Significance
The ubiquitin–proteasome system and autophagy are two major intracellular proteolytic pathways, and both remove misfolded and proteotoxic proteins from eukaryotic cells. This study describes the detailed regulatory pathway of proteasome degradation by autophagy for its own quality control. We discovered that a portion of inhibited proteasomes is actively sequestered into the aggresome, an insoluble fraction of the mammalian cell. The aggresome functions as a triage point for proteasome recovery and autophagic degradation. This mainly distinguishes proteasome quality control in mammals from that in other organisms. STUB1/CHIP E3 Ub ligase has a critical role in targeting inhibited proteasomes into the aggresome. These results provide strong insights into protein catabolism in various pathological conditions originating from impaired proteasomes.
Keywords: proteasome, aggresome, proteaphagy, ubiquitin, STUB1
Abstract
The 26S proteasome, a self-compartmentalized protease complex, plays a crucial role in protein quality control. Multiple levels of regulatory systems modulate proteasomal activity for substrate hydrolysis. However, the destruction mechanism of mammalian proteasomes is poorly understood. We found that inhibited proteasomes are sequestered into the insoluble aggresome via HDAC6- and dynein-mediated transport. These proteasomes colocalized with the autophagic receptor SQSTM1 and cleared through selective macroautophagy, linking aggresomal segregation to autophagic degradation. This proteaphagic pathway was counterbalanced with the recovery of proteasomal activity and was critical for reducing cellular proteasomal stress. Changes in associated proteins and polyubiquitylation on inhibited 26S proteasomes participated in the targeting mechanism to the aggresome and autophagosome. The STUB1 E3 Ub ligase specifically ubiquitylated purified human proteasomes in vitro, mainly via Lys63-linked chains. Genetic and chemical inhibition of STUB1 activity significantly impaired proteasome processing and reduced resistance to proteasomal stress. These data demonstrate that aggresomal sequestration is the crucial upstream event for proteasome quality control and overall protein homeostasis in mammals.
Intracellular proteolysis not only regulates various homeostatic pathways responding to environmental changes and nutritive conditions for survival but also contributes to protein quality control (PQC) by removing defective, misfolded, and potentially harmful proteins from the cell. Two machineries, the ubiquitin (Ub)–proteasome system (UPS) and autophagy–lysosome system (henceforth referred to as autophagy), are responsible for controlled protein degradation in eukaryotes. The UPS accounts for the majority of cellular proteolysis, continuously regulating the clearance of most short-lived proteins with polyubiquitin (polyUb) chains assembled in different linkage conformations (1). In contrast, autophagy operates at a basal level under normal conditions, but is stimulated during various pathological and physiological states (2). Cargoes subjected to autophagic degradation, including both proteins and nonproteinaceous components, provide energy and anabolic intermediates to the cell (3, 4).
Although bulky autophagy degrades numerous cytoplasmic entities in a nonselective manner (5, 6), under specific conditions, selected substrates can be degraded by cognate autophagy, which processes mitochondria (mitophagy), peroxisomes (pexophagy), microorganisms (xenophagy), ribosomes (ribophagy), and protein aggregates (aggrephagy). A prominent target selection mechanism in autophagy is the recognition of lysine 63 (Lys63)-linked polyUb chains on the substrates by autophagic receptors, such as SQSTM1, which contain the conserved Ub-associated domain (7). Most autophagic receptors have an additional LC3-interacting region; the target cargoes can be docked onto phosphatidylethanolamine-modified LC3 (LC3-II) on the expanding phagophore membrane, enveloped by an autophagosome, and eventually degraded in the autolysosomes. Notably, the enzymatic cascade attaching the lipid moiety at the C-terminal glycine of the cleaved LC3 protein in autophagy resembles the E1-, E2-, and E3-enzyme–mediated Ub-conjugating system. This suggests that complementation and feedback communication between these two catabolic systems in the cell might be based on the functional sharing of key components in their regulatory mechanisms (8, 9).
The 26S proteasome is composed of two structurally and functionally distinct protein complexes: the core particle (CP) and regulatory particle (RP). At the most downstream UPS level, the proteasome efficiently degrades target substrates into small peptides irreversibly, thereby ensuring protein homeostasis (proteostasis) in the cell. It is known that proteasomal activity is closely linked with cellular autophagic flux: for example, proteasome inhibition results in the compensatory autophagy induction (10–12). Conversely, proteasomal activity up-regulation leads to delayed autophagic flux, mainly due to the defective fusion between autophagosome and lysosome (13). If proteasomes are chronically inhibited, excess aberrant proteins concentrate into a proteinaceous inclusion body termed the aggresome. Aggresome formation is generally believed to represent a cytoprotective mechanism by which toxic aggregates are sequestered and their autophagic clearance is facilitated (14–16). Although a variety of proteins have been detected within aggresomes, the nature and fate of proteasomes found in the aggresome remain essentially unknown.
Several studies have focused on proteasome synthesis, assembly, and activity regulation, but our understanding of their destruction mechanism is incomplete as proteasomes are generally thought to be highly abundant and stable (17, 18). Only recently, studies on Arabidopsis and Saccharomyces cerevisiae have uncovered selective proteaphagic routes (19–24), and mammals appear to utilize a similar clearance process (25). In the current study, we discovered the adaptive response of mammalian cells upon proteasome inhibition, where purging inhibited proteasomes from the cell is preceded by its segregation into insoluble aggresomes via microtubule-based transport. Autophagy inhibition resulted in elevated proteasome levels in the insoluble fraction and a number of speckled puncta in the cytoplasm, indicating autophagic clearance of proteasomes in the aggresome. Targeting proteasomes to the aggresomes appeared to be associated with the attachment of Lys63-linked polyUb chains by STUB1. A biophysical screen identified a small molecule, which efficiently inhibited STUB1-mediated polyubiquitylation and aggresomal formation of inhibited proteasomes in vitro and in vivo. Considering the constant load of proteins being degraded and need for amino acids for de novo protein synthesis, nonfunctional proteasomes should be replaced to cope with proteotoxic stress on a global scale. Our findings demonstrate that STUB1-mediated proteasome ubiquitylation, aggresomal sequestration, and subsequent autophagic degradation are the major survival strategies for mammalian proteaphagy under proteotoxic stress.
Results
Inhibited Proteasomes Accumulate in the Insoluble Fraction and the Aggresome.
