Abstract
During breast cancer metastasis, the developmental process epithelial–mesenchymal transition (EMT) is abnormally activated. Transcriptional regulatory networks controlling EMT are well-studied; however, alternative RNA splicing also plays a critical regulatory role during this process. A comprehensive understanding of alternative splicing (AS) and the RNA binding proteins (RBPs) that regulate it during EMT and their impact on breast cancer remains largely unknown. In this study, we annotated AS in the breast cancer TCGA data set and identified an AS signature that is capable of distinguishing epithelial and mesenchymal states of the tumors. This AS signature contains 25 AS events, among which nine showed increased exon inclusion and 16 showed exon skipping during EMT. This AS signature accurately assigns the EMT status of cells in the CCLE data set and robustly predicts patient survival. We further developed an effective computational method using bipartite networks to identify RBP-AS networks during EMT. This network analysis revealed the complexity of RBP regulation and nominated previously unknown RBPs that regulate EMT-associated AS events. This study highlights the importance of global AS regulation during EMT in cancer progression and paves the way for further investigation into RNA regulation in EMT and metastasis.
Keywords: alternative splicing (AS), epithelial–mesenchymal transition (EMT), RNA-binding proteins (RBPs), breast cancer
INTRODUCTION
Alternative splicing (AS) is the process whereby a single gene transcript is spliced to generate numerous RNA isoforms with the potential to encode different proteins. It has been reported that nearly all human genes undergo AS, and alternatively spliced isoforms have been shown to perform distinct functional roles in the cell (Wang et al. 2008; Nilsen and Graveley 2010). The process of AS is regulated by various cis-elements that are located in the vicinity of variable exons of pre-mRNAs. These cis-elements are recognized by cognate RNA-binding proteins (RBPs) to influence splice site recognition by the spliceosome. Splicing-regulatory RBPs can have positive or negative effects on the inclusion of alternative exons and they frequently interact with one another in complexes to modulate splicing regulatory action (Fu and Ares 2014; Damianov et al. 2016; Ying et al. 2017). Therefore, the relationship between RBPs and their target splicing events is highly context-dependent, and deciphering the “splicing code” is a continuing challenge in the field (Barash et al. 2010). Developing computational methods to understand how and to what extent different groups of RBPs regulate specific splicing events is of great interest.
The epithelial–mesenchymal transition (EMT) is a developmental process where epithelial cells lose their cell-cell adhesions and apical-basal polarity while acquiring migratory and invasive properties characteristic of mesenchymal cells (Thiery and Sleeman 2006; Thiery et al. 2009). Numerous studies have linked abnormal activation of EMT to tumor metastasis, which remains the leading cause of death in cancer patients (Yang and Weinberg 2008). Genome-wide AS analysis revealed dynamic changes of AS during EMT (Shapiro et al. 2011; Yang et al. 2016). A handful of AS events and RBPs have also shown functional significance in EMT (Warzecha et al. 2009, 2010; Brown et al. 2011; Reinke et al. 2012; Lu et al. 2013; Huang et al. 2017; Li et al. 2018; Hu et al. 2020). Given the strong relationship between EMT and cancer metastasis, analysis of cancer patient data sets to study AS and regulatory RBPs during EMT could be clinically informative in finding drivers of tumor metastasis.
In this study, we made use of breast invasive carcinoma (BRCA) RNA-sequencing data sets available through The Cancer Genome Atlas (TCGA). We derived an AS signature of EMT that is capable of discriminating epithelial and mesenchymal tumor samples and faithfully predicting patient outcomes. In order to comprehensively address the combinatorial regulation of AS by RBPs during EMT, we developed a bipartite network to associate multiple RBPs with target splicing events. Through community detection, we identified novel clusters of RBPs involved in combinatorial regulation of AS. These clusters contain newly identified EMT-associated RBPs and splicing events that have the power to predict patient survival.
RESULTS
Identification of EMT-associated AS events in human breast cancer
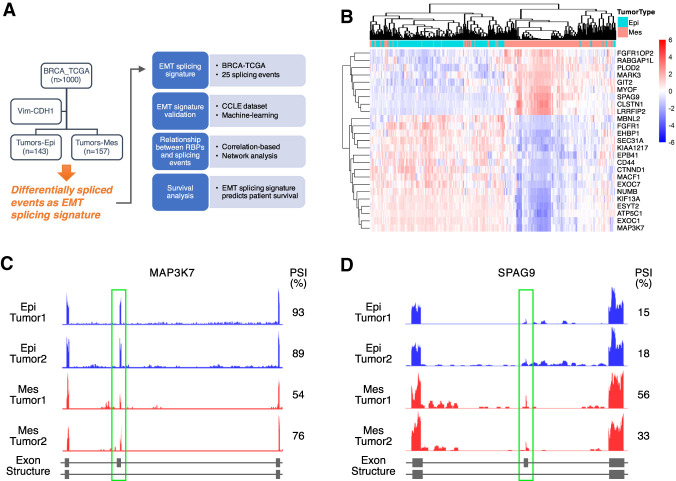
To identify important AS events that are associated with EMT in breast cancer, we first developed a method to differentiate between epithelial and mesenchymal tumors in TCGA-BRCA samples (Fig. 1A). We used E-cadherin (CDH1) and Vimentin (VIM), two genes highly expressed in epithelial and mesenchymal cell states, respectively, to separate tumor specimens into either epithelial or mesenchymal tumors. This method of characterizing EMT status was previously shown to be simple and robust (Park et al. 2008). Our rationale of choosing CDH1 and VIM is that down-regulation of E-cadherin is a hallmark of EMT which results in disassembly of the cell adherens junctions, and loss of E-cadherin is implicated in cancer progression and metastasis (Hirohashi 1998; Beavon 2000; Schmalhofer et al. 2009). In contrast, increased expression of the intermediate filament Vimentin is a predominant marker for mesenchymal cells (Leader et al. 1987; Satelli and Li 2011).
FIGURE 1.
