Significance
We examined recent whole-genome data of 53 organisms and found that the substantial differences in their genome sizes can be largely explained by the proportion of transposable elements (TEs) within them. TEs coexist with their host largely because CpG methylation suppresses their transcription. Genome expansion is therefore dependent to a large extent on the action of DNA methyltransferases, which evolved at roughly the same time as TEs. A long-term outcome of CpG methylation is an increase in C-to-T transition mutations both in the TEs and host DNA, which leads to a decreased proportion of CpG dinucleotides over evolutionary time. The survival of TEs in the host genome also provides extra DNA that may be repurposed for host regulatory functions.
Keywords: DNA methylation, transposable element, genome size
Abstract
Multicellular eukaryotic genomes show enormous differences in size. A substantial part of this variation is due to the presence of transposable elements (TEs). They contribute significantly to a cell’s mass of DNA and have the potential to become involved in host gene control. We argue that the suppression of their activities by methylation of the C–phosphate–G (CpG) dinucleotide in DNA is essential for their long-term accommodation in the host genome and, therefore, to its expansion. An inevitable consequence of cytosine methylation is an increase in C-to-T transition mutations via deamination, which causes CpG loss. Cytosine deamination is often needed for TEs to take on regulatory functions in the host genome. Our study of the whole-genome sequences of 53 organisms showed a positive correlation between the size of a genome and the percentage of TEs it contains, as well as a negative correlation between size and the CpG observed/expected (O/E) ratio in both TEs and the host DNA. TEs are seldom found at promoters and transcription start sites, but they are found more at enhancers, particularly after they have accumulated C-to-T and other mutations. Therefore, the methylation of TE DNA allows for genome expansion and also leads to new opportunities for gene control by TE-based regulatory sites.
Eukaryotic genomes contain much more DNA than necessary for the protein-coding and noncoding genes they contain, and they show as much as 64,000-fold variation in their sizes (1). Although the functional significance of these size differences remains enigmatic (2), much of the variability can be explained by the presence of repetitive DNA, particularly transposable elements (TEs), which were identified by Barbara McClintock many years ago (3). The human genome, for example, has three main classes of TEs that together make up more than 45% of human DNA: long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), and endogenous retroviruses (ERVs). These elements have inserted themselves and transposed in eukaryotic germlines in waves during evolution and have the potential to modify gene control in the host organism (4–6).
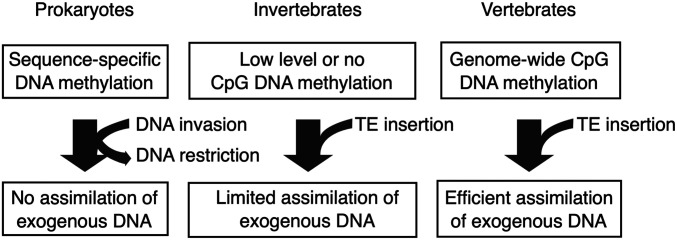
Thirty years ago, Bestor (7) proposed that an important function of DNA cytosine methylation was to silence the expression of TEs. Given the potentially lethal effects of ectopic expression of these elements, methylation would allow for the coexistence of TEs and the host in a type of host–parasite relationship. An important additional posit was that prokaryotic DNA methyltransferases began by protecting the host from foreign DNA integration but evolved into enzymes which allowed for the coexistence of foreign DNA within the host genome (Fig. 1). The transition from the relatively narrow and rare sequence specificities of prokaryotic DNA methyltransferases to eukaryotic enzymes recognizing the simple and frequent C–phosphate–G (CpG) dinucleotide therefore enabled the accommodation of TEs in the host. Bird and others (8), using methylation-sensitive restriction enzymes, subsequently found that invertebrates had either very little or highly compartmentalized regions of CpG methylation, whereas vertebrates had intergenic and far more widespread modification patterns (Fig. 1).
Fig. 1.
Model illustrating differing roles for DNA methylation in handling exogenous DNA. DNA methylation in prokaryotes is part of their restriction/modification system of host defense. Invertebrates can accommodate TE DNA to a limited extent due to low prevalence of DNA CpG methylation. Vertebrates, especially mammals, have extensive CpG methylation on a genomic scale and can tolerate high levels of TEs.
These insightful observations were made before the advent of whole-genome sequencing and did not fully appreciate that cytosine methylation was inherently strongly mutagenic (9). This is due to a much-increased rate of C-to-T transition mutations at methylation sites such as CpGs. In turn, this produces a strong decrease in the observed/expected (O/E) ratio of CpGs (a measure of the loss of CpG dinucleotides; Materials and Methods) in the DNA of organisms having CpG methylation. For example, human DNA shows a CpG O/E ratio of about 0.25, with methylated CpG sites having a half-life of about 35 million y in the germline (10). By measuring the underrepresentation of this dinucleotide in modern species, we can infer the prevalence of CpG DNA methylation in evolutionary time.
By examining the complete DNA sequences of 53 organisms, we can confirm the validity of Bestor’s original hypotheses and have uncovered some concepts, namely that the integration of TEs leads not only to genome expansion and methylation of the TE DNA but also to the methylation of the flanking host DNA. While the evolutionary driver for expansion remains unknown, there is a clear correlation between genome size and CpG underrepresentation, suggesting that DNA methylation led to substantial increases in DNA mass. We also confirmed earlier suggestions that TEs can contribute to the formation of new cis-regulatory DNA elements actually bound by transcription factors in living cells. However, in general, this contribution results in a modest number of binding events compared with those contributed by non-TE DNA and often requires that the TEs have undergone evolutionary alterations in the form of C-to-T and other mutations.
Results
Genome Size and the CpG O/E Ratio Are Negatively Correlated.