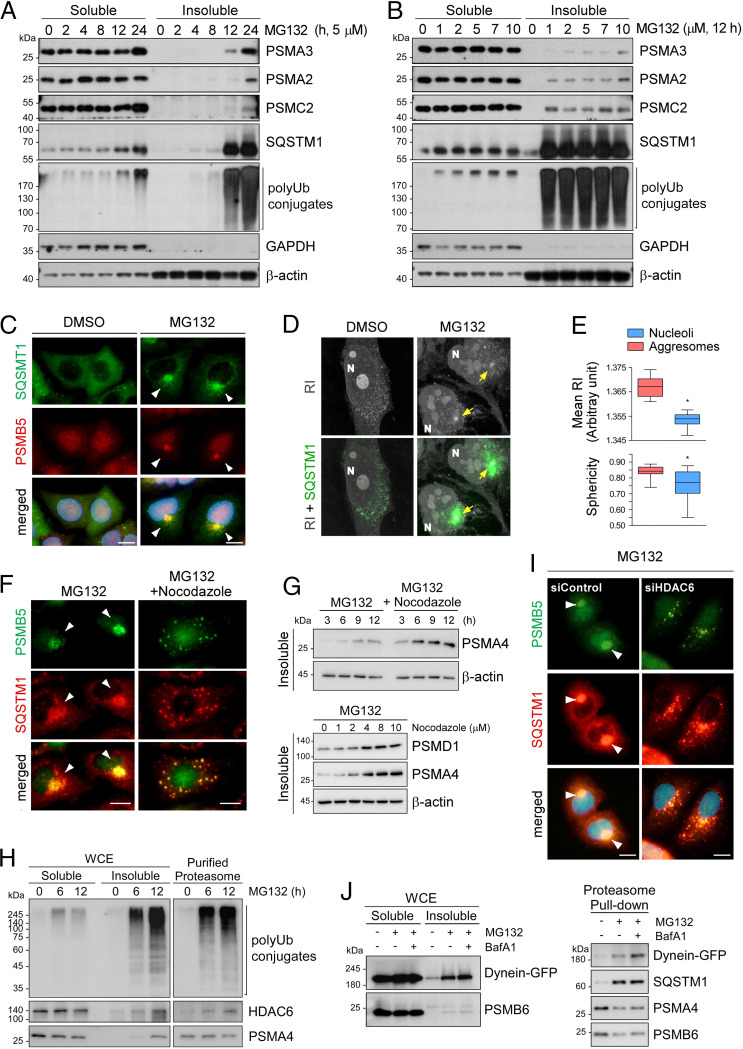
During our initial studies on proteasome homeostasis, autophagic induction (by amino acid or glucose starvation) resulted in reduced levels of proteasome subunits in whole-cell extracts (WCEs), but chemical or genetic inhibition of autophagy (via an bafilomycin A1 [BafA1] treatment or Atg5 knockout) did not alter this tendency (SI Appendix, Fig. S1A). These data suggest that, unlike in nitrogen-starved yeast and plants (19–21), bulk autophagy might not be the major route for 26S proteasome clearance in mammals. We examined various cellular stress conditions, including proteasome inhibition, to test whether they can induce selective autophagic proteasome degradation (proteaphagy), but the amount of proteasomes in the Triton X-100–soluble WCE fraction remained virtually unchanged (Fig. 1 A and B). In stark contrast, we noticed that the levels of both CP and RP subunits were significantly elevated in the insoluble fraction when the cells were treated with MG132 for more than 12 h. The relatively late response time is probably due to elaborate molecular and cellular processes. Prolonged proteasome inhibition also drastically increased the amounts of prominent autophagic receptor SQSTM1, its oligomeric forms, and total polyUb-conjugates in the insoluble fraction (Fig. 1 A and B and SI Appendix, Fig. S1 B–D).
Fig. 1.
Treatment with proteasome inhibitors led to the accumulation of proteasomes in the insoluble aggresome via microtubule-based transport. (A) Accumulation of proteasome subunits in the insoluble fraction of mouse embryonic fibroblasts (MEFs) after treatment with 5 μM MG132 for the indicated periods (0 to 24 h). Whole cell extracts (WCEs) were separated into Triton X-100–soluble (soluble) and pellet fractions (insoluble) and subjected to SDS/PAGE/immunoblotting (IB). (B) As in A, except that the cells were treated with various concentrations of MG132 for 12 h. (C) Representative images of A549 cells treated with MG132 (10 μM, 12 h). Immunofluorescent staining (IFS) with anti-SQSTM1 (green) and anti-PSMB5 (red) antibodies. Nuclei were counterstained with DAPI (blue). (D) Refractive index (RI)-based holotomographic images of MG132 (5 μM, 12 h)-treated cells. Distinct juxtanuclear inclusion bodies from RI images are colocalized with SQSTM1-positive signals (green) from IFS. (E) Quantitative analysis of tomographic images of the aggresomes (mean RI and sphericity) compared with those of nucleoli from the identical cells. A box-and-whisker plot with n = 29. (F) Inhibition of aggresome formation in A549 cells by cotreatment with nocodazole (2 μM) and MG132 (5 μM) for 12 h. (G, Top) As in F, except that WCEs were fractionated into detergent-soluble and -insoluble fractions. (Bottom) A549 cells were treated with 5 μM along with indicated final concentrations of nocodazole for 12 h. Insoluble fractions were fractionated from WCEs and analyzed by SDS/PAGE/IB. (H) Direct interaction between HDAC6 and a proteasome was stronger with MG132 treatment. HEK293 cells stably expressing biotin-tagged CP subunit PSMB2 were treated with MG132 (5 μM) for 6 h or 12 h. Human proteasomes were affinity-purified from WCEs and then analyzed by SDS/PAGE/IB. (I) A549 cells were transfected with 20 nM siRNA for silencing HDAC6 (siHDAC6) or with scrambled (control) siRNA (siControl) for 48 h and then treated with 5 μM MG132 for 12 h. The cells were fixed and subjected to co-IFS with anti-PSMB5 (green) and anti-SQSTM1 (red) antibodies. (Scale bars: 10 μm.) (J) GFP-tagged dynein was transiently overexpressed in HEK293-PSMB2-biotin cells in the presence of 5 μM MG132 for 12 h and/or 100 nM bafilomycin A1 (BafA1) for 4 h. WCEs (Left) were subjected to affinity purification of proteasomes with streptavidin (Right), followed by IB with indicated antibodies.
Under normal conditions, proteasomes are largely dispersed throughout the cytoplasm and nucleus (26, 27). To visualize and localize the accumulated proteasomes in the cell, we immunostained the CP subunit PSMB5 after treating the cells with 5 μM MG132 for 12 h. In yeast and plants, accumulated proteasomes were observed in either the vacuole or the cytoplasm (19–21). In contrast, the proteasome-positive signals in mammalian cells manifested as one ∼4-μm punctum mostly located near the nucleus (Fig. 1C and SI Appendix, Fig. S2 A and B), which resembles the proteasome storage granules (PSGs) generated in yeast and plants under carbon-deprived conditions (28, 29). These puncta strongly overlapped with immunostained SQSTM1, Ub, cystic fibrosis transmembrane conductance regulator-ΔF508 (misfolded protein), or vimentin in the MG132-treated cells (Fig. 1C and SI Appendix, Fig. S2 C–E). The characteristics and colocalization pattern of these proteins strongly indicated that proteasomal inclusions observed on the cytoplasmic side of the perinuclear region are aggresomes. Both RP and CP subunits showed similar enlarged juxtanuclear signals (SI Appendix, Fig. S2F).
Label-free holotomographic images, based on the differences in the refractive index (RI) among cellular organelles, uncovered a distinct juxtanuclear inclusion body with exceptionally high RI, concentration, and sphericity (Fig. 1 D and E and SI Appendix, Fig. S2G). When combined with fluorescence immunostaining, the RI images revealed that the aggresomal puncta were surrounded by an SQSTM1-positive inclusion body (Fig. 1D). Although the aggresome occupies a considerable volume near the nucleus, this rearrangement did not result in cell cycle arrest, mitotic failure, or apoptosis (SI Appendix, Fig. S3A). However, prolonged proteasome inhibitor treatment led to the significant transcriptional up-regulation of many proteasome subunit-, Ub-, and autophagy-related genes (SI Appendix, Fig. S3B). These results imply that sequestration might be linked to an autophagic clearance and compensatory de novo synthesis of active proteasomes for the maintenance of the cellular proteasome pool. Collectively, we confirm that inhibited 26S proteasomes accumulate into the aggresome with the autophagic receptor SQSTM1. We speculate that aggresomal segregation is crucial for the subsequent elimination of nonfunctional proteasomes or their recovery to the soluble and active state (see below).
Inhibited Proteasomes Are Delivered to the Aggresome through HDAC6- and Dynein-Mediated Transport.
