Abstract
Schaaf-Yang syndrome (SYS) is a neurodevelopmental disorder caused by truncating variants in the paternal allele of MAGEL2, located in the Prader-Willi critical region, 15q11-q13. Although the phenotypes of SYS overlap those of Prader-Willi syndrome (PWS), including neonatal hypotonia, feeding problems, and developmental delay/intellectual disability, SYS patients show autism spectrum disorder and joint contractures, which are atypical phenotypes for PWS. Therefore, we hypothesized that the truncated Magel2 protein could potentially produce gain-of-function toxic effects. To test the hypothesis, we generated two engineered mouse models; one, an overexpression model that expressed the N-terminal region of Magel2 that was FLAG tagged with a strong ubiquitous promoter, and another, a genome-edited model that carried a truncating variant in Magel2 generated using the CRISPR/Cas9 system. In the overexpression model, all transgenic mice died in the fetal or neonatal period indicating embryonic or neonatal lethality of the transgene. Therefore, overexpression of the truncated Magel2 could show toxic effects. In the genome-edited model, we generated a mouse model carrying a frameshift variant (c.1690_1924del; p(Glu564Serfs*130)) in Magel2. Model mice carrying the frameshift variant in the paternal or maternal allele of Magel2 were termed Magel2P:fs and Magel2M:fs, respectively. The imprinted expression and spatial distribution of truncating Magel2 transcripts in the brain were maintained. Although neonatal Magel2P:fs mice were lighter than wildtype littermates, Magel2P:fs males and females weighed the same as their wildtype littermates by eight and four weeks of age, respectively. Collectively, the overexpression mouse model may recapitulate fetal or neonatal death, which are the severest phenotypes for SYS. In contrast, the genome-edited mouse model maintains genomic imprinting and distribution of truncated Magel2 transcripts in the brain, but only partially recapitulates SYS phenotypes. Therefore, our results imply that simple gain-of-function toxic effects may not explain the patho-mechanism of SYS, but rather suggest a range of effects due to Magel2 variants as in human SYS patients.
Introduction
In 2013, the first four individuals with truncating variants in the paternal allele of MAGEL2 were reported, and later described as having Schaaf-Yang syndrome (SYS, OMIM#615547). The phenotypes of SYS patients overlap those of Prader-Willi syndrome (PWS, OMIM#176270), including neonatal hypotonia, feeding problems and developmental delay/intellectual disability (DD/ID) [1]. Additionally, SYS patients show autism spectrum disorder (ASD) and contractures of the small finger joints, which are atypical phenotypes for PWS [2].
PWS occurs as the result of absence of expression of paternal genes from chromosome 15q11.2-q13 [3, 4]. Chromosome 15q11.2-q13 contains paternal-only expressed genes encoding polypeptides (MKRN3, MAGEL2, NDN, NPAP1 and SNURF-SNRPN) [1]. It also contains snoRNAs (SNORD115, 116) which show paternal-only expression [5]. Recently, it was revealed that a paternal deletion of SNORD116 is responsible for PWS [6]. MAGEL2 is not expressed in patients with PWS. Therefore, a loss-of-function in MAGEL2 should be associated with PWS. Nevertheless, SYS patients generally show more severe phenotypes than typical PWS patients. Additionally, patients with a paternally inherited deletion including MAGEL2, but not SNRPN/SNORD116, have a milder phenotype than those with truncating variants in MAGEL2 [7, 8]. Thus, a gain-of-function mechanism in MAGEL2 was suggested as the pathological mechanism underlying SYS [9].
Human MAGEL2, and its mouse ortholog Magel2, are GC-rich, single-exon, maternally imprinted genes that are exclusively expressed from the unmethylated paternal allele. MAGEL2 and Magel2 encode putative proteins of 1249 and 1284 amino acids, respectively, which are highly homologous (Fig 1) [9, 10]. In SYS, more than half of the patients carry a truncating variant in nucleotides c.1990-1996, which is upstream of the region encoding the C-terminus of the proline-rich region in MAGEL2. Notably, c.1996dupC is the most common and severe variant in SYS patients [9]. In mice, Magel2 RNA is expressed at low levels throughout the brain, but shows the highest expression in hypothalamic regions, especially the paraventricular nucleus (PVN) and suprachiasmatic nucleus (SCN). A mouse model has been generated by inactivating Magel2 in C57BL/6 mice with the use of a lacZ knock-in allele with paternal inheritance [11, 12]. Magel2-null mice have reduced embryonic viability, but otherwise normal embryonic growth in survivors, followed by postnatal growth retardation. In their later development, they even showed more weight gain compared to littermates [11]. Such mild phenotypes in Magel2-null mice did not recapitulate those of SYS, but may represent those of patients with a deletion of the entire MAGEL2 gene.
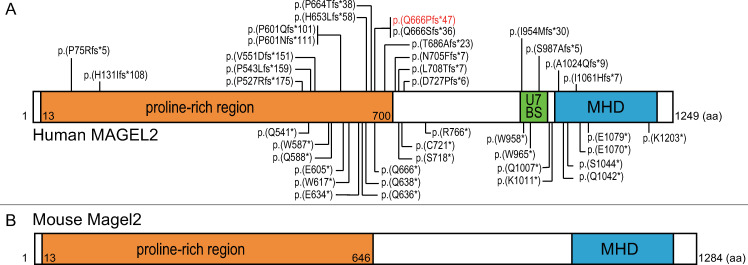
Fig 1. Schematic structure of the human MAGEL2 and mice Magel2.
