Significance
Malaria is a deadly disease affecting hundreds of millions of people. Cerebral malaria is a type of severe disease with a high mortality rate. However, the causes leading to cerebral malaria, likely including parasite and host factors, are still elusive. Here we discover and investigate a host gene (receptor transporter protein 4 [RTP4]) that can regulate host type I interferon responses and symptoms of experimental cerebral malaria. RTP4 also significantly affects West Nile virus load in the brains of infected mice, but not in the heart and the spleen. This study reveals important roles of RTP4 in antimalarial and antiviral immunity, particularly in brain infection and pathology. RTP4 is a potential target for therapy in some neurological diseases.
Keywords: Plasmodium yoelii, Plasmodium berghei, interferon, gene knockout, signaling
Abstract
Infection by malaria parasites triggers dynamic immune responses leading to diverse symptoms and pathologies; however, the molecular mechanisms responsible for these reactions are largely unknown. We performed Trans-species Expression Quantitative Trait Locus analysis to identify a large number of host genes that respond to malaria parasite infections. Here we functionally characterize one of the host genes called receptor transporter protein 4 (RTP4) in responses to malaria parasite and virus infections. RTP4 is induced by type I IFN (IFN-I) and binds to the TANK-binding kinase (TBK1) complex where it negatively regulates TBK1 signaling by interfering with expression and phosphorylation of both TBK1 and IFN regulatory factor 3. Rtp4−/− mice were generated and infected with malaria parasite Plasmodiun berghei ANKA. Significantly higher levels of IFN-I response in microglia, lower parasitemia, fewer neurologic symptoms, and better survival rates were observed in Rtp4−/− than in wild-type mice. Similarly, RTP4 deficiency significantly reduced West Nile virus titers in the brain, but not in the heart and the spleen, of infected mice, suggesting a specific role for RTP4 in brain infection and pathology. This study reveals functions of RTP4 in IFN-I response and a potential target for therapy in diseases with neuropathology.
Malaria parasite infection stimulates a complex and delicately regulated host immune response (1, 2). In addition to IFN-γ, which is a central cytokine in malaria control (3, 4), IFN-I (IFN-α and IFN-β) also plays important roles in malaria parasite infections (5). IFN-I inhibits both blood stages of some Plasmodium yoelii strains during early infection and liver stages of Plasmodium berghei (6–8). Although the mechanisms remain largely unknown, studies have also shown that IFN-I inhibits T cell activation and IFN-γ production (9–13). For example, IFNAR1 deficiency boosts inducible T cell costimulator signaling and accelerates humoral immune responses during nonlethal blood-stage infections (13). Additionally, IFN-I and IFN-γ were reported to contribute to dendritic cell death during malaria parasite infection (14). Therefore, chronically high levels of IFN-I may also negatively impact host immune responses and restrict humoral immunity against malaria parasite blood-stage development (5, 12–15). The effects of IFN-I on parasite control and host pathology likely depend on a combination of host and parasite factors as well as the timing of IFN production.
Several pathways of IFN-I production involving pattern recognition receptors such as Toll-like receptors (TLRs), cyclic GMP-AMP synthase (cGAS), melanoma differentiation-associated protein 5(MDA5), retinoic acid-inducible gene I (RIG-I), and their signaling adaptors (myeloid differentiation primary response 88 [MyD88], stimulator of interferon genes [STING], mitochondrial antiviral-signaling protein [MAVS], and TIR-domain-containing adapter-inducing interferon-β [TRIF]) have been shown to play important roles in IFN-I production during malaria parasite infections (6–8, 16–20). MDA5 was shown to detect parasite RNA and initiate IFN-I responses to control blood stage parasitemia of P. yoelii strains (6) and prevent development of P. berghei liver stages (8). cGAS and STING were reported to detect parasite DNA (pDNA) and stimulate IFN-I responses in Plasmodium falciparum (21, 22) and P. yoelii (7) infections. Various TLRs have also been implicated in IFN-I responses and antimalarial immunity (19, 23–26). However, the timing and level of the IFN-I response must be regulated to avoid adverse effects on the host. In fact, many molecules such as suppressor of cytokine signaling 1 (SOCS1), FOS-like antigen-1 (FOSL1), and adenylate kinase 3 (AK) have been identified as negative regulators of IFN-I production during malaria parasite infections (7, 27, 28). Polymorphisms in the genomes of individual parasite strains could lead to variations in the nature and quantity of pathogen-associated molecular patterns or danger-associated molecular patterns released after malaria infection. Parasite DNA, RNA, glycosylphosphatidylinositol, protein-DNA complex, and hemozoin have been reported to stimulate IFN and inflammatory responses (6, 8, 16–18, 21, 25, 29–31). However, the complex regulatory network and the dynamics of IFN-I pathways in response to malaria parasite infections are still poorly understood.
Receptor transporter protein 4 (RTP4) is a member of the RTP family known to promote cell-surface expression of a group of G-protein–coupled receptors (GPCRs) that have been reported to mediate pain relief, bitter taste sense, or smell sensing (odorant receptors [ORs]) (32–34). Additionally, RTP4 expression can be induced upon viral infection, indicating that it may affect virus replication (35–38). However, the role of RTP4 in immune responses remains to be investigated. In this report, we demonstrate that RTP4 negatively regulates the IFN-I response by affecting TBK1 and IRF3 phosphorylation. We generated Rtp4 knockout (KO or Rtp4−/−) mice and show that Rtp4−/− mice produce higher levels of IFN-I than wild-type (WT) mice after malaria parasite infection. Compared to WT mice, Rtp4−/− mice had significantly lower parasitemia and fewer malaria-induced neurosymptoms after malaria parasite infection and had lower viral titers in the brain after West Nile virus (WNV) infection. This study reveals important roles of RTP4 in regulating IFN-I responses and antimalarial/viral immunity.
Results
Genetic Screen of Host–Parasite Interaction Identifies Rtp4 as an IFN-Stimulated Gene.
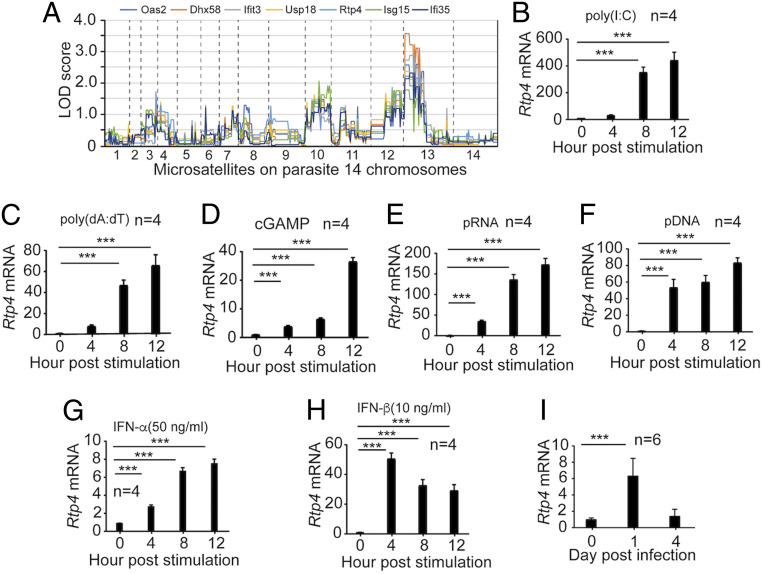
To investigate molecular mechanisms of host–parasite interaction, we previously performed Trans-species Expression Quantitative Trait Locus (Ts-eQTL) analysis to identify host genes that respond to malaria parasite infection (27). We identified several host gene clusters containing known IFN-stimulated genes (ISGs). Among the ISG clusters was cluster #243 that contained Rtp4 and many known ISGs or genes that function in IFN-I responses such as Oas2, Dhx58, Ifit3, Usp18, Isg15, and Ifi35 (Dataset S1). To further characterize the Rtp4 gene functionally, we first plotted the genome-wide pattern of logarithm of the odds (LOD) scores (GPLS) of selected ISGs in the cluster and showed similar GPLSs with a major LOD score peak on parasite chromosome 13 (Fig. 1A). The result suggests significant linkages of these ISGs to a parasite locus on chromosome 13 and potential functions of RTP4 in IFN-I pathways in response to malaria parasite infection. Microarray analysis (27) using RNA extracted from spleens of mice 4 d postinfection (p.i.) with P. yoelii parasites (17XNL or N67) showed higher Rtp4 expression in the splenic tissue of N67-infected mice than those infected with 17XNL (least squares mean [Lsmean]) = −0.141 for 17XNL-infected mice; Lsmean = 0.776 for N67-infected mice). Given that the N67 parasite induces a stronger IFN-I response than the 17XNL parasite (27), these results further support the possibility that Rtp4 may be induced by IFN-I during malaria parasite infection.
