Abstract
Background
The accurate annotation of protein functions is of great significance in elucidating the phenomena of life, treating disease and developing new medicines. Various methods have been developed to facilitate the prediction of these functions by combining protein interaction networks (PINs) with multi-omics data. However, it is still challenging to make full use of multiple biological to improve the performance of functions annotation.
Results
We presented NPF (Network Propagation for Functions prediction), an integrative protein function predicting framework assisted by network propagation and functional module detection, for discovering interacting partners with similar functions to target proteins. NPF leverages knowledge of the protein interaction network architecture and multi-omics data, such as domain annotation and protein complex information, to augment protein-protein functional similarity in a propagation manner. We have verified the great potential of NPF for accurately inferring protein functions. According to the comprehensive evaluation of NPF, it delivered a better performance than other competing methods in terms of leave-one-out cross-validation and ten-fold cross validation.
Conclusions
We demonstrated that network propagation, together with multi-omics data, can both discover more partners with similar function, and is unconstricted by the “small-world” feature of protein interaction networks. We conclude that the performance of function prediction depends greatly on whether we can extract and exploit proper functional information of similarity from protein correlations.
Keywords: Network propagation, Protein-protein interaction, Prediction of protein function
Background
Proteins are the main component of cells and play an essential role in nearly all cell functions such as composing cellular structure. Biological functions are performed by groups of interacting and functionally associated proteins, instead of individual proteins. The accurate characterization of protein functions is a key to understanding life at the molecular level and has a profound impact on biomedicine and pharmaceuticals.
Proteins of unknown function comprise a significant fraction of sequenced genomes [1]. Thus, how accurately unknown proteins are determined in their purposes has become one of the greatest challenges in the post-gene era. However, due to the inherent difficulty and high costs, experimental techniques to determine protein functions has been unable to meet the growing genomic sequence data. An increasing number of protein-protein interaction data urgently requires computational methods to predict protein functions.. A protein interaction network (PIN) can be modelled as an undirected graph, in which a vertex represents a protein and an edge denotes an interaction between a pair of proteins. Intuitively, numerous network-based [2–4] or graph-based [5, 6] approaches are applied to predict protein functions from PINs. These methods are based on the observation that proteins often possess similar or identical biochemical functions with their interaction partners in the PINs [2, 7]. Unfortunately, these methods are often plagued by noise and errors, resulting in biased outcomes and reduced confidence in PINs.
To provide an accurate prediction results, the integration of different types of biological data has become an important and popular strategy. A number of approaches have been developed to facilitate the prediction of protein functions by combining PPIs with multi-source biological data. Cozzetto et al. [8] proposed an effective method to deduce protein functions by integrating PINs with a wide variety of biological information, such as sequence, gene expression, etc. Zhang et al. [9] developed the domain context similarity for the prediction of protein functions using protein domain composition and PINs. As an improvement on Zhang’s method, two algorithms, named DCS (domain combination similarity) [10] and DSCP (domain combination similarity in context of protein complexes) were proposed to annotate unknown proteins by combining PINs with proteins’ domain information and protein complexes information. For the annotation of protein functions, the PON (protein overlap network) [11] was constructed using the protein domain information and PIN topology. Sarker et al. [12] initially reconstructed a protein-protein network based on PINs and protein domains, and then presented the GrAPFI method for the annotation of protein functions. INGA [13] and INGA 2.0 [14] web servers were developed to infer protein functions by combining protein interaction networks, domain assignments and sequence similarity. PANNZER2 [15] was another functional annotation web server based on sequence similarity practical. On the basis of the deep learning framework, Zhang proposed two methods: DeepGOA [16] and DeepFunc [17], for accurate prediction of protein functions. Normally, these methods are based network or neighbour-count. Multiple biological data is fused into these methods to improve the quality of the PINs. For example, Zhang, DCS and DSCP have improved neighbour-count-based methods with integrated protein domain data. The combinatorial theory was used in these three methods to calculate the functional similarity between proteins. Combining protein domain information with the topologies of PINs, the PON and GrAPFI method rebuilt protein interaction networks for the prediction of protein functions. Then, these two methods annotated unknown proteins according to their 1-layer neighbours in the constructed network based on neighbor count and link weights. In spite of the advances in these methods, it was a central challenge to the integration of multiple biological data categories within a single analysis framework.
In the context of functions prediction, most network analysis methods depended on the principle of ‘guilt by association’, which is based on observations that a protein shares many functional features with its direct interacting partners in PINs. A simple and generic method might be to characterize unknown proteins with functions of all direct neighbours in PINs. Nevertheless, such a straightforward way would potentially yield false positives that are linked to proteins by irrelevant interactions; it would also introduce false negatives that do not directly connect to proteins with known functions [18]. It is verified by our statistics on the yeast PINs. We investigated the shortest path length distribution of protein pairs with common functions and proteins pairs that do not share any function. The statistics results were shown in Fig. 1. Figure 1 shows that the proportion of protein pairs with co-annotation is higher than that of protein pairs with none-annotation when the distance is less than 3. This indicates that co-annotated proteins are closer to each other than non-co-annotated proteins. Figure 1 also reveals an interesting phenomenon that proteins seem to co-annotate with their level-3 or level-4 neighbours instead of direct interacting partners, due to the incompleteness and fault of the PINs.
Fig. 1.
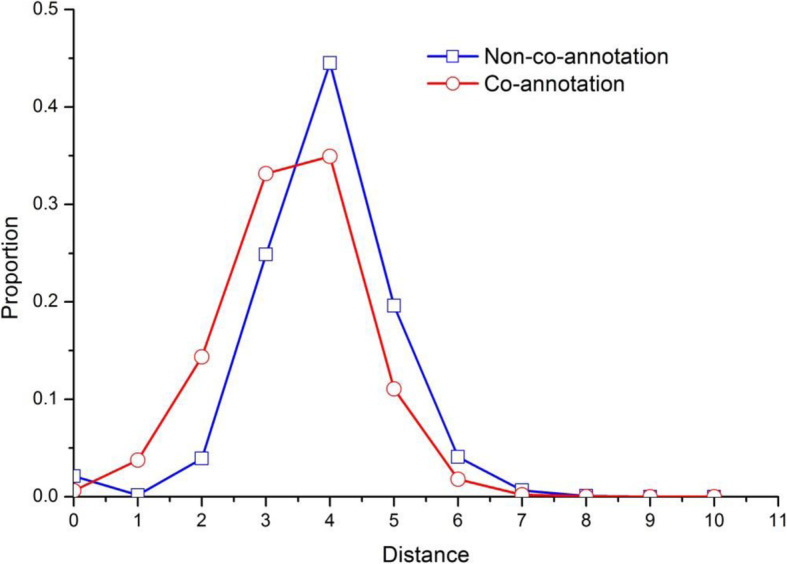
Distribution of shortest-path distances in the pairs of proteins sharing functions. This figure illustrates the shortest path length distribution of protein pairs with common functions and proteins pairs that do not share any function. The curve is plotted by short-path distances between proteins on the horizontal axis and the proportion of pairs of proteins on the vertical. The red round curve describes the relationship between pairs of proteins with common functions and the short path distances between them, and the blue rectangle curve depicts that of pairs of proteins without common functions
To clear this hurdle, as a proxy to a ‘functional distance’ between proteins, the short-path distance instead of Euclidean distance (i.e. the short-path distance between proteins is equal to 1) was adopted in some approaches to predict protein functions. However, most of proteins can arrive reach other proteins within a few steps because of the small-world feature of the PIN. Although these approaches can effectively suppress false negatives, it will also return many spurious functions by including irrelevant interactions. Network propagation provides us with a more refined approach by using the flow of information through network connections as a means to establish relationships between nodes [19]. There are various guises of network propagation, such as random walks on graphs [20], the Google PageRank search algorithm [21], heat diffusion processes [22], graph kernels [23], etc. In biological network, plenty of methods based on network propagation have been widely applied to essential proteins identification [24, 25], drug synergy prediction [26], tumors classification [27], disease associated genes identification [28, 29], microbe-disease associations inference [30] and protein functions prediction [31], which demonstrated that network propagation is a powerful data transformation method of broad utility in genetic research [18]. Additionally, the rationality of combining the protein domain, complex information and PINs for functions prediction is substantiated by the DCS and DSCP methods.
