Abstract
The wide prevalence and regulated expression of long noncoding RNAs (lncRNAs) highlight their functional roles, but the molecular basis for their activities and structure-function relationships remains to be investigated, with few exceptions. Among the relatively few lncRNAs conserved over significant evolutionary distances is the long intergenic noncoding RNA (lincRNA) Cyrano (orthologous to human OIP5-AS1), which contains a region of 300 highly conserved nucleotides within tetrapods, which in turn contains a functional stretch of 26 nt of deep conservation. This region binds to and facilitates the degradation of the microRNA miR-7, a short ncRNA with multiple cellular functions, including modulation of oncogenic expression. We probed the secondary structure of Cyrano in vitro and in cells using chemical and enzymatic probing, and validated the results using comparative sequence analysis. At the center of the functional core of Cyrano is a cloverleaf structure maintained over the >400 million years of divergent evolution that separates fish and primates. This strikingly conserved motif provides interaction sites for several RNA-binding proteins and masks a conserved recognition site for miR-7. Conservation in this region strongly suggests that the function of Cyrano depends on the formation of this RNA structure, which could modulate the rate and efficiency of degradation of miR-7.
Keywords: Cyrano, SHAPE, conservation, lincRNA, structure
INTRODUCTION
Long intergenic noncoding ribonucleic acids (lincRNAs) are defined as non-protein-encoding transcripts longer than 200 nt that do not overlap with protein-coding genes (Derrien et al. 2012; Ulitsky and Bartel 2013). At least 30,000 lincRNAs have been identified in the human genome from large-scale genomic analyses (GENCODE v29). These RNA polymerase II transcripts can localize to either the nucleus or cytoplasm and typically have very low abundance, often less than one copy per cell (Cabili et al. 2015; Kleaveland et al. 2018), yet are widely expressed and regulated in a temporal and tissue specific manner. LincRNAs evolve very rapidly, yet an increasing number of them have demonstrable biological functions in development and disease (Wapinski and Chang 2011), which includes, but is not limited to, the targeting of chromatin-remodeling enzymes to specific genomic loci (Ulitsky and Bartel 2013).
The structure, biological mechanism, and detailed molecular function of most lincRNAs remain unknown, with some exceptions (Li et al. 2016). While genome-wide mapping projects provide information regarding their protein, RNA, and DNA interactions, and polymorphisms, structure-function information for the majority of lincRNAs is especially limited (The ENCODE Project Consortium 2012). Although lincRNAs have a lower propensity for forming secondary structure relative to messenger RNAs (mRNAs) (Yang and Zhang 2015), structural probing of the lincRNAs NEAT1 (Lin et al. 2018), XIST (Pintacuda et al. 2017), MEG3 (Uroda et al. 2019), and HOTAIR (Somarowthu et al. 2015), for example, reveal that these transcripts can possess intricate secondary structures. A commonly stated but generally unproven hypothesis is that these secondary structures are functional, and that structural features are conserved despite rapid primary sequence divergence.
RNA secondary structure can be established through chemical and enzymatic probing techniques, as demonstrated decades ago for the ribosome (Stiegler et al. 1981). Prior to 2006, several chemical probing approaches were utilized to obtain structural information for every nucleotide (Ziehler and Engelke 2001); the development of selective 2′ hydroxyl acylation analyzed by primer extension (SHAPE) chemistry, which provides structural information on all nucleotides in a single experiment, greatly enhanced the technique of chemical probing (Wilkinson et al. 2006). The application of next generation sequencing (NGS) on probed RNA has enabled genome-wide structural evaluation of RNA transcripts both in vitro and in cells (Kwok 2016; Qian et al. 2019). However, the very low abundance of many lincRNAs makes their investigation in cells very challenging (Kwok et al. 2013), leading to the necessity for probing transcripts in vitro through nondenaturing purification of the RNA (Chillón et al. 2015). The shotgun secondary structure (3S) approach can be used for secondary structure validation (Novikova et al. 2013; Chillón et al. 2015), together with positional covariance (Rivas et al. 2017). Programs such as CMfinder (Yao et al. 2006), which is a motif discovery program driven by comparative sequence analysis, identifies structural motifs in RNAs based on covariation and evolutionary divergence.
Among the relatively small fraction of lincRNAs possessing primary sequence conservation over significant evolutionary distances is Cyrano (OIP5-AS1 is its human orthologue), present in a wide range of species from human to lamprey (Ulitsky et al. 2011; Mallory and Shkumatava 2015). While the lincRNA Cyrano was initially believed to be critical for embryonic development (this role was recently shown to be dispensable by Goudarzi et al. 2019), the transcript acts as a sponge for the RNA binding protein HuR (Kim et al. 2016) and appears to regulate cell proliferation and apoptosis in cancer cells through its association with several microRNAs, especially miR-7 (Smith et al. 2017; Kleaveland et al. 2018).
Most recently, Cyrano has been shown to target the microRNA miR-7 for degradation through TDMD (targeted RNA-directed microRNA degradation) (Kleaveland et al. 2018). MicroRNAs can regulate gene expression by binding to ncRNAs as well as mRNAs (Bartel 2004), and recruit Argonaute proteins to induce gene silencing (as part of the RNA-induced silencing complex [RISC]) (Hammond et al. 2001). The degradation of miR-7 by Cyrano prevents its repression of the circular RNA, Cdr1as (Kleaveland et al. 2018), which plays a key role in neuronal activity (Smith et al. 2017; Kleaveland et al. 2018).
Regulation of miR-7 by Cyrano occurs through a core of 26 bases, located within the Cyrano transcript, that are nearly perfectly conserved (96%) in species as divergent as zebrafish and human. Approximately 100 nt on either side of this 26-nt stretch are 40% identical between human and fish (Supplemental Fig. 1A), while a 300-nt region (which includes the 26-nt core region) is well-conserved between primates and tetrapods (Supplemental Fig. 1B; Ulitsky et al. 2011). The 26-nt core is complementary to microRNA miR-7 in all species for which sequences are available (Wienholds et al. 2005; Ulitsky et al. 2011), and mutations within the 26-nt miR-7-binding core resulted in significantly reduced miR-7 degradation in murine models (Kleaveland et al. 2018).
These observations prompted us to investigate whether this functional region of Cyrano, that we refer to as the 300-nt conserved core, would fold into a structure which might contribute to its miR-7 degradation. To date, no structural information has been reported for Cyrano RNA. We show here that the 300-nt core of Cyrano adopts an intricate secondary structure involving additional nucleotides just outside the conserved region, spanning ∼600 nt. The 26-nt miR-7 binding sequence is embedded within a cloverleaf domain that is conserved through positional covariation in species as distantly related as zebrafish and humans, which diverged 420 million years ago (Ahlberg and Milner 1994), and is recognized by miR-7 and by several RNA binding proteins. This structural conservation strongly suggests that the functions of the lincRNA Cyrano are dependent on the formation of this RNA structure.
