Abstract
tRNAs constitute the most highly modified class of RNA. Every tRNA contains a unique set of modifications, and Ψ55, m5U54, and m7G46 are frequently found within the elbow of the tRNA structure. Despite the abundance of tRNA modifications, we are only beginning to understand the orchestration of modification enzymes during tRNA maturation. Here, we investigated whether pre-existing modifications impact the binding affinity or catalysis by tRNA elbow modification enzymes. Specifically, we focused on the Escherichia coli enzymes TruB, TrmA, and TrmB which generate Ψ55, m5U54, and m7G46, respectively. tRNAs containing a single modification were prepared, and the binding and activity preferences of purified E. coli TrmA, TruB, and TrmB were examined in vitro. TruB preferentially binds and modifies unmodified tRNA. TrmA prefers to modify unmodified tRNA, but binds most tightly to tRNA that already contains Ψ55. In contrast, binding and modification by TrmB is insensitive to the tRNA modification status. Our results suggest that TrmA and TruB are likely to act on mostly unmodified tRNA precursors during the early stages of tRNA maturation whereas TrmB presumably acts on later tRNA intermediates that are already partially modified. In conclusion, we uncover the mechanistic basis for the preferred modification order in the E. coli tRNA elbow region.
Keywords: tRNA, RNA modification, pseudouridine, methylation, modification order
INTRODUCTION
Transfer RNA (tRNA) is the most highly and most diversely modified class of RNA, containing over 100 chemically distinct modifications and a median of twelve modifications per individual tRNA (Boccaletto et al. 2018). In some organisms, more than 1% of the genome encodes tRNA modification enzymes, highlighting the energetic cost all cells invest in tRNA modification (El Yacoubi et al. 2012) whereas in many organisms including humans the full set of tRNA modification enzymes is not even known yet (de Crecy-Lagard et al. 2019). The model organism Escherichia coli contains over 25 different modifications, with each tRNA containing between three and 13 modifications (Boccaletto et al. 2018).
Two clusters of tRNA modifications are evident in the tRNA tertiary structure. The first cluster of tRNA modifications is found within the tRNA anticodon loop. In general, these modifications are bulky, complex, and each modification is only found within a few isoacceptor tRNAs per organism. Several tRNA anticodon modifications have been shown to play important roles during translation (Ranjan and Rodnina 2016). The second cluster of tRNA modifications is found within the tRNA elbow, a region formed by long-range base-pairing interactions between the D and T loops.
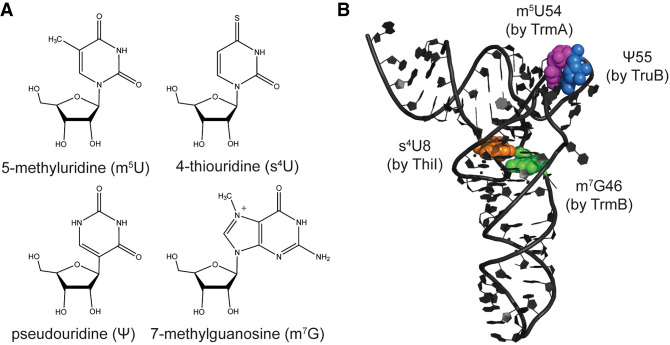
In contrast to anticodon loop modifications, tRNA elbow modifications are less diverse. For example, 5-methyluridine (m5U) 54 (also known as ribothymidine 54) and pseudouridine (Ψ) 55 are found within every tRNA in E. coli, where they are introduced by TrmA and TruB, respectively (Fig. 1A,B). Additionally, m5U54 and Ψ55 modifications are found widely within elongator tRNAs throughout all domains of life, including in humans, where these modifications are introduced by the TrmA and TruB homologs TRMT2A and PUS4 in the cytoplasm. Another abundant tRNA elbow modification is 7-methylguanosine (m7G) 46 (Fig. 1A,B), which is introduced by TrmB in E. coli and is frequently found within tRNAs containing short variable loops (Purta et al. 2005). TrmB is also widely conserved, with METTL1 as its human homolog. Within bacteria, 4-thiouridine (s4U) 8 is a common modification and is catalyzed by ThiI, which requires sulfur transfer by IscS (Fig. 1A,B; Kambampati and Lauhon 2000). tRNA elbow modifications are generally considered to be important for promoting tRNA structure and cellular stability (Alexandrov et al. 2006; Kimura and Waldor 2019); however, modifications within the tRNA elbow also have roles during translation. Specifically, E. coli TruB has been found to be important for the translation of consecutive arginine CGA codons (Urbonavicius et al. 2002), and Pseudomonas aeruginosa TrmB is important for the expression of genes enriched in phenylalanine and aspartate codons (Thongdee et al. 2019). In addition to the direct stabilizing effects of tRNA modifications on tRNA structure, two tRNA elbow modification enzymes, namely TruB and TrmA, have been described as tRNA chaperones that help tRNA to adopt its overall L-shaped fold independent of their modification activity (Keffer-Wilkes et al. 2016; LC Keffer-Wilkes, EF Soon, and U Kothe, in prep.).
FIGURE 1.
Structures of selected tRNA elbow modifications and their locations within E. coli tRNAPhe. (A) Structures of the 4-thiouridine, 7-methylguanosine, 5-methyluridine, and pseudouridine modifications. (B) The locations of these modifications within the tRNA elbow are highlighted as spheres within the crystal structure of unmodified tRNAPhe (PDB 3L0U; Byrne et al. 2010). The enzyme responsible for each modification is indicated next to the respective modification site.
Here, we are specifically focusing on the pseudouridine synthase TruB, and the two methyltransferases TrmA and TrmB. TruB utilizes a catalytic aspartate residue in conjunction with a conserved arginine and a second-shell aspartate residue to catalyze the isomerization of uridine to pseudouridine (Friedt et al. 2014). Specifically, the catalytic aspartate attacks the 2′ oxygen of the ribose to form a glycal intermediate and to break the N-C glycosidic bond. Upon rotation of the uracil base, a new C-C glycosidic bond is formed and the covalent bond to the catalytic aspartate is broken (Veerareddygari et al. 2016). TrmA and TrmB both use S-adenosylmethionine (SAM) as the methyldonor. During tRNA methylation by TrmA, a catalytic cysteine residue forms a covalent bond with C6 in the uracil ring enabling methyltransfer from SAM to the base. Subsequently, a catalytic glutamate residue acts a general base abstracting a proton from the uracil ring, allowing the covalent bond to the catalytic cysteine to be broken (Alian et al. 2008). The catalytic mechanism of TrmB methylating G46 is not known yet, but a conserved aspartate residue (D144 in E. coli) is essential for catalysis (Purta et al. 2005). It has been suggested that the catalytic aspartate acts as a general base deprotonating G46 such that the base can attack the SAM cofactor for methyltransfer (Tomikawa 2018).
Despite the abundance and importance of tRNA modifications, we are still in the early stages of understanding the multistep process through which tRNA is modified. From previous mechanistic studies, it is known that the majority of tRNA modification enzymes, including TrmA, TruB, TrmB, and ThiI, are active on in vitro transcribed, unmodified tRNA (Kambampati and Lauhon 2000; Wright et al. 2011; Thongdee et al. 2019; LC Keffer-Wilkes, EF Soon, and U Kothe, in prep.); however, completely unmodified tRNA may not be the preferred substrate of these enzymes in vivo. Recent reviews have emphasized that the order of tRNA modification is likely not random (Han and Phizicky 2018; Sokolowski et al. 2018; Barraud and Tisne 2019). In particular, five so-called “tRNA modification circuits” within the tRNA anticodon loop have been well-described where introduction of a certain modification strictly depends on the prior presence of another modification (Han and Phizicky 2018). Within the tRNA elbow, temperature-specific networks of tRNA modifications have been reported in Thermus thermophilus (Hori 2019). Additionally, a sequential formation of tRNA elbow modifications has been described recently for Saccharomyces cerevisiae system, wherein the modification of in vitro transcribed yeast tRNAPhe by yeast cell extract was monitored over time. Ψ55 was found to be fully introduced within the tRNA population prior to the appearance of any other modification, followed by m5U54 and m7G46, then m2G10, D16, and m5C49, and finally m1A58 (Barraud et al. 2019). This study provides strong evidence that modifications in the tRNA elbow region are not introduced randomly, but the molecular determinants of the sequential modification order remain unclear.