Inspired by these findings, we developed a network propagation-based method, named NPF, for prediction of protein functions. Our model initially simulates the random walk with restart algorithm and constructs a propagation network by integrating knowledge of the protein interaction network architecture, protein domains and protein complexes. This serves as the basis for us to detect functional modules with high coupling in the prediction of functions of unknown protein. To evaluate the performance of NPF, we apply our method and six other state-of-the-art methods for prediction of protein functions on yeast PINs. Experimental results demonstrated that NPF outperformed these competing methods, including Neighbourhood-counting (NC) [2], Zhang [9], DCS [10], DSCP [10], PON [11] and GrAPFI [12].
Methods
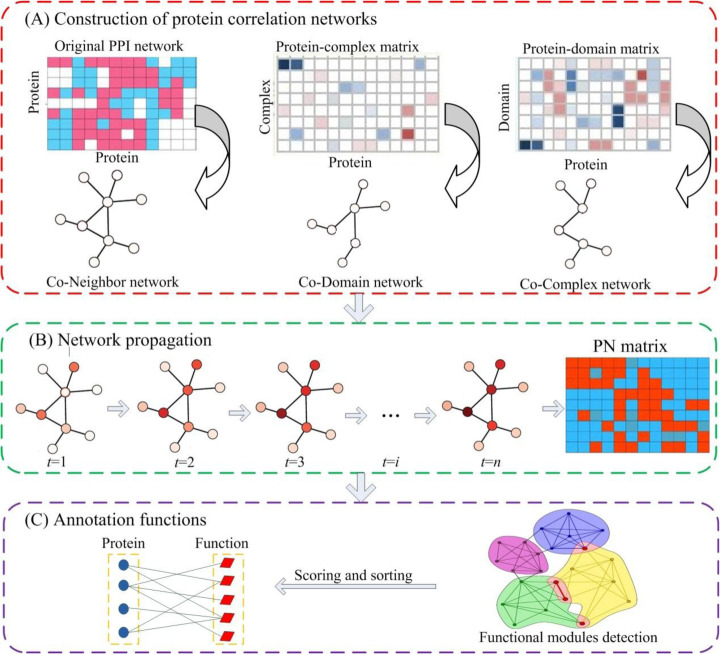
The NPF method is divided into three stages: (1) Constructing three protein-protein correlation networks by integrating knowledge of the protein interaction network architecture, protein-domain associations and protein-complex associations. (2) Building a propagation network by applying an improved random walk with restart algorithm to multiple protein correlation networks. (3) Detecting functional modules with high coupling in the propagation network and annotating functions for target proteins. The flowchart for the NPF method is shown in Fig. 2.
Fig. 2.
Flowchart of NPF method. a Three protein correlation networks Co_Neighbor, Co-Domain and Co-Complex are derived from original PIN, protein domain data, as well as protein complex information, respectively. b The propagation network PN is generated by running an improved random walk with restart algorithm on multiple functional similarity networks. The propagation process is illustrated at different steps until convergence. Changes in the color of nodes in the graph indicate the progress of the iterative process. c Annotation for target proteins. Taking the target node as the seed node, a highly cohesive functional module can be obtained. Functions of neighbors in the detected functional module are used to characterize the target node
Construction of multiple protein correlation networks
Biological functions are performed by a group of genes or proteins which are related to one or more cellular interactions, e.g. protein-protein interaction, co-regulation, co-expression or membership of a protein complex. Physical PINs directly indicate the cooperation of proteins to drive a biological process [32]. Moreover, computational approaches had successfully detected stable functional modules from co-expression networks [33]. We suspect that tightly interacting and functionally dependent proteins may co-express, co-regulate or share a common protein complex, etc. Therefore, we constructed multiple protein-protein correlation networks with integration of knowledge of protein interaction network architecture, protein domain annotation and protein complexes information.
Co-neighbor network
Molecular functions are performed by groups of proteins interacting to each other. Thus, a straightforward strategy is to annotate proteins for target proteins using knowledge of the protein interaction network architecture. In this study, we used the overlapping interacting partners between a pair of proteins as an estimate of their functional correlation. In the Co-Neighbor network, two proteins are connected if they have a physical interaction and link to one or more common proteins simultaneously. Given a pair of proteins pi and pj in the Co-Neighbor network, their correlation value was calculated as follow [34]:
| 1 |
where, N pi and N pj represents the set of direct neighbors of pi and pj respectively. Npi∩N pj is an intersection of N pi and N pj.
Co-domain network
Domains are sequential and structural motifs found independently in different proteins and play as the stable functional block of proteins. We now generalize the idea to construct a protein correlation network based on the protein domain annotation information. For a pair of proteins pi and pj, let M denotes the total number of domain categories in PINs, and let x and y represent the number of domain categories of pi and pj, respectively. Let z expresses the number of overlapping domain categories between pi and pj. Then, we measured the functional correlation between two proteins pi and pj in the Co-Domain network with the follow formula, which is an improvement of the Zhang method [9]:
| 2 |
Finally, the correlation score between pi and pj was obtained by the normalization processing, which was described as follows:
| 3 |
Co-complex network
Protein complexes consisting of molecular aggregations of proteins assembled by multiple protein interactions are fundamental units of macro-molecular organization and play crucial roles in integrating individual gene products to perform useful cellular functions. Studies [10] have revealed that if two proteins are consisted of the same protein complexes, they tend to perform the same or similar biological functions. As much, incorporating quality-controlled protein complexes and analysing functional associations are both essential for accurate function annotation. We therefore proposed to construct the protein correlation network Co-Complex, where the functional correlation between two proteins is measured using the Eq. (4) [35].
| 4 |
In Eq. (4), C pi and C pj represents the set of protein complexes in which pi and pj is involved respectively. Cpi∩C pj denotes the set of protein complexes containing both pi and pj.
Network propagation algorithm
The network propagation algorithm involved a random walk with restart process on multiple protein correlation networks to generate an aggregated protein functional similarity network with high confidence. This process considered the global connectivity patterns of the PIN for annotating target proteins. Moreover, this algorithm took into account the structural feature and modular feature of protein for measuring functional similarity by performing a two-step propagation operation. The output of the network propagation algorithm is a propagated protein functional matrix, which could be used as input for protein function prediction.