RESULTS
Human Cyrano hosts an SVA retrotransposable mobile element
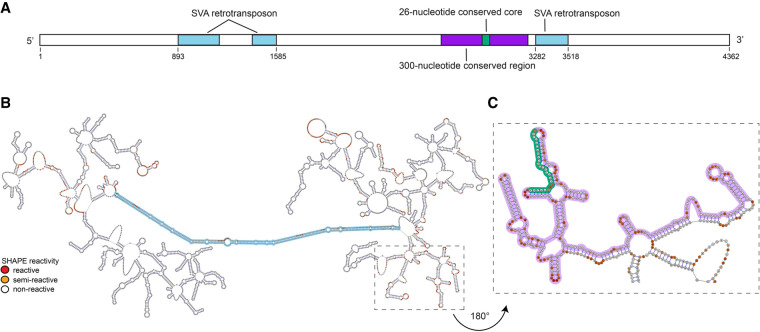
We used in vitro SHAPE (Wilkinson et al. 2006) to probe the secondary structure of the 300-nucleotide conserved core of Cyrano within a spliced 4.2 kilobase (kb) human Cyrano transcript (Fig. 1A); this splice variant comprises four exons and ends in a poly(A) tail (Ulitsky et al. 2011) and was targeted because it contains the most conserved region of Cyrano (Kleaveland et al. 2018). SHAPE probing revealed two important structural details: (i) Based on sparse SHAPE data restraints (∼1500 of 4200 nt probed), the secondary structure of the 4.2 kb transcript is proposed to possess a dumbbell shape, with two 2 kb domains separated by a 250 bp helical domain (Fig. 1B), and (ii) the 300-nt conserved core is located in one of the two domains; the structure adopted by the 300-nt conserved region does not fold into an independent domain, but is instead base-paired with neighboring nucleotides (Fig. 1C).
FIGURE 1.
The 4.2 kb human Cyrano transcript adopts a dumbbell-like secondary structure, as calculated by RNAstructure using sparse folding restraints provided by SHAPE chemical probing, with two structured domains separated by a long double helical retrotransposon insert. (A) Domain structure of Cyrano; (B) secondary structure of the complete 4.2 kb transcript; (C) expanded view of the secondary structure around the conserved miR-7 binding site. Regions corresponding to the SVA retrotransposable element are highlighted in blue; the 26-nt conserved region complementary to miR-7 is highlighted in green and its surrounding region, the 300-nt region conserved in tetrapods and the focus of this investigation, is highlighted in purple.
An NCBI BLASTn search (Altschul et al. 1990) revealed that the sequence corresponding to the 250 bp double helical region is a SINE VNTR Alu-like (SVA) retrotransposable mobile element, which is primate specific (Wang et al. 2005). A third region corresponding to an SVA insert was also identified, predicted to fold independently of the 250 bp helix. Using Cyrano as a query sequence in an NCBI BLASTn search, followed by an alignment with the Geneious software alignment tool (Kearse et al. 2012), we were able to locate and identify this SVA element in at least 19 other primate-specific Cyrano transcripts (Supplemental Fig. 2). The biological relevance, if any, of the conserved presence of the SVA element in primates remains to be elucidated. The presence of the SVA retrotransposable element simplified the structural analysis by separating human Cyrano into two independent domains of about 2 kb each (Fig. 1B). We focused our investigation on the region located between two of the fragmented SVA elements (nucleotides 1585 and 3282), which possesses the highest level of sequence conservation across the length of the transcript and contains the highly conserved 26-nt core fully complementary to miR-7.
In vitro structural probing of Cyrano
The 300-nt conserved core of human Cyrano was revealed to base-pair with neighboring nucleotides in the 4.2 kb transcript; using the 3S method, we established that a 600-nt region that encompasses the 26-nt sequence folds locally without requiring long-range interactions with the rest of the RNA (Fig. 2A). The SHAPE reactivity profiles of the 600-nt fragment, transcribed separately and compared to the same region within the full-length RNA transcript, are in very good agreement with each other, with a Pearson coefficient R of 0.74 (Supplemental Fig. 3D).
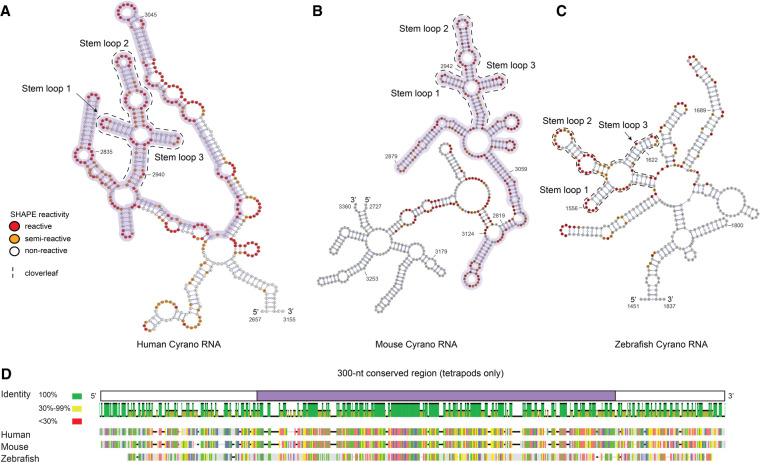
FIGURE 2.
(A) Secondary structure of the ∼600-nt fragment encompassing the stretch of human Cyrano (OIP5-AS1) recognized by miR-7, as calculated using RNAstructure with SHAPE restraints. (B) Secondary structure of the 600-nt fragment encompassing the same stretch of mouse Cyrano RNA. (C) Secondary structure of a 500-nt fragment including the comparable sequence of zebrafish Cyrano. The experimental SHAPE reactivity is mapped on each transcript. The 300-nt region, conserved only in tetrapods, is highlighted in purple. (D) Sequence alignment of human, mouse, and zebrafish Cyrano RNAs. Sequence identity is grouped in colored blocks: green: 100% identity, yellow: 30%–99%, and red: <30%.
The human Cyrano 600-nt domain forms an independently folded domain consisting of several helices connected by multihelical junctions (Fig. 2A). Nucleotides preceding the 5′-end of the 300-nt core constitute the central section of the independently folded domain, while nucleotides that follow complete the last helix of the structured domain. Thus, the 300-nt conserved sequence does not fold independently but requires about 100 flanking nucleotides on either side in order to form a stable secondary structure.
At the center of the 600-nt domain we observe an RNA secondary structure resembling a cloverleaf, ∼100 nt in length. This cloverleaf contains three stem–loops that originate from a common junction. A careful analysis of the SHAPE data plotted on the secondary structure of the human cloverleaf (Fig. 3A) demonstrate that the data match the structure very well. Internal loops, apical loops, bulges and some neighboring nucleotides, presumably experiencing partial conformational flexibility, are reactive, while helices generally are not, except for nucleotides neighboring single stranded regions. There are, however, two exceptions. Between the junction of the cloverleaf and the internal loop of stem–loop 2, we observed significant SHAPE reactivity for several AU base pairs. Since this short helical region is located between a three-way junction and an internal loop, it is plausible that this region is partly open, or “breathing” and the base pairs may be formed and yet still partly accessible to SHAPE reagents (Kenyon et al. 2014). We take this observation to suggest the presence of conformational exchange, or conformational dynamics, for this region of the RNA. The same conclusion can be drawn for the stem–loop around nucleotide 2835, bridging a multihelical junction and a large internal loop, and including three bulged nucleotides that interrupt two short helices, which is also the binding site for the FASTKD2 protein (described below).