Therefore, the goal of this study was to examine the impact of tRNA modification status on the in vitro binding and catalytic activity of E. coli tRNA elbow modification enzymes TruB, TrmA, and TrmB. Specifically, by conducting quantitative biochemical experiments, we aimed to not only identify the preferred tRNA substrate for these enzymes, but to also characterize the molecular basis for the ordered modification of tRNA in the elbow region. To accomplish this, we prepared in vitro transcribed tRNA that contained a single modification for use in enzyme binding and activity assays. Here, we report that TruB prefers to bind and modify tRNA that does not yet contain any modifications, whereas TrmA prefers to modify unmodified tRNA, but has a binding preference for tRNA already containing the Ψ55 modification. This proof-of-principle study suggests tRNA elbow modification enzymes are sensitive to the modification status of their substrate tRNAs, explaining why the order of tRNA elbow modification is not completely random.
RESULTS
In order to determine the effect of a pre-existing tRNA modification on the affinity and activity of another tRNA modification enzyme, we used in vitro transcribed E. coli tRNAPhe as a model substrate. Single-modified tRNAPhe was prepared by incubation with purified E. coli TrmA, TruB, TrmB, or ThiI and IscS. Each single-modified tRNA contained one modification at a level of at least 78% (see Materials and Methods). Unmodified tRNA as well as single-modified tRNAs were subsequently used in binding and activity assays in order to compare whether TrmA, TruB, or TrmB prefer binding and/or modifying tRNA with a certain modification status.
Binding and activity preferences of TruB
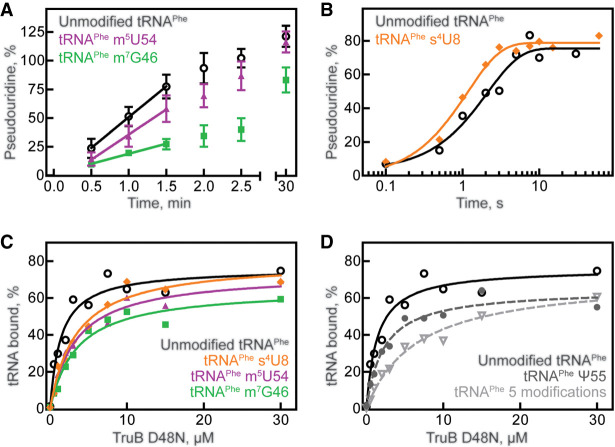
To assess whether TruB prefers to bind and modify unmodified tRNA or tRNA already containing a single modification, we prepared tritium-labeled tRNAPhe containing s4U8, m7G46, or m5U54. First, we compared the activity of TruB under multiple-turnover conditions with unmodified tRNA and tRNA containing m7G46 or m5U54 using tritium release assays. At a tRNA concentration of 600 nM (near its Michaelis constant, KM, 550 nM), 10 nM TruB has an initial velocity of approximately 4.7 nM s−1 for pseudouridylating unmodified tRNA (Table 1; Fig. 2A), as previously reported (Wright et al. 2011). Introduction of the adjacent m5U54 modification did not significantly affect the initial rate of TruB for tRNAPhe; however, incorporation of m7G46 into the tRNA lowered the initial velocity of pseudouridylation by TruB almost threefold to 1.6 nM s−1 (Table 1; Fig. 2A). The presence of s4U8 did not significantly affect single-turnover TruB rate (Table 2; Fig. 2B).
TABLE 1.
Average multiple-turnover initial velocities (v0, nM s−1) for TruB and TrmA modifying different single-modified tRNAs
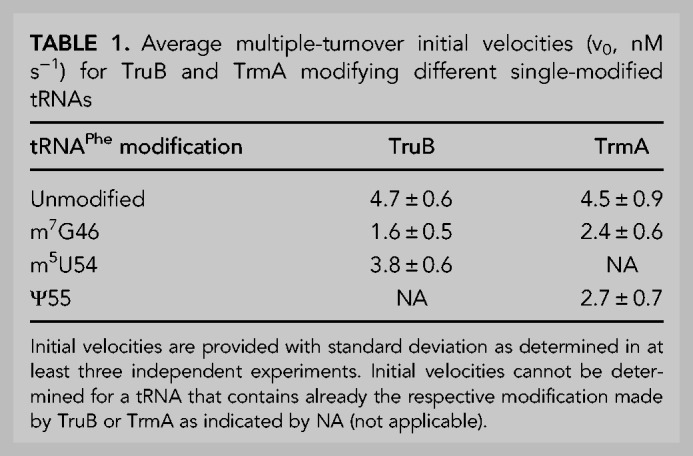
FIGURE 2.
tRNA modification and binding preferences of TruB. (A) Multiple-turnover modification by 10 nM TruB incubated with 600 nM unmodified tRNAPhe (open circles), tRNAPhe m5U54 (purple triangles), or tRNAPhe m7G46 (green squares). Time courses were performed at least in triplicate, and initial velocities were determined by linear regression (see Table 1). (B) Single-turnover modification by 5 µM TruB incubated with 1 µM unmodified tRNAPhe (open circles) or tRNAPhe s4U8 (orange diamonds). Here, we show one representative time course, but each time course was performed in triplicate and the apparent rate of each reaction was determined by fitting with a one-exponential equation, summarized in Table 2. Binding of catalytically inactive TruB D48N to 20 nM (C) unmodified tRNAPhe (open circles), tRNAPhe m5U54 (purple triangles), tRNAPhe m7G46 (green squares), tRNAPhe s4U8 (orange diamonds), or to (D) tRNAPhe Ψ55 (gray circles, dashed line), or tRNAPhe containing s4U8, Ψ32, Ψ38, m7G46, m5U54, and Ψ55 (light gray inverted triangles, dashed line), unmodified tRNAPhe (same as shown in C) determined by nitrocellulose filtration. One representative curve each of at least three replicates is shown. The data were fit with a hyperbolic equation to determine the dissociation constant, KD (see Table 3).
TABLE 2.
Average single-turnover apparent rates for TruB (kΨ, s−1), TrmA (kmethyl, s−1), and TrmB (kapp, s−1) modifying different single-modified tRNAs
In order to reveal whether the decreased speed of TruB modification for tRNA containing m7G46 was caused by a decrease in the binding affinity of TruB for this tRNA, dissociation constants (KD) were determined using nitrocellulose filtration assays. The catalytically inactive TruB D48N variant was used in order to measure the affinity of TruB for its substrate tRNA, rather than for its product, tRNAPhe Ψ55. As reported previously, the dissociation constant of TruB D48N for unmodified tRNA is 1.4 µM (Table 3; Fig. 2C; Wright et al. 2011) and thus slightly lower than the dissociation constant of TruB wild-type for pseudouridylated tRNA (2.4 µM) (Keffer-Wilkes et al. 2016). When m7G46 is present within the tRNA, the affinity of TruB D48N is reduced approximately twofold with a dissociation constant of about 3.1 µM (Table 3; Fig. 2C), in accordance with the decrease in TruB initial velocity for tRNA m7G46 (Table 1; Fig. 2A). Interestingly, the presence of either s4U8 or m5U54 also lowered the affinity of TruB D48N for tRNAPhe approximately twofold (Table 3; Fig. 2C), despite the fact these modifications did not significantly lower the initial velocity of TruB modification (Table 1; Fig. 2A,B). TruB D48N binding to tRNAPhe Ψ55, the product of TruB, was found to be similar to TruB D48N binding to its unmodified substrate tRNAPhe. In summary, the presence of s4U8, m7G46, or m5U54 (but not Ψ55) lowered the affinity of TruB D48N, highlighting that s4U8, m7G46, and m5U54 significantly and negatively affect TruB binding to tRNA (Table 3; Fig. 2D).
TABLE 3.
Average dissociation constants (KD, µM) for TruB, TrmA, and TrmB binding to differently modified tRNAs
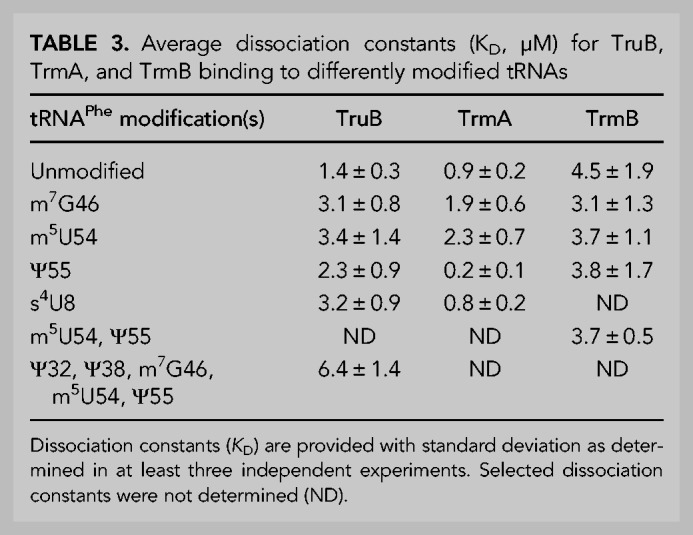
Next, we wanted to examine whether the presence of multiple modifications further decreased the affinity of TruB for tRNA. Therefore, [3H]tRNAPhe containing five modifications, namely two pseudouridines in the anticodon loop (Ψ32 and Ψ38 introduced by RluA and TruA), as well as m7G46, m5U54, and Ψ55 was prepared. The affinity of TruB for this tRNA was found to be further decreased compared to the single-modified tRNA variants with a KD of about 6.4 µM (Table 3; Fig. 2D), further suggesting TruB prefers to bind to tRNA prior to modification by other enzymes. Taken together, these results indicate that the binding of TruB to tRNA is sensitive to the presence of single s4U8, m7G46, and m5U54 modifications, with m7G46 additionally lowering the reaction velocity of pseudouridylation by TruB.