To gain insight into the transport of proteasomes to the aggresome, we cotreated the cells with nocodazole, a microtubule-disrupting drug, and MG132, which resulted in the extensive formation of speckled small puncta (<1 μm) of proteasomes throughout the cytoplasm (Fig. 1F). Under the same condition, a higher number of proteasomes accumulated in the insoluble fraction than with MG132 treatment (Fig. 1G and SI Appendix, Fig. S3C). We next tested whether HDAC6, an adaptor protein for polyUb and dynein motor complex, plays an active role in the translocation of proteasomes into aggresomes (16, 30). We observed that a significant amount of the endogenous HDAC6 protein was bound to affinity-purified mammalian proteasomes only in proteasome inhibitor-treated cells (Fig. 1H). Consistent with its direct interaction with inhibited proteasomes, endogenous HDAC6 was found to strongly colocalize with aggresome-sequestered proteasomes (SI Appendix, Fig. S4A). HDAC6 knockdown through small interfering RNA (siRNA) resulted in further proteasome accumulation in the insoluble fraction compared with that in the control siRNA group, and effectively abolished the formation of PSMB5- and SQSTM1-positive aggresomes (Fig. 1I and SI Appendix, Fig. S4 B and C). Considering that proteasome–dynein motor interaction strengthens in the presence of MG132 (Fig. 1J), mammalian proteasomes are expected to be sequestered and condensed into insoluble aggresomes through microtubule-based transport.
Proteasomes Deposited in the Aggresome Are Cleared by Autophagy.
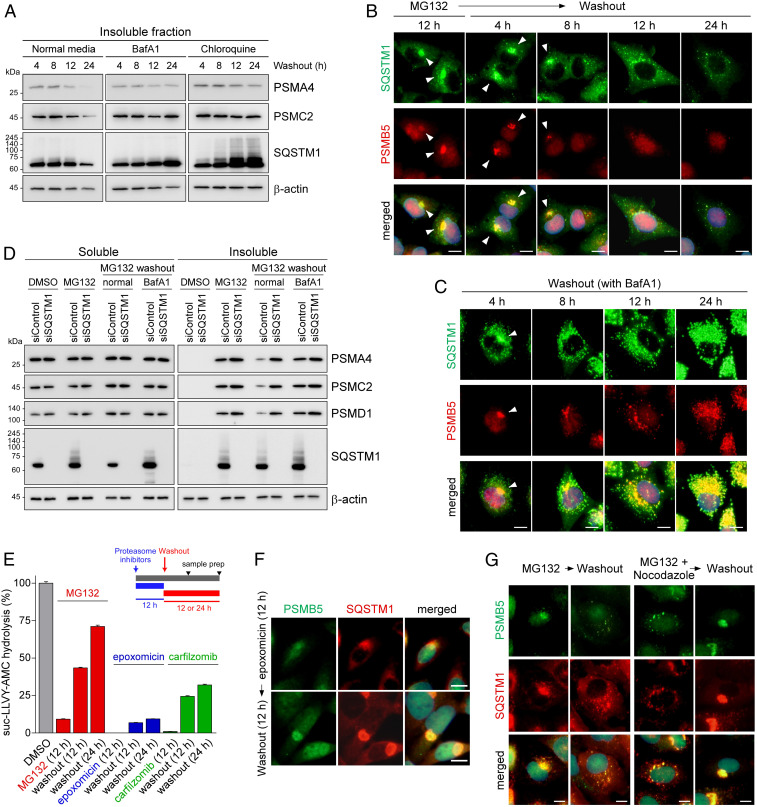
To examine the fate of proteasomes deposited in the aggresome, MG132-treated cells were returned to the inhibitor-free (normal) medium. The levels of both CP and RP subunits accumulated in the insoluble fraction gradually diminished during the normal washout process (Fig. 2A and SI Appendix, Fig. S4D). Correspondingly, the aggresomal structure dissipated from the outside and completely dissolved after ∼24 h of MG132 washout (Fig. 2B and SI Appendix, Fig. S2 A and B). A possible explanation for this observation is the reversion of inhibited proteasomes to active and soluble proteasomes in the cytosol. However, we found that cellular autophagic flux inhibition by adding either BafA1 or chloroquine led to a drastic delay in the elimination of proteasome subunits from the insoluble fraction during the washout process (Fig. 2A). Similarly, washing the drug out with a medium supplemented with autophagy inhibitors led to aggresome clearance failure and drastically increased the number of small cytoplasmic puncta (Fig. 2C), in sharp contrast to the results from normal washout (Fig. 2B). The proteasome-positive puncta colocalized with SQSTM1, and the lysosomal marker LAMP1 with the BafA1-containing washout media, both in the presence and absence of MG132, suggesting that the dissipating proteasomes are probably autolysosomes (SI Appendix, Fig. S4 E and F). These results strongly indicate that autophagic degradation is actively involved in the postaggresomal processing of inhibited proteasomes, while other mechanisms, such as recovery of aggresomal proteasomes and lagged-phase segregation, may still be active.
Fig. 2.
Inhibited proteasomes accumulated in the aggresome are cleared via the autophagy–lysosome system. (A) Washing out MG132 with either a normal medium or medium containing autophagy inhibitors, such as BafA1 (100 nM) and chloroquine (25 μM), for 4, 8, 12, or 24 h. HEK293 cells were treated with MG132 (5 μM) for 12 h to induce aggresome formation before the washout experiment. Immunoblotting analysis of a CP subunit (PSMA4), an RP subunit (PSMC2), SQSTM1, and β-actin was conducted after the isolation of the detergent-insoluble fraction of WCEs. (B and C) As in A, except that IFS analysis was performed with anti-PSMB5 (red) and anti-SQSTM1 (green) antibodies in the absence (B) or presence (C) of BafA1. (D) A549 cells were transfected with either siControl or siSQSTM1. After 24 h, cells were treated with 5 μM MG132 for 12 h and then washed out with either a normal medium or a medium containing BafA1 for additional 18 h. A detergent-soluble and -insoluble fraction was isolated and subject to SDS/PAGE/IB analysis. (E) The proteasome activity was measured using hydrolysis of fluorogenic suc-LLVY-AMC after treatment with 5 μM MG132, 0.5 μM epoxomicin, or 0.1 μM carfilzomib for 12 h (mean ± SD from three independent experiments). (F) Contrary to the results from MG132-treated samples, proteasome-positive aggresomes after treatment with irreversible proteasome inhibitor epoxomicin (100 nM, 12 h) did not dissipate even after 12 h inhibitor washout. (G) IFS images of A549 cells after 12 h washout following 12 h MG132 treatment (5 μM) or MG132 plus nocodazole (2 μM) cotreatments. (Scale bars: 10 μm.)
In agreement with the results of chemical inhibition of autophagy, a knockdown of SQSTM1 using siRNA caused a significant delay in proteasome clearance from the insoluble fraction after the washout, nearly as effectively as BafA1 treatment (Fig. 2D). Notably, cells deficient in SQSTM1 were unable to form the aggresome under proteasome inhibition conditions (SI Appendix, Fig. S4G). This finding implies that impaired autophagic flux may counter-regulate aggresomal assembly of inhibited proteasomes, but the underlying feedback mechanism is yet to be determined. Moreover, we observed that autophagosome formation suppression by ATG5 and ATG7 knockdown caused a significant accumulation of proteasome subunits in the washed-out cells, as seen after SQSTM1 knockdown (Fig. 2D and SI Appendix, Fig. S5A), further confirming the defective proteasome clearance when autophagy is impaired. We then up-regulated cellular autophagy by removing amino acids from the medium while washing out MG132. As shown in SI Appendix, Fig. S5B, significantly lower levels of proteasome components, SQSTM1, and LC3-II were observed in the insoluble fraction, as compared with the normal washout group. These results underscore the role of autophagy in the clearance of inhibited proteasomes from the aggresome.