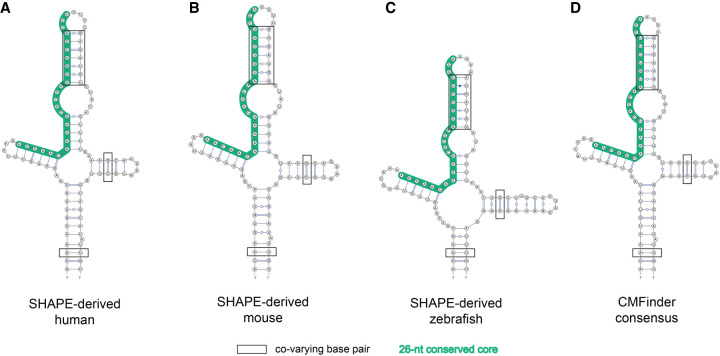
FIGURE 3.
The cloverleaf structures surrounding the miR-7 binding site (in green) of Cyrano, as calculated with RNAstructure constrained by SHAPE probing, rendered side by side, for (A) human, (B) mouse, and (C) zebrafish. The consensus motif discovered by CMfinder is shown in (D); covarying base pairs are boxed.
To further investigate the secondary structure of this region of the RNA, we used an orthogonal approach by performing S1 nuclease mapping, an enzyme which only cleaves single stranded nucleotides, and analyzed these 100 nt in the context of the full-length transcript. The results are consistent with the SHAPE-generated model and provide further support for the secondary structure obtained by SHAPE (Supplemental Fig. 4A).
To further validate the proposed cloverleaf structure, we used ScanFold (ScanFold-Scan and ScanFold-Fold) to fold the 600-nt region used in the 3S shotgun approach (Andrews et al. 2019). This sliding window approach, which folds the RNA sequentially, 120 nt at a time, revealed the same cloverleaf structural motifs identified by SHAPE, with a Z-score <1. The Stockholm format representation corresponding to these results can be found in Supplemental Table 2.
The cloverleaf region corresponds closely to the 100 nt of high conservation between primates and fish. Thus, we hypothesized that its structure might be conserved as well. To confirm this hypothesis, we probed an ∼600-nt sequence encompassing the fragment corresponding to the cloverleaf domain in mouse and zebrafish Cyrano (using the 3S approach) and also used comparative sequence analysis to identify and validate conserved motifs.
SHAPE probing, followed by secondary structure prediction with RNAstructure, revealed that both the mouse and zebrafish Cyrano RNAs are structured, despite significant (∼60%) sequence divergence (Fig. 2B–D; Supplemental Fig. 3A,D). Strikingly, the 100-nt cloverleaf structure observed in human Cyrano RNA is also observed in the mouse and zebrafish transcripts, with minor differences. In zebrafish, the first stem–loop possesses a 6-nt apical loop, whereas in humans and mice, the apical loop contains only 4 nt; the middle stem–loop possesses an internal loop, as in the human and mouse transcripts, but the internal loop contains 8 unpaired nucleotides, while 10 are observed in humans; the final stem–loop is 2 bp longer than in humans and mice, and possesses a 1-nt bulge. The junction bringing these three stem–loops together contains more unpaired nucleotides in zebrafish, especially 5′ to the first stem–loop. Significantly, the 26-nt conserved core is base-paired in a similar fashion in zebrafish, mouse, and human RNAs.
We further used covariation analysis (CMfinder) to query the presence of double helical regions, under the hypothesis that there would be evolutionary constraints on the RNA secondary structure, if the structure was functional. Sequence alignment from several species led to the identification of several double helices consistent with the SHAPE analysis, highlighting the concordance of chemical probing and phylogenetic analysis. The consensus secondary structures predicted by CMfinder are in agreement with our SHAPE-derived secondary structures (Fig. 3; Supplemental Fig. 4B–E; Supplemental Table 3). Most notable are the CMfinder results regarding the cloverleaf element identified in the SHAPE analysis.
MiR-7 Cyrano interaction
Cyrano was recently shown to direct the degradation of miR-7 (Kleaveland et al. 2018; Wightman et al. 2018) by nearly perfectly base-pairing with it (Supplemental Fig. 5A). Using electrophoretic mobility shift assays (EMSA), we confirmed that miR-7 RNA binds strongly to the cloverleaf structure within Cyrano (Kd < 100 nM), (Supplemental Fig. 5B), corroborating previously published in cell cross-linking data (Piwecka et al. 2017) by providing physical evidence of a direct interaction. We used IntaRNA, an RNA–RNA interaction prediction program, to predict the energy and folding architecture of the miR-7:Cyrano complex (Busch et al. 2008; Wright et al. 2014). The results revealed favorable minimum free energy for the complex, suggesting that the cloverleaf structure could be opened by miR-7 through a strand invasion mechanism, leaving the nucleotides complementary to the 26-nt conserved stretch single stranded (Supplemental Fig. 5C).
In cell structural probing of Cyrano
It is well known that the cellular environment can affect RNA folding through molecular crowding, high magnesium concentrations, post-transcriptional modifications and protein binding (Leamy et al. 2016; Watters et al. 2016). Thus, we performed SHAPE on human Cyrano expressed in eukaryotic cells to probe its conformation in the cellular environment.
We transfected HEK293 cells with human Cyrano RNA expression plasmid (endogenous Cyrano copy numbers are too low to generate a robust SHAPE signal), followed by treatment with the cell-permeable SHAPE adduct 1M7. The in cell results reveal a SHAPE reactivity profile highly similar to the in vitro data across the 600-nt fragment, suggesting the overall fold proposed in vitro accurately reflects cellular conditions (Supplemental Fig. 6). However, six regions, accounting for <10% of the RNA, have different SHAPE reactivity in vitro and in cell (Fig. 4A; Supplemental Fig. 6). We hypothesized that at least some of the regions where in cell SHAPE reactivity differs from in vitro SHAPE reactivity would correspond to protein binding sites, which could reduce access to the SHAPE reagents and/or alter the local RNA structure. This hypothesis is supported by the identification of protein binding sites near many of these regions.
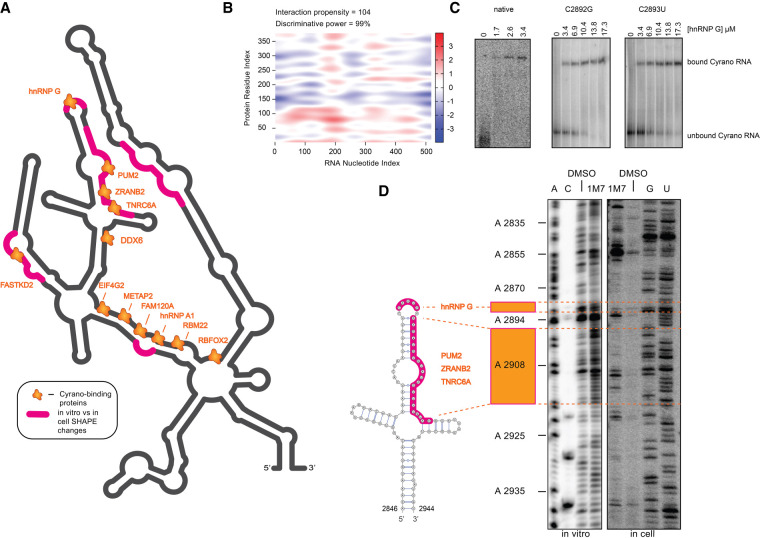
FIGURE 4.