(A) Human MAGEL2 contains a proline-rich region (residues 13–700), USP7 binding site (U7BS: residues 949–1004), and MAGE homolog domain (MHD: residues 1020–1219). Truncating variants reported previously are indicated by their positions (top; frameshift variants, bottom: nonsense variants). The mutation hotspot is located at nucleotides c.1990-1996. Over half of SYS patients carried c.1996dupC:p.(Q666Pfs*47) in MAGEL2 (in red text). (B) Mouse Magel2 contains proline-rich region (residues 13–646) and MHD (residues 1052–1251).
Therefore, we generated mouse models to test the hypothesis that the truncating MAGEL2 protein could potentially produce gain-of-function toxic effects. Assuming that mice carrying a truncating variant in Magel2 have a more severe phenotype than Magel2-null mice, we generated two types of mouse models: a transgenic mouse that overexpressed the N-terminal region of Magel2, and a genome-edited mouse expressing truncating Magel2 under the intrinsic promoter.
Materials and methods
Vector construction
pCAGGS1-Magel2-FLAG
We generated an overexpression model that overexpressed the N-terminal region of Magel2 (amino acid residues 1–437). We amplified a 1311bp fragment encoding the N-terminal region of Magel2 with a FLAG tag at the C-terminus by polymerase chain reaction (PCR). PCR was performed with mouse genomic DNA, AmpliTaq Gold 360 Master Mix (Thermo Fisher Scientific, Waltham, MA), and primers F1 and R1. Primers F1 and R1 contained recognition sites for EcoRI and XhoI, respectively. As an antibody specific to Magel2 protein was not available, we inserted a FLAG-tag at the C-terminus of the truncated Magel2 (S1 Fig). To express truncated Magel2 under the control of the CAG promoter, the product was subcloned into pCAGGS1 containing a modified chicken actin promoter with the CAG promoter, kindly provided by Dr. J. Miyazaki (Osaka University), using EcoRI and XhoI cloning sites (Fig 2A). The construct was named pCAGGS1-Magel2-FLAG. The construct was linearized by digestion with SalI and HindIII prior to microinjection into fertilized eggs.
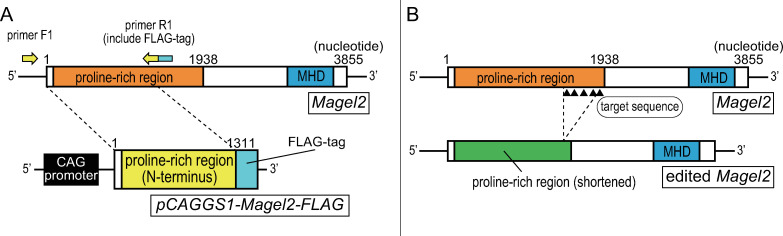
Fig 2. Strategies to generate transgenic mice.
(A) Strategy to generate the overexpression model which overexpresses the N-terminal region of Magel2 with FLAG-tag. (B) Strategy to generate genome-edited model which carries truncating variant in Magel2. Target sequence for CRISPR/Cas9 (5´-CCACAGGAGCTCCCGGTGCCACA-3´) is located on c.1702-1724, c.1720-1742, c.1810-1832, c.1882-1904 and c.1900-1922 (black triangles).
pX330-Magel2
We generated a genome-edited model that carried a truncating variant in intrinsic Magel2. We selected 5´-CCACAGGAGCTCCCGGTGCCACA-3´ as a target sequence for CRISPR/Cas9 which located on c.1702-1724, c.1720-1742, c.1810-1832, c.1882-1904 and c.1900-1922 in Magel2. Target sequences were located near the C-terminus of the proline-rich region in Magel2. The pX330 plasmid (Addgene plasmid #42230) carries both guide RNA and Cas9 expression unit. Magel2-CRISPR-F (5´-caccTGTGGCACCGGGAGCTCCTG-3´) and Magel2-CRISPR-R (5´-aaacCAGGAGCTCCCGGTGCCACA-3´) oligo DNAs were annealed and subcloned into pX330 with BbsI cloning site as described previously [13]. The plasmid was designated as pX330-Magel2.
p2color-Magel2
The p2color vector (RDB13948, RIKEN BRC, Tsukuba Japan) contains a multiple cloning site target site between RFP- and GFP-encoding DNA sequences. Magel2-screening-F (aattTGTGGCACCGGGAGCTCCTGTGGCACCGG) and Magel2-screening-R (ggccCCGGTGCCACAGGAGCTCCCGGTGCCACA) oligo DNAs were annealed and subcloned into the p2color vector with EcoRI/NotI cloning sites as described previously [13]. The plasmid was designated as p2color-Magel2. p2color-Magel2 was used for the RFP-GFP reporter assay which confirmed the cleavage activity of pX330-Magel2.
Western blotting
We transfected pCAGGS1-Magel2-FLAG into HEK293 cells using Lipofectamine 2000 (Thermo Fisher Scientific), and confirmed its expression by western blot analysis as previously described using primary antibodies against FLAG (diluted 1:1000; Cell Signaling Technology, Danvers, MA) and GAPDH (diluted 1:10,000; Cell Signaling Technology), and a horseradish peroxidase–conjugated secondary antibody (GE Healthcare, Little Chalfont, UK) [14].
