MALSU1-L0R8F8-mt-ACP module is necessary for establishing a specific intersubunit bridge (56); and a similar role has been proposed for the MTERF4-NSUN4 module (8). As proposed for bacterial ribosome biogenesis (46), these mitochondrial proteins may all act as part of a general quality control mechanism to follow the maturation state of the PTC as structural checkpoints, to prevent defective mtLSU particles from engaging in monosome formation and altered translation.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Corresponding author, Dr Antoni Barrientos, PhD (abarrientos@med.miami.edu).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Flavia Fontanesi for scientific discussions and comments on the manuscript and Zhen-Yuan Lin for analysis of the BioID samples. RiboMethSeq analysis of mitochondrial 16S rRNA was performed by Dr Virginie Marchand and Prof. Yuri Motorin from the Epitranscriptomics & RNA Sequencing (EpiRNA-Seq) core facility at the UMS2008/US40 of Université de Lorraine-CNRS-INSERM (Nancy, France).
Author contributions: P.M. generated the vast majority of the presented data. P.M. and A.B. designed the study, designed and performed experiments, analyzed results and wrote the paper. H.A., A.C.G. and E.A.S. developed the BioID studies, analyzed the data and prepared the corresponding figure. All authors read and edited the manuscript.
Contributor Information
Priyanka Maiti, Department of Neurology, University of Miami Miller School of Medicine, Miami, FL, USA.
Hana Antonicka, Montreal Neurological Institute and Department of Human Genetics, McGill University, Montreal, QC, Canada.
Anne-Claude Gingras, Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital and Department of Molecular Genetics, University of Toronto, Toronto, ON, Canada.
Eric A Shoubridge, Montreal Neurological Institute and Department of Human Genetics, McGill University, Montreal, QC, Canada.
Antoni Barrientos, Department of Neurology, University of Miami Miller School of Medicine, Miami, FL, USA; Department of Biochemistry and Molecular Biology, University of Miami Miller School of Medicine, Miami, FL, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIGMS-MIRA [R35GM118141 to A.B.]; Muscular Dystrophy Association Research Grant [MDA-381828 to A.B.]; Canadian Institutes of Health Research (CIHR) Foundation Grant [FDN 143301 to A.C.G.]; CIHR [MOP-133530 to E.A.S.]; American Heart Association postdoctoral fellowship [19POST34450174 to P.M.]. Funding for open access charge: NIGMS [R35GM118141 to A.B.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Amunts A., Brown A., Toots J., Scheres S.H.W., Ramakrishnan V.. The structure of the human mitochondrial ribosome. Science. 2015; 348:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Greber B.J., Bieri P., Leibundgut M., Leitner A., Aebersold R., Boehringer D., Ban N.. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 2015; 348:303–308. [DOI] [PubMed] [Google Scholar]
- 3. De Silva D., Fontanesi F., Barrientos A.. The DEAD-Box protein Mrh4 functions in the assembly of the mitochondrial large ribosomal subunit. Cell Metab. 2013; 18:712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maiti P., Kim H.J., Tu Y.T., Barrientos A.. Human GTPBP10 is required for mitoribosome maturation. Nucleic Acids Res. 2018; 13:11423–11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zeng R., Smith E., Barrientos A.. Yeast mitoribosome large subunit assembly proceeds by hierarchical incorporation of protein clusters and modules on the inner membrane. Cell Metab. 2018; 27:645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bogenhagen D.F., Ostermeyer-Fay A.G., Haley J.D., Garcia-Diaz M.. Kinetics and mechanism of mammalian mitochondrial ribosome assembly. Cell Rep. 2018; 22:1935–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rackham O., Busch J.D., Matic S., Siira S.J., Kuznetsova I., Atanassov I., Ermer J.A., Shearwood A.M., Richman T.R., Stewart J.B. et al.. Hierarchical RNA processing is required for mitochondrial ribosome assembly. Cell Rep. 2016; 16:1874–1890. [DOI] [PubMed] [Google Scholar]
- 8. Metodiev M.D., Spahr H., Loguercio Polosa P., Meharg C., Becker C., Altmueller J., Habermann B., Larsson N.G., Ruzzenente B.. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014; 10:e1004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim H.-J., Barrientos A.. MTG1 couples mitoribosome large subunit assembly and intersubunit bridge formation. Nucleic Acid Res. 2018; 46:8435–8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lavdovskaia E., Kolander E., Steube E., Mai M.M., Urlaub H., Richter-Dennerlein R.. The human Obg protein GTPBP10 is involved in mitoribosomal biogenesis. Nucleic Acids Res. 2018; 46:8471–8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tu Y.T., Barrientos A.. The human mitochondrial DEAD-box protein DDX28 resides in RNA granules and functions in mitoribosome assembly. Cell Rep. 2015; 10:854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Antonicka H., Shoubridge E.A.. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep. 2015; 10:920–932. [DOI] [PubMed] [Google Scholar]