Identification of EMT-associated AS events in human breast cancer. (A) Schematic of the analysis pipeline. (B) Heat map of the PSI values of the 25 significantly altered cassette exons in epithelial and mesenchymal samples. The PSI values were transformed into z-score (mean = 0 and standard deviation = 1) and plotted for each event. Red denotes higher inclusion of the cassette exon, while blue stands for higher skipping of that exon. Rows and columns are clustered based on the Pearson correlation coefficient. (C,D) Genome browser tracts of RNA-sequencing data showing alterations in AS between epithelial and mesenchymal samples. (C) Epithelial samples show higher inclusion of MAP3K7 exon 12 than mesenchymal samples. (D) Epithelial samples show lower inclusion of SPAG9 exon 24 than mesenchymal samples.
Tumor samples were scored as epithelial or mesenchymal tumors by plotting the difference in the expression between VIM and CDH1. For instance, a more mesenchymal tumor expresses relatively higher levels of VIM and lower levels of CDH1, resulting in a larger EMT score (Supplemental Table S1). As shown in Supplemental Figure S1A, tumors scoring one standard deviation below the mean were designated as epithelial tumors and those with an EMT score greater than one standard deviation above the mean were classified as mesenchymal tumors. This classification identified 143 epithelial specimens and 157 mesenchymal specimens out of a total of 1215 TCGA-BRCA samples.
To test whether this stratification creates a bias toward existing breast cancer subtypes, we plotted the distribution of tumors in each subtype (Supplemental Fig. S1B). Within the 300 epithelial and mesenchymal samples identified, 93 out of 157 mesenchymal samples and 129 out of 143 epithelial samples are associated with a known subtype within the database. Among the 93 mesenchymal samples, 66 were of the luminal A or B subtype (71.7%) and five were of basal subtype (5.4%). Among the 129 epithelial samples, 110 were of the luminal A or B subtype (85.3%) and six were of basal subtype (4.7%). While normal-like tumors tended to be more mesenchymal and luminal B tumors were generally more epithelial, our EMT classification did not display a significant bias toward epithelial luminal tumors and mesenchymal basal tumors.
Because the majority of AS events fall into the exon skipping category, also called skipped exons (SE), we focused on analyzing SE events of the above epithelial and mesenchymal tumors. We utilized exon junction reads and reads landing in variable exons extracted from the TCGA RNA-seq samples and calculated the Percent Spliced In (PSI) values of each of the AS events from a comprehensive list of 42,485 annotated SE events (Katz et al. 2010; Shen et al. 2014). We then calculated the differential PSI between tumors classified by epithelial or mesenchymal state. This approach led to the identification of 28 AS events that showed a change in average PSI greater than 20% between epithelial and mesenchymal, with an FDR less than 10−20. To avoid the confounding complications caused by transcription changes, we also eliminated splicing events with a change in gene expression greater than 1.5-fold between epithelial and mesenchymal tumors. This resulted in a final list of 25 AS events that showed significant differences in epithelial and mesenchymal tumors (Supplemental Table S2). Among them, EXOC7 and FGFR1 were previously reported to undergo isoform switching in EMT (Lu et al. 2013; Hopkins et al. 2017), supporting the validity of our 25 annotated AS events. Unsupervised clustering of the levels of these 25 AS events robustly separated the tumors into either epithelial or mesenchymal groups (Fig. 1B). Visualizing two representative examples, RNA sequencing read density plots showed that the degree of MAP3K7 exon 12 inclusion is decreased in mesenchymal tumors, whereas SPAG9 exon 24 inclusion preferentially occurs in mesenchymal tumors (Fig. 1C,D). These analyses reveal that the 25 splicing events exhibit significant differences between tumors that are epithelial or mesenchymal in nature.
Validation of the 25 AS signature in a CCLE breast cancer cell line data set
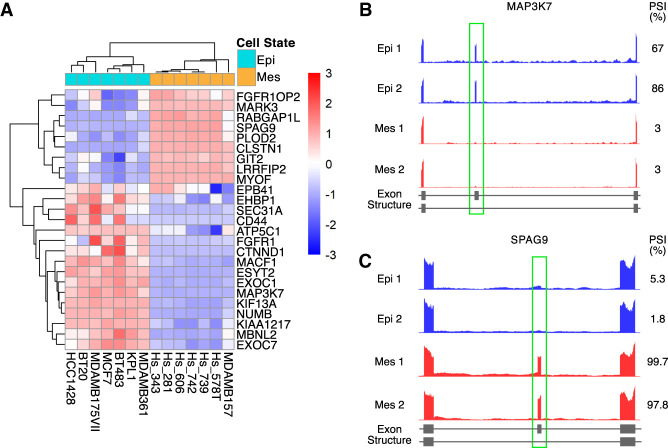
To assess whether the 25 AS signature can stratify independent EMT-associated cancer samples, we analyzed a different publicly available RNA sequencing database, the Cancer Cell Line Encyclopedia (CCLE). This database is composed of RNA sequencing data of 54 breast cancer cell lines. We used expression levels of CDH1 and VIM to derive their EMT scores and classified seven epithelial and seven mesenchymal cell lines (Supplemental Table S3). Comparison of PSI values in these 14 cell lines revealed that all of the AS events showed alternative splicing in the same direction as predicted, and 23 out of 25 AS events showed significant differences in PSI, with an absolute difference in PSI greater than 0.25 (Supplemental Table S4). As expected, PSI-based unsupervised hierarchical clustering analysis grouped all epithelial cells and mesenchymal cells as two separate clusters (Fig. 2A). Similar to what was observed in patient tumor specimens, the MAP3K7 and SPAG9 splicing events stratified the epithelial and mesenchymal breast cancer cell lines (Fig. 2B,C).
FIGURE 2.
Detection of the AS signature in the CCLE data set. (A) Heat map of the 25 EMT-associated AS events between epithelial (n = 7) and mesenchymal (n = 7) cell lines from the CCLE database. The heat map displays z-score transformed PSI values. The columns represent samples and the rows represent the 25 EMT-associated alternative splicing events. (B,C) Genome browser tracts of RNA-sequencing data showing alterations in alternative splicing between epithelial and mesenchymal cell lines. (B) Epithelial cell lines show higher inclusion of MAP3K7 exon 12 than mesenchymal cell lines. (C) Epithelial cell lines show lower inclusion of SPAG9 exon 24 than mesenchymal cell lines.