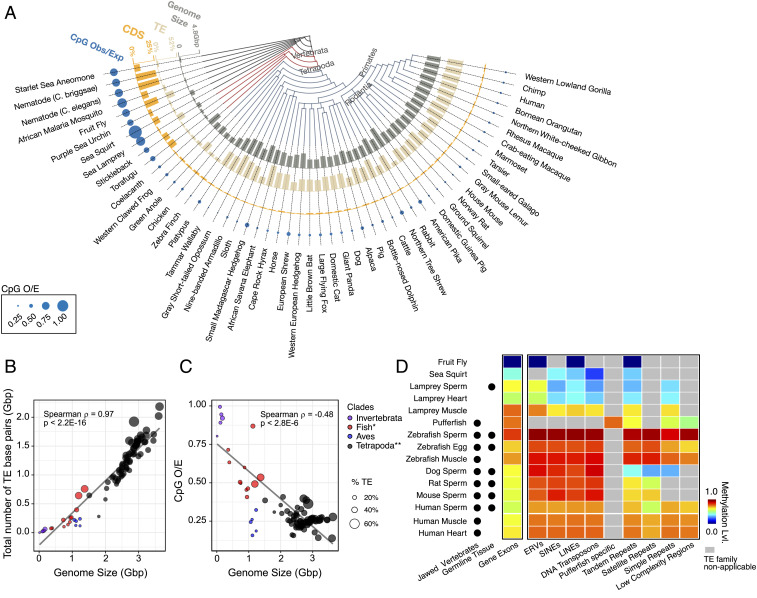
We used whole-genome DNA-sequencing data for invertebrates and vertebrates to assess the CpG O/E ratio, size of the genome, and percentage of the genome occupied by TEs (Fig. 2A). Invertebrates have small genomes, few TEs, higher percentages of coding sequence, and little CpG loss because they do not have strong intergenic CpG methylation (11). Fish and amphibians have intermediate-size genomes and relatively few TEs, but their lower CpG O/E ratio did show some CpG loss. Birds are strongly CpG-deficient even though they have relatively smaller genomes and a lower percentage of TEs. It has been suggested that birds may have lost substantial portions of their genomes during the transition to flight (2), and DNA methylation may have first allowed for a genome expansion before that loss. Most tetrapods show a two- to three-fold increase in genome size relative to fish, with a high percentage of TEs and fewer CpGs.
Fig. 2.
DNA methylation enabled genome expansion via TEs in higher-order vertebrates. (A) Genome size (billion bp), % TE, % coding sequence (CDS), and CpG O/E ratio, shown on a taxonomy tree. (B) Total number of TE bases versus genome size. (C) CpG O/E ratio versus genome size. *Fish includes Actinopterygii (ray-finned fishes), Chondrichthyes (cartilaginous fishes), and lamprey (a jawless fish) but not coelacanth. **Although part of Tetrapoda, birds (Aves) are colored separately. (D) Average methylation levels of gene exons and TEs in different organisms.
For the 53 organisms examined, Fig. 2B shows a positive linear relationship between genome size and TE content (Spearman’s ρ = 0.97, P < 2.2 × 10−16) and Fig. 2C shows an inverse relationship between the CpG O/E ratio and genome size (Spearman’s ρ = −0.48, P < 2.8 × 10−6). We also used data from whole-genome bisulfite sequencing of extant species to determine the distribution of CpG methylation (as opposed to CpG depletion) within them (SI Appendix, Fig. S1). Species with larger genomes have higher levels of intergenic DNA methylation, which is mostly attributable to the methylation of TEs (Fig. 2D). In invertebrates such as sea squirts and early jawless vertebrates such as lamprey, the TEs are less methylated relative to the genic regions. The ERVs are an exception; they are relatively more methylated compared with other TEs in early vertebrate evolution (Fig. 2D). In most vertebrates having a substantial TE content (Fig. 2A), we found that all TE families had equal or higher methylation than did gene exons (Fig. 2D). A likely explanation is that the TEs are able to play a role in the increase of genome size because transcriptional suppression by DNA methylation reduces their possible deleterious consequences to the host (Fig. 1).
Transposable Elements Can Initiate CpG Loss in Host DNA.
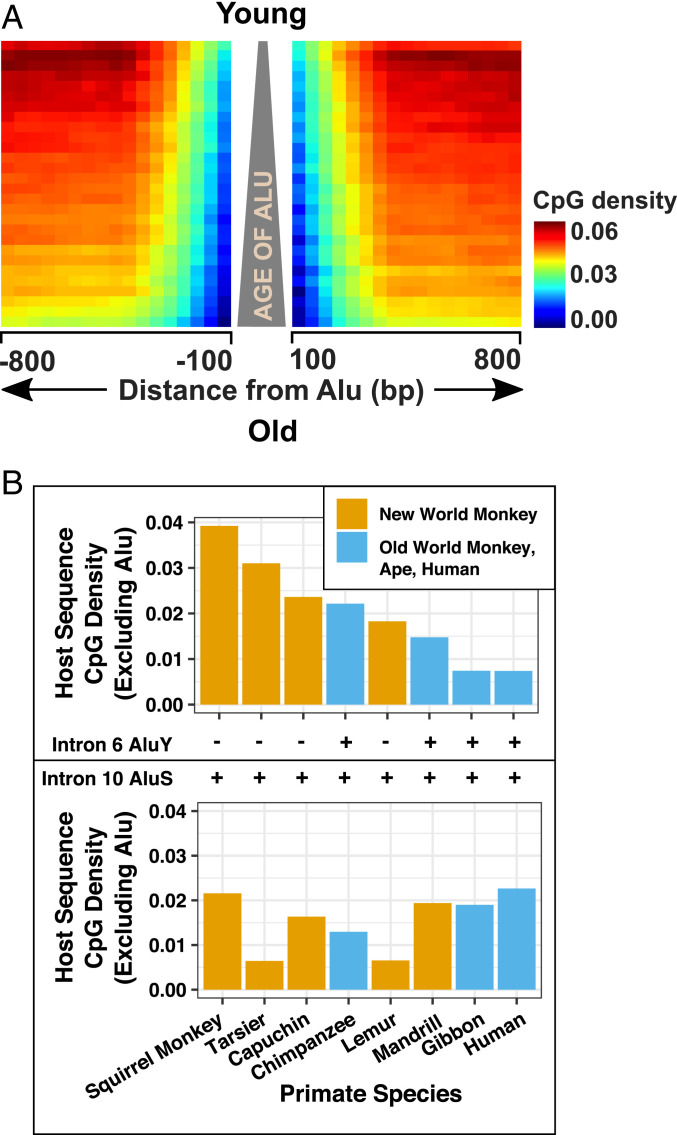
The data in Fig. 2 are consistent with the idea that TEs are likely responsible for both the expansion of the genome and its subsequent CpG depletion. We next asked whether the de novo methylation of a TE results in CpG loss not only in the TE but also in the surrounding host DNA. We used Alu elements as examples because unlike LINEs and ERVs, which are CpG-rich only in their promoter/long terminal repeat (LTR) regions, Alus are CpG-rich throughout their 280-bp sequences before their initial insertion or transposition (10, 12). We know that Alus can act as “methylation centers” after insertion into host DNA, whereby methylation subsequently spreads into the flanking DNA (13–15). To determine whether such methylation spreading might subsequently result in CpG loss in the flanking host DNA over evolutionary time, we arrayed Alu elements in the human genome according to their age and then generated a heatmap of the surrounding host CpG content (Fig. 3A). Evolutionarily older Alus show more CpG loss on their immediate flanks than younger ones.
