Abstract
Pyronaridine, tilorone, and quinacrine were recently identified by a machine learning model and demonstrated in vitro and in vivo activity against Ebola virus (EBOV) and represent viable candidates for drug repurposing. The target for these molecules was previously unknown. These drugs have now been docked into the crystal structure of the ebola glycoprotein and then experimentally validated in vitro using microscale thermophoresis to generate Kd values for tilorone (0.73 μM), pyronaridine (7.34 μM), and quinacrine (7.55 μM). These molecules were shown to bind with a higher affinity than the previously reported toremifene (16 μM). These three structures provide more insight into the structural diversity of ebola glycoprotein inhibitors which can be utilized in the discovery and design of additional inhibitors.
Keywords: Antiviral, ebola, glycoprotein, pyronaridine, tilorone, quinacrine
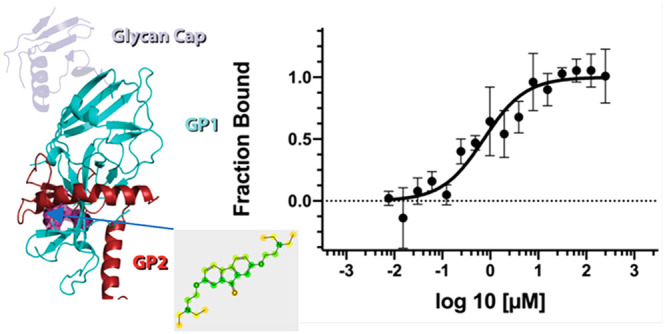
The most recent Ebola virus (EBOV) outbreak in the Democratic Republic of the Congo resulted in the deaths of over 2200 people according to the World Health Organization.1 While there is an approved vaccine for prevention of EBOV disease,2 we have no approved antivirals to treat patients, although there are several treatments that have reached the clinic3−6 with the most promising results for biologics only. One target of particular interest for antibody therapies7 is the glycoprotein which is composed of GP1 and GP2 subunits and is involved in attachment to the cell and entry.8 Earlier high throughput screens had identified benzodiazepine analogues as GP1 inhibitors which act early in viral entry.8,9 Several structurally diverse U.S. Food and Drug Administration approved drugs have also been identified including toremifene, benztropine, bepridil, paroxetine, sertraline, and ibuprofen10 which all bind in the same site at the tunnel entrance to the glycoprotein, destabilizing it and resulting in release of GP2, thus preventing fusion between virus and endosome.9 Several other research groups have identified small molecules that inhibit glycoprotein, suggesting that a wide array of molecules may bind.11−15 Others have used computational approaches to perform virtual screens to identify inhibitors of GP2.16 Recent efforts to identify small molecule drugs for EBOV have included using computational methods in the form of a Bayesian machine learning (ML) approach trained with EBOV inhibitors.17,18 This ML model enabled a virtual screen and selection of three compounds, tilorone, quinacrine, and pyronaridine tetraphosphate19 (Figure 1). All these drugs inhibited EBOV in vitro in HeLa cells (IC50’s of 1.14–1.48, 1.05–1.48, and 0.82–1.30 μM for tilorone, quinacrine, and pyronaridine tetraphosphate, respectively, across three strains of EBOV20) and in vivo in the mouse-adapted EBOV (ma-EBOV) efficacy model.21−24 Pyronaridine tetraphosphate also demonstrated significant activity in the guinea pig adapted model of EBOV infection20 and is currently used as an antimalarial in combination with artesunate (Pyramax). Most recently, pyronaridine was shown to be a potent lysosomotropic agent while artesunate which is also active against EBOV in vitro was not.25 Combining these two drugs had an additive effect on inhibiting EBOV replication in vitro and reduced cytotoxicity.25 Pursuit of the potential target of these computationally identified drugs pointed us to docking compounds in the glycoprotein structure and experimental validation.
Figure 1.
Molecular structures of compounds evaluated in this study.