Fig. 1.
RTP4 is induced by IFN-I, ligands of IFN-I response, and malaria parasite infection. (A) Plots of genome-wide pattern of LOD scores. The LOD scores were reported previously (27). The dashed lines indicate the boundaries of the parasite’s 14 chromosomes. (B–F) RTP4 mRNA transcript levels in MEFs (1 × 106) from WT mice after stimulation with various ligands for 4, 8, and 12 h. (B) RTP4 mRNA levels after stimulation with poly(I:C) (1.5 μg) for 4, 8, and 12 h; (C) with poly(dA:dT) (1 μg); (D) with cGAMP (4 μg); (E) with pRNA (5 μg); and (F) with pDNA (5 μg). (G and H) Stimulation of RTP4 expression by IFN-α and IFN-β. MEF cells (1 × 106) were stimulated with IFN-α (G, 100 ng) and IFN-β (H, 20 ng) for 4, 8, and 12 h, respectively. (I) RTP4 transcript levels from the spleens of WT mice p.i. with N67 parasites. Means and SD are from replicates (n) as indicated. One-way ANOVA: ***P < 0.001. All experiments were independently repeated with similar results.
Increased Rtp4 Expression after Stimulation with DNA/RNA Ligands or Parasite Infection.
We next investigated RTP4 expression in response to stimulation by ligands that can stimulate IFN-I production, pDNA, parasite RNA (pRNA), and malaria parasite infection. Mouse embryonic fibroblasts (MEFs) produced significantly higher RTP4 messenger RNA (mRNA) after stimulation with poly(I:C) or poly(dA:dT) at 8 and 12 h (Fig. 1 B and C). Similarly, treatment of MEFs with cGAMP, pRNA, and pDNA significantly increased RTP4 mRNA at 4, 8, and 12 h post stimulation (Fig. 1 D–F). Direct treatment of MEFs with IFN-α (50 ng/mL) and IFN-β (10 ng/mL) also significantly increased RTP4 expression (Fig. 1 G and H). In vivo, RTP4 mRNA levels significantly increased in the spleens of malaria parasite-infected mice at day 1, but not at day 4 p.i. (Fig. 1I). These results demonstrate that RTP4 can be induced by IFN-I and IFN-I pathways, including those induced by parasite infection.
RTP4 Is a Negative Regulator of IFN-I Responses In Vitro.
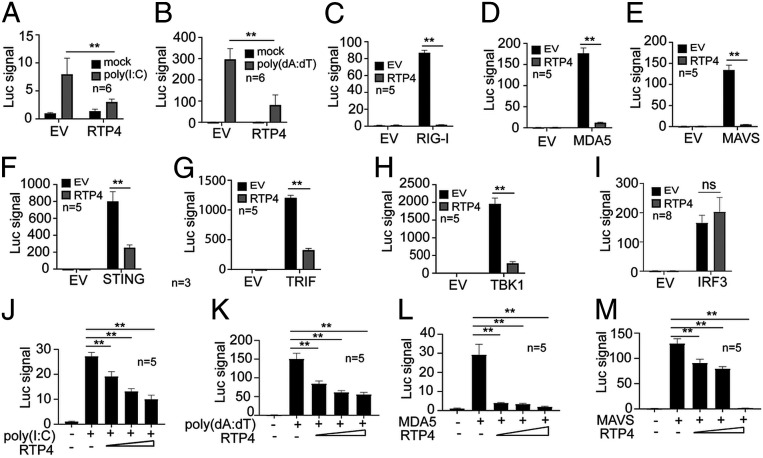
We first investigated the role of RTP4 in regulating IFN-I responses in vitro. Expression of RTP4 in human HEK 293T cells inhibited poly(I:C)- and poly(dA:dT)-stimulated IFN-β promoter activity (Fig. 2 A and B). Similarly, RTP4 expression significantly reduced IFN-β promotor activity stimulated by cotransfecting plasmids expressing RIG-I, MDA5, MAVS, STING, TRIF, or TBK1 (Fig. 2 C–H). In contrast, RTP4 expression did not reduce IFN-β promoter activity stimulated by cotransfection with a constitutively active phosphomimetic IRF3-5D mutant plasmid (Fig. 2I). RTP4 expression inhibited IFN-I responses stimulated by poly(I:C), poly(dA:dT), MDA5, and MAVS in a dose-dependent manner (Fig. 2 J–M). However, ectopic expression of RTP4 in 293T cells did not significantly affect NF-κB–mediated signaling after stimulation with poly(I:C) or poly(dA:dT) (SI Appendix, Fig. S1). Together, these results suggest that RTP4 acts as a negative regulator of IFN-I response downstream of MAVS, STING, and TRIF, and may affect the TBK1 complex.
Fig. 2.
RTP4 is a negative regulator of IFN-I response in vitro. (A and B) Luciferase signals (Luc signal) driven by IFN-β promoter after transfection of 2 × 105 293T cells with 100 ng empty plasmid vector (EV) or 100 ng plasmid encoding RTP4 after stimulation with poly(I:C) (500 ng) or poly(dA:dT) (125 ng). (C–I) Cotransfection of 2 × 105 293T cells with empty vector or plasmid (100 ng each) encoding the indicated proteins and RTP4. Luciferase signals were measured as in B. (J–M) Dose–responses of luciferase signals from 2 × 105 293T cells after cotransfection of the indicated genes with different amounts of plasmid (50, 100, and 200 ng) encoding RTP4. Mann–Whitney test, means and SD (n = 5 to 8); **P < 0.01; ns, not significant. All experiments were independently repeated with similar results.
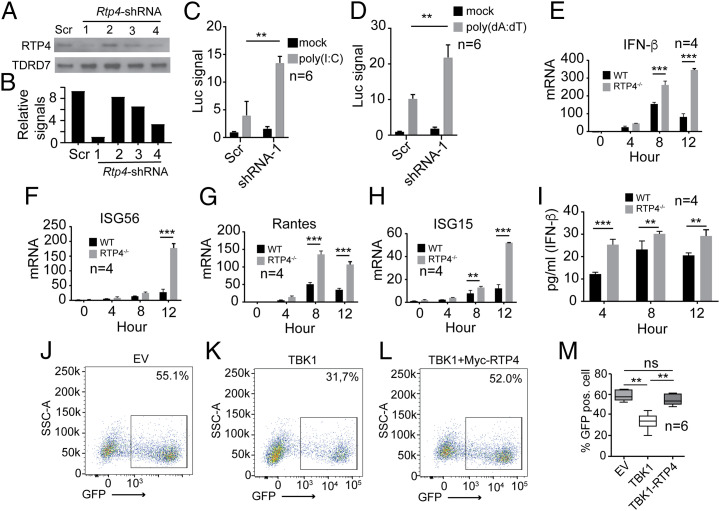
To further characterize RTP4 functions during IFN-I response, we designed four short-hairpin RNAs (shRNAs) to knock down RTP4 expression (Dataset S2). RTP4 shRNA #1 was the most potent, reducing RTP4 expression by 9.3-fold (Fig. 3 A and B). Knockdown of Rtp4 in 293T cells with shRNA #1 significantly increased the luciferase signal driven by the IFN-β promoter after poly(I:C) (Fig. 3C) or poly(dA:dT) stimulation (Fig. 3D). To confirm these results, we genetically disrupted the Rtp4 gene in 293T cells using CRISPR/Cas9 gene-editing technology and four gene-specific single-guide RNAs (sgRNAs) as described in Materials and Methods (SI Appendix, Fig. S2A and Dataset S2). Clonal cell lines with a disrupted Rtp4 gene were confirmed by PCR amplification (SI Appendix, Fig. S2B) and DNA sequencing (SI Appendix, Fig. S2C). WT and Rtp4−/− cells were stimulated with 2 μg poly(I:C) for 0, 4, 8, and 12 h, and mRNA was extracted for qPCR analysis of selected genes using primers listed in Dataset S2. Significantly higher mRNA transcripts for IFN-β and other ISGs such as ISG56, Rantes, and ISG15 were detected in Rtp4−/− cells at 8 and/or 12 h post stimulation with poly(I:C) (Fig. 3 E–H). In addition, IFN-β protein was more abundant in supernatants from Rtp4−/− cells compared to WT cells at 4, 8, and 12 h after poly(I:C) stimulation (Fig. 3I). These results demonstrate that RTP4 deficiency significantly increases expression of IFN-β and ISGs following immune stimulation in vitro, confirming that RTP4 is a negative regulator of IFN-I responses.