RFP-GFP reporter assay
We co-transfected HEK293T cells with pX330-Magel2 and p2color-Magel2 using Lipofectamine 2000 following the manufacturer’s protocol. In this assay, the GFP sequence is fused to the target site out of frame, and functional GFP is expressed only when CRISPR/Cas9 induces a double-strand break at the target site, whose repair by error-prone non-homologous end joining gives rise to indels that often result in a frameshift variant (S2 Fig) [15].
Mouse breeding and handling
C57BL/6N mice and ICR mice were purchased from Japan SLC, Inc. (Hamamatsu, Japan). Mice were kept in plastic cages under pathogen-free conditions in a room maintained at 23 ± 2°C under 12:12 light dark conditions. Mice were weaned at four weeks of age then housed at 1–7 mice per cage with food (Oriental Bio Service, Kyoto, Japan) and filtered water ad libitum. Mice used for weight measurement were housed at 4–5 mice per cage after weaning. Mice were euthanized at the appropriate time points with carbon dioxide followed by cervical dislocation. All experimental procedures conformed to the Regulations for Animal Experimentation at Nagoya City University, reviewed by the Institutional Laboratory Animal Care and Use Committee of Nagoya City University, and approved by the provost (Protocol Number: H27M-14, H28M-70, H29M-64).
Microinjection
Four-week-old C57BL/6N females were superovulated with 7.5 IU of pregnant mare serum gonadotropin and human chorionic gonadotropin and mated with 10-week-old C57BL/6N males. Pronuclear-stage eggs were injected with the linearized transgene, cultivated in KSOM overnight, and then transferred into the oviducts of 7-week-old pseudopregnant ICR females. On the 19th day of pregnancy, we sacrificed ICR females and performed cesarean section. Thus, we obtained live-born pups and stunted embryos.
Genomic DNA analysis
Genomic DNA was extracted from mouse tails or embryos with KAPA Mouse Genotyping Kit (Nippon Genetics Co., Ltd., Tokyo, Japan) according to manufacturer’s protocol. Mice were anesthetized with 1% isoflurane, and tail tips taken at four weeks of age. PCR on genomic DNA was performed with AmpliTaq Gold 360 Master Mix and primer pairs. The PCR products were separated using 3% agarose gel electrophoresis. The primers F2 (5´-CAGTATCAGGAGCACCAA-3´) and R2 (5´-ATCCTTGTAGTCCATAGGAC-3´) were used for the overexpression model (S3 Fig). As primer R2 was designed within FLAG-tag sequence, DNA from mice carrying the transgene were specifically amplified by PCR. The primers F3 (5´-CCAACTGTCTATCCCAAT-3´) and R3 (5´-TGCCAGAAGTGAGGAGGT-3´) were used for the genome-edited model (S4 Fig). In mice carrying indels in the C-terminus of the proline-rich region, the length of amplified DNA was shorter than those of wildtype. PCR products were sequenced with BigDye Terminator v3.1 Cycle Sequencing Kit and SeqStudio Genetic Analyzer (Thermo Fisher Scientific). Mice carrying the transgene were used for subsequent mating.
Reverse Transcription-PCR (RT-PCR) analysis
Total RNA was isolated from hypothalamus of neonatal mice (P10) using RNeasy Plus Mini Kit (Qiagen, Hilden, Germany). We removed genomic DNA from total RNA products by using recombinant DNase I (Takara Bio Inc., Shiga, Japan). Reverse transcription was performed using purified RNA and SuperScript IV Reverse Transcriptase (Invitrogen, Carlsbad, CA). All processes were performed following the manufacturer’s protocol. After reverse transcription, complementary DNA (cDNA) was amplified with primer F3 and R3, and AmpliTaq Gold 360 Master Mix. The RT-PCR products were separated using 3% agarose gel electrophoresis.
In situ hybridization (ISH)
ISH of Magel2 mRNA was performed on the brains of young-adult (8–9 weeks) male mice. DNA template for probe (targeted at bases c.1059-1679 of the Magel2 mRNA) was amplified from C57BL/6N mice DNA, with Magel2-probe primer pairs (5´-TGTACCACAAGCCCCCCA-3´ and 5´-GGGGCCTGGCCTTTGG-3´), and AmpliTaq Gold 360 Master Mix. The DNA template was subcloned into pGEM-T easy vector (Promega, Madison, WI, USA) following the manufacturer’s protocol (S5 Fig). We synthesized [35S]UTP-labeled Magel2 antisense strand probes with T7 RNA polymerase after DNA construct linearization by digestion with SalI. We also synthesized 35S-labeled Magel2 sense strand probes with SP6 RNA polymerase after DNA construct linearization by digestion with NcoI. The antisense strand and the sense strand were used for the cRNA probe and negative control, respectively. Cryosections (20 μm thick) were cut from freshly frozen mouse brains, and fixed in 4% formaldehyde in phosphate buffer (0.1 M, pH 7.4) with proteinase K (10 μg/mL). The sections were acetylated with acetic anhydride, dehydrated in ascending alcohol series, and air-dried. They were incubated in hybridization buffer (50% formamide, 0.3 M NaCl, 20 mM Tris-HCl, 10% dextran sulfate, Denhardt’s solution, 500 μg/mL yeast tRNA, 20 mM dithiothreitol, and 200 μg/mL salmon testis DNA) with the synthesized RNA probes for 12 hours at 55°C. After hybridization, they were washed with 50% formamide/2× standard sodium citrate (SSC) at 65°C and incubated with 1 μg/mL RNase A in RNase buffer (0.5 M NaCl, 10 mM Tris–HCl, and 1 mM EDTA, pH 8.0) for 30 min at 37°C. Subsequently, they were washed in 50% formamide/2× SSC at 65°C, rinsed with 2X SSC and 0.1X SSC, dehydrated in alcohol, and air-dried [16]. The slides were then stained with hematoxylin and eosin, and images captured with a CCD camera (OLYMPUS, Tokyo, Japan) connected to a stereomicroscope (Carl Zeiss, Oberkochen, Germany).