(A) Protein binding partners for human Cyrano (orange splashes) identified by eCLIP (ENCODE) or predicted by the RBP database, mapped onto the secondary structure of the 600-nt fragment of human Cyrano; regions where changes are observed when SHAPE reactivity in vitro and in cell were compared, are outlined in magenta. (B) CatRAPID heat map prediction of the amino-terminal domain (RRM) of hnRNP G and its potential binding site within Cyrano, which corresponds to the conserved cloverleaf structure. Red hotspots are indicative of likely protein–RNA interaction sites. (C) EMSA of the human cloverleaf structure of Cyrano probed with the RRM of hnRNP G, as well as two mutated forms of the RNA (which disrupt the hnRNP G binding site). (D) Representative gels showing differences in SHAPE reactivity between transcripts mapped in vitro and in cell for the cloverleaf domain of human Cyrano.
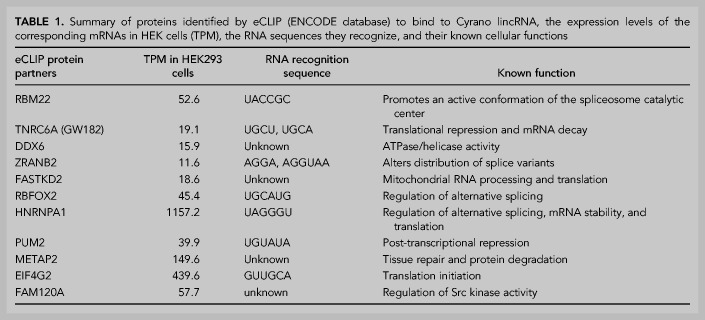
As was done on other lincRNAs (Lin et al. 2018), we used data available through the ENCODE project and the RBP database to identify protein binding partners of the lincRNA Cyrano (Smola et al. 2016). Enhanced cross-linking and immunoprecipitation (eCLIP) data sets found within the ENCODE database (Derrien et al. 2012; Van Nostrand et al. 2016) revealed at least 11 proteins (Table 1) that cross-link to the domain of Cyrano on which this investigation is focused. Since the eCLIP data in ENCODE corresponds to HepG2 and K562 cell-lines, we cross-checked each of the putative Cyrano protein binding partners’ expression levels in HEK293 cells; transcripts per kilobase million (TPM) scores are well above 1.0 for all 11 proteins (Human Protein Atlas; a TPM value of 1.0 is defined as a threshold for expression of the corresponding protein). The TPM scores of each protein, their RNA recognition sequences, and known functions, if available, are outlined in Table 1.
TABLE 1.
Summary of proteins identified by eCLIP (ENCODE database) to bind to Cyrano lincRNA, the expression levels of the corresponding mRNAs in HEK cells (TPM), the RNA sequences they recognize, and their known cellular functions
Interaction of Cyrano with hnRNP G
Using the RBP database (Cook et al. 2011) followed by catRAPID (Agostini et al. 2013) analysis, we identified several candidate interaction partners that might bind directly to, or in close proximity to the 26 nt complementary to miR-7 (Fig. 4B,C; Supplemental Table 4). We chose to examine the heterogeneous nuclear ribonucleoprotein (hnRNP) G (also known as RBMX) because of clear differences observed between in cell and in vitro SHAPE reactivity patterns for the region of the transcript where RMBX is predicted to bind (Fig. 4D). The apical loop of the middle stem–loop of the cloverleaf contains a 5′ CCAU 3′ sequence predicted by the RBP database to bind to the hnRNP G protein. This RNA recognition sequence was shown to be among the preferred RNA binding sequences of this protein (Moursy et al. 2014). hnRNP G has several roles in pre and post-transcriptional regulation (Soulard et al. 1993; Ravasi et al. 2006; Shin et al. 2008; Kanhoush et al. 2010). CatRAPID (Agostini et al. 2013), which estimates the binding propensity of a protein–RNA interaction, scored the amino-terminal RRM (RNA recognition motif) of hnRNP G as a strong interaction partner for this region of Cyrano (Fig. 4B).
We expressed and purified the amino-terminal RNA binding domain of hnRNP G and evaluated its binding affinity in vitro for the 100-nt cloverleaf subconstruct of Cyrano using EMSAs (Fig. 4C). We observed direct binding to be 0.83 ± 0.36 µM. Upon mutating nucleotides in the RBMX binding sites to 5′ GCAU 3′ or 5′ CUAU 3′, binding was reduced to 9.15 ± 1.20 and 10.6 ± 3.86 micromolar for each mutant, respectively, thus corroborating a specific interaction (Fig. 4C).
Tertiary structural modeling of the cloverleaf domain
To gain further insight into the structure of Cyrano, we modeled the 3D structure of the 100-nt human cloverleaf structure (Supplemental Fig. 7A,B). The prediction reveals a compact conformation stabilized by A-minor interactions formed by conserved and unpaired Adenines at the four-way junction, which brings stem–loop 2 into spatial proximity of the basal helix of the cloverleaf. The two outermost stem–loops (stem–loops 1 and 3), which are coaxially stacked, are located at the junction of the cloverleaf, seemingly inducing a V-like turn in the RNA structure. The stacking of the two stem–loops may make it possible for miR-7 to bind its complementary sequence (Supplemental Fig. 7C), as this region of the RNA is partially exposed to solution; the stacked helices may unwind with the aid of RISC-associated proteins, allowing miR-7 to access and bind to the 26-nt conserved region. Interestingly, both mouse and zebrafish adopt similar V-like turns at their junctions, despite sequence divergence from the human transcript (Supplemental Fig. 7D,E). Further investigation of the predicted tertiary structure using nuclear magnetic resonance (NMR) or X-ray crystallography could provide valuable experimental information to corroborate the model.
DISCUSSION
It is estimated that <5% of the approximately 30,000 transcribed lincRNAs in human are conserved over significant evolutionary distances (Ransohoff et al. 2018). Even in the case of conserved transcripts, sequence conservation is often limited, making comparative sequence analysis to establish secondary structure and identify functionally important domains difficult (Ulitsky and Bartel 2013; Eddy 2014; Rivas et al. 2017). Rapidly evolving regions of the genome within primates often map to these transcripts (Nitsche et al. 2015; Tavares et al. 2019). A common hypothesis is that RNA secondary structure is more conserved than sequence alone, leading to the frequently stated but generally untested hypotheses that structure, rather than sequence, is under evolutionary pressure in lincRNAs (Hawkes et al. 2016). If these hypotheses were true, then a structural understanding of lincRNAs would reveal functional correlations that might be unobservable from sequence analysis alone.
We show herein that the lincRNA Cyrano, one of few noncoding transcripts with recognizable orthologues over the evolutionary distances separating primates and zebrafish (Ulitsky et al. 2011), contains a conserved folded secondary structured element that is maintained over a long evolutionary history. The cloverleaf-shaped structure is ∼100 nt long and includes a 26-nt sequence nearly perfectly complementary to miR-7 in all organisms with recognizable orthologues from zebrafish to human. The cloverleaf can be modeled to fold into a compact structure stabilized by interhelical stacking and the likely formation of conserved A-minor interactions. Examination of sequence evolution reveals selective pressure to preserve the secondary and tertiary structure of this domain through compensatory mutations and by retaining nucleotides required to fold the four-way junction at the center of the cloverleaf (Supplemental Fig. 7). These compensatory base changes are few compared to the ribosome, which has a vastly longer evolutionary history. These observations support the notion that RNA secondary and perhaps higher order structure, rather than sequence alone, might be essential for the function of this lincRNA, as was observed for the lincRNA COOLAIR across divergent plant species (Hawkes et al. 2016).