Fig. 3.
Knockdown or knockout of RTP4 increases expression of IFN-β and ISGs. (A) Reduction of RTP4 protein expression after shRNA knockdown (2 μg each shRNA in 2 × 106 293T cells). Scr, shRNA with scrambled sequence; 1 to 4, four different shRNAs (see Dataset S2 for sequences). A plasmid encoding TDRD7 was used as internal control. (B) Scanned signals from A after subtraction from those of β-actin. (C) Luciferase signals (Luc signal) driven by IFN-β promoter in 2 × 105 293T cells transfected with RTP4-specific shRNA #1 or Scr shRNA (Mock) after poly(I:C) stimulation (500 ng). (D) Same as in C except stimulation with poly (dA:dT) (125 ng). (E–H) mRNA levels of IFN-β (E), ISG56 (F), Rantes (G), and ISG15 (H) in WT or Rtp4−/− 293T cells at 0, 4, 8, or 12 h after poly(I:C) stimulation (750 ng). mRNA levels were measured using RT-qPCR as described in Materials and Methods. (I) IFN-β protein in culture media in WT or Rtp4−/− 293T cells after poly(I:C) stimulation (750 ng). IFN-β was measured using an ELISA kit (PBL Assay Science). Means and SD from replicates (n) as indicated; two-way ANOVA: **P < 0.01; ***P < 0.001. (J–L) Representative images of flow cytometry counts of green fluorescent (GFP) positive cells infected with vesicular stomatitis virus (VSV). The 293T cells were transfected with empty EV, vector containing TBK1 gene (100 ng), or TBK1 vector plus vector containing myc-RTP4 gene (200 ng). After 24 h, the cells were infected with VSV-eGFP (MOI = 0.001) for 16 h and counted using flow cytometry. (M) Plot of percentages of GFP-positive cells. Mann–Whitney test (n = 6), mean + SD; **P < 0.01.
We next performed vesicular stomatitis virus (VSV) replication assays in vitro to evaluate the effects of RTP4 on TBK1-mediated inhibition of VSV replication. The 293T cells were cotransfected with plasmids encoding TBK1, MYC-tagged RTP4 (MYC-RTP4), or an empty vector control. After 24 h, the cells were infected with VSV-enhanced green fluorescent protein (eGFP) at a multiplicity of infection (MOI) of 0.001 plaque-forming units (PFU) per cell for 16 h and analyzed by flow cytometry. The results showed that RTP4 reversed TBK1-mediated inhibition of VSV replication in vitro (Fig. 3 J–M). These results further confirm that RTP4 negatively affects IFN-I response and antiviral activity.
RTP4 Binds to TBK1, but Not to MDA5 or RIG-I.
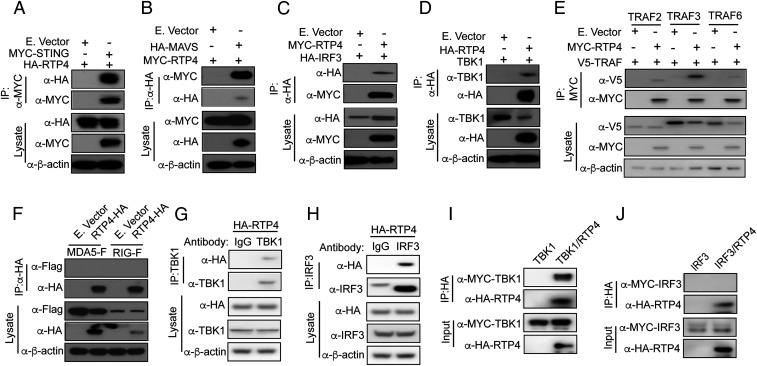
Our in vitro cotransfection results suggest that RTP4 may interact with the TBK1 complex. We next performed coimmunoprecipitation (co-IP) to investigate whether RTP4 binds to STING, TBK1, IRF3, and/or TRAFs. We transfected 293T cells with plasmids expressing HA-, MYC-, or V5-tagged proteins and pulled down the tagged proteins with antibodies against specific tags after stimulating cells with poly(I:C) for 24 h. The proteins were separated in sodium dodecyl sulfate/polyacrylamide gel electrophoresis gels, blotted to polyvinylidene difluoride membranes, and detected using anti-tag or anti-TBK1 antibodies. RTP4 was pulled down by STING (Fig. 4A), MAVS (Fig. 4B), and IRF3 (Fig. 4C). Similarly, RTP4 pulled down TBK1, TRAF2 (weak), TRAF3, and TRAF6 (weak), but not MDA5 or RIG-I (Fig. 4 D–F). We also used anti-TBK1 and anti-IRF3 antibodies to pull down endogenous TBK1 and IRF3, respectively. An anti-HA antibody was used to detect RTP4 after RTP4-HA plasmid transfection. Again, HA-tagged RTP4 was pulled down by both anti-TBK1 and anti-IRF3 antibodies (Fig. 4 G and H). These results suggest that RTP4 binds to the STING/TBK1/TRAFs/IRF3 complex. To identify the specific RTP4 interacting partners, we used a cell-free protein expression system (PURExpress, NEB) to synthesize HA- or MYC-tagged RTP4, TBK1, or IRF3 in vitro and anti-HA or anti-MYC antibodies to pull down RTP4. RTP4 pulled down TBK1 (Fig. 4I), but not IRF3 (Fig. 4J), suggesting that RTP4 interacts directly with TBK1.
Fig. 4.
Interaction of RTP4 with STING, MAVS, TBK1, and IRF3 in vitro. The 293T cells (1 × 106) were cotransfected with plasmids encoding RTP4 and STING, MAVS, TBK1, IRF3, or TRAFs (1 μg each) tagged with either HA, MYC, or V5. The tagged molecules were pulled down and detected using anti-tag antibodies. IP, indicating antibody used in immunoprecipitation; lysate, total proteins from cell lysis; β-actin was used as protein loading control. (A–F) The co-IP and protein expression levels as indicated for STING (A), MAVS (B), IRF3 (C), TBK1 (D), TRAF2, TRAF3, and TRAF6 (E), and MDA5/RIG-I (F). (G and H) 239T cells (1 × 107) were transfected with 5 μg of RTP4-HA plasmid only, and anti-TBK1 or anti-IRF3 mouse antibody was used to pull down TBK1 and IRF3, respectively. RTP4 was detected using anti-HA antibody. Mouse IgG was used as control for potential nonspecific binding in the pulldown experiments. Note: The band in the IgG lane in H is likely a IgG heavy chain. (I and J) HA- or MYC-tagged RTP4, TBK1, or IRF3 were synthesized in vitro using a cell-free protein expression system (PURExpress, NEB). Anti-HA or anti-MYC antibodies were then used to pull down RTP4. TBK1 and IRF3 were detected using anti-MYC (I, TBK1) and anti-HA (J, IRF3).
We also performed an immunofluorescence assay to determine RTP4 localization within 293T cells. RTP4 primarily localized to the cytoplasm and appeared to be associated with membrane-bound vesicles (SI Appendix, Fig. S3 A–E) even upon poly(I:C) stimulation (SI Appendix, Fig. S3 F–J). TBK1 localization was also cytoplasmic and might partially colocalize with RTP4 (SI Appendix, Fig. S3 E–J, yellow dots). However, we cannot rule out that some of the yellow dots are simply due to spatially overlapped signals, not actual colocalizations. Additional experiments using higher-resolution methods such as immune-electron microscopy may be necessary to provide a definitive answer on RTP4 and TBK1 colocalization.