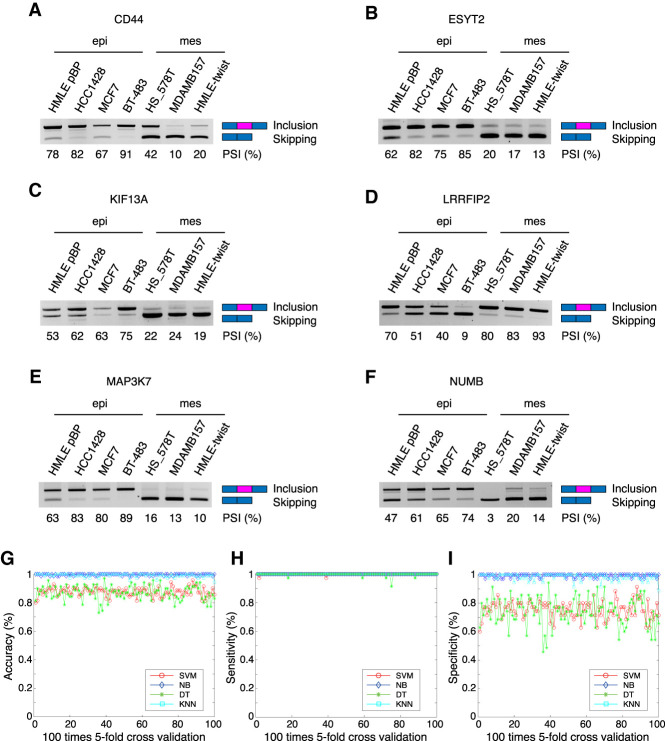
We next experimentally examined whether the computationally annotated AS events do, in fact, undergo isoform switching during EMT. Using semiquantitative PCR, we analyzed six out of the 25 AS events in four epithelial cells and three mesenchymal cells, including CCLE-derived lines and a pair of lines from an experimental EMT system where the human mammary epithelial cells (HMLE) transit into a mesenchymal state by overexpressing the transcription factor Twist (HMLE/Twist) (Mani et al. 2008). Each of the PCR reactions amplify both splice isoforms simultaneously, allowing for calculation of the PSI values. Not only were the AS events significantly altered in the experimental EMT system, but similar PSI distributions were also detected in breast cancer epithelial and mesenchymal cells (Fig. 3A–F).
FIGURE 3.
Experimental and computational validation of the AS signature. (A–F) Semiquantitative PCR validation of six out of 25 AS events in epithelial or mesenchymal cell lines from the CCLE database. (G–I) Prediction of epithelial or mesenchymal cell status from the CCLE database based on the EMT AS signature through machine learning methods (SVM, DT, KNN, and NB). (G) Accuracy. (H) Sensitivity. (I) Specificity distribution is shown for all four methods. All machine learning methods using the AS signature exhibit strong predictive power with average accuracies of SVM, DT, KNN, and NB at 87.81%, 87.47%, 99.13%, and 99.66%, respectively.
The 25 AS signature accurately predicts EMT status
If isoform switching of the 25 AS events is a general phenomenon during EMT, the PSI in each tumor or cell line should faithfully predict the epithelial or mesenchymal status. To test this idea, we used four widely used machine learning models, Support Vector Machine (SVM), Decision Tree (DT), K-nearest neighbor (KNN), and Naive Bayes (NB). We used fivefold cross-validation across 100 iterations and found that all machine learning methods exhibited strong predictive power (Supplemental Fig. S2A–C). A more stringent test was to use the 25 AS events derived from the BRCA-TCGA data set and predict the epithelial and mesenchymal states of the CCLE samples. This approach once again revealed high accuracy and sensitivity of the predictions, with the average accuracies of SVM, DT, KNN, and NB at 87.81%, 87.47%, 99.13%, and 99.66%, respectively (Fig. 3G–I). Considering both specificity and selectivity in tandem with accuracy, KNN exhibited the strongest predictive power. Thus, these results indicate that our AS signature can accurately and effectively distinguish epithelial and mesenchymal cell states and is a robust signature of EMT.
Community detection of RBP-AS bipartite networks reveals distinct clusters
The predictive capacity of the AS signature demonstrated that AS of this subset of events is consistent and robust during EMT. We hypothesized that a regulatory network tightly controls these EMT-associated AS events, which are likely functionally critical for EMT.
Since RBPs directly regulate AS, we focused on the relationship between RBPs and EMT-associated AS events by establishing correlation networks. We first analyzed the correlation between RBP gene expression and the EMT scores in each tumor sample bearing epithelial or mesenchymal states in the BRCA TCGA data set. This led to the identification of 22 RBPs that showed a significant positive or negative correlation (r ≥ 0.4 or r ≤ −0.4, respectively, P ≤ 10−10, log2 gene fold-change ≥ 1, Supplemental Table S5).
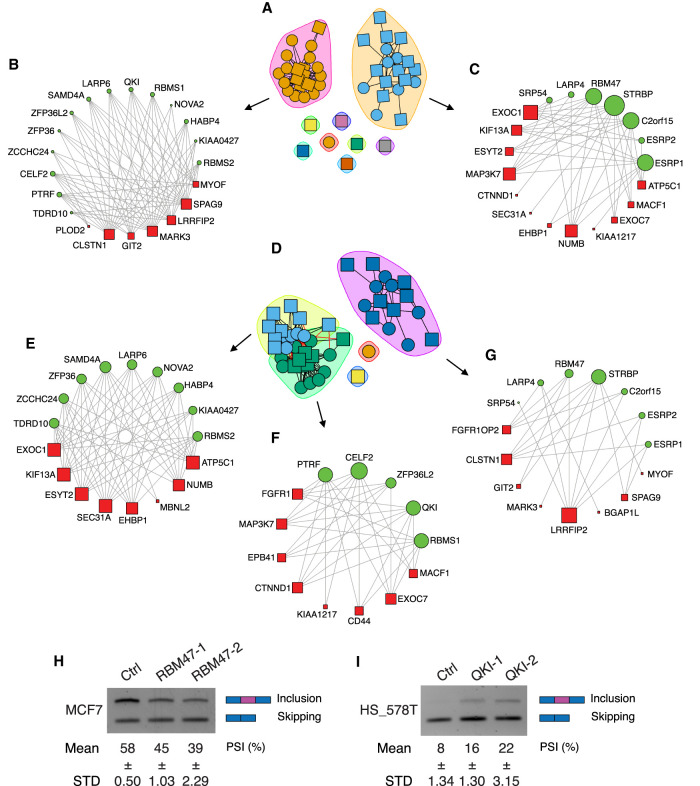
Bipartite networks contain two independent sets of nodes where connections, or edges, are only drawn between nodes in the two different sets and not within one set. To evaluate the relationships between RBPs and EMT-associated AS events, we constructed bipartite correlation networks using the 22 RBPs as one class of nodes and 25 AS events as the other class of nodes (Fig. 4). The RBP and AS nodes are connected when the absolute value of the Pearson correlation coefficient (PCC) of an RBP and AS node is equal to or greater than 0.4 (|PCC| ≥ 0.4). Following bipartite network construction, we applied a fast unipartite modularity maximization algorithm to detect communities within the network in order to identify groups of RBP-AS events that are more closely connected compared to the rest, as these communities are likely functionally significant. This annotation yielded two groups of RBPs based on their splicing activity: One group of RBPs promote exon inclusion (Fig. 4A) and the other group of RBPs promote exon skipping (Fig. 4D).