Furthermore, we found that significant proteasomal activity could be recovered after MG132 washout when measured with suc-LLVY-AMC as reporter substrates (Fig. 2E). Both inhibitor dissociation from proteasomes and transcriptional bounce-back mechanism are probably responsible for this recovery in activity. In contrast, when cells were treated with the irreversible inhibitors epoxomicin and carfilzomib (31, 32), there was much less recovery of proteasome function after the drugs were washed out (7.5- and 2.2-fold lower cellular proteasome activity, respectively, compared with that after MG132 washout; Fig. 2E). Furthermore, in stark contrast to MG132-treated cells, juxtanuclear aggresomes in the epoxomicin-treated cells were not cleared out, but increased in size after the washout (Fig. 2F). These results indicate that only a minor proportion of inhibited proteasomes in the cell can be concentrated in the aggresome, and defective proteasomes in the cytoplasm are still actively transported to the aggresome (lagged-phase segregation) until proteasomal activity is restored. These rationales are plausible owing to the abundance of proteasomes (>0.5% of the total cellular proteins) (17, 18). Consistent with this, in a washout experiment using cotreatment with MG132 and nocodazole, the cytoplasmic speckles in the nocodazole-treated cells appeared to be transported to the juxtanuclear aggresomes after ∼12 h of washout (Fig. 2G). Although MG132 alone did not affect cell viability, cotreatment with nocodazole and MG132 resulted in substantially elevated cytotoxicity (SI Appendix, Fig. S5C). Therefore, segregating inhibited proteasomes in the aggresome promotes cell survival during prolonged proteasome inhibition.
These results indicate that the spontaneous MG132 dissociation from the inhibited proteasome might be a slow process, and aggresomal sequestration of these proteasomes could be a faster responding mechanism in mammals than direct recovery to active status. In addition, these data establish aggresomal segregation as a cellular checkpoint, where decisions are made on whether to recover incompetent proteasomes (when the inhibitors are removed) or salvage them by autophagy (e.g., during chronic inhibition) as an effective survival strategy in mammals. Despite the existence of many mechanistic differences, this proteasome dynamic appears to be parallel to the observation in yeast and plants. For example, while PSGs reversibly accommodate inactive proteasomes under energy-limited conditions, more severe stress may result in sequestration of dysfunctional proteasomes in the insoluble protein deposit (IPOD)-like structure and subsequent degradation via autophagy (23, 33, 34).
Proteasome Structure and Interacting Proteins Are Actively Modified during the Sequestration Process.
To investigate the molecular mechanism underlying the translocation of inhibited proteasomes to the aggresome, we first performed a sucrose-gradient ultracentrifugation analysis to monitor proteasome assembly. Following immunoblotting analysis of the fractionated WCEs, we noticed that a significant proportion of the CP and RP subunits shifted from the free form to the 26S species (SI Appendix, Fig. S6A) in the presence of MG132. Increased binding of the CP and RP subcomplexes during MG132 treatment was also observed in size-exclusion chromatography (SI Appendix, Fig. S6B).
Next, we performed label-free quantitative mass spectrometry (MS) based on the intensity-based absolute quantification (iBAQ) algorithm, which uses the sum of peptide intensities of a protein of interest divided by the theoretically observable peptides numbers as an accurate proxy for protein amounts (35). Proteasomes and their associated proteins after prolonged MG132 treatment were affinity-purified from three independent cultures and subjected to the iBAQ analysis (SI Appendix, Fig. S7A). This method showed significant reproducibility (as evaluated by means of intragroup components and gene ontology clustering; SI Appendix, Table S1), without requiring pre- or posttreatment of samples, e.g., isotope labeling. According to our criteria (1.5-fold change and P < 0.05), 118 differentially associated proteins (DAPs) were identified; 81 and 87 proteins significantly changed their interaction with the proteasome after 6 and 12 h of MG132 treatment, respectively, compared to that in time 0 (SI Appendix, Fig. S7B). Most of the responding proteins, including the 70-kDa heat shock protein (HSP70) family, have previously been identified as proteasome-associated proteins (36), further validating this method (SI Appendix, Fig. S7C). We conducted an immunoblotting analysis of several DAPs under the same experimental conditions to verify the quantitative MS results, and observed that their levels were similar to that observed by quantitative MS (SI Appendix, Fig. S7D).
In the proteomic profile, we found that, among the 49 proteins shared between two time points, DAPs with >10-fold changes include E3 Ub ligases, proteasome adaptors, and autophagy-related proteins (SI Appendix, Fig. S7E). Multiple E3 Ub ligases, such as UBE3A, UBR4, and RNF181, accumulated during the treatment of proteasomes with MG132 for 6 h (SI Appendix, Fig. S7F). Components of the BAG6–BAT3 complex, which participate in the cytosolic PQC of misfolded proteins and proteasome substrates (37), were also significantly up-regulated (SI Appendix, Fig. S7G), suggesting that many of these DAPs reflect the cellular responsive mechanisms against long-term proteasome inhibition. In the native gel profile of purified MG132-treated proteasomes (SI Appendix, Fig. S7H), ECM29 and HDAC6 manifested a significant upshift into higher-molecular-weight species, which is consistent with the strong association of these proteins with inhibited proteasomes also suggesting structural remodeling (38, 39). The exact mechanism of the binding of these proteins to the nonfunctional proteasome remains to be determined. However, the biochemical and MS data strongly suggest that inhibition at the active site allosterically transforms the proteasome holoenzyme, such as a stronger CP–RP association and changes in various interacting proteins. In yeast and plant proteaphagy, the CP and RP are degraded at similar rates (20, 24). Therefore, the structural change of mammalian proteasomes may facilitate the sequestration of inhibited 26S proteasomes as well as subsequent autophagic degradation.
STUB1 Is Essential for Ubiquitylation and Aggresomal Formation of Inhibited Proteasomes.
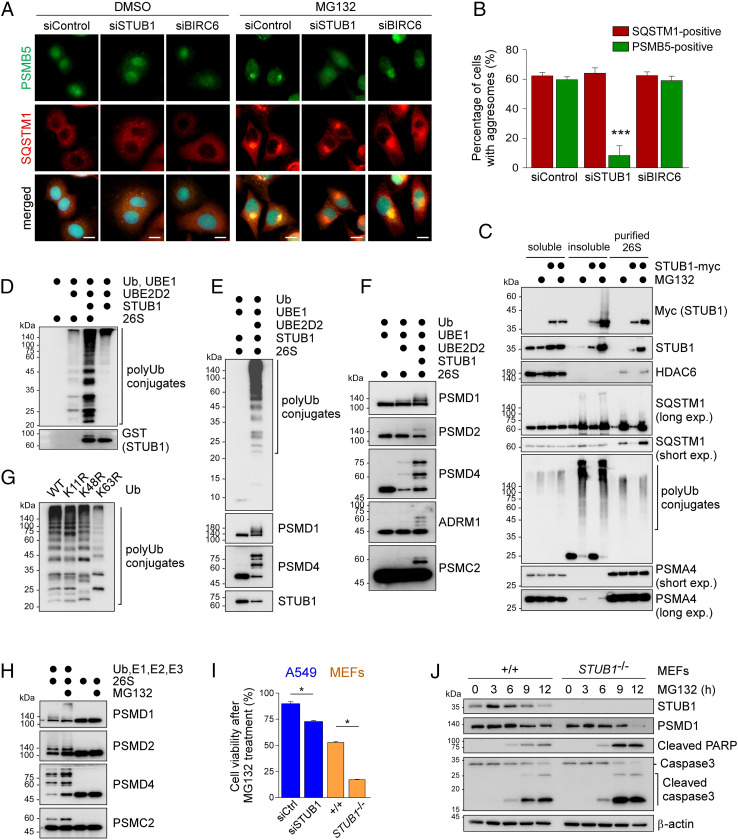
Due to the association of multiple E3 Ub ligases with inhibited proteasomes, we postulated that direct ubiquitylation might be involved in selective regulation during mammalian proteaphagy. To test this hypothesis, we used siRNA to knock down the E3 enzymes but found that silencing BIRC6 or UBE3A had little effect on cellular aggresome formation (Fig. 3 A and B and SI Appendix, Fig. S8A). Knocking down RNF181 or UBR4 delayed the formations of both PSMB5-positive and SQSTM1-positive aggresomes (SI Appendix, Fig. S8A). In contrast, we observed that STUB1, which recognizes and ubiquitylates HSP70 substrates responding to various kinds of cellular stress, is essential for the early phase of aggresomal segregation (Fig. 3 A and B). SQSTM1 localization in the aggresome of STUB1-deficient cells raised the possibility that STUB1-mediated polyubiquitylation might contribute to the selective transportation of inhibited proteasomes without affecting the formation of aggresomes per se. This hypothesis was supported by our findings that, upon MG132 treatment, proteasome subunits were less efficiently deposited in the insoluble fraction when STUB1 was knocked down (SI Appendix, Fig. S8B). A pulldown assay revealed a significantly increased interaction between inhibited proteasomes and STUB1, and HSP70 members such as HSPA1A and HSPA6 as well, in the presence of MG132 (Fig. 3C and SI Appendix, Figs. S7D and S9G). Although further studies are required to determine the possible contribution of other Ub E3 ligases in this process, our data suggest that STUB1 is a major regulator of proteasome ubiquitylation and aggresomal deposition.