Fig. 3.
Neighboring CpG density in flanking DNA is negatively correlated with the evolutionary age of SINEs (Alu). (A) More than a million Alu elements in the human genome are arrayed according to their evolutionary ages estimated by decreasing CpG density (defined as the number of CpGs per base pairs of DNA sequence). We used density rather than O/E ratios for this analysis because we assume GC content is relatively stable within short distances. Densities of 800-bp sequence flanking every Alu element were then calculated using a sliding 100-bp window and displayed as a heatmap. Details are described in Materials and Methods. (B) Reduced CpG density of TP53 intron 6 excluding an AluY which was inserted in Old World monkeys and apes, compared with New World monkeys which do not contain AluY; P = 0.057, Wilcoxon’s rank-sum test, two-tailed. No significant difference is seen in intron 10 of TP53 where a more ancient AluS insertion took place in the common ancestor of primates; P = 0.34, Wilcoxon’s rank-sum test, two-tailed.
To confirm this genome-wide analysis, we focused on a relatively recent AluY insertion into intron 6 of the TP53 gene which took place in Old World monkeys and apes after they had separated from New World monkeys (10). We studied eight primate species and confirmed a reduction in the CpG density of intron 6 in Old World monkeys and apes compared with New World monkeys (Fig. 3B). On the other hand, such a relationship was not seen in intron 10 of the same gene in which a more ancient AluS insertion took place in a common ancestor of both Old and New World monkeys (Fig. 3B). Collectively, these results suggest that an unmethylated, CpG-rich TE inserted into the germline is suppressed by DNA methylation, and that methylation can subsequently be spread into the surrounding DNA, leading eventually to the loss of CpG sites in neighboring DNA.
Evolutionarily Old TEs Are Found at Enhancers but Not at Transcription Start Sites.
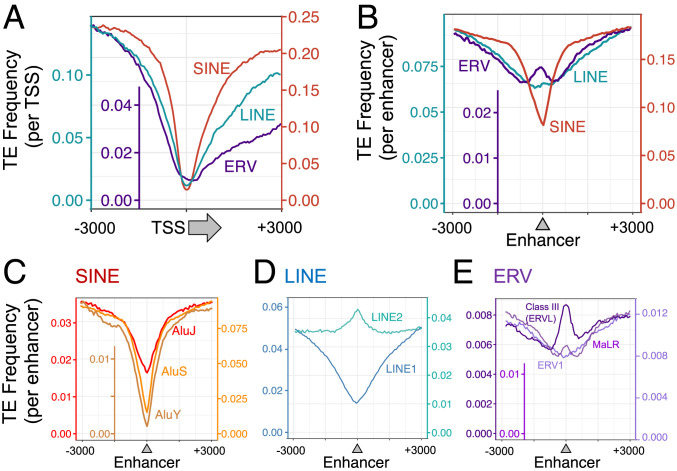
Next, we determined the distribution of TEs with respect to transcription start sites (TSSs) and enhancers (Fig. 4). An earlier study looked at the promoter as a broad region and found that 25% of such regions harbor TEs (16). After determining exact TSS locations (Ensembl release 87), we found that all three classes of TEs, irrespective of their evolutionary ages, were in fact strongly excluded from TSSs and that their frequency increased as a function of distance from a TSS (Fig. 4A). TE frequency is defined as the ratio of sequences originating from TEs found in TSSs or enhancers compared with host sequences found in all elements investigated. This relationship is consistent with our earlier report that fewer TEs are found at bidirectional promoter regions in which two proximal TSSs are oriented away from each other (17). Interestingly, the distribution curves showed decreased occupancy downstream of the TSS relative to upstream, with the asymmetry best exhibited by ERVs. A possible explanation for the asymmetry is that newly inserted TEs might interfere with transcription pausing or first-intron splicing (18). Alternatively, they might be too long to be accommodated in the 5′ untranslated region, therefore undergoing negative selection in these positions.
Fig. 4.
TEs are infrequently located at TSSs but can generate enhancers when mutated. (A) TEs (SINEs, LINEs, and ERVs) are almost never located at TSSs. The y axes represent the ratios of TE-generated TSSs compared with all TSSs and are unit-less. The color of the axes represents each class of TE and matches the color of the corresponding curve displayed. (B) Compared with TSSs, TEs are more frequently located in enhancers. (C–E) Young TEs are rarely seen bearing enhancer elements whereas older (mutated) TEs slowly evolved into enhancer elements.
In contrast to their absence at TSSs, ERVs were found at the centers of enhancers at similar frequencies to the surrounding host DNA; LINEs and SINEs were found at lower frequency (Fig. 4B). The presence of TEs at these locations was dependent on their evolutionary age, as assessed by the CpG O/E ratio. Young Alu families, as exemplified by AluY (19), were notably rare in enhancers relative to intermediate-age AluS and older AluJ family members (Fig. 4C). Because of the problem of mapping younger TEs that have not accumulated sufficient distinguishing mutations, we focused on mappable AluY copies to derive these data displayed in Fig. 4C.
Older LINE elements (LINE2) were slightly overrepresented in the centers of enhancers relative to the flanking DNA; younger (LINE1) sequences were less frequent (Fig. 4D). The same dependence on evolutionary age was seen with three classes of ERVs (Fig. 4E); the older class III (ERVL) family was overrepresented at enhancer centers relative to the younger class I and class II (ERVK) (Fig. 4E), likely due to the accumulation of C-to-T transitions or to having a lower CpG content when originally inserted. These data suggest that the likelihood that a TE will serve a regulatory function is increased by C-T and other mutations acquired over time.