Results
Generation of the Magel2 overexpression model
First, we transfected pCAGGS1-Magel2-FLAG into HEK293T cell and confirmed its expression by western blot analysis (S6 Fig). Second, pCAGGS1-Magel2-FLAG vector was injected into the pronuclei of fertilized oocytes to obtain mice that overexpressed the N-terminal region of Magel2 (amino acid residues 1–437) with a FLAG tag. We obtained 52 live-born pups and 29 stunted embryos from six litters. Although three of the 52 pups (5.8%) carried the transgene, two died immediately after birth and one exhibited a small body size and poor suck, and died at P13. None of the surviving 49 pups carried the transgene. Furthermore, 22 of 29 stunted embryos (75.9%) carried the transgene (Table 1). The probability of being transgene-positive was statistically significant between live-born pups and stunted embryos (P < 0.001, Pearson’s chi-squared test).
Table 1. Genotype distribution of live-born offspring and stunted embryos in the Magel2 overexpression model.
| Transgene-positive | Transgene-negative | Total | |
|---|---|---|---|
| Live-born pups | 3 (6.1%) | 49 | 52 |
| Stunted embryos | 22 (75.9%) | 7 | 29 |
Generation of the Magel2 genome-edited model
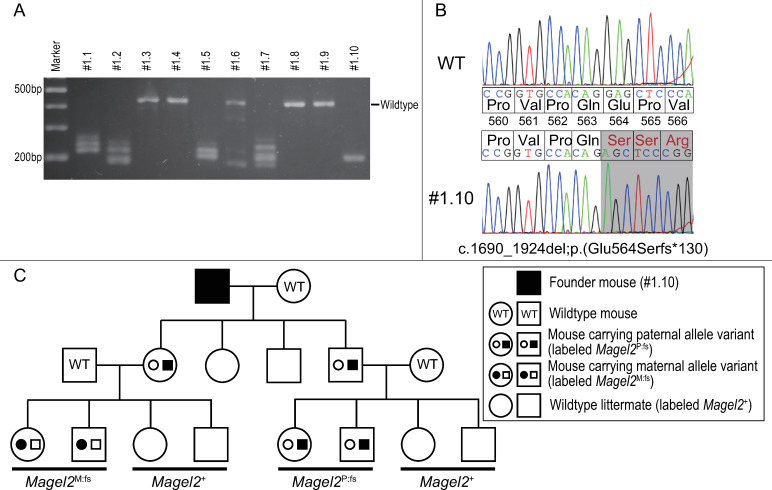
pX330-Magel2 was injected into the pronuclei of fertilized oocytes to obtain mice carrying frameshift variants in Magel2 target sites. From six litters, we obtained 24 pups, of which 20 survived and were genotyped. There were 12 unique Magel2 variants across the 20 pups (Fig 3A). We selected a male mouse carrying a homozygous frameshift variant in Magel2 (c.1690_1924del;p(Glu564Serfs*130)) as the founder mouse (Fig 3B). We then mated the founder mouse with wildtype females, and obtained mice carrying heterozygous frameshift variant in Magel2. Next, we mated affected males with wildtype females and obtained model mice carrying a paternal frameshift variant in Magel2, which were termed ‘Magel2P:fs’. We also mated wildtype males with affected females and obtained control mice carrying a maternal frameshift variant in Magel2, which were termed ‘Magel2M:fs’. Littermates without a variant in Magel2 were termed ‘Magel2+’ (Fig 3C). In Magel2P:fs mice, there were no obvious abnormality in physical findings, including contracture, which is a distinctive phenotype of human SYS patients.
Fig 3. Generation of a mouse model carrying a frameshift variant in Magel2.
(A) We obtained genome-edited mice carrying different variants in Magel2. We selected #1.10 as the founder mouse. (B) Comparison of the base sequence and amino acid residues in Magel2. Founder mouse #1.10 carried a homozygous frameshift variant in Magel2. (C) The pedigree of our mouse model. Mice carrying a variant in the paternal allele of Magel2 were termed ‘Magel2P:fs’. Mice carrying a variant in the maternal allele of Magel2 were termed ‘Magel2 M:fs’. Littermates not carrying a variant in Magel2 were termed ‘Magel2+’.