Binding and activity presence of TrmA
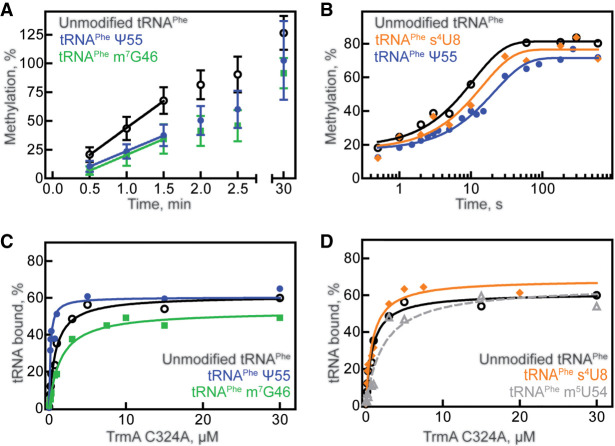
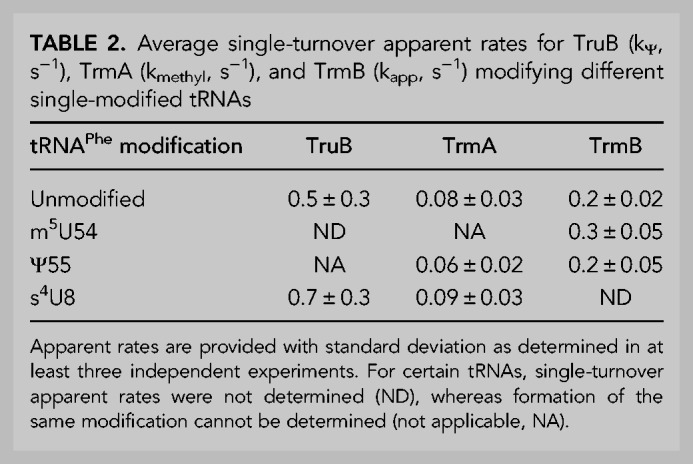
Similarly, we tested the impact of prior modifications on the TrmA enzyme using single-modified, tritium-labeled tRNAPhe in multiple-turnover tritium release assays. When acting on unmodified tRNAPhe, the initial velocity of TrmA-catalyzed methylation is about 4.5 nM s−1 (Table 1; Fig. 3A). The initial velocity of TrmA modification is reduced about twofold when either m7G46 or Ψ55 are present, to 2.4 nM s−1 and 2.7 nM s−1, respectively (Table 1; Fig. 3A). It is interesting that the initial velocity of TrmA is slower when Ψ55, the modification introduced by TruB, is present, because the affinity of TruB was reduced when the modification introduced by TrmA, m5U54, was present within tRNAPhe (Table 3; Fig. 2C), suggesting previous TruB modification negatively affects modification by TrmA and vice versa.
FIGURE 3.
tRNA modification and binding preferences for TrmA. (A) Multiple-turnover modification reaction of 10 nM TrmA incubated with 600 nM unmodified tRNAPhe (open circles), tRNAPhe m7G46 (green squares), or tRNAPhe Ψ55 (blue circles). Time courses were performed at least in triplicate. Initial velocities are summarized in Table 1. (B) Single-turnover modification by 5 µM TrmA and 50 µM SAM rapidly mixed with 1 µM unmodified tRNAPhe (open circles), tRNAPhe s4U8 (orange diamonds), or tRNAPhe Ψ55 (blue circles). Here, individual representative time courses are displayed, and the average apparent rates from three replicates is summarized in Table 2. Catalytically inactive TrmA C324A variant binding to 20 nM (C) unmodified tRNAPhe (open circles), tRNAPhe m7G46 (green squares), tRNAPhe Ψ55 (blue circles), or to (D) unmodified tRNAPhe (black circles, same as shown in C), tRNAPhe s4U8 (orange diamonds), or tRNAPhe m5U54 (gray triangles, dashed line). The average KD from at least three replicates for each experiment is summarized in Table 3.
To further examine the impact of the adjacent Ψ55 modification on catalysis by TrmA, the apparent rate of tRNA methylation by TrmA was compared for unmodified tRNAPhe and tRNAPhe Ψ55 using tritium release assays under single-turnover conditions in a quench-flow apparatus. In the absence and presence of the Ψ55 modification, the apparent rate was found to be about 0.08 and 0.06 s−1, respectively, suggesting that Ψ55 modification does not significantly affect catalysis by TrmA (Table 2; Fig. 3B). As observed with TruB, the presence of s4U8 also did not affect the apparent rate of TrmA (Table 2; Fig. 3B).
Next, we examined the influence of tRNA modification status on TrmA tRNA binding by nitrocellulose filter binding. Here, the TrmA C324A variant was used because this variant is deficient in catalysis and because it cannot form a covalent bond between enzyme and substrate (Kealey et al. 1994). The affinity of TrmA C324A for unmodified tRNAPhe was found to be about 0.9 µM (Table 3; Fig. 3C). The presence of the m7G46 modification lowered the affinity of TrmA C324A for tRNAPhe twofold to a dissociation constant of 1.9 µM (Table 3; Fig. 3C), consistent with the observation that this modification results in a reduction of the initial velocity of TrmA (Table 1; Fig. 3A). Similarly, we assessed the affinity of TrmA C324A for the product tRNA m5U54 and determined a reduced affinity compared to unmodified tRNA (Table 3; Fig. 3D). In other words, the affinity of TrmA C324A for tRNAPhe m7G46 is similar to the affinity of TrmA C324A for tRNA m5U54. The presence of s4U8 did not affect TrmA C324A binding (Table 3; Fig. 3D), in agreement with the lack of effect of this modification on TrmA activity (Table 2; Fig. 3A).
Interestingly, the presence of the Ψ55 modification strongly improved the affinity of TrmA C324A for tRNA Ψ55 almost fivefold compared to unmodified tRNAPhe (Table 3; Fig. 3C), contrasting the twofold decrease in TrmA activity for tRNAPhe Ψ55 (Table 1; Fig. 3A). Given that Ψ55 does not affect the rate constant of catalysis by TrmA (Table 2; Fig. 3B) and Ψ55 increases the affinity of TrmA for tRNAPhe (Table 3; Fig. 3C), the multiple-turnover activity of TrmA is surprisingly slow on tRNAPhe Ψ55 compared to unmodified tRNA (Table 1; Fig. 3A). As further discussed below, these findings suggest that Ψ55 must hinder another reaction step such as product release during tRNA modification by TrmA.
Binding and activity preferences of TrmB
Finally, we examined whether or not the methyltransferase TrmB is sensitive to the modification status of tRNA. In order to examine the single-turnover apparent rate for TrmB modifying tRNAs with different modification statuses, we measured radioactive methyl incorporation from tritium-labeled SAM into nonradioactive tRNA under single-turnover conditions. TrmB was found to modify tRNAPhe containing no modifications at a rate of about 0.2 sec−1, and the rate of modification was similar for tRNAPhe containing either m5U54 or Ψ55 (Table 2; Fig. 4A), suggesting TrmB does not have an activity preference for either unmodified or tRNA that is partially modified in the T loop.
FIGURE 4.