Our genome-wide TE analysis suggests that DNA methylation and the C-to-T mutation consequences are factors in how TEs can provide a source of host regulatory elements. The insertion of a TE has been proposed to speed up the process of enhancer creation by providing extra DNA containing preexisting regulatory sequences compatible with the host transcription factors (20). However, based on our findings, it seems more likely that insertion followed by de novo methylation of CpG sites in the germline causes C-to-T transitions that, along with other mutations, results in the generation of new regulatory elements both within the integrated TE and in the surrounding host DNA.
The Evolutionary Drive for TE-Derived Transcription Factor Binding Sites Is Dependent on the Genomic Context.
The presence of multiple transcription factor binding sites (TFBSs) in TE-derived DNA led to the hypothesis that TEs might provide a ready source of DNA that could be co-opted by the cell to help regulate gene expression (16, 21, 22), summarized in previous reviews (23, 24). We queried the potentials for each TE class to harbor binding motifs and compared these with the actual binding of TFs in living cells as measured by chromatin immunoprecipitation (ChIP). First, we found that the occupancy of TE-derived TFBSs is largely determined by genomic position and the local chromatin state. TE-derived binding motifs are less frequently (1.5%) bound by a TF relative to host-derived binding motifs (4.7%) (SI Appendix, Fig. S2). Second, TE-derived DNA close to gene promoters was more likely to have a bound TF (SI Appendix, Fig. S3), suggesting more frequent regulatory functions exerted by TE-derived DNA in gene proximity. Likewise, TE-derived sequences are less likely to account for the actual binding of TFs compared with host-derived sequences at virtually all distances to the promoters (SI Appendix, Fig. S3). Third, many active binding events mapped to TEs were associated with the inherent promoters of TEs. For example, analysis of TF enrichment (SI Appendix, Fig. S4A) identified RPC155/POLR3A, an RNA polymerase (RNAP) III subunit, as the only TF whose binding was increased in SINEs out of 148 TFs assayed in the ENCODE data (SI Appendix, Fig. S4 B and E). Likewise, TF-binding events mapped to LINEs and ERVs were best represented by RNAP II subunits and associated general transcription factors such as POLR2A, TAF1, and TBP. Fourth, CpG-rich, LTR-bearing ERVs were more likely to be bound by transcription initiation-associated TFs than were CpG-poor ERV fragments.
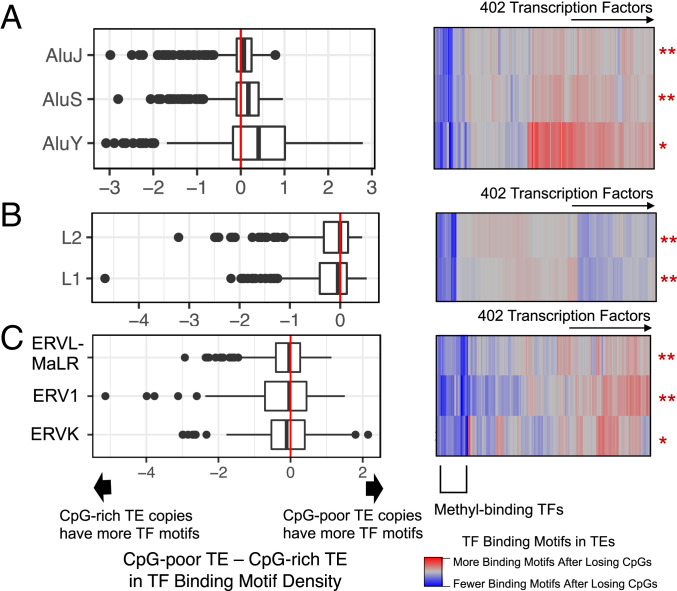
We compared CpG-rich copies and CpG-poor copies in each TE family and were able to identify a subset of TFs for which binding motifs would likely be gained after 5-methylcytosine deamination (Fig. 5). Gains were most notable in young TE families such as AluY (P = 1.1 × 10 −14, two-sided Wilcoxon test) (Fig. 5A). Older TE families such as AluJ (Fig. 5A), most LINEs (Fig. 5B), and most ERVs (Fig. 5C) tended to reach mutational homeostasis, where further CpG deamination does not lead to many more gains of binding motifs. This result is also supported by the existence of multiple TFBSs in LINEs and SINEs, but only in CpG-poor copies. The heatmaps (Fig. 5, Right) show that methyl-binding proteins such as MECP2, MBD2, and KAISO lose binding sites as a result of CpG loss, as expected.
Fig. 5.
Effect of CpG loss in TEs on their TF motif densities. Each dot in the graphs (Left) represents one TF. Rows of heatmaps correspond to TE subfamilies and columns correspond to TFs. CpG-rich and CpG-poor groups are not explicitly shown. In the heatmaps, each color shows the difference between the two groups for each TF. (A) CpG-poor SINEs are associated with more TF-binding motifs; P = 1.1 × 10−14 (AluY), 8.2 × 10−8 (AluS), and 6.2 × 10−4 (AluJ). **P < 1 × 10−4, *P < 0.01. (B) Comparison of CpG-rich and CpG-poor LINEs; P = 3.5 × 10−8 (L1) and 3.5 × 10−6 (L2). (C) Comparison of CpG-rich and CpG-poor ERVs; P = 0.011 (ERVL-MaLR), 0.006 (ERV1), and 0.004 (ERVK). Statistical significance was assessed using a two-sided Wilcoxon’s test.