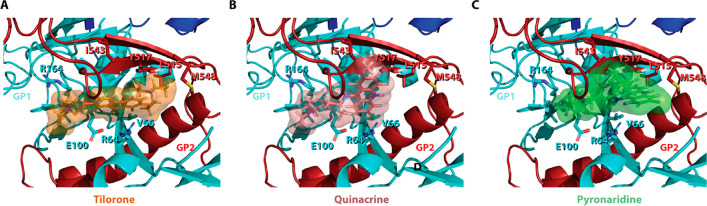
Tilorone had the best libdock score of 132.91 followed by pyronaridine (119.02) and quinacrine (111.56) (Figure 2). Binding energies for these docking poses were also calculated post ligand minimization but did not correlate with the libdock scores (data not shown). This may be due to the limitations of the CHARMM force fields used or other parameters, which may not have accurately estimated binding energy due to the complex π-system interactions and may require more robust calculations such as density functional theory. As the binding site used for docking is speculated based on the crystallographic position of toremifene, we also do not rule out the distinct possibility that the site of interaction may be different for one or all of these molecules. The main proposed electrostatic interaction between tilorone and quinacrine is a cation−π interaction with K64, with tilorone having a more idealized distance (Figure 3A and B). This differs with pyronaridine which has a π-stacking with Y517 (Figure 3C). Many studies have shown that cation−π interactions can enhance binding energies by 2–5 kcal/mol,26 and model calculations on benzene and toluene dimers (parallel and off center) suggest that these energies of interaction are in the range of 1–4 kcal/mol.27,28 This would suggest that these interactions may be comparable. The docking suggests that there are additional hydrophobic interactions that occur between tilorone and the EBOV GP protein. Tilorone and pyronaridine share interactions with residues L515 and M548. However, tilorone has additional substantial hydrophobic interactions with residues V66 and Y517. In addition, tilorone has nonpolar interactions with R164. Pyronaridine does have an additional interaction with I544 that is not shared with tilorone. Quinacrine shares some of the same interactions, specifically with I544 and V66 with a weaker cation−π interaction with R64 and nonpolar interactions with E100 and R164. All of these interactions are visualized in Figure 3.
Figure 2.
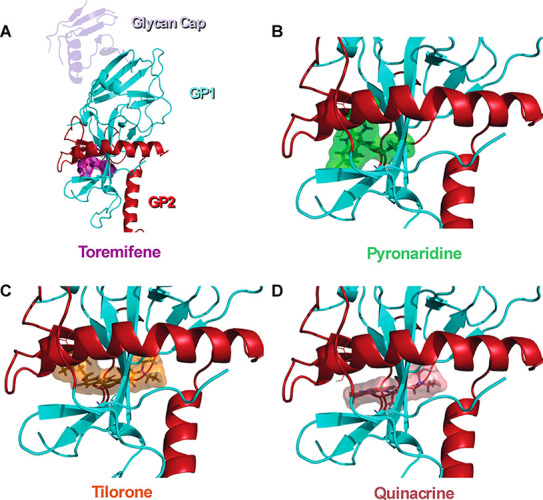
Docking of ligands in EBOV glycoprotein crystal structure showing lowest energy poses from the rigid docking (libdock). Cartoon representations of EBOV glycoprotein (GP1(Cyan)-GP2(Red)) with a glycan cap (purple). (A) Crystal structure of EBOV GP in complex with toremifene (PDB ID: 5JQ7; 2.69 Å). Top scoring docked poses of pyronaridine (B, libdock score: 119.016), tilorone (C, libdock score: 132.906), and quinacrine (D, libdock score: 111.558) in EBOV GP using libdock.
Figure 3.
Cartoon representation of the crystal structure of EBOV GP (PDB ID: 5JQ7; 2.69 Å) with side chains of the residues that form predicted interactions with the compounds tilorone (A), quinacrine (B), and pyronaridine (C) shown in a stick representation.