Birth rate of the genome-edited mouse model
To investigate the birth rate of Magel2P:fs and Magel2M:fs mice we mated affected males with wildtype females, and obtained 201 live-born pups from 27 litters. Eighty-five pups (42.3%) were Magel2P:fs. We also mated wildtype males with affected females and obtained 43 live-born pups from 6 litters. Twenty-two pups (51.2%) were Magel2M:fs. The birth rate of Magel2P:fs was less than expected, but there was no significant difference between Magel2P:fs and Magel2M:fs (P = 0.287, power 0.186, Pearson’s chi-squared test; Table 2).
Table 2. Genotype distribution of offspring born in the genome-edited model.
| Variant-positive | Variant-negative | Total | |
|---|---|---|---|
| Magel2P:fs | 85 (42.3%) | 116 | 201 |
| Magel2M:fs | 22 (51.2%) | 21 | 43 |
Expression of mRNA in Magel2
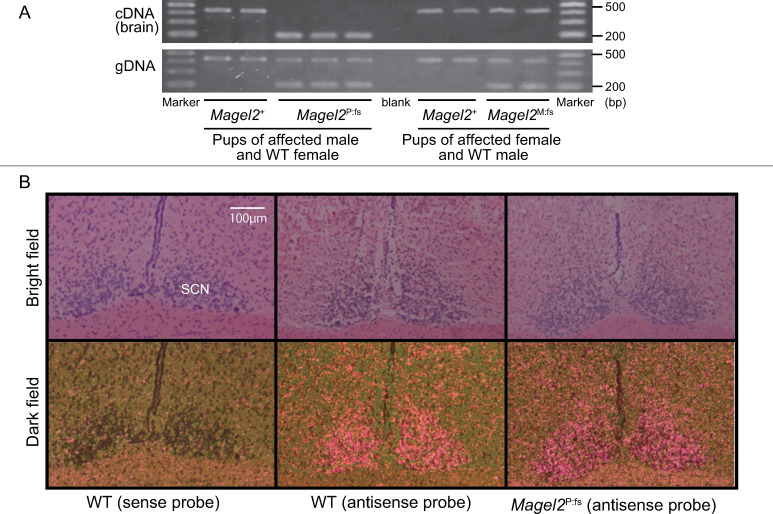
We performed RT-PCR on mRNA from newborn mouse brains. Magel2P:fs and Magel2M:fs mice expressed truncating Magel2 mRNA and normal Magel2 mRNA, respectively (Fig 4A). Thus, the paternal allele of Magel2 was expressed and the maternal allele of Magel2 was silenced.
Fig 4. Expression and distribution of Magel2 in the mouse brain.
(A) Expression of Magel2 transcripts in the neonatal mouse brain. Only the paternal allele of Magel2 is expressed in the brain. (B) Distribution of Magel2 transcripts in young-adult brains was similar in WT and Magel2P:fs mice. Magel2 mRNAs were expressed in the SCN of the hypothalamus in both groups. SCN: suprachiasmatic nucleus. Scale bar: 100 μm.
Localization of Magel2 mRNA in the young-adult mouse brain
We performed ISH on the brains of Magel2P:fs and wildtype young-adult mice. Magel2 mRNA was detected in the SCN and PVN of the hypothalamus in both groups (Fig 4B, S7 Fig). Thus, Magel2P:fs mice did not have an altered localization of Magel2 mRNA.
Body weight in the genome-edited mouse model
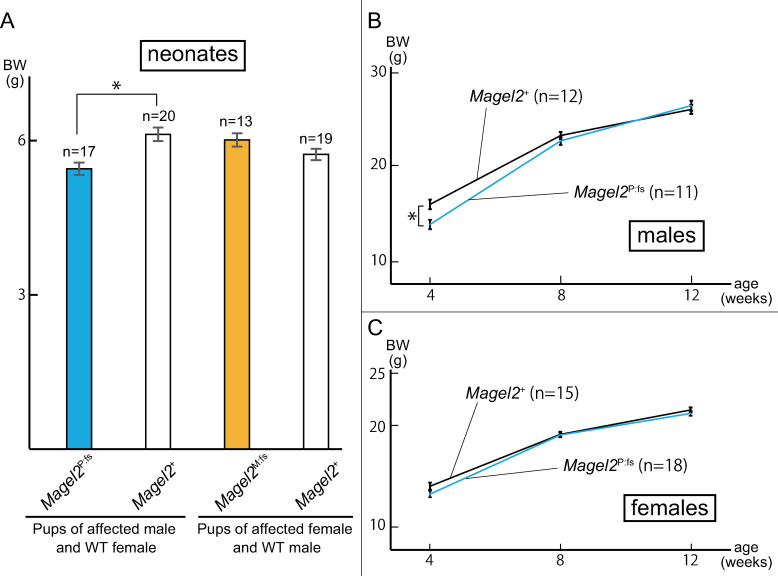
In neonates (P10), Magel2P:fs pups were lighter than Magel2+ (5.44 ± 0.12 g vs 6.11 ± 0.13 g, P = 0.0003, Welch’s t-test). By contrast, there was no difference between Magel2M:fs and Magel2+ pups (6.00 ± 0.13 g vs 5.72 ± 0.11 g, P = 0.058, Welch’s t-test; Fig 5A). For males, Magel2P:fs mice were lighter than their Magel2+ male littermates at four weeks of age (13.71 ± 0.49g vs 15.84 ± 0.50g, P = 0.0032, Welch’s t-test). By eight weeks of age however, the weight of Magel2P:fs males was similar to Magel2+ males (Fig 5B). For females, there was no difference between Magel2M:fs mice and Magel2+ female littermates at four weeks of age (13.84 ± 0.34 g vs 13.07 ± 0.31 g, P = 0.38, Welch’s t-test; Fig 5C).