At the first step of the network propagation algorithm, we established the transition matrix, H, based on the Co-Neighbor network. The transition probability from protein i to protein j was computed using the following equation:
| 5 |
Intuitively, we wish to calculate functional similarity between proteins by propagation that takes both structural feature and modular feature of proteins into account. These two features are derived from domain annotation and protein complex information, respectively. Therefore, we performed a two-step propagation operation to calculate functional similarity between the protein pi with other proteins by
| 6 |
| 7 |
where the parameter α ∈ [0, 1] balances between the propagation information and restart scores, VDit and VCit are two vectors at the t step to measure structural correlation and modular correlation between protein pi with the remaining proteins, respectively. Elements of the two vectors are initialized to 1/n (i.e., T, T). It was to note that it is possible to tune the functional similarity scores by defining two restart vectors RV_Di and RV_Ci by
| 8 |
| 9 |
In this study, we set α to 0.5 [36, 37]. When the propagation converges, we can obtain an adjacency matrix responding to the propagation network, which is formally described as follows:
| 10 |
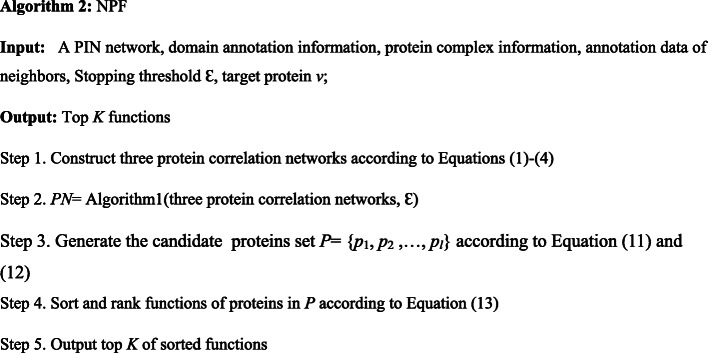
The overall framework of network propagation algorithm can be illustrated as the Algorithm 1. The proof of convergence on the Algorithm 1 can be found in Additional file 1.
Prediction of protein functions
Intuitively, interacting partners are helpful to characterize target proteins. However, members of the same functional module are often more densely connected than those across functional modules [38]. Therefore, at the final stage of our work, we threw out loosely connected neighbours and annotated target proteins with the remaining partners in the newly constructed propagation network. Given a target protein v, M_V is a module of the propagation network PN, which is composed of all neighbour nodes of v. The module fitness [39] was introduced to quantitative describe the cohesion of M_V.
| 11 |
where WDM _ Vin denotes the total weight of edges contained entirely by a group of proteins in the module M_V, WDM _ Vout denotes the total weight of edges that connect the group with the rest of the network. β is a positive real-valued parameter, controlling the size of the module. To simplify operation, we set β to 1. The aim of this stage was to determine a module starting from protein v such that the inclusion of a new neighbour or the elimination of one neighbor from the module would lower fM_V. Thus for this purpose, we introduced the concept of neighbour fitness. Given a v’s neighbour u, the neighbour fitness of u in reference to the module M_V was calculated as follows:
| 12 |
In eq. (12), M_V+ {u} and M_V-{u} represents the module obtained from M_V with neighbour u inside and outside, respectively.
First, neighbours of v were ranked in descending order according to the functional similarity to v. And then, all neighbours of v were visited and nodes with neighbour fitness greater than 0 were selected to form a candidate proteins set P = {p1, p2,…, pl}. Let F = {f1, f2,…, fm} be a list of functions of all proteins in P. The score of a candidate function fj in F can be calculated as follows:
| 13 |
where PN(v, u) represents the functional similarity between u and v in the newly constructed propagation network. If u contains function fj, then tuj = 1, otherwise tuj = 0. Finally, candidate functions were ranked in descending order according to their scores and TOP K of them were selected to characterize the target protein v. In this study, the parameter K was set to the number of functions of the protein with the greatest functional similarity to v in the propagation network PN. The Algorithm 2 gave the overall framework of the proposed NPF method.
Results
Experimental data
To test the performance of NPF, we applied our method and six competing methods to infer protein functions in the protein interaction network of Saccharomyces cerevisiae (Baker’s yeast), because of their completeness, convincement, and widespread used in function prediction algorithms as gold standard data. The PIN data is derived from BioGRID database [40], updated to Oct.28, 2017, which consists of 4113 proteins and 26,105 interactions among the proteins with self-interactions and repeated interactions removed. The BioGRID is an integrated network, which has been proven successful in tasks such as predicting disease genes [41].
The annotation data of proteins used for validation was downloaded from GO official website [42]. The GO system consists of three separate categories of annotations, namely molecular function, biological process and cellular component. This paper takes the biological process as an example to analyse the performance of NGF. The protein domain data was downloaded from Pfam database [43], which contains 1026 different types of domains associated with 2566 proteins in the BioGRID network. The benchmark protein complexes set was adopted from CYC2008 [44], which consists of 408 complexes involving 1600 proteins in the BioGRID dataset. The above four dataset were uniformly transformed to use the Ensemble Genomes Protein labelling system.
View of the constructed networks
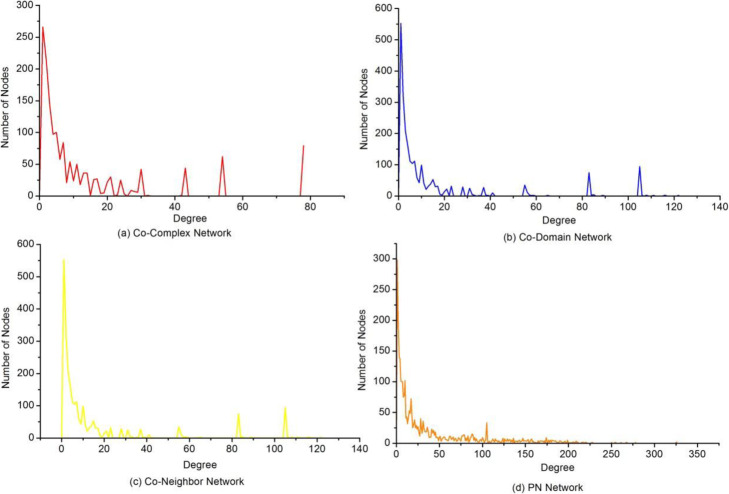
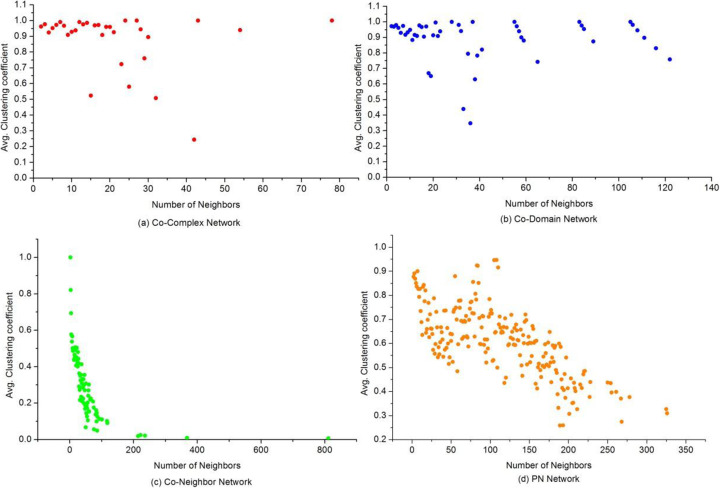
In order to better understand the behaviour of the proposed NPF method, we provided descriptive statistics on the constructed networks, including the Co-Neighbor network, Co-Domain network, Co-Complex network and PN (propagation network). Table 1 listed the basic statistics of the four constructed networks, such as size of networks, average degree etc. When considering network characteristics, the characteristic path length and the clustering coefficient are usually used to measure the network. Table 2 shows the topology features of the original PPI network and the constructed propagation network. The results indicate that the effect of the small-world characteristic on function prediction was improved through network propagation. Figures 3 and 4 depicted the distribution of degree and clustering coefficient in these four networks, respectively. Our statistics revealed reinforcing functional correlations or relationships between proteins in the PN. Therefore, it is reasonable to believe that network propagation is helpful to reduce the negative effect of false negative and improve the accuracy of prediction of protein functions.
