Abstract
Circadian clocks are endogenous oscillators that control ∼24-hour physiology and behaviors in virtually all organisms. The circadian oscillator comprises interconnected transcriptional and translational feedback loops, but also requires finely coordinated protein homeostasis including protein degradation and maturation. However, the mechanisms underlying the mammalian clock protein maturation is largely unknown. In this study, we demonstrate that necdin, one of the Prader-Willi syndrome (PWS)-causative genes, is highly expressed in the suprachiasmatic nuclei (SCN), the pacemaker of circadian clocks in mammals. Mice deficient in necdin show abnormal behaviors during an 8-hour advance jet-lag paradigm and disrupted clock gene expression in the liver. By using yeast two hybrid screening, we identified BMAL1, the core component of the circadian clock, and co-chaperone SGT1 as two necdin-interactive proteins. BMAL1 and SGT1 associated with the N-terminal and C-terminal fragments of necdin, respectively. Mechanistically, necdin enables SGT1-HSP90 chaperone machinery to stabilize BMAL1. Depletion of necdin or SGT1/HSP90 leads to degradation of BMAL1 through the ubiquitin–proteasome system, resulting in alterations in both clock gene expression and circadian rhythms. Taken together, our data identify the PWS-associated protein necdin as a novel regulator of the circadian clock, and further emphasize the critical roles of chaperone machinery in circadian clock regulation.
INTRODUCTION
Prader–Willi syndrome (PWS) is a genetic disorder induced by a deletion in the paternally-derived chromosome region 15q11–q13, which is a maternally imprinted region. The prevalence of PWS is estimated to be 1/10 000 to 1/25 000 in the general population (1). A number of genes have been mapped within the PWS region, including five protein-coding genes (MKRN3, MAGEL2, NDN, SNRPN-SNURF, C15orf2) and several antisense transcripts (2,3).
PWS is characterized by feeding problems and failure to thrive in early infancy, followed by growth delay, learning difficulties, hyperphagia and obesity, sleep abnormalities, behavioral problems and hypogonadism (3). Abnormalities in sleep and arousal are among the common features of PWS, which is characterized by excessive daytime sleepiness, defects in organization of rapid eye movement (REM) sleep and arousal, and sleep-disordered breathing (4). But the pathophysiologic mechanisms underlying such sleep disorders in PWS patients remain obscure.
Sleep is regulated by a homeostatic process as well as by circadian rhythms (5,6). A circadian rhythm is an endogenous oscillator that is maintained in an organism's tissues and cells and regulates daily changes in physiology, metabolism and behavior with an approximately 24-h daily period (7,8). The mammalian circadian clock is fundamentally based on a transcriptional-translational feedback loop. At the core of this molecular network are two transcription factors: circadian locomotor output cycles kaput (CLOCK) and brain and muscle aryl hydrocarbon receptor nuclear translocator-like 1 (BMAL1). They heterodimerize and bind to E-box elements (CACGTG) located in the promoters of a large number of clock-controlled genes. This mechanism drives the expression of these genes, including Period (Per1–3) and Cryptochrome (Cry1/2). The PER and CRY proteins form a complex and translocate to the nucleus to inhibit the transcriptional activity of the CLOCK-BMAL1 (9,10).
NDN, an intronless gene within the PWS region, encodes necdin, a protein with 321 amino acid residues (11). To study the expression pattern of necdin in the central nervous system, we searched the Allen Brain Atlas and found that this gene is highly expressed in the suprachiasmatic nucleus (SCN), the pacemaker of the mammalian circadian clock (6,10,12–14), implying that it should have a role in circadian rhythm regulation. In this study, we demonstrate that mice deficient in necdin show abnormal behaviors when subjected to an 8-hour advance jet-lag paradigm, accompanied by disrupted expression of clock genes in their livers. Further, we found that necdin interacts with BMAL1 and facilitates SGT1/HSP90 chaperone machinery to stabilize BMAL1. Disruption of necdin or SGT1 expression destabilizes BMAL1 by promoting its proteolytic degradation through the ubiquitin-proteasomal system, resulting in altered clock gene expression and disrupted circadian rhythms. These findings identify a novel mechanism involved in circadian clock regulation and provide evidence implicating necdin as a key endogenous regulator of the circadian clock in mammals.
MATERIALS AND METHODS
Sequences for siRNAs
The siRNA sequence of Negative Control was UUCUCC-GAACGUGUCACGUTT. The sense strand sequences of siRNA were CCUGC-ACACCAUGGAGUUUTT (#1), UCAUGAUCCUGAGCCUCAUTT(#2) and GGA-AGAAGCACUCCACCUUTT(#3) for necdin, and GCAGCUUUAAACAGAUUAU-TT(#1), CUGGUAUCAAAC AGAAUCUTT(#2) and GCAGAUGUAAAGAACC-UAUTT(#3) for Sgt1.
Antibodies and reagents
Antibodies against Necdin (ab18554), SGT1 (ab30931), BMAL1 (ab3350), HSP90 (ab13492), PER1 (ab136451), PER2 (ab180655), CRY1 (ab104736) were purchased from Abcam. Antibody against AVP (sc-390723) were obtained from Santa Cruz. Antibodies against Myc-tag (2276), Flag-tag (14793), HA-tag (3724), VIP (63269) were obtained from Cell Signaling Technology. Antibody against CRY2 were obtained from Invitrogen. Antibody against GRP (20073) were obtained from ImmunoStar. 17-AAG (HY-10211) and Geldanamycin (HY-15230) were purchased from MedChemExpress.
Animals
Necdin mice were generated by using CRISPR-Cas9 technology. Cas9 mRNA and two guide RNAs (gRNA) targeting the upstream and downstream regions of the mouse necdin gene were injected into C57BL/6 mouse oocytes, and a mouse with deletion of the entire necdin gene was used as a founder. Before behavioral tests, mice of the same sex were group-housed (3–5 animals per cage) under controlled conditions [temperature, 20 ± 2°C; relative humidity, 50–60%; 12:12-h light–dark (LD) cycle, lights on at 7:00 AM and lights off at 7:00 PM] and had free access to food and water. All procedures regarding the care and use of animals were approved by the Institutional Animal Care and Use Committee of Central South University of China.
The primers for genotyping were as following:
Ndn-WT forward (P1): 5′-CTTTCTCCAGG ACCTTCACATTTA-3′
Ndn-WT reverse (P2): 5′-GGGTCGCTCAGGTCCTTACTTTG-3′
Ndn-Mutant forward (P3): 5′-AAACAACTCATCATCATCATAAGG-3′
Ndn-Mutant reverse (P4): 5′-TTTGTAAAGGGTGCTAAGTGC-3′
Locomotor behavior
Mice aged 4–6 months were individually housed within cages equipped with running wheels and were allowed free access to food and water. Their locomotor activities were recorded as revolutions per 5-min interval. Mice were entrained to an initial LD cycle (light intensity ∼150 lx, lights on at 7:00 AM and lights off at 7:00 PM). After 2–3 weeks of activity recording in 12:12-h light–dark conditions, the mice were placed in constant darkness (DD) or constant light (LL) for ∼4 weeks.
These mice were then subjected to a light-induced phase shift at day 15 of DD. Animals in their home cages were moved to another room and exposed to a 10-min pulse of white light (∼150 lux) at circadian time (CT) 16, at which CT12 was designated as activity onset. The light-induced phase-shift amplitude was derived from regression lines drawn through the activity onset at least 7 days immediately before the day of stimulation and 7 days after reestablishment of a steady-state circadian period after stimulation.
The free-run period and fast Fourier transformation (FFT) were calculated using ClockLab software (Actimetrics, Evanston, IL, USA) in the Matlab environment. The free-run period was measured by a χ2 periodogram from days 10 through 25 under DD. Daily revolutions and FFT were determined by analyzing the activity of the last 10 days under LD and days 10–25 under DD. FFT circadian amplitude values represent the peak relative amplitude in the circadian range (18–30 h) normalized to a total variance of 100%.