We studied TF binding specific to each TE family and found increased binding of TRIM28/KAP1 was seen for ERVs, unlike for SINEs and LINES (SI Appendix, Fig. S4G). TRIM28 is known for its role in transcriptionally corepressing ERVs by binding to the ERV-targeting KRAB zinc-finger proteins (KZNFs) and subsequently mobilizing additional repressor proteins such as SETDB1, HP1, and the nucleosome remodeling and deacetylation (NuRD) complex (25). This binding is representative of TFs evolving to indirectly target TE sequences for their suppression, often in a non–sequence-specific way. Although ERVs are more likely to be bound by TFs than are SINEs and LINEs (SI Appendix, Fig. S4 B–D), many ERV-binding events are not sequence-specific; instead, they are associated with the local chromatin states of either active promoters or NuRD-mediated heterochromatinization. These results show diverse modes of TE-TFBS co-evolution, with some TFs evolving to target specific TE-associated genomic elements and chromatin states while others attracted to TFBS evolved from TE sequences. These distinct modes do not always follow a simple model of TE insertions delivering host fitness advantages by providing TFBSs and regulating host gene expression.
Host DNA Is More Likely to Harbor TFBSs than TE-Derived DNA.
The analysis above shows that some TEs can contribute to the generation of TFBSs, leading to the question of how often they actually participate in gene control networks relative to non-TE DNA. We calculated the TF-binding motifs and TFBSs (as identified by ChIP) in various genomic locations. Overall, we found that TEs are most common in intronic and intergenic regions and make up about 45% of total human DNA (Table 1). Known motifs for TFBSs were distributed almost equally in both TEs and non-TE DNA. However, bound (and therefore potentially functional) TFBSs in SINEs and LINEs were more frequent in introns than in intergenic DNA. On the other hand, TF binding in ERVs was more common in intergenic regions.
Table 1.
Occupied transcription factor binding sites are more prevalent in non-TE–derived DNA
| Region | SINE (1 × 106) | LINE (1 × 106) | ERV (1 × 106) | Non-TE DNA | |
| Bases (1×106) | Exon | 8 | 6 | 4 | 113 |
| Intron | 228 | 301 | 102 | 839 | |
| Intergenic | 161 | 332 | 162 | 643 | |
| Total | 397 | 638 | 268 | 1,595 | |
| % genome | 14 | 22 | 9 | 55 | |
| SINE + LINE + ERV | 45% | ||||
| Number of TF-binding motifs (1×106) | Exon | 2 | 1 | 1 | 29 |
| Intron | 63 | 52 | 20 | 173 | |
| Intergenic | 43 | 53 | 30 | 126 | |
| Total | 108 | 106 | 51 | 326 | |
| % genome | 18 | 18 | 9 | 55 | |
| SINE + LINE + ERV | 45% | ||||
| Number of TFBSs occupied in adult tissues (1×103) | Exon | 23 | 24 | 31 | 2,524 |
| Intron | 251 | 464 | 372 | 4,966 | |
| Intergenic | 150 | 337 | 487 | 3,578 | |
| Total | 425 | 825 | 890 | 11,068 | |
| % genome | 3 | 6 | 7 | 84 | |
| SINE + LINE + ERV | 16% | ||||
Transposable elements make up 45% of the human genome and provide a similar fraction of potential binding motifs but harbor only 16% of the actual TF binding. A small subset of TF motifs can overlap with multiple regions (e.g., both exons and introns) and are double-counted, causing their sums to be unequal to the genome-wide counts.
TEs contributed to 16% of the occupied TFBSs found in total cellular DNA (Table 1). ERVs were the largest contributor, associated with 7% of the occupied TFBSs. This is in contrast to the count of TF-binding motifs, in which TEs, and SINEs in particular, were found to harbor a similar (if not greater) number of binding motifs relative to non-TE–derived DNA (Table 1, Middle), consistent with SI Appendix, Fig. S2 and prior studies (26–28). Overall, therefore, TEs make up 45% of the genome yet contribute 16% of the occupied TFBSs, and TFBSs are two to six times more likely to be located in non-TE–derived DNA than TE-derived DNA genome-wide in somatic cells (SI Appendix, Table S1). As discussed above, this conclusion that TF binding is more common in non-TE sequences than TE sequences holds after correcting for the distance to the nearest TSSs (SI Appendix, Fig. S3).
Discussion
DNA methylation has roles in the control of gene expression at the levels of transcription initiation and elongation as well as in the function of regulatory elements such as promoters, enhancers, and insulators; those roles are relatively well understood (29). Its role as a suppressor of the transcription of TEs (30) is also widely accepted. In this paper, we suggest that the increase in eukaryotic genome size is a result of the interplay among TE insertion, DNA methylation, and 5-methylcytosine deamination. Although we focus on a simplified vision for the role of DNA methylation as primarily a defense mechanism, our model does not preclude other roles for DNA methylation in conferring selective evolutionary advantage. For example, Regev et al. (31) have argued that its role in gene control might have preceded its participation in TE suppression, thus contributing to genome evolution in additional ways.
The main features of our model are presented in Fig. 6. TEs are initially CpG-rich in their promoters and can insert and transpose while they have a high CpG O/E ratio. The insertion of a TE into the germline is potentially lethal to the host unless its transcription can be blocked by a process such as DNA cytosine methylation. Interestingly, they can insert widely in the genome but are almost completely excluded from host TSSs, suggesting that this might be immediately lethal to the host.
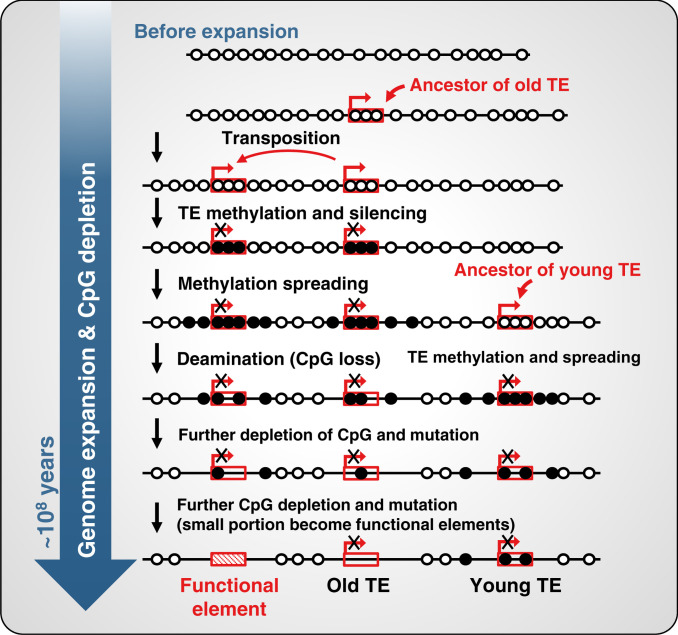
Fig. 6.