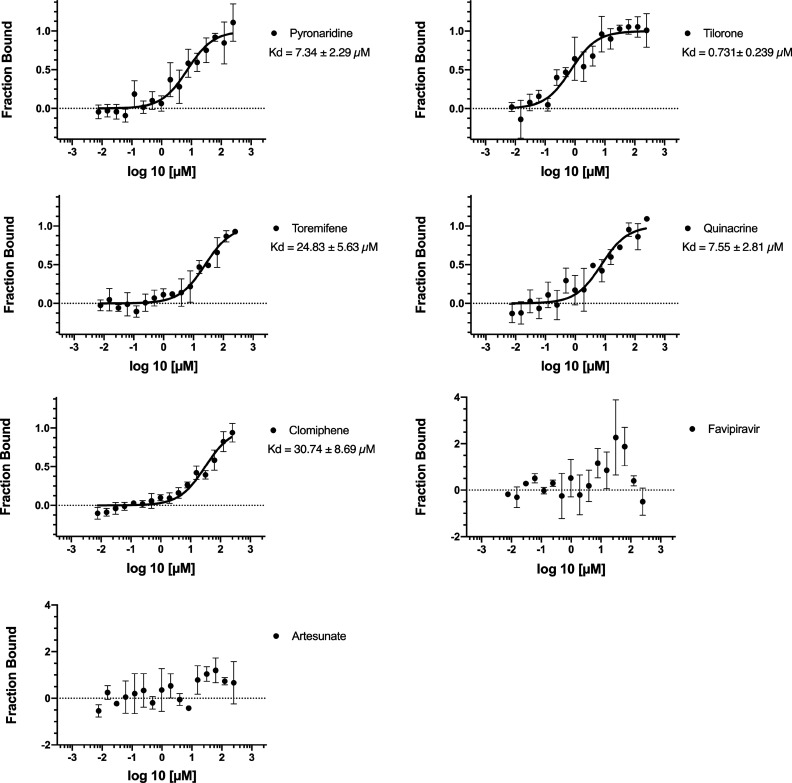
These docking predictions were then validated using microscale thermophoresis in order to calculate dissociation constants (Kd). The Kd values for pyronaridine (7.34 μM), tilorone (0.73 μM), and quinacrine (7.55 μM) were lower than that for the positive control toremifene (24.83 μM) (Figure 4) which is very similar to the literature value of 16 μM run under similar conditions.10 Additionally, we identified clomiphene (30.74 μM) (Figure 1), whereas favipiravir and artesunate did not bind to the EBOV glycoprotein (Figure 4). Favipiravir is a well-known EBOV RNA polymerase inhibitor and would not be expected to bind to the glycoprotein.29−32
Figure 4.
Ebola glycoprotein Kd values generated using microscale thermophoresis for test compounds.
We have previously demonstrated that artesunate does not inhibit lysotracker25 and is likely inhibiting EBOV via a different mechanism from that of pyronaridine with which it is used in combination as the drug Pyramax for malaria.33 The lack of inhibition of EBOV glycoprotein by artesunate provides further confirmation of this. Prior work on pyronaridine has demonstrated that it has promising in vitro activity against EBOV as well as excellent in vitro absorption, distribution, metabolism, and excretion (ADME) properties and a very long half-life across mice and humans.22 It also has statistically significant in vivo efficacy in mice and guinea pigs infected with EBOV.20 While the mechanism to date was unclear, it has been shown to inhibit lysostracker, suggesting a lysosomotropic effect.25 When this is coupled with the potential to bind to glycoprotein, this dual mechanism will likely prevent EBOV entry. Similarly, quinacrine is a small-molecule, orally bioavailable drug which has also been used clinically as an antimalarial. This also has very favorable ADME properties (apart from potent CYP2D6 inhibition), and it demonstrated in vivo activity in mice infected with EBOV.21 Tilorone is structurally different and is used in eastern Europe as an antiviral. It has excellent in vitro ADME properties, and it was recently demonstrated to have potent in vitro inhibition of EBOV as well as efficacy in mice infected with EBOV.23 Interestingly, this molecule did not demonstrate efficacy in the guinea pig EBOV model likely due to the significant species differences in metabolism when compared to the mouse model.20 Tilorone has also demonstrated in vitro inhibition of MERS,34 is under evaluation as a potential treatment for other coronaviruses such as SARS-CoV-2,24 and has shown a low μM IC50.35 This current work now provides further detail as to how these three molecules target EBOV glycoprotein as well as being lysosomotropic, enabling their previously reported blocking of viral entry. This information may aid in structure guided modification of these compounds and future X-ray crystallography. Such mechanistic insights may also aid drug discovery for other viruses (e.g., SARS-CoV-2). To date tilorone would appear to be the highest affinity compound for EBOV glycoprotein compared to the previously reported toremifene.10,15
In conclusion, the accumulated in vitro and in vivo data gathered for tilorone, quinacrine, and pyronaridine points to them sharing a common target or mechanism for the inhibition of EBOV.20−23,25,36 It would also appear all of these molecules block viral entry, are lysosomotropic, and are now identified to bind to the glycoprotein. Shedding further light on how these molecules work in vitro may provide further justification for clinically repurposing these compounds in future outbreaks.