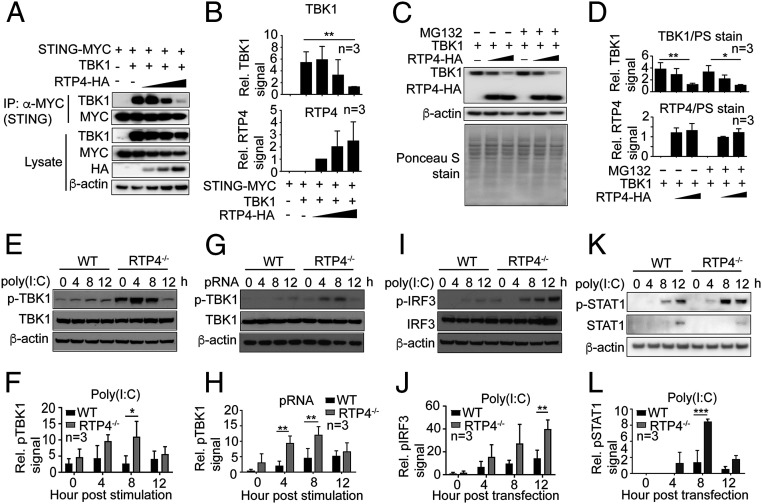
RTP4 Reduces TBK1 Binding to STING and Phosphorylation of TBK1 and IRF3.
We next investigated how RTP4 affects the functions of STING and the TBK1 complex to regulate IFN-I responses. Increasing (0.1, 1, or 2 μg) amounts of an RTP4-encoding plasmid transfected into 293T cells decreased TBK1 and STING expression and reduced binding of STING to TBK1 (Fig. 5 A and B). However, addition of proteasome inhibitor MG132 did not appear to affect TBK1 protein levels (Fig. 5 C and D). In addition, compared to WT cells, RTP4-deficient MEF cells had a higher level of phosphorylated TBK1 after stimulation with poly(I:C) (Fig. 5 E and F) or pRNA (Fig. 5 G and H). Similarly, IRF3 phosphorylation and protein expression was higher in the Rtp4−/− cells than in WT cells after poly(I:C) stimulation (Fig. 5 I and J), although the increase in IRF3 phosphorylation occurred later than that of TBK1. Moreover, Rtp4−/− cells had higher (significant at 8 h p.i.) STAT1 phosphorylation than WT cells after poly(I:C) stimulation (Fig. 5L). These results suggest that RTP4 can interfere with TBK1 and IRF3 protein expression and/or phosphorylation after activation of IFN-I responses by DNA or RNA, which may also affect expression of many ISGs or genes in IFN-I response pathways.
Fig. 5.
RTP4 impairs interaction of STING with TBK1 and phosphorylation of TBK1, IRF3, and STAT1. (A) The co-IP of STING and TBK1 in the presence of increasing amounts of RTP4 (50, 100, and 200 ng plasmid). The 293T cells (1 × 106) were transfected with plasmids encoding RTP4-HA, STING-MYC (500 ng), and TBK1 (500 ng). Proteins were pulled down using anti-MYC and detected with anti-TBK1 (dilution 1:1,000) or anti-MYC (dilution 1:1,000) antibodies. Protein expression in cell lysates was also detected using the same antibodies as well as anti-HA (dilution 1:1,000). (B) Plots of scanned signals of TBK1 and RTP4 from A after adjusting for protein loading (β-actin) and TBK1 or RTP4 protein expression in the lysates. (C and D) TBK1 and RTP4 protein levels by western blot (C) and plots (D) of scanned signals from C with or without MG132 treatment (10 μM) for 6 h. The 293T cells (1 × 106) were transfected with plasmids encoding TBK1 (500 ng) and RTP4-HA (0 ng, 500 ng, and 1.0 μg), and proteins were detected using anti-TBK1 and anti-HA. Ponceau S-stained total proteins and β-actin were used as protein loading controls. (E) Western blot detection of TBK1 protein expression and phosphorylation from 1 × 106 WT or Rtp4−/− cells after poly(I:C) (1.5 μg) stimulation. (F) Plot of scanned signals of TBK1 phosphorylation from E after adjusting for protein loading (β-actin) and TBK1 expression. (G and H) The same experiment as in E and F after pRNA (5 μg) stimulation. (I and J) The same experiment as in E and F but for pIRF3 levels. (K–L) The same experiment as in E and F but for pSTAT1 levels. Note: Proteins in E and I were from the same protein preparations, and only one β-actin loading control was performed. Two-way ANOVA, mean and SD (n = 3); *P < 0.05; **P < 0.01; ***P < 0.001. All of the experiments were independently repeated at least three times with similar results.
Higher IFN-I in Rtp4−/− Mice after Malaria Infection or Ligand Stimulations.
To better characterize RTP4 functions in vivo, we generated Rtp4 KO mice using CRISPR/Cas9. Two sgRNAs were designed to disrupt the Rtp4 gene by targeting both exons (SI Appendix, Fig. S4). Homozygous Rtp4 KO (Rtp4−/−) mice were obtained after backcrossing and genotyping offspring of mice heterozygous for the disrupted Rtp4 gene (SI Appendix, Fig. S5 A–D). Disruption of the Rtp4 gene did not have any observable adverse effects on mouse health or body weight gain (SI Appendix, Fig. S5E).
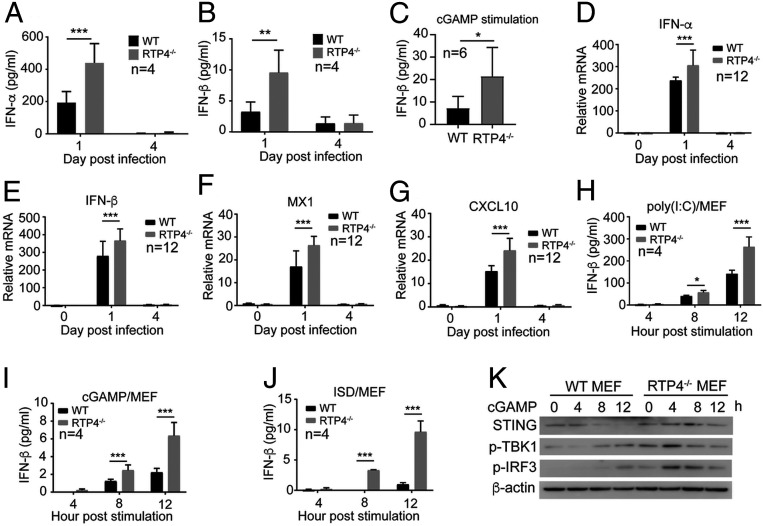
We next infected WT and Rtp4−/− mice with malaria parasites and measured serum IFN-α/β and mRNA transcripts in the spleen. Significantly higher levels of serum IFN-α and IFN-β were observed in Rtp4−/− mice compared to WT mice at day 1 p.i., but not at day 4 p.i. with N67 parasites (Fig. 6 A and B). Consistently, Rtp4−/− mice injected intravenously (i.v.) with cGAMP (50 μg/mouse) produced significantly higher IFN-β levels compared to WT mice (Fig. 6C). Additionally, mRNA levels for IFN-α/β and ISGs (MX1 and CXCL10) were significantly higher in the spleens of Rtp4−/− mice relative to WT mice at day 1 p.i. (Fig. 6 D–G). We also isolated and cultured MEFs from WT and Rtp4−/− mice and stimulated the cells with poly(I:C), cGAMP, or IFN stimulatory DNA (ISD) for 4, 8, and 12 h. Significantly higher IFN-β levels were detected in the supernatants of Rtp4−/− MEFs relative to WT MEFs at 8 and 12 h after stimulation with all three ligands (Fig. 6 H–J). Stimulation with cGAMP showed higher STING expression and higher levels of phosphorylation of TBK1 and IRF3 in Rtp4−/− MEFs relative to WT MEFs between 4 and 8 h after stimulation (Fig. 6K). Consistently, significantly higher fluorescent signals of phosphorylated IRF3 (pIRF3) were detected in the Rtp4−/− MEFs than in WT cells at 4 h post cGAMP stimulation (SI Appendix, Fig. S6 A and B).
Fig. 6.