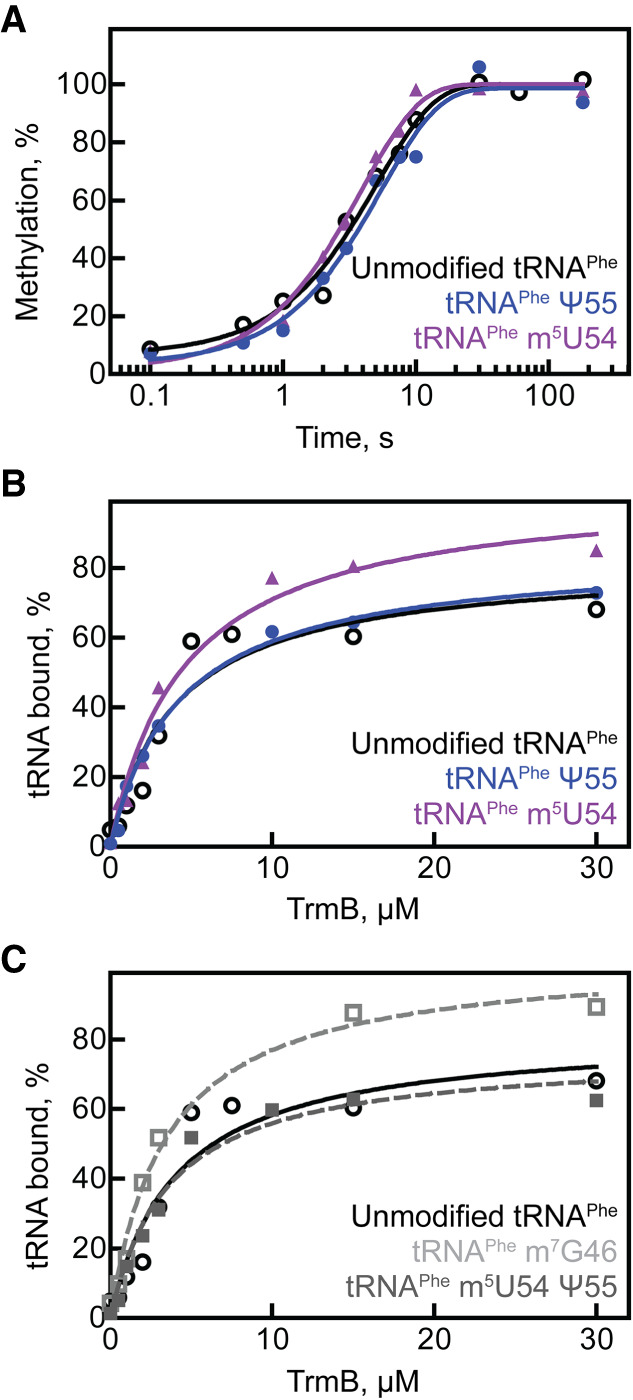
tRNA modification and binding preferences for TrmB. (A) Single-turnover modification by 5 µM TrmB and 50 µM [3H]SAM mixed with 1 µM unmodified tRNAPhe (open circles), tRNAPhe m5U54 (purple triangles), or tRNAPhe Ψ55 (blue circles). The average apparent rate of modification from three time courses (one representative each shown here) is reported in Table 2. TrmB binding to 20 nM (B) unmodified tRNAPhe (open circles), tRNAPhe m5U54 (purple triangles), tRNAPhe Ψ55 (blue circles), or to (C) unmodified tRNAPhe (black circles, same as shown in B), tRNAPhe m5U54 Ψ55 (gray squares, dashed line), or tRNAPhe m7G46 (open squares, dashed line). The experiments were repeated at least three times, and the average KD for each tRNA is summarized in Table 3.
Subsequently, we determined the affinity of TrmB for unmodified tRNA and tRNA containing either m5U54 or Ψ55 which may differ even though no changes in the modification rate were observed. Here, wild-type TrmB was used, but the cofactor SAM was omitted from the reaction such that TrmB is not capable of forming product. The affinity of TrmB for tRNAPhe containing either m5U54 or Ψ55 was similar to that of TrmB binding to unmodified tRNAPhe, characterized by a KD of approximately 3–4 µM (Table 3; Fig. 4B). Similarly, the presence of both m5U54 and Ψ55 modifications together did not impact the affinity of TrmB for tRNAPhe (Table 3; Fig. 4C). Like several other tRNA modification enzymes, TrmB was found to bind its product (tRNA m7G46) with a similar affinity as its substrate tRNA (Table 3; Fig. 4C). Our results point toward an insensitivity of TrmB binding and activity to the tRNA modification status in the T loop.
DISCUSSION
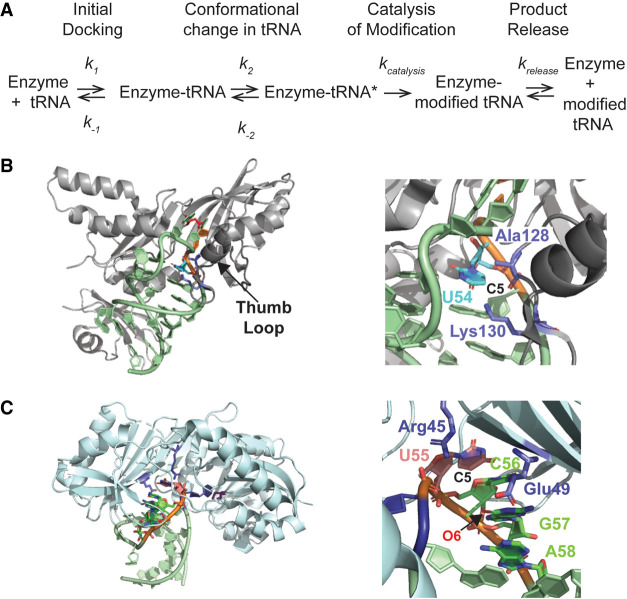
In this study, we examined the effects of single modifications within tRNAPhe on the activity and affinity of tRNA modification enzymes TruB, TrmA, and TrmB. We report here that the tRNA binding and modification activities of TruB and TrmA are sensitive to the modification status of the substrate tRNA. In contrast, none of the modifications examined here affected either tRNA binding or activity by TrmB. In the following, we will first discuss the molecular basis of how prior tRNA modifications affect (or do not affect) the molecular and kinetic mechanism of TruB, TrmA, and TrmB (Fig. 5A) before considering the cellular consequences and preferential modification order for tRNA maturation.
FIGURE 5.
Kinetic mechanism and tRNA interactions of TruB and TrmA. (A) Both TruB and TrmA have a two-step tRNA binding mechanism where the tRNA is first docked onto the enzyme (k1, k−1) followed by a conformational change that disrupts T and D arm interactions (k2, k−2) such that the enzyme gains access to its target uridine. Both binding steps are reversible and contribute to the dissociation constant. Subsequently, the tRNA is modified in one or more catalytic steps (kcatalysis), and finally the modified tRNA is released as the reaction product (krelease). All steps can contribute to the overall velocity (v0) of the enzyme. (B) Interaction of TruB with the T arm, in particular U54. (Left) Overview of TruB bound to the T arm (PDB ID 1K8W [Hoang and Ferre-D'Amare 2001]). U54 is shown in cyan, U55 in orange, and the catalytic aspartate 48 in red. The thumb loop is depicted in dark gray. (Right) Close-up view on the interaction of Lys130 and Ala128 (purple) in the thumb loop with U54. The backbone oxygen of Ala128 is 3.8 Å apart from C5 in U54 whereas the ζN of Lys130 is located 3.6 Å away from C5 of U54. The site of methylation by TrmA, C5 in U54, is indicated suggesting that the methyl group would cause a steric clash with Lys130 and Ala128. (C) Interaction of TrmA with the T arm, in particular U55. (Left) Overview of TrmA bound to the T arm (PDB ID: 3BT7 [Alian et al. 2008]). U54 is shown in blue, the catalytic cysteine in purple, and U55 in orange. (Right) Close-up, back view of U55 stacking with C56, G57, and A58 (green). The position of C5 of U55 which would be occupied by N1 in pseudouridine is indicated. Residues Arg45 and Glu49 (purple) are in proximity of U55, but not in direct contact with C5, that is, N1 in Ψ55 (3.1 Å between the ηN of Arg45 and C5 of U55, and 5.9 Å between the εO of Glu49 and C5 of U55). A pseudouridine would further strengthen the depicted base stacking interactions of U55–C56–G57–A58 in the T loop. Moreover, a water molecule could potentially form a hydrogen bonding bridge between N1 of pseudouridine and O6 of G57 (4.3 Å apart, labeled). All structural representations were prepared with the PyMOL software package.