Fig 5. Comparison of body weight of affected and WT mice.
(A) Mean weight ± SEM at P10. The weight of Magel2P:fs was reduced compared to Magel2+ (5.44 ± 0.12 g vs 6.11 ± 0.13 g, P = 0.0003). There was no difference between Magel2M:fs and Magel2+ mice (6.00 ± 0.13 g vs 5.72 ± 0.11 g, P = 0.058). Both sexes were included. (B, C) Mean weight ± SEM at four, eight and 12 weeks of age in both sexes. At four weeks, Magel2P:fs males were lighter than Magel2+ (13.71 ± 0.49 g vs 15.84 ± 0.50 g, P = 0.0032), but by eight weeks of age, there was no difference. In females, there was no difference between Magel2P:fs mice and Magel2+ at four, eight and 12 weeks of age.
Discussion
MAGEL2 is located in the PWS critical region and typically deleted in PWS patients. Therefore, loss-of-function phenotypes with MAGEL2 are likely to be included in typical PWS patients. However, most SYS patients with a truncating variant in MAGEL2 show more severe clinical features than PWS patients. Indeed, our previous study identified six patients with SYS with a truncating variant, including the common c.1996dupC, and all six patients showed severe intellectual disability and complication of joint contracture, which are atypical for PWS [17]. Thus, we hypothesized that the truncated Magel2 protein could potentially produce gain-of-function toxic effects, and we generated two types of mouse models expressing truncating mutations of Magel2.
First, we generated an overexpression model which expressed truncating Magel2 under the control of the CAG promoter, and found that all transgenic mice died in fetal or neonatal period. In normal mouse embryos, Magel2 is expressed in the hypothalamus, cerebral cortex, and spinal cord [18]. In contrast, our model mice expressed truncated Magel2 under the CAG promoter, which induced much higher expression levels ubiquitously. Therefore, we assumed that the expression of truncated Magel2 in various types of organs were responsible for fetal death. We tried to extract protein from the brain of dead mice in the overexpression model, and to detect truncated Magel2 protein with FLAG tag. However, we were not able to detect FLAG tag signal by western blot analysis as the extracted proteins were denatured. Therefore, we were not able to assess the patho-mechanism of fetal or neonatal death in the overexpression mouse model. Nevertheless, the overexpression model revealed the toxic effects of overexpression of truncated Magel2 in the fetal period. It is intriguing that Mejlachowics et al. reported that the c.1996delC variant in MAGEL2 was responsible for fetal death at 24 to 27 weeks of gestation in human [19], indicating the severe toxic effects of the specific truncated MAGEL2. Our overexpression model may, at least in some degree, recapitulate the most severe of the toxic effects of truncated MAGEL2.
Next, we generated a genome-edited mouse model carrying a frameshift variant in Magel2 (c.1690_1924del;p(Glu564Serfs*130)) with the CRISPR/Cas9 system. As the paternal allele of Magel2 is expressed, but the maternal allele is silenced, we classified the model mice into those carrying a paternal allele variant (Magel2P:fs) and those carrying a maternal allele variant (Magel2M:fs).
Kozlov et al. generated a mouse model with inactivate paternal Magel2 with the use of a lacZ knock-in allele, and Magel2 knockout model displayed 10% postnatal lethality [12]. We investigated the birth rate of Magel2P:fs and Magel2M:fs, but there was no difference between the two groups although statistical power was not high.
Imprinting regions, including Magel2, are governed by imprinting centers that regulate parent-of-origin epigenotypes and gene expression patterns [20]. Matarazo et al. reported the loss of imprinting and the expression of the maternal allele of Magel2 in a mouse model with a deletion of the paternal allele of Magel2, including its promoter [21]. We confirmed genome imprinting by RT-PCT on newborn mouse brain mRNA, and revealed that Magel2P:fs expressed truncated Magel2, and Magel2M:fs expressed full-length Magel2. That suggested that our mouse model maintained the genome imprinting mechanism of Magel2.
Magel2 expression is specifically localized to the SCN and PVN in the hypothalamus [12, 18]. We performed ISH to compare the distribution of Magel2 mRNA in wildtype and Magel2P:fs males. There was no difference in the distribution of Magel2 mRNA. Those results suggested that our model mice maintained the distribution of Magel2 mRNA in the brain. As we do not have a specific anti-Magel2 antibody, we could not assess the expression of the Magel2 protein.