Cyrano is known to speed up the degradation of miR-7 (Smith et al. 2017; Kleaveland et al. 2018), thus preventing repression of the circular RNA Cdr1as (Kleaveland et al. 2018). Repression of miR-7 expression by the complete lincRNA is significant, 45-fold, while a fragment corresponding to the cloverleaf exerts a more modest, three- to fourfold influence on miR-7 levels (Kleaveland et al. 2018). Our work shows that miR-7 binds to the conserved core structure of Cyrano. Perhaps the cloverleaf structure provides a regulatory role by controlling the loading of miR-7 onto this RNA through its secondary and three-dimensional structure. This hypothesis is supported by mutations made to the cloverleaf (Kleaveland et al. 2018); deletions and insertions within the cloverleaf domain lead to reduced regulation of miR-7 (Kleaveland et al. 2018). Secondary structural modeling of the deletions and insertions also reveals significant changes in the RNA structure; instead of the RNA adopting compact, V-shaped structure (which we propose is necessary for bringing RISC-associate proteins into proximity to the miR-7 site), we observe an elongated, rod-like conformation caused by the disruption of the three-way junction that induces the V-like turn (Supplemental Fig. 8). These changes lead to a 40% increase in relative miR-7 expression levels (Kleaveland et al. 2018). Interestingly, the mutant that maintains a secondary structure closest to that of our predicted cloverleaf shows only a smaller increase in miR-7 expression. Together, these results suggest a regulatory repressive role for the conserved structure described here.
In addition to intrinsic RNA structural features, lncRNA function is regulated by the proteins that bind to them. It is hypothesized that lncRNAs serve as scaffolds that assemble RNA binding proteins (RBP) to regulate gene expression (Wang and Chang 2011; Ulitsky and Bartel 2013). Cyrano was recently predicted to harbor binding sites for at least 37 RBPs (Li et al. 2014); this number is not surprising, given its length, and other lncRNAs, such as Xist and NEAT1, are predicted to bind to more than 30 RBPs as well (Li et al. 2014; Chu et al. 2015). In our study, most differences between the SHAPE reactivity profile in cells and in vitro can be attributed to interactions with proteins, as was also observed for Xist using SHAPE-MaP (Smola et al. 2015, 2016). Using both the experimental data stored in the ENCODE eCLIP database, and the theoretical predictions of the RBP database, we identified 24 RBPs that potentially bind to and regulate Cyrano, five of which (FUS, HuR, PUM2, SRSF1, and TNRC6) are known to bind to other lncRNAs (Li et al. 2014). Since several of these protein-binding regions are in close proximity to each other, it is possible that combinations of these protein binding events may be anti-cooperative or cooperative; binding of one protein partner may restrict or allow access of other proteins by altering the secondary structured domain required for binding (Becker et al. 2019).
Of the proteins identified to bind to Cyrano by eCLIP, TNRC6A, DDX6, FAM120A, and PUM2, are known to associate with Ago proteins (Frohn et al. 2012). DDX6 is an RNA helicase (Cordin et al. 2006), and TNRC6A functions in RNA degradation (Behm-Ansmant et al. 2006). PUM2 is implicated in mRNA stability and translation (Van Etten et al. 2012), and FAM120A binds to lncRNAs preferentially in the presence of microRNAs and Dicer (Frohn et al. 2012). Our tertiary structural model reveals that the binding sites of these proteins are all in close proximity to the miR-7 binding site. Further investigation of these proteins and their interaction with the Cyrano cloverleaf would provide a useful starting point for obtaining a more detailed understanding of how Cyrano regulates miR-7 degradation, but the presence of a helicase and an RNA degradation factor is highly suggestive of their role in regulation of miR-7 by this transcript, and of how the Cyrano structure brings key degradation factors in close proximity to their target.
MATERIALS AND METHODS
DNA template preparation
Cyrano DNA plasmids for zebrafish, human, and mouse were kindly provided by Dr. Alena Shkumatava (Unite de Genetique et Biologie due Developpement, Institut Curie). The plasmid for each species was transformed into DH5α cells and extracted using the Qiagen midi-prep kit. Plasmids were linearized with appropriate restriction enzymes according to manufacturer instructions (SalI : human, SacI : mouse, NotI : zebrafish). Linearization was confirmed on 1% agarose gels.
In order to isolate the DNA template corresponding to the 600-nt fragment, full length, cut DNA template was subject to polymerase chain reaction (PCR) with the Phusion polymerase (NEB) and with primers synthesized by IDT, containing the T7 promoter region (bold text on the forward [F] primer) and a poly(A) tail (bold text on the reverse [R] primer):
Human F: 5′-AAGCTTTAATACGACTCACTATAGGGGGATATATTCCAGCTGTAGTTGC-3′
Human R: 5′-TTTTTTTTTTTTTTTTCTCAGATTTTGACCCCACATTT-3′
Zebrafish F: 5′-AAGCTTTAATACGACTCACTATAGGGATCTGCTATAGAGCACTGTGA-3′
Zebrafish R: 5′-TTTTTTTTTTTTTTTTCCAGAATCGTGCAGCCCTAC-3′
Mouse F: 5′-AAGCTTTAATACGACTCACTATAGGGGTGGCACATTTCCATTTATAGTCT-3′
Mouse R: 5′-TTTTTTTTTTTTTTTTAGTGGCTCTCAGTGGGAA-3′
PCR was run according to the manufacturer's instructions. Following PCR, the DNA template was evaluated on a 1% agarose gel.
In vitro RNA transcription and purification
Full-length DNA plasmids for zebrafish and mouse Cyrano were transcribed in vitro with SP6 and T3 RNA polymerases, respectively (Messenger mega kit, Ambion). Full-length human Cyrano lincRNA, the 600-nt fragments corresponding to human, zebrafish, and mouse RNAs, and the 100-nt fragment were transcribed in vitro using in house-purified T7 RNA polymerase. RNA transcript lengths and purity were confirmed on a 1% agarose gel. Each RNA was purified by phenol:chloroform extraction, followed by precipitation with ethanol. When abortive transcripts were observed, RNA was purified on denaturing polyacrylamide gels, followed by electro-elution and buffer exchange against water. Mature miR-7 RNA was purchased from IDT. DNA template for the 100-nt fragment of human Cyrano was ordered from IDT and included the SHAPE linker, as previously described (Wilkinson et al. 2006).
In vitro 1M7/NMIA and S1 nuclease probing
1M7 was synthesized in house as described previously (Turner et al. 2013); NMIA was purchased from Sigma-Aldrich. SHAPE reactions were carried out as described (Wilkinson et al. 2006) and analyzed by both gel electrophoresis and capillary electrophoresis (described below); both methods gave consistent results. Enzymatic cleavage with S1 (NEB) was performed as described (Nilsen 2013). Modified or cleaved RNAs were reverse transcribed using the same primers used for SHAPE and the corresponding cDNA fragments were analyzed. At least three biological replicates were performed for in vitro probing, and two for SHAPE probing of RNA transcribed in cells. A table outlining the reagents and methods used to probe the RNAs can be found in Supplemental Table 1.