FIGURE 4.
Community detection of RBP-AS bipartite networks. (A) Subgroup A correlated with exon inclusion. (B) RBPs and associated exon inclusion events that promote EMT. (C) RBPs and associated exon inclusion events that inhibit EMT and promote epithelial state. (D) Subgroup B correlated with exon skipping. (E,F) RBPs and associated exon skipping events that promote EMT. (G) RBPs and associated exon skipping events that inhibit EMT. Node sizes are proportional to the number of related events. (H,I) Semiquantitative PCR validation of MAP3K7 exon 12 alternative splicing affected by RBP knockdown in MCF7 cells or HS_578T cells.
The majority of RBPs promoting exon inclusion shown in Figure 4A is further clustered into two networks (Fig. 4B,C). The network in Figure 4B represents those RBPs that promote exon inclusion and favors a mesenchymal AS pattern. Among these RBPs, QKI, ZCCHC24, and the CELF2 homolog CELF1 were previously reported to promote EMT (Chaudhury et al. 2016; Cieply et al. 2016; Yang et al. 2016), validating our computational output. In contrast to these RBPs that promote EMT-associated exon inclusion events (Fig. 4B), RBPs can also form a network with exon-inclusion events that favor an epithelial AS (Fig. 4C). Out of the seven RBPs, ESRP1, ESRP2, and RBM47 have been experimentally demonstrated to inhibit EMT through AS regulation, and EXOC7 exon inclusion promotes an epithelial cell state (Warzecha et al. 2010; Dittmar et al. 2012; Lu et al. 2013; Yang et al. 2016). These results show that our computationally derived RBP-AS networks are supported by experimental evidence.
In addition to the above RBPs that promote exon inclusion, the second group of RBPs promotes exon skipping (Fig. 4D). Clustering analysis separates RBPs into three networks. The first and second networks depict connections between RBPs and exon skipping events that favor a mesenchymal splicing pattern (Fig. 4E,F), while the third network connects RBPs and exon skipping events that favor an epithelial state (Fig. 4G). Noticeably, the majority of RBPs are connected to several AS events. Conversely, each AS event is regulated by a set of RBPs, promoting either the same splicing directions or opposite splicing directions, suggesting a combinatorial effect of RBPs through synergetic or antagonized fashions that tightly regulate AS events during EMT. For example, exon inclusion in MAP3K7 is one of the AS events favoring an epithelial state. Several RBPs including RBM47, STRBP, and C2orf15 promote the inclusion of MAP3K7 alternative exon (Fig. 4C), potentially promote an epithelial state. In contrast, a different group of RBPs, including PTRF, CELF2, and QKI, promote the skipping of the MAP3K7 variable exon, favoring a mesenchymal splicing pattern (Fig. 4F). These analyses reveal that the expression of different RBP groups act in concert to dictate the splicing direction of their gene targets, and that the combinatorial effect of RBP-AS networks promotes either an epithelial or a mesenchymal splicing pattern during EMT.
To experimentally validate our bioinformatic findings, we performed siRNA knockdown of RBPs and assessed their effects on regulating alternative splicing of their predicted downstream target, MAP3K7, whose exon 12 is preferentially included in epithelial cells and skipped in mesenchymal cells. We silenced the epithelial promoting RBM47 and mesenchymal promoting QKI by siRNAs in epithelial MCF7 and mesenchymal HS_578T cells, respectively, and examined the endogenous levels of MAP3K7 splice isoforms (Fig. 4H,I; Supplemental Fig. S3A,B). In agreement with our RBP network prediction, knocking down RBM47 promoted skipping of MAP3K7 exon 12, while knocking down QKI promoted inclusion of this exon. These results are further supported by previous findings that RBM47 promotes an epithelial state and that QKI accelerates EMT (Vanharanta et al. 2014; Pillman et al. 2018), illustrating the potential antagonizing role of these two RBPs on the same exon in different cell states.
AS levels predict patient survival
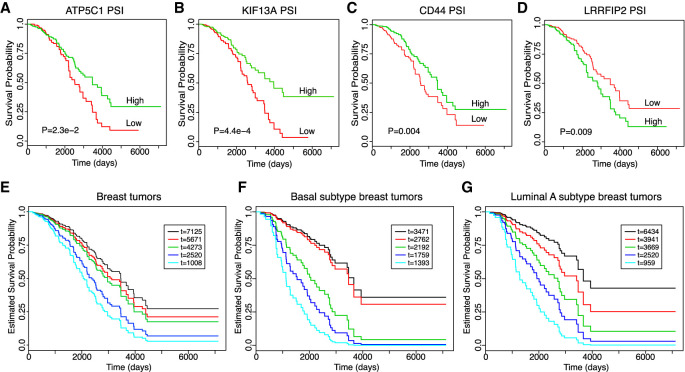
Since the AS signature was capable of distinguishing patient samples with regard to their EMT status and because abnormal activation of EMT is a hallmark for tumor metastasis, we next investigated the ability of this AS signature to predict patient survival using the TCGA-BRCA database. We found that the PSI values of four genes (ATP5C1, KIF13A, CD44, LRRFIP2) showed the most significant survival curve separation (Fig. 5A–D). Patients that had AS levels that favor a mesenchymal splicing pattern showed a worse survival prediction compared to those with the epithelial splicing pattern, indicating that promotion of EMT-associated splicing correlates with poorer patient outcomes. We also plotted the survival curves based on the gene expression level of the same four genes (Supplemental Fig. S4A–D). CD44 and ATP5C1 were not able to separate survival based on gene expression. Interestingly, gene expression levels of LRRFIP2 and KIF13A were able to separate survival curves, but the directionality of prediction was opposite between gene expression and alternative splicing levels. For example, high expression levels of KIF13A predicts poor survival while higher AS levels predicts better survival. These results indicate that gene expression and alternative splicing are independent measures in BRCA progression, further demonstrating the significance of alternative splicing in patient survival prediction.