For the 8-h advance jet-lag experiments, lights were turned off at 11:00 AM and turned on at 11:00 PM on the first day. Thereafter the animals were exposed to the new light–dark schedule for three weeks. The onset of activity for each cycle was defined as the occurrence of the first concentrated bout of activity after an extended period of rest.
Generation of Ndn−p/+m fibroblast cells
Briefly, we placed one male Ndn+p/−m mouse with two female WT mice in one cage. Once a plug was identified, the female mouse was transferred to a new cage and the time of embryo development was noted as 0.5 days. After the embryos developed for 13.5–15.5 days, the pregnant mouse was euthanized and each embryo was carefully transferred to a fresh dish. The embryos were dissected and the head, heart and liver were removed, with the head being used for genotyping. The fetal tissues were minced and digested with pre-chilled 0.25% trypsin-EDTA at 37°C for 10 min. The suspension was transferred to a 50 ml conical tube and the cells were spun down at 500 × g and 4°C for 5 min. Cell pellets from the tube were resuspended in complete culture media and the cells were cultured in a standard humidified 37°C, 5% CO2 incubator.
Cell culture and transfection
U2OS and HEK293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C in 5% CO2 incubators. Plasmid and siRNA transfections were performed with Lipofectamine 2000 (Invitrogen) reagents according to the manufacturer's protocol.
Co-immunoprecipitation
Cells were harvested with lysis buffer (150 mM NaCl, 1% NP-40, 2 mM EDTA, 50 mM Tris, pH 8.0, and protease inhibitor cocktail). Approximately 1 mg of whole cell lysate was incubated with 1 μg of the indicated antibody with constant agitation overnight at 4°C. 30 μl protein G agarose bead slurry (Sigma; P3296) was added to pull down the immunocomplexes. The beads were collected by centrifugation, washed extensively with lysis buffer 3–5 times, and boiled with 2× SDS loading buffer. The protein samples were then subjected to immunoblot analysis with the appropriate antibody.
Immunofluorescence staining
Mice were perfused intracardially with 4% paraformaldehyde, and brains were removed, post-fixed and cryoprotected in 25% sucrose overnight. Eight-micron-thick sections including the SCN region were collected and incubated with primary antibodies to necdin (diluted 1:500), AVP (diluted 1:300), VIP (diluted 1:300), GRP (diluted 1:300) or BMAL1 (diluted 1:200) overnight. The slices were then incubated in the dark with Alexa-conjugated labeled secondary antibodies. Nuclei were stained with DAPI. Slices were imaged using a confocal microscope (TCS SP5; Leica). Fluorescence intensities of BMAL1 were analyzed using fluorescence image analysis software (Lumina Vision, Mitani).
Ubiquitination assays
Cells were treated with MG132 (10 μM) for 6 h, and then lysed with ubiquitination buffer I (2% SDS, 10 mM Tris-base, pH 7.5, 150 mM NaCl) at 100°C for 10 min. The lysates were then diluted 10-fold with regular ubiquitination buffer II (1% Triton-100, 10 mM Tris-base, pH 7.5, 2 mM EDTA, 150 mM NaCl) and subjected to immunoprecipitation and western blot analysis.
RNA isolation, reverse transcription and RT-qPCR
Cells or tissues were lysed with TRIzol® reagent according to the manufacturer's instructions. Two micrograms of total RNA were reversed transcribed using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific; K1622), and 20 ng of total cDNA equivalents were then analyzed using Fast SYBR™ Green Master Mix (Thermo Scientific; 4385612) according to the manufacturer's instructions using a C1000 touch Thermal Cycler. Primers used for qPCR were: mNecdin forward 5′-GAGGTCCCCGACTGTGAGAT-3′ and reverse 5′- TGCAGGATTTTAGGGTCAA-CATC-3′; mBMAL1 forward 5′-TGACCCTCATG-GAAGGTTAGAA-3′ and reverse 5′-GGACATTGCATTGCA-TGTTGG-3′; mPer1 forward 5′-TGAAGCAAGACCG-GGAGAG-3′ and mPer1 reverse 5′-CACACACGCCGTCACATCA-3′; mPer2 forward 5′-GAAAGCTG-TCACCACCATAGAA-3′ and mPer2 reverse 5′-AACTCGCACTTCCTTTTCAGG-3′; mCry1 forward 5′-CACT-GGTTCCGAAAGGGACTC-3′ and mCry1 reverse 5′-CTGAAGCAAAAATC-GCCACCT-3′; mCry2 forward 5′-CACTGGTTCCGCAA-AGGACTA-3′ and mCry2 reverse 5′-CCACGGGTCGAGGATGTAGA-3′; mDbp forward 5′-GGAAACAGCAAGCCCAAAGAA-3′ and mDbp reverse 5′-CAGCGG-CGCAAAAAGACTC-3′; mDec1 forward 5′-ACGGAGACCTGTCAGGGATG-3′ and mDec1 reverse 5′-GGCAGTTTGTAAGTTTCCTTGC-3′; mDec2 forward 5′-TGTGTAAACCCAAAAGGAGCTT-3′ and mDec2 reverse 5′-TGTTCGG-GCAGTAAATCTTTCAG-3′. All reactions were performed in triplicate. The relative levels of gene mRNAs were normalized to the corresponding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels.
Real-time bioluminescence monitoring
U2OS cells stably expressing a luciferase under the control of Bmal1 promoter were a kind gift from Professor Eric Zhang (NIBS, Chinese Academy of Sciences). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, 100 ng/ml streptomycin and 5 ng/ml puromycin at 37°C in 5% CO2. A total of 6 × 105 cells were plated in 35-mm dishes for 24 h. The cells were treated with 0.1 μM dexamethasone (DEX, Sigma; D4902) for 2 h, and the medium was changed to phenol red-free DMEM supplemented with 10% FBS, 10 mM HEPES buffer, 0.1 mM luciferin (Promega, P1061), 100 units/ml penicillin and 100 ng/ml streptomycin. Bioluminescence was monitored continuously with a 32-channel LumiCycle (Actimetrics, Wilmette, IL, USA) and was analyzed by LumiCycle analysis software (Actimetrics, Wilmette, IL, USA).
Yeast two-hybrid screening assay
Necdin cDNA was cloned into a pGBKT7 plasmid and used as bait to screen a mouse embryo 11-day library (Clontech/ TaKaRa). Interacting clones were grown on a selection medium lacking the amino acids Leu and Trp and containing Aureobasidin A and X-α-Gal. Auxotrophic marker genes and the β-galactosidase assay were used to screen for positive clones. Plasmids of positive clones were isolated, amplified and sequenced. Interactions were further confirmed by retransforming the identified plasmids together with the bait. The identified DNA sequences were further characterized by BLAST analysis of the NCBI database (http://blast.ncbi.nlm.-nih.gov/Blast.cgi) to determine the identity of potential necdin-interacting proteins.
Western blotting
Cells or tissue samples were lysed in 2× SDS lysis buffer (2% SDS, 63 mM Tris–HCl, and 10% glycerol). Proteins in lysates were separated by SDS-PAGE, transferred to nitrocellulose membranes (PVDF), and immunoblotted with the corresponding antibodies overnight at 4°C. Membranes were then washed and incubated with horseradish peroxidase conjugate secondary antibodies. The proteins were visualized using the Pierce™ ECL Western Blotting Substrate kit (Thermo Scientific; 32106). Band intensities were quantified by ImageJ.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 6.0. All experiments were repeated at least three times and the distribution of data points is presented as mean ± s.e.m. Student's t-test for comparison of two conditions or ANOVAs were utilized with post hoc Bonferroni multiple-comparisons test for three or more conditions. P < 0.05 was considered significant.