Disruption of Rtp4 gene in MEF cells and mice increases IFN-I responses. (A and B) Detection of IFN-α (A) and IFN-β (B) protein levels in the blood of Rtp4−/− and WT mice at days 1 and 4 p.i. with N67 parasites. (C) IFN-β levels in Rtp4−/− and WT mice at 6 h after i.v. injection of cGAMP (50 μg/mice). (D–G) mRNA levels for IFN-α (D), IFN-β (E), MX1 (F), and CXCL10 (G) in the spleens of Rtp4−/− and WT mice at days 0, 1, and 4 p.i. with N67 parasites. (H–J) IFN-β levels in culture media after stimulation of MEF cells with poly(I:C) (H), cGAMP (I), and ISD (J) for 4, 8, and 12 h. IFN-α and IFN-β from mouse blood and supernatants of MEF cultures were measured using ELISA kits (PBL Assay Science). mRNA levels were measured using qPCR as described in Materials and Methods. Means and SD from replicates (n) as indicated; two-way ANOVA: *P < 0.05; **P < 0.01; ***P < 0.001. (K) Western blot detection of STING protein expression and levels of TBK1 and IRF3 phosphorylation in Rtp4−/− and WT MEF cells after cGAMP stimulation. All experiments were independently repeated with similar results.
Reduction in Brain Viral Load and Cerebral Malarial Symptoms in Mice Lacking Rtp4.
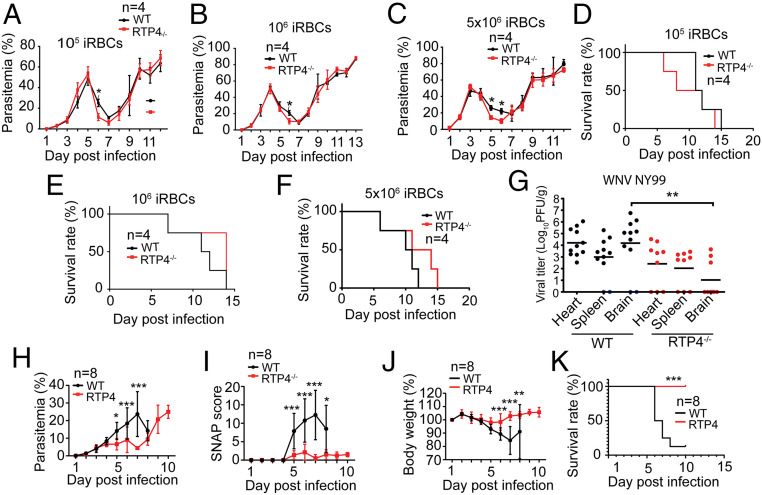
We next injected i.v. different dosages (1 × 105, 1 × 106, and 5 × 106) of N67-infected red blood cells (iRBCs) into WT and Rtp4−/− mice and evaluated parasite growth (parasitemia) and host survival. Significantly lower parasitemia was observed at day 6 p.i. in the Rtp4−/− mice for all three injection groups (dosages), although the parasitemia over the course of infection was similar in WT and Rtp4−/− mice (Fig. 7 A–C). No significant differences in host survival rate were observed between WT and Rtp4−/− mice (Fig. 7 D–F). These results are consistent with previous observations of infected mice in which higher levels of IFN-I at day 1 p.i. correlated with lower N67 parasitemia at day 6 p.i. (6). We also previously observed a subtle but significantly higher parasitemia at day 6 p.i. in mice deficient for MDA5 and MAVS (6). The elevated serum IFN-α/β levels at day 1 p.i. could contribute to the lower parasitemia at day 6 p.i. in N67-infected Rtp4−/− mice.
Fig. 7.
Deficiency of RTP4 significantly decreases parasitemia, cerebral malarial symptoms, and viral load in the brain. WT and Rtp4−/− mice (n = 4) were injected i.v. with N67-infected red blood cells at three different dosages (1 × 105, 1 × 106, and 5 × 106). Parasitemia and host survival rates were monitored by counting daily thin blood smears and pain scores. (A–C) Dynamics of parasitemia after infection of WT and Rtp4−/− mice with different dosages of P. yoelii N67 iRBCs. (D–F) Host survival curves of the infections in A–C. Parasitemia was quantified using Giemsa-stained smears of blood samples using a microscope. (G) Viral titers in the heart, spleen, and brain at day 7 p.i. with 103 PFUs of WNV. Each dot represents an individual mouse, and the data are pooled from three independent experiments. (H) Dynamics of parasitemia after infection of WT and Rtp4−/− mice with P. berghei ANKA iRBCs (1 × 106). (I) SNAP scores of infected WT and Rtp4−/− mice. (J) Body weights of infected WT and Rtp4−/− mice. (K) Host survival rates after P. berghei ANKA infection. Note: Only one mouse remained in the WT mouse group after day 8, and measurements for WT were not done. For malaria infections, means and SD from replicates (n) are as indicated; for WNV infections, means of 9 to 14 mice per group are indicated. Two-way ANOVA (A–C and H–J), logrank (D–F and K) , and one-way ANOVA (G): *P < 0.05; **P < 0.01; ***P < 0.001. F test compared variances for H–J with F = 1.53 and DFn = 9. All experiments were independently repeated with similar results.
The limited effect of RTP4 deficiency on N67 infection prompted us to investigate its potential effect on WNV infection because of the importance of IFN-I responses in controlling viral infections. WT and Rtp4−/− mice were inoculated subcutaneously with 103 PFUs of WNV via the rear foot pad, and viral titers in the brain, heart, and spleen were quantified at day 7 p.i. Significantly lower viral titers were found in the brains of Rtp4−/− mice relative to WT mice but not in the hearts or spleens (Fig. 7G). These results demonstrate that RTP4 may specifically modulate the host’s antiviral response within the brain.
We hypothesized a potential role for RTP4 in neurotropic infections based on the observation of lower WNV load in the brain of Rtp4−/− mice and tested WT and Rtp4−/− mice for susceptibility to P. berghei ANKA that causes neurologic symptoms and is considered an experimental cerebral malaria (ECM) parasite. Indeed, Rtp4−/− mice had significantly lower parasitemia, lower simple neuroassessment of asymmetric impairment (SNAP) scores for cerebral malaria (39), and a higher body weight relative to WT mice between days 5 and 8 p.i. (Fig. 7 H–J). Rtp4−/− mice also survived significantly longer than WT mice (Fig. 7K). Together, these results suggest that RTP4 may negatively affect outcomes of some neurotropic diseases, possibly through modulation of IFN-I responses, microglia activation, and parasite growth in the blood.
Improved IFN-I Response, Microglia Activation, and Brain Pathology in P. berghei-Infected Rtp4−/− Mice.
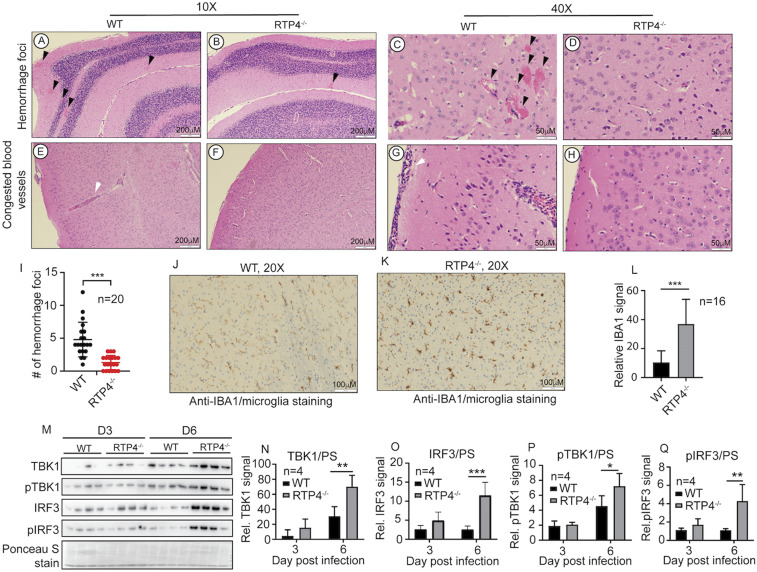
To investigate the mechanism underlying the improved cerebral malaria symptoms and host survival, we performed histochemistry to examine pathological changes in the brain, spleen, and the liver in WT and Rtp4−/− mice infected with P. berghei ANKA. Many hemorrhagic foci and blood vessels filled with iRBCs and host white blood cells were observed in brains of WT mice at day 6 p.i., particularly in the cerebellum, but were infrequent or absent in infected Rtp4−/− mice (Fig. 8 A–H). To quantitate the hemorrhagic foci, we examined the brain meningeal cortex from the cerebellum to the midbrain and enumerated hemorrhagic foci in 20 fields randomly selected from infected WT and Rtp4−/− mice. Significantly more hemorrhagic foci (average of 4.8 foci per microscopic field in WT vs. 1.3 foci in Rtp4−/− mice; P < 0.01) were observed in WT mice compared to Rtp4−/− mice (Fig. 8I). In contrast, no detectable pathological changes were observed in the spleen and the liver of P. berghei ANKA-infected WT and Rtp4−/− mice (SI Appendix, Fig. S7 A and B). The results show more tissue damage in the brain than in the spleen and liver of WT mice, similar to the higher viral loads in the brains of WNV-infected WT mice, and a specific effect of RTP4 on pathology related to brain infection.
