FIGURE 3.
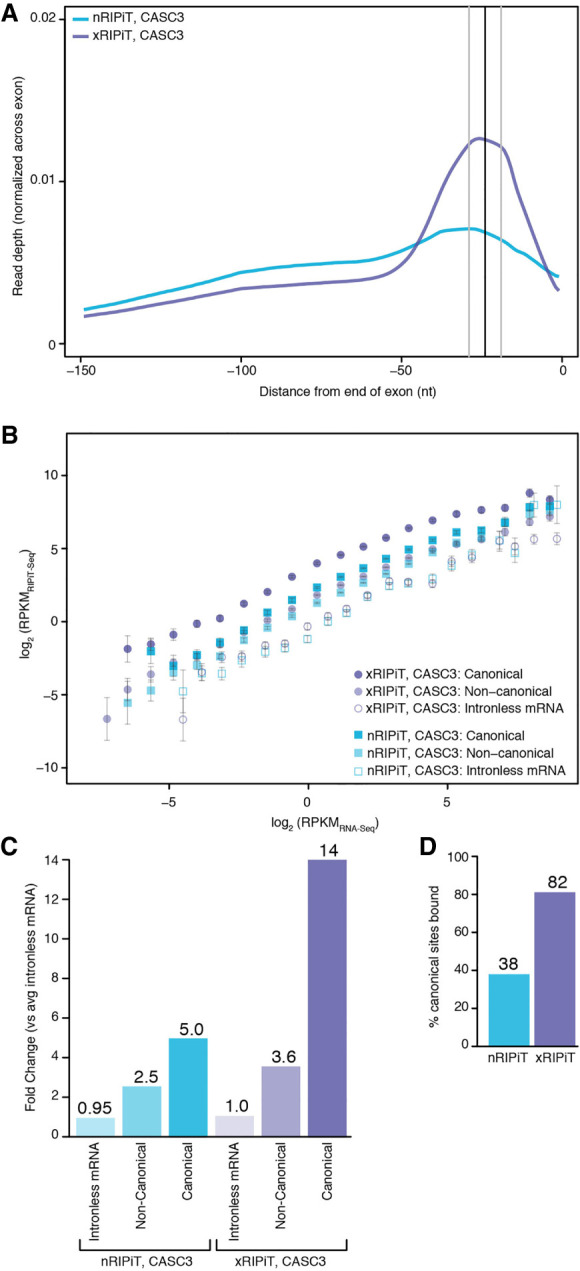
Formaldehyde crosslinking enhances RIPiT-seq signal for CASC3. (A) A meta-exon plot showing nRIPiT and xRIPiT read densities in the 150 nt window from the end of exons of protein-coding genes (excluding final exons). (B) Comparison of gene-level CASC3 read density (RPKMRIPiT-seq) in native RIPiT (nRIPiT, squares) and formaldehyde-crosslinked RIPiT (xRIPiT; circles) for canonical (darker-shaded shapes) and noncanonical regions (lighter-shaded shapes), and for intronless genes (empty shapes). Along the x-axis, genes are binned into twenty bins where each bin contains exons from genes within a twofold expression level range based on RNA-seq. Error bars represent the standard error of the mean signal in each bin. (C) A comparison of linear fit coefficients (or intercepts, in log space) of the six classes in B. Classes are labeled on the bottom. The coefficient for the average of the two intronless classes was set to 1 and all intercepts were adjusted accordingly. The fold-change as compared to the average of the two intronless classes is shown above each bar. (D) Percentage of all canonical EJC regions where read count is greater than or equal to twofold as compared to read counts on intronless genes of similar expression level.