RESULTS
Circadian behaviors and clock gene expression are altered in necdin-deficient mice
The SCN located in the hypothalamus is the pacemaker of the circadian clock in mammals. Using immunofluorescence staining, we found that necdin is highly expressed in the SCN (Supplementary Figure S1) and partly co-localizes with several SCN neuropeptides including arginine vasopressin (AVP), vasoactive intestinal polypeptide (VIP), gastrin releasing peptide (GRP) (Figure 1A). Nevertheless, necdin protein levels in the SCN were unchanged between the day and night (data not shown).
Figure 1.
Altered circadian behaviors and clock gene expression in necdin-deficient mice. (A) Expression of necdin (green), AVP (red), VIP and GRP (red) in the SCN using immunofluorescence staining. Scale bar, 100 μm. (B) Representative double-plotted actograms of wild-type (WT) and necdin KO mice subjected to an 8-h advanced jet-lag paradigm. The major activity onset during shift is labelled with red lines. (C) Statistics data of major activity onset plot from WT (n = 15) and necdin KO mice (n = 16). Data are presented as means ± s.e.m.; *P < 0.05; **P < 0.01; ***P < 0.005, Bonferroni post hoc analysis, two-way ANOVA. (D) Quantitative PCR analysis of the indicated clock genes in the livers from WT and Necdin KO mice taken at indicated ZTs of a day. Data are presented as means ± s.e.m.; *P < 0.05; **P < 0.01; ***P < 0.005, Bonferroni post hoc analysis, two-way ANOVA; n = 3 mice/genotype/time point. (E, F) Western blot analysis of liver nuclear extracts from WT and necdin KO mice over a 24 h circadian cycle (E) and the quantification of indicated protein levels (F); Data are presented as means ± s.e.m.; *P < 0.05; **P < 0.01; ***P < 0.005, Bonferroni post hoc analysis, two-way ANOVA; n = 3 mice/genotype/time point.
To elucidate the physiological function of necdin, we generated a mouse strain with a necdin-null allele (Supplementary Figure S2A and B). As necdin is a maternally imprinted gene, mice with defects in paternal necdin (necdin−p/+m) lack necdin expression (Supplementary Figure S2C and D). To address the function of necdin in circadian regulation, we monitored the wheel-running activity of necdin−p/+m mice and their wild-type (WT) littermate controls. Animals were housed individually in cages equipped with running wheels. After continuous monitoring of wheel running activities for 2–3 weeks under a 12:12-h light–dark (LD) schedule, mice were switched to constant darkness (DD) or constant light (LL) for 4 wk. Both WT and necdin−p/+m mice entrained to LD cycles and showed no significant differences in daily counts or amplitudes of locomotor rhythmicity (Supplementary Figure S3A). Under DD or LL conditions, both WT and necdin−p/+m mice showed similar free-running periods and fast Fourier transform (FFT) power levels (Supplementary Figure S3B, C and E). We also compared the phase shifts generated by exposure to a brief light pulse at CT16 (white light, ∼150 lux, 10 min) under DD conditions. In response to this treatment, we did not see any significant difference between WT and necdin−p/+m mice (WT, 121.0 ± 21.3 min, n = 6; necdin−p/+m, 113.7 ± 1.7 min, n = 6; P> 0.05) (Supplementary Figures S3A and S3D).
We then compared the adaptation of WT and necdin−p/+m mice to an 8-h advance jet-lag paradigm. WT mice gradually shifted their locomotor activity rhythms to the new LD schedule. Every subsequent day after the LD cycle advance, WT mice started their activity slightly earlier, and finally aligned it to the beginning of night after ∼11 days. In contrast, necdin−p/+m mice adapted more rapidly to the 8-h advance, requiring only ∼8 days for re-entrainment (Figure 1B, C). There was a significant difference in PS50 (50% phase-shift) values between the two strains (WT, 5.89 ± 0.11 days, n = 15; necdin−p/+m, 3.71 ± 0.12 days, n = 16; P < 0.05, Figure 1C). Although most necdin−p/+m mice showed a gradual shift to the new LD cycle, activity bouts clearly occurred between lights-off and the major activity onset during the shift; such bouts were absent in WT mice. Furthermore, Jet-lag treatment disrupted the behaviors in another two necdin−p/+m mice (Supplementary Figure S3F), implying an unstable central clock in these mice.
We also measured the expression levels of a panel of clock genes in the livers of WT and necdin−p/+m mice at different time points during a circadian cycle. As shown in Figure 1D, expression of the clock genes Per1/2, Cry1/2 and Dec1/2 showed a definite circadian rhythm in WT mice, the amplitude of which was significantly attenuated in necdin−p/+m mice. Necdin deficiency also led to significantly damped circadian amplitude of mCRY1 and mPER1 proteins in the liver nuclear extracts, whereas the patterns of mCRY2 and mPER2 in the liver nuclear extracts were largely intact (Figure 1E, F). Our data thus demonstrate an important role of necdin in the circadian regulation in mice.
Necdin interacts with BMAL1
To study the underlying mechanism by which necdin affects the circadian rhythm, we performed a yeast two hybrid screen using necdin as bait. As a result, BMAL1, a core component of the circadian clock, was identified as one of the necdin-interactive proteins. As shown in Figure 2A, only yeast colonies co-transformed with both pGBKT-necdin and pGADT7-Bmal1 plasmids grew and became blue on high-stringency plates (SD/-Trp/-Leu/AbA/ X-α-Gal), whereas no colonies grew when yeast was transformed with pGBKT7-necdin and pGADT7, or pGBKT7 and pGADT7-Bmal1.
Figure 2.
Necdin positively regulates BMAL1 protein levels. (A) Interaction of necdin and BMAL1 as assayed with a yeast two hybrid assay. The term pGBKT7-Ndn denotes necdin fused to a DNA-binding domain, whereas pGADT7-Bmal1 denotes BMAL1 fused to an activation domain. Only yeast colonies co-transformed with pGADT7-Bmal1 and pGBKT7-Ndn can grow and turn blue on a SD/-Leu/-Trp/AbA/X-α-Gal plate. (B) Interaction of necdin and BMAL1 as assayed by a co-immunoprecipitation (co-IP) assay in HEK293T cells expressing FLAG-necdin and HA-BMAL1. Antibodies against HA, FLAG were used to immunoprecipitate (IP) the cell lysate, and the immune complex was blotted with antibody against HA or FLAG, respectively. (C) Interaction of necdin and BMAL1 as assayed by a co-IP assay in U2OS cells. The cell lysate was immunoprecipitated with an antibody against necdin or control IgG, and the immune complex was blotted with antibodies against necdin or BMAL1, respectively. (D) Interaction of BMAL1 and full length or truncated necdin as assayed by a co-IP assay. HA-tagged BMAL1 and FLAG-tagged necdin fragments were expressed in HEK293T cells; the cell lysate was immunoprecipitated with FLAG antibody, and the immune complex was blotted with HA antibody. (E) Representative immunoblots (left) and statistics data of three independent experiments (right) from U2OS cells transfected with different doses of necdin or empty vector (EV). Data are presented as means ± s.e.m., *P < 0.05, post hoc Dunnett's t-test, one-way ANOVA. (F) Representative immunoblots (left) and statistics data of three independent experiments (right) from U2OS cells transfected with control siRNA (NC) or NDN siRNAs. Data are presented as means ± s.e.m.; *P < 0.05, post hoc Dunnett's t-test, one-way ANOVA. (G) Representative immunoblots (top) and statistics data of three independent experiments (bottom) from WT and necdin KO MEF cells. Data are presented as means ± s.e.m.; *P < 0.05, unpaired t-test. (H). Representative BMAL1 immunofluorescence in the SCN from WT and necdin KO mice taken at ZT0 and ZT12. (I) Immunoblots of BMAL1 and β-actin in the cortex, cerebellum, and liver from WT and necdin KO mice taken at the indicated ZTs of a day.
We further studied the necdin-BMAL1 interaction by using coimmunoprecipitation (co-IP). Only when HA-tagged BMAL1 and FLAG-tagged necdin were co-expressed in HEK293T cells, the FLAG antibody was able to pulldown HA-tagged BMAL1 and the HA antibody was able to pulldown FLAG-tagged necdin (Figure 2B). We also performed co-IP in U2OS cells with an antibody against necdin or control IgG. As shown in Figure 2C, BMAL1 was detected in the immunocomplex pulled down by necdin antibody, but not by control IgG, indicating that necdin interacts with BMAL1 endogenously.