Table 1.
Statistics of constructed networks
| Networks | Number of nodes | Number of edges | Average degree | Clustering coefficient | Connected components |
|---|---|---|---|---|---|
| Co-Neighbor | 2696 | 13,728 | 10.184 | 0.645 | 25 |
| Co-Domain | 2448 | 18,123 | 14.806 | 0.743 | 471 |
| Co-Complex | 1595 | 10,886 | 13.650 | 0.798 | 279 |
| PN | 3082 | 57,256 | 37.155 | 0.673 | 185 |
Table 2.
Comparison of the original network and the constructed network
| Networks | Number of nodes | Number of edges | Characteristic path length | Clustering coefficient |
|---|---|---|---|---|
| BioGRID | 4113 | 26,105 | 3.461 | 0.309 |
| PN | 3082 | 57,256 | 3.710 | 0.673 |
This table compares the topology features of the original network based on the BioGRID dataset and the constructed network PN
Fig. 3.
The distribution of degree in constructed networks. This Figure shows the distribution of degree in the four constructed networks. (a) Co-Complex network, (b) Co-Domain network, (c) Co-Neighbor network, (d) PN network
Fig. 4.
The distribution of clustering coefficient in constructed networks. This Figure shows the distribution of clustering coefficient in the four constructed networks. (a) Co-Complex network, (b) Co-Domain network, (c) Co-Neighbor network, (d) PN network
Assessment criteria
Two assessment criteria were adopted to compare function prediction performance of the NPF with six competing methods, including NC [2], ZhangDC [9], DCS [10], DSCP [10], PON [11] and GrAPFI [12]. The NC method is a classic protein function annotation method, which is only based on the PIN. Zhang and DCS inferred protein functions through protein domain composition and PINs, and DSCP extends the protein functional similarity definition in DCS by combining the domain compositions of both proteins and complexes including them. PON and GrAPFI constructed a protein correlation network and characterized unknown proteins by integrating PINs and protein domain information.
Proteins in PINs were divided into two categories: the training set and the testing set. In one round of cross validation, the functions of each protein in the testing set are predicted according to the proteins in the training set. The validation process is repeated multiple times until each protein has a chance to become a member of the testing set. The final performances were evaluated by the average of all rounds. The first assessment criterion was leave-one-out cross-validation [10] which put one target protein into the testing set and the rest of proteins into the training set per round. However, the leave-one-out cross-validation was often plagued by many unannotated proteins in the network. Another assessment criterion used in this study was ten-fold cross validation [45], in which the proteins set was randomly divided into ten subsets, a single subset was retained for the testing set, and the remaining nine subsets were used as the training set. The cross-validation process was then repeated ten times, with each of the ten subsets used exactly once as the testing set. The ten results from the folds were then averaged to produce the final performance.
To assess the quality of predicted functions, we matched inferred functions with actual functions of target proteins. Precision and Recall were the commonly used measures to test the performance of function prediction methods. Precision is the fraction of predicted functions that are matched with known proteins while Recall is the fraction of known functions that are matched with predicted functions. In this study, true positive (TP), false positive (FP) and false negative (FN) represents the number of matched predicted functions, incorrectly matched predicted functions and missing matched known functions, respectively. Therefore, these two measures can be defined as follows:
| 14 |
| 15 |
F-measure, as the harmonic mean of Precision and Recall, was another measure to evaluate the performance of a method synthetically, which was calculated as follows:
| 16 |
Leave-one-out cross-validation
First, the leave-one-out cross validation was applied to verify the quality of predicted functions inferred by our NPF methods, as well as a representative set of competing methods: NC, Zhang, DCS, DSCP, PON and GrAPFI. To ensure impartiality, we filtered out those GO terms whose number of annotated proteins is less than 10 or more than 200 proteins. After being processed by this step, the number of GO terms is 267. Out of all the 4113 proteins in the PINs, 2716 proteins were annotated. The average and median number of GO terms for these annotated protein was 2.1 and 2, respectively. The NPF method obtained 2146 functional modules for these 2716 training proteins on the BioGRID databases. The average size and fitness value of the detected functional modules is 13.48 and 0.5625, respectively.
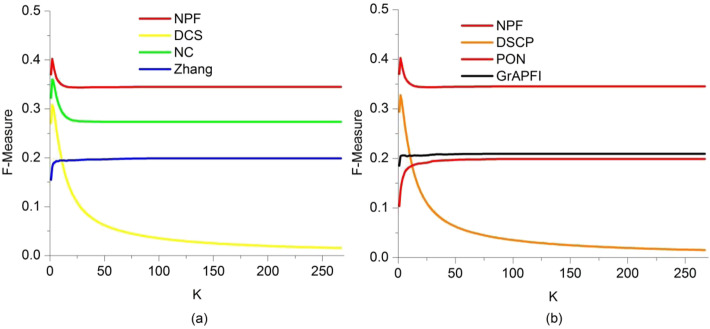
We first assessed the performance of NPF and six other competing methods on these target proteins by the average Precision, Recall and F-measure. The basic information about predicted functions by NPF and six other competing methods was presented in Table 3. In Table 3, MP was the number of proteins successfully matching at least one known function, while PMP represented the number of proteins perfectly matching the known functions, yet ZP was the number of proteins with zero-error prediction. MMP denoted the number of proteins completely mismatching the known functions. In other words, none of the predicted functions match the known functions. From Table 3, we can see that NPF contained the second-biggest number of perfect matching proteins (891) after NC (1428), while ZP of our method (885) is far more than NC’s (100). Figure 5 showed the overall comparison in terms of Precision, Recall and F-measure. It illustrated that NPF archives the largest value of Precision and F-measure, the second-largest value of Recall after NC. This is due to the maximum number of perfect matching proteins with NC. F-measure of NPF was 61.56, 109.41, 19.74, 11.53, 209.80 and 103.36% higher than NC, Zhang, DCS, DSCP, PON and GrAPFI, respectively.
Table 3.
Basic information of prediction by various algorithms
| Methods | MP | PMP | MMP | ZP |
|---|---|---|---|---|
| NPF | 1503 | 891 | 1213 | 885 |
| NC | 1945 | 1428 | 771 | 100 |
| Zhang | 727 | 421 | 1989 | 432 |
| DCS | 1269 | 743 | 1447 | 742 |
| DSCP | 1358 | 810 | 1358 | 799 |
| PON | 536 | 229 | 2180 | 277 |
| GrAPFI | 774 | 384 | 1942 | 432 |
This table shows the basic information of the results predicted by NPF, NC, Zhang, DCS, DSCP, PON and GrAPFI. MP is the number of proteins successfully matching at least one known function. PMP represents the number of proteins perfectly matching the known functions. MMP denotes the number of proteins completely mismatching the known functions. ZP is the number of proteins with zero-error prediction. That is, all the predicted functions in these proteins match the known functions
Fig. 5.
