Abstract
Besides lung drastic involvement, SARS-CoV-2 severely affected other systems including liver. Emerging epidemiological studies brought the attentions towards liver injury and impairment as a potential outcome of COVID19. Angiotensin-converting enzyme 2 (ACE2) and Transmembrane serine protease (TMPRSS2) are the main cell entry receptors of SARS-CoV-2. We have tested the ability of medications to regulate expression of SARS-CoV-2 receptors. Understanding that may reflect how such medications may affect the level of infectivity and permissibility of the liver following COVID-19. Using transcriptomic datasets, Toxicogenomic Project-Genomics Assisted Toxicity Evaluation System (Open TG-GATEs) and GSE30351, we have tested the ability of ninety common medications to regulate COVID-19 receptors expression in human primary hepatocytes. Most medications displayed a dose-dependent change in expression of receptors which could hint at a potentially more pronounced change with chronic use. The expression level of TMPRSS2 was increased noticeably with a number of medications such as metformin. Within the analgesics, acetaminophen revealed a dose-dependent reduction in expression of ACE2, while non-steroidal anti-inflammatory drugs had mixed effect on receptors expression. To confirm the observed effects on primary human hepatocytes, rat hepatocyte treatments data was obtained from DrugMatrix toxicogenomic database (GSE57805), which showed a similar ACE2 and TMPRSS2 expression pattern. Treatment of common co-morbidities often require chronic use of multiple medications, which may result in an additive increase in the expression of ACE2 and TMPRSS2. More research is needed to determine the effect of different medications on COVID-19 receptors.
Electronic supplementary material
The online version of this article (10.1007/s00204-020-02869-1) contains supplementary material, which is available to authorized users.
Keywords: ACE2, TMPRSS2, SARS-CoV-2, COVID-19, Liver, Medications, Metformin, Acetaminophen, Nsaids, Hepatocyte
Introduction
Besides the respiratory tissue, the SARS-CoV-2 infection has affected other systems such as gastrointestinal tract and liver. Emerging epidemiological studies have brought the attentions towards liver injury and liver impairment as a potential outcome of COVID19 infection. Liver impairment was reported in the previous coronavirus infections (Alsaad et al. 2018) affecting up to 60% percent of patients with SARS-COV-1 (Chau et al. 2004). Similarly, in COVID-19, more than half of patients developed abnormal levels of alanine aminotransferase and aspartate aminotransferase (AST) (Zhang et al. 2020). This liver abnormalities were not detected in pre-clinical stage of disease, and only started appearing with disease progression and appearance of infection symptoms. Moreover, higher liver dysfunction was observed with more severe presentations of COVID-19 and more admission to intensive care units (Guan et al. 2020; Huang et al. 2020). In the COVID-19, the resulting hepatic dysfunction is accompanied by abnormal coagulation and fibrinolytic pathways leading to worse outcomes (Tang et al. 2020; Thachil et al. 2020).
Liver impairment during COVID-19 infection could be due to several factors including the direct infection of liver cells with SARS-CoV-2 (Wang et al. 2020; Zhao et al. 2020), cytokine storm and inflammation, and drug-induced toxicity. To understand that, liver impairment needs to be correlated clinically with the course of the disease. Liver enzymes abnormalities early during infection could mostly be attributed to direct infection of liver cells; while late impairments could mostly be due to failure of other organs. Angiotensin-converting enzyme 2 (ACE2) and Transmembrane serine protease (TMPRSS2) are the main cell entry receptors of SARS-CoV-1 and SARS-CoV-2. Recently, several studies have shown the direct link between the number of receptors and the level of SARS-CoV-2 viral infection (Matsuyama et al. 2020; Monteil et al. 2020; Yang et al. 2007). We have tested the ability of ninety commonly used medications to regulate expression of SARS-CoV-2 receptors in liver cells. Understanding that may reflect how such medications may affect the level of infectivity and permissibility of the liver following COVID-19 infection (Boeckmans et al. 2020).