Fig. 3.
STUB1 is critical for the direct polyubiquitylation of inhibited proteasomes and for their aggresomal assembly. (A) Impaired aggresomal deposition of inhibited proteasomes when A549 cells were transfected with siSTUB1 but not when transfected with siControl or siBIRC6. The cells were analyzed by IFS with anti-PSMB5 (green) and anti-SQSTM1 (red) antibodies in the presence of DMSO (Upper) or MG132 (5 μM, 12 h; Lower). (B) Quantitation of A. Percentage of cells with aggresomes were counted and plotted as mean ± SD of three independent experiments with ∼500 cells (***P < 0.001, one-way ANOVA followed by the Bonferroni post hoc test). (C) Strong accumulation of STUB1 in the insoluble fraction and among purified proteasomes with MG132 treatment. Myc-tagged STUB1 was transfected into HEK293-PSMB2-biotin cells, which were then treated with MG132 (5 μM, 12 h). Soluble and insoluble fractions of WCEs were prepared and subjected to SDS/PAGE/IB along with affinity-purified proteasomes. (D) In vitro polyubiquitylation of human proteasomes by STUB1. The 26S proteasomes were affinity-purified and incubated with recombinant Ub, UBE1, UBE2D2, and STUB1 proteins in different combinations. The ubiquitylation of 26S proteasomes was monitored with SDS/PAGE/IB using anti-Ub antibodies. Note that autoubiquitylation of STUB1 produced distinct polyUb species of proteasome subunits. (E) As in D, except that the reaction lacks UBE2D2 and produces no ubiquitylation. (F) The RP subunits were decorated with polyUb chains only when all of the components were added in the reaction. (G) Different polyUb-conjugates were formed with UbK63R after in vitro ubiquitylation of 26S proteasomes, whereas UbK11R and UbK48R produced polyUb chains comparable to those of wild-type Ub. (H) In vitro polyubiquitylation reactions were conducted with recombinant proteins Ub, UBE1, UBE2D2, and STUB1 and purified proteasomes from MG132-treated cells. Proteasome subunits were examined by SDS/PAGE/IB with the indicated antibodies. (I) Comparison of cell survival in wild-type (+/+) and STUB1−/− MEFs and A549 cells, which were transfected with either siControl or siSTUB1 for 48 h. MG132 was administered at 5 μM concentration for 12 h. Values are presented as mean ± SD (n = 3; *P < 0.01, two-tailed Student’s t test). (J) The +/+ and STUB1−/− MEFs were treated with 5 μM MG132 for 12 h before harvesting the WCEs, which were subjected to immunoblotting with indicated antibodies.
To find whether STUB1 directly polyubiquitylates the 26S proteasome, we carried out an in vitro reconstitution assay with different combinations of recombinant wild-type Ub, UBE1 (as E1), UBE2D2 (E2), STUB1 (E3), and purified human 26S proteasomes. STUB1 generated extensive polyUb conjugates of 26S proteasomes, which is distinct from the autoubiquitylated species of STUB1 (Fig. 3D and SI Appendix, Fig. S8C). Moreover, we detected ubiquitylated species of PSMD1, PSMD2, PSMD4, ADRM1, and other RP subunits (Fig. 3 E and F). These were not observed if the reaction was carried out in the absence of either UBE2D2 or STUB1. This result from the control reactions lacking one or two key enzymes provided strong evidence that the polyUb modification of proteasomes does not originate from the E2 or E3 enzymes copurified with the proteasome. Many of these subunits are predicted to be ubiquitylated according to the cell-wide MS studies (40–42). Our study indicates that many subunits, including PSMD4, in the mammalian proteasome can be directly polyubiquitylated by STUB1, which is likely to participate in proteasome remodeling and interaction with the autophagosome.
To assess the topology of Ub chains that are assembled on the proteasomes by STUB1, we carried out similar in vitro reactions with mutant Ub, where UbK63R generated a much different polyUb pattern from that of wild-type Ub (Fig. 3G and SI Appendix, Fig. S8D). UbK11R and UbK48R also produced slightly altered polyubiquitylation patterns of proteasomes. Overall, the ubiquitin chains on the inhibited proteasome appeared to be mixed polymers mainly with Lys63 links. It is possible that STUB1 generates the nondegradable and heterogeneous forked Ub linkages (43) on the proteasome. After treating the cells with MG132, elevated levels of proteasome ubiquitylation were observed (Fig. 3H and SI Appendix, Fig. S8 E and F), but HSP70 inhibitors potently blocked this process (SI Appendix, Fig. S8 F and G). This could be because of the direct interaction between the proteasome and STUB1 in the presence of MG132 (Fig. 3C); however, whether HSP70 family members are involved is yet to be determined.
We then examined the fate of the cytoplasmic speckles formed in STUB1 knockdown cells after MG132 treatment. Unlike PSMB5-positive aggresomes in control cells, which dissolved after the 24-h drug washout under normal conditions, the proteasome speckles in the STUB1-knockdown cells showed much delayed reduction during the washout (SI Appendix, Fig. S8H). Therefore, STUB1 activity appears to be required for the proper segregation and disposal of inhibited proteasomes both in vitro and in vivo. Although other E3 Ub ligases deserve further investigation, our results collectively indicate that the direct ubiquitylation of 26S proteasomes by STUB1 is a major prerequisite for the selective segregation of nonfunctional proteasomes. The results also suggest that STUB1-regulated proteasome homeostasis might antagonize the cellular stress caused by aberrant UPS function. We found that genetic inactivation of STUB1 in A549 cells resulted in a significant increase in cytotoxicity only in the presence of MG132 and that this tendency was more prominent in STUB1-null (STUB1−/−) mouse embryonic fibroblasts (MEFs; Fig. 3I). Consistent with this observation, STUB1−/− MEFs elicited a stronger apoptotic response to MG132-mediated proteasomal stress as compared to wild-type cells (Fig. 3J).
A STUB1 Inhibitor Effectively Blocks Proteasome Polyubiquitylation and Aggresomal Sequestration.