FIGURE 5.
AS levels predict patient survival. (A–D) Kaplan-Meier survival plots of BRCA patients from TCGA stratified by the exon-inclusion level of four of the 25 EMT AS events. High PSI is 50% above the average PSI value while low PSI is 50% below the average PSI value. The “survdiff” function in R is used to compute P-values using the log-rank test. (E–G) Alternative splicing of the genes ATP5C1, KIF13A, CD44, and LRRFIP2 are used to predict the survival of five randomly selected patients from TCGA using the Cox proportional hazard model. The known patient survival times from TCGA are indicated in the inserted boxes as “t=” with the unit as days. The estimated survival probabilities over time by PSI values are plotted, and the color code of each patient survival curve corresponds to the same patient with indicated survival time from TCGA in the inserted box. Patients are randomly selected without specifying a breast cancer subtype (E), from the basal subtype (F), or from the luminal A subtype (G). Regardless of subtype classification, patients with the longer survival time in TCGA showed better predicted survival and vice versa.
We further examined whether composite PSI values are capable of predicting patient survival. We used patient tumor PSI values of the aforementioned four AS events as independent variables and applied them to the COX proportional hazard model (Cox 1972) to estimate the survival probability over time of a patient. We randomly selected five patients from the BRCA-TCGA data set and generated a putative survival curve for each patient (Fig. 5E). We then compared the predicted survival probability to the known survival time recorded in TCGA. Strikingly, the patient survival probabilities estimated from the PSI value-based COX model completely matched their known survival information. As shown in Figure 5E, the patient with shortest survival time (1008 d) has the worst predicted survival outcome (cyan line), whereas the patient with longest survival time (7125 d) is predicted with the best survival outcome (black line). The rank of predicted survival of all five patients is consistent with that of their known survival time. Moreover, we tested whether this prediction model holds true in different subtypes of breast cancer by comparing the survival of five randomly picked patients within the basal subtype (Fig. 5F) and the luminal A subtype (Fig. 5G). Once again, the survival probabilities were accurately predicted by the alternative splicing signature. In contrast, the gene expression levels of these four genes lack the ability to accurately estimate the survival probabilities (Supplemental Fig. S4E–G). Thus, these results indicate that the EMT-associated AS signature has strong predictive power for breast cancer patient survival.
DISCUSSION
Previous studies have focused on transcriptional regulation of EMT. However, the role of global AS regulation in EMT remains relatively uncharacterized, with little connection to clinical data sets. In this study, we attempted to characterize the AS differences between epithelial and mesenchymal tumors from TCGA. We derived an AS signature consisting of differentially spliced genes between epithelial and mesenchymal patient tumors. This signature is validated both experimentally and bioinformatically. We constructed bipartite networks of RBPs and the AS signature, revealing RBP-AS communities with a likelihood for functional connections. Our AS signature can accurately predict patient survival, suggesting its potential as a clinical prognostic factor. This study highlights the importance of global AS regulation during EMT and provides solid ground for further investigation into RNA regulation in EMT and metastasis.
Understanding the underlying mechanisms that regulate EMT is critical for developing new therapeutic approaches against metastasis. As AS plays a critical role in EMT and cancer progression, multiple attempts have been made at deciphering the global regulatory role of AS in EMT and metastasis. Recent studies have tried to understand AS regulation at the transcriptome level. However, efforts to explore AS changes during EMT have mostly focused on in vitro studies in cell lines with limited connections to human patient data sets. In this study, we utilized the human invasive breast cancer data set from TCGA in order to examine the AS changes between epithelial and mesenchymal breast tumors. We subdivided the data set into epithelial and mesenchymal tumor samples using an EMT gene expression signature. Our EMT gene expression signature utilizes CDH1 and VIM, two gene markers highly expressed in epithelial and mesenchymal cell states, respectively, which clearly and accurately separated patient tumor samples into epithelial or mesenchymal-like categories, demonstrating the simplicity and accuracy of our gene expression signature.
Through analysis of differential splicing between the epithelial and mesenchymal tumors, we identified an AS signature of 25 genes that showed a drastic change in splicing patterns between epithelial and mesenchymal tumors. These events are validated in a separate CCLE database both bioinformatically and experimentally. Several of these events have been shown in previous publications, by our laboratory and other groups, to not only occur during EMT, but also be functionally important for EMT (Warzecha et al. 2009, 2010; Brown et al. 2011; Reinke et al. 2012; Lu et al. 2013; Hopkins et al. 2017; Huang et al. 2017; Li et al. 2018; Hu et al. 2020). Our analysis highlights the clinical significance of AS in breast cancer and provides the impetus for further mechanistic investigations into how each individual splicing event contributes to EMT and cell plasticity.
From our analysis of epithelial and mesenchymal tumors, we extracted a list of RBPs that may play a critical role during EMT through correlation analysis and robust filtering. We linked splicing events with their regulators and tried to build a functionally interconnected regulation network that dictates EMT direction. Community detection methods on EMT associated splicing events identified subnetwork clusters that are positively or negatively associated with EMT. RBPs and their target AS events form highly connected communities which may indicate enrichment of specific biological functions. We tested our bioinformatic findings experimentally. MAP3K7 alternative exon 12 is a conserved tissue-specific alternative splicing event (Venables et al. 2012). It was recently reported that skipping of exon 12 generates a constitutively active MAP3K7 kinase important for cells to undergo EMT (Tripathi et al. 2019). Here, we have identified two splicing factors RBM47 and QKI that antagonize each other to promote an epithelial or mesenchymal isoform, respectively. Our analysis provides a new way of exploring the association between RBPs and AS events and provides an opportunity to further study global AS regulation in EMT.