Results and discussion
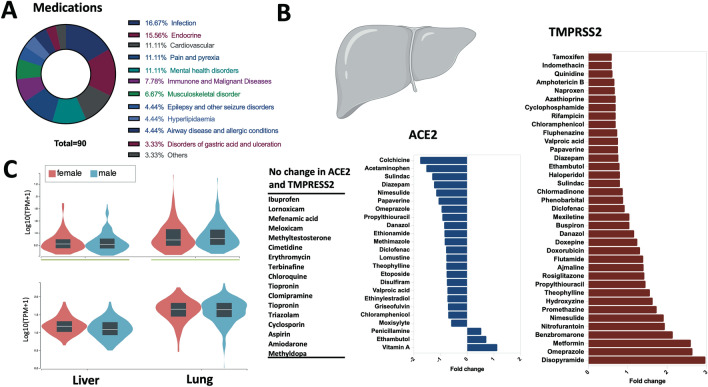
The effect of common medications on the expression of SARS-CoV-2 receptors was determined in vitro using human primary hepatocytes (Fig. 1a, b), which are known to express these receptors at a comparable level to bronchial lung tissue (Fig. 1c). Two doses, a high and a low of each medication, were tested. Most medications displayed a dose-dependent change in expression of entry genes which could hint at a potentially more pronounced change with chronic use of these medications. To confirm the observed effect of the medications on the expression of SARS-CoV-2 receptors in primary human hepatocytes, we have determined their expression in primary rat hepatocytes treated with part of the medications presented in Fig. 1b. Not all medications present in Fig. 1b were available in this dataset. A similar pattern of ACE2 and TMPRSS2 expression was observed following treatment of rat primary cells with all medications tested (Supplementary Fig. 1).
Fig. 1.
Effect of common medications on the expression levels of SARS-CoV-2 entry receptors in liver. a Represents the medication classes that their effect was tested for the expression levels of ACE2 or TMPRSS2 in the hepatocytes. b Shows selected medications with either no fold change, or a fold change of 0.5 or greater for ACE2 or TMPRSS2 expression levels. c Shows the baseline expression of ACE2 and TMPRSS2 in healthy liver and lung tissue of female and male extracted from the Genotype-Tissue Expression (GTEx) project
The expression level of TMPRSS2 was increased noticeably with a number of medications such as metformin, theophylline, and omeprazole (Fig. 1b). To our knowledge, this observed effect of these common medications on the liver expression of TMPRSS2 has not been reported previously. The increased in expression of TMPRSS2 will lead to an increase in priming of ACE2 and the virus spike protein and hence may increase the levels of infectivity (Wambier and Goren 2020). This transmembrane serine protease is known to be essential for activation of other viral infections with hepatic involvement such as hepatitis C (Esumi et al. 2015). This effect should, therefore, be considered when these medications are prescribed for individuals at high risk of hepatic infections.
Within the analgesics, acetaminophen revealed a dose-dependent reduction in expression level of ACE2, while the effect of non-steroidal anti-inflammatory drugs (NSAIDs) ranged between no expression change with ibuprofen and meloxicam and mixed effect of decreased ACE2 and increased TMPRSS2 expressions with diclofenac, naproxen, and nimesulide (Fig. 1b; Supplementary Table 1).
As for infection medications, griseofulvin decreased ACE2 expression, while nitrofurantoin caused dose-dependent increase in TMPRSS2. Interestingly, among medications used for COVID-19 infection, both erythromycin, which comes under the macrolide group of azithromycin, and hydroxychloroquine derivative displayed no change in expression of ACE2 and TMPRSS2. On the other hand, treatment with controversial corticosteroid medications like dexamethasone caused a slight decrease in ACE2 and an increase in TMPRSS2 expression (Supplementary Table 1).
Another interesting group of medications was anti-diabetics. In this group, metformin caused a dose-dependent increase in both of ACE2 and TMPRSS2; while rosiglitazone showed a decrease in ACE2 and an increase in TMPRSS2 (Fig. 1b; Supplementary Table 1).
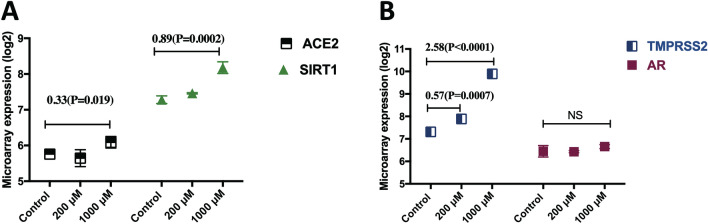
Metformin, a first-line treatment for type 2 diabetes, enhances the insulin secretion in response to glucose level elevation via the activation of SIRT1 (Yamamoto and Takahashi 2018). SIRT1 was shown to regulate ACE2 expression (Clarke et al. 2014), and hence the observed upregulation of ACE2 following metformin treatment could be due to Metformin induced SIRT1 expression (Fig. 2a). Another noticeable effect of metformin treatment was the increased expression of androgen-regulated gene TMPRSS2 by more than two-fold, with no significant change in the expression of androgen receptor. Therefore, metformin may increase TMPRSS2 through an androgen-independent mechanism (Fig. 2b).