Experimental Procedures
Materials
Pyronaridine tetraphosphate [4-[(7-chloro-2-methoxybenzo[b][1,5]naphthyridin-10-yl)amino]-2,6-bis(1-pyrrolidinylmethyl)phenol phosphate (1:4)]19 was purchased from BOC Sciences (Shirley, NY). Tilorone and quinacrine were purchased from Cayman Chemicals (Ann Arbor, MI). Toremifene and clomiphene were purchased from MedChemExpress (Monmouth Junction, NJ), and favipiravir was from TRC Canada (North York, ON, Canada). Zaire ebolavirus disulfide-linked glycoprotein heterodimer (GP1-GP2) was purchased from Novus Biologicals (Centennial, CO). According to the manufacturer, EBOV GP protein is purified from CHO-derived viral expression with previous internal verification of significant glycosylation.
Docking
Three EBOV inhibitors, tilorone, pyronaridine, and quinacrine, were docked into the EBOV glycoprotein structure (PDB: 5JQ7) at the same site as previously described for toremifene10 using Discovery Studio LibDock. Binding energies were calculated following a ligand minimization (rigid protein).
Microscale Thermophoresis
An amount of 200 μg of lyophilized protein was resuspended in RED-NHS second Generation labeling buffer (NanoTemper; Cambridge, MA). This was followed by the labeling of the primary amines using the RED-NHS dye according to the manufacturer’s protocol. Labeled protein was buffer exchanged into 10 mM MES, pH 5.0, 150 mM NaCl, 170 mM sodium malonate at pH 5.2 (MST buffer), which is similar to a buffer previously shown to be appropriate for EBOV GP,10 and then diluted to a final concentration of 1 μM. For each experimental compound, 16 independent stocks were made in DMSO using 2-fold serial dilution (10 mM initial concentration). The MST buffer used for a final dilution prior to MST was supplemented with 0.05% Tween 20 and 10 mM BME. The protein was diluted to 2.5 nM in the supplemented MST buffer, and 19.5 μL of this was combined with 0.5 μL of the compound stock and then mixed thoroughly. This resulted in 2-fold serial dilution testing series with the highest and lowest concentrations of 250 μM and 7.629 nM, respectively, with a consistent final DMSO concentration of 2.5%. These reactions were incubated for 20–30 min prior to transferring to standard Monolith NT.115 capillaries. Experiments were run at 20% excitation and high MST power at 23.0 °C on a Monolith NT.115Pico instrument (NanoTemper). Favipiravir and toremifene were also run as the negative and positive controls, respectively. Each experimental compound was run in quadruplicate.
The data were acquired with MO.Control 1.6.1 (NanoTemper Technologies).
Recorded data were analyzed with MO.Affinity Analysis 2.3 (NanoTemper
Technologies). The dissociation constant Kd quantifies the equilibrium of the reaction of the labeled molecule
A (concentration cA) with its target T
(concentration cT) to form the complex
AT (concentration cAT) and is defined
by the law of mass action as 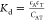 where all concentrations are “free”
concentrations. During the titration experiments, the concentration
of the labeled molecule A is kept constant and the concentration of
added target T is increased. These concentrations are known and can
be used to calculate the dissociation constant. The free concentration
of the labeled molecule A is the added concentration minus the concentration
of formed complex AT. The Kd is calculated
as
where all concentrations are “free”
concentrations. During the titration experiments, the concentration
of the labeled molecule A is kept constant and the concentration of
added target T is increased. These concentrations are known and can
be used to calculate the dissociation constant. The free concentration
of the labeled molecule A is the added concentration minus the concentration
of formed complex AT. The Kd is calculated
as 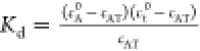 . The fraction of bound molecules x can be derived from Fnorm,
where Fnorm(A) is the normalized fluorescence
of only unbound labeled molecules A and Fnorm(AT) is the normalized fluorescence of complexes AT of labeled as
shown by the equation:
. The fraction of bound molecules x can be derived from Fnorm,
where Fnorm(A) is the normalized fluorescence
of only unbound labeled molecules A and Fnorm(AT) is the normalized fluorescence of complexes AT of labeled as
shown by the equation: 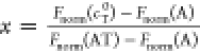 . The MST traces that showed aggregation
or outliers were removed from the data sets prior to Kd determination.