CpG methylation contributes to TE-mediated genome expansion and ultimately to CpG depletion by deamination and neofunctionalization of TEs in the expanded genome. The model depicts an early genome with no TEs and the unmethylated CpG sites shown as open circles and methylated CpGs as solid black circles. At this stage, the CpG O/E ratio is about 1. Insertion and transposition of a TE lead to its de novo methylation (shown as black circles) and silencing of the TE. Methylation can then spread into the flanking host DNA. Methylated CpGs have an enhanced mutation frequency relative to unmethylated CpGs and a half-life of about 35 million y in the primate germline (10). Over evolutionary time, this leads to an overall depletion of CpGs in the entire genome with the exception of CpG islands (11) and ultimately to the creation of new functional elements such as enhancers, depicted by the decreasing number of methylation sites and a decrease in CpG O/E ratio.
The evolution of prokaryotic DNA methylases into enzymes with the CpG recognition sequence allowed for the accommodation of silenced TEs in vertebrate genomes and therefore to massive genome expansion. A well-recognized consequence of TE methylation is the spread of methylation into the host DNA, which results eventually in the striking inverse correlation we found between the CpG O/E ratio and the genome size of the organism. The spread of methylation and rarely demethylation from TEs was previously demonstrated in locus-specific manner for Alu (10) and L1 (32) sequences through comparative analysis, underscoring their role in creating the epigenetic and eventually the genetic landscape of the mammalian germline.
The central role of cytosine methylation in repressing the transcription of evolutionarily young TEs (i.e., those more recently inserted) has been well-described (5, 33). The potentially lethal effects of inappropriate ERV expression have been suggested by the observation that a group of young ERVs is not demethylated even during the programmed genomic demethylation in preimplantation embryonic development (34, 35) and in primordial germ cell development (36). Further, mice have evolved a specialized Dnmt3c that targets ERVs during development (37). The DNA methylation at TEs in mammalian germlines has also been suggested to be guided by host factors including Piwi-interacting RNA (piRNA) (38, 39).
In synergy with DNA methylation-mediated TE silencing, other mechanisms may also contribute to the host’s tolerance of TEs on both the transcriptional and posttranscriptional level. Two notable candidates that act on the transcriptional level are KZNFs (40, 41) and the tumor suppressor protein p53, which both evolved roughly contemporaneously with the TE-mediated genome expansion and genome-wide DNA methylation in vertebrates (42, 43). KZNFs silence gene transcription by cooperating with the transcriptional corepressor TRIM28 and the NuRD complexes. ZNF91 and ZNF93 have undergone fast, recent evolution in primates to keep up with the accumulation of mutations that allow TEs to escape host suppression (44). The tumor suppressor p53 is known for its interaction with DNMT3A (45) and for cooperation with DNA methylation in the epigenetic silencing of transposable elements (46).
On the posttranscriptional level, an array of host factors has been identified which sense and respond to transposable element activation. RNA helicases such as MDA5 and RIG-I were found to target ERV transcripts activated from DNMT inhibitor administration (47). Host factors such as piRNA, ZAP, RNaseL, MOV10, and TREX1 suppress retrotransposition through sensing and degrading cytoplasmic viral RNA or complementary DNA. RNA editors such as APOBEC/AID enzymes enable posttranslational modification of TE transcripts, limiting its retrotransposition capacity. Viral suppression can also occur indirectly. For instance, enzymes that affect the level of the dNTP pool limit TE transcription and replication (48). These mechanisms have been summarized in previous reviews (49).
Although it is tempting to suggest that TEs become activated for the selective advantage of the host, the hypothesis that TE activity rewires regulatory networks (16, 22, 50) is complicated at several levels. There is a discrepancy between the motif provided by a TE and the actual TFBS occupancy (SI Appendix, Table S1). For example, Alu sequences are known to harbor a compendium of sequence-binding motifs for nuclear receptors such as RAR, VDR, and LXR (26, 27), but most of the occupancy by these TFs lies in non-TE genomic territory in the human soma (SI Appendix, Table S1). We saw a disproportionately smaller number of TFBSs in the TE-derived DNA (Table 1) relative to the 45% of TF motifs the human genome harbors. This is consistent with prior reports of a lack of direct evidence that TEs are conclusively used as cis-regulatory elements (51). The occupancy—and therefore the actual use—of motifs could depend on the developmental stage. Certain TFBSs are co-opted only when the epigenetic suppression is lifted, for example, by the global epigenetic remodeling occurring in early embryonic development. This is also consistent with earlier discoveries highlighting the more active use of TE-derived TFBSs in regulating stem cell renewal and differentiation (52, 53). Evolutionary age may play a significant role in the adoption of TE-derived TFBSs, because newly inserted TEs are not the most common starting material for TFBS generation. We have shown here that most of them need to be mutated to be optimized for such exaptation, which is consistent with previous observations in ERVs (54) and Alu elements (55) that C-to-T and other mutations are needed to complete TE co-option.
The emergence of whole-genome DNA methylation had profound implications as to how the genome evolved. DNA methylation will be the key to understanding how incremental evolution was replaced by a system of TEs and host DNA intricately interacting, coevolving, and contributing to regulatory innovations in greatly enlarged genomes. Our work suggests an unrecognized role for DNA methylation in enabling genome expansion and the increase in DNA mass.
Materials and Methods
Genome Data and Transposable Element Statistics.
The genome sequence and transcript annotation was retrieved from Ensembl release 87 (47 vertebrates) and Ensembl metazoa (6 invertebrates) release 38 (56). This diverse collection covers 43 tetrapods (including 38 mammals and 5 birds). The phylogenetic tree was obtained by pruning the National Center for Biotechnology Information taxonomy. We controlled the quality of the included assembly by requiring a minimum scaffold N50 of 200 kb. The CpG observed/expected ratio was calculated by the CpG density—which is N(CpG)/N, where N(CpG) is the number of CpG dinucleotides and N is the length of the genome—divided by the expected CpG density, N(C) × N(G)/(N × N), where N(C) is the number of cytosines and N(G) is the number of guanines. Transposable element coverage was estimated using annotation provided by the Ensembl database. A list of public methylome datasets reanalyzed in the study can be found in SI Appendix, Table S2. The chromatin accessibility in human early embryos was obtained from a recent study (57).