Impact of modifications on tRNA binding and catalysis by other modification enzymes
Of the tested enzymes, TruB is most sensitive to the presence of other modifications in tRNA. We observed that m7G46 decreases the initial reaction velocity of pseudouridine formation by TruB and decreases the affinity of TruB for tRNA (Tables 1, 3). In addition, the modifications m5U54 and s4U8 also decrease the affinity of TruB for tRNA although the formation of pseudouridine is not affected (Tables 1–3). Based on the structure of TruB with an unmodified T arm (PDB ID 1K8W), only m5U54 will be in contact with TruB whereas s4U8 and m7G46 are predicted to be distant from TruB (Hoang and Ferre-D'Amare 2001). Within the unmodified T arm in the TruB structure, U54 forms a base pair with A58 and is contacted by the thumb loop in TruB (residues 120 to 149). Specifically, the side chain of Lysine 130 and the backbone oxygen of Alanine 128 interact with each other and are in very close proximity of C5 of U54 (3.6 and 3.8 Å, respectively) leaving no space for an additional methyl group (Fig. 5B). The steric clash between the methyl group of m5U54 and Lysine 130 explains the decreased affinity of TruB for tRNA with m5U54 compared to unmodified tRNA and suggests that the thumb loop of TruB must undergo a conformational change to accommodate this modification. We speculate that within the kinetic mechanism of TruB, the presence of m5U54 could lead to a faster dissociation of tRNA compared to unmodified tRNA (k−1 and k−2, Fig. 5; Keffer-Wilkes et al. 2016). However, since the catalysis of pseudouridylation is the slow and rate-limiting step for TruB (Wright et al. 2011), which is not affected by m5U54 once bound, no change in the reaction velocity is observed.
As s4U8 and m7G46 are not in contact with TruB, their effects on TruB's activity must be indirect and communicated through the tRNA structure. Structural and functional evidence clearly shows that TruB must break interactions between the T and D arm to gain access to the target U55 (Hoang and Ferre-D'Amare 2001; Keffer-Wilkes et al. 2016). Both s4U8 and m7G46 stabilize the D arm in mature tRNA as a part of a strong network of tertiary interactions in tRNA in this region (Helm 2006). Notably, the methylation of G46 introduces a positive charge in the variable loop near the negatively charged backbone of the base of the D arm, thereby acting like a staple closely connecting the variable loop and the D arm. Similarly, s4U8 is also interacting with the D arm. By positioning and stabilizing the D arm, m7G46 and s4U8 may render it more difficult for TruB to disrupt the tRNA elbow structure, to displace the D arm, and to gain access to U55. Accordingly, we hypothesize that both modifications slow down the conformational change (k2) thereby decreasing the affinity of TruB for tRNA (Fig. 5A). In the case of m7G46, the initial velocity of pseudouridylation is also affected (Table 1). We can envision that the constrained location of the D arm may affect correct positioning of the T arm within the active site thereby slowing down catalysis.
The negative impact of s4U8 and m5U54 on TruB's affinity for tRNA (Table 3) is reminiscent of tRNA binding by ArcTGT, for which a negative correlation was observed between KM and tRNA melting temperature causing ArcTGT modification of unmodified tRNA to be quick in comparison to modification of tRNA containing all modifications except for its product, archaeosine (Nomura et al. 2016). This similarity between ArcTGT and TruB is notable since both enzymes disrupt the tRNA tertiary structure in order to access their target base (Hoang and Ferre-D'Amare 2001; Ishitani et al. 2003). A rigid tRNA elbow already containing one or more modifications may provide TruB and ArcTGT with a greater challenge in the accession of their target base. Likewise, Bacillus subtilis TrmK methylates A22 and also prefers to bind to completely unmodified tRNA compared to tRNA containing prior modifications (Degut et al. 2019).
Similar to pseudouridylation by TruB, tRNA methylation at U54 by TrmA is affected by tRNA modifications. We found that TrmA is sensitive to the presence of m7G46 and Ψ55, but not s4U8. The presence of m7G46 decreases methylation activity and affinity for tRNA. Again, this effect is most likely a result of the stabilization of the D arm by m7G46, which hinders the disruption of T and D arm interactions by TrmA (decreased k2), causing a higher dissociation constant. We also observed that the presence of Ψ55 decreases the initial velocity (v0) of methylation by TrmA, but without affecting the catalytic step (kmethyl) itself (Tables 1, 2). In contrast to the negative effect on multiple-turnover methylation, we surprisingly note that Ψ55 has a positive effect on the affinity of TrmA for tRNA. As we will further explain below, these observations for Ψ55 can only be explained if a forward reaction step other than catalysis is slowed down (k1, k2, or krelease), reducing the multiple-turnover activity, and if a reverse reaction step (e.g., k−2) is reduced even more, leading to tighter tRNA binding (Fig. 5A).
In the structure of TrmA bound to an unmodified T arm, U55 forms base stacking interactions with nucleotides 56–58, and the base of U55 is contacted by the side chain of Arginine 45 and the backbone of Glutamate 49 (Fig. 5C, PDB ID 3BT7; Alian et al. 2008). When U55 is converted to pseudouridine, N1 of pseudouridine will occupy the position of C5. None of TrmA's contacts with U55 involve this atom in the uracil ring such that it seems unlikely that TrmA residues directly sense the presence of Ψ55. However, TrmA can only gain access to U54 by imposing a structural rearrangement in the T loop including formation of the base stack between U55, C56, G57, and A58 (Alian et al. 2008). As pseudouridine forms stronger base stacking interactions than uridine (Davis 1995), the presence of Ψ55 will stabilize the T arm conformation as bound to TrmA. In addition, it is conceivable that a water molecule could possibly form a hydrogen bonding bridge between N1 of Ψ55 and O6 of G57, further rigidifying the base stacking interactions. The stabilization of the T arm conformation as bound to TrmA could reduce the dissociation of tRNA from TrmA (k−1 or k−2, Fig. 5C) explaining the lower dissociation constant of TrmA for tRNA containing Ψ55.
The initial velocity of tRNA methylation of TrmA, but not the catalytic rate, is reduced by Ψ55, despite tighter binding of TrmA to tRNA containing Ψ55. Thus surprisingly, despite tighter binding, the overall multiple-turnover reaction comprising all reaction steps is slower whereas the catalytic step is not directly affected. As mentioned, these observations can only be explained by slower tRNA binding (k1 or k2) or slower product release, limiting the turnover of TrmA. Slower product release could be a result of more stable binding of tRNA containing Ψ55 which should equally affect k−2 and krelease (Fig. 5A). The presence of Ψ55 itself will also stabilize tertiary interactions within the tRNA substrate, rendering it possibly more difficult for TrmA to gain access to U54 when disrupting the interaction between the D arm and the T arm (reduced k2), which may further contribute to the reduced multiple turnover rate of TrmA when Ψ55 is present.
Notably, TrmB is not affected in its activity or tRNA binding by the presence of m5U54 or Ψ55. Since no structure of TrmB bound to tRNA has been determined so far, we can only speculate whether and how TrmB interacts with other regions of the tRNA besides the variable loop. It may be that TrmB does not contact the T arm and that it does not affect the tertiary interaction between the D and T arm. Alternatively, TrmB may interact with the T arm, but this interaction may not be affected by T loop modifications.
Preferred order of tRNA modification
Based on our data, we hypothesize that there is a preferred order, although not a strict hierarchy, of tRNA modification in E. coli. Specifically, we propose that TruB and TrmA prefer to act earlier than TrmB and ThiI during tRNA maturation. Since TruB and TrmA are faster acting on tRNA lacking other modifications (Table 1–3), TruB and TrmA are likely to act during the relatively early stages of tRNA maturation within the cell prior to TrmB introducing m7G46 in the variable arm. This hypothesis is supported by our observations that m7G46 presence negatively affects tRNA modification by TruB and TrmA, but TrmB is unaffected by m5U54 and Ψ55. Accordingly, TrmB is hypothesized to preferably act after TrmA and TruB, but this does not mean that E. coli TrmB is acting late during tRNA modification as also observed in yeast (Barraud et al. 2019). Notably, a recent study uncovered that the presence of m7G46 is highly advantageous for the formation of the adjacent aminocarboxypropyluridine (acp3U47) modification in E. coli tRNA by the newly discovered YfiP/TapT enzyme (Meyer et al. 2020). Conversely, TrmB does not depend on the prior formation of acp3U47. Therefore, it is plausible that TruB and TrmA prefer to modify tRNA very early followed by TrmB, which methylates tRNA shortly after and before the likely late-acting TapT enzyme (Takakura et al. 2019; Meyer et al. 2020). The presence of the s4U8 modification did not affect the apparent rate of modification for either TrmA or TruB (Table 2; Figs. 2B, 3B); however, s4U8 presence did lower the affinity of TruB for tRNAPhe (Table 3; Fig. 2C). From these data, we cannot decisively predict temporally when ThiI is likely to act during tRNA maturation; however, it does seem probable that ThiI may act after TruB in order to provide TruB with an ideal substrate.