While our community detection analysis identified known EMT RBP regulators such as the ESRPs, QKI and RBM47 (Warzecha et al. 2010; Cieply et al. 2016; Yang et al. 2016), our networks contain numerous RBPs and AS events with no known functional connections to EMT. However, the magnitude of the node size in each community suggests putative RBPs or splicing events with dense connections have a higher likelihood of an important role. As an example, ZCCHC24 was one of the most significantly connected RBP nodes, and was recently shown to have highly labile expression during induction of pluripotency (Cieply et al. 2016). EMT has been associated with an increase in stemness and stem-cell phenotypes (Pradella et al. 2017), suggesting ZCCHC24 is a promising candidate for future study. LARP6 is another RBP node that we discovered to regulate mesenchymal splicing patterns. Interestingly, LARP6 regulates collagen synthesis and tissue fibrosis, phenotypes that are closely connected to EMT (Cai et al. 2010). It will be interesting as a future direction to investigate its role in alternative splicing regulation that impacts an EMT phenotype. Future investigations on the RBP-AS network have the potential to link previously undetected pathways with EMT progression and tumor metastasis.
AS has been shown to be a promising predictor of patient outcome (Shen et al. 2016). To test the predictive power of our AS signature derived from the patient TCGA data set, we utilized machine learning techniques to predict the EMT category of CCLE as another independent RNA sequencing data set. In all machine learning models, our 25-gene splicing signature has strong predictive power ranging from 87.47%–99.6% in the CCLE data set. Many of these splicing events are strong predictors of poor survival, while their gene expression levels are not as informative. Using only four out of the 25 AS signatures, we were able to accurately rank patient survival probabilities. For the same set of patients within the basal or luminal A subtype, using a combined signature of four alternative splicing events surpasses the predictive ability of using their gene expression levels. This showed that our analysis extracted meaningful alternative splicing information which cannot be replaced by traditional gene expression approaches. These results highlight the clinical relevance of this splicing signature. We speculate that this splicing signature will be a useful clinical predictor of tumor EMT status and patient survival.
MATERIALS AND METHODS
EMT score calculation
The EMT score is calculated as the gene expression difference between VIM and CDH1 (VIM-CDH1) and ranked in descending order. Samples whose EMT score was one standard deviation above the mean were classified as mesenchymal, while the samples with an EMT score one standard deviation below the mean were denoted as epithelial.
Analysis of alternative splicing
Raw junction reads in the PSI estimation were downloaded from Level 3 TCGA BRCA data from the GDC legacy archive (https://portal.gdc.cancer.gov/legacy-archive). Known alternative splicing events were classified using an annotated set of splicing events provided by the splicing analysis tool MISO downloaded from (http://miso.readthedocs.io/en/fastmiso/) (Katz et al. 2010). The percentage spliced in (PSI) values of each cassette exon were calculated using the following formula:
| (1) |
where I represents the exon inclusion reads which are from the upstream splice junction and the downstream splice junction. S represents the exon skipping reads which are from the skipping splice junction connecting the upstream exon to the downstream exon. LI is the effective length of inclusion isoform, LS is the effective length of skipping isoform, j is the junction length which is defined as j = 2 × (read length-anchor length), and i is the read length. When a gene contains multiple variable exons, the effective length of inclusion isoform (LI) is calculated as LI = m × (j − r + 1), and the junction length is denoted as j = m × (readLength − anchorLength). m represents the number of variable exons for isoform, such as m = 2 for a splicing event with two adjacent variable exons. For those events that have the same starting coordinates but different ending coordinates, only the one whose distance is largest is considered.
Identification of EMT-related AS events
The mean PSI for each gene from the epithelial or mesenchymal samples (TCGA or CCLE) is calculated. The absolute value of the difference between the average PSI for each gene is calculated (ΔPSI) (Supplemental Table S2). Significant AS events are selected by ΔPSI ≥ 0.2, FDR ≤ 10−20, and associated fold change of gene expression value ≤ 1.5. The known EMT associated splicing event CD44 is also included (Brown et al. 2011).
Validation of AS events through semiquantitative PCR
Briefly, RNA was extracted from cultured epithelial cell lines HMLE pBP, HCC1428, MCF7 and mesenchymal cell lines BT-578T, HS_578T, MDA-MB-157, and HMLE/Twist using the E.Z.N.A Total RNA Kit (Omega Bio-Tek). RNA concentration and purity were measured on a Nanodrop 2000 (Thermo Fisher Scientific).
cDNA was generated via reverse transcription using the GoScript Reverse Transcription System (Promega) with 1 µL GoScript RT and 250 ng of RNA in a total volume of 20 µL followed by incubation at 25°C for 5 min, 42°C for 30 min, and 70°C for 15 min. Semiquantitative RT-PCR assaying for splicing products was performed using Hot StarTaq DNA polymerase (Qiagen), and PCR cycles were run for 30 or fewer cycles. Primers for semiquantitative analysis were designed on constitutive exons flanking each variable exon. Semiquantitative PCR generates both exon inclusion and skipping products which were separated through agarose gel electrophoresis. PCR product intensity was measured using ImageJ image analysis software.
Identification of EMT-associated RBPs
The Pearson correlation value (r) is calculated between RBP expression and the EMT score of each tumor sample, then ranked in descending order. Significant RBPs are selected by r ≥ 0.4 or r ≤ −0.4, P ≤ 10−10, gene absolute fold change ≥ 1.5.
Machine-learning classification of EMT status
We evaluated how well AS events can distinguish epithelial or mesenchymal samples. We used fivefold cross-validation across 100 iterations using the machine learning methods support vector machine (SVM), decision tree (DT), K-nearest neighbor (KNN) and Naive Bayes. Epithelial and mesenchymal samples from TCGA were randomly divided into five equal sized samples with four samples randomly assigned as the training set, and the remaining sample designated as the test set. This process is repeated five times until each sample has been assigned as the test set (one run). The average of the five results from one run is then used as a single estimation. This process is repeated 100 times and plotted for accuracy, specificity and sensitivity. We repeated this process using RNA-seq data from CCLE breast cancer cell lines to further evaluate prediction performance, training the parameters using four TCGA subsets and using the CCLE data set as the test set. We used the fitcsvm function in Matlab R2018b for the SVM method and adopted the corresponding functions defined in Matlab for the DT, KNN and NB methods.