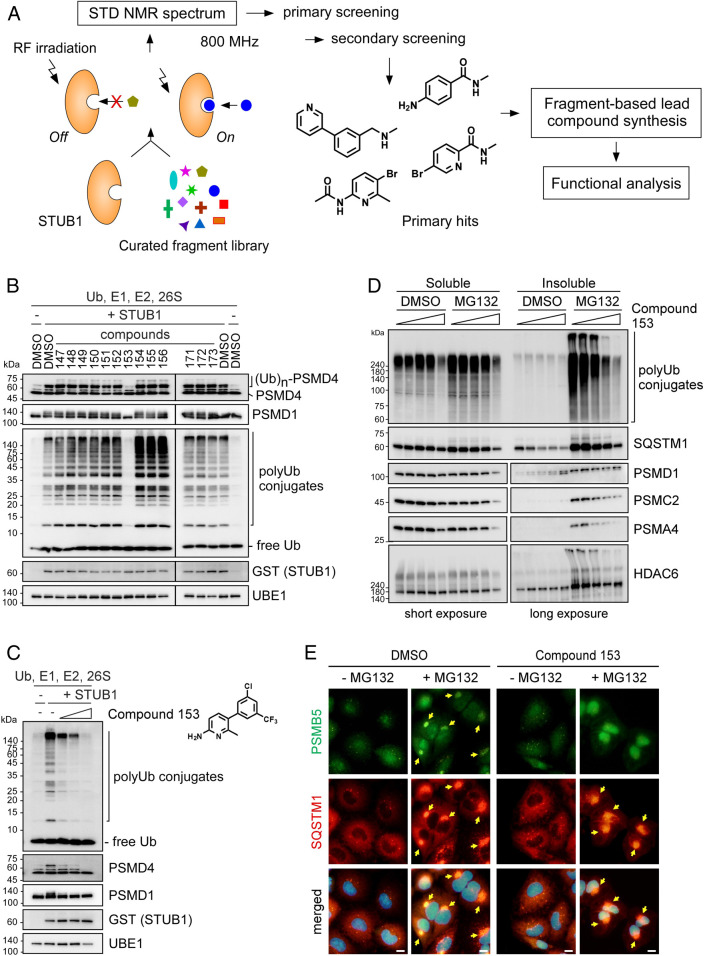
To further validate the function of STUB1 in proteasome ubiquitylation in the cell, we screened ∼500 curated fragments (<300 Da) based on NMR spectrum alteration induced by STUB1 interaction (Fig. 4A and SI Appendix, Fig. S9 A–D). Four hits, mostly having a six-membered ring, were identified as the true binding molecules of STUB1. Using the screened molecules as starting compounds, we synthesized various heterodimers and derivatives to strengthen the binding affinities. When the compounds were functionally analyzed in vitro, compound 153 virtually completely prevented ubiquitylation of purified human proteasomes (IC50 = 85.44 μM; Fig. 4 B and C). We further studied and confirmed that compound 153 effectively blocked the ubiquitylation of proteasome subunits, but did not influence proteasome activity or proteasomal DUB activity (Fig. 4C and SI Appendix, Fig. S9 E and F). Future studies on obtaining cocrystal structures of STUB1 with compound 153 will enable structure-guided optimization of the STUB1 inhibitors.
Fig. 4.
STUB1 inhibitors identified via biophysical screening specifically inhibit proteasome ubiquitylation in vitro and aggresome formation in vivo. (A) An overview of the STUB1 inhibitor identification process, which largely consists of the NMR-based fragment screenings using saturation transfer difference (STD), synthesis of lead compounds, and functional analysis for their inhibitory effects (both in vitro and in vivo). (B) Compounds (1 mM) derived from the screened fragments were subjected to in vitro ubiquitylation assays of the purified proteasomes. Compound 153 potently inhibited the ubiquitylation of proteasome subunits. (C) As in B, except that graded doses (0, 250, 500, and 1,000 μM) of compound 153 were used. Chemical structure of compound 153 is shown. (D) Reduced levels of proteasome subunits in the insoluble fraction of HEK293 cells after treatment with 5 μM MG132 with 0, 10, 25, 50, and 100 μM compound 153 for 12 h. Triton X-100–soluble and pellet fractions from WCEs were isolated and subjected to SDS/PAGE/IB analysis. For PSMD1, PSMC2, PSMA4, and HDAC6 from the soluble and insoluble fractions, short and long exposed images were shown respectively due to distinctively different protein levels. (E) Representative images of A549 cells treated with MG132 (5 μM, 12 h) in the absence or presence of compound 153 (75 μM, 12 h). IFS with anti-SQSTM1 (red) and anti-PSMB5 (green) antibodies. Nuclei were counterstained with DAPI (blue). Arrows indicate the aggresomes.
To determine whether suppression of STUB1 activity would affect aggresome formation in vivo, we treated HEK293 cells with compound 153 and MG132 simultaneously. Analysis of the insoluble fraction showed reduced levels of various proteasome subunits, along with polyUb conjugates, SQSTM1, and HDAC6, but little effects on these proteins in the soluble fraction (Fig. 4D). Consistently, compound 153 significantly reduced the levels of polyubiquitylated PSMD14 (SI Appendix, Fig. S9G). Furthermore, treatment with the STUB1 inhibitor significantly suppressed aggresomal sequestration of proteasomes after MG132 treatment, while SQSTM1-positive inclusion bodies were comparably observed (Fig. 4E and SI Appendix, Fig. S9H). The similarity between outcomes after treatments of STUB1 siRNA and compound 153 (Figs. 3A and 4E) signifies the effect of the small molecule and importance of STUB1 on proteasome quality control. When cells were cotreated with MG132 and 100 μM compound 153 for 12 h, synergistic cytotoxicity was observed, while the structurally closely related compound 154 had little effect on cell viability, regardless of the presence of MG132 (SI Appendix, Fig. S9I). Based on these data together with those of previous in vitro experiments, compound 153 appears to antagonize the upstream quality control cascade of nonfunctional proteasomes by blocking STUB1-mediated polyubiquitylation. Therefore, STUB1 may play a significant role in relieving cellular stress and promoting cell survival by mediating proteasome quality control.
Discussion
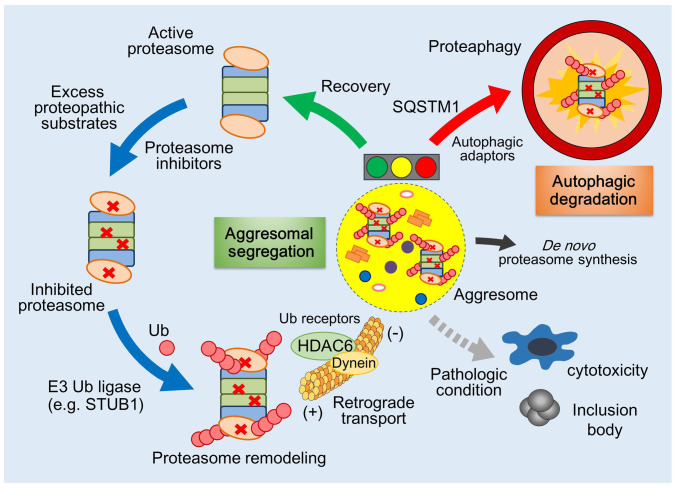
In this study, we report the pervasive but unique molecular mechanism of mammalian proteaphagy, which includes the extensive polyUb modification by STUB1, sequestration of inhibited proteasomes into the aggresome, and subsequent autophagic degradation (Fig. 5). However, the morphological and functional similarity between the aggresome (in mammals) and PSGs (in yeast and plants) suggests that the quality control mechanisms for nonfunctional proteasomes in eukaryotes are essentially conserved in evolution. The PSG may serve as a temporal reservoir of the proteasome pool, protecting them from autophagic degradation during specific stresses but dissolving to release functional proteasomes to the cell (24, 33). The irreversibly impaired proteasomes may be terminally sequestered into the perivacuolar IPOD for vacuolar or lysosomal degradation, but many studies also suggest that the IPODs function as a sorting compartment as well (22, 23, 29, 44). Although we do not present the precise mechanism, we propose that the aggresome also provides a transient platform for the recovery of proteasomes from their incompetent state. The aggresome-mediated sequestration of defective proteasomes in mammals (Fig. 1) may be the unique feature of mammalian proteasome quality control possibly because yeast and plants lack some of the key structural features or constituents of dynein motors (45–47).