In accordance with our suggestion that TruB and TrmA may act prior to TrmB during tRNA modification in E. coli, a similar preferred modification order was reported in a recent study of in vitro transcribed tRNAPhe modification in Saccharomyces cerevisiae cell extracts monitored by time-resolved NMR. Here, Ψ55 was the first modification to be introduced within tRNA, with both m5U54 and m7G46 appearing second within the tRNA where the introduction of m7G46 finished after the complete introduction of m5U54 (Barraud et al. 2019). Thus, in both E. coli and S. cerevisiae, TruB prefers to act prior to TrmB. But in yeast, it could be possible that TrmB and TrmA homologs act in parallel during tRNA maturation, which may be a result of species-specific differences between monomeric E. coli TrmB (Zhou et al. 2009) and the heterodimeric Trm8-Trm82 complex from yeast (Alexandrov et al. 2002).
Regarding tRNA modification by TruB and TrmA in E. coli, the most likely temporal order is not obvious because m5U54 hinders TruB binding and Ψ55 slows modification by TrmA (Table 1–3). On the other hand, Ψ55 presence increases the affinity of TrmA for tRNAPhe approximately fivefold (Table 3). The seemingly opposite effects of Ψ55 on TrmA are not without precedence as the apparent rate of U34 modification by CmoM is decreased when 1-methylguanosine 37 formed by TrmD is present within E. coli tRNAProUGG, despite 1-methylguanosine 37 lowering the affinity of CmoM for tRNAProUGG (Masuda et al. 2018). Interestingly, bulk tRNAs from yeast missing TruB's homolog, Pus4, have been found to lack m5U54 content, suggesting, at least in yeast, introduction of m5U54 actually relies on previous modification by TruB to a certain extent (Barraud et al. 2019). Accordingly, the yeast tRNA methyltransferase Trm2 may be differently affected by the presence of Ψ55 than its E. coli homolog TrmA.
Several reasons support our suggestion that both TruB and TrmA modify tRNA relatively early within the overall hierarchy of tRNA modifications. First, with TrmA and TruB as the only enzymes to modify all species of E. coli tRNA, it seems unlikely for these enzymes to rely on the previous introduction of another tRNA modification as a positive determinant as no other modification can be present within each of their substrate tRNAs. In contrast, it is possible for other modification enzymes to rely on either m5U54 or Ψ55 modifications as a determinant, considering that m5U54 and Ψ55 conversely are present within the tRNA substrates of every E. coli tRNA modification enzyme. Second, evidence for early tRNA modification by TrmA and TruB during tRNA maturation is provided by the observation that both m5U54 and Ψ55 appear prior to intron splicing and 5′ and 3′ end trimming when yeast tRNATyr was injected into Xenopus oocytes (Nishikura and De Robertis 1981). Similarly, yeast tRNAPhe contains m5U54 and Ψ55 prior to intron splicing (Etcheverry et al. 1979; Jiang et al. 1997). Third, considering that both TruB and TrmA are known tRNA chaperones (Keffer-Wilkes et al. 2016; LC Keffer-Wilkes, EF Soon, and U Kothe, in prep.), a scenario in which TruB and TrmA have evolved to interact with tRNA during the early stages of its maturation in order to quickly promote correct tRNA fold may be beneficial. In particular, the correct three-dimensional structure of tRNA promoted by the action of TruB and TrmA will allow the tRNA to be modified by other tRNA modification enzymes that recognize the entire tRNA structure (Grosjean et al. 1996).
Conclusion
In summary, our proof-of-principle study describes that TruB and TrmA prefer to act on unmodified tRNA compared to tRNA already containing a single modification whereas TrmB does not appear to have a preference for the modification status of tRNA. Our results corroborate recent findings in yeast (Barraud et al. 2019) and additionally provide mechanistic insight for why Ψ55 and m5U54 are likely to appear early during tRNA maturation. A recent review (Han and Phizicky 2018) hypothesized that tRNA modification circuits, that is, a strict order of modification, are most common within the anticodon loop because modifications can serve as additional sequence or structure determinants to ensure tRNA anticodon loop modification enzymes only modify their desired substrates, which is particularly critical considering the diversity of anticodon loop modification and its importance for translational fidelity. Taking our results together with other studies of tRNA elbow modification enzymes (Rider et al. 2009; Nomura et al. 2016; Barraud et al. 2019; Degut et al. 2019; Hori 2019; Meyer et al. 2020), it becomes clear that modifications in the tRNA elbow region do not occur in strict order where one modification is dependent on the other. However, there is still a preferred pathway of tRNA elbow modification and the modification enzymes do not act randomly on tRNA.
MATERIALS AND METHODS
Buffers and reagents
Experiments were performed in TAKEM4 buffer (50 mM Tris-HCl, pH 7.5, 70 mM NH4Cl, 30 mM KCl, 1 mM EDTA, 4 mM MgCl2). [C5-3H]UTP for in vitro transcription of [3H]tRNAPhe was purchased from Moraveck Biochemicals and radioactive adenosyl-l-methionine S-[methyl-3H] ([3H]SAM) was purchased from PerkinElmer. Nonradioactive SAM was obtained from New England Biolabs (NEB). All other chemicals were purchased from Fisher Scientific.
Protein expression and purification
To generate a plasmid encoding E. coli trmB, the forward primer, 5′-GCCAGAGCTAGCAAAAACGACGTCATTTCACCG-3′ and reverse primer, 5′-GCCACCGGATCCTTATTTCACCCTCTCGAACATTAAGTCCC-3′ were used to amplify the trmB open reading frame and clone into the pET28a(+) vector using the NheI and BamHI restriction sites, creating pET28a-TrmB. The gene sequence was confirmed by DNA sequencing (Genewiz) and the plasmid was transformed into BL21 (DE3) cells. Cells were grown in LB supplemented with 50 µg/mL kanamycin at 37°C and protein expression was induced at an OD600 of approximately 0.6 with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). After 3 h, cells were harvested by centrifugation at 5000g for 15 min, flash frozen, and stored at −80°C.
Wild-type TruB and TruB D48N were expressed from pET28a-TruB in BL21 (DE3) cells as previously described (Wright et al. 2011). For the expression of ThiI and IscS in BL21 (DE3) cells, plasmids pBH113 and pBH402 were obtained from Eugene Mueller (Mueller et al. 1998, 2001). TrmA wild-type and TrmA C324A proteins were overexpressed from the pCA24N(GFP minus)-TrmA (JW3937) plasmid in AG1 (ME5305) cells (Kitagawa et al. 2005; LC Keffer-Wilkes, EF Soon, and U Kothe, in prep.). Similarly, the tRNA pseudouridine synthases TruA and RluA were expressed using the respective pCA24N(GFP minus) plasmids (Wright et al. 2011).
All proteins were purified via their amino-terminal hexahistidine tag using nickel-sepharose and Superdex 75 chromatography as described (Wright et al. 2011). Protein concentrations were determined by absorbance at 280 nm using molar extinction coefficients of 35,870 M−1 cm−1 for TrmA, 20,860 M−1 cm−1 for TruB, and 27,960 M−1 cm−1 for TrmB (calculated using ProtParam [Gill and von Hippel 1989]), 63,100 M−1 cm−1 for ThiI and 25,400 M−1 cm−1 for IscS (as determined in Mueller et al. 2001). Concentrations were confirmed using comparative SDS-PAGE.
tRNA preparation
The E. coli tRNAPhe gene was amplified from the pCF0 plasmid (Sampson et al. 1989) for in vitro transcription. To produce internally tritium-labeled tRNA, [C5-3H]UTP was included in the reaction as previously described (Wright et al. 2011). Radioactive tRNAPhe transcripts were purified by Nucleobond Xtra Midi anion-exchange gravity columns (Macherey-Nagel) as described (Wright et al. 2011). Nonradioactive transcripts were purified by phenol-chloroform extraction to remove proteins followed by Superdex 75 (XK 26/100 column, GE Healthcare) size exclusion chromatography to remove unincorporated nucleotides. Subsequently, tRNAPhe was concentrated by isopropanol precipitation and resuspended in water. tRNAPhe concentration was determined by A260 measurements using a molar extinction coefficient of 500,000 M−1 cm−1 (Peterson and Uhlenbeck 1992). Scintillation counting was used to determine the specific activity of radioactive tRNA.