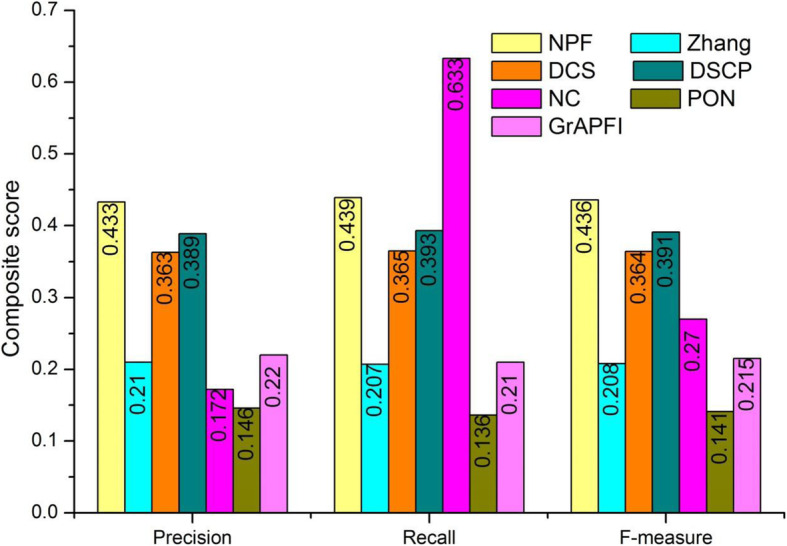
Overall comparisons of various methods. Numbers of each bar are the values for each score, including precision, recall and F-measure
To further investigate the performance of NPF and six other competing methods, we applied the Precision-Recall (PR) curve, whose vertical and horizontal coordination are the values of Precision and Recall, respectively. The PR curve is a standard for evaluation of the comprehensive performance of all methods in terms of different strategies of function selection. Predicted functions were ranked in descending order according to the values of functional similarity calculated by NPF, NC, PON and GrAPFI, respectively. Then, the top K functions were selected and annotated target proteins. The Parameter K changed from 1 to 267. As for the methods of Zhang, DSC and DSCP, top N (N < =K) proteins which had the highest similarity value with target proteins were selected and K functions in these fell out proteins were selected in turn to characterize target proteins. For a given target protein and the parameter K, the precision and recall values can be calculated according to the definition in Eqs. (14) and (15). The final PR curves of NPF and six other competing methods were drew according to the average precision and recall values over all target proteins. The PR curves of seven methods were illustrated in Fig. 4. Numbers in brackets represented the maximum F-measures for these seven methods. As shown in Fig. 6, NPF archived the first maximum F-measures in all methods. The PR curves of our method was above that of six other competing methods, which means that the NPF has a higher number of true positives and at the same time a smaller number of false positives when selecting different parameters. With the constant increase of K, the PR curve of NPF did not show drastic fluctuations. Even in the worst case, the precision value of NPF can still archive 0.248. However, the precision values of DSCP and DCS dropped sharply with the emergence of a large number of similar proteins.
Fig. 6.
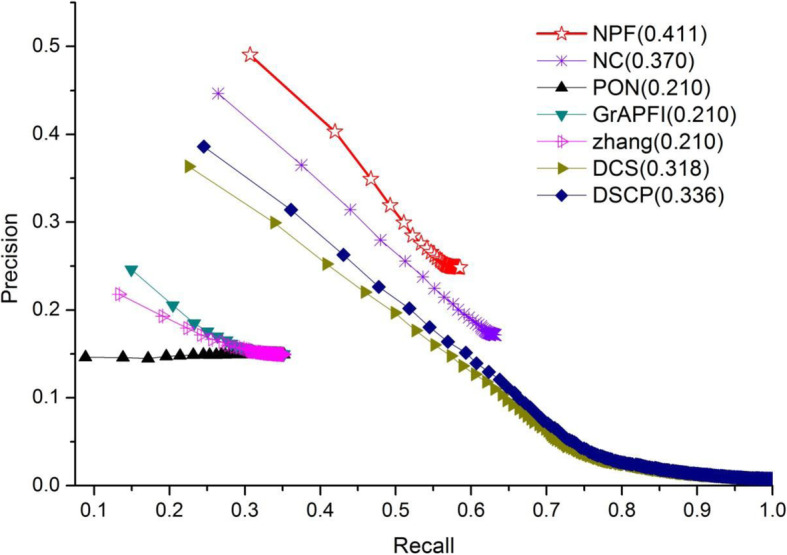
The precision-recall curves of NPF compared to six other competing methods. The figure denotes the precision-recall (PR) curves of NPF and six other competing methods (Zhang, DCS, DSCP, PON and GrAPFI) based on the average prediction performance over all testing protein. The vertical and horizontal coordination of the PR curves are the values of Precision and Recall, respectively. Numbers in brackets represent the maximum F-measures for these seven methods
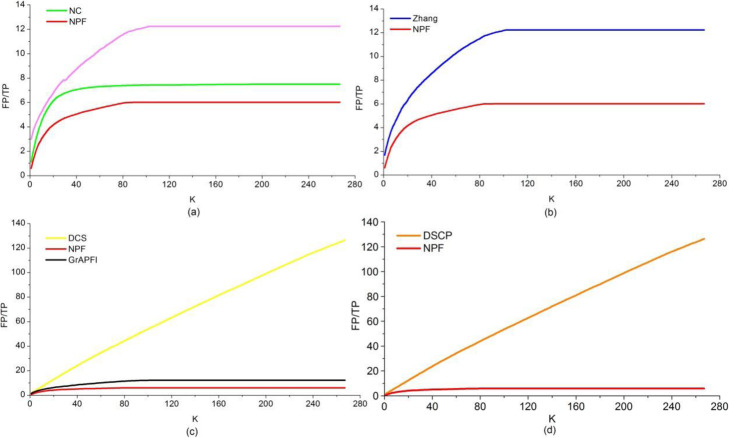
For overall comparison, we counted the number of true positive and false positive functions predicted by NPF and competing methods. A more valuable comparison between these methods was presented by plotting FP/TP curves as parameter K varies. Fig. 7 showed the FP/TP of our method and six other competing methods fluctuated under various value of the parameter K (ranging from 1 to 267). The smaller slope of the FP/TP curve of a method was, the lower the noise ratio was, which resulted in a greater predicted accuracy of the method. From this figure we can see that, FP/TP curve of NPF has consistently been covered with that of all other methods. That is, NPF generated the fewest false positives among all the methods when matching the same number of known functions.
Fig. 7.