In cell RNA structural probing with 1M7
An amount of 5 µg of human Cyrano plasmid, cloned into the CMV vector, was transfected into HEK293 cells. Plates were incubated at 37°C for 48 h, before washing and concentrating with PBS buffer. The cells were then immediately aliquoted equally into six different tubes, two containing DMSO, one containing fresh PBS, and three containing 260 mM 1M7 (final concentration 90 mM). The tubes were then incubated at 37°C in a shaker (220 rpm) for 8 min followed by treatment with TRIzol to halt the SHAPE reaction. Cells were treated with chloroform to separate and extract the RNA before purification using the Zymo RNA Easy Clean kit. The RNA was diluted to a final concentration of ∼10 µg/µL. The amount of cDNA was too low for detection by capillary electrophoresis. Therefore, we analyzed in cell SHAPE data using gel electrophoresis with two biological replicates. Approximately 8 µg of total RNA was used per SHAPE reaction. As a control, a reverse transcription reaction on 5S rRNA was also performed using a 5S specific primer on the DMSO, PBS, and 1M7 treated samples (data not shown).
Gel electrophoresis
S1 nuclease reactions for in vitro and in cell reactions were analyzed on 8% denaturing polyacrylamide gels. Gels were run for 5 h (at the 2 h mark, a new set of samples was loaded to gather information about nucleotides closer to the 3′-end more efficiently) at 70 Watts and 4°C. Gels were then dried under vacuum pressure at 80°C and exposed to a phosphor plate overnight. The phosphor plates were scanned using a Typhoon 600 imager, followed by analysis and SHAPE normalization with SAFA (Semi Automated Foot-printing Analysis) software and with GelAnalyzer2010 (Das et al. 2005; http://www.gelanalyzer.com, accessed July 29, 2010).
Capillary electrophoresis
Three primers (one each for human, mouse, and zebrafish Cyrano), labeled with four of the five dyes from the G5 dye set (NED, VIC, FAM, PET) were ordered from Life Technologies. Primers were diluted to 2 pmol/µL in water. The sequences of each primer were as follows:
Human: 5′-GCTGGTTTCCAGATACCTTATGG-3′,
Mouse: 5′-GTGGCACATTTCCATTTATAGTCT-3′,
Zebrafish: 5′-AATTAACCAAGGTGTGGATACTG-3′.
cDNA fragments were analyzed using a 3130xl Genetic Analyzer. SHAPE reactivity was analyzed and normalized using QuSHAPE (Karabiber et al. 2013).
Secondary structure prediction and image rendering
SHAPE reactivity data were incorporated as a SHAPE constraint file in the RNAstructure folding program, and the 20 lowest energy structures based on those constraints were generated (Reuter and Mathews 2010; Lorenz et al. 2016; Spasic et al. 2018). Structures were calculated with RNAstructure under default secondary structure options: 310.15 K, maximum loop size of 30, maximum percent energy difference of 10 and a minimum helix length of 3. The SHAPE intercept was set at −0.6 and the SHAPE slope was set at 1.8. No “hard” folding constraints were used during the calculation to avoid any bias. SHAPE and nuclease foot-printing data have been submitted to RMDB in RDAT file format.
The structure of Cyrano reported in this manuscript was selected based on a combination of lowest energy, best agreement with SHAPE reactivity, and agreement with comparative sequence analysis. Each structure image presented in this manuscript was rendered with VARNA (Darty et al. 2009). Higher energy structures can be provided upon request.
ScanFold analysis
We used the ScanFold webserver (https://mosslabtools.bb.iastate.edu/) to validate local motifs throughout the cloverleaf domain with the ScanFold-Scan and ScanFold-Fold analysis tools. The sequence of the RNA was uploaded in fasta format and default values were used for window size (120), step size (10), randomizations (30), temperature (37°C), and shuffle type (mono).
Comparative sequence analysis
CMfinder was used for phylogenetic analysis of Cyrano (Yao et al. 2006). The unaligned sequences evaluated by CMfinder were gathered from NCBI's Basic Local Alignment Search Tool (BLAST) (Altschul et al. 1990), corresponding to 51 different species. Because alignment methods generally fail when sequence conservation falls below 60% or is above 85%, not all of the sequences obtained from NCBI were used; redundant sequences were removed until ∼70% primary sequence conservation was obtained, before analysis with CMfinder. The sequences were then formatted to a FASTA file and run by the cmfinder.pl Perl script under default conditions. A list of these sequences can be provided upon request.
3D structure modeling
simRNA and RNAComposer (human/mouse, and zebrafish, respectively) were used to model the tertiary structure of the cloverleaf domain in human Cyrano using the documentation provided by the developers (Boniecki et al. 2015; Antczak et al. 2016; Magnus et al. 2016). No experimental or theoretical restraints were used to model the structure; the Stockholm format corresponding to the secondary structure predicted by SHAPE served as the starting input file. FARFAR energy was used to minimize the models (Cheng et al. 2015). Chimera was used to render the structural images (Pettersen et al. 2004). Additional details of the simulations, including relevant commands, configuration parameters, and RMSD and energy distribution plots corresponding to the Cyrano cloverleaf structure, can be found at: https://github.com/Dreycey/cyrano_simulations.
hnRNP G expression and purification
The amino-terminal RRM of hnRNP G (RBMX) (containing an amino-terminal 6×-Histidine-tag followed by a TEV cleavage site) was expressed in E coli using BL21 competent cells. The cells were harvested in buffer containing 20 mM Tris, 150 mM, 5 mM β-mercaptoethanol NaCl, pH 8, followed by purification of the protein on a nickel column. The His-tag was removed with TEV protease, followed by a second purification on the same nickel column. Protein fractions were evaluated by SDS page gel to ensure purity.
Electrophoretic mobility shift assays (EMSA)
Purified and dephosphorylated cloverleaf Cyrano RNA was rephosphorylated with γ32ATP. Varying concentrations of either miR-7 RNA or RBMX protein were incubated with the Cyrano-derived cloverleaf structure on ice in a binding mixture containing 0.05 M Tris-HCl, 0.05 M KCl, 0.2 M DTT, 1% Triton X-100 for 20 min, followed by the addition of 10% glycerol. Samples were loaded onto a 6% native polyacrylamide gel and run at 15 W for 1.5 h. Gels were dried and placed in a phosphor imager plate overnight. The plate was scanned with a phosphor scanner (Typhoon 9000) and autoradiograms were analyzed with ImageJ software.
Sequence of lincRNA Cyrano used in this study
Cyrano lincRNA:
Human: (4 Exons, 8706 base pairs) Ensembl ENST00000500949.6
Mouse (3 exons, 8300 base pairs): Ensembl ENSMUST00000153581.1
Zebrafish (3 exons, 4606 base pairs): Ensembl ENSDART00000139872.3
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Larry Ruzzo for his insight and guidance concerning CMfinder; Dr. Kate Godin for sequencing of the RNA plasmids; Dr. Maryam Yahyazee for preparation of the 1M7 reagent; Dr. Alena Shkumatava for plasmids and discussions; and Gregory P. Wolfe for manuscript and image editing. The work was supported by grants from the National Institutes of Health–National Institute of General Medical Sciences (NIH-NIGMS): RO1 GM103834 and R35 GM121487 (G.V.).