Partial modification of tRNAPhe
In vitro transcribed tRNAPhe was refolded in TAKEM4 buffer by heating to 65°C for 5 min and cooled to room temperature for 10 min. To prepare single-modified tRNAPhe Ψ55, tRNAPhe m5U54, and tRNAPhe m7G46, 1.6 µM of tritium-labeled or nonradioactive refolded tRNAPhe was incubated with 5 µM of purified TruB, TrmA, or TrmB enzyme for 2 h at 37°C in a 5 mL reaction. Methylation reactions included 50 µM SAM. To prepare [3H]tRNAPhe s4U8, 4 µM of folded [3H]tRNAPhe, 2 µM of purified ThiI, and 1 µM purified IscS was incubated with 40 µM pyridoxal-5′-phosphate, 4 mM ATP, 0.5 mM L-cysteine, and 1 mM DTT in a 10 mL reaction volume in TAKEM4 buffer for 2 h at 37°C. To prepare a [3H]tRNAPhe with five modifications, 5 µM [3H]tRNAPhe was incubated with 7.5 µM of each RluA, TruA, TruB, TrmA, and TrmB in the presence of 50 µM SAM in a 100 µL reaction. In all cases, enzymes were subsequently removed by phenol-chloroform extraction followed by ethanol precipitation of the tRNA. After resuspending modified tRNAPhe in water, tRNAPhe concentration was determined by A260 measurements and scintillation counting.
To quantify the level of modification for prepared [3H]tRNAPhe m5U54 and [3H]tRNAPhe Ψ55, 600 nM of the now single-modified tRNA was incubated with 5 µM TrmA or TruB, respectively. The end level of modification by the enzyme after 1 h at 37°C was determined in triplicate by tritium release assays in order to determine the percentage of the tRNA that had remained unmodified. Thereby, it was determined that [3H]tRNAPhe m5U54 was 78 ± 5% modified and [3H]tRNAPhe Ψ55 was 81 ± 4% modified. The Ψ55 content within the [3H]tRNAPhe with five modifications was similarly quantified, and Ψ55 was found to be present in 83 ± 3% of this preparation. To assess the level of modification within [3H]tRNAPhe m7G46 and nonradioactive tRNAPhe m5U54 and tRNAPhe Ψ55 preparations, small scale reactions with [3H]SAM or [3H]tRNAPhe were performed in parallel in triplicate to estimate the level of modification within each respective large-scale preparation. The respective tRNAPhe preparations contained 83 ± 5% of m7G46, 88 ± 2% of m5U54, and 89 ± 3% of Ψ55. Finally, to quantify the level of s4U8 within tRNA, modified tRNA was analyzed in triplicate on an [(N-acryloylamino)phenyl]mercuric chloride (APM) gel (Igloi 1988). In brief, 50 pmol of RNA was analyzed on a denaturing gel containing 10% acrylamide (19:1 acrylamide:bisacrylamide), 7 M urea, and 20 µM APM in Tris/Borate/EDTA (TBE) buffer. Following SYBR Green II staining, the ratio of thiolated tRNA retarded by APM to nonthiolated, faster migrating tRNA was determined using ImageJ software and [3H]tRNAPhe s4U8 was determined to be 83 ± 2% modified.
Tritium release assay to measure Ψ55 and m5U54 formation
Tritium release assays were performed as previously described to detect pseudouridylation by TruB and C5-methylation by TrmA (Wright et al. 2011; LC Keffer-Wilkes, EF Soon, and U Kothe, in prep.). First, tritium-labeled tRNAPhe was refolded as before. All reactions with TrmA included 50 µM SAM. For multiple-turnover reactions, 600 nM [3H]tRNAPhe was incubated with 10 nM enzyme. The initial velocity (v0) of the reaction was determined by linear fitting of the first 1.5 min of the reaction. For single-turnover reactions, enzyme and [3H]tRNAPhe were mixed in a Kintek quench-flow with final concentrations of 5 µM of enzyme and 1 µM of tRNA. Reactions were stopped by addition of 0.1 M hydrochloric acid after 0.1, 0.5, 1, 2, 3, 5, 7.5, 10, 30, 60, and 180 sec, and the level of tritium released was determined as previously described (Wright et al. 2011). To determine the apparent reaction rate (kapp), time courses were fit with a one-exponential equation:
where Y is the percentage of pseudouridine formation at a given time point, Y0 is the initial level of apparent pseudouridylation (due to tritium release from the tRNA in absence of enzyme), Y∞ is the endlevel of pseudouridylation, and t is the time.
For TruB and TrmA, the apparent reaction rate is concentration-independent and therefore directly reflects the rate constant of pseudouridylation (kΨ) and the rate constant of methylation (kmethyl), respectively (Wright et al. 2011; LC Keffer-Wilkes, EF Soon, and U Kothe, in prep.).
Methylation assay to measure m7G46 formation
To determine methylation by TrmB, TrmB and [3H]SAM were rapidly mixed in a quench-flow apparatus with nonradioactive tRNAPhe to final concentrations 5 µM, 50 µM, and 1 µM, respectively, and reactions were stopped by addition of 0.1 M HCl at the same timepoints as described above. A constant volume of each quenched sample was precipitated on Whatman paper disks presoaked with 5% (w/v) trichloracetic acid. To remove unincorporated [3H]SAM, disks were washed three times with 5% (w/v) trichloracetic acid for 5 min followed by a final wash in ethanol. After drying, paper disks were added to 4 mL EcoLite(+) scintillation cocktail (MP Biomedical), and the amount of [3H]methyl incorporated was determined by scintillation counting. The apparent rate of methylation was determined by fitting to a single exponential function, as stated above. Since percent tRNA methylated cannot be determined directed with this assay, the maximum amplitude determined by fitting was set to be 100%.
Nitrocellulose filtration to determine the affinity for tRNA
Refolded, uniformly tritium-labeled tRNAPhe (20–40 nM) was incubated with increasing concentrations of enzyme (0, 0.5, 1.0, 2.0, 3.0, 5.0, 7.5, 10.0, 15.0, 30.0 µM for TruB D48N and wild-type TrmB, and 0, 0.15, 0.2, 0.3, 0.75, 1.0, 3.0, 5.0, 15.0, 30.0 µM for TrmA C324A) in TAKEM4 buffer for 10 min at room temperature prior to filtration through a nitrocellulose membrane. The proportion of tRNA bound to enzyme was determined by scintillation counting as described in (Wright et al. 2011). The dissociation constant (KD) was determined by plotting percent RNA bound (Bound) as a function of enzyme concentration ([enzyme]) and fitting to a hyperbolic equation:
ACKNOWLEDGMENTS
We thank Saskia Funk for preparation of ThiI and IscS proteins, and the National BioResource Project (National Institute of Genetics, Japan) for the TrmA expression plasmid. In addition, we are grateful for the gift of ThiI and IscS expression plasmids by Dr. Eugene Mueller. This work was supported by the Natural Sciences and Engineering Research Council of Canada (Discovery Grant RGPIN-2014-05954).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.075473.120.