Fig. 2.
Metformin caused a dose-dependent effect on gene expression of SARS-CoV-2 entry genes in primary human hepatocytes. a Treatment with 1000 μM metformin for 24 h significantly increased expression of both ACE2 and SIRT1 (n = 2/group). b Metformin caused a significant dose-dependent increase in mRNA levels of TMPRSS2, but it did not change expression of androgen receptor (AR). All treatments (n = 2/group) were conducted for 24 h
Treatment of common co-morbidities such as cardiovascular and metabolic disorders often requires chronic use of multiple medications, which may result in an additive increase in the expression of ACE2 and TMPRSS2. For example, metformin is often prescribed along with one or more other medications such as gemfibrozil and amitriptyline; both medications were found to increase ACE2 and TMPRSS2 expression (Supplementary Table 1).
Our results are based on public gene expression datasets and thus they may or may not reflect changes in protein expression. Therefore, confirmatory experiments at the mRNA and protein levels are needed to support our findings. Given the fact that SARS-CoV-2 infection does not increase expressions of ACE2 and TMPRSS2 (Hoffmann et al. 2020; Saheb Sharif-Askari et al. 2020), the combined effect of chronic use of these medications could affect liver susceptibility to the SARS-CoV-2 infection. Although the increased risk of developing severe COVID-19 infections should not be correlated solely with the use of medications, data presented here suggest that we should be vigilant about the potential medication effects. Obviously, more elaborative research is needed to guide proper usage of these medications and suggest safer alternatives.
Methods
Bioinformatic analyses were conducted to evaluate the effect of different groups of medications on expression levels of ACE2 and TMPRSS2 gene signatures in primary human hepatocytes. Publicly available gene expression datasets available via Toxicogenomics Project-Genomics Assisted Toxicity Evaluation System (Open TG-GATEs) (Igarashi et al. 2015), National Center for Biotechnology Information Gene Expression Omnibus (NCIB GEO, https://www.ncbi.nlm.nih.gov/geo) and the European Bioinformatics Institute (EMBL-EBI, https://www.ebi.ac.uk) were used. All the selected studies used the Affymetrix microarray platforms. Medication treatments were extracted from TG-GATEs database using the high and middle concentrations. The data for chloroquine derivative medication were extracted from the GSE30351 dataset. The expression of ACE2 and TMPRSS2 was also analyzed in medication treated primary rat hepatocytes (Supplementary Fig. 1). The rat in-invitro treatments were obtained from DrugMatrix toxicogenomic database (GSE57805). Hybridization to the whole genome was performed for all included samples using the GPL570 human and RG230_2.0 rat GeneChip (Affymetrix, CA). The details for the in vitro treatments, the Open TG-GATEs and DrugMatrix project are publicly available and previously published (Gusenleitner et al. 2014; Igarashi et al. 2015). Further, the information on dose, duration and number of treatments are provided in the Supplementary Table 1 and Supplementary Fig. 1.
Orthogonal data were extracted using single value decomposition (SVD) (Gusenleitner et al. 2014; Igarashi et al. 2015; Kossenkov and Ochs 2010; Uehara et al. 2010). All the included data were evaluated for quality control (QC). The QC was performed at each step of GeneChip analysis and covered details on background signals, corner signals, and expression of housekeeping genes (Gusenleitner et al. 2014; Igarashi et al. 2015; Kossenkov and Ochs 2010; Uehara et al. 2010). The raw Affymetrix data were normalized and log transformed. Microarray data (CEL files) were pre-processed with Robust Multi-Array Average (RMA) technique using R software (Hughey and Butte 2015). Log-transformed normalized intensities were used in Linear Models for MicroArray data (LIMMA) analyses to identify differentially expressed genes between treated and control hepatocytes. The number of RMA normalized gene array included was 20, 606 genes. We used the default Benjamini-Horchberg correction for multiple testing (Dudoit et al. 2002; Smyth Gordon 2004). Statistical analyses were performed using R software (v 3.0.2) and Prism (v8; GraphPad Software). For all analyses, p values < 0.05 were considered significant.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
RH, NSA, FSA, QH, and TK conceived and designed the experiments; NSA, FSA, BAM, EA, analyzed the data. MA, SA, RH, and SAH revised the manuscript. All authors contributed to writing and revision of the manuscript.