Fig. 5.
Illustration of the proteasome homeostasis mechanism in mammals. Schematic representation of proteasome quality control mediating aggresomal segregation and autophagic degradation. When proteasomes are in a chronic state of inhibition, they are polyubiquitylated by STUB1 and transported to the detergent-insoluble aggresome by the HDAC6/dynein motor. Inhibited proteasomes deposited in the aggresome can be cleared by the autophagy–lysosome system while recovered proteasomes can be rescued from the aggresome and transported to the cytoplasm. Along with induction of de novo proteasome biogenesis, this proteasome homeostasis mechanism appears to be essential for the maintenance of the basal level of functional proteasomes in the cell. Although the in vivo significance of this pathway is largely undetermined, it seems plausible to predict that an imbalance between recovery and degradation of inhibited proteasomes results in the failed cellular adaptation to proteopathic stress, which would also be implicated in accumulated intracellular inclusions prevalent in neurodegenerative diseases.
For delivery to the aggresome, proteasomes need to be extensively decorated with polyUb moieties (Fig. 3), which are also key determinants in yeast for depositing misfolded proteins along with proteasomes to the juxtanuclear quality control compartments (JUNQ) (34, 48). The presence of proteasomes along with unprocessed substrates in the aggresome has been regarded as a collateral deposition, but our data imply that the aggresome may be an active player in PQC and act as a temporary storage place for excess amounts of proteins destined for proteasomal degradation. Therefore, the spatial exchange of proteasomes under inhibitory conditions is critical for not only proteasome dynamics but also global proteostasis. Notably, the components in the JUNQ were highly fluidic through the cytosolic compartments, while those in the IPODs showed less mobility (34). PolyUb chains on inhibited proteasomes may trigger spontaneous and reversible phase separation in the aggresome (49, 50). In contrast to the proteins in the insoluble aggregates, which lack structural integrity and intrinsic activity, proteins in liquid droplets are functional and can reversibly diffuse to the cytoplasm. If the inhibition is persistent, then the nonfunctional proteasomes are degraded by autophagy (Fig. 2). Therefore, our findings lead to the conclusion that UPS and autophagy are alternative pathways but tightly connected in a compensatory feedback circuit, potentially involving phase transition.
We observed that most of the proteasome-interacting proteins showed increased interaction when proteasomes were inhibited (SI Appendix, Fig. S7). Some of these DAPs are related with the PQC, likely reflecting the consequence of a cellular effort to eliminate defective polypeptides (by BAG-6) (37, 51) or cytosolic misfolded proteins (by UBR proteins) (52, 53). Previous studies have identified that Ub receptors Rpn10/PSMD4 and Cue5/Tollip are involved in proteaphagy in yeast and plants, respectively (19, 20). Similarly, our MS analysis identified that many Ub receptors associated with mammalian proteasomes, such as PSMD4, Tollip, UBQLN2, SQSTM1, and RAD23, were highly up-regulated (SI Appendix, Table S1). The role of these Ub receptors in mammalian proteaphagy remains to be further characterized. Using in vitro reconstitution and small-molecule inhibitors, we also demonstrated that STUB1 directly mediates polyubiquitylation of inhibited proteasomes. Therefore, the function of STUB1 as a pivotal E3 Ub ligase for degrading nonnative proteins expands to proteasome homeostasis. Structural remodeling or posttranslational modifications may enable STUB1 to specifically recognize nonfunctional proteasomes. Since many other E3 Ub ligases interact with proteasomes, it is possible that proteasome quality control is mediated by different E3 enzymes in other cell types. Overall, it appears that the upstream regulatory cascade governing proteaphagy starts from the inhibition of active sites and ends in the polyubiquitylation of proteasomes.
It is conceivable that the adaptive mechanism for proteasome homeostasis has strong pathological implications. Under mild stress conditions, excessive amounts of misfolded proteins may clog the translocation channel or entry pore of the RP, thereby accumulating incompetent proteasomes in the insoluble aggresome and initiating de novo proteasome synthesis as a compensatory mechanism. However, chronic stress may impair the proteasome–aggresome–autophagy circuit and result in the development of proteopathies gradually over time. Our study provides a framework for integrating proteasome quality control mechanisms in mammals and other eukaryotes, suggesting that maintaining a functional proteasome pool in the cell has cytoprotective effects upon numerous proteopathic challenges. Further understanding of this pathway will provide effective therapeutic strategies to reduce the levels of aberrant proteins in cells. Specifically, a strategy to improve this regulatory circuit could potentially attenuate drug resistance to proteasome inhibition therapy in patients with multiple myeloma.
Materials and Methods
Analysis of Soluble and Insoluble Fractions.
Cultured cells were washed with ice-cold PBS and lysed with either Triton X-100 buffer (200 mM KCl, 20 mM 4-[2-hydroxyethyl]-1-piperazineethanesulfonic acid-KOH, pH 7.9, 1 mM MgCl2, 1 mM EDTA, 1% of Triton X-100, and 10% of glycerol) or RIPA buffer (50 mM Tris⋅HCl, pH 8, 1% of Nonidet P-40, 0.5% of deoxycholate, 0.1% of sodium dodecyl sulfate [SDS], and 150 mM NaCl), all supplemented with protease inhibitors. The lysates were then centrifuged at 16,000 × g for 30 min at 4 °C. The supernatants were designated as a detergent-soluble fraction. The pellets were washed with lysis buffer and resuspended in lysis buffer supplemented with 1% of SDS, then were sonicated for 10 s with a microtip sonicator, heated at 100 °C for 10 min, and were analyzed as a detergent-insoluble fraction. Equal volumes of each insoluble and soluble fraction were boiled for 10 min in SDS sample buffer and analyzed by IB.
Purification and In Vitro Ubiquitylation of 26S Proteasomes.
Human proteasomes were purified as previously described (54). For ubiquitylation of the purified proteasomes, 50 μM Ub, 150 nM UBE1, and 1 μM UBE2D2 were incubated for 10 min at room temperature. Then, 500 nM recombinant STUB1 and 60 nM purified proteasome were added. The reactions were allowed to proceed for 4 h at 37 °C and stopped by the addition of SDS sample buffer.
Quantitative LC-MS/MS Analysis.
LC-MS/MS analysis on a Q Exactive Plus Hybrid Quadrupole-Orbitrap mass spectrometer was performed as previously described (55). For label-free quantification of proteasome-associated proteins, the iBAQ algorithm (= ∑ peak intensities of all peptides matching to a protein of interest/∑ the number of theoretically observable peptides of the protein of interest) was utilized as part of the MaxQuant platform. A DAP was considered statistically significant if its fold change was ≥1.5 and if it had a P value <0.05. For a comparison of proteomes, two-sided t tests were performed with a significance level of 5%. All statistical analyses were performed in the Perseus software.
Hydrogen-1 NMR Screening and Compound Synthesis.
In order to identify compounds that can bind to STUB1 protein, we employed 1H ligand-observed NMR screening methods like the saturation-transfer difference experiment with second-generation BIONET Premium Fragment Library. The STD NMR experiments were carried out at 293 K on an 800-MHz Bruker NMR spectrometer. Preliminary NMR screenings were performed with a mixture of five different fragments. For fragments presenting the binding potential, secondary STD NMR experiments were carried out. Detailed synthesis methods for compound 153 are available in SI Appendix.
Data Availability Statement.