REFERENCES
- Alexandrov A, Martzen MR, Phizicky EM. 2002. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8: 1253–1266. 10.1017/S1355838202024019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21: 87–96. 10.1016/j.molcel.2005.10.036 [DOI] [PubMed] [Google Scholar]
- Alian A, Lee TT, Griner SL, Stroud RM, Finer-Moore J. 2008. Structure of a TrmA–RNA complex: a consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc Natl Acad Sci 105: 6876–6881. 10.1073/pnas.0802247105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud P, Tisne C. 2019. To be or not to be modified: miscellaneous aspects influencing nucleotide modifications in tRNAs. IUBMB Life 71: 1126–1140. 10.1002/iub.2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud P, Gato A, Heiss M, Catala M, Kellner S, Tisne C. 2019. Time-resolved NMR monitoring of tRNA maturation. Nat Commun 10: 3373 10.1038/s41467-019-11356-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, et al. 2018. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46: D303–D307. 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne RT, Konevega AL, Rodnina MV, Antson AA. 2010. The crystal structure of unmodified tRNAPhe from Escherichia coli. Nucleic Acids Res 38: 4154–4162. doi:10.1093/nar/gkq133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DR. 1995. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res 23: 5020–5026. 10.1093/nar/23.24.5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crecy-Lagard V, Boccaletto P, Mangleburg CG, Sharma P, Lowe TM, Leidel SA, Bujnicki JM. 2019. Matching tRNA modifications in humans to their known and predicted enzymes. Nucleic Acids Res 47: 2143–2159. 10.1093/nar/gkz011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degut C, Roovers M, Barraud P, Brachet F, Feller A, Larue V, Al Refaii A, Caillet J, Droogmans L, Tisne C. 2019. Structural characterization of B. subtilis m1A22 tRNA methyltransferase TrmK: insights into tRNA recognition. Nucleic Acids Res 47: 4736–4750. 10.1093/nar/gkz230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Bailly M, de Crecy-Lagard V. 2012. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet 46: 69–95. 10.1146/annurev-genet-110711-155641 [DOI] [PubMed] [Google Scholar]
- Etcheverry T, Colby D, Guthrie C. 1979. A precursor to a minor species of yeast tRNASer contains an intervening sequence. Cell 18: 11–26. 10.1016/0092-8674(79)90349-0 [DOI] [PubMed] [Google Scholar]
- Friedt J, Leavens FM, Mercier E, Wieden HJ, Kothe U. 2014. An arginine-aspartate network in the active site of bacterial TruB is critical for catalyzing pseudouridine formation. Nucleic Acids Res 42: 3857–3870. 10.1093/nar/gkt1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182: 319–326. 10.1016/0003-2697(89)90602-7 [DOI] [PubMed] [Google Scholar]
- Grosjean H, Edqvist J, Straby KB, Giege R. 1996. Enzymatic formation of modified nucleosides in tRNA: dependence on tRNA architecture. J Mol Biol 255: 67–85. 10.1006/jmbi.1996.0007 [DOI] [PubMed] [Google Scholar]
- Han L, Phizicky EM. 2018. A rationale for tRNA modification circuits in the anticodon loop. RNA 24: 1277–1284. 10.1261/rna.067736.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M. 2006. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res 34: 721–733. 10.1093/nar/gkj471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang C, Ferre-D'Amare AR. 2001. Cocrystal structure of a tRNA Ψ55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell 107: 929–939. 10.1016/S0092-8674(01)00618-3 [DOI] [PubMed] [Google Scholar]
- Hori H. 2019. Regulatory factors for tRNA modifications in extreme-thermophilic bacterium Thermus thermophilus. Front Genet 10: 204 10.3389/fgene.2019.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igloi GL. 1988. Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry 27: 3842–3849. 10.1021/bi00410a048 [DOI] [PubMed] [Google Scholar]
- Ishitani R, Nureki O, Nameki N, Okada N, Nishimura S, Yokoyama S. 2003. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell 113: 383–394. 10.1016/S0092-8674(03)00280-0 [DOI] [PubMed] [Google Scholar]
- Jiang HQ, Motorin Y, Jin YX, Grosjean H. 1997. Pleiotropic effects of intron removal on base modification pattern of yeast tRNAPhe: an in vitro study. Nucleic Acids Res 25: 2694–2701. 10.1093/nar/25.14.2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambampati R, Lauhon CT. 2000. Evidence for the transfer of sulfane sulfur from IscS to ThiI during the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. J Biol Chem 275: 10727–10730. 10.1074/jbc.275.15.10727 [DOI] [PubMed] [Google Scholar]
- Kealey JT, Gu X, Santi DV. 1994. Enzymatic mechanism of tRNA (m5U54)methyltransferase. Biochimie 76: 1133–1142. 10.1016/0300-9084(94)90042-6 [DOI] [PubMed] [Google Scholar]
- Keffer-Wilkes LC, Veerareddygari GR, Kothe U. 2016. RNA modification enzyme TruB is a tRNA chaperone. Proc Natl Acad Sci 113: 14306–14311. 10.1073/pnas.1607512113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Waldor MK. 2019. The RNA degradosome promotes tRNA quality control through clearance of hypomodified tRNA. Proc Natl Acad Sci 116: 1394–1403. 10.1073/pnas.1814130116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. 2005. Complete set of ORF clones of Escherichia coli ASKA library (A Complete Set of E. coli K-12 ORF Archive): unique resources for biological research. DNA Res 12: 291–299. 10.1093/dnares/dsi012 [DOI] [PubMed] [Google Scholar]
- Masuda I, Takase R, Matsubara R, Paulines MJ, Gamper H, Limbach PA, Hou YM. 2018. Selective terminal methylation of a tRNA wobble base. Nucleic Acids Res 46: e37 10.1093/nar/gky013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B, Immer C, Kaiser S, Sharma S, Yang J, Watzinger P, Weiss L, Kotter A, Helm M, Seitz HM, et al. 2020. Identification of the 3-amino-3-carboxypropyl (acp) transferase enzyme responsible for acp3U formation at position 47 in Escherichia coli tRNAs. Nucleic Acids Res 48: 1435–1450. 10.1093/nar/gkz1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller EG, Buck CJ, Palenchar PM, Barnhart LE, Paulson JL. 1998. Identification of a gene involved in the generation of 4-thiouridine in tRNA. Nucleic Acids Res 26: 2606–2610. 10.1093/nar/26.11.2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller EG, Palenchar PM, Buck CJ. 2001. The role of the cysteine residues of ThiI in the generation of 4-thiouridine in tRNA. J Biol Chem 276: 33588–33595. 10.1074/jbc.M104067200 [DOI] [PubMed] [Google Scholar]
- Nishikura K, De Robertis EM. 1981. RNA processing in microinjected Xenopus oocytes. Sequential addition of base modifications in the spliced transfer RNA. J Mol Biol 145: 405–420. 10.1016/0022-2836(81)90212-6 [DOI] [PubMed] [Google Scholar]
- Nomura Y, Ohno S, Nishikawa K, Yokogawa T. 2016. Correlation between the stability of tRNA tertiary structure and the catalytic efficiency of a tRNA-modifying enzyme, archaeal tRNA-guanine transglycosylase. Genes Cells 21: 41–52. 10.1111/gtc.12317 [DOI] [PubMed] [Google Scholar]
- Peterson ET, Uhlenbeck OC. 1992. Determination of recognition nucleotides for Escherichia coli phenylalanyl-tRNA synthetase. Biochemistry 31: 10380–10389. 10.1021/bi00157a028 [DOI] [PubMed] [Google Scholar]
- Purta E, van Vliet F, Tricot C, De Bie LG, Feder M, Skowronek K, Droogmans L, Bujnicki JM. 2005. Sequence-structure-function relationships of a tRNA (m7G46) methyltransferase studied by homology modeling and site-directed mutagenesis. Proteins 59: 482–488. 10.1002/prot.20454 [DOI] [PubMed] [Google Scholar]
- Ranjan N, Rodnina MV. 2016. tRNA wobble modifications and protein homeostasis. Translation (Austin) 4: e1143076 10.1080/21690731.2016.1143076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider LW, Ottosen MB, Gattis SG, Palfey BA. 2009. Mechanism of dihydrouridine synthase 2 from yeast and the importance of modifications for efficient tRNA reduction. J Biol Chem 284: 10324–10333. 10.1074/jbc.M806137200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson JR, DiRenzo AB, Behlen LS, Uhlenbeck OC. 1989. Nucleotides in yeast tRNAPhe required for the specific recognition by its cognate synthetase. Science 243: 1363–1366. 10.1126/science.2646717 [DOI] [PubMed] [Google Scholar]
- Sokolowski M, Klassen R, Bruch A, Schaffrath R, Glatt S. 2018. Cooperativity between different tRNA modifications and their modification pathways. Biochim Biophys Acta Gene Regul Mech 1861: 409–418. 10.1016/j.bbagrm.2017.12.003 [DOI] [PubMed] [Google Scholar]
- Takakura M, Ishiguro K, Akichika S, Miyauchi K, Suzuki T. 2019. Biogenesis and functions of aminocarboxypropyluridine in tRNA. Nat Commun 10: 5542 10.1038/s41467-019-13525-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongdee N, Jaroensuk J, Atichartpongkul S, Chittrakanwong J, Chooyoung K, Srimahaeak T, Chaiyen P, Vattanaviboon P, Mongkolsuk S, Fuangthong M. 2019. TrmB, a tRNA m7G46 methyltransferase, plays a role in hydrogen peroxide resistance and positively modulates the translation of katA and katB mRNAs in Pseudomonas aeruginosa. Nucleic Acids Res 47: 9271–9281. 10.1093/nar/gkz702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomikawa C. 2018. 7-methylguanosine modifications in transfer RNA (tRNA). Int J Mol Sci 19: E4080 10.3390/ijms19124080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J, Durand JM, Bjork GR. 2002. Three modifications in the D and T arms of tRNA influence translation in Escherichia coli and expression of virulence genes in Shigella flexneri. J Bacteriol 184: 5348–5357. 10.1128/JB.184.19.5348-5357.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerareddygari GR, Singh SK, Mueller EG. 2016. The pseudouridine synthases proceed through a glycal intermediate. J Am Chem Soc 138: 7852–7855. 10.1021/jacs.6b04491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JR, Keffer-Wilkes LC, Dobing SR, Kothe U. 2011. Pre-steady-state kinetic analysis of the three Escherichia coli pseudouridine synthases TruB, TruA, and RluA reveals uniformly slow catalysis. RNA 17: 2074–2084. 10.1261/rna.2905811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Liu Q, Yang W, Gao Y, Teng M, Niu L. 2009. Monomeric tRNA (m7G46) methyltransferase from Escherichia coli presents a novel structure at the function-essential insertion. Proteins 76: 512–515. 10.1002/prot.22413 [DOI] [PubMed] [Google Scholar]