. The MST traces that showed aggregation
or outliers were removed from the data sets prior to Kd determination.
Acknowledgments
We would like to sincerely thank Dr. Peter Madrid, Dr. Jason Comer, Dr. Julie Dyall, Dr. Manu Anantpadma, and Dr. Robert Davey for many Ebola related discussions and collaborations. Dr. Ana Puhl Is gratefully acknowledged for discussions on MST.
Author Contributions
T.R.L. performed all experimental work and data analysis and helped draft the manuscript. S.E. designed the study and wrote the manuscript.
We kindly acknowledge NIH funding: R21TR001718 from NCATS as well as 1R43GM122196-01 and R44GM122196–02A1 “Centralized assay datasets for modelling support of small drug discovery organizations” from NIGMS.
The authors declare the following competing financial interest(s): S.E. is CEO of Collaborations Pharmaceuticals, Inc. T.R.L. is an employee at Collaborations Pharmaceuticals, Inc. Collaborations Pharmaceuticals, Inc. has obtained FDA orphan drug designations for pyronaridine, tilorone, and quinacrine for use against Ebola.
References
- Ilunga Kalenga O.; Moeti M.; Sparrow A.; Nguyen V. K.; Lucey D.; Ghebreyesus T. A. The Ongoing Ebola Epidemic in the Democratic Republic of Congo, 2018–2019. N. Engl. J. Med. 2019, 381, 373–383. 10.1056/NEJMsr1904253. [DOI] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration . First FDA-approved vaccine for the prevention of Ebola virus disease, marking a critical milestone in public health preparedness and response. https://www.fda.gov/news-events/press-announcements/first-fda-approved-vaccine-prevention-ebola-virus-disease-marking-critical-milestone-public-health (accessed 2020-04-05).
- Qiu X.; Wong G.; Audet J.; Bello A.; Fernando L.; Alimonti J. B.; Fausther-Bovendo H.; Wei H.; Aviles J.; Hiatt E.; Johnson A.; Morton J.; Swope K.; Bohorov O.; Bohorova N.; Goodman C.; Kim D.; Pauly M. H.; Velasco J.; Pettitt J.; Olinger G. G.; Whaley K.; Xu B.; Strong J. E.; Zeitlin L.; Kobinger G. P. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 2014, 514 (7520), 47–53. 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D.; Misasi J.; Mulangu S.; Stanley D. A.; Kanekiyo M.; Wollen S.; Ploquin A.; Doria-Rose N. A.; Staupe R. P.; Bailey M.; Shi W.; Choe M.; Marcus H.; Thompson E. A.; Cagigi A.; Silacci C.; Fernandez-Rodriguez B.; Perez L.; Sallusto F.; Vanzetta F.; Agatic G.; Cameroni E.; Kisalu N.; Gordon I.; Ledgerwood J. E.; Mascola J. R.; Graham B. S.; Muyembe-Tamfun J. J.; Trefry J. C.; Lanzavecchia A.; Sullivan N. J. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science 2016, 351 (6279), 1339–42. 10.1126/science.aad5224. [DOI] [PubMed] [Google Scholar]
- Sivapalasingam S.; Kamal M.; Slim R.; Hosain R.; Shao W.; Stoltz R.; Yen J.; Pologe L. G.; Cao Y.; Partridge M.; Sumner G.; Lipsich L. Safety, pharmacokinetics, and immunogenicity of a co-formulated cocktail of three human monoclonal antibodies targeting Ebola virus glycoprotein in healthy adults: a randomised, first-in-human phase 1 study. Lancet Infect. Dis. 2018, 18 (8), 884–893. 10.1016/S1473-3099(18)30397-9. [DOI] [PubMed] [Google Scholar]