To determine which domains in necdin might be involved in the interaction with BMAL1, we carried out co-IP using full length necdin or several truncated necdin fragments (amino acids 1–115, 1–170, 116–170, 190–325). As shown in Figure 2D, only full length and two N-terminal fragments (amino acids 1–170, 116–170) associated with BMAL1.
Necdin positively regulates BMAL1 protein levels
To study the functional consequences of the necdin-BMAL1 interaction, we first overexpressed different amounts of necdin in U2OS cells and measured the endogenous BMAL1 protein levels. As shown in Figure 2E, necdin dose-dependently increased endogenous BMAL1 protein levels. Conversely, knockdown of necdin with siRNAs significantly downregulated BMAL1 (Figure 2F). Moreover, BMAL1 protein levels were also significantly decreased in embryonic fibroblasts (MEFs) from Ndn−p/+m mice as compared with WT controls (Figure 2G), and the decrease was rescued by overexpression of necdin (Supplementary Figure S4A).
We also measured BMAL1 protein levels in a variety of tissues from WT and Ndn−p/+m mice. As shown in Figure 2H, BMAL1 levels in the SCN of Ndn−p/+m mice were significantly decreased as compared to WT controls as measured by immunofluorescence staining. Further, BMAL1 protein levels showed a circadian rhythm in the cortex (peak at ZT4, trough at ZT12, ratio of peak to trough was 1.9), cerebellum (peak at ZT0, trough at ZT16, ratio of peak to trough was 2) and liver (peak at ZT8, trough at ZT20, ratio of peak to trough was 2.7), which was significantly attenuated in Ndn−p/+m mice (Figure 2I and Supplementary Figure S5). Taken together, these data indicate that necdin positively regulates BMAL1 protein levels.
Depletion of necdin promotes BMAL1 degradation through the ubiquitin-proteasomal system
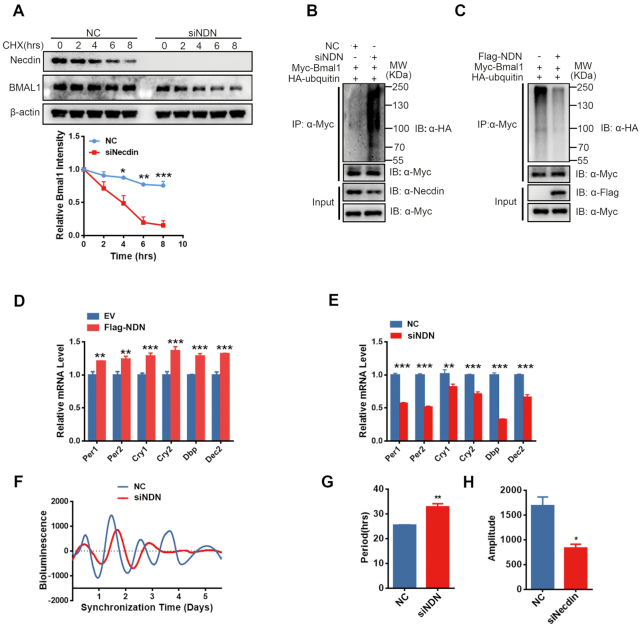
As overexpression or knockdown of necdin did not affect Bmal1 transcription (Supplementary Figure S6A, B), we speculated that necdin may regulate BMAL1 protein stability. Indeed, knockdown of necdin significantly decreased the half-life of BMAL1 (Figure 3A). Furthermore, overexpression of necdin markedly decreased, whereas knockdown of necdin increased the levels of ubiquitinylated BMAL1 (Figure 3B, C), indicating that depletion of necdin destabilizes BMAL1 through the ubiquitin-proteasomal system.
Figure 3.
Depletion of necdin promotes BMAL1 degradation through the ubiquitin proteasomal system. (A) Representative immunoblots (top) and statistics data of three independent experiments (bottom) from U2OS cells transfected with control siRNA (NC) or NDN siRNA and treated with 10 mg/ml cycloheximide (CHX) for 0–8 h. Data are presented as means ± s.e.m.; *P < 0.05; **P < 0.01; ***P < 0.005, Bonferroni post hoc analysis, two-way ANOVA. (B) Ubiquitination of BMAL1 in U2OS cells transfected with control siRNA (NC) or NDN siRNA. (C) Ubiquitination of BMAL1 in U2OS cells transfected with empty vector or necdin. (D, E) Quantitative PCR of the indicated clock genes in U2OS cells transfected with empty vector (EV) or necdin (D), or control siRNA (NC) or NDN siRNA (E). Data are presented as means ± s.e.m.; *P < 0.05; **P < 0.01; ***P < 0.005, Student's t-test, n = 3. (F) Representative bioluminescence signals from control (NC) or NDN-depleted (siNDN) U2OS cells stably expressing luciferase under the control of Bmal1 promoter. Cells were synchronized with 100 nM dexamethasone and recorded in a Luminometer. (G, H) Statistics data of the amplitudes (G) and periods (H) obtained from Bmal1:luciferase U2OS cells transfected with control siRNA (NC) or NDN siRNA. Data are presented as means ± s.e.m.; *P < 0.05; **P < 0.01; ***P < 0.005, Student's t-test, n = 6.
BMAL1 is one of the core components of the circadian clock. BMAL1 and CLOCK form a heterodimer that positively regulates the transcription of downstream genes through binding to the E-box elements on target gene promoters. We therefore measured the expression of several BMAL1-CLOCK target genes after overexpression or depletion of necdin in U2OS cells. As shown in Figure 3D and Figure 3E, overexpression of necdin increased, whereas depletion of necdin decreased the transcription of Per1/2, Cry1/2, Dbp and Dec2 in U2OS cells. We also measured the circadian rhythm of U2OS cells stably expressing luciferase under control of the Bmal1 promoter. Depletion of necdin significantly decreased the circadian amplitude (Control siRNA, 1690 ± 175.0; Necdin siRNA, 834.9 ± 76.1; P < 0.05; Figure 3F, G) and lengthened the circadian period (Control siRNA, 25.50 ± 0.06; Necdin siRNA, 32.83 ± 1.3; P < 0.01; Figure 3H).
Necdin bridges BMAL1 and SGT1 to form a complex
SGT1 is another necdin-interactive protein identified from the above-mentioned yeast two hybrid screening (Supplementary Figure S7A). We chose SGT1 for further study as SGT1 is a co-chaperone of HSP90, a protein which facilitates the maturation and stabilization of a wide range of client proteins and is a well-known regulator of the circadian clock in mammals, Arabidopsis and Drosophila (15–17).
We confirmed the interaction between necdin and SGT1 by a co-IP assay. Only when HA-tagged SGT1 and FLAG-tagged necdin were co-expressed in HEK293T cells, the FLAG antibody was able to pulldown HA-tagged SGT1 and the HA antibody was able to pulldown FLAG-tagged necdin (Figure 4A). We also performed co-IP in U2OS cells with an antibody against SGT1 or control IgG. As shown in Figure 4B, necdin was detected in the immunocomplex pulled-down by SGT1 antibody, but not by control IgG, indicating that necdin interacts with SGT1 endogenously.
Figure 4.