Alu CpG Densities.
We downloaded human repeat masker data (58, 59) and grouped 1,142,278 Alu elements by internal CpG density. For each group, we computed the fraction of CpGs in the ±800-bp flanking region using a 100-bp overlapping window. The CpG density, defined as the number of CpGs per base pair of DNA sequence, was then plotted as a heatmap. Analysis of TP53 intronic Alu insertion used the following genome assemblies and TP53 transcripts: ENST00000617185 (GRCh38, human), ENSPTRT00000016033 (Pan_tro_3.0, chimpanzee), ENSNLET00000012443 (Nleu_3.0, gibbon), ENSMLET00000060690 (Mleu.le_1.0, mandrill), ENSMICT00000052203 (Mmur_3.0, lemur), ENSSBOT00000024929 (SaiBol1.0, squirrel monkey), ENSCCAT00000047260 (Cebus_imitator-1.0, capuchin), and ENSTSYT00000028083 (Tarsius_syrichta-2.0.1, tarsier). Gibbon and squirrel monkey have extra Alu element insertion into intron 10 and the extra Alu elements were excluded from the host sequence CpG density calculation, in addition to the shared AluS insertion.
TEs around TSSs and Enhancers.
We studied transposable element frequency for each of the 100-bp windows evenly positioned from the transcription start sites. Transcription start sites were obtained by collapsing the TSSs of messenger RNA transcripts included in Ensembl release 87. For the enhancer analysis, we studied the 15-state chromHMM annotation generated from the Roadmap Epigenome Project (60). We considered a region to be an enhancer only if it was found to be either a strong enhancer or poised enhancer in more than 50 samples. We centered these regions and probed 3,000 bp upstream and downstream of the enhancer region and computed the frequency of observing transposable elements. To normalize by flanking region, we equalized the y axis of each plot using the last 300 bp from both ends for the repetitive element classes plotted in the same panel in order to highlight the relative depletion for each TE category.
TFBS-Generating Potentials.
TF-binding motifs were obtained by scanning the human genome sequence (GRCh37) using FIMO (61). We studied 402 core motifs included in the HOCOMOCO database (version 11) (62). CpG density was defined as the observed CpG over the expected CpG: N(CpG) × N/(N(C) × N(G)), where N(CpG), N(C), and N(G) are the number of CpG dinucleotides, number of cytosines, and number of guanines, respectively. High-CpG TEs were defined as having CpG density >0.3 and low-CpG TEs as having CpG density <0.2. Only SINEs greater than 200 bp in length and LINEs greater than 3,000 bp in length were included to avoid fragments. The overlap between TF-binding motifs and TEs was computed by BEDTools (63) and normalized by the length of the TEs. For TF-binding events, we collected narrow peaks for 508 ChIP-seq (ChIP sequencing) experiments of 148 TF binding sites in 84 cell lines from the ENCODE project. We computed the frequency of TE presence in the 3-kb flanking sequence centered on each TF binding site. Only mappable TFBSs were considered. Genome mappability was downloaded from the UCSC ENCODE data track (https://genome.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeMapability). We used a 50-mer track and excluded regions of mappability less than 0.5. TEs overlapping with nonmappable regions were excluded from the analysis. Only ChIP-seq peaks in mappable regions were included in the analysis to sidestep arbitrariness in placing multimapping reads. The TFBS potential of a TE was characterized by its enrichment at the TFBS apex normalized by the flanking genomic region (SI Appendix, Fig. S4A). In other words, for each TF, we computed the enrichment score of the TE in the center of the TFBS. The enrichment score was defined as the relative TE depletion of the TFBS center with respect to the flanking region. Different TFBSs were clustered using uniform manifold approximation and projection by TE enrichment.
TE Enrichment in the Human Genome.
We downloaded exon definitions and transcript definitions for humans from GENCODE database release 26. Exonic regions were merged from all of the exons from all isoform definitions. TE definitions were downloaded from RepeatMasker (58). For each 100-bp nonoverlapping window in the genome, we computed the density of TFBSs and compared it with the distance of the window to the transcription start site.
Supplementary Material
Acknowledgments
This work was supported by Grant R35 CA209859 from the National Cancer Institute. We thank David Nadziejka PhD for editing the manuscript and Jason Jones PhD, who was involved in the early development of some of the concepts in this manuscript.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921719117/-/DCSupplemental.
Data Availability.
All of the data were retrieved from public data repositories (Materials and Methods).