All protocols and source data are presented in the main text or the SI Appendix. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD019193. The plasmid constructs and reagents used in this study are available on request from the corresponding author.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Research Foundation of Korea (2019R1A2B5B02069530 to M.J.L., 2019R1A2C1005987 to J.H.L., 2017R1A6A3A11029936 to S.P., 2019R1A6A3A01094785 to W.H.C., 2016R1A2B3011389 to Y.T.K., and 2020R1A5A1019023 to Y.T.K., D.H., and M.J.L.), Korea Toray Science Foundation (800-20180524 to M.J.L.), and the Creative-Pioneering Researchers of Seoul National University (800-20160281 to M.J.L.). We gratefully acknowledge the Y. K. Kim (Korea University), J. Song (Yonsei University), and J. Y. Mun (Korea Brain Research Institute) laboratories for reagents and/or critical discussion.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD019193.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920327117/-/DCSupplemental.
References
- 1.Glickman M. H., Ciechanover A., The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 82, 373–428 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Rubinsztein D. C., Codogno P., Levine B., Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 11, 709–730 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stolz A., Ernst A., Dikic I., Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 16, 495–501 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Weidberg H., Shvets E., Elazar Z., Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 80, 125–156 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Youle R. J., Narendra D. P., Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 9–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galluzzi L. et al., Molecular definitions of autophagy and related processes. EMBO J. 36, 1811–1836 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pankiv S. et al., p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Korolchuk V. I., Mansilla A., Menzies F. M., Rubinsztein D. C., Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol. Cell 33, 517–527 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J. H., Park S., Kim E., Lee M. J., Negative-feedback coordination between proteasomal activity and autophagic flux. Autophagy 15, 726–728 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu W. K. et al., Macroautophagy and ERK phosphorylation counteract the antiproliferative effect of proteasome inhibitor in gastric cancer cells. Autophagy 6, 228–238 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Zhu K., Dunner K. Jr., McConkey D. J., Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene 29, 451–462 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kageyama S. et al., Proteasome dysfunction activates autophagy and the Keap1-Nrf2 pathway. J. Biol. Chem. 289, 24944–24955 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim E. et al., Dual function of USP14 deubiquitinase in cellular proteasomal activity and autophagic flux. Cell Rep. 24, 732–743 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Kawaguchi Y. et al., The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115, 727–738 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Kopito R. R., Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524–530 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Hao R. et al., Proteasomes activate aggresome disassembly and clearance by producing unanchored ubiquitin chains. Mol. Cell 51, 819–828 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka K., Ichihara A., Half-life of proteasomes (multiprotease complexes) in rat liver. Biochem. Biophys. Res. Commun. 159, 1309–1315 (1989). [DOI] [PubMed] [Google Scholar]
- 18.Russell S. J., Steger K. A., Johnston S. A., Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J. Biol. Chem. 274, 21943–21952 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Marshall R. S., Li F., Gemperline D. C., Book A. J., Vierstra R. D., Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in arabidopsis. Mol. Cell 58, 1053–1066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marshall R. S., McLoughlin F., Vierstra R. D., Autophagic turnover of inactive 26S proteasomes in yeast is directed by the ubiquitin receptor Cue5 and the Hsp42 chaperone. Cell Rep. 16, 1717–1732 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Waite K. A., De-La Mota-Peynado A., Vontz G., Roelofs J., Starvation induces proteasome autophagy with different pathways for core and regulatory particles. J. Biol. Chem. 291, 3239–3253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enenkel C., Proteasome dynamics. Biochim. Biophys. Acta 1843, 39–46 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Karmon O., Ben Aroya S., Spatial organization of proteasome aggregates in the regulation of proteasome homeostasis. Front. Mol. Biosci. 6, 150 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshall R. S., Vierstra R. D., Proteasome storage granules protect proteasomes from autophagic degradation upon carbon starvation. eLife 7, e34532 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Cohen-Kaplan V. et al., p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 113, E7490–E7499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pack C. G. et al., Quantitative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome. Nat. Commun. 5, 3396 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Asano S. et al., Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science 347, 439–442 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Peters L. Z., Hazan R., Breker M., Schuldiner M., Ben-Aroya S., Formation and dissociation of proteasome storage granules are regulated by cytosolic pH. J. Cell Biol. 201, 663–671 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters L. Z., Karmon O., Miodownik S., Ben-Aroya S., Proteasome storage granules are transiently associated with the insoluble protein deposit in Saccharomyces cerevisiae. J. Cell Sci. 129, 1190–1197 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Ouyang H. et al., Protein aggregates are recruited to aggresome by histone deacetylase 6 via unanchored ubiquitin C termini. J. Biol. Chem. 287, 2317–2327 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kisselev A. F., Callard A., Goldberg A. L., Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J. Biol. Chem. 281, 8582–8590 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Meng L. et al., Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl. Acad. Sci. U.S.A. 96, 10403–10408 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J., Breker M., Graham M., Schuldiner M., Hochstrasser M., AMPK regulates ESCRT-dependent microautophagy of proteasomes concomitant with proteasome storage granule assembly during glucose starvation. PLoS Genet. 15, e1008387 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaganovich D., Kopito R., Frydman J., Misfolded proteins partition between two distinct quality control compartments. Nature 454, 1088–1095 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwanhäusser B. et al., Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Besche H. C., Haas W., Gygi S. P., Goldberg A. L., Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry 48, 2538–2549 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minami R. et al., BAG-6 is essential for selective elimination of defective proteasomal substrates. J. Cell Biol. 190, 637–650 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehmann A., Niewienda A., Jechow K., Janek K., Enenkel C., Ecm29 fulfils quality control functions in proteasome assembly. Mol. Cell 38, 879–888 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Kleijnen M. F. et al., Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat. Struct. Mol. Biol. 14, 1180–1188 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Kim W. et al., Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng J. et al., A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Book A. J. et al., Affinity purification of the Arabidopsis 26 S proteasome reveals a diverse array of plant proteolytic complexes. J. Biol. Chem. 285, 25554–25569 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim H. T. et al., Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282, 17375–17386 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Specht S., Miller S. B., Mogk A., Bukau B., Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J. Cell Biol. 195, 617–629 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gicking A. M., Swentowsky K. W., Dawe R. K., Qiu W., Functional diversification of the kinesin-14 family in land plants. FEBS Lett. 592, 1918–1928 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Nakajima Y., Suzuki S., Environmental stresses induce misfolded protein aggregation in plant cells in a microtubule-dependent manner. Int. J. Mol. Sci. 14, 7771–7783 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicholas M. P. et al., Control of cytoplasmic dynein force production and processivity by its C-terminal domain. Nat. Commun. 6, 6206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu Z. C. et al., Ubiquitin orchestrates proteasome dynamics between proliferation and quiescence in yeast. Mol. Biol. Cell 28, 2479–2491 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun D., Wu R., Zheng J., Li P., Yu L., Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 28, 405–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yasuda S. et al., Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 578, 296–300 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Wang Q. et al., A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol. Cell 42, 758–770 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heck J. W., Cheung S. K., Hampton R. Y., Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl. Acad. Sci. U.S.A. 107, 1106–1111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szoradi T. et al., SHRED is a regulatory cascade that reprograms Ubr1 substrate specificity for enhanced protein quality control during stress. Mol. Cell 70, 1025–1037.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Choi W. H. et al., Open-gate mutants of the mammalian proteasome show enhanced ubiquitin-conjugate degradation. Nat. Commun. 7, 10963 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han D., Jin J., Woo J., Min H., Kim Y., Proteomic analysis of mouse astrocytes and their secretome by a combination of FASP and StageTip-based, high pH, reversed-phase fractionation. Proteomics 14, 1604–1609 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All protocols and source data are presented in the main text or the SI Appendix. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD019193. The plasmid constructs and reagents used in this study are available on request from the corresponding author.