- Mulangu S.; Dodd L. E.; Davey R. T. Jr.; Tshiani Mbaya O.; Proschan M.; Mukadi D.; Lusakibanza Manzo M.; Nzolo D.; Tshomba Oloma A.; Ibanda A.; Ali R.; Coulibaly S.; Levine A. C.; Grais R.; Diaz J.; Lane H. C.; Muyembe-Tamfum J. J.; Group P. W.; Sivahera B.; Camara M.; Kojan R.; Walker R.; Dighero-Kemp B.; Cao H.; Mukumbayi P.; Mbala-Kingebeni P.; Ahuka S.; Albert S.; Bonnett T.; Crozier I.; Duvenhage M.; Proffitt C.; Teitelbaum M.; Moench T.; Aboulhab J.; Barrett K.; Cahill K.; Cone K.; Eckes R.; Hensley L.; Herpin B.; Higgs E.; Ledgerwood J.; Pierson J.; Smolskis M.; Sow Y.; Tierney J.; Sivapalasingam S.; Holman W.; Gettinger N.; Vallee D.; Nordwall J.; Team P. C. S. A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. N. Engl. J. Med. 2019, 381 (24), 2293–2303. 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire E. O.; Schendel S. L.; Fusco M. L.; Gangavarapu K.; Gunn B. M.; Wec A. Z.; Halfmann P. J.; Brannan J. M.; Herbert A. S.; Qiu X.; Wagh K.; He S.; Giorgi E. E.; Theiler J.; Pommert K. B. J.; Krause T. B.; Turner H. L.; Murin C. D.; Pallesen J.; Davidson E.; Ahmed R.; Aman M. J.; Bukreyev A.; Burton D. R.; Crowe J. E. Jr.; Davis C. W.; Georgiou G.; Krammer F.; Kyratsous C. A.; Lai J. R.; Nykiforuk C.; Pauly M. H.; Rijal P.; Takada A.; Townsend A. R.; Volchkov V.; Walker L. M.; Wang C. I.; Zeitlin L.; Doranz B. J.; Ward A. B.; Korber B.; Kobinger G. P.; Andersen K. G.; Kawaoka Y.; Alter G.; Chandran K.; Dye J. M. Viral Hemorrhagic Fever Immunotherapeutic, C., Systematic Analysis of Monoclonal Antibodies against Ebola Virus GP Defines Features that Contribute to Protection. Cell 2018, 174 (4), 938–952.e13. 10.1016/j.cell.2018.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A.; Li B.; Mills D. M.; Panchal R. G.; Cardinale S. C.; Butler M. M.; Peet N. P.; Majgier-Baranowska H.; Williams J. D.; Patel I.; Moir D. T.; Bavari S.; Ray R.; Farzan M. R.; Rong L.; Bowlin T. L. Identification of a small-molecule entry inhibitor for filoviruses. J. Virol 2011, 85 (7), 3106–19. 10.1128/JVI.01456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J.; Zhao Y.; Fry E. E.; Stuart D. I. Target Identification and Mode of Action of Four Chemically Divergent Drugs against Ebolavirus Infection. J. Med. Chem. 2018, 61 (3), 724–733. 10.1021/acs.jmedchem.7b01249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Ren J.; Harlos K.; Jones D. M.; Zeltina A.; Bowden T. A.; Padilla-Parra S.; Fry E. E.; Stuart D. I. Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature 2016, 535 (7610), 169–172. 10.1038/nature18615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q.; Cheng H.; Xiong R.; Zhang G.; Du R.; Anantpadma M.; Davey R. A.; Rong L. Identification of Diaryl-Quinoline Compounds as Entry Inhibitors of Ebola Virus. Viruses 2018, 10 (12), 678. 10.3390/v10120678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Ma L.; Yi D.; Wang H.; Wang J.; Zhang Y.; Guo Y.; Li X.; Zhou J.; Shi Y.; Gao G. F.; Cen S. Novel cyclo-peptides inhibit Ebola pseudotyped virus entry by targeting primed GP protein. Antiviral Res. 2018, 155, 1–11. 10.1016/j.antiviral.2018.04.020. [DOI] [PubMed] [Google Scholar]