We measured the body weight of genome-edited model mice compared with their littermates (Magel2+). At P10, Magel2P:fs pups were statistically lighter than Magel2+. Although Magel2P:fs males were statistically lighter than Magel2+ males at four weeks of age, their body weight caught up with those of Magel2+ males at eight weeks of age. Furthermore, the body weight of Magel2P:fs females was similar to that of Magel2+ females at four weeks of age. Bischof et al. reported that Magel2-null mice exhibited neonatal growth retardation and excessive weight gain after weaning, and their growth abnormality was similar to PWS [11]. In contrast, it was reported that 97% of SYS patients exhibited poor suck in infancy, but only 22–41% exhibited excessive weight gain [9, 22]. Our mouse model showed growth retardation in neonates, but they did not show excessive weight gain after weaning. They may partially recapitulate human SYS phenotype in terms of characteristic growth abnormality.
The genome-edited mouse model did not show obvious abnormality in physical findings. In humans, genotype-phenotype association in MAGEL2 has been discussed previously. McCarthy et al. mentioned that c.1996dupC in MAGEL2 is the most common variant in SYS. Patients carrying c.1996dupC in MAGEL2 showed a higher prevalence of joint contractures, feeding difficulties, and severe ID/DD than patients carrying other variants in MAGEL2. They mentioned that the severity of SYS depended on the specific location of the truncating mutation. MAGEL2 and Magel2 are single exon genes, and mutations leading to a premature stop codon are predicted not to cause nonsense-mediated mRNA decay. The pathogenic effect of the truncated MAGEL2 protein may differ depending on the precise location of the mutation in MAGEL2 [9]. Our mouse model carrying the c.1690_1924del variant in paternal Magel2 may produce truncated Magel2 protein which shows a milder toxic effect.
Our genome-edited mouse model showed almost comparable or less severe phenotypes to previously reported Magel2 null mice, and failed to recapitulate the common phenotype of SYS. This may be due to the position of the variant we made and the wide clinical spectrum of human SYS patients. Thus, the gain-of-function hypothesis remains unsolved. Nevertheless, in our genome-edited mouse model, we showed the maintenance of imprinted expression and the distribution of the truncated Magel2 transcripts in the mouse brain.
There are several limitations of the study. First, although we analyzed Magel2 mRNA, we did not analyze Magel2 protein due to the lack of a specific anti-Magel2 antibody. Second, we were not able to investigate the patho-mechanism of fatal or neonatal death of the overexpression model because we were unable to obtain purified brain proteins. Third, the analyses of our mouse models are not exhaustive. It is known that human SYS patients have ASD and arthrogryposis, and Magel2-null mice have altered circadian rhythm, reduced motor activity, and increased adiposity [2, 12, 23]. Therefore, behavioral, anatomical and serological tests are required for our mouse model in the future.
Conclusion
We generated two types of mouse models carrying a truncating variant in Magel2. The overexpression model was embryonic or neonatal lethal, indicating toxic effects of overexpression of the truncated Magel2. The genome-edited model maintained genomic imprinting and distribution of truncated Magel2 transcripts in the brain, and only partially recapitulate SYS phenotypes. Our results suggest that not simple gain-of-function toxic effects, but rather varied effects due to the position and type of MAGEL2 variants, underlie the patho-mechanism of SYS.
Supporting information
(EPS)
(A) Scheme of the RFP-GFP reporter-based assay for measuring the activity of the CRISPR/Cas9 system. The CRISPR/Cas9 system induces double-strand breaks for target sequence, with frameshift variants incorporated after non-homologous end joining. (B) Robust EGFP signals were only seen in HEK293T cells co-transfected with both p2color-Magel2 and pX330-Magel2.
(EPS)
Primer R2 contains the sequence complementary to the terminus of truncated Magel2 and FLAG-tag sequence. DNA from transgenic mice is specifically amplified with primers F2 and R2.
(EPS)
In wildtype DNA, PCR with primers F3 and R3 was predicted to produce an amplicon of 438bp. An amplicon would be predicted to be shorter than that of wildtype when Magel2 was edited by CRISPR/Cas9 system.
(EPS)
Magel2 DNA (c.1059-1679) was subcloned into pGEM-T easy vector. Magel2 antisense strand for the cRNA probe and sense strand for negative control were synthesized by T7 and SP6 RNA polymerase, respectively.
(EPS)
(EPS)
Distributions of Magel2 transcripts in PVN of wild-type mice and Magel2p:fs mice are shown in middle and right panels, respectively. Left panels, sense controls. Scale bar: 100 μm.
(EPS)
(PDF)
Acknowledgments
We wish to thank all members of the laboratories of Department of Pediatrics and Neonatology, Nagoya City University Graduate School of Medical Sciences, for their assistance. We also acknowledge the assistance of the Research Equipment Sharing Center at the Nagoya City University.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This study was partly supported by JSPS KAKENHI Grant-in-Aid for Early-Career Scientists (JP18K15682). There was no additional external funding received for this study.