Fig. 8.
Brain pathological changes, microglia activation, and IFN-I response in mice infected with P. berghei ANKA. (A–D) Images showing hemorrhage foci at the cerebellum. (E–H) Blood vessels with iRBCs and host white cells from the cerebellum. (I) Numbers of hemorrhage foci from 20 randomly selected microscopic images from the cerebellum of P. berghei-infected WT and Rtp4−/− mice. Brain samples from mice at day 6 p.i. were fixed in 10% buffered formalin, embedded in paraffin, and sequential coronal sections of the whole brain were stained with hematoxylin and eosin (H&E) or antibodies as described. H&E-stained brain tissue sections were examined under a light microscope at 10× and 40× magnifications. (J and K) Anti-IBA1 staining (brown color) of microglia in the brain from cerebellar cortex of infected WT (J) and Rtp4−/− (K) mice, as described in Materials and Methods. (L) Quantitative microglia signals as shown in J and K were scanned from anti-IAB1–stained images (16 each) of brain tissues of WT and Rtp4−/− mice at day 6 p.i. with P. berghei using ImageJ. (M) Western blot of protein expression and phosphorylation of TBK1 and IRF3 in microglia purified from the whole brains of infected WT and Rtp4−/− mice (n = 4) at days 3 and 6 p.i. with P. berghei ANKA. Scanned signals of Ponceau S-stained total proteins as loading controls. (N–Q) Plots of scanned protein and phosphorylation signals of TBK1 and IRF3 from M. For I, L, and N–Q, two-way ANOVA, mean and SD: *P < 0.05; **P < 0.01; ***P < 0.001.
In a transcriptomic study of microglia from brains of mice infected with P. berghei ANKA, IFN-I was shown to activate microglia to produce inflammatory cytokines during the acute phase of ECM (40). Therefore, we stained brain sections from WT and Rtp4−/− mice for ionized calcium-binding adaptor molecule 1 (IBA1) as a specific marker of microglia. Significantly stronger IBA1-staining signals were observed in microglia of Rtp4−/− mice compared to those of WT mice at day 6 p.i. (Fig. 8 J–L). We next investigated IFN-I response by examining levels of protein expression and phosphorylation of TBK1 and IRF3 in microglia isolated from mice on days 3 and 6 p.i. with P. berghei ANKA. The results showed that microglia from Rtp4−/− mice had significantly higher protein and phosphorylation levels of TBK1 and IRF3 at 6 d p.i. (Fig. 8 M–Q), suggesting that the increased levels of pTBK1 and pIRF3 were largely due to increased protein levels. Consistently, significantly higher IFN-α and IFN-β mRNA levels were detected in microglia of infected Rtp4−/− mice than WT mice (SI Appendix, Fig. S6 C and D). These results suggest increased IFN-I responses in the brain of infected Rtp4−/− mice. Therefore, enhanced IFN-I responses and microglia activation may play a role in the protective effect of RTP4 deficiency during P. berghei ANKA infection.
Discussion
This study investigates functions of RTP4 in regulating IFN-I production during malaria parasite and viral infections. The Rtp4 gene clustered with various ISGs through Ts-eQTL analysis in response to malaria parasite infection (27). RTP4 was shown to be strongly up-regulated in individuals with moderate/severe laboratory-confirmed influenza (41). RTP4 is expressed at significantly lower levels in the STAT1-, STAT2-, and IRF9-deficient primary murine mixed glial cell cultures, and its expression can be induced by IFN-α (42). Elevated RTP4 expression was also observed in mice after treatment with IFN-α or poly(I:C) (37). These observations suggest that RTP4 expression can be induced by IFN-I after malaria parasite and viral infections and may play important roles in these infections. However, whether RTP4 can regulate IFN-I and host immune responses during infection remains to be determined. Here we demonstrate that RTP4 is a negative regulator of the IFN-I response. Overexpression of RTP4 in 239T cells significantly reduced IFN-β promoter activity after stimulation with ligands such as poly(I:C) and poly(dA:dT) or overexpression of STING, MAVS, RIG-I, and MDA5. In contrast, inhibition of Rtp4 expression or genetic disruption of Rtp4 increased IFN-I responses or production. We also generated Rtp4 genetic KO cell lines and mice to confirm its effects on IFN-I production and its role in malarial and viral infections. Mechanistically, we show that the inhibitory effect of RTP4 on the IFN-I response is mediated in part through inhibition of TBK1 and IRF3 protein expression and/or phosphorylation by its binding to TBK1 to limit production of IFN-I. RTP4 can be pulled down with many proteins in the TBK1 complex of IFN-I response pathways; however, co-IP experiments using in vitro-expressed proteins suggest that RTP4 binds to TBK1 to interfere with TBK1-mediated IFN-I responses. Together, these results demonstrate a function for RTP4 as a negative regulator of IFN-I pathways in addition to its reported function in protein transport (32–34). Our model for the functional roles of RTP4 in IFN-I responses is summarized in SI Appendix, Fig. S8A.
Our data suggest that RTP4 contributes to antimalaria parasite immunity by modulating host IFN-I responses that may affect early parasitemia during N67 infection. Significantly higher serum levels of IFN-α/β were observed in Rtp4−/− mice compared to WT mice at day 1 p.i. with N67 parasites. This increase in IFN-I may lead to a significantly lower parasitemia at day 6 p.i. in the Rtp4−/− mice. In WT mice, RTP4 helps to suppress IFN-I levels leading to higher parasitemia at day 6 p.i. Similarly, significant increases in day 6 parasitemia, but not in host survival rate, were observed in N67 infection of Mda5−/− and Mavs−/− mice, which could be linked to a decrease in IFN-I levels at day 1 p.i. (6, 7). Therefore, early differences in IFN-I levels may have a direct impact on later-stage parasite growth during N67 infection. RTP4 is likely part of the host regulatory circuit in limiting TBK1 signaling to avoid potential damage caused by continuous IFN production. Infections with malaria parasites affect a large number of genes (>1,000) and induce complicated networks of host responses that could be significantly linked to many parasite genomic loci (27). Rtp4 is just one of the genes that can affect the outcome of host immune responses, with some impact on early growth of the N67 parasite.