Necdin bridges BMAL1 to form a complex with SGT1. (A) Interaction of necdin and SGT1 as assayed by co-immunoprecipitation (co-IP) assay in HEK293T cells expressing FLAG-necdin and HA-SGT1. Antibodies against HA, FLAG were used to immunoprecipitate (IP) cell lysate, and the immune complex was blotted with antibody against HA or FLAG, respectively. (B) Interaction of necdin and BMAL1 as assayed by co-IP assay in U2OS cells. Cell lysate was immunoprecipitated with an antibody against SGT1 or control IgG, and the immune complex was blotted with antibodies against necdin or SGT1, respectively. (C) Interaction of BMAL1 and full length or truncated necdin as assayed by co-IP assay. HA-tagged SGT1 and FLAG-tagged necdin fragments were expressed in HEK293T cells, cell lysate was immunoprecipitated with FLAG antibody, and the immune-complex was blotted with HA antibody. A diagram indicating the interaction of necdin with BMAL1 and SGT1 is shown at the bottom. (D) An anti-necdin antibody was used to immunoprecipitate the cell lysate from WT and necdin-KO MEFs, and the immune complexes were blotted with the indicated antibodies. (E) Co-IP assay of HEK293T cells expressing Myc-BMAL1 and HA-SGT1. Antibodies against HA, Myc or control IgG were used to immunoprecipitate (IP) cell lysate, and the immune complex was blotted with antibody against HA or Myc, respectively. (F) An anti-Myc antibody or control IgG was used to immunoprecipitate cell lysate from HEK293T cells expressing FLAG-necdin, Myc-BMAL1 or HA-SGT1; immune-complexes were blotted with the indicated antibodies.
SGT1 bound to full-length necdin or to a C-terminal fragment (amino acids 190–325) of necdin in the co-IP assay (Figure 4C). We therefore speculate that necdin may bridge BMAL1 and SGT1 to form a complex (Figure 4C, bottom). Indeed, antibody against SGT1 was able to pull down BMAL1 in U2OS cells (Supplementary Figure S7B) and in WT MEF cells, but not in Ndn−p/+m MEF cells (Figure 4D). When HA-SGT1 and Myc-BMAL1 were co-expressed in HEK293T cells, HA antibody failed to pull down Myc-BMAL1 (Figure 4E). Similarly, Myc antibody was unable to pull down HA-SGT1 (Figure 4E). Yet, HA antibody pulled down Myc-BMAL1 when Myc-BMAL1, HA-SGT1 and FLAG-necdin were co-expressed in HEK293T cells (Figure 4F). These results indicate that necdin was required and sufficient for formation of a complex between it, BMAL1 and SGT1.
SGT1 regulates BMAL1 stability in a necdin-dependent manner
To investigate the effect of SGT1 on BMAL1, we knocked down SGT1 by siRNA in U2OS cells and measured the endogenous BMAL1 protein levels. As shown in Figure 5A, depletion of SGT1 significantly decreased BMAL1 protein levels. Moreover, knockdown of SGT1 decreased the half-life of BMAL1 (Figure 5B) and increased ubiquitination of BMAL1 (Figure 5C). Conversely, overexpression of SGT1 dose-dependently increased BMAL1 protein levels (Figure 5D). Nevertheless, necdin is required for SGT1 to stabilize BMAL1, as overexpression of SGT1 failed to increase the BMAL1 protein level in necdin-depleted U2OS cells (Figure 5E) or Ndn−p/+m MEF cells (Figure 5F).
Figure 5.
SGT1 regulates BMAL1 stability in a necdin-dependent manner. (A) The representative immunoblots (top) and statistics data of three independent experiments (bottom) from U2OS cells transfected with control siRNA (NC) or Sgt1 siRNAs. Data are presented as means ± s.e.m.; **P < 0.01, post hoc Dunnett's t-test, one-way ANOVA. (B) Representative immunoblots (top) and statistics data of three independent experiments (bottom) from U2OS cells transfected with control siRNA (NC) or Sgt1 siRNA and treated with 10 mg/ml CHX for 0–12 h. Data are presented as means ± s.e.m.; *P < 0.05; **P < 0.01; ***P < 0.005, Bonferroni post hoc analysis, two-way ANOVA. (C) The ubiquitination of BMAL1 in U2OS cells transfected with control siRNA (NC) or Sgt1 siRNA. (D) Representative immunoblots (top) and statistics data of three independent experiments (bottom) from U2OS cells transfected with different doses of NDN or empty vector (EV). Data are presented as means ± s.e.m.; *P < 0.05, post hoc Dunnett's t-test, one-way ANOVA. (E) SGT1 failed to increase the BMAL1 protein level in necdin-depleted (siNDN) U2OS cells. Representative immunoblots are shown at left; statistics data of three independent experiments are shown at right. Data are presented as means ± s.e.m.; *P < 0.05, unpaired t-test. (F) SGT1 failed to increase BMAL1 protein levels in necdin-KO MEF cells. Representative immunoblots are shown at left; statistics data of three independent experiments are shown at right. Data are presented as means ± s.e.m.; *P < 0.05, unpaired t-test. (G, H) Quantitative PCR of the indicated clock genes in U2OS cells transfected with empty vector (EV) or SGT (G), or control siRNA (NC) or NDN siRNA (H). Data are presented as means ± s.e.m.; *P < 0.05; **P < 0.01; ***P < 0.005, Student's t-test, n = 3. (I) Representative bioluminescence signals from control (NC) or Sgt1-depleted (siSgt1) U2OS cells stably expressing luciferase under the control of the Bmal1 promoter. Cells were synchronized with 100 nM dexamethasone and luminescence was recorded by a Luminometer.
As expected, overexpression of SGT1 increased, whereas depletion of Sgt1 decreased the transcription of Per1/2, Cry1/2, Dbp and Dec2 in U2OS cells (Figure 5G, H). Depletion of SGT1 in U2OS cells significantly decreased its circadian amplitude (Control siRNA, 2062 ± 184.93; Sgt1 siRNA, 920.3 ± 35.80; P < 0.05; Figure 5I and Supplementary Figure S8A) and lengthened its circadian period (Control siRNA, 24.17 ± 0.52; Sgt1 siRNA, 26.70 ± 0.35, P < 0.05; Figure 5I and Supplementary Figure S8B).
Necdin regulates BMAL1 stability in an SGT1- and HSP90-dependent manner
SGT1 is a co-chaperone for heat shock protein 90 (HSP90); among its effects, Schneider et al. have shown that HSP90 can regulate the stability of BMAL1 and circadian gene expression. Consistent with their data, HSP90 inhibitors 17-AAG and geldanamycin (GA) downregulated BMAL1 protein levels (Figure 6A, B). Furthermore, treatment with HSP90 inhibitors 17-AAG or GA decreased the transcription of Per1/2, Cry1, Dbp, and Dec2 in U2OS cells (Figure 6C, D). Moreover, treatment with 17-AAG or GA decreased the amplitude of the circadian rhythm (DMSO, 1691 ± 115.9; 17-AAG, 767.2 ± 83.10; GA, 293.7 ± 20.95; P < 0.05; Figure 6E and Figure 6F), and increased the circadian period (DMSO, 25.83 ± 0.20 h; 17-AAG, 33.10 ± 2.14 h; GA, 36.37 ± 0.23 h; P < 0.05; Figure 6E and G). Interestingly, overexpression of necdin in SGT1-depleted U2OS cells failed to increase BMAL1 protein levels (Figure 7A), indicating that SGT1 is required for necdin to stabilize BMAL1 protein. Furthermore, overexpression of necdin in GA-treated U2OS cells failed to increase the BMAL1 protein level (Figure 7B), indicating the requirement of HSP90 for necdin to stabilize BMAL1 protein.
Figure 6.
HSP90 regulates BMAL1 stability and clock gene expression. (A, B) Representative immunoblots (left) and statistics data of three independent experiments (right) from U2OS cells following treatment with different concentrations of 17-AAG (A) or GA (B). Data are presented as means ± s.e.m.; *P < 0.05; **P < 0.01, post hoc Dunnett's t-test, one way ANOVA. (C, D) Quantitative PCR of the indicated clock genes in U2OS cells following treatment with 17-AAG (C) or GA (D). Data are presented as means ± s.e.m.; *P < 0.05; **P < 0.01; ***P < 0.005, Student's t test, n = 3. (E) Representative bioluminescence signals from control (DMSO)-, 17-AAG- or GA-treated U2OS cells stably expressing luciferase under control of the Bmal1 promoter. Cells were synchronized with 100 nM dexamethasone and recorded by a Luminometer. (F, G) Statistics data of the amplitudes (F) and periods (G) obtained from Bmal1:luciferase U2OS cells treated with DMSO, 17-AAG or GA. Data are presented as means ± s.e.m.; *P < 0.05; **P < 0.01; ***P < 0.005, post hoc Dunnett's t-test, one way ANOVA. n = 6.