Bipartite network between RBPs and AS events and community detection
To construct an RBP-AS bipartite network, we represent the relationship between RBPs and AS events in the form of a nR × nA adjacency matrix, where nR is the number of the selected RBPs and nA is the total number of AS events. The matrix B is defined as follows:
| (2) |
The bipartite network is created by joining together pairs of RBPs and AS when Bij = 1.
We further applied modularity to measure the density of links inside communities as compared to links between communities (Blondel et al. 2008). The bipartite modularity is defined as follows:
| (3) |
where m is the number of links in the network, Bij is the weight of the edge between RBP i and isoform j, ki is the degree of RBP i, dj is the degree of AS event j, and Ci,Cj are the community indices of RBP i and isoform j, respectively. The δ-function δ (u, v) is defined as:
| (4) |
We ran the algorithm in R and adopted the “igraph” packages for the figures. Node sizes are proportional to the number of related events.
siRNA-mediated knockdown of RBP nodes
Briefly, 5 × 104 MCF7 or HS_578T cells were seeded into each well of a 24-well plate. Twenty-four hours after seeding, siRNAs against RBPs were added at a final concentration of 10 nM per well using 1 µL Lipofectamine RNAiMAX transfection reagent (Invitrogen). RNA was collected 72 h after transfection using the E.Z.N.A Total RNA Kit (Omega Bio-Tek).
Survival analysis of TCGA BRCA data
Kaplan-Meier survival plots of BRCA patients from TCGA stratified by the exon-inclusion level of four of the 25 EMT AS events were plotted. High or low PSI is defined by being 50% above or below the average PSI value, respectively.
The Cox proportional hazards model demonstrates that the hazard function h(t), which means the risk of death at time t. for an individual with a certain gene expression profile, is given by
| (5) |
where h0(t) is the baseline hazard when the PSI values of all four genes (ATP5C1, KIF13A, CD44 and LRRFIP2) are equal to zero. β = (β1, β2, · · · βp)T denotes the column vector of regression parameter. X = (x1, x2, · · · xp)T denotes the PSI value of p splicing events. The Cox partial likelihood is derived as follows:
| (6) |
where D denotes the set of indices of patient death and Rr is the set of indices of individuals at risk for death at time tr (Gui and Li 2005). We take the logarithm of the Cox partial likelihood as follows:
| (7) |
The normal maximum likelihood estimation method is then applied to calculate the unknown parameters β. This cox proportional hazard model is used to plot the predicted survival curve for any patient sample with known PSI values for the four splicing events. We picked five random data sets with different survival times and plotted their survival curves using this model.
We adopted the “survival” package in R to plot the predicted survival curve. All Kaplan-Meier curves are plotted using the “survfit” function, and the “survdiff” function is used compute P-values using the log-rank test.
DATA DEPOSITION
Processed TCGA BRCA Level 3 RNA-SeqV2 gene expression data were downloaded from the Genomic Data Commons (GDC) Legacy Archive (https://portal.gdc.cancer.gov/legacy-archive). CCLE data were downloaded from the CCLE database (https://portals.broadinstitute.org/ccle). All data generated or analyzed during this study (compiled tables of EMT scores, differential splicing analysis, EMT-related RBP analysis, and survival analysis from TCGA and CCLE) are included in this published article (and its Supplemental Information files). The original scripts generated during the current study are available from the corresponding author on reasonable request.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Natural Science Foundation of Shenzhen University (grant number 000393 and JCYJ 20170817100950436 to Y.Q.); and the U.S. National Institutes of Health (www.nih.gov) (grant number 5F30CA196118 to S.E.H., R01CA182467 and R35GM131876 to C.C.). C.C. is a Cancer Prevention and Research Institute of Texas (CPRIT, https://www.cprit.state.tx.us) Scholar in Cancer Research (RR160009). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Sarah Herzog for help with the experiments and editing the manuscript. We thank Rong Zheng for helpful discussions and designing RT-PCR primers.
Author contributions: C.C. conceived the project. Y.Q. and C.C. designed the experiments. Y.Q. performed the bioinformatics analyses. J.L. and S.E.H. helped in analyzing and organizing the data. Y.Q. and J.L. performed the revision experiments. M.D. performed the semi-qPCR validation and created the figures. Y.Q., J.L., S.E.H., and C.C. wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.074187.119.