Author contributions: A.N.J. performed and analyzed all in vitro and in cell SHAPE, nuclease foot-printing, and EMSA experiments with contributions from G.P., J.W., M.K., and N.F. A.N.J. carried out the comparative sequence analysis. T.P. prepared all plasmids used for cell transfection with contributions from N.F. Identification, expression, and purification of hnRNP G was performed by M.K. Tertiary structural modeling was performed by D.A. and A.N.J. A.N.J., G.P., T.P., and G.V. drafted the manuscript and conceived the project and experimental approaches.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.076117.120.
REFERENCES
- Agostini F, Zanzoni A, Klus P, Marchese D, Cirillo D, Tartaglia GG. 2013. catRAPID omics: a web server for large-scale prediction of protein–RNA interactions. Bioinformatics 29: 2928–2930. 10.1093/bioinformatics/btt495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlberg PE, Milner AR. 1994. The origin and early diversification of tetrapods. Nature 368: 507–514. 10.1038/368507a0 [DOI] [Google Scholar]
- Altschul SF, Gish W, Miller W, Lipman DJ, Myers EW. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Andrews RJ, Baber L, Moss WN. 2019. Mapping the RNA structural landscape of viral genomes. Methods 19: 30172-0 10.1016/j.ymeth.2019.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antczak M, Popenda M, Zok T, Sarzynska J, Ratajczak T, Tomczyk K, Adamiak RW, Szachniuk M. 2016. New functionality of RNAComposer: an application to shape the axis of miR160 precursor structure. Acta Biochim Pol 63: 737–744. 10.18388/abp.2016_1329 [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Becker WR, Jarmoskaite I, Vaidyanathan PP, Greenleaf WJ, Herschlag D. 2019. Demonstration of protein cooperativity mediated by RNA structure using the human protein PUM2. RNA 25: 702–712. 10.1261/rna.068585.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. 2006. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20: 1885–1898. 10.1101/gad.1424106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniecki MJ, Lach G, Dawson WK, Tomala K, Lukasz P, Soltysinski T, Rother KM, Bujnicki JM. 2015. SimRNA: a coarse-grained method for RNA folding simulations and 3D structure prediction. Nucleic Acids Res 44: e63 10.1093/nar/gkv1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, Backofen R, Richter AS. 2008. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 24: 2849–2856. 10.1093/bioinformatics/btn544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, Rinn JL, Raj A. 2015. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol 16: 20 10.1186/s13059-015-0586-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Chou FC, Das R. 2015. Modeling complex RNA tertiary folds with Rosetta. Methods Enzymol 553: 35–64. 10.1016/bs.mie.2014.10.051 [DOI] [PubMed] [Google Scholar]
- Chillón I, Marcia M, Legiewicz M, Liu F, Somarowthu S, Pyle AM. 2015. Native purification and analysis of long RNAs. Methods Enzymol 558: 3–37. 10.1016/bs.mie.2015.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Zhang Q, da Rocha S, Flynn R, Bharadwaj M, Calabrese J, Magnuson T, Heard E, Chang H. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161: 404–416. 10.1016/j.cell.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. 2011. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res 39: D301–D308. 10.1093/nar/gkq1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P. 2006. The DEAD-box protein family of RNA helicases. Gene 367: 17–37. 10.1016/j.gene.2005.10.019 [DOI] [PubMed] [Google Scholar]
- Darty K, Denise A, Ponty Y. 2009. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics 25: 1974–1975. 10.1093/bioinformatics/btp250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. 2005. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11: 344–354. 10.1261/rna.7214405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2014. Computational analysis of conserved RNA secondary structure in transcriptomes and genomes. Annu Rev Biophys 43: 433–456. 10.1146/annurev-biophys-051013-022950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohn A, Eberl HC, Stohr J, Glasmacher E, Rudel S, Heissmeyer V, Mann M, Meister G. 2012. Dicer-dependent and -independent Argonaute2 protein interaction networks in mammalian cells. Mol Cell Proteomics 11: 1442–1456. 10.1074/mcp.M112.017756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi M, Berg K, Pieper LM, Schier AF. 2019. Individual long non-coding RNAs have no overt functions in zebrafish embryogenesis, viability and fertility. eLife 8: e40815 10.7554/eLife.40815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150. 10.1126/science.1064023 [DOI] [PubMed] [Google Scholar]
- Hawkes EJ, Hennelly SP, Novikova IV, Irwin JA, Dean C, Sanbonmatsu KY. 2016. COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Rep 16: 3087–3096. 10.1016/j.celrep.2016.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhoush R, Beenders B, Perrin C, Moreau J, Bellini M, Penrad-Mobayed M. 2010. Novel domains in the hnRNP G/RBMX protein with distinct roles in RNA binding and targeting nascent transcripts. Nucleus 1: 109–122. 10.4161/nucl.1.1.10857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabiber F, McGinnis JL, Favorov OV, Weeks KM. 2013. QuShape: rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA 19: 63–73. 10.1261/rna.036327.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon J, Prestwood L, Lever A. 2014. Current perspectives on RNA secondary structure probing. Biochem Soc Trans 42: 1251–1255. 10.1042/BST20140084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Abdelmohsen K, Yang X, De S, Grammatikakis I, Noh JH, Gorospe M. 2016. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Res 44: 2378–2392. 10.1093/nar/gkw017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleaveland B, Shi CY, Stefano J, Bartel DP. 2018. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell 174: 350–362. 10.1016/j.cell.2018.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok CK. 2016. Dawn of the in vivo RNA structurome and interactome. Biochem Soc Trans 44: 1395–1410. 10.1042/BST20160075 [DOI] [PubMed] [Google Scholar]
- Kwok CK, Ding Y, Tang Y, Assmann SM, Bevilacqua PC. 2013. Determination of in vivo RNA structure in low-abundance transcripts. Nat Commun 4: 2971 10.1038/ncomms3971 [DOI] [PubMed] [Google Scholar]
- Leamy KA, Assmann SM, Mathews DH, Bevilacqua PC. 2016. Bridging the gap between in vitro and in vivo RNA folding. Q Rev Biophys 49: e10 10.1017/S003358351600007X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-H, Liu S, Zheng L-L, Wu J, Sun W-J, Wang Z-L, Zhou H, Qu L-H, Yang J-H. 2014. Discovery of protein–lncRNA interactions by integrating large-scale CLIP-Seq and RNA-Seq datasets. Front Bioeng Biotechnol 2: 88 10.3389/fbioe.2014.00088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhu H, Luo Y. 2016. Understanding the functions of long non-coding RNAs through their higher-order structures. Int J Mol Sci 17: 702 10.3390/ijms17050702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Schmidt BF, Bruchez MP, McManus CJ. 2018. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res 46: 3742–3752. 10.1093/nar/gky046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Hofacker IL, Stadler PF. 2016. RNA folding with hard and soft constraints. Algorithms Mol Biol 11: 8 10.1186/s13015-016-0070-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus M, Boniecki MJ, Dawson W, Bujnicki JM. 2016. SimRNAweb: a web server for RNA 3D structure modeling with optional restraints. Nucleic Acids Res 44: W315–W319. 10.1093/nar/gkw279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Shkumatava A. 2015. LncRNAs in vertebrates: advances and challenges. Biochimie 117: 3–14. 10.1016/j.biochi.2015.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moursy A, Allain FHT, Cléry A. 2014. Characterization of the RNA recognition mode of hnRNP G extends its role in SMN2 splicing regulation. Nucleic Acids Res 42: 6659–6672. 10.1093/nar/gku244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW. 2013. RNA structure determination using nuclease digestion. Cold Spring Harb Protoc 4: 379–382. 10.1101/pdb.prot072330 [DOI] [PubMed] [Google Scholar]