Figure 7.
Necdin regulates BMAL1 stability in a SGT1- and HSP90-dependent manner (A) Necdin failed to increase BMAL1 protein levels in Sgt1-depleted (siSgt1) U2OS cells. Representative immunoblots are shown at top; statistics data of three independent experiments are shown at bottom; data are presented as means ± s.e.m.; *P < 0.05, unpaired t test. (B) Necdin failed to increase BMAL1 protein levels in U2OS cells treated with geldanamycin (GA). Representative immunoblots are shown at top; statistics data of three independent experiments are shown at bottom; data are presented as means ± s.e.m.; *P < 0.05, unpaired t-test.
DISCUSSION
Roles of PWS-associated proteins in the circadian clock
PWS-related sleep problems may arise from a variety of mechanisms. Defects in REM sleep may be due to disrupted sleep homeostatic regulation, whereas sleep-disordered breathing may be a consequence of obesity (18). REM sleep defects and/or sleep-disordered breathing may lead to excessive daytime sleepiness (19), which may also be attributable to dysfunctional circadian clocks.
Magel2 is the first PWS-associated gene that is reported to be involved in sleep regulation. Kozlov et al. found that mice deficient in Magel2 showed markedly reduced circadian amplitude of activity and increased daytime activity. They further showed that orexin, a critical wake-promoting protein, is substantially reduced in the lateral hypothalamus of Magel2-deficient mice (20). These data indicate that Magel2 deficiency may be responsible for, at least partly, the sleep homeostatic dysregulation in PWS patients. Nevertheless, Devos et al. found that Magel2 is able to repress the activity of the CLOCK-BMAL1 heterodimer in a Per2-luciferase assay, implying a potential role in circadian regulation (21).
Magel2 and necdin belong to the so-called MAGE protein family (22). Interestingly, Xu and colleagues showed that mice lacking MAGED1, a MAGE family member, leads to a shortened period and altered rest-activity bouts. They further identified MAGED1 as an RORα-binding protein that regulates the expression of core clock genes such as Bmal1, Rev-erbα and E4bp4 (23).
In this study, we showed that the PWS-associated protein necdin interacts with the core circadian component BMAL1. Necdin stabilized BMAL1 through SGT1-HSP90 chaperone machinery. Deficiency in necdin led to altered clock gene expression and circadian behavior, which may underlie part of the circadian dysfunction in PWS patients.
The roles of HSP90 and protein quality control in circadian clocks
In addition to the interconnected transcriptional and translational feedback loops, the circadian clock is also regulated by numerous post-transcriptional and post-translational processes. The protein homeostasis process, involving the synthesis, folding, maturation, and turnover of polypeptides, has been recognized as being essential for circadian regulation (24).
To prevent mis-folding and facilitate folding, a wide range of molecular chaperones function in cells to stabilize and/or help a non-native protein acquire its native conformation (25,26). In particular, the HSP90-related protein quality control pathway has been reported to participate in circadian control in flies, plants and mammals (15–17).
HSP90 functions downstream of the ribosome-bound HSP70 in the structural maturation and conformational regulation of numerous molecules (27). In Arabidopsis, HSP90 stabilizes the circadian clock-associated F-box protein ZEITLUPE. Further studies indicate that GIGANTEA acts as a co-chaperone of HSP90 to facilitate the maturation of ZEITLUPE (28). In Drosophila, pharmacologically or genetically reduced HSP90 levels increase behavioral variability of circadian locomotor activity (17).
In mammals, Schneider et al. indicated that inhibition of the ATP-dependent chaperone activity of HSP90 impairs circadian rhythmicity of cultured mouse fibroblasts. Inhibition of HSP90 shortened the half-life of BMAL1 and blunted expression of rhythmic BMAL1-CLOCK target genes (15).
In this study, we found that necdin may enable SGT1-HSP90 chaperone machinery to stabilize BMAL1. Although necdin levels did not show circadian variation in the SCN or liver, we speculate that the circadian rhythm of BMAL1 protein levels may be controlled in part by the oscillation of HSP90, as levels of two forms of HSP90 (HSP86 and HSP84) in the mouse SCN displayed an overt circadian rhythm with a peak at night and a trough during the day (29).
The mechanism underlying how necdin affects protein stability and function
Necdin is involved in a variety of physiological functions, including, adipogenesis, myogenesis as well as neuronal differentiation, survival and migration (30,31). Deficiency in necdin leads to hypothalamic dysfunction and central apnea, which mimic some symptoms of' PWS (32).
Necdin may regulate its target genes/proteins at either the transcriptional or post-translational levels. Although necdin has no transcriptional activity, it may interact with transcription activators/repressors to regulate transcription indirectly. Necdin may enhance target gene expression by relieving the repression of some transcriptional suppressors, such as Bmi1, Msx and EID-1 (33–35). Necdin may also down-regulate gene expression by suppressing the activity of certain transactivators, such as p53, Sp1, E2F1, ARNT2:SIM1 and ARNT2:HIF1α (36–39). Furthermore, necdin has been shown to regulate PPARγ expression through an epigenetic mechanism, by affecting both the recruitment of MeCP2 (methyl CpG binding protein 2) and HP-1α co-repressor to the Pparγ promoter as well as the H3K27 dimethylation status at the exon 5 locus (40).
Necdin may antagonize transcription repressors or activators at the post-translational level. Yoshikawa and colleagues found that necdin is able to recruit Sirt1 and consequently facilitate the de-acetylation of Foxo1 or p53, leading to diminished transcription activity of these two factors (41,42).
Necdin may also regulate the ubiquitination and stability of proteins with which it interacts. In this scenario, necdin stabilizes PGC-1α and leptin receptor, but promotes the degradation of HIF-1α, PIAS1 and CCAR1 (43–46). Although the underlying mechanism is largely unknown, Wijesuriya et al. demonstrated that necdin stabilizes the leptin receptor through interaction with MAGEL2 as well as with RNF41 and USP8 (47).
Necdin may also enhance the cytoplasmic retention of proteins such as EID-1 and NEFA. Interestingly, cytoplasmic retention of EID-1 relieves its transcriptional repression, whereas enhanced cytoplasmic retention of NEFA potentiates its effect on the caffeine-evoked elevation of cytosolic Ca2+ levels (48).
Our study indicates that necdin may serve as a scaffold protein, facilitating SGT1-HSP90 chaperone machinery to stabilize the core circadian clock component BMAL1. Nevertheless, we cannot totally exclude the possibility that necdin may also regulate the BMAL1 protein level through other mechanisms. It will be intriguing to study whether necdin also affects the translation of BMAL1 protein.
Post-translational regulation of BMAL1
The transcription of Bmal1 is positively regulated by RORα, and antagonized by Rev-Erbα (10). In addition, BMAL1 protein is subjected to extensive post-translational modifications, such as acetylation, ubiquitination, sumoylation and phosphorylation (49–52). Interestingly, the acetylation and deacetylation of BMAL1 is catalyzed by its partner CLOCK and Sirt1, respectively. Although necdin is able to recruit Sirt1 to de-acetylate Foxo1 and p53, it is unlikely that this pathway explains the effect of necdin on BMAL1. First, BMAL1 acetylation does not affect its stability (53), whereas necdin positively regulates BMAL1 protein levels. Second, BMAL1 acetylation facilitates recruitment of CRY1 to CLOCK-BMAL1, thereby promoting transcriptional repression (53). In contradistinction, necdin correlates positively with the transcriptional activity of BMAL1.