- 13. Jourdain A.A., Koppen M., Wydro M., Rodley C.D., Lightowlers R.N., Chrzanowska-Lightowlers Z.M., Martinou J.C.. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab. 2013; 17:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He J., Cooper H.M., Reyes A., Di Re M., Kazak L., Wood S.R., Mao C.C., Fearnley I.M., Walker J.E., Holt I.J.. Human C4orf14 interacts with the mitochondrial nucleoid and is involved in the biogenesis of the small mitochondrial ribosomal subunit. Nucleic Acids Res. 2012; 40:6097–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Uchiumi T., Ohgaki K., Yagi M., Aoki Y., Sakai A., Matsumoto S., Kang D.. ERAL1 is associated with mitochondrial ribosome and elimination of ERAL1 leads to mitochondrial dysfunction and growth retardation. Nucleic Acids Res. 2010; 38:5554–5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kotani T., Akabane S., Takeyasu K., Ueda T., Takeuchi N.. Human G-proteins, ObgH1 and Mtg1, associate with the large mitochondrial ribosome subunit and are involved in translation and assembly of respiratory complexes. Nucleic Acids Res. 2013; 41:3713–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hirano Y., Ohniwa R.L., Wada C., Yoshimura S.H., Takeyasu K.. Human small G proteins, ObgH1, and ObgH2, participate in the maintenance of mitochondria and nucleolar architectures. Genes Cells. 2006; 11:1295–1304. [DOI] [PubMed] [Google Scholar]
- 18. Datta K., Fuentes J.L., Maddock J.R.. The yeast GTPase Mtg2p is required for mitochondrial translation and partially suppresses an rRNA methyltransferase mutant, mrm2. Mol. Biol. Cell. 2005; 16:954–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sato A., Kobayashi G., Hayashi H., Yoshida H., Wada A., Maeda M., Hiraga S., Takeyasu K., Wada C.. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells. 2005; 10:393–408. [DOI] [PubMed] [Google Scholar]
- 20. Scott J.M., Ju J., Mitchell T., Haldenwang W.G.. The Bacillus subtilis GTP binding protein obg and regulators of the sigma(B) stress response transcription factor cofractionate with ribosomes. J. Bacteriol. 2000; 182:2771–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kint C., Verstraeten N., Hofkens J., Fauvart M., Michiels J.. Bacterial Obg proteins: GTPases at the nexus of protein and DNA synthesis. Crit. Rev. Microbiol. 2014; 40:207–224. [DOI] [PubMed] [Google Scholar]
- 22. Lin B., Skidmore J.M., Bhatt A., Pfeffer S.M., Pawloski L., Maddock J.R.. Alanine scan mutagenesis of the switch I domain of the Caulobacter crescentus CgtA protein reveals critical amino acids required for in vivo function. Mol. Microbiol. 2001; 39:924–934. [DOI] [PubMed] [Google Scholar]
- 23. Tan J., Jakob U., Bardwell J.C.. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 2002; 184:2692–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. King M.P., Attardi G.. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989; 246:500–503. [DOI] [PubMed] [Google Scholar]
- 25. Morita E., Arii J., Christensen D., Votteler J., Sundquist W.I.. Attenuated protein expression vectors for use in siRNA rescue experiments. BioTechniques. 2012; 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Enriquez J.A., Attardi G.. Analysis of aminoacylation of human mitochondrial tRNAs. Methods Enzymol. 1996; 264:183–196. [DOI] [PubMed] [Google Scholar]
- 27. Leary S.C., Sasarman F.. Oxidative phosphorylation: synthesis of mitochondrially encoded proteins and assembly of individual structural subunits into functional holoenzyme complexes. Methods Mol. Biol. 2009; 554:143–162. [DOI] [PubMed] [Google Scholar]
- 28. Bourens M., Barrientos A.. A CMC1-Knockout reveals translation-independent control of human mitochondrial Complex IV biogenesis. EMBO Rep. 2017; 8:477–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J.. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951; 193:265–275. [PubMed] [Google Scholar]
- 30. Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970; 227:680–685. [DOI] [PubMed] [Google Scholar]
- 31. Szczepanowska K., Maiti P., Kukat A., Hofsetz E., Nolte H., Senft K., Becker C., Ruzzenente B., Hornig-Do H.T., Wibom R. et al.. CLPP coordinates mitoribosomal assembly through the regulation of ERAL1 levels. EMBO J. 2016; 35:2566–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marchand V., Blanloeil-Oillo F., Helm M., Motorin Y.. Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA. Nucleic Acids Res. 2016; 44:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pichot F., Marchand V., Ayadi L., Bourguignon-Igel V., Helm M., Motorin Y.. Holistic optimization of bioinformatic analysis pipeline for detection and quantification of 2′-O-methylations in RNA by RiboMethSeq. Front. Genet. 2020; 11:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Couzens A.L., Knight J.D., Kean M.J., Teo G., Weiss A., Dunham W.H., Lin Z.Y., Bagshaw R.D., Sicheri F., Pawson T. et al.. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci. Signal. 2013; 6:rs15. [DOI] [PubMed] [Google Scholar]