Funding
This research has been financially supported by Tissue Injury and Repair (TIR) group operational grant (Grant code: 150317); COVID-19 research grant; seed grant (Grant code: 2001090275); and by collaborative research grant (Grant code: 2001090278) to RH, University of Sharjah, UAE; and by a Sandooq Al Watan Applied Research & Development grant to RH; and by Prince Abdullah Ben Khalid Celiac Disease Research Chair, under the Vice Deanship of Research Chairs, King Saud University, Riyadh, Kingdom of Saudi Arabia.
Compliance with ethical standards
Conflict of interest
The authors have no conflicts of interest to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alsaad KO, et al. Histopathology of Middle East respiratory syndrome coronovirus (MERS-CoV) infection—clinicopathological and ultrastructural study. Histopathology. 2018;72:516–524. doi: 10.1111/his.13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmans J, Rodrigues RM, Demuyser T, Piérard D, Vanhaecke T, Rogiers V. COVID-19 and drug-induced liver injury: a problem of plenty or a petty point? Arch Toxicol. 2020 doi: 10.1007/s00204-020-02734-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau TN, et al. SARS-associated viral hepatitis caused by a novel coronavirus: report of three cases. Hepatology. 2004;39:302–310. doi: 10.1002/hep.20111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke NE, Belyaev ND, Lambert DW, Turner AJ. Epigenetic regulation of angiotensin-converting enzyme 2 (ACE2) by SIRT1 under conditions of cell energy stress. Clin Sci (Lond) 2014;126:507–516. doi: 10.1042/cs20130291. [DOI] [PubMed] [Google Scholar]
- Dudoit S, Yang YH, Callow MJ, Speed TP. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat Sin. 2002;12:111–139. [Google Scholar]
- Esumi M, et al. Transmembrane serine protease TMPRSS2 activates hepatitis C virus infection. Hepatology. 2015;61:437–446. doi: 10.1002/hep.27426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W-J, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusenleitner D, Auerbach SS, Melia T, Gómez HF, Sherr DH, Monti S. Genomic models of short-term exposure accurately predict long-term chemical carcinogenicity and identify putative mechanisms of action. PLoS ONE. 2014;9:e102579. doi: 10.1371/journal.pone.0102579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughey JJ, Butte AJ. Robust meta-analysis of gene expression using the elastic net. Nucleic Acids Res. 2015;43:e79–e79. doi: 10.1093/nar/gkv229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi Y, Nakatsu N, Yamashita T, Ono A, Ohno Y, Urushidani T, Yamada H. Open TG-GATEs: a large-scale toxicogenomics database. Nucleic Acids Res. 2015;43:D921–D927. doi: 10.1093/nar/gku955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossenkov AV, Ochs MF. Matrix factorisation methods applied in microarray data analysis. Int J Data Min Bioinform. 2010;4:72–90. doi: 10.1504/ijdmb.2010.030968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S, et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci. 2020;117:7001. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif-Askari NS, Sharif-Askari FS, Alabed M, Temsah M-H, Al Heialy S, Hamid Q, Halwani R. Airways expression of SARS-CoV-2 receptor, ACE2, and TMPRSS2 is lower in children than adults and increases with smoking and COPD. Mol Ther Methods Clin Dev. 2020;18:1–6. doi: 10.1016/j.omtm.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth Gordon K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18:844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thachil J, et al. ISTH interim guidance on recognition and management of coagulopathy in COVID-19. J Thromb Haemost. 2020 doi: 10.1111/jth.14810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Ono A, Maruyama T, Kato I, Yamada H, Ohno Y, Urushidani T. The Japanese toxicogenomics project: application of toxicogenomics. Mol Nutr Food Res. 2010;54:218–227. doi: 10.1002/mnfr.200900169. [DOI] [PubMed] [Google Scholar]
- Wambier CG, Goren A. SARS-COV-2 infection is likely to be androgen mediated. J Am Acad Dermatol. 2020 doi: 10.1016/j.jaad.2020.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. SARS-CoV-2 infection of the liver directly contributes to hepatic impairment in patients with COVID-19. J Hepatol. 2020 doi: 10.1016/j.jhep.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Takahashi Y. The essential role of SIRT1 in hypothalamic-pituitary axis. Front Endocrinol (Lausanne) 2018 doi: 10.3389/fendo.2018.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X-H, et al. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp Med. 2007;57:450–459. [PubMed] [Google Scholar]
- Zhang C, Shi L, Wang F-S. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol Hepatol. 2020;5:428–430. doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, et al. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell. 2020 doi: 10.1007/s13238-020-00718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.