The effect of necdin deficiency on Per2 expression
It should be noted that Per2 mRNA level was only subtly downregulated in the livers from necdin−p/+m mice, although BMAL1 protein levels were significantly damped. The oscillation of Per2 in necdin-deficient mice may be driven by the system cues, such as body temperature and feeding. In a seminar study, Kornmann et al. repressed hepatic Bmal1 transcription by overexpressing Rev-erba in the liver. Although this manipulation damped the rhythmic transcription of most genes, the expression of Per2 oscillated robustly (54,55).
Consistent with the mRNA levels, the circadian rhythm of mPER2 from the liver nuclear extracts were also largely intact in necdin−p/+m mice. PER1, PER2, CRY1 and CRY2 proteins form a repressor complex with a molecular mass of ∼2 MDa. Among them, PER2 may be the rate-limiting component for the formation of this repressor complex. Schibler et al. speculate that CRY proteins and PER1 may be produced in excess, and the proteins that are not incorporated into the repressosome complex may be rapidly degraded (56). Indeed, necdin-deficiency did not alter the pattern of mCRY2 in the liver nuclear extracts, in spite of the significant downregulation of Cry2 mRNA levels.
In conclusion, we have demonstrated that the PWS protein necdin regulates BMAL1 stability and circadian rhythm amplitudes through the SGT1-HSP90 pathway. Our data uncovered a possible mechanism underlying the sleep disorders observed in PWS patients, and further highlighted an important role for protein homeostasis in circadian control. It will be intriguing to identify co-chaperones/ chaperones that control other components of the circadian clock.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Edwards Sharman for language editing.
Contributor Information
Renbin Lu, Center for Medical Genetics, School of Life Sciences, Central South University, Changsha 410078, Hunan, P. R. China; Hunan Key Laboratory of Animal Models for Human Diseases, Changsha 410078, Hunan, P. R. China.
Yufan Dong, Center for Medical Genetics, School of Life Sciences, Central South University, Changsha 410078, Hunan, P. R. China.
Jia-Da Li, Center for Medical Genetics, School of Life Sciences, Central South University, Changsha 410078, Hunan, P. R. China; Hunan Key Laboratory of Animal Models for Human Diseases, Changsha 410078, Hunan, P. R. China; Hunan Key Laboratory of Medical Genetics, Changsha 410078, Hunan, P. R. China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [81770780, 81728013, 31972913 to J.-D.L.]; Key Research and Development Programs from Hunan Province [2018DK2010, 2018DK2013 to J.-D.L.]; Guangdong Key Project in ‘Development of new tools for diagnosis and treatment of Autism’ [2018B030335001 to J.-D.L.]; Strategic Priority Research Program of Central South University [ZLXD2017004 to J.-D.L.]; Fundamental Research Funds for the Central Universities of Central South University[1053320171214 to R.L.]. Funding for open access charge: National Natural Science Foundation of China [81770780, 81728013, 31972913 to J.-D.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Goldstone A.P. Prader-Willi syndrome: advances in genetics, pathophysiology and treatment. Trends Endocrinol. Metab. 2004; 15:12–20. [DOI] [PubMed] [Google Scholar]
- 2. Murrell A. Cross-talk between imprinted loci in Prader-Willi syndrome. Nat. Genet. 2014; 46:528–530. [DOI] [PubMed] [Google Scholar]
- 3. Cassidy S.B., Schwartz S., Miller J.L., Driscoll D.J.. Prader-Willi syndrome. Genet. Med. 2012; 14:10–26. [DOI] [PubMed] [Google Scholar]
- 4. Nixon G.M., Brouillette R.T.. Sleep and breathing in Prader-Willi syndrome. Pediatr. Pulmonol. 2002; 34:209–217. [DOI] [PubMed] [Google Scholar]
- 5. Franken P. A role for clock genes in sleep homeostasis. Curr. Opin. Neurobiol. 2013; 23:864–872. [DOI] [PubMed] [Google Scholar]
- 6. Chong S.Y., Ptacek L.J., Fu Y.H.. Genetic insights on sleep schedules: this time, it's PERsonal. Trends Genet. 2012; 28:598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bass J., Takahashi J.S.. Circadian integration of metabolism and energetics. Science. 2010; 330:1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Panda S. Circadian physiology of metabolism. Science. 2016; 354:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schibler U. The daily rhythms of genes, cells and organs. Biological clocks and circadian timing in cells. EMBO Rep. 2005; 6(Suppl. 1):S9–S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takahashi J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017; 18:164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nakada Y., Taniura H., Uetsuki T., Inazawa J., Yoshikawa K.. The human chromosomal gene for necdin, a neuronal growth suppressor, in the Prader-Willi syndrome deletion region. Gene. 1998; 213:65–72. [DOI] [PubMed] [Google Scholar]
- 12. Hastings M.H., Maywood E.S., Brancaccio M.. Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 2018; 19:453–469. [DOI] [PubMed] [Google Scholar]
- 13. Top D., Young M.W.. Coordination between differentially regulated circadian clocks generates rhythmic behavior. Cold Spring Harb. Perspect. Biol. 2018; 10:a033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Okamura H., Doi M., Fustin J.M., Yamaguchi Y., Matsuo M.. Mammalian circadian clock system: Molecular mechanisms for pharmaceutical and medical sciences. Adv. Drug. Deliv. Rev. 2010; 62:876–884. [DOI] [PubMed] [Google Scholar]
- 15. Schneider R., Linka R.M., Reinke H.. HSP90 affects the stability of BMAL1 and circadian gene expression. J. Biol. Rhythms. 2014; 29:87–96. [DOI] [PubMed] [Google Scholar]
- 16. Kim T.S., Kim W.Y., Fujiwara S., Kim J., Cha J.Y., Park J.H., Lee S.Y., Somers D.E.. HSP90 functions in the circadian clock through stabilization of the client F-box protein ZEITLUPE. PNAS. 2011; 108:16843–16848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hung H.C., Kay S.A., Weber F.. HSP90, a capacitor of behavioral variation. J. Biol. Rhythms. 2009; 24:183–192. [DOI] [PubMed] [Google Scholar]
- 18. Miller J., Wagner M.. Prader-Willi syndrome and sleep-disordered breathing. Pediatr. Ann. 2013; 42:200–204. [DOI] [PubMed] [Google Scholar]
- 19. Camfferman D., McEvoy R.D., O’Donoghue F., Lushington K.. Prader Willi Syndrome and excessive daytime sleepiness. Sleep Med. Rev. 2008; 12:65–75. [DOI] [PubMed] [Google Scholar]
- 20. Kozlov S.V., Bogenpohl J.W., Howell M.P., Wevrick R., Panda S., Hogenesch J.B., Muglia L.J., Van Gelder R.N., Herzog E.D., Stewart C.L.. The imprinted gene Magel2 regulates normal circadian output. Nat. Genet. 2007; 39:1266–1272. [DOI] [PubMed] [Google Scholar]
- 21. Devos J., Weselake S.V., Wevrick R.. Magel2, a Prader-Willi syndrome candidate gene, modulates the activities of circadian rhythm proteins in cultured cells. J. Circadian Rhythms. 2011; 9:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barker P.A., Salehi A.. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J. Neurosci. Res. 2002; 67:705–712. [DOI] [PubMed] [Google Scholar]
- 23. Wang X., Tang J., Xing L., Shi G., Ruan H., Gu X., Liu Z., Wu X., Gao X., Xu Y.. Interaction of MAGED1 with nuclear receptors affects circadian clock function. EMBO J. 2010; 29:1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hirano A., Fu Y.H., Ptacek L.J.. The intricate dance of post-translational modifications in the rhythm of life. Nat. Struct. Mol. Biol. 2016; 23:1053–1060. [DOI] [PubMed] [Google Scholar]