REFERENCES
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. 2010. Deciphering the splicing code. Nature 465: 53–59. 10.1038/nature09000 [DOI] [PubMed] [Google Scholar]
- Beavon IR. 2000. The E-cadherin-catenin complex in tumour metastasis: structure, function and regulation. Eur J Cancer 36: 1607–1620. 10.1016/S0959-8049(00)00158-1 [DOI] [PubMed] [Google Scholar]
- Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. 2008. Fast unfolding of communities in large networks. J Stat Mech 2008: P10008 10.1088/1742-5468/2008/10/P10008 [DOI] [Google Scholar]
- Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C. 2011. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest 121: 1064–1074. 10.1172/JCI44540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Fritz D, Stefanovic L, Stefanovic B. 2010. Binding of LARP6 to the conserved 5′ stem-loop regulates translation of mRNAs encoding type I collagen. J Mol Biol 395: 309–326. 10.1016/j.jmb.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A, Cheema S, Fachini JM, Kongchan N, Lu G, Simon LM, Wang T, Mao S, Rosen DG, Ittmann MM, et al. 2016. CELF1 is a central node in post-transcriptional regulatory programmes underlying EMT. Nat Commun 7: 13362 10.1038/ncomms13362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieply B, Park JW, Nakauka-Ddamba A, Bebee TW, Guo Y, Shang X, Lengner CJ, Xing Y, Carstens RP. 2016. Multiphasic and dynamic changes in alternative splicing during induction of pluripotency are coordinated by numerous RNA-binding proteins. Cell Rep 15: 247–255. 10.1016/j.celrep.2016.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DR. 1972. Regression models and life-tables. J R Stat Soc B (Methodol) 34: 187–202. 10.1007/978-1-4612-4380-9_37 [DOI] [Google Scholar]
- Damianov A, Ying Y, Lin C-H, Lee J-A, Tran D, Vashisht AA, Bahrami-Samani E, Xing Y, Martin KC, Wohlschlegel JA, et al. 2016. Rbfox proteins regulate splicing as part of a large multiprotein complex LASR. Cell 165: 606–619. 10.1016/j.cell.2016.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Jiang P, Park JW, Amirikian K, Wan J, Shen S, Xing Y, Carstens RP. 2012. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol Cell Biol 32: 1468–1482. 10.1128/MCB.06536-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-D, Ares M Jr. 2014. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 15: 689–701. 10.1038/nrg3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J, Li H. 2005. Penalized Cox regression analysis in the high-dimensional and low-sample size settings, with applications to microarray gene expression data. Bioinformatics 21: 3001–3008. 10.1093/bioinformatics/bti422 [DOI] [PubMed] [Google Scholar]
- Hirohashi S. 1998. Inactivation of the E-cadherin-mediated cell adhesion system in human cancers. Am J Pathol 153: 333–339. 10.1016/S0002-9440(10)65575-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins A, Coatham ML, Berry FB. 2017. FOXC1 regulates FGFR1 isoform switching to promote invasion following TGFβ-induced EMT. Mol Cancer Res 15: 1341–1353. 10.1158/1541-7786.MCR-17-0185 [DOI] [PubMed] [Google Scholar]
- Hu X, Harvey SE, Zheng R, Lyu J, Grzeskowiak CL, Powell E, Piwnica-Worms H, Scott KL, Cheng C. 2020. The RNA-binding protein AKAP8 suppresses tumor metastasis by antagonizing EMT-associated alternative splicing. Nat Commun 11: 486 10.1038/s41467-020-14304-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zhang J, Harvey SE, Hu X, Cheng C. 2017. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev 31: 2296–2309. 10.1101/gad.305862.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7: 1009–1015. 10.1038/nmeth.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader M, Collins M, Patel J, Henry K. 1987. Vimentin: an evaluation of its role as a tumour marker. Histopathology 11: 63–72. 10.1111/j.1365-2559.1987.tb02609.x [DOI] [PubMed] [Google Scholar]
- Li J, Choi PS, Chaffer CL, Labella K, Hwang JH, Giacomelli AO, Kim JW, Ilic N, Doench JG, Ly SH, et al. 2018. An alternative splicing switch in FLNB promotes the mesenchymal cell state in human breast cancer. Elife 7: e37184 10.7554/eLife.37184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Liu J, Liu S, Zeng J, Ding D, Carstens RP, Cong Y, Xu X, Guo W. 2013. Exo70 isoform switching upon epithelial-mesenchymal transition mediates cancer cell invasion. Dev Cell 27: 560–573. 10.1016/j.devcel.2013.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao M-J, Eaton N, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133: 704–715. 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463. 10.1038/nature08909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-M, Gaur AB, Lengyel E, Peter ME. 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22: 894–907. 10.1101/gad.1640608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillman KA, Phillips CA, Roslan S, Toubia J, Dredge BK, Bert AG, Lumb R, Neumann DP, Li X, Conn SJ, et al. 2018. miR-200/375 control epithelial plasticity-associated alternative splicing by repressing the RNA-binding protein quaking. EMBO J 37: e99016 10.15252/embj.201899016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradella D, Naro C, Sette C, Ghigna C. 2017. EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer 16: 8 10.1186/s12943-016-0579-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke LM, Xu Y, Cheng C. 2012. Snail represses the splicing regulator epithelial splicing regulatory protein 1 to promote epithelial-mesenchymal transition. J Biol Chem 287: 36435–36442. 10.1074/jbc.M112.397125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satelli A, Li S. 2011. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci 68: 3033–3046. 10.1007/s00018-011-0735-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalhofer O, Brabletz S, Brabletz T. 2009. E-cadherin, β-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev 28: 151–166. 10.1007/s10555-008-9179-y [DOI] [PubMed] [Google Scholar]
- Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, Gertler FB. 2011. An EMT–driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet 7: e1002218 10.1371/journal.pgen.1002218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Park JW, Lu Z, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y. 2014. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci 111: E5593–E5601. 10.1073/pnas.1419161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Wang Y, Wang C, Wu YN, Xing Y. 2016. SURVIV for survival analysis of mRNA isoform variation. Nat Commun 7: 1–11. 10.1038/ncomms11548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. 2006. Complex networks orchestrate epithelial–mesenchymal transitions. Nat Rev Mol Cell Biol 7: 131 10.1038/nrm1835 [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RYJ, Nieto MA. 2009. Epithelial-mesenchymal transitions in development and disease. Cell 139: 871–890. 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Tripathi V, Shin J-H, Stuelten CH, Zhang YE. 2019. TGF-β-induced alternative splicing of TAK1 promotes EMT and drug resistance. Oncogene 38: 3185–3200. 10.1038/s41388-018-0655-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanharanta S, Marney CB, Shu W, Valiente M, Zou Y, Mele A, Darnell RB, Massagué J. 2014. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. Elife 3: e02734 10.7554/eLife.02734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Vignal E, Baghdiguian S, Fort P, Tazi J. 2012. Tissue-specific alternative splicing of Tak1 is conserved in deuterostomes. Mol Biol Evol 29: 261–269. 10.1093/molbev/msr193 [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. 2009. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 33: 591–601. 10.1016/j.molcel.2009.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S, Guo W, Xing Y, Carstens RP. 2010. An ESRP-regulated splicing programme is abrogated during the epithelial–mesenchymal transition. EMBO J 29: 3286–3300. 10.1038/emboj.2010.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. 2008. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 14: 818–829. 10.1016/j.devcel.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Yang Y, Park JW, Bebee TW, Warzecha CC, Guo Y, Shang X, Xing Y, Carstens RP. 2016. Determination of a comprehensive alternative splicing regulatory network and combinatorial regulation by key factors during the epithelial-to-mesenchymal transition. Molecular and Cell Biol 36: 1704–1719. 10.1128/MCB.00019-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y, Wang X-J, Vuong CK, Lin C-H, Damianov A, Black DL. 2017. Splicing activation by Rbfox requires self-aggregation through its tyrosine-rich domain. Cell 170: 312–323.e10. 10.1016/j.cell.2017.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.