References
- 1.Schaaf CP, Gonzalez-Garay ML, Xia F, Potocki L, Gripp KW, Zhang B, et al. Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism. Nat Genet. 2013;45:1405–1408. 10.1038/ng.2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fountain MD, Aten E, Cho MT, Juusola J, Walkiewicz MA, Ray JW, et al. The phenotypic spectrum of Schaaf-Yang syndrome: 18 new affected individuals from 14 families. Genet Med. 2017;19:45–52. 10.1038/gim.2016.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler MG. Prader-Willi syndrome: current understanding of cause and diagnosis. Am J Med Genet. 1990;35:319–332. 10.1002/ajmg.1320350306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holm VA, Cassidy SB, Butler MG, Hanchett JM, Greenswag LR, Whitman BY, et al. Prader-Willi syndrome: consensus diagnostic criteria. Pediatrics. 1993;91:398–402. [PMC free article] [PubMed] [Google Scholar]
- 5.Cavaillé J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci U S A. 2000;97:14311–14316. 10.1073/pnas.250426397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet. 2008;40:719–721. 10.1038/ng.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanber D, Giltay J, Wieczorek D, Zogel C, Hochstenbach R, Caliebe A, et al. A paternal deletion of MKRN3, MAGEL2 and NDN does not result in Prader-Willi syndrome. Eur J Hum Genet. 2009;17:582–590. 10.1038/ejhg.2008.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buiting K, Di Donato N, Beygo J, Bens S, von der Hagen M, Hackmann K, et al. Clinical phenotypes of MAGEL2 mutations and deletions. Orphanet J Rare Dis. 2014;9:40 10.1186/1750-1172-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarthy J, Lupo PJ, Kovar E, Rech M, Bostwick B, Scott D, et al. Schaaf-Yang syndrome overview: Report of 78 individuals. Am J Med Genet A. 2018;176:2564–2574. 10.1002/ajmg.a.40650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tacer KF, Potts PR. Cellular and disease functions of the Prader-Willi Syndrome gene MAGEL2. Biochem J. 2017;474:2177–2190. 10.1042/BCJ20160616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bischof JM, Stewart CL, Wevrick R. Inactivation of the mouse Magel2 gene results in growth abnormalities similar to Prader-Willi syndrome. Hum Mol Genet. 2007;16:2713–2719. 10.1093/hmg/ddm225 [DOI] [PubMed] [Google Scholar]
- 12.Kozlov SV, Bogenpohl JW, Howell MP, Wevrick R, Panda S, Hogenesch JB, et al. The imprinted gene Magel2 regulates normal circadian output. Nat Genet. 2007;39:1266–1272. 10.1038/ng2114 [DOI] [PubMed] [Google Scholar]
- 13.Hasegawa Y, Hoshino Y, Ibrahim AE, Kato K, Daitoku Y, Tanimoto Y, et al. Generation of CRISPR/Cas9-mediated bicistronic knock-in ins1-cre driver mice. Exp Anim. 2016;65:319–327. 10.1538/expanim.16-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Negishi Y, Miya F, Hattori A, Johmura Y, Nakagawa M, Ando N, et al. A combination of genetic and biochemical analyses for the diagnosis of PI3K-AKT-mTOR pathway-associated megalencephaly. BMC Med Genet. 2017;18:4 10.1186/s12881-016-0363-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, et al. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol. 2013;31:251–258. 10.1038/nbt.2517 [DOI] [PubMed] [Google Scholar]
- 16.Kumamoto N, Matsuzaki S, Inoue K, Hattori T, Shimizu S, Hashimoto R, et al. Hyperactivation of midbrain dopaminergic system in schizophrenia could be attributed to the down-regulation of dysbindin. Biochem Biophys Res Commun. 2006;345:904–909. 10.1016/j.bbrc.2006.04.163 [DOI] [PubMed] [Google Scholar]
- 17.Negishi Y, Ieda D, Hori I, Nozaki Y, Yamagata T, Komaki H, et al. Schaaf-Yang syndrome shows a Prader-Willi syndrome-like phenotype during infancy. Orphanet J Rare Dis. 2019;14:277 10.1186/s13023-019-1249-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S, Walker CL, Wevrick R. Prader–Willi syndrome transcripts are expressed in phenotypically significant regions of the developing mouse brain. Gene Expr Patterns. 2003;3:599–609. 10.1016/s1567-133x(03)00113-3 [DOI] [PubMed] [Google Scholar]
- 19.Mejlachowicz D, Nolent F, Maluenda J, Ranjatoelina-Randrianaivo H, Giuliano F, Gut I, et al. Truncating mutations of MAGEL2, a gene within the Prader-Willi locus, are responsible for severe arthrogryposis. Am J Hum Genet. 2015;97:616–620. 10.1016/j.ajhg.2015.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brant JO, Riva A, Resnick JL, Yang TP. Influence of the Prader-Willi syndrome imprinting center on the DNA methylation landscape in the mouse brain. Epigenetics. 2014;9:1540–1556. 10.4161/15592294.2014.969667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matarazzo V, Muscatelli F. Natural breaking of the maternal silence at the mouse and human imprinted Prader-Willi locus: A whisper with functional consequences. Rare Dis. 2013;1:e27228 10.4161/rdis.27228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleinendorst L, Pi Castan G, Caro-Llopis A, Boon EMJ, van Haelst MM. The role of obesity in the fatal outcome of Schaaf-Yang syndrome: Early onset morbid obesity in a patient with a MAGEL2 mutation. Am J Med Genet A. 2018;176:2456–2459. 10.1002/ajmg.a.40486 [DOI] [PubMed] [Google Scholar]
- 23.Fountain MD, Tao H, Chen CA, Yin J, Schaaf CP. Magel2 knockout mice manifest altered social phenotypes and a deficit in preference for social novelty. Genes Brain Behav. 2017;16:592–600. 10.1111/gbb.12378 [DOI] [PMC free article] [PubMed] [Google Scholar]