The FP/TP curves of various methods. This Figure depicts the FP/TP of our method and other competing methods fluctuate under various value of the parameter K. The vertical and horizontal coordination of the curve are the values of FP/TP and K, respectively. a Shows the FP/TP curves of NC and NPF. b Shows the FP/TP curves of Zhang and NPF. c Shows the FP/TP curves of DCS, GrAPFI and NPF. d Shows the FP/TP curves of DSCP and NPF
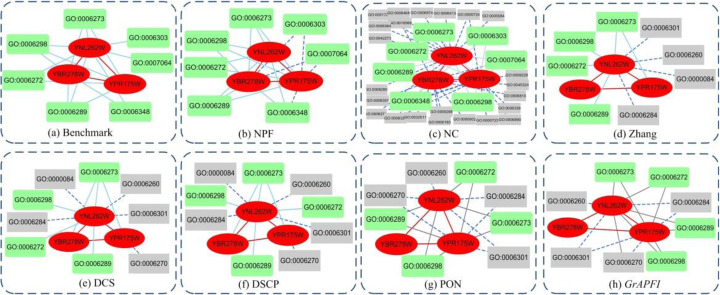
To further analyze the difference between NPF and six other competing methods, we selected YNL262W, YBR278W and YPR175W as examples and inferred proteins using the seven methods. Table 4 listed the basic information of these target proteins, including degree, number of domains and number of involving complexes. Figure 8 showed the predicted functions by various methods and the benchmark set. In Fig. 8, red elliptic nodes were target proteins, and red edges represented interactions between target proteins. Green round rectangle and grey rectangle nodes represented matched functions and false matched functions, respectively. Solid edges and dash edges between proteins and functions denoted correct and false associations. Table 5 showed the description of seven known functions of the three selected proteins. Take the protein YBR278W as an example, which does not contain any domains. For the three domain-based methods Zhang, PON and GrAPFI, no one function was inferred, let alone matched a known function. DCS and DSCP generated two predicted functions with one function matched by including neighbors or complex members for calculation of domain context similarities. The NC method annotated the protein YBR278W with functions of its all neighbors. Although the method successfully matched five functions, it introduced a large number of false-positive functions. Out of seven functions predicted by NPF, five functions were matched with known functions. This is due to the fact that we discovered more partners with similar functions through network propagation and got rid of some functionally unrelated proteins by detecting functional modules with high coupling. The example exhibited the highest predicting accuracy of NPF, compared to the results archived by other competing methods.
Table 4.
Basic information of selected target proteins
| Proteins | Degree | Number of domains | Number of complexes |
|---|---|---|---|
| YNL262W | 13 | 2 | 1 |
| YBR278W | 9 | 0 | 1 |
| YPR175W | 9 | 1 | 1 |
This table shows the basic information of three target proteins. The second column represents the number of its direct neighbors in the original PINs, while the third column is the number of domains it contains. The last column denotes the number of complexes involved
Fig. 8.
Functions of three selected proteins predicted by various methods. Red elliptic nodes denote target proteins, and red edges represent interactions between them. Green round rectangle and gray rectangle nodes represent matched functions and false matched functions respectively. Solid edges and dash edges between proteins and functions denote correct and false associations. (a) Benchmark results (b)-(h) is the result generated by NPF, Zhang, DCS, DSCP, PON and GrAPFI, respectively
Table 5.
Description of selected GO Terms
| GO Term | Description |
|---|---|
| GO:0006272 | Leading strand elongation, which is continuous as it proceeds in the same direction as the replication fork. |
| GO:0006273 | Lagging strand elongation proceeds by discontinuous synthesis of short stretches of DNA, known as Okazaki fragments, from RNA primers; these fragments are then joined by DNA ligase. |
| GO:0006289 | Nucleotide excision repair recognizes a wide range of substrates, including damage caused by UV irradiation and chemicals. |
| GO:0006298 | The mismatch repair system promotes genomic fidelity by repairing base-base mismatches, insertion-deletion loops and heterologies generated during DNA replication and recombination. |
| GO:0006303 | The repair of a double-strand break in DNA in which the two broken ends are re-joined with little or no sequence complementarity. |
| GO:0006348 | Chromatin silencing at telomere means the repression of transcription of telomere DNA by altering the structure of chromatin. |
| GO:0007064 | Mitotic sister chromatid cohesion. The cell cycle process in which the sister chromatids of a replicated chromosome are joined along the entire length of the chromosome. |
The underscored text represents the name of GO Term
Ten-fold cross validation
In the previous section, we applied the leave-one-out cross-validation to exhibit the NPF’s improvement on function prediction compared to the state-of-the-art methods. However, in real-world applications, there are usually much more unknown proteins than just one. To do this we adopted the ten-fold validation to verify the validity of our method on PINs with less function information. The entire set of proteins was divided into ten equal sets randomly, nine of which were used for training and the remaining part was used for testing. The process is repeated 1000 times, each time using another testing set. We ran the functional annotation methods of NPF, Zhang, DCS, DSCP, PON and GrAPFI on PINs to get mean values and standard deviations of precision, recall and F-measure, as shown in Table 6. Additionally, predicted functions were ranked in descending order according to the values obtained by various method and the top K functions were selected to annotate target proteins. A more valuable comparison between these methods was presented by plotting PR curves and F-measure curves as the parameter K varies using the ten-fold validation. Figures 9 and 10 illustrated the PR curves and F-measure curves of various methods, respectively. Table 6, Figs. 9 and 10 exhibited the performance improvement of NPF compared to six other competing methods. Therefore, NPF seemed to be an effective method for characterizing unknown proteins.
Table 6.
The prediction results using ten-fold validation
| Methods | Precison | Recall | F-measure | |||
|---|---|---|---|---|---|---|
| mean value | standard deviations | mean value | standard deviations | mean value | standard deviations | |
| NPF | 0.424 | 0.025 | 0.429 | 0.022 | 0.426 | 0.022 |
| NC | 0.176 | 0.014 | 0.610 | 0.023 | 0.273 | 0.018 |
| Zhang | 0.198 | 0.019 | 0.196 | 0.019 | 0.197 | 0.019 |
| DCS | 0.352 | 0.025 | 0.354 | 0.027 | 0.353 | 0.025 |
| DSCP | 0.378 | 0.027 | 0.382 | 0.028 | 0.380 | 0.027 |
| PON | 0.139 | 0.017 | 0.129 | 0.016 | 0.134 | 0.016 |
| GrAPFI | 0.219 | 0.018 | 0.209 | 0.018 | 0.214 | 0.018 |
Fig. 9.
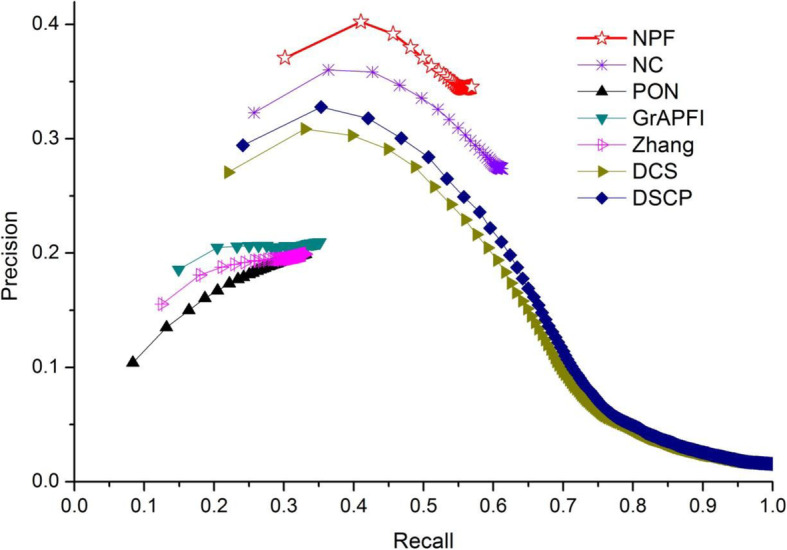
The precision-recall curves of various methods using ten-fold validation. This Figure shows the PR curves of NPF and six other methods using ten-fold validation. The entire set of proteins is divided into ten equal sets randomly, nine of which are used for training and the remaining part is used for testing. The process is repeated 1000 times, each time using another testing set
Fig. 10.