- 25. Wolff S., Weissman J.S., Dillin A.. Differential scales of protein quality control. Cell. 2014; 157:52–64. [DOI] [PubMed] [Google Scholar]
- 26. Bukau B., Weissman J., Horwich A.. Molecular chaperones and protein quality control. Cell. 2006; 125:443–451. [DOI] [PubMed] [Google Scholar]
- 27. Hartl F.U., Bracher A., Hayer-Hartl M.. Molecular chaperones in protein folding and proteostasis. Nature. 2011; 475:324–332. [DOI] [PubMed] [Google Scholar]
- 28. Cha J.Y., Kim J., Kim T.S., Zeng Q., Wang L., Lee S.Y., Kim W.Y., Somers D.E.. GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat. Commun. 2017; 8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fukuyama T., Doi M., Matsuo M., Nishinaga H., Miyake S., Okamura H.. Circadian expression of 86- and 84-kDa heat shock proteins in the mouse suprachiasmatic nucleus. Biomed. Res. 2008; 29:93–98. [DOI] [PubMed] [Google Scholar]
- 30. Kubota Y., Osawa M., Jakt L.M., Yoshikawa K., Nishikawa S.. Necdin restricts proliferation of hematopoietic stem cells during hematopoietic regeneration. Blood. 2009; 114:4383–4392. [DOI] [PubMed] [Google Scholar]
- 31. Takazaki R., Nishimura I., Yoshikawa K.. Necdin is required for terminal differentiation and survival of primary dorsal root ganglion neurons. Exp. Cell Res. 2002; 277:220–232. [DOI] [PubMed] [Google Scholar]
- 32. Muscatelli F., Abrous D.N., Massacrier A., Boccaccio I., Le Moal M., Cau P., Cremer H.. Disruption of the mouse Necdin gene results in hypothalamic and behavioral alterations reminiscent of the human Prader-Willi syndrome. Hum. Mol. Genet. 2000; 9:3101–3110. [DOI] [PubMed] [Google Scholar]
- 33. Minamide R., Fujiwara K., Hasegawa K., Yoshikawa K.. Antagonistic interplay between necdin and Bmi1 controls proliferation of neural precursor cells in the embryonic mouse neocortex. PLoS One. 2014; 9:e84460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kuwajima T., Taniura H., Nishimura I., Yoshikawa K.. Necdin interacts with the Msx2 homeodomain protein via MAGE-D1 to promote myogenic differentiation of C2C12 cells. J. Biol. Chem. 2004; 279:40484–40493. [DOI] [PubMed] [Google Scholar]
- 35. Bush J.R., Wevrick R.. The Prader-Willi syndrome protein necdin interacts with the E1A-like inhibitor of differentiation EID-1 and promotes myoblast differentiation. Differentiation. 2008; 76:994–1005. [DOI] [PubMed] [Google Scholar]
- 36. Taniura H., Matsumoto K., Yoshikawa K.. Physical and functional interactions of neuronal growth suppressor necdin with p53. J. Biol. Chem. 1999; 274:16242–16248. [DOI] [PubMed] [Google Scholar]
- 37. Matsumoto K., Taniura H., Uetsuki T., Yoshikawa K.. Necdin acts as a transcriptional repressor that interacts with multiple guanosine clusters. Gene. 2001; 272:173–179. [DOI] [PubMed] [Google Scholar]
- 38. Taniura H., Taniguchi N., Hara M., Yoshikawa K.. Necdin, a postmitotic neuron-specific growth suppressor, interacts with viral transforming proteins and cellular transcription factor E2F1. J. Biol. Chem. 1998; 273:720–728. [DOI] [PubMed] [Google Scholar]
- 39. Friedman E.R., Fan C.M.. Separate necdin domains bind ARNT2 and HIF1alpha and repress transcription. Biochem. Biophys. Res. Commun. 2007; 363:113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhu N.L., Wang J., Tsukamoto H.. The Necdin-Wnt pathway causes epigenetic peroxisome proliferator-activated receptor gamma repression in hepatic stellate cells. J. Biol. Chem. 2010; 285:30463–30471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hasegawa K., Yoshikawa K.. Necdin regulates p53 acetylation via Sirtuin1 to modulate DNA damage response in cortical neurons. J. Neurosci. 2008; 28:8772–8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hasegawa K., Kawahara T., Fujiwara K., Shimpuku M., Sasaki T., Kitamura T., Yoshikawa K.. Necdin controls Foxo1 acetylation in hypothalamic arcuate neurons to modulate the thyroid axis. J. Neurosci. 2012; 32:5562–5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hasegawa K., Yasuda T., Shiraishi C., Fujiwara K., Przedborski S., Mochizuki H., Yoshikawa K.. Promotion of mitochondrial biogenesis by necdin protects neurons against mitochondrial insults. Nat. Commun. 2016; 7:10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moon H.E., Ahn M.Y., Park J.A., Min K.J., Kwon Y.W., Kim K.W.. Negative regulation of hypoxia inducible factor-1alpha by necdin. FEBS Lett. 2005; 579:3797–3801. [DOI] [PubMed] [Google Scholar]
- 45. Gur I., Fujiwara K., Hasegawa K., Yoshikawa K.. Necdin promotes ubiquitin-dependent degradation of PIAS1 SUMO E3 ligase. PLoS One. 2014; 9:e99503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Francois S., D’Orlando C., Fatone T., Touvier T., Pessina P., Meneveri R., Brunelli S.. Necdin enhances myoblasts survival by facilitating the degradation of the mediator of apoptosis CCAR1/CARP1. PLoS One. 2012; 7:e43335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wijesuriya T.M., De Ceuninck L., Masschaele D., Sanderson M.R., Carias K.V., Tavernier J., Wevrick R.. The Prader-Willi syndrome proteins MAGEL2 and necdin regulate leptin receptor cell surface abundance through ubiquitination pathways. Hum. Mol. Genet. 2017; 26:4215–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Taniguchi N., Taniura H., Niinobe M., Takayama C., Tominaga-Yoshino K., Ogura A., Yoshikawa K.. The postmitotic growth suppressor necdin interacts with a calcium-binding protein (NEFA) in neuronal cytoplasm. J. Biol. Chem. 2000; 275:31674–31681. [DOI] [PubMed] [Google Scholar]
- 49. Nakahata Y., Kaluzova M., Grimaldi B., Sahar S., Hirayama J., Chen D., Guarente L.P., Sassone-Corsi P.. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008; 134:329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gossan N.C., Zhang F., Guo B., Jin D., Yoshitane H., Yao A., Glossop N., Zhang Y.Q., Fukada Y., Meng Q.J.. The E3 ubiquitin ligase UBE3A is an integral component of the molecular circadian clock through regulating the BMAL1 transcription factor. Nucleic Acids Res. 2014; 42:5765–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cardone L., Hirayama J., Giordano F., Tamaru T., Palvimo J.J., Sassone-Corsi P.. Circadian clock control by SUMOylation of BMAL1. Science. 2005; 309:1390–1394. [DOI] [PubMed] [Google Scholar]
- 52. Sanada K., Okano T., Fukada Y.. Mitogen-activated protein kinase phosphorylates and negatively regulates basic helix-loop-helix-PAS transcription factor BMAL1. J. Biol. Chem. 2002; 277:267–271. [DOI] [PubMed] [Google Scholar]
- 53. Hirayama J., Sahar S., Grimaldi B., Tamaru T., Takamatsu K., Nakahata Y., Sassone-Corsi P.. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007; 450:1086–1090. [DOI] [PubMed] [Google Scholar]
- 54. Kornmann B., Schaad O., Bujard H., Takahashi J.S., Schibler U.. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007; 5:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schibler U., Gotic I., Saini C., Gos P., Curie T., Emmenegger Y., Sinturel F., Gosselin P., Gerber A., Fleury-Olela F et al.. Clock-Talk: interactions between central and peripheral circadian oscillators in mammals. Cold Spring Harb. Symp. Quant. Biol. 2015; 80:223–232. [DOI] [PubMed] [Google Scholar]
- 56. Aryal R.P., Kwak P.B., Tamayo A.G., Gebert M., Chiu P.L., Walz T., Weitz C.J.. Macromolecular assemblies of the mammalian circadian clock. Mol. Cell. 2017; 67:770–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.