- Shaikh F.; Zhao Y.; Alvarez L.; Iliopoulou M.; Lohans C.; Schofield C. J.; Padilla-Parra S.; Siu S. W. I.; Fry E. E.; Ren J.; Stuart D. I. Structure-Based in Silico Screening Identifies a Potent Ebolavirus Inhibitor from a Traditional Chinese Medicine Library. J. Med. Chem. 2019, 62 (6), 2928–2937. 10.1021/acs.jmedchem.8b01328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yermolina M. V.; Wang J.; Caffrey M.; Rong L. L.; Wardrop D. J. Discovery, synthesis, and biological evaluation of a novel group of selective inhibitors of filoviral entry. J. Med. Chem. 2011, 54 (3), 765–81. 10.1021/jm1008715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Ren J.; Fry E. E.; Xiao J.; Townsend A. R.; Stuart D. I. Structures of Ebola Virus Glycoprotein Complexes with Tricyclic Antidepressant and Antipsychotic Drugs. J. Med. Chem. 2018, 61 (11), 4938–4945. 10.1021/acs.jmedchem.8b00350. [DOI] [PubMed] [Google Scholar]
- Singleton C. D.; Humby M. S.; Yi H. A.; Rizzo R. C.; Jacobs A. Identification of Ebola Virus Inhibitors Targeting GP2 Using Principles of Molecular Mimicry. J. Virol. 2019, 93 (15), e00676-19. 10.1128/JVI.00676-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid P. B.; Chopra S.; Manger I. D.; Gilfillan L.; Keepers T. R.; Shurtleff A. C.; Green C. E.; Iyer L. V.; Dilks H. H.; Davey R. A.; Kolokoltsov A. A.; Carrion R. Jr.; Patterson J. L.; Bavari S.; Panchal R. G.; Warren T. K.; Wells J. B.; Moos W. H.; Burke R. L.; Tanga M. J. A systematic screen of FDA-approved drugs for inhibitors of biological threat agents. PLoS One 2013, 8 (4), e60579 10.1371/journal.pone.0060579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid P. B.; Panchal R. G.; Warren T. K.; Shurtleff A. C.; Endsley A. N.; Green C. E.; Kolokoltsov A. A.; Davey R. A.; Manger I. D.; Gilfillan L.; Bavari S.; Tanga M. J. Evaluation of Ebola Virus Inhibitors for Drug Repurposing. ACS Infect. Dis. 2015, 1, 317–326. 10.1021/acsinfecdis.5b00030. [DOI] [PubMed] [Google Scholar]
- Ekins S.; Freundlich J.; Clark A.; Anantpadma M.; Davey R.; Madrid P. Machine learning models identify molecules active against Ebola virus in vitro. F1000Research 2015, 4, 1091. 10.12688/f1000research.7217.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane T. R.; Massey C.; Comer J. E.; Freiberg A. N.; Zhou H.; Dyall J.; Holbrook M. R.; Anantpadma M.; Davey R. A.; Madrid P. B.; Ekins S. Pyronaridine Tetraphosphate Efficacy Against Ebola Virus Infection in Guinea Pig. Antiviral Res. 2020, 181, 104863. 10.1016/j.antiviral.2020.104863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane T. R.; Comer J. E.; Freiberg A. N.; Madrid P. B.; Ekins S. Repurposing Quinacrine Against Ebola Virus Infection In vivo. Antimicrob. Agents Chemother. 2019, 63, e01142–19. 10.1128/AAC.01142-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane T. R.; Massey C.; Comer J. E.; Anantpadma M.; Freundlich J. S.; Davey R. A.; Madrid P. B.; Ekins S. Repurposing The Antimalarial Pyronaridine Tetraphosphate To Protect Against Ebola Virus Infection. PLoS Neglected Trop. Dis. 2019, 13, e0007890 10.1371/journal.pntd.0007890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S.; Lingerfelt M. A.; Comer J. E.; Freiberg A. N.; Mirsalis J. C.; O’Loughlin K.; Harutyunyan A.; McFarlane C.; Green C. E.; Madrid P. B. Efficacy of Tilorone Dihydrochloride against Ebola Virus Infection. Antimicrob. Agents Chemother. 2018, 62 (2), e01711–17. 10.1128/AAC.01711-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S.; Lane T. R.; Madrid P. B. Tilorone: a Broad-Spectrum Antiviral Invented in the USA and Commercialized in Russia and beyond. Pharm. Res. 2020, 37 (4), 71. 10.1007/s11095-020-02799-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane T. R.; Dyall J.; Mercer L.; Goodin C.; Foil D. H.; Zhou H.; Postnikova E.; Liang J. Y.; Holbrook M. R.; Madrid P. B.; Ekins S.. Repurposing Pyramax for the Treatment of Ebola Virus Disease: Additivity of the Lysosomotropic Pyronaridine and Non-Lysosomotropic Artesunate. bioRxiv, April 27, 2020, ver. 1. 10.1101/2020.04.25.061333. [DOI]