- 35. Teo G., Liu G., Zhang J., Nesvizhskii A.I., Gingras A.C., Choi H.. SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J. Proteomics. 2014; 100:37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr., Jungkamp A.C., Munschauer M. et al.. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiang M., Datta K., Walker A., Strahler J., Bagamasbad P., Andrews P.C., Maddock J.R.. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 2006; 188:6757–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim D.I., Jensen S.C., Noble K.A., Kc B., Roux K.H., Motamedchaboki K., Roux K.J.. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell. 2016; 27:1188–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roux K.J., Kim D.I., Raida M., Burke B.. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012; 196:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Antonicka H., Lin Z.-Y., Janer A., Weraarpachai W., Gingras A.C., Shoubridge E.A.. A high-density human mitochondrial proximity interaction network. 2020; bioRxiv doi:02 April 2020, preprint: not peer reviewed 10.1101/2020.04.01.020479. [DOI] [PubMed]
- 41. Brown A., Amunts A., Bai X.C., Sugimoto Y., Edwards P.C., Murshudov G., Scheres S.H., Ramakrishnan V.. Structure of the large ribosomal subunit from human mitochondria. Science. 2014; 46:718–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rorbach J., Boesch P., Gammage P.A., Nicholls T.J., Pearce S.F., Patel D., Hauser A., Perocchi F., Minczuk M.. MRM2 and MRM3 are involved in biogenesis of the large subunit of the mitochondrial ribosome. Mol. Biol. Cell. 2014; 25:2542–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Motorin Y., Marchand V.. Detection and analysis of RNA ribose 2′-O-methylations: challenges and solutions. Genes (Basel). 2018; 9:642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee K.W., Bogenhagen D.F.. Assignment of 2′-O-methyltransferases to modification sites on the mammalian mitochondrial large subunit 16 S ribosomal RNA (rRNA). J. Biol. Chem. 2014; 289:24936–24942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pintard L., Bujnicki J.M., Lapeyre B., Bonnerot C.. MRM2 encodes a novel yeast mitochondrial 21S rRNA methyltransferase. EMBO J. 2002; 21:1139–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Feng B., Mandava C.S., Guo Q., Wang J., Cao W., Li N., Zhang Y., Zhang Y., Wang Z., Wu J. et al.. Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol. 2014; 12:e1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Britton R.A. Role of GTPases in bacterial ribosome assembly. Annu. Rev. Microbiol. 2009; 63:155–176. [DOI] [PubMed] [Google Scholar]
- 48. Koripella R.K., Sharma M.R., Risteff P., Keshavan P., Agrawal R.K.. Structural insights into unique features of the human mitochondrial ribosome recycling. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:8283–8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bugl H., Fauman E.B., Staker B.L., Zheng F., Kushner S.R., Saper M.A., Bardwell J.C., Jakob U.. RNA methylation under heat shock control. Mol. Cell. 2000; 6:349–360. [DOI] [PubMed] [Google Scholar]
- 50. Arai T., Ishiguro K., Kimura S., Sakaguchi Y., Suzuki T., Suzuki T.. Single methylation of 23S rRNA triggers late steps of 50S ribosomal subunit assembly. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E4707–E4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brown A., Rathore S., Kimanius D., Aibara S., Bai X.C., Rorbach J., Amunts A., Ramakrishnan V.. Structures of the human mitochondrial ribosome in native states of assembly. Nat. Struct. Mol. Biol. 2017; 24:866–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Maeder C., Draper D.E.. A small protein unique to bacteria organizes rRNA tertiary structure over an extensive region of the 50 S ribosomal subunit. J. Mol. Biol. 2005; 354:436–446. [DOI] [PubMed] [Google Scholar]
- 53. Fung S., Nishimura T., Sasarman F., Shoubridge E.A.. The conserved interaction of C7orf30 with MRPL14 promotes biogenesis of the mitochondrial large ribosomal subunit and mitochondrial translation. Mol. Biol. Cell. 2013; 24:184–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rathore A., Chu Q., Tan D., Martinez T.F., Donaldson C.J., Diedrich J.K., Yates J.R. 3rd, Saghatelian A.. MIEF1 microprotein regulates mitochondrial translation. Biochemistry. 2018; 57:5564–5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hauser R., Pech M., Kijek J., Yamamoto H., Titz B., Naeve F., Tovchigrechko A., Yamamoto K., Szaflarski W., Takeuchi N. et al.. RsfA (YbeB) proteins are conserved ribosomal silencing factors. PLoS Genet. 2012; 8:e1002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li X., Sun Q., Jiang C., Yang K., Hung L.W., Zhang J., Sacchettini J.C.. Structure of ribosomal silencing factor bound to Mycobacterium tuberculosis ribosome. Structure. 2015; 23:1858–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.