The F-measure curves of various methods using ten-fold validation. This Figure depicts the F-measure of seven methods fluctuate under various value of the parameter K. The vertical and horizontal coordination of the curve are the values of F-measure and K, respectively. a Shows the F-measure curves of NC, DCS, Zhang and NPF. b Shows the F-measure curves of DSCP, GrAPFI, PON and NPF
Discussions
The accurate annotation of protein functions is the key to understanding life at the molecular level and plays an important role in disease treatment, new drug development. Limited by the quality of protein interaction data generated by high-throughput technologies, methods that infer protein functions in terms of protein interactions may not work well [10]. A popular optimization scheme for the problem is to infer protein functions by combining PINs with multiple biological data. Despite the advances in these methods, designing efficient algorithms to fuse these multi-source biological data remains challenging. Additionally, the topology of the PINs, such as the “small world”, is also one of the factors that affect the prediction performances. Here, we presented the NPF, a network propagation-based method to annotate functions for target proteins. To overcome the problem of incomplete and false interaction data, we constructed a propagation network by integrating knowledge of the protein interaction network architecture, protein-domain associations and protein-complex associations. By propagating functional similarities across the networks, we can obtain more functionally relevant interacting partners to characterize the target proteins, which effectively free from the constraints of the “small-world” characteristic. Additionally, we take out those redundant function-independent partners by forming functional modules with high cohesion. Comprehensive comparisons among the state-of-the-art methods and our method have been made in terms of the leave-one-out cross-validation and the ten-fold cross validation. Experimental results demonstrated that our method outperforms other competing methods. Specially, DSCP used the same kind of data as NPF, yet NPF outperformed DSCP. There are two reasons to believe that NPF probably come out much better in the comparison. On the one hand, NPF can discover more neighbors with similar functions through network propagation; on the other hand, NPF predicted functions using multiple neighbors, not just the closest neighbors. In my opinion, proteins may be involved in different functional modules to perform multiple biological functions. Based on these results, we can conclude that the network propagation is useful for the study of protein interaction networks.
Conclusions
In this study, we proposed a novel protein functions annotation method based on network propagation, named NPF, which incorporates the topology of PINS and multiple biological data, such as domain annotation information, protein complexes information. Furthermore, we guarantee the NPF against false functions by detecting functional modules based on the neighbour fitness. Experimental comparison results between NPF and six state-of-the-art methods on yeast PINs showed that NPF significantly outperforms other competing methods. In our future study, we will take the hierarchical structure of GO Terms into account for further improvement of the performance of function prediction.
Supplementary information
Additional file 1 Algorithm convergence. This file provides the proof of the Algorithm 1 convergence about the effect of parameter α and ∂ on the speed of convergence.
Acknowledgements
Not applicable.
Abbreviations
- PIN
Protein interaction network
- NPF
Network propagation for functions prediction
- DCS
Domain combination similarity
- DSCP
Domain combination similarity in context of protein complexes
- PON
Protein overlap network
- INGA
Interaction network go annotator
- PANNZER
Protein ANNotation with Z-scoRE
- NC
Neighbourhood-counting
- PN
Propagation network
- GO
Gene ontology
- TP
True positive
- FP
False positive
- FN
False negative
Authors’ contributions
BHZ, ZHZ and LW designed the NPF method; SH, YCL and LW analysed data for the work; BHZ and MPJ drafted and revised the manuscript. The authors read and approved the final manuscript.
Funding
This work was supported in part by the National Natural Science Foundation of China (61772089, 61873221, 61672447), Natural Science Foundation of Hunan Province (No. 2019JJ40325, No. 2018JJ3566, No. 2018JJ3565, No. 2018JJ4058), National Scientific Research Foundation of Hunan Province (19A048), Major Scientific and Technological Projects for collaborative prevention and control of birth defects in Hunan Province (2019SK1010), Hunan Provincial Key Laboratory of Industrial Internet Technology and Security (2019TP1011), and Hunan Provincial Key Laboratory of Nutrition and Quality Control of Aquatic Animals (2018TP1027).
The funding body played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
Publicly available datasets were analysed in this study. This data and the NGF program can be found here: https://github.com/husaiccsu/NPF.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Bihai Zhao and Zhihong Zhang contributed equally to this work.
Contributor Information
Meiping Jiang, Email: meipingjiang123@163.com.
Lei Wang, Email: wanglei@xtu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12859-020-03663-7.
References
- 1.Shumilin IA, Cymborowski M, Chertihin O, et al. Identification of unknown protein function using metabolite cocktail screening. Structure. 2012;20(10):1715–1725. doi: 10.1016/j.str.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwikowski B, Uetz P, Fields S. A network of protein–protein interactions in yeast. Nat Biotechnol. 2000;18(12):1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- 3.Bogdanov P, Singh AK. Molecular function prediction using neighborhood features. IEEE/ACM Trans Comput Biol Bioinformatics. 2009;7(2):208–217. doi: 10.1109/TCBB.2009.81. [DOI] [PubMed] [Google Scholar]
- 4.Cho YR, Zhang A. Predicting protein function by frequent functional association pattern mining in protein interaction networks. IEEE Trans Inf Technol Biomed. 2009;14(1):30–36. doi: 10.1109/TITB.2009.2028234. [DOI] [PubMed] [Google Scholar]
- 5.Vazquez A, Flammini A, Maritan A, et al. Global protein function prediction from protein-protein interaction networks. Nat Biotechnol. 2003;21(6):697–700. doi: 10.1038/nbt825. [DOI] [PubMed] [Google Scholar]
- 6.Nabieva E, Jim K, Agarwal A, et al. Whole-proteome prediction of protein function via graph-theoretic analysis of interaction maps. Bioinformatics. 2005;21(suppl_1):i302–i310. doi: 10.1093/bioinformatics/bti1054. [DOI] [PubMed] [Google Scholar]
- 7.Tang XW, Xiao Q, Yu K. Breast Cancer candidate gene detection through integration of subcellular localization data with protein-protein interaction networks. IEEE Trans Nanobioscience. 2020;19(3):556–561. doi: 10.1109/TNB.2020.2990178. [DOI] [PubMed] [Google Scholar]