- Dougherty D. A. The cation-pi interaction. Acc. Chem. Res. 2013, 46 (4), 885–93. 10.1021/ar300265y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipot C.; Jaffe R.; Maigret B.; Pearlman D. A.; Kollman P. A. Benzene Dimer: A Good Model for π–π Interactions in Proteins? A Comparison between the Benzene and the Toluene Dimers in the Gas Phase and in an Aqueous Solution. J. Am. Chem. Soc. 1996, 118 (45), 11217–11224. 10.1021/ja961379l. [DOI] [Google Scholar]
- Tsuzuki S.; Honda K.; Uchimaru T.; Mikami M.; Tanabe K. Origin of attraction and directionality of the pi/pi interaction: model chemistry calculations of benzene dimer interaction. J. Am. Chem. Soc. 2002, 124 (1), 104–12. 10.1021/ja0105212. [DOI] [PubMed] [Google Scholar]
- Oestereich L.; Ludtke A.; Wurr S.; Rieger T.; Munoz-Fontela C.; Gunther S. Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antiviral Res. 2014, 105, 17–21. 10.1016/j.antiviral.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Smither S. J.; Eastaugh L. S.; Steward J. A.; Nelson M.; Lenk R. P.; Lever M. S. Post-exposure efficacy of oral T-705 (Favipiravir) against inhalational Ebola virus infection in a mouse model. Antiviral Res. 2014, 104, 153–5. 10.1016/j.antiviral.2014.01.012. [DOI] [PubMed] [Google Scholar]
- Guedj J.; Piorkowski G.; Jacquot F.; Madelain V.; Nguyen T. H. T.; Rodallec A.; Gunther S.; Carbonnelle C.; Mentre F.; Raoul H.; de Lamballerie X. Antiviral efficacy of favipiravir against Ebola virus: A translational study in cynomolgus macaques. PLoS Med. 2018, 15 (3), e1002535 10.1371/journal.pmed.1002535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer J. E.; Escaffre O.; Neef N.; Brasel T.; Juelich T. L.; Smith J. K.; Smith J.; Kalveram B.; Perez D. D.; Massey S.; Zhang L.; Freiberg A. N. Filovirus Virulence in Interferon alpha/beta and gamma Double Knockout Mice, and Treatment with Favipiravir. Viruses 2019, 11 (2), 137. 10.3390/v11020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft S. L.; Duparc S.; Arbe-Barnes S. J.; Craft J. C.; Shin C. S.; Fleckenstein L.; Borghini-Fuhrer I.; Rim H. J. Review of pyronaridine anti-malarial properties and product characteristics. Malar. J. 2012, 11, 270. 10.1186/1475-2875-11-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S.; Madrid P. B. Tilorone: A Broad-Spectrum Antiviral For Emerging Viruses. Antimicrob. Agents Chemother. 2020, 64, e00440-20. 10.1128/AAC.00440-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon S.; Ko M.; Lee J.; Choi I.; Byun S. Y.; Park S.; Shum D.; Kim S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020, 64, e00819–20. 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantpadma M.; Lane T.; Zorn K. M.; Lingerfelt M. A.; Clark A. M.; Freundlich J. S.; Davey R. A.; Madrid P.; Ekins S. Ebola Virus Bayesian Machine Learning Models Enable New In Vitro Leads. ACS Omega 2019, 4, 2353–2361. 10.1021/acsomega.8b02948. [DOI] [PMC free article] [PubMed] [Google Scholar]