Although RTP4 deficiency had a limited effect on parasitemia in mice infected with N67 parasites, Rtp4−/− mice had dramatically reduced neurological symptoms following infection with P. berghei ANKA parasites. Significantly lower SNAP scores and longer host survival times were observed in Rtp4−/− mice compared to WT mice after P. berghei ANKA infection. Additionally, significant reduction in hemorrhage foci in the brain of Rtp4−/− mice was observed compared to WT mice, with no obvious pathological differences in the liver and spleen between WT and Rtp4−/− mice. These results were similar to the observation of a significant reduction in WNV load in the brain of Rtp4−/− mice compared to that of WT mice, but not in the heart or the spleen. These results suggest that RTP4 may have specific functions in the brain or the central nervous system (CNS) in some infections. Our observations are consistent with previous reports of increased RTP4 expression in the brains of mice infected with chikungunya virus and Newcastle disease virus (36, 43). RTP4 mRNA was also significantly up-regulated in the hypothalamus after morphine administration (44), suggesting a specific function of RTP4 in the brain. The protective effects of RTP4 deficiency after P. berghei ANKA infection could be due to alteration in IFN-I responses in the brain because levels of protein and phosphorylation of TBK1 and IRF3 as well as mRNAs of IFN-α and IFN-β increased in microglia of infected Rtp4−/− mice at day 6 p.i. However, we cannot rule out that the improvement in SNAP scores and better host survival was also associated with the reduced parasitemia in these mice. Interestingly, the effects of RTP4 on TBK1 and IRF3 are different in in vitro-transfected 293T cells and in microglia from P. berghei ANKA-infected mice. RTP4 mostly affects TBK1 and IRF3 phosphorylation in 239T or MEF cells, but influences TBK1 and IRF3 protein levels in microglia from infected mice, suggesting potentially different regulating mechanisms in vitro and in vivo. RTP4 overexpression has been reported to inhibit yellow fever virus replication in a dose-dependent manner in STAT1-deficient human fibroblasts (35) and to moderately inhibit human norovirus replication in HG23 cells (38). The inhibitory effects of RTP4 appear to conflict with our observation of RTP4 being an inhibitor of IFN-I response. However, the inhibitory effects of RTP4 on replications of yellow fever virus and human norovirus were observed in STAT1-deficient cells and in HG23 cells, respectively. These effects could be mediated through other unknown mechanisms such as G-protein–mediated pathways or by directly inhibiting viral replication.
Both IFN-I and IFN-II have been shown to contribute to ECM pathology in P. berghei ANKA infection (45). Whereas Ifn-γR1−/− mice were fully resistant to ECM after sporozoite or merozoite infection, Ifnar−/− mice were partially protected with reduced microvascular pathology that was associated with reduced T cell responses in the brain. Additionally, transcriptome analysis of microglia from the brain of mice infected with P. berghei ANKA suggests that IFN-β activates microglia to produce inflammatory cytokines during the acute phase of ECM (40). Our observation of higher levels of microglia activation in the Rtp4−/− mice suggests potential roles of RTP4 and IFN-β in regulating microglia responses to P. berghei ANKA infection. In contrast, injections of a recombinant human IFN-α prevented death by cerebral malaria, with decreased parasitemia, less sequestered iRBCs and leukocytes in cerebral vessels, reduced ICAM-1 expression in brain endothelial cells, and increased levels of IFN-γ (46). Therefore, the roles of IFN-I in brain pathology and host survival after P. berghei ANKA infection are likely time and dosage dependent, which requires further investigation. It is possible that IFN-α and IFN-β have different effects on host responses and disease pathology. Nonetheless, our observations of the specific effects of RTP4 on P. berghei ANKA and WNV infections may open up important research opportunities for studying the potential roles of RTP4 on diseases affecting the brain and CNS. It will be interesting to investigate the effects of RTP4 on other neurotropic pathogens (e.g., Eastern equine encephalitis virus and Toxoplama gondii), and even extend these findings into studies of neurodegenerative diseases such as Alzheimer’s disease.
Increased levels of RTP4 transcript were detected in the spleens of N67-infected mice, suggesting activation of gene expression after malaria infection. RTP4 transcripts also increased in MEFs after stimulation with poly(I:C), poly(dA:dT), cGAMP, pRNA, pDNA, and IFN-α/β in vitro. These results confirm that Rtp4 is an ISG that is induced by IFN-I response and/or IFN-I. However, we still do not know the exact signaling pathways leading to increased RTP4 expression. Similar to serum IFN-α/β levels, RTP4 transcript was down-regulated at day 4 p.i. with N67 parasites. Again, the mechanism of RTP4 down-regulation requires further investigation. Elucidation of the mechanisms regulating RTP4 expression may help to design strategies to modulate IFN-I responses and improve parasite clearance and disease management.
RTP4 was initially characterized as a chaperone protein that helps transport GPCR to the plasma membrane (32). It binds to opioid receptors, participates in the proper folding of the μ-δ heterodimer of the receptor, and helps translocate the proteins out of the Golgi apparatus to the cell surface (32, 44). In a recent study, RTP4 expression was found to specifically regulate μ-opioid receptor abundance at the cell surface (44). Both RTP3 and RTP4 are colocalized with bitter taste receptors (TAS2Rs) and improve TAS2R16 plasma membrane localization (33). Additionally, overexpression of RTP4 decreased ubiquitination of the receptors, preventing degradation of the proteins. Other members of the RTP family, RTP1 and RTP2, have also been shown to play significant roles in the translocation of GPCR odorant receptors to the plasma membrane and promote the functioning of ORs (34). A search of the functional protein association network database STRING (https://string-db.org) shows two groups of proteins interacting with RTP4 (SI Appendix, Fig. S8B): those encoded by ISGs (IFIT1, IFIT3, IFI44, OASL2, and USP18) and by olfactory receptor genes (OLFR1444, OLFR66, OLF4522, OLFR414, and OLFR569). Our results further show that RTP4 inhibits IFN-I responses, in addition to its known function of enhancing specific GPCR expression. The combined effects of inhibiting IFN-I response and enhancing specific GPCR expression can influence the capacity to fight infections in the brain (SI Appendix, Fig. S8C). It is not clear what the functional connections for RTP4 in GPCR transport are and what role this connection plays in regulating IFN-I responses. The induction of RTP4 expression after malaria parasite/viral infections and the subsequent effects on IFN-I response will likely influence its function as a chaperone in transporting GPCRs to the plasma membrane. Whereas increased RTP4 expression can inhibit IFN-I responses, elevated RTP4 expression may also enhance the activities of pathways mediated by specific GPCRs. RTP4 is located on the membrane of many cellular organelles, and its functions are likely dependent on its cellular localizations. Additionally, it is not known whether other members of the RTP family can also regulate the IFN-I response or can compensate for loss of RTP4. RTP1 and RTP2 were not detected in the spleen, and RTP3 expression levels did not change in Rtp4−/− mice with or without parasite infections (data not shown). Despite many unanswered questions about RTP4’s functions, our current study demonstrates an important function for RTP4 in regulating host immune responses to malaria parasite and viral infections, which may be explored for disease treatment and management. Inhibition of RTP4 expression may help reduce parasite or virus replication and help to alleviate symptoms of cerebral malaria.
Materials and Methods
Malaria Parasites and Mice.
Procedures for infection of mice with P. yoelii nigeriensis N67 or P. berghei ANKA parasites were as reported previously (27, 47). Briefly, parasites were thawed from frozen stocks and injected into naive C57BL/6 mice to initiate infection. An inoculum containing 1 × 105 to 5 × 106 iRBCs from the donor mice suspended in 100 µL sterile phosphate-buffered saline, pH 7.4, was injected i.v. into experimental mice (n = 3 to 6 mice per clone or strain). Inbred female C57BL/6 mice, aged 6 to 8 wk old, were obtained from Charles River Laboratory, Jackson Laboratory, or National Institute of Allergy and Infectious Diseases/Taconic repository, NIH. Parasitemia was monitored by microscopic examination of Giemsa-stained thin tail blood smears. All experiments, including those in SI Appendix, Supplementary Methods, were independently repeated with similar results. Additional methods can be found in SI Appendix.
Study Approval.
All animal procedures for this study were performed in accordance with the protocol approved (approval #LMVR11E) by the Institutional Animal Care and Use Committee (IACUC) at NIAID and Rockefeller University’s IACUC following the guidelines of the Public Health Service Policy on Humane Care and Use of Laboratory Animals and Association for Assessment and Accreditation of Laboratory Animal Care. All mice were maintained under pathogen-free conditions.
Supplementary Material
Acknowledgments
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH; by Public Health Service Awards F32 AI133910 (to A.W.A.) and R01 AI091707 (to C.M.R.); by grants (R01CA101795 and 1U54CA210181) from National Cancer Institute, the NIH (to R.F.W.). We thank Bradley Otterson of the NIH library for editing, William M. Schneider for scientific input and manuscript editing, and Corrine Quirk for assistance with mouse colonies.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006492117/-/DCSupplemental.
Data Availability.
All data are available in the figures, SI Appendix, SI Appendix figures, and Datasets S1 and S2.