References
- 1.Hidalgo O. et al., Is there an upper limit to genome size? Trends Plant Sci. 22, 567–573 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Canapa A., Barucca M., Biscotti M. A., Forconi M., Olmo E., Transposons, genome size, and evolutionary insights in animals. Cytogenet. Genome Res. 147, 217–239 (2015). [DOI] [PubMed] [Google Scholar]
- 3.McClintock B., The significance of responses of the genome to challenge. Science 226, 792–801 (1984). [DOI] [PubMed] [Google Scholar]
- 4.Bestor T. H., Sex brings transposons and genomes into conflict. Genetica 107, 289–295 (1999). [PubMed] [Google Scholar]
- 5.Yoder J. A., Walsh C. P., Bestor T. H., Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13, 335–340 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Lee E. et al.; Cancer Genome Atlas Research Network , Landscape of somatic retrotransposition in human cancers. Science 337, 967–971 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bestor T. H., DNA methylation: Evolution of a bacterial immune function into a regulator of gene expression and genome structure in higher eukaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 326, 179–187 (1990). [DOI] [PubMed] [Google Scholar]
- 8.Bird A. et al., Studies of DNA methylation in animals. J. Cell Sci. Suppl. 19, 37–39 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Coulondre C., Miller J. H., Farabaugh P. J., Gilbert W., Molecular basis of base substitution hotspots in Escherichia coli. Nature 274, 775–780 (1978). [DOI] [PubMed] [Google Scholar]
- 10.Yang A. S. et al., The rate of CpG mutation in Alu repetitive elements within the p53 tumor suppressor gene in the primate germline. J. Mol. Biol. 258, 240–250 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Zemach A., McDaniel I. E., Silva P., Zilberman D., Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916–919 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Deininger P., Alu elements: Know the SINEs. Genome Biol. 12, 236 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magewu A. N., Jones P. A., Ubiquitous and tenacious methylation of the CpG site in codon 248 of the p53 gene may explain its frequent appearance as a mutational hot spot in human cancer. Mol. Cell. Biol. 14, 4225–4232 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yates P. A., Burman R. W., Mummaneni P., Krussel S., Turker M. S., Tandem B1 elements located in a mouse methylation center provide a target for de novo DNA methylation. J. Biol. Chem. 274, 36357–36361 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Egger G., Liang G., Aparicio A., Jones P. A., Epigenetics in human disease and prospects for epigenetic therapy. Nature 429, 457–463 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Jordan I. K., Rogozin I. B., Glazko G. V., Koonin E. V., Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 19, 68–72 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Takai D., Jones P. A., Origins of bidirectional promoters: Computational analyses of intergenic distance in the human genome. Mol. Biol. Evol. 21, 463–467 (2004). [DOI] [PubMed] [Google Scholar]
- 18.van de Lagemaat L. N., Medstrand P., Mager D. L., Multiple effects govern endogenous retrovirus survival patterns in human gene introns. Genome Biol. 7, R86 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price A. L., Eskin E., Pevzner P. A., Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res. 14, 2245–2252 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourque G. et al., Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 18, 1752–1762 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Britten R. J., Davidson E. H., Gene regulation for higher cells: A theory. Science 165, 349–357 (1969). [DOI] [PubMed] [Google Scholar]
- 22.Feschotte C., Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 9, 397–405 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chuong E. B., Elde N. C., Feschotte C., Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 18, 71–86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeh D. W., Zeh J. A., Ishida Y., Transposable elements and an epigenetic basis for punctuated equilibria. BioEssays 31, 715–726 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Iyengar S., Farnham P. J., KAP1 protein: An enigmatic master regulator of the genome. J. Biol. Chem. 286, 26267–26276 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polak P., Domany E., Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics 7, 133 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gombart A. F., Saito T., Koeffler H. P., Exaptation of an ancient Alu short interspersed element provides a highly conserved vitamin D-mediated innate immune response in humans and primates. BMC Genomics 10, 321 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui F., Sirotin M. V., Zhurkin V. B., Impact of Alu repeats on the evolution of human p53 binding sites. Biol. Direct 6, 2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones P. A., Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Ohtani H., Liu M., Zhou W., Liang G., Jones P. A., Switching roles for DNA and histone methylation depend on evolutionary ages of human endogenous retroviruses. Genome Res. 28, 1147–1157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regev A., Lamb M. J., Jablonka E., The role of DNA methylation in invertebrates: Developmental regulation or genome defense? Mol. Biol. Evol. 15, 880–891 (1998). [Google Scholar]
- 32.Grandi F. C. et al., Retrotransposition creates sloping shores: A graded influence of hypomethylated CpG islands on flanking CpG sites. Genome Res. 25, 1135–1146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh C. P., Chaillet J. R., Bestor T. H., Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20, 116–117 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Guo H. et al., The DNA methylation landscape of human early embryos. Nature 511, 606–610 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Göke J. et al., Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell 16, 135–141 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Tang W. W. C. et al., A unique gene regulatory network resets the human germline epigenome for development. Cell 161, 1453–1467 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barau J. et al., The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 354, 909–912 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Aravin A. A. et al., A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuramochi-Miyagawa S. et al., DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22, 908–917 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rowe H. M. et al., De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development 140, 519–529 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quenneville S. et al., The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Rep. 2, 766–773 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imbeault M., Helleboid P.-Y., Trono D., KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543, 550–554 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Belyi V. A. et al., The origins and evolution of the p53 family of genes. Cold Spring Harb. Perspect. Biol. 2, a001198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jacobs F. M. J. et al., An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516, 242–245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y. A. et al., DNA methyltransferase-3a interacts with p53 and represses p53-mediated gene expression. Cancer Biol. Ther. 4, 1138–1143 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Levine A. J., The p53 protein plays a central role in the mechanism of action of epigentic drugs that alter the methylation of cytosine residues in DNA. Oncotarget 8, 7228–7230 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishak C. A., Classon M., De Carvalho D. D., Deregulation of retroelements as an emerging therapeutic opportunity in cancer. Trends Cancer 4, 583–597 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Gramberg T. et al., Restriction of diverse retroviruses by SAMHD1. Retrovirology 10, 26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodier J. L., Restricting retrotransposons: A review. Mob. DNA 7, 16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang T. et al., Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl. Acad. Sci. U.S.A. 104, 18613–18618 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Souza F. S. J., Franchini L. F., Rubinstein M., Exaptation of transposable elements into novel cis-regulatory elements: Is the evidence always strong? Mol. Biol. Evol. 30, 1239–1251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunarso G. et al., Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 42, 631–634 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Kano H. et al., L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 23, 1303–1312 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emera D., Wagner G. P., Transposable element recruitments in the mammalian placenta: Impacts and mechanisms. Brief. Funct. Genomics 11, 267–276 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Su M., Han D., Boyd-Kirkup J., Yu X., Han J. J., Evolution of Alu elements toward enhancers. Cell Rep. 7, 376–385 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Zerbino D. R. et al., Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu J. et al., Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature 557, 256–260 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Smit A. F. A., Riggs A. D., MIRs are classic, tRNA-derived SINEs that amplified before the mammalian radiation. Nucleic Acids Res. 23, 98–102 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smit A. F., Riggs A. D., Tiggers and DNA transposon fossils in the human genome. Proc. Natl. Acad. Sci. U.S.A. 93, 1443–1448 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kundaje A. et al.; Roadmap Epigenomics Consortium , Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grant C. E., Bailey T. L., Noble W. S., FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kulakovskiy I. V. et al., HOCOMOCO: Towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-seq analysis. Nucleic Acids Res. 46, D252–D259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data were retrieved from public data repositories (Materials and Methods).