- Nitsche A, Rose D, Fasold M, Reiche K, Stadler PF. 2015. Comparison of splice sites reveals that long noncoding RNAs are evolutionarily well conserved. RNA 21: 801–812. 10.1261/rna.046342.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova I, Dharap A, Hennelly SP, Sanbonmatsu KY. 2013. 3S: shotgun secondary structure determination of long non-coding RNAs. Methods 63: 170–177. 10.1016/j.ymeth.2013.07.030 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pintacuda G, Young AN, Cerase A. 2017. Function by structure: spotlights on Xist long non-coding RNA. Front Mol Biosci 4: 90 10.3389/fmolb.2017.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Jara CAC, Fenske P, et al. 2017. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357: eaam8526 10.1126/science.aam8526 [DOI] [PubMed] [Google Scholar]
- Qian X, Zhao J, Yeung PY, Zhang QC, Kwok CK. 2019. Revealing lncRNA structures and interactions by sequencing-based approaches. Trends in Biochem Sci 44: 33–52. 10.1016/j.tibs.2018.09.012 [DOI] [PubMed] [Google Scholar]
- Ransohoff JD, Wei Y, Khavari PA. 2018. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 19: 143–157. 10.1038/nrm.2017.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, et al. 2006. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res 16: 11–19. 10.1101/gr.4200206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH. 2010. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11: 129 10.1186/1471-2105-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E, Clements J, Eddy SR. 2017. Lack of evidence for conserved secondary structure in long noncoding RNAs. Nat Methods 14: 45–48. 10.1038/nmeth.4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin KH, Kim RH, Kim RH, Kang MK, Park NH. 2008. hnRNP G elicits tumor-suppressive activity in part by upregulating the expression of Txnip. Biochem Biophys Res Commun 372: 880–885. 10.1016/j.bbrc.2008.05.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KN, Starmer J, Miller SC, Sethupathy P, Magnuson T. 2017. Long noncoding RNA moderates microRNA activity to maintain self-renewal in embryonic stem cells. Stem Cell Rep 9: 108–121. 10.1016/j.stemcr.2017.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Calabrese JM, Weeks KM. 2015. Detection of RNA–protein interactions in living cells with SHAPE. Biochem 54: 6867–6875. 10.1021/acs.biochem.5b00977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, Lee DM, Calabrese JM, Weeks KM. 2016. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc Natl Acad Sci 113: 10322–10327. 10.1073/pnas.1600008113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somarowthu S, Legiewicz M, Liu F, Marcia M, Liu F, Pyle AM. 2015. HOTAIR forms an intricate and modular secondary structure. Mol Cell 58: 353–361. 10.1016/j.molcel.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulard M, Valle VD, Siomi MC, Pinol-Roma S, Codogno P, Bauvy C, Bellini M, Lacroix JC, Monod G, Dreyfuss G. 1993. hnRNP G: sequence and characterization of a glycosylated RNA-binding protein. Nucleic Acids Res 21: 4210–4217. 10.1093/nar/21.18.4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spasic A, Assmann SM, Bevilacqua PC, Mathews DH. 2018. Modeling RNA secondary structure folding ensembles using SHAPE mapping data. Nucleic Acids Res 46: 314–323. 10.1093/nar/gkx1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler P, Carbon P, Zuker M, Ebel JP, Ehresmann C. 1981. Structural organization of the 16S ribosomal RNA from E. coli. Topography and secondary structure. Nucleic Acids Res 9: 2153–2172. 10.1093/nar/9.9.2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares RCA, Pyle AM, Somarowthu S. 2019. Phylogenetic analysis with improved parameters reveals conservation in lncRNA structures. J Mol Biol 431: 1592–1603. 10.1016/j.jmb.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner R, Shefer K, Ares M Jr. 2013. Safer one-pot synthesis of the ‘SHAPE’ reagent 1-methyl-7-nitroisatoic anhydride (1m7). RNA 19: 1857–1863. 10.1261/rna.042374.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. 2013. lincRNAs: genomics, evolution, and mechanisms. Cell 154: 26–46. 10.1016/j.cell.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. 2011. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550. 10.1016/j.cell.2011.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uroda T, Anastasakou E, Rossi A, Teulon J-M, Pellequer J-L, Annibale P, Pessey O, Inga A, Chillón I, Marcia M. 2019. Conserved pseudoknots in lncRNA MEG3 are essential for stimulation of the p53 pathway. Mol Cell 75: 982–995. 10.1016/j.molcel.2019.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten J, Schagat TL, Hrit J, Weidmann CA, Brumbaugh J, Coon JJ, Goldstrohm AC. 2012. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J Biol Chem 287: 36370–36383. 10.1074/jbc.M112.373522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nyugen TB, Surka C, Elkins K, et al. 2016. Robust transcriptome-wide discovery of RNA binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13: 508–514. 10.1038/nmeth.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. 2011. Molecular mechanisms of long noncoding RNAs. Mol Cell 43: 904–914. 10.1016/j.molcel.2011.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xing J, Grover D, Hedges DJ, Han K, Walkter JA, Batzer MA. 2005. SVA elements: a hominid-specific retrotransposon family. J Mol Biol 354: 994–1007. 10.1016/j.jmb.2005.09.085 [DOI] [PubMed] [Google Scholar]
- Wapinski O, Chang HY. 2011. Long noncoding RNAs and human disease. Trends Cell Biol 21: 354–361. 10.1016/j.tcb.2011.04.001 [DOI] [PubMed] [Google Scholar]
- Watters KE, Abbott TR, Lucks JB. 2016. Simultaneous characterization of cellular RNA structure and function with in-cell SHAPE-Seq. Nucleic Acids Res 44: e12 10.1093/nar/gkv879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, De Bruijn E, Horvitz HR, Kauppinen S, Plasterk RHA. 2005. MicroRNA expression in zebrafish embryonic development. Science 309: 310–311. 10.1126/science.1114519 [DOI] [PubMed] [Google Scholar]
- Wightman FF, Giono LE, Fededa JP, De La Mata M. 2018. Target RNAs strike back on microRNAs. Front Genet 9: 435 10.3389/fgene.2018.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1: 1610–1616. 10.1038/nprot.2006.249 [DOI] [PubMed] [Google Scholar]
- Wright PR, Georg J, Mann M, Sorescu DA, Richter AS, Lott S, Kleinkauf R, Hess WR, Backofen R. 2014. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res 42: 119–123. 10.1093/nar/gku359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J-R, Zhang J. 2015. Human long noncoding RNAs are substantially less folded than messenger RNAs. Mol Biol Evol 32: 970–977. 10.1093/molbev/msu402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Weinberg Z, Ruzzo WL. 2006. CMfinder—a covariance model based RNA motif finding algorithm. Bioinformatics 22: 445–452. 10.1093/bioinformatics/btk008 [DOI] [PubMed] [Google Scholar]
- Ziehler WA, Engelke DR. 2001. Probing RNA structure with chemical reagents and enzymes. Curr Protoc Nucleic Acid Chem 10.1002/0471142700.nc0601s00 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.