References
- 1.Stevenson M. M., Riley E. M., Innate immunity to malaria. Nat. Rev. Immunol. 4, 169–180 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Riley E. M., Wahl S., Perkins D. J., Schofield L., Regulating immunity to malaria. Parasite Immunol. 28, 35–49 (2006). [DOI] [PubMed] [Google Scholar]
- 3.King T., Lamb T., Interferon-γ: The Jekyll and Hyde of malaria. PLoS Pathog. 11, e1005118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lacerda-Queiroz N. et al., Mechanism of splenic cell death and host mortality in a Plasmodium yoelii malaria model. Sci. Rep. 7, 10438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mooney J. P., Wassmer S. C., Hafalla J. C., Type I interferon in malaria: A balancing act. Trends Parasitol. 33, 257–260 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Wu J. et al., Strain-specific innate immune signaling pathways determine malaria parasitemia dynamics and host mortality. Proc. Natl. Acad. Sci. U.S.A. 111, E511–E520 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu X. et al., Cross-regulation of two type I interferon signaling pathways in plasmacytoid dendritic cells controls anti-malaria immunity and host mortality. Immunity 45, 1093–1107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liehl P. et al., Host-cell sensors for Plasmodium activate innate immunity against liver-stage infection. Nat. Med. 20, 47–53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haque A. et al., Type I interferons suppress CD4+ T-cell-dependent parasite control during blood-stage Plasmodium infection. Eur. J. Immunol. 41, 2688–2698 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Haque A. et al., Type I IFN signaling in CD8- DCs impairs Th1-dependent malaria immunity. J. Clin. Invest. 124, 2483–2496 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montes de Oca M. et al., Type I interferons regulate immune responses in humans with blood-stage Plasmodium falciparum infection. Cell Rep. 17, 399–412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zander R. A. et al., Type I interferons induce T regulatory 1 responses and restrict humoral immunity during experimental malaria. PLoS Pathog. 12, e1005945 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebina I. et al., IFNAR1-Signalling obstructs ICOS-mediated humoral immunity during non-lethal blood-stage Plasmodium infection. PLoS Pathog. 12, e1005999 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura T., Kimura K., Yui K., Yoshida S., Reduction of conventional dendritic cells during Plasmodium infection is dependent on activation induced cell death by type I and II interferons. Exp. Parasitol. 159, 127–135 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Loughland J. R. et al., Plasmacytoid dendritic cells appear inactive during sub-microscopic Plasmodium falciparum blood-stage infection, yet retain their ability to respond to TLR stimulation. Sci. Rep. 7, 2596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gowda N. M., Wu X., Gowda D. C., TLR9 and MyD88 are crucial for the development of protective immunity to malaria. J. Immunol. 188, 5073–5085 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao X. et al., Increased CD40 expression enhances early STING-mediated type I interferon response and host survival in a rodent malaria model. PLoS Pathog. 12, e1005930 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X., Gowda N. M., Kumar S., Gowda D. C., Protein-DNA complex is the exclusive malaria parasite component that activates dendritic cells and triggers innate immune responses. J. Immunol. 184, 4338–4348 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gazzinelli R. T., Kalantari P., Fitzgerald K. A., Golenbock D. T., Innate sensing of malaria parasites. Nat. Rev. Immunol. 14, 744–757 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Baccarella A., Fontana M. F., Chen E. C., Kim C. C., Toll-like receptor 7 mediates early innate immune responses to malaria. Infect. Immun. 81, 4431–4442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma S. et al., Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity 35, 194–207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallego-Marin C. et al., Cyclic GMP-AMP synthase is the cytosolic sensor of Plasmodium falciparum genomic DNA and activates type I IFN in malaria. J. Immunol. 200, 768–774 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gazzinelli R. T., Denkers E. Y., Protozoan encounters with toll-like receptor signalling pathways: Implications for host parasitism. Nat. Rev. Immunol. 6, 895–906 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y. et al., TLR4 and TLR9 signals stimulate protective immunity against blood-stage Plasmodium yoelii infection in mice. Exp. Parasitol. 170, 73–81 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Coban C. et al., Toll-like receptor 9 mediates innate immune activation by the malaria pigment hemozoin. J. Exp. Med. 201, 19–25 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mockenhaupt F. P. et al., Toll-like receptor (TLR) polymorphisms in African children: Common TLR-4 variants predispose to severe malaria. Proc. Natl. Acad. Sci. U.S.A. 103, 177–182 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J. et al., Genome-wide analysis of host-Plasmodium yoelii interactions reveals regulators of the type I interferon response. Cell Rep. 12, 661–672 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai B., Wu J., Yu X., Su X.-z et al., FOSL1 inhibits type I interferon responses to malaria and viral infections by blocking TBK1 and TRAF3/TRIF interactions. mBio 8, e02161-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schofield L. et al., Glycosylphosphatidylinositol toxin of Plasmodium up-regulates intercellular adhesion molecule-1, vascular cell adhesion molecule-1, and E-selectin expression in vascular endothelial cells and increases leukocyte and parasite cytoadherence via tyrosine kinase-dependent signal transduction. J. Immunol. 156, 1886–1896 (1996). [PubMed] [Google Scholar]
- 30.Krishnegowda G. et al., Induction of proinflammatory responses in macrophages by the glycosylphosphatidylinositols of Plasmodium falciparum: Cell signaling receptors, glycosylphosphatidylinositol (GPI) structural requirement, and regulation of GPI activity. J. Biol. Chem. 280, 8606–8616 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Togbe D. et al., Murine cerebral malaria development is independent of toll-like receptor signaling. Am. J. Pathol. 170, 1640–1648 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Décaillot F. M., Rozenfeld R., Gupta A., Devi L. A., Cell surface targeting of mu-delta opioid receptor heterodimers by RTP4. Proc. Natl. Acad. Sci. U.S.A. 105, 16045–16050 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behrens M. et al., Members of RTP and REEP gene families influence functional bitter taste receptor expression. J. Biol. Chem. 281, 20650–20659 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Saito H., Kubota M., Roberts R. W., Chi Q., Matsunami H., RTP family members induce functional expression of mammalian odorant receptors. Cell 119, 679–691 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Schoggins J. W. et al., A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nair S. R., Abraham R., Sundaram S., Sreekumar E., Interferon regulated gene (IRG) expression-signature in a mouse model of chikungunya virus neurovirulence. J. Neurovirol. 23, 886–902 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Hoyo-Becerra C. et al., Rapid regulation of depression-associated genes in a new mouse model mimicking interferon-α-related depression in Hepatitis C virus infection. Mol. Neurobiol. 52, 318–329 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Dang W. et al., IRF-1, RIG-I and MDA5 display potent antiviral activities against norovirus coordinately induced by different types of interferons. Antiviral Res. 155, 48–59 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Riggle B. A. et al., MRI demonstrates glutamine antagonist-mediated reversal of cerebral malaria pathology in mice. Proc. Natl. Acad. Sci. U.S.A. 115, E12024–E12033 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Capuccini B. et al., Transcriptomic profiling of microglia reveals signatures of cell activation and immune response, during experimental cerebral malaria. Sci. Rep. 6, 39258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davenport E. E., Antrobus R. D., Lillie P. J., Gilbert S., Knight J. C., Transcriptomic profiling facilitates classification of response to influenza challenge. J. Mol. Med. (Berl.) 93, 105–114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W. et al., Type I interferon-regulated gene expression and signaling in murine mixed glial cells lacking signal transducers and activators of transcription 1 or 2 or interferon regulatory factor 9. J. Biol. Chem. 292, 5845–5859 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patiño D. A. G., The contribution of mast cells to antiviral immune responses: Characterization and functional definition of receptor transporter protein 4 (RTP4). PhD thesis, Free University of Berlin. (2013). Available at https://d-nb.info/1034527789/34.
- 44.Fujita W. et al., Regulation of an opioid receptor chaperone protein, RTP4, by morphine. Mol. Pharmacol. 95, 11–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palomo J. et al., Type I interferons contribute to experimental cerebral malaria development in response to sporozoite or blood-stage Plasmodium berghei ANKA. Eur. J. Immunol. 43, 2683–2695 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Vigário A. M. et al., Recombinant human IFN-alpha inhibits cerebral malaria and reduces parasite burden in mice. J. Immunol. 178, 6416–6425 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Li J. et al., Linkage maps from multiple genetic crosses and loci linked to growth-related virulent phenotype in Plasmodium yoelii. Proc. Natl. Acad. Sci. U.S.A. 108, E374–E382 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the figures, SI Appendix, SI Appendix figures, and Datasets S1 and S2.