- 8.Cozzetto D, Buchan DWA, Bryson K, et al. Protein function prediction by massive integration of evolutionary analyses and multiple data sources. BMC Bioinformatics. 2013;14(Suppl 3):S1. doi: 10.1186/1471-2105-14-S3-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang S, Chen H, Liu K, et al. Inferring protein function by domain context similarities in protein-protein interaction networks. BMC Bioinformatics. 2009;10(1):395. doi: 10.1186/1471-2105-10-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng W, Wang J, Cai J, et al. Improving protein function prediction using domain and protein complexes in PPI networks. BMC Syst Biol. 2014;8(1):35. doi: 10.1186/1752-0509-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang S, Zheng D, Standley DM, et al. A novel function prediction approach using protein overlap networks. BMC Syst Biol. 2013;7(1):61. doi: 10.1186/1752-0509-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarker B, Rtichie DW, Aridhi S. International Conference on Complex Networks and their Applications. Cham: Springer; 2018. Exploiting complex protein domain networks for protein function annotation; pp. 598–610. [Google Scholar]
- 13.Piovesan D, Giollo M, Leonardi E, et al. INGA: protein function prediction combining interaction networks, domain assignments and sequence similarity. Nucleic Acids Res. 2015;43(W1):W134–W140. doi: 10.1093/nar/gkv523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piovesan D, Tosatto SCE. INGA 2.0: improving protein function prediction for the dark proteome. Nucleic Acids Res. 2019;47(W1):W373–W378. doi: 10.1093/nar/gkz375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Törönen P, Medlar A, Holm L. PANNZER2: a rapid functional annotation web server. Nucleic Acids Res. 2018;46(W1):W84–W88. doi: 10.1093/nar/gky350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang F, Song H, Zeng M, et al. A deep learning framework for gene ontology annotations with sequence - and network-based information. IEEE/ACM Trans Comput Biol Bioinform. 2020;10.1109/TCBB.2020.2968882. 10.1109/TCBB.2020.2968882. [published online ahead of print, 2020 Jan 23]. [DOI] [PubMed]
- 17.Zhang FH, Song H, Zeng M, et al. DeepFunc: a deep learning framework for accurate prediction of protein functions from protein sequences and interactions. Proteomics. 2019;19(12):1900019. doi: 10.1002/pmic.201900019. [DOI] [PubMed] [Google Scholar]
- 18.Cowen L, Ideker T, Raphael BJ, et al. Network propagation: a universal amplifier of genetic associations. Nat Rev Genet. 2017;18(9):551. doi: 10.1038/nrg.2017.38. [DOI] [PubMed] [Google Scholar]
- 19.Martiniano HFMC, Asif M, Vicente AM, et al. International Meeting on Computational Intelligence Methods for Bioinformatics and Biostatistics. Cham: Springer; 2018. Network propagation-based semi-supervised identification of genes associated with autism Spectrum disorder; pp. 239–248. [Google Scholar]
- 20.Tong H, Faloutsos C, Pan J, et al. Random walk with restart: fast solutions and applications. Knowl Inf Syst. 2008;14(3):327–346. [Google Scholar]
- 21.Haveliwala TH. Topic-sensitive pagerank: a context-sensitive ranking algorithm for web search. IEEE Trans Knowl Data Eng. 2013;15(4):784–796. [Google Scholar]
- 22.Lebowitz JL, Krapivsky PL, Redner S, Ben-Naim E. A kinetic view of statistical physics. J Stat Phys. 2011;143(4):831–832. [Google Scholar]
- 23.Borgwardt KM, Ong CS, Schonauer S, et al. Protein function prediction via graph kernels. Bioinformatics. 2005;21(1):47–56. [DOI] [PubMed]
- 24.Zhao BH, Zhao YL, Zhang XX, et al. An iteration method for identifying yeast essential proteins from heterogeneous network. BMC Bioinformatics. 2019;20(1):1–13. Article number: 355 (2019). [DOI] [PMC free article] [PubMed]
- 25.Zhao BH, Han X, Liu XR, et al. A novel method to predict essential proteins based on diffusion distance networks. IEEE Access. 2020:29385–94.
- 26.Li H, Li T, Quang D, et al. Network propagation predicts drug synergy in cancers. Cancer Res. 2018;78(18):5446–5457. doi: 10.1158/0008-5472.CAN-18-0740. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W, Ma J, Ideker T. Classifying tumors by supervised network propagation. Bioinformatics. 2018;34(13):i484–i493. doi: 10.1093/bioinformatics/bty247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gottlieb A, Magger O, Berman I, et al. PRINCIPLE: a tool for associating genes with diseases via network propagation. Bioinformatics. 2011;27(23):3325–3326. doi: 10.1093/bioinformatics/btr584. [DOI] [PubMed] [Google Scholar]
- 29.Qian Y, Besenbacher S, Mailund T, et al. Identifying disease associated genes by network propagation. BMC Syst Biol. 2014;8(S1):S6. doi: 10.1186/1752-0509-8-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng L, Zhou D, Liu W, et al. Prioritizing human microbe-disease associations utilizing a node-information-based link propagation method. IEEE Access. 2020:31341–9.
- 31.Peng W, Li M, Chen L, et al. Predicting protein functions by using unbalanced random walk algorithm on three biological networks. IEEE/ACM Trans Comput Biol Bioinformatics. 2017;14(2):360–369. doi: 10.1109/TCBB.2015.2394314. [DOI] [PubMed] [Google Scholar]
- 32.Liang L, Chen V, Zhu K, et al. Integrating data and knowledge to identify functional modules of genes: a multilayer approach. BMC Bioinformatics. 2019;20(1):225. doi: 10.1186/s12859-019-2800-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stuart JM, Segal E, Koller D, et al. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302(5643):249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 34.Chua HN, Sung W, Wong L, et al. Exploiting indirect neighbours and topological weight to predict protein function from protein-protein interactions. Bioinformatics. 2006;22(13):1623–1630. doi: 10.1093/bioinformatics/btl145. [DOI] [PubMed] [Google Scholar]
- 35.Zhao BH, Hu S, Li XY, et al. An efficient method for protein function annotation based on multilayer protein networks. Hum Genomics. 2016;10(33):1–15. doi: 10.1186/s40246-016-0087-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang TH, Sicotte H, Tian Z, et al. Robust and efficient identification of biomarkers by classifying features on graphs. Bioinformatics. 2008;24(18):2023–2029. doi: 10.1093/bioinformatics/btn383. [DOI] [PubMed] [Google Scholar]
- 37.Vanunu O, Magger O, Ruppin E, et al. Associating genes and protein complexes with disease via network propagation. PLoS Comput Biol. 2010;6(1):e1000641. [DOI] [PMC free article] [PubMed]
- 38.Hartwell LH, Hopfield JJ, Leibler S, et al. From molecular to modular cell biology. Nature. 1999;402(6761):C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 39.Lancichinetti A, Fortunato S, Kertész J. Detecting the overlapping and hierarchical community structure in complex networks. New J Phys. 2009;11(3):033015. [Google Scholar]
- 40.Oughtred R, Stark C, Breitkreutz B, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019. [DOI] [PMC free article] [PubMed]
- 41.Huang JK, Carlin DE, Yu MK, et al. Systematic evaluation of molecular networks for discovery of disease genes. Cell Syst. 2018;6(4):484–495. doi: 10.1016/j.cels.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bateman A, Coin L, Durbin R, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32(suppl 1):D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pu S, Wong J, Turner B, et al. Up-to-date catalogues of yeast protein complexes. Nucleic Acids Res. 2009;37:825–831. doi: 10.1093/nar/gkn1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreno-Torres JG, Sáez JA, Herrera F. Study on the impact of partition-induced dataset shift on k-fold cross-validation. IEEE Trans Neural Netw Learn Syst. 2012;23(8):1304–1312. doi: 10.1109/TNNLS.2012.2199516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Algorithm convergence. This file provides the proof of the Algorithm 1 convergence about the effect of parameter α and ∂ on the speed of convergence.
Data Availability Statement
Publicly available datasets were analysed in this study. This data and the NGF program can be found here: https://github.com/husaiccsu/NPF.